
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Phys., 20 November 2023
Sec. Soft Matter Physics
Volume 11 - 2023 | https://doi.org/10.3389/fphy.2023.1251146
This article is part of the Research TopicReviews in Soft Matter PhysicsView all 6 articles
Lipid bilayers—the main matrix of cell membranes—are a paradigm of soft molecular assemblies whose properties have been evolutionarily optimized to satisfy the functional requirements of cells. For instance, lipid bilayers must be rigid enough to serve as the protective barrier between cells and their environment, yet fluid enough to enable the diffusion of proteins and molecular clusters necessary for biological functions. Inspired by their biological multifunctionality, lipid membranes have also been used as a central design element in many practical applications including artificial cells, drug nanocarriers, and biosensors. Whether biological or synthetic, lipid membranes often involve molecular or nanoscopic additives that modulate the membrane properties through various mechanisms. Hence, how lipid membranes respond to additives has justifiably drawn much attention in recent years. This review summarizes findings and observations on different classes of additives and their effects on structural, thermodynamic, elastic, and dynamical membrane properties that are central to biological function or synthetic membrane performance. The review primarily focuses on phospholipids as a major component of cell membranes and a widely used lipid type in synthetic membrane designs.
Lipid bilayers are the primary structure of the plasma membranes of living cells, playing a crucial role as the barrier between the cytosol and the extracellular environment and the first line of cellular defense against pathogens and foreign particles. These 3–4 nm thick membranes are formed of dynamic, fluid self-assemblies of lipids—amphiphilic molecules characterized by hydrophilic heads and hydrophobic fatty acid tails [1]. The properties of lipid membranes dictate various cellular functions, including molecular transport, protein recruitment, signal transduction, and maintaining a stable intracellular environment for biochemical reactions. This multifunctionality has inspired the use of synthetic and biomimetic lipid membranes in numerous practical applications, including artificial cells [2–6], molecular and therapeutic nanocarriers [7–10], as well as biosensing and biosorting platforms [11–13]. Whether biological or synthetic, lipid membranes have to meet important yet often contradictory requirements. For instance, they should maintain reasonable mechanical integrity while also providing a fluid environment for membrane components to diffuse and interact. Therefore, understanding the physical properties of lipid membranes is central to every aspect of their biological function and their practical applications.
Lipids constitute a large class of molecules that vary in their head group chemistry, tail length, and tail unsaturation (see Figure 1) [16]. In cells, lipid chemical structures have evolved to meet different functional requirements, and accordingly they exist in varying concentrations in the membranes of different organelles and across different cellular tissues [15, 17, 18]. In fact, mammalian membranes include over 1,000 unique lipid species which play various roles in biological function. In this review article, we primarily focus on phospholipids, which constitute a major lipid component of mammalian plasma cell membranes and are widely used in synthetic lipid membrane applications. Phospholipids are characterized by a phosphate headgroup and two hydrophobic fatty acid chains. Although over half of the phospholipids in the plasma membrane have a phosphatidylcholine (PC) headgroup, the small fraction that do not are still required in vital cell processes and play a critical role in locally modifying the membrane properties [17]. This diversity of chemical structures has positioned phospholipids as attractive molecular candidates in numerous applications requiring synthetic membranes with biomimetic functionality.
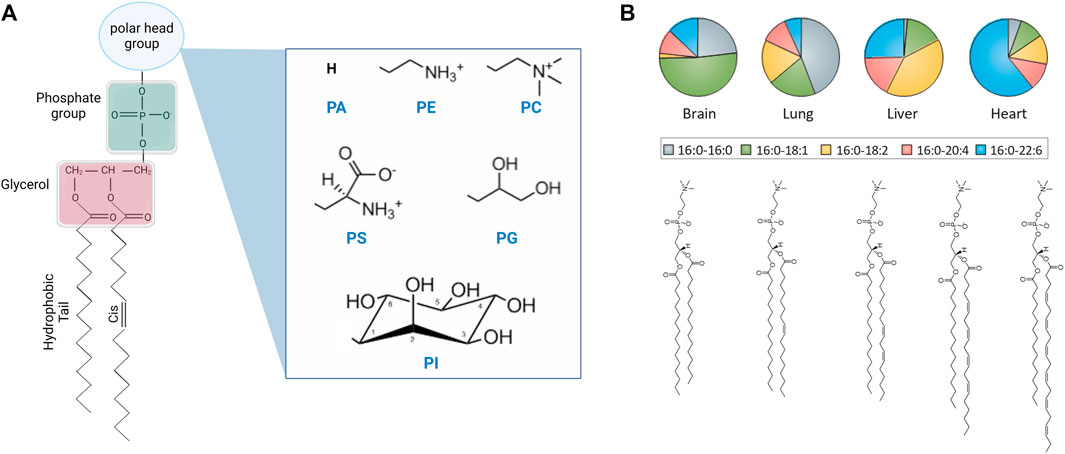
FIGURE 1. Chemical structure of lipid molecules and their relative abundance in liposomal cell extracts. (A) Schematic of the chemical structure of a phospholipid with different headgroups, including phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylglycerol (PG), and phosphatidylinositol (PI) headgroups (created with BioRender.com). (B) Schematic of saturated and unsaturated PC lipids and their abundance in the lipidome of different mouse tissues [14] (reprinted with permission from Harayama et. al [15]. Copyright (2018) Nature). Lipids are typically described by their headgroup chemistry, the number of carbon atoms in their hydrocarbon fatty acid chains, and the number and position of double bonds per chain, e.g., 16:0–16:0 PC corresponds to a saturated phosphatidylcholine lipid with 16 carbon atoms and zero double bonds in both tails. The chain description of the form X:Y ω-Z represents the chain length and saturation where X is the number of carbon atoms in the chain, Y is the number of double bonds, and Z is the position of the last double bond along the chain.
Besides phospholipids, biological and synthetic lipid membranes typically host other molecules or additives that impart specific functions. For example, cholesterol—an abundant molecular component in mammalian plasma cell membranes—is known to regulate membrane compartmentalization and cell signaling, two essential processes in cell survival [19, 20]. In synthetic and liposomal membranes, cholesterol is often used as a membrane stabilizer and a regulator for lateral lipid organization [21–24]. Membrane proteins are another functionally critical constituent of cell membranes and synthetic membrane mimics. How proteins interact with their host lipid membranes has direct effects on human health and disease [25, 26] and on the functional properties of synthetic membranes and artificial cell communication [27, 28]. In addition to biological molecules, cell membranes are often exposed to small synthetic additives, such as small drug molecules, diagnostic nanoparticles, and environmental cytotoxins [29–31]. Interestingly, studies show compelling evidence that cell membranes often adapt to changes in molecular uptake or environmental conditions by modifying their lipid composition to maintain specific membrane properties that are necessary for function [32–34], a process known as homeoviscous adaptation [35–37]. Replicating this adaptive behavior in artificial cells or utilizing it in molecular biosensing or therapeutic approaches would be transformative. Therefore, understanding how lipid membranes respond to additives is necessary in uncovering adaptive cellular mechanisms and emulating them in the design of lipidic membranes as configurable soft materials.
Importantly, such applications strongly rely on the mode of incorporation of different additives in lipid membranes and the magnitude of their perturbation of the membrane physical properties. Knowledge of the molecular mechanisms that underly the interaction of additives with their host membranes is central to the functional assignment of additives and their informed use in artificial and biomimetic membrane systems. To describe these mechanisms and identify emerging rules for soft matter interactions of molecular and nanoscopic additives, we will discuss the partitioning of different types of additives into lipid membranes and their affinity to different lipid phases within the lateral lipid organization [38–40]. This in turn will be mapped to trends of additive-induced effects on the structural, thermodynamic, or elastic properties of phospholipid membranes. The obtained trends will elucidate general design rules that could accelerate the development of artificial membrane technologies with targeted functionality and tunability.
To this end, we will summarize how different classes of molecular and nanoscopic additives—including sterols, fatty acids, proteins/peptides, drug molecules, polymers, and nanoparticles—affect the membrane structure, function, and dynamics and how these properties can be obtained using various characterization techniques. In each section, we will discuss the effects of these additives on physical membrane descriptors associated with biological functions or synthetic membrane performance and that are commonly ascribed to molecular self-assemblies. This includes structural properties described by the molecular packing (expressed in terms of the area per lipid (AL) [41] or average area per molecule [42]) and the membrane thickness (typically expressed in terms of the hydrophobic thickness of the fatty acid chain region (
Cholesterol (Chol) is an abundant component of mammalian plasma membranes and a common additive in synthetic and liposomal membranes. Accordingly, cholesterol has drawn much attention across various fields of research [61–63] due to its distinct role in membrane structure, rigidity, and fluidity [64, 65]. How cholesterol modulates physical membrane properties is largely determined by where and how it incorporates into the lipid membrane. In membranes with compatible lipid tail lengths, nuclear magnetic resonance (NMR) and neutron diffraction studies show that cholesterol orients itself in an upright position against lipid tails (see Figure 2A), i.e., parallel to the bilayer normal [66, 71]. However, when the hydrophobic thickness of the host lipid membrane becomes smaller than the hydrophobic thickness of cholesterol, cholesterol starts to exhibit tilted orientations and can even sequester between the two membrane leaflets in very thin lipid membranes [66, 72–74]. The dependence of cholesterol’s orientation on bilayer composition has also been observed in molecular dynamics (MD) simulations on membranes formed of saturated lipids (i.e., lipids with no double bonds in the fatty acid chains) and unsaturated lipids (with one or more double bonds in the fatty acid chains) [75–77]. For example, MD simulations show that cholesterol segregates neatly into the hydrocarbon region of SOPC (18:0–18:1) membranes up to a concentration of around 30 mol%, but above this threshold cholesterol is driven towards the center of the bilayer and its tilt angle relative to the bilayer normal increases (Figure 2B) [68]. Similarly, in membranes composed of polyunsaturated DAPC (di20:4 ω-6), doped with 10 mol% DMPC (14:0–14:0), a non-negligible fraction of cholesterol is found to reside between the monolayer leaflets (i.e., perpendicular to the bilayer normal) in DAPC-rich domains, whereas an upright orientation of cholesterol is observed in the DMPC-rich regions [78]. This is in line with cholesterol’s stronger affinity to saturated hydrocarbon chains compared to polyunsaturated lipids. Indeed, other studies on DAPC-cholesterol membranes show that while only 5 mol% of fully saturated DMPC (14:0–14:0) is required to pull cholesterol back up into its upright position, it takes 50 mol% of POPC (16:0–18:1) to achieve the same effect [79, 80].
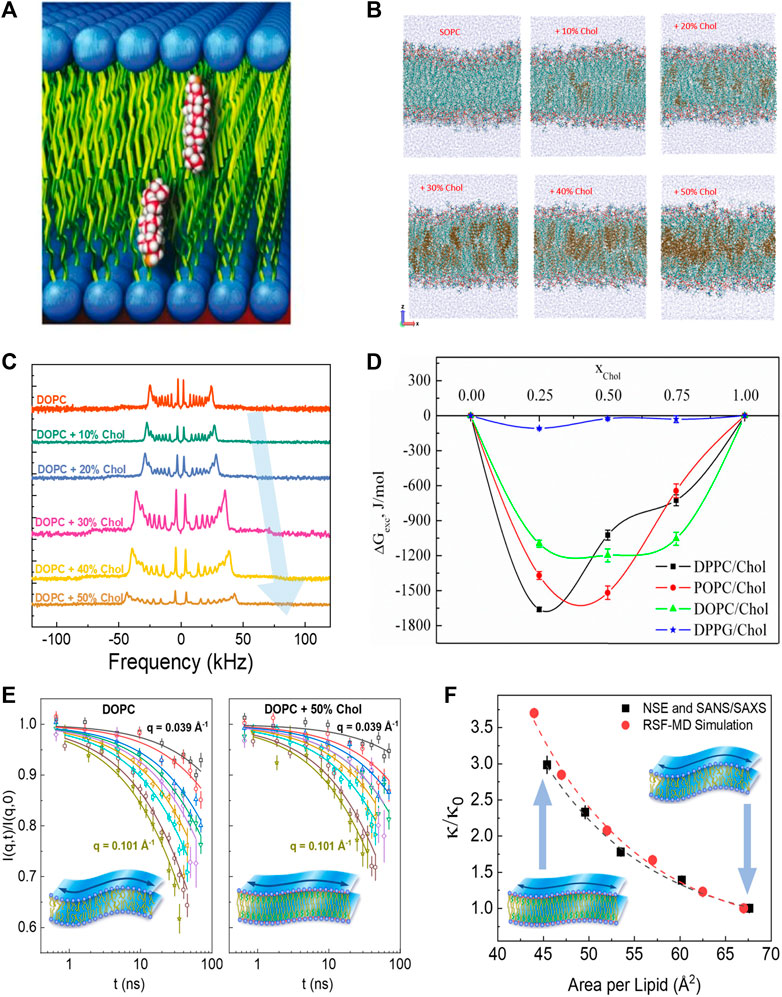
FIGURE 2. Effects of cholesterol on membrane structure and dynamics. (A) Schematic of cholesterol’s upright orientation in lipid membranes, with its hydroxyl group near the lipid-water interface [66] (reproduced with permission from Kinnun et al. [67]. Copyright (2021) Frontiers). (B) MD simulations of SOPC bilayers with increasing cholesterol concentration (reprinted with permission from Ivanova et al [68] Copyright (2023) MDPI). (C) Solid-state 2H NMR spectra illustrating cholesterol-induced acyl-chain ordering in multilamellar dispersion of DOPC, indicated by the increase in quadrupolar splitting with increasing cholesterol content (reprinted with permission from Chakraborty et al [69]. Copyright (2020) National Academy of Sciences). (D) Excess free energy of mixing (
The upright positioning of cholesterol along lipid tails induces tighter molecular packing or condensing of fluid phospholipid membranes, a phenomenon explained by the umbrella model whereby the phospholipid headgroups shield the non-polar cholesterol from unfavorable interactions with the aqueous medium [81]. Although this condensing effect has been observed across various phospholipid membranes with different degrees of chain unsaturation [82–84], the extent to which cholesterol affects molecular packing depends on the degree of lipid chain unsaturation and the position of the double bond along the acyl chain [85, 86]. Notably, the ordering of lipids by cholesterol often results in an increased thickness of the lipid membrane, as observed by numerous techniques including small angle neutron and X-ray scattering (SANS/SAXS) [82], neutron and X-ray diffraction [84, 87], and 2H-NMR [88, 89]. For example, 2H-NMR studies on lipid membranes with varying degrees of chain unsaturation show an increase in quadrupolar splitting with increasing cholesterol content, indicating greater acyl chain ordering (see example in Figure 2C) [89]. Similar studies have illustrated that cholesterol-induced thickening of membranes decreases with lipid chain unsaturation, such that DMPC (14:0–14:0) membranes exhibit the strongest thickening with cholesterol, followed by POPC (16:0–18:1) and DOPC (18:1–18:1) membranes [90]. These observations are in agreement with scattering experiments [69, 91–93] as well as 2H-NMR studies and MD simulations [68, 69, 94]. Importantly, the area-per-molecule values calculated from the membrane thickness agree well with direct measurements of molecular packing in Langmuir monolayer studies [75–77, 95–100]. These observations are consistent with the excess Gibbs free energy,
Differences in cholesterol-lipid affinity are a critical factor in the incorporation of cholesterol into various phospholipid membranes, especially in synthetic or liposomal phospholipid membranes requiring high cholesterol concentrations for additional stability. In such applications, cholesterol loading into the membrane is dictated by cholesterol’s solubility limit, which strongly depends on the phospholipid headgroup, e.g., phosphatidylcholine (PC) vs phosphatidylethanolamine (PE) or phosphatidylserine (PS) [81, 102, 103]. These observations highlight differences in cholesterol interactions with phospholipids of various chain or headgroup structures and are confirmed by a range of techniques including SANS [104], NMR [105], fluorescence microscopy [106–108] Langmuir isotherms [109, 110] and MD simulations [111]. This differential affinity of cholesterol to lipids with saturated vs unsaturated tails or PC vs PE headgroups has important consequences on cholesterol partitioning across different lipid phases in mixed lipid membranes [112–114]. Such concepts have been frequently used in model lipid membranes mimicking the lateral organization of cell membranes [108, 115–117], of high importance in artificial cell designs mimicking signaling pathways. Specifically, in lipid membranes containing saturated and unsaturated PC lipids, cholesterol was initially thought to preferentially partition into regions rich in saturated lipids, resulting in lipid phase separation into a liquid-ordered (LO) phase that is rich in saturated lipids and cholesterol and a liquid-disordered (LD) phase that is predominantly formed by unsaturated and/or short chain lipids [118, 119]. However, recent MD simulations suggest a different partitioning mechanism, in which saturated lipids form highly ordered domains surrounded by regions of unsaturated lipids and cholesterol [60]. Nonetheless, and regardless of the partitioning mechanism, the introduction of cholesterol in phospholipid membranes containing saturated and unsaturated lipids typically results in lipid phase separation into domains with different packing states, with tighter lipid packing in the LO domains and looser packing in the LD regions.
As expected, such changes in molecular packing due to cholesterol have important implications in the fluidity and permeability of lipid membranes [120–123], with consequences both in biological function and in applications [124, 125]. For example, liposomes rich in cholesterol have been regularly used in drug delivery applications where the addition of cholesterol results in reduced leakage, higher retention, and extended release of the drug load [126, 127]. Cholesterol also plays a significant role in solute partitioning into lipid membranes depending on lipid-cholesterol interactions (discussed earlier) with more pronounced effects for large solutes, membranes with less chain unsaturation, and membranes with smaller phospholipid headgroups [128]. In cell membranes, cholesterol-induced molecular packing can significantly impact the folding of membrane proteins and resultant biological function [129]. Similarly, changes in lipid packing by cholesterol have been associated with changes in the elastic membrane properties. For example, micropipette aspiration studies [130] have demonstrated that the inclusion of cholesterol in lipid membranes leads to an increase in the area stretch modulus, as corroborated by recent MD simulations [131, 132]. Interestingly, similar correlations between molecular packing and the area compressibility modulus,
Despite the well-documented effects of cholesterol on lipid packing and lateral compressibility in common phospholipid membranes (particularly PC lipids), its effects on the bending rigidity of these membranes is still a topic of current debate. For example, earlier studies using diffuse X-ray scattering, flicker spectroscopy, and electrodeformation have reported that cholesterol has different stiffening effects on membranes with varying degrees of chain unsaturation [134, 135]. In these studies, cholesterol was found to significantly stiffen saturated DMPC (14:0–14:0) membranes, moderately stiffen mono-unsaturated SOPC (18:0–18:1) membranes, and have no effect on the bending rigidity of di-monounsaturated DOPC (18:1–18:1) membranes. However, a recent study using neutron spin-echo (NSE) spectroscopy, 2H-NMR relaxometry, and MD simulations showed that the bending fluctuations of DOPC membranes experience significant slowdown on nanosecond timescales with the addition of cholesterol (Figure 2E) [57]. More importantly, the observed increase in the bending rigidity modulus
In liposomal applications, while cholesterol has been frequently used in stabilizing liposomes and reducing premature drug release [139, 140], it tends to rapidly exchange with cell membranes and lipoproteins—eventually resulting in compromised liposomal stability. An alternative workaround is to use synthetic molecules formed by the conjugation of sterols to lipid acyl chains, also known as sterol modified lipids (SMLs) [141]. As intended, SMLs prevent the exchange of free sterols out of liposomal membranes while maintaining the structural and mechanical properties of the liposome [141]. Studies of SMLs in saturated DMPC (14:0–14:0) and DPPC (16:0–16:0) lipid bilayers using NMR, Langmuir pressure-area isotherms, SANS, and neutron reflectometry show an increase in membrane thickness and concomitant decrease in the mean molecular area with the addition of SMLs [142, 143]. More importantly, liposomes containing SMLs show remarkable decrease in leakage over extended time compared to traditional liposomes [141], further emphasizing the interdependence of molecular packing, stability, and leakage in lipid membranes.
In addition to cholesterol, there is a considerable number of related sterols that are similar in chemical structure but differ in the number of double bonds, planar roughness, or alkyl chain (see Figure 3A). The sterol group originates from the triterpenoid squalene, which is the precursor molecule to sterols found in higher order organisms [146]. For example, ergosterol, sitosterol and stigmasterol are commonly found in plants and fungi [147] and are then taken up into mammalian cells through dietary means. Understanding how such sterols affect the physical properties of phospholipid membranes is necessary for gaining a full grasp of their biological role and potential applications [148]. Ergosterol, for example, is known to induce lipid ordering in fungal membranes leading to denser packing, reduced lateral diffusion, increased membrane thickness, and higher membrane stiffness due to the conformational restrictions on adjacent lipids [149–151]. However, X-ray diffraction studies by Hung et al. show that the condensing effect of ergosterol on PC lipid membranes strongly depends on lipid chain unsaturation and the sterol tilt angle, resulting in different condensing and membrane thickening effects compared to cholesterol [152].
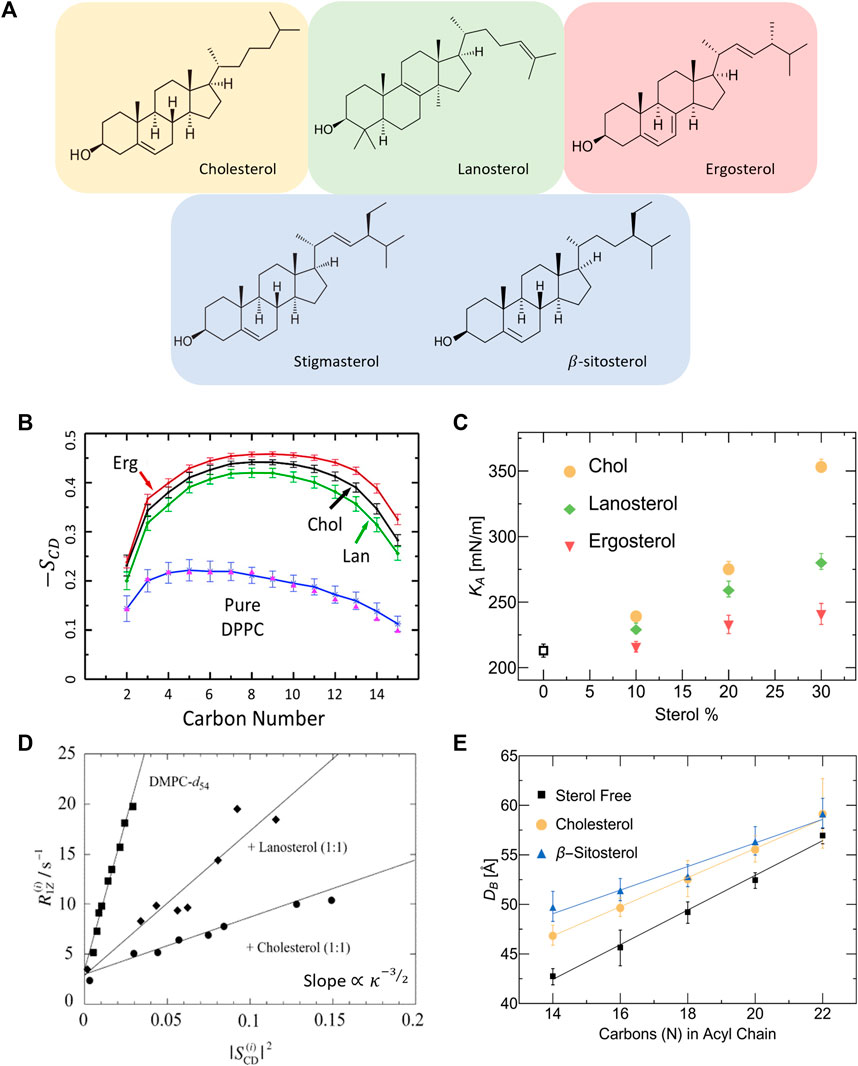
FIGURE 3. Effect of different sterols on PC lipid membranes. (A) Schematic of the chemical structures of sterols found in animal cells (tan), fungi and protozoa (red), plants (blue), and the precursor molecule to animal and fungal steroids, lanosterol (green). (B) Simulation results on DPPC show that ergosterol and lanosterol respectively induce stronger and weaker ordering of DPPC chains compared to cholesterol, illustrated by the order parameter
Differences in sterol effects on lipid membranes have also been observed in MD simulations on saturated DMPC and DPPC membranes, showing that ergosterol orders the lipid chains to a larger extent than cholesterol while lanosterol (the parent sterol) shows a weaker ordering effect (see Figure 3B) [153, 154]. Interestingly, micropipette aspiration studies on the three sterols in monounsaturated POPC (16:0–18:1) membranes show that the area expansion modulus increases significantly with the addition of cholesterol and to a lesser extent with lanosterol and ergosterol (see Figure 3C) [148]. Remarkably, these effects are commensurate with the respective sterol-induced ordering of lipids. The effects of sterols on membrane properties have also been investigated in several NMR studies reporting that, compared to cholesterol, ergosterol causes increased ordering of the hydrocarbon chains of DMPC [148, 153, 155] and DPPC [154] up to 30 mol% sterol concentration. In comparison, lanosterol was found to result in reduced condensing [77] and stiffening effects [145, 148] compared to cholesterol in DMPC membranes (see Figure 3D). For details of NMR investigations of lipid membrane with various sterols, the reader is referred to other focused reviews [156].
