- 1Key Laboratory of Tea Quality and Safety Control, Ministry of Agriculture, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, China
- 4College of Plant Protection, Anhui Agricultural University, Hefei, China
- 5College of Life Sciences, Huaibei Normal University, Huaibei, China
Carboxylesterases (CXEs) belong to a family of metabolic enzymes. Some CXEs act as odorant-degrading enzymes (ODEs), which are reportedly highly expressed in insect olfactory organs and participate in the rapid deactivation of ester pheromone components and plant volatiles. The tea geometrid Ectropis obliqua Prout produces sex pheromones consisting of non-ester functional compounds but relies heavily on acetic ester plant volatiles to search for host plants and locate oviposition sites. However, studies characterizing putative candidate ODEs in this important tea plant pest are still relatively scarce. In the present study, we identified 35 candidate EoblCXE genes from E. obliqua chemosensory organs based on previously obtained transcriptomic data. The deduced amino acid sequences possessed the typical characteristics of the insect CXE family, including oxyanion hole residues, the Ser-Glu-His catalytic triad, and the Ser active included in the conserved pentapeptide characteristic of esterases, Gly-X-Ser-X-Gly. Phylogenetic analyses revealed that the EoblCXEs were diverse, belonging to several different insect esterase clades. Tissue- and sex-related expression patterns were studied via reverse-transcription and quantitative real-time polymerase chain reaction analyses (RT- and qRT-PCR). The results showed that 35 EoblCXE genes presented a diversified expression profile; among these, 12 EoblCXEs appeared to be antenna-biased, two EoblCXEs were non-chemosensory organ-biased, 12 EoblCXEs were ubiquitous, and nine EoblCXEs showed heterogeneous expression levels among different tissues. Intriguingly, two EoblCXE genes, EoblCXE7 and EoblCXE13, were not only strongly localized to antennal sensilla tuned to odorants, such as the sensilla trichodea (Str I and II) and sensilla basiconica (Sba), but were also expressed in the putative gustatory sensilla styloconica (Sst), indicating that these two CXEs might play multiple physiological roles in the E. obliqua chemosensory processing system. This study provides the first elucidation of CXEs in the chemosensory system of a geometrid moth species and will enable a more comprehensive understanding of the functions of insect CXEs across lepidopteran species.
Introduction
The sophisticated olfactory system, particularly the peripheral chemical signal coding, is essential for insects to find mates, locate food, and avoid predators (Dweck et al., 2013; Tauxe et al., 2013; Strauch et al., 2014; Li and Liberles, 2015). Biologically important odorants are generally perceived sensitively and specifically in the multiporous sensilla hairs on insect antennae (Meijerink and van Loon, 1999; Pophof et al., 2005; Park et al., 2013; Sun L. et al., 2014). It is well established that at least three major classes of molecules are involved in this process: odorant-binding proteins (OBPs), odorant receptors (ORs), and odorant-degrading enzymes (ODEs). In brief, airborne odorants enter the hydrosoluble sensillum lymph through the sensilla pores, bind to OBPs, activate ORs and trigger signal transduction cascades and olfactory coding; odorants are then rapidly removed from the vicinity of the ORs by ODEs to restore the sensitivity of the sensory neuron (Rützler and Zwiebel, 2005; Vogt, 2005; Pelosi et al., 2006; Leal, 2013).
The highly sensitive odorant signal transduction pathway of insects represents an excellent model that researchers can use to develop new environmentally friendly pest-management strategies through targeting key molecules and screening biologically active compounds for behavioral control. Previous functional reports regarding OBPs and ORs indeed led to the rapid discovery of high-efficiency pest repellents and attractants. For example, compounds that are behaviorally active in the mirid bug Adelphocoris lineolatus were successfully screened via studies on the interaction between antenna-enriched AlinOBP10 and its putative ligands (Sun et al., 2013). In the aphid alarm pheromone EBF perception pathway, ApisOBP3 and ApisOBP7 as well as ApisOR5 were proven to be potentially crucial targets for aphid repellent screening (Sun et al., 2012; Zhang R. et al., 2017). However, compared with OBPs and ORs, similar reports on ODEs appear to be rare. Given that the rapid degradation of redundant odorants can rescue the sensitivity of odorant sensory neurons, putative genes encoding insect ODEs that are highly expressed in the chemosensory system should be identified, and their potential roles in odorant degradation deserve thorough exploration.
Carboxylesterases (CXEs) belong to the α/β-fold hydrolase superfamily and are widely distributed in insects and other organisms. CXEs commonly include a conserved catalytic triad (Ser-His-Glu) and specifically catalyze the hydrolysis of ester bonds in various substrates (Oakeshott et al., 1999, 2005). Because most insect species, including hemipteran bugs and lepidopteran moths, utilize aliphatic esters as intraspecific sex pheromones and ovipositional stimulants (Ando et al., 2004; Millar, 2005; Pan et al., 2015), many antennae-biased CXEs have been identified, and their activities associated with sex pheromone and odorant degradation have been assessed (Vogt, 2005; Jacquin-Joly and Maïbèche-Coisne, 2009). The first CXE subfamily of ODEs, known as Apol-SE (Vogt and Riddiford, 1981), or ApolPDE (Ishida and Leal, 2005), was isolated from the giant silk moth, Antheraea polyphemus. Subsequently, genes encoding putative antennal esterases were cloned and described across insect species using a polymerase chain reaction (PCR) strategy. These genes included two other CXEs, ApolODE and Apol-IE, in A. polyphemus (Ishida and Leal, 2002); Mbra-EST from the cabbage armyworm, Mamestra brassicae (Maïbèche-Coisne et al., 2004); D-AP1, a honeybee homolog of CXE in Apis mellifera L. (Kamikouchi et al., 2004); Slit-EST and Snon-EST from the Egyptian armyworm, Spodoptera littoralis, and the Mediterranean corn borer, Sesamia nonagrioides (Merlin et al., 2007); and PjapPDE, cloned from the Japanese beetle, Popillia japonica (Ishida and Leal, 2008). Furthermore, through expressed sequence tag (EST) and RNA-Seq analyses, diverse CXE genes have been identified from various insect species, such as Epiphyas postvittana (Jordan et al., 2008), Spodoptera littoralis (Durand et al., 2010b), Agrotis ipsilon (Gu et al., 2013), Sesamia inferens (Zhang Y. N. et al., 2014), Chilo suppressalis (Liu et al., 2015; Xia et al., 2015), and Spodoptera litura (Zhang et al., 2016).
Convincing evidence obtained through biochemical characterization and enzyme kinetic activity analyses showed that Apol-SE/ApolPDE displays expression specific to male antennal sensilla and exhibits rapid catalytic activity toward the acetate sex pheromone component E6Z11-16:OAc (Vogt et al., 1985; Prestwich et al., 1986; Klein, 1987; Ishida and Leal, 2005). In vitro functional analyses and potential hydrolyzed substrates of CXEs have also been documented in other insect species, particularly lepidopteran moths, whose main sex pheromone components are acetate esters (Ishida and Leal, 2008; Durand et al., 2010a, 2011; He et al., 2014a,b,c, 2015). Additionally, an extracellular carboxylesterase esterase-6 (EST-6) of Drosophila melanogaster has been demonstrated to be a potential ODE for both the sex pheromone ester cis-vaccenyl acetate (CVA) and other bioactive volatile esters, such as pentyl acetate (Chertemps et al., 2012, 2015). All of the available data support potential roles of CXEs in degrading either sex pheromones or host plant odorants containing ester functional groups.
The tea geometrid Ectropis obliqua Prout is a common pest of the tea plant, Camellia sinensis (L.), and causes serious economic damage to tea production (Ye et al., 2014; Zhang G. H. et al., 2014). Multiple electrophysiological and behavioral studies show that E. obliqua relies heavily on chemical cues to locate host plants, oviposition sites and conspecific mates. Furthermore, larval infection of tea plants strongly induces the release of several kinds of host volatiles with ester functional groups, and these ester compounds can in turn regulate the ovipositional preference of E. obliqua adult females (Sun X. L. et al., 2014). Hence, studies on the molecular mechanism of ester odorant degradation are particularly important for the identification of potential target genes mediating oviposition signal inactivation and the development of ODE-based strategies in geometrid pest management.
In this study, we identified putative genes encoding CXEs by analyzing the BLASTX annotations of transcriptomic data. The phylogenetic relationships between the candidate CXEs and homologs in other Lepidoptera species were further analyzed. Finally, the tissue expression patterns of the identified CXEs were investigated in olfactory organs (particularly in the different antennal sensilla) and non-olfactory organs, and potential functional differentiation was discussed.
Materials and Methods
Insect Rearing and Tissue Collection
The tea geometrid E. obliqua was collected from the Yuhang tea plantation in Zhejiang Province, China. Phylogenetic identity analysis and laboratory colony construction were performed according to Zhang G. H. et al. (2014). The pupae were sexed, and male and female individuals were raised separately until eclosion. Adult moths of different sexes were maintained in different cages and fed a 10% honey solution on water-soaked cotton.
