
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol., 04 February 2016
Sec. Respiratory Physiology and Pathophysiology
Volume 7 - 2016 | https://doi.org/10.3389/fphys.2016.00021
This article is part of the Research TopicBiologics for the treatment of respiratory disease: A focus on translational researchView all 4 articles
MicroRNAs (miRNAs) are small non-coding RNA molecules that modulate expression of the majority of genes by inhibiting protein translation. Growing literature has identified functional roles for miRNAs across a broad range of biological processes. As such, miRNAs are recognized as potential disease biomarkers and novel targets for therapies. While several miRNA-targeted therapies are currently in clinical trials (e.g., for the treatment of hepatitis C virus infection and cancer), no therapies have targeted miRNAs in respiratory diseases in the clinic. In this mini-review, we review the current knowledge on miRNA expression and function in respiratory diseases, intervention strategies to target miRNA function, and considerations specific to respiratory diseases. Altered miRNA expression profiles have been reported in a number of respiratory diseases, including asthma, chronic obstructive pulmonary disease, cystic fibrosis, and idiopathic pulmonary fibrosis. These include alterations in isolated lung tissue, as well as sputum, bronchoalveolar lavage fluids and peripheral blood or serum. The observed alterations in easily accessible body fluids (e.g., serum) have been proposed as new biomarkers that may inform disease diagnosis and patient management. In a subset of studies, miRNA-targeted interventions also improved disease outcomes, indicating functional roles for altered miRNA expression in disease pathogenesis. In fact, direct administration of miRNA-targeting molecules to the lung has yielded promising results in a number of animal models. The ability to directly administer compounds to the lung holds considerable promise and may limit potential off-target effects and side effects caused by the systemic administration required to treat other diseases.
MicroRNAs (miRNAs) are small non-coding RNA molecules that inhibit protein translation from target mRNAs. More than 1000 unique miRNAs are present in the human genome (Lewis et al., 2005; Berezikov, 2011) and miRNAs are thought to modulate expression of >60% of genes (Friedman et al., 2009). MiRNAs function across a wide variety of biological processes and are increasingly recognized as biomarkers for disease diagnosis and potential therapeutic targets for treatment. In respiratory diseases, there is emerging evidence that altered miRNA expression modulates disease processes and ultimately disease pathogenesis. In this mini-review, we will briefly summarize the current understanding of how miRNAs are processed and function, their roles in respiratory diseases and techniques available to modulate miRNA function in vivo.
The mechanisms regulating miRNA expression, processing and function have been extensively reviewed (Ha and Kim, 2014) and a schematic of miRNA processing and function is presented in Figure 1. MiRNAs are commonly encoded either within the introns of protein-coding genes or as independent genes, and transcribed by RNA polymerase II (Lee et al., 2004). After transcription, mature miRNAs are generated through a multi-step process (Lee et al., 2002). First, the primary transcript (pri-miRNA) is processed by the nuclear RNase III Drosha-DGCR8 (DiGeorge syndrome critical region gene 8) complex, into a ~65 nucleotide hairpin precursor miRNA (pre-miRNA; Denli et al., 2004). Pre-miRNAs are exported from the nucleus into the cytoplasm by exportin 5 and cleaved by the RNase III enzyme Dicer into a double-stranded miRNA-miRNA* duplex approximately 22 nucleotides (nt) in length (Hutvágner et al., 2001; Ketting et al., 2001). This miRNA duplex is unwound by helicases and a mature miRNA is incorporated into the RNA-induced silencing complex (RISC; Bartel, 2004). The RISC complex binds target mRNA sequences through partially complementary binding with the incorporated miRNA (Bartel, 2004; Weinmann et al., 2009).
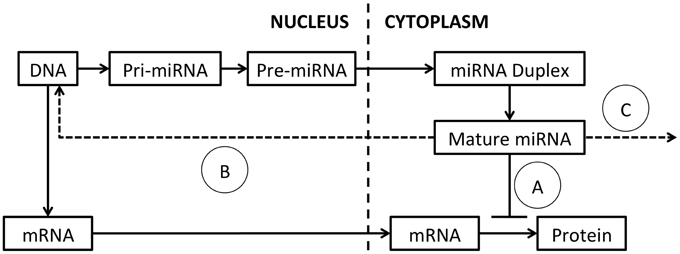
Figure 1. MiRNA processing and function. The primary miRNA transcript (pri-miRNA) is transcribed from DNA and excised by Drosha, to produce the pre-miRNA. The pre-miRNA is exported to the cytoplasm by exportin-5 and spliced by Dicer to generate a miRNA duplex. The duplex is unwound by helicases and a mature single-stranded miRNA is assembled into the RISC complex. (A) MiRNA typically modulate target mRNA translation by complementary binding, within the 3′ UTR, 5′ UTR or coding region. New evidence suggests that miRNAs also function by (B) binding of DNA promoters, acting as RNA decoys or through direct binding of target mRNAs. (C) Mature miRNAs can also be released in exosomes and act on distant cells.
MiRNAs typically modulate target mRNA levels by binding to the 3′ untranslated region (UTR) of mRNA transcripts (Hammond et al., 2001). This occurs through complementary binding of the highly specific seed sequence at the 5′ end of a miRNA to the target mRNA (Mallory et al., 2004). Sequence complementarity between the rest of the miRNA and the target mRNA is often quite low, allowing individual miRNAs to target multiple mRNA sequences and making the prediction of miRNA targets difficult (Lewis et al., 2003; Mallory et al., 2004; Lim et al., 2005; Liu, 2008). MiRNA:mRNA interactions repress protein translation and/or reduce target mRNA stability, resulting in decreased protein translation from the target mRNA.
New studies are adding increased complexity to our understanding of how miRNAs function. In addition to binding target sites within the 3′-UTR of mRNA transcripts, miRNA can also bind within the 5′-UTR (Lytle et al., 2007), or coding regions (Forman et al., 2008). Further, a subset of miRNAs increase target expression through a number of mechanisms (Vasudevan et al., 2007), including binding DNA promoters (Place et al., 2008), acting as RNA decoys (Eiring et al., 2010) or through binding of target mRNAs (Ørom et al., 2008). MiRNAs can also be released from expressing cells and detected in exosomes in the circulation allowing communication between neighboring and distant cell populations (Mitchell et al., 2008; Liu et al., 2010a; Redis et al., 2012). As miRNAs are single-stranded RNA molecules, they may also activate immune cells following binding to Toll-like receptors (TLRs) either as part of their function, or as an unintended consequence following administration (Fabbri et al., 2012; Lehmann et al., 2012).