Other changes in sterol chemistry are also known to result in non-trivial changes in membrane structure and dynamics which can affect basic cellular functions. For example, plant sterols have been found to increase the bilayer thickness in a similar fashion to cholesterol [149]. Stigmasterol and sitosterol were specifically found to increase lateral packing density and bilayer thickness in both DMPC (14:0–14:0) and POPC (16:0–18:1) membranes [147]. By comparing the effects of cholesterol and β-sitosterol, SANS measurements show that they have different condensing effects on membranes with varying chain lengths, with β-sitosterol causing a more pronounced thickening (or condensing) effect in lipid membranes with shorter chains (Figure 3E) [144]. Importantly for synthetic membrane applications, small differences in sterol structures have been found to result in drastic differences in their solubility limits in phospholipid membranes containing saturated and unsaturated PC lipids [157]. This can potentially influence the degree to which different sterols can be used in modifying membrane properties. These observations point to the importance of understanding the effects of sterol structure on the properties of membranes with different phospholipid compositions and sterol content.
Free Fatty Acids (FFAs) are known as an important energy source for cells and cellular tissues. They are classified according to their aliphatic chain length and degree of chain unsaturation, i.e., saturated fatty acids (SFAs) with no double bonds, unsaturated fatty acids (UFAs) containing one double bond, and polyunsaturated fatty acids (PUFAs) containing two or more double bonds. For a detailed review on natural and synthetic fatty acids, we refer to Ibarguren et al. [158]. FFAs are primarily taken in through diet and are known to perturb the membranes of cells they interact with [159]. For instance, the high intake of oleic acid (18:1 ω-9) in olive-oil rich Mediterranean diet has been linked to a reduction in blood pressure through regulation of cell membrane properties and protein signaling [160]. In this review, we focus on the physical effects of FFAs on lipid membranes studied with various techniques including differential scanning calorimetry (DSC) [161–164], fluorescence spectroscopy [165, 166], electron spin resonance [167, 168], light scattering [169], NMR [170], and other approaches. These studies report that the interaction and incorporation of FFAs with phospholipid membranes happens within minutes [171, 172]. More importantly, structural differences in the chain length, degree of unsaturation, location of double bond, and cis/trans isomerization can significantly change the membrane properties [173, 174].
For example, long-chain saturated fatty acids increase the gel-to-fluid phase transition temperature of phospholipid bilayers (also known as the melting temperature,
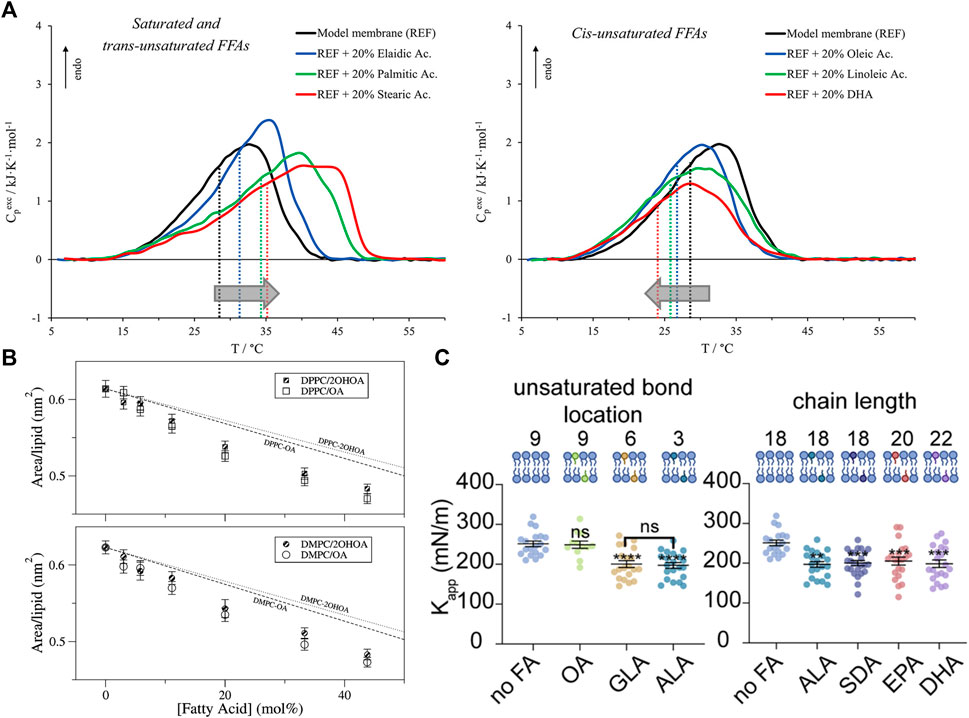
FIGURE 4. Effects of free fatty acids (FFAs) on PC lipid membranes. (A) DSC and DPH fluorescence measurements show that the inclusion of FFAs in DMPC:DPPS:DOPC membranes causes changes in membrane transition temperature towards the melting point of the respective FFA, with saturated and trans-unsaturated FFAs causing an increase in the transition temperature (left) and opposite effects for unsaturated FFAs (right). Measurements were run at a scan rate of 0.5 °C/min (reprinted with permission from Saitta et al. [178]. Copyrights (2020) American Chemical Society). (B) Studies of oleic acid (OA) embedded in bilayers composed of DMPC and DPPC show favorable interaction of OA with the host membrane indicated by lower area per lipid values compared to the theoretical prediction of ideal mixing (reprinted with permission from Cerezo et al. [179]. Copyright (2011) American Chemical Society). (C) Micropipette aspiration studies show that the membrane area compressibility
As stated earlier, FFA-induced changes in membrane phase transitions are typically associated with a change in the membrane fluidity. For example, DPH fluorescence anisotropy studies show that increasing the degree of unsaturation of incorporated FFAs leads to decreased anisotropy and decreased resistance to detergent, indicating membrane fluidization [181]. But, in the presence of cholesterol, EPA has little to no effect on membrane fluidity compared to DHA [182]. Interestingly, X-ray diffraction studies have shown that neither DHA nor EPA have an effect on the headgroup-headgroup thickness of POPC bilayers but an increase in electron density in the hydrocarbon region was observed with EPA indicating tighter packing [183]. Increased packing was also found for DMPC or DPPC monolayers with free oleic acid based on area per lipid values that are lower than the weighted area average, indicating favorable FFA-lipid interactions [179] (Figure 4B). Here we note that changes in membrane fluidity are not always directly correlated with changes in membrane permeability. For example, earlier studies showed that the effect of FFAs (namely, monoglycerides) have on membrane permeability is due to both induced acyl chain disorder as well as their specific interaction with the lipid headgroup [184]. Naturally, the effects that FFAs have on the bilayer would depend on the ordered state of bilayer and the incorporated FFA. For example, the addition of myristic acid to DMPC (14:0–14:0) membranes was found to slightly reduce picosecond dynamics in the gel state but no differences were observed above
Due to the high degree of chain conformations that PUFAs can explore, they can locally and globally modify membrane structural properties, thus impacting membrane elasticity [183]. For example, flicker spectroscopy shows that oleic acid decreases the bending rigidity modulus
Interestingly, in comparing FFA effects on phospholipid membranes with PE headgroups Langer and coworkers argue that the partitioning of FFAs into lipid membranes depends more on the molecular packing than on the nature of the lipid headgroups [187]. For instance, due to their much smaller polar headgroup relative to their fatty acid chains, PE lipids have a negative spontaneous curvature and thus they self-assemble into non-bilayer structures—typically resulting in hexagonal phases unless paired with other lipids or sterols. X-ray diffraction studies on C18 FAs (oleic, elaidic, and stearic acid) show that oleic acid (OA) causes concentration dependent alterations of the lipid self-assembly. More specifically, OA was found to induce a reduction of up to 20°C–23°C in the bilayer-to-hexagonal transition temperature of POPE (16:0–18:1) and DOPE (18:1–18:1) while elaidic and stearic acids did not markedly alter the membrane morphology. The above effects in PE membranes [188] as well as physiological cells [160] are attributed to the different molecular shape of OA with respect to their congeners, elaidic and stearic acids.
Other forms of fatty acids used in physiological processes include squalene typically utilized in the production of human sebum [189] and amine-conjugated free fatty acids used as molecular messengers in cells [190]. For instance, squalene, the precursor to sterols [146], was found via DSC to lower the main transition temperature and the fluid-bilayer to hexagonal phase transition in 95:5 SOPE:POPC mixtures [191]. On the other hand, N-acyltaurines (NATs) have a fatty acid chain linked to a taurine headgroup and thus have a large headgroup to chain ratio, resulting in conical molecular geometry and a positive spontaneous curvature [192]. In a recent publication, Prakash et al. found that NATs easily intercalate into the membrane and can fluidize the membrane up to a maximum miscibility of approximately 50 mol% [193]. Other molecules like dioleoyl-glycerol (DOG), a molecule similar to DOPC (18:1–18:1) but lacking the phosphocholine headgroup motif, can significantly modify lipid packing in their host membranes. For example, when introduced in DOPC membranes, DOG resides in the center of the bilayer and interdigitates between the two membrane leaflets, resulting in an increase in the order parameter and membrane thickness, inducing curvature strain [192]. The absence of the headgroup in DOG can also induce curvature strain and create stress and packing defects in membranes. A molecule similar to DOG but lacking one of the oleic acid chains is monoolein (1-oleoyl-rac-glycerol), which due to its simple structure can adapt to complex membrane morphologies [194]. Indeed, monoolein was found to greatly increase the permeability of EggPC membranes as much as, if not more than, unsaturated FFAs which due to its conical shape [184].
Proteins are a major component of plasma cell membranes and an important design element in artificial cells and synthetic cell membrane mimics. Membrane proteins are mainly divided into two categories: peripheral proteins which adhere to the membrane surface and integral proteins that span partial or full membrane thickness (see Figure 5A). The focus of this section is to highlight the effects proteins (and peptides) on membrane properties including membrane thickness, intrinsic curvature, and elastic moduli [198]. Nevertheless, it is imperative to emphasize that the functions of membrane proteins are simultaneously regulated by their lipid environment [199–202]. For example, the lipid headgroup and fatty acid structure as well as cholesterol content of lipid membranes can significantly affect the binding affinity of proteins to membranes [203–205], their partitioning into the membrane-water interface [206, 207], and their folding into stable conformational states [208–210]. Other studies have shown that the activity of mechanosensitive proteins like Piezo1 [175], TRPV4 [186], and MscL [211] can be altered by the presence of PUFAs, becoming activated or inactivated depending on the fatty acid identity. Knowing that the activity of proteins is dictated by their conformational state, numerous examples have illustrated that conformational changes in membrane proteins tightly depend on the membrane material properties. Recent simulations by Soubias and coworkers show that the thickness of lipid membranes, determined by the cholesterol content, closely regulates the active state of G-protein-coupled receptor (GPCR) rhodopsin [212]. Similarly, the activity of mechanosensitive ion channels—which are responsible for regulating intracellular pressure—is closely determined by the elasticity and molecular packing of its immediate lipid membrane environment (see Figure 5B) [213]. Perozo et al. have shown that changes in the membrane intrinsic curvature by the external addition of lyso-PC lipids plays a critical role in the conformations of MscL, generating significant asymmetry in the transbilayer pressure profile that can trap the channel in a fully open state [214]. More importantly, using electron paramagnetic resonance spectroscopy and spin labelling, they found that changes in the channel conformation are highly dynamic and require multiple transitions in the transmembrane helix domain of the channel protein [215].
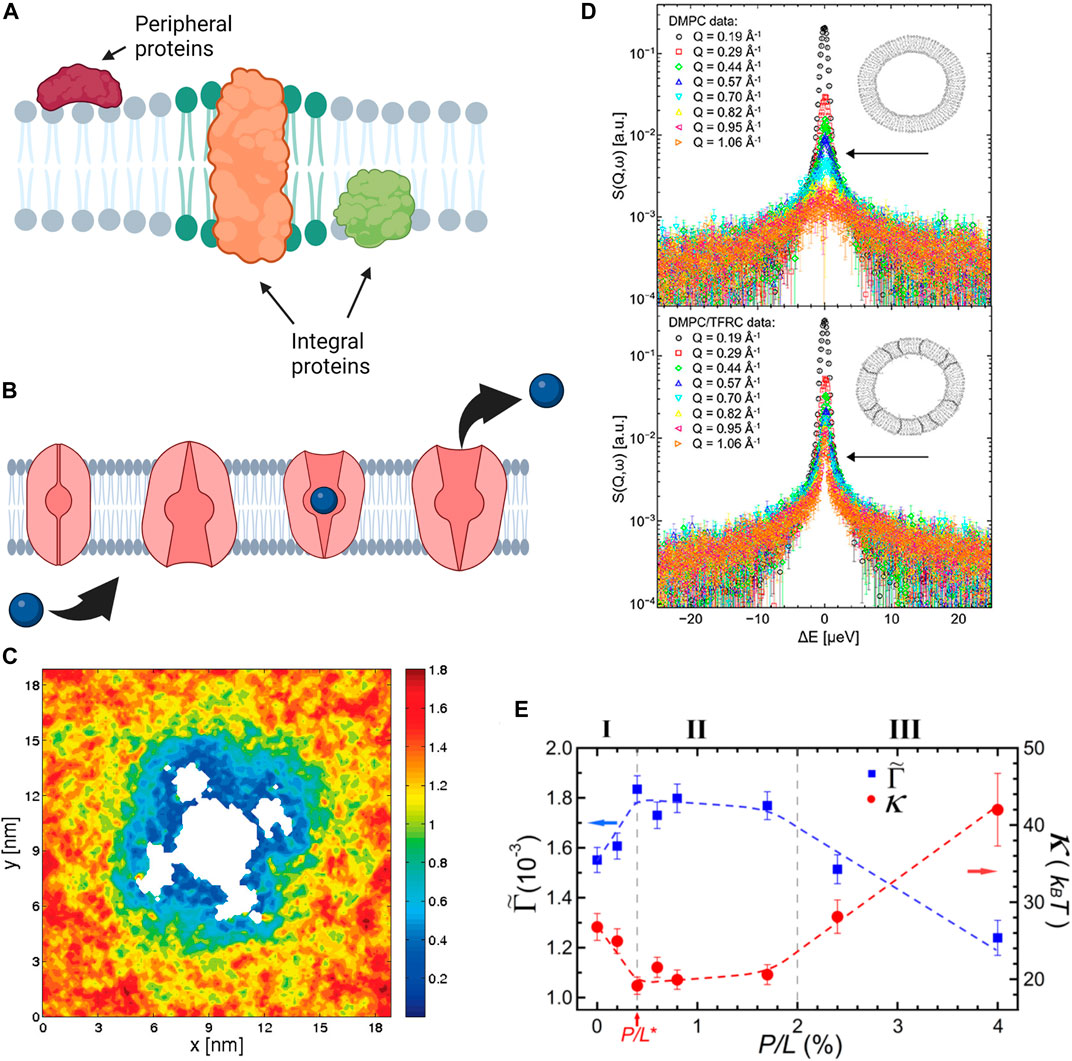
FIGURE 5. Effects of proteins and antimicrobial peptides on membrane structure, mechanics, and dynamics. (A) Schematic of protein incorporation into lipid membranes, showing peripheral proteins that bind to the membrane surface and integral proteins that partially or fully insert within lipid membranes. (B) The function of integral proteins such as the transport properties of ion channels are affected by local material properties of the membrane. (A,B) were created with BioRender.com. (C) MD simulations demonstrating the mobility of DOPC lipids around Kv1.2 voltage gated ion channel. Cooler colors correspond to reduced diffusion, showing that lipid mobility in the immediate channel vicinity is significantly reduced in comparison to the bulk lipids (reprinted with permission from Niemela et al. [195]. Copyright (2010) American Chemical Society). (D) Quasi-elastic neutron scattering spectra of DMPC membranes (top) and DMPC membranes with Transferrin Receptor protein TRFC (bottom) indicates that the presence of the protein significantly slows the diffusion of membrane lipids (reprinted with permission from Ebersberger et al. [196]. Copyright (2020) Frontiers). (E) Neutron spin-echo results on DOPC vesicles with melittin, a pore forming peptide, illustrating the changes in the bending rigidity
Therefore, understanding the synergy of proteins and lipid membranes and their effects on structural and dynamical membrane properties is key to biological function [216, 217]. Emulating these functions often results in useful technologies when translated to synthetic lipid platforms, such as liposomes [218–220], droplet interface bilayers (DIBs) [221–223], and supported lipid bilayers [11, 224, 225]. For instance, alpha hemolysin (αHL), a protein that forms transmembrane pores, disrupts the membrane structure and eventually causes membrane rupture due to ion transport [226, 227]. While this is detrimental for cells, it nonetheless enables a variety of applications requiring transport of ions and molecules across the membrane, including biosensing [228], activation of genetic circuits [229], and engineered AND gates for controlled cargo release [230].
Equally important to advanced membrane applications is the binding of proteins to specific lipid domains for controlled functionality, as in the case of raft-forming membranes with coexisting LO and LD phases [60, 231, 232]. In fact, the selective partitioning of proteins into lipid domains with specific headgroup and fatty acid structures [233, 234] is a functional membrane feature that is of high interest in synthetic biology and technological membrane applications [235–237]. For example, the HIV GAG protein favors disordered lipids with unsaturated fatty acids on both chains, yet the addition of cholesterol which increases lipid packing was found to also increase GAG binding [204]. On the other hand, palmitoylated proteins such as those implicated in T-cell signaling [238–240] have a preferential interaction to the LO phase whereas de-palmitoylated proteins have stronger interactions with the LD phase due to the fatty acid residue acting as an anchor into the membrane [241]. As Lorent and Levental point out, while a handful of proteins have been identified to preferentially bind to or insert into LO domains, the general mechanism of protein partitioning is yet to be established in complex in vitro or in vivo systems [242]. Indeed, this is the topic of ongoing investigations which will have far-reaching implications both in biological function and in constantly evolving membrane technologies.
One of the consequences of lipid-protein interactions is their effect on membrane phase transitions, i.e., their ability to influence the formation or disruption of distinct raft-like lipid domains that serve as a functional platform for various membrane processes [243]. For example, recent studies show that upon membrane association, α-helices of proteins are driven together by lipophobic effects which reduce the conformational freedom of lipids surrounding the α-helix complex, preventing them from taking part in gel-fluid phase transitions [244, 245]. Consequently, these bound proteins affect the diffusion of lipids within the membrane by acting as diffusion barriers [246]. Because lipid-protein interactions are highly dynamic in nature and span a wide range of timescales down to picosecond (ps) and nanosecond (ns) timescales, a full understanding of their underlying mechanism is still missing. However, the development and application of high-resolution techniques has started to shed light on the molecular nature of these interactions and their effects on lipid membrane properties. For instance, NMR studies on rhodopsin − a protein receptor responsible for dim light vision—show that increasing the mole fraction of rhodopsin in DMPC membranes causes an increase in the orientational order parameter of DMPC lipids and a corresponding increase in the measured gel-to-fluid transition [247]. Other studies using electron spin resonance (ESR) spectroscopy have been performed to inspect the effect of protein inclusions on the mobility and rotational dynamics of lipids within host membranes [248]. ESR spectra reveal that the SecA protein found in Escherichia coli slows lipid mobility and eliminates the LO-LD transition [249].
Neutron spectroscopy and MD simulations are also regularly used in dynamic studies of lipid-protein interactions due to the compatibility of the length and time scales that they can access. For instance, MD simulations of voltage-gated ion channel, Kv1.2, embedded in POPC membranes show that the inclusion of protein significantly impacts the mobility of the lipids surrounding it (Figure 5C) [195]. Accordingly, the protein and the neighboring lipids form a dynamical complex which diffuses as a single unit within the membrane plane. Similarly, neutron spectroscopy studies have demonstrated that the inclusion of transferrin receptor protein (TFRC) in DMPC membranes (Figure 5D) leads to restricted lateral lipid mobility in the vicinity of the protein, as well as long-ranged lipid dynamics [196]. In more recent neutron spectroscopy studies, Kelley and coworkers showed that channel-forming peptides, gramicidin (dimer-forming) and alamethicin (membrane-spanning), have markedly different effects on nanosecond bending and thickness fluctuations of DMPC lipid membranes [250]. Their findings demonstrate that peptides cannot be simply treated as rigid objects in terms of their effects on membrane fluctuations. These examples illustrate the complexity of protein-membrane interactions and their manifestation on different spatial and temporal scales, from local dynamics to collective fluctuations.
An important class of peptides is antimicrobial peptides (AMPs) which target specific lipid compositions found in bacterial membranes, and thus have various uses in therapeutic, clinical, and agricultural applications. These peptides can destroy dormant antibiotic-resistant bacteria by lysing their membranes, leading to bacterial death [251]. As such, AMPs have gained increasing interest as a potential antibacterial treatment, especially with the rise of antibiotic-resistant bacteria. Their interactions with lipid membranes have been studied using numerous techniques, including most recently MD simulations [252, 253]. These studies show that depending on peptide concentration, lipid composition, and membrane mechanical properties, the peptide can change state from surface-bound to pore-forming [254]. Pore formation is dominated by the ability of the peptide to generate curvature strain [255]. In addition to pore formation, AMPs can modulate membranes in a detergent-like process, i.e., by solubilizing the membrane. An example of this was observed on NH125 which binds to the outer lipid monolayer through electrostatic forces. When it flips to the inner leaflet, it solubilizes the membrane leading to complete membrane destabilization and eventually cell death [251]. Other AMPs, including melittin [256] and alamethicin [257], have been observed to reduce the bilayer thickness with increasing peptide concentration until a critical value, corresponding to the onset of pore formation state [254]. These observations are consistent with neutron spectroscopy studies showing that melittin alters the bending rigidity of DOPC membranes in a concentration dependent manner (Figure 5E) [197]. At low melittin concentrations, the adsorption of melittin to the membrane surface disrupts lipid packing and causes a significant decrease in the bending rigidity. At a critical concentration, melittin forms pores in the membrane and the bending rigidity starts to exhibit a slight increase. At higher melittin concentrations, the membrane bending rigidity increases significantly due to repulsive interpore interactions. Additionally, melittin displays an affinity to membranes displaying lower area compressibility moduli [258]. Not surprisingly, membrane budding and leakage due to melittin decreases upon the addition of cholesterol [259, 260] given the stiffening role of cholesterol described earlier. However, the maximum leakage correlates with intrinsic curvature of the specific lipid species within the membrane [259]. In addition, melittin has been shown to inhibit the gel-to-fluid phase transition in DMPC lipid membranes, while simultaneously enhancing lateral lipid mobility and chain flexibility [260]. In comparison, the introduction of alamethicin causes significant lipid disordering up to a critical concentration required pore formation, after which alamethicin stabilizes pore edges and reintroduces order [257].
