
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 04 April 2017
Sec. Redox Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.00191
Singlet oxygen generated in a type II photodynamic action, due to its limited lifetime (1 μs) and reactive distance (<10 nm), could regulate live cell function nanoscopically. The genetically-encoded protein photosensitizers (engineered fluorescent proteins such as KillerRed, TagRFP, and flavin-binding proteins such as miniSOG, Pp2FbFPL30M) could be expressed in a cell type- and/or subcellular organelle-specific manner for targeted protein photo-oxidative activation/desensitization. The newly emerged active illumination technique provides an additional level of specificity. Typical examples of photodynamic activation include permanent activation of G protein-coupled receptor CCK1 and photodynamic activation of ionic channel TRPA1. Protein photosensitizers have been used to photodynamically modulate major cellular functions (such as neurotransmitter release and gene transcription) and animal behavior. Protein photosensitizers are increasingly used in photon-driven nanomanipulation in cell physiology research.
Photodynamic action as a physiological curiosity has a long history, dating back to more than a century ago (for an early review on this topic, please see Blum, 1932). Investigation of photodynamic modulation of cellular physiology, however, has been rather limited until the recent past. A number of technological advances in photodynamic research have been made in the past few years. The newly renovated photodynamic modulation is now poised to be used on a much wider scale in physiological research.
A typical photodynamic action involves light, light-absorbing organic molecule (photosensitizer, S), and oxygen. Singlet oxygen is generated in a Type II photodynamic action. The photosensitizer (S) after absorption of a photon (hυ) of appropriate wavelength is excited from ground state (S) to the singlet excited state (1S). The 1S then undergoes a physical process named intersystem crossing (isc), to reach the triplet excited state (3S). If the triplet state is sufficiently long-lived, its excitation energy can be transferred to the ground state molecular oxygen (1), to generate the delta singlet oxygen (1Δg)(1O2). The singlet oxygen so generated can react with cellular components (A), to trigger the full-scale cellular photodynamic responses (Cui and Matthews, 1998; Cui et al., 2012; Dai et al., 2012) (Scheme 1).
SCHEME 1
Uncontrolled photodynamic action is detrimental as noted in porphyria patients (Kaestner et al., 2004; Norman, 2005), but measured photodynamic action has been utilized for major clinical advances. Concentrated generation of singlet oxygen at mega doses is cytocidal in different patterns (Agostinis et al., 2011; Krammer and Verwanger, 2012; Bacellar et al., 2015; Abrahamse and Hamblin, 2016). Singlet oxygen could trigger apoptosis, for example, by oxidizing multiple proteins (such as Bcl-2, Bcl-XL, BAX, BID) in the apoptosis pathway (Oleinick and Evans, 1998; Xue et al., 2001; Usuda et al., 2002, 2003; Chiu et al., 2005; Wan et al., 2008; Liu et al., 2011). Due to such cytocidal effects of singlet oxygen, photodynamic action has been found to be effective in the clinical treatments of both cancers and non-malignant lesions (Kennedy et al., 1990; Bown et al., 2002; Mittra and Singerman, 2002; Brown et al., 2004; Szeimies et al., 2010; Agostinis et al., 2011; Bown, 2013; Huggett et al., 2014; Craig et al., 2015; Abrahamse and Hamblin, 2016; Liu et al., 2016; Newman, 2016). On the other hand, it has been found that controlled doses of singlet oxygen could modulate cellular signaling in different cell types such as glandular cells with proven high specificity (Matthews and Cui, 1989, 1990a,b; al-Laith et al., 1993; Cui and Kanno, 1997; Cui et al., 1997, 2000, 2003, 2012; Cui and Matthews, 1998; Hashikura et al., 2001; Cui and Guo, 2002a,b; An et al., 2003; Wang et al., 2003; Krammer and Verwanger, 2012; Bacellar et al., 2015). One particular noted case is the photodynamic activation of CCK1 receptors in rat pancreatic acinar cells.
Singlet oxygen generated in photodynamic action with the sulphonated aluminum phthalocyanine (SALPC), has been found to activate rather permanently the CCK1 receptor in rat pancreatic acinar cells (Cui and Kanno, 1997; An et al., 2003; Cui et al., 2012), but desensitizes the α1 adrenergic receptor in rat hepatocytes (Cui et al., 2000) and other G protein-coupled receptors (GPCR).
CCK-CCK receptors play important roles in both gastrointestinal (GI) and central nervous system (CNS) functions (Cawston and Miller, 2010; Yu and Smagghe, 2014), due to the wide-spread distribution of both CCK1 and CCK2 receptors (Miller and Gao, 2008; Dockray and Burdyga, 2011). Of the group A GPCR receptors, CCK1 is unique in that it can be permanently activated by photodynamic oxidation (Cui and Kanno, 1997; An et al., 2003; Cui et al., 2012). It may be noted that ligand (agonist CCK)-induced cytosolic calcium oscillations disappeared immediately after washout of CCK (Figure 1A), but the photodynamically-induced, ligand-independent calcium oscillations persisted well after photodynamic action (duration: 1 min) had completed (Figure 1B).
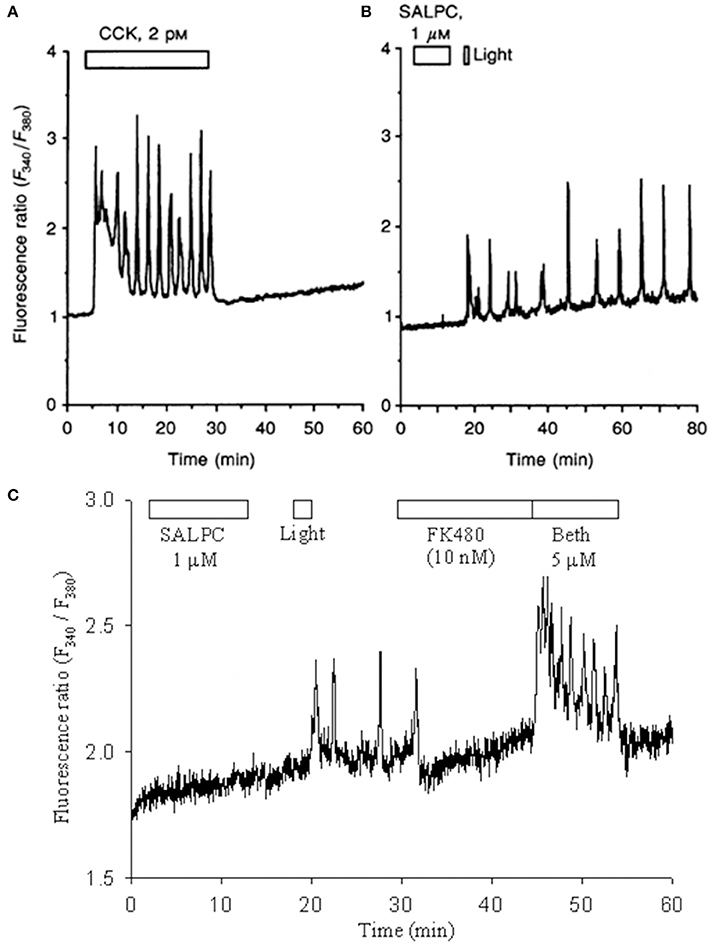
Figure 1. SALPC photodynamic action triggers permanent activation of CCK1 receptors and persistent calcium oscillations in rat pancreratic acinar cells. Fura-2 AM-loaded rat pancreatic acini were perifused, CCK (A), sulphonated aluminium phthalocyanine (SALPC, B,C), CCK1R antagonist FK480 (C), muscarinic agonist bethanechol (Beth, C), light illumination (λ > 580 nm) (55,000 lux, B; 53,000 lux or 72 mW/cm2, C) were applied, as indicated by the horizontal bars. Note that CCK-induced calcium oscillations ceased immediately after wash-out of CCK (A), but photodynamically-induced calcium oscillations persisted even after photodynamic action (B,C). The photodynamically-induced calcium oscillations were completely blocked by the CCK1R antagonist FK480, then stimulation with bethanechol still induced new regular calcium oscillations (C). Reproduced from Cui and Kanno (1997) and An et al. (2003).
An early hint for irreversible activation of CCK1 receptor was noted as early as 1980 by the Jamieson group. A photoaffinity probe to label the CCK1 receptor, CCK octapeptide (Asp-Tyr-(SO3H)-Met-Gly-Trp-Met-Asp-Phe-NH2, CCK-8) analog 2-nitro-5-azidobenzoyl-Gly-Asp-Tyr-(SO3H)-Met-Gly-Trp-Met-Asp-Phe-NH2 (NAB-Gly-CCK-8), was found to elicit irreversible secretion after UV irradiation of guinea pig pancreatic acini, although whether such secretion was mediated by the CCK1 receptor was not at that time verified (Galardy et al., 1980). A series of works by us have clearly delineated the irreversible nature of oxidative activation of CCK1 receptor (Matthews and Cui, 1989, 1990a,b; al-Laith et al., 1993; Cui et al., 1997, 2012; Cui and Kanno, 1997; An et al., 2003).
Photodynamic action with the photosensitizer sulphonated aluminum phthalocyanine (SALPC) was initially found to stimulate amylase secretion and regulate cytosolic signaling in the freshly isolated rat pancreatic acini (as reviewed in Cui and Matthews, 1998). In those experiments, the freshly isolated pancreatic acini were perifused, exposed to SALPC briefly (10 min), subsequent light illumination (2 min) then triggered persistent calcium oscillations which were completely blocked by CCK1 antagonist FK480 (10 nM) (An et al., 2003) (Figure 1C). After the blockade of calcium oscillations with FK480 (10 nM), the muscarinic agonist bethanechol (Beth) still triggered robust new calcium oscillations (An et al., 2003) (Figure 1C). These data indicated that after photodynamically-triggered calcium oscillations, pancreatic acinar cells remained perfectly healthy (An et al., 2003; Cui et al., 2012). Here the photodynamic action was restricted to the plasma membrane (Cui and Kanno, 1997; An et al., 2003; Cui et al., 2012).
