- 1College of Science, Wuhan University of Science and Technology, Wuhan, China
- 2College of Energy Engineering, Huanghuai University, Zhumadian, China
In this paper, a new method for obtaining the basic reproduction number is proposed, called the path analysis method. Compared with the traditional next-generation method, this method is more convenient and less error-prone. We develop a general model that includes most of the epidemiological characteristics and enumerate all disease transmission paths. The path analysis method is derived by combining the next-generation method and the disease transmission paths. Three typical examples verify the effectiveness and convenience of the method. It is important to note that the path analysis method is only applicable to epidemic models with bilinear incidence rates. The Volterra-type Lyapunov function is given to prove the global stability of the system. The simulations prove the correctness of our conclusions.
1 Introduction
Research on the epidemic compartment model began with Kermack–McKendrick’s SIR [1] system. It took the Black Death as the research object and had only one infected population during the illness period. The advantage of the SIR system is that it only needs to focus on the total number of patients per unit time [2–4]. With the development of medical sciences, it is found that some patients have already been infected before they develop symptoms. Statistics show that most infectious diseases have an asymptomatic infected population, such as COVID-19 [5], SARS [6], and Ebola [7]. Therefore, scholars proposed the SIR [8–11] model with two infected populations: asymptomatic and symptomatic populations. The asymptomatic population is transformed into a symptomatic population by a certain percentage after a latent period.
In recent years, researchers have developed more complex high-dimensional models based on the transmission characteristics. In [12], the
The basic reproduction number [18–23] is one of the most important indicators of the infectious disease compartment model. Its basic form is
The study of stability is one of the most important subjects in the infectious disease model. Many studies [27–35] give the methods for proving the local and global stabilities of the singularities. Lyapunov’s second method and Lasalle’s invariance principle are the most common methods for proving global stability. However, they are not easy to operate because there is no general way to construct a suitable Lyapunov function. In the Lyapunov function toolbox, linear-, quadratic-, and Volterra-type functions are three frequently used functions applied to biological systems. These functions are as follows:
where
In summary, most researchers introduce their models, then calculate the basic reproduction number, and prove the stability of the equilibrium point. These processes are similar but require tedious calculations. Is it possible to obtain a basic reproduction number with universal applicability by building a general model containing the main features? This paper develops a model with
2 Model and method
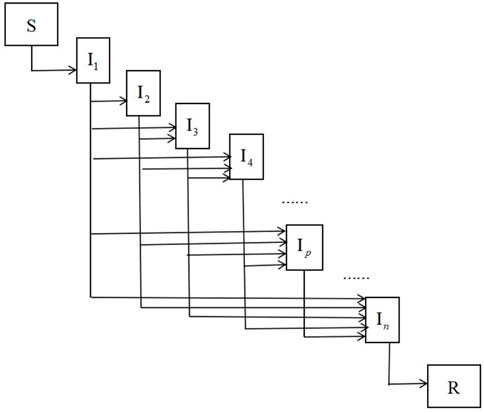
Individuals are divided into three categories, susceptible (
Here, the next-generation method [38] is used to calculate the basic reproduction number. We rewrite system 1 as (
where
where
Hence,
where
Here, the basic reproduction number consists of
3 Application examples
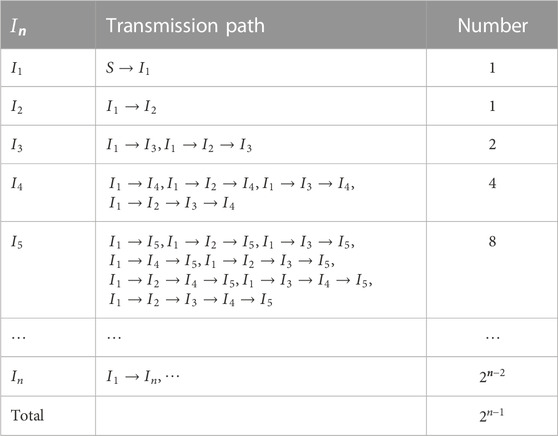
For high-dimensional epidemic model, it is cumbersome and error-prone to derive the basic reproduction number using the next-generation method. In this section, we use the path analysis method of Section 2 to directly give the basic reproduction numbers for three bilinear compartment models without any calculation.
