
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Phys., 06 September 2022
Sec. Biophysics
Volume 10 - 2022 | https://doi.org/10.3389/fphy.2022.963716
This article is part of the Research TopicDynamical Systems, PDEs and Networks for Biomedical Applications: Mathematical Modeling, Analysis and SimulationsView all 18 articles
G-protein signaling pathways mediate communication across cell membranes. The first steps of this communication occur at the cell membrane, where upon receiving an external signal –the binding of an agonist ligand– the membrane-embedded G-Protein Coupled Receptor adopts a conformation recognized by a cytoplasmatic G protein. Whereas specialized GPCRs sense protons from the extracellular milieu, thus acting as pH sensors in specialized cells, accumulating evidence suggests that pH sensitivity might be common to distinct GPCRs. In this perspective article we discuss general principles of protonation-coupled protein conformational dynamics and how these apply to GPCRs. To dissect molecular interactions that might govern the protonation-coupled conformational dynamics of GPCRs, we use graph-based algorithms to compute graphs of hydrogen bond networks. We find that the internal H-bond networks contain sites where structural rearrangements upon protonation change could be transmitted throughout the protein. Proton binding to bulk-exposed clusters of titratable protein sidechains ensures the pH sensing mechanism is robust.
G-Protein Coupled Receptors (GPCRs) mediate communication between eukaryotic cells and their environments. Signals sensed by GPCRs can be of external origin, such as light or odors, or endogenous, such as hormones or neurotransmitters. Humans have >750 GPCRs [1], and about 30–35% of the drugs target GPCRs that respond to endogenous signals (endo-GPCRs)—though just a minority (10%) of the known GPCRs are used as targets [2–4]. As endo-GPCRs are significantly conserved between human and mouse, and are expressed preferentially in the brain [1], they are of tremendous interest in expanding the repertoire of GPCRs used for therapeutics [4]. Moreover, since the majority of the GPCRs that are currently targeted by approved drugs couple to cytoplasmatic G proteins Gs or Gi [4], in the future G proteins might also become drug targets [5, 6], e.g., by using ligands that can modulate interactions between G proteins and their upstream/downstream interaction partners and thus impact signal transduction pathways.
The general principles of how GPCRs interact with G proteins to effect cell signaling are illustrated in Figure 1A. Upon binding of an agonist the GPCR changes conformation and binds to the G protein (Figure 1A). Some class B GPCRs alter their ligand specificity upon interactions with receptor activity-modifying proteins, RAMPS, which are single-pass transmembrane proteins with an extracellular domain [7]. Both GPCRs and G proteins are dynamic [8, 9]. GPCRs can couple to lipids [10, 11], can bind sodium ions [12, 13], and change protonation during their function [14, 15]. Indeed, recent data were interpreted to suggest that “proton-sensing and H+-gated agonism are recurring features of GPCR signaling biology” [16]; however it remains unclear how GPCRs couple proton binding with protein conformation. Mechanisms for protonation-coupled function have been dissected for microbial rhodopsins –which, similar to GPCRs, are seven-helical membrane proteins, and pass through discrete intermediate conformations during their reaction cycles. Here, we rely on GPCRs and microbial rhodopsin structures to dissect interactions potentially important for protonation-coupled conformational change. We computed H-bond networks using the graph-based algorithms Bridge [17, 18] and C-Graphs [19] and as H-bond criterion a distance of ≤3.5 Å between the donor and acceptor heavy atoms; the graph computations included all H-bonding protein sidechains, and water-mediated bridges between sidechains with up to three water molecules in a bridge. Thus, nodes of an H-bond graph are H-bonding protein sidechains, and edges, sidechain-sidechain H-bonds or water-mediated bridges between sidechains. We suggest that GPCRs that couple proton binding with conformational change might use mechanisms similar to other membrane proteins (Figure 1B).
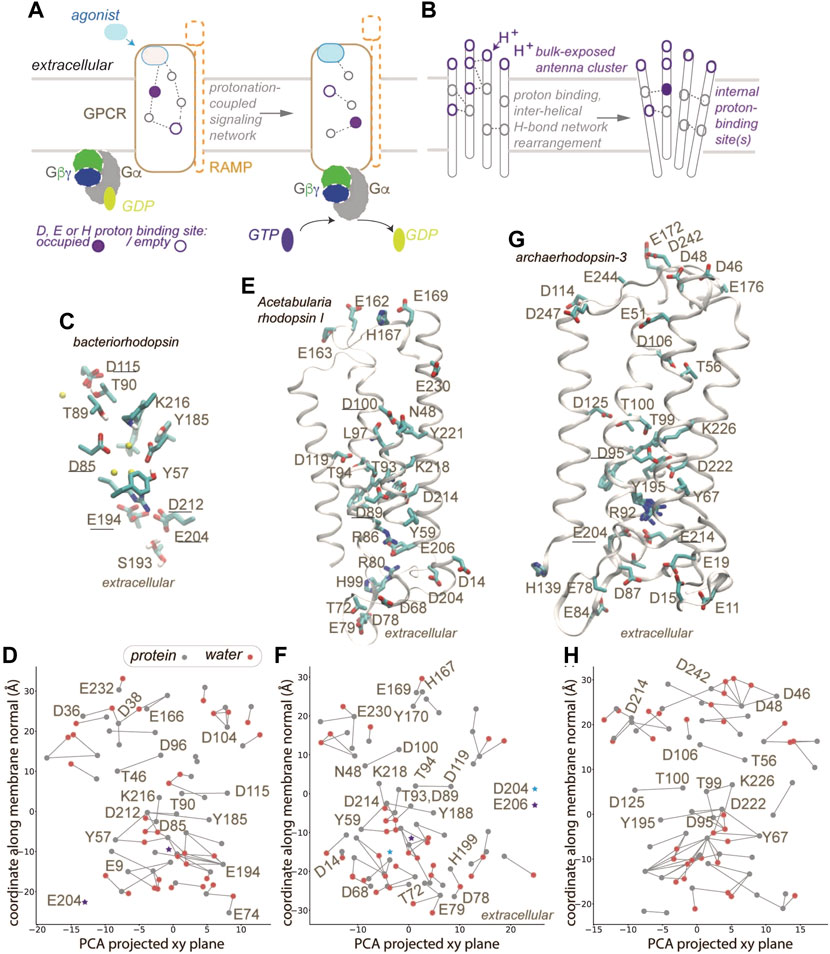