AMPs are also dependent on lipid headgroup composition. For example, Zhao et al. demonstrated that temporin B and L, two related AMPs, readily insert into membranes composed of anionic PG headgroups and thus vesiculate SOPC:POPG liposomes but have no effect on pure SOPC (18:0–18:1) liposomes [261]. This suggests that negatively charged lipids reduce repulsion forces between peptides, inducing aggregation and leading to pore formation to relax leaflet asymmetry [261]. Another example can be found in the lysing mechanism of MSI-78 [262], a synthetic peptide mimicking the magainin family, as well as LL-37 [263] which were found via DSC to induce a positive curvature strain by increasing the transition temperature corresponding to the phase transition from a fluid bilayer to an inverted-hexagonal phase of POPE membranes. These changes can also be observed in shifts of the
Furthermore, AMPs can modulate lipid distribution within the membrane plane or across the two membrane leaflets. In general, many antimicrobial compounds have multiple charged domains which can sequester oppositely charged lipids to induce lipid phase separation, thus resulting in phase boundary defects that can change the membrane permeability or alter the membrane stability and subsequently affecting bacterial function [265]. For example, aurein—a 13-amino acid antimicrobial peptide in the frog Litoria genus that exhibits high antibiotic efficacy—is found to induce significant lateral segregation in initially uniform lipid bilayers composed of zwitterionic lipids and anionic lipids. Interestingly, reduced lipid diffusion in the fluid phase was observed even at low aurein concentrations, making the membrane prone to additional stresses and defects that change membrane properties and impede membrane-related biological processes [266]. In asymmetric membranes with different leaflet compositions, AMPs have been observed to cause lipid flip-flop between the two membrane leaflets. In a recent study, using time-resolved SANS and selective lipid deuteration, Nguyen et al. illustrated that melittin significantly accelerates lipid flip-flop, resulting in complete scrambling of pre-formed asymmetric vesicles within a couple of hours [267]. Similar findings were observed in asymmetric membranes containing antimicrobial frog peptides L18W-PGLa and magainin 2, further relating lipid flip-flop to peptide translocation and membrane leakage [268]. These studies shed light on the mechanisms of AMP interactions with lipid membranes, illustrating how AMPs assume their antimicrobial potency with potential applications in clinical, agricultural, and food industries [269, 270].
Drug-lipid interactions have a wide range of applications in various scientific disciplines from synthetic chemistry to pharmacology. These interactions often alter membrane structure and mechanics and subsequently dictate the effectiveness or design of the administered drug molecules, whether natural or synthetic [7, 271]. Among drug molecules, those produced by life forms are referred to as biogenic compounds. Examples of biogenic compounds include neurotransmitters and hormones, which play a major physiological role and therefore mimicking or modifying their chemical structures can be of benefit in future therapeutic approaches.
For example, melatonin—a hormone produced in the pineal gland of the human brain [272]—is generally used as supplement for regulating sleep but has many other therapeutic uses of relevance to health including the suppression of inflammations and tumors, cardio- and nervous system protection, and antioxidant activity [272–274]. Structurally, melatonin is known to partition into membranes at the interface between the headgroup and tail group regions of the bilayer, generally resulting in an increase in the area per lipid, a decrease in the bilayer thickness, and a subsequent increase in membrane permeability [275]. Indeed, melatonin is known to exhibit a high partition coefficient or association constant in lipid membranes, enabling it to permeate multilamellar vesicles and intracellular membranes [276]. Recent studies combining confocal microscopy, SANS, and DSC show that the incorporation of melatonin in phospholipid membranes, with compositions similar to pulmonary membranes, results in the stabilization of lipid domains implicated in membrane functions [277]. In phospholipid monolayers, melatonin is found to decrease the packing density in saturated DPPC (16:0–16:0) monolayers [278, 279] and di-unsaturated DOPC (18:1–18:1) monolayers [280], accompanied with a decrease in the area compressibility modulus
Other changes in membrane properties have also been observed with similar lipophilic neurohormones, which readily partition into lipid membranes through non-specific lipid interactions [282, 283]. For example, N-acetylserotonin is reported to increase the fluidity and area per molecule in phospholipid membranes, whereas serotonin is found to reduce the order parameter of the LD phase but increases the order parameter of the LO phase in a POPC:POPS mixture [279]. On the other hand, all-atom MD simulations of POPC:POPS mixtures found that natural psychedelics increase the area per lipid and decrease bilayer thickness [284]. These simulations also found that the hallucinogenic bufotenine causes the largest decrease in chain order parameter and largest modification of structural properties. In another study, low concentrations of psilocin, a psychoactive hallucinogen, were observed to decrease the transition temperature of DPPC:DPPS membranes in a manner similar to anesthetics [285]. In the same study, simulations of psilocin in a POPC:POPS mixture found an increase of 3–4
Other forms of biogenic compounds include phytochemicals, which are small natural bioactive molecules derived from plants and are part of the plant’s immune system. Various phytochemicals have found pharmacological use and are known to play a protective role against several diseases and infections [286, 287]. Phenolic phytochemicals are particularly promiscuous in modifying cell functions and membrane protein activity due to their ability to readily partition into or permeate through lipid membranes. To explore this mechanism, recent MD simulations investigated five bioactive phenols of reported medicinal value—namely, curcumin from turmeric, EGCG from green tea, capsaicin from chili peppers, genistein from soybeans, and resveratrol from grapes [288]. Findings from these studies show that these compounds alter the membrane properties by localizing to the lipid-water interface, resulting in changes in the lateral pressure profiles. More importantly, the induced changes in membrane properties result in similar effects on membrane proteins suggesting that the mechanism underlying the biological activity of phenolic phytochemicals is due to their propensity in modifying membrane properties, rather than specific protein binding. Correlating changes in the physical membrane properties induced by such molecules paves the way to a better understanding of their effect on human health and informing future designs of therapeutic agents with optimized membrane interactions.
Pharmaceutical molecules are used across the world for various maladies, such as moderating blood pressure and pain relief [289, 290]. While some of these drug compounds target specific receptor proteins, others such as anti-depressants may directly affect the membrane properties [291]. Synthetically produced drugs have highly complex chemical structures, each with different functional groups that interact with the membrane in various ways. Here, we relate the effects of drugs in modifying membrane properties to their partitioning into lipid membranes [292]. As summarized below, literature on common classes of pharmaceutical compounds points to common observations of increased membrane fluidity, decreased molecular packing, and an overall softening of the membrane. However, deviations from these general observations can occur depending on whether a pharmaceutical compound resides in the membrane hydrophobic core or partitions to the membrane-water interface [293, 294]. For example, azithromycin, a well-known synthetic antioxidant, has been shown to be effective in inhibiting viral entry across cell membranes [295] and preventing bacterial growth [296, 297]. In membranes, azithromycin interacts with the polar lipid headgroups of phospholipids and disrupts lipid order. AFM and optical microscopy studies show that it alters lateral lipid organization and the formation of lipid domains [298, 299]. These observations align with the propensity of azithromycin to decrease the membrane bending rigidity and area compressibility [299].
Other drug classes, like sartans which are used for hypertension [289], display varied interactions with lipid membranes depending on the lipid headgroup and acyl chain length. Many of these compounds reduce and broaden the main phase transition temperature in PC lipid membranes, indicating an induced decrease in lipid order [300, 301]. These findings align with observed changes in the lipid tilt angle [302] and with X-ray scattering studies reporting an increase in the area per lipid and a decrease in the membrane thickness [303]. This is also in agreement with other studies reporting amplified sartan-induced thermal fluctuations that uncouple the bilayer stacks [304, 305], most likely due to a softening of the membrane as expected from the reduction in the area per lipid.
Nonsteroid anti-inflammatory drugs (NSAIDs) are another class of commonly-used, over-the-counter medications for pain, inflammation, and fever [290]. To reach their target, the cyclooxygenase enzyme, NSAIDs must diffuse through cell membranes which can interfere with the membrane properties. For instance, aspirin [306]—the most common NSAID—has been shown to broaden and suppress the main phase transition of lipid membranes, resulting in overall membrane fluidization [307]. Scanning electron microscopy (SEM) studies show that red blood cells become blebby when exposed to aspirin or related compounds [308], suggesting membrane softening. Neutron spin-echo (NSE) measurements confirm that aspirin reduces the bending rigidity and increases lipid diffusion in DMPC membranes [307, 309]. Further studies using micropipette aspiration have demonstrated that aspirin-related compounds reduce the apparent area compressibility and bending rigidity in SOPC vesicles in a concentration dependent manner, concomitant with a decrease in the bilayer thickness and membrane tension [310].
Other studies on ibuprofen [311–313] and indomethacin [311] also show a shift in the main phase transition in DMPC lipid membranes to lower temperatures, indicating reduced lipid cooperativity typical of increased disorder and fluidity [311, 313]. Similar to aspirin, ibuprofen and indomethacin have also been demonstrated to lower the bending rigidity and area compressibility modulus of their host membranes [309, 314]. Analogous observations were reported on oxicams, another type of NSAIDs [315, 316]. These effects of NSAIDs on lipid membranes extend beyond the types discussed above. For example, acemetacin significantly reduces the cooperativity and phase transition in host lipid membranes [317]. Other NSAIDs like indomethacin influence phase coexistence in DPPC:Chol membranes and show preference to ordered domains induced by cholesterol [318]. Supporting studies found that indomethacin and acemetacin can influence chain packing in planar bilayers and destabilize the gel phase [317]. Similarly, aescin—another anti-inflammatory agent—lowers the main phase transition temperature of DMPC and simultaneously increases the area per lipid [319].
Studies on trifluoperazine (TFP), an antipsychotic, also show that it reduces the main phase transition temperature of DPPC membranes and completely eliminates the pretransition at concentrations as low as 1:100 TFP:DPPC [320]. Another antidepressant, fluoxetine (commonly known as Prozac) also broadens and shifts the main transition of DMPC membranes to lower temperatures and at a concentrations of 10 mM it completely suppresses the phase transition; however, less pronounced effects were observed in membranes with longer acyl chains like DPPC (16:0–16:0) and DSPC (18:0–18:0) [321]. All of these studies provide excellent examples of how pharmaceutical compounds modulate the physical properties of membranes in a way that is consistent with their partitioning mechanism and with structure-property relations of soft molecular assemblies.
In the very start of the 20th century, Meyer [38] and Overton [39] established a rule in predicting anesthetic strength. It was quite simple—the anesthetic potency of a specific chemical structure correlates linearly with partitioning from an aqueous to an organic phase. This correlation improves if the oil phase is substituted by octanol or even a lipid bilayer [322]. However, anesthetics vary wildly in chemical size, structure, and functional groups; how could such a diversity in molecules result in similar effects? One of the hypotheses is that anesthetics have a non-specific indirect mechanism of action via the membrane itself, in line with the pressure reversal effect [323, 324]. Other hypotheses support a direct effect on membrane receptors, whose function can also be significantly altered by local membrane properties as discussed earlier [325, 326].
In a dedicated review article on the lipid-centric vs protein-centric mechanism of action by anesthetics, Eckenhoff [327] drew examples to the fact that the effects seen on membrane properties were detectable at concentrations far above clinical concentrations, with the caveat that these model cell membranes may be poor approximations to real, highly complex biological systems. For instance, a commonly observed characteristic of anesthetics is their fluidizing effect, resulting in the reduction of the main phase transition at clinical concentrations in membrane mimics. In living systems, however, the plasma cell membrane is composed of hundreds of lipid species, proteins, and other molecules that prohibit a well-defined phase transition, rendering this descriptor of anesthetics invalid. Other studies have reported that clinical concentrations of volatile anesthetics like isoflurane inhibit the activity of voltage-gated sodium channels without affecting lipid bilayer properties, pointing to direct interactions with the channel protein instead of membrane-mediated interactions [328]. On the other hand, Cantor proposed a mechanism of action via changes in the lateral pressure profile of the membrane whereby anesthetic molecules modify the lateral pressure across the depth of the membrane, adjusting the preferred conformation of the protein [329]. These types of density profile calculations have become readily available in recent years due to increased computing power and as such MD simulations have become an invaluable tool for studying these systems.
For instance, MD simulations on general inhalation anesthetics, like desflurane and methoxyflurane, show that they induce a significant increase in the area per lipid (
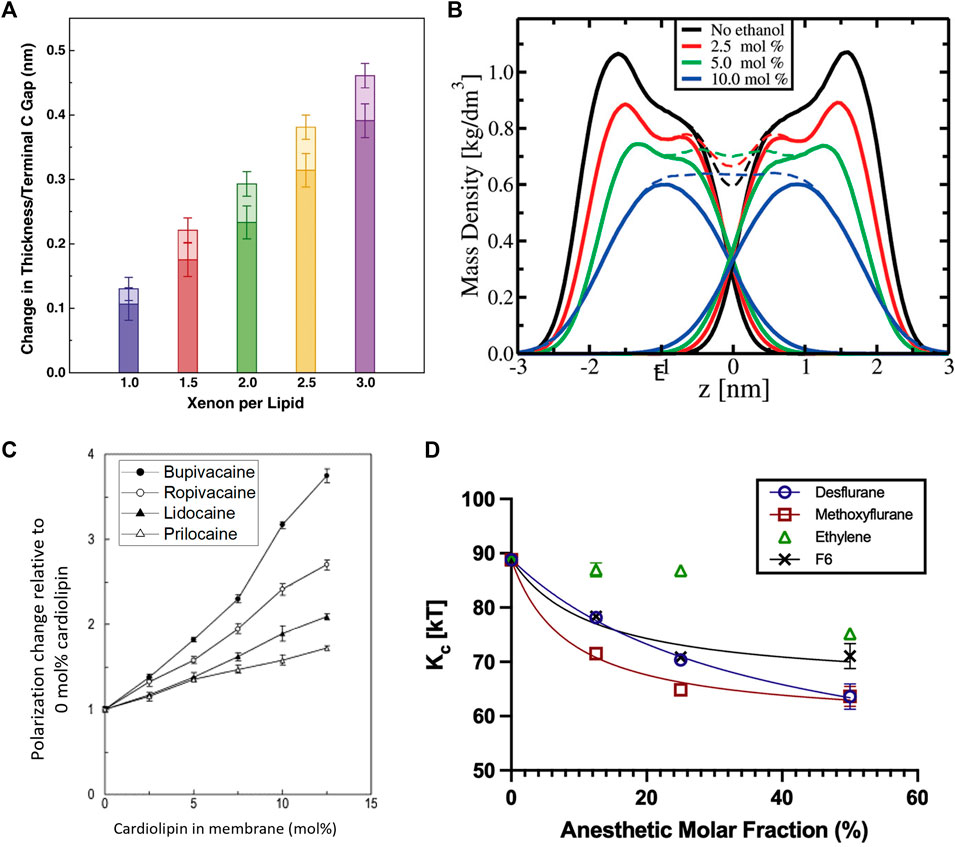
FIGURE 6. Effects of anesthetics on the thickness, mass density, fluidity, and mechanics of lipid membranes. (A) Effect of Xenon on simulated DOPC membranes showing that increasing amounts of Xenon increase the bilayer thickness with the majority of the effect coming from an increased gap between the terminal methyl groups and with some contributions due to chain ordering. Addition of bulk pressure had little to no effect on the area per lipid but was noted to increase the bilayer thickness counterintuitively at the highest pressures and concentrations (reprinted with permission from Booker et al. [332]. Copyright (2013) Elsevier). (B) Effect of ethanol on the mass density of simulated POPC bilayers, indicating a reduction in the bilayer thickness by inducing chain interdigitation—seen by the lack of a dip at the
Not surprisingly, anesthetic-induced changes in molecular packing correlate with changes in the membrane fluidity. MD simulations illustrate that these additives induce similar effects to the chain order parameter as lipid melting, explaining the observed increase in fluidity [338]. Indeed, tetracaine [339] as well as sevoflurane and diethylether [331] are all found to fluidize lipid membranes, with a reduction of this effect upon application of high pressure (typically inducing higher lipid order). Notably, the effect that local anesthetics have on lipid membranes depends on lipid composition. For example, a study on biomimetic membranes with four different local anesthetics show that bupivacaine has the strongest effect on the membrane fluidity due to its high lipophilicity compared to the other anesthetics, especially in the presence of cardiolipin—an abundant lipid in mitochondrial membranes (Figure 6C) [334]. In fact, the striking disruption of cardiolipin (CL) membranes by bupivacaine can result in heavy leakage and complete rupture of cardiolipin vesicles [340]. At concentrations below clinical use, DPH fluorescence anisotropy studies show that lidocaine can fluidize pure DPPC liposomes but had a particularly strong impact on anionic lipids with a PS, PA, or PG headgroup with cardiolipin being affected the strongest [341]. The lipid dependence of the fluidizing effects of anesthetics was also observed in a study on dibucaine in various mixtures of POPC:POPE:CL, showing that the diffusion coefficient varies with lipid composition with an obvious dependence on cardiolipin [342]. Ethanol was found to have similar fluidizing effects on membranes, with a significant enhancement of lipid diffusion on short timescales [343]. This increase in membrane fluidity is commensurate with the observed increase in membrane thermal fluctuations and a reduction in the bending rigidity with ethanol [333] and other inhalational anesthetics [330] such as desflurane, methoxyflurane, and ethylene (Figure 6D).
As expected, anesthetic-induced changes in lipid packing also influence the thermodynamic properties of lipid membranes. For example, tetracaine, a local anesthetic which fluidizes lipid membranes, is found to lower the membrane gel-fluid transition temperature [344], with similar results found for lidocaine [345, 346] and dibucaine [342, 346]. Analogous observations were reported in DSC studies on alphaxalone, a general steroid anesthetic [347]. On the other hand, Xenon exhibits a reversible effect on the melting transition. It lowers the transition temperature over time, but upon venting heat capacity profiles recover to typical profiles of the pure membrane, as corroborated by simulations [332]. Anesthetics were also found to disrupt the phase transition of membranes into ordered-disordered lipid domains. In fact, the potency of lidocaine, dibucaine, and tetracaine has been correlated with their ability to disturb lipid domains and change membrane organization [348]. For example, due to the amphiphilic nature of tetracaine, it has the ability to significantly solubilize membranes and to further disrupt membrane structure by inducing pore formation and micron-scale tubules [337, 339]. These studies highlight the importance of the interaction of anesthetics with lipid membranes and their modulation of membrane properties in ways that can significantly influence cellular processes, potentially explaining their mode of action through modification of the membrane physical properties.
Straight chain alkanes and alkanols are chemically similar to the free fatty acids, discussed in a previous section, but they lack the carboxylic acid group of free fatty acids; yet one can still draw similarities in their effects. Notably, the polar hydroxyl group in alkanols gives them an amphiphilic character, in contrast to straight chain alkanes which are purely non-polar. This causes alkanes and alkanols to partition differently into lipid membranes depending on the polar chemistry as well as chain length, and thus leads to different effects on the host membrane. Alcohols generally incorporate in membranes with their hydroxyl group near the lipid headgroup and the chain intercalated with the lipid acyl chain [349], but those with short chains, such as ethanol [350–352], tend to partition to the lipid glycerol moiety while the shortest, methanol, is unable to penetrate at all into the chain region due to its comparably high polarity [353]. Ethanol has been found to displace water molecules from the phosphate headgroups of lipid membranes, thus increasing the headgroup area and decreasing bilayer thickness [354, 355]. Additionally, gas chromatography has demonstrated that ethanol has a partition coefficient in membranes with PG and PS headgroups nearly double that of PC headgroups [356]. Similar changes in membrane structure have been reported for short chain alcohols [357, 358] with a dependence on the alcohol concentration and chain length [359]. The membrane modifying potency of alcohols increases logarithmically with each additional methylene group and a cutoff effect occurs approximately when the length of the alkanol becomes longer than half of the acyl chain length [360, 361] corresponding to the most pronounced changes in membrane thickness [349]. Notably, a recent high-resolution SANS study of co-solvents on DMPC lipid membranes demonstrated that tetrahydrofuran (THF) exhibits two partition coefficients describing the fraction of THF that partitions to the membrane-water interface (or the lipid headgroup region) and the fraction of THF residing in the hydrophobic core of the membrane [362]. This two partition constant model was necessary for fitting the collected data, emphasizing the importance of accurately measuring solvent partitioning in lipid membranes for various applications.
Thickness changes may also occur by alkanol partitioning into the LD phase of lipid membranes [363]. It should be noted that short alkanols, such as ethanol [343], have an affinity towards disordered domains with a partition coefficient that is 3–4 times larger than the LO phase [364]. In contrast, longer alkanols prefer the ordered phase [363]. More interestingly, recent studies show that these differential effects of ethanol cause an increase in the interfacial tension of LO-LD domains, resulting in larger domain sizes with important implications in biofuel production [364]. SANS measurements [365] and simulations [366] have shown that longer chains have a condensing effect on DMPC (14:0–14:0) membranes, reducing the area per lipid. Other studies [367–370] have yielded contrasting results, showing a decrease in thickness for longer alkanol chain lengths. Among these structural effects, it was also found that both linear and branched alcohols are able to induce an interdigitated phase at a critical concentration in saturated PC membranes [367–370]. This is attributed to the disruption of the hydrogen bonding between the headgroups, leading to decreased packing and interdigitation [371, 372]. Ethanol, in particular, can form non-bilayer globular structures from POPC [333] and DPPC [373] above concentrations of ∼15% mol. Gurotvenko and Anwar point out that even though these concentrations are not found in typical physiological conditions, epithelial tissues come in contact with a localized, high concentration of ethanol when consuming hard alcohol [333].
In addition, alkanols and n-alkanes are known to generally broaden the main phase transition in phospholipid membranes [374]. However, the chain length plays an important role in the thermodynamics of lipid phase transitions. For instance, short to medium chains such as ethanol [358, 369], octane [375], and decane [375] decrease the main transition temperature while longer chains such as dodecane [282] and tetradecanol [358] increase the transition temperature. Fluorescence measurements explain these changes by assuming that alcohols have their own effective transition temperature, which is distinct from their respective bulk melting point, and thus act as a quasi-lipid when embedded in lipid membranes [376]. Similarly, n-alkanes exhibit dependence on the specific lipid headgroup but induce little to no observable change in the main transition temperature [375]. For PE membranes, alkanes reduce the transition temperature,
Furthermore, fluorescence studies [378] and simulations [343, 351, 366] show that ethanol increases the diffusion of membrane lipids, while longer chain alcohols tend to greatly reduce translational diffusion [366]. Complementary studies by 2H-NMR [379], fluorescence assays [380], and simulations [358] have also reported observations of alcohols inducing order or disorder in lipid membranes depending on chain length. Notably, the addition of cholesterol to PC lipid membranes modifies the extent of chain disordering by ethanol [381]. Micropipette aspiration studies on SOPC (18:0–18:1) bilayers [355] and MD simulations on DMPC (14:0–14:0) membranes [358] both reported a reduction in the bending rigidity and membrane tension in the presence of ethanol [343, 366]. However, Ly and Longo found that alcohols with increasing chain length cause a larger decrease in the bending rigidity [357], in agreement with trends in the area compressibility modulus predicted by Traube’s rule for alcohol surfactants at the water interface. Similar to ethanol, short chain alkanols have been found to decrease the bilayer interfacial tension, reducing the tension by up to 50% at membrane rupture [357]. For medium length alkanols, simulations show a decrease in the area compressibility modulus with increasing concentration of octanol [211] whereas long chain alkanes can stiffen [358] the membrane at high enough concentrations. Overall, short chains perturb the membrane until reaching a chain length “cut-off” where they then instead reinforce the membrane, increasing order, bilayer thickness, and bending rigidity [382].
