
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Phys., 16 June 2022
Sec. Optics and Photonics
Volume 10 - 2022 | https://doi.org/10.3389/fphy.2022.894797
This article is part of the Research TopicAdvances in Optical Imaging Techniques for Microscopy and NanotechnologyView all 7 articles
Lipid droplets are the major organelles for fat storage in a cell and analyzing lipid droplets in Caenorhabditis elegans (C. elegans) can shed light on obesity-related diseases in humans. In this work, we propose to use a label free scattering-based method, namely dark field microscopy, to visualize the lipid droplets with high contrast, followed by deep learning to perform automatic segmentation. Our method works through combining epi-illumination dark field microscopy, which provides high spatial resolution, with asymmetric illumination, which computationally rejects multiple scattering. Due to the raw data’s high quality, only 25 images are required to train a Convolutional Neural Network (CNN) to successfully segment lipid droplets in dense regions of the worm. The performance is validated on both healthy worms as well as those in starvation conditions, which alter the size and abundance of lipid droplets. Asymmetric illumination substantially improves CNN accuracy compared with standard dark field imaging from 70% to be 85%, respectively. Meanwhile, standard segmentation methods such as watershed and DIC object tracking (DICOT) failed to segment droplets due to the images’ complex label-free background. By successfully analyzing lipid droplets in vivo and without staining, our method liberates researchers from dependence on genetically modified strains. Further, due to the “open top” of our epi-illumination microscope, our method can be naturally integrated with microfluidic chips to perform large scale and automatic analysis.
As a model organism, Caenorhabditis elegans has the advantage of a short life span, easy maintenance, and many shared genes with humans [1]. Thus, since its introduction to the model organism research community, C. elegans has been under extensive study to gain knowledge of neuroscience, behavior, and disease [2]. In particular, with the prevalence of obesity in the world, C. elegans has become a popular animal model for studying the regulation of lipid metabolism and obesity-related diseases. Further, many genes related to the metabolic diseases are shared with humans [3]. The lipid droplets, as the major fat storage organelles, exist mostly in the intestine and epidermal layer of the animal. They are generally round and their sizes range from a few hundred nanometers to a few microns [4]. During the development and growth of C. elegans, lipid droplets undergo morphological changes according to the dietary regulation and gene mutation [5]. Lipid droplet morphology can thus provide a vast amount of information that can lead to detection of new genes and pathways, through studying the diverse morphology and chemical content of lipid droplets. Currently this is done through exogenous or genetically-encoded staining of lipid droplets and manual analysis, which is laborious and time-consuming[6]. Therefore, large scale screening systems, including both image acquisition and automatic analysis, will be beneficial to the gene identification and life-time monitoring of C. elegans.
Current visualization of lipid droplets is still accomplished through fluorescence microscopy and electron microscopy due to the small and dense package of the lipid droplets [7,8]. Traditional differential interference contrast (DIC) microscopy utilizing oil-immersion condensers could visualize the lipid droplets. However, this method is polarization dependent and would not work with the plastic material of the microfluidic chip which is often used in large scale screening or time-lapse monitoring. In the fluorescence-based methods, often confocal microscopy is preferred if individual droplets are of interest. Nile Red dye or genetically encoded fluorescent proteins are used to create contrast. However, the Nile Red is not specific and not every strain of C. elegans has a matched genetic fluorescent protein [4]. Further the genetically modified C. elegans are not available to everyone, thus limiting capacities of many research labs.
A label-free method that does not depend on polarization and staining is therefore preferred. Recently both coherent and spontaneous Raman imaging has been used to study the lipid diversity [9–11]. However due to its slow speed, Raman microscopy would not be a good screening method. Another line of label-free methods is phase microscopy. Recently, there is a strong push to expand its application to image thick samples such as C. elegans[12–14]. However, current resolution and contrast are still not enough to visualize individual lipid droplets due to their dense packing within the C. elegans. Phase imaging methods are generally wide-field illumination and imaging, resulting in optical sectioning that is significantly worse compared to confocal fluorescence [15,16]. This issue becomes pressing in thick samples, where out-of-plane structures can dominate the image quality. Perhaps due to this reason, reported work on using phase microscopy to analyze lipid droplets is scarce. Recently, we have developed an epi-illumination dark field microscope which can visualize the unlabeled and individual lipid droplets in living C. elegans [17]. The system has a transverse resolution of 260 nm and lateral resolution of 520nm, which is comparable to a fluorescence microscope of equivalent numerical aperture. Thus we hope to use this system as a basis to perform lipid droplet analysis in vivo.
