- 1 Department of Biological Science, National University of Singapore, Singapore
- 2 Natural Sciences and Science Education, National Institute of Education, Nanyang Technological University, Singapore
The swamp eel, Monopterus albus, is an obligatory air-breathing teleost which can undergo long period of emersion, has high environmental and tissue ammonia tolerance, and can survive in brackish water. We obtained a cDNA sequence of glutamate dehydrogenase (gdh), which consisted of a 133-bp 5′ UTR, a complete coding sequence region spanning 1629 bp and a 3′ UTR of approximately 717 bp, from the liver, intestine, and brain of M. albus. The translated Gdh amino acid sequence had 542 residues, and it formed a monophyletic clade with Bostrychus sinensis Gdh1a, Tetraodon nigroviridis Gdh1a, Chaenocephalus aceratus Gdh1a, Salmo salar Gdh1a1 and Gdh1a2, and O. mykiss Gdh1a. One day of exposure to terrestrial conditions or 75 mmol l−1 NH4Cl, but not to water at salinity 20, resulted in a significant increase in mRNA expression of gdh1a and Gdh amination activity in the liver of M. albus. However, exposure to brackish water, but not to terrestrial conditions or 75 mmol l−1 NH4Cl, led to a significant increase in the mRNA expression of gdh1a and Gdh amination activity in the intestine. By contrast, all the three experimental conditions had no significant effects on the mRNA expression of gdh1a in the brain of M. albus, despite a significant decrease in the Gdh amination activity in the brain of fish exposed to 75 mmol l−1 NH4Cl for 6 days. Our results indicate for the first time that the mRNA expression of gdh1a was differentially up-regulated in the liver and intestine of M. albus in response to ammonia toxicity and salinity stress, respectively. The increases in mRNA expression of gdh1a and Gdh amination activity would probably lead to an increase in glutamate production in support of increased glutamine synthesis for the purpose of ammonia detoxification or cell volume regulation under these two different environmental conditions.
Introduction
The swamp eel, Monopterus albus (Zuiew) belongs to Order Synbranchiformes, Family Synbranchidae, and is commonly found in muddy ponds, swamps, canals, and rice fields in India, Malaysia, Indonesia, and Southern China (Rainboth, 1996). It is an obligate air breather and its gills are vastly atrophied to become a fold of skin within the opercular chamber (Graham, 1997). As an air breather, M. albus can survive on land for an extended period. Since no water is available to flush branchial or cutaneous surfaces during emersion, ammonia excretion becomes inefficient leading to the accumulation of ammonia within the body (Ip et al., 2004b). During drought, ammonia excreted into a small volume of external medium would increase in concentration which may reach to levels that would impede endogenous ammonia excretion. For fish living in the rice field, it may encounter high levels of environmental ammonia (Ip et al., 2004a) during the application of ammonium sulfate fertilizers (Freney et al., 1981). Since the environmental salinity may increase when the habitat dries up during drought or during high tide in swamps located at the river mouth, M. albus may also encounter hyperosmotic stress.
Ammonia is toxic to fishes as it affects many cellular processes (Ip et al., 2001, 2004a,b; Chew et al., 2006; Ip and Chew, 2010), but some tropical air-breathing teleosts can ameliorate ammonia toxicity through a variety of adaptations (see Ip et al., 2001, 2004a,b; Chew et al., 2006 for reviews). During 6 days of emersion (Tay et al., 2003) or 40 days of aestivation in mud (Chew et al., 2005), M. albus demonstrates high tissue ammonia tolerance, suppresses endogenous ammonia production, and detoxifies endogenous ammonia to glutamine in extra-cranial tissues. Furthermore, M. albus has high tolerance of acute ammonia toxicity and detoxifies exogenous ammonia to glutamine (Tng et al., 2009). After 6 days of exposure to 75 mmol l−1 NH4Cl, ammonia (6.5–15.2 μmol g−1), and glutamine (9.4–17.1 μmol g−1) accumulate to very high levels in the brain, muscle, liver, and intestine (Ip et al., 2004c). Hence, glutamine can be regarded as a temporary product of ammonia detoxification in M. albus during emersion or ammonia exposure. The fate of glutamine during prolonged exposure to or recovery from emersion or environmental ammonia is unclear at present, but one of the advantages of producing glutamine is that it can be stored momentarily in the body and used for the synthesis of other molecules, e.g., purines and pyrimidines, when the adverse environmental conditions subside. Besides, glutamine can also act as an intracellular osmolyte for the purpose of cell volume regulation. Recently, Tok et al. (2009) reported that M. albus switched from hyperosmotic hyperionic regulation in freshwater to a combination of osmoconforming and hypoosmotic hypoionic regulation to survive in brackish water (salinity 25). Salinity stress resulted in relatively large increases in the plasma osmolality, [Na+] and [Cl−] in M. albus. This in turn led to the accumulation of organic osmolytes and inorganic ions for cell volume regulation, with the hepatic glutamine content increased to a phenomenal level of >30 μmol g−1.
Glutamine synthesis is catalyzed by glutamine synthetase (GS) and requires glutamate as a substrate. Although transamination reactions can generate glutamate, the key enzyme regulating ammonia and glutamate levels in vertebrates is glutamate dehydrogenase (GDH; Shoemaker and Haley, 1993), which is involved in several metabolic pathways (Hudson and Daniel, 1993; Thatcher and Storey, 2001). GDH catalyzes both glutamate formation in the amination direction using ammonia and α-ketoglutarate as substrates and glutamate degradation in the deamination direction (Smith et al., 1975; Hudson and Daniel, 1993). To date, there is a dearth of information on how adverse conditions would affect the mRNA expression of gdh in various tissues of M. albus and in fish in general. Therefore, this study was undertaken to clone and sequence the cDNA of gdh from the liver, brain, and intestine of M. albus. Efforts were then made to determine whether exposure to terrestrial conditions, environmental ammonia (75 mmol l−1 NH4Cl), or brackish water (salinity 20) would induce changes in the mRNA expression of gdh in these three organs.
Materials and Methods
Fish
Specimens of M. albus (150–400 g body mass) were purchased locally from a fish farm in Singapore. They were maintained in plastic aquaria in freshwater (salinity 1) at 25°C under a 12 h:12 h dark:light regime in the laboratory and water was changed daily. No attempt was made to separate the sexes. Fish were acclimated to laboratory conditions for at least 1 week before experimentation, during which they were fed live guppy. Food was withdrawn 48 h prior to experiments, which gave sufficient time for the gut to be emptied of all food and waste products. Fish were not fed throughout the experimental period. Procedures adopted in this study were approved by the Institutional Animal Care and Use Committee of the National University of Singapore (IACUC 021/10).
Exposure of Fish to Experimental Conditions and Collection of Samples
Fish were exposed to control or experimental conditions individually in plastic tanks (50 cm length × 30 cm width × 10 cm height) with free access to air. A total of 12 fish were immersed in 10 volumes (w/v) of freshwater and they served as controls. Water was changed daily. For terrestrial conditions, fish were kept in plastic aquaria tanks containing a thin film (100 ml) of freshwater. These tanks were rinsed and the freshwater was replenished daily. For ammonia exposure, a total of 8 fish were immersed in 10 volumes of freshwater containing 75 mmol l−1 NH4Cl (pH 7), which was also changed daily. For salinity stress, 4 fish were immersed in 10 volumes (w/v) of water of various salinities prepared by mixing freshwater with an appropriate quantity of seawater, and the pH was adjusted to 7. The ambient salinity was raised progressively through a 4-day period from freshwater (day 0) to salinity 5 (day 1), salinity 10 (day 2), salinity 15 (day 3), and salinity 20 (day 4). At the end of the exposure periods (day 1, 4, and 6 for control fish, N = 4 each; day 1 and 6 for fish exposed to terrestrial or ammonia conditions, N = 4 each; day 4 for fish exposed to salinity stress, N = 4), fish were killed with a strong blow to the head, and samples of the liver, intestine, and brain were excised, frozen in liquid nitrogen, and stored at −80°C until analysis.
