
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 11 April 2017
Sec. Vascular Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.00214
 Jie Zhang1†
Jie Zhang1† Jialing Qi1†
Jialing Qi1† Shuilong Wu2
Shuilong Wu2 Lijiao Peng2
Lijiao Peng2 Yunwei Shi3
Yunwei Shi3 Jinxian Yang4
Jinxian Yang4 Zhenhua Yin1
Zhenhua Yin1 Yu Gao2
Yu Gao2 Chengniu Wang1
Chengniu Wang1 Jie Gong5
Jie Gong5 Haijun Zhang6
Haijun Zhang6 Jingjing Zhang2*
Jingjing Zhang2* Dong Liu3*
Dong Liu3*The monolayer of endothelial cells (ECs) lining the intima of all blood vessel wall forms a semipermeable barrier that regulates tissue-fluid homeostasis, transport of nutrients, and migration of blood cells across the barrier. A number of signaling pathways and molecules mediate endothelial permeability, which plays important roles in a variety of the physiological and pathological conditions. Fatty acid binding proteins (FABPs) are able to bind various hydrophobic molecules, such as long-chain fatty acids, prostaglandins and eicosanoids. FABP4, a member of the family of FABPs, plays an important role in maintenance of glucose and lipid homeostasis as well as angiogenesis. In the present study, we found that fabp11a, the ortholog of mammalian FABP4, was highly expressed in developing brain vessels of zebrafish. Knockout of fabp11a gene caused hemorrhage in zebrafish brain. Morpholino mediated fabp11a gene knockdown phenocopied the hemorrhage in mutants. Furthermore, we demonstrated permeability of brain vessels in fabp11a mutant is significantly higher than that of control. In addition, COX and LOX inhibition partially rescued the brain vessel integrity defects caused by fabp11a loss-of-function, suggesting the integrity defect was relevant to the Fatty Acid function.
The vascular system of vertebrates composed of a well-organized and hierarchical network of blood vessels, including arteries, veins, and capillaries. This network overspreads throughout every tissue and organ of the body and is adapt to the physiological function in local microenvironment (Risau, 1997; Carmeliet and Tessier-Lavigne, 2005; Larrivee et al., 2009). The development of vascular system undergoes two sequential processes, vasculogenesis and angiogenesis (Risau, 1997; Carmeliet and Tessier-Lavigne, 2005; Larrivee et al., 2009). Vasculogenesis is a de novo formation of the blood vessels, whereas angiogenesis is the outgrowth of endothelial cells (ECs) to form new branches from a pre-existing vasculature (Risau and Flamme, 1995; Risau, 1997). The monolayer of endothelium lining the intima of all blood vessel wall forms a semipermeable barrier that regulates tissue-fluid homeostasis, transport of nutrients, and migration of blood cells across the barrier. A number of signaling pathways and molecules mediate endothelial permeability, which is important in a variety of the physiological and pathological conditions (Komarova et al., 2017).
Fatty acid binding protein 4 (FABP4, adipocyte-FABP, aP2), plays an important role in maintenance of glucose and lipid homeostasis as well as inflammation through its actions in adipocytes and macrophages (Kuwano et al., 1993; Li and Wilson, 1997), belongs to a family of intracellular FABPs. There are 9 highly conserved FABP family members which are cytosolic proteins with small molecular weight around 15 kDa. FABP members are expressed specifically in different tissues with some overlaps (Hertzel and Bernlohr, 2000). FABPs function in binding to a variety of hydrophobic ligands, such as long-chain fatty acids, prostaglandins, leukotrienes, and eicosanoids (Haunerland and Spener, 2004; Makowski and Hotamisligil, 2005). Compared with other pan-endothelial cell markers such as CD31, the expression of FABP4 is restricted to microvascular and small vascular ECs, which contributes to angiogenic responses, such as cell proliferation and migration (Elmasri et al., 2012). Recent studies in mice have shown that endothelial FABP4 are involved in some angiogenesis-related processes, such as EC survival, migration, and angiogenic sprouting (Elmasri et al., 2012). However, the developmental role of FABP4 in vasculature remains largely unknown.
The zebrafish (Danio rerio) has been shown by large amounts of studies as a powerful in vivo vertebrate model system for the study of vasculogenesis and angiogenesis (Weinstein, 2002). Combined with the transgenic methods to label the vascular ECs with fluorescent proteins, it allows high-resolution imaging of blood vessel in live embryos (Lawson and Weinstein, 2002; Wang et al., 2016). Recently, arising reverse genetic tools, such the antisense morpholino oligonucleotide (MO)-based gene knockdown strategies (Nasevicius and Ekker, 2000; Eisen and Smith, 2008) and genomic editing techniques (Huang et al., 2011; Zhu et al., 2011; Bedell et al., 2012; Gupta et al., 2012; Chang et al., 2013; Hwang et al., 2013), have been widely applied in manipulating the gene expression and function in zebrafish (Beis and Stainier, 2006). In this study, we examined the role of Fabp4 in blood vessel development in zebrafish. Current study provides new insight into the role of fatty acid binding protein in blood vessel.
