
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 23 March 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.00168
This article is part of the Research Topic Phytohormones and the Regulation of Stress Tolerance in Plants: Current Status and Future Directions View all 28 articles
 Yi Wang1,2†
Yi Wang1,2† Xiaolu Wang1,2†
Xiaolu Wang1,2† Chao Wang1,2†
Chao Wang1,2† Fan Peng1,2
Fan Peng1,2 Ruijiao Wang1,2
Ruijiao Wang1,2 Xue Xiao1,2
Xue Xiao1,2 Jian Zeng3
Jian Zeng3 Houyang Kang1,2
Houyang Kang1,2 Xing Fan1,2
Xing Fan1,2 Lina Sha1,2
Lina Sha1,2 Haiqin Zhang1,2
Haiqin Zhang1,2 Yonghong Zhou1,2*
Yonghong Zhou1,2*Different intra- or interspecific wheat show different interactions of Cd/Zn. Normally, Zn has been/being widely utilized to reduce the Cd toxicity. In the present study, the DPW seedlings exhibited strong Cd tolerance. Zn and Cd mutually inhibited their uptake in the roots, showed antagonistic Cd/Zn interactions. However, Zn promoted the Cd transport from the roots to shoots, showed synergistic. In order to discover the interactive molecular responses, a transcriptome, including 123,300 unigenes, was constructed using RNA-Sequencing (RNA-Seq). Compared with CK, the expression of 1,269, 820, and 1,254 unigenes was significantly affected by Cd, Zn, and Cd+Zn, respectively. Only 381 unigenes were co-induced by these three treatments. Several metal transporters, such as cadmium-transporting ATPase and plant cadmium resistance 4, were specifically regulated by Cd+Zn. Other metal-related unigenes, such as ABC transporters, metal chelator, nicotianamine synthase (NAS), vacuolar iron transporters (VIT), metal-nicotianamine transporter YSL (YSL), and nitrate transporter (NRT), were regulated by Cd, but were not regulated by Cd+Zn. These results indicated that these transporters participated in the mutual inhibition of the Cd/Zn uptake in the roots, and also participated in the Cd transport, accumulation and detoxification. Meanwhile, some unigenes involved in other processes, such as oxidation-reduction, auxin metabolism, glutathione (GSH) metabolism nitrate transport, played different and important roles in the detoxification of these heavy metals.
In plants, non-essential heavy metals cause toxicity and eventually inhibit plant growth and development (Balen et al., 2011). Cadmium (Cd), a heavy metal widespread in the environment, damages the photosynthetic apparatus, affects the respiratory and nitrogen metabolism, and alters the balance of water and nutrient uptake (Herbette et al., 2006; Balen et al., 2011). Cd absorbed by plants can be introduced into the food chain (McLaughlin et al., 1999). Consumption, either directly or indirectly, of these parts could be a human health concern (Grant et al., 2008). Therefore, the Cd concentrations in several safe cereal grains were limited below 0.2 mg/Kg. In contrast, zinc (Zn) is an essential metal for plant growth. It has been/being widely utilized to reduce the Cd toxicity, although the effect varies with genotypes, the dose and duration of the Zn and Cd exposure (Rizwan et al., 2016). Excess Zn also limits the plant growth and causes a strong toxicity (Zhao et al., 2005; Wang et al., 2009). Plants therefore need to prevent damage from non-essential metals and ensure the proper homeostasis of essential metals (Lin and Aarts, 2012).
Cd and Zn, co-existed in the soil, cause various synergistic and antagonistic interactions according to the species, external metal concentrations, tissues, and developmental stages (Cataldo et al., 1983; Nan et al., 2002; Hassan et al., 2005; Sun et al., 2005; Balen et al., 2011; Cherif et al., 2011; Tkalec et al., 2014). In soybean, the uptake of Cd/Zn exhibits competitive inhibition (Cataldo et al., 1983). In durum and bread wheat, Cd and Zn mutually inhibit their uptake in the roots, stems and leaves (Hart et al., 2002, 2005). In tomato (Cherif et al., 2011) and Lemna minor (Balen et al., 2011), Zn inhibits the Cd uptake. However, the Cd/Zn interactions are not always antagonistic. Synergistic interactions were observed in both wheat and corn under field conditions (Nan et al., 2002). In tobacco, Zn promotes the Cd uptake, while Cd inhibits the Zn uptake in the roots and leaves (Tkalec et al., 2014). In rice, Zn increases the Cd concentration in the shoots, but inhibits the Cd uptake in the roots (Hassan et al., 2005). However, all these studies focused on the transport and biochemical responses by measuring plant growth, metal concentration, pigment content, and antioxidant content (Cataldo et al., 1983; Nan et al., 2002; Hassan et al., 2005; Sun et al., 2005; Cherif et al., 2011; Balen et al., 2011; Tkalec et al., 2014). Meanwhile, studies revealed the changes of transcriptomic profiles were focused on Cd or Zn alone (Herbette et al., 2006; Di Baccio et al., 2011; Lin et al., 2013). The changes of transcriptomic profiles for Cd/Zn interactions were not revealed.
Transcriptomes of Triticum turgidum (2n = 4x = 28, AABB) and common wheat (2n = 6x = 42, AABBDD) using RNA-sequencing (RNA-Seq) have been reported (Duan et al., 2012; Schreiber et al., 2012; Krasileva et al., 2013). Recently, the transcriptomic profiles of the developing starchy endosperm and the grain filling of bread wheat, and the dwarfism of dwarf Polish wheat were revealed using RNA-Seq (Pont et al., 2011; Pellny et al., 2012; Wang et al., 2016a). Although the molecular responses to Cd or Zn in plants have been widely investigated (Herbette et al., 2006; Di Baccio et al., 2011; Lin et al., 2013), the similar study in wheat using RNA-Seq is not processed. Based on genetic analysis and taxonomical classification, Polish wheat (2n = 4x = 28, AABB, Triticum polonicum L.) presents a low level of genetic similarity with T. durum, T. turgidum, and T. aestivum (Wang et al., 2013; Michalcová et al., 2014). Due to the high thousand kernel weights and high Zn, Fe, and Cu concentrations in the seeds (Wiwart et al., 2013), and the dwarfing gene (Kang et al., 2012), Polish wheat has attracted the interest of producers and breeders (Wiwart et al., 2013). Dwarf polish wheat (DPW, Triticum polonicum L.) which collected from Tulufan, Xingjiang, China, shows high tolerance to Cd and Zn. Therefore, it is a desirable material for studying Cd/Zn interactions. Previously proteomic study revealed that many proteins mainly participated in sucrose, glutathione (GSH), S-adenosyl-l-methionine (SAM), organic acids metabolisms and oxidation-reduction process were response to the Cd/Zn interactions on two days after treatments (Wang et al., 2016b). However, results of transtriptomic and proteomic analysis are very low overlay. It is interesting to investigate that what kinds of genes response to the Cd/Zn interactions when prolonged the treated time. In the present study, our aims are therefore to investigate the transcriptome responses under Cd, Zn and Cd+Zn stresses, finally reveal the molecular mechanisms of Cd/Zn interactions in the DPW roots on 5 days after treatments.
