- 1Department of Animal Genetics and Breeding, College of Animal Science and Technology, Yangzhou University, Yangzhou, China
- 2Department of Poultry Genetics and Breeding, Poultry Institute, Chinese Academy of Agricultural Sciences, Yangzhou, China
Chicken reticuloendotheliosis virus (REV) causes the atrophy of immune organs and immuno-suppression. The pathogenic mechanisms of REV are poorly understood. The aim of this study was to use RNA sequencing to analyse the effect of REV on immunity and cell proliferation in chicken lymphocytes from peripheral blood in vitro. Overall, 2977 differentially expressed genes (DEGs) were examined from cells between infected with REV or no; 56 DEGs related to cell proliferation and 130 DEGs related to immunity were identified. MTT, Q-PCR, and FCM indicated that REV reduced the number of lymphocytes by inhibiting the transition of S/G1 phase through FOXO and p53 pathways. Similarly, REV infection would destroy the immune defense of lymphocytes through MAPK-AP1 via Toll-like receptor-, NOD-like receptor-, and salmonella infection pathways to reduce the secretion of IL8 and IL18. In addition, the reduction of lymphocytes also might be responsible for the lower levels of IL8 and IL18, and the rescue of lymphocytes would been activated still through FOXO and p53 pathways. Moreover, the immune response for REV in lymphocytes would activate by up-regulating the expression of NOD1, MYD88, and AP1 through Toll-like receptor-/NOD-like receptor/salmonella-MAPK-AP1 pathways. These results indicate that REV could affect lymphocytes from peripheral blood by inhibit the cell proliferation and the immune system. It also was revealed that NOD1, MYD88, and AP1 were the key genes to activate the immune response through Toll-like receptor-/NOD-like receptor/salmonella-MAPK-AP1 pathways. These findings establish the groundwork and provide new clues for deciphering the molecular mechanism underlying REV infection in chickens.
Introduction
Chicken reticuloendotheliosis is a serious infectious oncosis caused by reticuloendotheliosis virus (REV) infected lymphocytes or reticuloendothelial cells (Witter et al., 1979). Many studies have been reported on the epidemiology, vaccines, and the pathogenesis of REV (García et al., 2003; Fadly and Garcia, 2006; Cheng et al., 2007; Li and Cui, 2007). REV causes the atrophy of immune organs (thymus and bursa of Fabricius) leading to immuno-suppression (Witter et al., 1981; Nazerian et al., 1982). Previous studies have demonstrated a change in IL-2 secretion. T lymphocytes in the thymus and spleen were damaged by REV causing a reduction in the secretion levels of IL-2 (Hrdlicková et al., 1994). A decrease in IL-2 secretion further reduced the number of T cells, and caused serious immune dysfunction when secreting positive immune cytokines to influence the differentiation of T cells into TH and CTL cells, which have different functions (Li and Liu, 1999; Kim et al., 2004).
Although a certain studies on the mechanism of REV pathogenesis and immunity have focused on genome and proteome levels (Li et al., 2016; Xue et al., 2017), these processes are still poorly understood on gene expression level. This study had systematically identified the global genes and pathways by RNA sequencing in chicken lymphocytes after REV infection. The present study investigated the effect of REV on chicken blood lymphocytes.
Materials and Methods
Animals and Ethics Statement
Experimental chickens at 21 day were from a pure line of SPF Rugao chicken from the Poultry Institute, Chinese Academy of Agricultural Sciences. Blood was collected from the wing vein, using citric acid dextrose as an anticoagulant for subsequent lymphocytes derived. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People's Republic of China (2006).
All experimental procedures were performed in accordance with the Administration Act of Experimental Animals using and care in Jiangsu Province (#115th Jiangsu Province Government Notice in 2008). All of the animal experimental operations were approved and guided by the Animal Care and Use Committee of Yangzhou University.
The Standard Biosecurity or Institutional Safety Procedures
All experiments with the REV viruses in vivo and vitro were conducted in a key biosafety Laboratory of Animal Genetics and Breeding of Molecular Design of Jiangsu Province.
The half lethal dose and the minimum lethal dose of biological agents involved in the experiment were tested by the test center of Yangzhou University (Certificate Number: 150000002432), the doses of biological agents used in experiment are within half the amount of lethal dose, normal animals do not die, which were approved by the laboratory management of Yangzhou University.
Lymphocytes Culture and REV Infection
Lymphocytes were derived from chicken blood according the protocol of the chicken blood lymphocyte separation medium kit. Cells (5 × 105/ ml) were incubated with RPMI-1640 medium containing 10% FBS in 10- cm dishes for 24 h, and then infected with reticuloendotheliosis virus strain HA1101 (REV, GenBank accession number: KF305089.1) with 105 TCID50/0.1 ml coming from the Key Laboratory of Jiangsu Preventive Veterinary Medicine. Cells were harvested for use in RNA sequencing, FCM, Q-PCR, and ELISA after infection for 36 h. Another set of eight independent replicates were subjected to the same treatment for MTT analysis. Other studies in lymphocytes were carried out in triplicate.
RNA Sequencing Analysis
Total RNA was isolated from REV infected or uninfected cells using Trizol reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions and dissolved in RNase-free water at a final concentration, 2.0 μg/μl. RNA sequencing was performed by Anno-road Genomics Company (Beijing, China), and data extraction was carried out following the standard protocol (Liao et al., 2013; Love et al., 2014). Genes were considered differentially expressed genes (DEGs) only when the fold-change in abundance for all comparisons exceeded 2.0, with a P-value < 0.05. Using the Euclidian metric, average linkage hierarchical clustering was performed based on DEGs. In heat-maps, the color of features was determined by log2 (reference/sample).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
Based on the DEGs, Gene Ontology enrichment analysis was performed using the GOEAST software toolkit. The significance level of GO term enrichment was set as FDR-adjusted p-value < 0.05 by the Yekutieli method. Enriched KEGG pathways with DEGs were identified by a hypergeometric test using R packages (P < 0.01, FDR adjusted). Pathways with < 3 known chicken genes were discarded. Graphical pathway maps were downloaded from the KEGG FTP server, and DEGs were then highlighted in them according to the coordinate description in the XML files at the KEGG FTP server, using Perl GD, XML::Parser and XML::LibXML modules.