Total RNA Extraction, cDNA Synthesis, PCR, and Gene Sequencing
The total RNA was extracted from the liver, intestine, and brain, using the chaotropic extraction protocol described by Whitehead and Crawford (2005), and purified using RNeasy Mini Kit (QIAGEN Inc., Hilden, Germany). RNA quality was checked electrophoretically and quantified at 260 nm using a 105.810-UVS Hellma traycell (Hellma GmbH & Co., KG, Müllheim, Baden-Württemberg, Germany) adapted to a Shimadzu UV-1601 UV–VIS recording spectrophotometer (Shimadzu Corporation, Nakagyo-ku, Kyoto, Japan). The total RNA (1 μg) from each sample was then reverse transcribed using RevertAid™ M-MuLV Reverse Transcriptase and oligo(dT)18 primer from the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas Inc., Hanover, MD, USA). PCR was performed on the cDNA obtained using the forward primers 1, 2, and 3 and the reverse primers 4, 5, 6, and 7 (Table 1). The primer sequences for 1 and 4 were obtained from Hirata et al. (2003) while the primer sequences for 2, 3, 5, 6, and 7 were obtained from Peh (2008). Each PCR was carried out in 25 μl reaction volumes, containing 10× Dreamtaq Buffer (2.5 μl; Fermentas Inc.), dNTPs (10 μmol l−1, 0.5 μl), MgCl2 (25 mmol l−1, 0.5 μl), Dreamtaq polymerase (5 IU μl−1, 0.125 μl; Fermentas Inc.), forward primer and reverse primers (10 μmol l−1, 1.25 μl each), and cDNA template (0.5 μl), using a Veriti® Fast 96-well Thermal Cycler (Applied Biosystems Inc., Foster City, CA, USA). The following thermocycling program was used: 95°C for 3 min, followed by 40 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C and 1 cycle of final extension at 72°C for 10 min. The PCR products obtained were separated by 1% agarose gel electrophoresis and the band of expected size was excised from the gel. The PCR product was purified from the gel slice by the centrifugation method of Wizard SV gel and PCR clean-up system kit (Promega Corporation, Madison, WI, USA). Purified PCR products were subjected to cycle sequencing using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc.) and purified by the ethanol/sodium acetate precipitation. Purified products were automatically sequenced using the Prism™ 3130XL Genetic Analyzer (Applied Biosystems Inc.).
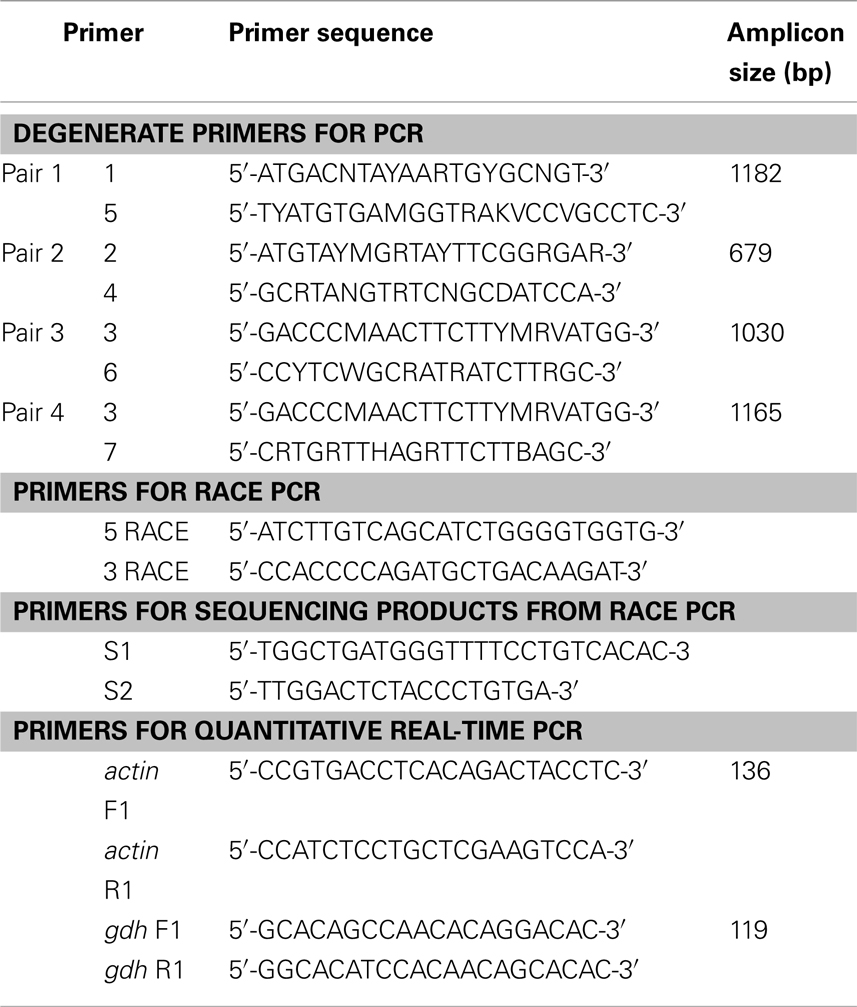
Table 1. Primers for PCR or RACE PCR to amplify glutamate dehydrogenase (gdh), or for quantitative real-time PCR on actin (reference gene) and gdh1a from the liver, intestine, and brain of Monopterus albus.
RACE PCR and Cloning
Total RNA (1 μg) was reverse transcribed into cDNA for RACE PCR using SMARTer™ RACE cDNA Amplification kit (Clontech Laboratories, Mountain View, CA, USA). RACE PCR was performed using the Advantage® 2 PCR kit (Clontech Laboratories) with gene-specific gdh RACE primers (Table 1). Thermocycling conditions were: 5 cycles at 94°C for 30 s, 72°C for 4 min, continued with 5 cycles at 94°C for 30 s, 70°C for 30 s, 72°C for 4 min, and then followed by 30 cycles at 94°C for 30 s, 68°C for 30 s, 72°C for 4 min. RACE PCR products were separated by 1% agarose gel electrophoresis and the distinct bands observed were excised from the gel. The RACE PCR products were purified from the gel slices and quantified as stated above.
RACE PCR products were cloned using pGEM Easy T/A cloning kit (Promega Corporation), transformed into Escherichia coli JM109 strain, and plated onto Luria–Bertani agar with ampicillin, 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside and isopropyl-beta-thio galactopyranoside. Subsequently, white colonies were selected and grown overnight in LB broth with ampicillin. The plasmids were extracted using the resin-based plasmid miniprep kit (Axygen Biosciences, Union City, CA, USA) and quantified by spectrophotometry. Cloned gdh RACE PCR products were then sequenced.