Seeds of DPW were sterilized with 1% NaOCl. After germination at room temperature for 5 days, the seedlings with plastic foam support grown on distilled water for 3 days and then were cultured in nutrient solution (Hoagland's Modified Basal Salt Mixture, MP Biomedicals, USA) in a growth chamber at 25°C with a relative humidity 70% under a 16-h-light/8-h-dark cycle. Per 50 plants were cultured in a container which contained 8 l nutrient solution with pH 6.0. The nutrient solution was refreshed every 5 days. Two-leaf seedlings were stressed with control (CK, null), 40 μM CdSO4 (Cd), 800 μM ZnCl2 (Zn, the Zn concentration of arable soil varies from 25 to 150 mg/Kg), and 40 μM CdSO4+ 800 μM ZnCl2 (Cd+Zn). On 5 days after treatments, the roots collected from 15 plants (15 plants per biological replicate, three biological replicates) were snap frozen in liquid nitrogen and stored at −80°C for RNA-Seq.
On 5 days after treatments, the leaves and roots were collected from 20 plants (20 plants per biological replicate, three biological replicates). The roots were successively washed with 0.1 μM EDTA and ddH2O. The length of the longest root and leaf per plant were measured. Their fresh and dry weights were also determined. The percentage of leaf or root dry weight was calculated as (leaf or root dry weight of 20 plants)/(total dry weight of 20 plants) ×100%; the percentage of leaf or root fresh weight was calculated as (leaf or root fresh weight of 20 plants)/(total fresh weight of 20 plants) × 100%. After weighing, all tissues were dried at 80°C for 2 days to measure metal concentration. At the same time, the percentages of water content and dry weight were calculated. All data analysis (student's t-test) was performed with SPSS 20.0 and figures were drawn with Sigmaplot 12.0.
The Cd and Zn concentrations were measured as described by Wang et al. (2014). Reference standard solutions of Cd and Zn were purchased from the Fisher Scientific Ltd. (Shanghai, China). All data analysis (student's t-test) was performed with SPSS 20.0 and figures were drawn with Sigmaplot 12.0.
Total RNA of each sample (null, Cd, Zn and Cd+Zn) was isolated using the E.Z.N.A.® Total RNA Kit II (Omega, USA). The RNA was checked for quality on 1% agarose gels and the NanoPhotometer® spectrophotometer (Implen, Germany) and the RNA 6000 Nano Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, USA). The Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, Shanghai, China) was used to measure RNA concentration.
mRNA was purified from total RNA using poly-T oligo-attached magnetic beads (Life Technologies, USA) and transcribed to cDNA using random oligonucleotides and M-MuLV Reverse Transcriptase (RNase H−) (TaKaRa, Dalian, China). NEBNext adaptor oligonucleotides (Illumia, USA) were ligated to 3′ ends of cDNA fragments. Then, 200-bp cDNA fragments were purified using the AMPure XP beads system (Beckman Coulter, USA). Ten cycles of PCR amplifications were performed to enrich cDNA fragments using the NEB Universal PCR primer and Index primer (Illumia, USA). The PCR products were purified using the AMPure XP beads system and quantified using the Agilent Bioanalyzer 2100 system. Finally, the four-coded samples were clustered by a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumia, USA), and then sequenced on an Illumina Hiseq 2000 platform.
Adapter reads containing poly-N and low-quality reads were removed using Novogene-written perl scripts to product clean reads. The paired-end clear reads generated contigs using Trinity (V2012-10-15) (Grabherr et al., 2011) with minimum K-mer coverage was 2, and other parameters were default.
The putative unigene function was annotated using a series of databases, including BLASTx against the NCBI NR and NT, Swiss-Prot databases, the Kyoto Encyclopedia of Genes and Genomes (KEGG), Ortholog database (KO) and Clusters of Orthologous Groups of proteins (KOG/COG) database, with an E-value cutoff of 10-6, hidden Markov models scan (hmmscan) against the protein family (Pfam) (Eddy, 2011), and Blast2GO against Gene Ontology (GO) (Götz et al., 2008). Functional categories of putative unigenes were grouped using the GO database, KEGG database, and KOG database.
Clean reads were aligned against reference transcript sequences to produce a read count using the RSEM package (Li and Dewey, 2011). The read counts of each unigene were converted into RPKM values to normalize the gene expression (Mortazavi et al., 2008). Differentially expressed genes (DEGs) were calculated using the DEseq method (Ander and Huber, 2010).
qPCR and data analysis were performed as described by Wang et al. (2015). Twelve differential expressed genes were validated and their primers were listed in STable 1. Actin (Wang et al., 2015) was used to standardize transcript levels in each sample. The ΔΔCt method was used to normalize the relative expression of each gene using the software of Bio-Rad CFX manager v. 1.6.541.1028. The student's t-test (P < 0.05) was conducted for the evaluation of significance of mean values.
Compared with control (CK), Cd did not inhibit the root and shoot growth after 5 days of treatment (Figures 1A,B). Excess Zn and Zn + Cd slightly reduced the root length (Figure 1A) and significantly (P < 0.05) inhibited the shoot growth (Figure 1B). Compared with CK, Cd significantly reduced the fresh root weight percentage (Figure 1D), but did not affect the dry root weight percentage (Figure 1C) and the dry and fresh leaf weight percentage (Figures 1E,F). Zn and Cd+Zn significantly increased the fresh root weight percentage (Figure 1D), but did not affect the leaf weight percentage (Figures 1E,F). The results described above indicated that metal stresses obviously affected the plant growth.
