
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol., 18 December 2017
Sec. Exercise Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.01080
Over the last couple of decades, research has focused on attempting to understand the genetic influence on sports performance. This has led to the identification of a number of candidate genes which may help differentiate between elite and non-elite athletes. One of the most promising genes in that regard is ACTN3, which has commonly been referred to as “a gene for speed”. Recent research has examined the influence of this gene on other performance phenotypes, including exercise adaptation, exercise recovery, and sporting injury risk. In this review, we identified 19 studies exploring these phenotypes. Whilst there was large variation in the results of these studies, as well as extremely heterogeneous cohorts, there is overall a tentative consensus that ACTN3 genotype can impact the phenotypes of interest. In particular, the R allele of a common polymorphism (R577X) is associated with enhanced improvements in strength, protection from eccentric training-induced muscle damage, and sports injury. This illustrates that ACTN3 is more than just a gene for speed, with potentially wide-ranging influence on muscle function, knowledge of which may aid in the future personalization of exercise training programmes.
ACTN3 is a gene that encodes for alpha-actinin-3, a protein expressed only in type-II muscle fibers (North et al., 1999). A common polymorphism in this gene is R577X (rs1815739), where a C-to-T base substitution results in the transformation of an arginine base (R) to a premature stop codon (X). X allele homozygotes are deficient in the alpha-actinin-3 protein, which is associated with a lower fast-twitch fiber percentage (Vincent et al., 2007), but does not result in disease (MacArthur and North, 2004). The XX genotype frequency differs across ethnic groups, with approximately 25% of Asians, 18% of Caucasians, 11% of Ethiopians, 3% of Jamaican and US African Americans, and 1% of Kenyans and Nigerians possessing the XX genotype (Yang et al., 2007; MacArthur et al., 2008; Scott et al., 2010). ACTN3 genotype is associated with speed and power phenotypes. Yang et al. (2003) reported that elite sprint athletes had significantly higher frequencies of the R allele than controls, a finding that has been replicated multiple times in speed, power and strength athletes (Druzhevskaya et al., 2008; Roth et al., 2008; Eynon et al., 2009; Ahmetov et al., 2011; Cieszczyk et al., 2011; Kikuchi et al., 2016; Papadimitriou et al., 2016; Weyerstraß et al., 2017; Yang et al., 2017), although these findings are not unequivocal (Scott et al., 2010; Gineviciene et al., 2011; Sessa et al., 2011). Whilst Yang et al. (2003) found a trend toward an increased XX genotype frequency in endurance athletes vs. controls, this relationship is less robust, with most studies reporting a lack of association between XX genotype and endurance performance (Lucia et al., 2006; Saunders et al., 2007; Döring et al., 2010; Kikuchi et al., 2016). In addition, whilst Kenyan and Ethiopian endurance runners are highly successful (Wilber and Pitsiladis, 2012), the frequency of the XX genotype within this group is very low at 8% (Ethiopian) and 1% (Kenyan) (Yang et al., 2007). As such, the general consensus is that ACTN3 X allele likely does not modify elite endurance athlete status (Vancini et al., 2014).
Much of the attention on ACTN3 has focused on the robust relationship with the R allele and strength/power phenotype, with a number of reviews further exploring this relationship (Eynon et al., 2013; Ma et al., 2013; Ahmetov and Fedotovskaya, 2015). Indeed, a number of papers referenced ACTN3 as a “gene for speed” (MacArthur and North, 2004; Chan et al., 2008; Berman and North, 2010). However, emerging evidence suggests that this polymorphism may impact a number of other traits, including exercise recovery, injury risk, and training adaptation (Delmonico et al., 2007; Pimenta et al., 2012; Massidda et al., 2017). The purpose of this mini-review is to further explore these potential relationships, as an increased understanding of the role played by ACTN3 on these traits may lead to improvements in the utilization of genetic information in exercise training.
Over the last 20 or so years, the consistent underlying impact of genetics on exercise adaptation has been well explored (Bouchard et al., 2011; Bouchard, 2012). Whilst it is clear that genetics has an undoubted influence on both exercise performance (Guth and Roth, 2013) and adaptation (Mann et al., 2014), fewer studies examine the influence of individual single nucleotide polymorphisms (SNPs) (Delmonico et al., 2007), or a combination of SNPs (Jones et al., 2016), on this process. In this section, we explore the evidence regarding the impact of ACTN3 on the post-exercise adaptive response.
Following a structured literature search, we found five studies that examined the influence of ACTN3 on exercise adaptation to a standardized training programme (Table 1). Four of these studied resistance training (Clarkson et al., 2005a; Delmonico et al., 2007; Pereira et al., 2013; Erskine et al., 2014), and one focused on aerobic training (Silva et al., 2015). An additional study (Mägi et al., 2016), monitored changes in VO2peak over a five-year period in elite skiers, with no significant ACTN3 genotype differences. However, the exercise intervention in this study was not controlled, and so we did not include it within Table 1. There was considerable variation in the findings. For resistance training, two studies reported that the RR genotype was associated with the greatest increase in strength (Pereira et al., 2013) and power (Delmonico et al., 2007) following resistance training. One study reported no effect of ACTN3 genotype on training adaptations following resistance training (Erskine et al., 2014). Another reported greater improvement in one-repetition maximum (1RM) in X allele carriers compared to RR genotypes (Clarkson et al., 2005a). A further study utilized ACTN3 within a 15-SNP total genotype score (TGS), finding that individuals with a higher number of power alleles (such as ACTN3 R) exhibited greater improvements following high-intensity resistance training compared to low-intensity resistance training (Jones et al., 2016). However, because subjects could have the ACTN3 XX genotype and still be classed as those who would best respond to high-intensity training (due to the possession of a higher number of alleles in other power-associated SNPs), we did not include this study within Table 1.
