
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 20 April 2017
Sec. Integrative Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.00230
 Bolin Cai1,2,3
Bolin Cai1,2,3 Zhenhui Li1,2,3
Zhenhui Li1,2,3 Manting Ma1,2,3
Manting Ma1,2,3 Zhijun Wang1,2,3
Zhijun Wang1,2,3 Peigong Han1,2,3
Peigong Han1,2,3 Bahareldin A. Abdalla1,2,3
Bahareldin A. Abdalla1,2,3 Qinghua Nie1,2,3*
Qinghua Nie1,2,3* Xiquan Zhang1,2,3
Xiquan Zhang1,2,3Long non-coding RNAs (lncRNAs) play important roles in epigenetic regulation of skeletal muscle development. In our previous RNA-seq study (accession number GSE58755), we found that lncRNA-Six1 is an lncRNA that is differentially expressed between White Recessive Rock (WRR) and Xinghua (XH) chicken. In this study, we have further demonstrated that lncRNA-Six1 is located 432 bp upstream of the gene encoding the protein Six homeobox 1 (Six1). A dual-luciferase reporter assay identified that lncRNA-Six1 overlaps the Six1 proximal promoter. In lncRNA-Six1, a micropeptide of about 7.26 kDa was found to play an important role in the lncRNA-Six1 in cis activity. Overexpression of lncRNA-Six1 promoted the mRNA and protein expression level of the Six1 gene, while knockdown of lncRNA-Six1 inhibited Six1 expression. Moreover, tissue expression profiles showed that both the lncRNA-Six1 and the Six1 mRNA were highly expressed in chicken breast tissue. LncRNA-Six1 overexpression promoted cell proliferation and induced cell division. Conversely, its loss of function inhibited cell proliferation and reduced cell viability. Similar effects were observed after overexpression or knockdown of the Six1 gene. In addition, overexpression or knockdown of Six1 promoted or inhibited, respectively, the expression levels of muscle-growth-related genes, such as MYOG, MYHC, MYOD, IGF1R, and INSR. Taken together, these data demonstrate that lncRNA-Six1 carries out cis-acting regulation of the protein-encoding Six1 gene, and encodes a micropeptide to activate Six1 gene, thus promoting cell proliferation and being involved in muscle growth.
Meat production from chicken muscle is one of the most important factors in determining the economic value of poultry. Numerous processes regulate muscle growth, including genetics, environment, and nutrition, but genetics play the least understood of these critical roles (Scanes et al., 1984).
Some non-coding RNAs are regulators of muscle growth and development, but are not strictly subject to normal genetic analyses (Kallen et al., 2013; Mousavi et al., 2013; Luo et al., 2014). Therefore, these RNAs have been underrepresented in genetic studies, especially those concerning chicken muscle development. Protein-encoding genes only account for a small portion (2%) of the genome, and yet 70 to 90% of the genome is transcribed at some point during development. The result is a large long non-coding RNA (lncRNA) transcriptome in mammals (Lee, 2012).
LncRNAs have been found both in the nucleus and in the cytoplasm, with sizes ranging from 200 bp to more than 100 kb (Cabili et al., 2011; Djebali et al., 2012). LncRNAs are involved in diverse aspects of cell biology and regulation, such as genomic imprinting, X chromosome inactivation, chromatin modifications, nuclear trafficking, transcriptional interference, and activation. These RNAs can regulate gene expression via epigenetic alterations, as well as transcriptional and post-transcriptional processing (Kino et al., 2010; Lee and Bartolomei, 2013; Zhao et al., 2013). LncRNA is an important regulatory factor for biology, but little is known about lncRNA in chicken research. Using next generation sequencing, 281 new intergenic lncRNAs were first systematically identified in chicken embryo skeletal muscle. Further analysis found that these lncRNAs were less conserved than protein-encoding genes, but slightly more conserved than random non-coding sequences (Li et al., 2012). In another study, 1,336 lncRNAs were differentially expressed in the preadipocytes of Jinghai Yellow chicken at different stages, for which the number of differentially expressed genes decreased with differentiation, demonstrating that the early stage might be most important for chicken preadipocyte differentiation (Zhang et al., 2017). LncRNA-αGT is a nuclear, non-coding transcript involved in the stage-specific activation of the chicken adult αD globin gene. Research has shown that lncRNA-αGT is required for full activation of the adult αD gene and maintenance of transcriptionally active chromatin, and could maintain adult α-globin gene expression by promoting an active chromatin structure (Arriaga-Canon et al., 2014). Although, more and more lncRNAs have been found by high-throughput sequencing, the mechanism of lncRNA regulation involved in muscle growth in chicken is still poorly understood.
Six1 (Sine oculis homeobox 1) is an SO (Sine oculis) ortholog in vertebrates and is a member of the Sine oculis homeobox family of highly conserved transcription factors. Six1 was first identified as a regulator of Drosophila visual system development (Cheyette et al., 1994). Subsequently, a role for Six1 in vertebrate development of sensory systems, skeletal muscle, craniofacial organs, and kidneys has been found (Xu et al., 2003; Stierwald et al., 2004; Nonomura et al., 2010; Gordon et al., 2012; Sato et al., 2012). Six1 is expressed in numerous tissues and especially in skeletal muscle (Boucher et al., 1996; Wu et al., 2011; Wang et al., 2014). This gene regulates skeletal muscle development and transformation of muscle fiber types throughout the embryonic to postnatal stages (Ozaki et al., 2001; Laclef et al., 2003; Grifone et al., 2004, 2005; Hetzler et al., 2014; O'Brien et al., 2014).
Some lncRNAs regulate gene expression primarily in cis, where both the regulatory RNA and the target gene are transcribed from the same locus (Guil and Esteller, 2012). WRR is a broiler chicken with a fast growth rate, which exhibits a different growth performance from the XH chicken (a Chinese native breed with a slow growth rate) at 7 weeks of age (Ouyang et al., 2015). In our previous study, two chicken breeds, WRR and XH, of 7 weeks in age were used for RNA-seq analysis, and a large population of lncRNAs that were differentially expressed between WRR and XH chickens were identified. This was suggestive that these lncRNAs might affect chicken growth. Both Six1 and lncRNA-Six1 are differentially expressed between WRR and XH chicken, and lncRNA-Six1 is upstream of Six1 in the chicken genome.
To study the role of lncRNA-Six1 in chicken skeletal muscle growth, we analyzed its molecular function and expression level, and performed an analysis of Six1 promoter activity to identify the relationship between lncRNA-Six1 and Six1. Additionally, biological function and tissue expression profiles were also analyzed for Six1.
All animal experiments were carried out in strict accordance with the regulations for the Administration of Laboratory Animals of Guangdong Province. These experiments were handled in compliance with, and were approved by, the Institutional Animal Care and Use Committee at the South China Agricultural University (Guangzhou, China) with approval number SCAU#0011. All efforts were made to minimize animal suffering.
Six 7-week-old XH female chickens were obtained from the Avian Farm of the South China Agriculture University (Guangzhou, China). Chickens were euthanized, and organs and tissues were collected after rapid dissection, then immediately frozen in liquid nitrogen and stored at −80°C. The samples included the cerebrum, cerebellum, hypothalamus, pituitary, heart, liver, spleen, lung, kidney, muscular stomach, glandular stomach, breast muscle, leg muscle, abdominal fat, and subcutaneous fat.
Total RNA was extracted from each tissue using Trizol reagent (TaKaRa, Japan) following the manufacturer's protocol. The integrity and concentration of all obtained RNA samples were assayed by 1.5% agarose gel electrophoresis, and evaluated by ultraviolet spectroscopy using the 260/280 nm ratio.