Natural polymers are present in almost all living organisms ranging from the extracellular matrix of animals and plant cell walls [1] to glucans and chitin that give fungi their structure [383] to the lipopolysaccharide layer of Gram-negative bacteria [384]. These biopolymers do exert effects on lipid membranes under various conditions. However, in their free form, they do not typically insert into the membrane but rather form coatings or mats in the membrane vicinity. Monolayer compression isotherms of DOPC on an aqueous subphase containing chitosan or hyaluronic acid display minor changes in molecular packing and monolayer compressibility, indicating minimal changes to the monolayer properties [385]. Therefore, despite the biological relevance of free biopolymers, we will not discuss them as a membrane additive. Instead, we focus here on bioderived or synthetic polymers that are conjugated to lipids or sterols or those that are designed to insert directly into lipid membranes, acting as a true additive.
Due to their versatile properties, synthetic polymers play an essential role in everyday life and have recently been incorporated in various designs of lipid membranes, often referred to as “polymer-lipid hybrid membranes”. These hybrid membranes are attractive candidates for numerous applications such as controlled drug delivery [386, 387], biosensors [388–391], and artificial cells [392, 393]. Earlier studies on lipid-polymer hybrids, specially block copolymers, show that the thickness, permeability, and bending rigidity of membranes can be changed by controlling the properties of incorporated polymer, including the block length and hydrophilic to hydrophobic block ratio [394, 395]. Optimizing these properties is critical for designing successful targeted drug delivery approaches [396]. This affords adaptable polymer-liposome a great potential in the design of stable and long lasting controlled-release nanocarriers as a hybrid alternative to fluid liposomes and rigid polymersomes [397–399].
For instance, PEO-b-PCL-b-PEO, an amphiphilic triblock copolymer structured with a hydrophobic middle block and long hydrophilic end blocks, was shown to remarkably enhance the stability of DPPC (16:0–16:0) vesicles by increasing the pressure required for the onset of membrane lysis [400]. The membrane thickness and size polydispersity can be additionally altered by modifying the hydrophilic block fraction [401]. Other polymers including polyethylene glycol (PEG) [402–405] and polybutadiene-b-poly(ethylene oxide) (PB-b-PEO) have also been used in stable biocompatible drug delivery technologies (Figure 7A) [408], including most recent mRNA vaccines. Their incorporation into liposomes allows cyclic dosage, tunable release of both hydrophilic and hydrophobic drugs, and controlled release of therapeutic agents over extended periods. Polymer-lipid hybrid membranes also have widespread applications in medical devices and biomaterials. Polymers composed of 2-methacryloyloxyethyl phosphorylcholine (MPC) units are exceptional biomaterials for the design of artificial cell membrane structures with compatible bio-interfaces between artificial and biological systems [410]. For example, synthetic polymer vesicle nanoreactors composed of oligo(aspartic acid)-b-poly(propylene oxide)—with a negatively charged surface—are favorably permeable to cationic and neutral compounds and act as a synthetic molecular channel when inserted into lipid membranes [411]. This suggests that imparting the vesicle surface with anionic charge enhances the permeability and usage in biomedical materials and artificial cells.
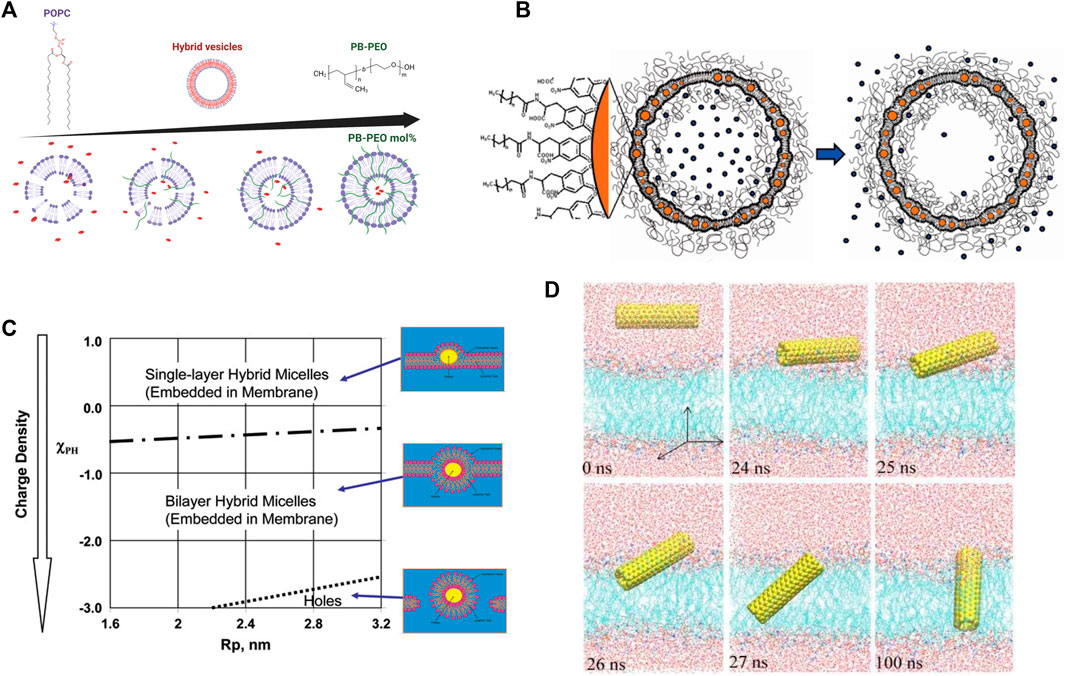
FIGURE 7. Effects of polymers and nanoparticles (NPs) on lipid membranes. (A) Addition of PB-PEO decreases the permeability of POPC lipid vesicles indicating a greater potential as drug carriers. Figure is adapted from Lim et al. [408], copyright (2013) MDPI, and illustrated with BioRender.com. (B) Drug delivery nanocarrier designs composed of a liposome functionalized with polymers for stability and containing embedded nanoparticles for controlled release of cargo upon application of alternating magnetic field (reprinted with permission from Amstad et al. [409]. Copyright (2011) American Chemical Society). (C) Schematic of the interactions of NPs with a lipid membrane as a function of NP radius and surface charge (reprinted with permission from Ginzburg et al. [406]. Copyright (2007) American Chemical Society). (D) Simulations of the insertion of a pristine carbon nanotube (CNT) into a lipid bilayer, illustrating that CNTs quickly angle themselves after contacting lipid-water interface to insert within the lipid membranes (reprinted with permission from Gao et al. [407]. Copyright (2019) MDPI).
Knowledge of how these polymers change the structure and dynamics of the lipid membrane is necessary to improve these applications and generate controllable membrane platforms. Previous studies have predicted that the insertion or grafting of polymer chains in lipid membranes can significantly impact the elasticity of lipid membranes and subsequent membrane-protein interactions [412]. These concepts were recently illustrated in liposomal studies, where the introduction of block-copolymers in liposomal membranes was shown to regulate membrane elasticity for optimal folding of mechanosensitive membrane proteins [413] and the insertion rates of natively folded peptides [414]. The inclusion of polymers in membranes has also been shown to impact membrane phase transitions. For example, the biocompatible polymers polyisobutylene (PIB) and poly(ethylene oxide) (PEO) completely disrupt the typical behavior of DPPC monolayers at the air/water interface, indicating a disruption to lipid packing [64, 415]. In DOPC, n-alkyl PEO was found to reduce the bending rigidity via transformation of the lamellar structure [416]. Other forms of amphiphilic polymers have also been used to generate transient pores in lipid membranes [417], aiding in the design of hybrid lipid membranes with controllable transport properties.
Cell membranes frequently interact with small nanoscopic particles, or nanoparticles (NPs). For example, NPs present in the atmosphere from industrial processes or environmental pollution are known to partition into pulmonary membranes, causing lung complications and airway irritation [418]. A practical way to model the effect that various NPs have on the pulmonary surfactant, the monolayer lining the alveoli, is through the use of Langmuir lipid monolayers. This method has shown that silica NPs increase the area per lipid at any given surface pressure and simultaneously reduce the surface pressure required for collapse in DPPC:DOPC:Chol monolayers [419]. A similar result was seen for carbon NPs in DPPC:DPPG, but the degree to which the carbon NPs affect the monolayer depends on the value of the surface pressure at the time when the NPs are deposited [420]. Other studies on saturated and unsaturated lipid monolayers composed of PC and PG headgroups show that cationic and anionic silver NPs have different effects depending on the lipid composition, charge mismatch, and surface pressure during NP insertion [421]. In a similar study on model PS monolayer composed of DPPC:DOPC, the addition of nanoparticles was found to change the surface pressure response to cyclic compression cycles simulating the respiratory rhythm. This study found that the total harmonic distortion increases in a pure DOPC monolayer, but in a DPPC:DOPC system NPs decrease the overall distortion, leading to a more linear response between area compression and surface pressure [422].
Nanoparticles also have novel uses in drug delivery applications, which still have shortcomings in controlled cargo release and exhibit a great increase in permeability around the lipid melting transition temperature. To circumvent these pitfalls, magnetic nanoparticles allow for controlled release of the drug by using timed magnetic field pulses as an external stimulus. This results in local magnetic heating in the vicinity of the NPs, raising the local membrane temperature beyond the melting transition, thereby increasing the permeability and releasing the encapsulated drug (Figure 7B) [409]. The incorporation of NPs in lipid membranes requires judicious surface functionalization; for instance, hydrophobic magnetic NPs are typically intended to heat their local lipid environment whereas hydrophilic NPs must heat the bulk aqueous medium to achieve the same release efficiency [409]. Hydrophobic cobalt ferrite NPs coated with a shell of oleic acid easily partition into the hydrocarbon region of a liposomal membrane. Interestingly, these particles cause slow release of cargo right after the application of a magnetic field due to pore formation but induce quicker release after a few hours due to increased membrane permeability [423]. On the other hand, superparamagnetic iron oxide NPs in bare, silica-coated, or charge-functionalized forms did not have any significant effect on lipid order, membrane fluidity, or phase transition when either incubated or encapsulated into PC liposomes [424]. These findings are important in understanding the biocompatibility of NPs and guiding future NP designs.
Liposomal leakage by nanoparticles is mainly due to the interactions between the lipid molecules and the functional group on the NP surface, with stronger effects of headgroup interactions compared to chain interactions. For example, a NP with a positive surface charge will interact much more strongly with negatively charged lipids compared to a NP with negative charge [425]. However, the NP itself plays a key role in membrane destabilization, i.e., free functional groups alone have no effect on leakage but can result in significant leakage when decorating a membrane-embedded NP. For example, in comparison to melittin, a pore forming protein discussed earlier, a single nanoparticle can cause the same degree of disruption that requires hundreds of melittin proteins [425].
Nanoparticle size plays a key role in NP partitioning into membranes and the induced disruption to the membrane structure. MD simulations show that a single gold NP alters the structural properties and fluidity of the membrane on a short and long range in a way that correlates with particle size [426]. Isothermal calorimetry studies show that the interaction of small NPs with DPPC:DPTAP liposomes is an entropically driven process favoring individual NP binding to the surface, while larger NPs interact through an enthalpically driven process which favors NP aggregation [427]. This aggregation may lead to structural changes such as fusion of neighboring or encapsulated liposomes together [428]. Complementary TEM studies demonstrate that NP incorporation into membrane is size limited (maximum size of ∼5 nm) [409], consistent with theoretical results [429] from a simple geometrical model which predicts whether a nanoparticle would partition into a membrane or cause micellar formation. Including the charge density of the NP into the model shows that, besides size, the charge strength on the NP surface is another determining factor in membrane partitioning or micellization (Figure 7C) [406]. In drug delivery liposomes, the NP size and surface functionalization should be optimized with the applied magnetic field strength [428], frequency, application time, and pulse sequence [409] to control the leakage rate and amount [423].
Other forms of environmental NP irritants include fullerenes, highly hydrophobic spherical nanoparticles composed of 60 carbon atoms with a diameter of approximately 1 nm. This class of molecules has a plethora of uses, both industrial and therapeutic [430, 431]. However, it is known to damage the plasma membrane through lipid peroxidation [432] which is an important health factor in the use of engineered nanomaterials in consumer applications. Fullerenes tend to sit off the bilayer center and have minimal changes on the local area per lipid or lipid chain stretching [433, 434]. However, due to this preferred displacement from the center of the bilayer, fullerene inclusion in membranes induces leaflet asymmetry and differential stresses that result in changes to the overall area compressibility, bilayer thickness, and bending rigidity [434]. Moreover, solid state NMR and FTIR studies show that fullerenes have an affinity towards lipids with an anionic headgroup, like those abundantly present in bacterial membranes, and therefore disturb chain packing and increase fluidity [435]. In monolayers, fullerenes were found to aggregate and greatly affect the stability and surface tension of monolayers formed of DPPC, a major component of the pulmonary surfactant [436].
Carbon nanotubes (CNTs) are another form of carbon-based nanoparticles and have been used as a synthetic analogue for porin, a protein involved in molecular exchange and transport across cell membranes. Short CNTs (10–20 nm in length) span the membrane (Figure 7D) and form pore sizes that can be engineered from tenths of a nanometer up to 10 nm in width, allowing for stable nanofluidic channels that have applications in transport, sensing, and filtration [437, 438]. The length and surface functionalization of CNTs can significantly impact the properties of their immediate lipid membrane environment, i.e., the annular lipid shell surrounding each CNT [439]. At high CNT densities, lipid-mediated interactions facilitate CNT diffusion and drive CNTs to assemble into clusters of 2D hexagonal arrays spaced apart by the annular lipid shells [440]. Other simulations found that while CNTs had virtually no effect on the structural properties of the membrane they cause slight ordering in neighboring lipid tails [407].
The interaction of lipid membranes with small molecules and nanoscopic additives is a common phenomenon in biological and synthetic membranes, offering a plethora of possibilities for tuning membrane properties either through biological adaptation or through sophisticated engineering of molecules and nanoparticles. Whether biological or synthetic, how additives modulate the physical properties of lipid membranes is necessary to understanding the role of membranes as evolved and adaptive soft materials, and refining their designs for practical applications and advanced technologies. To date, numerous studies using experimental, theoretical, or computational approaches have contributed significant knowledge about the mechanisms by which different additives partition into lipid membranes and their subsequent effects on the membrane structural, elastic, thermodynamic, and dynamic properties. Nonetheless, a major gap still exists in combining these effects into unified physical laws or design rules to realize the potential of lipid membranes as functional platforms beyond trial-and-error approaches.
This review sheds light on the interdependence of partitioning of additives into phospholipid membranes and resultant physical membrane properties. By comparing findings across seemingly disparate classes of additives and phospholipid architectures, a more complete picture of molecular partitioning and structure-property relations starts to emerge. For example, a common observation is that additives that partition to the membrane-water interface, i.e., to the headgroup region of lipid membranes, tend to disrupt lipid packing and subsequently lower the phase transition temperatures, decrease the bending moduli, and increase lipid mobility. In contrast, additives that reside in the hydrophobic region of the lipid membrane in a way that induces higher lipid chain order generally have opposite effects. Put together, this shows a consistent picture in which additive-induced changes to the membrane structure are often coupled with changes to the physical membrane properties in an interdependent pattern. These observations point to the potential of establishing structure-property relations that unify the effects of additives and the design principles of lipid membranes as soft molecular assemblies. This interdependence is yet to be systematically investigated.
More importantly, exploring structure-property relations should be done across multiple spatiotemporal scales to identify emergent membrane properties that dictate function across different scales. Given the soft and highly dynamic nature of lipid membranes, how they respond to additives on fast and slow timescales could transform the nature of membrane-based technologies. Current and future investigations in these directions will help accelerate the development of artificial lipid membranes with functions that can be specifically tuned for therapeutic or technological purposes. With the advancement of characterization techniques, synthesis approaches, and membrane functionalization methods, lipid membranes and their interactions with biological and synthetic additives will continue to push the boundaries in technologies that improve human health, advance synthetic biology platforms, and further our knowledge of soft materials and their various applications.
TK and NBM contributed equally to collecting and summarizing existing literature. RA reviewed and edited the article. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Alberts B, Bray B, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 4th Edn, Garland Science (2002). p. 1616.
2. Xu C, Hu S, Chen X. Artificial cells: from basic science to applications. Mater Today (2016) 19:516–32. doi:10.1016/j.mattod.2016.02.020
3. Pohorille A, Deamer D. Artificial cells: Prospects for biotechnology. Trends Biotechnol (2002) 20:123–8. doi:10.1016/S0167-7799(02)01909-1
4. Siontorou CG, Nikoleli G-P, Nikolelis DP, Karapetis SK. Artificial lipid membranes: Past, present, and future. Membranes (2017) 7:38. doi:10.3390/membranes7030038
5. Elani Y, Trantidou T, Wylie D, Dekker L, Polizzi K, Law RV, et al. Constructing vesicle-based artificial cells with embedded living cells as organelle-like modules. Sci Rep (2018) 8:4564. doi:10.1038/s41598-018-22263-3
6. Vance JA, Devaraj NK. Membrane mimetic chemistry in artificial cells. J Am Chem Soc (2021) 143:8223–31. doi:10.1021/jacs.1c03436
7. Peetla C, Stine A, Labhasetwar V. Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery. Mol Pharm (2009) 6:1264–76. doi:10.1021/mp9000662
8. Narain A, Asawa S, Chhabria V, Patil-Sen Y. Cell membrane coated nanoparticles: Next-generation therapeutics. Nanomedicine (2017) 12:2677–92. doi:10.2217/nnm-2017-0225
9. Jain S, Jain V, Mahajan SC. Lipid based vesicular drug delivery systems. Adv Pharm (2014) 2014:1–12. doi:10.1155/2014/574673
10. Carvalho PM, Makowski M, Domingues MM, Martins IC, Santos NC. Lipid membrane-based therapeutics and diagnostics. Arch Biochem Biophys (2021) 704:108858. doi:10.1016/j.abb.2021.108858
11. Sackmann E. Supported membranes: Scientific and practical applications. Science (1996) 271:43–8. doi:10.1126/science.271.5245.43
12. Ottova AL, Ti Tien H. Self-assembled bilayer lipid membranes: from mimicking biomembranes to practical applications. Bioelectrochemistry Bioenerg (1997) 42:141–52. doi:10.1016/S0302-4598(96)05098-2
13. Zhang Z, Huang X, Qian Y, Chen W, Wen L, Jiang L. Engineering smart nanofluidic systems for artificial ion channels and ion pumps: From single-pore to multichannel membranes. Adv Mater (2020) 32:1904351. doi:10.1002/adma.201904351
14. Harayama T, Eto M, Shindou H, Kita Y, Otsubo E, Hishikawa D, et al. Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cel Metab (2014) 20:295–305. doi:10.1016/j.cmet.2014.05.019
15. Harayama T, Riezman H. Understanding the diversity of membrane lipid composition. Nat Rev Mol Cel Biol (2018) 19:281–96. doi:10.1038/nrm.2017.138
16. Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res (2005) 46:839–61. doi:10.1194/jlr.E400004-JLR200
17. van Meer G, Voelker D, Feigenson G. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cel Biol (2008) 9:112–24. doi:10.1038/nrm2330
18. Lorent J, Levental KR, Ganesan L, Rivera-Longsworth G, Sezgin E, Doktorova M, et al. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat Chem Biol (2020) 16:644–52. doi:10.1038/s41589-020-0529-6
19. Yeagle PL. Modulation of membrane function by cholesterol. Biochimie (1991) 73:1303–10. doi:10.1016/0300-9084(91)90093-G
20. Simons K, Ikonen E. How cells handle cholesterol. Science (2000) 290:1721–6. doi:10.1126/science.290.5497.1721
21. Raffy S, Teissié J. Control of lipid membrane stability by cholesterol content. Biophys J (1999) 76:2072–80. doi:10.1016/s0006-3495(99)77363-7
22. Veatch SL, Keller SL. Seeing spots: Complex phase behavior in simple membranes. Biochim Biophys Acta (BBA) - Mol Cel Res (2005) 1746:172–85. doi:10.1016/j.bbamcr.2005.06.010
23. Feigenson GW. Phase diagrams and lipid domains in multicomponent lipid bilayer mixtures. Biochim Biophys Acta (BBA) - Biomembranes (2009) 1788:47–52. doi:10.1016/j.bbamem.2008.08.014
24. Xu X, London E. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry (2000) 39:843–9. doi:10.1021/bi992543v
25. Gasanov SE, Dagda RK, Rael ED. Snake venom cytotoxins, phospholipase A2s, and Zn2+-dependent metalloproteinases: Mechanisms of action and pharmacological relevance. J Clin Toxicol (2014) 4:1000181. doi:10.4172/2161-0495.1000181
26. Wang R, Kniazeva M, Han M. Peroxisome protein transportation affects metabolism of branched-chain fatty acids that critically impact growth and development of C. elegans. PloS one (2013) 8:e76270. doi:10.1371/journal.pone.0076270
27. Aufinger L, Simmel FC. Establishing communication between artificial cells. Chem – A Eur J (2019) 25:12659–70. doi:10.1002/chem.201901726
28. Lentini R, Yeh Martín N, Mansy SS. Communicating artificial cells. Curr Opin Chem Biol (2016) 34:53–61. doi:10.1016/j.cbpa.2016.06.013
29. Monticelli L, Salonen E, Ke PC, Vattulainen I. Effects of carbon nanoparticles on lipid membranes: A molecular simulation perspective. Soft Matter (2009) 5:4433–45. doi:10.1039/B912310E
30. Rossi G, Barnoud J, Monticelli L. Polystyrene nanoparticles perturb lipid membranes. J Phys Chem Lett (2014) 5:241–6. doi:10.1021/jz402234c
31. Roiter Y, Ornatska M, Rammohan AR, Balakrishnan J, Heine DR, Minko S. Interaction of nanoparticles with lipid membrane. Nano Lett (2008) 8:941–4. doi:10.1021/nl080080l
32. Levental KR, Malmberg E, Symons JL, Fan YY, Chapkin RS, Ernst R, et al. Lipidomic and biophysical homeostasis of mammalian membranes counteracts dietary lipid perturbations to maintain cellular fitness. Nat Commun (2020) 11:1339. doi:10.1038/s41467-020-15203-1
33. Budin I, de Rond T, Chen Y, Chan LJG, Petzold CJ, Keasling JD. Viscous control of cellular respiration by membrane lipid composition. Science (2018) 362:1186–9. doi:10.1126/science.aat7925
34. Leveille CL, Cornell CE, Merz AJ, Keller SL. Yeast cells actively tune their membranes to phase separate at temperatures that scale with growth temperatures. Proc Natl Acad Sci (2022) 119:e2116007119. doi:10.1073/pnas.2116007119
35. Ernst R, Ejsing CS, Antonny B. Homeoviscous adaptation and the regulation of membrane lipids. J Mol Biol (2016) 428:4776–91. doi:10.1016/j.jmb.2016.08.013
36. Hazel JR. Thermal adaptation in biological membranes: Is homeoviscous adaptation the explanation? Annu Rev Physiol (1995) 57:19–42. doi:10.1146/annurev.ph.57.030195.000315
37. Sinensky M. Homeoviscous adaptation—a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc Natl Acad Sci (1974) 71:522–5. doi:10.1073/pnas.71.2.522
38. Meyer H. Zur Theorie der Alkoholnarkose: Erste Mittheilune. Welche Eigenschaft der Anästhetica bedingt ihre narkotische Wirkung? Archiv für Experimentelle Pathologie und Pharmakologie (1899) 42:109–18. doi:10.1007/BF01834479
