- 1Muscle Research and Therapeutics, Baker Heart and Diabetes Institute, Melbourne, VIC, Australia
- 2Department of Diabetes, Monash University, Melbourne, VIC, Australia
- 3Department of Pathology, University of Melbourne, Melbourne, VIC, Australia
- 4Organogenesis and Cancer Programme, Peter MacCallum Cancer Centre, Melbourne, VIC, Australia
- 5Sir Peter MacCallum Department of Oncology, University of Melbourne, Melbourne, VIC, Australia
- 6Department of Anatomy and Developmental Biology, and Biomedicine Discovery Institute, Monash University, Clayton, VIC, Australia
- 7Department of Physiology, University of Melbourne, Melbourne, VIC, Australia
- 8Department of Biochemistry and Molecular Biology, Monash University, Clayton, VIC, Australia
- 9Department of Neurology, University of Washington School of Medicine, Seattle, WA, United States
The integrative control of diverse biological processes such as proliferation, differentiation, apoptosis and metabolism is essential to maintain cellular and tissue homeostasis. Disruption of these underlie the development of many disease states including cancer and diabetes, as well as many of the complications that arise as a consequence of aging. These biological outputs are governed by many cellular signaling networks that function independently, and in concert, to convert changes in hormonal, mechanical and metabolic stimuli into alterations in gene expression. First identified in Drosophila melanogaster as a powerful mediator of cell division and apoptosis, the Hippo signaling pathway is a highly conserved regulator of mammalian organ size and functional capacity in both healthy and diseased tissues. Recent studies have implicated the pathway as an effector of diverse physiological cues demonstrating an essential role for the Hippo pathway as an integrative component of cellular homeostasis. In this review, we will: (a) outline the critical signaling elements that constitute the mammalian Hippo pathway, and how they function to regulate Hippo pathway-dependent gene expression and tissue growth, (b) discuss evidence that shows this pathway functions as an effector of diverse physiological stimuli and (c) highlight key questions in this developing field.
Introduction
The fine control of biological processes such as cell division, terminal differentiation, cell death (apoptosis) and metabolism in response to changes in the external environment is essential for biological organisms to maintain homeostasis and function appropriately (Purvis and Lahav, 2013). These processes are mediated by intracellular signaling networks including the Transforming-Growth Factor beta (TGF-β), Notch, WNT and Insulin-PI3K-mTOR signaling pathways that act in isolation to control the expression of sub-sets of genes, and as an orchestrated network that collectively influences biological phenotypes (Bedinger and Adams, 2015; Bray, 2016; Hata and Chen, 2016; Masuda and Ishitani, 2017). Recent evidence also implicates the relatively less-well-characterized Hippo pathway as a major signaling pathway governing the cellular response to diverse physiological stimuli (Harvey et al., 2013; Meng et al., 2016). The role of the Hippo pathway in Drosophila (Richardson and Portela, 2017), during early mammalian developmental stages (Sasaki, 2017), or in specific tissues such as the heart (He et al., 2017; Zhang and Del Re, 2017), intestine (Gregorieff and Wrana, 2017), and liver (Patel et al., 2017) have recently been described. In this review, we aim to provide the reader with an outline of the critical elements that form the core Hippo signaling pathway in mammals, and to describe the experimental evidence supporting a vital role for this pathway as a major regulator of tissue growth and differentiation in response to three main forms of external regulation: hormonal, mechanical and metabolic stimuli. We will use mammalian nomenclature in general, unless discussing specific results in other organisms.
The Hippo Signaling Pathway Negatively Regulates the Activity of YAP and TAZ
First identified from genetic loss-of-function screens in Drosophila Melanogaster, the Hippo signaling pathway has emerged as a powerful regulator of organ size and cell fate across species (Harvey et al., 2013). To date, over 40 proteins have been implicated as members of this diverse pathway; however, the capacity for many of these elements to control Hippo signaling activity is highly context-dependent (Plouffe et al., 2016). Despite this, most stimuli that influence the Hippo pathway converge at a common level to activate, or inhibit, the core Hippo pathway kinases, Mammalian Ste-20 like kinase 1 and 2 (MST1/2) and/or mitogen-activated protein kinase kinase kinase kinases 1-4, 6, and 7 (MAP4K) (Tapon et al., 2002; Harvey et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003; Li et al., 2015; Meng et al., 2015; Zheng et al., 2015; Figure 1). The biochemical regulation of these proteins has been studied mainly with MST1/2, where activation of the kinase is dependent on phosphorylation by the TAO family kinases (TAO1-3) in an activation loop at Thr180/183, respectively (Praskova et al., 2004; Boggiano et al., 2011; Poon et al., 2011). This phosphorylation event is critical for increased catalytic activity and, in the case of MST1/2, the formation of a complex with the adaptor protein WW-domain containing 1 (SAV1) (Pantalacci et al., 2003; Wu et al., 2003; Callus et al., 2006). This interaction appears exclusive to MST1/2 since MAP4K do not interact with SAV1 in vitro (Meng et al., 2015; Zheng et al., 2015). Upon activation, MST1/2 or MAP4K can phosphorylate the C-terminal hydrophobic motif of the NDR family kinases Large Tumour Suppressor kinase 1 and 2 (LATS1/2Thr1079) and/or the related Nuclear dbf-2 related (NDR) kinases 1/2 (Chan et al., 2005; Hergovich et al., 2009; Li et al., 2015; Meng et al., 2015; Tang et al., 2015; Zheng et al., 2015). Phosphorylated LATS1/2 and/or NDR1/2 then undergo auto-phosphorylation in an activation loop (Chan et al., 2005). MST1/2 also binds the adaptor proteins MOB1A/1B (MOB) leading to phosphorylation on two N-terminal residues; Thr12 and Thr35 (Lai et al., 2005; Praskova et al., 2008). MST1/2 phosphorylation results in activation of MOB and a conformational change that favors binding to LATS1/2 or NDR1/2 kinases (Praskova et al., 2008). When active, the LATS/MOB or NDR/MOB complexes suppress the activity of the transcriptional co-activators Yes-associated protein (YAP1) and transcriptional co-activator with PDZ-binding motif (WWTR1/TAZ) (Huang et al., 2005; Zhao et al., 2007; Lei et al., 2008; Zhang et al., 2015).
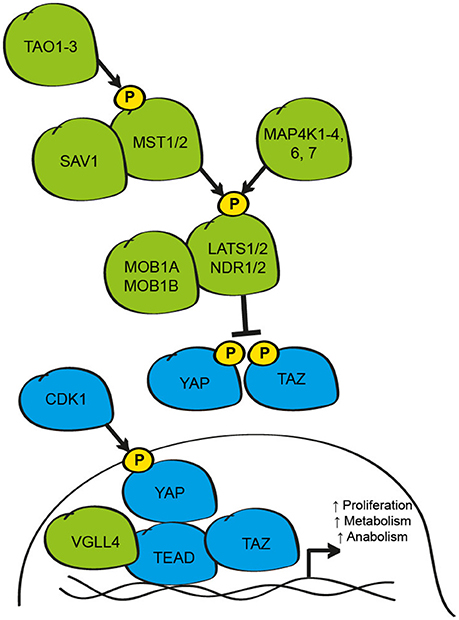
Figure 1. Schematic of the core elements of the mammalian Hippo signaling pathway. The core Hippo pathway kinases (TAO1-3, MST1/2, MAP4K1-4, 6, 7, LATS1/2, and NDR1/2), plus the adaptor proteins (SAV1 and MOB1A/B), function to inhibit the activity of the transcriptional co-activators YAP and TAZ/WWTR1 by phosphorylation at critical serine residues that results in cytoplasmic retention and/or protein degradation. When active, YAP and TAZ bind to TEAD transcription factors to regulate proliferative, metabolic and anabolic gene expression. YAP-TEAD signaling is limited by competitive interaction with VGLL-4. During mitosis, YAP activity can be enhanced by phosphorylation by CDK1. Elements inhibitory to YAP and TAZ activity are shown in green; elements that active/enhance YAP and TAZ activity are shown in blue.
YAP and TAZ are the primary effectors of the Hippo pathway in mammals, and are the homologs of the Drosophila gene Yorkie (Yki) (Huang et al., 2005). While the activity of these proteins can be influenced transcriptionally, YAP and TAZ are mainly regulated by post-translational modifications, in particular by phosphorylation on critical serine residues in a consensus motif (HXRXXS), by LATS1/2 and/or NDR1/2 (Zhao et al., 2007; Lei et al., 2008; Zhang et al., 2015). The importance of this mechanism was highlighted by studies where single and multiple point mutations of critical serine residues (Ser61, 109, 127, 164, and 381; mutated to alanine to create phosphorylation-resistant mutations) were introduced into the human YAP protein (Zhao et al., 2007). Using these mutant YAP proteins, the authors demonstrated that these serine residues are directly phosphorylated by activated LATS1/2 to limit the activity of YAP (Zhao et al., 2007). Similar findings have been reported for TAZ, where LATS phosphorylates Ser66, 89, 117, and 311 (Lei et al., 2008). Of these residues, the best studied are YAPSer127/TAZSer89 which influence sub-cellular localization and interaction with 14-3-3 binding proteins, and YAPSer381/TAZSer311 which function as priming sites for a second phosphorylation event at Ser384 by casein kinase 1γ/δ and the subsequent ubiquitination and degradation of YAP or TAZ by the E3 ligase, SCFβ−TRCP (Zhao et al., 2007, 2010; Liu et al., 2010). While the precise function of the other Serine residues remains unclear, these likely also influence YAP and TAZ activity since mutation of all 5 serine residues results in greater activation of the proteins than mutation of individual residues, at least in cultured cells (Zhao et al., 2009).