A PARIS Kit (Ambion, Life Technologies, USA) was used to harvest the cytoplasmic and nuclear cell lysates. Fresh cultured cells were placed on ice, after being collected and washed once in PBS. Subsequently, cells were resuspended in 500 μL ice-cold Cell Fractionation Buffer and incubated on ice for 10 min. After centrifugation for 5 min at 4°C and 500 × g, the cytoplasmic fraction was carefully aspirated away from the nuclear pellet; thereafter, the nuclear pellet was washed in ice-cold Cell Fractionation Buffer and lysed in Cell Disruption Buffer. Samples were split for RNA isolation, following the manufacturer's protocol.
The PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan) was used to synthesize cDNA. The iTaq Universal SYBR Green Supermix Kit (Toyobo, Japan) was used for cDNA quantification, according to the manufacturer's protocol, in a BioRad CFX96 Real-Time Detection instrument. The chicken β-actin gene was used as an internal control. Data analysis was carried out using the comparative 2−ΔΔCT method (Livak and Schmittgen, 2001).
Primers were designed using Premier Primer 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) or OLIGO Primer Analysis Software Version 7 (Molecular Biology Insights, USA), and synthesized by Sangon Biotech (Shanghai, China). The major primers used in this study are listed in Table 1. Of these primers, lncRNA-Six1 5′ RACE-outer, lncRNA-Six1 5′ RACE-inner and lncRNA-Six1-3′ RACE were used to clone the full-length of chicken lncRNA-Six1. The other 7 primers (lncRNA-Six1-ORF-1, lncRNA-Six1-ORF-2, lncRNA-Six1-ORF-3, lncRNA-Six1-ORF-4, lncRNA-Six1-ORF-5, lncRNA-Six1-ORF-6, and lncRNA-Six1-ORF-7) were used for the open reading frame (ORF) amplification of lncRNA-Six1, and pSDS-β-actin was used as a positive control. Moreover, primers pSDS-lncRNA-Six1 and pSDS-Six1 were used to amplify the coding sequences, and then construct the overexpression vector. The last 4 primers (pGL3-basic-P1, pGL3-basic-P2, pGL3-basic-P3, and pGL3-basic-P4) were used for the dual-luciferase reporter assay for the promoter region of Six1. Primers for RT-quantitative PCR (qPCR) are shown in Table 2. The siRNAs used for the knockdown of Six1 were synthesized by Guangzhou RiboBio (Guangzhou, China) and are listed in Table 3. LncRNA-Six1 is an RNA molecule present in the cytoplasm and nucleus. The siRNA and ASO that were used for the specific knockdown of lncRNA-Six1 in the cytoplasm and nucleus, respectively, were designed and synthesized by Guangzhou RiboBio (Guangzhou, China), and are listed in Table 3.
The partial lncRNA-Six1 sequence was obtained from our previous lncRNA-seq data (accession number GSE58755). RACE PCR was performed to obtain the full-length sequence of the lncRNA-Six1. Total RNA from breast muscle tissue was used as the template for nested-PCR reactions using a SMARTer RACE cDNA Amplification Kit (Clontech, Osaka, Japan), following the manufacturer's instructions. The products of the RACE PCR were cloned into the pJET 1.2/blunt cloning vector (CloneJET PCR Cloning Kit; Fermentas, Glen Burnie, MD, USA) and sequenced by Sangon Biotech (Shanghai, China).
For lncRNA-Six1 overexpression plasmid construction, the full-length sequence of lncRNA-Six1 was amplified by PCR, and cloned into the expression plasmid pSDS-20218 (SiDanSai, Shanghai, China) by using BsaI restriction enzyme. Six1 overexpression constructs were generated by amplifying the Six1 coding sequence, which was then subcloned into the overexpression plasmid vector, pSDS-20218 (SiDanSai, Shanghai, China). Seven ORFs of lncRNA-Six1 were also amplified and cloned into pSDS-20218 (SiDanSai, Shanghai, China). A region from the GFP gene was amplified and cloned into pSDS-20218 as a negative control, and was named pSDS-Control.
Luciferase reporter vectors including different sized Six1 promoter fragments were constructed from the chicken genome using XhoI and HindIII restriction sites. The PCR products were excised with XhoI and HindIII restriction endonucleases and ligated into plasmid vector pGL3-Basic (Promega, Wisconsin, USA). The recombinant constructs were named pGL3-basic-P1 (−2247/+216), pGL3-basic-P2 (−1595/+216), pGL3-basic-P3 (−1129/+216), and pGL3-basic-P4 (−495/+216). These were numbered relative to the first base of the initiation codon of the Six1 gene.
DF-1 cells were cultured in DMEM (Gibco, USA) supplemented with 10% (v/v) fetal bovine serum (Hyclone, USA) and 0.2% penicillin/streptomycin (Invitrogen, USA). QM-7 cells were cultured in high-glucose M199 medium (Gibco, USA) with 10% fetal bovine serum, 10% tryptose phosphate broth solution (Sigma, USA), and 0.2% penicillin/streptomycin. All cells were cultured at 37°C in a 5% CO2, humidified atmosphere. In this study, DF-1 cells were used to study the function of lncRNA-Six1, lncRNA-Six1-ORF-2, and Six1 in chicken genome. For QM-7 cells, were used to study the influence of Six1 on muscle growth.
All transient transfections used Lipofectamine 3000 reagent (Invitrogen, USA) following the manufacturer's protocol. The concentrations used for plasmid transfections were as follows: 6-well plate, 2.5 μg/well; 12-well plate, 1 μg/well; 24-well plate, 0.5 μg/well; 48-well plate, 0.25 μg/well; 96-well plate, 0.1 μg/well. For siRNAs and ASO, a concentration of 100 nM were used. si-lncRNA-Six1 and ASO-lncRNA-Six1 were co-transfected for interference with lncRNA-Six1, and named as si-ASO-lncRNA-Six1 in this study. The PRL-TK vector (Promega, USA) was used as an internal control plasmid for Renilla luciferase expression and was co-transfected with the pGL3 luciferase reporter vectors. pSDS-Control and pGL3 basic were used as the empty vector control plasmids. In this study, except for Cell Counting Kit (CCK)-8 assays, all cells were harvested or analyzed after 48 h transfection.
The recombinant plasmids (pGL3-basic-P1, pGL3-basic-P2, pGL3-basic-P3, and pGL3-basic-P4), used to analyze the Six1 promoter region, were co-transfected with pRL-TK in DF-1 cells. The pGL3-basic was also co-transfected with pRL-TK as a control. Firefly and Renilla luciferase activities were measured at 48 h post-transfection using a Dual-GLO Luciferase Assay System Kit (Promega, USA), following the manufacturer's instructions. Luminescence was measured using a Fluorescence/Multi-Detection Microplate Reader (BioTek, USA) and firefly luciferase activities were normalized to Renilla luminescence in each well.
DF-1 cellular proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer with phenylmethanesulfonyl fluoride protease inhibitor. Western blot analysis was performed as previously reported (Feng et al., 2016). The antibodies used for Western blots were as follows: Flag tag polyclonal antibody (20543-1-AP; Proteintech, USA; 1:1,000), goat anti-rabbit IgG (H&L)-HRP (BS13278; Bioworld, USA; 1:5,000), rabbit anti-Six1 (sc-9127; Santa Cruz, USA; 1:100), myogenin antibody (orb6492; Biorbyt, UK; 1:100), B103 (DHSB, USA; 0.5 μg/ml), goat anti-rabbit IgG-HRP (BA1054; Boster, China; 1:5,000), peroxidase-goat anti-mouse IgG (BA1051; Boster, China; 1:2,500), and HRP-conjugated monoclonal mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (KC-5G5; Kangchen, China; 1:10,000).