All animal experimentation was carried out in accordance with the NIH Guidelines for the care and use of laboratory animals (http://oacu.od.nih.gov/regs/index.htm) and ethically approved by the Administration Committee of Experimental Animals, Jiangsu Province, China [Approval ID: SYXK (SU) 2007–0021]. Care and breeding of zebrafish was carried out essentially as we previously described (Wang et al., 2016). AB and transgenic zebrafish lines: Tg(kdrl:EGFP) and Tg(gata1:DsRed) used were described in our previous work (Wang et al., 2016).
The zebrafish fabp11a exons information is got from Ensembl (http://asia.ensembl.org/Danio_rerio/Transcript/Summary?db=core;g=ENSDARG00000017299;r=19:32579389-32581848;t=ENSDART00000021798). The protein molecular weight is calculated using Protein Molecular Weight Calculator (http://www.sciencegateway.org/tools/proteinmw.htm). And the phylogenetic tree was built using PhyML software (Guindon et al., 2010). The protein sequences alignment were performed using T-coffee and edited by Jalview (Waterhouse et al., 2009; Di Tommaso et al., 2011).
Total RNA of zebrafish embryos was isolated with Trizol (Invitrogen) and stored at –80°C. The total amount of extracted RNA was measured by Nanodrop. Genomic cDNA was then synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche). The PCR amplifications were carried out in a total volume of 20 or 50 μl with specific primers (Supplementary Table 1) and Advantage 2 Polymerase Kit (Clontech). The primers for Real-time PCR analysis of fabp11a was described in our previous work (Qi et al., 2016). Quantitative PCR was carried out in triplicate using the FastStart Universal SYBR Green Master Mix (Roche Applied Science) on a real-time PCR detection system (StepOne™ Real-Time PCR Systems).
The procedure for whole-mount in situ hybridization was carried out as described previously (Huang et al., 2013). The antisense RNA probe used was described in our previous work (Qi et al., 2016).
Morpholino (Gene Tools) were resuspended in DNase/RNase-free distilled water at a stock concentration of 1 mM according to the manufacturer's instruction and stored at −20°C. MOs were diluted to 0.3 mM and injected into one-cell stage embryos. The sequences of the morpholino targeting mfsd2aa used was (named: mfsd2aa-MO): 5′-ACCATTTTCCCGAATAATGATGCTC-3′. The sequence of morpholino targeting fabp11a was listed in previous work (Qi et al., 2016).
Rapamycin, L-NAME and SB203580 LY294002, inhibitors of m-TOR, eNOS and P38 respectively, were purchased from Sigma-Aldrich. Rapamycin dissoveled in DMSO and was used at a final concentration of 10 nM. L-NAME and SB203580 were dissolved in tank water and were used at a final concentration of 10 uM. The inhibitors of Cyclooxygenase (COX) indomethoinc and Lornoxican were used at the final concentration 28 nM and 16 nM respectively. The inhibitor of Lipoxygenase (LOX) nordihyroguaria-retic acid (NDGA) was used at the final concentration 14 uM. O-dianisidine staining for globin was completed as described previously (Jiang et al., 2009).
For confocal imaging, Tg(kdrl:EGFP) or Tg(gata1:DsRed) transgenic lines were used to investigate the blood vessel development. Embryos were anesthetized with egg water containing 0.16 mg/mL tricaine and 1% 1-phenyl-2 thiourea (Sigma), and mounted in 0.6% low melting agarose. Confocal images was taken with a Leica TCS-SP5 LSM. Analysis was performed using Imaris software. 10 kD and 40 kD dextran was injected into the heart, and fluorescence was visualized by confocal imaging. For the results of in situ hybridization and Tg(gata1:DsRed) and zebrafish embryos, images were acquired with an Olympus DP71 camera on an Olympus stereomicroscope MVX10.
Statistical analysis was performed using GraphPad Prism® version 6.0c. Fisher's exact test, χ2-test and Student's t-test were used (P < 0.05).
In zebrafish, fabp11a gene was identified as the ortholog of mammalian FABP4 (Liu et al., 2007; Karanth et al., 2008). A number of closely related syntenic genes in the region of the zebrafish fabp11a locus were identified to be conserved with human FABP4, confirming that zebrafish fabp4 is the orthologous gene of mammalian FABP4 (Liu et al., 2007). The zebrafish fabp11a gene localizes on chromosome 19, and is composed of 4 exons spanning around 2.46 KB (Figure 1A). According to the sequence information, it is predicted to encode a 134 amino acid protein (NCBI Reference Sequence: NP_001004682.1, gi: 52219194), with low molecular weight of around 15.12 KDa. Multi-alignment analyses indicated that zebrafish Fabp11a and other vertebrate FABP4 proteins have orthologous relationship (Supplementary Table 2). The sequences of FABP4 from different species of vertebrates showed significant high similarities (Figure 1B). The phylogenetic analysis of selected vertebrate FABP4 proteins shows that zebrafish (Danio rerio) Fabp11a is mostly closed to Mus musculus FABP4, followed by human FABP4 (Figure 1C). These results indicate that FABP4 protein is highly conserved during the evolution of vertebrate, suggesting the functional importance of FABP4.
