- 1State Key Laboratory of Advanced Electromagnetic Technology, School of Electrical and Electronic Engineering, Huazhong University of Science and Technology, Wuhan, Hubei, China
- 2Wuhan National High Magnetic Field Center, Wuhan, China
- 3Department of Dermatology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Recently, it is been shown that cold atmospheric pressure plasmas Cold Atmospheric Plasma effectively inactivate the 2019-nCoV virus. Despite this promising finding, the precise mechanism of this inactivation remains unclear due to the limited number of studies conducted on the subject. Consequently, this paper focuses on the spike protein, a crucial part of the novel coronavirus, and the various reactive oxygen and nitrogen species (RONS) generated by the plasma. The study employs reactive molecular dynamics simulation and ReaxFF potential to explore the reactions between the spike protein molecules and different reactive oxygen nitrogen species (including H2O2, OH, O, O3, HOONO, and 1O2). The findings suggest that when a single RONS interacts with the spike protein, 1O2 and HOONO have the most potent ability to sever the spike protein. Additionally, the combined effect of long-lived and short-lived RONS presents a more potent decomposition impact.
1 Introduction
Coronaviruses (CoVs) represent a group of enveloped viruses containing large, single-stranded positive RNA genomes [1]. Over the past 2 decades, they have been responsible for two major pandemics: Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) [2]. SARS-CoV, primarily found in bats, is thought to potentially trigger future outbreaks [3]. In 2020, a new coronavirus was officially designated as SARS-CoV-2 [4]. This virus spreads more rapidly among the population than either MERS-CoV or SARS-CoV [5, 6]. Comprising four structural proteins (spike protein, envelope protein, membrane protein, and nucleocapsid protein) along with genetic genomic RNA [7], SARS-CoV-2 relies heavily on its spike protein for infection transmission. This protein consists of two subunits, S1 and S2. The S1 subunit’s receptor binding domain attaches to the host receptor, followed by the S2 subunit fusing with the cell membrane [8]. Disrupting the spike protein could significantly help in diminishing the virus’s activity.
The term “plasma” was first introduced by Irving Langmuir in 1927 to describe ionized gas [9]. Initial applications of plasma technology primarily revolved around engineering fields such as nuclear fusion and plasma etching [10, 11]. Over the past 2 decades, cold atmospheric pressure plasma (CAP) has become a cornerstone of plasma biomedicine, with widespread use in cancer therapy, treatment of wounds and skin diseases, and disinfection of medical devices [12–14]. Numerous recent studies have explored the efficacy of discharge plasma in neutralizing coronaviruses, including SARS-CoV-2 [15–17]. These findings indicate that discharge plasma holds potential for environmental decontamination and disinfection, reducing the transmission of infectious agents like SARS-CoV-2. Nonetheless, further research is required to determine the practicality and safety of CAP implementation in real-world scenarios.
Cold atmospheric pressure plasma (CAP) is a complex mixture composed of reactive oxygen nitrogen species (RONS) (e.g., O, OH, H2O2, NO, NO2, ONOO−), ultraviolet light (UV), electrons, ions, and electric fields [14, 18–20]. Current experimental detection methods primarily demonstrate the ability of Cold Atmospheric Plasma (CAP) to eliminate bacteria, fungi, viruses, and other microorganisms [21, 22]. Additionally, some studies have explored the impact of nitro-oxidative stress on membranes and proteins [23–25]. However, elucidating the microscopic mechanisms behind CAP’s effects remains challenging. Molecular dynamics (MD) simulation technology [26], as a complement to experimental research, can simulate the physical and chemical reactions between various active substances and biological structures, leading to its growing application in plasma biomedicine.
In molecular dynamics (MD) simulations, simulations based on quantum mechanics, such as “ab initio MD,” primarily include Born-Oppenheimer MD (BOMD) and Car-Parrinello MD (CPMD) [27], both of which are founded on density functional theory (DFT). CPMD is often used in plasma medicine-related simulation studies, for instance, in the investigation of OH radical-induced DNA-Base damage [28]. This method is known for its precision and the capacity to examine reactions involving the breakage or formation of covalent bonds. However, it demands significant computational resources and typically only permits simulation of systems with around 100 atoms on a picosecond (ps) scale.
Classical MD simulations utilize force fields to define potential interatomic interactions, which are categorized into reactive and non-reactive types. Non-reactive force fields are suitable for movements of systems with a large number of atoms on a nanosecond (ns) scale. This approach has been applied to study the effects of lipid peroxidation products on cell membrane structures and the transmembrane transport of RONS [29]. Nonetheless, it cannot depict the breaking and forming of bonds during reactions. Therefore, when the specific reaction processes between RONS particles and biomolecules need to be investigated, reactive force fields are employed [30].
Employing reaction-based approaches, such as ReaxFF, to simulate chemical bond breakage and formation can help uncover the reaction mechanisms between active substances and biological structures. ReaxFF is a force field widely used for reactive systems. It was initially developed for studying complex reactions in Hydrocarbons. Remarkably, ReaxFF’s outcomes align well with thermal formation, geometric data, and quantum chemical data concerning bond dissociation in various hydrocarbon molecules. This demonstrates ReaxFF’s ability to reproduce the energies corresponding to both non-reactive and reactive behaviors of these compounds. The currently prevalent ReaxFF is an extensively expanded and improved reactive force field [31–35].
The expansion and improvement of ReaxFF in other fields also demonstrate its wide range of applications, such as in the field of plasma medicine. Utilizing ReaxFF, Bogaerts A et al. employed MD simulations to reveal that reactive oxygen species (ROS) can react with bacterial cell walls to achieve a bactericidal effect [36]. Recently, Shuhui Y. et al. Conducted MD simulations using ReaxFF, uncovering the reaction mechanism between OH radicals and bacterial biofilms [37]. Furthermore, numerous studies have employed ReaxFF-based MD simulations to investigate the microscopic principles governing interactions between active substances and biological structures, such as cell membranes and DNA. Inspired by these findings, MD simulations can be employed to observe the process of CAP inactivating SARS-CoV-2 and investigate its microscopic inactivation mechanism. Simultaneously, the results derived from these simulations can be validated and complemented by macroscopic experimental observations.
RONS, a pivotal component of CAP, plays a crucial role in its ability to neutralize bacteria and viruses [38]. The spike protein, which is fundamental to the initial infection process in host cells, has significant impact on the virus’s activity [39]. Consequently, selecting the spike protein of SARS-CoV-2 to examine its interaction with RNOS provides a clear picture of how CAP deactivates SARS-CoV-2. This study employs molecular dynamics simulation to explore the intricate process of a single diverse RONS within CAP interacting with the SARS-CoV-2 spike protein, as well as the interaction between both short-lived and long-lived RONS with the spike protein simultaneously, and compared with the existing experimental results. This will shed light on the microscopic mechanisms by which CAP neutralizes SARS-CoV-2.