Phylogenetic Analysis
The translated Gdh sequence from M. albus was aligned with 25 other Gdh protein sequences using ClustalX version 2.0.12 (Larkin et al., 2007). The sequences used for comparison and their respective accession number in either GenBank or Ensembl databases were as follows: Oncorhynchus mykiss Gdh1a (AAM73777.1) and Gdh1b (AAM73775.1), Danio rerio Gdh1a (NP_955839.2) and Gdh1b (NP_997741.1), Salmo salar Gdh1a1 (CAD58714.1), Gdh1a2 (CAD89353.1), and Gdh1b (CAD58715.1), Tribolodon hakonensis Gdh1a (BAD83654.1), Chaenocephalus aceratus Gdh1a (P82264.1), Litopenaeus vannamei Gdh (ACC95446.1), Xenopus laevis GDH1 (NP_001087023.1), X. (Silurana) tropicalis GDH1 (NP_001011138.1), Gallus gallus GDH1 (P00368.1), Rattus norvegicus GDH1 (NP_036702.1), Mus musculus GDH1 (NP_032159.1), Bos taurus GDH1 (AAI03337.1), Homo sapiens GLUD1 (NP_005262.1) and GLUD2 (NP_036216.2), Takifugu rubripes Gdh1a (ENSTRUP00000000720) and Gdh1b (ENSTRUP00000009100), Tetraodon nigroviridis Gdh1a (ENSTNIP00000008014) and Gdh1b (ENSTNIP00000016349), and Taeniopygia guttata GDH1 (ENSTGUP00000005951). Protein sequences for B. sinensis Gdh1a and Gdh1b were obtained from Peh (2008). A number of the current gene and protein names in the databases do not reflect co-orthologous relationships to the human gene or paralogous relationships. Therefore, the author has attempted to reflect these relationships by renaming (where applicable) the protein names presented in the phylogenetic tree following the method suggested by Taylor et al. (2001). The PHYLIP package (version 3.5) was then used to carry out neighbor-joining phylogenetic analysis (Saitou and Nei, 1987), with a bootstrapping resampling option to assess the support for nodes (100 pseudoreplicates). The Pacific white shrimp L. vannamei was specified as the outgroup and the input order of species was randomized where possible.
Quantitative Real-Time PCR (qPCR)
The primers used for qPCR (Table 1) were designed using PerlPrimer version 1.1.18 (Marshall, 2004). Primers for the housekeeping gene actin (Table 1) were designed based on the M. albus actin sequence (AY647143.1) deposited in GenBank. All primer pairs produced amplicons of the predicted size. The PCR products from each primer pair were subjected to 2% agarose gel electrophoresis to confirm amplification specificity. The bands were excised from the gel and PCR products purified, quantified, and then sequenced as described above.
Total RNA samples (N = 4 for each organ and each condition) were subjected to DNase treatment using deoxyribonuclease I (Sigma-Aldrich Co. St, Louis, MO, USA). Total RNA (1 μg) from each treated sample was then reverse transcribed using random hexamer primers and the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas Inc.). The qPCR reactions were performed in triplicates using 1 ng of cDNA template, 0.4 μmol l−1 of each primer, and 5 μl of Fast SYBR® Green Master Mix (Applied Biosystems Inc.) in a volume of 10 μl. Reactions were analyzed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems Inc.) and cycling conditions were: 95°C for 20 s, 50 cycles at 95°C for 3 s followed by 60°C for 30 s. A melt curve analysis was carried out after each run with the following conditions: 95°C for 15 s, increasing temperature from 60 to 95°C in 1 h, ending with 95°C for 15 s. No primer dimers were detected. Relative standard curves were generated by 10-fold serial dilutions of cDNA from the liver, intestine, and brain of M. albus exposed to freshwater conditions. Amplification efficiencies of gdh1a and actin ranged between 85 and 90%. The amplification efficiencies of the target (gdh1a) and reference gene (actin) must be approximately equal for the ΔΔCt calculation to be valid (Livak and Schmittgen, 2001). Thus, plots of the log cDNA dilution versus ΔCt were made and the absolute values of the slopes (−0.05 for liver; 0.03 for intestine; 0.06 for brain) were verified to be close to zero. ΔΔCt calculation for relative quantification of target gene with reference to actin was applied to obtain the fold change in gene expression. All data were normalized to the abundance of actin mRNA. Results were expressed in fold change but statistical analyses were performed after log2 transformation of the fold change values.
Determination of Gdh Activity
Tissue samples (N = 4 for each organ and each condition) were homogenized three times in 5 volumes (w/v) of ice-cold extraction buffer containing 50 mmol l−1 imidazole-HCl (pH 7.0), 50 mmol l−1 sodium fluoride, 3 mmol l−1 EGTA, and 3 mmol l−1 EDTA at 24,000 r.p.m. for 20 s each with a 10-s off interval using an Ultra-Turrax homogenizer. The homogenates were centrifuged at 10,000 × g and 4°C for 20 min. The supernatant obtained were passed through a 5-ml Econo-Pac 10DG desalting column (Bio-Rad Laboratories, Hercules, CA, USA) equilibrated with 50 mmol l−1 imidazole-HCl (pH 7.0) and eluted with the same buffer. The resulting eluents were used for the determination of Gdh activities. The dilution involved was corrected by monitoring the change in protein concentrations of the sample before and after passing through the column. Protein was determined according to the method of Bradford (1976). Bovine gamma globulin was used as a standard for comparison. The amination activity of GDH was determined according to Peng et al. (1994), and was expressed as micromole NADH oxidized per minute per gram tissue.
Statistical Analyses
Results were presented as mean ± SD. Student’s t-test and one-way analysis of variance (ANOVA) followed by multiple comparison of means by the Tukey test were used to evaluate differences between means where applicable. Differences were regarded as statistically significant at p < 0.05.
Results
gdh Nucleotide and the Translated Gdh Sequences
At the beginning, partial fragments of gdh were obtained from the liver, intestine, and brain of control M. albus kept in freshwater or fish exposed to terrestrial conditions, environmental ammonia, or salinity stress through PCR using four pairs of gdh primers. There was no variation in the partial gdh sequences between tissues or between conditions. Furthermore, there was no sequence variation among the 5′ RACE products or the 3′ RACE products generated from the cDNA of the three organs. The complete gdh gene sequence (GenBank accession no.: JF694445) obtained from the liver of M. albus consisted of a 133-bp 5′ UTR, a complete coding sequence (CDS) region spanning 1629 bp and a 3′ UTR of approximately 717 bp (Figure 1). The same complete sequence was obtained for gdh from the intestine and brain of M. albus. In addition, there was no variation in the complete gdh sequences from all three tissues of fish exposed to terrestrial conditions, environmental ammonia, or salinity stress. The translated Gdh amino acid sequence obtained from the liver of M. albus had 542 residues (Figure 1), and a comparison with those of other animals revealed that it is closely related to Gdh1a (Table 2). From the alignment of the deduced M. albus Gdh1a with Gdh from C. aceratus, O. mykiss, T. hakonensis, X. laevis, and H. sapiens, it is evident that Gdh is highly conserved across species (Figure 2).

Figure 1. The complete nucleotide sequence and the corresponding translated amino acid sequence of the complete coding sequence (CDS) of glutamate dehydrogenase 1a (gdh1a; GenBank accession no. JF694445) from the liver of Monopterus albus. “*” Indicates the stop codon. The start and the end of the CDS are indicated in boldface type, and the priming positions of the RACE primers used are underlined and indicated in boldface type. Pentameric motifs corresponding to AU-rich elements (AREs) are highlighted in gray.

Figure 2. The alignment of the translated amino acid sequence of glutamate dehydrogenase 1a (Gdh1a) from the liver of Monopterus albus and the amino acid sequences of Gdh in Chaenocephalus aceratus (P82264.1), Oncorhynchus mykiss (NP_001166000.1), Tribolodon hakonensis (BAD83654.1), Xenopus laevis (NP_001087023.1), and Homo sapiens (NP_005262.1). Identical residues in the alignment are indicated by “*”; similar amino acids in the alignment are indicated by “:”; dissimilar amino acids in the alignment are indicated by “.”. Residues involved in adenine binding domain are boxed; residues contributing to the antenna domain are shaded gray.