For large scale screening purposes or for time-lapse monitoring, it would be desirable to have the images automatically analyzed. Edge detection, Hough transform and granulometry-based digital sieves have been used to segment or analyze lipid droplets [18,19]. However they are all developed based on fluorescence images with nearly perfect contrast. In the label-free images of lipid droplets in vivo, including those generated by the dark field microscope used here, the multiple scattering in the thick sample degrades the image contrast and creates subtle textures in the images, not to mention the lack of specificity means that the images in general are complex. Traditional image analysis methods are therefore not well suited to automated lipid droplet segmentation using unlableled C. elegans images.
In this work, we propose to solve those issues through two strategies, one is to improve the image contrast through adding asymmetrical illumination to our previously developed epi-illumination dark field microscopy. Asymmetrical illumination has been shown previously to yield differential phase contrast (DPC) images which have improved optical sectioning [20–22]. However, to date it is mostly explored in cases of either small NA forward illumination or the collection of forward scattering signals. Here, we apply the asymmetry to epi-illumination with a large NA and to the collection of back scattered signals. In this way, we can maintain the high spatial resolution while removing the multiply-scattered background signal from the image. The second strategy is in the image segmentation side, where we utilize deep learning rather than traditional image processing methods to perform the image segmentation of lipid droplets in the dense regions. Due to the improved image quality from the asymmetric illumination, our training requires a small dataset of only 25 images. With this, we have achieved automatic image analysis of unlabeled lipid droplets with analysis accuracy of around 85%, while the traditional methods basically fail to segment. We validate our approach through recovering changes in lipid number and size due to starvation and feeding conditions in living C. elegans. Since our method is not fluorescence-dependent, we can analyze any strain of the C. elegans, without requiring exogenous labels or genetically-encoded fluorophores.
The schematic diagram of the system is shown in Figure 1A. The detailed description of the imaging system was described previously [17]. Briefly, the system utilizes a 60X, NA 1.49 oil-immersion objective (Nikon, Japan) and a ring source to provide the epi-illumination. The exposure time of data in this paper is 80 ms with a sCMOS camera (pco.edge 4.2, PCO GmbH, Germany). The back scattered light is collected to form the image. In the conjugate plane of the ring source, a ring-shaped metal mask is used to block the light reflected from the sample for dark-field imaging. The ring source consists of 32 green LEDs with a central wavelength of 531 nm and a line-width of about 30 nm. In order to generate an asymmetric illumination pattern, the LED ring is divided into 4 groups, with each group having eight LEDs. The adjacent two groups are used to form top and bottom or left and right illumination patterns as shown in Figure 1B. If the worm body is largely horizontal in the field of view, we will use horizontally asymmetrical illumination scheme. Otherwise, we will use vertically asymmetric scheme. While multi-axis asymmetric illumination imaging has been previously demonstrated [23], for simplicity in this study we utilize only a single axis to create each image.
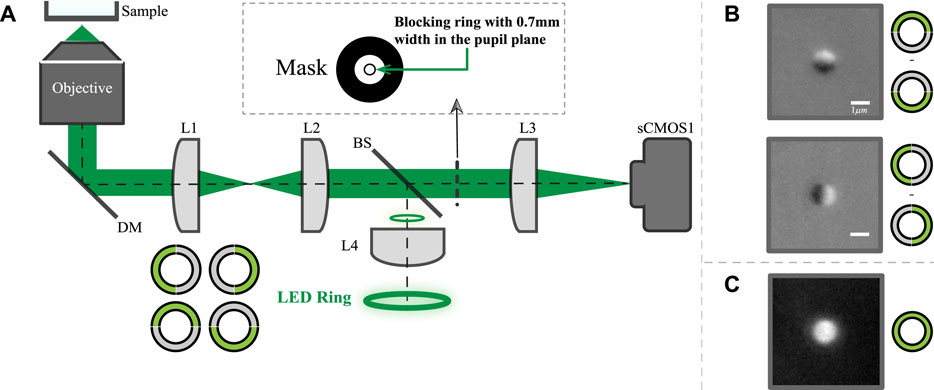
FIGURE 1. The proposed imaging system layout and exemplary images. (A) Schematic layout of the proposed system where both the illumination and detection is on one side of the objective. (B) images of 1 μm bead with vertically or horizontally asymmetrical illumination; (C) Symmetric illumination dark field image of the same bead.