2 Methods and setup
2.1 Principle of ReaxFF potential
In this study, ReaxFF MD within Materials Studio is employed to investigate the interactions between various RONS and the SARS-CoV-2 spike protein, thereby elucidating the mechanism of CAP-mediated inactivation of the novel coronavirus. ReaxFF MD combines the ReaxFF chemical reaction force field with molecular dynamics simulations [31]. This reactive force field models the relationship between bond distance and bond order, as well as the connection between bond order and bond energy. This approach effectively simulates the breaking of bonds between atoms [32].
The crucial sequence, BO’ij, can be determined using the subsequent equation:
where
ReaxFF not only retains accuracy comparable to quantum mechanics but also requires minimal computational resources. Moreover, this method is applicable to systems comprising various macromolecules [40]. The overall system energy in ReaxFF can be expressed as:
Where
ReaxFF is designed to depict the stability and geometry of non-conjugated, conjugated, and radical-containing compounds, as well as the dissociation and formation of chemical bonds in hydrocarbon compounds. Its accuracy has been validated through numerous experiments [33].
2.2 Simulation model
Figure 1 presents a schematic representation of SARS-CoV-2 infecting cells through the spike protein [7]. The SARS-CoV-2 spike protein is a type I membrane protein, which can be divided into two segments: the receptor-binding fragment, responsible for recognizing and binding to the host cell receptor, and the fusion fragment, responsible for fusing with the host cell membrane [41]. The complete spike protein’s immense structure and molecular weight make it unsuitable for calculations, and its three monomers share similar structural and chemical properties [42]. To simplify the calculations, we extracted the TM domain from the fusion fragment of spike protein monomers for ReaxFF MD simulation. TM domain is a transmembrane domain anchored in the viral membrane and its chemical structure and structural model are depicted in Figure 2. The amino acid sequence, extending from the amino-terminal to the carboxy terminal, is WLGFIAGLIAIVLVTILLSST.
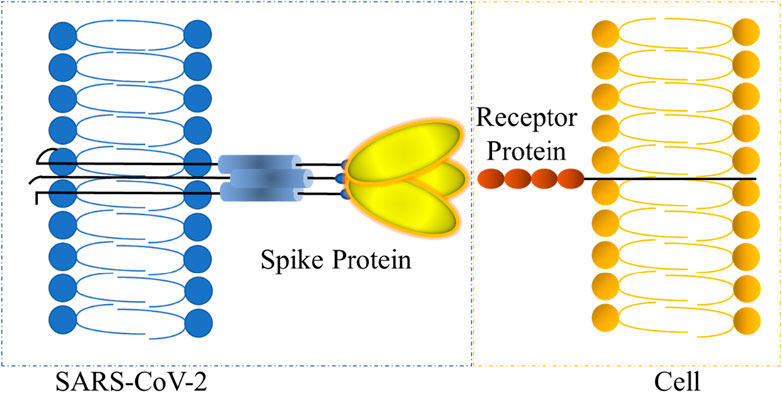
FIGURE 1. SARS-CoV-2 infects cells through the spike protein. The left is a partial map of SARS-CoV-2, and the right is a partial map of cells.
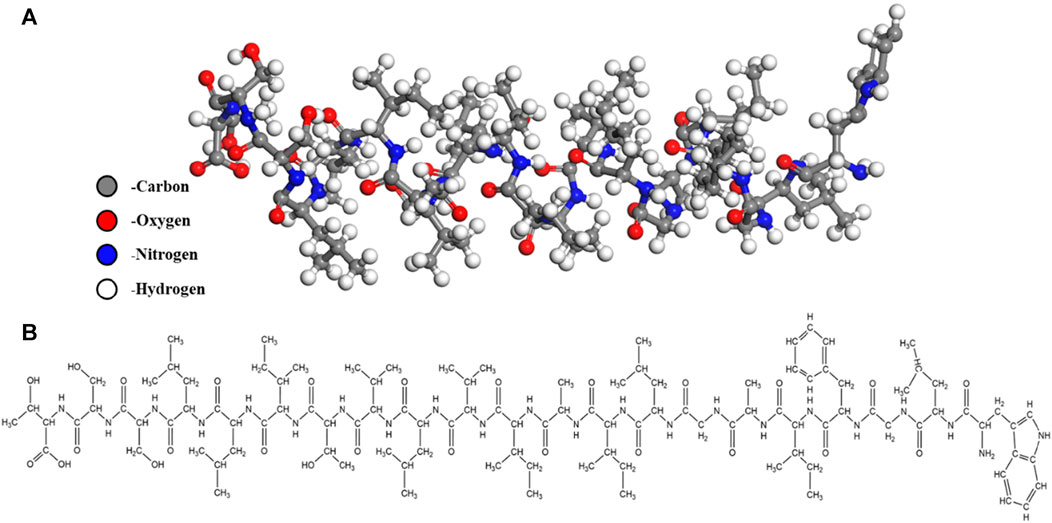
FIGURE 2. Diagram of the 7LC8 single-chain structure used in the simulation. (A) The ball-and-stick model of 7LC8 (B) The structural formula of 7LC8.
2.3 Simulation parameters
This paper delves into the reactions between varying concentrations of major RONS and SARS-CoV-2 spike protein molecules, disregarding the self-reaction among the active substances. Furthermore, as the specific reaction involving the O atom generated by CAP and the spike protein molecule has been extensively discussed in our previous paper [43], we will not elaborate on it. Instead, we will only refer to its simulation results during the result comparison phase.
Numerous studies have indicated that plasma generates varying types and amounts of RONS under different operating conditions, which can somewhat influence sterilization efficiency [44–47]. Consequently, we employed six distinct RONS, including H2O2, OH, O, O3, HOONO, and 1O2, set 1, 2, 3, 4, and 5 active particles as reactive particles to be introduced into the model system, simulating different concentrations of reactants. Although CAPs generally produce lower concentrations of reactive particles than these [48, 49], such increases are deemed acceptable after extensive experience with MD simulations to ensure adequate reaction efficiency within the system.
Initially, we positioned the geometrically optimized spike protein molecules and active particles within a 25 Å × 25 Å × 25Å box using the Amorphous Cell Calculation function and applied periodic boundary conditions to maintain a constant system density of 0.25 g/cm3. Subsequently, we employed the Geometry Optimization function in the software to optimize its geometry, enabling rapid energy minimization. Since geometric optimization does not consider thermodynamic factors such as temperature and pressure, the molecular configuration remains distant from the actual molecular state, necessitating kinetic optimization. We then set the ambient temperature to room temperature (300K) [50] and subjected the system to the COMPASSII force field, using the “macroscopic canonical ensemble” (i.e., constant temperature and volume) and the Berendsen temperature control mechanism [51] (i.e., assuming the system and a constant-temperature external heat bath were coupled, maintaining the simulated reaction process temperature at around 300K). After performing hundreds of thousands of calculations, kinetically optimized molecules were obtained.