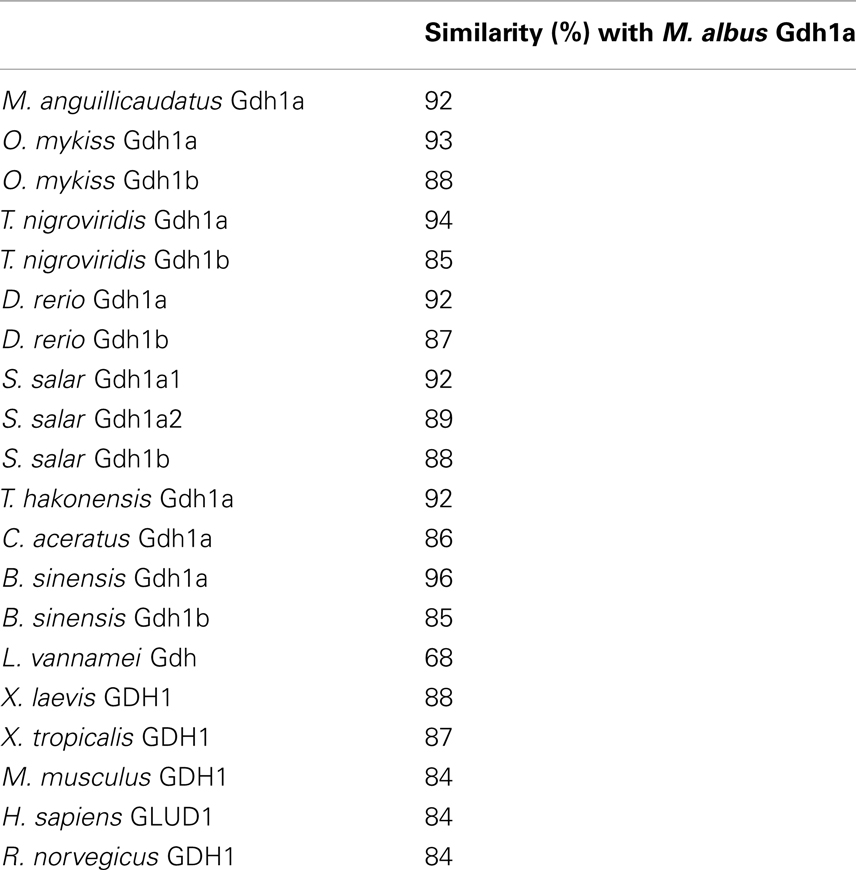
Table 2. Percentage similarity of GDH/Gdh from various organisms with Monopterus albu s Gdh1a (GenBank accession no. JF694445) obtained using Cluster W multiple alignment.
Phylogenetic Analysis
Monopterus albus Gdh1a formed a monophyletic clade with B. sinensis Gdh1a, T. nigroviridis Gdh1a, T. rubripes Gdh1a, C. aceratus Gdh1a, S. salar Gdh1a1 and Gdh1a2, and O. mykiss Gdh1a (Figure 3). Danio rerio Gdh1a and Gdh1b, T. hakonensis Gdh1a, B. sinensis Gdh1b, T. nigroviridis Gdh1b, S. salar Gdh1b, and O. mykiss Gdh1b diverged from M. albus Gdh1a (Figure 3) and generally exhibited lower sequence identity scores (<90%) with M. albus Gdh1a.
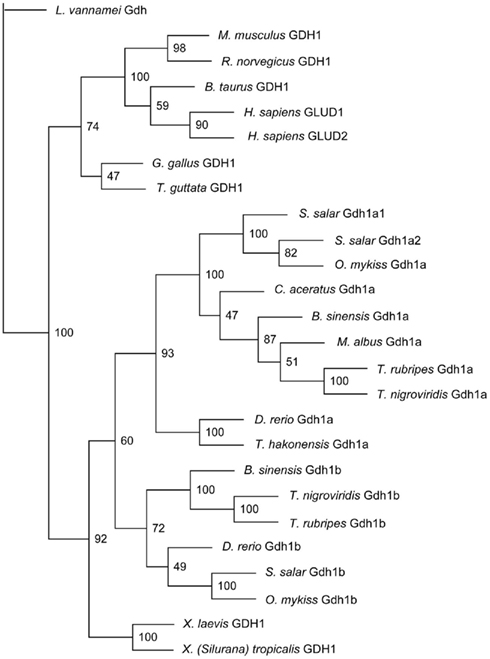
Figure 3. The phylogenetic tree of several vertebrate glutamate dehydrogenase (Gdh) protein sequences and Monopterus albus Gdh1a sequence. Litopenaeus vannamei Gdh sequence was used as the outgroup. Bootstrap values are indicated at the nodes of tree branches. The sequences used in the tree and their respective accession number in either GenBank or Ensembl databases were presented in the Section “Materials and Methods.”
Effects of Various Environmental Conditions on the MRNA Expression of Gdh1a in the Liver, Intestine, and Brain
The mRNA expression of gdh1a increased significantly by 2.6 and 3.2-fold in the liver of M. albus exposed to terrestrial conditions or 75 mmol l−1 NH4Cl, respectively, for 1 day (Figure 4A), but it returned to the control level after 6 days of exposure to both conditions (Figure 4B). There was no significant change in the mRNA expression of gdh1a in the liver of fish exposed to brackish water for 1 day (Figure 4C). Exposure of fish to terrestrial conditions or 75 mmol l−1 NH4Cl did not result in any significant changes in the mRNA expression of gdh1a in the intestine (Figures 5A,B). In contrast, there was a significant increase (2.5-fold) in the mRNA expression of gdh1a in the intestine of fish exposed to brackish water for 1 day (Figure 5C). On the other hand, the mRNA expression of gdh1a remained unchanged in the brain of M. albus exposed to all three experimental conditions (Figure 6).
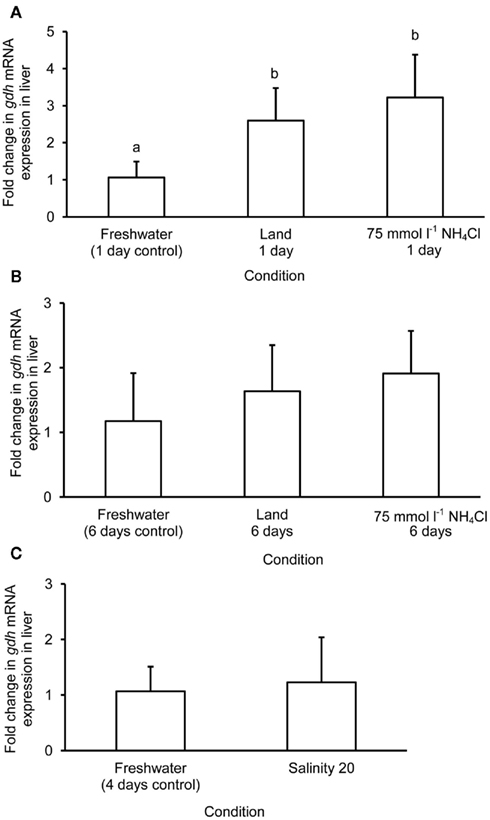
Figure 4. Effects of (A) 1 day of exposure to freshwater (control), terrestrial conditions, or 75 mmol l−1 NH4Cl, (B) 6 days of exposure to freshwater (control), terrestrial conditions (land), or 75 mmol l−1 NH4Cl, and (C) 4 days of exposure to freshwater (control) or 1 day of exposure to salinity 20 after a 3-day progressive increase in salinity from freshwater on the mRNA expression (fold change) of glutamate dehydrogenase 1a (gdh1a) in the liver of Monopterus albus. Results represent mean + SD (N = 4). Statistical analysis was applied after log2 transformation of the individual fold change values. Means not sharing the same letter are significantly different, p < 0.05.