Validation by Quantitative Polymerase Chain Reaction (Q-PCR)
Primer information is listed in Table S1. Each 20 μl PCR mixture contained 10 μl of the 2 × iQ™ SYBR Green Supermix, 0.5 μl (10 mM) of each primer and 1 μl of cDNA. Mixtures were incubated in a Real-Time PCR Detection System (ABI7500, Carlsbad, California, USA). A melting curve was constructed to verify that only a single PCR product was amplified. Samples were assayed in triplicate with standard deviations of threshold cycle (CT) values not exceeding 0.5 on a within-run basis. Correlation analysis for gene expression between the two methods was performed.
Screen of Related DEGs
Based on DEGs, gene ontology (GO) enrichment analysis was performed using the GOEAST software toolkit, and the significance level of GO term enrichment was set as a FDR-adjusted p-value < 0.05 by the Yekutieli method. DEG-related focus traits were screened.
Cell Proliferation Assay
Lymphocytes in six-well culture dishes were infected with REV or were uninfected, in RPMI1640 medium containing 10% FBS for 48 h, and then collected by centrifugation at 500 × g for 5 min for MTT detection. For the MTT assay, 20 μl of MTT (5 mg/ml) was added to each well and incubated for 4 h at 37°C. After removing the supernatant, formazan crystals were dissolved in 200 μl DMSO and the absorbance was measured at 490 nm. Each group had eight repeat wells to ensure the accuracy of the experiment.
Flow Cytometry (FCM) Assay
The same samples in the cell proliferation assay were used for the cell cycle assay by FCM. Cells were digested by trypsin, collected after 500 × g centrifugation for 5 min and washed with ice-cold PBS. The cell pellet was suspended with 70% ethanol at 4°C overnight, washed with PBS, then incubated with 180 μ g/ml RNase A at 37°C for 15 min. For FCM, 50 μg/ml propidium iodide (final concentration) was added for 15 min staining in the dark at 37°C. Data were plotted and analyzed using FCS Software (De Novo Software, Los Angeles, CA).
ELISA for Immune Factors
The cell medium from REV infected or uninfected groups were used for the detection of TNF2, IL8, and IL18, and the assay was performed using a specific ELISA kit (B&D, Minnesota, USA) according to the manufacturer's instructions.
Statistical Analyses
Statistical differences between groups were evaluated using the Student's t-test. P < 0.05 (*) or < 0.01 (**) was considered significant. Data are represented as the mean ± SD.
Results
Differentially Expressed Gene Profiles in Lymphocytes after Infection with REV
Using the lymphocytes from peripheral blood infected with REV or not, gene expression profiles were examined using RNA sequencing. 2977 known genes, 1981 down-regulated, and 996 up-regulated (Infection vs. Control), were detected as differentially expressed genes (DEGs) with consistent fold changes ≥2.0 (Table S2). Based on the 2977 known DEGs, a cluster analysis of all six samples was performed by Cluster 3.0 software, three samples in same one group were respectively polymerized (Figure 1A). in addition, some DEGs were selected to examine the relative expression using Q-PCR. As shown in Figure 1B, the fold-changes of 26 genes by RNA-sequencing and Q-PCR were highly correlated (r = 0.9014, P < 0.01).
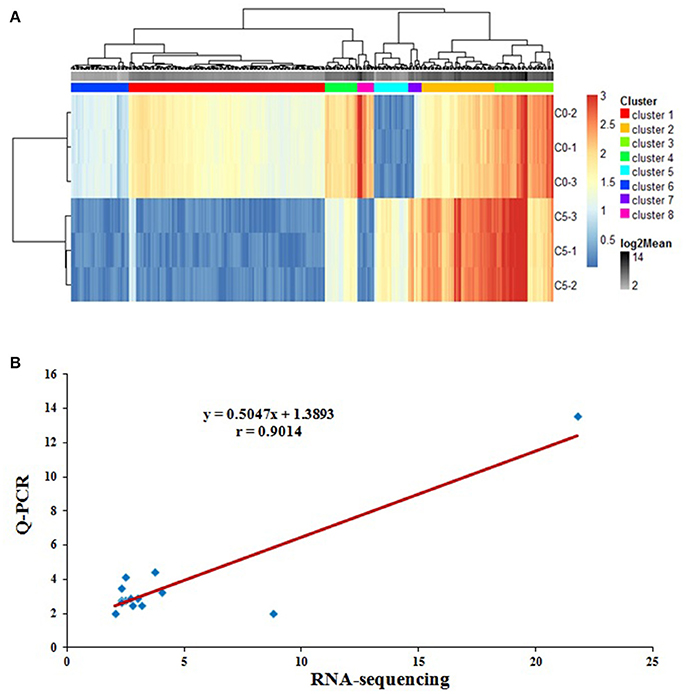
Figure 1. The quality analysis of RNA-sequencing. (A) Based on the 2977 known DEGs from RNA sequencing between REV infected or uninfected cells for 36 h, the heat-map of all six samples using Cluster 3.0 software demonstrated that three samples in same group were respectively polymerized; (B) Correlation analysis of the fold-changes of 26 DEGs by RNA-sequencing and Q-PCR to validate the results from RNA sequencing. The data was highly correlated (r = 0.9014, P < 0.01), and the results from RNA sequencing were accurate, n = 26.