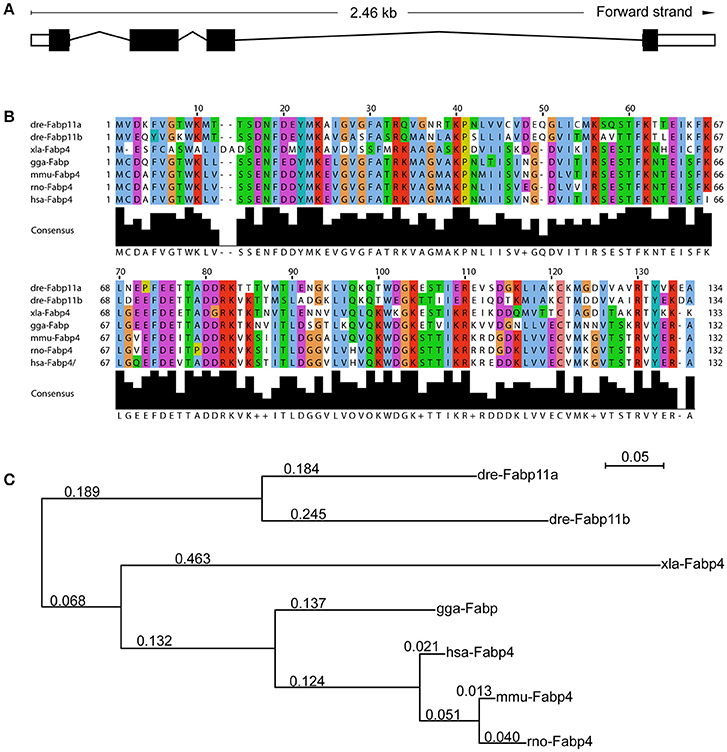
Figure 1. Functional domains of Fabp11a are highly conserved during vertebrates. (A) The fabp11a gene is composed of 4 exons spanning around 2.46 KB. (B) Alignment of Fabp11a motor domain amino acid residue sequences of Danio rerio (NP_001004682.1), Danio rerio (AAI65498.1) Fabp11b, Gallus gallus FABP4 (NP_989621.1), Xenopus laevis FABP4 (NP_001089252.1), Mus musculus FABP4 (CAJ18597.1), Rattus norvegicus FABP4 (AAH84721.1), and Homo sapiens FABP4 (CAG33184.1). The sequences are retrieved from NCBI Protein sequence database. (C) Phylogenetic tree of amino acid sequences generated using the PhyML software.
After in silicon analysis of zebrafish Fabp11a, we carried out the whole-mount in situ hybridization experiments to analyze the temporal and spatial expression of fabp11a during zebrafish blood vessel development. We did not detect the expression of fabp11a in zebrafish blood vessels at stages earlier than 24 hpf (Figure 2A). fabp11a mRNA was discovered to emerge in mid-cerebral vein (MCeV) at 24 hpf (Figures 2B,B'). From 30 hpf, fabp11a was present in part of primordial hindbrain channel (PHBC), where central arteries (CtAs) initiate to branch, and gradually expanded to the entire PHBC and CtAs at 48 hpf (Figures 2C–E”). Additionally, we also observed that fabp11a was expressed in anterior cerebral Vein (ACeV) and metencephalic artery (MtA) at 72 hpf (Figures 2F–F”).
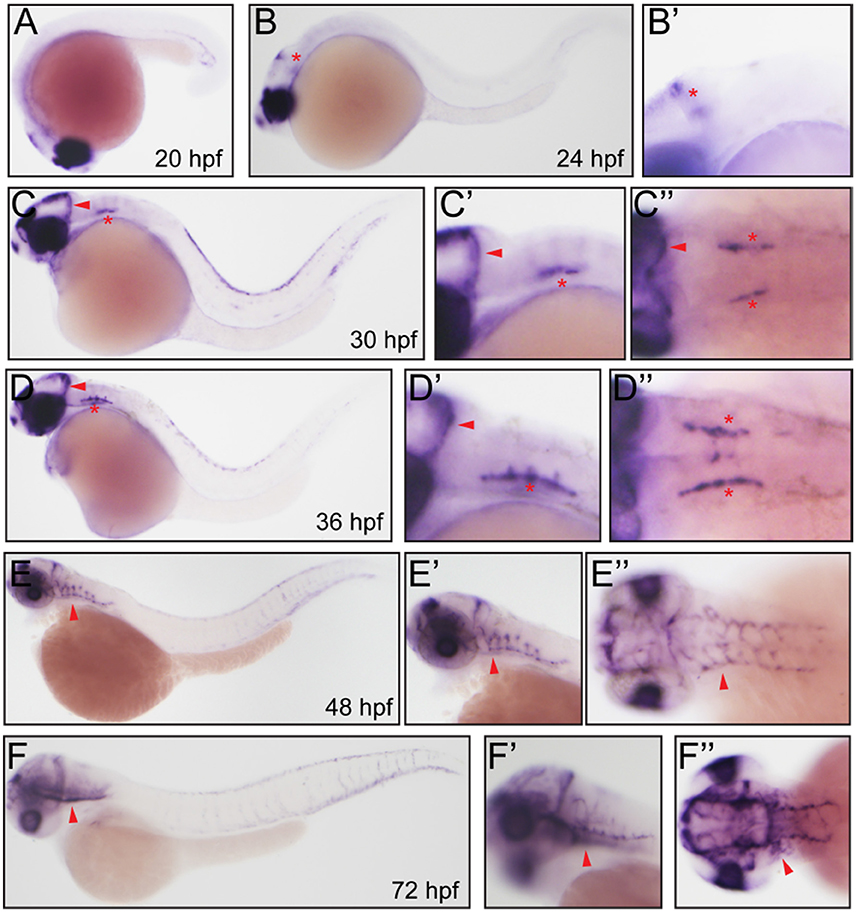
Figure 2. Whole mount in situ hybridization analysis of fabp11a in zebrafish embryos. (A) 20 hpf, lateral view. (B) 24 hpf, lateral view. Red asterisk indicates mid-cerebral vein (MCeV). (B') 24 hpf, lateral view. Red asterisk indicates mid-cerebral vein (MCeV). (C) 30 hpf, lateral view. Red arrowhead indicates MCeV; Red asterisk indicates primordial hindbrain channel (PHBC). (C') 30 hpf, lateral view. Red arrowhead indicates MCeV; Red asterisk indicates PHBC. (C”) 30 hpf, dorsal view. Red arrowhead indicates MCeV; Red asterisks indicate PHBC. (D) 36 hpf, lateral view. Red arrowhead indicates MCeV; Red asterisk indicates primordial hindbrain