Singlet oxygen in the cellular milieu has a short lifetime (μs) (Cui and Matthews, 1998; Bovis et al., 2012; Kim et al., 2014), therefore has a limited effective diffusion distance of <10 nm (Moan and Berg, 1991; Cui and Matthews, 1998; Dougherty et al., 1998; Nowis et al., 2005; Cui et al., 2012). Singlet oxygen generated in photodynamic action is, therefore, effective only at the site of generation, or at the site of photosensitizer localization in the cell.
Due to their intrinsic physicochemical properties, photosensitizers of different chemical classes tend to accumulate preferentially at specific subcellular sites such as the plasma membrane, the endoplasmic reticulum (ER), lysosomes, mitochondria, to modulate cellular activities from their different subcellular locations after photodynamic action (Theodossiou et al., 2006; Allison and Sibata, 2010; Agostinis et al., 2011). Hematoporphyrin derivative (HPD) monomers tend to accumulate at the mitochondria, and HPD oligomers at the plasma membrane (Scourides et al., 1987). Mono-aspartyl chlorin e6 (MACE) after endocytosis localizes to lysosomes (Berg and Moan, 1997). Phthalocyanines tend to accumulate at mitochondria (Peng et al., 1991). Benzoporphyrin derivative (BpD) is localized to the Golgi apparatus (Rosenkranz et al., 2000). Protoporphyrin IX (PPIX) precursor ALA-synthesized PPIX distributes to the plasma membrane, lysosomes and mitochondria (Kennedy et al., 1990).
Interestingly, photodynamic action at different cellular sites triggers cell death by distinct pathways. Photodynamic action at mitochondria and lysosomes triggers apoptosis; photodynamic action at the ER elicits autophagy; whereas photodynamic action at the plasma membrane induces necrosis (Almeida et al., 2004; Bacellar et al., 2015; Abrahamse and Hamblin, 2016). As mentioned above, SALPC photodynamic action at the plasma membrane induced permanent activation of CCK1 receptor (Cui and Kanno, 1997; An et al., 2003; Cui et al., 2012), photodynamic action with Victoria Blue VO at mitochondria reduced the oscillatory frequency of receptor-mediated calcium oscillations (Cui and Guo, 2002a,b).
It is recognized that the specific localization of chemical photosensitizers are only relative, distribution in multiple subcellular sites are quite common (Kessel, 1997, 2004, 2012; Kessel et al., 1997; Oleinick and Evans, 1998). The plasma membrane-localized photosensitizer MCP, for example, is also found in the ER and lysosomes; photosensitizer SnOPA is present in the plasma membrane, but also in the ER, lysosomes and elsewhere (for an in-depth review, see Kessel, 2004).
Mutated fluorescent proteins or engineered flavin-binding proteins have significantly enhanced photosensitivity compared with their parental proteins. Such genetically-encoded protein photosensitizers can be targeted precisely to subcellular organelles after tagging with signal sequences or after fusion with target proteins, resulting in high-precision spatially-controlled photodynamic action, or targeted protein oxidation. Locally-expressed protein photosensitizers after absorption of photons at specified wavelength generate the highly reactive singlet oxygen. Singlet oxygen as noted above has limited diffusion distance (<10 nm), therefore modulates target proteins or subcellular organelles nanoscopically.
Protein photosensitizers are either fluorescent protein variants such as KillerRed, KillerOrange, TagRFP, or flavin-binding proteins such as miniSOG and variants miniSOGQ102L/V, and Pp2FbFPL30M. The miniSOGQ102L is also named by some as singlet oxygen protein photosensitizer (SOPP). The chromophores (fluorophores) of KillerRed, KillerOrange, TagRFP are QYG, QWG, MYG, respectively, whereas miniSOG, miniSOGQ102L/V, Pp2FbFPL30M all share the same chromophore of FMN (Figure 2).
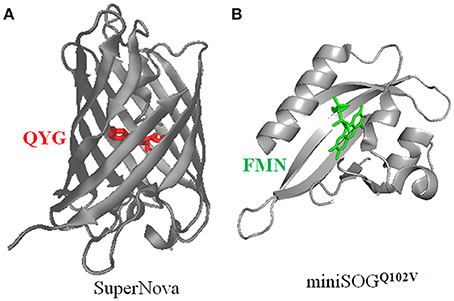
Figure 2. Three dimensional structures of SuperNova (A), miniSOGQ102V (B), with their respective chromophores highlighted. (A) The full amino acid sequence of SuperNova is obtained from PDB database (3WCK). The three dimensional structure of SuperNova is from PDB in pdb format, input to VMD graphics. The chromophore of Gln65-Tyr66-Gly67 (Takemoto et al., 2013) in SuperNova is highlighted in red. (B) The full amino acid sequence of miniSOGQ102V is from Rodríguez-Pulido et al. (2016). The sequence is put into the protein structure website Swiss-model, three-dimensional model is then obtained after a build-model step. The chromophore FMN in miniSOGQ102V is highlighted in green. Model building similar to Mironova et al. (2013).
The prototypical protein photosensitizer KillerRed is derived from the jellyfish chromoprotein anm2CP (239 residues, MWt 27 kD) (Bulina et al., 2006a,b; Pletnev et al., 2009), with point mutations of Thr145Asp, Cys161Gly (Shagin et al., 2004). KillerRed is composed of 11 anti-parallel β-sheets which form a barrel structure, with a central chromophore of Q65-Y66-G67 (see Figure 2) (Roy et al., 2010). An aqueous central channel/pore exists in the KillerRed structure, which is composed of the chromophore Q65-Y66-G67 and residues Ile142, Leu143, Pro144, Ile199, Ile200, Thr201. The excited chromophore can transfer its excitation energy to ground state molecular oxygen which has reached the chromophore region by diffusion through this channel; the generated ROS also exit KillerRed via the same channel (Carpentier et al., 2009; Pletnev et al., 2009; Serebrovskaya et al., 2009; Roy et al., 2010). The ROS quantum yield of KillerRed is more than 1000 times of EGFP (Bulina et al., 2006a,b; Carpentier et al., 2009; Pletnev et al., 2009). The current consensus is that KillerRed undergoes a Type I photodynamic action to generate superoxide anion, although it was previously thought to generate singlet oxygen by a Type II photodynamic action (Pletnev et al., 2009; Serebrovskaya et al., 2009; Shu et al., 2011; Vegh et al., 2011; Kim et al., 2014). A singlet oxygen-generating capacity cannot be completely ruled out, however (Roy et al., 2010; Petrova et al., 2016). Since KillerRed tends to dimerize, a monomeric mutant, Supernova, has been reported (Figure 2), which has the following 6 mutations compared with KillerRed: G3V, N145S, L160T, F162T, L172K, M204T (Takemoto et al., 2013). Supernova is believed to have similar photochemical properties as the parental KillerRed (Takemoto et al., 2013). The basic characteristics of KillerRed are listed in Table 1 (Bulina et al., 2006a,b; Lukyanov et al., 2010). Further mutations (G5C, Y68W, D119S, N147S, F179L, Y223H, E237Q) of KillerRed result in a blue-shifted KillerOrange, the photosensitization properties of KillerOrange remain to be investigated (Pletneva et al., 2015; Sarkisyan et al., 2015).
KillerRed fused with signal sequences can be targeted to the mitochondria in 293T cells (Bulina et al., 2006b), HEK293 cells, in the body wall muscle cells (Shibuya and Tsujimoto, 2012) and neurons of C. elegans (Williams et al., 2013). KillerRed can also be targeted to the lysosomes (Serebrovskaya et al., 2014), Golgi apparatus (Jarvela and Linstedt, 2014), the plasma membrane, mitochondria, or chromosomes (Shirmanova et al., 2013) in Hela cells. KillerRed has also been targeted to the plasma membrane in C. elegans neurons (Williams et al., 2013), zebrafish neurons and cardiomyocytes (Lee et al., 2010; Teh et al., 2010), or targeted to chromosomes in DU145 cells (Waldeck et al., 2011). The fusion protein TRF1-KillerRed has been used to target the telomeres (Sun et al., 2015). KillerRed fusion proteins (laminB1-KillerRed/histone2A-KillerRed) have been used to determine the spatial localization of chromosomal genes in the cell nucleus (Waldeck et al., 2013). The histone fusion protein H2B-KillerRed can be used to exert photodynamic blockade of cell division (Serebrovskaya et al., 2011; Shirmanova et al., 2015). Light irradiation of mitochondria-localized KillerRed (Mito-KillerRed) has been used to prune neuronal dendritic spines in defined dendritic regions of cultured neurons via the induction of caspase-3 activity (Ertuerk et al., 2014). Work done with larval zebrafish expressing KillerRed in the habenula afferent neurons from the ventral-lateral forebrain has helped to confirm that the habenula region is important for avoidance learning and helpless behavior (Lee et al., 2010).
TagRFP is derived from the Entacmaea quadricolor fluorescent protein TurboRFP (a random mutant of eqFP578), with mutations of R162E, Q166D, S180N, F198V, F200Y at the hydrophilic interface (Merzlyak et al., 2007). TagRFP has a central chromophore of M63-Y64-G65 (Subach et al., 2010). Light irradiated-TagRFP generates only 1O2, with a quantum yield of 0.004, but does not produce superoxide anion (Ragas et al., 2011). TagRFP has a high fluorescent quantum yield (Φfluo 0.48) and is widely used for fluorescent imaging (Merzlyak et al., 2007; Khrenova et al., 2015; Manoharan et al., 2015) (see Table 1). But when using TagRFP as a fluorescent probe for imaging, care must be taken that no undue photodamage is induced in either live cells or fixed tissue sections. Light illumination (532 nm, 40 mWatt/cm2) of TagRFP-expressing E. coli has been found to result in bacterial cell death (Ruiz-González et al., 2012).