At 48 h after transfection, DF-1 cells were incubated at 37°C for 2 h in the presence of 50 μM EdU (RiboBio, China). The cells were then fixed in 4% paraformaldehyde for 30 min and neutralized using 2 mg/mL glycine solution. The cells were permeabilized by adding 0.5% Triton X-100. A solution containing EdU (Apollo Reaction Cocktail; RiboBio, China) was added and the cells were incubated at room temperature for 30 min. The nuclear stain Hoechst 33342 was then added and incubation was continued for another 30 min. The number of EdU-stained cells was determined using images of three randomly selected fields obtained with a fluorescence microscope (TE2000-U; Nikon, Japan).
Flow cytometry analysis was performed on a BD AccuriC6 flow cytometer (BD Biosciences, USA) and data was processed using FlowJo7.6 software. The cell cultures in growth media were collected after a 48 h transfection and fixed in 70% ethanol overnight at −20°C. Propidium iodide (Sigma, USA) (50 μg/mL) containing 10 μg/mL RNase A (Takara, USA) was added and incubated for 30 min at 37°C in the dark and then the samples were sent for detection.
For the cell growth assays, cells were seeded in a 96-well plate. After being transfected with (a) pSDS-lncRNA-Six1, (b) si-ASO-lncRNA-Six1, (c) pSDS-Six1, or (d) si-Six1, the proliferation of the cell culture was monitored at 24 h, 48 h, 72 h, and 96 h using the TransDetect CCK (TransGen Biotech, Beijing, China). Every 24 h, 10 μL of CCK solution was added and the absorbance at 450 nm was measured using a Model 680 Microplate Reader (Bio-Rad) after a 1 h incubation.
DF-1 cells were seeded in 12-well plates. After a density of 50% confluence was reached, cells were transfected. Subsequently, a linear wound was generated by scratching the monolayer of cells with a 1 mL pipette tip, when the cells reached a density of 100% confluence. The proliferation and migration of cells were captured at 0, 24, 48, and 72 h by using a microscope (TE2000-U; Nikon, Japan). The width of the scratches was measured by Nano Measurer software. The formula to calculate the cell migration rate was as follows: (W0h – W × h) × 100%/ W0h, where W0h represents the mean wound width at 0 h, and W × h represents the mean wound width at 24, 48, and 72 h.
After transfection for 48 h, cells were fixed in 1% formaldehyde for 10 min at 37°C, and quenched with 125 mM glycine for 5 min. Samples were then washed twice with cold PBS and placed in SDS lysis buffer. After sonication process [(5 s pulse + 5 s interval) × 25% of maximum power, for 14 times], chromatin was immunoprecipitated with the DYKDDDDK Tag (D6W5B) rabbit monoclonal antibody (14,793; Cell Signaling, USA; 1:50), as previously reported (Harms et al., 2015). The relative quantity of the immunoprecipitated factor was calculated by qPCR.
All results are presented as the mean ± S.E.M, based on at least three independent experiments for each treatment. Statistical significance of differences between means was assessed by performing an unpaired Student's t-test and P < 0.05 or less was considered significant.
We identified the 5′ and 3′ ends of lncRNA-Six1 by RACE analysis (Figure 1A and Supplementary Data 1). Predictions of the identity of lncRNA-Six1 by the Coding Potential Calculator suggested a low coding potential and low evolutionary conservation consistent with a non-coding RNA (Kong et al., 2007; Figure 1B). In order to verify this prediction, we analyzed the coding ability of seven potential ORFs of lncRNA-Six1 by Western blot. We found that only ORF-2 generated a micropeptide (7.26 kDa), suggestive that lncRNA-Six1 is an lncRNA with low protein-encoding potential (Figure 1C and Supplementary Data 2). The National Center for Biotechnology Information's Basic Local Alignment Search Tool (BLAST) showed that the lncRNA-Six1 was 2,350 bp long, located on chromosome 5 and spanned from 54,483,370 to 54,487,175, and comprised two exons. The micropeptide was also located at chromosome 5 from 54,484,978 to 54,485,172, and the 3′ end of lncRNA-Six1 was separated from Six1 by 432 bp (Figure 1D). We also investigated the intracellular localization of lncRNA-Six1, the RT-PCR results confirmed that lncRNA-Six1 is an RNA molecule present in the cytoplasm and nucleus (Figure 1E).
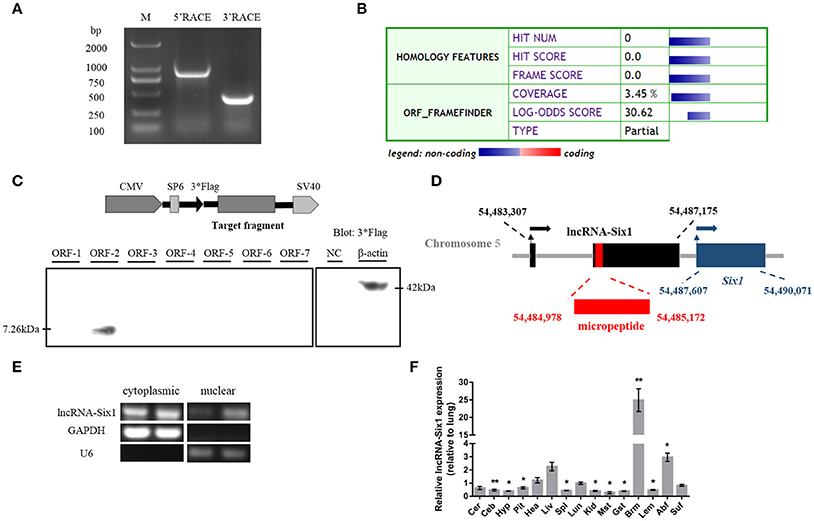
Figure 1. Identification of lncRNA-Six1. (A) Results of 5′ RACE and 3′ RACE. M, DL2000 Marker; 5′ RACE product, 806 bp; 3′ RACE product, 365 bp. (B) Analysis obtained from the Coding Potential Calculator (http://cpc.cbi.pku.edu.cn/) based on evolutionary conservation and ORF attributes. (C) Western blot analysis of the coding ability of lncRNA-Six1. The possible ORF of lncRNA-Six1 was cloned into the eukaryotic expression vector pSDS-20218 with a 3*Flag tag. Untransfected DF-1 cells were used as a negative control (NC) and DF-1 cells transfected with β-actin were used as a positive control. The upper panel shows the model target fragment in the pSDS vector. The lower left panel is the blot of ORFs 1–7, and the lower right panel is the blot of the NC and β-actin. (D) A schematic image of the locus for lncRNA-Six1 (black), micropeptide (red) and Six1 (blue). Coordinates are listed according to the Gallus_gallus-5.0 reference Annotation Release 103. Thin arrows represent the transcription start site. Thick arrows represent the direction of transcription. (E) RT-PCR detection of lncRNA-Six1 in the cytoplasmic and nuclear fractions of chicken primary myoblasts. GAPDH and U6 serve as cytoplasmic and nuclear localization controls, respectively. (F) XH chicken tissue expression profiles of lncRNA-Six1. The horizontal axis and vertical axis indicate different tissues and their relative expression values (mean ± S.E.M). Cer, cerebrum; Ceb, cerebellum; Hyp, hypothalamus; Pit, pituitary; Hea, heart; Liv, liver; Spl, spleen; Lun, lung; Kid, kidney; Mst, muscular stomach; Gst, glandular stomach; Brm, breast muscle; Lem, leg muscle; Abf, abdominal fat; Suf, subcutaneous fat. In panel (F), the results are shown as the mean ± S.E.M. and the data were representative of three independent assays from six female XingHua chickens. Statistical significance of differences between means was assessed using an unpaired Student's t-test. (* P < 0.05; ** P < 0.01.)
We measured lncRNA-Six1 expression from different tissues from 7-week-old XH chickens using RT-qPCR. LncRNA-Six1 was expressed in all 15 tissues, and breast muscle had the highest levels (Figure 1F). This suggested that lncRNA-Six1 may function in chicken skeletal muscle development.