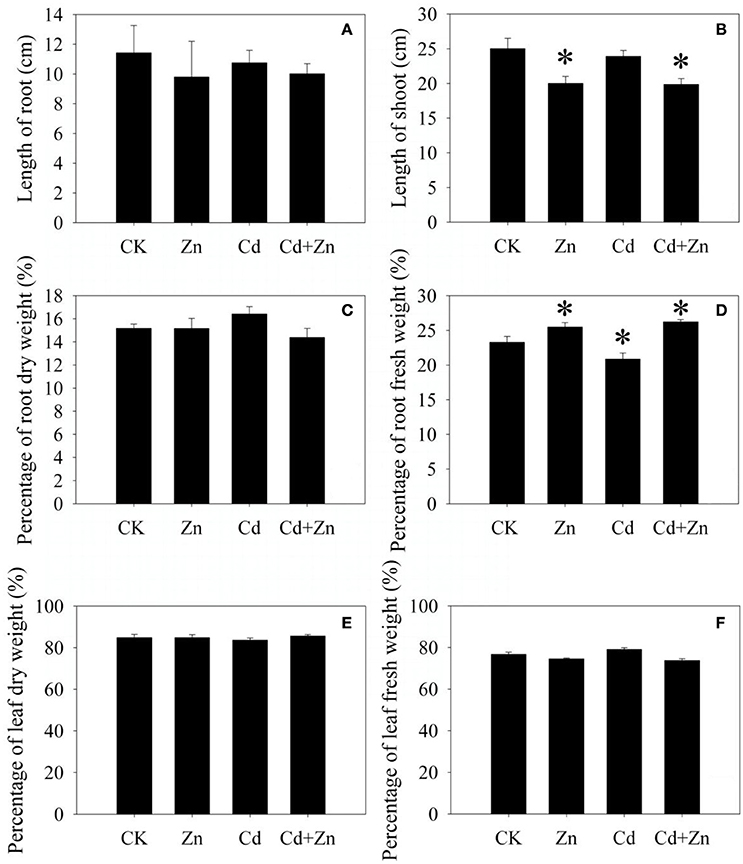
Figure 1. Growth of DPW exposed to Cd, Zn and Cd+Zn. (A) root length; (B) shoot length; (C) weight percentages of dry roots; (D) weight percentages of fresh roots; (E) weight percentages of dry leaves; (F) weight percentages of fresh leaves. Values were means ± standard error (three biological replicates); asterisk represented significant difference (P < 0.05).
After 5 days of treatments, an accumulation of Cd was not observed in all investigated samples which were unexposed to Cd (CK and Zn, Figures 2A,B). The Cd concentration in the roots under Cd+Zn stress was significantly lower (P < 0.01) than that under Cd stress (Figure 2A). However, opposite result was observed in the shoots (Figure 2B). These results indicated that Zn inhibited the Cd uptake in the roots, but promoted the Cd transport from the roots to shoots.
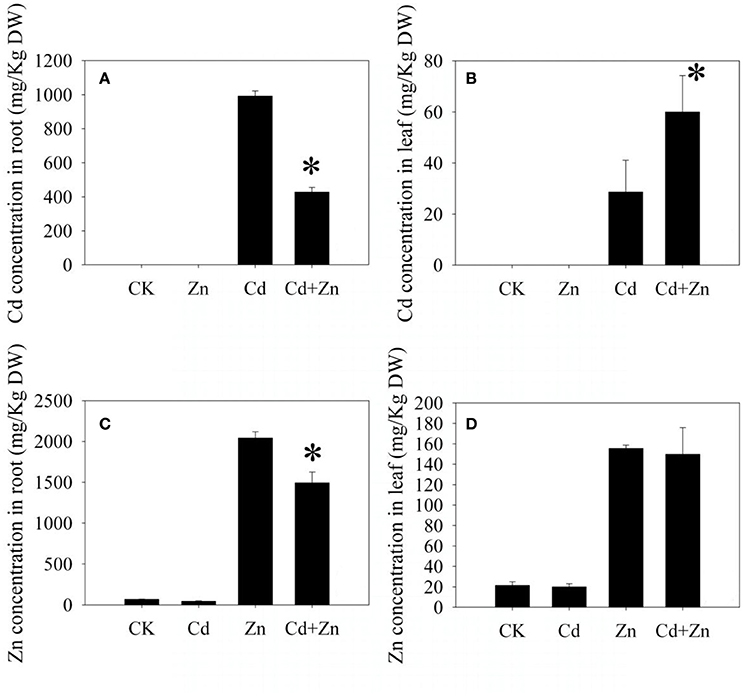
Figure 2. Metal concentrations in the roots and leaves. (A,B) Cd concentrations; (C,D) Zn concentrations. Values were means ± standard error (three biological replicates); asterisk represented significant difference (P < 0.05).
The Zn concentration was higher in the roots than that in the leaves (Figures 2C,D). In the roots, the Zn concentration under Cd+Zn stress was significantly lower (P < 0.01) than that under Zn stress (Figure 2C). In the leaves, the Zn concentrations were similar between Zn stress and Cd+Zn stress (Figure 2D). Thus, Cd only inhibited the Zn uptake in the roots.
RNA-Seq generated 18.35 Gb nucleotides. All raw read sequences were deposited to the NCBI Sequence Read Archive (SRA) database with accession numbers SRR2973581, SRR2973582, SRR2973583, and SRR2973584. Approximately 123,300 unigenes that varied from 201 bp to 16,390 bp (mean length was 660 bp, N50 value was 870 bp) were assembled. Amino acid (AA) sequences of 76,395 (61.96%) unigenes were predicted.
Though Blastx against several public databases, 84,709 (68.70%) unigenes were annotated. Among these annotated unigenes, 63,221 unigenes were functionally classified in GO; 13,637 unigenes were classified into 26 KOG categories; 10,576 unigenes were functionally classified in KEGG. All data of sequences and functional annotation were deposited to the NCBI Transcriptome Shotgun Assembly (TSA) database with accession number GEDP00000000.
Cd and Zn mutually inhibited their uptake in the roots, Zn promoted the Cd transport from the roots to shoots (Figure 2), and these treatments also affected growth (Figure 1). Theoretically, some unigenes participated in the interactions should be regulated by these treatments, which were revealed using RNA-Seq.
Compared with CK, the expression of 1,269, 1,254, and 820 unigenes was changed by Cd, Cd+Zn, and Zn, respectively (Figure 3). Among these DEGs, the expression of 381 unigenes mainly participated in several basic processes were co-changed by Cd, Zn, and Cd+Zn (STable 2), suggesting that there were differential molecular responses to Cd, Zn, and Cd+Zn stresses.
The remaining DEGs were arranged into 6 subgroups (Figure 3).
(1) The expression of 763 DEGs, which consisted of 96 down- and 667 up-regulated unigenes, was changed by Cd, but was not affected by Zn and Cd+Zn (log2fold-changes of CK/Cd were >1 or <−1 with P values below 1.00E-05, log2fold-changes of CK/Zn and CK/Cd+Zn varied from 1 to −1, Figure 3, STable 3). These DEGs mainly participated in glutathione (GSH) metabolism (4 down- and 7 up-), oxidation-reduction process (1 down- and 12 up-), carbohydrate metabolism (3 down- and 19 up-), metal transport (9 up-), nitrate metabolism (3 up- and 1 down-), and metal chelation (2 up-) (Table 1).