For the tissue-specific expression profile analysis of E. obliqua adults, approximately 500 antennae, three abdomens, and 300 legs of both male and female adults 1–3 days after emergence were dissected and collected. Two biological replicates were prepared for RT-PCR, and two additional biological replicates were prepared for qRT-PCR. All of the specimens were immediately stored at −80°C until use.
RNA Extraction and cDNA Synthesis
Total RNA from each specimen was extracted with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The integrity of the total RNA was examined through 1.2% agarose electrophoresis, and the purity was assessed using a NanoDrop™ instrument (Wilmington, DE, USA). First-strand cDNA was synthesized from 2 μg of RNA using a FastQuant RT kit with gDNA Eraser (TianGen, Beijing, China) according to the manufacturer's instructions.
Identification of Candidate EoblCXEs and Sequence Analysis
Candidate EoblCXEs were identified through keyword screening of the BLASTX annotations of transcriptomic data from adult E. obliqua chemosensory organs, including the antennae, legs, wings and proboscises. The TBLASTN program was also applied using the previously identified S. littoralis CXEs (Durand et al., 2010b) as the query. The open reading frames (ORFs) of genes were predicted using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The theoretical isoelectric points and molecular weights of the deduced proteins were calculated using the ExPASy tool (http://web.expasy.org/compute_pi/). Homology searches were performed with BLAST (http://blast.ncbi.nlm.nih.gov/). Catalytic residues were predicted by searching the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/structure/cdd/cdd.shtml). Putative N-terminal signal peptides were predicted using the SignalP 4.0 program (http://www.cbs.dtu.dk/services/SignalP/) (Brunak et al., 2010).
Phylogenetic Analysis
The amino acid sequences of EoblCXEs and CXEs from other species were aligned using ClustalX 2.0 (Larkin et al., 2007). A neighbor-joining tree was constructed using the program MEGA 6.0 with the Jones–Taylor–Thornton (JTT) amino acid substitution model (Tamura et al., 2013). Node support was assessed using a bootstrapping procedure with 1,000 replicates, uniform rates, and pairwise deletion of data gaps. The protein names and accession numbers corresponding to the genes used for construction of the phylogenetic tree are listed in Table S1.
Reverse-Transcription PCR
The tissue-specific expression of EoblCXEs was determined via reverse-transcription PCR (RT-PCR) using ExTaq DNA polymerase (TaKaRa, Dalian, China). The E. obliqua glyceraldehyde-3-phosphate dehydrogenase (EoblGAPDH, GenBank accession no. KT991373) reference gene was employed as an internal control to normalize target gene expression in order to correct for sample-to-sample variation. The specific primers used for amplification are listed in Table S2.
The experiment was performed according to a previous report (Sun et al., 2017b): each reaction of 50 μL contained 1 μL of 200 ng/μL (200 ng) single-stranded cDNA, 5 μL of 10 × ExTaq buffer, 4 μL of deoxyribonucleoside triphosphates (dNTPs), 2 μL of each primer and 0.25 U of ExTaq DNA polymerase. The PCR conditions were as follows: initial denaturation at 94°C for 4 min followed by 40 cycles of 94°C for 30 s, 55–65°C for 30 s, and 72°C for 30 s and a final elongation step at 72°C for 10 min. After PCR, the products were analyzed in 1.5% agarose gels. To check reproducibility, each RT-PCR run for each sample was performed with two biological replicates and three technical replicates. The relative expression levels of the EoblCXE genes in different tissues were calculated using the ratio of RT-PCR band intensity between the target gene and the internal reference gene, EoblGAPDH, using Bio-Rad Quantity One 4.6.2 software (Zhang et al., 2013).
Quantitative Real-Time PCR
Based on the RT-PCR results, 18 EoblCXEs were randomly selected to conduct quantitative real-time PCR (qRT-PCR). The experiment was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA), and each reaction was conducted in a 20-μL reaction mixture containing 10 μL of 2 × SYBR Green PCR Master Mix (TaKaRa, Dalian, Liaoning, China), 0.8 μL of each primer (10 μM), 0.4 μL of ROX Reference Dye II, 2 μL of sample cDNA (200 ng), and 6.0 μL of sterilized H2O. The qPCR cycling parameters were as follows: 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 31 s. Subsequently, the fluorescence was measured using a 55–95°C melting curve to detect a single gene-specific peak and to confirm the absence of primer dimer peaks; single, discrete peaks were detected for all primers tested.
The primers employed for qPCR (Table S3) were designed using the Beacon Designer 7.90 program (PREMIER Biosoft International). The reference gene EoblGAPDH was found to be expressed at a similar level in different tissues and was used as an internal control to normalize target gene expression in order to correct for sample-to-sample variation (Sun et al., 2017a). The amplification efficiency for the target and reference genes was assessed using gradient dilution templates to examine the variation of ΔCT (CT, Target gene − CT, reference gene) with template dilution (Livak and Schmittgen, 2001). The absolute values of the slopes of all lines obtained from template dilution plots (log cDNA dilution vs. ΔCT) were close to zero, indicating that the efficiency for EoblCXEs was similar to that for EoblGAPDH. Non-template reactions (replacing cDNA with sterilized H2O) were performed as negative controls. To check the reproducibility of the qPCR assays, each reaction for each sample was performed with three technical replicates and two biological replicates.
Comparative analyses of target gene expression between different tissues were performed using one-way nested analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) test. The relative mRNA expression levels between males and females of 10 antennae-biased EoblCXEs were compared with Student's t-test. All analyses were performed using SPSS Statistics 18.0 software (SPSS Inc., Chicago, IL, USA).
Fluorescence in Situ Hybridization
Based on the observed tissue expression patterns and the results of phylogenetic analyses, two EoblCXE genes, EoblCXE7, and EoblCXE13, were selected for fluorescence in situ hybridization assays. Biotin-labeled antisense or sense RNA probes were transcribed from the linearized recombinant pGEM-T vector using a biotin RNA Labeling Mix (SP6/T7) (Roche, Mannheim, Germany) following the recommended protocols. RNA probes were subsequently fragmented to an average length of approximately 400 bp via incubation in carbonate buffer (80 mM NaHCO3, 120 mM Na2CO3, pH 10.2).
The experiment was performed following a reported protocol (Wang et al., 2017). The antennae of both male and female 1–3-day-old moths were dissected, embedded with Tissue-Tek optimal cutting temperature (O.C.T.) compound (Sakura Finetek, Torrance, CA, USA) and rapidly frozen at −60°C. Sections (12 μm) were prepared using a Cryostar NX50 cryostat (Thermo Scientific, San Jose, CA, USA) at −20°C, thaw-mounted on SuperFrost Plus microscope slides (Fisher Scientific, Pittsburgh, PA, USA), and air-dried at room temperature for 15 min. After a series of fixing and washing procedures, the tissue sections were covered with 100 μL of hybridization solution containing biotin-labeled antisense RNA probes and incubated at 60°C for at least 16 h. After hybridization, the slides were washed twice for 20 min in 0.2 × saline-sodium citrate (SSC) at 60°C and treated with 1% blocking reagent (Roche, Basel, Switzerland) in TBST for 30 min at room temperature. Biotin-labeled probes were detected via incubation with streptavidin-horseradish peroxidase (HRP) (Perkin Elmer, Boston, MA, USA) diluted 1:100 in TBS with 0.03% Triton X-100 and 1% blocking reagent at 37°C for 1 h. After three 5-min washes in TBS with 0.05% Tween 20 (Sigma, Louis, MO, USA), the biotin-labeled probes were detected with the TSA Plus Fluorescein System (Perkin Elmer). Images were captured via laser scanning microscopy (LSM) using a Zeiss LSM880 confocal microscope (Zeiss, Oberkochen, Germany). Photoshop CS5 (Adobe Systems, San Jose, CA, USA) was employed to adjust the brightness or contrast of the figures.