To unveil functions for lncRNA-Six1, we performed lncRNA-Six1 overexpression and knockdown experiments to assess its role in cell proliferation and viability. The plasmid pSDS-lncRNA-Six1 was constructed for lncRNA-Six1 overexpression, and siRNA and ASO for lncRNA-Six1 knockdown (si-ASO-lncRNA-Six1) were designed and synthesized. After 48 h of transfection, the relative RNA expressions of lncRNA-Six1 were detected (Figures 2A,B). LncRNA-Six1 overexpression significantly promoted cell proliferation, as judged by EdU incorporation (Figures 2C,E). Conversely, proliferation was significantly inhibited after lncRNA-Six1 knockdown (Figures 2D,F). LncRNA-Six1 overexpression also resulted in a slight decrease in the number of G0/G1 cells, and a slightly increase in the number S phase cells. LncRNA-Six1 knockdown reversed this trend, and G0/G1 cells increased significantly and S phase cells decreased significantly (Figures 2G,H). In addition, we also used the CCK-8 assay and found that lncRNA-Six1 overexpression promoted an increase in cell number. However, with lncRNA-Six1 interference, the opposite was true (Figures 2I,J). Together, these data were indicative that lncRNA-Six1 has a positive regulatory effect on cell growth and proliferation, and induces cell division.
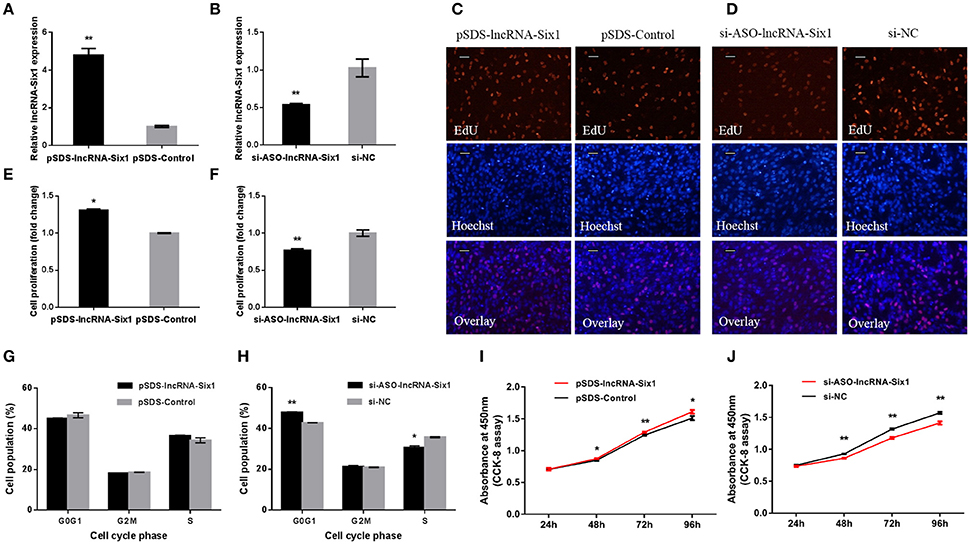
Figure 2. lncRNA-Six1 promotes cell proliferation and induces cell division. (A,B) lncRNA-Six1 relative expression determined by RT-qPCR in DF-1 cells after transfection with the listed nucleic acids. (C,D) EdU proliferation assays for the cells in (A,B) 48 h after transfection, Scale bar, 5 μm. (E,F) the numbers of proliferative cells were also counted. (G,H) Cell cycle analysis of DF-1 cells 48 h after transfection using propidium iodide staining for DNA content. (I,J) DF-1 cell growth curves following transfection of the indicated plasmids over time. In all panels, data are presented as the mean ± S.E.M. of three biological replicates with unpaired Student's t-test. (* P < 0.05; ** P < 0.01) vs. NC, negative control.
In order to further understand the molecular mechanism of lncRNA-Six1, we predicted its cis-regulated target genes on the UCSC Genome Browser (http://genome.ucsc.edu/) (The differentially expressed genes at 100 Kb upstream or downstream of differentially expressed lncRNAs were selected as the potential cis target genes of lncRNAs) (Li et al., 2017; Figure 3A). The results showed that the lncRNA-Six1 is located upstream of Six1, which suggests that Six1 may be a potential cis-regulatory target gene for lncRNA-Six1.
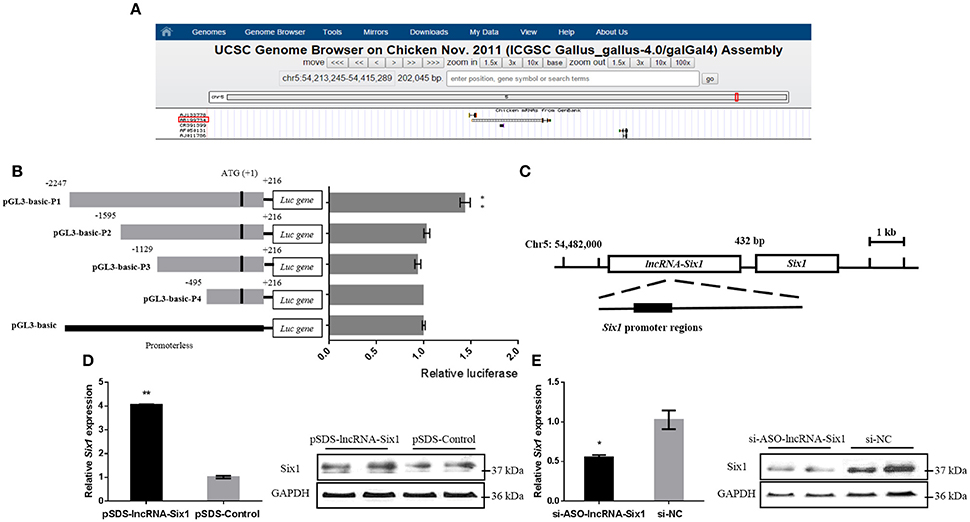
Figure 3. Six1 is a targeted cis-regulatory gene of lncRNA-Six1. (A) UCSC Genome Browser on Chicken Nov. 2011 (ICGSC Gallus_gallus-4.0/galGal4) assembly showing the positional relationship between lncRNA-Six1 and Six1. (B) Dual-luciferase reporter assay using the clones' upstream regions of the chicken Six1 promoter. (C) Model of lncRNA-Six1 cis-regulated Six1. (D) The mRNA and protein expression levels of Six1 from pSDS-lncRNA-Six1 transfected DF-1 cells. (E) Knockdown of the Six1 gene in transfected DF-1 cells. In all panels, data are presented as means ± S.E.M. of three independent experiments with unpaired Student's t-test. (* P < 0.05; ** P < 0.01.) vs. NC, negative control.
To investigate whether lncRNA-Six1 targeted regulate Six1, we cloned individual segments from the Six1 promoter in front of firefly luciferase. These constructs were used to transiently transfect DF-1 cells to determine the region required for Six1 promoter activity. Deletions extending from −1595 to +216, −1129 to +216, and −495 to +216 showed no significant difference in luciferase activities compared with the pGL3-basic control group. However, a significant increase of luciferase activity was observed with the longest construct (−2247 to +216) when compared with the region from −1595 to +216. This was indicative that an essential region of the Six1 promoter was located at −2247 to −1595 bp (Figure 3B). Interestingly, the 3′ end of lncRNA-Six1 was separated from Six1 by 432 bp and the whole RNA overlaps the key promoter regions of Six1 (Figure 3C). This was suggestive that this lncRNA is the regulatory factor of Six1.