The variation between studies is likely due to heterogeneity at baseline between genotypes, and differences in exercise prescription. Given the prevalence of the R allele in elite speed-power and strength athletes (Yang et al., 2003; Vincent et al., 2007), it is speculatively considered that R allele carriers would respond best to speed-power and strength training (Kikuchi and Nakazato, 2015). However, as illustrated here, there is perhaps a paucity of data to support this position. Nevertheless, there are some potential molecular mechanisms that could underpin this proposition. Norman et al. (2014) reported that mammalian target of rapamycin (mTOR) and p70S6k phosphorylation was greater in R allele carriers than XX genotypes following sprint exercise. Both mTOR and p70S6k regulate skeletal muscle hypertrophy (Bodine et al., 2001; Song et al., 2005), providing mechanistic support for the belief that hypertrophy, and hence strength and power improvements, should be greater in R allele carriers following resistance training. In addition, Ahmetov et al. (2014) reported that testosterone levels were higher in male and female athletes with at least one R allele compared to XX genotypes. Whilst the direction of this association is not clear, it again supplies a possible mechanism explaining why R allele carriers may experience greater training-induced strength improvements.
A single study examined the impact of this polymorphism on the magnitude of VO2 improvements following endurance training (Silva et al., 2015). Here, VO2 scores at baseline were greater in XX genotypes, but following training this difference was eliminated, indicating that RR genotypes had a greater percentage improvement following training. The population in this cohort were police recruits. Given that the X allele is potentially associated with elite endurance athlete status (Yang et al., 2003), it is not clear whether these results would be mirrored in elite endurance athletes. Clearly, further work is required to fully understand what relationship, if any, exists between ACTN3 and improvements in aerobic capacity following training.
ACTN3 R577X has also been associated with exercise-induced muscle damage; here, increased muscle damage will likely reduce speed of recovery, suggesting a potential modifying effect of this polymorphism on between-session recovery. Of the eight studies identified that examined the impact of this polymorphism on post-exercise muscle damage (Table 2), six reported that that the X allele and/or the XX genotype was associated with higher levels of markers associated with muscle damage (Vincent et al., 2010; Djarova et al., 2011; Pimenta et al., 2012; Belli et al., 2017; Del Coso et al., 2017a,b). One study found no effect of the polymorphism (Clarkson et al., 2005b), and one found that RR genotypes experienced a greater exercise-induced reduction in force compared to XX genotypes (Venckunas et al., 2012). An additional study (Del Coso et al., 2017c) examined the impact of ACTN3 as part of a TGS on creatine kinase (CK) response following a marathon race. Within this TGS, the R allele was considered protective against increased CK concentrations. The results indicated that those athletes with a higher TGS, and therefore greater genetic protection, had a lower CK response to the marathon. Whilst not direct evidence of the R allele's protective effect, as it is possible that the other SNPs used in the TGS conveyed this effect, it nevertheless strengthens the supporting argument.
The increase in post-exercise muscle damage is likely due to structural changes associated with this polymorphism. Alpha-actinin-3 is expressed only in fast-twitch muscle fibers, and X allele homozygotes are alpha-actinin-3 deficient; instead, they upregulate production of alpha-actinin-2 in these fast-twitch fibers (MacArthur et al., 2007; Seto et al., 2011). Both alpha-actinin-3 (encoded for by ACTN3) and alpha-actinin-2 are major structural components of the Z-disks within muscle fibers (Beggs et al., 1992). The Z-disk itself is vulnerable to injury during eccentric contractions (Friden and Lieber, 2001), and knock-out mouse models illustrates these Z-disks are less stable during contraction with increased alpha-actinin-2 concentrations (Seto et al., 2011). A number of the studies in Table 2 exclusively utilized eccentric contractions, whilst others focused on prolonged endurance events that include running, which incorporates eccentric contractions as part of the stretch shortening cycle with each stride (Komi, 2000).
The overall consensus of these studies is that the X allele, and/or the XX genotype, is associated with greater markers of muscle damage following exercise that has an eccentric component; either through direct eccentric muscle action (Vincent et al., 2010), from sport-specific training (Pimenta et al., 2012), or from a competitive event requiring eccentric contractions (Belli et al., 2017; Del Coso et al., 2017a,b). However, there are a number of weaknesses to these studies, potentially limiting the strength of these findings. The overall subject number is modest, with a total of 376 (mean 47) across all eight studies; indeed, the study with the greatest number of subjects, Clarkson et al. (2005b), reported no modifying effect of this polymorphism on post-exercise muscle damage. The total number of XX genotypes was also low, with 85 reported across the studies. This is partly a function of the lower prevalence (~18%) of this genotype, but again the study with the largest number (n = 48) of XX genotypes found no effect of this polymorphism (Clarkson et al., 2005b). It is clear that, in order to increase the robustness of this association, further work with greater subject numbers is required.