Results
Identification and Sequence Characteristics of Candidate EoblCXEs
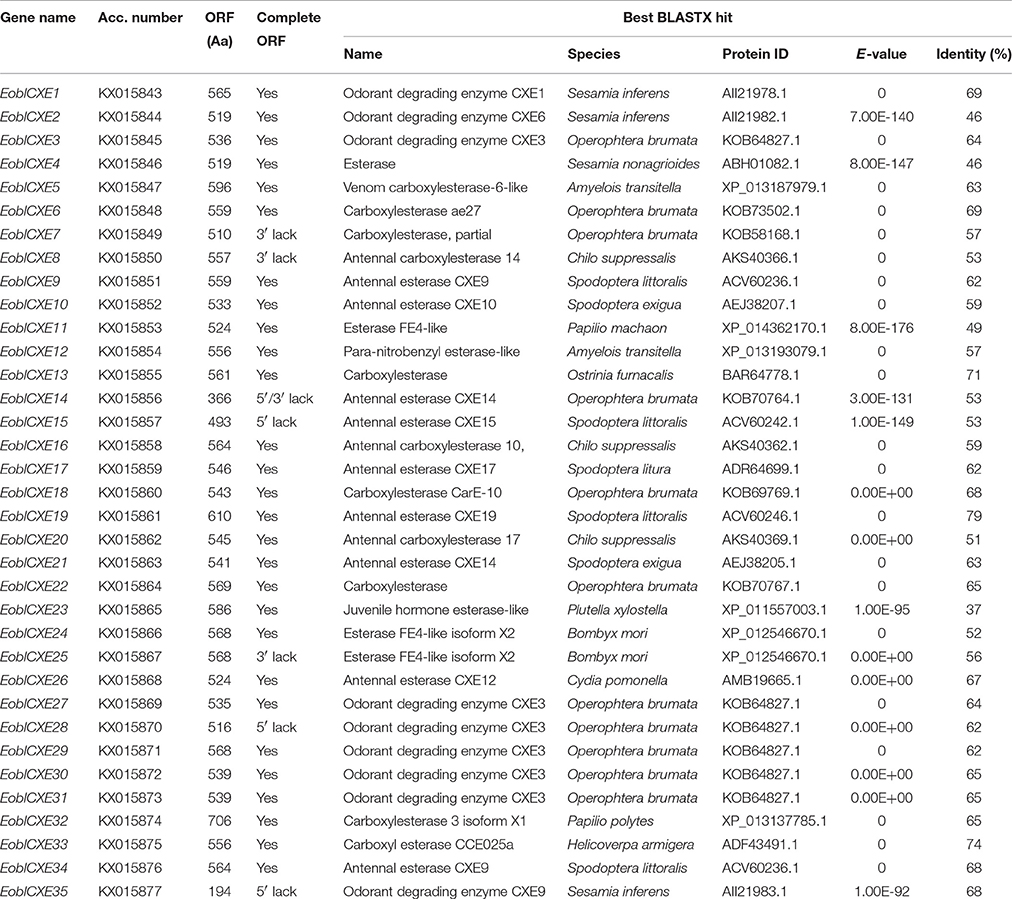
Thirty-five candidate EoblCXEs were identified in the chemosensory organs of E. obliqua. Of these 35 candidates, 28 EoblCXEs possessed full-length open reading frames (ORFs), and seven lacked either the 5′ or 3′ region (Table 1). The candidate sequences were designated EoblCXE1-35 according to their presumptive orthologs in other insects, particularly S. littoralis, S. exigua and S. inferens, and were deposited in the GenBank database under sequential accession numbers from KX015843 to KX015877 (Table 1).
The amino acid identity among the 35 candidate EoblCXEs ranged from 13 to 98% (Table S4). The 28 full-length EoblCXEs exhibited an average coding region length of 1650 bp and encoded 519 to 706 amino acids. Their predicted theoretical isoelectric points ranged from 4.95 to 8.80, and their calculated molecular masses ranged from 58.10 to 74.89. Putative N-terminal signal peptide prediction showed that 16 of the 28 sequences displayed typical sequence cleavage sites. Multiple sequence alignments revealed that all 28 full-length EoblCXEs displayed a conserved sequence motif including the oxyanion hole residues, the catalytic triad (Ser-Glu-His), and the Ser active site in the conserved pentapeptide Gly-X-Ser-X-Gly, characteristic of esterases (Table 2).
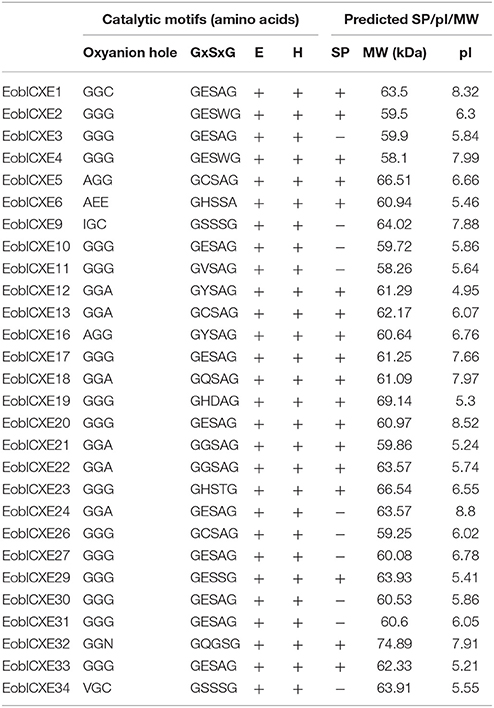
Table 2. Motif analysis and biochemical characteristics of the 28 putative EoblCXEs with full-length sequences.
Phylogenetic Analyses
To define the putative functions of the candidate EoblCXEs, phylogenetic analyses were performed (Figure 1). The results revealed that the insect esterases could be divided into 10 major clades: mitochondrial and cytosolic esterases, dipteran microsomal α-esterases, cuticular and antennal esterases, ß-esterases and pheromone esterases, lepidopteran juvenile hormone esterases (JHE), non-lepidopteran JHE, moth antennal esterases, neuroligins, neuroreceptors, and gliotactins (Oakeshott et al., 2005; Durand et al., 2010b).
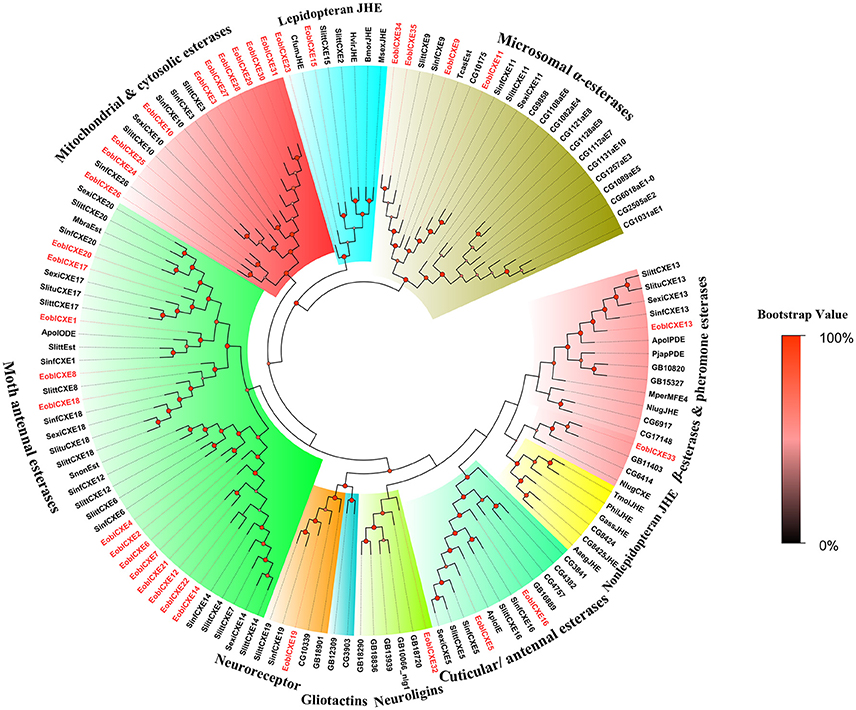
Figure 1. Phylogenetic tree of insect carboxylesterases (CXEs). The tree was constructed with MEGA 6.0 using the neighbor-joining method. The values at the nodes are the results of bootstrapping with 1,000 replicates. EoblCXEs are shown in red. The accession numbers are given in Table S1 and alignment document was included in Supplementary Material.
EoblCXEs were generally distributed in eight different clades: EoblCXE13 and 33, along with the pheromone-degrading enzymes Apol-PDE and Pjap-PDE, clustered with the ß-esterase and pheromone esterase group; EoblCXE5 and 16 were distributed within a clade of cuticular and antennal esterases; and EoblCXE19, EoblCXE32, two EoblCXEs (EoblCXE15 and 23), four EoblCXEs (EoblCXE9, 11, 34, and 35), and 10 EoblCXEs (EoblCXE3, 10, 24–31) were assigned to neuroreceptors, neuroligins, lepidopteran JHE, dipteran microsomal α-esterases and mitochondrial and cytosolic esterases, respectively. The moth antennal esterases exhibited the greatest abundance of EoblCXEs (13 EoblCXEs, including EoblCXE1, 2, 4, 6–8, 12, 14, 17, 18, and 20–22), whereas no EoblCXEs clustered into the non-lepidopteran JHE or gliotactin clades (Figure 1).