In [37], system (2) has two populations with infection capability, which are called asymptomatic
Therefore, the basic reproduction number is as follows:
System (3) with nine dimensions has been developed in [25] to depict the transmission of COVID-19. The first equation reveals that
shown as
The basic reproduction number is as follows:
In [13], an epidemic model (4) incorporating quarantine was built to predict the COVID-19 trend in the United Kingdom. The first equation shows that the quarantine
The basic reproduction number is
4 Global stability analysis
4.1 Global stability analysis of the disease-free equilibrium point
Theorem 4.1:. The disease-free equilibrium point of system (1) is
where
When
4.2 Global stability analysis of the endemic equilibrium point
Theorem 4.2:. When
Therefore, when
We denote
Differentiating
By calculation, we can get
Finally, we get
According to Lyapunov’s second method, the endemic equilibrium point is globally stable.
5 Model simulation
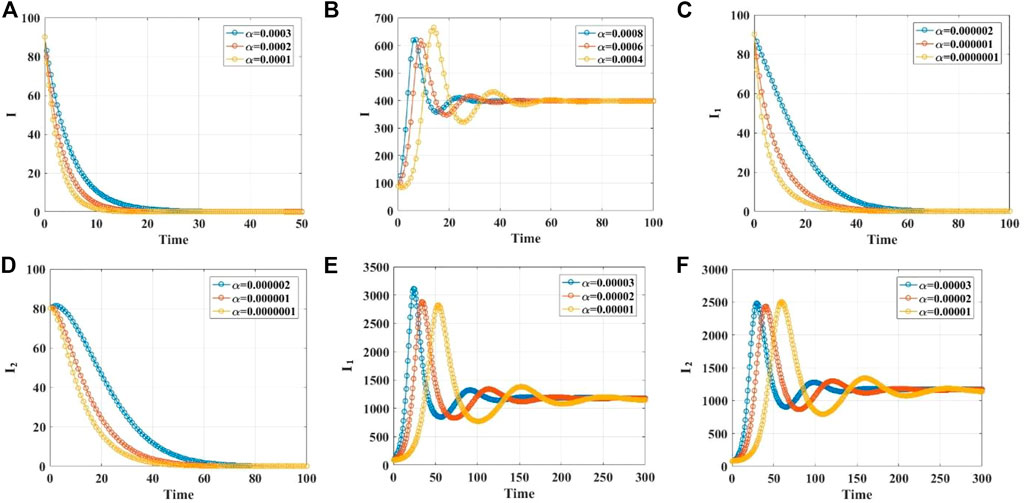
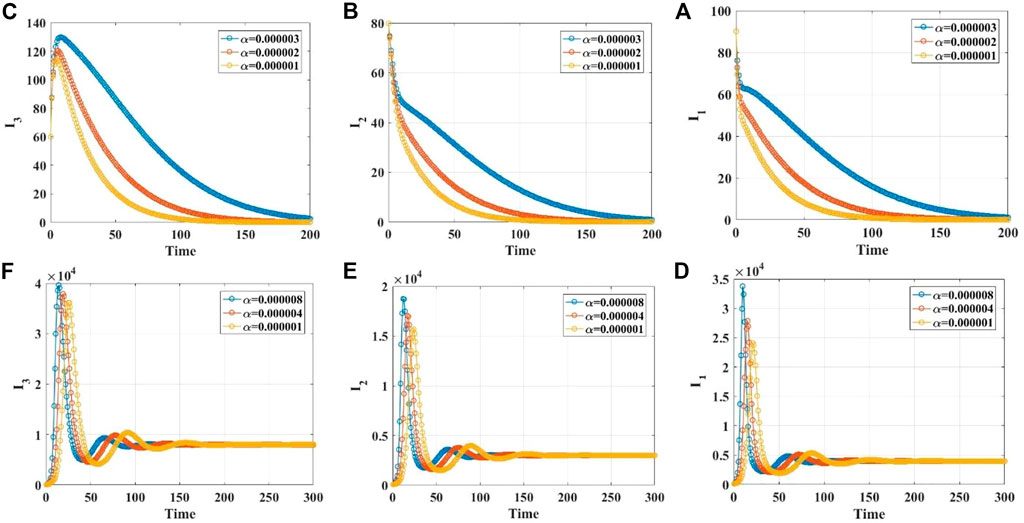
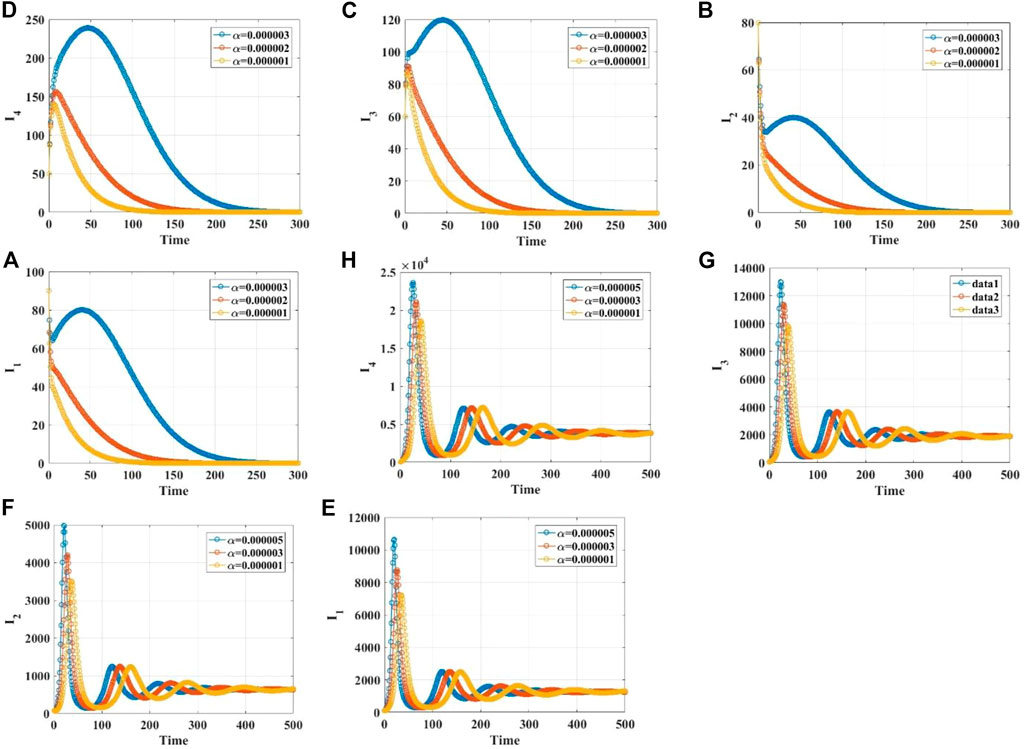
We demonstrate the stabilities of the disease-free and endemic equilibrium points with 1, 2, 3, and 4 infected populations through simulations. Supplementary Material S1 gives the values of the parameters in different cases. When the infection rate
6 Conclusion and discussions
This paper constructs a general epidemic system with bilinear incidence rates. It contains
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
YZ: conceptualization, methodology, software, and writing—original draft preparation. YD: visualization, investigation and supervision. MG: writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 12271418).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2023.1158814/full#supplementary-material
References
1. Kermack WO, McKendrick AG. Contributions to the mathematical theory of epidemics. II.—the problem of endemicity[J]. Proc R Soc Lond Ser A, containing Pap a Math Phys character (1932) 138(834):55–83.
2. Beretta E, Takeuchi Y. Global stability of an SIR epidemic model with time delays. J Math Biol (1995) 33(3):250–60. doi:10.1007/BF00169563
3. Korobeinikov A. Lyapunov functions and global stability for SIR and SIRS epidemiological models with non-linear transmission. Bull Math Biol (2006) 68:615–26. doi:10.1007/s11538-005-9037-9