GO analysis based on the 2977 DEGs was performed, the enriched GO-terms (P < 0.05) were selected (Table S3), including defense response to bacterium, inflammatory response, cell cycle, cell proliferation, etc. By GO-term analysis, 56 known DEGs related to cell cycle (Table S4), and 130 known DEGs related to immunity (Table S5) were screened. After KEGG pathway analysis, 14 metabolic pathways were screened (Table S6). As expected, some well-known pathways were enriched, which affect immune responses (Toll-like receptor-, NOD-like receptor-, and salmonella infection signaling pathways), cell proliferation or apoptosis pathways (cell cycle- and FOXO signaling), glyco- and lipid- metabolism pathways (Insulin signaling).
REV Infection Inhibited the Cell Proliferation of Lymphocyte
Cell proliferation detection by MTT showed that lymphocyte numbers were significantly reduced in REV infected cells compared with controls (P < 0.01) (Figure 2A). Compared with the controls, the results by FCM showed that the percentage of REV infected cells was significantly lower (P < 0.01) in S phase and higher (P < 0.01) in the G1 phase (Figure 2B).
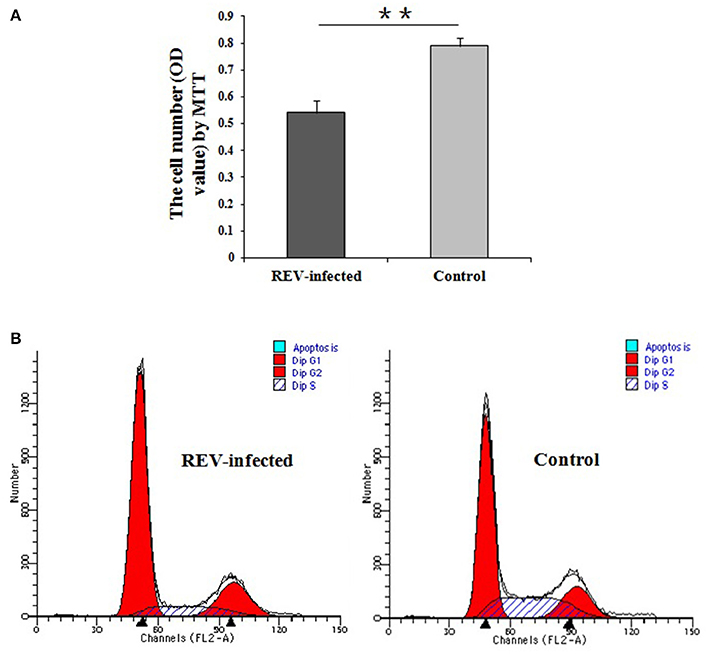
Figure 2. Detection of cell proliferation and cell cycle of lymphocytes infected with REV for 36 h. (A) Comparative analysis of cell number in REV infected or uninfected groups by MTT. The cell numbers were significantly reduced in REV infected cells compared with controls (P < 0.01). Data are means ± SEM (n = 8); (B) Cell cycle analysis by FCM. The cell percentage in S phase was significantly lower (P < 0.01) and in G1 phase was higher (P < 0.01) in REV infected cells compared to the controls. The G1/S transition in lymphocytes was inhibited by REV infection. ** P < 0.01.
In FOXO signaling pathway (including TGFβ, Insulin, IL10, and MAPK pathways) (Figure S1A), SMAD3 (the key factor in TGFβ pathway) and FOXO (including FOXO1, FOXO3, and FOXO4, the common key factor in Insulin-, IL10-, and MAPK pathways) were enriched, and would regulate the expression of CCNB2, CCNB3, CCND1, CCND2, CCND3, CCNG2, and CDKN1B, which involves in the cell cycle. In p53 signaling pathway (Figure S1B), CCNA2, CCND1, CCND2, CCND3, CCNE2, CCNH, CDKN1B, CDK6, CDK7, and GADD45A were enriched to regulate cell cycle. In addition, SIAH1 and WIPI2 were enriched in p53 negative feedback of p53 pathway. By Q-PCR, SMAD3, FOXO1, FOXO3, FOXO4, CCND1, CCND2, CCNE2, and CDKN1B were significantly down-regulated (P < 0.01) (Figure 3A). These results strongly supported that REV would inhibit the cell proliferation of lymphocyte from chicken peripheral blood through p53 and FOXO pathways.
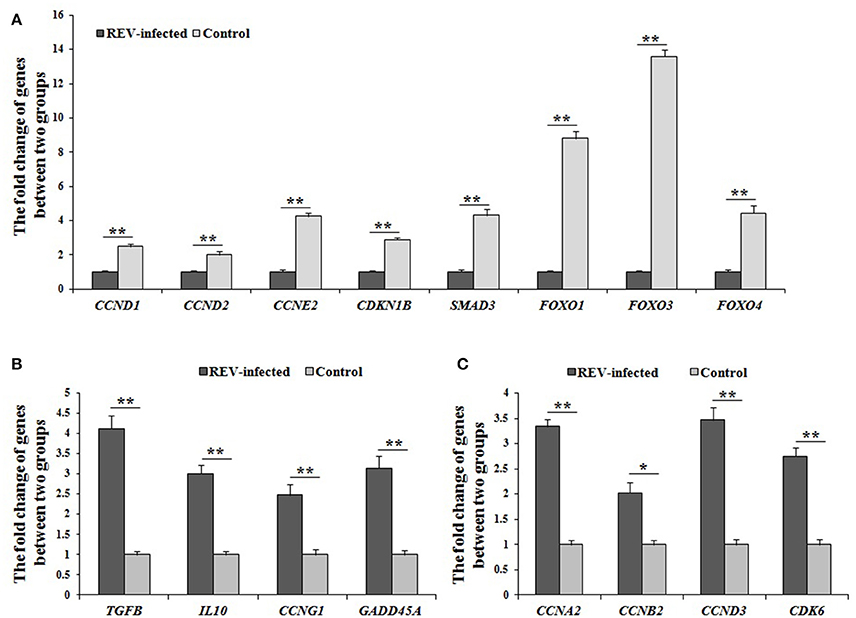
Figure 3. The differential expression of genes related to cell cycle by Q-PCR in REV infected cells. (A) The expression levels of CCND1, CCND2, CCNE2, CDKN1B, SMAD3, FOXO1, FOXO3, and FOXO4 were significantly down-regulated (P < 0.01) in infected cells compared with controls; (B) The expression levels of TGFB, IL10, CCNG1, and GADD45A were significantly up-regulated (P < 0.05 or P < 0.01) in infected cells compared with controls; (C) The expression levels of CCNA2, CCNB2, CCND3, and CDK6 were significantly up-regulated (P < 0.05 or P < 0.01) in infected cells compared with controls. Data are means ± SEM (n = 3). *P < 0.05, **P < 0.01.