Counter to inhibitory phosphorylation by LATS1/2, in mitotically active cells during the G2-M phase of the cell cycle, YAP and TAZ activity is enhanced by phosphorylation by cyclin-dependent kinase 1 (CDK1) at multiple threonine and serine residues; findings that may explain the efficacy of CDK inhibitors under certain circumstances (Yang et al., 2013; Zhao et al., 2014; Pegoraro et al., 2015). Independent of direct inhibition by LATS1/2, the activity of YAP and TAZ can also be influenced by physical retention of the proteins in the cytosol by the Angiomotin family proteins (AMOT, AMOT1L, and AMOT2L) in a LATS1/2 dependent- or independent-manner (Chan et al., 2011; Paramasivam et al., 2011; Wang et al., 2011; Zhao et al., 2011), by disruption of protein-protein interactions necessary for transcriptional activity due to phosphorylation by kinases such as AMP-activated protein kinase (AMPK1) (DeRan et al., 2014; Mo et al., 2015; Wang et al., 2015), interaction with tyrosine kinases including YES and Src (Vassilev et al., 2001), the STRIPAK PP2A phosphatase complex (Ribeiro et al., 2010), methylation by the methyltransferase SET-7 at lysine 494 (Oudhoff et al., 2013), sumolyation by the promyelocytic leukemia protein (PML) (Lapi et al., 2008) and acetylation by the nuclear acetyltransferases CREB binding protein (CBP) and p300 (Hata et al., 2012). Collectively, the co-ordinated influence of these post-translational modifications functions to limit the activity of YAP and/or TAZ.
When Hippo pathway activity is low, YAP and TAZ accumulate predominantly in the nucleus of the cell where they can interact with transcription factors to regulate gene expression including p73, Tbx-5, Smad proteins, FoxO, and Runx (Strano et al., 2001; Ferrigno et al., 2002; Zaidi et al., 2004; Murakami et al., 2005; Varelas et al., 2010b; Rosenbluh et al., 2012; Shao et al., 2014). At this level, Hippo signaling may intersect with a number of other signaling pathways to control subsets of genes during specific cellular contexts (Alarcon et al., 2009; Azzolin et al., 2014). However, the most significant interacting partners of YAP and TAZ are the TEA domain family member transcription factors, (TEAD 1-4; Scalloped (Sd) in Drosophila) that are essential under most conditions for YAP and TAZ to promote growth (Vassilev et al., 2001; Wu et al., 2008; Zhang et al., 2008; Zhao et al., 2008). The interaction between YAP and TEAD, and TAZ and TEAD is mediated via a C-terminal binding domain in YAP/TAZ and an N-terminal binding domain in TEAD (Vassilev et al., 2001; Zhao et al., 2008). Solving the crystal structure of the YAP-TEAD and TAZ-TEAD complexes has identified the critical regions and residues necessary for this interaction (Chen et al., 2010; Li et al., 2010; Kaan et al., 2017). These studies support previous genetic and bio-chemical studies in vitro and in vivo, as well as genomic studies of individuals carrying mutations in TEAD1 at Tyr421 who develop Sveinsson's Chorioretinal atrophy, a rare autosomal-dominant disease that leads to degeneration of the photoreceptor cells in the eye (Fossdal et al., 2004).
While the over-expression of YAP or TAZ results in TEAD-dependent gene expression, the over-expression of TEAD or Sd alone leads to the depression of many of these target genes (Koontz et al., 2013). This fascinating observation lead to the demonstration that YAP binding to TEAD results in the de-repression of many of these genes, which under basal conditions are suppressed by TEAD in complex with the Vestigal-like 4 protein (VGLL-4; Tgi in Drosophila) (Koontz et al., 2013). In these elegant studies, it was demonstrated that under Hippo pathway active conditions, where YAP activity is suppressed, a TEAD/VGLL-4 complex represses the expression of target genes. When the Hippo pathway is inhibited, YAP physically competes with VGLL-4 for TEAD binding, resulting in de-repression of target genes (Koontz et al., 2013). In addition to the activation of YAP and TAZ, CDK1 also phosphorylates VGLL-4 during mitosis to limit binding to TEAD and repression of target genes (Zeng et al., 2017). While Yki/Sd binding occurs mainly in the promoter region of target genes (Oh et al., 2013), active YAP-TEAD and TAZ-TEAD complexes bind mainly to distal enhancer regions where they interact with elements of the transcriptional machinery such as the Mediator complex to influence gene expression (Galli et al., 2015; Kim et al., 2015; Zanconato et al., 2015).
Control of Organ Size by YAP and TAZ
The first identified, and best studied, role of the Hippo pathway is the regulation of organ size. This was initially demonstrated during the development of Drosophila melanogaster imaginal discs, where loss of upstream elements, or activation of Yki results in a profound over-growth phenotype of epithelial tissues, caused by ectopic cell proliferation, increased progression through the cell cycle and impaired apoptosis (Tapon et al., 2002; Harvey et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003; Huang et al., 2005; Li et al., 2015; Meng et al., 2015; Zheng et al., 2015). Subsequent studies have shown that YAP and TAZ function in a similar manner during the early stages of mammalian embryonic development, and in the specialized development of a number of mammalian tissues, demonstrating the conserved nature of this function for the Hippo pathway (Morin-Kensicki et al., 2006; Camargo et al., 2007; Dong et al., 2007). While the functional output and requirement for TEAD is conserved across species (Hilman and Gat, 2011), the magnitude and complexity of the genes controlled by YAP-TEAD and TAZ-TEAD complexes in mammals differs in tissue- and temporal-specific manners (Meng et al., 2016). Thus, it is likely that YAP and TAZ influence mammalian cell proliferation and apoptosis by controlling the expression of tissue-specific gene signatures, rather than a common subset of target genes. However, recent reports demonstrate in Drosophila and a number of mammalian cell types, that Yki/YAP activation is associated with the upregulation of genes encoding Hippo pathway upstream kinases and adaptor proteins, including Neurofibromin 2 (NF2) and LATS2 (Dai et al., 2015; Moroishi et al., 2015; Park G. S. et al., 2016). The induction of these genes forms a negative feedback loop that acts to limit the transcriptional activity of YAP and TAZ, and is likely a critical mechanism to ensure proper development of mammalian tissues indicating that some subsets of target genes are likely common between mammalian tissues (Park G. S. et al., 2016).
In addition to the well-described developmental role for YAP and TAZ, a number of studies now implicate these genes as critical gate-keepers of stem cell expansion and differentiation in adult tissues. The first example of this in mammals was in the mouse intestine where activation of YAP leads to the rapid proliferation of the undifferentiated stem/progenitor cell population (Camargo et al., 2007). Subsequent studies have demonstrated similar findings in the progenitor cells of the skin, intestine, brain and skeletal muscle (Cao et al., 2008; Gee et al., 2011; Schlegelmilch et al., 2011; Beverdam et al., 2013; Tremblay et al., 2014; Sun et al., 2017). Concurrently, appropriate regulation of tissue homeostasis and regeneration requires the subsequent down-regulation of YAP for terminal differentiation to proceed, as demonstrated in genetic mouse models expressing active YAP mutants, or lacking upstream signaling elements (Cao et al., 2008; Lian et al., 2010; Judson et al., 2012; Tremblay et al., 2014; Sun et al., 2017). While YAP and TAZ function in a redundant manner in many settings, this appears more complex during the differentiation of skeletal muscle cells where YAP inhibits, while TAZ promotes terminal differentiation and regeneration (Jeong et al., 2010; Watt et al., 2010; Park G. H. et al., 2014; Mohamed et al., 2016; Sun et al., 2017). These findings have broad implications for regenerative medicine and suggest that appropriate modulation of YAP and/or TAZ activity may be an approach that can be utilized to treat degenerative conditions. However, the efficacy of such an approach is likely to be tissue- and context-dependent, as demonstrated in the mammalian heart where YAP activation can enhance the regenerative capacity of the heart, but only at early stages of post-natal life (Xin et al., 2013). Caution is also warranted with strategies aimed at modulating YAP activity in tissues such as the skin which display profound phenotypes upon activation and inhibition of YAP-TEAD activity in the adult (Schlegelmilch et al., 2011). However, since not all tissue types display phenotypes in response to loss of YAP or TAZ under basal conditions e.g., the intestine or mammary gland (Cai et al., 2010; Chen et al., 2014), targeting this pathway in a tissue-specific manner may still offer great promise for the treatment of a number of conditions.