FIGURE 1. GPCR activation and protonation-coupled conformational dynamics with H-bond networks. (A) Agonist and G-protein binding may associate with protonation-coupled formation of a continuous H-bond network inside the GPCR. (B) Schematic representation of a membrane protein with a bulk-exposed proton antenna cluster and an internal proton-binding site. (C,D) Close view of the internal H-bond network (C) and H-bond graph (D) computed for bacteriorhodopsin, pdb 7z09 [39]. Gray and red nodes represent, respectively, protein sidechains and water oxygen atoms; only selected nodes are labeled. (E,F) Molecular graphics (E) and H-bond graph (F) computed for Acetabularia rhodopsin I, pdb 5awz [40]. (G,H) Molecular graphics (G) and H-bond graph computed for archaeorhodopsin-3, pdb 6gux [41]. Only some helices are displayed in panels E,G. Groups underlined in panels C,E,G were implicated in proton binding [40, 41, 78]. Molecular graphics included in Figure 1 and Figure 2 were prepared with Visual Molecular Dynamics, VMD [79]. H-bond graphs in panels D, E, H were computed with C-Graphs [19] using structures aligned with OPM [80].
Analyses of H-bonds in static experimental structures of GPCRs are commonly used to formulate hypotheses about reaction mechanisms. As summarized below, the view of internal H-bond networks of GPCRs depends drastically on the resolution at which structures were solved, and on the internal number of water molecules.
The structure of the (inactive) visual rhodopsin GPCR from squid presented a remarkable internal protein-water H-bond network hypothesized to mediate propagation of structural change during receptor activation [20]. Much of this H-bond network remains present in structures of the early intermediates batho and lumi [19]. Moreover, some of the internal water molecules of squid rhodopsin are conserved in structures of jumping-spider rhodopsin-1 and adenosine A2A receptor (AA2AR), suggesting conserved roles of water molecules in propagating structural change [19].
A difficulty with identifying protein-water H-bond networks of GPCRs based on static structures is that the number of internal water molecules tends to depend on the resolution: GPCR structures solved at resolutions of 2 Å or higher typically have at least 30 internal water molecules, some of which are found at conserved sites [21]. A dataset of 63 GPCR structures solved at a resolution of 2.5 Å or better, and with at least 10 internal water molecules, were found to host a conserved, core protein-water H-bond network that inter-connects functionally important regions of GPCRs [21]; within this dataset, structures solved at resolution of at least 2.3 Å and with more internal water molecules had additional local H-bond clusters [21]. In stark contrast with the extended H-bond network of the inactive squid rhodopsin structure, two recent structures of G-protein bound GPCRs solved at resolutions of 2.9–3.15 Å [22, 23], which lack internal water molecules, have only small, localized H-bond clusters of up to 3-4 protein sidechains [24]. This suggests that both the resolution and internal water content need to be accounted for in hypotheses about putative roles of H-bond networks for conformational couplings of GPCRs [21, 24].
GPCRs that change protonation during function include bovine rhodopsin –two internal Glu groups, one at the ligand binding site (E3.28 in the standard Ballesteros-Weinstein numbering scheme for class A GPCRs), and one at a conserved functional motif (E3.49) [14]; the M2 muscarinic receptor –two conserved carboxylic groups, D2.50 and D3.32, might change protonation [25]; the calcium receptor –whose activity might be modulated by proton binding to carboxylic sidechains [26]; the μ-opioid receptor –the propensity of the ligand to H-bond to H6.52 could explain pH sensitivity [27]; AA2ARs –ligand binding depends on pH-sensitive interactions of E169 and H264 at the extracellular side [28, 29].
Other GPCRs signal changes in extracellular pH to ensure cell homeostasis, i.e., their biological function is to sense pH, and they have been implicated in disease conditions associated with acidic pH –cancer, inflammatory disease, and ischemia [30, 31]. As no three-dimensional structures have been solved for these canonical pH-sensing GPCRs, their reaction mechanism remains elusive. Central roles for proton sensing have been assigned to H-bonding extracellular His sidechains [32], or to a triad of internal carboxylic groups [33]. How the protonation change of external His sidechains would cause receptor structural changes, and how protons from the extracellular bulk would make their way to internal carboxylic groups, are key open questions. As summarized below, we suggest that mechanisms used by microbial rhodopsins, and by other proton-binding membrane proteins, provide clues about common principles of action for protonation-coupled membrane proteins.