(2) 172 Zn-induced DEGs, which consisted of 36 down- and 136 up-regulated unigenes, were not affected by Cd and Cd+Zn (log2fold_changes of CK/Zn were >1 or <−1 with P-values below 1.00E-05, log2fold-changes of CK/Cd, and CK/Cd+Zn varied from 1 to −1, Figure 3, STable 4). Among them, several noteworthy DEGs were correspondingly grouped into carbohydrate metabolism (3 down- and 8 up-), GSH metabolism (4 up-), and oxidation-reduction process (3 up-) (Table 1).
(3) The expression of 42 DEGs was changed by Cd and Zn, but was not changed by Cd+Zn (log2-fold changes of CK/Cd and CK/Zn were >1 or <−1 with P-values below 1.00E-05, log2fold-changes of CK/Cd+Zn varied from 1 to −1, Figure 3, STable 5), which suggested that the expression of these DEGs was mutually suppressed by Cd and Zn under Cd+Zn stress. Among these unigenes, several noteworthy DEGs participated in GSH and carbohydrate metabolism, respectively (Table 1).
(4) The expression of 83 DEGs which included 51 down- and 32 up-regulated unigenes was changed by Cd and Cd+Zn, but was not changed by Zn (log2−fold changes of CK/Cd and CK/Cd+Zn were >1 or <−1 with P-values below 1.00E-05, log2fold-changes of CK/Zn varied from 1 to −1, Figure 3, STable 6), suggesting that these DEGs were specifically Cd-induced. Among these unigenes, some noteworthy unigenes participated in carbohydrate metabolism (2 up-), oxidation-reduction process (peroxidase 15 and catalase isozyme 2), and nitrate transport (nitrate transporter 1.5) (Table 1).
(5) The expression of 225 DEGs, consisting of 174 down- and 51 up-regulated unigenes, was affected by Zn and Cd+Zn, but was not affected by Cd alone (log2-fold changes of CK/Zn and CK/Cd+Zn were >1 or <−1 with P-values below 1.00E-05, log2fold-changes of CK/Cd varied from 1 to −1, Figure 3, STable 7), suggesting that these DEGs were specifically Zn-induced. Among them, noteworthy DEGs mainly participated in oxidation-reduction process (3 up-), carbohydrate metabolism (2 down-), GSH metabolism (1 up-) and nitrate metabolism (1 up-) (Table 1).
(6) Additionally, the expression of 565 DEGs which consisted of 95 down- and 470 up-regulated unigenes was affected by Cd+Zn, but was not affected by Cd and Zn (log2-fold changes of CK/Cd+Zn were >1 or <−1 with P-values below 1.00E-05, log2fold-changes of CK/Zn and CK/Cd varied from 1 to −1, Figure 3, STable 8), suggesting that these DEGs specifically responded to Cd+Zn. Among these DEGs, some noteworthy DEGs participated in carbohydrate metabolism (2 down- and 9 up-), metal transport (2 up-) and nitrate transport (2 down-) (Table 1).
To validate different expression levels that resulted from RNA-Seq, the expression of 12 DEGs were normalized (STable 1). As shown in Figure 4, compared with CK, two DEGs were significantly (P < 0.01) regulated by Cd and Cd+Zn, but were not regulated by Zn (Figure 4A). Two DEGs were significantly (P < 0.01) regulated by Zn and Cd+Zn, but were not regulated by Cd (Figure 4A). Two DEGs were significantly (P < 0.01) regulated by Cd+Zn, but were not regulated by Cd and Zn (Figure 4A). Two DEGs were significantly (P < 0.01) regulated by Cd, but were not regulated by Zn and Cd+Zn (Figure 4B). Two DEGs were significantly (P < 0.01) regulated by Zn, but were not regulated by Cd and Cd+Zn (Figure 4B). Two DEGs were significantly (P < 0.01) regulated by Cd and Zn, but were not regulated by Cd+Zn (Figure 4B). These results were similar with the differential expression resulted from RNA-Seq, suggesting that DEGs resulted from RNA-Seq were credibly used to analyze the molecular responses to Cd, Zn, and Cd+Zn.
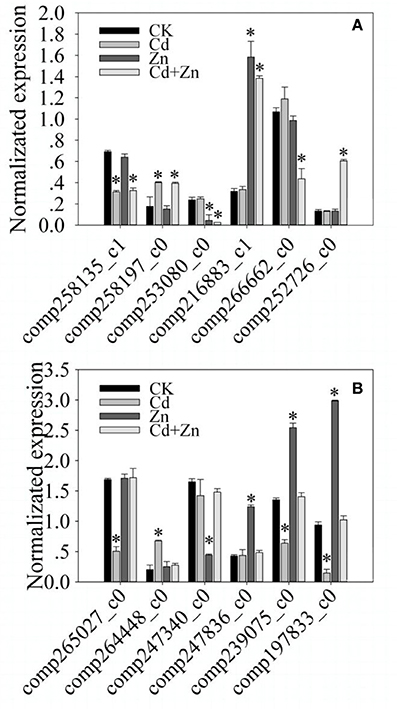
Figure 4. qRT-PCR validation of the expression of randomly selected 12 DEGs that resulted from RNA-Seq. (A) compared with CK, comp258135_c1 was down induced by Cd and Cd+Zn; comp258197_c0 was up induced by Cd and Cd+Zn; comp253080_c0 was down induced by Zn and Cd+Zn; comp216883_c1 was up induced by Zn and Cd+Zn; comp26662_c0 was specifically down induced by Cd+Zn; comp252726_c0 was specifically up induced by Cd+Zn. (B) comp265027_c0 was down induced by Cd; comp264448_c0 was up induced by Cd; comp247340_c0 was down induced by Zn; comp247836_c0 was up induced by Zn; comp239075_c0 and comp197833_c0 were down induced by Cd, but was up induced by Zn. Bars represented standard errors of three biological replicates. Asterisks represented significant differences between treatments and CK.
Cd inhibits the growth of bread wheat (Sun et al., 2005) and durum wheat (Hart et al., 2005; Koleva-Valkova et al., 2012). However, it also stimulates or does not inhibit the plant growth of other types of bread wheat (Stolt et al., 2003; Zhao et al., 2005; Lin et al., 2007). In the present study, although DPW accumulated 992.29 ± 29.83 mg/Kg (dry weight, DW) Cd in the roots and 40.82 ± 13.70 mg/Kg (DW) Cd in the shoots, the lengths of root and shoot were not affected (Figures 1A,B), which validated that DPW seedlings had stronger Cd tolerance than other types of durum and bread wheat (Hart et al., 2002, 2005). Meanwhile, the growth was inhibited by Zn and Zn+Cd stresses when the leaf and root accumulated high Zn concentrations (Figure 1), which indicated that excess Zn could cause the toxicity in wheat seedling (Zhao et al., 2005).