Key roles for miRNAs in normal lung development and respiratory diseases have been extensively reviewed (Plank et al., 2013; Rupani et al., 2013; Booton and Lindsay, 2014). These include extensive reviews of specific respiratory diseases including asthma (Tay et al., 2014), chronic obstructive pulmonary disease (COPD) (De Smet et al., 2015), cystic fibrosis (CF) (Sonneville et al., 2015), and idiopathic pulmonary fibrosis (IPF) (Pandit et al., 2011), as well as lung cancer (Lin et al., 2010; Rusek et al., 2015). For the purpose of this mini-review, we will briefly summarize recent findings on miRNA expression and function in each of these diseases (Table 1).
Many studies have performed miRNA profiling on samples from patients with asthma. Profiling of bronchial epithelial cells identified 60–200 differentially expressed miRNAs (Jardim et al., 2012; Solberg et al., 2012), and isolated exosomes from bronchial alveolar lavage fluids exhibited changes in 24 miRNAs (Levänen et al., 2013). Profiling of peripheral blood samples revealed alterations in miRNA expression patterns, with miR-192 notably decreased (Yamamoto et al., 2012), and alterations in miR-124, -26a, let-7a, and let-7d (Panganiban et al., 2012). Profiling of circulating lymphocytes also identified upregulation of miR-221 and miR-485-3p in asthmatic children compared to healthy controls (as well as in an ovalbumin (OVA)-induced mouse model of asthma; Liu et al., 2012). Profiling of airway T cells also revealed increased levels of miR-19a in patients with asthma, with functional roles in TH2 cytokine production (Simpson et al., 2014).
A growing number of studies have also demonstrated alterations and functional roles for miRNAs in mouse models of allergic airways disease. A profiling study using three murine models of allergic inflammation identified a number of altered miRNAs, including miR-29b, -29c, -146b, -223, -483, -574-5p, -672, and -690 (Garbacki et al., 2011). Deep sequencing of mouse lungs revealed dynamic changes in miRNA expression following OVA-induced allergic airway inflammation and demonstrated high levels of let-7 family members in OVA-challenged lungs (Polikepahad et al., 2010). Inhibition of let-7 family members reduced the allergic phenotype and IL-13 expression (Polikepahad et al., 2010). MiR-21 expression is increased in several models of experimental asthma (Lu et al., 2009) and mir-21 gene deletion reduced TH2 responses and decreased eosinophilia in an ovalbumin (OVA)-induced asthma model (Lu et al., 2011). Inhibition of miR-106a also reduced features of disease including AHR, inflammation and fibrosis (Sharma et al., 2009, 2012). Mir-155-deficient mice also have decreased asthma disease severity (Malmhäll et al., 2014), which was attributed to altered T cell function (Okoye et al., 2014). Inhibition of miR-221 in a mouse model of asthma also suppresses airway inflammation (Qin et al., 2012).
Our group demonstrated that antagomir-mediated inhibition of miR-126 significantly reduced airway hyperreactivity, eosinophil recruitment, mucus hypersecretion, and TH2 cell activation (Mattes et al., 2009). However, while inhibition of miR-126 reduced eosinophil infiltration in chronic asthma models, it failed to inhibit inflammation and airway remodeling (Collison et al., 2011a). Expression of miR-145, miR-21, and let-7b were increased after house dust-mite (HDM) exposure and inhibition of miR-145 (but not miR-21 or let-7b) suppressed airways inflammation (Collison et al., 2011b). Recently, we also demonstrated that inhibition of miR-9 restored steroid sensitivity and dampened airways inflammation in an otherwise steroid-resistant airways disease model (Li et al., 2015).
Respiratory bacterial and viral infections are also associated with asthma onset and disease exacerbations (reviewed in Falsey and Walsh, 2000; Hansbro et al., 2004; Friedlander and Busse, 2005; Piedimonte, 2013; Starkey et al., 2013; Choroszy-Król et al., 2014; Leigh and Proud, 2015). For example, Haemophilus influenzae bacterial infection is linked to both exacerbations and disease severity in patients with asthma (Wood et al., 2010). Importantly, key roles for miRNA functions are recognized in responses to respiratory infections [reviewed for virus (Globinska et al., 2014) and bacterial infections (Staedel and Darfeuille, 2013)]. For example, inhibition of miR-328 promoted clearance of H. influenzae, even in the context of steroid-induced immunosuppression (Tay et al., 2015). Thus, modulation of miRNA function may impact asthma severity by altering the course of respiratory infections and miRNAs may also serve as therapeutic targets for the management of infection-induced asthma exacerbation.
A growing number of studies have also assessed miRNA expression in COPD patients. As smoking is a major contributing factor for COPD development, many studies have also assessed impacts of cigarette smoke exposure alone. However, these studies have not yet assessed functional impacts in COPD.
Cigarette smoke reduced global miRNA expression, in human alveolar macrophages isolated from smokers compared to non-smokers (Graff et al., 2012; Gross et al., 2014) and lung tissue from smoke-exposed mice or rats (Izzotti et al., 2009a,b). In isolated bronchial airway epithelium from smokers, 28 miRNAs were differentially expressed, including miR-218 (Schembri et al., 2009). Profiling of miRNAs from different lung regions in smokers with COPD revealed subsets of miRNAs that correlated with regional emphysema severity (Christenson et al., 2013). In a comparison of lung tissue from smokers with COPD, to smokers without COPD, miRNA profiling revealed increased levels of 57 miRNAs, with 13 decreased (Ezzie et al., 2012). Further, altered miRNA expression in lung tissue was detectable between COPD patients based on emphysema severity (Savarimuthu Francis et al., 2014).
MiRNA expression has been proposed as an accessible biomarker of COPD disease. The ratio of serum miR-21 to -181a was predictive for COPD in asymptomatic heavy smokers (Xie et al., 2014). Decreased serum levels of miR-20a, -28-3p, -34c-5p, and -100, and increased miR-7 were also detected in COPD patients (Akbas et al., 2012). Similarly, altered miRNA expression in sputum (Van Pottelberge et al., 2011) and exhaled breath condensates (Pinkerton et al., 2013) have been detected in patients with COPD.