39. Overton CE. Studien über die Narkose: Zugleich ein Beitrag zur allgemeinen Pharmakologie. Gustav Fischer (1901).
40. Hannesschlaeger C, Horner A, Pohl P. Intrinsic membrane permeability to small molecules. Chem Rev (2019) 119:5922–53. doi:10.1021/acs.chemrev.8b00560
41. Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta (BBA)-Reviews Biomembranes (2000) 1469:159–95. doi:10.1016/S0304-4157(00)00016-2
42. Smaby JM, Momsen MM, Brockman HL, Brown RE. Phosphatidylcholine acyl unsaturation modulates the decrease in interfacial elasticity induced by cholesterol. Biophys J (1997) 73:1492–505. doi:10.1016/S0006-3495(97)78181-5
43. Saffman P, Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci (1975) 72:3111–3. doi:10.1073/pnas.72.8.3111
44. Hughes BD, Pailthorpe BA, White LR. The translational and rotational drag on a cylinder moving in a membrane. J Fluid Mech (1981) 110:349–72. doi:10.1017/S0022112081000785
45. Petrov EP, Schwille P. Translational diffusion in lipid membranes beyond the saffman-delbrück approximation. Biophys J (2008) 94:L41–3. doi:10.1529/biophysj.107.126565
46. Sakuma Y, Kawakatsu T, Taniguchi T, Imai M. Viscosity landscape of phase-separated lipid membrane estimated from fluid velocity field. Biophys J (2020) 118:1576–87. doi:10.1016/j.bpj.2020.01.009
47. Fitzgerald JE, Venable RM, Pastor RW, Lyman ER. Surface viscosities of lipid bilayers determined from equilibrium molecular dynamics simulations. Biophys J (2023) 122:1094–104. doi:10.1016/j.bpj.2023.01.038
48. Nagle J, Scott H. Lateral compressibility of lipid mono-and bilayers. Theory of membrane permeability. Biochim Biophys Acta (BBA)-Biomembranes (1978) 513:236–43. doi:10.1016/0005-2736(78)90176-1
49. Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv Colloid Interf Sci (2014) 208:225–34. doi:10.1016/j.cis.2014.03.003
50. Faizi HA, Frey SL, Steinkühler J, Dimova R, Vlahovska PM. Bending rigidity of charged lipid bilayer membranes. Soft Matter (2019) 15:6006–13. doi:10.1039/C9SM00772E
51. Doktorova M, Harries D, Khelashvili G. Determination of bending rigidity and tilt modulus of lipid membranes from real-space fluctuation analysis of molecular dynamics simulations. Phys Chem Chem Phys (2017) 19:16806–18. doi:10.1039/C7CP01921A
52. Nagao M, Kelley EG, Ashkar R, Bradbury R, Butler PD. Probing elastic and viscous properties of phospholipid bilayers using neutron spin echo spectroscopy. J Phys Chem Lett (2017) 8:4679–84. doi:10.1021/acs.jpclett.7b01830
53. Helfrich W. Elastic properties of lipid bilayers: Theory and possible experiments. Z Naturforsch C (1973) 28:693–703. doi:10.1515/znc-1973-11-1209
54. Zilker A, Engelhardt H, Sackmann E. Dynamic reflection interference contrast (RIC-) microscopy: A new method to study surface excitations of cells and to measure membrane bending elastic moduli. J De Physique (1987) 48:2139–51. doi:10.1051/jphys:0198700480120213900
55. Milner ST, Safran S. Dynamical fluctuations of droplet microemulsions and vesicles. Phys Rev A (1987) 36:4371–9. doi:10.1103/PhysRevA.36.4371
56. Evans EA. Bending resistance and chemically induced moments in membrane bilayers. Biophys J (1974) 14:923–31. doi:10.1016/S0006-3495(74)85959-X
57. Goodrich FC. Interaction in mixed monolayers In: Proceeding of 2nd International Congress on Surface Activity. editor JH Schulman. London: Butterworths (1957).
58. Lee A. Lipid phase transitions and phase diagrams I. Lipid phase transitions. Biochim Biophys Acta (BBA)-Reviews Biomembranes (1977) 472:237–81. doi:10.1016/0304-4157(77)90018-1
59. Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science (2006) 313:1948–51. doi:10.1126/science.1130279
60. Sodt AJ, Sandar ML, Gawrisch K, Pastor RW, Lyman E. The molecular structure of the liquid-ordered phase of lipid bilayers. J Am Chem Soc (2014) 136:725–32. doi:10.1021/ja4105667
61. Mouritsen OG, Zuckermann MJ. What's so special about cholesterol? Lipids (2004) 39:1101–13. doi:10.1007/s11745-004-1336-x
62. Narwal V, Deswal R, Batra B, Kalra V, Hooda R, Sharma M, et al. Cholesterol biosensors: A review. Steroids (2019) 143:6–17. doi:10.1016/j.steroids.2018.12.003
63. Large DE, Abdelmessih RG, Fink EA, Auguste DT. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv Drug Deliv Rev (2021) 176:113851. doi:10.1016/j.addr.2021.113851
64. Filippov A, Orädd G, Lindblom G. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J (2003) 84:3079–86. doi:10.1016/S0006-3495(03)70033-2
65. Needham D, McIntosh T, Evans E. Thermomechanical and transition properties of dimyristoylphosphatidylcholine/cholesterol bilayers. Biochemistry (1988) 27:4668–73. doi:10.1021/bi00413a013
66. Marquardt D, Kučerka N, Wassall SR, Harroun TA, Katsaras J. Cholesterol's location in lipid bilayers. Chem Phys Lipids (2016) 199:17–25. doi:10.1016/j.chemphyslip.2016.04.001
67. Kinnun JJ, Scott HL, Ashkar R, Katsaras J. Biomembrane structure and material properties studied with neutron scattering. Front Chem (2021) 9:642851. doi:10.3389/fchem.2021.642851
68. Ivanova N, Chamati H. The effect of cholesterol in SOPC lipid bilayers at low temperatures. Membranes (2023) 13:275. doi:10.3390/membranes13030275
69. Chakraborty S, Doktorova M, Molugu TR, Heberle FA, Scott HL, Dzikovski B, et al. How cholesterol stiffens unsaturated lipid membranes. Proc Natl Acad Sci (2020) 117:21896–905. doi:10.1073/pnas.2004807117
70. Jurak M. Thermodynamic aspects of cholesterol effect on properties of phospholipid monolayers: Langmuir and Langmuir–blodgett monolayer study. The J Phys Chem B (2013) 117:3496–502. doi:10.1021/jp401182c
71. Mojumdar E, Groen D, Gooris G, Barlow D, Lawrence M, Deme B, et al. Localization of cholesterol and fatty acid in a model lipid membrane: A neutron diffraction approach. Biophys J (2013) 105:911–8. doi:10.1016/j.bpj.2013.07.003
72. Harroun TA, Katsaras J, Wassall SR. Cholesterol hydroxyl group is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry (2006) 45:1227–33. doi:10.1021/bi0520840
73. Harroun TA, Katsaras J, Wassall SR. Cholesterol is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry (2008) 47:7090–6. doi:10.1021/bi800123b
74. Marquardt D, Heberle FA, Greathouse DV, Koeppe RE, Standaert RF, Van Oosten BJ, et al. Lipid bilayer thickness determines cholesterol's location in model membranes. Soft Matter (2016) 12:9417–28. doi:10.1039/C6SM01777K
75. Chou T-H, Chang C-H. Thermodynamic behavior and relaxation processes of mixed DPPC/cholesterol monolayers at the air/water interface. Colloids Surf B: Biointerfaces (2000) 17:71–9. doi:10.1016/S0927-7765(99)00102-2
76. Wydro P, Hac-Wydro K. Thermodynamic description of the interactions between lipids in ternary Langmuir monolayers: The study of cholesterol distribution in membranes. J Phys Chem B (2007) 111:2495–502. doi:10.1021/jp066950+
77. Sabatini K, Mattila J-P, Kinnunen PK. Interfacial behavior of cholesterol, ergosterol, and lanosterol in mixtures with DPPC and DMPC. Biophys J (2008) 95:2340–55. doi:10.1529/biophysj.108.132076
78. Risselada HJ, Marrink SJ. The molecular face of lipid rafts in model membranes. Proc Natl Acad Sci (2008) 105:17367–72. doi:10.1073/pnas.0807527105
79. Kucerka N, Marquardt D, Harroun TA, Nieh MP, Wassall SR, de Jong DH, et al. Cholesterol in bilayers with PUFA chains: Doping with DMPC or POPC results in sterol reorientation and membrane-domain formation. Biochemistry (2010) 49:7485–93. doi:10.1021/bi100891z
80. Lyu D, Zhang L, Zhang Y. Effects of cholesterol on bilayers with various degrees of unsaturation of their phospholipid tails under mechanical stress. RSC Adv (2020) 10:11088–94. doi:10.1039/D0RA00624F
81. Huang J, Feigenson GW. A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers. Biophys J (1999) 76:2142–57. doi:10.1016/S0006-3495(99)77369-8
82. Kučerka N, Pencer J, Nieh M-P, Katsaras J. Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles. Eur Phys J E (2007) 23:247–54. doi:10.1140/epje/i2007-10202-8
83. Kučerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J, Sachs JN. The effect of cholesterol on short-and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering. Biophys J (2008) 95:2792–805. doi:10.1529/biophysj.107.122465
84. Hung W-C, Lee M-T, Chen F-Y, Huang HW. The condensing effect of cholesterol in lipid bilayers. Biophys J (2007) 92:3960–7. doi:10.1529/biophysj.106.099234
85. Martinez-Seara H, Róg T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M, Reigada R. Interplay of unsaturated phospholipids and cholesterol in membranes: Effect of the double-bond position. Biophys J (2008) 95:3295–305. doi:10.1529/biophysj.108.138123
86. Keller F, Heuer A. Chain ordering of phospholipids in membranes containing cholesterol: What matters? Soft Matter (2021) 17:6098–108. doi:10.1039/D1SM00459J
87. Karmakar S, Raghunathan VA. Structure of phospholipid-cholesterol membranes: An x-ray diffraction study. Phys Rev E (2005) 71:061924. doi:10.1103/PhysRevE.71.061924
88. Leftin A, Molugu TR, Job C, Beyer K, Brown MF. Area per lipid and cholesterol interactions in membranes from separated local-field 13C NMR spectroscopy. Biophys J (2014) 107:2274–86. doi:10.1016/j.bpj.2014.07.044
89. Trouard TP, Nevzorov AA, Alam TM, Job C, Zajicek J, Brown MF. Influence of cholesterol on dynamics of dimyristoylphosphatidylcholine bilayers as studied by deuterium NMR relaxation. J Chem Phys (1999) 110:8802–18. doi:10.1063/1.478787
90. Petrache HI, Dodd SW, Brown MF. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys J (2000) 79:3172–92. doi:10.1016/S0006-3495(00)76551-9
91. Kučerka N, Nieh M-P, Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta (BBA) - Biomembranes (2011) 1808:2761–71. doi:10.1016/j.bbamem.2011.07.022
92. Marquardt D, Heberle FA, Pan J, Cheng X, Pabst G, Harroun TA, et al. The structures of polyunsaturated lipid bilayers by joint refinement of neutron and X-ray scattering data. Chem Phys Lipids (2020) 229:104892. doi:10.1016/j.chemphyslip.2020.104892
93. Lewis-Laurent A, Doktorova M, Heberle FA, Marquardt D. Vesicle Viewer: Online analysis of small angle scattering from lipid vesicles. Biophys J (2021) 120. doi:10.1016/j.bpj.2021.09.018
94. Kelley EG, Butler PD, Ashkar R, Bradbury R, Nagao M. Scaling relationships for the elastic moduli and viscosity of mixed lipid membranes. Proc Natl Acad Sci (2020) 117:23365–73. doi:10.1073/pnas.2008789117
95. Miyoshi T, Kato S. Detailed analysis of the surface area and elasticity in the saturated 1, 2-diacylphosphatidylcholine/cholesterol binary monolayer system. Langmuir (2015) 31:9086–96. doi:10.1021/acs.langmuir.5b01775
96. Worthman L, Nag K, Davis PJ, Keough K. Cholesterol in condensed and fluid phosphatidylcholine monolayers studied by epifluorescence microscopy. Biophys J (1997) 72:2569–80. doi:10.1016/S0006-3495(97)78900-8
97. Kim K, Kim C, Byun Y. Preparation of a dipalmitoylphosphatidylcholine/cholesterol Langmuir− blodgett monolayer that suppresses protein adsorption. Langmuir (2001) 17:5066–70. doi:10.1021/la0102096
98. Lancelot E, Grauby-Heywang C. Comparison of the interaction of dihydrocholesterol and cholesterol with sphingolipid or phospholipid Langmuir monolayers. Colloids Surf B: Biointerfaces (2007) 59:81–6. doi:10.1016/j.colsurfb.2007.04.017
99. Duncan SL, Larson RG. Comparing experimental and simulated pressure-area isotherms for DPPC. Biophys J (2008) 94:2965–86. doi:10.1529/biophysj.107.114215
100. Herculano RD, Pavinatto FJ, Caseli L, D'Silva C, Oliveira ON. The lipid composition of a cell membrane modulates the interaction of an antiparasitic peptide at the air–water interface. Biochim Biophys Acta (BBA)-Biomembranes (2011) 1808:1907–12. doi:10.1016/j.bbamem.2011.03.012
101. Mills TT, Huang J, Feigenson GW, Nagle JF. Effects of cholesterol and unsaturated DOPC lipid on chain packing of saturated gel-phase DPPC bilayers. Biophys J (2009) 96:126–39. doi:10.4149/gpb_2009_02_126
102. Huang J, Buboltz JT, Feigenson GW. Maximum solubility of cholesterol in phosphatidylcholine and phosphatidylethanolamine bilayers. Biochim Biophys Acta (BBA) - Biomembranes (1999) 1417:89–100. doi:10.1016/S0005-2736(98)00260-0
103. Garg S, Castro-Roman F, Porcar L, Butler P, Bautista PJ, Krzyzanowski N, et al. Cholesterol solubility limit in lipid membranes probed by small angle neutron scattering and MD simulations. Soft Matter (2014) 10:9313–7. doi:10.1039/C4SM01219D
104. Heberle FA, Doktorova M, Goh SL, Standaert RF, Katsaras J, Feigenson GW. Hybrid and nonhybrid lipids exert common effects on membrane raft size and morphology. J Am Chem Soc (2013) 135:14932–5. doi:10.1021/ja407624c
105. Veatch SL, Soubias O, Keller SL, Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc Natl Acad Sci (2007) 104:17650–5. doi:10.1073/pnas.0703513104
106. Konyakhina TM, Wu J, Mastroianni JD, Heberle FA, Feigenson GW. Phase diagram of a 4-component lipid mixture: DSPC/DOPC/POPC/chol. Biochim Biophys Acta (BBA)-Biomembranes (2013) 1828:2204–14. doi:10.1016/j.bbamem.2013.05.020
107. Veatch SL, Keller SL. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J (2003) 85:3074–83. doi:10.1016/S0006-3495(03)74726-2
108. Zhao J, Wu J, Heberle FA, Mills TT, Klawitter P, Huang G, et al. Phase studies of model biomembranes: Complex behavior of DSPC/DOPC/cholesterol. Biochim Biophys Acta (BBA) - Biomembranes (2007) 1768:2764–76. doi:10.1016/j.bbamem.2007.07.008
109. Smaby JM, Brockman HL, Brown RE. Cholesterol’s interfacial interactions with sphingomyelins and-phosphatidylcholines: hydrocarbon chain structure determines the magnitude of condensation. Biochemistry (1994) 33:9135–142. doi:10.1021/bi00197a016
110. Albrecht O, Gruler H, Sackmann E. Pressure-composition phase diagrams of cholesterol/lecithin, cholesterol/phosphatidic acid, and lecithin/phosphatidic acid mixed monolayers: A Langmuir film balance study. J Colloid Interf Sci (1981) 79:319–38. doi:10.1016/0021-9797(81)90084-9
111. Heberle FA, Feigenson GW. Phase separation in lipid membranes. Cold Spring Harbor Perspect Biol (2011) 3:a004630. doi:10.1101/cshperspect.a004630
112. Nyholm TKM, Jaikishan S, Engberg O, Hautala V, Slotte JP. The affinity of sterols for different phospholipid classes and its impact on lateral segregation. Biophys J (2019) 116:296–307. doi:10.1016/j.bpj.2018.11.3135
113. Shaw TR, Wisser KC, Schaffner TA, Gaffney AD, Machta BB, Veatch SL. Chemical potential measurements constrain models of cholesterol-phosphatidylcholine interactions. Biophys J (2023) 122:1105–117. doi:10.1016/j.bpj.2023.02.009
114. Kato A, Tsuji A, Yanagisawa M, Saeki D, Juni K, Morimoto Y, et al. Phase separation on a phospholipid membrane inducing a characteristic localization of DNA accompanied by its structural transition. J Phys Chem Lett (2010) 1:3391–5. doi:10.1021/jz101376m
115. Veatch SL, Keller SL. Organization in lipid membranes containing cholesterol. Phys Rev Lett (2002) 89:268101. doi:10.1103/PhysRevLett.89.268101
116. Heberle FA, Petruzielo RS, Pan J, Drazba P, Kučerka N, Standaert RF, et al. Bilayer thickness mismatch controls domain size in model membranes. J Am Chem Soc (2013) 135:6853–9. doi:10.1021/ja3113615
117. Feigenson GW. Phase boundaries and biological membranes. Annu Rev Biophys Biomol Struct (2007) 36:63–77. doi:10.1146/annurev.biophys.36.040306.132721
118. Mesquita RM, Melo E, Thompson TE, Vaz WL. Partitioning of amphiphiles between coexisting ordered and disordered phases in two-phase lipid bilayer membranes. Biophys J (2000) 78:3019–25. doi:10.1016/S0006-3495(00)76840-8
119. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science (2010) 327:46–50. doi:10.1126/science.1174621
120. Presti FT. The role of cholesterol in regulating membrane fluidity. In: Membrane Fluidity in Biology: Cellular Aspects. editors RC Aloia, and JM Boggs. Academic Press (1985) 4. p. 97–145.
121. Kaddah S, Khreich N, Kaddah F, Charcosset C, Greige-Gerges H. Cholesterol modulates the liposome membrane fluidity and permeability for a hydrophilic molecule. Food Chem Toxicol (2018) 113:40–8. doi:10.1016/j.fct.2018.01.017
122. Szabo G. Dual mechanism for the action of cholesterol on membrane permeability. Nature (1974) 252:47–9. doi:10.1038/252047a0
123. Galla HJ, Hartmann W, Theilen U, Sackmann E. On two-dimensional passive random walk in lipid bilayers and fluid pathways in biomembranes. J Membr Biol (1979) 48:215–36. doi:10.1007/BF01872892
124. Papahadjopoulos D, Cowden M, Kimelberg H. Role of cholesterol in membranes effects on phospholipid-protein interactions, membrane permeability and enzymatic activity. Biochim Biophys Acta (BBA) - Biomembranes (1973) 330:8–26. doi:10.1016/0005-2736(73)90280-0
125. Srivastava RC, Sahney R, Upadhyay S, Gupta RL. Membrane permeability based cholesterol sensor — a new possibility. J Membr Sci (2000) 164:45–9. doi:10.1016/S0376-7388(99)00201-X
126. Briuglia M-L, Rotella C, McFarlane A, Lamprou DA. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv Transl Res (2015) 5:231–42. doi:10.1007/s13346-015-0220-8
127. De Gier J, Mandersloot JG, Van Deenen LLM. Lipid composition and permeability of liposomes. Biochim Biophys Acta (BBA) - Biomembranes (1968) 150:666–75. doi:10.1016/0005-2736(68)90056-4
128. Wennberg CL, van der Spoel D, Hub JS. Large influence of cholesterol on solute partitioning into lipid membranes. J Am Chem Soc (2012) 134:5351–61. doi:10.1021/ja211929h
129. Tripathy M, Srivastava A. Lipid packing in biological membranes governs protein localization and membrane permeability. Biophys J (2023) 122:2727–43. doi:10.1016/j.bpj.2023.05.028
130. Needham D, Nunn RS. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys J (1990) 58:997–1009. doi:10.1016/S0006-3495(90)82444-9
131. Saeedimasine M, Montanino A, Kleiven S, Villa A. Role of lipid composition on the structural and mechanical features of axonal membranes: A molecular simulation study. Sci Rep (2019) 9:8000. doi:10.1038/s41598-019-44318-9
132. Doktorova M, LeVine MV, Khelashvili G, Weinstein H. A new computational method for membrane compressibility: Bilayer mechanical thickness revisited. Biophys J (2019) 116:790–502. doi:10.1016/j.bpj.2019.07.039
133. Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J (2000) 79:328–39. doi:10.1016/s0006-3495(00)76295-3
134. Pan J, Tristram-Nagle S, Nagle JF. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys Rev E (2009) 80:021931. doi:10.1103/physreve.80.021931
135. Gracià RS, Bezlyepkina N, Knorr RL, Lipowsky R, Dimova R. Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter (2010) 6:1472–82. doi:10.1039/B920629A
136. Khondker A, Alsop RJ, Dhaliwal A, Saem S, Moran-Mirabal JM, Rheinstädter MC. Membrane cholesterol reduces polymyxin B nephrotoxicity in renal membrane analogs. Biophys J (2017) 113:2016–28. doi:10.1016/j.bpj.2017.09.013
137. Kirby C, Clarke J, Gregoriadis G. Effect of the cholesterol content of small unilamellar liposomes on their stability in vivo and in vitro. Biochem J (1980) 186:591–8. doi:10.1042/bj1860591
138. Nickels JD, Cheng X, Mostofian B, Stanley C, Lindner B, Heberle FA, et al. Mechanical properties of nanoscopic lipid domains. J Am Chem Soc (2015) 137:15772–80. doi:10.1021/jacs.5b08894
139. Ruwizhi N, Aderibigbe BA. The efficacy of cholesterol-based carriers in drug delivery. Molecules (2020) 25:4330. doi:10.3390/molecules25184330
140. Hu DR, Qian ZY. Cholesterol in drug delivery systems. In: Cholesterol: From Chemistry and Biophysics to the Clinic. editors AN Bukiya, and AM Dopico Academic Press (2022). p. 797–824.