More importantly, after overexpression of lncRNA-Six1, the mRNA and protein levels of Six1 increased, while inhibition of lncRNA-Six1 down-regulated Six1 mRNA and protein (Figures 3D,E). Therefore, we confirmed that Six1 is a direct target gene of lncRNA-Six1 in chickens.
To further investigate whether the micropeptide was involved in the cis-regulation of Six1, the relative mRNA expression of the lncRNA-Six1-ORF-2 was detected after overexpression or interference of lncRNA-Six1 (Figures 4A,B). The intracellular localization and tissue expression profiles of lncRNA-Six1-ORF-2 were also analyzed (Figures 4C,D). The results showed that lncRNA-Six1-ORF-2 was present in the cytoplasm and nucleus, and highly expressed in breast muscle. Overexpression of lncRNA-Six1-ORF-2 was performed to study its physiological effects (Figure 4E). A cell cycle analysis after overexpression of lncRNA-Six1-ORF-2 was carried out (Figure 4F). Moreover, we also used the scratch-wound assay and found that lncRNA-Six1-ORF-2 overexpression promoted cell proliferation and migration (Figures 4G,H).
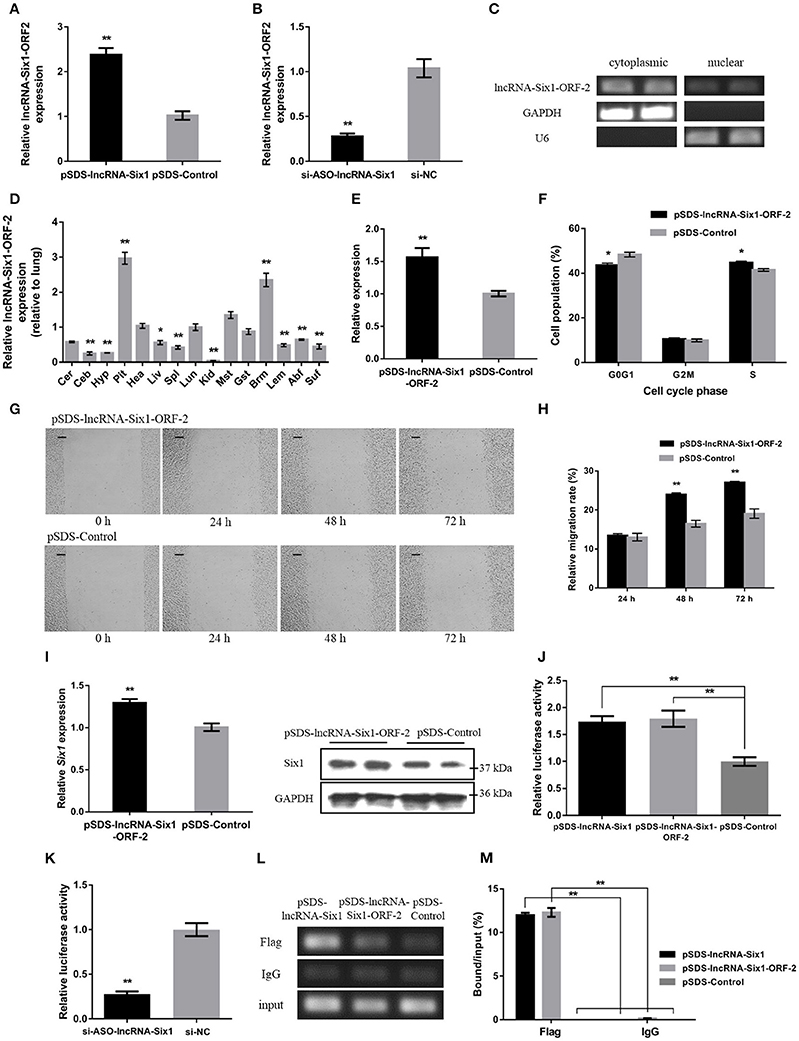
Figure 4. The micropeptide is a key factor required for lncRNA-Six1 to act in cis. (A,B) The mRNA expression levels of lncRNA-Six1-ORF-2 from pSDS-lncRNA-Six1 or si-ASO-lncRNA-Six1 transfected DF-1 cells. (C) RT-PCR detection of lncRNA-Six1-ORF-2 in the cytoplasmic and nuclear fractions of chicken primary myoblast. (D) Tissue expression profiles of the lncRNA-Six1-ORF-2 in XH chickens. The abbreviations for the tissue names are as described in Figure 1. (E) LncRNA-Six1-ORF-2 mRNA expression levels after overexpressing lncRNA-Six1-ORF-2 in DF-1 cells. (F) Cell cycle analysis of DF-1 cells after lncRNA-Six1-ORF-2 overexpression. (G) Representative images of the scratch-wound assay at 0, 24, 48, and 72 h after overexpression of the lncRNA-Six1-ORF-2 in DF-1 cells. Scale bar, 37.5 μm. (H) Quantification of the scratch-wound assay at 24, 48, and 72 h after overexpression of the lncRNA-Six1-ORF-2. (I) The mRNA and protein expression levels of Six1 from pSDS- lncRNA-Six1-ORF-2 transfected DF-1 cells. (J, K) Dual-luciferase reporter assay of the Six1 promoter activity after overexpression of lncRNA-Six1 and lncRNA-Six1-ORF-2, or interference of lncRNA-Six1. (L) Chromatin immunoprecipitation PCR analysis of the binding capacity of lncRNA-Six1 and the micropeptide to the Six1 promoter region. (M) Chromatin immunoprecipitation qPCR analysis of the binding capacity of lncRNA-Six1 and the micropeptide to the Six1 promoter region. In all panels, results are expressed as the mean ± S.E.M. of three independent experiments with unpaired Student's t-test. (* P < 0.05; ** P < 0.01.) vs. NC, negative control.
The mRNA and protein levels of Six1 were significantly increased after overexpression of lncRNA-Six1-ORF-2 (Figure 4I). Meanwhile, a dual-luciferase reporter assay was developed to measure the Six1 promoter activity, after co-transfection with pSDS-lncRNA-Six1, pSDS-lncRNA-Six1-ORF-2, and si-ASO-lncRNA-Six1 with pGL3-basic-P1 (Figures 4J,K). We found that the relative luciferase activity was significantly increased with the overexpression of lncRNA-Six1 or lncRNA-Six1-ORF-2, while the knockdown of lncRNA-Six1 caused the relative luciferase activity to be decreased significantly. In the chromatin immunoprecipitation assay, it was found also that the micropeptide was enriched by the Six1 promoter region (Figures 4L,M). These results indicate that the micropeptide may be a key factor required for lncRNA-Six1 to act in cis.
We examined tissue expression profiles for Six1 and found high expression in breast and leg muscle (Figure 5A). Six1 overexpression and knockdown in DF-1 and QM-7 cells verified the positive regulatory function of Six1 in myogenic proliferation (Figures 5B,C). In DF-1 cells, overexpression of Six1 significantly promoted cell proliferation (Figures 5D,F). This also resulted in a significant decrease in the number of cells in G0/G1, and a significant increase in the number of S-phase cells (Figure 5H). However, cell proliferation was inhibited after interference of Six1 expression (Figures 5E,G), and resulted in a larger number of G0/G1 cells and fewer S phase cells (Figure 5I).