In addition to the role of the pathway as a regulator of cell division and differentiation, growing evidence supports a role for the Hippo pathway as a mediator of cell size in post-mitotic tissues such as skeletal muscle and the heart, where activation of YAP directly, or by inhibition of LATS2, causes hypertrophy by influencing the rates of protein synthesis, without altering cell number (Matsui et al., 2008; Goodman et al., 2015; Watt et al., 2015). As in epithelial cells, the ability for YAP to regulate basal tissue size in striated muscle appears mostly dependent on TEAD (Watt et al., 2015) providing further evidence for the critical role for the YAP-TEAD and TAZ-TEAD complexes as the primary effectors of Hippo signaling in mammals.
Integration of Hormonal Signaling by the Hippo Pathway
One common approach that cells utilize to modulate intra-cellular responses to changes in the external environment is the synthesis and release of soluble factors such as hormones, growth factors and diffusible molecules including insulin and glucose (Bedinger and Adams, 2015). While genetic studies were instrumental for elucidating the critical intracellular signaling proteins that regulate YAP and TAZ activity, defining how the Hippo pathway is activated by external stimuli to respond to changes in the environment has proved more elusive. Studies exploring this area demonstrated that soluble factors including lysophosphatidic acid (LPA), Sphingosine-1-phosphate (S1P), epinephrine, estrogen, and glucagon can activate, or inhibit, YAP and TAZ via the Hippo pathway by their cognate G-protein coupled receptors (Miller et al., 2012; Yu et al., 2012; Zhou et al., 2015; Figure 2). When activated by such soluble factors, these receptors recruit and couple with their associated G-protein sub-units to engage the small GTPases RHOA and RAC1 leading to alterations in LATS1/2 activity (Yu et al., 2012; Plouffe et al., 2016). In most cases, stimulation of Gαq/11, Gα12/13, and Gαi/o subunits is reported to activate YAP and TAZ, while Gαs subunits inhibit; however, this is likely dependent on many factors including receptor recycling rate, G-subunit protein levels, and expression of downstream signaling effector proteins meaning that individual sub-units could both activate, or inhibit, YAP and TAZ depending on the cellular context (Yu et al., 2012; Plouffe et al., 2016). Of note, activating mutations in G protein sub-units are observed in a number of cancer sub-types that display increased YAP protein (Yu et al., 2012; Feng et al., 2014; Zhou et al., 2015).
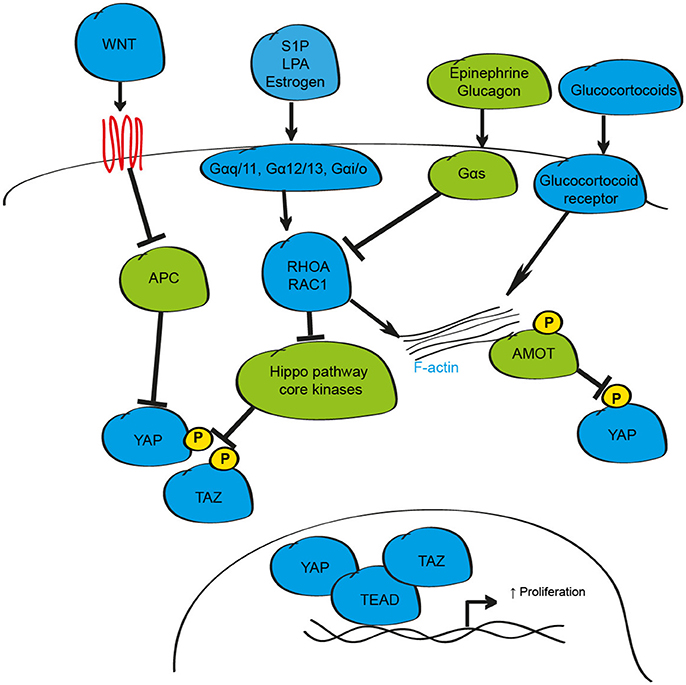
Figure 2. Schematic depicting the hormonal regulation of the mammalian Hippo signaling pathway. In response to hormonal stimuli including S1P, LPA, Estrogen, Epinephrine and Glucagon, the activity of YAP and TAZ can be enhanced via the GPCR G-proteins Gαq/11, Gα12/13, or Gαi/o, or inhibited via Gαs signaling axis. When active, these stimuli influence YAP and TAZ phosphorylation/localization through modulation of F-actin/RHOA/RAC1. YAP can also be phosphorylated and sequestered by the AMOT family of proteins. In addition to regulation by GPCR signaling, YAP, and TAZ activity is altered by secreted proteins such as WNT and by glucocorticoids.
A number of soluble signaling proteins that act independently of the G-protein coupled receptor signaling pathways have also been proposed to influence cell proliferation, differentiation and inflammation through YAP and TAZ including Glucocorticoids, Epidermal Growth Factor Receptor (EGFR), Insulin-like growth factor-1, WNT, TGF-β, Bone Morphogenic Proteins (BMPs) and the cytokines leukemia inhibitory factor (LIF) and Interlukin-6 (IL6) (Alarcon et al., 2009; Tamm et al., 2011; Strassburger et al., 2012; Reddy and Irvine, 2013; Azzolin et al., 2014; Haskins et al., 2014; Park H. W. et al., 2015; Taniguchi et al., 2015; Sorrentino et al., 2017). The importance of these interactions with Hippo signaling in many cell and tissue types, especially during non-pathological conditions, has yet to be demonstrated. However, a growing body of evidence in multiple tissues supports the suggestion that interactions at multiple levels with the WNT signaling pathway may influence YAP and TAZ activity to alter tissue growth in multiple settings. The interaction between these pathways is complex and likely context-dependent, but can include transcriptional regulation of YAP expression, effects on YAP and TAZ protein stability and localization, and modulation of Hippo pathway target gene expression (Varelas et al., 2010a; Heallen et al., 2011; Azzolin et al., 2012, 2014; Rosenbluh et al., 2012). In addition to WNT-mediated control of Hippo signaling, YAP-TEAD, and TAZ-TEAD may also influence WNT activity through transcriptional regulation of WNT signaling pathway elements (Heallen et al., 2011).
Hippo Signaling and Mechano-Transduction
In addition to soluble molecules, regulation of physiological function in the local microenvironment is highly dependent on changes in mechanical properties derived from alterations in cell shape, fluid shear-stress or cell-cell contact (Finch-Edmondson and Sudol, 2016). Recent experimental evidence highlights a role for YAP and TAZ as critical effectors of mechanical signals (Figure 3). The first findings linking changes in physical dynamics to Hippo signaling came from studies showing that as cells divide and make contact with neighboring cells, YAP phosphorylation increased and was re-localized to the cytosol of the cell (Zhao et al., 2007). Growing evidence supports the conclusion that in addition, and in isolation of cell-cell contact, changes in mechanical properties of the ECM, cell geometry and polymerisation of the F-actin cytoskeleton are essential inputs that influence the activity of YAP and TAZ (Dupont et al., 2011; Wada et al., 2011; Aragona et al., 2013; Bertrand et al., 2014).
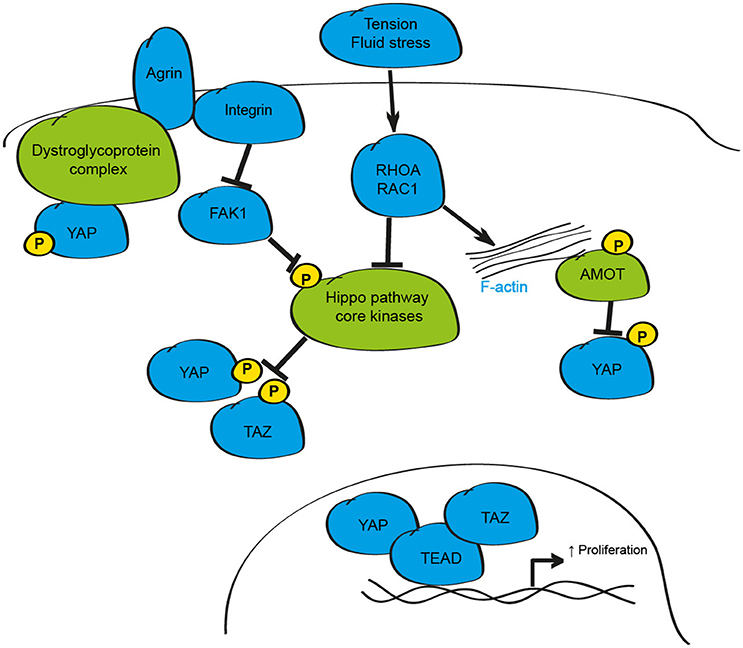
Figure 3. Schematic depicting the mechanical regulation of the mammalian Hippo signaling pathway. Changes in mechanical tension, shear stress, and cell-cell contact can alter YAP and TAZ activity through the F-actin/RHOA/RAC1 axis. Additionally, the proteoglycan Agrin modulates YAP and TAZ activity via Integrin/FAK-mediated activation of the Hippo core kinases and by modulation of the dystroglyoprotein complex.