The flavin-binding protein miniSOG is derived from the LOV2 domain of A. thaliana phototropin 2 with a Cys426Gly mutation, with further mutations S24G, I387M, N390S, S394T, F470L surrounding the chromophore (FMN) binding site (Shu et al., 2011). The resultant miniSOG is composed of 2 α-helix, interspersed with 5 β-sheets, with the chromophore FMN located between the α-helix and β-sheets (Pietra, 2014) (see Figure 2). Point mutation Cys426Gly facilitates transfer of FMN excitation energy absorbed from a photon to ground state O2 instead of to covalent bonding with Cys, significantly enhancing its 1O2 quantum yield (Shu et al., 2011). Initial report with anthracene-9, 10-dipropionic acid (ADPA) as the 1O2 probe obtained a quantum yield of 0.476 (Shu et al., 2011). Subsequent work has found that ADPA is also oxidized by other ROS (Ruiz-González et al., 2012). Direct measurement of 1O2 phosphorescence at 1275 nm, and the use of uric acid or PNS as 1O2 probes revealed a much lower quantum yield of 0.03 (Ruiz-González et al., 2012; Pimenta et al., 2013). The basic spectroscopic characteristics of miniSOG are listed in Table 1 (Shu et al., 2011; Ryumina et al., 2013; Wingen et al., 2014; Westberg et al., 2015).
Photosensitizer miniSOG can be targeted to the plasma membrane, mitochondria or chromosomes in Hela cells (as fusion proteins miniSOG-mem, miniSOG-mito, H2B-miniSOG) (Ryumina et al., 2013). In C. elegans motor neurons, miniSOG-TOMM-20 is targeted to mitochondrial outer membrane, whereas miniSOG-COX8a is targeted to mitochondrial matrix (Qi et al., 2012). miniSOG can be expressed as a fusion protein with the SDHC subunit (mev-1) of the mitochondrial respiratory chain complex II (succinate:ubiquinone oxidoreductase), to photodynamically inactivate with high specificity the respiratory chain complex II (on mitochondrial inner membrane) in C. elegans, without any damage toward its immediate neighbors complex I (NADH:ubiquinone oxidoreductase) or complex IV (cytochrome C oxidase) (Wojtovich et al., 2016). Photosensitizer miniSOG has also been fusion-expressed with vesicular SNARE proteins vesicule-associated membrane protein 2 (VAMP2) or synaptophysin 1 (SYP1), to target the small synaptic vesicles in cultured rat hippocampal neurons (Lin et al., 2013) or C. elegans neurons (Lin et al., 2013). miniSOG could be fusion-expressed with the synaptic active zone protein Munc13, to the pre-synaptic active zone in C. elegans neurons (Zhou et al., 2013).
Pp2FbFPL30M is derived from the LOV domain of the flavin-binding protein Pp2FbFP from Pseudomonas sputita, with a further mutation of L30M (Torra et al., 2015). Measurement of phosphorescence at 1275 nm found that the 1O2 quantum yield of Pp2FbFPL30M was 0.09 (Torra et al., 2015). The spectroscopic characteristics of Pp2FbFPL30M) are listed in Table 1 (Ryumina et al., 2013; Wingen et al., 2014).
miniSOGQ102L is also named SOPP, as mentioned above. In comparison with the parental miniSOG, in SOPP the FMN-binding glutamine is mutated to leucine (Q102L). This mutation reduces the hydrogen bond between Q102 and FMN, diminishing electron transfer, but enhancing energy transfer, with the net result of a much enhanced 1O2 quantum yield of 0.25 (Westberg et al., 2015, 2016). Another mutant, miniSOGQ102V (Figure 2), has an even higher 1O2 quantum yield of 0.39 (Rodríguez-Pulido et al., 2016). When miniSOGQ102L is expressed at the plasma membrane (by fusion with a PH domain) in C. elegans epithelial cells or in the cholinergic neurons, blue light illumination induces worm paralysis and neuronal injury, at efficiency higher than with miniSOG as the photosensitizer (Xu and Chisholm, 2016). The basic properties of miniSOGQ102L are listed in Table 1. A new version of miniSOG, miniSOG2, involves seven point mutations: G22S, G40P, Q44R, R57H, L84F, H85R, M89I. Mutations R57H, Q44R, G40P, L84F directly interacting with the chromophore FMN are likely responsible for the red-shifted excitation and emission spectra, together with enhanced singlet oxygen generation, but the quantum yield remains to be measured (Makhijani et al., 2017).
Subcellular targeting of protein photosensitizers has been done as mentioned above in different cell types, such as targeted KillerRed expression in Hela cells (Shirmanova et al., 2013; Jarvela and Linstedt, 2014; Serebrovskaya et al., 2014), HEK293 cells (Bulina et al., 2006b), DU145 cells (Waldeck et al., 2011), and miniSOG expression in Hela cells (Ryumina et al., 2013) or in cultured hippocampal neurons (Lin et al., 2013).
To target-express a protein photosensitizer, the photosensitizer gene needs to be fused with a subcellular localization sequence (SLS), to localize the protein photosensitizer to the desired subcellular compartments. KillerRed, for example, could be targeted to the plasma membrane (Bulina et al., 2006b; Teh et al., 2010) (plasma membrane LS, PMLS) with the N-terminal (20 residues) sequence of neuromodulin (Skene and Virág, 1989), or with the PH Delta1 sequence (Fujii et al., 1999; Bulina et al., 2006b). KillerRed can be targeted to mitochondria with the double sequences of MTS1 and MTS2 (Yang and Yang, 2011; Shibuya and Tsujimoto, 2012). Mitochondrial targeting sequence (MTS) can be derived from human cytochrome C oxygenase VIII subunit (Rizzuto et al., 1989, 1995), or from members of the respiratory chain complexes (Wojtovich et al., 2016). For lysosomal targeting, the C-terminal cytosolic tail sequence of the lysosomal-associated membrane protein II (LIMP II) could be used (Tabuchi et al., 2000). GTPase Rab7A sequence has been used to target KillerRed to lysosomes (Serebrovskaya et al., 2011; Ryumina et al., 2016). ER targeting could use the ER localization sequence (MLLSVPLLLGLLGLAVA) of calreticulin (Fliegel et al., 1989) and the ER retaining sequence KDEL (Munro and Pelham, 1987). The human β-1,4-galactotransferase N-terminal sequence can be used to target TagRFP to the Golgi apparatus (Shaner et al., 2008). Fusion proteins miniSOG-H2B, miniSOG-VAMP2, SYP1-miniSOG as mentioned above target miniSOG to Hela cell chromosomes (Ryumina et al., 2013), and to the small synaptic vesicles, respectively (Lin et al., 2013). KillerRed-TRF1 targets to the telomeres (Sun et al., 2015). The complete amino acid sequence of protein photosensitizers are listed in Table 2.
For whole organism studies, it is routine to place the desired gene under cell type- or tissue-specific promoters. Place the protein photosensitizer gene or gene construct under a suitable promoter, the sensitizer could then be expressed in that tissue (neuron, muscle, for example) only, in model animals C. elegans, zebrafish or others at the desired subcellular compartments (Serebrovskaya et al., 2011, 2014; Kobayashi et al., 2013; Williams et al., 2013). Cell-type specific viral vectors are also useful for in vivo injections. Engineered Muller cell-specific adenovirus variant containing the KillerRed gene, ShH10-KillerRed, for example, has been used to target the mouse retinal Muller cells. This study has confirmed the essential roles of Muller cells in visual perception and in normal retinal structure formation (Byrne et al., 2013). In this regard some technical strategies for gene delivery (Kaestner et al., 2015a; El-Shamayleh et al., 2016) and past works on tissue-specific expression of fluorescent protein sensors may be considered (Akemann et al., 2013; Kaestner et al., 2015b).
Other than subcellularlly-defined expression of protein photosensitizers, photodynamic action could be further spatially-delimited with ultra-structurally distinct point illumination. The recently emerged active illumination (AI) technology can be done at single or dual wavelengths, in multiple cells or cellular regions, simultaneously with imaging. The illumination light spot could vary in size, shape, and light intensity, with the spot size down to the theoretical diffraction limit (0.2 × 0.2 μm) (Shkryl et al., 2012). Active or selective illumination is made possible due to the invention of digital micromirror arrays, which can in real-time control the angle of each micromirror in the array (Shkryl et al., 2012).
Such selective illumination has been used to un-cage calcium or inositol 1, 4, 5-triphosphate (IP3) at multiple cellular sites simultaneously (Shkryl et al., 2012). Selective dual-wavelength illumination (390, 510 nm) could open or close, in tandem, designer photosensitive potassium channels in neurons (Janovjak et al., 2010). It has been shown that after selective irradiation of cultured vascular endothelial cells, localized photodynamic action readily triggered focused cell death in the irradiated areas (Feine et al., 2012). In channelrhodopsin 2 (ChR2)-expressing C. elegans, selective illumination of dendrites of sensory neurons stimulated cytosolic calcium increase, leading to enhanced worm activities (Cho and Sternberg, 2014).
Photodynamic modulation of cellular functions with protein photosensitizers is outlined above. Works in the following areas have emerged which are of particular significance in cellular physiology.
The earliest example of photodynamic modulation of native ionic channels is illustrated by the photodynamic blockade, with photosensitizers Rose Bengal and Eosin Y, of voltage-gated sodium channels (Nav) in the squid giant axons. In addition, photodynamic action also slowed or disrupted Nav inactivation (Oxford et al., 1977). Photodynamic action with Rose Bengal has also been found to inhibit other voltage-gated channels as well (Nav, Kv, Cav) in the isolated frog (Rana pipiens) atrial cardiomyocyte. After photodynamic action, Nav inactivation was significantly slowed; Cav inactivation was also inhibited (Tarr and Valenzeno, 1991). Photodynamic action with Rose Bengal was found to inhibit Cav, Kv, KCa channels in rat anterior pituitary cells GH3 (Valenzeno and Tarr, 1997).