We found six studies examining the association between ACTN3 genotype and sports injury risk (Table 3). Three of these examined ankle sprains (Kim et al., 2014; Shang et al., 2015; Qi et al., 2016), with one each for non-contact injuries (Iwao-Koizumi et al., 2015), professional soccer players (Massidda et al., 2017), and exertional rhabdomyolysis (ER) (Deuster et al., 2013). Whilst ER is strongly related to increased CK following exercise (Clarkson and Ebbeling, 1988; Brancaccio et al., 2010), because it requires medical treatment we classified it as an injury. Of these papers, five reported a protective effect of the R allele and/or the RR genotype against injury (Deuster et al., 2013; Kim et al., 2014; Shang et al., 2015; Qi et al., 2016; Massidda et al., 2017). Specifically, Deuster et al. (2013) found that XX genotypes were almost three times more likely to be ER patients than R allele carriers. Qi et al. (2016) reported a significantly lower frequency of the RR genotype in a group of ankle sprain patients vs. controls. Kim et al. (2014) found that XX genotypes were 4.7 times more likely to suffer an ankle injury than R allele carriers in their cohort of ballerinas. Shang et al. (2015) reported the R allele as significantly under-represented in a cohort of military recruits reporting ankle sprains. Finally, Massidda et al. (2017) demonstrated that XX genotypes were 2.6 times more likely to suffer an injury than RR genotypes, and that these injuries were more likely to be of increased severity. Only one study (Iwao-Koizumi et al., 2015) reported that the R allele was associated with an increased risk (OR = 2.52) of a muscle injury compared to X allele carriers in a female cohort.
Regarding ER, the likely mechanism is similar to that discussed in the post-exercise muscle damage section; increased damage at the Z-disk during exercise. For ankle sprains, the mechanism is potentially related to muscle function. R allele carriers tend to have greater levels of muscle mass (MacArthur and North, 2007), and specifically type-II fibers (Vincent et al., 2007), indicating that both the RX and RR genotypes tend to have increased strength capabilities (Pimenta et al., 2013). For other soft-tissue injury types, again, the decreased potential of damage at the Z-disk likely reduces injury risk. This would be particularly true for eccentric contractions; given the importance of this contraction type in the etiology of hamstring injuries, this could be a further causative mechanism (Askling et al., 2003), alongside that of reduced muscle strength (Yamamoto, 1993).
Alongside the modifying role of ACTN3 on muscle strength and injury risk, emerging evidence suggests this SNP may also impact flexibility and muscle stiffness. Two studies reported an association between RR genotype and a decreased flexibility score in the sit-and-reach test (Zempo et al., 2016; Kikuchi et al., 2017). Conversely, Kim et al. (2014) reported that XX genotypes had decreased flexibility in the same test. This lack of consensus is largely due to the small total study number, with greater clarity expected as research in the area evolves. It also mirrors the lack of consensus as to whether flexibility increases or decreases risk of injury (Gleim and McHugh, 1997), indicating the complex, multifactorial nature of injuries and their development (Bahr and Holme, 2003).
In summary, it appears that the R allele of ACTN3 is somewhat protective against injuries. The mechanisms underpinning this are likely varied, and related to a combination of the modifying effects of this SNP on both strength (particularly eccentric strength), exercise-induced muscle damage, and flexibility.
The results of this mini-review indicate that, aside from its established role in sporting performance, the ACTN3 R577X polymorphism also potentially modifies exercise adaption, exercise recovery, and exercise-associated injury risk. As this polymorphism directly influences both muscle structure and muscle fiber phenotype, this is perhaps unsurprising, and points to the potential use of knowledge of this polymorphism in the development of personalized training programmes. However, it is important to consider the limitations surrounding many of these studies. The subject numbers in the considered studies tended to be low, with large heterogeneity between study cohorts, ranging from untrained subjects to professional sports people, as well as differences in sex. Both of these aspects will impact the study findings; the effect of this polymorphism may be smaller in untrained subjects, for example, whereas in elite, well-trained athletes, who are likely closer to their genetic ceiling, the effect may be greater. The low subject numbers are troubling due to the relatively low XX genotype frequency, which is ~18% in Caucasian cohorts, and even lower in African and African-American cohorts. As such, XX genotypes are considerably under-represented across the considered research.
The above limitations indicate further work is required to fully understand the impact of this polymorphism on these phenotypes. That said, there is some consistency between trials, allowing speculative guidelines to be developed for the use of genetic information in the development of personalized training. XX genotypes potentially have increased muscle damage following exercise that includes an eccentric component (Pimenta et al., 2012; Belli et al., 2017; Del Coso et al., 2017a,b). This information may, consequently, be used to guide between-session recovery, and during the competitive season recovery times post-competition. For example, in an elite soccer club, ACTN3 genotype could be utilized alongside other well-established markers to determine training intensity in the days following a match, with players genetically predisposed to increased muscle damage either having a longer recovery period, or increased recovery interventions such as cold-water immersion. In addition, recent research has illustrated the positive impact of Nordic Hamstring Exercises on hamstring injury risk (van der Horst et al., 2015), making these exercises increasingly common in professional sports teams. These exercises have a large eccentric component, upon which this polymorphism may have a direct effect. As such, it would be expected that XX genotypes would have increased muscle soreness and damage following these exercises, potentially impacting the timing of their use within a training programme.