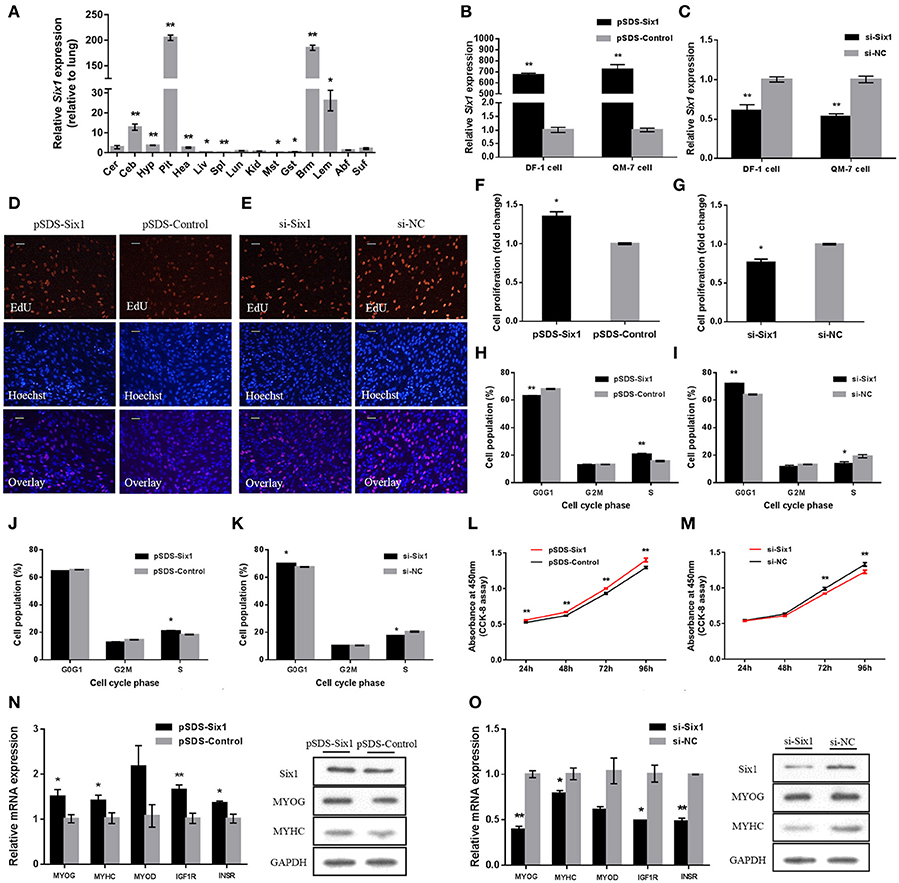
Figure 5. Six1 promotes the growth of chicken skeletal muscle. (A) Tissue expression profiles of Six1 in XH chickens. The abbreviations for the tissue names are as described in Figure 1. (B,C) Six1 mRNA levels in (B) transfected and (C) knockdown DF-1 and QM-7 cells. (D,E) Proliferation of transfected DF-1 cells was assessed by EdU incorporation. Scale bar, 5μm. (F,G) Proliferation rates of DF-1 cells transfected with pSDS-Six1 and with si-Six1. (H–K) Both DF-1 and QM-7 cells, respectively, were collected for cell cycle analysis 48h after transfection. (L,M) Cell growth was measured following the transfection of pSDS-Six1 and si-Six1. (N,O) mRNA and protein levels of some related genes induced by Six1 overexpression and siRNA interference in DF-1 cells. In all panels, results are expressed as the mean ± S.E.M. of three replicates with unpaired Student's t-test. (* P < 0.05; ** P < 0.01.) vs. NC, negative control.
We found similar results for transfected QM-7 myoblasts. Six1 overexpression significantly reduced the number of cells that progressed to G0/G1 and increased the number of S phase cells (Figure 5J). These observations coincided with enhanced cell viability (Figure 5L). Conversely, Six1 knockdown significantly inhibited myoblast proliferation and reduced cell viability (Figures 5K,M).
To identify the role of Six1 in the chicken skeletal muscle growth, the mRNA expression level of growth-related genes, such as MYOG, MYHC, MYOD, IGF1R, and INSR, were detected by qPCR after overexpression or knockdown of Six1. Furthermore, the protein levels of MYOG and MYHC were also detected by Western blot. The results showed that overexpression of Six1 could promote the upregulation of growth-related genes (Figure 5N). In contrast, growth-related gene expression was inhibited after interference with Six1 (Figure 5O). This data demonstrated that Six1 promoted the expression of muscle-growth-related genes and further induced chicken muscle growth.
With the development of genome research, it has been found that the transcription of the animal genome is not only ubiquitous, but also incredibly complex. In the past, lncRNA was considered to be part of the “noise” of the transcribed genome, without any function. However, there is growing evidence that lncRNAs can serve as versatile regulators of diverse aspects of biology by a variety of mechanisms (Rinn et al., 2007). LncRNAs are a class of RNA generally considered to be non-coding with low sequence conservation. However, for some lncRNAs, low protein-encoding potential have been shown. A minimum ORF length is generally used to discriminate between protein-encoding genes and lncRNAs (Karapetyan et al., 2013). For example, the FANTOM consortium originally considered the protein-encoding gene to have an ORF of at least 300 nucleotides (100 amino acids) (Okazaki et al., 2002), and this result is highly consistent with subsequent studies using more sophisticated methods of identification (Frith et al., 2006a,b). Another alternative approach to discriminating lncRNAs from protein-encoding genes is to assess putative ORFs for similarity to known proteins or protein domains, since such homology provides indirect evidence of function as an mRNA (Dinger et al., 2008). In the present study, we identified a lncRNA (lncRNA-Six1) that was differentially expressed between WRR and XH chickens. For this lncRNA, the evolutionary conservation was analyzed and ORF attributes were predicted by the Coding Potential Calculator. We found a micropeptide of about 7.26 kDa using Western bolt assays, which demonstrated that lncRNA-Six1 is an lncRNA. LncRNA-Six1 is present in the cytoplasm and nucleus, and showed a higher expression level in breast muscle.
There is evidence that lncRNAs function during muscle growth. A muscle-specific mouse lncRNA (linc-MD1) is a competitive endogenous RNA for miR-133 and miR-135 targets. This lncRNA controls the expression of MEF2C and MAML1, which are transcription factors that activate late-differentiation muscle genes; thus, affecting myoblast differentiation (Cesana et al., 2011). Moreover, Linc-YY1, a dynamically regulated factor during myogenesis in vitro and in vivo, was discovered from the promoter of the transcription factor Yin Yang 1 (YY1) gene (Zhou et al., 2015). The study found that Linc-YY1 can activate the gene expression of YY1 in trans by interacting with YY1; thus, altering myogenic differentiation and affecting the regeneration of impaired muscle. The MyoD upstream non-coding RNA (MUNC) is specifically expressed in skeletal muscle and can act directly or indirectly on multiple promoters to increase myogenic gene expression, affecting myoblast differentiation in myogenesis (Mueller et al., 2015). LncRNA-Six1 showed a higher expression level in breast muscle, which is suggestive that lncRNA-Six1 may have a role in chicken muscle growth.
LncRNAs are primarily cis-acting regulatory elements and have been described for numerous regulators, including Xist, lincRNA-p21, Kcnq1ot1, and ANRIL (Clemson et al., 1996; Pandey et al., 2008; Yap et al., 2010; Dimitrova et al., 2014). In our study, using an in silico analysis, we identified lncRNA-Six1 upstream of the Six1 gene, which itself was also differentially expressed. Interestingly, lncRNA-Six1 was found to overlap the Six1 promoter. Moreover, the relative luciferase activity was significantly increased with the overexpression of lncRNA-Six1, while the knockdown of lncRNA-Six1 caused the relative luciferase activity to be decreased significantly, suggesting it regulates via a cis mechanism.