When grown in isolation on stiff substrates, or under conditions where cells can spread, YAP and TAZ are active and localize to the nucleus (Dupont et al., 2011; Wada et al., 2011; Aragona et al., 2013). However, in soft matrices, or when cultured in small surface areas so that they are compressed, YAP and TAZ are inhibited and re-localize to the cytosol (Dupont et al., 2011; Aragona et al., 2013). These effects were proposed to be mediated by Hippo pathway dependent- and independent-mechanisms; however, as in the case of soluble factors, redundancy with kinases such as NDR1/2 was not tested, and so the precise mechanism of action remains unclear. Despite this, studies assessing the mechanisms downstream of mechanical cues collectively demonstrate that mechanical stimuli influence YAP and TAZ activity through alterations in F-actin polymerisation, and by changes in the activity of the small GTPases RHOA and RAC1 (Dupont et al., 2011; Fernandez et al., 2011; Sansores-Garcia et al., 2011; Aragona et al., 2013; Plouffe et al., 2016). Disruption of the F-actin-RHOA/RAC1-YAP/TAZ pathway has been implicated in the specification of cell identify during development, in the activation and differentiation of various cells and in the maintenance of vertebrate three-dimensional tissue structure and tension, as well as in response to fluid flow stresses to influence the stability of blood vessels during development and the formation atherosclerotic plaques in APOE−/− mice, demonstrating the significance of this mode of regulation to a number of cellular processes and disease-relevant settings (Sansores-Garcia et al., 2011; Calvo et al., 2013; Liu et al., 2015; Porazinski et al., 2015; Wang K. C. et al., 2016; Wang L. et al., 2016; Nakajima et al., 2017; Totaro et al., 2017).
The critical pathways that are required to relay external mechanical forces in the extracellular matrix to influence Hippo signaling in mammalian tissues have only recently been identified. Studies in liver and heart cells suggest that Agrin, a proteoglycan that regulates formation and maintenance of the neuromuscular junction, and the dystroglycoprotein-complex (DGC), a multiprotein complex that links the extra-cellular matrix and actin cytoskeleton, may both influence Hippo signaling in mammalian tissues (Bassat et al., 2017; Chakraborty et al., 2017; Morikawa et al., 2017). While Agrin, via Integrin/PAK1 signaling, regulates YAP activity through the core Hippo kinases (Chakraborty et al., 2017), the DGC appears to limit YAP activity by physical interaction of phosphorylated YAP through the DGC component Dag1 (Morikawa et al., 2017). Recent evidence in cardiomyocytes suggests that Agrin-dependent activation of YAP is also mediated by disruption of the DGC complex in settings of cardiomyocyte regeneration providing further evidence for this axis as a regulator of YAP activity (Bassat et al., 2017). Given the critical role of Agrin-Integrin and DGC protein complexes during development, and in neurodegenerative and neuromuscular diseases, further study of these mechanistic links may offer exciting strategies that can be exploited to modulate YAP and TAZ activity in a range of disease states.
Cellular Metabolism and the Hippo Pathway
Integration of tissue-specific, and whole-body, metabolic signaling responses is essential to ensure homeostasis under basal conditions. Furthermore, the dysregulation of metabolic pathways in many tissues from lipid metabolism toward anaerobic glycolysis (the Warburg effect) is an established event in the development and progression of many cancers, and in the activation of stem cell populations (Agathocleous and Harris, 2013; Zhou et al., 2017). Recent studies highlight a number of essential metabolic pathways that appear to function via modulation of YAP and TAZ (Figure 4).
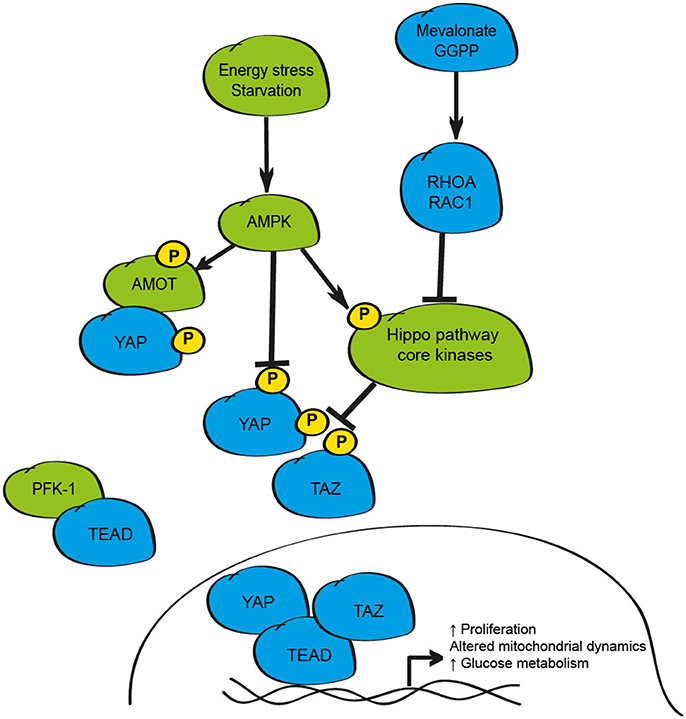
Figure 4. Schematic depicting the metabolic regulation of the mammalian Hippo signaling pathway. Energy stress as observed in settings of glucose withdrawal/starvation inhibits YAP activity via activation of AMPK leading to phosphorylation of AMOT, YAP, or disruption of the YAP/TEAD complex. Metabolic pathways including the mevalonate/HMD CoA reductase-GGPP pathway also influence YAP and TAZ activity via RHOA/RAC1. Activity of this pathway can also be limited by disruption of YAP-TEAD binding by PFK-1.
The first link between the Hippo pathway and cellular metabolism was the demonstration that the inhibition of the mevalonate/HMG-CoA reductase pathway, a critical mediator of cholesterol and isoprenoids production, was sufficient to promote cytoplasmic retention, and to impair the transcriptional activity of YAP and TAZ in breast cancer cells (Sorrentino et al., 2014; Wang et al., 2014). The mevalonate pathway acts to enhance YAP and TAZ activity via geranylgeranyl pyrophosphate (GGPP) and RHOA suggesting that the small GTPases RHOA and RAC1 may form common intercellular mediators of YAP/TAZ function in response to metabolic, hormonal and mechanical stimuli (Sorrentino et al., 2014; Wang et al., 2014). The significance of these findings in non-cancerous cell types is less clear; yet inhibition of HMG-CoA reductase by statin exposure in developing mouse embryos prevents blastocyst formation and impairs YAP activity highlighting a role for this axis during early mammalian development (Alarcon and Marikawa, 2016).
The activity of YAP and TAZ can also be impacted by the cellular concentrations of glucose and glucose-responsive pathways (Adler et al., 2013; DeRan et al., 2014; Enzo et al., 2015; Mo et al., 2015; Wang et al., 2015). Energy stress, as induced by culturing cells in glucose-free conditions, results in inhibition of YAP activity in mouse hepatocytes in vivo as demonstrated by starvation/re-feeding experiments (Wang et al., 2015). While these studies collectively demonstrate that glucose withdrawal inhibits YAP activity, the underlying mechanisms reported differ between studies and include AMPK1-mediated phosphorylation of AMOTL1, direct phosphorylation of YAP on multiple serine residues by AMPK1, disruption of the YAP-TEAD complex by AMPK1 phosphorylation at Ser94, and physical interaction between TEADs and phosphofructokinase-1 (PFK-1), a rate-limiting enzyme in the glycolysis pathway (Adler et al., 2013; DeRan et al., 2014; Enzo et al., 2015; Mo et al., 2015; Wang et al., 2015).
In addition to being responsive to changes in cellular metabolism, the Hippo pathway appears to control key metabolic components to influence cell proliferation under certain conditions including transcriptional control of glucose transporters, gluconeogenic gene expression, amino acid transporters, and elements of the glutamine metabolism pathways (Hansen et al., 2015; Wang et al., 2015; Cox et al., 2016; Park Y. Y. et al., 2016; Hu et al., 2017). Further to the effect of these metabolism-induced changes on cell division, the manipulation of YAP and TAZ can also influence mitochondrial dynamics, through enhancing the rates of mitochondrial fusion and fission, by transcriptional up-regulation of genes such as Opa-1 and Mfn2, critical elements controlling these vital cellular processes (Nagaraj et al., 2012; von Eyss et al., 2015). These findings highlight the interplay between Hippo signaling and multiple metabolic processes and demonstrate further mechanisms of growth control by this pathway that could be exploited in settings such as cancer, metabolic disease or regenerative medicine.