Tissue- and Sex-Related Expression Patterns of Candidate EoblCXE Genes
To clarify whether candidate EoblCXEs could function in chemosensory organs with physiological roles in odorant degradation, the tissue- and sex-related expression profiles of the 35 EoblCXE genes were determined via RT-PCR. As shown in Figure 2, the EoblCXE genes displayed four general expression patterns: 12 EoblCXE genes (EoblCXE2, 5, 7, 10, 13, 15–17, 19, 20, 22, and 24) were strongly expressed in olfactory organ antennae; two EoblCXEs (EoblCXE14 and 23) were non-chemosensory organ biased; 12 EoblCXEs (EoblCXE1, 3, 4, 6, 9, 11, 12, 18, 27, 29, 30, and 34) were ubiquitous, and their expression levels were comparable among the tested tissues; and nine EoblCXEs (EoblCXE8, 21, 26, 35, 25, 28, and 31–33) exhibited heterogeneous expression profiles.
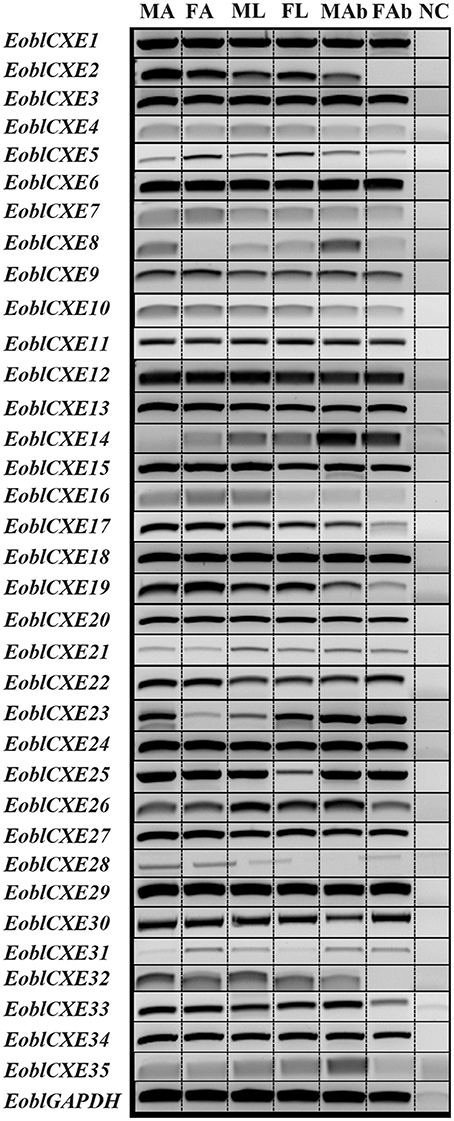
Figure 2. Tissue-related expression patterns of EoblCXE genes, as revealed via RT-PCR. The EoblGAPDH gene was used as a positive control, and NC (no cDNA template) was used as a negative control. MA, male antennae; FA, female antennae; ML, male legs; FL, female legs; MAB, male abdomen; FAB, female abdomen. The original image of this is shown in Figure S2.
To confirm the RT-PCR results, 18 candidate EoblCXE genes randomly selected from all expression patterns were quantified through qRT-PCR assays. The qRT-PCR results are shown in Figure 3. Similar to the RT-PCR results, EoblCXE2, 5, 7, 10, 13, 15, 20, 22, and 24 were strongly expressed in moth antennae, whereas EoblCXE14 and 23 were primarily expressed in the abdomen (a non-chemosensory organ), and EoblCXE6 was ubiquitously expressed within different tissues. However, the qRT-PCR and RT-PCR results were somewhat contradictory; the qRT-PCR results showed that EoblCXE3 was highly expressed in the abdomens of both sexes, whereas EoblCXE12 and 26 were highly expressed in the antennae and legs, respectively.
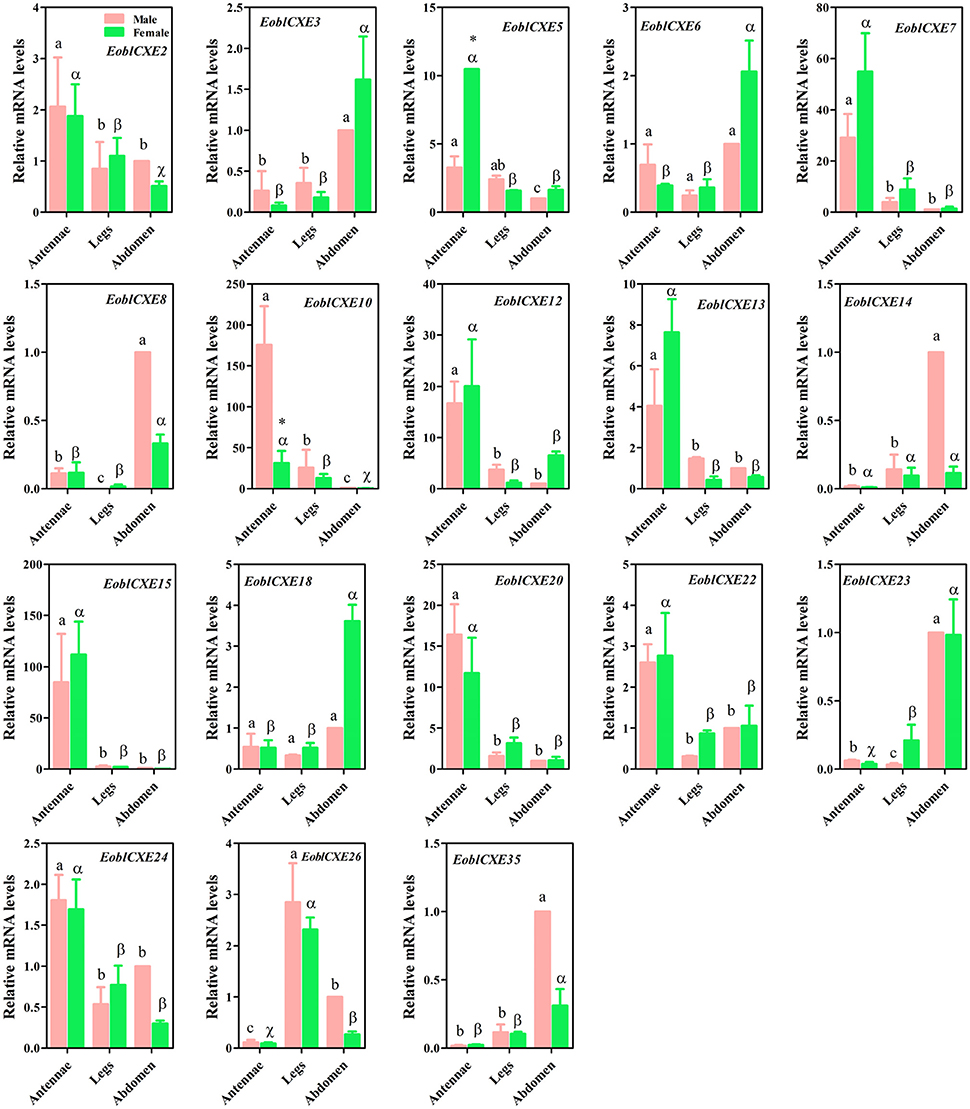
Figure 3. Relative mRNA levels of EoblCXE genes in different tissues of male and female E. obliqua adults, as revealed via qRT-PCR. The reference gene EoblGAPDH was found to be expressed at a similar level in different tissues and was employed as an internal control to normalize target gene expression in order to correct for sample-to-sample variation (Sun et al., 2017a). The amplification efficiency for the target and reference genes were assessed using gradient dilution templates to examine the variation of ΔCT (CT, Target gene − CT, reference gene) with template dilution (Livak and Schmittgen, 2001). The absolute values of the slopes of all lines from the template dilution plots (log cDNA dilution vs. ΔCT) were close to zero, indicating that the efficiency for EoblCXEs was similar to that for EoblGAPDH. The fold changes are relative to the transcript levels in the male abdomen. The error bars represent the standard error, and different letters (a, b and c for male; α, ß, and χ for females) above each bar denote significant differences (P < 0.05). The t and p-values in Student's t-test are shown in Table S5 and the asterisk indicates significantly different relative expression levels between male and female antennae.
To dissect the putatively different roles of EoblCXEs in olfaction between male and female moths, sex-biased expression profiles were determined for the antennae-enriched EoblCXE genes. Eight of the 10 tested EoblCXE genes (EoblCXE2, 5, 7, 10, 12, 13, 15, 20, 22, and 24) exhibited comparable expression levels between the sexes. In contrast, EoblCXE5 and 10 were highly expressed in female and male antennae, respectively.
Cellular Localization of EoblCXE7 and EoblCXE13 within Different Antennal Sensilla
EoblCXE7 and EoblCXE13 were strongly labeled on the sensilla sides of both male and female antennae but differed from each other in their cellular localization in the different types of sensilla found in each sex (Figures 4, 5). EoblCXE7 exhibited a similar localization in the antennae of male and female moths (Figures 4A–F). The antisense EoblCXE7 probe clearly labeled the base of the sensilla trichodea (Str I for male and Str II for female moths), sensilla basiconica (Sba) and sensilla styloconica (Sst) (Figures 4A–C). The scale sides of the antennae were not labeled in either sex. Compared with EoblCXE7, EoblCXE13 showed different expression patterns between male and female antennal sensilla (Figures 5A–I). The antisense EoblCXE13 probe was restricted to the base of Str I rather than Sba or Sst in male moths (Figures 5A–D). In contrast, strong labeling was detected not only in Str II but also in Sba and Sst in female moths (Figures 5E–I). The sense probes of EoblCXE7 and EoblCXE13 produced no positive signals (Figure S1).