Interestingly, recent studies have shown that lncRNAs could also form micropeptide by encoding short ORFs, thus, directly regulating a large variety of functions. Myoregulin (MLN), which is a micropeptide encoded by an annotated lncRNA, was discovered to inhibit the pump activity of sarcoplasmic reticulum Ca2+-ATPase (SERCA), affecting exercise performance and Ca2+ handling in muscle (Anderson et al., 2015). In mice, a peptide, DWORF (dwarf open reading frame), of 34 amino acids was discovered in a putative muscle-specific lncRNA. DWORF can enhance muscle performance by activating the same calcium pump, and may prove to be useful in improving the cardiac muscle function of mammals with heart disease (Nelson et al., 2016). Another recent study identified a small polypeptide (small regulatory polypeptide of amino acid response, SPAR) encoded by the lncRNA LINC00961, which can inhibit amino acid-induced mTORC1 activation and muscle regeneration in skeletal muscle (Matsumoto et al., 2017). In our study, we have identified a micropeptide encoded by lncRNA-Six1. LncRNA-Six1-ORF-2 was present in the cytoplasm and nucleus, and highly expressed in breast muscle. Overexpression of lncRNA-Six1-ORF-2 promoted cell proliferation and migration, and the micropeptide was enriched by the Six1 promoter region. We speculate that this micropeptide plays an important role in lncRNA-Six1 in cis activity.
In vertebrates, Six1 is a regulator of skeletal muscle development and its mRNA is localized to regions of myogenesis in mouse embryos (Oliver et al., 1995; Buckingham and Rigby, 2014). In zebrafish, the Six1a ortholog drives proliferation and differentiation of zebrafish embryonic muscle fiber precursor cells, and its knockdown results in apoptosis (O'Brien et al., 2014). Six1 also plays a key role in the transcriptional regulation of the myogenic regulatory factor (MRF) gene family (Fougerousse et al., 2002; Grifone et al., 2005). For in situ hybridization, no MyoD- and MyoG-expressing cells are detected in Six1−/− limb buds at E11.5 (Laclef et al., 2003). During the differentiation of C2C12 cells, Six1 was down-regulated and the same down-regulation in expression was shown in Myf5, MyoD, and MyoG (Wu et al., 2012). Moreover, the expression of MyHCIIB or MyHCIIX was never observed by immunohistochemistry in Sol fibers, when Six1 could not be detected in peripheral nuclei of these fibers (Grifone et al., 2004). IGF1R and INSR are the insulin receptors and have been found to be involved in the regulation of animal growth. A study showed that Six1, IGF1R, and INSR are essential for the correct initiation and up-regulation of SRY, and affect mammalian gonad development (Eggers et al., 2014). Similarly, our results are indicative that Six1 has a high expression level in chicken muscle tissue. Six1 overexpression also significantly promoted proliferation of myoblast cell lines. Furthermore, overexpression of Six1 could promote the expression of MYOG, MYHC, MYOD, IGF1R, and INSR. These genes were down-regulated with knockdown of Six1.
In this study, we have confirmed that lncRNA-Six1 locates in the upstream of Six1 and overlaps the Six1 proximal promoter. Overexpression or knockdown of lncRNA-Six1 promoted or inhibited, respectively, the mRNA and protein expression level of Six1. These data demonstrate that Six1 is a direct cis-acting target of lncRNA-Six1. In addition, a micropeptide of about 7.26 kDa encoded by lncRNA-Six1 was found to be required for lncRNA-Six1 to act in cis. This micropeptide was enriched by the Six1 promoter region to regulate Six1 expression. Overexpression of lncRNA-Six1 or Six1 promoted cell proliferation and cell division, while the opposite effects were observed after knockdown of lncRNA-Six1 or Six1. Moreover, overexpression or knockdown of Six1 promoted or inhibited, respectively, the expression of muscle-growth-related genes. Taken together, the results of our study demonstrate that lncRNA-Six1 carries out cis-acting regulation of the protein-encoding Six1 gene. A micropeptide encoded by lncRNA-Six1 binds with the promoter of Six1 and activates Six1 to promote cell proliferation and cell division, and induce the expression of muscle-growth-related genes in chicken.
BC: Performed research, analyzed data, wrote paper. ZL: Analyzed data and participated in its design. MM: Constructed vectors. ZW and PH: Analyzed data. BA: Reviewed the manuscript. QN: Conceived of the study, and participated in its design and coordination. XZ: Participated in the design of the study.
This research was supported by the Program for New Century Excellent Talents in University (NCET-13-0803), the Natural Scientific Foundation of China (31571269), the Foundation for High-level Talents in Higher Education in Guangdong, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00230/full#supplementary-material
Anderson, D. M., Anderson, K. M., Chang, C. L., Makarewich, C. A., Nelson, B. R., McAnally, J. R., et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606. doi: 10.1016/j.cell.2015.01.009
Arriaga-Canon, C., Fonseca-Guzmán, Y., Valdes-Quezada, C., Arzate-Mejía, R., Guerrero, G., and Recillas-Targa, F. (2014). A long non-coding RNA promotes full activation of adult gene expression in the chicken alpha-globin domain. Epigenetics 9, 173–181. doi: 10.4161/epi.27030
Boucher, C. A., Carey, N., Edwards, Y. H., Siciliano, M. J., and Johnson, K. J. (1996). Cloning of the human SIX1 gene and its assignment to chromosome 14. Genomics 33, 140–142. doi: 10.1006/geno.1996.0172
Buckingham, M., and Rigby, P. W. (2014). Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238. doi: 10.1016/j.devcel.2013.12.020
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. doi: 10.1101/gad.17446611
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Cheyette, B. N., Green, P. J., Martin, K., Garren, H., Hartenstein, V., and Zipursky, S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977–996. doi: 10.1016/0896-6273(94)90308-5
Clemson, C. M., McNeil, J. A., Willard, H. F., and Lawrence, J. B. (1996). XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275. doi: 10.1083/jcb.132.3.259
Dimitrova, N., Zamudio, J. R., Jong, R. M., Soukup, D., Resnick, R., Sarma, K., et al. (2014). LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 54, 777–790. doi: 10.1016/j.molcel.2014.04.025
Dinger, M. E., Pang, K. C., Mercer, T. R., and Mattick, J. S. (2008). Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4:e1000176. doi: 10.1371/journal.pcbi.1000176
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Eggers, S., Ohnesorg, T., and Sinclair, A. (2014). Genetic regulation of mammalian gonad development. Nat. Rev. Endocrinol. 10, 673–683. doi: 10.1038/nrendo.2014.163
Feng, M., Dai, M., Xie, T., Li, Z., Shi, M., and Zhang, X. (2016). Innate immune responses in ALV-J infected chicks and chickens with hemangioma in vivo. Front. Microbiol. 7:786. doi: 10.3389/fmicb.2016.00786
Fougerousse, F., Durand, M., Lopez, S., Suel, L., Demignon, J., Thornton, C., et al. (2002). Six and Eya expression during human somitogenesis and MyoD gene family activation. J. Muscle Res. Cell Motil. 23, 255–264. doi: 10.1023/A:1020990825644
Frith, M. C., Bailey, T. L., Kasukawa, T., Mignone, F., Kummerfeld, S. K., Madera, M., et al. (2006a). Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 3, 40–48. doi: 10.4161/rna.3.1.2789
Frith, M. C., Forrest, A. R., Nourbakhsh, E., Pang, K. C., Kai, C., Kawai, J., et al. (2006b). The abundance of short proteins in the mammalian proteome. PLoS Genet. 2:e52. doi: 10.1371/journal.pgen.0020052