The traditional dark field image can also be obtained through turning all LEDs on or simply adding up the pair of the asymmetrical illuminated images. Representative top-bottom and left-right dark-field DPC images of a single 1 μm bead are shown in Figure 1B, while Figure 1C shows the traditional dark field image of the same bead.
For C. elegans, the body length is about 1 mm and the moving speed could be up to 0.5 mm/s, however the field of view of the present imaging system is only about 177μm × 177 μm. Thus, in order to view a freely-moving worm with submicron resolution, a high speed and high precision tracking platform will be necessary. In this work, for simplicity, the worm is imaged after anesthetization.
There are two steps of the processing: preprocessing and segmentation of the image.
Since images are generated when part of the ring is on, we have two kinds of data. One is the differential phase image, Idpc, through subtraction. The other is the epi-illumination dark field microscopy data, Idm, through summation. The formulas can be described below:
where the subscripts L, R, T and B represent left, right, top and bottom halves respectively. In our system, the camera collects the back scattered signals from subcellular structures, which is typically weak. The background light, which arises from out-of-plane scattering, reflections within the optical system, and other stray light sources, can be substantial compared to the signals from the sample. Thus a background image collected for each half-ring is subtracted from the raw images before computing Idpc and Idm.
Three methods, two traditional machine-learning based and one deep learning based, are used. The first is the traditional thresholding method. For differential phase type images, DIC-object-tracking (DICOT) method, which is thresholding based, uses a modified Gaussian filter called scaling of Gaussian (SoG) and has been shown to achieve reasonable results on the yolk granules inside the C. elegans embryo [24]. The method can be downloaded (https://github.com/Self-OrganizationLab/DICOT_GUI) and has parameters such as the kernel size and sigma which define the estimated object size and the variation of the Gaussian filter. In our DPC image, we optimized the kernel size to be 3 μm and sigma to be 0.1 μm. The second method is the watershed algorithm, which is edge detection based. Through searching the ridges (high intensity region) and valleys (low intensity region) in an image, the algorithm accomplishes segmentation. A custom Matlab script is used to perform the watershed segmentation and can be found at https://github.com/pipi8jing/pytorch-unet-dropletsegmentation.
The third method utilizes deep learning, which has achieved tremendous success on a wide variety of image segmentation and classification problems in recent years [25,26]. However, typically deep learning requires substantial amounts of training data (from thousands to millions of images). In this work, the ground truth is generated through manual annotation and the sizes of the training data set are 25 and 20 images for DPC and DF cases respectively. The sizes of the test data for DPC and DF based analysis are 10 and 3 respectively. For DF based analysis, due to the poorer performance, we annotated only 3 images for the testing.
The CNN architecture for segmentation is based on the classic U-Net[27]. Compared with the original U-Net structure, the downsampling is achieved through a 2 × 2 pixel convolution [28]. A dropout layer (p = 0.2) before each downsampling and upsampling process is used to prevent overfitting. The loss function is defined as:
where f(xi), yi are the output and target image respectively. The network was trained using the Adam optimizer and a learning rate of 0.001. The convolutional kernel were initialized using a truncated normal distribution with a standard deviation of 0.02 and a mean of zero. The bias were initialized as zeros. The training is finished after a total of 3,000 mini-batch (size = 8) iterations. The network was implemented using the Pytorch framework (version 1.10.0) and on a desktop computer with a NVIDIA GeForce RTX 3090 (NVidia, Santa Clara, CA, United States) and 16 GB RAM.