However, the confusing results were also found in this study. As the starting factors of TGFβ-, IL10-, MAPK-, and p53- pathways, the expressions of TGFB, TGFBR1, TGFBR2, IL10, Grb2, CCNG1, and GADD45A were significantly up-regulated in REV infected cells compared with controls, and the expression of TGFB, IL10, CCNG1, and GADD45A by Q-PCR was same with the data by RNA-sequencing (Figure 3B). In addition, the expression levels of CCNA2, CCNB2, CCND3, and CDK6 were also significantly up-regulated (P < 0.05 or P < 0.01) in REV infected cells compared with controls (Figure 3C).
The Immune Damage and Response Synchronously Occurs by MAPK-AP1 Pathway in Lymphocytes for REV
As lymphocytes were used, it is reasonable to assume that DEGs in the current study contributed to immune responses after REV infection, and the representative immune factors also were screened. Some genes involved in ILs and their receptors were differentially expressed by RNA sequencing. The expression levels of IL8L1, IL15, IL16, IL18, IL1R, IL2R, and IL18R were significantly down-regulated in infected cells. Similarly, several genes of the TNF super-family (TNFSF8, TNFSF10, TNFSF13B, and TNFSF15) also were found from RNA sequencing data. Except for TNFSF10, their expression levels were down-regulated by REV. Moreover, TLR2A, TLR4, TLR7, TLR15 genes also were identified as DEGs, and their mRNA levels were significantly down-regulated in infected cells.
Combined with the results of the KEGG analysis, the identified DEGs related to immune defense were enriched in salmonella infection-, NOD-like receptor-, and Toll-like receptor pathways. In the Toll-like receptor pathway (Figure S2A), a decrease in TLR2A, TLR4, and TLR7 through MAPK-AP-1 pathway, down-regulated the transcript abundance of IL8 to reduce chemotaxis, T cell stimulation and antiviral effects. For NOD-like receptor pathway (Figure S2B), the transcript abundance of IL8, IL18, and IFN were regulated MAPK-AP-1 pathway and reduced the immune actions of pro-inflammatory cytokines and chemokines. In salmonella infection pathway (Figure S2C), IL8 and IL18 genes down-regulated the expression of through MAPK-AP-1 pathway. Among the factors, TNF2 gene was identified, and had significantly lower mRNA and protein levels (P < 0.01, P < 0.05, respectively) (Figure 4A). Similarly, IL8 and IL18 genes also had significantly lower mRNA and protein levels (P < 0.01) in infected cells compared with controls (Figure 4B). Moreover, TLR2A, TLR4, and TLR7 genes also had significantly lower mRNA and protein levels (P < 0.01) in infected cells compared with controls (Figure 4C). Overall the above results, REV would reduce the secretion of IL8 and/or IL18 through three (salmonella infection-, NOD-like receptor-, and Toll-like receptor pathways), which would together in MAPK-AP1 pathway.
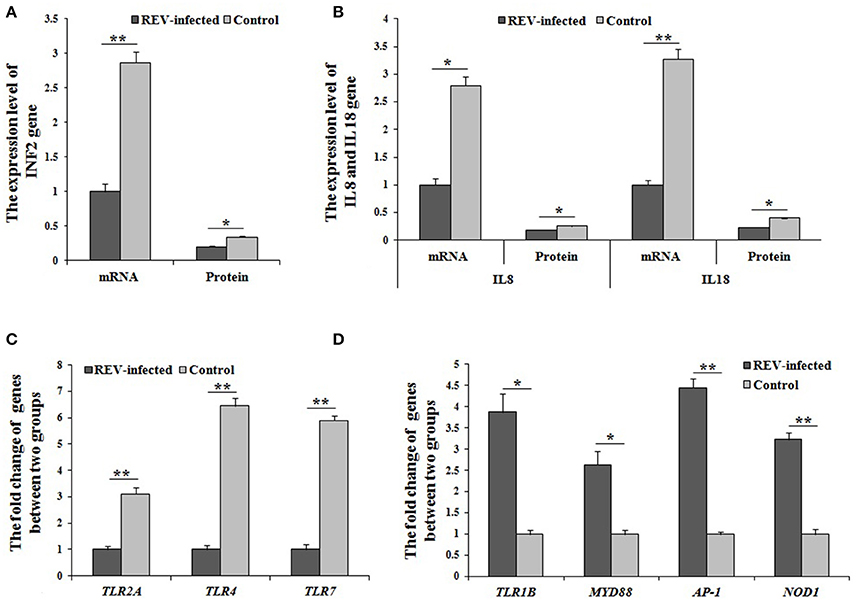
Figure 4. The mRNA or protein levels of genes related to immunity by Q-PCR or ELISA. (A,B) The mRNA and protein levels of TNF2, IL8, and IL18 were significantly lower (P < 0.05 or P < 0.01) in infected cells compared with controls; The expression levels of cluster of differentiation (CD) genes were significantly up-regulated (P < 0.05 or P < 0.01) in infected cells compared with controls; (C) The mRNA levels of TLR2A, TLR4, and TLR7 were significantly changed (P < 0.01) between the two groups; (D) The mRNA levels of TLR1B, MYD88, AP-1, and NOD1 were significantly changed (P < 0.05 or P < 0.01) between the two groups. Data are means ± SEM (n = 3).