Figures 3A, B display the molecular structure diagrams before and after optimization. Upon comparison, it is evident that the spatial structure becomes more three-dimensional. The enthalpy change of the system throughout the optimization process is illustrated in Figure 3C. Enthalpy is a thermodynamic energy state function of an object, and its magnitude can characterize the stability of the entire system: higher enthalpy indicates higher total energy and less stability, whereas lower enthalpy suggests lower total energy and relative stability. Following geometric and kinetic optimization, the enthalpy of the entire system decreased from the initial −600 to −800 kcal/mol, approximately 33% lower, exhibiting a saturation trend. This indicates that the system’s energy has been substantially reduced, and its stability has improved. The optimized structure is used as the initial state for MD simulation, the reaction force field is selected as ReaxFF6.0, the ensemble is selected as NVT, the temperature is set at 300 K, the reaction time step is set at 1fs, and the reaction lasts 200 ps, repeated the simulations for each types and concentration 50 times.
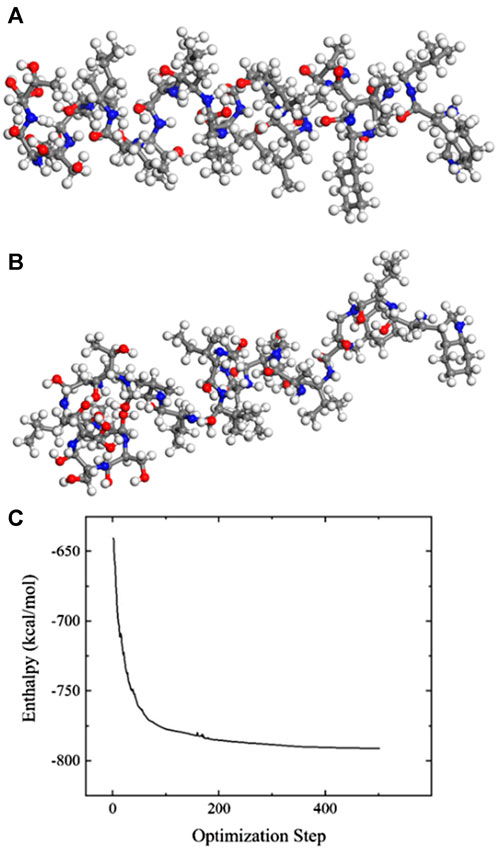
FIGURE 3. Spike protein molecular optimization results. (A) Spatial structure before optimization, (B) Spatial structure after optimization, (C) Enthalpy change during optimization.
3 Simulation results
Numerous RONS exist in CAP. To evaluate the effectiveness of distinct RONS in degrading the spike protein, we conducted molecular dynamics simulations employing individual reactants, including H2O2, OH, O, O3, HOONO, and 1O2 interacting with the spike protein. In our prior publication [43], we explored and analyzed the scenario involving O atoms as the reactant and summarized the relevant principles. The molecular structures of these RONS, represented as ball-and-stick models, can be found in Figure 4.
FIGURE 4. Ball-and-stick model of different RONS produced by CAP. (A) H2O2 (B) OH (C) O (D) O3 (E) HOONO (F) 1O2.
Initially, when a RONS as reactive particles to be introduced into the model system, the groups undergoing hydrogen abstraction were tallied, with the results displayed in Figure 5. Upon comparing these outcomes, it became evident that the methyl group (—CH3) possesses the highest likelihood of hydrogen abstraction, ranging between 50% and 60%. This was attributable to the greater prevalence of these groups in the spike protein’s molecular chain, as well as their location on the periphery, making them more likely to interact with reactive species. Conversely, the hydroxyl group (—OH) demonstrated the lowest probability of hydrogen abstraction, at less than 5%. This was because the spike protein’s molecular chain contained very few hydroxyl groups, and they possess a higher bond energy (464 kJ/mol), rendering them more stable than C-H (414 kJ/mol) and N-H (389 kJ/mol) bonds. Generally, the hydrogen abstraction probability of amino groups was lower than that of carbon groups. Although the N-H bond energy was lower than that of the C-H bond, there were fewer amino groups, and their positions were closer to the molecular chain’s center. The oxygen group exhibited the lowest probability of hydrogen abstraction.
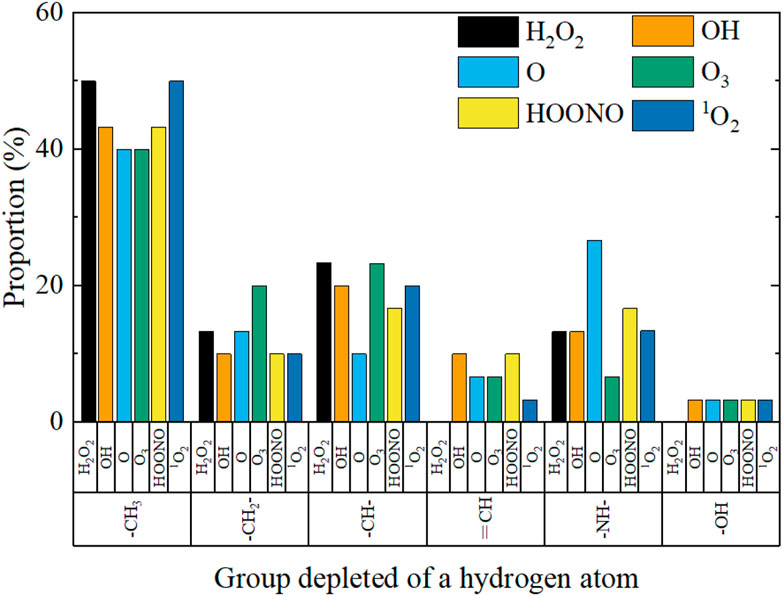
FIGURE 5. Statistics on the hydrogen abstraction of different groups during the reaction between plasma RONS and spike protein.
The results were imported into Pymol, and we generated a comparative diagram showcasing the surface structure of SARS-CoV-2 spike protein molecules before and after the reaction, as presented in. This image highlighted the varying efficiency of different RONS in destroying the spike protein. Comparing Figures 6A–G revealed that the extent of damage inflicted on the spike protein molecules varied depending on the specific active substance involved.