A number of individual miRNAs have also been assessed in samples isolated from COPD patients. MiR-146a induction following ex vivo cytokine activation is reduced in fibroblasts from COPD patients that smoke, compared to fibroblasts from smokers without COPD (Sato et al., 2010). MiR-199a-5p was decreased in peripheral blood monocytes from symptomatic COPD patients (Hassan et al., 2014), with decreases also observed in circulating regulatory T cells (Chatila et al., 2014). However, levels of miR-199-5p were increased in total lung tissues (Mizuno et al., 2012). In mouse models, miR-135b was increased in lungs from smoke-exposed mice, which required interleukin 1 (IL-1)-R1 expression and acted as a negative regulator of IL-1 activation and signaling (Halappanavar et al., 2013). In lungs from smoke-exposed mice and COPD patients, miR-144 and -101 were also both increased, and suppressed expression of their target CF transmembrane conductance regulator (CFTR) (Hassan et al., 2012a).
Cystic fibrosis is caused by mutations in the CFTR gene. Despite this simple cause, significant heterogeneity exists between CF patients, suggesting potential roles for epigenetic regulation, including miRNA alterations (Cutting, 2010).
Most work on miRNA function in CF has assessed direct impacts on CFTR expression. Increased miR-101 and -494 repressed CFTR expression in cell lines in vitro (Megiorni et al., 2011). Further, miR-138 overexpression increased levels of CFTR (via suppression of its target SIN3 transcription regulator family member A, in CF airway epithelial cells (Ramachandran et al., 2012). MiR-145, -223, and -494 were increased in CF bronchial brushings, and directly regulated CFTR expression in vitro (Oglesby et al., 2013). MiR-509-3p and -494 were also increased in primary cultured airway epithelia from CF patients, vs. non-CF controls, and cooperatively repressed CFTR expression (Ramachandran et al., 2013).
Several studies have also assessed miRNA-mediated regulation of inflammation in CF patients. MiR-17 was decreased in CF bronchial brushings, resulting in increased expression of its pro-inflammatory target, IL-8 (Oglesby et al., 2015). MiR-126 was downregulated in bronchial epithelial cells from CF patients, with significant increases in target of myb protein 1, which modulates inflammatory responses (Oglesby et al., 2010). MiR-155 was increased in mutant CFTR epithelial cell lines, contributing to increased pro-inflammatory IL-8 release (Bhattacharyya et al., 2011). Finally, elevated miR-145 levels negatively correlated with its target SMAD family member 3 in nasal epithelial cells from CF patients, potentially regulating downstream transforming growth factor, beta 1 (TGF-β1) inflammatory pathways (Megiorni et al., 2013).
In addition to direct effects on CFTR and inflammation, bacterial infections worsen CF disease symptoms and contribute to long-term airway remodeling. Thus, impacts of miRNA function on anti-bacterial immunity may also regulate disease progression and shape disease management strategies.
IPF is a chronic fibrosing interstitial lung disease with unknown causes. Initial profiling of IPF lungs revealed that 10% of lung-expressed miRNAs were altered, compared to normal controls (Pandit et al., 2010). Profiling of circulating serum miRNAs in patients with IPF revealed altered expression of miR-21, -155, and -101-3p associated with clinical features of disease (Li et al., 2014). MiR-21 expression was increased in the serum of IPF patients, and levels correlated with decreases in lung function (Li et al., 2013). Let-7 was also significantly downregulated in IPF lungs and may contribute to fibrosis through regulation of high mobility group A2 (Pandit et al., 2010).
Little is known about miRNA function in IPF and importantly, much of the functional data has been inferred from chemically-induced mouse models of lung fibrosis. MiR-29 was reduced in mouse models, as seen in IPF lung samples (Pandit et al., 2010), and knockdown in human fetal lung fibroblasts increased fibrosis-associated gene expression (Cushing et al., 2011). MiR-21 expression was increased in myofibroblasts from IPF lungs and inhibition in mouse fibrosis models dampened disease severity (Liu et al., 2010b). MiR-155 was increased in mouse fibrosis models (Pottier et al., 2009), and may modulate fibrosis through regulation of angiotensin II type I receptor, which is increased in IPF lungs (Königshoff et al., 2007). MiR-200 family members were reduced in mouse models of lung fibrosis and restoration of miR-200c inhibited fibrosis (Yang et al., 2012). MiR-26a was decreased in fibrosis models and inhibition caused pulmonary fibrosis, while overexpression repressed fibrotic disease (Liang et al., 2014). MiR-326 was also decreased in mouse fibrosis models and human IPF lung samples, and administration of miR-326 mimics inhibited TGF-β expression and dampened fibrosis (Das et al., 2014). MiR-486-5p was decreased in lung tissues of patients with IPF and overexpression of miR-486-5p limited fibrosis in mouse models (Ji et al., 2015).
The impacts of miRNA dysregulation on cancer initiation and progression have been extensively studied and reviewed (Hayes et al., 2014; Lin and Gregory, 2015). A summary of the proposed roles for miRNAs in cancer is beyond the scope of this mini-review and we direct readers to reviews that have summarized miRNA functions in lung cancers specifically (Lin et al., 2010; Rusek et al., 2015).
The studies described have revealed changes in miRNA expression in respiratory diseases and key functions across a range of disease processes. Interestingly, miRNA-targeted interventions modified disease pathogenesis in pre-clinical disease models, for a small subset of the miRNAs identified in each disease.
Based on similar findings in other fields, many biopharmaceutical companies are now targeting miRNAs using novel therapeutics. The first drug to inhibit a specific miRNA (miR-122) entered Phase II clinical trials in 2010 for the treatment of hepatitis C virus infection (Janssen et al., 2013). The first drug to increase a specific miRNA (miR-34 mimic; MRX34) entered Phase I clinical trials in 2013, for the treatment of advanced hepatocellular carcinoma (Bouchie, 2013). However, no clinical trials have yet assessed the impacts of miRNA-targeted strategies in respiratory diseases.
One approach to alter miRNA function in vivo is through administration of double-stranded synthetic miRNA oligonucleotides, termed mimics, which have been used extensively in cancer studies (Henry et al., 2011). These double-stranded molecules are processed by the endogenous miRNA processing machinery, integrated into the RISC complex and dampen target mRNA translation. A main limitation of this approach is the effective delivery of mimic molecules into target cells.