Based on the identified salmonella infection-, NOD-like receptor-, and Toll-like receptor pathways related to immunity, some important in immune genes were screened as DEGs by RNA-sequencing. In Toll-like receptor pathway, TLR1B, MYD88, and AP-1 were significantly up-regulated (P < 0.05 or P < 0.01) in REV infected cells compared with control cells by Q-PCR (Figure 4D). For the NOD-like receptor pathway, NOD1 and AP-1 were significantly up-regulated (P < 0.01) in REV infected cells compared with control cells by Q-PCR (Figure 4D). In the salmonella infection signaling pathway, MYD88 and AP-1 were significantly up-regulated (P < 0.05 or P < 0.01) in REV infected cells compared with control cells by Q-PCR (Figure 4D).
Discussion
Chicken REV infected lymphocytes or reticuloendothelial cells, and caused the atrophy of immune organs (thymus and bursa of Fabricius) leading to immuno-suppression (Witter et al., 1981; Nazerian et al., 1982). Many studies of REV regarding epidemiology, vaccines, and the mechanism of pathogenesis and immunity have been reported (García et al., 2003; Fadly and Garcia, 2006; Cheng et al., 2007; Li and Cui, 2007), however is still lacking the systematically undertaken research on molecular regulation mechanism and immune response for REV in chicken lymphocytes from peripheral blood. The objective of this study was to investigate the effect of REV on chicken blood lymphocytes by RNA sequencing. Potential candidate related DEGs were defined from the data of RNA sequencing using different RNA samples (n = 3 dishes), with a required fold change in expression of ≥2.0. The cluster analysis of six RNA samples was performed, three samples in the same group were respectively polymerized, so the comparability among groups was believed. To deeply confirm the obtained data by RNA sequencing, 156 tests of 26 related DEGs were performed between two groups by Q-PCR, and the relationship on fold-changes of gene expression by two methods were significantly correlated (r = 0.9014), indicating that the accuracy of data from RNA sequencing was verified in this study.
It was reported that REV infection induced a splenic suppressor cell population which cytostatically inhibited the proliferation of cytotoxic cells capable of lysing REV-T tumor cells (Bose, 1984). A reduction of lymphocytes with REV infected in vitro was found in this study. Moreover, the results of FCM revealed that the G1/S phase transition was interdicted to inhibit cell proliferation of lymphocytes. Genetically, the FOXO and p53 pathways, which regulate cell proliferation (Zaballos and Santisteban, 2013; Liu et al., 2016; Yi et al., 2016; Tang et al., 2017), were enriched by KEGG analysis. Further, SMAD3 and FOXO (including FOXO1, FOXO3, and FOXO4) were involved in FOXO pathway (including TGFβ, Insulin, IL10, and MAPK pathways), and would regulate the expression of CCNB2, CCNB3, CCND1, CCND2, CCND3, CCNG2, and CDKN1B in this study. Similarly, CCNA2, CCND1, CCND2, CCND3, CCNE2, CCNH, CDKN1B, CDK6, CDK7, and GADD45A were involved in p53 pathway. In addition, SIAH1 and WIPI2 were enriched in p53 negative feedback. In overall consideration of above results and known information (Lee et al., 2012; Hill et al., 2014; Zhao et al., 2015; Vezzali et al., 2016), genes (SMAD3, FOXO1, FOXO3, FOXO4, CCND1, CCND2, CCNE2, CDKN1B) were validated by Q-PCR, and their expressions were significantly down-regulated (P < 0.05 or P < 0.01). So it was strongly supported that REV would inhibit the cell proliferation of lymphocyte, and the key genes (SMAD3, FOXO1, FOXO3, FOXO4, CCND1, CCND2, CCNE2, CDKN1B) and the pathways (p53 and FOXO) were considered with the important role in inhibiting cell proliferation of lymphocyte from chicken peripheral blood for REV infection in this study. However, the expression levels of the starting factors (TGFB, TGFBR1, TGFBR2, IL10, Grb2, CCNG1, and GADD45A) in FOXO and p53 pathways, and genes related to cell cycle (CCNA2, CCNB2, CCND3, CDK6) were up-regulated (P < 0.05 or P < 0.01) in REV infected cells compared with controls by RNA-sequencing and Q-PCR. So it was speculated that the rescue of lymphocyte would activate through FOXO and p53 pathways.
This reduction of lymphocytes caused serious immune dysfunction in secreting immune cytokines that influence the differentiation of T cells into TH and CTL cells (Li and Liu, 1999; Kim et al., 2004). After infected by REV, DEGs contributed to immune responses also were screened, including ILs, ILRs, TNFs, IFNs, and TLRs, and the expression levels of these genes were significantly down-regulated in infected cells. It was indicated that the immune defense was destroyed for REV infection, and this view was same as previous reported (Witter et al., 1981; Nazerian et al., 1982). Combined with the results of the KEGG analysis, the identified DEGs related to immune responses were enriched in Toll-like receptor-, NOD-like receptor- and salmonella infection signaling pathways, which involved in the process of immune responses (Wong et al., 2009; Coutermarsh-Ott et al., 2016; Huang et al., 2016; Yin et al., 2017). For three enriched pathways (salmonella infection, NOD-like receptor and Toll-like receptor) would directly connect MAPK pathway and AP-1 in this study. Many studies have reported on the MAPK signaling pathway and AP-1 in the regulation of immune factor secretion (Shi et al., 2014; Jiang et al., 2015; Lanna et al., 2017). Moreover, TLR2, TLR4, TLR7, and IL8 were enriched in Toll-like receptor pathway, IL8 and IL18 were enriched in the NOD-like receptor pathway, and TLR4, IL8, and IL18 were enriched in the salmonella infection pathway. By Q-PCR, TLR2, TLR4, TLR7, IL8, and IL18 were significantly down-regulated in REV infected cells compared with controls. Further, the protein levels of TNF2, IL8, and IL18 were also detected, and were significantly lower for REV infection. Considered to the function of TLR2, TLR4, TLR7, TNF2, IL8, and IL18 (Briend et al., 2017; Sanguinete et al., 2017; Zhang et al., 2017), it was revealed that the immune defense would been inhibited for REV infection through salmonella infection, NOD-like receptor, Toll-like receptor and MAPK-AP1 pathways. Together all, REV infection would decrease the secretion of immune factors for the reduction of lymphocytes and inhibition of immune defense.