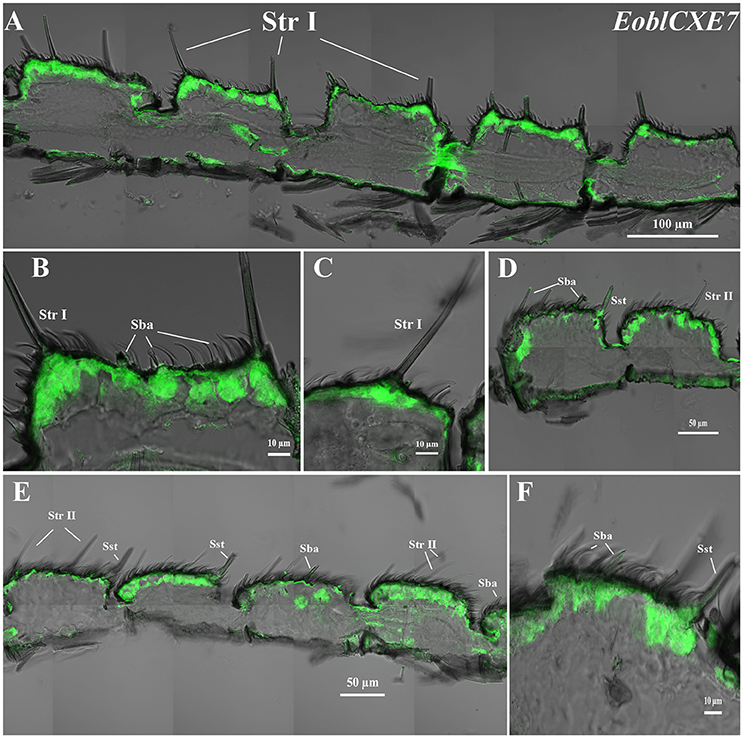
Figure 4. Localization of the EoblCXE7 gene in different antennal sensilla of both sexes, as revealed via fluorescence in situ hybridization. (A–C) male antennae; (D–F) female antennae; Str I, sensilla trichodea I; Str II, sensilla trichodea II; Sba, sensilla basiconica; Sst, sensilla styloconica.
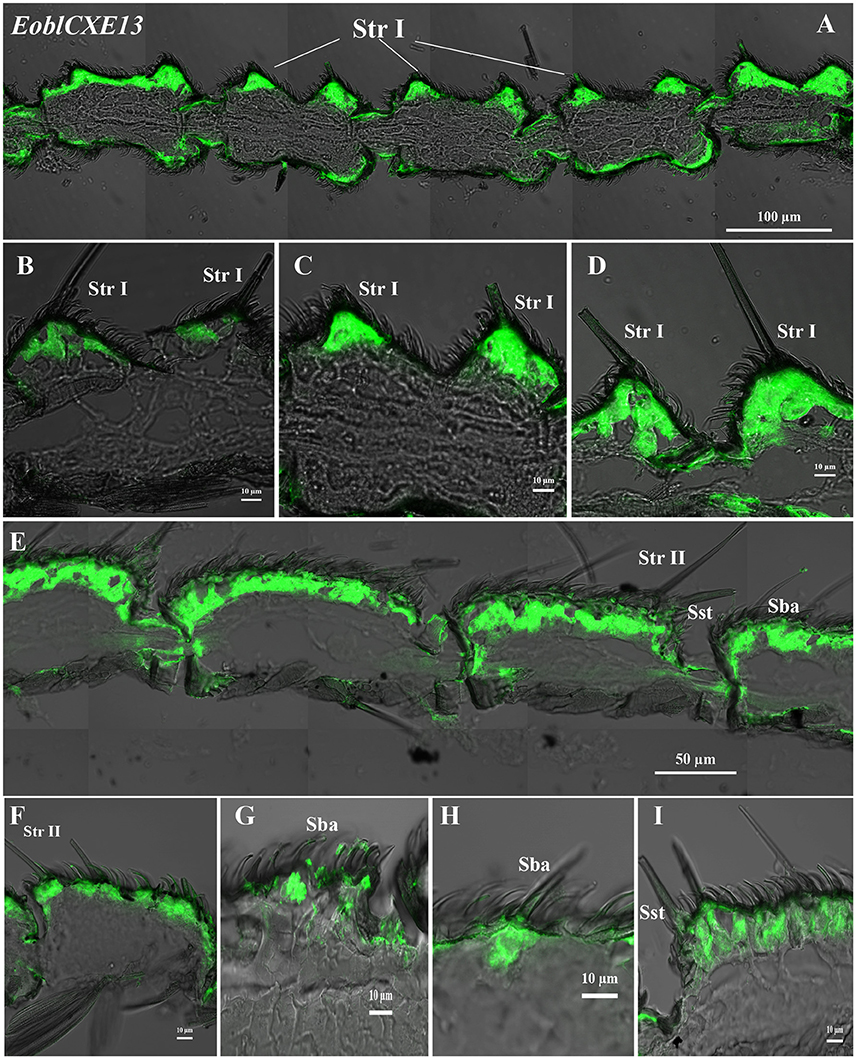
Figure 5. Localization of the EoblCXE13 gene in different antennal sensilla of both sexes, as revealed via fluorescence in situ hybridization. (A–D) male antennae; (E–I) female antennae; Str I, sensilla trichodea I; Str II, sensilla trichodea II; Sba, sensilla basiconica; Sst, sensilla styloconica.
Discussion
In the present study, we identified and characterized 35 candidate genes encoding EoblCXEs from the chemosensory organs of the moth E. obliqua through transcriptomic analysis, including 28 full-length sequences. Phylogenetic analyses and tissue- and sex-related expression profiling showed that EoblCXEs exhibited diverse sequence structures, multiple subfamily clades and distinct expression patterns, suggesting potential differentiation of physiological functions among EoblCXEs. As expected, we demonstrated that 12 EoblCXE genes were highly expressed in E. obliqua antennae, particularly EoblCXE7 and EoblCXE13, which were strongly localized to the olfactory sensilla of both sexes. These results were consistent with the previously reported proposition that CXEs function specifically in insect olfaction and are involved in olfactory signal termination and maintenance of the sensitivity of the olfactory sensilla (Vogt and Riddiford, 1981; Vogt et al., 1985; Ishida and Leal, 2005; Vogt, 2005; Durand et al., 2010a,b, 2011; Chertemps et al., 2012, 2015).
Gene sequence identification represents the first step in elucidating the potential physiological functions of insect CXEs. Due to the lack of genomic information for E. obliqua, we identified putative CXEs through a transcriptomic approach. The number of putative CXE genes (35) identified in E. obliqua (Table 1) was comparable to those found in insect species with available genome data, including D. melanogaster (35 genes), A. gambiae (51 genes), A. aegypti (49 genes) and A. mellifera (24 genes). However, this number was significantly greater than the number of CXE genes identified in moth species that utilize ester compounds as intraspecific sex pheromones, including 20 from S. littoralis (Merlin et al., 2007; Durand et al., 2010b), 24 from S. litura (Zhang et al., 2016), 20 from S. inferens (Zhang Y. N. et al., 2014), 19 from C. suppressalis (Liu et al., 2015), 17 from A. ipsilon (Gu et al., 2013), and 30 from Cnaphalocrocis medinalis (Zhang Y. X. et al., 2017).
Insect CXEs expressed in the olfactory system are mainly related to ester odorant degradation, particularly that of lepidopteran moth sex pheromones (Vogt and Riddiford, 1981; Vogt et al., 1985; He et al., 2015); however, E. obliqua females produce sex pheromones containing unsaturated hydrocarbons and enantiomers of epoxy hydrocarbons rather than acetate esters (Yang et al., 2016a). Thus, it is particularly interesting that the number of CXE genes from E. obliqua was significantly higher than that in species that utilize ester compounds as intraspecific sex pheromones. We speculate that this situation might be attributed to either the use of a different sequencing strategy or the fact that E. obliqua adults depend on the detection of multiple odorants with ester functional groups to find host plants and egg-laying sites. Our use of a high-throughput RNA-sequencing approach in the chemosensory organs (adult antennae of both sexes, legs, wings and proboscises) enabled us to identify as many CXE genes from E. obliqua chemosensory organs as possible. The tea plant, which is the most preferred host of E. obliqua, releases large quantities of ester compounds, particularly under attack by E. obliqua caterpillars; these ester compounds, such as (Z)-3-hexenyl hexanoate and (Z)-3-hexenyl acetate, can in turn regulate the host searching and ovipositional preferences of E. obliqua female adults (Wang, 2010; Sun X. L. et al., 2014). Hence, we propose that some EoblCXEs might act as candidate ODEs and potentially exhibit crucial physiological functions in the degradation of tea plant volatiles with ester functional groups rather than the degradation of sex pheromone components produced by E. obliqua females. This inference corresponds well to the tissue- and sex-related expression patterns found in this study, in which only EoblCXE10 of the antennae-enriched EoblCXE genes displayed a male-specific expression pattern; the other EoblCXEs were either highly abundant in female antennae or were not sex-biased (Figure 3); however, even if then, this inference remains to be supported by the in vitro biochemical enzymatic experiment.