In order to use the training data more efficiently, we have rotated Idpc images generated from vertical illumination 90° so that all images appear to utilize the same horizontal differential contrast. To quantify the accuracy, we have used dice, precision and recall, defined as follows:
where true/false positive/negative are calculated at the pixel level in each image. After the segmentation by the CNN,there are still holes and irregular boundaries in the binary result. In order to facilitate the morphology analysis of lipid droplets for different physiology states, we used morphological operators in ImageJ (https://imagej.nih.gov/ij/download.html) to fill holes within the binary image, by simply clicking the dilation and then the erosion buttons without need for parameter tuning.
C. elegans were anesthetized with 0.05 mM of levamisole for 15 minutes at an ambient temperature of 21°C immediately prior to imaging. After anesthetization the worms were placed on coverslip-bottomed Petri dishes for observation.
Lipid droplets are not only well inside the body of C. elegans (within
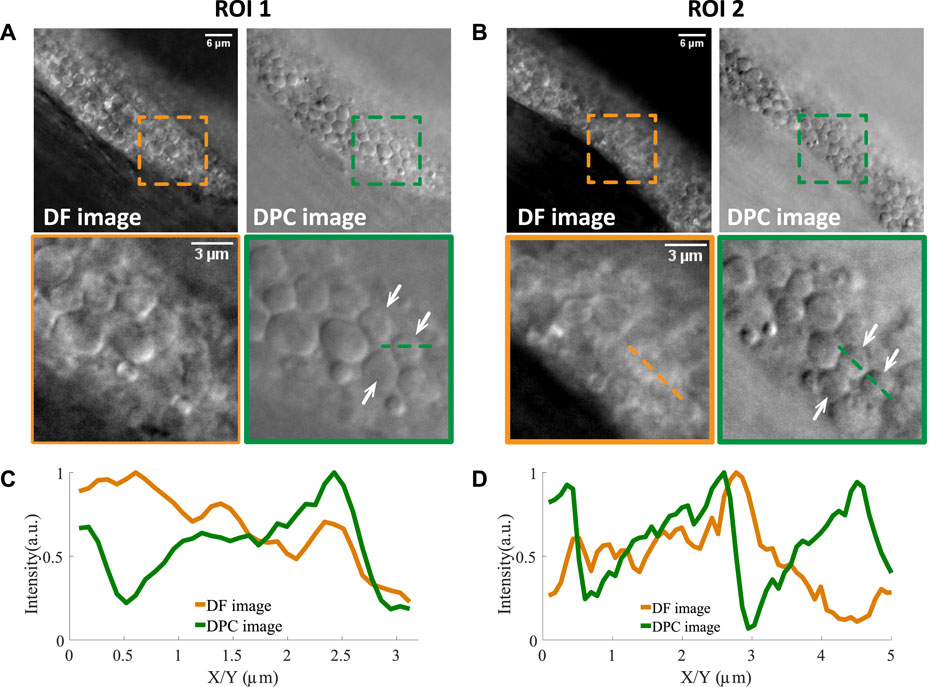
FIGURE 2. Two examples of lipid droplets in dense regions imaged by normal dark field microscopy and asymmetrically illuminated dark field microscopy. (A,B) Two regions of interest and their zoom-in versions where lipid droplets pointed by the white arrows can be resolved in the DPC image and not in the DF image. The line profiles of two such lipid droplets are shown in (C,D). Scale bars in (A,B) represent 6 μm.
In the raw dark field images (labeled as DF images), one can see that there are still many lipid droplets with low contrast. Two examples are shown in (A) and (B), indicated by white arrows. Then with asymmetrical illumination, the image of the same field of view can be obtained (labeled as DPC images). One can see that the shape of lipid droplets becomes more obvious since its boundary is more clearly defined and the intensity within the droplet area is more uniform. The background of the DPC image is also cleaner compared to DF image. The line profile of two lipid droplets (indicated by the dotted lines in the zoomed ROIs) are shown in Figures 2C,D. One can see that both edges of the droplets are obvious in the DPC image while only one edge is clearly visible in the dark field images, due to the out-of-focus signals. However, in the DPC images, the out-of-focus light is eliminated through the subtraction operation (as shown in Eq. 1) and the edges can be revealed with higher contrast. This increased contrast would facilitate the automatic image analysis as we demonstrate below.