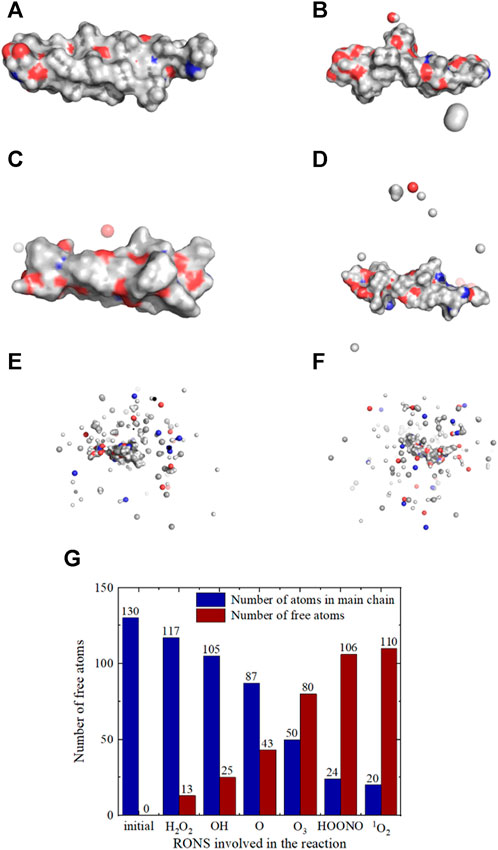
FIGURE 6. Surface structure of SARS-CoV-2 spike protein molecules after reactions with different RONS, and statistics on the destruction of spike protein molecules by RONS.(A) The structure after reacting with an H2O2, (B) The structure after reacting with an OH, (C) The structure after reaction with an O, (D) The structure after reaction with a O3, (E) The structure after reaction with a HOONO, (F) The structure after reaction with an 1O2, (G) The statistics of damage to spike protein molecules caused by different RONS.
During simulation, each C/N/O/H atom within the spike protein is named to observe the specific reaction processes between various particles and the spike protein. The study investigates the reaction mechanisms of the particles with the spike protein, explaining the representative reaction processes involved. Additionally, these representative reaction processes are identified and illustrated in the figures below.
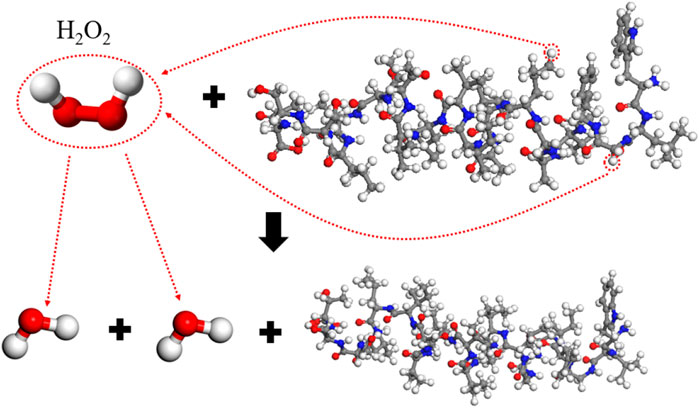
The effect of a single H2O2 molecule is the weakest, with 13 free atoms and a main chain integrity of 90.0%. By analyzing the reaction process, the reaction mechanism is illustrated as in Figure 7. When H2O2 reacts, it decomposes into two hydroxyl radicals, which each perform a hydrogen abstraction reaction on the main chain, resulting in two molecules of H2O and a largely intact main chain. In specific reactions, as mentioned before, the probability of hydrogen abstraction differs among various groups; for example, in a set of reactions, the two hydrogen abstractions occur on the -CH2- group of 3-Gly (glycine at position three in the amino acid sequence) and the -CH3 group of 8-Leu (leucine). Theoretically, the hydrogen abstraction efficiency of H2O2 should be double that of OH, but since H2O2 is relatively stable, in 50 simulations, only 30 reacted, which makes the reaction probability lower than that of OH. Therefore, the impact of H2O2 on the spike protein is slightly lower than that of OH.
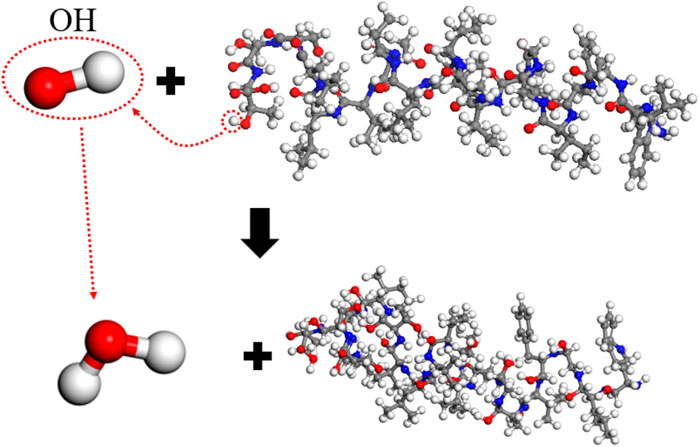
The effect of a single hydroxyl radical (OH) is similar to that of H2O2, resulting in 25 free atoms and a main chain integrity of 80.8%. The reaction mechanism, as analyzed, is depicted in Figure 8. The OH radical abstracts a hydrogen atom from the spike protein, subsequently becoming a stable water molecule, while a small number of atoms detach from the spike protein. Most hydrogen abstraction reactions break C-H bonds, reacting with the free hydrogen atom, but in some cases, the abstraction may occur on -OH groups, such as the removal of a hydrogen atom from the -OH group in 21-Thr at the end of the molecular chain. This indicates that the probability of hydrogen abstraction is somewhat related to the group’s position. OH has a higher reactivity than H2O2; during simulations, hydrogen abstraction occurs in all 50 groups, making its overall effect stronger.
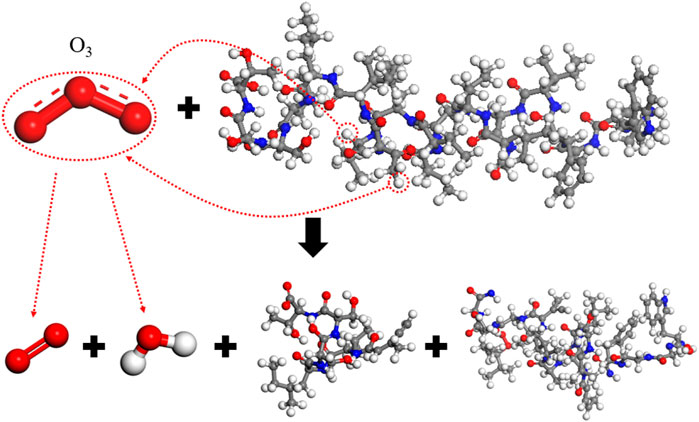
A single ozone (O3) molecule causes the spike protein molecular chain to break, resulting in 80 free atoms and a main chain integrity of 38.5%. The reaction mechanism, as analyzed, is presented in Figure 9. In the reaction with O3, hydrogen abstraction also occurs, removing two hydrogen atoms, with the end products being O2 and H2O. This leads to the breakage of the spike protein molecular chain and a significant impact on the overall structure. For example, during the reaction, hydrogen is first abstracted from the -CH- group in 14-Val and the -CH3 group in 15-Thr, which then leads to a substantial number of breaks in the surrounding C-O, C-C, and C-N bonds, compromising the integrity of the spike protein.