One strategy to improve cell targeting is the coupling of mimics to antibody-coated nanoparticles, as used in neuroblastoma (Liu et al., 2011). Another approach (used in the current MRX34 clinical trial), is encapsulation of mimic molecules in liposome-based delivery system to facilitate target cell uptake (Bouchie, 2013).
An alternative approach that stably increases miRNA levels is delivery of miRNA expression vectors (Stegmeier et al., 2005). This approach can yield stable, long-term miRNA expression and provides the potential to target vectors to specific cell types or enforce cell-type specific miRNA expression using specific promoters.
To inhibit target miRNAs, oligonucleotide inhibitors with complementary sequences to the miRNA of interest can be used (anti-miRs). These molecules are often comprised of locked nucleic acid backbones with specific modifications to increase stability in vivo (Lennox and Behlke, 2011). One specific category of inhibitor, termed antagomirs, is further modified with a 2′-O-methyl linkage and phosphorothioate modification to improve binding efficiency and prevent nuclease degradation (Krützfeldt et al., 2005). Anti-miR treatments interfere with target miRNA function, increasing translation of miRNA-targeted mRNAs. Antagomir-mediated miRNA-silencing in vivo is dose-dependent and can last for several weeks after a single administration (Krützfeldt et al., 2005).
Another strategy to inhibit miRNA function is the use of “miRNA sponges.” MiRNA sponges are transgenes that encode RNA molecules comprised of multiple tandem miRNA target sites (Ebert et al., 2007). The repeated target sequences compete with endogenous mRNA targets for RISC binding, reducing the impact of a specific miRNA on its intended target (Ebert et al., 2007). This approach has been used extensively in vitro and in preclinical animal models. However, this approach also requires delivery of a transgene-encoding vector to achieve stable inhibition (Gentner et al., 2009).
There are a number of considerations and issues to consider when modulating miRNA expression in general, and more specifically in lung disease (reviewed in Hassan et al., 2012b).
Individual miRNAs modulate expression of multiple mRNA targets and interfering with single miRNAs can have broad effects on multiple cellular pathways. For this reason, therapies targeting individual miRNAs can have broader impacts than traditional single-molecule/single-target approaches. Further, compared to short-interfering RNA approaches, miRNA targeting may have broader effects on more mRNAs, but more moderate effects on each individual mRNA (Baker, 2010). This can be beneficial, as individual miRNAs often regulate multiple genes in a related gene network. However, the alteration of multiple downstream targets also increases the likelihood of undesired side effects, particularly if systemic drug delivery is used.
The simplest approaches to modulate miRNA levels are through administration of “naked” nucleic acids (either mimic or anti-miR). This approach provides the advantage of being able to tailor dosing and withdraw treatment if complications arise. In respiratory diseases, direct tissue administration by aerosol provides targeted delivery to lung tissue, with minimal dissemination into the circulation and systemic tissues. This is likely to limit side effects, compared to systemic delivery. However, this also means that these therapies will only be effective if altering miRNA function specifically within the lung is sufficient to dampen disease processes. Even with direct administration into the lung, efficient delivery to target cell populations remains difficult, as seen in siRNA-based therapies (Lam et al., 2012). For example, we observe cell type-specific differences in antagomir distribution following intranasal administration in vivo, with efficient uptake in myeloid cell populations and poor targeting of lymphocytes, which was also replicated in vitro (Plank et al., 2015).
More complex treatment strategies requiring stable expression and viral delivery systems (e.g., sustained miRNA delivery or miRNA sponges) add additional complications relating to the expression vector itself. Viral delivery platforms have been associated with direct toxicity, increased risks associated with genomic integration events and the inability to discontinue treatment if problems do arise (Ibrahim et al., 2011). A further drawback, specific to miRNA modulation using viral delivery systems, is toxicity induced by the overwhelming of the exportin-5 pathway following enforced miRNA expression (Castanotto et al., 2007).
Increasing numbers of publications have identified differences in miRNA expression and functional roles for miRNAs across a range of respiratory diseases. MiRNAs have been proposed as biomarkers of disease and potential novel therapeutic targets for treatment. In respiratory diseases, direct administration of miRNA-targeting drugs into the lungs may allow tissue-specific targeting, while limiting side effects resulting from systemic delivery. Continued mechanistic studies are required to optimize drug delivery systems and demonstrate acceptable efficacy and safety for translation into the clinic. While miRNA-targeting approaches hold promise, no studies have yet assessed impacts in human respiratory diseases.
SM, MP, HT, AC, and PF wrote and edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by project grants from the National Health and Medical Research Council (NHMRC) of Australia and the Hunter Medical Research Institute (HMRI). SM is supported by fellowships from the Canadian Institutes of Health Research (CIHR), the University of Newcastle and the HMRI.
