
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 30 March 2012
Sec. Vascular Physiology
Volume 3 - 2012 | https://doi.org/10.3389/fphys.2012.00068
SIRT1 is an NAD-dependent deacetylase that regulates stress response pathways. By deacetylating transcription factors and co-factors, SIRT1 modulates metabolism, inflammation, hypoxic responses, circadian rhythms, cell survival, and longevity. Since SIRT1 plays a key role in regulating pathways involved in cardiovascular diseases and metabolic diseases cancer, the regulation of SIRT1 has received intense scrutiny. The post-transcriptional regulation of SIRT1 is mediated by two classes of molecules, RNA-binding proteins (RBPs) and non-coding small RNAs. MicroRNAs (miRNAs) are short non-coding RNAs that regulate target gene expression in a post-transcriptional manner. More than 16 miRNAs modulate SIRT1 expression, including miR-34a. miR-34a induces colon cancer apoptosis through SIRT1, and miR-34a also promotes senescence in endothelial cells via SIRT1. This review describes the impact of miRNAs on SIRT1. The background of SIRT1 and miRNAs will be summarized, followed by the mechanism by which several key miRNAs alter SIRT1 levels, and how the RBP HuR regulates SIRT1. MicroRNA regulation of SIRT1 might affect a wide variety of pathways in humans, from metabolic diseases such as diabetes to cardiovascular diseases and cancer.
Sir2 was originally identified as the silencing information regulator 2 in S. cerevisiae (Kennedy et al., 1995). In yeast, Sir2 is recognized as a regulator of lifespan (Kaeberlein et al., 1999). Sir2 is a histone deacetylase (Imai et al., 2000), and it promotes longevity by its deacetylase activity. The human orthologs of Sir2 include seven members, SIRT1-7. SIRT1 is a highly conserved nicotinamide adenine dinucleotide (NAD)+-dependent protein deacetylase, that regulates chromatin remodeling, stress responses, DNA repair, insulin regulation, lifespan (Guarente and Picard, 2005). The diverse effects of SIRT1 reflect its ability to deacetylate a variety of transcriptional factors, such as p53, forkhead box O (FoxO), NF-κB, and peroxisome proliferators activated receptor gamma coactivator-1α (PGC-1α; Brunet et al., 2004; Yeung et al., 2004; Rodgers et al., 2005).
SIRT1 plays an important role in cancer (Liu et al., 2009). SIRT1 expression is increased in human cancers such as prostate cancer, colon cancer, acute myeloid leukemia, and some skin cancers (Bradbury et al., 2005; Hida et al., 2007; Huffman et al., 2007; Stunkel et al., 2007). SIRT1 might act as a tumor promoter in these diseases by interacting with and inhibiting p53 (van Leeuwen and Lain, 2009). SIRT1 also represses expression of tumor suppressor proteins and DNA repair proteins. But SIRT1 expression is decreased in other cancers, including ovarian cancer, glioblastoma, and bladder carcinoma (Deng, 2009). SIRT1 might serve as a tumor suppressor in these diseases by blocking oncogenic pathways. For example, SIRT1 limits β-catenin signaling in colon cancer, and in breast cancer BRCA1 signaling interacts with the SIRT1 pathway (Mullan et al., 2006; Firestein et al., 2008). Thus SIRT1 can serve as a tumor promoter or tumor suppressor, depending on the oncogenic pathways specific to particular tumors.
SIRT1 also regulates metabolism and modulates metabolic diseases like diabetes (Lee and Kemper, 2010). Cellular studies showed that SIRT1 modulates fat accumulation, regulates mitochondrial biogenesis, and activates fatty acid oxidation. Mouse studies have revealed important physiological effects of SIRT1. SIRT1 affects metabolism of mice during caloric restriction (Haigis and Guarente, 2006). Transgenic mice over-expressing SIRT1 are protected from some pathological conditions including insulin resistance and glucose tolerance (Banks et al., 2008). Moreover, mice treated with SRT1720, a specific synthetic activator of SIRT1, are protected from diet-induced obesity and insulin resistance (Feige et al., 2008). Taken together, these in vivo results suggest that SIRT1 regulates mammalian metabolism.
SIRT1 also protects the host against obesity. SIRT1 transgenic mice were protected from high-fat diet-induced obesity and liver steatosis (Banks et al., 2008; Pfluger et al., 2008). Conversely SIRT1 heterozygous mice fed with high-fat diet gain body weight, exhibit liver steatosis and inflammation in fat (Xu et al., 2010). These mice data indicate that SIRT1 regulates systemic metabolism.
The physiological impact of SIRT1 upon aging in mammals is controversial. The original proposal that SIRT1 regulates mammalian aging came from two sources. Studies of the SIRT1 homolog Sir2 show that Sir2 prolongs longevity in yeast (Hekimi and Guarente, 2003). Additionally, prolongation of survival by caloric restriction also increases SIRT1 activity (Bordone and Guarente, 2005). Despite these tantalizing hints about the relation of SIRT1 and longevity, a conclusive link between the two has not yet been established. For example, moderate expression of SIRT1 attenuates age-dependent incidence of cardiac hypertrophy and dysfunction by inducing resistance to oxidative stress in mice heart; however high level of SIRT1 increases heart dysfunction (Alcendor et al., 2007).
SIRT1 controls angiogenesis, in part by regulating key transcription factors and radicals in response to stress (Potente and Dimmeler, 2008). FoxO1 and FoxO3a are transcription factors in endothelial cells that inhibit endothelial cells migration and tube formation (Potente et al., 2005). Loss of SIRT1 increases acetylation of FoxO1, a negative regulator of angiogenesis, thereby limiting angiogenic activity in endothelial cells (Potente et al., 2007). Furthermore, SIRT1 also increases expression of Kruppel-like factor 2 (KLF2), a transcription factor in endothelial cells that activates expression of vasculoprotective genes (Gracia-Sancho et al., 2010). Thus SIRT1 controls transcription factors such as FOXO and KLF2 that are crucial regulators of endothelial cells (Figure 1).