141. Huang Z, Szoka FC. Sterol-modified phospholipids: Cholesterol and phospholipid chimeras with improved biomembrane properties. J Am Chem Soc (2008) 130:15702–12. doi:10.1021/ja8065557
142. Shaghaghi M, Keyvanloo A, Huang Z, Szoka FC, Thewalt JL. Constrained versus free cholesterol in DPPC membranes: A comparison of chain ordering ability using deuterium NMR. Langmuir (2017) 33:14405–13. doi:10.1021/acs.langmuir.7b03299
143. Foglia F, Barlow DJ, Szoka FC, Huang Z, Rogers SE, Lawrence MJ. Structural studies of the monolayers and bilayers formed by a novel cholesterol-phospholipid chimera. Langmuir (2011) 27:8275–81. doi:10.1021/la200739y
144. Gallová J, Uhríková D, Kučerka N, Teixeira J, Balgavý P. Hydrophobic thickness, lipid surface area and polar region hydration in monounsaturated diacylphosphatidylcholine bilayers: SANS study of effects of cholesterol and β-sitosterol in unilamellar vesicles. Biochim Biophys Acta (BBA)-Biomembranes (2008) 1778:2627–32. doi:10.1016/j.bbamem.2008.08.009
145. Martinez GV, Dykstra EM, Lope-Piedrafita S, Brown MF. Lanosterol and cholesterol-induced variations in bilayer elasticity probed by 2H NMR relaxation. Langmuir (2004) 20:1043–6. doi:10.1021/la036063n
146. Bloch KE. Sterol, structure and membrane function. Crit Rev Biochem (1983) 14:47–92. doi:10.3109/10409238309102790
147. Hodzic A, Rappolt M, Amenitsch H, Laggner P, Pabst G. Differential modulation of membrane structure and fluctuations by plant sterols and cholesterol. Biophys J (2008) 94:3935–44. doi:10.1529/biophysj.107.123224
148. Henriksen J, Rowat A, Brief E, Hsueh Y, Thewalt J, Zuckermann M, et al. Universal behavior of membranes with sterols. Biophys J (2006) 90:1639–49. doi:10.1529/biophysj.105.067652
149. Bernsdorff C, Winter R. Differential properties of the sterols cholesterol, ergosterol, β-sitosterol, trans-7-dehydrocholesterol, stigmasterol and lanosterol on DPPC bilayer order. J Phys Chem B (2003) 107:10658–64. doi:10.1021/jp034922a
150. Pencer J, Nieh MP, Harroun TA, Krueger S, Adams C, Katsaras J. Bilayer thickness and thermal response of dimyristoylphosphatidylcholine unilamellar vesicles containing cholesterol, ergosterol and lanosterol: A small-angle neutron scattering study. Biochim Biophys Acta (BBA)-Biomembranes (2005) 1720:84–91. doi:10.1016/j.bbamem.2005.10.017
151. Ermakova E, Zuev Y. Effect of ergosterol on the fungal membrane properties. All-atom and coarse-grained molecular dynamics study. Chem Phys Lipids (2017) 209:45–53. doi:10.1016/j.chemphyslip.2017.11.006
152. Hung W-C, Lee MT, Chung H, Sun YT, Chen H, Charron N, et al. Comparative study of the condensing effects of ergosterol and cholesterol. Biophys J (2016) 110:2026–33. doi:10.1016/j.bpj.2016.04.003
153. Cournia Z, Ullmann GM, Smith JC. Differential effects of cholesterol, ergosterol and lanosterol on a dipalmitoyl phosphatidylcholine membrane: A molecular dynamics simulation study. J Phys Chem B (2007) 111:1786–801. doi:10.1021/jp065172i
154. Czub J, Baginski M. Comparative molecular dynamics study of lipid membranes containing cholesterol and ergosterol. Biophys J (2006) 90:2368–82. doi:10.1529/biophysj.105.072801
155. Urbina JA, Pekerar S, Patterson J, Montez B, Oldfield E. Molecular order and dynamics of phosphatidylcholine bilayer membranes in the presence of cholesterol, ergosterol and lanosterol: A comparative study using 2H-13C-and 31P-NMR spectroscopy. Biochim Biophys Acta (BBA)-Biomembranes (1995) 1238:163–76. doi:10.1016/0005-2736(95)00117-L
156. Dufourc EJ. Sterols and membrane dynamics. J Chem Biol (2008) 1:63–77. doi:10.1007/s12154-008-0010-6
157. Stevens MM, Honerkamp-Smith AR, Keller SL. Solubility limits of cholesterol, lanosterol, ergosterol, stigmasterol, and β-sitosterol in electroformed lipid vesicles. Soft Matter (2010) 6:5882–90. doi:10.1039/C0SM00373E
158. Ibarguren M, López DJ, Escribá PV. The effect of natural and synthetic fatty acids on membrane structure, microdomain organization, cellular functions and human health. Biochim Biophys Acta (BBA)-Biomembranes (2014) 1838:1518–28. doi:10.1016/j.bbamem.2013.12.021
159. Lapshina E, Zavodnik I, Bryszewska M. Effect of free fatty acids on the structure and properties of erythrocyte membrane. Scand J Clin Lab Invest (1995) 55:391–7. doi:10.3109/00365519509104978
160. Teres S, Barceló-Coblijn G, Benet M, Álvarez R, Bressani R, Halver JE, et al. Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc Natl Acad Sci (2008) 105:13811–6. doi:10.1073/pnas.0807500105
161. Eliasz A, Chapman D, Ewing D. Phospholipid phase transitions. Effects of n-alcohols, n-monocarboxylic acids, phenylalkyl alcohols and quatenary ammonium compounds. Biochim Biophys Acta (BBA)-Biomembranes (1976) 448:220–33. doi:10.1016/0005-2736(76)90238-8
162. Mabrey S, Sturtevant JM. Incorporation of saturated fatty acids into phosphatidylcholine bilayers. Biochim Biophys Acta (BBA)-Lipids Lipid Metab (1977) 486:444–50. doi:10.1016/0005-2760(77)90094-7
163. Kantor HL, Prestegard JH. Fusion of phosphatidylcholine bilayer vesicles: Role of free fatty acid. Biochemistry (1978) 17:3592–7. doi:10.1021/bi00610a027
164. Schullery SE, Seder TA, Weinstein DA, Bryant DA. Differential thermal analysis of dipalmitoylphosphatidylcholine-fatty acid mixtures. Biochemistry (1981) 20:6818–24. doi:10.1021/bi00527a012
165. Usher J, Epand R, Papahadjopoulos D. The effect of free fatty acids on the thermotropic phase transition of dimyristoyl glycerophosphocholine. Chem Phys Lipids (1978) 22:245–53. doi:10.1016/0009-3084(78)90031-2
166. Klausner RD, Kleinfeld A, Hoover R, Karnovsky M. Lipid domains in membranes: Evidence derived from structural perturbations induced by free fatty acids and lifetime heterogeneity analysis. J Biol Chem (1980) 255:1286–95. doi:10.1016/S0021-9258(19)86027-1
167. Marsh D, Seddon JM. Gel-to-inverted hexagonal (Lβ-HII) phase transitions in phosphatidylethanolamines and fatty acid-phosphatidylcholine mixtures, demonstrated by 31P-NMR spectroscopy and X-ray diffraction. Biochim Biophys Acta (BBA)-Biomembranes (1982) 690:117–23. doi:10.1016/0005-2736(82)90245-0
168. Podo F, Blasie J. Proton magnetic relaxation studies of mixed phosphatidylcholine/fatty acid and mixed phosphatidylcholine bimolecular bilayers. Biochim Biophys Acta (BBA)-Biomembranes (1976) 419:1–18. doi:10.1016/0005-2736(76)90368-0
169. Kremer J, Wiersema P. Exchange and aggregation in dispersions of dimyristoyl phosphatidylcholine vesicles containing myristic acid. Biochim Biophys Acta (BBA)-Biomembranes (1977) 471:348–60. doi:10.1016/0005-2736(77)90041-4
170. Pauls KP, MacKay AL, Bloom M. Deuterium nuclear magnetic resonance study of the effects of palmitic acid on dipalmitoylphosphatidylcholine bilayers. Biochemistry (1983) 22:6101–9. doi:10.1021/bi00295a010
171. Mally M, Peterlin P, Svetina S. Partitioning of oleic acid into phosphatidylcholine membranes is amplified by strain. J Phys Chem B (2013) 117:12086–94. doi:10.1021/jp404135g
172. Ibarguren M, López DJ, Encinar JA, González-Ros JM, Busquets X, Escribá PV. Partitioning of liquid-ordered/liquid-disordered membrane microdomains induced by the fluidifying effect of 2-hydroxylated fatty acid derivatives. Biochim Biophys Acta (BBA)-Biomembranes (2013) 1828:2553–63. doi:10.1016/j.bbamem.2013.06.014
173. Wang C, Harris WS, Chung M, Lichtenstein AH, Balk EM, Kupelnick B, et al. n−3 Fatty acids from fish or fish-oil supplements, but not α-linolenic acid, benefit cardiovascular disease outcomes in primary- and secondary-prevention studies: a systematic review. Am J Clin Nutr (2006) 84:5–17. doi:10.1093/ajcn/84.1.5
174. Epand RM, Epand RF, Ahmed N, Chen R. Promotion of hexagonal phase formation and lipid mixing by fatty acids with varying degrees of unsaturation. Chem Phys Lipids (1991) 57:75–80. doi:10.1016/0009-3084(91)90051-C
175. Romero LO, Massey AE, Mata-Daboin AD, Sierra-Valdez FJ, Chauhan SC, Cordero-Morales JF, et al. Dietary fatty acids fine-tune Piezo1 mechanical response. Nat Commun (2019) 10:1200. doi:10.1038/s41467-019-09055-7
176. De Santis A, Varela Y, Sot J, D’Errico G, Goñi FM, Alonso A. Omega-3 polyunsaturated fatty acids do not fluidify bilayers in the liquid-crystalline state. Sci Rep (2018) 8:16240–13. doi:10.1038/s41598-018-34264-3
177. Stillwell W, Wassall SR. Docosahexaenoic acid: Membrane properties of a unique fatty acid. Chem Phys Lipids (2003) 126:1–27. doi:10.1016/S0009-3084(03)00101-4
178. Saitta F, Motta P, Barbiroli A, Signorelli M, La Rosa C, Janaszewska A, et al. Influence of free fatty acids on lipid membrane–Nisin interaction. Langmuir (2020) 36:13535–44. doi:10.1021/acs.langmuir.0c02266
179. Cerezo J, Zuniga J, Bastida A, Requena A, Ceron-Carrasco JP. Atomistic molecular dynamics simulations of the interactions of oleic and 2-hydroxyoleic acids with phosphatidylcholine bilayers. J Phys Chem B (2011) 115:11727–38. doi:10.1021/jp203498x
180. Jacobs ML, Faizi HA, Peruzzi JA, Vlahovska PM, Kamat NP. EPA and DHA differentially modulate membrane elasticity in the presence of cholesterol. Biophys J (2021) 120:2317–29. doi:10.1016/j.bpj.2021.04.009
181. Onuki Y, Morishita M, Chiba Y, Tokiwa S, Takayama K. Docosahexaenoic acid and eicosapentaenoic acid induce changes in the physical properties of a lipid bilayer model membrane. Chem Pharm Bull (2006) 54:68–71. doi:10.1248/cpb.54.68
182. Mason RP, Jacob RF, Shrivastava S, Sherratt SC, Chattopadhyay A. Eicosapentaenoic acid reduces membrane fluidity, inhibits cholesterol domain formation, and normalizes bilayer width in atherosclerotic-like model membranes. Biochim Biophys Acta (BBA)-Biomembranes (2016) 1858:3131–40. doi:10.1016/j.bbamem.2016.10.002
183. Sherratt SC, Mason RP. Eicosapentaenoic acid and docosahexaenoic acid have distinct membrane locations and lipid interactions as determined by X-ray diffraction. Chem Phys Lipids (2018) 212:73–9. doi:10.1016/j.chemphyslip.2018.01.002
184. Muranushi N, Takagi N, Muranishi S, Sezaki H. Effect of fatty acids and monoglycerides on permeability of lipid bilayer. Chem Phys Lipids (1981) 28:269–79. doi:10.1016/0009-3084(81)90013-X
185. Sebastian B, Tobias U. The influence of additives on the nanoscopic dynamics of the phospholipid dimyristoylphosphatidylcholine. Biochim Biophys Acta (BBA) - Biomembranes (2011) 1808:199–208. doi:10.1016/j.bbamem.2010.10.012
186. Caires R, Sierra-Valdez FJ, Millet JR, Herwig JD, Roan E, Vásquez V, et al. Omega-3 fatty acids modulate TRPV4 function through plasma membrane remodeling. Cel Rep (2017) 21:246–58. doi:10.1016/j.celrep.2017.09.029
187. Langner M, Isac T, Hui S. Interaction of free fatty acids with phospholipid bilayers. Biochim Biophys Acta (BBA)-Biomembranes (1995) 1236:73–80. doi:10.1016/0005-2736(95)00037-4
188. Funari SS, Barceló F, Escribá PV. Effects of oleic acid and its congeners, elaidic and stearic acids, on the structural properties of phosphatidylethanolamine membranes. J Lipid Res (2003) 44:567–75. doi:10.1194/jlr.M200356-JLR200
189. Pappas A, Johnsen S, Liu J-C, Eisinger M. Sebum analysis of individuals with and without acne. Dermato-endocrinology (2009) 1:157–61. doi:10.4161/derm.1.3.8473
190. Ezzili C, Otrubova K, Boger DL. Fatty acid amide signaling molecules. Bioorg Med Chem Lett (2010) 20:5959–68. doi:10.1016/j.bmcl.2010.08.048
191. Lohner K, Degovics G, Laggner P, Gnamusch E, Paltauf F. Squalene promotes the formation of non-bilayer structures in phospholipid model membranes. Biochim Biophys Acta (BBA)-Biomembranes (1993) 1152:69–77. doi:10.1016/0005-2736(93)90232-O
192. Vamparys L, Gautier R, Vanni S, Bennett WD, Tieleman DP, Antonny B, et al. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys J (2013) 104:585–93. doi:10.1016/j.bpj.2012.11.3836
193. Prakash SA, Sengan M, John ML, Veerappan A, Kamlekar RK. Interaction of N-acyltaurines with phosphatidylcholines. Biochim Biophys Acta (BBA)-Biomembranes 1865, 184103 (2023). doi:10.1016/j.bbamem.2022.184103
194. Kulkarni CV, Wachter W, Iglesias-Salto G, Engelskirchen S, Ahualli S. Monoolein: A magic lipid? Phys Chem Chem Phys (2011) 13:3004–21. doi:10.1039/C0CP01539C
195. Niemela PS, Miettinen MS, Monticelli L, Hammaren H, Bjelkmar P, Murtola T, et al. Membrane proteins diffuse as dynamic complexes with lipids. J Am Chem Soc (2010) 132:7574–5. doi:10.1021/ja101481b
196. Ebersberger L, Schindler T, Kirsch SA, Pluhackova K, Schambony A, Seydel T, et al. Lipid dynamics in membranes slowed down by transmembrane proteins. Front Cel Dev Biol (2020) 8:579388. doi:10.3389/fcell.2020.579388
197. Lee J-H, Choi SM, Doe C, Faraone A, Pincus PA, Kline SR. Thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides. Phys Rev Lett (2010) 105:038101. doi:10.1103/physrevlett.105.038101
198. Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Biophys Biomol Struct (2007) 36:107–30. doi:10.1146/annurev.biophys.36.040306.132643
199. Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta (BBA) - Biomembranes (2004) 1666:62–87. doi:10.1016/j.bbamem.2004.05.012
200. Brown MF. Soft matter in lipid–protein interactions. Annu Rev Biophys (2017) 46:379–410. doi:10.1146/annurev-biophys-070816-033843
201. Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature (2009) 459:379–85. doi:10.1038/nature08147
202. White SH, Ladokhin AS, Jayasinghe S, Hristova K. How membranes shape protein structure. J Biol Chem (2001) 276:32395–8. doi:10.1074/jbc.R100008200
203. Zou Y, Pan R, Ruan Q, Wan Z, Guo J, Yang X. Interaction of soybean 7s globulin peptide with cell membrane model via isothermal titration calorimetry, quartz crystal microbalance with dissipation, and Langmuir monolayer study. J Agric Food Chem (2018) 66:4913–22. doi:10.1021/acs.jafc.8b00414
204. Dick RA, Goh SL, Feigenson GW, Vogt VM. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci (2012) 109:18761–6. doi:10.1073/pnas.1209408109
205. Guixà-González R, Albasanz JL, Rodriguez-Espigares I, Pastor M, Sanz F, Martí-Solano M, et al. Membrane cholesterol access into a G-protein-coupled receptor. Nat Commun (2017) 8:14505. doi:10.1038/ncomms14505
206. Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol (1996) 3:842–8. doi:10.1038/nsb1096-842
207. Marx DC, Fleming KG. Local bilayer hydrophobicity modulates membrane protein stability. J Am Chem Soc (2021) 143:764–72. doi:10.1021/jacs.0c09412
208. White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct (1999) 28:319–65. doi:10.1146/annurev.biophys.28.1.319
209. Fleming KG. Energetics of membrane protein folding. Annu Rev Biophys (2014) 43:233–55. doi:10.1146/annurev-biophys-051013-022926
210. Popot J-L, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem (2000) 69:881–922. doi:10.1146/annurev.biochem.69.1.881
211. Melo MN, Arnarez C, Sikkema H, Kumar N, Walko M, Berendsen HJC, et al. High-throughput simulations reveal membrane-mediated effects of alcohols on MscL gating. J Am Chem Soc (2017) 139:2664–71. doi:10.1021/jacs.6b11091
212. Soubias O, Sodt AJ, Teague WE, Hines KG, Gawrisch K. Physiological changes in bilayer thickness induced by cholesterol control GPCR rhodopsin function. Biophys J (2023) 122:973–83. doi:10.1016/j.bpj.2022.11.2937
213. Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature (2016) 534:347–51. doi:10.1038/nature17964
214. Perozo E, Kloda A, Cortes DM, Martinac B. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol (2002) 9:696–703. doi:10.1038/nsb827
215. Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature (2002) 418:942–8. doi:10.1038/nature00992
216. Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature (2007) 450:376–82. doi:10.1038/nature06265
217. Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cel Biol (2008) 9:99–111. doi:10.1038/nrm2328
218. Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature (2001) 409:387–90. doi:10.1038/35053176
219. Watanabe M, Tomita T, Yasuda T. Membrane-damaging action of staphylococcal alpha-toxin on phospholipid-cholesterol liposomes. Biochim Biophys Acta (BBA)-Biomembranes (1987) 898:257–65. doi:10.1016/0005-2736(87)90065-4
220. Tomita T, Watanabe M, Yasuda T. Influence of membrane fluidity on the assembly of Staphylococcus aureus alpha-toxin, a channel-forming protein, in liposome membrane. J Biol Chem (1992) 267:13391–7. doi:10.1016/S0021-9258(18)42223-5
221. Tsofina L, Liberman E, Babakov A. Production of bimolecular protein-lipid membranes in aqueous solution. Nature (1966) 212:681–3. doi:10.1038/212681a0
222. Bayley H, Cronin B, Heron A, Holden MA, Hwang WL, Syeda R, et al. Droplet interface bilayers. Mol BioSystems (2008) 4:1191–208. doi:10.1039/B808893D
223. Funakoshi K, Suzuki H, Takeuchi S. Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis. Anal Chem (2006) 78:8169–74. doi:10.1021/ac0613479
224. König BW, et al. Neutron reflectivity and atomic force microscopy studies of a lipid bilayer in water adsorbed to the surface of a silicon single crystal. Langmuir (1996) 12:1343–50. doi:10.1021/la950580r
225. Tamm LK, McConnell HM. Supported phospholipid bilayers. Biophys J (1985) 47:105–13. doi:10.1016/S0006-3495(85)83882-0
226. Menestrina G. Ionic channels formed by Staphylococcus aureus alpha-toxin: Voltage-dependent inhibition by divalent and trivalent cations. J Membr Biol (1986) 90:177–90. doi:10.1007/BF01869935
227. Gouaux JE, Braha O, Hobaugh MR, Song L, Cheley S, Shustak C, et al. Subunit stoichiometry of staphylococcal alpha-hemolysin in crystals and on membranes: A heptameric transmembrane pore. Proc Natl Acad Sci (1994) 91:12828–31. doi:10.1073/pnas.91.26.12828
228. Kawano R, Tsuji Y, Kamiya K, Kodama T, Osaki T, Miki N, et al. A portable lipid bilayer system for environmental sensing with a transmembrane protein. PLoS One (2014) 9:e102427. doi:10.1371/journal.pone.0102427
229. Manzer ZA, Ghosh S, Jacobs ML, Krishnan S, Zipfel WR, Piñeros M, et al. Cell-free synthesis of a transmembrane mechanosensitive channel protein into a hybrid-supported lipid bilayer. ACS Appl Bio Mater (2021) 4:3101–12. doi:10.1021/acsabm.0c01482
230. Hilburger CE, Jacobs ML, Lewis KR, Peruzzi JA, Kamat NP. Controlling secretion in artificial cells with a membrane and gate. ACS Synth Biol (2019) 8:1224–30. doi:10.1021/acssynbio.8b00435
231. Sengupta P, Baird B, Holowka D. Lipid rafts, fluid/fluid phase separation, and their relevance to plasma membrane structure and function. Seminars in cell & developmental biology (2007) 18 (5): 583–90. doi:10.1016/j.semcdb.2007.07.010
232. Brown DA, London E. Structure and function of sphingolipid-and cholesterol-rich membrane rafts. J Biol Chem (2000) 275:17221–4. doi:10.1074/jbc.R000005200
233. Sengupta P, Hammond A, Holowka D, Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim Biophys Acta (BBA)-Biomembranes (2008) 1778:20–32. doi:10.1016/j.bbamem.2007.08.028
234. Sengupta P, Seo AY, Pasolli HA, Song YE, Johnson MC, Lippincott-Schwartz J. A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat Cel Biol (2019) 21:452–61. doi:10.1038/s41556-019-0300-y
235. Czogalla A, Franquelim HG, Schwille P. DNA nanostructures on membranes as tools for synthetic biology. Biophys J (2016) 110:1698–707. doi:10.1016/j.bpj.2016.03.015
236. Chabanon M, Stachowiak JC, Rangamani P. Systems biology of cellular membranes: A convergence with biophysics. WIREs Syst Biol Med (2017) 9:e1386. doi:10.1002/wsbm.1386
237. Aumiller WM, Davis BW, Keating CD. Phase separation as a possible means of nuclear compartmentalization. In: R Hancock, and KW Jeon, editors. International review of cell and molecular biology Academic Press (2014) 307. p. 109–49.