Membrane proteins are commonly thought to rely on internal H-bond networks to couple protein conformation with a change in protonation, typically of a carboxylate and/or His sidechains, as these moieties titrate in a pH range relevant to biology [34, 35]. Protonation change at internal sites of membrane proteins involves H-bond paths that transiently inter-connect proton donor and acceptor pairs. In the bacteriorhodopsin proton pump (Figures 1C,D 2A), a few internal carboxylic groups, including D85, D96 and E194/E204 (Figures 1C,D) change protonation during bacteriorhodopsin’s function. In the resting state, the primary proton acceptor D85 H-bonds with T89 and water, protonated D115 with T90, D212 with Y57 and Y185, and the proton-release site E194-E204 with S193. At the cytoplasmic side, a cluster of carboxylic groups might function as a proton-collecting antenna [36–38] that picks up protons and delivers them to an internal carboxylic group [37, 38] (Figure 2A).
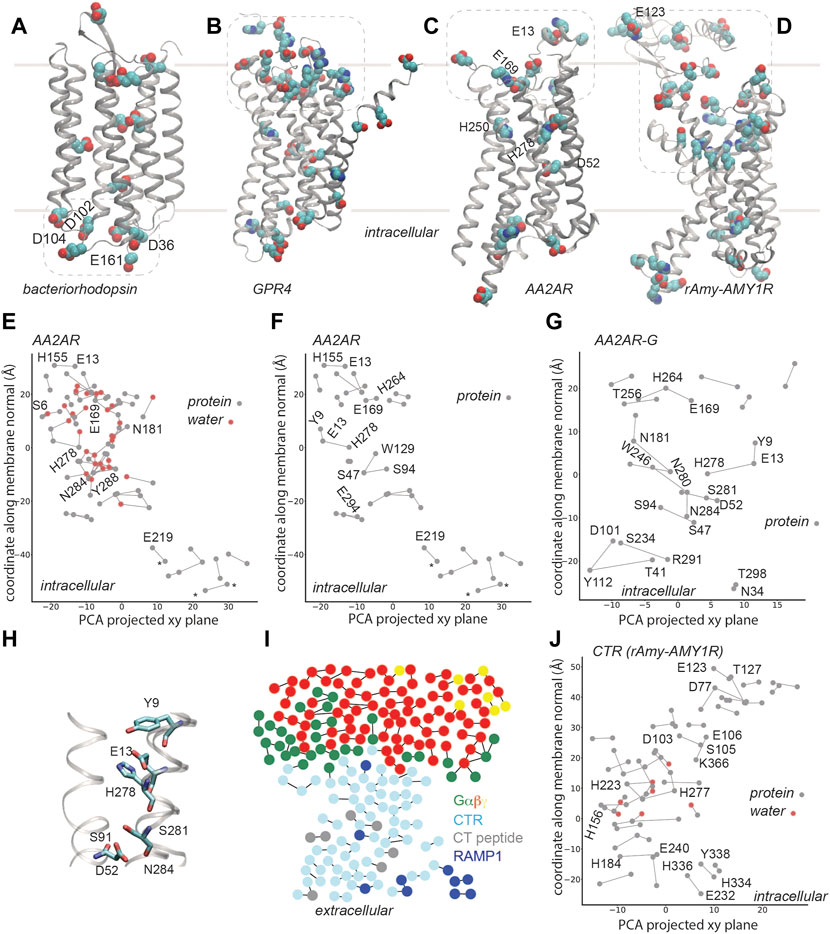
FIGURE 2. H-bond networks of GPCRs. Dotted lines indicate selected clusters of bulk-exposed carboxylic and His groups. (A) Carboxylic groups of bacteriorhodopsin, based on pdb 5zim [81]; Asp/Glu groups labeled are part of the proton-collecting antenna [37, 38]. (B) Carboxylic and His groups of human GPR4 from model AF-P46093-F1-model_v2, UniProt entry P46093 [61]. (C,D) Carboxylic and His groups of AA2AR (panel B, pdb 5nm4, 1.7Å resolution [63]) and of rAmy-AMY1R (pdb 7tyf [72]). (E–G) C-Graphs H-bond graphs computed with water bridges (E) vs. for protein sidechains (F) for AA2AR based on pdb 5 nm4, and for G-protein bound AA2AR, pdb 5g53, 3.4 Å resolution [82] (G). H-bond networks of AA2ARs were reported in Refs. [19, 21]. In panels (E,F), stars indicate groups of a synthetic construct. (H) Selected H-bonds of AA2AR. (I) Bridge2 H-bond graph of the CTR-G protein complex, pdb 6e3y, 3.3 Å resolution [70]. (J) C-Graphs H-bond graph for the CTR, pdb 7tyf, 2.2 Å resolution [72]. Panels (E–G,J) use structures with the receptor domain oriented along the membrane normal using VMD and OPM [80].
The protein-water H-bond graph computed from a high-resolution structure (1.05 Å) of bacteriorhodopsin [39] contains 79 H-bonding sidechains and water molecules, and 73 H-bonds between these groups (Figure 1D); when we computed the H-bond graph of the same structure without water molecules, only 19 H-bonds remained. We obtained similar results for a 1.6 Å resolution structure of Acetabularia rhodopsin I [40] (Figure 1E), whose H-bond graph has about 3.7-fold more H-bonds when both protein sidechains and water molecules are included (Figure 1F), than when water was excluded. Likewise, for the 1.3 Å resolution structure of archaerhodopsin-3 [41] (Figure 1G) the protein-water H-bond graph has about 3.8-fold more H-bonds than when (Figure 1H), vs. 24 H-bonds when only sidechains were included in computations.