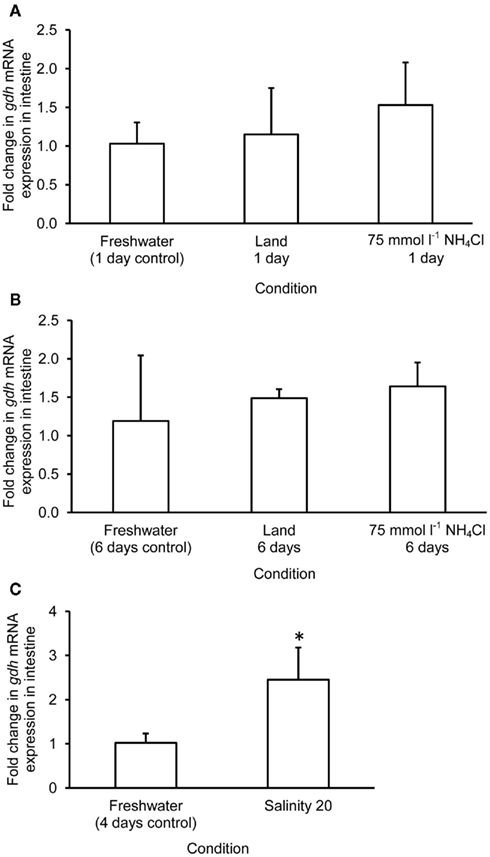
Figure 5. Effects of (A) 1 day of exposure to freshwater (control), terrestrial conditions, or 75 mmol l−1 NH4Cl, (B) 6 days of exposure to freshwater (control), terrestrial conditions (land), or 75 mmol l−1 NH4Cl, and (C) 4 days of exposure to freshwater (control) or 1 day of exposure to salinity 20 after a 3-day progressive increase in salinity from freshwater on the mRNA expression (fold change) of glutamate dehydrogenase 1a (gdh1a) in the intestine of Monopterus albus. Results represent mean + SD (N = 4). Statistical analysis was applied after log2 transformation of the individual fold change values. *Significantly different from corresponding control, p < 0.05.
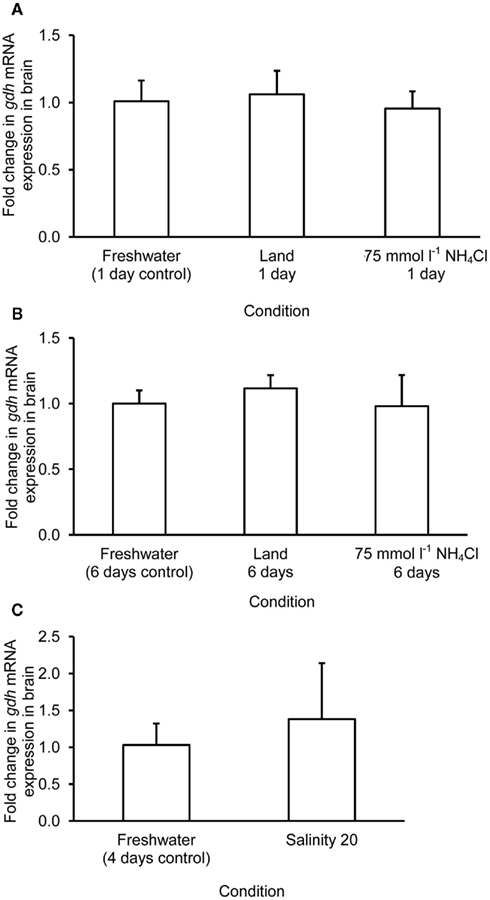
Figure 6. Effects of (A) 1 day of exposure to freshwater (control), terrestrial conditions, or 75 mmol l−1 NH4Cl, (B) 6 days of exposure to freshwater (control), terrestrial conditions (land), or 75 mmol l−1 NH4Cl, and (C) 4 days of exposure to freshwater (control) or 1 day of exposure to salinity 20 after a 3-day progressive increase in salinity from freshwater on the mRNA expression (fold change) of glutamate dehydrogenase 1a (gdh1a) in the brain of Monopterus albus. Results represent mean + SD (N = 4). Statistical analysis was applied after log2 transformation of the individual fold change values.
Effects of Various Environmental Conditions on the Amination Activity of Gdh from the Liver, Intestine, and Brain
There was a significant increase in the amination activity of Gdh from the liver of M. albus after 1 day of exposure to terrestrial conditions or 75 mmol l−1 NH4Cl, but the activity returned to the control level on day 6 (Table 3). One day or 6 days of exposure to terrestrial conditions or 75 mmol l−1 NH4Cl had no significant effect on the intestinal Gdh amination activity. As for the brain, there were no changes in the amination activity of Gdh except for fish exposed to 75 mmol l−1 NH4Cl for 6 days whereby the activity decreased significantly (Table 3). Exposure to brackish water had no significant effects on the amination activity of Gdh from the liver and brain of M. albus, but resulted in a significant increase (threefold) in the intestinal Gdh amination activity (Table 4).
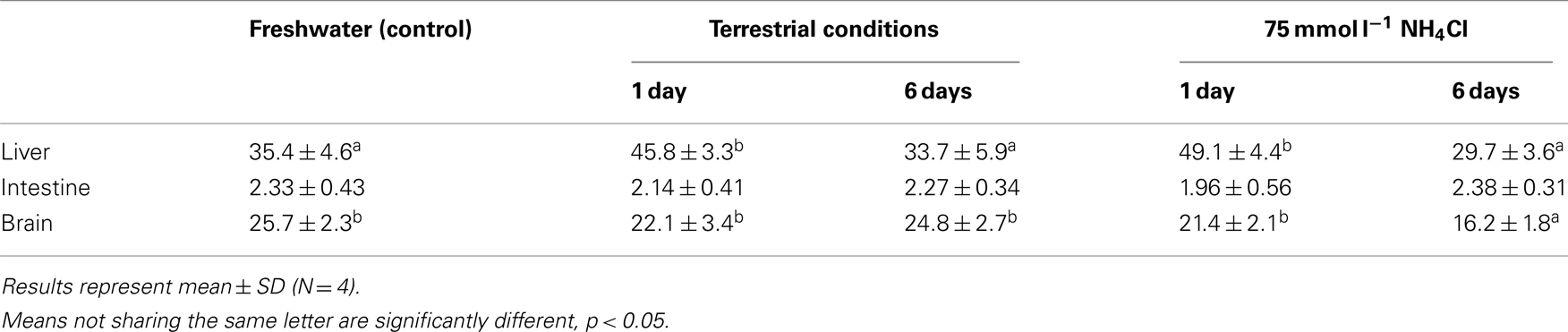
Table 3. The amination activity (μmol NADH oxidized min− 1 g− 1 tissue) of glutamate dehydrogenase from the liver, intestine, and brain of Monopterus albu s kept in freshwater (control) or after 1 day or 6 days of exposure to terrestrial conditions or 75 mmol l− 1 NH4Cl.