Kv channels expressed in cell lines have been found to be inhibited by photodynamic action with porphyrins (photosensitizer) conjugated to subtype-specific monoclonal antibodies. Photodynamic action with anti-Kv4.2 mAb-porphyrin conjugates was found to facilitate photoablation of Kv4.2, but not of Kv4.3 or Kv2.1 (Sack et al., 2013). Similarly photodynamic action (laser light at 473 nm, 350 mW/cm2) with channel-binding photosensitizer FITC-cAMP (bound to CNBD in mHCN2) was found to inhibit mouse potassium channel mHCN2 expressed in Xenopus laevis oocytes. Photodynamic modulation of mHCN2 in the closed state decreased subsequently induced Ih current significantly. In contrast, photodynamic action enhanced the Iinst current and delayed channel inactivation when photosensitization was executed in the open channels (Gao et al., 2014). These latter two photodynamically-induced changes were determined to be due to oxidation or cross-linking of the His434 residue located at the cytosolic side of the S6 segment, because H434A mutation abolished the delay in channel deactivation, and the generation of Iinst (Gao et al., 2014). Interestingly, when miniSOG was fused to the C terminal end (C terminal end of CNBD) of mHCN2, light irradiation was found to exert (on mHCN2) effects similar to those observed with the chemical photosensitizer FITC-cAMP (Gao et al., 2014).
In contrast to the inhibitory effect on the voltage-gated channels Nav and Kv, photo-oxidation has been found to activate the calcium-permeant sensory channels TRPA1 and TRPV1 (Hill and Schaefer, 2009; Babes et al., 2016). Photodynamic action with photosensitizers acridine orange (490 nm), or hypericin (590 nm) was found to activate TRPA1 expressed in HEK293 cells (Hill and Schaefer, 2009). Photodynamic action with photosensitizer protoporphyrin IX was found to drastically activate both TRPA1 and TRPV1. Purified human TRPA1 inserted in artificial lipid bilayers was found to be activated only after photodynamic action with protoporphyrin IX and blue light (Babes et al., 2016) (Figure 3). Similar works involving the protein photosensitizers both ex vivo and in vivo are eagerly awaited.
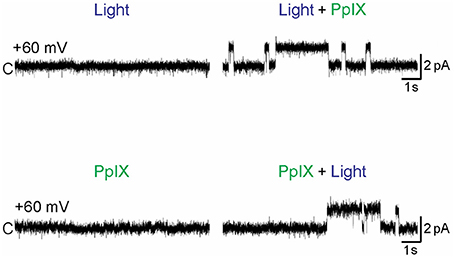
Figure 3. Photodynamic activation of TRPA1. Purified human TRPA1 reconstituted into artificial lipid (1,2-diphytanoyl-snglycero-3-phosphocholine: cholesterol was 9:1) bilayers with both the N- and C-terminals facing the cytosol were voltage-clamped at + 60 mV. PPIX (1 μM) was added to the cytosolic side, laser light (405 nm) was applied (at 0.45 mW/mm2). Single channel currents were measured using Patch-a-Patch in symmetrical K+ solution (150 mM KCl, 10 mM HEPES) under light illumination alone (Light), protoporphyrin IX alone (PPIX), or PPIX plus light (Light + PPIX) as indicated. Scale bars indicate 2 pA current and 1 s. Only simultaneous presence of light and PPIX (photodynamic action) led to channel opening. From Babes et al. (2016).
The SNARE complex proteins and associated synaptic active zone proteins are essential for regulated neurotransmitter release. Protein photosensitizer miniSOG fused with the v-SNARE proteins VAMP2 or synaptophysin (miniSOG-VAMP2, SYP1-miniSOG) was expressed in neonatal rat hippocampal slices or in cultured hippocampal and cortical neurons. Transgenic C. elegans worms with panneuronal expression of miniSOG-VAMP2 were also made. The fused miniSOG (miniSOG-VAMP2, SYP1-miniSOG) was found to localize to the synaptic vesicles, but without any effect on transmitter release or on animal behavior in the dark. Upon illumination with blue light (480 nm), neurotransmitter release in cultured neurons or in hippocampal slices were completely blocked, with the inhibition lasting for >1 h (Lin et al., 2013). In C. elegans panneuronally expressing miniSOG-VAMP2, light irradiation similarly led to marked inhibition of spontaneous neurotransmitter release, with reduced movement and worm paralysis (Lin et al., 2013). Light irradiation (480 nm) of C. elegans expressing miniSOG fused to the pre-synaptic active zone protein Munc13 led to similar acute inhibition of neurotransmitter release (Zhou et al., 2013) (Figure 4).
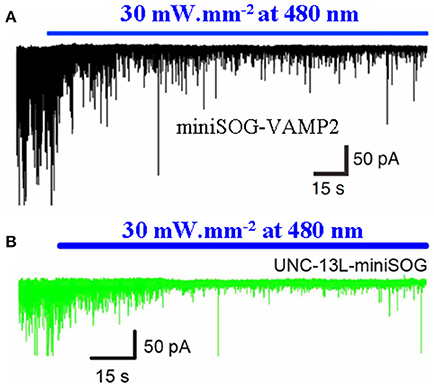
Figure 4. Photodynamic inhibition of neurotransmitter release in C. elegans with miniSOG-VAMP2 or UNC-13L-miniSOG. The protein photosensitizer miniSOG was fusion-expressed with the vSNARE vesicular associated membrane protein 2 (VAMP2) (A) or with the active zone protein UNC-13-L (Munc 13) (B) in C. elegans neurons under neuron-specific promoter. The transgenic worms were illuminated with blue light (480 nm, 30 mW/mm2), and postsynaptic current was recorded from ventral medial wall body muscle with whole cell configuration. Note the rapid decrease in excitatory postsynaptic current upon blue light illumination. Adapted from Lin et al. (2013) and Zhou et al. (2013).
Another place of interest that has been investigated is the chromosomal tip, the telomeres. The photosensitizer KillerRed could be fusion-expressed with the telomere repeat binding factor 1 (KillerRed-TRF1). The spatially-defined generation of singlet oxygen after photodynamic action at the telomeres was found to result in telomere abnormalities (telomere associations, shortened or complete loss of telomeres), leading to fastened cell senescence or cell death in cultured cancer cells HeLa, U2OS and IMR90 (Sun et al., 2015) (Figure 5A). KillerRed fusion expressed with the chromosomal protein histone H2B (H2B-KR-KR) in adherent HeLa-Kyoto cells after green light illumination was found to induce acute wide spread damages to genomic DNA, leading to non-separation of chromosomes, and to blockade of cell division and proliferation. H2B-KR-KR target expressed in specific tissues (under control of tissue-specific promoters) in Xenopus embryos was found, after green light illumination (540–580 nm 120 mW/cm2; 525 nm, 45 mW/cm2), to retard organogenesis. Therefore, H2B-KR-KR photodynamic action could be used to study cell division, organism development, organogenesis or carcinogenesis in a cell-specific manner in vivo (Serebrovskaya et al., 2011) (Figure 5B). Protein photosensitizers tandem-KillerRed and miniSOG target-expressed at chromatin (H2B-tKR or H2B-miniSOG) in HeLa-Kyoto cells after brief light irradiation (H2B-tandem KillerRed expressing cells with green light 540–580 nm for 15 min at 200 mW/cm2; H2B-miniSOG for 5 min with blue light 465–495 nm at 65 mW/cm2) was able to induce single strand breaks but only miniSOG induced double strand breaks of the genomic DNA, leading to DNA damage response and cell senescence (Petrova et al., 2016). Most interestingly, germline C. elegans expressing Histone-mSOG after exposure to blue light has been found to produce progenies with inheritable phenotypes (Noma and Jin, 2015). KillerRed fusion-expressed either with the peripheral nuclear protein lamin B1 (KRed-Lamin B1) or with the diffusely distributed chromosomal protein H2A (H2A-KRed) was used to identify gene damages after photodynamic action. The extent of photo-oxidative damage was used to delineate the spatial localization of the respective genes and the gene-carrying chromosomes in the nucleus (Waldeck et al., 2013). KillerRed fused to a tet-repressor (tetR-KR) or a transcription activator (TA-KR) have been used to target oxidative stress to hetero- or euchromatin respectively in U2OS cells (Lan et al., 2014).
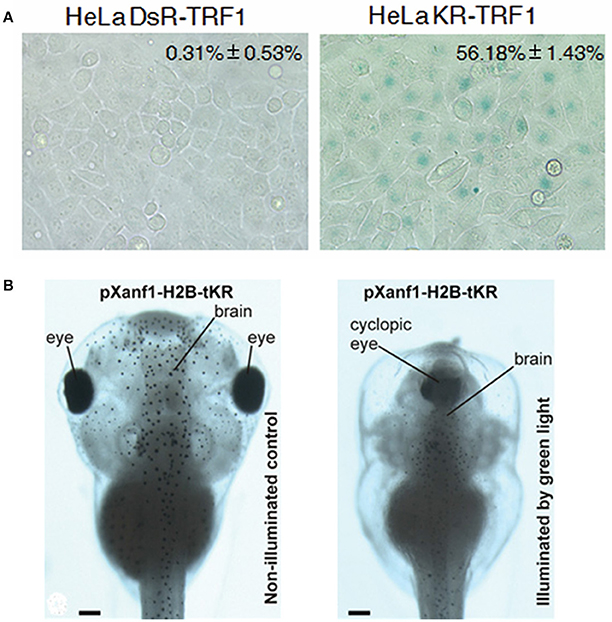
Figure 5. Photodynamic transformation of cell fate and zebrafish development with KillerRed fusion-expressed with TRF1 at the telomere (KR-TRF1) in HeLa cells or with H2B under the promoter pXanf1 in X. laevis. (A) KillerRed (KR) was fusion-expressed with telomere repeat binding factor 1 (TRF1) to Hela cell telomeres (with the non-photosensitizer DsRed used as control), repeated light exposure (1.5 mW/cm2, 10 min in each passage for 30 passages) induced fastened cell senescence as indicated by enhanced β–galactosidase activity (percentage cells with SA-β-gal staining). The negative control fluorescent protein DsR showed no such effect. (B) KillerRed was fusion-expressed with core histone H2B under the control of forebrain promoter pXanf1, the transgenic embryos were illuminated at the early midneurula stages with green LED (525 nm, 45 mW/cm2, 1 h). Such photodynamic treatment severely retarded the development of the forebrain, sometimes resulted in a completely cyclopic phenotype, similar to phenotypes with suppressed pXanf1 expression (illuminated by green light). The tadpoles developed from transgenic X. laevis embryos not illuminated showed normal phenotype (non-illuminated control). Scale bars in (B) 100 μm. Adapted from Sun et al. (2015) and Serebrovskaya et al. (2011).