Since Cd and Zn mutually inhibited their uptake in the roots (Figure 2), the Cd/Zn interactions in the DPW roots were antagonistic, which was same as the antagonists in bread and durum wheat (Hart et al., 2002, 2005; Sun et al., 2005). However, Zn promoted the Cd transport from the roots to shoots (Figure 2B), Cd did not affect Zn transport (Figure 2D), which indicated that the Cd/Zn interactions in the DPW leaves were synergistic. This result was different from that Zn inhibited the Cd transport from the roots to shoots on 2 days after treatments with the same metal treated concentrations (Wang et al., 2016b).
Our previous study revealed that various proteins participated in the Cd/Zn interactions on 2 days after treatment (Wang et al., 2016b). In the present study, the transcriptomic changes in the roots also indicated that some genes were involved in the Cd/Zn interactions (Figure 3, STables 2–8). On 5 days after treatments, the expression of 1,269, 1,254, and 820 unigenes was individually changed by Cd, Cd+Zn, and Zn. However, only 381 unigenes were co-regulated by these treatments (Figure 3). In addition to the specific unigenes individually induced by Cd, Zn, and Cd+Zn, the remaining DEGs classified into different subgroups were considered to participate in the Cd/Zn interactions. These results indicated that Cd, Zn, and Cd+Zn induced differential molecular responses, which ultimately resulted in the differential molecular responses for Zn and Cd stresses (Lin and Aarts, 2012). In the following discussion, some DEGs involved in several important processes were described.
Heavy metal transporters play important roles in the metal uptake, transport and distribution. In yeast, the expression of cadmium-transporting ATPase which is a cadmium-specific efflux pump enhanced Cd resistance by extruding intercellular Cd (Adle et al., 2007; Adle and Lee, 2008). In the DPW roots, the expression of cadmium-transporting ATPase and plant cadmium resistance 4 was specifically up-regulated by Cd+Zn (Table 1), suggesting that Cd might be extruded from roots, finally resulted in that the Cd concentration in the roots under Cd+Zn stress was significantly lower than that under Cd stress (Figure 2A). Therefore, Zn can enhance the Cd resistance by reducing the Cd accumulation (Rizwan et al., 2016).
Except of metal efflux pumps, other heavy metal transporters also play important roles in heavy metal detoxification. In Arabidopsis, AtABC25 (Kim D. Y. et al., 2006) and AtABCC1-3 (Bovet et al., 2003, 2005; Park et al., 2012; Brunetti et al., 2015) transported Cd into the vacuoles to increase Cd tolerance. Overexpression of a metal chelator, metallothionein (MT), increased Cd content in the roots and enhanced Cd tolerance (Sekhar et al., 2011). In this study, 6 ABC transporters (B, C, and D family members, Table 1) and a MT were up-induced by Cd, suggesting that most of Cd accumulated by root might be sequestrated into the vacuoles so that DPW seedling exhibited strong Cd tolerance under Cd stress (Figure 1). When these regulations were not observed under Cd+Zn stress (Table 1), the Cd accumulations in the roots should be reduced (Bovet et al., 2003, 2005; Sekhar et al., 2011; Park et al., 2012; Brunetti et al., 2015), finally resulted in that the Cd concentration in the roots under Cd stress was higher than that under Cd+Zn stress (Figure 2A). Meanwhile, most of Cd accumulated by the roots was not sequestrated into the vacuoles, but was uploaded into the xylems and then transported into the shoots (Figure 2B, Kim et al., 2007). Thus, these ABC transporters and MT participated in the Cd/Zn interactions for the Cd transport and accumulation.
In planta, the high accumulation of Fe in the shoots under Cd stress could alleviate Cd toxicity (Wu et al., 2012). Overexpression of nicotianamine synthase (NAS), vacuolar iron transporters (VIT), and metal-nicotianamine transporter YSL (YSL) increased the Fe content in the roots and shoots (Kim S. A. et al., 2006; Ishimaru et al., 2010; Wu et al., 2012). In the present study, Cd stress through up regulating NAS1, VIT, and YSL12 (Table 1) to increase the Fe content in the DPW roots and shoots (data not shown), which could alleviate Cd toxicity (Wu et al., 2012), and then enhanced the Cd tolerance in DPW seedlings (Figure 1). However, these regulations were not observed under Zn and Cd+Zn stresses (Table 1), suggesting that these up regulations were activated when the roots accumulating a certain amount of Cd.
Cd inhibits the nitrate assimilation and transport in planta (Sanita di Toppi and Gabbrielli, 1999; Li et al., 2010). Cd regulates several nitrate-related genes, such as glutamate dehydrogenase and nitrate transporter (NRT) (Chaffei et al., 2004; Li et al., 2010), and also affects leaf nitrogen remobilization and root nitrogen storage (Chaffei et al., 2004). Down-regulation of AtNRT2.8 reduced the Cd accumulation in the roots and increased it in the shoots. Thus, AtNRT2.8-regulated nitrate distribution controls the Cd uptake and transport (Li et al., 2010). In the present study, Cd alone up regulated glutamate dehydrogenase and two NRTs (Table 1), suggesting that Cd might regulate the nitrate assimilation and transport in the DPW roots (Chaffei et al., 2004). Meanwhile, two high affinity nitrate transporters (HANTs) were specifically down-regulated by Cd+Zn (Table 1). HANTs participate in the nitrate uptake (Cerezo et al., 2001; Li et al., 2007). The inhibition of nitrate uptake reduces the Cd uptake and other essential metals (Mao et al., 2014). Thus, down regulation of HANTs might mutually inhibit the Cd/Zn uptake under Cd+Zn stress (Table 1). Due to different nitrate-related genes were induced, Cd and Cd+Zn might regulate different nitrate metabolism.
Although these heavy metal and nitrate transporters were not observed at proteomic level on two days after treatments (Wang et al., 2016b), unigenes participated in glutathione (GSH) metabolism, antioxidant enzymes and cell wall composition were induced at both transcriptomic (Table 1) and proteomic level (Wang et al., 2016b). GSH is a substrate for phytochelatin synthesis and crucial for detoxification of heavy meals (Freeman et al., 2004; Yadav, 2009). Formation of GSH-Cd or Zn complexes and then sequestration into the vacuoles is another mechanism of detoxification (Seth et al., 2012; Jozefczak et al., 2015). Genes involved in GSH metabolism, such as glutathione S-transferase (GST), hydroxyacylglutathione hydrolase (HGH), and glutaredoxin (Grx), were differentially regulated by heavy metals (Di Baccio et al., 2011; Lin et al., 2013; Jozefczak et al., 2015). As detoxifying enzymes present in all aerobic organisms, GSTs catalyze the nucleophilic attack of the sulfur atom of the tripeptide GSH on the electrophilic group of the substrate (Adamis et al., 2004), and also transport compound of GSH-cytotoxic substrates into the vacuoles for detoxification (Kumar et al., 2013). In this study, several unigenes of GSH metabolism, such as Grx, HGH, lactoyglutathione lyase, S-formylglutathione hydrolase, disulfide isomerase-like 1-4, and 5 GSTs, were induced and grouped into different interactions of Cd/Zn (Table 1), suggesting that GSH metabolism played different roles in the Cd/Zn interactions (Wang et al., 2016b).