Based on the identified salmonella infection-, NOD-like receptor-, Toll-like receptor, and MAPK-AP1 pathways, TLR1B, MYD88, NOD1, AP-1, which had the important role in regulation the immune action (Buchholz and Stephens, 2008; Ramasamy et al., 2012; Limoge et al., 2017; Wang et al., 2017), were significantly up-regulated in REV infected cells compared with control cells by RNA-sequencing and Q-PCR. In addition, CD40 and CD80 also were significantly up-regulated in REV infected cells compared with control cells by two methods.
Among them, NOD1 and AP-1 were involved in NOD-like receptor pathway. TLR1B, MYD88, CD40, CD80, and AP-1 were involved in Toll-like receptor pathway. MYD88 and AP-1 were involved in salmonella infection pathway. NOD1, TLR1B, MYD88, AP-1. These results revealed that the immune response for REV would been activated by up-regulating the expression levels of genes (TLR1B, MYD88, NOD1, and AP-1) through salmonella infection-, NOD-like receptor-, Toll-like receptor, and MAPK-AP1 pathways.
In summary, we report the effect of REV infection on cell proliferation and immune response in chicken peripheral blood lymphocytes in vitro. REV infection inhibited the G1/S transition to reduce lymphocyte through p53 and FOXO signaling pathways, and destroyed through salmonella infection-, NOD-like receptor-, Toll-like receptor, and MAPK-AP1 pathways in lymphocytes. On the other hand, the immune response would activate by up-regulating the expression of TLR1B, MYD88, NOD1, and AP-1. In addition, the immune inhibition of REV was rescued in lymphocytes by activating cell proliferation through p53 and FOXO pathways. Our findings will help to establish the groundwork (Figure 5) and provide new clues for deciphering the molecular mechanisms underlying the pathogenesis of REV in chicken blood lymphocytes. The field of our view was only focus on the transcriptional level of genes, so the additional studies of the post-translational effects were required to complement these mRNA expression analyses.
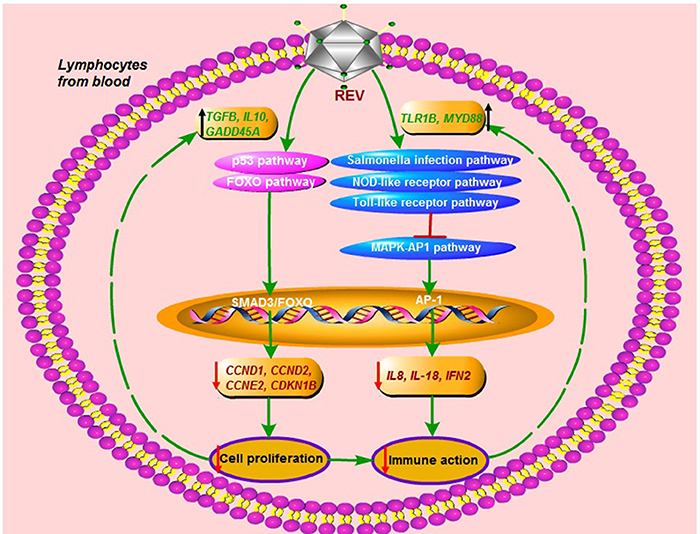
Figure 5. The immune destroy and regulatory mechanism in lymphocytes from chicken peripheral blood for REV infection.
Author Contributions
GChang, GChen, and YB conceived the project. GChang and YB designed all the experiments. YB, LX, LQ, SW, and XL performed the experiments. YB, YaniZ, YC, YangZ, and QX conducted the bioinformatics analyses. YB contributed to the manuscript preparation.
Funding
The research was supported by the National & Science Technology Pillar Program of China (2015BAD03B03), the Project funded of Priority Academic Program Development of Jiangsu Higher Education Institutions, the National natural science foundation of China (Grant No. 31372297), and the Science and Technology Innovation Foundation of Yangzhou University (2016CXJ067).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Kun Qian (Key Laboratory of Jiangsu Preventive Veterinary Medicine, Yangzhou University, Yangzhou, China) and Zhizhong Cui (Shandong Agriculture University, Taian, China) for providing the REV strain HA1101.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00004/full#supplementary-material
Table S1. The specific primers for Q-PCR in this study.
Table S2. The differentially expressed gene (DEG) profiles (fold change ≥ 2.0).
Table S3. The enriched GO-terms (based on the 2977 DEGs).
Table S4. The DEGs related to cell cycle and cell proliferation.
Table S5. The DEGs related to immune.
Table S6. The enriched pathways, based on the 2977 DEGs.
Figure S1. The enriched pathways involved in the cell cycle in this study.
Figure S2. The enriched pathways involved in the immunity in this study.
Abbreviations
CCNs, Cyclins; CD, Cluster of differentiation; CDK, Cyclin-dependent kinases; CT, Threshold cycle; DEGs, Differentially expressed genes; FCM, Flow cytometry; FOXO, Forkhead box O; IFN, Interferon; GO, Gene ontology; IL, Interleukin; KEGG, Kyoto encyc lopedia of genes and genomes; MAPK, Mitogen-activated protein kinase; MYD88, Myeloid differentiation primary response gene 88; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; NOD, Nucleotide-binding oligomer-zation domain; Q-PCR, Quantitative polymerase chain reaction; REV, Reticuloendotheliosis virus; TGF, Transforming growth factor; TLR, Toll-like receptor; TNF, Tumor necrosis factor.