The above examples showcase targeted photonanomanipulation in subcellular organelle-based events at the plasma membrane, synaptic vesicles and at chromosomes. Numerous other subcellular sites can be similarly modulated with this light-controlled nanomanipulation technique.
In conclusion, the emergence of protein photosensitizers with enhanced singlet oxygen quantum yield has made possible the fast-track progress in photodynamic nanomanipulation of cellular physiology, mainly in C. elegans and zebrafish, whole organism works in mammalian animals remain to be expanded. The targeted expression after insertion of signaling sequence, fusion expression of target proteins, or promoter-driven tissue-specific expression, have made possible highly confined generation of singlet oxygen, with targeted nanoscopical protein oxidative activation or inactivation. Of the three major photopharmacological techniques available today—optogenetics involving channelrhodopsin and other photosensory domain-containing proteins spatial configurational-responsive to the absorption of a photon, photochromic ligands of receptors or ionic channels involving the photon-driven configurational changes of small molecule ligands, and photodynamic modulation involving photosensitizer-generated singlet oxygen, the protein photosensitizers are unique in that they could be repetitively irradiated (used) rather like an enzyme molecule to generate multiple copies of singlet oxygen molecules but not merely repetitive configuration changes in the light-absorbing molecules themselves. This makes possible multiple-hit oxidative activation or desensitization of receptors and channels important in cellular physiology, with the use of attenuated lasers and LED, or with cheap halogen cold light sources. In the future, more extensive works may be done with this photonanomanipulation technique both in the elucidation of basic cellular functions such as gene transcription, protein synthesis, transport, degradation, or in the elucidation of cell-type specific functions in the central nervous system circuits or in the innervation hubs (ganglia) and pathways (nerve fiber wirings) of the peripheral nervous system. Such photonanomanipulations in situ could also be used to study the intrinsic long-term repair mechanisms with high spatial and temporal resolutions.
HJ and YL wrote the initial drafts. ZC conceived the idea of the review and finalized the last version of the article. All authors checked and approved the submitted version.
Work in the authors' laboratory has been supported by grants from The Natural Science Foundation of China (Nos. 31670856, 31270892) and The National Basic Research Program (973 Program) from The Ministry of Science and Technology of China (No. 2011CB809101).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abrahamse, H., and Hamblin, M. R. (2016). New photosensitizers for photodynamic therapy. Biochem. J. 473, 347–364. doi: 10.1042/BJ20150942
Agostinis, P., Berg, K., Cengel, K. A., Foster, T. H., Girotti, A. W., Gollnick, S. O., et al. (2011). Photodynamic therapy of cancer: an update. CA Cancer J. Clin. 61, 250–281. doi: 10.3322/caac.20114
Akemann, W., Sasaki, M., Mutoh, H., Imamura, T., Honkura, N., and Knoepfel, T. (2013). Two-photon voltage imaging using a genetically encoded voltage indicator. Sci. Rep. 3:2231. doi: 10.1038/srep02231
al-Laith, M., Matthews, E. K., and Cui, Z. J. (1993). Photodynamic drug action on isolated rat pancreatic acini. Mobilization of arachidonic acid and prostaglandin production. Biochem. Pharmacol. 46, 567–573. doi: 10.1016/0006-2952(93)90539-9
Allison, R. R., and Sibata, C. H. (2010). Oncologic photodynamic therapy photosensitizers: a clinical review. Photodiagnosis Photodyn. Ther. 7, 61–75. doi: 10.1016/j.pdpdt.2010.02.001
Almeida, R. D., Manadas, B. J., Carvalho, A. P., and Duarte, C. B. (2004). Intracellular signaling mechanisms in photodynamic therapy. Biochim. Biophys. Acta 1704, 59–86. doi: 10.1016/j.bbcan.2004.05.003
An, Y. P., Xiao, R., Cui, H., and Cui, Z. J. (2003). Selective activation by photodynamic action of cholecystokinin receptor in the freshly isolated rat pancreatic acini. Br. J. Pharmacol. 139, 872–880. doi: 10.1038/sj.bjp.0705295
Babes, A., Sauer, S. K., Moparthi, L., Kichko, T., Neacsu, C., Namer, B., et al. (2016). Photosensitization in porphyrias and photodynamic therapy involves TRPA1 and TRPV1. J. Neurosci. 36, 5264–5278. doi: 10.1523/JNEUROSCI.4268-15.2016
Bacellar, I. O., Tsubone, T. M., Pavani, C., and Baptista, M. S. (2015). Photodynamic efficiency: from molecular photochemistry to cell death. Int. J. Mol. Sci. 16, 20523–20559. doi: 10.3390/ijms160920523
Berg, K., and Moan, J. (1997). Lysosomes. Microtubules as targets for photochemotherapy of cancer. Photochem. Photobiol. 65, 403–409. doi: 10.1111/j.1751-1097.1997.tb08578.x
Bovis, M. J., Woodhams, J. H., Loizidou, M., Scheglmann, D., Bown, S. G., and MacRobert, A. J. (2012). Improved in vivo delivery of m-THPC via pegylated liposomes for use in photodynamic therapy. J. Control Release 157, 196–205. doi: 10.1016/j.jconrel.2011.09.085
Bown, S. G. (2013). Photodynamic therapy for photochemists. Philos. Trans. A Math. Phys. Eng. Sci. 371:20120371. doi: 10.1098/rsta.2012.0371
Bown, S. G., Rogowska, A. Z., Whitelaw, D. E., Lees, W. R., Lovat, L. B., Ripley, P., et al. (2002). Photodynamic therapy for cancer of the pancreas. Gut 50, 549–557. doi: 10.1136/gut.50.4.549
Brown, S. B., Brown, E. A., and Walker, I. (2004). The present and future role of photodynamic therapy in cancer treatment. Lancet Oncol. 5, 497–508. doi: 10.1016/S1470-2045(04)01529-3
Bulina, M. E., Chudakov, D. M., Britanova, O. V., Yanushevich, Y. G., Staroverov, D. B., Chepurnykh, T. V., et al. (2006a). A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95–99. doi: 10.1038/nbt1175
Bulina, M. E., Lukyanov, K. A., Britanova, O. V., Onichtchouk, D., Lukyanov, S., and Chudakov, D. M. (2006b). Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nature Protoc 1, 947–953. doi: 10.1038/nprot.2006.89
Byrne, L. C., Khalid, F., Lee, T., Zin, E. A., Greenberg, K., Visel, M., et al. (2013). AAV-mediated, optogenetic ablation of Muller glia leads to structural and functional changes in the mouse retina. PLoS ONE 8:e76075. doi: 10.1371/journal.pone.0076075
Carpentier, P., Violot, S., Blanchoin, L., and Bourgeois, D. (2009). Structural basis for the phototoxicity of the fluorescent protein KillerRed. FEBS Lett. 583, 2839–2842. doi: 10.1016/j.febslet.2009.07.041
Cawston, E. E., and Miller, L. J. (2010). Therapeutic potential for novel drugs targeting the type 1 cholecystokinin receptor. Br. J. Pharmacol. 159, 1009–1021. doi: 10.1111/j.1476-5381.2009.00489.x
Chiu, S. M., Xue, L. Y., Azizuddin, K., and Oleinick, N. L. (2005). Photodynamic therapy-induced death of HCT 116 cells: apoptosis with or without Bax expression. Apoptosis 10, 1357–1368. doi: 10.1007/s10495-005-2217-0
Cho, J. Y., and Sternberg, P. W. (2014). Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell 156, 249–260. doi: 10.1016/j.cell.2013.11.036
Craig, R. A., McCoy, C. P., Gorman, S. P., and Jones, D. S. (2015). Photosensitisers-the progression from photodynamic therapy to anti-infective surfaces. Expert Opin. Drug Deliv. 12, 85–101. doi: 10.1517/17425247.2015.962512
Cui, Z. J., and Guo, L. L. (2002a). Assessing physiological concentrations of endogenous substances in situ by inducing calcium oscillations in vitro. Case of liver. Acta Pharmacol. Sin. 23, 27–32.
Cui, Z. J., and Guo, L. L. (2002b). Photodynamic modulation by Victoria Blue BO of phenylephrine-induced calcium oscillations in freshly isolated rat hepatocytes. Photochem. Photobiol. Sci. 1, 1001–1005. doi: 10.1039/b208215m
Cui, Z. J., Habara, Y., and Satoh, Y. (2000). Photodynamic modulation of adrenergic receptors in the isolated rat hepatocytes. Biochem. Biophys. Res. Commun. 277, 705–710. doi: 10.1006/bbrc.2000.3742
Cui, Z. J., Habara, Y., Wang, D. Y., and Kanno, T. (1997). A novel aspect of photodynamic action: induction of recurrent spikes in cytosolic calcium concentration. Photochem. Photobiol. 65, 382–386. doi: 10.1111/j.1751-1097.1997.tb08574.x
Cui, Z. J., Han, Z. Q., and Li, Z. Y. (2012). Modulating protein activity and cellular function by methionine residue oxidation. Amino Acids 43, 505–517. doi: 10.1007/s00726-011-1175-9
Cui, Z. J., and Kanno, T. (1997). Photodynamic triggering of calcium oscillation in the isolated rat pancreatic acini. J. Physiol. 504, 47–55. doi: 10.1111/j.1469-7793.1997.047bf.x
Cui, Z. J., and Matthews, E. K. (1998). Photodynamic modulation of cellular function. Acta Pharmacol. Sin. 19, 297–303.