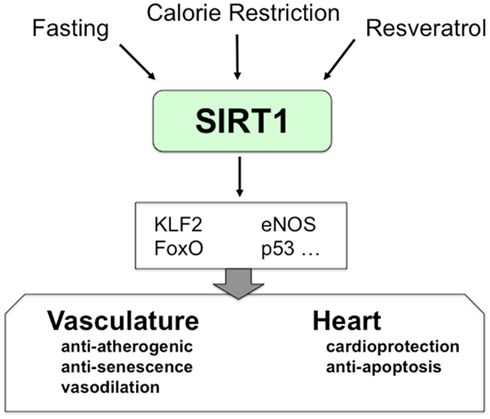
Figure 1. The role of SIRT1 in vasculature. SIRT1 mediates important cellular processes in cardiovascular systems. Calorie restriction and resveratrol enhance SIRT1 expression and activity. SIRT1 modulates cellular responses in cardiovascular systems by regulating the activity of several transcription factors.
Two interesting studies indicate the link between SIRT1 and endothelial function. First, SIRT1 regulates eNOS and NO synthesis. Calorie restriction induces SIRT1 and eNOS expression in male mice, and this effect is attenuated in eNOS knockout mice (Nisoli et al., 2005). Second, SIRT1 deacetylates and activates endothelial nitric oxide synthase (eNOS; Mattagajasingh et al., 2007). Since eNOS synthesizes NO, which maintains vascular homeostasis, these data support the beneficial role of SIRT1 in the vascular endothelium.
Finally, SIRT1 protects smooth muscle by decreasing angiotensin I signaling: SIRT1 decreases expression of the angiotensin receptor, which in turn limits reactive oxygen species (Li et al., 2011). Thus SIRT1 acts within endothelial cells and smooth muscle cells to protect the vasculature.
MicroRNAs (miRNAs) are non-coding ∼23 nt RNAs that control gene expression by suppressing translation or reducing stability of their target mRNAs (Bartel, 2004). MiRNAs modulate a variety of important functions including proliferation, differentiation, apoptosis, and senescence in animals and plants (Ambros, 2004; He and Hannon, 2004; Croce, 2008).
The biogenesis of miRNAs consists of several processing steps. Most miRNAs are transcribed into long primary transcripts (pri-miRNAs) by RNA polymerase II. Pri-miRNAs are cleaved into stem–loop structural miRNA precursors (pre-miRNAs) by the microprocessor complex, composed of RNase III enzymes Drosha and DGCR8. These ∼70 nt pre-miRNAs are exported to the cytoplasm by exportin-5 and cleaved by RNase III enzyme Dicer to generate duplex mature miRNAs. One or both single strands of mature miRNAs are incorporated into Argonaute (Ago2) in a miRNA–protein complex (RISC), which binds to the 3′ untranslated region (3′UTR) of target mRNAs. The binding of mature miRNA to the 3′UTR of target mRNAs depends on the interaction of the 6- to 8-nt seed sequence at the 5′ ends of miRNA, and miRNA response elements in the target mRNA.
Several algorithms are available to predict potential target genes of miRNAs (Sethupathy et al., 2006; Hofacker, 2007). These methods are incorporated into miRNA target prediction programs on line such as Targetscan1, miRanda2, and PicTar3. The algorithms are based upon seed sequence of mRNA, and identify hundreds of potential target genes. Using these methods, several miRNA have been predicted to regulate SIRT1, as discussed below.
Several endogenous and exogenous regulators of SIRT1 have been identified. The protein encoded by deleted in breast cancer 1 (DBC1) is a natural inhibitor of SIRT1 activity. DBC1 forms a complex with SIRT1 to act as a tumor suppressor, but knockout of DBC1 increases SIRT1 activity, promoting tumorigenesis (Kim et al., 2008; Zhao et al., 2008). Resveratrol was the small molecule first identified to regulate SIRT1 activity and to extend life span. After the effect of resveratrol upon SIRT1 was characterized, high throughput screening was used to find other small molecule activators of SIRT. For example, SRT1720 was found to improve insulin sensitivity and decrease blood glucose levels (Milne et al., 2007), although its effects on SIRT1 are still questionable (Pacholec et al., 2010).
Under environmental stress, cells respond by activating sets of genes to maintain homeostasis. Although the role of SIRT1 in mediating stress responses is well established, the factors that control SIRT1 expression are less well understood. The following section reviews recent studies demonstrating the post-transcriptional regulation of SIRT1 by miRNAs and RNA-binding proteins (RBPs).
More than 16 miRNAs regulate SIRT1 expression and activity (Table 2). Among these miRNAs, miR-34a has been the most studied (Table 1).
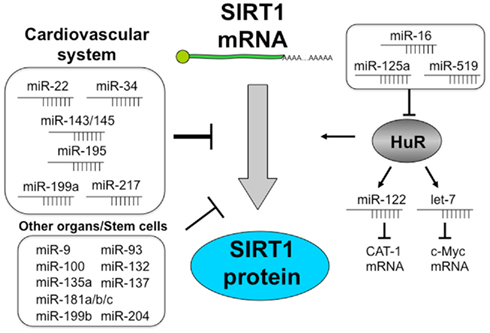
Figure 2. Regulation of SIRT1 by miRNAs. Schema shows that miRNAs regulate SIRT1 expression and function directly or through HuR. miR-22, miR-34, miR-143/145, miR-195, miR-199a, and miR-217 are expressed well in vascular tissues and control SIRT1 protein. SIRT1 is regulated by more than 15 miRNAs. miR-16, miR-125a, and miR-519 decrease HuR expression. In contrast, HuR controls the post-transcriptional regulation of miRNAs such as miR-122 and let-7. Moreover HuR binds to the 3′UTR of SIRT1 and changes SIRT1 mRNA stability.