Focusing on sporting injuries, the general consensus from the studies found is that the X allele increased the risk of ankle injuries (Kim et al., 2014; Shang et al., 2015; Qi et al., 2016) and general sporting injury (Massidda et al., 2017). Again, this information could guide training interventions. In this case, X allele carriers might undertake increased general strengthening exercises and neuromuscular training targeting injury risk reduction. Furthermore, knowledge of this information could increase athlete motivation to undertake these exercises (Goodlin et al., 2015).
Finally, maximizing the training response is crucial, both to elite athletes looking to improve by fractions of a second, and to beginners looking to decrease their risk of disease. Increasingly, there is evidence that polymorphisms, including ACTN3 R577X, can impact this adaptive process (Delmonico et al., 2007; Pereira et al., 2013). If further research replicates these early findings, then again, this information could be used in the development of training programmes. Regarding ACTN3, at present it appears that R allele carriers potentially exhibit greater increases in strength and power following high-load resistance training (Delmonico et al., 2007). As such, Kikuchi and Nakazato (2015) speculate that R allele carriers should prioritize high-load, low-repetition resistance training if improvements in muscle strength are required, and high intensity interval (HIT) training to specifically elicit improvements in VO2max.
There is a clear, undoubted impact of genetics on both sporting performance and exercise adaptation. In this regard, one of the most well-studied genes is ACTN3, which has been reliably shown to impact speed-power and strength phenotypes. However, emerging research indicates that this polymorphism may also impact other exercise associated variables, including training adaptation, post-exercise recovery, and exercise-associated injuries; this research is summarized in Figure 1. This information is important, not just because it illustrates the wide-ranging impact SNPs can have, but also because it represents an opportunity to personalize, and therefore enhance, training guidelines. At present, there are no best-practice guidelines pertaining to the use of genetic information in both elite sport and the general public. However, sports teams have been using genetic information for over 10 years (Dennis, 2005), and continue to do so. Consequently, the development of these guidelines represents an important step from lab to practice. Clearly, further research is required to fully develop these guidelines, and at present such information is speculative. Nevertheless, the use of genetic information represents an opportunity to enhance training prescription and outcomes in exercisers of all abilities.
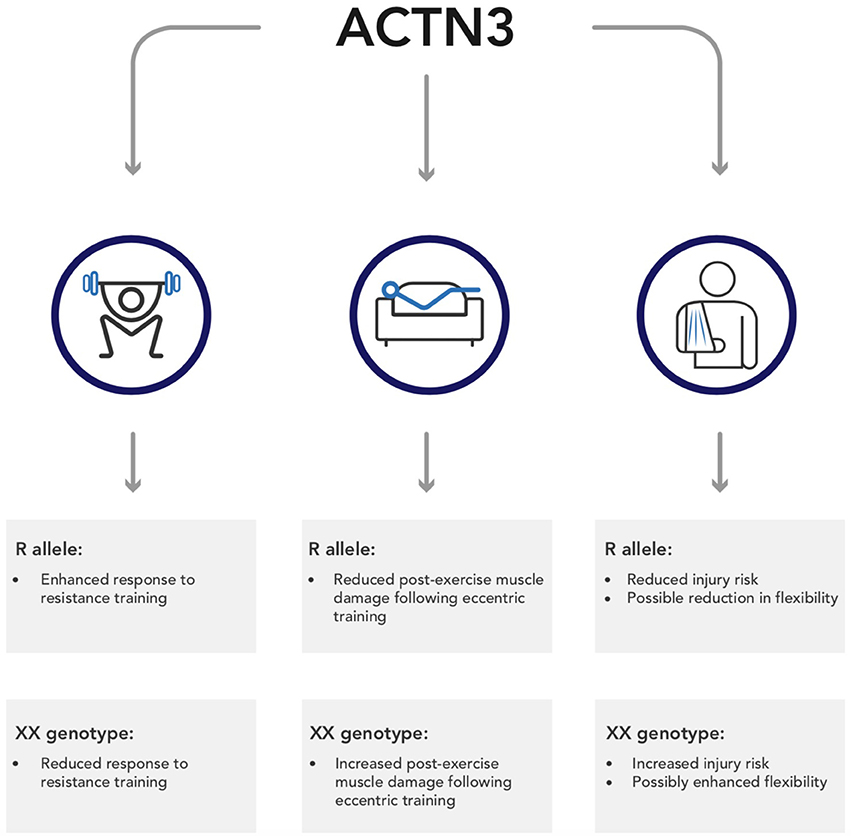
Figure 1. A summary of the potential wider implications of ACTN3 genotype on outcomes from exercise.
CP: Conceived the idea for this manuscript, and wrote the initial draft. JK: Provided feedback on the initial draft, and made valuable changes to the manuscript, as well as providing direction. All the authors made contributions in drafting the manuscript and have approved the final version.
CP is an employee of DNAFit Ltd., a genetic testing company. He received no payment for the production of this article, which was completed as part of his Professional Doctorate studies at the University of Central Lancashire.
The other author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Both authors would like to thank Tshepo Mofokeng for his assistance in the design of Figure 1.