channel (PHBC). (D') 36 hpf, lateral view. Red arrowhead indicates MCeV; Red asterisk indicates PHBC. (D”) 36 hpf, dorsal view. Red arrowhead indicates MCeV; Red asterisks indicate PHBC. (E,E') 48 hpf, lateral view. Red arrowhead indicates brain vessels. (E”) 48 hpf, dorsal view. Red arrowhead indicates brain vessels. (F,F') 72 hpf, lateral view. Red arrowhead indicates brain vessels. (F”) 72 hpf, dorsal view. Red arrowhead indicates brain vessels.
To uncover the biological function of fabp11a in zebrafish brain vessels, we did the confocal imaging analysis of fabp11a mutants, which were generated using CRISPR-associated (Cas) 9 system (CRISPR/Cas9), as previously described (Qi et al., 2016), on Tg(kdrl:EGFP) line. We did not find obvious morphological defects of the brain vessels, including MCeV, PHBC, and CtAs in fabp11a mutants (Figure 3A). However, we found that around 22% of the fabp11a mutants displayed hemorrhage in the head (Figure 3B). The leaked blood cell were evidently observed in the brain after O-dianisidine staining (Figure 3C) or in Tg(gata1:dsRed) line (Figure 3D), in which red blood cell is labeled with red florescence. Morpholino mediated fabp11a gene knock down phenocopied the hemorrhage in mutants (Figures S1A–D”), which confirmed that the hemorrhage phenotype was caused by fabp11a loss-of-function. Furthermore, we demonstrated that the hemorrhage phenotype could be rescued by co-injection of fabp11a mRNA together with fabp11a MO (Figure S1E).
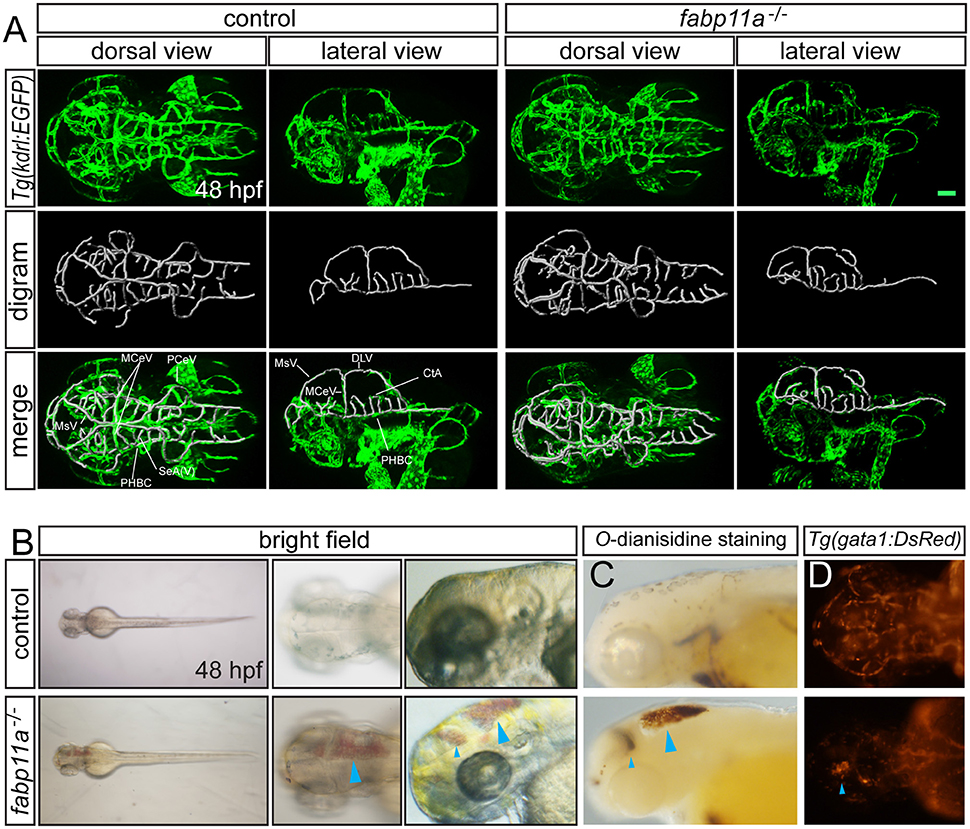
Figure 3. fabp11a knockout leads to zebrafish brain hemorrhage. (A) Confocal imaging analysis of brain vascular morphology in control and fabp11a mutants Tg(kdrl:EGFP) embryos at 48 hpf. (B) Microscopy analysis of control embryos and fabp11a mutants in bright field at 48 hpf. Blue arrowheads indicate hemorrhage in zebrafish head. (C) Microscope analysis of control embryos and fabp11a mutants with O-dianisidine staining in bright field at 48 hpf. Blue arrowheads indicate hemorrhage in zebrafish head. (D) Fluorescent microscopy imaging of Tg(gata1:DsRed) embryos at 48 hpf. Scale bar: 20 μm.