With the DF or DPC images, we first processed them with the traditional image segmentation methods, namely, watershed and DICOT(more details can be found in Section 2). The raw images and the ground truth (generated through manual annotation) are shown in Figure 3A and the segmentation results in Figure 3B, where the ground truth is labeled in yellow, and the watershed results (edges of lipid droplets) are overlaid in sepia, while the DICOT results (brightest point in a lipid droplet) are black dots. One can see that due to the dense structure and complex textures of the droplets, the intensity variations along the edges and inside the lipid droplets cause the standard segmentation results to not be usable. In the image shown in Figure 3A, there are 73 lipid droplets in total. The watershed method gives out only 7 correctly. For DICOT method, the most obvious problem is that the same particle can be detected multiple times (a few examples indicated by the green arrows in Figure 3B). Further, it can only be used to count particles roughly without giving a complete morphological segmentation. We believe the primary reason for this is that, unlike fluorescence images with low backgrounds, our label-free microscope has a strong, complex background that varies across the image, and even within a single lipid droplet (More examples of the irregular intensity variation can be found in the Supplementary Figure S1). Thus, traditional methods which are mostly based on local intensity values, have difficulty accurately segmenting the complex label-free image.
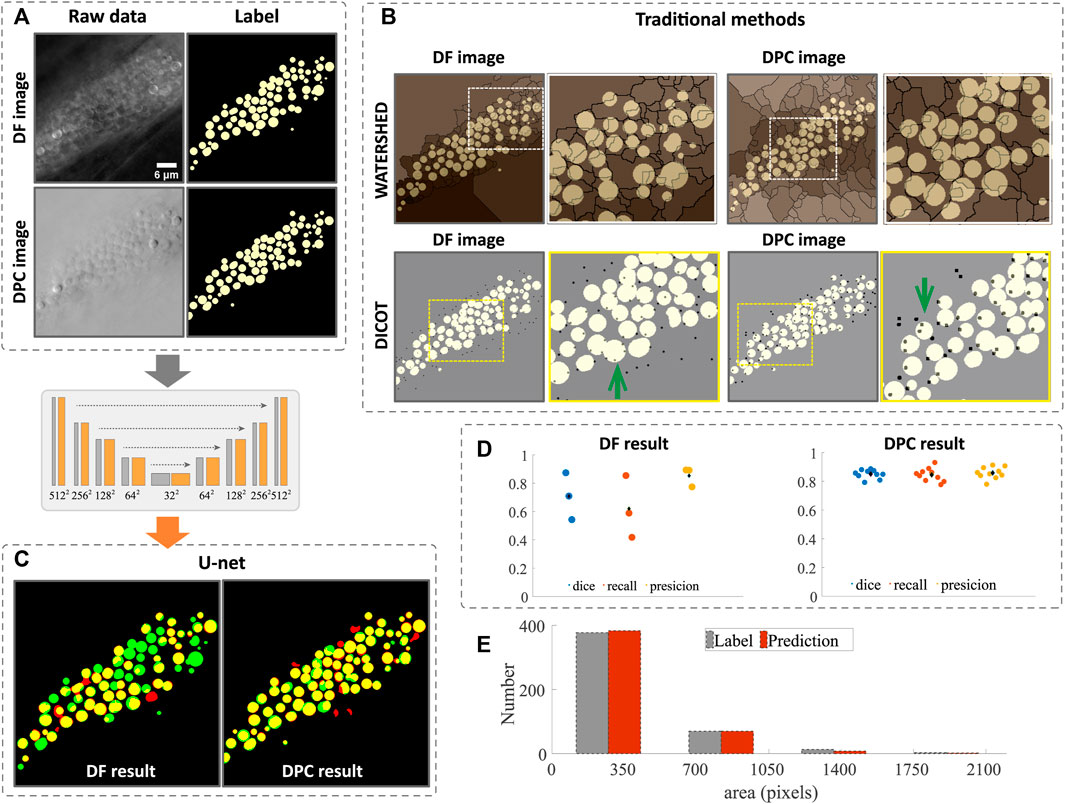
FIGURE 3. Image segmentation and morphology analysis of lipid droplet in dense regions. (A) raw data of DF and DPC images and their ground truth annotated manually. (B) segmentation by traditional methods and the ground truth is shown in white and the edges (by watershed) and bright peaks (by DIOCT) of the lipid droplets are shown in black. One can see that many edges or the peaks are in the wrong places. The segmentation results using CNN are shown in (C) and is significantly better. The quantitative measure of the DL segmentation shown in (D) suggests that the accuracy for DPC case is around 85% and 70% for DF case. The area and number of the lipid droplets are analyzed from DPC segmentation and is shown (E) along with the analysis based on manual segmentation. Scale bars in (B) represent 6 μm.