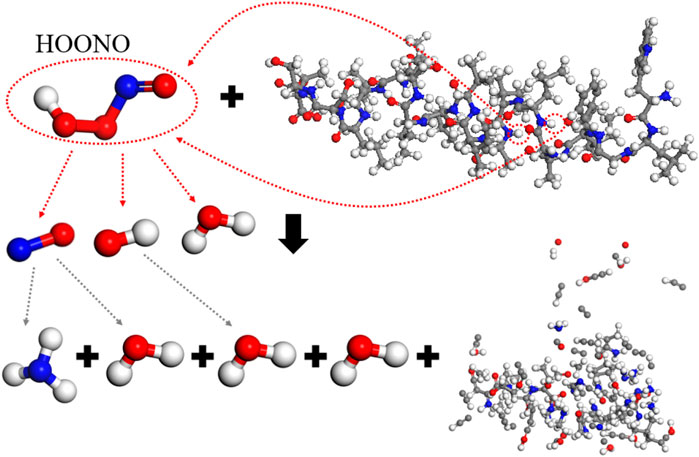
A single HOONO molecule causes extensive breakage of bonds in spike protein molecules, resulting in 106 free atoms and maintaining only 18.5% of the main chain’s integrity. Analysis of the reaction process (Figure 10) reveals that at the onset, HOONO produces NO, OH, and H2O. Both NO and OH undergo redox reactions with the spike protein, eventually transforming HOONO into NH3 and H2O, leading to the disintegration of the spike protein into smaller molecules and compromising its integrity. For example, HOONO first triggers a hydrogen abstraction reaction at the -NH- groups of 8-Leu and 11-Ile, forming H2O, NO, and OH. These then react further with the spike protein.
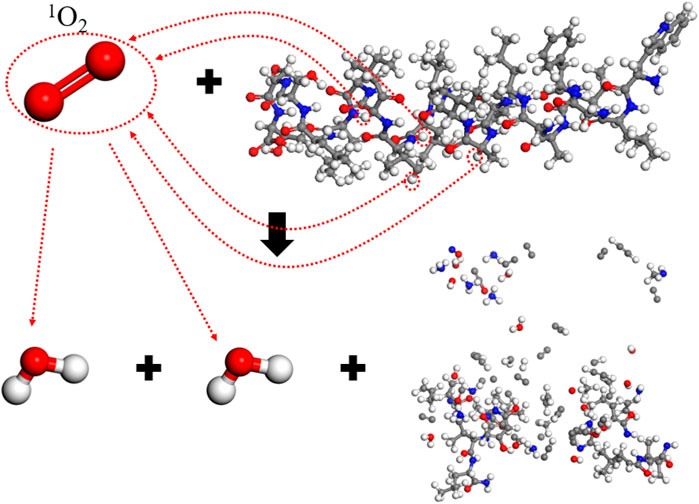
The effect of a single 1O2 molecule is similar to HOONO, with 110 free atoms and a main chain integrity of 15.4%. 1O2 can directly react with electron-rich double bonds without forming free radical intermediates. Due to the low activation energy required, singlet oxygen oxidation reactions are very rapid. Analysis of the reaction process, as shown in Figure 11, indicates that 1O2 reacts with the spike protein, resulting in two H2O molecules and the disintegration of the spike protein into many smaller molecules, damaging its integrity. During the 1O2 reaction, four hydrogen atoms are abstracted from the spike protein, causing extensive bond breakage around the dehydrogenated groups.
To visually compare the extent of damage caused by various substances to the spike protein molecule at different concentrations, we displayed the number of unbound atoms in the SARS-CoV-2 spike protein molecule after reacting with different amounts of RONS in Figure 12. The effect of H2O2 is the weakest, with damage to the spike protein reaching saturation at about 4 molecules. Although OH abstracts fewer hydrogen atoms, its reaction probability is relatively higher than H2O2, also reaching saturation at 4 molecules. The reaction products of O3 are similar to those of atomic oxygen, reaching saturation in damage at 3 molecules. HOONO and 1O2 cause the most significant damage, reaching saturation earlier, with spike protein damage saturating at just two molecules.
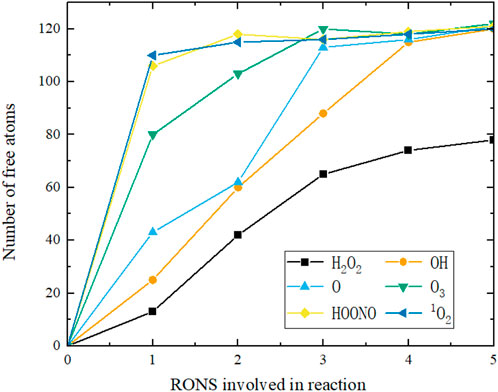
FIGURE 12. The number of free atoms of the SARS-CoV-2 spike protein molecule reacted with different amounts of RONS indicates the degree of damage to the spike protein molecule (the more free atoms, the stronger the damage of the spike protein molecule).
During the reaction between RONS and the spike protein, the number of free atoms breaking away from the main chain initially increased rapidly and then reached a saturation point. The growth in the number of free atoms indicates the ongoing reaction, and reaching the limit signifies the reaction’s endpoint. The saturation time represents the time taken for the reaction to complete. Figure 13 captures the variation in the number of free atoms over time due to a single 1O2, where the red dotted line shows that the saturation time for the number of free atoms was 73 ps, indicating that the reaction was complete at 73 ps with no further increase in free atoms. Figure 14 displays the saturation times for different RONS reactions. The reactions involving HOONO, 1O2, and O3 reach saturation more quickly compared to those with H2O2, OH, and O. This suggests that the reactions with H2O2, OH, and O are slower, taking approximately twice as long to complete compared to those with O3, HOONO, and 1O2.
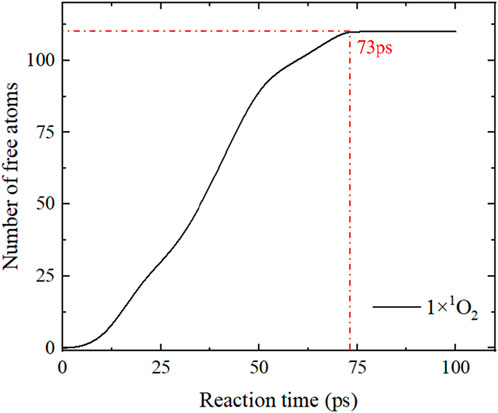
FIGURE 13. The number of free atoms dissociated from the protein backbone gradually increases with the reaction time (during the reaction between a single 1O2 and the SARS-CoV-2 spike protein); the red dotted line marks the saturation time of the number of free atoms.