4. Wei W, Xu W, Song Y, Liu J. Bifurcation and basin stability of an SIR epidemic model with limited medical resources and switching noise. Solitons & Fractals (2021) 152:111423. doi:10.1016/j.chaos.2021.111423
5. Lei S, Jiang F, Su W, Chen C, Chen J, Mei W, et al. Clinical characteristics and outcomes of patients undergoing surgeries during the incubation period of COVID-19 infection. EClinicalMedicine (2020) 21:100331. doi:10.1016/j.eclinm.2020.100331
6. Chan-Yeung M, Xu RH. Sars: Epidemiology. Respirology (2003) 8:S9–S14. doi:10.1046/j.1440-1843.2003.00518.x
7. Eichner M, Dowell SF, Firese N. Incubation Period of Ebola hemorrhagic virus subtype zaire. Osong Public Health Res Perspect (2011) 2(1):3–7. doi:10.1016/j.phrp.2011.04.001
8. Schwartz IB, Smith HL. Infinite subharmonic bifurcation in an SEIR epidemic model. J Math Biol (1983) 18:233–53. doi:10.1007/bf00276090
9. Buonomo B, Lacitignola D. On the dynamics of an SEIR epidemic model with a convex incidence rate. Ricerche di matematica (2008) 57:261–81. doi:10.1007/s11587-008-0039-4
10. Wang X, Tao Y, Song X. Pulse vaccination on SEIR epidemic model with nonlinear incidence rate. Appl Math Comput (2009) 210(2):398–404. doi:10.1016/j.amc.2009.01.004
11. Efimov D, Ushirobira R. On an interval prediction of COVID-19 development based on a SEIR epidemic model. Annu Rev Control (2021) 51:477–87. doi:10.1016/j.arcontrol.2021.01.006
12. Wang L, Wang J, Zhao H, Shi YY, Wang K, Wu P, et al. Modelling and assessing the effects of medical resources on transmission of novel coronavirus (COVID-19) in Wuhan, China. Math Biosci Eng (2020) 17(4):2936–49. doi:10.3934/mbe.2020165
13. Nadim SS, Ghosh I, Chattopadhyay J. Short-term predictions and prevention strategies for COVID-19: A model-based study. Appl Math Comput (2021) 404:126251. doi:10.1016/j.amc.2021.126251
14. Das P, Nadim SS, Das S, et al. Dynamics of COVID-19 transmission with comorbidity: A data driven modelling based approach. Nonlinear Dyn (2021) 106:1197–211. doi:10.1007/s11071-021-06324-3
15. Péni T, Csutak B, Szederkényi G, Rost G. Nonlinear model predictive control with logic constraints for COVID-19 management. Nonlinear Dyn (2020) 102:1965–86. doi:10.1007/s11071-020-05980-1