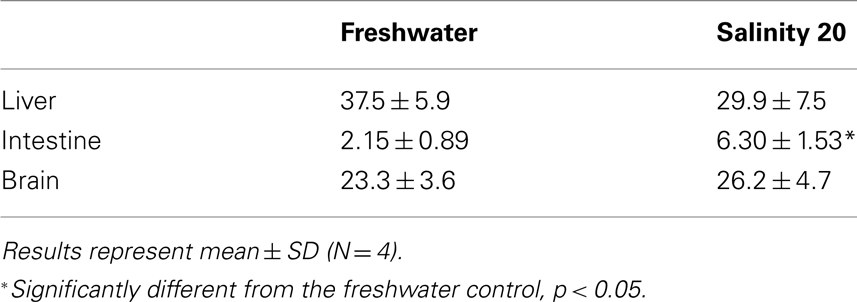
Table 4. The amination activity (μmol NADH oxidized min−1 g−1 tissue) of glutamate dehydrogenase from the liver, intestine, and brain of Monopterus albu s kept in freshwater (control) or in water at salinity 20 for 1 day after a 3-day progressive increase in salinity from freshwater.
Discussion
Expression of Gdh1a in the Liver, Intestine, and Brain of M. albus
Two forms of GDH, GLUD1 (ubiquitous), and GLUD2 (nerve tissue specific) are expressed in human (Mavrothalassitis et al., 1988; Shashidharan et al., 1994, 1997). The hibernating Richardson’s ground squirrel also expresses two distinct forms of GDH (Thatcher and Storey, 2001). Similarly, two forms of gdh have been identified in D. rerio (Woods et al., 2005) and B. sinensis (Peh, 2008), and multiple isoforms of gdh in O. mykiss (AAM73775.1, AAM73776.1, AAM73777.1) and S. salar (CAD89353.1, CAD58714.1, CAD58715.1, CAD58716.1) can be found in GenBank. In contrast, only one form of gdh is expressed in the gills of T. hakonensis. In addition, the presence of only one form of Gdh in the liver of C. aceratus (Ciardiello et al., 2000) implies the expression of a single gdh. In this study, we identified only gdh1a from the liver, intestine, and brain of M. albus, but the possibility of the presence of other gdh isoforms, especially in other organs, cannot be ignored. The translated Gdh1a of M. albus had high sequence identity, and was grouped together, with those of T. nigroviridis, T. Rubripes, B. sinensis, C. aceratus, S. salar, and O. mykiss, and by coincidence, these fishes are capable of inhabiting waters of high salinities.
In general, the Gdh amino acid sequences of M. albus and other fishes, and those of amphibians and mammals are highly conserved. The activator of GDH, ADP, is noted to have varying stimulatory effects for different GDH proteins or isoforms (Cho et al., 1995; Ciardiello et al., 2000; Plaitakis and Zaganas, 2001; Plaitakis et al., 2003). An adenine binding domain consisting of 14 consecutive amino acid residues exists within the ADP binding site in GDH, in which the Tyr187 residue is shown to be essential for GDH activation by ADP (Yoon et al., 2002). In M. albus, the amino acids contributing to the adenine binding domain and the antenna domain were identical to those in T. hakonensis and C. aceratus, but differed from that of O. mykiss Gdh1b and X. laevis GDH1. The GDH antenna domain, present only in GDH from multicellular animals, is apparently involved in allosteric regulation (Banerjee et al., 2003). Variations in the amino acid sequences in this region may confer selective advantages, allowing the GDH isoform to cater to the metabolic needs of an organism (Banerjee et al., 2003; Choi et al., 2007). Hence, it is probable that the allosteric regulation of Gdh1a in M. albus would bear some similarity with those in T. hakonensis and C. aceratus.
Differential Regulation of gdh1a mRNA Expression in the Liver, Intestine, and Brain of M. albus Exposed to Terrestrial Conditions or Environmental Ammonia
The mRNA expression of gdh1a in the liver of M. albus exposed to terrestrial conditions or 75 mmol l−1 NH4Cl for 1 day increased significantly, but returned to control levels on day 6. GDH is generally crucial to the regulation of amino acid catabolism and ammonia production. Excess amino acids from diet proteins are preferentially degraded, and their carbon skeletons can be channeled directly into the tricarboxylic acid cycle or converted to glucose through gluconeogenesis in the liver. The first step in amino acid catabolism in the liver involves the removal of the α-amino nitrogen as ammonia, and many amino acids are oxidatively deaminated by GDH after transamination (Campbell, 1991). Since M. albus were unfed during the experimental period, it is unlikely that the initial increase in gdh1a mRNA expression was to facilitate glutamate catabolism. In fact, it has been established that M. albus suppresses amino acid catabolism and detoxifies ammonia to glutamine during terrestrial and ammonia exposure (Tay et al., 2003; Ip et al., 2004c). Hence, the initial increase in gdh1a mRNA expression could be a response to increase glutamate production to support the increased synthesis of glutamine catalyzed by GS. It has been demonstrated in mammals that hepatic GDH mRNA expression is up-regulated in the pericentral zones, resulting in increases in GDH activity and consequently glutamate production in support of the increased detoxification of ammonia to glutamine after feeding (Boon et al., 1999). In fish, such as the scale-less carp Gymnocypris przewalskii (Wang et al., 2003) and the air-breathing catfish Clarias batrachus (Saha et al., 2000), hepatic Gdh activities are up-regulated following exposure to ammonia. Thus, the initial up-regulation of hepatic gdh1a mRNA expression in the liver of M. albus exposed to terrestrial conditions or ammonia could be an adaptation to increase glutamate synthesis in preparation of the detoxification of ammonia to glutamine.
Tay et al. (2003) and Ip et al. (2004c) reported that there were significant changes in the Gdh activity in the liver of M. albus after 6 days of exposure to terrestrial conditions and environmental ammonia, respectively, but they did not examine effects of a shorter period of exposure to these conditions. Results obtained from this study confirmed that an increase in Gdh amination activity occurred in the liver of M. albus after 1 day of exposure to terrestrial conditions or environmental ammonia. However, both hepatic gdh1a mRNA expression and hepatic Gdh amination activity returned to control level on day 6. Post-transcriptional control through the regulation of mRNA stability and translation are important in mediating gene expressions (Ross, 1995; Mitchell and Tollervey, 2000, 2001; Guhaniyogi and Brewer, 2001). An AU-rich element (ARE) is defined as a region with frequent adenine and uridine bases in the 3′ UTR of mRNA. It usually targets the mRNA for degradation (Chen and Shyu, 1995; Wilson and Brewer, 1999; Barreau et al., 2005), although there are a few instances where ARE-binding proteins may stabilize the mRNA transcript (Schroeder et al., 2003; Barreau et al., 2005). Present in the 3′ UTR of gdh1a from M. albus are random ATTTA pentamers (Figure 1) that corresponds to AUUUA pentamers, a type of Class I AREs (Chen and Shyu, 1995). The existence of these AREs in M. albus suggested that increases in gdh1a mRNA transcripts in response to 1 day of exposure to terrestrial conditions or 75 mmol l−1 NH4Cl were probably transient, corroborating the observation that the mRNA expression of gdh1a mRNA returned to control level on day 6.