Akbas, F., Coskunpinar, E., Aynaci, E., Oltulu, Y. M., and Yildiz, P. (2012). Analysis of serum micro-RNAs as potential biomarker in chronic obstructive pulmonary disease. Exp. Lung Res. 38, 286–294. doi: 10.3109/01902148.2012.689088
Baker, M. (2010). RNA interference: homing in on delivery. Nature 464, 1225–1228. doi: 10.1038/4641225a
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Berezikov, E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. doi: 10.1038/nrg3079
Bhattacharyya, S., Balakathiresan, N. S., Dalgard, C., Gutti, U., Armistead, D., Jozwik, C., et al. (2011). Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J. Biol. Chem. 286, 11604–11615. doi: 10.1074/jbc.M110.198390
Booton, R., and Lindsay, M. A. (2014). Emerging role of MicroRNAs and long noncoding RNAs in respiratory disease. Chest 146, 193–204. doi: 10.1378/chest.13-2736
Bouchie, A. (2013). First microRNA mimic enters clinic. Nat. Biotechnol. 31:577. doi: 10.1038/nbt0713-577
Castanotto, D., Sakurai, K., Lingeman, R., Li, H., Shively, L., Aagaard, L., et al. (2007). Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 35, 5154–5164. doi: 10.1093/nar/gkm543
Chatila, W. M., Criner, G. J., Hancock, W. W., Akimova, T., Moldover, B., Chang, J. K., et al. (2014). Blunted expression of miR-199a-5p in regulatory T cells of patients with chronic obstructive pulmonary disease compared to unaffected smokers. Clin. Exp. Immunol. 177, 341–352. doi: 10.1111/cei.12325
Choroszy-Król, I., Frej-Madrzak, M., Hober, M., Sarowska, J., and Jama-Kmiecik, A. (2014). Infections caused by Chlamydophila pneumoniae. Adv. Clin. Exp. Med. 23, 123–126. doi: 10.17219/acem/37035
Christenson, S. A., Brandsma, C. A., Campbell, J. D., Knight, D. A., Pechkovsky, D. V., Hogg, J. C., et al. (2013). miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome Med. 5, 114. doi: 10.1186/gm519
Collison, A., Herbert, C., Siegle, J. S., Mattes, J., Foster, P. S., and Kumar, R. K. (2011a). Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm. Med. 11:29. doi: 10.1186/1471-2466-11-29
Collison, A., Mattes, J., Plank, M., and Foster, P. S. (2011b). Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J. Allergy Clin. Immunol. 128, 160–167.e4. doi: 10.1016/j.jaci.2011.04.005
Cushing, L., Kuang, P. P., Qian, J., Shao, F., Wu, J., Little, F., et al. (2011). miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 45, 287–294. doi: 10.1165/rcmb.2010-0323OC
Cutting, G. R. (2010). Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann. N. Y. Acad. Sci. 1214, 57–69. doi: 10.1111/j.1749-6632.2010.05879.x
Das, S., Kumar, M., Negi, V., Pattnaik, B., Prakash, Y. S., Agrawal, A., et al. (2014). MicroRNA-326 regulates profibrotic functions of transforming growth factor-beta in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 50, 882–892. doi: 10.1165/rcmb.2013-0195OC
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F., and Hannon, G. J. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235. doi: 10.1038/nature03049
De Smet, E. G., Mestdagh, P., Vandesompele, J., Brusselle, G. G., and Bracke, K. R. (2015). Non-coding RNAs in the pathogenesis of COPD. Thorax 70, 782–791. doi: 10.1136/thoraxjnl-2014-206560
Ebert, M. S., Neilson, J. R., and Sharp, P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726. doi: 10.1038/nmeth1079
Eiring, A. M., Harb, J. G., Neviani, P., Garton, C., Oaks, J. J., Spizzo, R., et al. (2010). miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140, 652–665. doi: 10.1016/j.cell.2010.01.007
Ezzie, M. E., Crawford, M., Cho, J. H., Orellana, R., Zhang, S., Gelinas, R., et al. (2012). Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 67, 122–131. doi: 10.1136/thoraxjnl-2011-200089
Fabbri, M., Paone, A., Calore, F., Galli, R., Gaudio, E., Santhanam, R., et al. (2012). MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–E2116. doi: 10.1073/pnas.1209414109
Falsey, A. R., and Walsh, E. E. (2000). Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13, 371–384. doi: 10.1128/CMR.13.3.371-384.2000
Forman, J. J., Legesse-Miller, A., and Coller, H. A. (2008). A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. U.S.A. 105, 14879–14884. doi: 10.1073/pnas.0803230105
Friedlander, S. L., and Busse, W. W. (2005). The role of rhinovirus in asthma exacerbations. J. Allergy Clin. Immunol. 116, 267–273. doi: 10.1016/j.jaci.2005.06.003
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Garbacki, N., Di Valentin, E., Huynh-Thu, V. A., Geurts, P., Irrthum, A., Crahay, C., et al. (2011). MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS ONE 6:e16509. doi: 10.1371/journal.pone.0016509
Gentner, B., Schira, G., Giustacchini, A., Amendola, M., Brown, B. D., Ponzoni, M., et al. (2009). Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods 6, 63–66. doi: 10.1038/nmeth.1277
Globinska, A., Pawelczyk, M., and Kowalski, M. L. (2014). MicroRNAs and the immune response to respiratory virus infections. Expert Rev. Clin. Immunol. 10, 963–971. doi: 10.1586/1744666X.2014.913482
Graff, J. W., Powers, L. S., Dickson, A. M., Kim, J., Reisetter, A. C., Hassan, I. H., et al. (2012). Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE 7:e44066. doi: 10.1371/journal.pone.0044066
Gross, T. J., Powers, L. S., Boudreau, R. L., Brink, B., Reisetter, A., Goel, K., et al. (2014). A microRNA processing defect in smokers' macrophages is linked to SUMOylation of the endonuclease DICER. J. Biol. Chem. 289, 12823–12834. doi: 10.1074/jbc.M114.565473
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524. doi: 10.1038/nrm3838
Halappanavar, S., Nikota, J., Wu, D., Williams, A., Yauk, C. L., and Stampfli, M. (2013). IL-1 receptor regulates microRNA-135b expression in a negative feedback mechanism during cigarette smoke-induced inflammation. J. Immunol. 190, 3679–3686. doi: 10.4049/jimmunol.1202456
Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R., and Hannon, G. J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150. doi: 10.1126/science.1064023