238. Dinic J, Riehl A, Adler J, Parmryd I. The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Sci Rep (2015) 5:10082. doi:10.1038/srep10082
239. Kabouridis PS. Lipid rafts in T cell receptor signalling (Review). Mol Membr Biol (2006) 23:49–57. doi:10.1080/09687860500453673
240. Balagopalan L, Kortum RL, Coussens NP, Barr VA, Samelson LE. The linker for activation of T cells (LAT) signaling hub: from signaling complexes to microclusters. J Biol Chem (2015) 290:26422–9. doi:10.1074/jbc.R115.665869
241. Lin X, Gorfe AA, Levental I. Protein partitioning into ordered membrane domains: Insights from simulations. Biophys J (2018) 114:1936–44. doi:10.1016/j.bpj.2018.03.020
242. Lorent JH, Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids (2015) 192:23–32. doi:10.1016/j.chemphyslip.2015.07.022
243. Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature (2014) 510:48–57. doi:10.1038/nature13474
244. Jähnig F. Thermodynamics and kinetics of protein incorporation into membranes. Proc Natl Acad Sci (1983) 80:3691–5. doi:10.1073/pnas.80.12.3691
245. Lemmon MA, Engelman DM. Specificity and promiscuity in membrane helix interactions. Q Rev Biophys (1994) 27:157–218. doi:10.1017/S0033583500004522
246. Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annu Rev Biophys Biomol Struct (1997) 26:373–99. doi:10.1146/annurev.biophys.26.1.373
247. Bienvenue A, Bloom M, Davis J, Devaux P. Evidence for protein-associated lipids from deuterium nuclear magnetic resonance studies of rhodopsin-dimyristoylphosphatidylcholine recombinants. J Biol Chem (1982) 257:3032–8. doi:10.1016/S0021-9258(19)81069-4
248. Borbat P, Costa-Filho A, Earle K, Moscicki J, Freed . Electron spin resonance in studies of membranes and proteins. Science (2001) 291:266–9. doi:10.1126/science.291.5502.266
249. Keller RC, Snel MM, de Kruijff B, Marsh D. SecA restricts, in a nucleotide-dependent manner, acyl chain mobility up to the center of a phospholipid bilayer. FEBS Lett (1995) 358:251–4. doi:10.1016/0014-5793(94)01439-8
250. Kelley EG, Butler PD, Nagao M. Collective dynamics in lipid membranes containing transmembrane peptides. Soft Matter (2021) 17:5671–81. doi:10.1039/D1SM00314C
251. Kim W, Fricke N, Conery AL, Fuchs BB, Rajamuthiah R, Jayamani E, et al. NH125 kills methicillin-resistant Staphylococcus aureus persisters by lipid bilayer disruption. Future Med Chem (2016) 8:257–69. doi:10.4155/fmc.15.189
252. Lipkin R, Lazaridis T. Computational studies of peptide-induced membrane pore formation. Phil Trans R Soc B: Biol Sci (2017) 372:20160219. doi:10.1098/rstb.2016.0219
253. Khandelia H, Ipsen JH, Mouritsen OG. The impact of peptides on lipid membranes. Biochim Biophys Acta (BBA)-Biomembranes (2008) 1778:1528–36. doi:10.1016/j.bbamem.2008.02.009
254. Chen F-Y, Lee M-T, Huang HW. Evidence for membrane thinning effect as the mechanism for peptide-induced pore formation. Biophys J (2003) 84:3751–8. doi:10.1016/s0006-3495(03)75103-0
255. Mishra A, Gordon VD, Yang L, Coridan R, Wong GC. HIV TAT forms pores in membranes by inducing saddle-splay curvature: Potential role of bidentate hydrogen bonding. Angew Chem Int Edition (2008) 47:2986–9. doi:10.1002/anie.200704444
256. Dempsey CE. The actions of melittin on membranes. Biochim Biophys Acta (BBA)-Reviews Biomembranes (1990) 1031:143–61. doi:10.1016/0304-4157(90)90006-X
257. Wang KF, Nagarajan R, Camesano TA. Antimicrobial peptide alamethicin insertion into lipid bilayer: A QCM-D exploration. Colloids Surf B: Biointerfaces (2014) 116:472–81. doi:10.1016/j.colsurfb.2014.01.036
258. Yu Y, Vroman JA, Bae SC, Granick S. Vesicle budding induced by a pore-forming peptide. J Am Chem Soc (2010) 132:195–201. doi:10.1021/ja9059014
259. Allende D, Simon S, McIntosh TJ. Melittin-induced bilayer leakage depends on lipid material properties: Evidence for toroidal pores. Biophys J (2005) 88:1828–37. doi:10.1529/biophysj.104.049817
260. Sharma VK, Mamontov E, Anunciado DB, O'Neill H, Urban VS. Effect of antimicrobial peptide on the dynamics of phosphocholine membrane: Role of cholesterol and physical state of bilayer. Soft Matter (2015) 11:6755–67. doi:10.1039/C5SM01562F
261. Zhao H, Rinaldi AC, Di Giulio A, Simmaco M, Kinnunen PK. Interactions of the antimicrobial peptides temporins with model biomembranes. Comparison of temporins B and L. Biochemistry (2002) 41:4425–36. doi:10.1021/bi011929e
262. Hallock KJ, Lee D-K, Ramamoorthy A. MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys J (2003) 84:3052–60. doi:10.1016/S0006-3495(03)70031-9
263. Henzler Wildman KA, Lee D-K, Ramamoorthy A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry (2003) 42:6545–58. doi:10.1021/bi0273563
264. Powers J-PS, Tan A, Ramamoorthy A, Hancock RE. Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry (2005) 44:15504–13. doi:10.1021/bi051302m
265. Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta (BBA)-Biomembranes (2009) 1788:289–94. doi:10.1016/j.bbamem.2008.08.023
266. Sharma VK, Qian S. Effect of an antimicrobial peptide on lateral segregation of lipids: A structure and dynamics study by neutron scattering. Langmuir (2019) 35:4152–60. doi:10.1021/acs.langmuir.8b04158
267. Nguyen MHL, DiPasquale M, Rickeard BW, Doktorova M, Heberle FA, Scott HL, et al. Peptide-induced lipid flip-flop in asymmetric liposomes measured by small angle neutron scattering. Langmuir (2019) 35:11735–44. doi:10.1021/acs.langmuir.9b01625
268. Marx L, Frewein MPK, Semeraro EF, Rechberger G, Lohner K, Porcar L, et al. Antimicrobial peptide activity in asymmetric bacterial membrane mimics. Faraday Discuss (2021) 232:435–47. doi:10.1039/d1fd00039j
269. Lei J, Sun L, Huang S, Zhu C, He J, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res (2019) 11:3919–31.
270. Huan Y, Kong Q, Mou H, Yi H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front Microbiol (2020) 11:582779. doi:10.3389/fmicb.2020.582779
271. Seddon AM, Casey D, Law RV, Gee A, Templer RH, Ces O. Drug interactions with lipid membranes. Chem Soc Rev (2009) 38:2509–19. doi:10.1039/B813853M
272. Hardeland R, Cardinali DP, Srinivasan V, Spence DW, Brown GM, Pandi-Perumal SR. Melatonin—a pleiotropic, orchestrating regulator molecule. Prog Neurobiol (2011) 93:350–84. doi:10.1016/j.pneurobio.2010.12.004
273. Acuna Castroviejo D, C. Lopez L, Escames G, Lopez A, A. Garcia J, J. Reiter R. Melatonin-mitochondria interplay in health and disease. Curr Top Med Chem (2011) 11:221–40. doi:10.2174/156802611794863517
274. Kopustinskiene DM, Bernatoniene J. Molecular mechanisms of melatonin-mediated cell protection and signaling in health and disease. Pharmaceutics (2021) 13:129. doi:10.3390/pharmaceutics13020129
275. Drolle E, Kučerka N, Hoopes M, Choi Y, Katsaras J, Karttunen M, et al. Effect of melatonin and cholesterol on the structure of DOPC and DPPC membranes. Biochim Biophys Acta (BBA)-Biomembranes (2013) 1828:2247–54. doi:10.1016/j.bbamem.2013.05.015
276. Costa EJX, Lopes RH, Lamy-Freund MT. Permeability of pure lipid bilayers to melatonin. J Pineal Res (1995) 19:123–6. doi:10.1111/j.1600-079X.1995.tb00180.x
277. Bolmatov D, McClintic WT, Taylor G, Stanley CB, Do C, Collier CP, et al. Deciphering melatonin-stabilized phase separation in phospholipid bilayers. Langmuir (2019) 35:12236–45. doi:10.1021/acs.langmuir.9b01534
278. Choi Y, Attwood SJ, Hoopes MI, Drolle E, Karttunen M, Leonenko Z. Melatonin directly interacts with cholesterol and alleviates cholesterol effects in dipalmitoylphosphatidylcholine monolayers. Soft Matter (2014) 10:206–13. doi:10.1039/C3SM52064A
279. Robinson M, Turnbull S, Lee BY, Leonenko Z. The effects of melatonin, serotonin, tryptophan and NAS on the biophysical properties of DPPC monolayers. Biochim Biophys Acta (BBA)-Biomembranes (2020) 1862:183363. doi:10.1016/j.bbamem.2020.183363
280. De Lima VR, Caro MSB, Munford ML, Desbat B, Dufourc E, Pasa AA, et al. Influence of melatonin on the order of phosphatidylcholine-based membranes. J Pineal Res (2010) 49:169–75. doi:10.1111/j.1600-079x.2010.00782.x
281. Dies H, Cheung B, Tang J, Rheinstädter MC. The organization of melatonin in lipid membranes. Biochim Biophys Acta (BBA) - Biomembranes (2015) 1848:1032–40. doi:10.1016/j.bbamem.2015.01.006
282. Postila PA, Róg T. A perspective: Active role of lipids in neurotransmitter dynamics. Mol Neurobiol (2020) 57:910–25. doi:10.1007/s12035-019-01775-7
283. Peters GH, Wang C, Cruys-Bagger N, Velardez GF, Madsen JJ, Westh P. Binding of serotonin to lipid membranes. J Am Chem Soc (2013) 135:2164–71. doi:10.1021/ja306681d
284. Zohairi F, Khandelia H, Zanjani AAH. Interaction of psychedelic tryptamine derivatives with a lipid bilayer. Chem Phys Lipids (2023) 251:105279. doi:10.1016/j.chemphyslip.2023.105279
285. Nguyen TQT, Lund FW, Zanjani AAH, Khandelia H. Magic mushroom extracts in lipid membranes. Biochim Biophys Acta (BBA)-Biomembranes (2022) 1864:183957. doi:10.1016/j.bbamem.2022.183957
286. Khatun R, Singh S, Dubey NK, Prasad Das A. In: S Pati, T Sarkar, and D Lahiri, editors. Recent Frontiers of Phytochemicals. Amsterdam, Netherlands: Elsevier (2023). p. 383–95.
287. Martel J, Ojcius DM, Ko Y-F, Young JD. Phytochemicals as prebiotics and biological stress inducers. Trends Biochem Sci (2020) 45:462–71. doi:10.1016/j.tibs.2020.02.008
288. Ingólfsson HI, Thakur P, Herold KF, Hobart EA, Ramsey NB, Periole X, et al. Phytochemicals perturb membranes and promiscuously alter protein function. ACS Chem Biol (2014) 9:1788–98. doi:10.1021/cb500086e
289. Kaschina E, Steckelings UM, Unger T. Hypertension and the renin-angiotensin-aldosterone system. In: I Huhtaniemi, and L Martini. editors Encyclopedia of endocrine diseases Elsevier (2018). p. 505–10.
290. Vonkeman HE, van de Laar MA. Seminars in Arthritis and Rheumatism. Amsterdam, Netherlands: Elsevier (2023). 294–312.
291. Kapoor R, Peyear TA, Koeppe RE, Andersen OS. Antidepressants are modifiers of lipid bilayer properties. J Gen Physiol (2019) 151:342–56. doi:10.1085/jgp.201812263
292. Hill MW. The effect of anaesthetic-like molecules on the phase transition in smectic mesophases of dipalmitoyllecithin I. The normal alcohol up to C= 9 and three inhalation anaesthetics. Biochim Biophys Acta (BBA)-Biomembranes (1974) 356:117–24. doi:10.1016/0005-2736(74)90299-5
293. Bae S-J, Kitamura S, Herbette LG, Sturtevant JM. The effects of calcium channel blocking drugs on the thermotropic behavior of dimyristoylphosphatidylcholine. Chem Phys Lipids (1989) 51:1–7. doi:10.1016/0009-3084(89)90059-5
294. Giaginis C, Tsantili-Kakoulidou A. Alternative measures of lipophilicity: from octanol–water partitioning to IAM retention. J Pharm Sci (2008) 97:2984–3004. doi:10.1002/jps.21244
295. Du X, Zuo X, Meng F, Han C, Ouyang W, Han Y, et al. Direct inhibitory effect on viral entry of influenza A and SARS-CoV-2 viruses by azithromycin. Cel Prolif (2021) 54:e12953. doi:10.1111/cpr.12953
296. Hoffmann N, Lee B, Hentzer M, Rasmussen TB, Song Z, Johansen HK, et al. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob Agents Chemother (2007) 51:3677–87. doi:10.1128/aac.01011-06
297. Tateda K, Comte R, Pechere JC, Kohler T, Yamaguchi K, Van Delden C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother (2001) 45:1930–3. doi:10.1128/aac.45.6.1930-1933.2001
298. Berquand A, Mingeot-Leclercq M-P, Dufrene Y. Real-time imaging of drug–membrane interactions by atomic force microscopy. Biochim Biophys Acta (BBA)-Biomembranes (2004) 1664:198–205. doi:10.1016/j.bbamem.2004.05.010
299. Fa N, Lins L, Courtoy P, Dufrêne Y, Van Der Smissen P, Brasseur R, et al. Decrease of elastic moduli of DOPC bilayers induced by a macrolide antibiotic, azithromycin. Biochim Biophys Acta (BBA)-Biomembranes (2007) 1768:1830–8. doi:10.1016/j.bbamem.2007.04.013
300. Fotakis C, Christodouleas D, Zoumpoulakis P, Kritsi E, Benetis NP, Mavromoustakos T, et al. Comparative biophysical studies of sartan class drug molecules losartan and candesartan (CV-11974) with membrane bilayers. J Phys Chem B (2011) 115:6180–92. doi:10.1021/jp110371k
301. Ntountaniotis D, Mali G, Grdadolnik SG, Maria H, Skaltsounis AL, Potamitis C, et al. Thermal, dynamic and structural properties of drug AT1 antagonist olmesartan in lipid bilayers. Biochim Biophys Acta (BBA)-Biomembranes (2011) 1808:2995–3006. doi:10.1016/j.bbamem.2011.08.001
302. Kiriakidi S, Chatzigiannis C, Papaemmanouil C, Tzakos AG, Mavromoustakos T. Exploring the role of the membrane bilayer in the recognition of candesartan by its GPCR AT1 receptor. Biochim Biophys Acta (BBA)-Biomembranes (2020) 1862:183142. doi:10.1016/j.bbamem.2019.183142
303. Potamitis C, Chatzigeorgiou P, Siapi E, Viras K, Mavromoustakos T, Hodzic A, et al. Interactions of the AT1 antagonist valsartan with dipalmitoyl-phosphatidylcholine bilayers. Biochim Biophys Acta (BBA)-Biomembranes (2011) 1808:1753–63. doi:10.1016/j.bbamem.2011.02.002
304. Fotakis C, Megariotis G, Christodouleas D, Kritsi E, Zoumpoulakis P, Ntountaniotis D, et al. Comparative study of the AT1 receptor prodrug antagonist candesartan cilexetil with other sartans on the interactions with membrane bilayers. Biochim Biophys Acta (BBA)-Biomembranes (2012) 1818:3107–20. doi:10.1016/j.bbamem.2012.08.009
305. Liossi ΑS, Ntountaniotis D, Kellici TF, Chatziathanasiadou MV, Megariotis G, Mania M, et al. Exploring the interactions of irbesartan and irbesartan–2-hydroxypropyl-β-cyclodextrin complex with model membranes. Biochim Biophys Acta (BBA)-Biomembranes (2017) 1859:1089–98. doi:10.1016/j.bbamem.2017.03.003
306. Weissmann G. Aspirin. Scientific Am (1991) 264:84–90. stable/24936757. doi:10.1038/scientificamerican0191-84
307. Sharma V, Mamontov E, Ohl M, Tyagi M. Incorporation of aspirin modulates the dynamical and phase behavior of the phospholipid membrane. Phys Chem Chem Phys (2017) 19:2514–24. doi:10.1039/C6CP06202D
308. Suwalsky M, Belmar J, Villena F, Gallardo MJ, Jemiola-Rzeminska M, Strzalka K. Acetylsalicylic acid (aspirin) and salicylic acid interaction with the human erythrocyte membrane bilayer induce in vitro changes in the morphology of erythrocytes. Arch Biochem Biophys (2013) 539:9–19. doi:10.1016/j.abb.2013.09.006
309. Sharma V, Nagao M, Rai DK, Mamontov E. Membrane softening by nonsteroidal anti-inflammatory drugs investigated by neutron spin echo. Phys Chem Chem Phys (2019) 21:20211–8. doi:10.1039/C9CP03767E
310. Zhou Y, Raphael RM. Effect of salicylate on the elasticity, bending stiffness, and strength of SOPC membranes. Biophys J (2005) 89:1789–801. doi:10.1529/biophysj.104.054510
311. Sharma V, Mamontov E, Tyagi M. Effects of NSAIDs on the nanoscopic dynamics of lipid membrane. Biochim Biophys Acta (BBA)-Biomembranes (2020) 1862:183100. doi:10.1016/j.bbamem.2019.183100
312. Moreno MM, Garidel P, Suwalsky M, Howe J, Brandenburg K. The membrane-activity of ibuprofen, diclofenac, and naproxen: A physico-chemical study with lecithin phospholipids. Biochim Biophys Acta (BBA)-Biomembranes (2009) 1788:1296–303. doi:10.1016/j.bbamem.2009.01.016
313. Boggara MB, Krishnamoorti R. Small-angle neutron scattering studies of phospholipid− NSAID adducts. Langmuir (2010) 26:5734–45. doi:10.1021/la903854s
314. Boggara MB, Faraone A, Krishnamoorti R. Effect of pH and ibuprofen on the phospholipid bilayer bending modulus. J Phys Chem B (2010) 114:8061–6. doi:10.1021/jp100494n
315. Nunes C, Brezesinski G, Lima JL, Reis S, Lúcio M. Effects of non-steroidal anti-inflammatory drugs on the structure of lipid bilayers: Therapeutical aspects. Soft Matter (2011) 7:3002–10. doi:10.1039/C0SM00686F
316. Kyrikou I, Hadjikakou S, Kovala-Demertzi D, Viras K, Mavromoustakos T. Effects of non-steroid anti-inflammatory drugs in membrane bilayers. Chem Phys Lipids (2004) 132:157–69. doi:10.1016/j.chemphyslip.2004.06.005
317. Lúcio M, Bringezu F, Reis S, Lima JL, Brezesinski G. Binding of nonsteroidal anti-inflammatory drugs to DPPC: Structure and thermodynamic aspects. Langmuir (2008) 24:4132–9. doi:10.1021/la703584s
318. Zhou Y, Plowman SJ, Lichtenberger LM, Hancock JF. The anti-inflammatory drug indomethacin alters nanoclustering in synthetic and cell plasma membranes. J Biol Chem (2010) 285:35188–95. doi:10.1074/jbc.M110.141200
319. Sreij R, Prévost S, Dargel C, Dattani R, Hertle Y, Wrede O, et al. Interaction of the saponin aescin with ibuprofen in DMPC model membranes. Mol Pharm (2018) 15:4446–61. doi:10.1021/acs.molpharmaceut.8b00421
320. Hendrich AB, Wesołowska O, Michalak K. Trifluoperazine induces domain formation in zwitterionic phosphatidylcholine but not in charged phosphatidylglycerol bilayers. Biochim Biophys Acta (BBA)-Biomembranes (2001) 1510:414–25. doi:10.1016/S0005-2736(00)00373-4
321. Momo F, Fabris S, Stevanato R. Interaction of fluoxetine with phosphatidylcholine liposomes. Biophysical Chem (2005) 118:15–21. doi:10.1016/j.bpc.2005.06.006
322. Cantor RS. Breaking the meyer-overton rule: Predicted effects of varying stiffness and interfacial activity on the intrinsic potency of anesthetics. Biophys J (2001) 80:2284–97. doi:10.1016/S0006-3495(01)76200-5
323. Johnson FH, Flagler EA. Hydrostatic pressure reversal of narcosis in tadpoles. Science (1950) 112:91–2. doi:10.1126/science.112.2899.91.b
324. Johnson SM, Miller KW. Antagonism of pressure and anaesthesia. Nature (1970) 228:75–6. doi:10.1038/228075b0
325. Mashour GA, Forman SA, Campagna JA. Mechanisms of general anesthesia: from molecules to mind. Best Pract Res Clin Anaesthesiology (2005) 19:349–64. doi:10.1016/j.bpa.2005.01.004
326. Tsuchiya H, Mizogami M. Interaction of local anesthetics with biomembranes consisting of phospholipids and cholesterol: Mechanistic and clinical implications for anesthetic and cardiotoxic effects. Anesthesiology Res Pract (2013) 2013, 297141–18. doi:10.1155/2013/297141
327. Eckenhoff RG. Promiscuous ligands and attractive cavities: how do the inhaled anesthetics work?. Mol Interventions (2001) 1:258–68.