The first identified miRNA that regulates SIRT1 is miR-34a (Yamakuchi et al., 2008). miR-34a was discovered as a tumor suppressor gene in neuroblastoma (Welch et al., 2007). Deletion of chromosome 1p where miR-34a is located on was found in 30% of advanced stage neuroblastomas (Versteeg et al., 1995; Calin et al., 2004). Since these initial reports, low levels of miR-34a expression were reported in prostate cancer, hepatocellular carcinoma, chronic lymphocytic leukemia (CLL), colon cancer, and ovarian cancer (Bommer et al., 2007; Chang et al., 2007; Tazawa et al., 2007; Fujita et al., 2008; Li et al., 2009; Corney et al., 2010).
One mechanism by which miR-34a expression decreases in cancers is due to aberrant CpG methylation of the promoter of miR-34a (Lodygin et al., 2008). Another potential molecule that alters miR-34a expression in cancer is p53. More than half of all human cancers have mutations in p53 tumor suppressor gene. p53 directly increases miR-34a expression (Bommer et al., 2007; Chang et al., 2007; Tazawa et al., 2007; Fujita et al., 2008; Li et al., 2009; Corney et al., 2010).
Reduced expression of miR-34a alters cellular function in cancer cells. Ectopic expression of miR-34a induced cell cycle arrest in the G1 phase and apoptosis in several cancer cells and many potential targets of miR-34a have been identified (Bommer et al., 2007; Chang et al., 2007; He et al., 2007; Tarasov et al., 2007). However, the precise molecular targets of miR-34a were unknown.
We discovered that miR-34a regulates SIRT1, using a colon cancer cell line, HCT116 as a model system. The first clue to SIRT1 as a target of miR-34a came from prediction algorithms. The next evidence for a link between miR-34a and SIRT1 came from experiments in which treatment of HCT116 with adriamycin increased p53 protein, induced miR-34a expression, and decreased SIRT1. (However, adriamycin did not alter the expression of miR-34a and SIRT1 in HCT116 cells that lack p53). These results suggested that p53 induced miR-34a expression, which decreased SIRT1 protein expression. miR-34a inhibition of SIRT1 increased acetylated p53 and expression of p53 transcriptional targets, p21 and PUMA, which regulate the cell cycle and apoptosis. Since this initial report, SIRT1 regulation by miR-34a has been reported in colorectal cancer, prostate cancer, hepatocarcinoma, glioma, and pancreatic cancer (Fujita et al., 2008; Pogribny et al., 2009; Kojima et al., 2010; Luan et al., 2010; Akao et al., 2011; Pramanik et al., 2011).
In the cardiovascular system, miR-34a also plays an important role in regulating cellular senescence and angiogenesis through SIRT1. We have shown that miR-34a induces senescence and cell cycle arrest in endothelial cells. Overexpression of miR-34a inhibits SIRT1 expression and endothelial senescence (Ito et al., 2010). miR-34a expression in heart and spleen are higher in older mice than those in younger mice. Conversely SIRT1 expression decreased with age. Our data suggested that miR-34a is a direct regulator of endothelial senescence. miR-34a also inhibits endothelial progenitor cells (EPC) mediated angiogenesis by induction of senescence (Zhao et al., 2010). miR-34a also modulates downstream targets of SIRT1 such as FoxO1, enhancing senescence. In murine ESCs, miR-34a targets SIRT1 and contributes to pluripotency of ESCs. Thus SIRT1 plays an important role in the cardiovascular system, and miR-34a in turn modulates SIRT1 expression.
miR-34a also influences cellular metabolism through its effects upon SIRT1. SIRT1 directly interacts with PGC-1α and deacetylates PGC-1α, suggesting that SIRT1 maintains metabolic homeostasis by modulating the activity of PGC-1α (Nemoto et al., 2005). SIRT1 also suppresses a key adipogenic transcription factor, PPARγ, and promotes fat mobilization in white adipocytes (Picard et al., 2004). The liver is a key metabolic organ that regulates lipid metabolism and glucose homeostasis. Hepatocyte-specific deletion of SIRT1 impairs PPARα signaling and fatty acid β-oxidation (Purushotham et al., 2009). In addition, liver-specific knockout of SIRT1 leads to hepatic inflammation and endoplasmic reticulum (ER) stress in the liver upon high-fat diet feeding. In hepatocyte, miR-34a regulates SIRT1. Farnesoid X receptor (FXR), one of the key regulatory factors in liver metabolism, inhibits miR-34a in HepG2 cells and hepatocyte-specific knockout of FXR increases miR-34a expression in liver. In fatty livers of diet-induced obese mice and ob/ob mice, the level of miR-34a is elevated and SIRT1 is decreased (Lee et al., 2010). In pancreas, miR-34a overexpression inhibits insulin secretion (Lovis et al., 2008). These studies suggested that miR-34a strongly affects glucose and insulin homeostasis through SIRT1.