The H-bond graphs above of microbial rhodopsins have H-bonds that are common at proton-binding sites of membrane transporters and receptors, including GPCRs [42, 43]. In class A GPCRs, the conserved D2.50 is paired with S7.46 [21, 44, 45]. In bacteriorhodopsin, for which proton binding has been studied extensively, including with Fourier Transform Infrared Spectroscopy [38, 46–50], Nuclear Magnetic Resonance [51], time-resolved serial femtosecond crystallography [52, 53], and computations [54–56], the internal proton donor D96 H-bonds to T46, and D115, which remains protonated throughout the reaction cycle, H-bonds to T90 (Figure 1D). Interhelical H-bonds of the corresponding Asp are present in Acetabularia rhodopsin-1 and archaerhodopsin-3, though in the former D100 (corresponding to bacteriorhodopsin D96) H-bonds to an Asn (N48) instead of a Ser/Thr. More generally, Asp/Glu-Ser/Thr and Asp-Asn H-bonds are rather common in membrane transporters and receptors [42]. Similarly, bulk-exposed proton antenna clusters that deliver/release a proton to/from an internal site (Figures 1B, 2A), as proposed for bacteriorhodopsin, have also been discussed for cytochrome c oxidase and for photosystem II [36–38, 44, 57–59].
The AlphaFold [60] model deposited for the pH-sensing GPR4 [61] suggests that a large cluster of Asp, Glu, and His groups is located at the extracellular site, where pH must be sensed, and several carboxylic and histidine sidechains, at the core of the receptor (Figure 2B). Although details of the predicted structural model might be debatable, we suggest that, qualitatively, the overall arrangement with a bulk-exposed carboxylic-histidine cluster and several internal titratable sidechains resembles that of bacteriorhodopsin (Figures 1B, 2A,2B) and other microbial rhodopsins (Figures 1E–H).
Given the lack of experimental structures for pH-sensing GPCRs, below we further dissect internal H-bond networks of AA2AR as a class A receptor for which activation upon proton binding was recently discovered [16], and the calcitonin receptor (CTR), as a class B GPCR involved in cell signaling paths likely to depend on pH; as proton sensing appears to be rather common to GPCRs, studies of H-bond networks of GPCRs susceptible of pH sensitivity could inform on general principles of function.
AA2ARs are targeted by one of the most consumed substances in the world, caffeine, and by drugs against Parkinson’s disease [62]. Very recently, experimental data were interpreted to suggest that acidic pH can activate AA2AR [16], but it remains unclear how AA2ARs may couple proton binding to protein conformational change for receptor activation.
AA2AR (Figure 2C) contains relatively few internal carboxylic and His sidechains; there are, however, numerous other charged and polar sidechains within the transmembrane region of AA2AR, such that its protein-water H-bond graph computed for the highest-resolution structure [63] has no fewer than 82 H-bonds (Figure 2E). An extended H-bond cluster with 19 H-bonding protein sidechains and water molecules extends across >20 Å along the membrane normal, from S6 (1.32) to N181; a few other relatively large protein-water H-bond clusters are present, such that the entire receptor is spanned by H-bond clusters. H250 (6.52) and H278 (7.42) are located in the ligand binding site [64], with H278 part of an H-bond cluster with Y9 (1.35) and E13 (1.39) (Figures 2F,G). In an active-like structure, D52 (2.50) is H-bonded to S91 (3.39), S281 (7.46) and N284 (7.49) (Figure 2H), thus being part of a network that connects several GPCR motifs involved in ligand binding and receptor activation. For detailed analyses and discussions of GPCR H-bond networks of GPCRs, including of the adenosine A2A receptor we refer the reader to [21].
Without water molecules, the H-bond graph of the same AA2AR structure consists of just 29 H-bonds, and the largest H-bond cluster has 4 H-bonding sidechains (compare Figure 2E with Figure 2F). Likewise, the H-bond graph obtained for AA2AR in the AA2AR-G complex has 21 H-bonds, with local H-bond clusters of up to 4 H-bonding sidechains (Figure 2G). This finding is compatible with the aforementioned observations on the chemokine [23] and cannabinoid [24] receptors bound to G proteins, with the graph computations presented above for microbial rhodopsins (Figure 1), and with computations suggesting important roles of water-mediated interactions in GPCR activation [21, 65, 66]. That is, internal water molecules might be needed to establish extended connections throughout active-like, G-protein bound GPCRs. Moreover, the H-bond networks at the extracellular side, where bulk water molecules could visit the receptor at least transiently, are likely dynamic and could rearrange upon binding of an agonist, including –in the recently proposed acid sensor AA2AR [16]- upon proton binding.
Internal His, Asp and Glu of AA2AR mentioned above are conserved in the non-proton-sensing adenosine A1 and A2B receptors, which suggests that details of the intra-molecular interactions of titratable groups might determine pH sensitivity. Further studies that integrate high-resolution static structures with spectroscopy, site-directed mutagenesis, and computation, will be needed to unravel mechanisms of proton binding to adenosine receptors.
The CTR is among the best-studied class B GPCRs that are regulated by RAMPs. By itself, the CTR has high affinity for calcitocin (CT), being implicated in bone homeostasis. In complex with one of the three RAMPs, CTR forms amylin receptors, AMY1-3R, which have high affinity for the peptide amylin (Amy) and are involved in the control of food intake [7]; in addition to Amy, AMY1R binds the neuropeptide calcitonin gene-related peptide [7] and could be involved in migraines [67]. Whether and how pH impacts CTRs is unclear, though observations of an increased expression of the CTR at acidic pH [68] and of pH-dependent interactions between calcitonin and model membranes [69], suggest that CTR could be among GPCRs whose functioning is influenced by pH.