To investigate the permeability of cerebrovasculature after the loss of fabp11a, micro-injection of 10,000 and 40,000 MW Rhodamine-conjugated dextrans into the arterial vessel at 3 dpf followed by live imaging were performed. As a result, fluorescent dextran was observed being confined within vessels of control embryos (Figures 4A–A”). In contrast, dextran extravasated from cerebrovessels as evidenced by fluorescence within the brain parenchyma of fabp11a mutant larvae (Figures 4B–B”). In addition we measured the fluorescence intensity of extra- and intra-vascular and used E/I = extravascular fluorescence intensity /Intravascular fluorescence intensity to quantify the permeability of the selected vessels, where the fabp11a is expressed highly and found that permeability in fabp11a mutant is significantly higher than that of control (Figure 4C). These results suggest that fabp11a regulates brain vessel integrity in zebrafish.
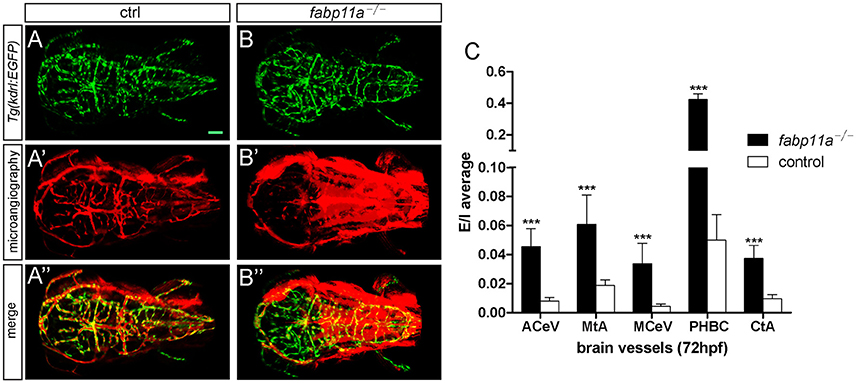
Figure 4. Zebrafish fabp11a mutants present disrupted brain vessel integrity. (A–B”) Confocal imaging analysis of fabp11a mutants and control Tg(kdrl:EGFP) embryos intracardiac injected with Rhodamine-conjugated dextran at 72 hpf. (C) Quantification of the permeability of the selected vessels in fabp11a mutants and control Tg(kdrl:EGFP) embryos using E/I = extravascular fluorescence intensity /Intravascular fluorescence intensity. T-test, ***P < 0.001, scale bar: 20 μm.
A number of signaling was implicated to the role of FABP4 in angiogenesis. To understand the potential mechanisms, through which fabp11a regulates the brain vessel integrity, we did a serials of experiments to investigate whether these pathways got involved (Figures S2A–D). Pharmacological inhibition of mTOR signaling with rapamycin resulted in significant reduction of branching angiogenesis of brain vessels but not the hemorrhage in head (Figure S2B). We also found that L-NAME (eNOS inhibitor) and SB203580 (P38 inhibitor) treatment induced the similar phenotypes with those caused by mTOR inhibition (Figures S2C,D). Through RT-PCR analysis, we showed fabp11a was downregulated in the rapamycin, L-NAME, and SB203580 treated embryos (Figure S2E).
The role of fatty acids (FA) on blood-brain barrier (BBB) permeability is well documented. It has been reported that in rat cerebral microvessels, inhibition of COX by indomethacin and LOX by nordihyroguaria-retic acid could effectively block the endothelial barrier defects caused by arachidonic acid (AA) (Easton and Fraser, 1998). AA is a precursor for the formation of various bioactive molecules including prostaglandins, such as PGE2, and leukotrienes. Since FABPs are capable of binding a variety of hydrophobic ligands, such as long-chain fatty acids, eicosanoids, leukotrienes and prostaglandins (Haunerland and Spener, 2004; Makowski and Hotamisligil, 2005), we examined whether COX inhibition by indomethacin and Lornoxican treatment and LOX inhibition by nordihyroguaria-retic acid treatment could rescue the brain vessel integrity defects caused by fabp11a deficiency. As a result, we found that treatment of indomethacin/Lornoxican, nordihyroguaria-retic acid separately, and their combination significantly reduced the ratio of brain vessel integrity defects caused by fabp11a deficiency (Figure 5A). Additionally, we discovered that the expression of cox2 was significantly upregulated (Figure 5C); meanwhile lox was slightly upregulated (Figure 5D), in fabp11a knockout embryos. In contrast, the expression of cox1 was not significantly affected (Figure 5B).