It should be noted that insect CXE genes belong to a multigene family that encodes sequence-divergent and functionally diverse proteins (Oakeshott et al., 1999; Tsubota and Shiotsuki, 2010). The 35 candidate EoblCXEs, which show average amino acid identities lower than 30%, fall into at least 10 different subclades; seven of these clades, which contain 33 EoblCXEs, possess clearly conserved functional characteristic features of the α/β-hydrolase structure, such as the Ser-Glu-His catalytic triad and the nucleophilic elbow surrounding the active-site serine residue (Gly-X-Ser-X-Gly) (Figure 1, Table 2). These features suggest that most EoblCXEs are catalytically active and participate in the degradation of diverse biologically important compounds. Other CXEs fall into the neuroligin, neuroreceptor, or gliotactin clades, including EoblCXE19 in the neuroreceptor clade and EoblCXE32 among the neuroligins, and lack the crucial residue Ser responsible for catalytic activity (Table 2); thus, these CXEs are considered to be catalytically inactive and are mainly involved in neurological and developmental functions related to sensory processing (Biswas et al., 2008; Durand et al., 2017).
Ten EoblCXEs belonging to the dipteran microsomal α-esterase or mitochondrial and cytosolic esterase clade lack a predicted signal peptide, indicating that they are intracellular esterases. These clades (particularly the α-esterases) are well-known for their involvement in the detoxification of insecticides and xenobiotics and the digestion of dietary esters (Newcomb et al., 1997; Campbell et al., 2003; Liang et al., 2007; Tang et al., 2011; Yang et al., 2016b; Gong et al., 2017). An orthologous gene of CXE10 has been functionally studied in two closely related Spodoptera species, S. littoralis (SlittCXE10) and S. exigua (SexiCXE10) (Durand et al., 2010a; He et al., 2015). Both SlittCXE10 and SexiCXE10 are reportedly expressed in the olfactory sensilla and preferentially degrade an ester plant volatile, (Z)-3-hexenyl acetate. EoblCXE10 shares high amino acid identity with SlittCXE10 and SexiCXE10 and is therefore considered an orthologous gene in E. obliqua. Its tissue expression was found to be restricted to the antennae of adults, particularly adult males, and resembled the distribution of its orthologs in Spodoptera species (Figure 3), implying a similar role in the degradation of host plant volatile compounds.
Approximately half of the candidate EoblCXEs clustered in the moth antennal esterase or ß-esterase and pheromone esterase clade and presented both a catalytically active Ser residue and a predicted signal peptide (Table 2). These CXEs represented typical secreted or extracellular esterases that could be secreted into the sensillum lymph surrounding the sensory neurons or into the hemolymph filling the antennal lumen, indicating potential roles in the degradation of odorants and the maintenance of OSN sensitivity. Indeed, functional reports regarding several members of these clades, such as ApolPDE (Vogt and Riddiford, 1981; Ishida and Leal, 2005), PjapPDE (Ishida and Leal, 2008), SexiCXE13 (He et al., 2014a), SexiCXE14 (He et al., 2014c), and SlittCXE7 (Durand et al., 2011), indicate that they play crucial roles in the degradation of insect sex pheromones and biologically important plant volatiles.
To gain further insight into the physiological roles of the EoblCXEs involved in hydrolyzing tea plant volatiles in E. obliqua olfaction, we selected EoblCXE7 and EoblCXE13 for a fluorescence in situ hybridization assay, not only because EoblCXE7 and EoblCXE13 are comparably expressed in the antennae of both E. obliqua sexes but also because EoblCXE13 clusters with ApolPDE, PjapPDE and SexiCXE13, while EoblCXE7 falls into the same clade as SlittCXE7 and SexiCXE14; therefore, these genes provide a useful model for comparative analyses of the functional evolution of CXE orthologs across lepidopteran species. In contrast to SlittCXE7 (Durand et al., 2011), the localization of EoblCXE7 and EoblCXE13 at the sensillum level is more complex. EoblCXE7 exhibits a similar cellular localization at Str and Sba between the sexes, whereas EoblCXE13 is extensively expressed at multiple sensilla of female E. obliqua but is mainly restricted to Str I in males. Interestingly, (Z)-3-hexenyl acetate, a substrate of both CXE13 and CXE7 in Spodoptera species (Durand et al., 2011; He et al., 2014a), can attract both virgin males and mated females in E. obliqua. These results suggested that EolbCXE7 and EoblCXE13 might function in the E. obliqua olfactory system; however, whether the functions of EolbCXE7 and EoblCXE13 resemble those of their orthologs SlittCXE7 and SexiCXE13 as candidate ODEs in the degradation of (Z)-3-hexenyl acetate remains to be further confirmed. Additionally, EoblCXE7 and EoblCXE13 were highly expressed at Sst, a putative gustatory sensillum in lepidopteran species (Zenker et al., 2011; Tang et al., 2015), and we therefore cannot rule out the possibility that both of these EoblCXEs might function in the gustatory system.
In summary, the present study provides the first identification and characterization of the expression patterns of candidate CXE genes in E. obliqua, a common lepidopteran insect pest of the Geometridae, which will aid the development of new pest management techniques using CXE as potential targets for the disruption of insect foraging behavior.
Author Contributions
LS, QianW, and YoZ conceived and designed the experimental plan. LS, QianW, and QiW preformed the experiments. LS, QianW, YuZ, MT, HG, JF, QX, and YaZ analyzed the data. LS and QianW drafted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31501652), Zhejiang Provincial Natural Science Foundation of China (LQ16C140003), Central public-interest Scientific Institution Basal Research Fund (1610212016015), China National Basic Research Program (2012CB114104), Research Foundation of State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201514 and SKLOF201719) and The Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2015-TRICAAS). We thank master student Tengfei Mao for his help in the qPCR, and undergraduate students Mengyao Ou, Xuanxuan Yue, and Hongyue Li for their help in insect rearing and tissue collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2017.01085/full#supplementary-material
Figure S1. Sense probe control for in situ hybridization with biotin-labeled probes.
Figure S2. Original image of Figure 2.
Table S1. Accession numbers and full names of the insect CXEs used in the phylogenetic analysis.
Table S2. Primers employed in the RT-PCR assay.
Table S3. Primers used in the qPCR assay.
Table S4. Amino acid identities of EoblCXEs.
Table S5. The t and p values of Student's t-test.
References
Ando, T., Inomata, S. I., and Yamamoto, M. (2004). “Lepidopteran sex pheromones,” in The Chemistry of Pheromones and Other Semiochemicals I, ed S. Schulz (Berlin; Heidelberg: Springer), 51–96.