A structure of the human(h) CTR bound to Gαsβγ and to salmon calcitocin (sCT) was initially solved with cryo-EM at a resolution of 4.1 Å [70] and then refined to a resolution of 3.3 Å [71]. Although some ambiguity remained in the CTR-CT interactions, the structure suggested polar receptor-ligand contacts and a number of H-bonds within the CTR and at the CTR-G protein interface [71]. The transmembrane region of the refined CTR structure contains several local H-bond clusters, but they are localized, with at most 4-5 sidechains in a cluster, and most of the H-bond graph consists of singular H-bonds (Figure 2I). Such a picture is compatible with the findings for AA2AR and other GPCRs, which need internal water molecules to establish extended H-bond clusters.
Cryo-EM structures solved were recently at resolutions of 2.0–3.3 Å for ligand-bound Gs-CTR and Gs-AMY1-3R [72]. Protein-water H-bonds were suggested to help stabilize the active conformation of the receptor, and to contribute to the binding of sCT [72]. The H-bond graph we computed for the CTR domain contains 51 sidechain-sidechain and water-mediated H-bonds; the largest H-bond clusters contain two water molecules each and 5-6 protein sidechains (Figure 2J). Several Asp, Glu, and His sidechains are part of the internal H-bond network, and thus of interest for potential couplings between protonation change and protein conformation. A large cluster of carboxylic and His sidechains that faces the extracellular bulk (Figure 2D) could couple protonation change with ligand binding.
Protonation is of general importance for the post-translational regulation of protein function [35], and a number of GPCRs bind protons during function [25, 73] or function as pH sensors implicated in cancer [32]. H-bond graphs of GPCRs and microbial rhodopsins suggest that GPCRs might share with other protonation-coupled membrane proteins common principles of how protonation change couples with protein conformation (Figure 1B). In the future, given the rapid pace at which GPCR structures are solved, H-bond graph computations could help identify H-bond networks and sites where the H-bond network is interrupted –which could guide, e.g., the placing of internal water molecules in GPCR structures. H-bond graphs of high-resolution structures of GPCRs trapped in distinct intermediates could help interpret spectroscopic fingerprints for putative proton-binding sites of GPCRs. We anticipate that future studies will capitalize on improvements in cryo-EM structure solving [74], computational modeling of GPCR conformational intermediates [75], and public repositories for GPCRs [76, 77].
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
A-NB designed and performed research, wrote the original draft and edited the manuscript. MA-P performed research and edited the manuscript.
Open-access publication funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—491111487. We thank the Central Library of the Forschungszentrum Jülich for making the open access publication possible. MA-P acknowledges financial support in part from the DFG Research Unit FOR2518 “Functional Dynamics of Ion Channels and Transporters—DynIon” Project P6. A-NB acknowledges financial support in part from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Sklodowska-Curie grant agreement No 860592, Innovative Training Network “Proton and proton-coupled transport”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A (2003) 100:4903–8. doi:10.1073/pnas.0230374100
2. Wise A, Gearing K, Rees S. Target validation of G-protein coupled receptors. Drug Discov Today (2002) 7:235–46. doi:10.1016/s1359-6446(01)02131-6
3. Hauser A, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: New agents, targets and indications. Nat Rev Drug Discov (2017) 16:829–42. doi:10.1038/nrd.2017.178
4. Sriram K, Insel PAG. Protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol Pharmacol (2018) 93:251–8. doi:10.1124/mol.117.111062
5. Höller C, Freissmuth M, Nanoff CG. G proteins as drug targets. CMLS Cel Mol Life Sci (1999) 55:257–70. doi:10.1007/s000180050288
6. Nubbemeyer B, Pepanian A, George AAP, Imhof D. Strategies towards targeting Gαi/s proteins: Scanning of protein‐protein interaction sites to overcome inaccessibility. ChemMedChem (2021) 16:1697–716. doi:10.1002/cmdc.202100039
7. Gingell JJ, Simms J, Barwell J, Poyner DR, Watkins HA, Pioszak AA, et al. An allosteric role for receptor activity-modifying proteins in defining GPCR pharmacology. Cell Discov (2016) 2:16012. doi:10.1038/celldisc.2016.12
8. Deupi X, Kobilka B. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (2010) 25:293–303. doi:10.1152/physiol.00002.2010
9. Alhadeff R, Vorobyov I, Yoon HW, Warshel A. Exploring the free-energy landscape of GPCR activation. Proc Natl Acad Sci U S A (2018) 115:10327–32. doi:10.1073/pnas.1810316115
10. Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids (1994) 73:159–80. doi:10.1016/0009-3084(94)90180-5
11. Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr Opin Struct Biol (2011) 21:802–7. doi:10.1016/j.sbi.2011.09.007
12. Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci (2014) 39:233–44. doi:10.1016/j.tibs.2014.03.002
13. Selent J, Sanz F, Pastor M, De Fabritiis G. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. Plos Comput Biol (2010) 6:e1000884. doi:10.1371/journal.pcbi.1000884
14. Mahalingam M, Martínez-Mayorga K, Broen MF, Vogel R. Two protonation switches control rhodopsin activation in membranes. Proc Natl Acad Sci U S A (2008) 105:17795–800. doi:10.1073/pnas.0804541105
15. Madathil S, Fahmy K. Lipid protein interactions couple protonation to conformation in a conserved cytosolic domain of G Protein-Coupled Receptors. J Biol Chem (2009) 284:28801–9. doi:10.1074/jbc.m109.002030
16. Kapolka NJ, Rowe JB, Taghon GJ, Morgan WM, OShea CR, Isom DG. Proton-gated coincidence detection is a common feature of GPCR signaling. Proc Natl Acad Sci U S A (2021) 118:e2100171118. doi:10.1073/pnas.2100171118
17. Siemers M, Lazaratos M, Karathanou K, Guerra F, Brown LS, Bridge BA-N. Bridge: A graph-based algorithm to analyze dynamic H-bond networks in membrane proteins. J Chem Theor Comput (2019) 15:6781–98. doi:10.1021/acs.jctc.9b00697
18. Siemers M, Bondar A-N. Interactive interface for graph-based analyses of dynamic H-bond networks: Application to spike protein S. J Chem Inf Model (2021) 61:2998–3014. doi:10.1021/acs.jcim.1c00306
19. Bertalan E, Lesca E, Schertler GFX, Bondar A-N. C-Graphs tool with graphical user interface to dissect conserved hydrogen-bond networks: Applications to visual rhodopsins. J Chem Inf Model (2021) 61:5692–707. doi:10.1021/acs.jcim.1c00827
20. Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature (2008) 453:363–7. doi:10.1038/nature06925
21. Bertalan É, Lešnik S, Bren U, Bondar A-N. Protein-water hydrogen-bond networks of G protein-coupled receptors: Graph-based analyses of static structures and molecular dynamics. J Struct Biol X (2020) 212:107634. doi:10.1016/j.jsb.2020.107634
22. Hua T, Li X, Wu L, Iliopoulos-Tsoutsouvas C, Yuxia W, Wu M, et al. Activation and signaling mechanisms revealed by cannabinoid receptor-Gi complex structures. Cell (2020) 180:1–11. doi:10.1016/j.cell.2020.01.008
23. Isaikina P, Tsai C-J, Dietz N, Pamula F, Grahl A, Goldie KN, et al. Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist. Sci Adv (2021) 7:eabg8685. doi:10.1126/sciadv.abg8685
24. Bondar A-N. Graphs of hydrogen-bond networks to dssect protein conformational dynamics. J Phys Chem B (2022) 126:3973–84. doi:10.1021/acs.jpcb.2c00200
25. Vickery ON, Carvalheda CA, Zaidi SA, Pisliakov AV, Katritch V, Zachariae U. Intracellular transfer of Na+ in an active-state G-Protein-Coupled Receptor. Structure (2018) 26:171–80.e2. doi:10.1016/j.str.2017.11.013
26. Quinn SJ, Bai M, Brown EM. pH sensing by the calcium-sensing receptor. J Biol Chem (2004) 279:37241–9. doi:10.1074/jbc.m404520200
27. Meyer J, del Vecchio G, Steitz V, Massaly N, Stein C. Modulation of µ-opioid receptor activation by acidic pH is dependent on ligand structure and an ionizable amino acid residue. Br J Pharmacol (2019) 176:4510–20. doi:10.1111/bph.14810
28. Carpenter B, Lebon G. Human adenosine A2A receptor: Molecular mechanism of ligand binding and activation. Front Pharmacol (2017) 8:898. doi:10.3389/fphar.2017.00898
29. Cao R, Giorgietti A, Bauer A, Neumaier B, Rosetti G, Carloni P. Role of extracellular loops and membrane lipids for ligand recognition in the neuronal adenosine receptor type 2A: An enhanced sampling simulation study. Molecules (2018) 23:2616. doi:10.3390/molecules23102616
30. Klatt W, Wallner S, Brochhausen C, Stolwijk JA, Schreml S. Expression profiles of proton-sensing G-protein coupled receptors in common skin tumors. Sci Rep (2020) 10:15327. doi:10.1038/s41598-020-71700-9
31. Silva PHI, Wagner CA. Physiological relevance of proton-activated GPCrs. Pflugers Arch - Eur J Physiol (2022) 474:487–504. doi:10.1007/s00424-022-02671-1
32. Ludwig M-G, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, et al. Proton-sensing G-protein-coupled receptors. Nature (2003) 425:93–8. doi:10.1038/nature01905
33. Rowe JB, Kapolka NJ, T GJ, Morgan WM, Isom DG. The evolution and mechanism of GPCR proton sensing. J Biol Chem (2020) 296:100167–13. doi:10.1074/jbc.ra120.016352
34. Thurlkill RA, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci (2006) 15:1214–8. doi:10.1110/ps.051840806
35. Schönichen A, Webb BA, Jacobson MP, Barber DL. Considering protonation as a posttranslational modification regulating protein structure and function. Annu Rev Biophys (2013) 42:289–314. doi:10.1146/annurev-biophys-050511-102349