328. Herold KF, Sanford RL, Lee W, Schultz MF, Ingólfsson HI, Andersen OS, et al. Volatile anesthetics inhibit sodium channels without altering bulk lipid bilayer properties. J Gen Physiol (2014) 144:545–60. doi:10.1085/jgp.201411172
329. Cantor RS. The lateral pressure profile in membranes: A physical mechanism of general anesthesia. Biochemistry (1997) 36:2339–44. doi:10.1021/bi9627323
330. Zizzi EA, Cavaglià M, Tuszynski JA, Deriu MA. Alteration of lipid bilayer mechanics by volatile anesthetics: Insights from μs-long molecular dynamics simulations. iScience (2022) 25:103946. doi:10.1016/j.isci.2022.103946
331. Hantal G, Fábián B, Sega M, Jójárt B, Jedlovszky P. Effect of general anesthetics on the properties of lipid membranes of various compositions. Biochim Biophys Acta (BBA)-Biomembranes (2019) 1861:594–609. doi:10.1016/j.bbamem.2018.12.008
332. Booker RD, Sum AK. Biophysical changes induced by xenon on phospholipid bilayers. Biochim Biophys Acta (BBA)-Biomembranes (2013) 1828:1347–56. doi:10.1016/j.bbamem.2013.01.016
333. Gurtovenko AA, Anwar J. Interaction of ethanol with biological membranes: the formation of non-bilayer structures within the membrane interior and their significance. J Phys Chem B (2009) 113:1983–92. doi:10.1021/jp808041z
334. Tsuchiya H, Ueno T, Mizogami M, Takakura K. Local anesthetics structure-dependently interact with anionic phospholipid membranes to modify the fluidity. Chem. Biol. Interact. (2010) 183:19–24. doi:10.1016/j.cbi.2009.10.006
335. Moskovitz Y, Yang H. Modelling of noble anaesthetic gases and high hydrostatic pressure effects in lipid bilayers. Soft Matter (2015) 11:2125–38. doi:10.1039/C4SM02667E
336. Hauet N, Artzner F, Boucher F, Grabielle-Madelmont C, Cloutier I, Keller G, et al. Interaction between artificial membranes and enflurane, a general volatile anesthetic: DPPC-enflurane interaction. Biophys J (2003) 84:3123–37. doi:10.1016/S0006-3495(03)70037-X
337. Takeda K, Okuno H, Hata T, Nishimoto M, Matsuki H, Kaneshina S. Interdigitation and vesicle-to-micelle transformation induced by a local anesthetic tetracaine in phospholipids bilayers. Colloids Surf B: Biointerfaces (2009) 72:135–40. doi:10.1016/j.colsurfb.2009.03.026
338. Zapata-Morin PA, Sierra-Valdez F, Ruiz-Suárez J. The interaction of local anesthetics with lipid membranes. J Mol Graphics Model (2014) 53:200–5. doi:10.1016/j.jmgm.2014.08.001
339. Hu S, Zhao T, Li H, Cheng D, Sun Z. Effect of tetracaine on dynamic reorganization of lipid membranes. Biochim Biophys Acta (BBA)-Biomembranes (2020) 1862:183351. doi:10.1016/j.bbamem.2020.183351
340. Önyüksel H, Sethi V, Weinberg GL, Dudeja PK, Rubinstein I. Bupivacaine, but not lidocaine, disrupts cardiolipin-containing small biomimetic unilamellar liposomes. Chem Biol Interact (2007) 169:154–9. doi:10.1016/j.cbi.2007.06.002
341. Tsuchiya H, Mizogami M. Membrane interactivity of charged local anesthetic derivative and stereoselectivity in membrane interaction of local anesthetic enantiomers. Local Reg Anesth (2008) 1:1–9. doi:10.2147/lra.s3876
342. Lopes S, Ivanova G, de Castro B, Gameiro P. Cardiolipin and phosphatidylethanolamine role in dibucaine interaction with the mitochondrial membrane. Biochim Biophys Acta (BBA)-Biomembranes (2019) 1861:1152–61. doi:10.1016/j.bbamem.2019.02.011
343. Terama E, Ollila OHS, Salonen E, Rowat AC, Trandum C, Westh P, et al. Influence of ethanol on lipid membranes: from lateral pressure profiles to dynamics and partitioning. J Phys Chem B (2008) 112:4131–9. doi:10.1021/jp0750811
344. Böttner M, Winter R. Influence of the local anesthetic tetracaine on the phase behavior and the thermodynamic properties of phospholipid bilayers. Biophys J (1993) 65:2041–6. doi:10.1016/s0006-3495(93)81254-2
345. Yi Z, Nagao M, Bossev DP. Effect of charged lidocaine on static and dynamic properties of model bio-membranes. Biophysical Chem (2012) 160:20–7. doi:10.1016/j.bpc.2011.08.007
346. Hata T, Matsuki H, Kaneshina S. Effect of local anesthetics on the bilayer membrane of dipalmitoylphosphatidylcholine: Interdigitation of lipid bilayer and vesicle–micelle transition. Biophysical Chem (2000) 87:25–36. doi:10.1016/S0301-4622(00)00175-7
347. Mavromoustakos T, Theodoropoulou E, Yang D-P. The use of high-resolution solid-state NMR spectroscopy and differential scanning calorimetry to study interactions of anaesthetic steroids with membrane. Biochim Biophys Acta (BBA)-Biomembranes (1997) 1328:65–73. doi:10.1016/S0005-2736(97)00078-3
348. Kinoshita M, Chitose T, Matsumori N. Mechanism of local anesthetic-induced disruption of raft-like ordered membrane domains. Biochim Biophys Acta (BBA)-General Subjects (2019) 1863:1381–9. doi:10.1016/j.bbagen.2019.06.008
349. Aagaard TH, Kristensen MN, Westh P. Packing properties of 1-alkanols and alkanes in a phospholipid membrane. Biophysical Chem (2006) 119:61–8. doi:10.1016/j.bpc.2005.09.003
350. Barry JA, Gawrisch K. Direct NMR evidence for ethanol binding to the lipid-water interface of phospholipid bilayers. Biochemistry (1994) 33:8082–8. doi:10.1021/bi00192a013
351. Chanda J, Bandyopadhyay S. Perturbation of phospholipid bilayer properties by ethanol at a high concentration. Langmuir (2006) 22:3775–81. doi:10.1021/la053398r
352. Holte LL, Gawrisch K. Determining ethanol distribution in phospholipid multilayers with MAS− NOESY spectra. Biochemistry (1997) 36:4669–74. doi:10.1021/bi9626416
353. Patra M, Salonen E, Terama E, Vattulainen I, Faller R, Lee BW, et al. Under the influence of alcohol: The effect of ethanol and methanol on lipid bilayers. Biophys J (2006) 90:1121–35. doi:10.1529/biophysj.105.062364
354. Stetter FW, Hugel T. The nanomechanical properties of lipid membranes are significantly influenced by the presence of ethanol. Biophys J (2013) 104:1049–55. doi:10.1016/j.bpj.2013.01.021
355. Ly HV, Block DE, Longo ML. Interfacial tension effect of ethanol on lipid bilayer rigidity, stability, and area/molecule: A micropipet aspiration approach. Langmuir (2002) 18:8988–95. doi:10.1021/la026010q
356. Nizza DT, Gawrisch K. A layer model of ethanol partitioning into lipid membranes. Gen Physiol Biophys (2009) 28:140–5. doi:10.4149/gpb_2009_02_140
357. Ly HV, Longo ML. The influence of short-chain alcohols on interfacial tension, mechanical properties, area/molecule, and permeability of fluid lipid bilayers. Biophys J (2004) 87:1013–33. doi:10.1529/biophysj.103.034280
358. Griepernau B, Leis S, Schneider MF, Sikor M, Steppich D, Böckmann RA. 1-Alkanols and membranes: A story of attraction. Biochim Biophys Acta (BBA)-Biomembranes (2007) 1768:2899–913. doi:10.1016/j.bbamem.2007.08.002
359. Klacsová M, Bulacu M, Kučerka N, Uhríková D, Teixeira J, Marrink S, et al. The effect of aliphatic alcohols on fluid bilayers in unilamellar DOPC vesicles—a small-angle neutron scattering and molecular dynamics study. Biochim Biophys Acta (BBA)-Biomembranes (2011) 1808:2136–46. doi:10.1016/j.bbamem.2011.04.010
360. Ingólfsson HI, Andersen OS. Alcohol's effects on lipid bilayer properties. Biophys J (2011) 101:847–55. doi:10.1016/j.bpj.2011.07.013
361. Zapata-Morin PA, Sierra-Valdez F, Ruiz-Suárez J. The cut-off effect of n-alcohols in lipid rafts: A lipid-dependent phenomenon. J Mol Graphics Model 101, 107732 (2020). doi:10.1016/j.jmgm.2020.107732
362. Tan L, Smith MD, Scott HL, Yahya A, Elkins JG, Katsaras J, et al. Modeling the partitioning of amphiphilic molecules and co-solvents in biomembranes. J Appl Crystallogr (2022) 55:1401–12. doi:10.1107/s1600576722008998
363. Ludwig J, Maibaum L. Effect of alcohol on the phase separation in model membranes. Chem Phys Lipids (2020) 233:104986. doi:10.1016/j.chemphyslip.2020.104986
364. Tan L, Scott HL, Smith MD, Pingali SV, O’Neill HM, Morrell-Falvey JL, et al. Amphiphilic Co-solvents modulate the structure of membrane domains. ACS Sust Chem Eng (2023) 11:1598–609. doi:10.1021/acssuschemeng.2c06876
365. Uhrıková D, Kucerka N, Islamov A, Kuklin A, Gordeliy V, Balgavý P. Small-angle neutron scattering study of the lipid bilayer thickness in unilamellar dioleoylphosphatidylcholine vesicles prepared by the cholate dilution method: n-Decane effect. Biochim Biophys Acta (BBA)-Biomembranes (2003) 1611:31–4. doi:10.1016/S0005-2736(02)00705-8
366. Griepernau B, Böckmann RA. The influence of 1-alkanols and external pressure on the lateral pressure profiles of lipid bilayers. Biophys J (2008) 95:5766–78. doi:10.1529/biophysj.108.142125
367. Rowe ES, Campion JM. Alcohol induction of interdigitation in distearoylphosphatidylcholine: fluorescence studies of alcohol chain length requirements. Biophys J (1994) 67:1888–95. doi:10.1016/s0006-3495(94)80671-x
368. Löbbecke L, Cevc G. Effects of short-chain alcohols on the phase behavior and interdigitation of phosphatidylcholine bilayer membranes. Biochim Biophys Acta (BBA)-Biomembranes (1995) 1237:59–69. doi:10.1016/0005-2736(95)00076-F
369. Tran R, Ho S, Dea P. Effects of ethanol on lipid bilayers with and without cholesterol: The distearoylphosphatidylcholine system. Biophysical Chem (2004) 110:39–47. doi:10.1016/j.bpc.2004.01.005
370. Pillman HA, Blanchard G. Effects of ethanol on the organization of phosphocholine lipid bilayers. J Phys Chem B (2010) 114:3840–6. doi:10.1021/jp910897t
371. Nambi P, Rowe ES, McIntosh TJ. Studies of the ethanol-induced interdigitated gel phase in phosphatidylcholines using the fluorophore 1, 6-diphenyl-1, 3, 5-hexatriene. Biochemistry (1988) 27:9175–82. doi:10.1021/bi00426a015
372. Roth LG, Chen CH. Thermodynamic elucidation of ethanol-induced interdigitation of hydrocarbon chains in phosphatidylcholine bilayer vesicles. J Phys Chem (1991) 95:7955–9. doi:10.1021/j100173a073
373. Mou J, Yang J, Huang C, Shao Z. Alcohol induces interdigitated domains in unilamellar phosphatidylcholine bilayers. Biochemistry (1994) 33:9981–5. doi:10.1021/bi00199a022
374. Pope J, Dubro D. The interaction of n-alkanes and n-alcohols with lipid bilayer membranes: A 2H-NMR study. Biochim Biophys Acta (BBA)-Biomembranes (1986) 858:243–53. doi:10.1016/0005-2736(86)90329-9
375. Hishida M, Endo A, Nakazawa K, Yamamura Y, Saito K. Effect of n-alkanes on lipid bilayers depending on headgroups. Chem Phys Lipids (2015) 188:61–7. doi:10.1016/j.chemphyslip.2015.05.002
376. Lee A. Interactions between anesthetics and lipid mixtures. Normal alcohols. Biochemistry (1976) 15:2448–54. doi:10.1021/bi00656a031
377. Hornby A, Cullis P. Influence of local and neutral anaesthetics on the polymorphic phase preferences of egg yolk phosphatidylethanolamine. Biochim Biophys Acta (BBA)-Biomembranes (1981) 647:285–92. doi:10.1016/0005-2736(81)90256-X
378. Hossain M, Blanchard G. Effects of ethanol and n-butanol on the fluidity of supported lipid bilayers. Chem Phys Lipids (2021) 238:105091. doi:10.1016/j.chemphyslip.2021.105091
379. Westerman P, Pope J, Phonphok N, Doane J, Dubro D. The interaction of n-alkanols with lipid bilayer membranes: A 2H-NMR study. Biochim Biophys Acta (BBA)-Biomembranes (1988) 939:64–78. doi:10.1016/0005-2736(88)90048-X
380. Ho C, Stubbs CD. Effect of n-alkanols on lipid bilayer hydration. Biochemistry (1997) 36:10630–7. doi:10.1021/bi9703150
381. Barry JA, Gawrisch K. Effects of ethanol on lipid bilayers containing cholesterol, gangliosides, and sphingomyelin. Biochemistry (1995) 34:8852–60. doi:10.1021/bi00027a037
382. Nguyen MH, Dziura D, DiPasquale M, Castillo SR, Kelley EG, Marquardt D. Investigating the cut-off effect of n-alcohols on lipid movement: A biophysical study. Soft Matter (2023) 19:5001–15. doi:10.1039/D2SM01583H
383. Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N. The fungal cell wall: Candida, cryptococcus, and Aspergillus species. Front Microbiol (2020) 10:2993. doi:10.3389/fmicb.2019.02993
384. Bertani B, Ruiz N Function and biogenesis of lipopolysaccharides. EcoSal Plus (2018) 8. doi:10.1128/ecosalplus.esp-0001-2018
385. Ładniak A, Jurak M, Wiącek AE. The effect of chitosan/TiO2/hyaluronic acid subphase on the behaviour of 1, 2-dioleoyl-sn-glycero-3-phosphocholine membrane. Biomater Adv (2022) 138:212934. doi:10.1016/j.bioadv.2022.212934
386. Porras-Gomez M, Leal C. Lipid-based liquid crystalline films and solutions for the delivery of cargo to cells. Liquid Crystals Rev (2019) 7:167–82. doi:10.1080/21680396.2019.1666752
387. Kang M, Kim H, Leal C. Self-organization of nucleic acids in lipid constructs. Curr Opin Colloid Interf Sci (2016) 26:58–65. doi:10.1016/j.cocis.2016.09.006
388. Rideau E, Dimova R, Schwille P, Wurm FR, Landfester K. Liposomes and polymersomes: A comparative review towards cell mimicking. Chem Soc Rev (2018) 47:8572–610. doi:10.1039/C8CS00162F
389. Le Meins J-F, Schatz C, Lecommandoux S, Sandre O. Hybrid polymer/lipid vesicles: State of the art and future perspectives. Mater Today (2013) 16:397–402. doi:10.1016/j.mattod.2013.09.002
390. Schulz M, Binder WH. Mixed hybrid lipid/polymer vesicles as a novel membrane platform. Macromolecular Rapid Commun (2015) 36:2031–41. doi:10.1002/marc.201500344
391. Kim M, Porras-Gomez M, Leal C. Graphene-based sensing of oxygen transport through pulmonary membranes. Nat Commun (2020) 11:1103. doi:10.1038/s41467-020-14825-9
392. Garni M, Wehr R, Avsar SY, John C, Palivan C, Meier W. Polymer membranes as templates for bio-applications ranging from artificial cells to active surfaces. Eur Polym J (2019) 112:346–64. doi:10.1016/j.eurpolymj.2018.12.047
393. Cook AB, Novosedlik S, van Hest JCM. Complex coacervate materials as artificial cells. Acc Mater Res (2023) 4:287–98. doi:10.1021/accountsmr.2c00239
394. Renggli K, Baumann P, Langowska K, Onaca O, Bruns N, Meier W. Selective and responsive nanoreactors. Adv Funct Mater (2011) 21:1241–59. doi:10.1002/adfm.201001563
395. Le Meins J-F, Sandre O, Lecommandoux S. Recent trends in the tuning of polymersomes’ membrane properties. Eur Phys J E (2011) 34:14–7. doi:10.1140/epje/i2011-11014-y
396. Yoo J-W, Doshi N, Mitragotri S. Adaptive micro and nanoparticles: Temporal control over carrier properties to facilitate drug delivery. Adv Drug Deliv Rev (2011) 63:1247–56. doi:10.1016/j.addr.2011.05.004
397. Discher DE, Ahmed F. Polymersomes. Annu Rev Biomed Eng (2006) 8:323–41. doi:10.1146/annurev.bioeng.8.061505.095838
398. Guo X, Szoka FC. Chemical approaches to triggerable lipid vesicles for drug and gene delivery. Acc Chem Res (2003) 36:335–41. doi:10.1021/ar9703241
399. Bochicchio S, Lamberti G, Barba AA. Polymer–lipid pharmaceutical nanocarriers: Innovations by new formulations and production technologies. Pharmaceutics (2021) 13:198. doi:10.3390/pharmaceutics13020198
400. Chen D, Santore MM. Hybrid copolymer–phospholipid vesicles: Phase separation resembling mixed phospholipid lamellae, but with mechanical stability and control. Soft Matter (2015) 11:2617–26. doi:10.1039/C4SM02502D
401. Perera RM, Gupta S, Li T, Van Leeuwen CJ, Bleuel M, Hong K, et al. Nanoscale lipid/polymer hybrid vesicles: Effects of triblock copolymer composition and hydrophilic weight fraction. ACS Appl Polym Mater (2022) 4:8858–68. doi:10.1021/acsapm.2c01272
402. Nunes SS, Fernandes RS, Cavalcante CH, da Costa César I, Leite EA, Lopes SCA, et al. Influence of PEG coating on the biodistribution and tumor accumulation of pH-sensitive liposomes. Drug Deliv Transl Res (2019) 9:123–30. doi:10.1007/s13346-018-0583-8
403. Allen TM, Hansen C, Rutledge J. Liposomes with prolonged circulation times: factors affecting uptake by reticuloendothelial and other tissues. Biochim Biophys Acta (BBA)-Biomembranes (1989) 981:27–35. doi:10.1016/0005-2736(89)90078-3
404. Mayer LD, Cullis PR, Bally MB. Designing therapeutically optimized liposomal anticancer delivery systems: Lessons from conventional liposomes. In: DD Lasic, and D Papahadjopoulos, editors Med Appl liposomes. Elsevier (1998). p. 231–57.
405. Parr MJ, Masin D, Cullis PR, Bally MB. Accumulation of liposomal lipid and encapsulated doxorubicin in murine lewis lung carcinoma: The lack of beneficial effects by coating liposomes with poly (ethylene glycol). J Pharmacol Exp Ther (1997) 280:1319–27.
406. Ginzburg VV, Balijepalli S. Modeling the thermodynamics of the interaction of nanoparticles with cell membranes. Nano Lett (2007) 7:3716–22. doi:10.1021/nl072053l
407. Gao Y, Mao D, Wu J, Wang X, Wang Z, Zhou G, et al. Carbon nanotubes translocation through a lipid membrane and transporting small hydrophobic and hydrophilic molecules. Appl Sci (2019) 9:4271. doi:10.3390/app9204271
408. Lim SK, De Hoog H-P, Parikh AN, Nallani M, Liedberg B. Hybrid, nanoscale phospholipid/block copolymer vesicles. Polymers (2013) 5:1102–14. doi:10.3390/polym5031102
409. Amstad E, Kohlbrecher J, Müller E, Schweizer T, Textor M, Reimhult E. Triggered release from liposomes through magnetic actuation of iron oxide nanoparticle containing membranes. Nano Lett (2011) 11:1664–70. doi:10.1021/nl2001499
410. Iwasaki Y, Ishihara K. Cell membrane-inspired phospholipid polymers for developing medical devices with excellent biointerfaces. Sci Tech Adv Mater (2012) 13:064101. doi:10.1088/1468-6996/13/6/064101
411. Nishimura T, Hirose S, Sasaki Y, Akiyoshi K. Substrate-sorting nanoreactors based on permeable peptide polymer vesicles and hybrid liposomes with synthetic macromolecular channels. J Am Chem Soc (2019) 142:154–61. doi:10.1021/jacs.9b08598
412. Marsh D, Bartucci R, Sportelli L. Lipid membranes with grafted polymers: Physicochemical aspects. Biochim Biophys Acta (BBA) - Biomembranes (2003) 1615:33–59. doi:10.1016/S0005-2736(03)00197-4
413. Jacobs ML, Boyd MA, Kamat NP. Diblock copolymers enhance folding of a mechanosensitive membrane protein during cell-free expression. Proc Natl Acad Sci (2019) 116:4031–6. doi:10.1073/pnas.1814775116
414. Steinkühler J, Jacobs ML, Boyd MA, Villaseñor CG, Loverde SM, Kamat NP. PEO-b-PBD diblock copolymers induce packing defects in lipid/hybrid membranes and improve insertion rates of natively folded peptides. Biomacromolecules (2022) 23:4756–65. doi:10.1021/acs.biomac.2c00936
415. Schulz M, Glatte D, Meister A, Scholtysek P, Kerth A, Blume A, et al. Hybrid lipid/polymer giant unilamellar vesicles: Effects of incorporated biocompatible PIB–PEO block copolymers on vesicle properties. Soft Matter (2011) 7:8100–10. doi:10.1039/C1SM05725A
416. De Mel JU, Gupta S, Willner L, Allgaier J, Stingaciu LR, Bleuel M, et al. Manipulating phospholipid vesicles at the nanoscale: A transformation from unilamellar to multilamellar by an n-Alkyl-poly (ethylene oxide). Langmuir (2021) 37:2362–75. doi:10.1021/acs.langmuir.0c03302
417. Binder WH. Polymer-induced transient pores in lipid membranes. Angew Chem Int Edition (2008) 47:3092–5. doi:10.1002/anie.200800269
418. Colvin VL. The potential environmental impact of engineered nanomaterials. Nat Biotechnol (2003) 21:1166–70. doi:10.1038/nbt875
419. Guzmán E, Ferrari M, Santini E, Liggieri L, Ravera F. Effect of silica nanoparticles on the interfacial properties of a canonical lipid mixture. Colloids Surf B: Biointerfaces (2015) 136:971–80. doi:10.1016/j.colsurfb.2015.11.001
420. Hu J, Li X, Li M, Shang Y, He Y, Liu H. Real-time monitoring of the effect of carbon nanoparticles on the surface behavior of DPPC/DPPG Langmuir monolayer. Colloids Surf B: Biointerfaces (2020) 190:110922. doi:10.1016/j.colsurfb.2020.110922
421. Bothun G, Ganji N, Khan I, Xi A, Bobba C. Anionic and cationic silver nanoparticle binding restructures net-anionic PC/PG monolayers with saturated or unsaturated lipids. Langmuir (2017) 33:353–60. doi:10.1021/acs.langmuir.6b02003
422. Guzmán E, Liggieri L, Santini E, Ferrari M, Ravera F. DPPC–DOPC Langmuir monolayers modified by hydrophilic silica nanoparticles: Phase behaviour, structure and rheology. Colloids Surf A: Physicochemical Eng Aspects (2012) 413:174–83. doi:10.1016/j.colsurfa.2011.12.059
423. Nappini S, Bonini M, Ridi F, Baglioni P. Structure and permeability of magnetoliposomes loaded with hydrophobic magnetic nanoparticles in the presence of a low frequency magnetic field. Soft Matter (2011) 7:4801–11. doi:10.1039/C0SM01264E
424. Santhosh PB, Drašler B, Drobne D, Kreft ME, Kralj S, Makovec D, et al. Effect of superparamagnetic iron oxide nanoparticles on fluidity and phase transition of phosphatidylcholine liposomal membranes. Int J Nanomedicine (2015) 10:6089–103. doi:10.2147/IJN.S89679
425. Moghadam BY, Hou W-C, Corredor C, Westerhoff P, Posner JD. Role of nanoparticle surface functionality in the disruption of model cell membranes. Langmuir (2012) 28:16318–26. doi:10.1021/la302654s
426. Mhashal AR, Roy S. Effect of gold nanoparticle on structure and fluidity of lipid membrane. PLoS One (2014) 9:e114152. doi:10.1371/journal.pone.0114152
427. Chen Y, Bothun GD. Cationic Gel-phase liposomes with “decorated” anionic SPIO nanoparticles: Morphology, colloidal, and bilayer properties. Langmuir (2011) 27:8645–52. doi:10.1021/la2011138
428. Chen Y, Bose A, Bothun GD. Controlled release from bilayer-decorated magnetoliposomes via electromagnetic heating. ACS Nano (2010) 4:3215–21. doi:10.1021/nn100274v
429. Wi HS, Lee K, Pak HK. Interfacial energy consideration in the organization of a quantum dot–lipid mixed system. J Phys Condens Matter (2008) 20:494211. doi:10.1088/0953-8984/20/49/494211
430. Wilson SR. Nanomedicine: fullerene and carbon nanotube biology. Perspect fullerene nanotechnology. In: E Ōsawa, editor Springer (2002). p. 155–63.
431. Partha R, Conyers JL. Biomedical applications of functionalized fullerene-based nanomaterials. Int J Nanomedicine (2009) 4:261–75. doi:10.2147/IJN.S5964
432. Sayes CM, Gobin AM, Ausman KD, Mendez J, West JL, Colvin VL. Nano-C60 cytotoxicity is due to lipid peroxidation. Biomaterials (2005) 26:7587–95. doi:10.1016/j.biomaterials.2005.05.027
433. Li L, Davande H, Bedrov D, Smith GD. A molecular dynamics simulation study of C60 fullerenes inside a dimyristoylphosphatidylcholine lipid bilayer. J Phys Chem B (2007) 111:4067–72. doi:10.1021/jp064982r
434. Wong-Ekkabut J, Baoukina S, Triampo W, Tang IM, Tieleman DP, Monticelli L. Computer simulation study of fullerene translocation through lipid membranes. Nat Nanotechnol (2008) 3:363–8. doi:10.1038/nnano.2008.130
435. Brisebois PP, Arnold AA, Chabre YM, Roy R, Marcotte I. Comparative study of the interaction of fullerenol nanoparticles with eukaryotic and bacterial model membranes using solid-state NMR and FTIR spectroscopy. Eur Biophys J (2012) 41:535–44. doi:10.1007/s00249-012-0809-5
436. Nisoh N, Karttunen M, Monticelli L, Wong-Ekkabut J. Lipid monolayer disruption caused by aggregated carbon nanoparticles. RSC Adv (2015) 5:11676–85. doi:10.1039/C4RA17006G
437. Li Z, Misra RP, Li Y, Yao YC, Zhao S, Zhang Y, et al. Breakdown of the Nernst–Einstein relation in carbon nanotube porins. Nat Nanotechnol (2023) 18:177–83. doi:10.1038/s41565-022-01276-0
438. Sullivan K, Zhang Y, Lopez J, Lowe M, Noy A. Carbon nanotube porin diffusion in mixed composition supported lipid bilayers. Sci Rep (2020) 10:11908. doi:10.1038/s41598-020-68059-2
439. Vögele M, Köfinger J, Hummer G. Molecular dynamics simulations of carbon nanotube porins in lipid bilayers. Faraday Discuss (2018) 209:341–58. doi:10.1039/C8FD00011E
Keywords: sterols, drug molecules, proteins and peptides, nanoparticles, molecular packing, bending rigidity, fluidity, phase transitions
Citation: Kumarage T, Morris NB and Ashkar R (2023) The effects of molecular and nanoscopic additives on phospholipid membranes. Front. Phys. 11:1251146. doi: 10.3389/fphy.2023.1251146
Received: 30 June 2023; Accepted: 18 October 2023;
Published: 20 November 2023.
Edited by:
Yun Liu, National Institute of Standards and Technology (NIST), United StatesReviewed by:
Shuo Qian, Oak Ridge National Laboratory (DOE), United StatesCopyright © 2023 Kumarage, Morris and Ashkar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rana Ashkar, YXNoa2FyQHZ0LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.