Biswas, S., Russell, R. J., Jackson, C. J., Vidovic, M., Ganeshina, O., Oakeshott, J. G., et al. (2008). Bridging the synaptic gap: neuroligins and neurexin I in Apis mellifera. PLoS ONE 3:e3542. doi: 10.1371/journal.pone.0003542
Brunak, S., Nielsen, H. V., Heijne, G. V., Nordahl Petersen, T., and Von Heijine, G. (2010). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Campbell, P. M., Robin, G. C., Court, L. N., Dorrian, S. J., Russell, R. J., and Oakeshott, J. G. (2003). Developmental expression and gene/enzyme identifications in the alpha gene cluster of Drosophila melanogaster. Insect Mol. Biol. 12, 459–471. doi: 10.1046/j.1365-2583.2003.00430.x
Chertemps, T., François, A., Durand, N., Rosell, G., Dekker, T., Lucas, P., et al. (2012). A carboxylesterase, esterase-6, modulates sensory physiological and behavioral response dynamics to pheromone in Drosophila. BMC Biol. 10:56. doi: 10.1186/1741-7007-10-56
Chertemps, T., Younus, F., Steiner, C., Durand, N., Coppin, C. W., Pandey, G., et al. (2015). An antennal carboxylesterase from Drosophila melanogaster, esterase 6, is a candidate odorant-degrading enzyme toward food odorants. Front. Physiol. 6:35. doi: 10.3389/fphys.2015.00315
Durand, N., Carot-Sans, G., Bozzolan, F., Rosell, G., Siaussat, D., Debernard, S., et al. (2011). Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS ONE 6:e29147. doi: 10.1371/journal.pone.0029147
Durand, N., Carot-Sans, G., Chertemps, T., Bozzolan, F., Party, V., Renou, M., et al. (2010a). Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PLoS ONE 5:e15026. doi: 10.1371/journal.pone.0015026
Durand, N., Carot-Sans, G., Chertemps, T., Montagné, N., Jacquin-Joly, E., Debernard, S., et al. (2010b). A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol. Biol. 19, 87–97. doi: 10.1111/j.1365-2583.2009.00939.x
Durand, N., Chertemps, T., Bozzolan, F., and Maïbèche, M. (2017). Expression and modulation of neuroligin and neurexin in the olfactory organ of the cotton leaf worm Spodoptera littoralis. Insect Sci. 24, 210–221. doi: 10.1111/1744-7917.12312
Dweck, H. K. M., Ebrahim, S. A. M., Kromann, S., Bown, D., Hillbur, Y., Sachse, S., et al. (2013). Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 23, 2472–2480. doi: 10.1016/j.cub.2013.10.047
Gong, Y. H., Ai, G. M., Li, M., Shi, X. Y., Diao, Q. Y., and Gao, X. W. (2017). Functional characterization of carboxylesterase gene mutations involved in Aphis gossypii resistance to organophosphate insecticides. Insect Mol. Biol. 26, 702–714. doi: 10.1111/imb.12331
Gu, S. H., Wu, K. M., Guo, Y. Y., Pickett, J., Field, L., Zhou, J. J., et al. (2013). Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics 14:636. doi: 10.1186/1471-2164-14-636
He, P., Li, Z. Q., Liu, C. C., Liu, S. J., and Dong, S. L. (2014a). Two esterases from the genus Spodoptera degrade sex pheromones and plant volatiles. Genome 57, 201–208. doi: 10.1139/gen-2014-0041
He, P., Zhang, J., Li, Z. Q., Zhang, Y. N., Yang, K., and Dong, S. L. (2014b). Functional characterization of an antennal esterase from the noctuid moth, Spodoptera exigua. Arch. Insect Biochem. Physiol. 86, 85–99. doi: 10.1002/arch.21164
He, P., Zhang, Y. N., Li, Z. Q., Yang, K., Zhu, J. Y., Liu, S. J., et al. (2014c). An antennae-enriched carboxylesterase from Spodoptera exigua displays degradation activity in both plant volatiles and female sex pheromones. Insect Mol. Biol. 23, 475–486. doi: 10.1111/imb.12095
He, P., Zhang, Y.-N., Yang, K., Li, Z. Q., and Dong, S. L. (2015). An antenna-biased carboxylesterase is specifically active to plant volatiles in Spodoptera exigua. Pest. Biochem. Phys. 123, 93–100. doi: 10.1016/j.pestbp.2015.03.009
Ishida, Y., and Leal, W. S. (2002). Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus. Insect Biochem. Mol. Biol. 32, 1775–1780. doi: 10.1016/S0965-1748(02)00136-4
Ishida, Y., and Leal, W. S. (2005). Rapid inactivation of a moth pheromone. Proc. Natl. Acad. Sci. U.S.A. 102, 14075–14079. doi: 10.1073/pnas.0505340102
Ishida, Y., and Leal, W. S. (2008). Chiral discrimination of the Japanese beetle sex pheromone and a behavioral antagonist by a pheromone-degrading enzyme. Proc. Natl. Acad. Sci. U.S.A. 105, 9076–9080. doi: 10.1073/pnas.0802610105
Jacquin-Joly, E., and Maïbèche-Coisne, M. (2009). “Molecular mechanisms of sex pheromone reception in Lepidoptera,” in Short Views on Insect Molecular Biology, ed R. Chandrasekar (International Book Mission), 147–158.
Jordan, M. D., Stanley, D., Marshall, S. D. G., De Silva, D., Crowhurst, R. N., Gleave, A. P., et al. (2008). Expressed sequence tags and proteomics of antennae from the tortricid moth, Epiphyas postvittana. Insect Mol. Biol. 17, 361–373. doi: 10.1111/j.1365-2583.2008.00812.x
Kamikouchi, A., Morioka, M., and Kubo, T. (2004). Identification of honeybee antennal proteins/genes expressed in a sex- and/or caste selective manner. Zool. Sci. 21, 53–62. doi: 10.2108/0289-0003(2004)21[53:IOHAGE]2.0.CO;2
Klein, U. (1987). Sensillum-lymph proteins from antennal olfactory hairs of the moth Antheraea polyphemus (Saturniidae). Insect Biochem. 7, 1193–1204. doi: 10.1016/0020-1790(87)90093-X
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li, Q., and Liberles, S. D. (2015). Aversion and attraction through olfaction. Curr. Biol. 25, R120–R129. doi: 10.1016/j.cub.2014.11.044
Liang, P., Cui, J. Z., Yang, X. Q., and Gao, X. W. (2007). Effects of host plants on insecticide susceptibility and carboxylesterase activity in Bemisia tabaci biotype B and greenhouse whitefly, Trialeurodes vaporariorum. Pest Manag. Sci. 63, 365–371. doi: 10.1002/ps.1346
Liu, S., Gong, Z. J., Rao, X. J., Li, M. Y., and Li, S. G. (2015). Identification of putative carboxylesterase and glutathione s-transferase genes from the antennae of the Chilo suppressalis (Lepidoptera: Pyralidae). J. Insect Sci. 15, 1–9. doi: 10.1093/jisesa/iev082
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maïbèche-Coisne, M., Merlin, C., François, M. C., Queguiner, I., Porcheron, P., and Jacquin-Joly, E. (2004). Putative odorant-degrading esterase cDNA from the moth Mamestra brassicae: cloning and expression patterns in male and female antennae. Chem. Senses 29, 381–390. doi: 10.1093/chemse/bjh039
Meijerink, J., and van Loon, J. J. A. (1999). Sensitivities of antennal olfactory neurons of the malaria mosquito, Anopheles gambiae, to carboxylic acids. J. Insect Physiol. 45, 365–373. doi: 10.1016/S0022-1910(98)00135-8
Merlin, C., Rosell, G., Carot-Sans, G., François, M. C., Bozzolan, F., Pelletier, J., et al. (2007). Antennal esterase cDNAs from two pest moths, Spodoptera littoralis and Sesamia nonagrioides, potentially involved in odourant degradation. Insect Mol. Biol. 16, 73–81. doi: 10.1111/j.1365-2583.2006.00702.x
Newcomb, R. D., Campbell, P. M., Ollis, D. L., Cheah, E., Russell, R. J., and Oakeshott, J. G. (1997). A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. U.S.A. 94, 7464–7468. doi: 10.1073/pnas.94.14.7464
Oakeshott, J. G., Claudianos, C., Campbell, P., Newcomb, R., and Russell, R. J. (2005). “Biochemical genetics and genomics of insect esterases,” in Comprehensive Molecular Insect Science, eds I. G. Lawrence, I. Kostas and S. G. Sarjeet (Amsterdam: Elsevier), 309–381.