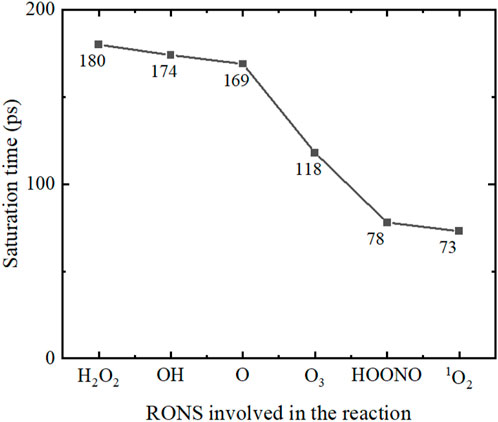
FIGURE 14. The saturation times of the reaction between different RONS and the SARS-CoV-2 spike protein.
Plasma can simultaneously produced a variety of high-density RONS, and these RONS also reacted and transformed each other, so Figure 15 systematically compared the number of free atoms and saturation time of different RONS (and their combinations). The reactivity of RONS particles is demonstrated by their interaction with the spike protein, where 2×HOONO, 2×1O2, and 2×O3 show greater potency compared to 2×H2O2, 2×OH, and 2×O in terms of the number of free atoms and reaction saturation time. Notably, combinations of different RONS particles exhibit enhanced effectiveness in reacting with the spike protein. For instance, the combination of H2O2+HOONO and H2O2+1O2, despite reaching saturation in spike protein disruption and having a less noticeable increase in free atoms, demonstrates a significantly faster reaction speed and shorter saturation time compared to 2×HOONO and 2×1O2.
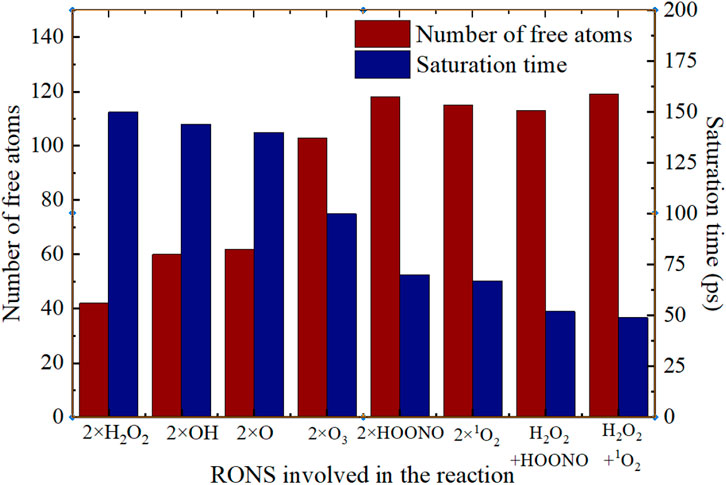
FIGURE 15. The number of free atoms and the saturation time of the reactions between different RONS (two or a combination) and the spike protein.
4 Discussions
This paper analyzes the application of molecular dynamics simulations to evaluate the effectiveness of various RONS in degrading the spike protein of SARS-CoV-2. The simulations consider individual reactants, including H2O2, OH, O, O3, HOONO, and 1O2, and their interactions with the spike protein. Previous experiments have studied the modifications of amino acids by Cold Atmospheric Plasma (CAP) [52]. Similar to experimental results, it is observed that dehydrogenation reactions are more likely to occur in branched-chain amino acids such as Valine (Val), Leucine (Leu), and Isoleucine (Ile). Furthermore, the protein structure is altered due to RONS-induced modifications, impacting the overall stability of the protein.
The degree of damage to the spike protein varies with different active substances. H2O2 and OH show weaker destructive capabilities, causing no significant structural impact on the spike protein. The reaction product of a single O3 molecule is similar to that of a single O atom; however, after O3 abstracts a hydrogen atom, it causes a breakage of chemical bonds surrounding the abstracted group, resulting in more severe damage to the spike protein. Both single HOONO and 1O2 cause substantial destruction to the integrity of the spike protein, as these particles abstract more hydrogen atoms, triggering chain reactions that disrupt molecular structures, leading to the cleavage of the spike protein, with less than 20% integrity remaining after the reaction.
A visual comparison of the damage caused to the spike protein by various substances at different concentrations shows that the destruction of the spike protein reaches saturation at different levels for each substance. When four H2O2 molecules are involved in the reaction, saturation is achieved in terms of spike protein damage. The effect of OH is slightly stronger than H2O2, also reaching saturation with four OH molecules. Both O3 and O atoms reach saturation with three molecules involved in the reaction. HOONO and 1O2 cause the most severe damage to the spike protein, reaching saturation with the involvement of just two particles.
The efficacy of disinfection and sterilization is largely due to the combined action of long-lasting and transient reactive particles in plasma. Though HOONO has a short half-life of about 1 μs, it is continually regenerated in plasma by the longer-lasting H2O2 (with a half-life of approximately 104 s), H+, and NO2− (lasting days). Similarly, 1O2 with a half-life of around 3 μs, like HOONO, is continuously produced through reactions involving H2O2 and HOONO, or by electron collision with O2, especially in environments like plasma-activated water or air plasma-treated liquids [53–55]. The combinations of H2O2+HOONO and H2O2+1O2 decrease the reaction saturation time (Figure 15), underscoring the combined potency of long-lasting and transient active particles in plasma for effectively neutralizing the virus spike protein.
5 Conclusion
The comprehensive molecular dynamics simulations revealed the varied effectiveness of different RONS, including H2O2, OH, O, O3, HOONO and 1O2, in interacting with and degrading the spike protein of SARS-CoV-2. Each RONS displayed distinct impacts, with H2O2 showing the least effect and HOONO and 1O2 causing the most significant damage to the spike protein’s structure.
The study found that the damage to the spike protein reached saturation at different concentrations for each reactant. This saturation point is important as it indicates the concentration at which maximum disruption of the spike protein can be achieved. Additionally, the time required to reach saturation, or the reaction speed, varied among the different RONS, providing a metric for determining the necessary concentration of RONS particles to inactivate the virus.
The study also emphasized the synergistic effects of various RONS, highlighting that a mix of transient and long-lasting active particles in plasma can significantly accelerate the virus inactivation process. This indicates plasma’s potential as a key tool in disinfection and sterilization processes to combat the spread of SARS-CoV-2.