Ahmetov, I. I., Donnikov, A. E., and Trofimov, D. Y. (2014). ACTN3 genotype is associated with testosterone levels of athletes. Biol. Sport 31, 105–108. doi: 10.5604/20831862.1096046
Ahmetov, I. I., Druzhevskaya, A. M., Lyubaeva, E. V., Popov, D. V., Vinogradova, O. L., and Williams, A. G. (2011). The dependence of preferred competitive racing distance on muscle fibre type composition and ACTN3 genotype in speed skaters. Exp. Physiol. 96, 1302–1310. doi: 10.1113/expphysiol.2011.060293
Ahmetov, I. I., and Fedotovskaya, O. N. (2015). Current progress in sports genomics. Adv. Clin. Chem. 70, 247–314. doi: 10.1016/bs.acc.2015.03.003
Askling, C., Karlsson, J., and Thorstensson, A. (2003). Hamstring injury occurrence in elite soccer players after preseason strength training with eccentric overload. Scand. J. Med. Sci. Sports 13, 244–250. doi: 10.1034/j.1600-0838.2003.00312.x
Bahr, R., and Holme, I. (2003). Risk factors for sports injuries—a methodological approach. Br. J. Sports Med. 37, 384–392. doi: 10.1136/bjsm.37.5.384
Beggs, A. H., Byers, T. J., Knoll, J. H., Boyce, F. M., Bruns, G. A., and Kunkel, L. M. (1992). Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J. Biol. Chem. 267, 9281–9288.
Belli, T., Crisp, A. H., and Verlengia, R. (2017). Greater muscle damage in athletes with ACTN3 R577X (RS1815739) gene polymorphism after an ultra-endurance race: a pilot study. Biol. Sport 34, 105–110. doi: 10.5114/biolsport.2017.64583
Berman, Y., and North, K. N. (2010). A gene for speed: the emerging role of α-actinin-3 in muscle metabolism. Physiology 25, 250–259. doi: 10.1152/physiol.00008.2010
Bodine, S. C., Stitt, T. N., Gonzalez, M., Kline, W. O., Stover, G. L., Bauerlein, R., et al. (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019. doi: 10.1038/ncb1101-1014
Bouchard, C. (2012). Genomic predictors of trainability. Exp. Physiol. 97, 347–352. doi: 10.1113/expphysiol.2011.058735
Bouchard, C., Sarzynski, M. A., Rice, T. K., Kraus, W. E., Church, T. S., Sung, Y. J., et al. (2011). Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J. Appl. Physiol. 110, 1160–1170. doi: 10.1152/japplphysiol.00973.2010
Brancaccio, P., Lippi, G., and Maffulli, N. (2010). Biochemical markers of muscular damage. Clin. Chem. Lab. Med. 48, 757–767. doi: 10.1515/CCLM.2010.179
Chan, S., Seto, J. T., MacArthur, D. G., Yang, N., North, K. N., and Head, S. I. (2008). A gene for speed: contractile properties of isolated whole EDL muscle from an α-actinin-3 knockout mouse. Am. J. Physiol. Cell Physiol. 295, C897–C904. doi: 10.1152/ajpcell.00179.2008
Cieszczyk, P., Eider, J., Ostanek, M., Arczewska, A., Leonska-Duniec, A., Sawczyn, S., et al. (2011). Association of the ACTN3 R577X polymorphism in polish power-orientated athletes. J. Hum. Kinet. 28, 55–61. doi: 10.2478/v10078-011-0022-0
Clarkson, P. M., Devaney, J. M., Gordish-Dressman, H., Thompson, P. D., Hubal, M. J., Urso, M., et al. (2005a). ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J. Appl. Physiol. 99, 154–163. doi: 10.1152/japplphysiol.01139.2004
Clarkson, P. M., and Ebbeling, C. (1988). Investigation of serum creatine kinase variability after muscle-damaging exercise. Clin. Sci. 75, 257–261. doi: 10.1042/cs0750257
Clarkson, P. M., Hoffman, E. P., Zambraski, E., Gordish-Dressman, H., Kearns, A., Hubal, M., et al. (2005b). ACTN3 and MLCK genotype associations with exertional muscle damage. J. Appl. Physiol. 99, 564–569. doi: 10.1152/japplphysiol.00130.2005
Del Coso, J., Salinero, J. J., Lara, B., Gallo-Salazar, C., Areces, F., Puente, C., et al. (2017a). ACTN3 X-allele carriers had greater levels of muscle damage during a half-ironman. Eur. J. Appl. Physiol. 117, 151–158. doi: 10.1007/s00421-016-3507-7
Del Coso, J., Valero, M., Salinero, J. J., Lara, B., Díaz, G., Gallo-Salazar, C., et al. (2017b). ACTN3 genotype influences exercise-induced muscle damage during a marathon competition. Eur. J. Appl. Physiol. 117, 409–416. doi: 10.1007/s00421-017-3542-z
Del Coso, J., Valero, M., Salinero, J. J., Lara, B., Gallo-Salazar, C., and Areces, F. (2017c). Optimum polygenic profile to resist exertional rhabdomyolysis during a marathon. PLoS ONE 12:e0172965. doi: 10.1371/journal.pone.0172965
Delmonico, M. J., Kostek, M. C., Doldo, N. A., Hand, B. D., Walsh, S., Conway, J. M., et al. (2007). Alpha-actinin-3 (ACTN3) R577X polymorphism influences knee extensor peak power response to strength training in older men and women. J. Gerontol. A Biol. Sci. Med. Sci. 62, 206–212. doi: 10.1093/gerona/62.2.206
Dennis, C. (2005). Rugby team converts to give gene tests a try. Nature 434:260. doi: 10.1038/434260a
Deuster, P. A., Contreras-Sesvold, C. L., O'Connor, F. G., Campbell, W. W., Kenney, K., Capacchione, J. F., et al. (2013). Genetic polymorphisms associated with exertional rhabdomyolysis. Eur. J. Appl. Physiol. 113, 1997–2004. doi: 10.1007/s00421-013-2622-y
Djarova, T., Watson, G., Basson, A., Grace, J., Cloete, J., and Ramakoaba, A. (2011). ACTN3 and TNF gene polymorphism association with C-reactive protein, uric acid, lactate and physical characteristics in young African cricket players. Afr. J. Biochem. Res. 5, 22–27.