Computer algorithms predict a set of miRNA that regulate SIRT1 (Table 2), and many of these have been verified in cells. A miRNA profiling study revealed that miR-217 was significantly upregulated in old human umbilical vein endothelial cells (HUVEC; PDL44) compared to young endothelial cells (PDL8; Menghini et al., 2009). Computational target prediction identified homology between miR-217 and 3′UTR of human SIRT1 mRNA. Overexpression of miR-217 suppresses SIRT1 expression in young endothelial cells. In contrast, inhibition of miR-217 increased SIRT1 expression and promoted cellular senescence in older HUVEC. Alteration of miR-217 controls SIRT1 level, which deacetylates FoxO1. Confirming these in vitro studies, miR-217 level is negatively associated with SIRT1 expression in human atherosclerotic plaque from patients underwent carotid endarterectomy. These results suggest that miR-217 plays an important role in senescence during the development of atherosclerosis.
In cardiomyocytes, SIRT1 is controlled by miR-195. The free fatty acid palmitate, upregulates miR-195 expression which inhibits SIRT1 and promotes apoptosis (Zhu et al., 2011). Another cardiac miRNA, miR-199a, limits expression of both SIRT1 and HIF-1α (Rane et al., 2009). Hypoxia or cardiac ischemia decreases miR-199a, permitting an increase in SIRT1 in cardiac myocytes. SIRT1 in turn downregulates prolyl hydroxylase 2 (PHD2), which stabilizes HIF-1α and hypoxia signaling. Thus miR-199 increases HIF-1α in two ways: first by regulating HIF-1α directly, second by a SIRT1–PHD2–HIF-1 pathway.
Two miRNAs control SIRT1 in pancreas and adipose tissue, critical organs that regulate glucose metabolism. One miRNA is miR-9, which modulates SIRT1 level in pancreatic β-cells (Ramachandran et al., 2011). Previous studies showed that SIRT1 affects insulin secretion by downregulation of UCP2 (Bordone et al., 2006). SIRT1 conditional transgenic mice (BESTO mice) exhibit a significant alteration of insulin secretion in response to glucose uptake, indicating a critical role of SIRT1 in the control of insulin secretion (Moynihan et al., 2005). Ramachandran et al. showed that glucose-stimulated insulin secretion is accompanied by changes in miR-9, which is mediated by SIRT1. In adipocytes, miR-132 affects SIRT1 regulation. Adipose tissue secretes a series of pro-inflammatory mediators that contribute to metabolic disorders. Serum deprivation induces the production of chemokines such as IL-8 and MCP-1 in adipocytes. In low serum condition, miR-132 expression is upregulated. Overexpression of miR-132 decreases SIRT1 expression, which blocks deacetylation of p65 NFκB, causing induction of IL-8 and MCP-1.
In cancer cells, senescence inhibits proliferation and induces apoptosis. Since SIRT1 is a major protein that regulates cellular senescence, miRNAs targeting SIRT1 have the potential to control cancer progression.
The miR-200 family is important regulator of tumor progression and metastasis (Brabletz and Brabletz, 2010). The miR-200 family consist of two clusters; (1) miR-200b, miR-200a, and miR-429, and (2) miR-200c and miR-141. Loss of miR-200a expression is associated with epithelial to mesenchymal transformation, in part via SIRT1 upregulation in breast cancer (Eades et al., 2011). miR-449 expression is decreased in Gastrin knockout mice, that are achlorhydric and tend to develop antral hyperplasia and gastric adenomas later (Bou Kheir et al., 2011). In human gastric cancers, miR-449 level is also down-regulated. One of miR-449 target is SIRT1, although miR-449 has a number of other targets of cell cycle related genes. Since the seed sequence of miR-449 is the same as that of miR-34a, target genes of miR-449 and miR-34a overlap. Increased miR-449 inhibits cellular proliferation and induces senescence in gastric cancer cell line, SNU638. Xu et al. (2011) performed miRNA profiling of senescent fibroblasts, showing that several miRNAs including miR-22, miR-34a, and miR-125a-5p are involved in cellular senescence in human diploid fibroblasts. Among them, miR-22 is highly expressed in senescent fibroblast, but relatively lower in cancers. Overexpression of miR-22 suppresses the proliferation in fibroblasts and human breast cancer cell line MCF7. SIRT1 is identified as a direct target of miR-22 as well as CDK6 and Sp1. Moreover synthetic miR-22 delivery inhibits breast tumor growth and metastasis in mice model of breast cancer. Tumor over-expressing miR-22 shows high number of senescent tumor cells.
These miRNAs described above are examples of tumor suppressor miRNAs targeting SIRT1. However, the role of SIRT1 in tumorigenesis is complex (Deng, 2009). Some studies show that SIRT1 acts as a tumor suppressor protein. Mice that are double heterozygous for SIRT1 and p53 (Sirt1+/−;p53+/−) develop tumors in multiple tissues such as primarily sarcomas, lymphomas, teratomas, and carcinomas (Wang et al., 2008). When SIRT1 was activated by resveratrol, the frequency of tumorigenesis was reduced. Another study showed that SIRT1 is a tumor promoter. The interaction of SIRT1 and p53 leads to deacetylation of p53 (Lys382 residue). SIRT1 overexpression inhibits p53 dependent cell cycle arrest and apoptosis (Luo et al., 2001; Vaziri et al., 2001). In contrast, suppression of SIRT1 activity increases p53 dependent transcriptional activity (Lain et al., 2008). These data indicate that SIRT1 deacetylates and inactivates p53 function, enhancing tumorigenesis. Thus the effects of SIRT1 upon tumors depend in part upon the type of tumor and the p53 status of the tumor.
SIRT1 regulates many transcriptional factors by their deacetylation, as described above. Therefore it is likely that SIRT1 controls expression of miRNAs by activating their transcriptional factors. One example of this is SIRT1 regulation of a brain specific miRNA, miR-134 (Gao et al., 2010). SIRT1 modulates synaptic plasticity and memory formation via a miRNA-mediated mechanism. SIRT1 suppresses miR-134 transcription in normal condition by a repressor complex containing transcription factor YY1. Loss of SIRT1 activity results in the increase of miR-134, which downregulates CREB and BDNF expression, impairing synaptic plasticity.