Plants have established effective antioxidative systems to protect cells against damage from metal-induced oxidative threats (Di Baccio et al., 2011). Previous studies revealed that many Cd or Zn- induced genes participated in the defense against oxidative stress (Di Baccio et al., 2011; Lin et al., 2013). In this study, 13 peroxidases (POD) and 3 aldehyde dehydrogenases that removed the toxic aldehydes from lipid peroxidation were regulated by Cd, but were not regulated by Zn and Cd+Zn stresses (Table 1). Three genes (two ubiquinol oxidase 1as and NADPH: quinone oxidoreductase 1) were up-regulated by Zn but were not induced by Cd and Cd+Zn stresses (Table 1). Catalase (CAT) isozyme 2 was specifically up-regulated by Cd (Table 1). These results confirmed that antioxidant enzymes play different and important roles in adaptive response to Cd, Zn, or Cd+Zn stresses (Qiu et al., 2008; Zeng et al., 2011).
The plant cell wall is mainly composed of cellulose and polysaccharides (Cosgrove, 2005). It can be modified by Cd (Li et al., 2015; Shi et al., 2015). Therefore, modification of cell wall composition is associated with the Cd exclusion in the roots (Zhu et al., 2012). Additionally, exogenous glucose also alleviates the Cd toxicity by fixing Cd in the cell wall and sequestering it into the vacuoles (Shi et al., 2015). In this study, many carbohydrate (including glucose, cellulose, callose and mannose) metabolism-related genes were differentially regulated by Cd, Zn, or Cd+Zn (Table 1), which suggested that cellulose, glucose and polysaccharides play different roles in the Cd and Zn fixation, exclusion and sequestration in the roots (Li et al., 2015).
YW, XW, CW, and YZ conceived and designed research, and wrote the manuscript. YW, XW, CW, XX, FP, and RW conducted experiments. YW, XW, JZ, HK, XF, LS, and HZ analyzed data. All authors read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the National Natural Science Foundation of China (No. 31671688 and 31470305), Bureau of Science and Technology and Bureau of Education of Sichuan Province, China.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00168/full#supplementary-material
Adamis, P. D. B., Gomes, D. S., Pinto, M. L., Panek, A. D., and Eleutherio, E. C. (2004). The role of glutathione transferases in cadmium stress. Toxicol. Lett. 154, 81–88. doi: 10.1016/j.toxlet.2004.07.003
Adle, D. J., and Lee, J. (2008). Expressional control of a cadmium-transporting P1B-type ATPase by a metal sensing degradation signal. J. Biol. Chem. 283, 31460–31468. doi: 10.1074/jbc.M806054200
Adle, D. J., Sinani, D., Kim, H., and Lee, J. (2007). A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 947–955. doi: 10.1074/jbc.M609535200
Ander, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Balen, B., Tkalec, M., Šikić, S., Tolić, S., Cvjetko, P., Pavlica, M., et al. (2011). Biochemical responses of Lemna minor experimentally exposed to cadmium and zinc. Ecotoxicology 20, 915–826. doi: 10.1007/s10646-011-0633-1
Bovet, L., Eggmann, T., Meylan-Bettex, M., Polier, J., Kammer, P., Marin, E., et al. (2003). Transcript levels of AtMRPs after cadmium treatment: induction of AtMRP. Plant Cell Environ. 26, 371–381. doi: 10.1046/j.1365-3040.2003.00968.x
Bovet, L., Feller, U., and Martinoia, E. (2005). Possible involvement of plant ABC transporters in cadmium detoxification: a cDNA submicroarray approach. Environ. Int. 31, 263–267. doi: 10.1016/j.envint.2004.10.011
Brunetti, P., Zanella, L., De Paolis, A., Di Litta, D., Cecchetti, V., Falasca, G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin- mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66, 3815–3829. doi: 10.1093/jxb/erv185
Cataldo, D. A., Garland, T. R., and Wildung, R. E. (1983). Cadmium uptake kinetics in intact soybean plants. Plant Physiol. 73, 844–848. doi: 10.1104/pp.73.3.844
Cerezo, M., Tillard, P., Filleur, S., Munos, S., Daniel-Vedele, F., and Gojon, A. (2001). Major alterations of the regulation of root NO3- uptake are associated with the mutaton of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127, 262–271. doi: 10.1104/pp.127.1.262
Chaffei, C., Pageau, K., Suzuki, A., Gouia, H., Ghorbel, M., and Masclaux-Daubresse, C. (2004). Cadmium toxicity induced changes in nitrogen management in Lycopersicon esculentum leading to a metabolic safeguard through an amino acid storage strategy. Plant Cell Physiol. 45, 1681–1693. doi: 10.1093/pcp/pch192
Cherif, J., Mediouni, C., Ammar, W. B., and Jemal, F. (2011). Interactions of zinc and cadmium toxicity in their effects on growth and in antioxidative systems in tomato plants (Solanum lycopersicum). J. Environ.Sci. 23, 837–844. doi: 10.1016/S1001-0742(10)60415-9
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Di Baccio, D., Galla, G., Bracci, T., Andreucci, A., Barcaccia, G., Tognetti, R., et al. (2011). Transcriptome analysis of Populus × euramericana clone I-214 leaves exposed to excess zinc. Tree Physiol. 31, 1293–1308. doi: 10.1093/treephys/tpr106
Duan, J., Xia, C., Zhao, G., Jia, J., and Kong, X. (2012). Optimizing de novo common wheat transcriptome assembly using short-read RNA-Seq data. BMC Genomics 13:392. doi: 10.1186/1471-2164-13-392
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Freeman, J. L., Persans, M. W., Nieman, K., Albrecht, C., Peer, W., Pickering, I. J., et al. (2004). Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Cell 16, 2176–2191. doi: 10.1105/tpc.104.023036