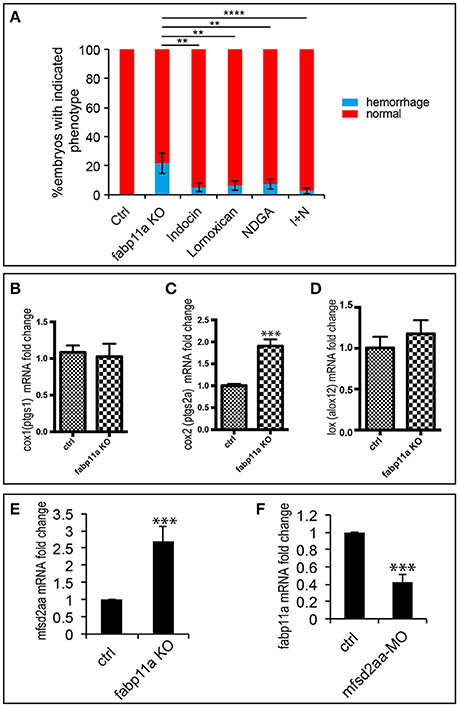
Figure 5. COX and LOX inhibition partially rescued the brain vessel integrity defects caused by fabp11a loss-of-function. (A) Percentage of embryos with hemorrhage phenotype in each group: control, fabp11a knockout, Indocin treated, Lornoxican treated, nordihyroguaria-retic acid (NDGA) treated, and Indocin and NDGA jointly (I+N) treated larvae. Fisher's exact test, **P < 0.01, ****P < 0.0001. (B–D) Real-time PCR analysis of expression of cox1, cox2, and lox in control and fabp11a knockout embryos. T-test, ***P < 0.001. (E) Real-time PCR analysis of mfsd2aa expression in control and fabp11a knockout embryos T-test, ***P < 0.001. (F) Real-time PCR analysis of fabp11a expression in control embryos and mfsd2aa morphants. T-test, ***P < 0.001.
During the process of lysophosphatidylcholine (LPC) synthesis, Mfsd2a could transport DHA and other needed fatty acids (Ben-Zvi et al., 2014; Nguyen et al., 2014). Given mfsd2aa zebrafish morphants present disruption of the blood vessel integrity (Guemez-Gamboa et al., 2015), we reasoned that mfsd2a is involved in the effects of FABP4 on vascular permeability. We did the expression analysis of mfsd2aa in the fabp11a mutants and fabp11a in mfsd2aa morphants by Real-time PCR, and found that mfsd2aa was upregulated in the fabp11a mutants (Figure 5E). In contrast, fabp11a was downregulated in mfsd2aa morphants (Figure 5F). However, either knockdown or upregulation of mfsd2aa in the fabp11a mutants failed to rescue the blood vessel integrity defects (Figure S3).
Fabp11a is highly expressed in the developing brain vessels of zebrafish. Currently, we revealed that fabp11a loss-of-function caused hemorrhage in the head and impaired brain vessel integrity for the first time.
Recently, accumulating data indicates that FABP4 was involved in the regulation of angiogenesis. Elmasri et al. reported that FABP4, which is expressed in the microvascular ECs in several normal tissues and organs, functions as a target of the VEGF/VEGFR2 signaling pathway and is a positive regulator during the mice and human endothelial cell (EC) proliferation (Elmasri et al., 2009). Consistently, It was also demonstrated that deficiency of FABP4 in human umbilical vein endothelial cells (HUVECs) and aortic rings from FABP4−/− mice leads to attenuation of angiogenic responses, such as endothelial cell migration, proliferation, apoptosis, and morphogenesis through modulation of several pathways, including p38, eNOS and SCF/c-kit signaling (Elmasri et al., 2009, 2012; Ghelfi et al., 2013). It was also found that EC-FABP4 is strongly regulated by the mTORC1 pathway and its inhibitor rapamycin in HUVECs (Elmasri et al., 2012). Later on, extended studies from the same lab showed that FABP4 is required for the retinal development but not for normal vascular development (Saint-Geniez et al., 2014). In addition, Harjes and his colleagues showed that DLL4-NOTCH could bind NICD to FABP4 promoter regions to regulate FABP4 gene expression directly, and endothelial NOTCH pathway is necessary for the FABP4 response to VEGFA signaling (Harjes et al., 2014). Currently we showed that chemical inhibition of p38 MAPK, eNOS and mTORC1 signaling did not cause hemorrhage in zebrafish. These results suggest that those are not the pathways responsible for brain vessel integrity defects caused by fabp11a deficiency. We also showed that COX and LOX inhibition partially rescued the brain vessel integrity defect caused by fabp11a loss-of-function, suggesting the integrity defect was relevant to the fatty acid function. We have shown that mfsd2aa was upregulated in the fabp11a mutants. However, neither knockdown nor upregulation of mfsd2aa in the fabp11a mutants could rescue the blood vessel integrity defects. These data suggests that mfsd2aa and fabp11a regulate vascular integrity in a different pathway. The upregulation of mfsd2aa in response to the fabp11a deficiency might be a compensational effect of brain vessel integrity defects.