A number of miRNAs regulate SIRT1 expression and activity. Some of these miRNAs are controlled by stress-induced transcriptional factors. As shown in miR-34a, p53 upregulates miR-34a expression. SIRT1, the target of miR-34a, deacetylates and destabilize p53, decreasing p53 activity (Luo et al., 2001; Vaziri et al., 2001). Therefore p53 increases miR-34a expression, then downregulates SIRT1 level, which increases p53 expression and activity. The increase of p53 produces more miR-34a. This is a positive loop, p53–miR-34a–SIRT1 (Yamakuchi and Lowenstein, 2009). This study of miR-134 indicates the possibility that SIRT1 alters some miRNAs expression, suggesting that another interesting loop: miRNA–SIRT1–miRNA. These studies suggest the complexities of the miRNA–SIRT1 network.
The RBP HuR (Hu antigen R, ELAVL1) regulates SIRT1 in a post-transcriptional manner. HuR is a member of ELAVL1family of RBPs (Hinman and Lou, 2008; Katsanou et al., 2009; Papadaki et al., 2009). HuR binds to AU-rich elements (AREs) in the 3′UTR of mRNAs (Caput et al., 1986; Shaw and Kamen, 1986; Szabo et al., 1991). HuR controls the stability of thousands of mRNA, including SIRT1.
HuR regulates SIRT1 expression by direct and indirect pathways. HuR directly binds to ARE in the SIRT1 mRNA, which stabilizes SIRT1 (Abdelmohsen et al., 2007). When HuR is silenced, SIRT1 mRNA levels are decreased; in contrast, overexpression of HuR increases SIRT1 mRNA. Interestingly, senescent fibroblasts exhibit low level of SIRT1 and HuR compared to young fibroblasts. One possible explanation is that repression of HuR by cellular senescence reduces SIRT1 expression in fibroblasts. In fact, knockdown of HuR decreases the proliferation rate. Under oxidative stress, HuR dissociated from the SIRT1 mRNA, causing its rapid degradation. Furthermore the checkpoint kinase 2 (Chk2) phosphorylates HuR, which inhibits the binding of SIRT1 and HuR. Three phosphorylation sites (two serine residues and one threonine residue) by Chk2 are identified on HuR. These results clearly suggest that RBP, HuR, controls the alteration of SIRT1 expression.
In addition to HuR binding directly to SIRT1 mRNA, HuR also regulates SIRT1 expression through indirect pathways that involve microRNA. The profile of microRNA expression is altered in senescent fibroblasts, with older cells expressing higher levels of several microRNA species, including miR-519 (Marasa et al., 2010). miR-519 inhibits HuR levels, which leads to decreased SIRT1 expression and senescence. Others have confirmed that miR-519 represses HuR translation, thereby altering HuR regulation of mRNA stability (Abdelmohsen et al., 2008).
SIRT1 signaling is complex, and controversies about the role of SIRT1 in longevity and cancer persist. Does SIRT1 really extend lifespan? Does SIRT1 accelerate or slow cancer progression? Regulation of SIRT1 adds to its complexity. Two major pathways for post-transcriptional regulations of SIRT1 exist, RBPs and miRNAs. A discrete set of miRNAs regulates SIRT1 expression, and different miRNA regulate SIRT1 in a cell specific manner. A major regulator of SIRT1 is miR-34a, which suppresses SIRT1 expression in specific tissues and cancers, including colon cancer. The RBP HuR is also involved in regulation of SIRT1 expression. The interaction between HuR and miRNAs is interesting and is an important area of future study.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I thank Dr. Charles J. Lowenstein for helpful advice and discussions. This work was supported by Scientist Development Grant 835446N from American Heart Association.
Abdelmohsen, K., Pullmann, R. Jr., Lal, A., Kim, H. H., Galban, S., Yang, X., Blethrow, J. D., Walker, M., Shubert, J., Gillespie, D. A., Furneaux, H., and Gorospe, M. (2007). Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25, 543–557.
Abdelmohsen, K., Srikantan, S., Kuwano, Y., and Gorospe, M. (2008). miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U.S.A. 105, 20297–20302.
Akao, Y., Noguchi, S., Iio, A., Kojima, K., Takagi, T., and Naoe, T. (2011). Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 300, 197–204.
Alcendor, R. R., Gao, S., Zhai, P., Zablocki, D., Holle, E., Yu, X., Tian, B., Wagner, T., Vatner, S. F., and Sadoshima, J. (2007). Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 100, 1512–1521.
Aranha, M. M., Santos, D. M., Solá, S., Steer, C. J., and Rodrigues, C. M. (2011). miR-34a regulates mouse neural stem cell differentiation. PLoS One 6, e21396. doi:10.1371/journal.pone.0021396
Banks, A. S., Kon, N., Knight, C., Matsumoto, M., Gutierrez-Juarez, R., Rossetti, L., Gu, W., and Accili, D. (2008). SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8, 333–341.
Bommer, G. T., Gerin, I., Feng, Y., Kaczorowski, A. J., Kuick, R., Love, R. E., Zhai, Y., Giordano, T. J., Qin, Z. S., Moore, B. B., MacDougald, O. A., Cho, K. R., and Fearon, E. R. (2007). p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307.
Bordone, L., and Guarente, L. (2005). Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 6, 298–305.