Concluding Remarks
Here, we summarize evidence supporting an essential role for the Hippo signaling pathway as a critical element integrating hormonal, mechanical and metabolic stimuli during the growth and differentiation of mammalian tissues during development, in the post-natal environment, and in the progression of specific disease states. Together, these findings suggest that targeting the Hippo pathway may be beneficial in particular clinical settings, with inhibition of the pro-proliferative/anti-apoptotic function of YAP and TAZ during the growth of cancer cells of particular interest. Potential approaches that could be used to achieve this goal include inhibitors of the mevalonate pathway (Statins, Bisphosphonates and Geranylgeranyl-transferase inhibitors), RHO GTPases (ROCK inhibitors), or small molecules that disrupt the interaction between YAP and TEAD, such as Verteporfin; many of which are used in clinical practice for other conditions (Dupont et al., 2011; Liu-Chittenden et al., 2012; Sorrentino et al., 2014; Wang et al., 2014). However, studies using these agents have typically limited their analyses to a single cell/tissue type and so the consequences on whole-body physiology are not clear. A greater understanding of this will be critical for the successful development of therapeutics targeting YAP and TAZ; particularly in light of the detrimental effects of inhibiting basal levels of YAP activity in tissues such as skeletal muscle and skin (Schlegelmilch et al., 2011; Watt et al., 2015). Targeting elements of the Hippo pathway using lower effective doses of agents, but as part of a combinatorial therapeutic approach, may therefore be an alternative means to inhibit the Hippo pathway while limiting potential detrimental consequences on other tissues. Such approaches appear to be effective in a number of cell types (Lin et al., 2015; Li et al., 2016; Zhao et al., 2017).
A major limitation of studies investigating the function of the Hippo pathway is the use of genetic manipulation whereby pathway members are exogenously over-expressed or completely deleted from a target tissue; yet complete deletion of pathway members in healthy or diseased tissues is not commonly observed (the exception being the NF2 gene in select cancers; Harvey et al., 2013). Consequently, assessing the function of Hippo pathway members in specific tissues at physiological and appropriate patho-physiological levels using more elegant approaches such as genome editing with CRISPR/CAS9, where the introduction of point-mutations, protein domain deletions, and activation of genes at the endogenous locus is possible may reveal more relevant information about the role of this pathway in the progression of disease pathologies.
While the significance of the Hippo pathway in many aspects of mammalian biology is becoming increasingly clear, evidence of a role for the Hippo pathway in cellular processes such as the response to hypoxia, autophagy and the unfolded protein response is limited to various tumor cell types, tissue culture models, or specific tissues (Liang et al., 2014; Yan et al., 2014; Wu et al., 2015). Future work will be required to explore how significant alterations in the activity of the Hippo pathway are to these critical cellular functions in non-cancerous tissues if we are to understand the significance of the Hippo pathway in these processes.
In summary, the published literature to date implicates the Hippo signaling pathway as a mediator of tissue growth and function in response to diverse physiological cues. While our understanding of the requirement for the Hippo pathway in mammals has expanded in recent years, the consequence of altered Hippo signaling in specific tissues on whole-body physiology, and the effect of such changes upon disease progression remains unexplored. Future studies aimed at understanding the integrative nature of the Hippo pathway on human physiology will be required to reveal the extent that this pathway influences biological function and the implications of targeting this pathway for clinical benefit.
Author Contributions
KW, KH, and PG wrote and edited the manuscript.
Funding
The work related to this project was partially supported by a Diabetes Australia general grant awarded to KW (Y16G-WATK), a project grant (1099588) awarded to KH and PG from the Australian National Health and Medical Research Council (NHMRC). KH and PG hold Senior Research Fellowships from the NHMRC. The Baker Heart and Diabetes Institute is supported in part by the Operational Infrastructure Support Program of the Victorian Government.
Conflict of Interest Statement
The handling Editor declared a shared affiliation, though no other collaboration, with the authors KW and PG.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adler, J. J., Johnson, D. E., Heller, B. L., Bringman, L. R., Ranahan, W. P., Conwell, M. D., et al. (2013). Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc. Natl. Acad. Sci. U.S.A. 110, 17368–17373. doi: 10.1073/pnas.1308236110
Agathocleous, M., and Harris, W. A. (2013). Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol. 23, 484–492. doi: 10.1016/j.tcb.2013.05.004
Alarcon, C., Zaromytidou, A. I., Xi, Q., Gao, S., Yu, J., Fujisawa, S., et al. (2009). Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139, 757–769. doi: 10.1016/j.cell.2009.09.035
Alarcon, V. B., and Marikawa, Y. (2016). Statins inhibit blastocyst formation by preventing geranylgeranylation. Mol. Hum. Reprod. 22, 350–363. doi: 10.1093/molehr/gaw011
Aragona, M., Panciera, T., Manfrin, A., Giulitti, S., Michielin, F., Elvassore, N., et al. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059. doi: 10.1016/j.cell.2013.07.042
Azzolin, L., Panciera, T., Soligo, S., Enzo, E., Bicciato, S., Dupont, S., et al. (2014). YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170. doi: 10.1016/j.cell.2014.06.013
Azzolin, L., Zanconato, F., Bresolin, S., Forcato, M., Basso, G., Bicciato, S., et al. (2012). Role of TAZ as mediator of Wnt signaling. Cell 151, 1443–1456. doi: 10.1016/j.cell.2012.11.027
Bassat, E., Mutlak, Y. E., Genzelinakh, A., Shadrin, I. Y., Baruch Umansky, K., Yifa, O., et al. (2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547, 179–184. doi: 10.1038/nature22978
Bedinger, D. H., and Adams, S. H. (2015). Metabolic, anabolic, and mitogenic insulin responses: a tissue-specific perspective for insulin receptor activators. Mol. Cell. Endocrinol. 415, 143–156. doi: 10.1016/j.mce.2015.08.013
Bertrand, A. T., Ziaei, S., Ehret, C., Duchemin, H., Mamchaoui, K., Bigot, A., et al. (2014). Cellular microenvironments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J. Cell Sci. 127(Pt 13), 2873–2884. doi: 10.1242/jcs.144907
Beverdam, A., Claxton, C., Zhang, X., James, G., Harvey, K. F., and Key, B. (2013). Yap controls stem/progenitor cell proliferation in the mouse postnatal epidermis. J. Invest. Dermatol. 133, 1497–1505. doi: 10.1038/jid.2012.430
Boggiano, J. C., Vanderzalm, P. J., and Fehon, R. G. (2011). Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21, 888–895. doi: 10.1016/j.devcel.2011.08.028
Bray, S. J. (2016). Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735. doi: 10.1038/nrm.2016.94
Cai, J., Zhang, N., Zheng, Y., de Wilde, R. F., Maitra, A., and Pan, D. (2010). The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383–2388. doi: 10.1101/gad.1978810
Callus, B. A., Verhagen, A. M., and Vaux, D. L. (2006). Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 273, 4264–4276. doi: 10.1111/j.1742-4658.2006.05427.x
Calvo, F., Ege, N., Grande-Garcia, A., Hooper, S., Jenkins, R. P., Chaudhry, S. I., et al. (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646. doi: 10.1038/ncb2756
Camargo, F. D., Gokhale, S., Johnnidis, J. B., Fu, D., Bell, G. W., Jaenisch, R., et al. (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060. doi: 10.1016/j.cub.2007.10.039
Cao, X., Pfaff, S. L., and Gage, F. H. (2008). YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 22, 3320–3334. doi: 10.1101/gad.1726608
Chakraborty, S., Njah, K., Pobbati, A. V., Lim, Y. B., Raju, A., Lakshmanan, M., et al. (2017). Agrin as a mechanotransduction signal regulating YAP through the Hippo pathway. Cell Rep. 18, 2464–2479. doi: 10.1016/j.celrep.2017.02.041
Chan, E. H., Nousiainen, M., Chalamalasetty, R. B., Schafer, A., Nigg, E. A., and Sillje, H. H. (2005). The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076–2086. doi: 10.1038/sj.onc.1208445