Döring, F. E., Onur, S., Geisen, U., Boulay, M. R., Pérusse, L., Rankinen, T., et al. (2010). ACTN3 R577X and other polymorphisms are not associated with elite endurance athlete status in the Genathlete study. J. Sports Sci. 28, 1355–1359. doi: 10.1080/02640414.2010.507675
Druzhevskaya, A. M., Ahmetov, I. I., Astratenkova, I. V., and Rogozkin, V. A. (2008). Association of the ACTN3 R577X polymorphism with power athlete status in Russians. Eur. J. Appl. Physiol. 103, 631–634. doi: 10.1007/s00421-008-0763-1
Erskine, R. M., Williams, A. G., Jones, D. A., Stewart, C. E., and Degens, H. (2014). The individual and combined influence of ACE and ACTN3 genotypes on muscle phenotypes before and after strength training. Scand. J. Med. Sci. Sports 24, 642–648. doi: 10.1111/sms.12055
Eynon, N., Duarte, J. A., Oliveira, J., Sagiv, M., Yamin, C., Meckel, Y., et al. (2009). ACTN3 R577X polymorphism and Israeli top-level athletes. Int. J. Sports Med. 30, 695–698. doi: 10.1055/s-0029-1220731
Eynon, N., Hanson, E. D., Lucia, A., Houweling, P. J., Garton, F., North, K. N., et al. (2013). Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 43, 803–817. doi: 10.1007/s40279-013-0059-4
Friden, J., and Lieber, R. L. (2001). Eccentric exercise-induced injuries to contractile and cytoskeletal muscle fibre components. Acta Physiol. Scand. 171, 321–326. doi: 10.1046/j.1365-201x.2001.00834.x
Gineviciene, V., Pranculis, V., Jakaitiene, V., Milasius, V., and Kucinskas, V. (2011). Genetic variation of the human ACE and ACTN3 gene and their association with functional muscle properties in Lithuanian elite athletes. Medicina 47, 284–290.
Gleim, G. W., and McHugh, M. P. (1997). Flexibility and its effects on sports injury and performance. Sports Med. 24, 289–299. doi: 10.2165/00007256-199724050-00001
Goodlin, G. T., Roos, A. K., Roos, T. R., Hawkins, C., Beache, S., Baur, S., et al. (2015). Applying personal genetic data to injury risk assessment in athletes. PLoS ONE 10:e0122676. doi: 10.1371/journal.pone.0122676
Guth, L. M., and Roth, S. M. (2013). Genetic influence on athletic performance. Curr. Opin. Pediatr. 25, 653–658. doi: 10.1097/MOP.0b013e3283659087
Iwao-Koizumi, K., Ota, T., Hayashida, M., Yonetani, Y., Nakata, K., Kinoshita, K., et al. (2015). The ACTN3 gene is a potential biomarker for the risk of non-contact sports injury in female athletes. J. Mol. Biomark. Diagn. S6:2. doi: 10.4172/2155-9929.S6-002
Jones, N., Kiely, J., Suraci, B., Collins, D. J., de Lorenzo, D., Pickering, C., et al. (2016). A genetic-based algorithm for personalized resistance training. Biol. Sport 33, 117–126. doi: 10.5604/20831862.1198210
Kikuchi, N., Miyamoto-Mikami, E., Murakami, H., Nakamura, T., Min, S. K., Mizuno, M., et al. (2016). ACTN3 R577X genotype and athletic performance in a large cohort of Japanese athletes. Eur. J. Sport Sci. 16, 694–701. doi: 10.1080/17461391.2015.1071879
Kikuchi, N., and Nakazato, K. (2015). Effective utilization of genetic information for athletes and coaches: focus on ACTN3 R577X polymorphism. J. Exerc. Nutrition Biochem. 19, 157–164. doi: 10.5717/jenb.2015.15093001
Kikuchi, N., Zempo, H., Fuku, N., Murakami, H., Sakamaki-Sunaga, M., Okamoto, T., et al. (2017). Association between ACTN3 R577X Polymorphism and trunk flexibility in 2 different cohorts. Int. J. Sports Med. 38, 402–406. doi: 10.1055/s-0042-118649
Kim, J. H., Jung, E. S., Kim, C. H., Youn, H., and Kim, H. R. (2014). Genetic associations of body composition, flexibility and injury risk with ACE, ACTN3 and COL5A1 polymorphisms in Korean ballerinas. J. Exerc. Nutrition Biochem. 18, 205–214. doi: 10.5717/jenb.2014.18.2.205
Komi, P. V. (2000). Stretch-shortening cycle: a powerful model to study normal and fatigued muscle. J. Biomech. 33, 1197–1206. doi: 10.1016/S0021-9290(00)00064-6
Lucia, A., Gomez-Gallego, F., Santiago, C., Bandres, F., Earnest, C., Rabadan, M., et al. (2006). ACTN3 genotype in professional endurance cyclists. Int. J. Sports Med. 27, 880–884. doi: 10.1055/s-2006-923862
Ma, F., Yang, Y., Li, X., Zhou, F., Gao, C., Li, M., et al. (2013). The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS ONE 8:e54685. doi: 10.1371/journal.pone.0054685