Meanwhile, deep learning based segmentation, even using only 25 annotated images in the training, results in much improved segmentation with most of the lipid droplets labeled successfully. As one can see in Figure 3C, some droplets that failed to segment by the traditional methods are successfully segmented by the CNN network. In Figure 3C, the ground truth annotations are colored green, while the deep network results are in red, such that areas of agreement are colored yellow. Comparing the DF or DPC image based segmentation, one can see that the latter gives a markedly better result. The quantitative measures such as dice, recall and precision are computed and plotted in Figure 3D for both cases. One can see that these numbers are all higher for the DPC image based segmentation. Note that the image analysis is performed on 2D data due to the focus of this paper on assessing different imaging modes for droplet segmentation. The microscope itself is capable of collecting 3D images by z-scanning, and overall performance of the deep learning may be further improved by considering 3D information, but this is beyond the scope of this article. More segmentation results for images at different z-slices can be found in the Supplementary Figure S2.
With the segmentation accomplished, we can perform bulk morphology analysis. Figure 3E shows the size distribution of the lipid droplets analyzed from the CNN segmented results compared to the manual analysis, with the two methods agreeing closely, showing that both area and number of droplets are accurately predicted. Although the CNN method still has some mistakes compared to the ground truth, these could be improved with a larger training set size. Nevertheless, the segmentation shown here is accurate enough to determine bulk morphological parameters.
In order to demonstrate the utility of the system, we applied the imaging and analysis pipeline to analyze the effect of starvation on lipid droplets in C. elegans (strain shg366), which have the same phenotype as wild-type. Three starvation statuses were studied: starved for 12 h, restricted diet and normal diet. The representative imaging results (with individual planes selected by the operator as those where the lipid droplets area appears largest) are shown in Figure 4A. One can easily see that the densities of lipid droplets in the starvation states are relatively smaller compared to the normal diet case. This implies that in the hungry state, the nematodes will consume their stored energy, resulting in a decrease in the density of lipid droplets. This is consistent with previous reports [29]. Based on the DL segmentation results, we can analyze the morphology and the result is shown in Figure 4B. One can see that the quantitative analysis is consistent with the visual inspection. The advantages of automatic analysis, however, is that one can easily analyze a large amount of data, obtain a time course of the changes due to the starvation or other types of stimulus, and more quantitatively compare morphological changes without tedious manual annotation.