As for the intestine of M. albus, the mRNA expression of gdh1a and Gdh amination activity remained unchanged after 1 day or 6 days of exposure to terrestrial conditions or 75 mmol l−1 NH4Cl, which is in agreement with previous reports that 6 days of emersion or ammonia exposure have no significant effect on the intestinal Gdh activity in M. albus (Tay et al., 2003; Ip et al., 2004c). By contrast, Peh et al. (2010) reported that there were significant increases in the ammonia content, and amination and deamination activities and protein abundance of Gdh, in the intestine of B. sinensis exposed to 15 mmol l−1 NH4Cl in seawater (pH 7.0). The excess glutamate formed in the intestine was apparently channeled into other amino acids and/or transported to other organs. However, when B. sinensis was exposed to a much higher concentration (30 mmol l−1) of NH4Cl in water at salinity 50 (pH. 7.0), the magnitude of increase in ammonia content in the intestine was less prominent, and there were no changes in activities and kinetic properties of intestinal Gdh. Therefore, Peh et al. (2010) proposed that intestinal Gdh could be involved in the defense against ammonia toxicity during exposure to ammonia in a hyperosmotic medium, whereby drinking was essential for osmoregulation. Since M. albus would refrain from imbibing water in a freshwater environment, the lack of effects of ammonia exposure on the mRNA expression of gdh1a and on the Gdh amination activity in its intestine is in support of the proposition of Peh et al. (2010).
One day or 6 days of terrestrial or ammonia exposure had no significant effects on the mRNA expression of gdh1a in the brain of M. albus. Indeed, there was no change in the Gdh amination activity from the brain of M. albus exposed to terrestrial conditions. However, in agreement with the report of Ip et al. (2004c), our results indicated that 6 days of exposure to ammonia led to a significant decrease in brain Gdh activity in M. albus. Hence, it is logical to deduce that a change in the protein expression of Gdh at the translation level could have occurred in the brain of fish after 6 days of exposure to 75 mmol l−1 NH4Cl.
Up-Regulation of gdh1a mRNA Expression in the Intestine of M. albus Exposed to Brackish Water
It has been reported recently that M. albus exposed to brackish water accumulates glutamine as a major organic osmolyte for cell volume regulation (Tok et al., 2009). The activity and protein abundance of GS increase significantly in the liver and muscle of fish exposed to brackish water. Increased production of glutamine requires an increased supply of glutamate, and indeed our results indicated that a significant increase in gdh1a mRNA expression occurred in the intestine of M. albus exposed to brackish water, which corroborates the findings of increased intestinal Gdh amination activity in this and previous (Tok et al., 2009) studies. Since the juvenile marble goby, Oxyeleotris marmorata, also up-regulates intestinal Gdh activity and protein abundance during seawater acclimation (Chew et al., 2010), it can be concluded that the intestines of some euryhaline fishes have an important osmoregulatory function of upregulating glutamate synthesis in response to salinity stress. The excess glutamate could be transported to other organs in support of increased glutamine synthesis for cell volume regulation.
Conclusion
The liver, intestine, and brain of M. albus expressed gdh1a, the translated amino acid of which had the greatest sequence identity with O. mykiss Gdh1a, S. salar Gdh1a1, B. sinensis Gdh1a, and T. hakonensis Gdh1a. The mRNA expression of gdh1a was differentially up-regulated in the liver and intestine of M. albus in response to ammonia toxicity (terrestrial conditions or environmental ammonia) and salinity stress (brackish water), respectively. It is probable that the corresponding increases in Gdh amination activities in these two organs would lead to increases in the production of glutamate to support increased glutamine synthesis for ammonia detoxification or cell volume regulation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported in part by the Singapore Ministry of Education through a grant (R154-000-470-112) administered to Yuen K. Ip.
References
Banerjee, S., Schmidt, T., Fang, J., Stanley, C. A., and Smith, T. J. (2003). Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation. Biochemistry 42, 3446–3456.
Barreau, C., Paillard, L., and Osborne, H. B. (2005). AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150.
Boon, L., Geerts, W. J. C., Lamers, A. J. W. H., and Van Noorden, C. J. F. (1999). High protein diet induces pericentral glutamate dehydrogenase and ornithine aminotransferase to provide sufficient glutamate for pericentral detoxification of ammonia in rat liver lobules. Histochem. Cell Biol. 111, 445–452.
Bradford, M. M. (1976). A rapid and sensitive method of the quantitation of microgram quantities of protein utilizing the principle of protein- dye binding. Anal. Biochem. 72, 248–254.
Campbell, J. W. (1991). “Excretory nitrogen metabolism,” in Comparative Animal Physiology, Environmental and Metabolic Animal Physiology, 4th Edn, ed. C. L. Prosser (New York: Wiley-Liss Inc.), 277–324.
Chen, C. Y., and Shyu, A. B. (1995). AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470.
Chew, S. F., Gan, J., and Ip, Y. K. (2005). Nitrogen metabolism and excretion in the swamp eel, Monopterus albus, during 6 or 40 days of estivation in mud. Physiol. Biochem. Zool. 78, 620–629.
Chew, S. F., Tng, Y. Y. M., Wee, N. L. J., Tok, C. Y., Wilson, J. M., and Ip, Y. K. (2010). Intestinal osmoregulatory acclimation and nitrogen metabolism in juveniles of the freshwater marble goby exposed to seawater. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 180, 511–520.
Chew, S. F., Wilson, J. M., Ip, Y. K., and Randall, D. J. (2006). “Nitrogenous excretion and defense against ammonia toxicity,” in Fish Physiology, Vol. 21, The Physiology of Tropical Fishes, eds A. Val, V. Almedia-Val, and D. J. Randall (New York: Academic Press), 307–395.
Cho, S., Lee, J., and Choi, S. Y. (1995). Two soluble forms of glutamate dehydrogenase isoproteins from bovine brain. Eur. J. Biochem. 233, 340–346.
Choi, M. M., Kim, E. A., Yang, S. J., Choi, S. Y., Cho, S. W., and Huh, J. W. (2007). Amino acid changes within antenna helix are responsible for different regulatory preferences of human glutamate dehydrogenase isozymes. J. Biol. Chem. 282, 19510–19517.
Ciardiello, M. A., Camardella, L., Carratore, V., and di Prisco, G. (2000). L-Glutamate dehydrogenase from the antarctic fish Chaenocephalus aceratus Primary structure, function and thermodynamic characterisation: relationship with cold adaptation. Biochim. Biophys. Acta 1543, 11–23.
Freney, J. R., Denmead, O. T., Watanabe, I., and Craswell, E. T. (1981). Ammonia and nitrous oxide losses following applications of ammonium sulfate to flooded rice. Aust. J. Agric. Res. 32, 37–45.
Graham, J. B. (1997). Air-breathing Fishes: Evolution, Diversity and Adaptation. London: Academic Press.
Guhaniyogi, J., and Brewer, G. (2001). Regulation of mRNA stability in mammalian cells. Gene 265, 11–23.
Hirata, T., Kaneko, T., Ono, T., Nakazato, T., Furukawa, N., Hasegawa, S., Wakabayashi, S., Shigekawa, M., Chang, M. H., Romero, M. F., and Hirose, S. (2003). Mechanism of acid adaptation of a fish living in a pH 3.5 lake. Am. J. Physiol. Regul. Integr. Comp. Physiol. 284, R1199–R1212.
Hudson, R. C., and Daniel, R. M. (1993). L-glutamate dehydrogenases: distribution, properties and mechanism. Comp. Biochem. Physiol. 106, 767–792.
Ip, Y. K., and Chew, S. F. (2010). Ammonia production, excretion, toxicity, and defense in fish: a review. Front. Physiol. 1:134. doi: 10.3389/fphy.2010.00134
Ip, Y. K., Chew, S. F., and Randall, D. J. (2001). “Ammonia toxicity, tolerance and excretion,” in Fish Physiology, Vol. 19, eds P. A. Wright, and P. M. Anderson (New York: Academic Press), 109–148.