Oakeshott, J. G., Claudianos, C., Russell, R. J., and Robin, G. C. (1999). Carboxyl/cholinesterases: a case study of the evolution of a successful multigene family. Bioessays 21, 1031–1042. doi: 10.1002/(SICI)1521-1878(199912)22:1<1031::AID-BIES7>3.0.CO;2-J
Pan, H., Lu, Y. H., Xiu, C., Geng, H., Cai, X., Sun, X., et al. (2015). Volatile fragrances associated with flowers mediate host plant alternation of a polyphagous mirid bug. Sci. Rep. 5:14805. doi: 10.1038/srep14805
Park, K. C., Mcneill, M., Unelius, C. R., Oh, H. W., and Suckling, D. M. (2013). Characterization of olfactory receptor neurons for pheromone candidate and plant volatile compounds in the clover root weevil, Sitona lepidus. J. Insect Physiol. 59, 1222–1234. doi: 10.1016/j.jinsphys.2013.10.002
Pelosi, P., Zhou, J., Ban, L., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Pophof, B., Stange, G., and Abrell, L. (2005). Volatile organic compounds as signals in a plant-herbivore system: electrophysiological responses in olfactory sensilla of the moth Cactoblastis cactorum. Chem. Senses 30, 51–68. doi: 10.1093/chemse/bji001
Prestwich, G. D., Vogt, R. G., and Riddiford, L. M. (1986). Binding and hydrolysis of radiolabeled pheromone and several analogs by male-specific antennal proteins of the moth Antheraea polyphemus. J. Chem. Ecol. 12, 323–333. doi: 10.1007/BF01020559
Rützler, M., and Zwiebel, L. J. (2005). Molecular biology of insect olfaction: recent progress and conceptual models. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 191, 777–790. doi: 10.1007/s00359-005-0044-y
Strauch, M., Lüdke, A., Münch, D., Laudes, T., Galizia, C. G., Martinelli, E., et al. (2014). More than apples and oranges–detecting cancer with a fruit fly's antenna. Sci. Rep. 4:3576. doi: 10.1038/srep03576
Sun, L., Gu, S. H., Xiao, H. J., Zhou, J. J., Guo, Y. Y., Liu, Z. W., et al. (2013). The preferential binding of a sensory organ specific odorant binding protein of the alfalfa plant bug Adelphocoris lineolatus AlinOBP10 to biologically active host plant volatiles. J. Chem. Ecol. 39, 1221–1231. doi: 10.1007/s10886-013-0333-9
Sun, L., Mao, T. F., Zhang, Y. X., Wu, J. J., Bai, J. H., Zhang, Y. N., et al. (2017a). Characterization of candidate odorant-binding proteins and chemosensory proteins in the tea geometrid Ectropis obliqua Prout (Lepidoptera: Geometridae). Arch. Insect Biochem. Physiol. 94:e21383. doi: 10.1002/arch.21383
Sun, L., Wang, Q., Yang, S., Wang, Q., Zhang, Z., Khashaveh, A., et al. (2017b). Functional analysis of female-biased odorant binding protein 6 for volatile and nonvolatile host compounds in Adelphocoris lineolatus (Goeze). Insect Mol. Biol. 26, 601–615 doi: 10.1111/imb.12322
Sun, L., Xiao, H. J., Gu, S. H., Guo, Y. Y., Liu, Z. W., and Zhang, Y. J. (2014). Perception of potential sex pheromones and host-associated volatiles in the cotton plant bug, Adelphocoris fasciaticollis (Hemiptera: Miridae): morphology and electrophysiology. Appl. Entomol. Zool. 49, 43–57. doi: 10.1007/s13355-013-0223-1
Sun, X. L., Wang, G. C., Gao, Y., Zhang, X. Z., Xin, Z. J., and Chen, Z. M. (2014). Volatiles emitted from tea plants infested by Ectropis obliqua larvae are attractive to conspecific moths. J. Chem. Ecol. 40, 1080–1089. doi: 10.1007/s10886-014-0502-5
Sun, Y. F., De Biasio, F., Qiao, H. L., Iovinella, I., Yang, S. X., Ling, Y., et al. (2012). Two odorant-binding proteins mediate the behavioral response of aphids to the alarm pheromone E-ß-farnesene and structural analogues. PLoS ONE 7:e32759. doi: 10.1371/journal.pone.0032759
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, B., Sun, J., Zhou, X., Gao, X., and Liang, P. (2011). The stability and biochemical basis of fufenozide resistance in a laboratory-selected strain of Plutella xylostella. Pest. Biochem Phys. 101, 80–85. doi: 10.1016/j.pestbp.2011.08.003
Tang, Q. B., Hong, Z. Z., Cao, H., Yang, F. M., and Zhao, X. (2015). Characteristics of morphology, electrophysiology, and central projections of two sensilla styloconica in Helicoverpa assulta larvae. Neuroreport 26:703. doi: 10.1097/WNR.0000000000000413
Tauxe, G. M., Macwilliam, D., Boyle, S. M., Guda, T., and Ray, A. (2013). Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell 155, 1365–1379. doi: 10.1016/j.cell.2013.11.013
Tsubota, T., and Shiotsuki, T. (2010). Genomic analysis of carboxyl/cholinesterase genes in the silkworm Bombyx mori. BMC Genomics 11:377. doi: 10.1186/1471-2164-11-377
Vogt, R. G. (2005). “Molecular basis of pheromone detection in insects,” in Comp Insect Physiol Biochem Pharmacol Mol Biol, eds L. I. Gilbert, K. Iatrou and S. Gill (London: Elsevier), 753–804.
Vogt, R. G., and Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Vogt, R. G., Riddiford, L. M., and Prestwich, G. D. (1985). Kinetic properties of a sex pheromone-degrading enzyme: the sensillar esterase of Antheraea polyphemus. Proc. Natl. Acad. Sci. U.S.A 82, 8827–8831. doi: 10.1073/pnas.82.24.8827
Wang, G. C. (2010). Ecological Function of Tea Plant Volatiles Induced by Three Herbivores. PhD, Chinese Academy of Agricultural Sciences.
Wang, S. N., Shan, S., Zheng, Y., Peng, Y., Lu, Z. Y., Yang, Y. Q., et al. (2017). Gene structure and expression characteristic of a novel odorant receptor gene cluster in the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae). Insect Mol. Biol. 26, 420–431. doi: 10.1111/imb.12306
Xia, Y. H., Zhang, Y. N., Hou, X. Q., Li, F., and Dong, S. L. (2015). Large number of putative chemoreception and pheromone biosynthesis genes revealed by analyzing transcriptome from ovipositor-pheromone glands of Chilo suppressalis. Sci. Rep. 5:7888. doi: 10.1038/srep07888
Yang, Y., Zhang, L., Guo, F., Long, Y., Wang, Y., and Wan, X. (2016a). Reidentification of sex pheromones of tea geometrid Ectropis obliqua Prout (Lepidoptera: Geometridae). J. Econ. Entomol. 109, 167–175. doi: 10.1093/jee/tov282
Yang, Y., Zhang, Y., Yang, B., Fang, J., and Liu, Z. W. (2016b). Transcriptomic responses to different doses of cycloxaprid involved in detoxification and stress response in the whitebacked planthopper, Sogatella furcifera. Entomol. Exp. Appl. 158, 248–257. doi: 10.1111/eea.12406
Ye, G. Y., Xiao, Q., Chen, M., Chen, X. X., Yuan, Z. J., Stanley, D. W., et al. (2014). Tea: biological control of insect and mite pests in China. Biol. Control 68, 73–91. doi: 10.1016/j.biocontrol.2013.06.013
Zenker, M. M., Penz, C., Paris, M. D., and Specht, A. (2011). Proboscis morphology and its relationship to feeding habits in noctuid moths. J. Insect Sci. 11:42. doi: 10.1673/031.011.0142
Zhang, G. H., Yuan, Z. J., Zhang, C. X., Yin, K. S., Tang, M. J., Guo, H. W., et al. (2014). Detecting deep divergence in seventeen populations of tea geometrid (Ectropis obliqua Prout) in China by COI mtDNA and cross-breeding. PLoS ONE 9:e99373. doi: 10.1371/journal.pone.0099373
Zhang, R., Wang, B., Grossi, G., Falabella, P., Liu, Y., Yan, S., et al. (2017). Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 27, 55–61. doi: 10.1016/j.cub.2016.10.013
Zhang, Y. N., Jin, J. Y., Jin, R., Xia, Y. H., Zhou, J. J., Deng, J. Y., et al. (2013). Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (walker). PLoS ONE 8:e69715. doi: 10.1371/journal.pone.0069715
Zhang, Y. N., Li, J. B., He, P., Sun, L., Li, Z. Q., Fang, L. P., et al. (2016). Molecular identification and expression patterns of carboxylesterase genes based on transcriptome analysis of the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). J. Asia Pac. Entomol. 19, 989–994. doi: 10.1016/j.aspen.2016.07.020
Zhang, Y. N., Xia, Y. H., Zhu, J. Y., Li, S. Y., and Dong, S. L. (2014). Putative pathway of sex pheromone biosynthesis and degradation by expression patterns of genes identified from female pheromone gland and adult antenna of Sesamia inferens (Walker). J. Chem. Ecol. 40, 439–451. doi: 10.1007/s10886-014-0433-1
Zhang, Y. X., Wang, W. L., Li, M. Y., Li, S. G., and Liu, S. (2017). Identification of putative carboxylesterase and aldehyde oxidase genes from the antennae of the rice leaffolder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). J. Asia Pac. Entomol. 20, 907–913. doi: 10.1016/j.aspen.2017.06.001
Keywords: Ectropis obliqua, carboxylesterases (CXEs), odorant-degrading enzymes, phylogenetic analyses, expression patterns, fluorescence in situ hybridization
Citation: Sun L, Wang Q, Wang Q, Zhang Y, Tang M, Guo H, Fu J, Xiao Q, Zhang Y and Zhang Y (2017) Identification and Expression Patterns of Putative Diversified Carboxylesterases in the Tea Geometrid Ectropis obliqua Prout. Front. Physiol. 8:1085. doi: 10.3389/fphys.2017.01085
Received: 18 August 2017; Accepted: 08 December 2017;
Published: 18 December 2017.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
Pei Liang, China Agricultural University, ChinaThomas Chertemps, Université Pierre et Marie Curie, France
Copyright © 2017 Sun, Wang, Wang, Zhang, Tang, Guo, Fu, Xiao, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Sun, bGlhbmdzdW5AdHJpY2Fhcy5jb20=
Yanan Zhang, eW56aGFuZ19pbnNlY3RAMTYzLmNvbQ==
Yongjun Zhang, eWp6aGFuZ0BpcHBjYWFzLmNu
†These authors have contributed equally to this work and co-first authors.
 Liang Sun
Liang Sun Qian Wang2,3†
Qian Wang2,3† Yanan Zhang
Yanan Zhang Yongjun Zhang
Yongjun Zhang