From an experimental point of view, these simulation results can provide insights into the potential mechanisms of action for plasma-generated RONS against the spike protein. This knowledge can help design experiments to validate the efficacy of RONS in inactivating SARS-CoV-2, either in vitro or in vivo. Moreover, understanding the interactions between RONS and the spike protein can help identify the most effective RONS and their optimal concentrations for disinfection and environmental decontamination.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
ST: Writing–original draft, Data curation, Methodology. HZ: Data curation, Methodology, Writing–original draft. DL: Writing–review and editing, Funding acquisition, Methodology, Project administration. SZ: Writing–review and editing, Investigation, Software, Validation. HC: Formal Analysis, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 52277149) and the Interdisciplinary Program of Wuhan National High Magnetic Field Center (Grant No. WHMFC202144), Huazhong University of Science and Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lu G, Liu D. SARS-like virus in the Middle East: a truly bat-related coronavirus causing human diseases. Protein Cell (2012) 3:803–5. doi:10.1007/s13238-012-2811-1
2. Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Engl J Med (2003) 348:1967–76. doi:10.1056/NEJMoa030747
3. Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol (2019) 17:181–92. doi:10.1038/s41579-018-0118-9
4. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature (2020) 579:270–3. doi:10.1038/s41586-020-2012-7
5. Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol (2020) 79:104212. doi:10.1016/j.meegid.2020.104212
6. Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, et al. A new coronavirus associated with human respiratory disease in China. Nature (2020) 579:265–9. doi:10.1038/s41586-020-2008-3
7. Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta (Bba) - Mol Basis Dis (2020) 1866:165878. doi:10.1016/j.bbadis.2020.165878
8. Lau YL, Peiris JM. Pathogenesis of severe acute respiratory syndrome. Curr Opin Immunol (2005) 17:404–10. doi:10.1016/j.coi.2005.05.009
9. Langmuir I. Oscillations in ionized gases. Proc Natl Acad Sci (1928) 14:627–37. doi:10.1073/pnas.14.8.627
10. Ichimaru S. Nuclear fusion in dense plasmas. Rev Mod Phys (1993) 65:255–99. doi:10.1103/RevModPhys.65.255
11. Team JET. Fusion energy production from a deuterium-tritium plasma in the JET tokamak. Nucl Fusion (1992) 32:187–203. doi:10.1088/0029-5515/32/2/I01
12. Wu E, Nie L, Liu D, Lu X, Ostrikov KK. Plasma poration: transdermal electric fields, conduction currents, and reactive species transport. Free Radic Biol Med (2023) 198:109–17. doi:10.1016/j.freeradbiomed.2023.02.011
13. Liu D, Szili EJ, Ostrikov KK. Plasma medicine: opportunities for nanotechnology in a digital age. Plasma Process Polym (2020) 17:2000097. doi:10.1002/ppap.202000097
14. Liu D, Zhang Y, Xu M, Chen H, Lu X, Ostrikov KK. Cold atmospheric pressure plasmas in dermatology: sources, reactive agents, and therapeutic effects. Plasma Process Polym (2020) 17:1900218. doi:10.1002/ppap.201900218
15. Gao H, Wang G, Chen B, Zhang Y, Liu D, Lu X, et al. Atmospheric-pressure non-equilibrium plasmas for effective abatement of pathogenic biological aerosols. Plasma Sourc Sci Technol (2021) 30:053001. doi:10.1088/1361-6595/abf51b
16. Filipić A, Gutierrez-Aguirre I, Primc G, Mozetič M, Dobnik D. Cold plasma, a new hope in the field of virus inactivation. Trends Biotechnol (2020) 38:1278–91. doi:10.1016/j.tibtech.2020.04.003
17. Assadi I, Guesmi A, Baaloudj O, Zeghioud H, Elfalleh W, Benhammadi N, et al. Review on inactivation of airborne viruses using non-thermal plasma technologies: from MS2 to coronavirus. Environ Sci Pollut Res (2022) 29:4880–92. doi:10.1007/s11356-021-17486-3
18. Lee YJ, Kim SW, Jung MH, Kim KS, Suh DS, Kim KH, et al. Plasma-activated medium inhibits cancer stem cell-like properties and exhibits a synergistic effect in combination with cisplatin in ovarian cancer. Free Radic Biol Med (2022) 182:276–88. doi:10.1016/j.freeradbiomed.2022.03.001
19. Miebach L, Freund E, Clemen R, Weltmann K, Metelmann HR, von Woedtke T, et al. Conductivity augments ROS and RNS delivery and tumor toxicity of an argon plasma jet. Free Radic Biol Med (2022) 180:210–9. doi:10.1016/j.freeradbiomed.2022.01.014
20. Zou X, Xu M, Pan S, Gan L, Zhang S, Chen H, et al. Plasma activated oil: fast production, reactivity, stability, and wound healing application. ACS Biomater Sci Eng (2019) 5:1611–22. doi:10.1021/acsbiomaterials.9b00125
21. Lan C, Zhu H, Wang S, Nie L, Liu D, Shi Q, et al. Disinfection of viruses with cold atmospheric-pressure plasmas: sources, mechanisms, and efficacy. Plasma Process Polym (2023):e2300183. doi:10.1002/ppap.202300183
22. Li J, Lan C, Nie L, Liu D, Lu X. Distributed plasma-water-based nitrogen fixation system based on cascade discharge: generation, regulation, and application. Chem Eng J (2023) 478:147483. doi:10.1016/j.cej.2023.147483
23. Abduvokhidov D, Yusupov M, Shahzad A, Attri P, Shiratani M, Oliveira MC, et al. Unraveling the transport properties of RONS across nitro-oxidized membranes. Biomolecules (2023) 13:1043. doi:10.3390/biom13071043
24. Lin A, Razzokov J, Verswyvel H, Privat-Maldonado A, De Backer J, Yusupov M, et al. Oxidation of innate immune checkpoint CD47 on cancer cells with non-thermal plasma. Cancers (2021) 13:579. doi:10.3390/cancers13030579
25. Razzokov J, Fazliev S, Erkinova D, Mamatkulov S, Chen Z. Understanding the effect of nitrosylation on dynamics of human epidermal growth factor: a µs simulation study. J Phys D: Appl Phys (2022) 55:475201. doi:10.1088/1361-6463/ac9148
26. Li Y, Tan S, Liu D, Zhang Y. Molecular dynamics simulation research on the interaction between plasma and living organisms: a comprehensive review. Plasma Process Polym (2023):e2300119. doi:10.1002/ppap.202300119
27. Iftimie R, Minary P, Tuckerman ME. Ab initio molecular dynamics: concepts, recent developments, and future trends. Proc Natl Acad Sci USA (2005) 102:6654–9. doi:10.1073/pnas.0500193102