References
Bose, H. R. Jr. (1984). Reticuloendotheliosis virus and disturbance in immune regulation. Microbiol. Sci. 1, 107–112.
Briend, E., Ferguson, G. J., Mori, M., Damera, G., Stephenson, K., and Karp, N. A. (2017). IL-18 associated with lung lymphoid aggregates drives IFNγ production in severe COPD. Respir. Res. 18:159. doi: 10.1186/s12931-017-0641-7
Buchholz, K. R., and Stephens, R. S. (2008). The cytosolic pattern recognition receptor NOD1 induces inflammatory interleukin-8 during Chlamydia trachomatis infection. Infect. Immun. 76, 3150–3155. doi: 10.1128/IAI.00104-08
Cheng, Z., Shi, Y., Zhang, L., Zhu, G., Diao, X., and Cui, Z. (2007). Occurrence of reticuloendo- theliosis in Chinese partridge. J. Vet. Med. Sci. 69, 1295–1298. doi: 10.1292/jvms.69.1295
Coutermarsh-Ott, S., Eden, K., and Allen, I. C. (2016). Beyond the inflammasome: regulatory NOD-like receptor modulation of the host immune response following virus exposure. J. Gen. Virol. 97, 825–838. doi: 10.1099/jgv.0.000401
Fadly, A., and Garcia, M. C. (2006). Detection of reticuloendotheliosis virus in live virus vaccines of poultry. Dev. Biol. 126, 301–305.
García, M., Narang, N., Reed, W. M., and Fadly, A. M. (2003). Molecular characterization of reticuloendotheliosis virus insertions in the genome of field and vaccine strains of fowl poxvirus. Avian Dis. 47, 343–354. doi: 10.1637/0005-2086(2003)047[0343:MCORVI]2.0.CO;2
Hill, R., Kalathur, R. K., Callejas, S., Colaço, L., Brandão, R., Serelde, B., et al. (2014). A novel phosphatidylinositol 3-kinase (PI3K) inhibitor directs a potent FOXO-dependent, p53- independent cell cycle arrest phenotype characterized by the differential induction of a subset of FOXO-regulated genes. Breast Cancer Res. 16, 482. doi: 10.1186/s13058-014-0482-y
Hrdlicková, R., Nehyba, J., and Humphries, E. H. (1994). V-rel induce expression of three avian immuno-regulatory surface receptors more efficiently than c-rel. J. Virol. 68, 308–319.
Huang, H., Liu, A., Wu, H., Ansari, A. R., Wang, J., Huang, X., et al. (2016). Transcriptome analysis indicated that Salmonella lipopoly-saccharide-induced thymocyte death and thymic atrophy were related to TLR4-FOS/JUN pathway in chicks. BMC Genomics. 17:322. doi: 10.1186/s12864-016-2674-6
Jiang, Y., Yu, L., and Wang, M. H. (2015). N-trans-feruloyltyramine inhibits LPS-induced NO and PGE2 production in RAW 264.7 macrophages: involvement of AP-1 and MAP kinase signalling pathways. Chem. Biol. Interact. 235, 56–62. doi: 10.1016/j.cbi.2015.03.029
Kim, Y., Brown, T. P., and Pantin-Jackwood, M. J. (2004). The effects of cyclophosphamide treatment on the pathogenesis of subgroup J avian leukosis virus (ALV-J) infection in broiler chickens with Marek's disease virus exposure. J. Vet. Sci. 5, 49–58.
Lanna, A., Gomes, D. C., Muller-Durovic, B., McDonnell, T., Escors, D., Gilroy, D. W., et al. (2017). A sestrin-dependent Erk-Jnk-p38 MAPK activation complex inhibits immunity during aging. Nat. Immunol. 18, 354–363. doi: 10.1038/ni.3665
Lee, J. Y., Jeong, W., Kim, J. H., Kim, J., Bazer, F. W., Han, J. Y., et al. (2012). Distinct expression pattern and post-transcriptional regulation of cell cycle genes in the glandular epithelia of avian ovarian carcinomas. PLoS ONE 7:e51592. doi: 10.1371/journal.pone.0051592
Li, G. X., and Liu, Z. G. (1999). Study on the kinetic alterations of tumor necrosis factor of SPF chickens infected with reticuloendotheliosis virus. Prog. Vet. Med. 20, 50–54.
Li, Y., Cui, S., Cui, Z., Chang, S., and Zhao, P. (2016). Genome analysis and pathogenicity of reticuloendotheliosis virus isolated from a contaminated vaccine seed against infectious bursal disease virus: first report in China. J. Gen. Virol. 97, 2809–2815. doi: 10.1099/jgv.0.000588
Li, Y. P., and Cui, Z. Z. (2007). Pathogenicity and genomic sequence comparison of a chicken infectious anemia virus field isolate. Wei Sheng Wu Xue Bao Chinese 47, 894–898.