Gordon, B. S., Delgado, D. D., White, J. P., Carson, J. A., and Kostek, M. C. (2012). Six1 and Six1 cofactor expression is altered during early skeletal muscle overload in mice. J. Physiol. Sci. 62, 393–401. doi: 10.1007/s12576-012-0214-y
Grifone, R., Laclef, C., Spitz, F., Lopez, S., Demignon, J., Guidotti, J. E., et al. (2004). Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 24, 6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004
Grifone, R., Demignon, J., Houbron, C., Souil, E., Niro, C., Seller, M. J., et al. (2005). Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132, 2235–2249. doi: 10.1242/dev.01773
Guil, S., and Esteller, M. (2012). Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19, 1068–1075. doi: 10.1038/nsmb.2428
Harms, M. J., Lim, H. W., Ho, Y., Shapira, S. N., Ishibashi, J., Rajakumari, S., et al. (2015). PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29, 298–307. doi: 10.1101/gad.252734.114
Hetzler, K. L., Collins, B. C., Shanely, R. A., Sue, H., and Kostek, M. C. (2014). The homoeobox gene SIX1 alters myosin heavy chain isoform expression in mouse skeletal muscle. Acta Physiol. 210, 415–428. doi: 10.1111/apha.12168
Kallen, A. N., Zhou, X. B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52, 101–112. doi: 10.1016/j.molcel.2013.08.027
Karapetyan, A. R., Buiting, C., Kuiper, R. A., and Coolen, M. W. (2013). Regulatory Roles for Long ncRNA and mRNA. Cancers 5, 462–490. doi: 10.3390/cancers5020462
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 3:r68. doi: 10.1126/scisignal.2000568
Kong, L., Zhang, Y., Ye, Z. Q., Liu, X. Q., Zhao, S. Q., Wei, L., et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349. doi: 10.1093/nar/gkm391
Laclef, C., Hamard, G., Demignon, J., Souil, E., Houbron, C., and Maire, P. (2003). Altered myogenesis in Six1-deficient mice. Development 130, 2239–2252. doi: 10.1242/dev.00440
Lee, J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439. doi: 10.1126/science.1231776
Lee, J. T., and Bartolomei, M. S. (2013). X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323. doi: 10.1016/j.cell.2013.02.016
Li, T., Wang, S., Wu, R., Zhou, X., Zhu, D., and Zhang, Y. (2012). Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 99, 292–298. doi: 10.1016/j.ygeno.2012.02.003
Li, Z., Ouyang, H., Zheng, M., Cai, B., Han, P., Abdalla, B. A., et al. (2017). Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression profiles reveals the potential role of LncRNAs in skeletal muscle development of the chicken. Front. Physiol. 7:687. doi: 10.3389/fphys.2016.00687
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, W., Wu, H., Ye, Y., Li, Z., Hao, S., Kong, L., et al. (2014). The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 5, e1347. doi: 10.1038/cddis.2014.289
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. doi: 10.1038/nature21034
Mousavi, K., Zare, H., Dell'Orso, S., Grontved, L., Gutierrez-Cruz, G., Derfoul, A., et al. (2013). eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617. doi: 10.1016/j.molcel.2013.07.022
Mueller, A. C., Cichewicz, M. A., Dey, B. K., Layer, R., Reon, B. J., Gagan, J. R., et al. (2015). MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol. 35, 498–513. doi: 10.1128/MCB.01079-14
Nelson, B. R., Makarewich, C. A., Anderson, D. M., Winders, B. R., Troupes, C. D., Wu, F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. doi: 10.1126/science.aad4076
Nonomura, K., Takahashi, M., Wakamatsu, Y., Takano-Yamamoto, T., and Osumi, N. (2010). Dynamic expression of Six family genes in the dental mesenchyme and the epithelial ameloblast stem/progenitor cells during murine tooth development. J. Anat. 216, 80–91. doi: 10.1111/j.1469-7580.2009.01167.x
O'Brien, J. H., Hernandez-Lagunas, L., Artinger, K. B., and Ford, H. L. (2014). MicroRNA-30a regulates zebrafish myogenesis through targeting the transcription factor six1. J. Cell Sci. 127, 2291–2301. doi: 10.1242/jcs.143677
Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., et al. (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573. doi: 10.1038/nature01266
Oliver, G., Mailhos, A., Wehr, R., Copeland, N. G., Jenkins, N. A., and Gruss, P. (1995). Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121, 4045–4055.
Ouyang, H., He, X., Li, G., Xu, H., Jia, X., Nie, Q., et al. (2015). Deep sequencing analysis of miRNA Expression in breast muscle of fast-growing and slow-growing broilers. Int. J. Mol. Sci. 16, 16242–16262. doi: 10.3390/ijms160716242
Ozaki, H., Watanabe, Y., Takahashi, K., Kitamura, K., Tanaka, A., Urase, K., et al. (2001). Six4, a putative myogenin gene regulator, Is not essential for mouse embryonal development. Mol. Cell. Biol. 10, 3343–3350. doi: 10.1128/MCB.21.10.3343-3350.2001
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246. doi: 10.1016/j.molcel.2008.08.022
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Sato, S., Ikeda, K., Shioi, G., Nakao, K., Yajima, H., and Kawakami, K. (2012). Regulation of Six1 expression by evolutionarily conserved enhancers in tetrapods. Dev. Biol. 368, 95–108. doi: 10.1016/j.ydbio.2012.05.023
Scanes, C. G., Harvey, S., Marsh, J. A., and King, D. B. (1984). Hormones and growth in poultry. Poult. Sci. 63, 2062–2074. doi: 10.3382/ps.0632062
Stierwald, M., Yanze, N., Bamert, R. P., Kammermeier, L., and Schmid, V. (2004). The Sine oculis/Six class family of homeobox genes in jellyfish with and without eyes: development and eye regeneration. Dev. Biol. 274, 70–81. doi: 10.1016/j.ydbio.2004.06.018
Wang, H., Jin, H., Liu, H., Sun, L., Li, X., Yang, C., et al. (2014). Molecular cloning and expression pattern of duck Six1 and its preliminary functional analysis in myoblasts transfected with eukaryotic expression vector. Indian J. Biochem. Biophys. 51, 271–281.
Wu, W., Ren, Z., Wang, Y., Chao, Z., Xu, D., and Xiong, Y. (2011). Molecular characterization, expression patterns and polymorphism analysis of porcine Six1 gene. Mol. Biol. Rep. 38, 2619–2632. doi: 10.1007/s11033-010-0403-9
Wu, W., Ren, Z., Chen, C., Liu, Y., Zhang, L., Chao, Z., et al. (2012). Subcellular localization of different regions of porcine Six1 gene and its expression analysis in C2C12 myoblasts. Mol. Biol. Rep. 39, 9995–10002. doi: 10.1007/s11033-012-1868-5
Xu, P. X., Zheng, W., Huang, L., Maire, P., Laclef, C., and Silvius, D. (2003). Six1 is required for the early organogenesis of mammalian kidney. Development 130, 3085–3094. doi: 10.1242/dev.00536
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674. doi: 10.1016/j.molcel.2010.03.021
Zhang, T., Zhang, X., Han, K., Zhang, G., Wang, J., Xie, K., et al. (2017). Analysis of long noncoding RNA and mRNA using RNA sequencing during the differentiation of intramuscular preadipocytes in chicken. PLoS ONE 12:e0172389. doi: 10.1371/journal.pone.0172389
Zhao, X., Tang, Z., Zhang, H., Atianjoh, F. E., Zhao, J. Y., Liang, L., et al. (2013). A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16, 1024–1031. doi: 10.1038/nn.3438
Keywords: lncRNA-Six1, cis-acting, micropeptide, Six1, muscle growth
Citation: Cai B, Li Z, Ma M, Wang Z, Han P, Abdalla BA, Nie Q and Zhang X (2017) LncRNA-Six1 Encodes a Micropeptide to Activate Six1 in Cis and Is Involved in Cell Proliferation and Muscle Growth. Front. Physiol. 8:230. doi: 10.3389/fphys.2017.00230
Received: 16 January 2017; Accepted: 31 March 2017;
Published: 20 April 2017.
Edited by:
Kevin I. Watt, Baker IDI Heart and Diabetes Institute, AustraliaReviewed by:
Ling Lian, China Agricultural University, ChinaCopyright © 2017 Cai, Li, Ma, Wang, Han, Abdalla, Nie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghua Nie, bnFpbmdodWFAc2NhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.