Chan, S. W., Lim, C. J., Chong, Y. F., Pobbati, A. V., Huang, C., and Hong, W. (2011). Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 286, 7018–7026. doi: 10.1074/jbc.C110.212621
Chen, L., Chan, S. W., Zhang, X., Walsh, M., Lim, C. J., Hong, W., et al. (2010). Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 24, 290–300. doi: 10.1101/gad.1865310
Chen, Q., Zhang, N., Gray, R. S., Li, H., Ewald, A. J., Zahnow, C. A., et al. (2014). A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 28, 432–437. doi: 10.1101/gad.233676.113
Cox, A. G., Hwang, K. L., Brown, K. K., Evason, K., Beltz, S., Tsomides, A., et al. (2016). Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol. 18, 886–896. doi: 10.1038/ncb3389
Dai, X., Liu, H., Shen, S., Guo, X., Yan, H., Ji, X., et al. (2015). YAP activates the Hippo pathway in a negative feedback loop. Cell Res. 25, 1175–1178. doi: 10.1038/cr.2015.101
DeRan, M., Yang, J., Shen, C. H., Peters, E. C., Fitamant, J., Chan, P., et al. (2014). Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 9, 495–503. doi: 10.1016/j.celrep.2014.09.036
Dong, J., Feldmann, G., Huang, J., Wu, S., Zhang, N., Comerford, S. A., et al. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133. doi: 10.1016/j.cell.2007.07.019
Dupont, S., Morsut, L., Aragona, M., Enzo, E., Giulitti, S., Cordenonsi, M., et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183. doi: 10.1038/nature10137
Enzo, E., Santinon, G., Pocaterra, A., Aragona, M., Bresolin, S., Forcato, M., et al. (2015). Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 34, 1349–1370. doi: 10.15252/embj.201490379
Feng, X., Degese, M. S., Iglesias-Bartolome, R., Vaque, J. P., Molinolo, A. A., Rodrigues, M., et al. (2014). Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 25, 831–845. doi: 10.1016/j.ccr.2014.04.016
Fernandez, B. G., Gaspar, P., Bras-Pereira, C., Jezowska, B., Rebelo, S. R., and Janody, F. (2011). Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337–2346. doi: 10.1242/dev.063545
Ferrigno, O., Lallemand, F., Verrecchia, F., L'Hoste, S., Camonis, J., Atfi, A., et al. (2002). Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-β/Smad signaling. Oncogene 21, 4879–4884. doi: 10.1038/sj.onc.1205623
Finch-Edmondson, M., and Sudol, M. (2016). Framework to function: mechanosensitive regulators of gene transcription. Cell. Mol. Biol. Lett. 21:28. doi: 10.1186/s11658-016-0028-7
Fossdal, R., Jonasson, F., Kristjansdottir, G. T., Kong, A., Stefansson, H., Gosh, S., et al. (2004). A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration). Hum. Mol. Genet. 13, 975–981. doi: 10.1093/hmg/ddh106
Galli, G. G., Carrara, M., Yuan, W. C., Valdes-Quezada, C., Gurung, B., Pepe-Mooney, B., et al. (2015). YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol. Cell 60, 328–337. doi: 10.1016/j.molcel.2015.09.001
Gee, S. T., Milgram, S. L., Kramer, K. L., Conlon, F. L., and Moody, S. A. (2011). Yes-associated protein 65 (YAP) expands neural progenitors and regulates Pax3 expression in the neural plate border zone. PLoS ONE 6:e20309. doi: 10.1371/journal.pone.0020309
Goodman, C. A., Dietz, J. M., Jacobs, B. L., McNally, R. M., You, J. S., and Hornberger, T. A. (2015). Yes-Associated Protein is up-regulated by mechanical overload and is sufficient to induce skeletal muscle hypertrophy. FEBS Lett. 589, 1491–1497. doi: 10.1016/j.febslet.2015.04.047
Gregorieff, A., and Wrana, J. L. (2017). Hippo signalling in intestinal regeneration and cancer. Curr. Opin. Cell Biol. 48, 17–25. doi: 10.1016/j.ceb.2017.04.005
Hansen, C. G., Ng, Y. L., Lam, W. L., Plouffe, S. W., and Guan, K. L. (2015). The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 25, 1299–1313. doi: 10.1038/cr.2015.140
Harvey, K. F., Pfleger, C. M., and Hariharan, I. K. (2003). The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467. doi: 10.1016/S0092-8674(03)00557-9
Harvey, K. F., Zhang, X., and Thomas, D. M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257. doi: 10.1038/nrc3458
Haskins, J. W., Nguyen, D. X., and Stern, D. F. (2014). Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci. Signal. 7:ra116. doi: 10.1126/scisignal.2005770
Hata, A., and Chen, Y.-G. (2016). TGF-β signaling from receptors to smads. Cold Spring Harb. Perspect. Biol. 8:a022061. doi: 10.1101/cshperspect.a022061
Hata, S., Hirayama, J., Kajiho, H., Nakagawa, K., Hata, Y., Katada, T., et al. (2012). A novel acetylation cycle of transcription co-activator Yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents. J. Biol. Chem. 287, 22089–22098. doi: 10.1074/jbc.M111.334714
He, J., Bao, Q., Yan, M., Liang, J., Zhu, Y., Wang, C., et al. (2017). The role of Hippo/yes-associated protein signalling in vascular remodelling associated with cardiovascular disease. Br. J. Pharmacol. doi: 10.1111/bph.13806. [Epub ahead of print].
Heallen, T., Zhang, M., Wang, J., Bonilla-Claudio, M., Klysik, E., Johnson, R. L., et al. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461. doi: 10.1126/science.1199010
Hergovich, A., Kohler, R. S., Schmitz, D., Vichalkovski, A., Cornils, H., and Hemmings, B. A. (2009). The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 19, 1692–1702. doi: 10.1016/j.cub.2009.09.020
Hilman, D., and Gat, U. (2011). The evolutionary history of YAP and the hippo/YAP pathway. Mol. Biol. Evol. 28, 2403–2417. doi: 10.1093/molbev/msr065
Hu, Y., Shin, D.-J., Pan, H., Lin, Z., Dreyfuss, J. M., Camargo, F. D., et al. (2017). YAP suppresses gluconeogenic gene expression through PGC1α. Hepatology 66, 2029–2041. doi: 10.1002/hep.29373
Huang, J., Wu, S., Barrera, J., Matthews, K., and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434. doi: 10.1016/j.cell.2005.06.007
Jeong, H., Bae, S., An, S. Y., Byun, M. R., Hwang, J. H., Yaffe, M. B., et al. (2010). TAZ as a novel enhancer of MyoD-mediated myogenic differentiation. FASEB J. 24, 3310–3320. doi: 10.1096/fj.09-151324
Judson, R. N., Tremblay, A. M., Knopp, P., White, R. B., Urcia, R., De Bari, C., et al. (2012). The Hippo pathway member Yap plays a key role in influencing fate decisions in muscle satellite cells. J. Cell Sci. 125(Pt 24), 6009–6019. doi: 10.1242/jcs.109546
Kaan, H. Y. K., Chan, S. W., Tan, S. K. J., Guo, F., Lim, C. J., Hong, W., et al. (2017). Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex. Sci. Rep. 7:2035. doi: 10.1038/s41598-017-02219-9
Kim, M., Kim, T., Johnson, R. L., and Lim, D. S. (2015). Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 11, 270–282. doi: 10.1016/j.celrep.2015.03.015
Koontz, L. M., Liu-Chittenden, Y., Yin, F., Zheng, Y., Yu, J., Huang, B., et al. (2013). The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell 25, 388–401. doi: 10.1016/j.devcel.2013.04.021
Lai, Z. C., Wei, X., Shimizu, T., Ramos, E., Rohrbaugh, M., Nikolaidis, N., et al. (2005). Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120, 675–685. doi: 10.1016/j.cell.2004.12.036
Lapi, E., Di Agostino, S., Donzelli, S., Gal, H., Domany, E., Rechavi, G., et al. (2008). PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol. Cell 32, 803–814. doi: 10.1016/j.molcel.2008.11.019
Lei, Q. Y., Zhang, H., Zhao, B., Zha, Z. Y., Bai, F., Pei, X. H., et al. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28, 2426–2436. doi: 10.1128/MCB.01874-07
Li, H., Huang, Z., Gao, M., Huang, N., Luo, Z., Shen, H., et al. (2016). Inhibition of YAP suppresses CML cell proliferation and enhances efficacy of imatinib in vitro and in vivo. J. Exp. Clin. Cancer Res. 35, 134. doi: 10.1186/s13046-016-0414-z
Li, S., Cho, Y. S., Yue, T., Ip, Y. T., and Jiang, J. (2015). Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discov 1:15038. doi: 10.1038/celldisc.2015.38
Li, Z., Zhao, B., Wang, P., Chen, F., Dong, Z., Yang, H., et al. (2010). Structural insights into the YAP and TEAD complex. Genes Dev. 24, 235–240. doi: 10.1101/gad.1865810
Lian, I., Kim, J., Okazawa, H., Zhao, J., Zhao, B., Yu, J., et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106–1118. doi: 10.1101/gad.1903310
Liang, N., Zhang, C., Dill, P., Panasyuk, G., Pion, D., Koka, V., et al. (2014). Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J. Exp. Med. 211, 2249–2263. doi: 10.1084/jem.20140341
Lin, L., Sabnis, A. J., Chan, E., Olivas, V., Cade, L., Pazarentzos, E., et al. (2015). The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47, 250–256. doi: 10.1038/ng.3218