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, I. A., Adiconis, X., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grant, C. A., Clarke, J. M., Duguid, S., and Chaney, R. L. (2008). Selection and breeding of plant cultivars to minimize cadmium accumulation. Sci. Total Environ. 390, 301–310. doi: 10.1016/j.scitotenv.2007.10.038
Hart, J., Welch, R. M., Norvell, W. A., Clarke, J. M., and Kochian, L. V. (2005). Zinc effects on cadmium accumulation and partitioning in near-isogenic lines of durum wheat that differ in grain cadmium concentration. New Phytol. 167, 391–401. doi: 10.1111/j.1469-8137.2005.01416.x
Hart, J., Welch, R. M., Norvell, W. A., and Kochian, L. V. (2002). Transport interactions between cadmium and zinc in roots of bread and durum wheat seedlings. Physiol. Plant. 116, 73–78. doi: 10.1034/j.1399-3054.2002.1160109.x
Hassan, M. J., Zhang, G., Wu, F., Wie, K., and Chen, Z. (2005). Zinc alleviates growth inhibition and oxidative stress caused by cadmium in rice. J. Plant Nutr. Soil Sci. 168, 255–261. doi: 10.1002/jpln.200420403
Herbette, S., Taconnat, L., Hugouvieux, V., Piette, L., Magniette, M., Cuine, S., et al. (2006). Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88, 1751–1765. doi: 10.1016/j.biochi.2006.04.018
Ishimaru, Y., Masuda, H., Bashir, K., Inoue, H., Tsukamoto, T., Takahashi, M., et al. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62, 379–390. doi: 10.1111/j.1365-313X.2010.04158.x
Jozefczak, M., Bohler, S., Schat, H., Horemans, N., Guisez, Y., Remans, T., et al. (2015). Both the concentration and redox state of glutathione and ascorbate influence the sensitively of Arabidopsis to cadmium. Ann. Bot. 116, 601–612. doi: 10.1093/aob/mcv075
Kang, H. Y., Lin, L. J., Song, Z. J., Yuan, J. Y., Zhong, M. Y., Zhang, H. Q., et al. (2012). Identification, fine mapping and characterization of Rht-dp, a recessive wheat dwarfing (reduced height) gene derived from Triticum polonicum. Genes Genomics 34, 509–515. doi: 10.1007/s13258-012-0022-z
Kim, D. Y., Bovet, L., Kushnir, S., Noh, E. W., Martinoia, E., and Lee, Y. (2006). AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 140, 922–932. doi: 10.1104/pp.105.074146
Kim, D. Y., Bovet, L., Masayoshi, M., Enrico, M., and Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Kim, S. A., Punshon, T., Lanzirotti, A., Li, L., Alonso, J. M., Ecker, J. R., et al. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314, 1295–1298. doi: 10.1126/science.1132563
Koleva-Valkova, L., Vasilev, A., Cuypers, A., and Vangronsveld, J. (2012). Comparative study of cadmium and zinc toxic effects on the cell redox status of durum wheat plants. Agrarni Nauki 4, 39–46.
Krasileva, K. V., Buffalo, V., Bailey, P., Pearce, S., Ayling, S., Tabbit, F., et al. (2013). Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 14:R66. doi: 10.1186/gb-2013-14-6-r66
Kumar, S., Asif, M. H., Chakrabarty, D., Tripathi, R. D., Dubey, R. S., and Trivedi, P. K. (2013). Expression of a rice Lambda class of glutathione S-transferase, OsGST2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J. Hazard. Mater. 248–249, 228–237. doi: 10.1016/j.jhazmat.2013.01.004
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, J. Y., Fu, Y. L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Li, T., Tap, Q., Shohag, M. J. I., Yang, X., Sparks, D. L., and Liang, T. (2015). Root cell wall polysaccharides are involved in cadmium hyperaccumulation in Sedum alfredii. Plant Soil 389, 387–399. doi: 10.1007/s11104-014-2367-3
Li, W., Wang, Y., Okamoto, M., Crawford, N., Siddiqi, M., and Glass, A. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Lin, C. Y., Trinh, N. N., Fu, S. F., Hsiung, Y. C., Chia, L. C., Lin, C. W., et al. (2013). Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol. Biol. 81, 507–522. doi: 10.1007/s11103-013-0020-9
Lin, R., Wang, X., Luo, Y., Du, W., Guo, H., and Yin, D. (2007). Effects of soil cadmium on growth, oxidative stress and antioxidant system in wheat seedlings (Triticum aestivum L.). Chemosphere 69, 89–98. doi: 10.1016/j.chemosphere.2007.04.041
Lin, Y. F., and Aarts, M. G. (2012). The molecular mechanism of zinc and cadmium stress response in plants. Cell. Mole. Life Sci. 69, 3187–3206. doi: 10.1007/s00018-012-1089-z
Mao, Q. Q., Guan, M. Y., Lu, K. X., Du, S. T., Fan, S. K., Ye, Y. Q., et al. (2014). Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in Arabidopsis. Plant Physiol. 166, 934–944. doi: 10.1104/pp.114.243766
McLaughlin, M. J., Parker, D. R., and Clarke, J. M. (1999). Metal and micronutrients – food safety issues. Field Crops Res. 60, 143–163. doi: 10.1016/S0378-4290(98)00137-3
Michalcová, V., Dušinskŷ, R., Sabo, M., Beyroutiová, M. A., Hauptvogel, P., Ivaničocá, Z., et al. (2014). Taxonomical classification and origin of Kamut® wheat. Plant Syst. Evol. 300, 1749–1757. doi: 10.1007/s00606-014-1001-4
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nan, Z., Li, J., Zhang, J., and Cheng, G. (2002). Cadmium and zinc interactions and their transfer in soil-crop system under actual field conditions. Sci. Total Environ. 285, 187–195. doi: 10.1016/S0048-9697(01)00919-6
Park, J., Song, W. Y., Ko, D., Eom, Y., Hansen, T. H., Schiller, M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Pellny, T. K., Lovegrove, A., Freeman, J., Tosi, P., Love, C. G., Paul Knox, J., et al. (2012). Cell walls of developing wheat starchy endosperm comparison of composition and RNA-Seq transcriptome. Plant Physiol. 158, 612–627. doi: 10.1104/pp.111.189191
Pont, C., Murat, F., Confolcnt, C., Balzcrguc, S., and Salsc, J. (2011). RNA-Seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol. 12:R119. doi: 10.1186/gb-2011-12-12-r119