MacArthur, D. G., and North, K. N. (2004). A gene for speed? The evolution and function of α-actinin-3. Bioessays 26, 786–795. doi: 10.1002/bies.20061
MacArthur, D. G., and North, K. N. (2007). ACTN3: a genetic influence on muscle function and athletic performance. Exerc. Sport Sci. Rev. 35, 30–34. doi: 10.1097/JES.0b013e31802d8874
MacArthur, D. G., Seto, J. T., Chan, S., Quinlan, K. G., Raftery, J. M., Turner, N., et al. (2008). An Actn3 knockout mouse provides mechanistic insights into the association between α-actinin-3 deficiency and human athletic performance. Hum. Mol. Genet. 17, 1076–1086. doi: 10.1093/hmg/ddm380
MacArthur, D. G., Seto, J. T., Raftery, J. M., Quinlan, K. G., Huttley, G. A., Hook, J. W., et al. (2007). Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat. Genet. 39, 1261–1265. doi: 10.1038/ng2122
Mägi, A., Unt, E., Prans, E., Raus, L., Eha, J., Veraksitš, A., et al. (2016). The association analysis between ACE and ACTN3 genes polymorphisms and endurance capacity in young cross-country skiers: longitudinal study. J. Sports Sci. Med. 15, 287–294.
Mann, T. N., Lamberts, R. P., and Lambert, M. I. (2014). High responders and low responders: factors associated with individual variation in response to standardized training. Sports Med. 44, 1113–1124. doi: 10.1007/s40279-014-0197-3
Massidda, M., Voisin, S., Culigioni, C., Piras, F., Cugia, P., Xu, Y., et al. (2017). ACTN3 R577X polymorphism is associated with the incidence and severity of injuries in professional football players. Clin. J. Sport Med. doi: 10.1097/JSM.0000000000000487. [Epub ahead of print].
Norman, B., Esbjörnsson, M., Rundqvist, H., Österlund, T., Glenmark, B., and Jansson, E. (2014). ACTN3 genotype and modulation of skeletal muscle response to exercise in human subjects. J. Appl. Physiol. 116, 1197–1203. doi: 10.1152/japplphysiol.00557.2013
North, K. N., Yang, N., Wattanasirichaigoon, D., Mills, M., Easteal, S., and Beggs, A. H. (1999). A common nonsense mutation results in α-actinin-3 deficiency in the general population. Nat. Genet. 21, 353–354. doi: 10.1038/7675
Papadimitriou, I. D., Lucia, A., Pitsiladis, Y. P., Pushkarev, V. P., Dyatlov, D. A., Orekhov, E. F., et al. (2016). ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: a multi-cohort study. BMC Genomics 17:285. doi: 10.1186/s12864-016-2462-3
Pereira, A., Costa, A. M., Izquierdo, M., Silva, A. J., Bastos, E., and Marques, M. C. (2013). ACE I/D and ACTN3 R/X polymorphisms as potential factors in modulating exercise-related phenotypes in older women in response to a muscle power training stimuli. Age 35, 1949–1959. doi: 10.1007/s11357-012-9461-3
Pimenta, E. M., Coelho, D. B., Cruz, I. R., Morandi, R. F., Veneroso, C. E., de Azambuja Pussieldi, G., et al. (2012). The ACTN3 genotype in soccer players in response to acute eccentric training. Eur. J. Appl. Physiol. 112, 1495–1503. doi: 10.1007/s00421-011-2109-7
Pimenta, E. M., Coelho, D. B., Veneroso, C. E., Coelho, E. J., Cruz, I. R., Morandi, R. F., et al. (2013). Effect of ACTN3 gene on strength and endurance in soccer players. J. Strength Cond. Res. 27, 3286–3292. doi: 10.1519/JSC.0b013e3182915e66
Qi, B., Liu, J. Q., and Liu, G. L. (2016). Genetic association between ACTN3 polymorphism and risk of non-acute ankle sprain. Genet. Mol. Res. 15. doi: 10.4238/gmr15048962
Roth, S. M., Walsh, S., Liu, D., Metter, E. J., Ferrucci, L., and Hurley, B. F. (2008). The ACTN3 R577X nonsense allele is under-represented in elite-level strength athletes. Eur. J. Hum. Genet. 16, 391–394. doi: 10.1038/sj.ejhg.5201964
Saunders, C. J., September, A. V., Xenophontos, S. L., Cariolou, M. A., Anastassiades, L. C., Noakes, T. D., et al. (2007). No association of the ACTN3 gene R577X polymorphism with endurance performance in Ironman Triathlons. Ann. Hum. Genet. 71, 777–781. doi: 10.1111/j.1469-1809.2006.00385.x
Scott, R. A., Irving, R., Irwin, L., Morrison, E., Charlton, V., Austin, K., et al. (2010). ACTN3 and ACE genotypes in elite Jamaican and US sprinters. Med. Sci. Sports Exerc. 42, 107–112. doi: 10.1249/MSS.0b013e3181ae2bc0
Sessa, F., Chetta, M., Petito, A., Franzetti, M., Bafunno, V., Pisanelli, D., et al. (2011). Gene polymorphisms and sport attitude in Italian athletes. Genet. Test. Mol. Biomark. 15, 285–290. doi: 10.1089/gtmb.2010.0179