Liao, Y., Smyth, G. K., and Shi, W. (2013). The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. doi: 10.1093/nar/gkt214
Limoge, M., Safina, A., Beattie, A., Kapus, L., Truskinovsky, A. M., and Bakin, A. V. (2017). Tumor-fibroblast interactions stimulate tumor vascularization by enhancing cytokine-driven production of MMP9 by tumor cells. Oncotarget 8, 35592–35608. doi: 10.18632/oncotarget.16022
Liu, Y. H., Liu, G. H., Mei, J. J., and Wang, J. (2016). The preventive effects of hyperoside on lung cancer in vitro by inducing apoptosis and inhibitingproliferation through Caspase-3 and p53 signaling pathway. Biomed. Pharmacother. 83, 381–391. doi: 10.1016/j.biopha.2016.06.035
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Nazerian, K., Witter, R. L., Crittenden, L. B., Noori-Dalloii, M. R., and Kung, H. J. (1982). An IgM-producing B lymphoblastoid cell line established from lymphomas induced by a non- defective reticuloendotheliosis virus. J. Gen. Virol. 58, 351–360. doi: 10.1099/0022-1317-58-2-351
Ramasamy, K. T., Reddy, M. R., Verma, P. C., and Murugesan, S. (2012). Expression analysis of turkey (Meleagris gallopavo) toll-like receptors and molecular characterization of avian specific TLR15. Mol. Biol. Rep. 39, 8539–8549. doi: 10.1007/s11033-012-1709-6
Sanguinete, M. M. M., Oliveira, P. H., Martins-Filho, A., Micheli, D. C., Tavares-Murta, B. M., Murta, E. F. C., et al. (2017). Serum IL-6 and IL-8 correlate with prognostic factors in ovarian cancer. Immunol. Invest. 46, 677–688. doi: 10.1080/08820139.2017.1360342
Shi, Q., Cao, J., Fang, L., Zhao, H., Liu, Z., Ran, J., et al. (2014). Geniposide suppresses LPS-induced nitric oxide, PGE2 and inflammatory cytokine by downregulating NF-κB, MAPK and AP-1 signaling pathways in macrophages. Int. Immunol. Pharmacol. 20, 298–306. doi: 10.1016/j.intimp.2014.04.004
Tang, M., Zhao, Y., Liu, N., Chen, E., Quan, Z., Wu, X., et al. (2017). Overexpression of HepaCAM inhibits bladder cancer cell proliferation and viability through the AKT/ FOXO pathway. J. Cancer Res. Clin. Oncol. 143, 793–805. doi: 10.1007/s00432-016-2333-y
Vezzali, R., Weise, S. C., Hellbach, N., Machado, V., Heidrich, S., and Vogel, T. (2016). The FOXG1/FOXO/SMAD network balances proliferation and differentiation of cortical progenitors and activates Kcnh3 expression in mature neurons. Oncotarget 7, 37436–37455. doi: 10.18632/oncotarget.9545
Wang, Y., Liu, Q., Chen, D., Guan, J., Ma, L., Zhong, G., et al. (2017). Chlamydial lipoproteins stimulate toll-like receptors 1/2 mediated inflammatory responses through MyD88-dependent pathway. Front. Microbiol. 8:78. doi: 10.3389/fmicb.2017.00078
Witter, R. L., Lee, L. F., Bacon, L. D., and Smith, E. J. (1979). Depression of vaccinal immunity to Marek's disease by infection with reticuloendotheliosis virus. Infect. Immun. 26, 90–98.
Witter, R. L., Smith, E. J., and Crittenden, L. B. (1981). Tolerance, viral shedding, and neoplasia in chicken infected with non-defective reticuloendotheliosis viruses. Avian Dis. 25, 374–394. doi: 10.2307/1589930
Wong, J. P., Christopher, M. E., Viswanathan, S., Karpoff, N., Dai, X., Das, D., et al. (2009). Activation of toll-like receptor signaling pathway for protection against influenza virus infection. Vaccine 27, 3481–3483. doi: 10.1016/j.vaccine.2009.01.048
Xue, M., Zhao, Y., Hu, S., Shi, X., Cui, H., and Wang, Y. (2017). Analysis of the spleen proteome of chickens infected with reticuloendotheliosis virus. Arch. Virol. 162, 1187–1199. doi: 10.1007/s00705-016-3180-5
Yi, W., Xiao, E., Ding, R., Luo, P., and Yang, Y. (2016). High expression of fibronectin is associated with poor prognosis, cell proliferation and malignancy via the NF-κB/p53-apoptosis signaling pathway in colorectal cancer. Oncol. Rep. 36, 3145–3153. doi: 10.3892/or.2016.5177
Yin, Y., Zhou, Z., Liu, W., Chang, Q., Sun, G., and Dai, Y. (2017). Vascular endothelial cells senescence is associated with NOD-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome activation via reactive oxygen species (ROS)/thioredoxin-interacting protein (TXNIP) pathway. Int. J. Biochem. Cell Biol. 84, 22–34. doi: 10.1016/j.biocel.2017.01.001
Zaballos, M. A., and Santisteban, P. (2013). FOXO1 controls thyroid cell proliferation in response to TSH and IGF-I and is involved in thyroid tumorigenesis. Mol. Endocrinol. 27, 50–62. doi: 10.1210/me.2012-1032
Zhang, A., Xu, J., Lai, H., Huang, W., Fang, N., and Chen, R. (2017). Age-related changes and distribution of T cell markers (CD3 and CD4) and toll-like receptors(TLR2, TLR3, TLR4 and TLR7) in the duck lymphoid organs. Immunobiology 222, 857–864. doi: 10.1016/j.imbio.2017.01.004
Keywords: REV, chicken lymphocyte, DGEs and pathway, cell proliferation, immune response
Citation: Bi Y, Xu L, Qiu L, Wang S, Liu X, Zhang Y, Chen Y, Zhang Y, Xu Q, Chang G and Chen G (2018) Reticuloendotheliosis Virus Inhibits the Immune Response Acting on Lymphocytes from Peripheral Blood of Chicken. Front. Physiol. 9:4. doi: 10.3389/fphys.2018.00004
Received: 18 October 2017; Accepted: 03 January 2018;
Published: 23 January 2018.
Edited by:
Sami Dridi, University of Arkansas, United StatesReviewed by:
Paweł Tomasz Maćkowiak, Poznan University of Life Sciences, PolandLisa Bielke, The Ohio State University, United States
Copyright © 2018 Bi, Xu, Qiu, Wang, Liu, Zhang, Chen, Zhang, Xu, Chang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guobin Chang, Z2hjaGVuQHl6dS5lZHUuY24=
Guohong Chen, cGFzc2lvbmNnYkAxNjMuY29t
 Yulin Bi1
Yulin Bi1 Guohong Chen
Guohong Chen