Bordone, L., Motta, M. C., Picard, F., Robinson, A., Jhala, U. S., Apfeld, J., McDonagh, T., Lemieux, M., McBurney, M., Szilvasi, A., Easlon, E. J., Lin, S. J., and Guarente, L. (2006). Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 4, e31. doi:10.1371/journal.pbio.0040031
Bou Kheir, T., Futoma-Kazmierczak, E., Jacobsen, A., Krogh, A., Bardram, L., Hother, C., Grønbæk, K., Federspiel, B., Lund, A. H., and Friis-Hansen, L. (2011). miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol. Cancer 18, 10–29.
Brabletz, S., and Brabletz, T. (2010). The ZEB/miR-200 feedback loop – a motor of cellular plasticity in development and cancer? EMBO Rep. 11, 670–677.
Bradbury, C. A., Khanim, F. L., Hayden, R., Bunce, C. M., White, D. A., Drayson, M. T., Craddock, C., and Turner, B. M. (2005). Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia 19, 1751–1759.
Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E., Mostoslavsky, R., Cohen, H. Y., Hu, L. S., Cheng, H. L., Jedrychowski, M. P., Gygi, S. P., Sinclair, D. A., Alt, F. W., and Greenberg, M. E. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015.
Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M., and Croce, C. M. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004.
Caput, D., Beutler, B., Hartog, K., Thayer, R., Brown-Shimer, S., and Cerami, A. (1986). Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. U.S.A. 83, 1670–1674.
Chang, T. C., Wentzel, E. A., Kent, O. A., Ramachandran, K., Mullendore, M., Lee, K. H., Feldmann, G., Yamakuchi, M., Ferlito, M., Lowenstein, C. J., Arking, D. E., Beer, M. A., Maitra, A., and Mendell, J. T. (2007). Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752.
Corney, D. C., Hwang, C. I., Matoso, A., Vogt, M., Flesken-Nikitin, A., Godwin, A. K., Kamat, A. A., Sood, A. K., Ellenson, L. H., Hermeking, H., and Nikitin, A. Y. (2010). Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 16, 1119–1128.
Deng, C. X. (2009). SIRT1, is it a tumor promoter or tumor suppressor? Int. J. Biol. Sci. 5, 147–152.
Eades, G., Yao, Y., Yang, M., Zhang, Y., Chumsri, S., and Zhou, Q. (2011). miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J. Biol. Chem. 286, 25992–26002.
Feige, J. N., Lagouge, M., Canto, C., Strehle, A., Houten, S. M., Milne, J. C., Lambert, P. D., Mataki, C., Elliott, P. J., and Auwerx, J. (2008). Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 8, 347–358.
Firestein, R., Blander, G., Michan, S., Oberdoerffer, P., Ogino, S., Campbell, J., Bhimavarapu, A., Luikenhuis, S., de Cabo, R., Fuchs, C., Hahn, W. C., Guarente, L. P., and Sinclair, D. A. (2008). The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE 3, e2020. doi:10.1371/journal.pone.0002020
Fujita, Y., Kojima, K., Hamada, N., Ohhashi, R., Akao, Y., Nozawa, Y., Deguchi, T., and Ito, M. (2008). Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem. Biophys. Res. Commun. 377, 114–119.
Gao, J., Wang, W. Y., Mao, Y. W., Graff, J., Guan, J. S., Pan, L., Mak, G., Kim, D., Su, S. C., and Tsai, L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Gracia-Sancho, J., Villarreal, G. Jr., Zhang, Y., and Garcia-Cardena, G. (2010). Activation of SIRT1 by resveratrol induces KLF2 expression conferring an endothelial vasoprotective phenotype. Cardiovasc. Res. 85, 514–519.
Haigis, M. C., and Guarente, L. P. (2006). Mammalian sirtuins – emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921.
He, L., and Hannon, G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531.
He, L., He, X., Lim, L. P., de Stanchina, E., Xuan, Z., Liang, Y., Xue, W., Zender, L., Magnus, J., Ridzon, D., Jackson, A. L., Linsley, P. S., Chen, C., Lowe, S. W., Cleary, M. A., and Hannon, G. J. (2007). A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134.
Hekimi, S., and Guarente, L. (2003). Genetics and the specificity of the aging process. Science 299, 1351–1354.
Hida, Y., Kubo, Y., Murao, K., and Arase, S. (2007). Strong expression of a longevity-related protein, SIRT1, in Bowen’s disease. Arch. Dermatol. Res. 299, 103–106.
Hinman, M. N., and Lou, H. (2008). Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65, 3168–3181.
Hofacker, I. L. (2007). RNA consensus structure prediction with RNAalifold. Methods Mol. Biol. 395, 527–544.
Huffman, D. M., Grizzle, W. E., Bamman, M. M., Kim, J. S., Eltoum, I. A., Elgavish, A., and Nagy, T. R. (2007). SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 67, 6612–6618.
Imai, S., Armstrong, C. M., Kaeberlein, M., and Guarente, L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800.
Ito, T., Yagi, S., and Yamakuchi, M. (2010). MicroRNA-34a regulation of endothelial senescence. Biochem. Biophys. Res. Commun. 398, 735–740.
Kaeberlein, M., McVey, M., and Guarente, L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580.
Katsanou, V., Milatos, S., Yiakouvaki, A., Sgantzis, N., Kotsoni, A., Alexiou, M., Harokopos, V., Aidinis, V., Hemberger, M., and Kontoyiannis, D. L. (2009). The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol. Cell. Biol. 29, 2762–2776.
Kennedy, B. K., Austriaco, N. R. Jr., Zhang, J., and Guarente, L. (1995). Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80, 485–496.
Kim, J. E., Chen, J., and Lou, Z. (2008). DBC1 is a negative regulator of SIRT1. Nature 451, 583–586.
Kojima, K., Fujita, Y., Nozawa, Y., Deguchi, T., and Ito, M. (2010). MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate 70, 1501–1512.