16. Batabyal S, Batabyal A. Mathematical computations on epidemiology: A case study of the novel coronavirus (SARS-CoV-2). Theor Biosciences (2021) 140:123–38. doi:10.1007/s12064-021-00339-5
17. Biswas SK, Ghosh JK, Sarkar S, Ghosh U. COVID-19 pandemic in India: A mathematical model study. Nonlinear Dyn (2020) 102:537–53. doi:10.1007/s11071-020-05958-z
18. Ojo MM, Peter OJ, Goufo EFD, Panigoro HS, Oguntolu FA. Mathematical model for control of tuberculosis epidemiology. J Appl Math Comput (2023) 69(1):69–87. doi:10.1007/s12190-022-01734-x
19. Yin Q, Wang Z, Xia C, Bauch CT. Impact of co-evolution of negative vaccine-related information, vaccination behavior and epidemic spreading in multilayer networks. Commun Nonlinear Sci Numer Simulation (2022) 109:106312. doi:10.1016/j.cnsns.2022.106312
20. Ojo MM, Peter OJ, Goufo EFD, Nisar KS. A mathematical model for the co-dynamics of COVID-19 and tuberculosis. Mathematics Comput Simulation (2023) 207:499–520. doi:10.1016/j.matcom.2023.01.014
21. Fan J, Yin Q, Xia C, et al. Epidemics on multilayer simplicial complexes[J]. Proc R Soc A (2022) 478(2261):20220059.
22. James Peter O, Ojo MM, Viriyapong R, Abiodun Oguntolu F. Mathematical model of measles transmission dynamics using real data from Nigeria. J Difference Equations Appl (2022) 28(6):753–70. doi:10.1080/10236198.2022.2079411
23. Wang Z, Xia C, Chen Z, Chen G. Epidemic propagation with positive and negative preventive information in multiplex networks. IEEE Trans cybernetics (2020) 51(3):1454–62. doi:10.1109/TCYB.2019.2960605
24. D'Arienzo M, Coniglio A. Assessment of the SARS-CoV-2 basic reproduction number, R0, based on the early phase of COVID-19 outbreak in Italy. Biosafety and health (2020) 2(2):57–9. doi:10.1016/j.bsheal.2020.03.004
25. Amouch M, Karim N. Modeling the dynamic of COVID-19 with different types of transmissions. J Chaos, Solitons Fractals (2021) 150:111188. doi:10.1016/j.chaos.2021.111188
26. Ghosh JK, Biswas SK, Sarkar S, Ghosh U. Mathematical modelling of COVID-19: A case study of Italy. Math Comput Simulation (2022) 194:1–18. doi:10.1016/j.matcom.2021.11.008
27. Lv R, Li H, Sun Q, et al. Stability analysis and optimal control of a time-delayed panic-spreading model[J]. Front Phys (2022) 1026.