Hansbro, P. M., Beagley, K. W., Horvat, J. C., and Gibson, P. G. (2004). Role of atypical bacterial infection of the lung in predisposition/protection of asthma. Pharmacol. Ther. 101, 193–210. doi: 10.1016/j.pharmthera.2003.10.007
Hassan, F., Nuovo, G. J., Crawford, M., Boyaka, P. N., Kirkby, S., Nana-Sinkam, S. P., et al. (2012a). MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE 7:e50837. doi: 10.1371/journal.pone.0050837
Hassan, T., Carroll, T. P., Buckley, P. G., Cummins, R., O'Neill, S. J., McElvaney, N. G., et al. (2014). miR-199a-5p silencing regulates the unfolded protein response in chronic obstructive pulmonary disease and alpha1-antitrypsin deficiency. Am. J. Respir. Crit. Care Med. 189, 263–273. doi: 10.1164/rccm.201306-1151OC
Hassan, T., McKiernan, P. J., McElvaney, N. G., Cryan, S. A., and Greene, C. M. (2012b). Therapeutic modulation of miRNA for the treatment of proinflammatory lung diseases. Expert Rev. Anti Infect. Ther. 10, 359–368. doi: 10.1586/eri.11.175
Hayes, J., Peruzzi, P. P., and Lawler, S. (2014). MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20, 460–469. doi: 10.1016/j.molmed.2014.06.005
Henry, J. C., Azevedo-Pouly, A. C., and Schmittgen, T. D. (2011). MicroRNA replacement therapy for cancer. Pharm. Res. 28, 3030–3042. doi: 10.1007/s11095-011-0548-9
Hutvágner, G., McLachlan, J., Pasquinelli, A. E., Bálint, E., Tuschl, T., and Zamore, P. D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. doi: 10.1126/science.1062961
Ibrahim, A. F., Weirauch, U., Thomas, M., Grünweller, A., Hartmann, R. K., and Aigner, A. (2011). MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 71, 5214–5224. doi: 10.1158/0008-5472.CAN-10-4645
Izzotti, A., Calin, G. A., Arrigo, P., Steele, V. E., Croce, C. M., and De Flora, S. (2009b). Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 23, 806–812. doi: 10.1096/fj.08-121384
Izzotti, A., Calin, G. A., Steele, V. E., Croce, C. M., and De Flora, S. (2009a). Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 23, 3243–3250. doi: 10.1096/fj.09-135251
Janssen, H. L., Reesink, H. W., Lawitz, E. J., Zeuzem, S., Rodriguez-Torres, M., Patel, K., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694. doi: 10.1056/NEJMoa1209026
Jardim, M. J., Dailey, L., Silbajoris, R., and Diaz-Sanchez, D. (2012). Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am. J. Respir. Cell Mol. Biol. 47, 536–542. doi: 10.1165/rcmb.2011-0160OC
Ji, X., Wu, B., Fan, J., Han, R., Luo, C., Wang, T., et al. (2015). The Anti-fibrotic effects and mechanisms of MicroRNA-486-5p in pulmonary fibrosis. Sci. Rep. 5:14131. doi: 10.1038/srep14131
Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J., and Plasterk, R. H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. doi: 10.1101/gad.927801
Königshoff, M., Wilhelm, A., Jahn, A., Sedding, D., Amarie, O. V., Eul, B., et al. (2007). The angiotensin II receptor 2 is expressed and mediates angiotensin II signaling in lung fibrosis. Am. J. Respir. Cell Mol. Biol. 37, 640–650. doi: 10.1165/rcmb.2006-0379TR
Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689. doi: 10.1038/nature04303
Lam, J. K., Liang, W., and Chan, H. K. (2012). Pulmonary delivery of therapeutic siRNA. Adv. Drug Deliv. Rev. 64, 1–15. doi: 10.1016/j.addr.2011.02.006
Lee, Y., Jeon, K., Lee, J. T., Kim, S., and Kim, V. N. (2002). MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21, 4663–4670. doi: 10.1093/emboj/cdf476
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Lehmann, S. M., Krüger, C., Park, B., Derkow, K., Rosenberger, K., Baumgart, J., et al. (2012). An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835. doi: 10.1038/nn.3113
Leigh, R., and Proud, D. (2015). Virus-induced modulation of lower airway diseases: pathogenesis and pharmacologic approaches to treatment. Pharmacol. Ther. 148, 185–198. doi: 10.1016/j.pharmthera.2014.12.005
Lennox, K. A., and Behlke, M. A. (2011). Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 18, 1111–1120. doi: 10.1038/gt.2011.100
Levänen, B., Bhakta, N. R., Torregrosa Paredes, P., Barbeau, R., Hiltbrunner, S., Pollack, J. L., et al. (2013). Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 131, 894–903. doi: 10.1016/j.jaci.2012.11.039
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. doi: 10.1016/j.cell.2004.12.035
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798. doi: 10.1016/S0092-8674(03)01018-3
Li, J. J., Tay, H. L., Maltby, S., Xiang, Y., Eyers, F., Hatchwell, L., et al. (2015). MicroRNA-9 regulates steroid-resistant airway hyperresponsiveness by reducing protein phosphatase 2A activity. J. Allergy Clin. Immunol. 136, 462–473. doi: 10.1016/j.jaci.2014.11.044
Li, P., Li, J., Chen, T., Wang, H., Chu, H., Chang, J., et al. (2014). Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int. J. Mol. Med. 33, 1554–1562. doi: 10.3892/ijmm.2014.1712
Li, P., Zhao, G. Q., Chen, T. F., Chang, J. X., Wang, H. Q., Chen, S. S., et al. (2013). Serum miR-21 and miR-155 expression in idiopathic pulmonary fibrosis. J. Asthma 50, 960–964. doi: 10.3109/02770903.2013.822080
Liang, H., Xu, C., Pan, Z., Zhang, Y., Xu, Z., Chen, Y., et al. (2014). The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol. Ther. 22, 1122–1133. doi: 10.1038/mt.2014.42
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. doi: 10.1038/nature03315
Lin, P. Y., Yu, S. L., and Yang, P. C. (2010). MicroRNA in lung cancer. Br. J. Cancer 103, 1144–1148. doi: 10.1038/sj.bjc.6605901
Lin, S., and Gregory, R. I. (2015). MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333. doi: 10.1038/nrc3932
Liu, F., Qin, H. B., Xu, B., Zhou, H., and Zhao, D. Y. (2012). Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Mol. Med. Report 6, 1178–1182. doi: 10.3892/mmr.2012.1030
Liu, G., Friggeri, A., Yang, Y., Milosevic, J., Ding, Q., Thannickal, V. J., et al. (2010b). miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 207, 1589–1597. doi: 10.1084/jem.20100035