Liu, C. Y., Zha, Z. Y., Zhou, X., Zhang, H., Huang, W., Zhao, D., et al. (2010). The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ−TrCP E3 ligase. J. Biol. Chem. 285, 37159–37169. doi: 10.1074/jbc.M110.152942
Liu, F., Lagares, D., Choi, K. M., Stopfer, L., Marinkovic, A., Vrbanac, V., et al. (2015). Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 308, L344–357. doi: 10.1152/ajplung.00300.2014
Liu-Chittenden, Y., Huang, B., Shim, J. S., Chen, Q., Lee, S. J., Anders, R. A., et al. (2012). Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305. doi: 10.1101/gad.192856.112
Masuda, T., and Ishitani, T. (2017). Context-dependent regulation of the β-catenin transcriptional complex supports diverse functions of Wnt/β-catenin signaling. J. Biochem. 161, 9–17. doi: 10.1093/jb/mvw072
Matsui, Y., Nakano, N., Shao, D., Gao, S., Luo, W., Hong, C., et al. (2008). Lats2 is a negative regulator of myocyte size in the heart. Circ. Res. 103, 1309–1318. doi: 10.1161/CIRCRESAHA.108.180042
Meng, Z., Moroishi, T., and Guan, K. L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17. doi: 10.1101/gad.274027.115
Meng, Z., Moroishi, T., Mottier-Pavie, V., Plouffe, S. W., Hansen, C. G., Hong, A. W., et al. (2015). MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6:8357. doi: 10.1038/ncomms9357
Miller, E., Yang, J., DeRan, M., Wu, C., Su, A. I., Bonamy, G. M., et al. (2012). Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 19, 955–962. doi: 10.1016/j.chembiol.2012.07.005
Mo, J. S., Meng, Z., Kim, Y. C., Park, H. W., Hansen, C. G., Kim, S., et al. (2015). Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 17, 500–510. doi: 10.1038/ncb3111
Mohamed, A., Sun, C., De Mello, V., Selfe, J., Missiaglia, E., Shipley, J., et al. (2016). The Hippo effector TAZ (WWTR1) transforms myoblasts and TAZ abundance is associated with reduced survival in embryonal rhabdomyosarcoma. J. Pathol. 240, 3–14. doi: 10.1002/path.4745
Morikawa, Y., Heallen, T., Leach, J., Xiao, Y., and Martin, J. F. (2017). Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 547, 227–231. doi: 10.1038/nature22979
Morin-Kensicki, E. M., Boone, B. N., Howell, M., Stonebraker, J. R., Teed, J., Alb, J. G., et al. (2006). Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell. Biol. 26, 77–87. doi: 10.1128/MCB.26.1.77-87.2006
Moroishi, T., Park, H. W., Qin, B., Chen, Q., Meng, Z., Plouffe, S. W., et al. (2015). A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284. doi: 10.1101/gad.262816.115
Murakami, M., Nakagawa, M., Olson, E. N., and Nakagawa, O. (2005). A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl. Acad. Sci. U.S.A. 102, 18034–18039. doi: 10.1073/pnas.0509109102
Nagaraj, R., Gururaja-Rao, S., Jones, K. T., Slattery, M., Negre, N., Braas, D., et al. (2012). Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 26, 2027–2037. doi: 10.1101/gad.183061.111
Nakajima, H., Yamamoto, K., Agarwala, S., Terai, K., Fukui, H., Fukuhara, S., et al. (2017). Flow-dependent endothelial YAP regulation contributes to vessel maintenance. Dev. Cell 40, 523–536 e526. doi: 10.1016/j.devcel.2017.02.019
Oh, H., Slattery, M., Ma, L., Crofts, A., White, K. P., Mann, R. S., et al. (2013). Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 3, 309–318. doi: 10.1016/j.celrep.2013.01.008
Oudhoff, M. J., Freeman, S. A., Couzens, A. L., Antignano, F., Kuznetsova, E., Min, P. H., et al. (2013). Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell 26, 188–194. doi: 10.1016/j.devcel.2013.05.025
Pantalacci, S., Tapon, N., and Leopold, P. (2003). The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927. doi: 10.1038/ncb1051
Paramasivam, M., Sarkeshik, A., Yates, J. R. III., Fernandes, M. J., and McCollum, D. (2011). Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell 22, 3725–3733. doi: 10.1091/mbc.E11-04-0300
Park, G. H., Jeong, H., Jeong, M. G., Jang, E. J., Bae, M. A., Lee, Y. L., et al. (2014). Novel TAZ modulators enhance myogenic differentiation and muscle regeneration. Br. J. Pharmacol. 171, 4051–4061. doi: 10.1111/bph.12755
Park, G. S., Oh, H., Kim, M., Kim, T., Johnson, R. L., Irvine, K. D., et al. (2016). An evolutionarily conserved negative feedback mechanism in the Hippo pathway reflects functional difference between LATS1 and LATS2. Oncotarget 7, 24063–24075. doi: 10.18632/oncotarget.8211
Park, H. W., Kim, Y. C., Yu, B., Moroishi, T., Mo, J. S., Plouffe, S. W., et al. (2015). Alternative Wnt Signaling Activates YAP/TAZ. Cell 162, 780–794. doi: 10.1016/j.cell.2015.07.013
Park, Y. Y., Sohn, B. H., Johnson, R. L., Kang, M. H., Kim, S. B., Shim, J. J., et al. (2016). Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 63, 159–172. doi: 10.1002/hep.28223
Patel, S. H., Camargo, F. D., and Yimlamai, D. (2017). Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology 152, 533–545. doi: 10.1053/j.gastro.2016.10.047
Pegoraro, S., Ros, G., Ciani, Y., Sgarra, R., Piazza, S., and Manfioletti, G. (2015). A novel HMGA1-CCNE2-YAP axis regulates breast cancer aggressiveness. Oncotarget 6, 19087–19101. doi: 10.18632/oncotarget.4236
Plouffe, S. W., Meng, Z., Lin, K. C., Lin, B., Hong, A. W., Chun, J. V., et al. (2016). Characterization of Hippo pathway components by gene inactivation. Mol. Cell 64, 993–1008. doi: 10.1016/j.molcel.2016.10.034
Poon, C. L., Lin, J. I., Zhang, X., and Harvey, K. F. (2011). The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 21, 896–906. doi: 10.1016/j.devcel.2011.09.012
Porazinski, S., Wang, H., Asaoka, Y., Behrndt, M., Miyamoto, T., Morita, H., et al. (2015). YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature 521, 217–221. doi: 10.1038/nature14215
Praskova, M., Khoklatchev, A., Ortiz-Vega, S., and Avruch, J. (2004). Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381(Pt 2), 453–462. doi: 10.1042/BJ20040025
Praskova, M., Xia, F., and Avruch, J. (2008). MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311–321. doi: 10.1016/j.cub.2008.02.006
Purvis, J. E., and Lahav, G. (2013). Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956. doi: 10.1016/j.cell.2013.02.005
Reddy, B. V., and Irvine, K. D. (2013). Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell 24, 459–471. doi: 10.1016/j.devcel.2013.01.020
Ribeiro, P. S., Josue, F., Wepf, A., Wehr, M. C., Rinner, O., Kelly, G., et al. (2010). Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39, 521–534. doi: 10.1016/j.molcel.2010.08.002
Richardson, H. E., and Portela, M. (2017). Tissue growth and tumorigenesis in Drosophila: cell polarity and the Hippo pathway. Curr. Opin. Cell Biol. 48, 1–9. doi: 10.1016/j.ceb.2017.03.006
Rosenbluh, J., Nijhawan, D., Cox, A. G., Li, X., Neal, J. T., Schafer, E. J., et al. (2012). β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457–1473. doi: 10.1016/j.cell.2012.11.026
Sansores-Garcia, L., Bossuyt, W., Wada, K., Yonemura, S., Tao, C., Sasaki, H., et al. (2011). Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30, 2325–2335. doi: 10.1038/emboj.2011.157
Sasaki, H. (2017). Roles and regulations of Hippo signaling during preimplantation mouse development. Dev. Growth Differ. 59, 12–20. doi: 10.1111/dgd.12335
Schlegelmilch, K., Mohseni, M., Kirak, O., Pruszak, J., Rodriguez, J. R., Zhou, D., et al. (2011). Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144, 782–795. doi: 10.1016/j.cell.2011.02.031
Shao, D., Zhai, P., Del Re, D. P., Sciarretta, S., Yabuta, N., Nojima, H., et al. (2014). A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nat. Commun. 5:3315. doi: 10.1038/ncomms4315
Sorrentino, G., Ruggeri, N., Specchia, V., Cordenonsi, M., Mano, M., Dupont, S., et al. (2014). Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 16, 357–366. doi: 10.1038/ncb2936
Sorrentino, G., Ruggeri, N., Zannini, A., Ingallina, E., Bertolio, R., Marotta, C., et al. (2017). Glucocorticoid receptor signalling activates YAP in breast cancer. Nat. Commun. 8:14073. doi: 10.1038/ncomms14073
Strano, S., Munarriz, E., Rossi, M., Castagnoli, L., Shaul, Y., Sacchi, A., et al. (2001). Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 276, 15164–15173. doi: 10.1074/jbc.M010484200
Strassburger, K., Tiebe, M., Pinna, F., Breuhahn, K., and Teleman, A. A. (2012). Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev. Biol. 367, 187–196. doi: 10.1016/j.ydbio.2012.05.008
Sun, C., De Mello, V., Mohamed, A., Ortuste Quiroga, H. P., Garcia-Munoz, A., Al Bloshi, A., et al. (2017). Common and distinctive functions of the Hippo effectors Taz and Yap in skeletal muscle stem cell function. Stem Cells. 35, 1958–1972. doi: 10.1002/stem.2652