Cui, Z. J., Zhou, Y. D., Satoh, Y., and Habara, Y. (2003). A physiological role for protoporphyrin IX photodynamic action in the rat Harderian gland? Acta Physiol. Scand. 179, 149–154. doi: 10.1046/j.1365-201X.2003.01177.x
Dai, T., Fuchs, B. B., Coleman, J. J., Prates, R. A., Astrakas, C., St Denis, T. G., et al. (2012). Concepts and principles of photodynamic therapy as an alternative antifungal discovery platform. Front. Microbiol. 3:120. doi: 10.3389/fmicb.2012.00120
Dockray, G. J., and Burdyga, G. (2011). Plasticity in vagal afferent neurones during feeding and fasting: mechanisms and significance. Acta Physiol. (Oxf). 201, 313–321. doi: 10.1111/j.1748-1716.2010.02219.x
Dougherty, T. J., Gomer, C. J., Henderson, B. W., Jori, G., Kessel, D., Korbelik, M., et al. (1998). Photodynamic therapy. J. Natl. Cancer Inst. 90, 889–905. doi: 10.1093/jnci/90.12.889
El-Shamayleh, Y., Ni, A. M., and Horwitz, G. D. (2016). Strategies for targeting primate neural circuits with viral vectors J. Neurophysiol. 116, 122–134. doi: 10.1152/jn.00087.2016
Ertuerk, A., Wang, Y. Y., and Sheng, M. (2014). Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J. Neurosci. 34, 1672–1688. doi: 10.1523/JNEUROSCI.3121-13.2014
Feine, I., Pinkas, I., Salomon, Y., and Scherz, A. (2012). Local oxidative stress expansion through endothelial cell - a key role for gap junction intercellular communication. PLoS ONE 7:e41633. doi: 10.1371/journal.pone.0041633
Fliegel, L., Burns, K., MacLennan, D. H., Reithmeier, R. A., and Michalak, M. (1989). Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 264, 21522–21528.
Fujii, M., Ohtsubo, M., Ogawa, T., Kamata, H., Hirata, H., and Yagisawa, H. (1999). Real-time visualization of PH domain-dependent translocation of phospholipase C-delta 1 in renal epithelial cells (MDCK): response to hypo-osmoticstress. Biochem. Biophys. Res. Commun. 254, 284–291. doi: 10.1006/bbrc.1998.9936
Galardy, R. E., Hull, B. E., and Jamieson, J. D. (1980). Irreversible photoactivation of a pancreatic secretagogue receptor with cholecystokinin COOH-terminal octapeptides. J. Biol. Chem. 255, 3148–3155.
Gao, W. H., Su, Z. C., Liu, Q. L., and Zhou, L. (2014). State-dependent and site-directed photodynamic transformation of HCN2 channel by singlet oxygen. J. Gen. Physiol. 143, 633–644. doi: 10.1085/jgp.201311112
Hashikura, S., Satoh, Y., Cui, Z. J., and Habara, Y. (2001). Photodynamic action inhibits compound 48/80-induced exocytosis in rat peritoneal mast cells. Jpn. J. Vet. Res. 49, 239–247. doi: 10.14943/jjvr.49.3.239
Hill, K., and Schaefer, M. (2009). Ultraviolet light and photosensitising agents activate TRPA1 via generation of oxidative stress. Cell Calcium 45, 155–164. doi: 10.1016/j.ceca.2008.08.001
Huggett, M. T., Jermyn, M., Gillams, A., Illing, R., Mosse, S., Novelli, M., et al. (2014). Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br. J. Cancer 110, 1698–1704. doi: 10.1038/bjc.2014.95
Janovjak, H., Szobota, S., Wyart, C., Trauner, D., and Isacoff, E. Y. (2010). A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat. Neurosci. 13, 1027–1032. doi: 10.1038/nn.2589
Jarvela, T., and Linstedt, A. D. (2014). Isoform-specific tethering links the Golgi ribbon to maintain compartmentalization. Mol. Biol. Cell 25, 133–144. doi: 10.1091/mbc.E13-07-0395
Kaestner, L., Juzeniene, A., and Moan, J. (2004). Erythrocytes–the house elves of photodynamic therapy. Photochem. Photobiol. Sci. 3, 981–989. doi: 10.1039/B403127J
Kaestner, L., Scholz, A., and Lipp, P. (2015a). Conceptual and technical aspects of transfection and gene delivery. Bioorg. Med. Chem. Lett. 25, 1171–1176. doi: 10.1016/j.bmcl.2015.01.018
Kaestner, L., Tian, Q., Kaiser, E., Xian, W., Müller, A., Oberhofer, M., et al. (2015b). Genetically encoded voltage indicators in circulation research. Int. J. Mol. Sci. 16, 21626–21642. doi: 10.3390/ijms160921626
Kennedy, J. C., Pottier, R. H., and Pross, D. C. (1990). Photodynamic therapy with endogenous protoporphyrin IX: basic principles and present clinical experience. J. Photochem. Photobiol. B Biol. 6, 143–148. doi: 10.1016/1011-1344(90)85083-9
Kessel, D. (1997). Subcellular localization of photosensitizing agents introduction. Photochem. Photobiol. 65, 387–388. doi: 10.1111/j.1751-1097.1997.tb08575.x
Kessel, D. (2004). Correlation between subcellular localization and photodynamic efficacy. J. Porphyr. Phthalocyanines 8, 1009–1014. doi: 10.1142/S1088424604000374
Kessel, D. (2012). Subcellular targets for photodynamic therapy: implications for initiation of apoptosis and autophagy. J. Natl. Compr. Canc. Netw. 10, S56–S59.
Kessel, D., Luo, Y., Deng, Y., and Chang, C. K. (1997). The role of subcellular localization in initiation of apoptosis by photodynamic therapy. Photochem. Photobiol. 65, 422–426. doi: 10.1111/j.1751-1097.1997.tb08581.x
Khrenova, M., Topol, I., Collins, J., and Nemukhin, A. (2015). Estimating orientation factors in the FRET theory of fluorescent proteins: the TagRFP-KFP pair and beyond. Biophys. J. 108, 126–132. doi: 10.1016/j.bpj.2014.11.1859
Kim, S., Tachikawa, T., Fujitsuka, M., and Majima, T. (2014). Far-red fluorescence probe for monitoring singlet oxygen during photodynamic therapy. J. Am. Chem. Soc. 136, 11707–11715. doi: 10.1021/ja504279r
Kobayashi, J., Shidara, H., Morisawa, Y., Kawakami, M., Tanahashi, Y., Hotta, K., et al. (2013). A method for selective ablation of neurons in C. elegans using the phototoxic fluorescent protein, KillerRed. Neurosci. Lett. 548, 261–264. doi: 10.1016/j.neulet.2013.05.053
Krammer, B., and Verwanger, T. (2012). Molecular response to hypericin-induced photodamage. Curr. Med. Chem. 19, 793–798. doi: 10.2174/092986712799034842
Lan, L., Nakajima, S., Wei, L. Z., Sun, L. X., Hsieh, C. L., Sobol, R. W., et al. (2014). Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 42, 2330–2345. doi: 10.1093/nar/gkt1233
Lee, A., Mathuru, A. S., Teh, C., Kibat, C., Korzh, V., Penney, T. B., et al. (2010). The habenula prevents helpless behavior in larval zebrafish. Curr. Biol. 20, 2211–2216. doi: 10.1016/j.cub.2010.11.025
Lin, J. Y., Sann, S. B., Zhou, K., Nabavi, S., Proulx, C. D., Malinow, R., et al. (2013). Optogenetic inhibition of synaptic release with chromophore-assisted light inactivation (CALI). Neuron 79, 241–253. doi: 10.1016/j.neuron.2013.05.022
Liu, C., Hu, M., Ma, D., Lei, J., and Xu, J. (2016). Photodynamic inactivation of antibiotic-resistant bacteria and biofilms by hematoporphyrin monomethyl ether. Lasers Med. Sci. 31, 297–304. doi: 10.1007/s10103-015-1859-6
Liu, L., Zhang, Z. Z., and Xing, D. (2011). Activation of Bax by lysosomal photodamage induces cell death via mitochondrial apoptotic pathway. Free Radic. Biol. Med. 51, 53–68. doi: 10.1016/j.freeradbiomed.2011.03.042
Lukyanov, K. A., Serebrovskaya, E. O., Lukyanov, S., and Chudakov, D. M. (2010). Fluorescent proteins as light-inducible photochemical partners. Photochem. Photobiol. Sci. 9, 1301–1306. doi: 10.1039/c0pp00114g
Makhijani, K., To, T. L., Ruiz-González, R., Lafaye, C., Royant, A., and Shu, X. K. (2017). Precision optogenetic tool for selective single- and multiple-cell ablation in a live animal model system. Cell Chem. Biol. 24, 110–119. doi: 10.1016/j.chembiol.2016.12.010
Manoharan, G. B., Enkvist, E., Kasari, M., Viht, K., Zenn, M., Prinz, A., et al. (2015). FRET-based screening assay using small-molecule photoluminescent probes in lysate of cells overexpressing RFP-fused protein kinases. Anal. Biochem. 481, 10–17. doi: 10.1016/j.ab.2015.04.009
Matthews, E. K., and Cui, Z. J. (1989). Photodynamic action of rose bengal on isolated rat pancreatic acini: stimulation of amylase release. FEBS Lett. 256, 29–32. doi: 10.1016/0014-5793(89)81711-9
Matthews, E. K., and Cui, Z. J. (1990a). Photodynamic action of sulphonated aluminium phthalocyanine (SALPC) on AR4-2J cells, a carcinoma cell line of rat exocrine pancreas. Br. J. Cancer 61, 695–701. doi: 10.1038/bjc.1990.157
Matthews, E. K., and Cui, Z. J. (1990b). Photodynamic action of sulphonated aluminium phthalocyanine (SALPC) on isolated rat pancreatic acini. Biochem. Pharmacol. 39, 1445–1457. doi: 10.1038/bjc.1990.157
Merzlyak, E. M., Goedhart, J., Shcherbo, D., Bulina, M. E., Shcheglov, A. S., Fradkov, A. F., et al. (2007). Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 4, 555–557. doi: 10.1038/nmeth1062
Miller, L. J., and Gao, F. (2008). Structural basis of cholecystokinin receptor binding and regulation. Pharmacol. Ther. 119, 83–95. doi: 10.1016/j.pharmthera.2008.05.001
Mironova, K. E., Proshkina, G. M., Ryabova, A. V., Stremovskiy, O. A., Lukyanov, S. A., Petrov, R. V., et al. (2013). Genetically encoded immunophotosensitizer 4D5scFv-miniSOG is a highly selective agent for targeted photokilling of tumor cells in vitro. Theranostics 3, 831–840. doi: 10.7150/thno.6715
Mittra, R. A., and Singerman, L. J. (2002). Recent advances in the management of age-related macular degeneration. Optom. Vis. Sci. 79, 218–224. doi: 10.1097/00006324-200204000-00008
Moan, J., and Berg, K. (1991). The photodegradation of porphyrins in cells can be used to estimate the lifetime of singlet oxygen. Photochem. Photobiol. 53, 549–553. doi: 10.1111/j.1751-1097.1991.tb03669.x
Munro, S., and Pelham, H. R. (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907. doi: 10.1016/0092-8674(87)90086-9
Newman, D. K. (2016). Photodynamic therapy: current role in the treatment of chorioretinal conditions. Eye 30, 202–210. doi: 10.1038/eye.2015.251
Noma, K., and Jin, Y. (2015). Optogenetic mutagenesis in Caenorhabditis elegans. Nat. Commun. 6:8868. doi: 10.1038/ncomms9868
Norman, R. A. (2005). Past and future: porphyria and porphyrins. Skinmed 4, 287–292. doi: 10.1111/j.1540-9740.2005.03706.x
Nowis, D., Makowski, M., Stoklosa, T., Legat, M., Issat, T., and Golab, J. (2005). Direct tumor damage mechanisms of photodynamic therapy. Acta Biochim. Pol. 52, 339–352.