36. Sacks V, Marantz Y, Aagaard A, Checover S, Nachliel E, Gutman M. The dynamic feature of the proton collecting antenna of a protein surface. Biochim Biophys Acta - Bioenerg (1998) 1365:232–40. doi:10.1016/s0005-2728(98)00073-5
37. Checover S, Marantz Y, Nachliel E, Gutman M, Pfeiffer M, Tittor J, et al. Dynamics of the proton transfer reaction on the cytoplasmic surface of bacteriorhodopsin. Biochemistry (2001) 40:4281–92. doi:10.1021/bi002574m
38. Riesle J, Oesterhelt D, Dencher NA, Heberle J. D38 is an essential part of the proton translocation pathway in bacteriorhodopsin. Biochemistry (1996) 35:6635–43. doi:10.1021/bi9600456
39. Borshchevskiy V, Kovalev K, Round E, Efremov R, Astashkin R, Bourenkov G, et al. True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins. Nat Struct Mol Biol (2022) 29:440–50. doi:10.1038/s41594-022-00762-2
40. Furuse M, Tamogami J, Hosaka T, Kikukawa T, Shinya N, Hato M, et al. Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum. Acta Crystallogr D Biol Crystallogr (2015) 71:2203–16. doi:10.1107/s1399004715015722
41. Bada Juarez JF, Judge PJ, Adam S, Axford D, Vinals J, Birch J, et al. Structures of the archaerhodopsin-3 transporter reveal that disordering of internal water networks underpins receptor sensitization. Nat Commun (2021) 12:629. doi:10.1038/s41467-020-20596-0
42. Lazaratos M, Siemers M, Brown LS, Bondar A-N. Conserved hydrogen-bond motifs of membrane transporters and receptors. Biochim Biophys Acta - Biomembranes (2022) 1864:183896. doi:10.1016/j.bbamem.2022.183896
43. Bondar A-N, Lemieux HJ. Reactions at biomembrane interfaces. Chem Rev (2019) 119:6162–83. doi:10.1021/acs.chemrev.8b00596
44. Bondar A-N. Proton-binding motifs of membrane-bound proteins: From bacteriorhodopsin to spike protein S. Front Chem (2021) 9:685761. doi:10.3389/fchem.2021.685761
45. Bondar A-N. Mechanisms of long-distance allosteric couplings in proton-binding membrane transporters. Adv Protein Chem Struct Biol (2022) 128:199–239. doi:10.1016/bs.apcsb.2021.09.002
46. Gerwert K, Hess B, Soppa J, Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A (1989) 86:4943–7. doi:10.1073/pnas.86.13.4943
47. Balashov SP, Govindjee R, Imasheva ES, Misra S, Ebrey TG, Feng Y, et al. The two pKa's of aspartate-85 and control of thermal isomerization and proton release in the arginine-82 to lysine mutant of bacteriorhodopsin. Biochemistry (1995) 34:8820–34. doi:10.1021/bi00027a034
48. Dioumaev AK, Brown LS, Needleman R, Lanyi JK. Fourier Transform infrared spectra of a late intermediate of the bacteriorhodopsin photocycle suggest transient protonation of asp-212. Biochemistry (1999) 38:10070–8. doi:10.1021/bi990873+
49. Metz G, Siebert F, Engelhardt M. Asp85 is the only internal aspartic acid that gets protonated in the M intermediate and the purple-to-blue transition of bacteriorhodopsin. A solid-state 13C CP-MAS NMR investigation. FEBS Lett (1992) 303:237–41. doi:10.1016/0014-5793(92)80528-o
50. Kandori H, Belenki M, Herzfeld J. Vibrational frequency and dipolar orientation of the protonated Schiff base in bacteriorhodopsin before and after photoisomerization. Biochemistry (2002) 41:6026–31. doi:10.1021/bi025585j
51. Ni QZ, Can Tv ED, Belenky M, Griffin RG, Herzfeld J. Primary transfer step in the light-driven ion pump bacteriorhodopsin: An irreversible U-turn revealed by Dynamic Nuclear Polarization-Enhanced Magic Angle Spinning NMR. J Am Chem Soc (2018) 140:4085–91. doi:10.1021/jacs.8b00022
52. Nango E, Royant A, Kubo M, Nakane T, Wickstrand C, Kimura T, et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science (2016) 354:1552–7. doi:10.1126/science.aah3497
53. Weinert T, Skopintsev P, James D, Dworkowski F, Panepucci E, Kekilli D, et al. Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography. Science (2019) 365:61–5. doi:10.1126/science.aaw8634
54. Bondar A-N, Elstner M, Suhai S, Smith JC, Fischer S. Mechanism of primary proton transfer in bacteriorhodopsin. Structure (2004) 12:1281–8. doi:10.1016/j.str.2004.04.016
55. Hayashi S, Ohmine I. Proton transfer in bacteriorhodopsin: Structure, excitation, IR spectra, and potential energy surface analyses by an ab initio QM/MM method. J Phys Chem B (2000) 104:10678–91. doi:10.1021/jp001508r
56. Goyal P, Ghosh N, Phatak P, Clemens M, Gaus M, Elstner M, et al. Proton storage site in bacteriorhodopsin: New insights from quantum mechanics/molecular mechanics simulations of microscopic pKa and infrared spectra. J Am Chem Soc (2012) 133:14981–97. doi:10.1021/ja201568s
57. Ädelroth P, Brzezinski P. Surface-mediated proton-transfer reactions in membrane-bound proteins. Biochim Biophys Acta - Bioenerg (2004) 1655:102–15. doi:10.1016/j.bbabio.2003.10.018
58. Shutova T, Klimov VV, Andersson B, Samuelsson G. A cluster of carboxylic groups in PsbO protein is involved in proton transfer from the water oxidizing complex of Photosystem II. Biochim Biophys Acta - Bioenerg (2007) 1767:434–40. doi:10.1016/j.bbabio.2007.01.020