Liu, J. (2008). Control of protein synthesis and mRNA degradation by microRNAs. Curr. Opin. Cell Biol. 20, 214–221. doi: 10.1016/j.ceb.2008.01.006
Liu, X. Q., Song, W. J., Sun, T. M., Zhang, P. Z., and Wang, J. (2011). Targeted delivery of antisense inhibitor of miRNA for antiangiogenesis therapy using cRGD-functionalized nanoparticles. Mol. Pharm. 8, 250–259. doi: 10.1021/mp100315q
Liu, Y., Xiang, X., Zhuang, X., Zhang, S., Liu, C., Cheng, Z., et al. (2010a). Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am. J. Pathol. 176, 2490–2499. doi: 10.2353/ajpath.2010.090777
Lu, T. X., Hartner, J., Lim, E. J., Fabry, V., Mingler, M. K., Cole, E. T., et al. (2011). MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 187, 3362–3373. doi: 10.4049/jimmunol.1101235
Lu, T. X., Munitz, A., and Rothenberg, M. E. (2009). MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 182, 4994–5002. doi: 10.4049/jimmunol.0803560
Lytle, J. R., Yario, T. A., and Steitz, J. A. (2007). Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. U.S.A. 104, 9667–9672. doi: 10.1073/pnas.0703820104
Mallory, A. C., Reinhart, B. J., Jones-Rhoades, M. W., Tang, G., Zamore, P. D., Barton, M. K., et al. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. doi: 10.1038/sj.emboj.7600340
Malmhäll, C., Alawieh, S., Lu, Y., Sjostrand, M., Bossios, A., Eldh, M., et al. (2014). MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J. Allergy Clin. Immunol. 133, 1429–1438, 1438.e1-7. doi: 10.1016/j.jaci.2013.11.008
Mattes, J., Collison, A., Plank, M., Phipps, S., and Foster, P. S. (2009). Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. U.S.A. 106, 18704–18709. doi: 10.1073/pnas.0905063106
Megiorni, F., Cialfi, S., Cimino, G., De Biase, R. V., Dominici, C., Quattrucci, S., et al. (2013). Elevated levels of miR-145 correlate with SMAD3 down-regulation in cystic fibrosis patients. J. Cyst. Fibros. 12, 797–802. doi: 10.1016/j.jcf.2013.03.007
Megiorni, F., Cialfi, S., Dominici, C., Quattrucci, S., and Pizzuti, A. (2011). Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS ONE 6:e26601. doi: 10.1371/journal.pone.0026601
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. doi: 10.1073/pnas.0804549105
Mizuno, S., Bogaard, H. J., Gomez-Arroyo, J., Alhussaini, A., Kraskauskas, D., Cool, C. D., et al. (2012). MicroRNA-199a-5p is associated with hypoxia-inducible factor-1alpha expression in lungs from patients with COPD. Chest 142, 663–672. doi: 10.1378/chest.11-2746
Oglesby, I. K., Bray, I. M., Chotirmall, S. H., Stallings, R. L., O'Neill, S. J., McElvaney, N. G., et al. (2010). miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J. Immunol. 184, 1702–1709. doi: 10.4049/jimmunol.0902669
Oglesby, I. K., Chotirmall, S. H., McElvaney, N. G., and Greene, C. M. (2013). Regulation of cystic fibrosis transmembrane conductance regulator by microRNA-145, -223, and -494 is altered in DeltaF508 cystic fibrosis airway epithelium. J. Immunol. 190, 3354–3362. doi: 10.4049/jimmunol.1202960
Oglesby, I. K., Vencken, S. F., Agrawal, R., Gaughan, K., Molloy, K., Higgins, G., et al. (2015). miR-17 overexpression in cystic fibrosis airway epithelial cells decreases interleukin-8 production. Eur. Respir. J. 46, 1350–1360. doi: 10.1183/09031936.00163414
Okoye, I. S., Czieso, S., Ktistaki, E., Roderick, K., Coomes, S. M., Pelly, V. S., et al. (2014). Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc. Natl. Acad. Sci. U.S.A. 111, E3081–E3090. doi: 10.1073/pnas.1406322111
Ørom, U. A., Nielsen, F. C., and Lund, A. H. (2008). MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471. doi: 10.1016/j.molcel.2008.05.001
Pandit, K. V., Corcoran, D., Yousef, H., Yarlagadda, M., Tzouvelekis, A., Gibson, K. F., et al. (2010). Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 182, 220–229. doi: 10.1164/rccm.200911-1698OC
Pandit, K. V., Milosevic, J., and Kaminski, N. (2011). MicroRNAs in idiopathic pulmonary fibrosis. Transl. Res. 157, 191–199. doi: 10.1016/j.trsl.2011.01.012
Panganiban, R. P., Pinkerton, M. H., Maru, S. Y., Jefferson, S. J., Roff, A. N., and Ishmael, F. T. (2012). Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am. J. Clin. Exp. Immunol. 1, 154–165.
Piedimonte, G. (2013). Respiratory syncytial virus and asthma: speed-dating or long-term relationship? Curr. Opin. Pediatr. 25, 344–349. doi: 10.1097/MOP.0b013e328360bd2e
Pinkerton, M., Chinchilli, V., Banta, E., Craig, T., August, A., Bascom, R., et al. (2013). Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J. Allergy Clin. Immunol. 132, 217–219. doi: 10.1016/j.jaci.2013.03.006
Place, R. F., Li, L. C., Pookot, D., Noonan, E. J., and Dahiya, R. (2008). MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U.S.A. 105, 1608–1613. doi: 10.1073/pnas.0707594105
Plank, M., Maltby, S., Mattes, J., and Foster, P. S. (2013). Targeting translational control as a novel way to treat inflammatory disease: the emerging role of MicroRNAs. Clin. Exp. Allergy 43, 981–999. doi: 10.1111/cea.12135
Plank, M. W., Maltby, S., Tay, H. L., Stewart, J., Eyers, F., Hansbro, P. M., et al. (2015). MicroRNA expression is altered in an ovalbumin-induced asthma model and Targeting miR-155 with antagomirs reveals cellular specificity. PLoS ONE 10:e0144810. doi: 10.1371/journal.pone.0144810
Polikepahad, S., Knight, J. M., Naghavi, A. O., Oplt, T., Creighton, C. J., Shaw, C., et al. (2010). Proinflammatory role for let-7 microRNAS in experimental asthma. J. Biol. Chem. 285, 30139–30149. doi: 10.1074/jbc.M110.145698
Pottier, N., Maurin, T., Chevalier, B., Puissegur, M. P., Lebrigand, K., Robbe-Sermesant, K., et al. (2009). Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS ONE 4:e6718. doi: 10.1371/journal.pone.0006718
Qin, H. B., Xu, B., Mei, J. J., Li, D., Liu, J. J., Zhao, D. Y., et al. (2012). Inhibition of miRNA-221 suppresses the airway inflammation in asthma. Inflammation 35, 1595–1599. doi: 10.1007/s10753-012-9474-1