Oleinick, N. L., and Evans, H. H. (1998). The photobiology of photodynamic therapy: cellular targets and mechanisms. Radiat. Res. 150, S146–S156. doi: 10.2307/3579816
Oxford, G. S., Pooler, J. P., and Narahashi, T. (1977). Internal and external application of photodynamic sensitizers on squid giant-axons. J. Membr. Biol. 36, 159–173. doi: 10.1007/BF01868149
Peng, Q., Farrants, G. W., Madslien, K., Bommer, J. C., Moan, J., and Danielsen, H. E. (1991). Subcellular localization, redistribution and photobleaching of sulfonated aluminum phthalocyanines in a human melanoma cell line. Int. J. Cancer 49, 290–295. doi: 10.1002/ijc.2910490225
Petrova, N. V., Luzhin, A. V., Serebrovskaya, E. O., Ryumina, A. P., Velichko, A. K., Razin, S. V., et al. (2016). Inducing cellular senescence in vitro by using genetically encoded photosensitizers. Aging (Albany. NY) 8, 2449–2462. doi: 10.18632/aging.101065
Pietra, F. (2014). Molecular dynamics simulation of dioxygen pathways through mini singlet oxygen generator (miniSOG), a genetically encoded marker and killer protein. Chem. Biodivers. 11, 1883–1891. doi: 10.1002/cbdv.201400125
Pimenta, F. M., Jensen, R. L., Breitenbach, T., Etzerodt, M., and Ogilby, P. R. (2013). Oxygen-dependent photochemistry and photophysics of “‘miniSOG,” a protein-encased flavin. Photochem. Photobiol. 89, 1116–1126. doi: 10.1111/php.12111
Pletnev, S., Gurskaya, N. G., Pletneva, N. V., Lukyanov, K. A., Chudakov, D. M., Martynov, V. I., et al. (2009). Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed. J. Biol. Chem. 284, 32028–32039. doi: 10.1074/jbc.M109.054973
Pletneva, N. V., Pletnev, V. Z., Sarkisyan, K. S., Gorbachev, D. A., Egorov, E. S., Mishin, A. S., et al. (2015). Crystal structure of phototoxic orange fluorescent proteins with a tryptophan-based chromophore. PLoS ONE 10:e0145740. doi: 10.1371/journal.pone.0145740
Qi, Y. B., Garren, E. J., Shu, X., Tsien, R. Y., and Jin, Y. (2012). Photo-inducible cell ablation in Caenorhabditis elegans usingthe genetically encoded singlet oxygen-generating protein miniSOG. Proc. Natl. Acad. Sci. U.S.A. 109, 7499–7504. doi: 10.1073/pnas.1204096109
Ragas, X., Cooper, L. P., White, J. H., Nonell, S., and Flors, C. (2011). Quantification of photosensitized singlet oxygen production by a fluorescent protein. Chemphyschem 12, 161–165. doi: 10.1002/cphc.201000919
Rizzuto, R., Brini, M., Pizzo, P., Murgia, M., and Pozzan, T. (1995). Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr. Biol. 5, 635–642. doi: 10.1016/S0960-9822(95)00128-X
Rizzuto, R., Nakase, H., Darras, B., Francke, U., Fabrizi, G. M., Mengel, T., et al. (1989). A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues. J. Biol. Chem. 264, 10595–10600.
Rodríguez-Pulido, A., Cortajarena, A. L., Torra, J., Ruiz-González, R., Nonell, S., and Flors, C. (2016). Assessing the potential of photosensitizing flavoproteins as tags for correlative microscopy. Chem. Commun. 52, 8405–8408. doi: 10.1039/C6CC03119F
Rosenkranz, A. A., Jans, D. A., and Sobolev, A. S. (2000). Targeted intracellular delivery of photosensitizers to enhance photodynamic efficiency. Immunol. Cell Biol. 78, 452–464. doi: 10.1046/j.1440-1711.2000.00925.x
Roy, A., Carpentier, P., Bourgeois, D., and Field, M. (2010). Diffusion pathways of oxygen species in the phototoxic fluorescent protein KillerRed. Photochem. Photobiol. Sci. 9, 1342–1350. doi: 10.1039/c0pp00141d
Ruiz-González, R., White, J. H., Agut, M., Nonell, S., and Flors, C. (2012). A genetically-encoded photosensitiser demonstrates killing of bacteria by purely endogenous singlet oxygen. Photochem. Photobiol. Sci. 11, 1411–1413. doi: 10.1039/c2pp25126d
Ryumina, A. P., Serebrovskaya, E. O., Shirmanova, M. V., Snopova, L. B., Kuznetsova, M. M., Turchin, I. V., et al. (2013). Flavoprotein miniSOG as a genetically encoded photosensitizer for cancer cells. Biochim. Biophys. Acta 1830, 5059–5067. doi: 10.1016/j.bbagen.2013.07.015
Ryumina, A. P., Serebrovskaya, E. O., Staroverov, D. B., Zlobovskaya, O. A., Shcheglov, A. S., Lukyanov, S. A., et al. (2016). Lysosome-associated miniSOG as a photosensitizer for mammalian cells. BioTechniques 61, 92–94. doi: 10.2144/000114445
Sack, J. T., Stephanopoulos, N., Austin, D. C., Francis, M. B., and Trimmer, J. S. (2013). Antibody-guided photoablation of voltage-gated potassium currents. J. Gen. Physiol. 142, 315–324. doi: 10.1085/jgp.201311023
Sarkisyan, K. S., Zlobovskaya, O. A., Gorbachev, D. A., Bozhanova, N. G., Sharonov, G. V., Staroverov, D. B., et al. (2015). KillerOrange, a genetically encoded photosensitizer activated by blue and green light. PLoS ONE 10:e0145287. doi: 10.1371/journal.pone.0145287
Scourides, P. A., Bohmer, R. M., Kaye, A. H., and Morstyn, G. (1987). Nature of the tumor-localizing components of hematoporphyrin derivative. Cancer Res. 47, 3439–3445.