Seto, J. T., Lek, M., Quinlan, K. G., Houweling, P. J., Zheng, X. F., Garton, F., et al. (2011). Deficiency of α-actinin-3 is associated with increased susceptibility to contraction-induced damage and skeletal muscle remodeling. Hum. Mol. Genet. 20, 2914–2927. doi: 10.1093/hmg/ddr196
Shang, X., Li, Z., Cao, X., Xie, C., Gu, M., Chen, P., et al. (2015). The association between the ACTN3 R577X polymorphism and noncontact acute ankle sprains. J. Sports Sci. 33, 1775–1779. doi: 10.1080/02640414.2015.1012098
Silva, M. S., Bolani, W., Alves, C. R., Biagi, D. G., Lemos, J. R. Jr., da Silva, J. L., et al. (2015). Elimination of influences of the ACTN3 R577X variant on oxygen uptake by endurance training in healthy individuals. Int. J. Sports Physiol. Perform. 10, 636–641. doi: 10.1123/ijspp.2014-0205
Song, Y. H., Godard, M., Li, Y., Richmond, S. R., Rosenthal, N., and Delafontaine, P. (2005). Insulin-like growth factor I-mediated skeletal muscle hypertrophy is characterized by increased mTOR-p70S6K signaling without increased Akt phosphorylation. J. Investig. Med. 53, 135–142. doi: 10.2310/6650.2005.00309
Vancini, R. L., Pesquero, J. B., Fachina, R. J., dos Santos Andrade, M., Borin, J. P., Montagner, P. C., et al. (2014). Genetic aspects of athletic performance: the African runners phenomenon. Open Access J. Sports Med. 5, 123–127. doi: 10.2147/OAJSM.S61361
van der Horst, N., Smits, D. W., Petersen, J., Goedhart, E. A., and Backx, F. J. (2015). The preventive effect of the Nordic hamstring exercise on hamstring injuries in amateur soccer players: a randomized controlled trial. Am. J. Sports Med. 43, 1316–1323. doi: 10.1177/0363546515574057
Venckunas, T., Skurvydas, A., Brazaitis, M., Kamandulis, S., Snieckus, A., and Moran, C. N. (2012). Human alpha-actinin-3 genotype association with exercise-induced muscle damage and the repeated-bout effect. Appl. Physiol. Nutr. Metab. 37, 1038–1046. doi: 10.1139/h2012-087
Vincent, B., De Bock, K., Ramaekers, M., Van den Eede, E., Van Leemputte, M., Hespel, P., et al. (2007). ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol. Genom. 32, 58–63. doi: 10.1152/physiolgenomics.00173.2007
Vincent, B., Windelinckx, A., Nielens, H., Ramaekers, M., Van Leemputte, M., Hespel, P., et al. (2010). Protective role of α-actinin-3 in the response to an acute eccentric exercise bout. J. Appl. Physiol. 109, 564–573. doi: 10.1152/japplphysiol.01007.2009
Weyerstraß, J., Stewart, K., Wesselius, A., and Zeegers, M. (2017). Nine genetic polymorphisms associated with power athlete status–a meta-analysis. J. Sci. Med. Sport. doi: 10.1016/j.jsams.2017.06.012. [Epub ahead of print].
Wilber, R. L., and Pitsiladis, Y. P. (2012). Kenyan and Ethiopian distance runners: what makes them so good? Int. J. Sports Physiol. Perform. 7, 92–102. doi: 10.1123/ijspp.7.2.92
Yamamoto, T. (1993). Relationship between hamstring strains and leg muscle strength. A follow-up study of collegiate track and field athletes. J. Sports Med. Phys. Fitness 33, 194–199.
Yang, N., MacArthur, D. G., Gulbin, J. P., Hahn, A. G., Beggs, A. H., Easteal, S., et al. (2003). ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 73, 627–631. doi: 10.1086/377590
Yang, N., MacArthur, D. G., Wolde, B., Onywera, V. O., Boit, M., Lau, S. Y., et al. (2007). The ACTN3 R577X polymorphism in East and West African athletes. Med. Sci. Sports Exerc. 39, 1985–1988. doi: 10.1249/mss.0b013e31814844c9
Yang, R., Shen, X., Wang, Y., Voisin, S., Cai, G., Fu, Y., et al. (2017). ACTN3 R577X gene variant is associated with muscle-related phenotypes in Elite Chinese sprint/power athletes. J. Strength Cond. Res. 31, 1107–1115. doi: 10.1519/JSC.0000000000001558
Keywords: ACTN3, genetics, adaptation, recovery, injury, personalized, genetic testing
Citation: Pickering C and Kiely J (2017) ACTN3: More than Just a Gene for Speed. Front. Physiol. 8:1080. doi: 10.3389/fphys.2017.01080
Received: 11 October 2017; Accepted: 08 December 2017;
Published: 18 December 2017.
Edited by:
Kimberly Huey, Drake University, United StatesReviewed by:
Rudy Valentine, Iowa State University, United StatesCopyright © 2017 Pickering and Kiely. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Craig Pickering, Y3JhaWdAZG5hZml0LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.