Lain, S., Hollick, J. J., Campbell, J., Staples, O. D., Higgins, M., Aoubala, M., McCarthy, A., Appleyard, V., Murray, K. E., Baker, L., Thompson, A., Mathers, J., Holland, S. J., Stark, M. J., Pass, G., Woods, J., Lane, D. P., and Westwood, N. J. (2008). Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 13, 454–463.
Lee, J., and Kemper, J. K. (2010). Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany N.Y.) 2, 527–534.
Lee, J., Padhye, A., Sharma, A., Song, G., Miao, J., Mo, Y. Y., Wang, L., and Kemper, J. K. (2010). A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 285, 12604–12611.
Li, L., Gao, P., Zhang, H., Chen, H., Zheng, W., Lv, X., Xu, T., Wei, Y., Liu, D., and Liang, C. (2011). SIRT1 inhibits angiotensin II-induced vascular smooth muscle cell hypertrophy. Acta Biochim. Biophys. Sin. (Shanghai) 43, 103–109.
Li, N., Fu, H., Tie, Y., Hu, Z., Kong, W., Wu, Y., and Zheng, X. (2009). miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 275, 44–53.
Liu, T., Liu, P. Y., and Marshall, G. M. (2009). The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 69, 1702–1705.
Lizé, M., Pilarski, S., and Dobbelstein, M. (2010). E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 17, 452–458.
Lodygin, D., Tarasov, V., Epanchintsev, A., Berking, C., Knyazeva, T., Korner, H., Knyazev, P., Diebold, J., and Hermeking, H. (2008). Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600.
Lovis, P., Roggli, E., Laybutt, D. R., Gattesco, S., Yang, J. Y., Widmann, C., Abderrahmani, A., and Regazzi, R. (2008). Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes 57, 2728–2736.
Luan, S., Sun, L., and Huang, F. (2010). MicroRNA-34a: a novel tumor suppressor in p53-mutant glioma cell line U251. Arch. Med. Res. 41, 67–74.
Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. (2001). Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107, 137–148.
Marasa, B. S., Srikantan, S., Martindale, J. L., Kim, M. M., Lee, E. K., Gorospe, M., and Abdelmohsen, K. (2010). MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany N.Y.) 2, 333–343.
Mattagajasingh, I., Kim, C. S., Naqvi, A., Yamamori, T., Hoffman, T. A., Jung, S. B., DeRicco, J., Kasuno, K., and Irani, K. (2007). SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 14855–14860.
Menghini, R., Casagrande, V., Cardellini, M., Martelli, E., Terrinoni, A., Amati, F., Vasa-Nicotera, M., Ippoliti, A., Novelli, G., Melino, G., Lauro, R., and Federici, M. (2009). MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 120, 1524–1532.
Milne, J. C., Lambert, P. D., Schenk, S., Carney, D. P., Smith, J. J., Gagne, D. J., Jin, L., Boss, O., Perni, R. B., Vu, C. B., Bemis, J. E., Xie, R., Disch, J. S., Ng, P. Y., Nunes, J. J., Lynch, A. V., Yang, H., Galonek, H., Israelian, K., Choy, W., Iffland, A., Lavu, S., Medvedik, O., Sinclair, D. A., Olefsky, J. M., Jirousek, M. R., Elliott, P. J., and Westphal, C. H. (2007). Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716.
Moynihan, K. A., Grimm, A. A., Plueger, M. M., Bernal-Mizrachi, E., Ford, E., Cras-Meneur, C., Permutt, M. A., and Imai, S. (2005). Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2, 105–117.
Mullan, P. B., Quinn, J. E., and Harkin, D. P. (2006). The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25, 5854–5863.
Nemoto, S., Fergusson, M. M., and Finkel, T. (2005). SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J. Biol. Chem. 280, 16456–16460.
Nisoli, E., Tonello, C., Cardile, A., Cozzi, V., Bracale, R., Tedesco, L., Falcone, S., Valerio, A., Cantoni, O., Clementi, E., Moncada, S., and Carruba, M. O. (2005). Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 310, 314–317.
Pacholec, M., Bleasdale, J. E., Chrunyk, B., Cunningham, D., Flynn, D., Garofalo, R. S., Griffith, D., Griffor, M., Loulakis, P., Pabst, B., Qiu, X., Stockman, B., Thanabal, V., Varghese, A., Ward, J., Withka, J., and Ahn, K. (2010). SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351.
Papadaki, O., Milatos, S., Grammenoudi, S., Mukherjee, N., Keene, J. D., and Kontoyiannis, D. L. (2009). Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J. Immunol. 182, 6779–6788.
Pfluger, P. T., Herranz, D., Velasco-Miguel, S., Serrano, M., and Tschop, M. H. (2008). Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. U.S.A. 105, 9793–9798.
Picard, F., Kurtev, M., Chung, N., Topark-Ngarm, A., Senawong, T., Machado De Oliveira, R., Leid, M., McBurney, M. W., and Guarente, L. (2004). Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429, 771–776.
Pogribny, I. P., Muskhelishvili, L., Tryndyak, V. P., and Beland, F. A. (2009). The tumor-promoting activity of 2-acetylaminofluorene is associated with disruption of the p53 signaling pathway and the balance between apoptosis and cell proliferation. Toxicol. Appl. Pharmacol. 235, 305–311.
Potente, M., and Dimmeler, S. (2008). Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle 7, 2117–2122.
Potente, M., Ghaeni, L., Baldessari, D., Mostoslavsky, R., Rossig, L., Dequiedt, F., Haendeler, J., Mione, M., Dejana, E., Alt, F. W., Zeiher, A. M., and Dimmeler, S. (2007). SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 21, 2644–2658.