28. Wu Y, Mundy CJ, Colvin ME, Car R. On the mechanisms of OH radical induced DNA-base damage: a comparative quantum chemical and Car−Parrinello molecular dynamics study. J Phys Chem A (2004) 108:2922–9. doi:10.1021/jp0363592
29. Van Der Paal J, Neyts EC, Verlackt CCW, Bogaerts A. Effect of lipid peroxidation on membrane permeability of cancer and normal cells subjected to oxidative stress. Chem Sci (2016) 7:489–98. doi:10.1039/C5SC02311D
30. Neyts EC, Yusupov M, Verlackt CC, Bogaerts A. Computer simulations of plasma–biomolecule and plasma–tissue interactions for a better insight in plasma medicine. J Phys D: Appl Phys (2014) 47:293001. doi:10.1088/0022-3727/47/29/293001
31. Van Duin ACT, Dasgupta S, Lorant F, Goddard WA. ReaxFF: a reactive force field for hydrocarbons. J Phys Chem A (2001) 105:9396–409. doi:10.1021/jp004368u
32. Senftle TP, Hong S, Islam MM, Kylasa SB, Zheng Y, Shin YK, et al. The ReaxFF reactive force-field: development, applications and future directions. Npj Comput Mater (2016) 2:15011–4. doi:10.1038/npjcompumats.2015.11
33. Monti S, Corozzi A, Fristrup P, Joshi K L, Kyung Shin Y, Oelschlaeger P, et al. Exploring the conformational and reactive dynamics of biomolecules in solution using an extended version of the glycine reactive force field. Phys Chem Chem Phys (2013) 15:15062–77. doi:10.1039/C3CP51931G
34. Neyts EC, van Duin ACT, Bogaerts A. Insights in the plasma-assisted growth of carbon nanotubes through atomic scale simulations: effect of electric field. J Am Chem Soc (2012) 134:1256–60. doi:10.1021/ja2096317
35. Weismiller MR, Van Duin ACT, Lee J, Yetter RA. ReaxFF reactive force field development and applications for molecular dynamics simulations of ammonia borane dehydrogenation and combustion. J Phys Chem A (2010) 114:5485–92. doi:10.1021/jp100136c
36. Bogaerts A, Yusupov M, Van der Paal J, Verlackt CCW, Neyts EC. Reactive molecular dynamics simulations for a better insight in plasma medicine. Plasma Process Polym (2014) 11:1156–68. doi:10.1002/ppap.201400084
37. Yang S, Zhao T, Cui J, Han Z, Zou L, Wang X, et al. Molecular dynamics simulations of the interaction between OH radicals in plasma with poly- β -1–6-N-acetylglucosamine. Plasma Sci Technol (2020) 22:125401. doi:10.1088/2058-6272/abb454
38. Kye-Nam L, Kwang-Hyun P, Won-Tae J, Yeon-Hee L. Sterilization of bacteria, yeast, and bacterial endospores by atmospheric-pressure cold plasma using helium and oxygen. J Microbiol (2006) 44:269–75.
39. Wigginton KR, Pecson BM, Sigstam T, Bosshard F, Kohn T. Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity. Environ Sci Technol (2012) 46:12069–78. doi:10.1021/es3029473
40. Chenoweth K, Van Duin ACT, Goddard WA. ReaxFF reactive force field for molecular dynamics simulations of hydrocarbon oxidation. J Phys Chem A (2008) 112:1040–53. doi:10.1021/jp709896w
41. Zhang J, Xiao T, Cai Y, Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol (2021) 50:173–82. doi:10.1016/j.coviro.2021.08.010
42. Tu Y, Tang W, Yu L, Liu Z, Liu Y, Xia H, et al. Inactivating SARS-CoV-2 by electrochemical oxidation. Sci Bull (2021) 66:720–6. doi:10.1016/j.scib.2020.12.025
43. Yang Z, Xiao A, Liu D, Shi Q, Li Y. Damage of SARS-CoV-2 spike protein by atomic oxygen of cold atmospheric plasma: a molecular dynamics study. Plasma Process Polym (2023) 20:2200242. doi:10.1002/ppap.202200242
44. Zhang X, Zhou R, Bazaka K, Liu Y, Zhou R, Chen G, et al. Quantification of plasma produced OH radical density for water sterilization. Plasma Process Polym (2018) 15:1700241. doi:10.1002/ppap.201700241
45. Liu F, Sun P, Bai N, Tian Y, Zhou H, Wei S, et al. Inactivation of bacteria in an aqueous environment by a direct-current, cold-atmospheric-pressure air plasma microjet. Plasma Process Polym (2010) 7:231–6. doi:10.1002/ppap.200900070
46. Fang Z, Shao T, Wang R, Yang J, Zhang C. Influences of oxygen content on characteristics of atmospheric pressure dielectric barrier discharge in argon/oxygen mixtures. Eur Phys J D (2016) 70:79. doi:10.1140/epjd/e2016-60438-9
47. Graves DB. Reactive species from cold atmospheric plasma: implications for cancer therapy. Plasma Process Polym (2014) 11:1120–7. doi:10.1002/ppap.201400068
48. Tian W, Kushner MJ. Atmospheric pressure dielectric barrier discharges interacting with liquid covered tissue. J Phys D: Appl Phys (2014) 47:165201. doi:10.1088/0022-3727/47/16/165201
49. Zhao J, Nie L. Five gaseous reactive oxygen and nitrogen species (RONS) density generated by microwave plasma jet. Phys Plasmas (2019) 26:073503. doi:10.1063/1.5092840
50. Yusupov M, Neyts EC, Khalilov U, Snoeckx R, van Duin ACT, Bogaerts A. Atomic-scale simulations of reactive oxygen plasma species interacting with bacterial cell walls. New J Phys (2012) 14:093043. doi:10.1088/1367-2630/14/9/093043
51. Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys (1984) 81:3684–90. doi:10.1063/1.448118
52. Takai E, Kitamura T, Kuwabara J, Ikawa S, Yoshizawa S, Shiraki K, et al. Chemical modification of amino acids by atmospheric-pressure cold plasma in aqueous solution. J Phys D: Appl Phys (2014) 47:285403. doi:10.1088/0022-3727/47/28/285403
53. Li Y, Nie L, Liu D, Kim S, Lu X. Plasma-activated chemical solutions and their bactericidal effects. Plasma Process Polym (2022) 19:2100248. doi:10.1002/ppap.202100248
54. Ma M, Zhang Y, Lv Y, Sun F. The key reactive species in the bactericidal process of plasma activated water. J Phys D: Appl Phys (2020) 53:185207. doi:10.1088/1361-6463/ab703a
Keywords: molecular dynamics simulation, RONS, plasma, spike protein, coronavirus
Citation: Tan S, Zhu H, Liu D, Zhang S and Chen H (2024) Molecular mechanism of cleavage of SARS-CoV-2 spike protein by plasma generated RONS. Front. Phys. 12:1357639. doi: 10.3389/fphy.2024.1357639
Received: 18 December 2023; Accepted: 22 January 2024;
Published: 13 February 2024.
Edited by:
Yuantao Zhang, Shandong University, ChinaReviewed by:
Jamoliddin Razzokov, Tashkent Institute of Irrigation and Agricultural Mechanization Engineers (TIIAME), UzbekistanNikola Skoro, University of Belgrade, Serbia
Copyright © 2024 Tan, Zhu, Liu, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawei Liu, bGl1ZHdAaHVzdC5lZHUuY24=; Song Zhang, emhhbmdzb25nNzY3QDE2My5jb20=
 Shihao Tan
Shihao Tan Haiwei Zhu1
Haiwei Zhu1 Dawei Liu
Dawei Liu Song Zhang
Song Zhang Hongxiang Chen
Hongxiang Chen