Serebrovskaya, E. O., Edelweiss, E. F., Stremovskiy, O. A., Lukyanov, K. A., Chudakov, D. M., and Deyev, S. M. (2009). Targeting cancer cells by using an antireceptor antibody-photosensitizer fusion protein. Proc. Natl. Acad. Sci. U.S.A. 106, 9221–9225. doi: 10.1073/pnas.0904140106
Serebrovskaya, E. O., Gorodnicheva, T. V., Ermakova, G. V., Solovieva, E. A., Sharonov, G. V., Zagaynova, E. V., et al. (2011). Light-induced blockage of cell division with a chromatin-targeted phototoxic fluorescent protein. Biochem. J. 435, 65–71. doi: 10.1042/BJ20101217
Serebrovskaya, E. O., Ryumina, A. P., Boulina, M. E., Shirmanova, M. V., Zagaynova, E. V., Bogdanova, E. A., et al. (2014). Phototoxic effects of lysosome-associated genetically encoded photosensitizer KillerRed. J. Biomed. Opt. 19:071403. doi: 10.1117/1.JBO.19.7.071403
Shagin, D. A., Barsova, E. V., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Labas, Y. A., et al. (2004). GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol. Biol. Evol. 21, 841–850. doi: 10.1093/molbev/msh079
Shaner, N. C., Lin, M. Z., McKeown, M. R., Steinbach, P. A., Hazelwood, K. L., Davidson, M. W., et al. (2008). Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551. doi: 10.1038/nmeth.1209
Shibuya, T., and Tsujimoto, Y. (2012). Deleterious effects of mitochondrial ROS generated by KillerRed photodynamic action in human cell lines and C. elegans. J. Photochem. Photobiol. B Biol. 117, 1–12. doi: 10.1016/j.jphotobiol.2012.08.005
Shirmanova, M. V., Serebrovskaya, E. O., Lukyanov, K. A., Snopova, L. B., Sirotkina, M. A., Prodanetz, N. N., et al. (2013). Phototoxic effects of fluorescent protein KillerRed on tumor cells in mice. J. Biophotonics 6, 283–290. doi: 10.1002/jbio.201200056
Shirmanova, M., Yuzhakova, D., Snopova, L., Perelman, G., Serebrovskaya, E., Lukyanov, K., et al. (2015). Towards PDT with genetically encoded photosensitizer KillerRed: a comparison of continuous and pulsed laser regimens in an animal tumor model. PLoS ONE 10:e0144617. doi: 10.1371/journal.pone.0144617
Shkryl, V. M., Maxwell, J. T., and Blatter, L. A. (2012). A novel method for spatially complex diffraction-limited photoactivation and photobleaching in living cells. J. Physiol. 590, 1093–1100. doi: 10.1113/jphysiol.2011.223446
Shu, X., Lev-Ram, V., Deerinck, T. J., Qi, Y., Ramko, E. B., Davidson, M. W., et al. (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9:e1001041. doi: 10.1371/journal.pbio.1001041
Skene, J. H., and Virág, I. (1989). Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J. Cell Biol. 108, 613–624. doi: 10.1083/jcb.108.2.613
Subach, O. M., Malashkevich, V. N., Zencheck, W. D., Morozova, K. S., Piatkevich, K. D., Almo, S. C., et al. (2010). Structural characterization of acylimine-containing blue and redchromophores in mTagBFP and TagRFP fluorescent proteins. Chem. Biol. 17, 333–341. doi: 10.1016/j.chembiol.2010.03.005
Sun, L., Tan, R., Xu, J., LaFace, J., Gao, Y., Xiao, Y., et al. (2015). Targeted DNA damage at individual telomeres disrupts their integrity and triggers cell death. Nucleic Acids Res. 43, 6334–6347. doi: 10.1093/nar/gkv598
Szeimies, R. M., Stockfleth, E., Popp, G., Borrosch, F., Brüning, H., Dominicus, R., et al. (2010). Long-term follow-up of photodynamic therapy with a self-adhesive 5-aminolaevulinic acid patch: 12 months data. Br. J. Dermatol. 162, 410–414. doi: 10.1111/j.1365-2133.2009.09377.x
Tabuchi, N., Akasaki, K., and Tsuji, H. (2000). Two acidic amino acid residues, Asp(470) and Glu(471), contained in the carboxyl cytoplasmic tail of a major lysosomal membrane protein, LGP85 / LIMP II, are important for its accumulation in secondary lysosomes. Biochem. Biophys. Res. Commun. 270, 557–563. doi: 10.1006/bbrc.2000.2448
Takemoto, K., Matsuda, T., Sakai, N., Fu, D., Noda, M., Uchiyama, S., et al. (2013). SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci. Rep. 3:2629. doi: 10.1038/srep02629
Tarr, M., and Valenzeno, D. P. (1991). Modification of cardiac ionic currents by photosensitiser-generated reactive oxygen. J. Mol. Cell. Cardiol. 23, 639–649. doi: 10.1016/0022-2828(91)90055-Q
Teh, C., Chudakov, D. M., Poon, K. L., Mamedov, I. Z., Sek, J. Y., Shidlovsky, K., et al. (2010). Optogenetic in vivo cell manipulation in KillerRed-expressing zebrafish transgenics. BMC Dev. Biol. 10:110. doi: 10.1186/1471-213X-10-110
Theodossiou, T. A., Noronha-Dutra, A., and Hothersall, J. S. (2006). Mitochondria are a primary target of hypericin phototoxicity: synergy of intracellular calcium mobilisation in cell killing. Int. J. Biochem. Cell Biol. 38, 1946–1956. doi: 10.1016/j.biocel.2006.05.009
Torra, J., Burgos-Caminal, A., Endres, S., Wingen, M., Drepper, T., and Gensch, T. (2015). Singlet oxygen photosensitisation by the fluorescent protein Pp2FbFP L30M, a novel derivative of Pseudomonas putida flavin-binding Pp2FbFP. Photochem. Photobiol. Sci. 14, 280–287. doi: 10.1039/c4pp00338a
Usuda, J., Azizuddin, K., Chiu, S. M., and Oleinick, N. L. (2003). Association between the photodynamic loss of Bcl-2 and the sensitivity to apoptosis caused by phthalocyanine photodynamic therapy. Photochem. Photobiol. 78, 1–8. doi: 10.1562/0031-8655(2003)0780001ABTPLO2.0.CO2
Usuda, J., Chiu, S. M., Azizuddin, K., Xue, L. Y., Lam, M., Nieminen, A. L., et al. (2002). Promotion of photodynamic therapy-induced apoptosis by the mitochondrial protein Smac/DIABLO: dependence on Bax. Photochem. Photobiol. 76, 217–223. doi: 10.1562/0031-8655(2002)0760217POPTIA2.0.CO2
Valenzeno, D. P., and Tarr, M. (1997). GH3 cells, ionic currents and cell killing: photomodification sensitized by Rose Bengal. Photochem. Photobiol. 68, 519–526. doi: 10.1111/j.1751-1097.1998.tb02508.x
Vegh, R. B., Solntsev, K. M., Kuimova, M. K., Cho, S., Liang, Y., Loo, B. L., et al. (2011). Reactive oxygen species in photochemistry of the red fluorescent protein “KillerRed”. Chem. Commun. 47, 4887–4889. doi: 10.1039/c0cc05713d
Waldeck, W., Mueller, G., Glatting, K. H., Hotz-Wagenblatt, A., Diessl, N., Chotewutmonti, S., et al. (2013). Spatial localization of genes determined by intranuclear DNA fragmentation with the fusion proteins lamin KRED and histone KRED und visible light. Intl J Med Sci 10, 1136–1148. doi: 10.7150/ijms.6121
Waldeck, W., Mueller, G., Wiessler, M., Tóth, K., and Braun, K. (2011). Positioning effects of KillerRed inside of cells correlate with DNA strand breaks after activation with visible Light. Int. J. Med. Sci. 8, 97–105. doi: 10.7150/ijms.8.97
Wan, Q. L., Liu, L., Xing, D., and Chen, Q. (2008). Bid is required in NPe6-PDT-induced apoptosis. Photochem. Photobiol. 84, 250–257. doi: 10.1111/j.1751-1097.2007.00248.x
Wang, N., Liu, Y., Xie, M. X., and Cui, Z. J. (2003). Changes in plasma membrane protein structure after photodynamic action in freshly isolated rat pancreatic acini. An FTIR study. J. Photochem. Photobiol. B Biol. 71, 27–34. doi: 10.1016/j.jphotobiol.2003.07.001
Westberg, M., Bregnhoj, M., Banerjee, C., Blazquez-Castro, A., Breitenbach, T., and Ogilby, P. R. (2016). Exerting better control and specificity with singlet oxygen experiments in live mammalian cells. Methods 109, 81–91. doi: 10.1016/j.ymeth.2016.07.001
Westberg, M., Holmegaard, L., Pimenta, F. M., Etzerodt, M., and Ogilby, P. R. (2015). Rational design ofan efficient, genetically encodable, proteinencased singlet oxygen photosensitizer. J. Am. Chem. Soc. 137, 1632–1642. doi: 10.1021/ja511940j
Williams, D. C., Bejjani, R. E., Ramirez, P. M., Coakley, S., Kim, S. A., Lee, H., et al. (2013). Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep. 5, 553–563. doi: 10.1016/j.celrep.2013.09.023
Wingen, M., Potzkei, J., Endres, S., Casini, G., Rupprecht, C., Fahlke, C., et al. (2014). The photophysics of LOV-based fluorescent proteins–new tools for cell biology. Photochem. Photobiol. Sci. 13, 875–883. doi: 10.1039/c3pp50414j
Wojtovich, A. P., Wei, A. Y., Sherman, T. A., Foster, T. H., and Nehrke, K. (2016). Chromophore-assisted light inactivation of mitochondrial electron transport chain complex II in Caenorhabditis elegans. Sci. Rep. 6:29695. doi: 10.1038/srep29695
Xu, S., and Chisholm, A. D. (2016). Highly efficient optogenetic cell ablation in C. elegans using membrane-targeted miniSOG. Sci. Rep. 6, 21271–21283. doi: 10.1038/srep21271
Xue, L. Y., Chiu, S. M., and Oleinick, N. L. (2001). Photochemical destruction of the Bcl-2 oncoprotein during photodynamic therapy with the phthalocyanine photosensitizer Pc 4. Oncogene 20, 3420–3427. doi: 10.1038/sj.onc.1204441
Yang, J. Y., and Yang, W. Y. (2011). Spatiotemporally controlled initiation of Parkin-mediated mitophagy within single cells. Autophagy 7, 1230–1238. doi: 10.4161/auto.7.10.16626
Yu, N., and Smagghe, G. (2014). CCK(-like) and receptors: structure and phylogeny in a comparative perspective. Gen. Comp. Endocrinol. 209, 74–81. doi: 10.1016/j.ygcen.2014.05.003
Keywords: photosensitization, protein photosensitizer, photonanomanipulation, singlet oxygen, calcium oscillation, pancreatic acinar cells, photopharmacology, ligand-independent
Citation: Jiang HN, Li Y and Cui ZJ (2017) Photodynamic Physiology—Photonanomanipulations in Cellular Physiology with Protein Photosensitizers. Front. Physiol. 8:191. doi: 10.3389/fphys.2017.00191
Received: 12 December 2016; Accepted: 14 March 2017;
Published: 04 April 2017.
Edited by:
Murugesan Velayutham, University of Pittsburgh School of Medicine, USAReviewed by:
Lars Kaestner, Saarland University, GermanyCopyright © 2017 Jiang, Li and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zong Jie Cui, empjdWlAYm51LmVkdS5jbg==
†Equal first authors.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.