Ramachandran, S., Karp, P. H., Jiang, P., Ostedgaard, L. S., Walz, A. E., Fisher, J. T., et al. (2012). A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U.S.A. 109, 13362–13367. doi: 10.1073/pnas.1210906109
Ramachandran, S., Karp, P. H., Osterhaus, S. R., Jiang, P., Wohlford-Lenane, C., Lennox, K. A., et al. (2013). Post-transcriptional regulation of cystic fibrosis transmembrane conductance regulator expression and function by microRNAs. Am. J. Respir. Cell Mol. Biol. 49, 544–551. doi: 10.1165/rcmb.2012-0430OC
Redis, R. S., Calin, S., Yang, Y., You, M. J., and Calin, G. A. (2012). Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol. Ther. 136, 169–174. doi: 10.1016/j.pharmthera.2012.08.003
Rupani, H., Sanchez-Elsner, T., and Howarth, P. (2013). MicroRNAs and respiratory diseases. Eur. Respir. J. 41, 695–705. doi: 10.1183/09031936.00212011
Rusek, A. M., Abba, M., Eljaszewicz, A., Moniuszko, M., Niklinski, J., and Allgayer, H. (2015). MicroRNA modulators of epigenetic regulation, the tumor microenvironment and the immune system in lung cancer. Mol. Cancer 14, 34. doi: 10.1186/s12943-015-0302-8
Sato, T., Liu, X., Nelson, A., Nakanishi, M., Kanaji, N., Wang, X., et al. (2010). Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am. J. Respir. Crit. Care Med. 182, 1020–1029. doi: 10.1164/rccm.201001-0055OC
Savarimuthu Francis, S. M., Davidson, M. R., Tan, M. E., Wright, C. M., Clarke, B. E., Duhig, E. E., et al. (2014). MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genomics 15:88. doi: 10.1186/1471-2164-15-88
Schembri, F., Sridhar, S., Perdomo, C., Gustafson, A. M., Zhang, X., Ergun, A., et al. (2009). MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 106, 2319–2324. doi: 10.1073/pnas.0806383106
Sharma, A., Kumar, M., Ahmad, T., Mabalirajan, U., Aich, J., Agrawal, A., et al. (2012). Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. J. Appl. Physiol. 113, 459–464. doi: 10.1152/japplphysiol.00001.2012
Sharma, A., Kumar, M., Aich, J., Hariharan, M., Brahmachari, S. K., Agrawal, A., et al. (2009). Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc. Natl. Acad. Sci. U.S.A. 106, 5761–5766. doi: 10.1073/pnas.0808743106
Simpson, L. J., Patel, S., Bhakta, N. R., Choy, D. F., Brightbill, H. D., Ren, X., et al. (2014). A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol. 15, 1162–1170. doi: 10.1038/ni.3026
Solberg, O. D., Ostrin, E. J., Love, M. I., Peng, J. C., Bhakta, N. R., Hou, L., et al. (2012). Airway epithelial miRNA expression is altered in asthma. Am. J. Respir. Crit. Care Med. 186, 965–974. doi: 10.1164/rccm.201201-0027OC
Sonneville, F., Ruffin, M., Guillot, L., Rousselet, N., Le Rouzic, P., Corvol, H., et al. (2015). New insights about miRNAs in cystic fibrosis. Am. J. Pathol. 185, 897–908. doi: 10.1016/j.ajpath.2014.12.022
Staedel, C., and Darfeuille, F. (2013). MicroRNAs and bacterial infection. Cell. Microbiol. 15, 1496–1507. doi: 10.1111/cmi.12159
Starkey, M. R., Nguyen, D. H., Kim, R. Y., Nair, P. M., Brown, A. C., Essifie, A. T., et al. (2013). Programming of the lung in early life by bacterial infections predisposes to chronic respiratory disease. Clin. Obstet. Gynecol. 56, 566–576. doi: 10.1097/GRF.0b013e3182993a0c
Stegmeier, F., Hu, G., Rickles, R. J., Hannon, G. J., and Elledge, S. J. (2005). A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 102, 13212–13217. doi: 10.1073/pnas.0506306102
Tay, H. L., Kaiko, G. E., Plank, M., Li, J., Maltby, S., Essilfie, A. T., et al. (2015). Antagonism of miR-328 increases the antimicrobial function of macrophages and neutrophils and rapid clearance of non-typeable Haemophilus influenzae (NTHi) from infected lung. PLoS Pathog. 11:e1004549. doi: 10.1371/journal.ppat.1004549
Tay, H. L., Plank, M., Collison, A., Mattes, J., Kumar, R. K., and Foster, P. S. (2014). MicroRNA: potential biomarkers and therapeutic targets for allergic asthma? Ann. Med. 46, 633–639. doi: 10.3109/07853890.2014.958196
Van Pottelberge, G. R., Mestdagh, P., Bracke, K. R., Thas, O., van Durme, Y. M., Joos, G. F., et al. (2011). MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 183, 898–906. doi: 10.1164/rccm.201002-0304OC
Vasudevan, S., Tong, Y., and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934. doi: 10.1126/science.1149460
Weinmann, L., Höck, J., Ivacevic, T., Ohrt, T., Mütze, J., Schwille, P., et al. (2009). Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136, 496–507. doi: 10.1016/j.cell.2008.12.023
Wood, L. G., Simpson, J. L., Hansbro, P. M., and Gibson, P. G. (2010). Potentially pathogenic bacteria cultured from the sputum of stable asthmatics are associated with increased 8-isoprostane and airway neutrophilia. Free Radic. Res. 44, 146–154. doi: 10.3109/10715760903362576
Xie, L., Wu, M., Lin, H., Liu, C., Yang, H., Zhan, J., et al. (2014). An increased ratio of serum miR-21 to miR-181a levels is associated with the early pathogenic process of chronic obstructive pulmonary disease in asymptomatic heavy smokers. Mol. Biosyst. 10, 1072–1081. doi: 10.1039/c3mb70564a
Yamamoto, M., Singh, A., Ruan, J., Gauvreau, G. M., O'Byrne, P. M., Carlsten, C. R., et al. (2012). Decreased miR-192 expression in peripheral blood of asthmatic individuals undergoing an allergen inhalation challenge. BMC Genomics 13, 655. doi: 10.1186/1471-2164-13-655
Keywords: microRNA, respiratory diseases, non-coding RNA, antagomir, mimic
Citation: Maltby S, Plank M, Tay HL, Collison A and Foster PS (2016) Targeting MicroRNA Function in Respiratory Diseases: Mini-Review. Front. Physiol. 7:21. doi: 10.3389/fphys.2016.00021
Received: 26 November 2015; Accepted: 15 January 2016;
Published: 04 February 2016.
Edited by:
Yu Ru Kou, National Yang-Ming University, TaiwanReviewed by:
Kunihiro Sakuma, Toyohashi University of Technology, JapanCopyright © 2016 Maltby, Plank, Tay, Collison and Foster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven Maltby, c3RldmVuLm1hbHRieUBuZXdjYXN0bGUuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.