In our previous study, we have shown that mutation of fabp11 causes dramatic defects in zebrafish eye development (Qi et al., 2016). However, the hemorrhagic phenotypes describe in this study were found in both the fabp11a mutants with or without eye defects. This suggests independent manners of Fabp11a mutation in affecting hemorrhagic phenotypes and the eye defects. It was demonstrated that fabp11a was highly enriched in zebrafish developing brain vessel. However, FABP4 mRNA is not detectable in adult mouse brain and brain vessels (Elmasri et al., 2009). This might be due to the differences of the developmental stages when the expression level of FABP4 was investigated. To date, there is no evidence to show that FABP4 knockout leads to hemorrhage in the head and brain vessel integrity defects in mice. A possible explanation is that this phenotype in mice is non-lethal and not examined so far. If this is not the case, in mice another paralog of FABP4 may play the role, as fabp11a of zebrafish, in regulation of vascular permeability.
DL and JinZ designed the experiments, and wrote the manuscript. JieZ, JQ, SW, LP, YS, JY, ZY, YG, CW, JG, and HZ did the experiments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by grants from the National Natural Science Foundation of China (31370824, 31400918, 81570447), the Special Support Plan of Excellent Young Talents, Guangdong, China (YueRencaiBan [2015] 8) and Initial Program of Excellent Young Teachers in Colleges and Universities in Guangdong Province, China (No. [2016] 6: YQ2015081).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00214/full#supplementary-material
Bedell, V. M., Wang, Y., Campbell, J. M., Poshusta, T. L., Starker, C. G., Krug, R. G. II., et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118. doi: 10.1038/nature11537
Beis, D., and Stainier, D. Y. (2006). In vivo cell biology: following the zebrafish trend. Trends Cell Biol. 16, 105–112. doi: 10.1016/j.tcb.2005.12.001
Ben-Zvi, A., Lacoste, B., Kur, E., Andreone, B. J., Mayshar, Y., Yan, H., et al. (2014). Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509, 507–511. doi: 10.1038/nature13324
Carmeliet, P., and Tessier-Lavigne, M. (2005). Common mechanisms of nerve and blood vessel wiring. Nature 436, 193–200. doi: 10.1038/nature03875
Chang, N., Sun, C., Gao, L., Zhu, D., Xu, X., Zhu, X., et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472. doi: 10.1038/cr.2013.45
Di Tommaso, P., Moretti, S., Xenarios, I., Orobitg, M., Montanyola, A., Chang, J. M., et al. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17. doi: 10.1093/nar/gkr245
Easton, A. S., and Fraser, P. A. (1998). Arachidonic acid increases cerebral microvascular permeability by free radicals in single pial microvessels of the anaesthetized rat. J. Physiol. 507(Pt 2), 541–547.
Eisen, J. S., and Smith, J. C. (2008). Controlling morpholino experiments: don't stop making antisense. Development 135, 1735–1743. doi: 10.1242/dev.001115
Elmasri, H., Ghelfi, E., Yu, C. W., Traphagen, S., Cernadas, M., Cao, H., et al. (2012). Endothelial cell-fatty acid binding protein 4 promotes angiogenesis: role of stem cell factor/c-kit pathway. Angiogenesis 15, 457–468. doi: 10.1007/s10456-012-9274-0
Elmasri, H., Karaaslan, C., Teper, Y., Ghelfi, E., Weng, M., Ince, T. A., et al. (2009). Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J. 23, 3865–3873. doi: 10.1096/fj.09-134882
Ghelfi, E., Yu, C. W., Elmasri, H., Terwelp, M., Lee, C. G., Bhandari, V., et al. (2013). Fatty acid binding protein 4 regulates VEGF-induced airway angiogenesis and inflammation in a transgenic mouse model: implications for asthma. Am. J. Pathol. 182, 1425–1433. doi: 10.1016/j.ajpath.2012.12.009
Guemez-Gamboa, A., Nguyen, L. N., Yang, H., Zaki, M. S., Kara, M., Ben-Omran, T., et al. (2015). Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat. Genet. 47, 809–813. doi: 10.1038/ng.3311
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gupta, A., Christensen, R. G., Rayla, A. L., Lakshmanan, A., Stormo, G. D., and Wolfe, S. A. (2012). An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods 9, 588–590. doi: 10.1038/nmeth.1994
Harjes, U., Bridges, E., McIntyre, A., Fielding, B. A., and Harris, A. L. (2014). Fatty acid-binding protein 4, a point of convergence for angiogenic and metabolic signaling pathways in endothelial cells. J. Biol. Chem. 289, 23168–23176. doi: 10.1074/jbc.M114.576512
Haunerland, N. H., and Spener, F. (2004). Fatty acid-binding proteins–insights from genetic manipulations. Prog. Lipid Res. 43, 328–349. doi: 10.1016/j.plipres.2004.05.001