Ip, Y. K., Chew, S. F., Wilson, J. M., and Randall, D. J. (2004a). Defences against ammonia toxicity in tropical air-breathing fishes exposed to high concentrations of environmental ammonia: a review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 174, 565–575.
Ip, Y. K., Chew, S. F., and Randall, D. J. (2004b). Five tropical fishes, six different strategies to defend against ammonia toxicity on land. Physiol. Biochem. Zool. 77, 768–782.
Ip, Y. K., Tay, A. S. L., Lee, K. H., and Chew, S. F. (2004c). Strategies for surviving high concentrations of environmental ammonia in the swamp eel Monopterus albus. Physiol. Biochem. Zool. 77, 390–405.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408.
Marshall, O. J. (2004). PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20, 2471–2472.
Mavrothalassitis, G., Tzimagiorgis, G., Mitsialis, A., Zannis, V. I., Plaitakis, A., Papamatheakis, J., and Moschonas, N. K. (1988). Isolation and characterization of cDNA clones encoding human liver glutamate dehydrogenase: evidence for a small gene family. Proc. Natl. Acad. Sci. U.S.A. 85, 3494–3498.
Mitchell, P., and Tollervey, D. (2000). mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10, 193–198.
Peh, W. Y. X. (2008). Osmoregulation and Excretory Nitrogen Metabolism in the Euryhaline Four-Eyed Sleeper Bostrychus Sinensis Exposed to Salinity and Ammonia Stresses. Master’s thesis, National University of Singapore, Singapore.
Peh, W. Y. X., Chew, S. F., Ching, B. Y., Loong, A. M., and Ip, Y. K. (2010). Roles of intestinal glutamate dehydrogenase and glutamine synthetase in environmental ammonia detoxification in the euryhaline four-eyed sleeper, Bostrychus sinensis. Aquat. Toxicol. 98, 91–98.
Peng, K. W., Chew, S. F., and Ip, Y. K. (1994). Free amino acids and cell volume regulation in the sipunculid Phascolosoma arcuatum. Physiol. Zool. 67, 580–597.
Plaitakis, A., Spanaki, C., Mastorodemos, V., and Zaganas, I. (2003). Study of structure–function relationships in human glutamate dehydrogenases reveals novel molecular mechanisms for the regulation of the nerve tissue-specific (GLUD2) isoenzyme. Neurochem. Int. 43, 401–410.
Plaitakis, A., and Zaganas, I. (2001). Regulation of human glutamate dehydrogenases: implications for glutamate, ammonia and energy metabolism in brain. J. Neurosci. Res. 66, 899–908.
Rainboth, W. J. (1996). Fishes of the Cambodian Mekong: FAO Species Identification Field Guide for Fishery Purposes. Rome: FAO.
Saha, N., Dutta, S., and Haussinger, D. (2000). Changes in free amino acid synthesis in the perfused liver of an air-breathing walking catfish, Clarias batrachus infused with ammonium chloride: a strategy to adapt under hyperammonia stress. J. Exp. Zool. 286, 13–23.
Saitou, N., and Nei, M. (1987). The neighbour-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4, 405–425.
Schroeder, J. M., Liu, W., and Curthoys, N. P. (2003). pH-responsive stabilization of glutamate dehydrogenase mRNA in LLC-PK1-F+ cells. Am. J. Physiol. Renal Physiol. 285, F258–F265.
Shashidharan, P., Clarke, D. D., Ahmed, N., Moschonas, N., and Plaitakis, A. (1997). Nerve tissue-specific human glutamate dehydrogenase that is thermolabile and highly regulated by ADP. J. Neurochem. 68, 1804–1811.
Shashidharan, P, Michaelides, T. M., Robakis, N. K., Kretsovali, A., Papamatheakis, J., and Plaitakis, A. (1994). Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J. Biol. Chem. 269, 16971–16976.
Shoemaker, M. T., and Haley, B. E. (1993). Identification of a guanine binding domain peptide of the GTP binding site of glutamate dehydrogenase: isolation with metal-chelate affinity chromatography. Biochemistry 32, 1883–1890.
Smith, E. L., Austen, B. M., Blumenthal, K. M., and Nyc, J. F. (1975). “Glutamate dehydrogenases,” in The Enzymes, Vol. 11, ed. P. D. Boyer (New York: Academic Press), 293–367.
Tay, A. S. L., Chew, S. F., and Ip, Y. K. (2003). The swamp eel Monopterus albus reduces endogenous ammonia production and detoxifies ammonia to glutamine during aerial exposure. J. Exp. Biol. 206, 2473–2486.
Taylor, J. S., Van de Peer, Y., Braasch, I., and Meyer, A. (2001). Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1661–1679.
Thatcher, B. J., and Storey, K. B. (2001). Glutamate dehydrogenase from liver of euthermic and hibernating Richardson’s ground squirrels: evidence for two distinct enzyme forms. Biochem. Cell Biol. 79, 11–19.
Tng, Y. Y. M., Chew, S. F., Wee, N. L. J., Wong, F. K., Wong, W. P., Tok, C. Y., and Ip, Y. K. (2009). Acute ammonia toxicity and the protective effects of methionine sulfoximine on the swamp eel, Monopterus albus. J. Exp. Zool. 311A, 676–688.
Tok, C. Y., Chew, S. F., Peh, W. Y. X., Loong, A. M., Wong, W. P., and Ip, Y. K. (2009). Glutamine accumulation and up-regulation of glutamine synthetase activity in the swamp eel, Monopterus albus (Zuiew), exposed to brackish water. J. Exp. Biol. 212, 1248–1258.
Wang, Y. S., Gonzalez, R. J., Marjorie, L. P., Grosell, M., Zhang, C. G., Feng, Q., Du, J. Z., Walsh, P. J., and Wood, C. M. (2003). Unusual physiology of scale-less carp, Gymnocypris przewalskii, in Lake Qinghai: a high altitude alkaline saline lake. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 134, 409–421.
Whitehead, A., and Crawford, D. L. (2005). Variation in tissue-specific gene expression among natural populations. Genome Biol. 6, R13.
Wilson, G. M., and Brewer, G. (1999). The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62, 257–291.
Woods, I. G., Wilson, C., Friedlander, B., Chang, P., Reyes, D. K., Nix, R., Kelly, P. D., Chu, F., Postlethwait, J. H., and Talbot, W. S. (2005). The zebrafish map defines ancestral vertebrate chromosomes. Genome Res. 15, 1307–1314.
Keywords: ammonia, glutamate, glutamate dehydrogenase, Monopterus albus, nitrogen metabolism, osmoregulation, mRNA expression, swamp eel
Citation: Tok CY, Chew SF and Ip YK (2011) Gene cloning and mRNA expression of glutamate dehydrogenase in the liver, brain, and intestine of the swamp eel, Monopterus albus (Zuiew), exposed to freshwater, terrestrial conditions, environmental ammonia, or salinity stress. Front. Physio. 2:100. doi: 10.3389/fphys.2011.00100
Received: 06 September 2011; Accepted: 24 November 2011;
Published online: 13 December 2011.
Edited by:
Hans O. Poertner, Alfred Wegener Institute for Polar and Marine Research, GermanyReviewed by:
Pung P. Hwang, Academia Sinica, TaiwanJ. F. Staples, University of Western Ontario, Canada
Wayne R. Fitzgibbon, Medical University of South Carolina, USA
Copyright: © 2011 Tok, Chew and Ip. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Yuen K. Ip, Department of Biological Sciences, National University of Singapore, Kent Ridge, Singapore 117543. e-mail:ZGJzaXB5a0BudXMuZWR1LnNn
 Chia Y. Tok1
Chia Y. Tok1