Qiu, R. L., Zhao, X., Tang, Y. T., Yu, F. M., and Hu, P. J. (2008). Antioxidative response to Cd in a newly discovered cadmium hyperaccumulator, Arabis paniculata F. Chemosphere 74, 6–12. doi: 10.1016/j.chemosphere.2008.09.069
Rizwan, M., Ali, S., Abbas, T., Zia-ur-Rehman, M., Hannan, F., Keller, C., et al. (2016). Cadmium minimization in wheat: a critical review. Ecotoxicol. Environ. Saf. 130, 43–53. doi: 10.1016/j.ecoenv.2016.04.001
Sanita di Toppi, L., and Gabbrielli, R. (1999). Response to cadmium in higher plants. Environ. Exp. Bot. 41, 105–130. doi: 10.1016/S0098-8472(98)00058-6
Schreiber, A. W., Hayden, M. J., Forrest, K. L., Kong, S., Langridge, P., and Baumann, U. (2012). Transcriptome-scale homoeolog-specific transcript assemblies of bread wheat. BMC Genomics 13:492. doi: 10.1186/1471-2164-13-492
Sekhar, K., Priyanka, B., Reddy, V. D., and Rao, K. V. (2011). Metallothionein 1 (CcMT1) of pigeonpea (Cajanus cajan, L.) confers enhanced tolerance to copper and cadmium in Escherichia coli and Arabidopsis thaliana. Environ. Exp. Bot. 72, 131–139. doi: 10.1016/j.envexpbot.2011.02.017
Seth, C. S., Remans, T., Keunen, E., Jozefczak, M., Glelen, H., Opdenakker, K., et al. (2012). Phytoextraction of toxic metals: a central role for glutathione. Plant Cell Environ. 35, 334–346. doi: 10.1111/j.1365-3040.2011.02338.x
Shi, Y. Z., Zhu, X. F., Wang, J. X., Li, G. X., and Zheng, S. J. (2015). Glucose alleviates cadmium toxicity by increasing cadmium fixation in roots cell wall and sequestration into vacuole in Arabidopsis. J. Integr. Plant Biol. 10, 830–837. doi: 10.1111/jipb.12312
Stolt, J. P., Sneller, F. E. C., Bryngelsson, T., Lundborg, T., and Schat, H. (2003). Phytochelatin and cadmium accumulation in wheat. Environ. Exp. Bot. 49, 21–28. doi: 10.1016/S0098-8472(02)00045-X
Sun, Q., Wang, X. R., Ding, S. M., and Yuan, X. F. (2005). Effects of interactions between cadmium and zinc on phytochelation and glutathione production in wheat (Triticum aestivum L.). Environ. Toxicol. 20, 195–201. doi: 10.1002/tox.20095
Tkalec, M., Štefanić, P. P., Cvjetko, P., Šikić, S., Pavlica, M., and Balen, B. (2014). The effects of cadmium-zinc interactions on biochemical response in tobacco seedlings and adult plants. PLoS ONE 9:e87582. doi: 10.1371/journal.pone.0087582
Wang, C., Zhang, S. H., Wang, P. F., Hou, J., Zhang, W. J., Li, W., et al. (2009). The effect of excess Zn on mineral nutrition and antioxidative response in rapeseed seedling. Chemosphere 75, 1468–1476. doi: 10.1016/j.chemosphere.2009.02.033
Wang, Y., Wang, C., Zhang, H., Yue, Z., Liu, X., and Ji, W. (2013). Genetic analysis of wheat (Triticum aestivum L.) and related species with SSR markers. Genet. Resour. Crop Evol. 60, 1105–1117. doi: 10.1007/s10722-012-9907-6
Wang, Y., Wang, X., Gu, M., Kang, H., Zeng, J., Fan, X., et al. (2015). Cloning and characterization of four novel SnRK2 genes from Triticum polonicum. Biol. Plant. 59, 211–219. doi: 10.1007/s10535-015-0501-6
Wang, Y., Wang, X., Wang, C., Wang, R., Peng, F., Xiao, X., et al. (2016a). Proteomic profiling of the interactions of Cd/Zn in the roots of dwarf polish wheat (Triticum polonicum L.). Front. Plant Sci. 7:1378. doi: 10.3389/fpls.2016.01378
Wang, Y., Xiao, X., Wang, X., Zeng, J., Kang, H., Fan, X., et al. (2016b). RNA-Seq and iTRAQ reveal the dwarfing mechanism of dwarf polish wheat (Triticum polonicum L.). Int. J. Biol. Sci. 12, 653–666. doi: 10.7150/ijbs.14577
Wang, Y., Xiao, X., Zhang, T., Kang, H., Zeng, J., Fan, X., et al. (2014). Cadmium treatment alters the expression of five genes at the Cda1 loucs in two soybean cultivars (Glycine Max (L.) Merr). Sci. World J. 2014:979750. doi: 10.1155/2014/979750
Wiwart, M., Suchowilska, E., Kandler, W., Sulyok, M., Groenwald, P., and Krska, R. (2013). Can polish wheat (Triticum polonicum L.) be an interesting gene source for breeding wheat cultivars with increased resistance to Fusarium head blight? Genet. Res. Crop Evol. 60, 2359–2373. doi: 10.1007/s10722-013-0004-2
Wu, H., Cheng, C., Du, J., Liu, H., Cui, Y., Zhang, Y., et al. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis–enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158, 790–800. doi: 10.1104/pp.111.190983
Yadav, S. K. (2009). Heavy metals toxicity in plants: an overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S. Afr. J. Bot. 76, 167–179. doi: 10.1016/j.sajb.2009.10.007
Zeng, X. W., Qiu, R. L., Ying, R. R., Tang, Y. T., Tang, L., and Fang, X. H. (2011). The differentially-expressed proteome in Zn/Cd hyperaccumulator Arabis paniculata Franch. in response to Zn and Cd. Chemosphere 82, 321–328. doi: 10.1016/j.chemosphere.2010.10.030
Zhao, Z., Zhu, Y., and Cai, Y. (2005). Effects of zinc on cadmium uptake by spring wheat (Triticum aestivum, L.): long-time hydroponic study and short-time 109 Cd tracing study. J. Zhejiang Univ. Sci. A 6, 643–648. doi: 10.1007/BF02856167
Keywords: dwarf polish wheat, RNA-Seq, cadmium, zinc, interaction
Citation: Wang Y, Wang X, Wang C, Peng F, Wang R, Xiao X, Zeng J, Kang H, Fan X, Sha L, Zhang H and Zhou Y (2017) Transcriptomic Profiles Reveal the Interactions of Cd/Zn in Dwarf Polish Wheat (Triticum polonicum L.) Roots. Front. Physiol. 8:168. doi: 10.3389/fphys.2017.00168
Received: 05 September 2016; Accepted: 07 March 2017;
Published: 23 March 2017.
Edited by:
Sheo Mohan Prasad, Allahabad University, IndiaReviewed by:
Thomas J. Bach, University of Strasbourg, FranceCopyright © 2017 Wang, Wang, Wang, Peng, Wang, Xiao, Zeng, Kang, Fan, Sha, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Zhou, WmhvdXloQHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.