Hertzel, A. V., and Bernlohr, D. A. (2000). The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol. Metab. 11, 175–180. doi: 10.1016/S1043-2760(00)00257-5
Huang, P., Xiao, A., Zhou, M., Zhu, Z., Lin, S., and Zhang, B. (2011). Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700. doi: 10.1038/nbt.1939
Huang, Y., Wang, x., Wang, X., Xu, M., Liu, M., and Liu, D. (2013). Nonmuscle myosin II-B (myh10) expression analysis during zebrafish embryonic development. Gene Expr. Patterns 13, 265–270. doi: 10.1016/j.gep.2013.04.005
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi: 10.1038/nbt.2501
Jiang, Q., Liu, D., Sun, S., Hu, J., Tan, L., Wang, Y., et al. (2009). Critical role of connexin43 in zebrafish late primitive and definitive hematopoiesis. Fish Physiol. Biochem. 36, 945–951. doi: 10.1007/s10695-009-9371-1
Karanth, S., Denovan-Wright, E. M., Thisse, C., Thisse, B., and Wright, J. M. (2008). The evolutionary relationship between the duplicated copies of the zebrafish fabp11 gene and the tetrapod FABP4, FABP5, FABP8, and FABP9 genes. FEBS J. 275, 3031–3040. doi: 10.1111/j.1742-4658.2008.06455.x
Komarova, Y. A., Kruse, K., Mehta, D., and Malik, A. B. (2017). Protein interactions at endothelial junctions and signaling mechanisms regulating endothelial permeability. Circ. Res. 120, 179–206. doi: 10.1161/CIRCRESAHA.116.306534
Kuwano, K., Bosken, C. H., Pare, P. D., Bai, T. R., Wiggs, B. R., and Hogg, J. C. (1993). Small airways dimensions in asthma and in chronic obstructive pulmonary disease. Am. Rev. Respir. Dis. 148, 1220–1225. doi: 10.1164/ajrccm/148.5.1220
Larrivee, B., Freitas, C., Suchting, S., Brunet, I., and Eichmann, A. (2009). Guidance of vascular development: lessons from the nervous system. Circ. Res. 104, 428–441. doi: 10.1161/CIRCRESAHA.108.188144
Lawson, N. D., and Weinstein, B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318. doi: 10.1006/dbio.2002.0711
Li, X., and Wilson, J. W. (1997). Increased vascularity of the bronchial mucosa in mild asthma. Am. J. Respir. Crit. Care Med. 156, 229–233. doi: 10.1164/ajrccm.156.1.9607066
Liu, R. Z., Saxena, V., Sharma, M. K., Thisse, C., Thisse, B., Denovan-Wright, E. M., et al. (2007). The fabp4 gene of zebrafish (Danio rerio)–genomic homology with the mammalian FABP4 and divergence from the zebrafish fabp3 in developmental expression. FEBS J. 274, 1621–1633. doi: 10.1111/j.1742-4658.2007.05711.x
Makowski, L., and Hotamisligil, G. S. (2005). The role of fatty acid binding proteins in metabolic syndrome and atherosclerosis. Curr. Opin. Lipidol. 16, 543–548. doi: 10.1097/01.mol.0000180166.08196.07
Nasevicius, A., and Ekker, S. C. (2000). Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26, 216–220. doi: 10.1038/79951
Nguyen, L. N., Ma, D., Shui, G., Wong, P., Cazenave-Gassiot, A., Zhang, X., et al. (2014). Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509, 503–506. doi: 10.1038/nature13241
Qi, J., Dong, Z., Shi, Y., Wang, X., Qin, Y., Wang, Y., et al. (2016). NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 26, 1349–1352. doi: 10.1038/cr.2016.134
Risau, W., and Flamme, I. (1995). Vasculogenesis. Annu. Rev. Cell Dev. Biol. 11, 73–91. doi: 10.1146/annurev.cb.11.110195.000445
Saint-Geniez, M., Ghelfi, E., Liang, X., Yu, C., Spencer, C., Abend, S., et al. (2014). Fatty acid binding protein 4 deficiency protects against oxygen-induced retinopathy in mice. PLoS ONE 9:e96253. doi: 10.1371/journal.pone.0096253
Wang, X., Ling, C. C., Li, L., Qin, Y., Qi, J., Liu, X., et al. (2016). MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 110, 140–150. doi: 10.1093/cvr/cvw023
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Weinstein, B. (2002). Vascular cell biology in vivo: a new piscine paradigm? Trends Cell Biol. 12, 439–445. doi: 10.1016/S0962-8924(02)02358-9
Keywords: FABP4, fabp11a, vessel permeability, knockout, zebrafish
Citation: Zhang J, Qi J, Wu S, Peng L, Shi Y, Yang J, Yin Z, Gao Y, Wang C, Gong J, Zhang H, Zhang J and Liu D (2017) Fatty Acid Binding Protein 11a Is Required for Brain Vessel Integrity in Zebrafish. Front. Physiol. 8:214. doi: 10.3389/fphys.2017.00214
Received: 01 February 2017; Accepted: 24 March 2017;
Published: 11 April 2017.
Edited by:
Jincai Luo, Peking University, ChinaCopyright © 2017 Zhang, Qi, Wu, Peng, Shi, Yang, Yin, Gao, Wang, Gong, Zhang, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Zhang, amluZ2ppbmcuemhhbmdAbGl2ZS5jb20=
Dong Liu, bGl1ZG9uZ3RvbUBnbWFpbC5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.