Potente, M., Urbich, C., Sasaki, K., Hofmann, W. K., Heeschen, C., Aicher, A., Kollipara, R., DePinho, R. A., Zeiher, A. M., and Dimmeler, S. (2005). Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 115, 2382–2392.
Pramanik, D., Campbell, N. R., Karikari, C., Chivukula, R., Kent, O. A., Mendell, J. T., and Maitra, A. (2011). Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 10, 1470–1480.
Purushotham, A., Schug, T. T., Xu, Q., Surapureddi, S., Guo, X., and Li, X. (2009). Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9, 327–338.
Ramachandran, D., Roy, U., Garg, S., Ghosh, S., Pathak, S., and Kolthur-Seetharam, U. (2011). Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. FEBS J. 278, 1167–1174.
Rane, S., He, M., Sayed, D., Vashistha, H., Malhotra, A., Sadoshima, J., Vatner, D. E., Vatner, S. F., and Abdellatif, M. (2009). Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 104, 879–886.
Rodgers, J. T., Lerin, C., Haas, W., Gygi, S. P., Spiegelman, B. M., and Puigserver, P. (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118.
Saunders, L. R., Sharma, A. D., Tawney, J., Nakagawa, M., Okita, K., Yamanaka, S., Willenbring, H., and Verdin, E. (2010). miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging 2, 415–431.
Schonrock, N., Humphreys, D. T., Preiss, T., and Götz, J. (2012). Target gene repression mediated by miRNAs miR-181c and miR-9 both of which are down-regulated by amyloid-β. J. Mol. Neurosci. 46, 324–335.
Sethupathy, P., Megraw, M., and Hatzigeorgiou, A. G. (2006). A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods 3, 881–886.
Shaw, G., and Kamen, R. (1986). A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46, 659–667.
Strum, J. C., Johnson, J. H., Ward, J., Xie, H., Field, J., Hester, A., Alford, A., and Waters, K. M. (2009). MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 23, 1876–1884.
Stunkel, W., Peh, B. K., Tan, Y. C., Nayagam, V. M., Wang, X., Salto-Tellez, M., Ni, B., Entzeroth, M., and Wood, J. (2007). Function of the SIRT1 protein deacetylase in cancer. Biotechnol. J. 2, 1360–1368.
Szabo, A., Dalmau, J., Manley, G., Rosenfeld, M., Wong, E., Henson, J., Posner, J. B., and Furneaux, H. M. (1991). HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and sex-lethal. Cell 67, 325–333.
Tarantino, C., Paolella, G., Cozzuto, L., Minopoli, G., Pastore, L., Parisi, S., and Russo, T. (2010). miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 24, 3255–3263.
Tarasov, V., Jung, P., Verdoodt, B., Lodygin, D., Epanchintsev, A., Menssen, A., Meister, G., and Hermeking, H. (2007). Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6, 1586–1593.
Tazawa, H., Tsuchiya, N., Izumiya, M., and Nakagama, H. (2007). Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 15472–15477.
Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L., and Weinberg, R. A. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159.
Versteeg, R., Caron, H., Cheng, N. C., van der Drift, P., Slater, R., Westerveld, A., Voute, P. A., Delattre, O., Laureys, G., and Van Roy, N. (1995). 1p36: every subband a suppressor? Eur. J. Cancer 31A, 538–541.
Wang, R. H., Sengupta, K., Li, C., Kim, H. S., Cao, L., Xiao, C., Kim, S., Xu, X., Zheng, Y., Chilton, B., Jia, R., Zheng, Z. M., Appella, E., Wang, X. W., Ried, T., and Deng, C. X. (2008). Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–323.
Welch, C., Chen, Y., and Stallings, R. L. (2007). MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26, 5017–5022.
Xu, D., Takeshita, F., Hino, Y., Fukunaga, S., Kudo, Y., Tamaki, A., Matsunaga, J., Takahashi, R. U., Takata, T., Shimamoto, A., Ochiya, T., and Tahara, H. (2011). miR-22 represses cancer progression by inducing cellular senescence. J. Cell Biol. 193, 409–424.
Xu, F., Gao, Z., Zhang, J., Rivera, C. A., Yin, J., Weng, J., and Ye, J. (2010). Lack of SIRT1 (mammalian sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology 151, 2504–2514.
Yamakuchi, M., Ferlito, M., and Lowenstein, C. J. (2008). miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426.
Yamakuchi, M., and Lowenstein, C. J. (2009). MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle 8, 712–715.
Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A., and Mayo, M. W. (2004). Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380.
Zhao, T., Li, J., and Chen, A. F. (2010). MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am. J. Physiol. Endocrinol. Metab. 299, E110–E116.
Zhao, W., Kruse, J. P., Tang, Y., Jung, S. Y., Qin, J., and Gu, W. (2008). Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590.
Keywords: microRNA, SIRT1, miR-34a, RNA-binding proteins, HuR
Citation: Yamakuchi M (2012) MicroRNA regulation of SIRT1. Front. Physio. 3:68. doi: 10.3389/fphys.2012.00068
Received: 30 November 2011; Paper pending published: 23 December 2011;
Accepted: 10 March 2012; Published online: 30 March 2012.
Edited by:
Yu Wang, The University of Hong Kong, Hong KongReviewed by:
Sanjukta Chakraborty, Texas A&M Health Science Center, USACopyright: © 2012 Yamakuchi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Munekazu Yamakuchi, Department of Medicine, Aab Cardiovascular Research Institute, University of Rochester School of Medicine and Dentistry, 601 Elmwood Avenue, Box CVRI, Rochester, NY 14642, USA. e-mail:bXVuZWthenVfeWFtYWt1Y2hpQHVybWMucm9jaGVzdGVyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.