28. Zhang W, Ma X, Zhang Y, et al. Dynamical models of acute respiratory illness caused by human adenovirus on campus[J]. Front Phys (2022) 10:1325.
29. Wang L, Jin Z, Wang H. A switching model for the impact of toxins on the spread of infectious diseases. J Math Biol (2018) 77:1093–115. doi:10.1007/s00285-018-1245-7
30. Wei F, Xue R. Stability and extinction of SEIR epidemic models with generalized nonlinear incidence. Math Comput Simulation (2020) 170:1–15. doi:10.1016/j.matcom.2018.09.029
31. Pérez Á GC, Avila-Vales E, García-Almeida GE. Bifurcation analysis of an SIR model with logistic growth, nonlinear incidence, and saturated treatment. Complexity (2019) 2019:1–21. doi:10.1155/2019/9876013
32. Peter OJ, Panigoro HS, Ibrahim MA, et al. Analysis and dynamics of measles with control strategies: A mathematical modeling approach[J]. Int J Dyn Control (2023) 1–15.
33. Kammegne B, Oshinubi K, Babasola O, Peter OJ, Longe OB, Ogunrinde RB, et al. Mathematical modelling of the spatial distribution of a COVID-19 outbreak with vaccination using diffusion equation. Pathogens (2023) 12(1):88. doi:10.3390/pathogens12010088
34. Peter OJ, Yusuf A, Ojo MM, Kumar S, Kumari N, Oguntolu FA. A mathematical model analysis of meningitis with treatment and vaccination in fractional derivatives. Int J Appl Comput Math (2022) 8(3):117. doi:10.1007/s40819-022-01317-1
35. Ojo MM, Benson TO, Peter OJ, Goufo EFD. Nonlinear optimal control strategies for a mathematical model of COVID-19 and influenza co-infection. Physica A: Stat Mech its Appl (2022) 607:128173. doi:10.1016/j.physa.2022.128173
36. Melese AS, Makinde OD, Obsu LL. Mathematical modelling and analysis of coffee berry disease dynamics on a coffee farm. Math Biosciences Eng (2022) 19(7):7349–73. doi:10.3934/mbe.2022347
37. Ottaviano S, Sensi M, Sottile S. Global stability of SAIRS epidemic models. Nonlinear Anal Real World Appl (2022) 65:103501. doi:10.1016/j.nonrwa.2021.103501
38. Van den Driessche P, Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math biosciences (2002) 180(1-2):29–48. doi:10.1016/s0025-5564(02)00108-6
39. Meskaf A, Khyar O, Danane J, Allali K. Global stability analysis of a two-strain epidemic model with non-monotone incidence rates. Solitons & Fractals (2020) 133:109647. doi:10.1016/j.chaos.2020.109647
40. Safi MA, Garba SM. Global stability analysis of SEIR model with holling type II incidence function[J]. Comput Math Methods Med (2012) 2012.
41. Li CL, Cheng CY, Li CH. Global dynamics of two-strain epidemic model with single-strain vaccination in complex networks. Nonlinear Analysis: Real World Applications (2023) 69:103738. doi:10.1016/j.nonrwa.2022.103738
42. Khan Z A, Alaoui A L, Zeb A, Tilioua M, Djilali S. Global dynamics of a SEI epidemic model with immigration and generalized nonlinear incidence functional. Results in Physics (2021) 27:104477. doi:10.1016/j.rinp.2021.104477
Keywords: path analysis method, basic reproduction number, transmission paths, Lyapunov functions, stability
Citation: Zhou Y, Ding Y and Guo M (2023) Path analysis method in an epidemic model and stability analysis. Front. Phys. 11:1158814. doi: 10.3389/fphy.2023.1158814
Received: 04 February 2023; Accepted: 09 March 2023;
Published: 23 March 2023.
Edited by:
Chengyi Xia, Tiangong University, ChinaReviewed by:
Qianqian Zheng, Xuchang University, ChinaOlumuyiwa James Peter, University of Medical Sciences, Ondo, Nigeria
Guodong Zhang, South-Central University for Nationalities, China
Copyright © 2023 Zhou, Ding and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhou, emhvdXlvbmdlZHVAMTI2LmNvbQ==
 Yong Zhou
Yong Zhou Yiming Ding1
Yiming Ding1