59. Lorch S, Capponi S, Pieront F, Bondar A-N. Dynamic carboxylate/water networks on the surface of the PsbO subunit of photosystem II. J Phys Chem B (2015) 119:12172–81. doi:10.1021/acs.jpcb.5b06594
60. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature (2021) 596:583–9. doi:10.1038/s41586-021-03819-2
61. The UniProt Consortium UniProt: A hub for protein information. Nucleic Acid Res (2015) 43:D204–D212. doi:10.1093/nar/gku/989
62. Jacobson KA, Gao ZG, Matricon P, Eddy MT, Carlsson J. Adenosine A2A receptor antagonists: From caffeine to selective non-xanthines. Br J Pharmacol (2020) 179:3496–511. doi:10.1111/bph.15103
63. Weinert T, Olieric N, Cheng R, Brünle S, James D, Ozerov D, et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat Commun (2017) 8:542. doi:10.1038/s41467-017-00630-4
64. Jespers W, Schiedel AC, Heitman LH, Cooke RM, van Westen GJP, Gloriam DE, et al. Structural mapping of adenosine receptor mutations: Ligand binding and signaling mechanisms. Trends Pharmacol Sci (2018) 39:75–89. doi:10.1016/j.tips.2017.11.001
65. Yuan S, Filipek S, Palczewski K, Vogel H. Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat Commun (2014) 5:4733. doi:10.1038/ncomms5733
66. Lee Y, Kim S, Choi S, Hyeon C. Ultraslow water-medated transmembrane interactions regulate the activation of A2A adenosine receptor. Biophys J (2016) 111:1180–91. doi:10.1016/j.bpj.2016.08.002
67. Walker CS, Eftekhari S, Bower RL, Wilderman A, Insel PA, Edvinsson L, et al. A second trigeminal CGRP receptor: Function and expression of the AMY1 receptor. Ann Clin Transl Neurol (2015) 2:595–608. doi:10.1002/acn3.197
68. Biskobing DM, Fan D. Acid pH increases carbonic anhydrase II and calcitonin receptor mRNA expression in mature osteoclasts. Calcif Tissue Int (2000) 67:178–83. doi:10.1007/s00223001107
69. Micelli S, Melelo D, Piciarelli V, Gallucci E. Effect of pH-variation on insertion and ion channel formation of human calcitonin into planar lipid bilayers. Front Biosci (2006) 11:2035–44. doi:10.2741/1945
70. Liang Y-L, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature (2017) 546:118–23. doi:10.1038/nature22327
71. dal Maso E, Glukhova A, Zhu Y, Garcia-Nafria J, Tate CG, Atanasio S, et al. The molecular control of calcitonin receptor signaling. ACS Pharmacol Transl Sci (2019) 2:31–51. doi:10.1021/acsptsci.8b00056
72. Cao J, Belousoff J, Liang Y-L, Johnson RM, Josephs TM, Fletcher MM, et al. A structural basis for amylin receptor phenotype. Science (2022) 375:eabm9609. doi:10.1126/science.abm9609
73. Zhang XC, Cao C, Zhou Y, Zhao Y. Proton transfer-mediated GPCR activation. Protein Cell (2015) 6:12–7. doi:10.1007/s13238-014-0106-4
74. Garcia-Nafria J, Tate CG. Structure determination of GPCRs: Cryo-EM compared with X-ray crystallography. Biochem Soc Trans (2021) 49:2345–55. doi:10.1042/bst20210431
75. Del Alamo S, Sala D, McHaorab HS, Miller J. Sampling alternative conformational states of transporters and receptors with AlphaFold2. ELife (2022) 11:e75751. doi:10.7554/elife.75751
76. Rodriguez-Espigares I, Torrens-Fontanals M, Tiemann JKS, Aranda-Garcia D, Ramirez-Anguita JM, Stepniewski TM, et al. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nat Methods (2020) 17:777–87. doi:10.1038/s41592-020-0884-y
77. Kooistra AJ, Mordalski S, Pándy-Szekeres P, Esguerra M, Mamyrbekov A, Munk C, et al. GPCRdb in 2021: Integrating GPCR sequence, structure and function. Nucleic Acids Res (2021) 49:D335–D43. doi:10.1093/nar/gkaa1080
78. Lanyi JK. Bacteriorhodopsin. Int Rev Cytol (1999) 187:161–202. doi:10.1016/s0074-7696(08)62418-3
79. Humphrey W, Dalke W, Schulten K. VMD: Visual molecular dynamics. J Mol Graph (1996) 14:33–8. doi:10.1016/0263-7855(96)00018-5
80. Lomize M, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res (2011) 40:D370–6. doi:10.1093/nar/gkr703
81. Hasegawa N, Jonotsuka H, Miki K, Takeda K. X-ray structure analysis of bacteriorhodopsin at 1.3Å resolution. Sci Rep (2018) 8:13123. doi:10.1038/s41598-018-31370-0
Keywords: G-protein coupled receptor (GPCR), G protein, pH, hydrogen bond (H-bond), graph theory—graph algorithms
Citation: Bondar A-N and Alfonso-Prieto M (2022) Hydrogen-bond networks for proton couplings in G-Protein coupled receptors. Front. Phys. 10:963716. doi: 10.3389/fphy.2022.963716
Received: 07 June 2022; Accepted: 09 August 2022;
Published: 06 September 2022.
Edited by:
André H. Erhardt, Weierstrass Institute for Applied Analysis and Stochastics (LG), GermanyReviewed by:
Stefan Gahbauer, University of California, San Francisco, United StatesCopyright © 2022 Bondar and Alfonso-Prieto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana-Nicoleta Bondar, bmJvbmRhckBmaXppY2EudW5pYnVjLnJv, YS5ib25kYXJAZnotanVlbGljaC5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.