Tamm, C., Bower, N., and Anneren, C. (2011). Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J. Cell Sci. 124(Pt 7), 1136–1144. doi: 10.1242/jcs.075796
Tang, F., Gill, J., Ficht, X., Barthlott, T., Cornils, H., Schmitz-Rohmer, D., et al. (2015). The kinases NDR1/2 act downstream of the Hippo homolog MST1 to mediate both egress of thymocytes from the thymus and lymphocyte motility. Sci. Signal. 8:ra100. doi: 10.1126/scisignal.aab2425
Taniguchi, K., Wu, L. W., Grivennikov, S. I., de Jong, P. R., Lian, I., Yu, F. X., et al. (2015). A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519, 57–62. doi: 10.1038/nature14228
Tapon, N., Harvey, K. F., Bell, D. W., Wahrer, D. C., Schiripo, T. A., Haber, D., et al. (2002). Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478. doi: 10.1016/S0092-8674(02)00824-3
Totaro, A., Castellan, M., Battilana, G., Zanconato, F., Azzolin, L., Giulitti, S., et al. (2017). YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 8:15206. doi: 10.1038/ncomms15206
Tremblay, A. M., Missiaglia, E., Galli, G. G., Hettmer, S., Urcia, R., Carrara, M., et al. (2014). The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell 26, 273–287. doi: 10.1016/j.ccr.2014.05.029
Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C., and Halder, G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920. doi: 10.1038/ncb1050
Varelas, X., Miller, B. W., Sopko, R., Song, S., Gregorieff, A., Fellouse, F. A., et al. (2010a). The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell 18, 579–591. doi: 10.1016/j.devcel.2010.03.007
Varelas, X., Samavarchi-Tehrani, P., Narimatsu, M., Weiss, A., Cockburn, K., Larsen, B. G., et al. (2010b). The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev. Cell 19, 831–844. doi: 10.1016/j.devcel.2010.11.012
Vassilev, A., Kaneko, K. J., Shu, H., Zhao, Y., and DePamphilis, M. L. (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229–1241. doi: 10.1101/gad.888601
von Eyss, B., Jaenicke, L. A., Kortlever, R. M., Royla, N., Wiese, K. E., Letschert, S., et al. (2015). A MYC-Driven Change in Mitochondrial Dynamics Limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell 28, 743–757. doi: 10.1016/j.ccell.2015.10.013
Wada, K., Itoga, K., Okano, T., Yonemura, S., and Sasaki, H. (2011). Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907–3914. doi: 10.1242/dev.070987
Wang, K. C., Yeh, Y. T., Nguyen, P., Limqueco, E., Lopez, J., Thorossian, S., et al. (2016). Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc. Natl. Acad. Sci. U.S.A. 113, 11525–11530. doi: 10.1073/pnas.1613121113
Wang, L., Luo, J.-Y., Li, B., Tian, X. Y., Chen, L. J., Huang, Y., et al. (2016). Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 540, 579–582. doi: 10.1038/nature20602
Wang, W., Huang, J., and Chen, J. (2011). Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 286, 4364–4370. doi: 10.1074/jbc.C110.205401
Wang, W., Xiao, Z. D., Li, X., Aziz, K. E., Gan, B., Johnson, R. L., et al. (2015). AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 17, 490–499. doi: 10.1038/ncb3113
Wang, Z., Wu, Y., Wang, H., Zhang, Y., Mei, L., Fang, X., et al. (2014). Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc. Natl. Acad. Sci. U.S.A. 111, E89–E98. doi: 10.1073/pnas.1319190110
Watt, K. I., Judson, R., Medlow, P., Reid, K., Kurth, T. B., Burniston, J. G., et al. (2010). Yap is a novel regulator of C2C12 myogenesis. Biochem. Biophys. Res. Commun. 393, 619–624. doi: 10.1016/j.bbrc.2010.02.034
Watt, K. I., Turner, B. J., Hagg, A., Zhang, X., Davey, J. R., Qian, H., et al. (2015). The Hippo pathway effector YAP is a critical regulator of skeletal muscle fibre size. Nat. Commun. 6:6048. doi: 10.1038/ncomms7048
Wu, H., Wei, L., Fan, F., Ji, S., Zhang, S., Geng, J., et al. (2015). Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat. Commun. 6:6239. doi: 10.1038/ncomms7239
Wu, S., Huang, J., Dong, J., and Pan, D. (2003). hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456. doi: 10.1016/S0092-8674(03)00549-X
Wu, S., Liu, Y., Zheng, Y., Dong, J., and Pan, D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398. doi: 10.1016/j.devcel.2008.01.007
Xin, M., Kim, Y., Sutherland, L. B., Murakami, M., Qi, X., McAnally, J., et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. U.S.A. 110, 13839–13844. doi: 10.1073/pnas.1313192110
Yan, L., Cai, Q., and Xu, Y. (2014). Hypoxic conditions differentially regulate TAZ and YAP in cancer cells. Arch. Biochem. Biophys. 562, 31–36. doi: 10.1016/j.abb.2014.07.024
Yang, S., Zhang, L., Liu, M., Chong, R., Ding, S. J., Chen, Y., et al. (2013). CDK1 phosphorylation of YAP promotes mitotic defects and cell motility and is essential for neoplastic transformation. Cancer Res. 73, 6722–6733. doi: 10.1158/0008-5472.CAN-13-2049
Yu, F. X., Zhao, B., Panupinthu, N., Jewell, J. L., Lian, I., Wang, L. H., et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791. doi: 10.1016/j.cell.2012.06.037
Zaidi, S. K., Sullivan, A. J., Medina, R., Ito, Y., van Wijnen, A. J., Stein, J. L., et al. (2004). Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 23, 790–799. doi: 10.1038/sj.emboj.7600073
Zanconato, F., Forcato, M., Battilana, G., Azzolin, L., Quaranta, E., Bodega, B., et al. (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227. doi: 10.1038/ncb3216
Zeng, Y., Stauffer, S., Zhou, J., Chen, X., Chen, Y., and Dong, J. (2017). Cyclin-dependent kinase 1 (CDK1)-mediated mitotic phosphorylation of the transcriptional co-repressor Vgll4 inhibits its tumor-suppressing activity. J. Biol. Chem. 292, 15028–15038. doi: 10.1074/jbc.M117.796284
Zhang, L., Ren, F., Zhang, Q., Chen, Y., Wang, B., and Jiang, J. (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387. doi: 10.1016/j.devcel.2008.01.006
Zhang, L., Tang, F., Terracciano, L., Hynx, D., Kohler, R., Bichet, S., et al. (2015). NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr. Biol. 25, 296–305. doi: 10.1016/j.cub.2014.11.054
Zhang, Y., and Del Re, D. P. (2017). A growing role for the Hippo signaling pathway in the heart. J. Mol. Med. 95, 465–472. doi: 10.1007/s00109-017-1525-5
Zhao, B., Kim, J., Ye, X., Lai, Z. C., and Guan, K. L. (2009). Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 69, 1089–1098. doi: 10.1158/0008-5472.CAN-08-2997
Zhao, B., Li, L., Lu, Q., Wang, L. H., Liu, C. Y., Lei, Q., et al. (2011). Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63. doi: 10.1101/gad.2000111
Zhao, B., Li, L., Tumaneng, K., Wang, C. Y., and Guan, K. L. (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ−TRCP. Genes Dev. 24, 72–85. doi: 10.1101/gad.1843810
Zhao, B., Wei, X., Li, W., Udan, R. S., Yang, Q., Kim, J., et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761. doi: 10.1101/gad.1602907
Zhao, B., Ye, X., Yu, J., Li, L., Li, W., Li, S., et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971. doi: 10.1101/gad.1664408
Zhao, X., Wang, X., Fang, L., Lan, C., Zheng, X., Wang, Y., et al. (2017). A combinatorial strategy using YAP and pan-RAF inhibitors for treating KRAS-mutant pancreatic cancer. Cancer Lett. 402, 61–70. doi: 10.1016/j.canlet.2017.05.015
Zhao, Y., Khanal, P., Savage, P., She, Y. M., Cyr, T. D., and Yang, X. (2014). YAP-induced resistance of cancer cells to antitubulin drugs is modulated by a Hippo-independent pathway. Cancer Res. 74, 4493–4503. doi: 10.1158/0008-5472.CAN-13-2712
Zheng, Y., Wang, W., Liu, B., Deng, H., Uster, E., and Pan, D. (2015). Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev. Cell 34, 642–655. doi: 10.1016/j.devcel.2015.08.014
Zhou, W., Liotta, L. A., and Petricoin, E. F. (2017). The warburg effect and mass spectrometry-based proteomic analysis. Cancer Genomics Proteomics 14, 211–218. doi: 10.21873/cgp.20032
Keywords: hippo signaling pathway, YAP, TAZ, integrative physiology, cell signaling
Citation: Watt KI, Harvey KF and Gregorevic P (2017) Regulation of Tissue Growth by the Mammalian Hippo Signaling Pathway. Front. Physiol. 8:942. doi: 10.3389/fphys.2017.00942
Received: 11 August 2017; Accepted: 07 November 2017;
Published: 24 November 2017.
Edited by:
Joon (Kyungjoon) Lim, La Trobe University, AustraliaReviewed by:
Giovanni Sorrentino, École Polytechnique Fédérale de Lausanne, SwitzerlandCatherine Coirault, Institut National de la Santé et de la Recherche Médicale, France
Copyright © 2017 Watt, Harvey and Gregorevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin I. Watt, a2V2aW4ud2F0dEBiYWtlci5lZHUuYXU=
 Kevin I. Watt
Kevin I. Watt Kieran F. Harvey3,4,5,6
Kieran F. Harvey3,4,5,6