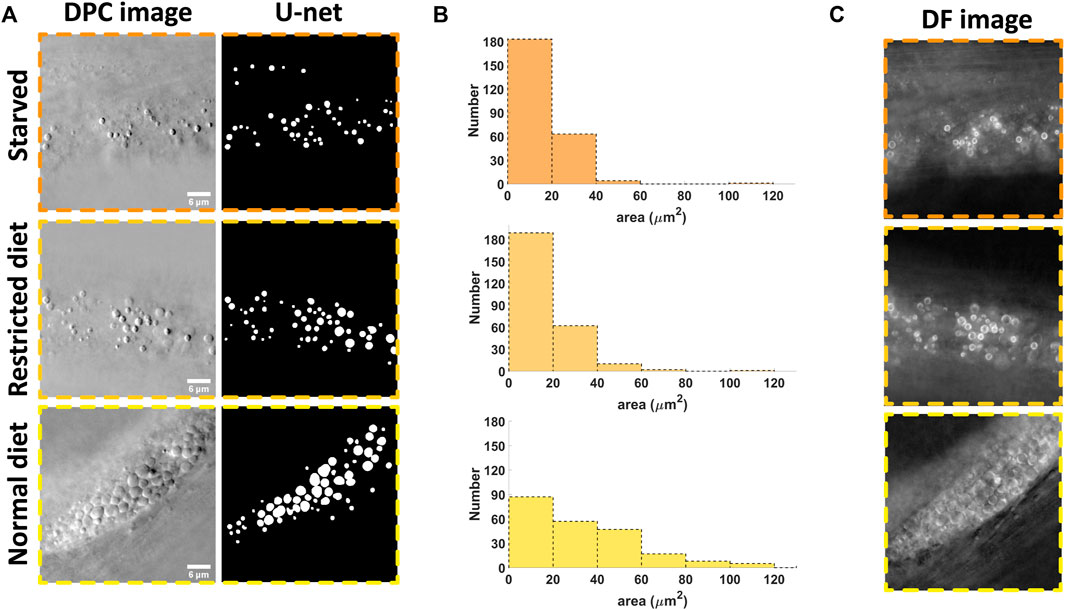
FIGURE 4. Results of the starvation experimentation where the number and size of the lipid droplets are analyzed. (A) example of the raw data and the segmented results for three different state: starved, restricted diet and normal diet; (B) The morphology analysis based on 5 images for each case; (C) In starved state, the lipid droplets become bright ring-like in the dark field image, signifying the strong variations in refractive index compared to those with normal state. Scale bars in (A) represent 6 μm.
Note that with our illumination scheme, the dark field and DPC images can be generated from the same set of data (with Eqs 1, 2). Interestingly, the dark field image could give unique information. As shown in Figure 4C, we can see that the edges of the lipid droplets in the starving state become either very bright or dark while, they are less so in the normal state. This may be caused by changes in the chemical composition inside the droplets, resulting in changes in the refractive index [11]. This also implies that the abnormal lipid droplets are more obvious in the dark field images while the edges of normal lipid droplets are more obvious in DPC images. These two kind of images could be combined in the future to analyze the effect of certain stimuli in more detail and address the heterogeneity of responses.
In this work, we have developed a novel pipeline for in vivo lipid droplet analysis where the visualization and automatic image segmentation are two necessary steps. Through combining epi-illumination dark-field and asymmetrical illumination, we have improved the optical sectioning and thus improved the image contrast of the lipid droplets. Even in the densely packed area, the boundary of most lipid droplets are clear as shown in Figures 2–4. For the automatic image segmentation, we have taken advantage of the well-validated U-net CNN architecture and applied it to our problem. Due to the high quality of the data, a miniscule training data set of only 25 images was used to obtain a segmentation accuracy around 85%, while the traditional methods such as DICOT or watershed failed. While with more data the accuracy can be further improved, the results shown here clearly demonstrate the proof-of-concept while at the same time demonstrating that the asymmetric illumination provides a distinct advantage to segmentation compared with the raw dark-field images. In both cases, we can also improve the image contrast further through deconvolution. Since our illumination configuration is rather different from other DPC work [30,31], a new theory about the image formation and optical transfer function would need to be developed in order to create a suitable theoretical inverse filter. Alternatively, as we have previously shown that our image system is nearly linear (compared to the typical bilinearity of phase imaging systems), we could use a measured point spread function to perform the deconvolution. Right now our morphology analysis of lipid droplets is based on representative 2D images. Incorporating 3D data and multiple FOVs within a single worm may improve CNN-based segmentation and allow more complete and statistically powerful lipid droplet morphological parameters. These would be directions of future work.
For lipid droplets, other than the morphology, the chemical content are also important. We can combine our method and Raman spectroscopy (for example, Coherent anti-Stokes Raman spectroscopy (CARS)) to obtain the chemical makeup of individual lipid droplets with improved speed and cleaner background. As we have shown in the starvation experiment, the morphology of lipid droplets exhibits unique features when in different starvation states. In a hypothetical future experiment one could envision using the dark field DPC image to guide a Raman excitation beam and obtain the specific chemical makeup of various lipid droplets of interest while saving the acquisition time of the Raman measurements compared to creating a full Raman image.
Further, as we have discussed in the introduction, our differential phase image is created through asymmetrical illumination, not through polarization means (as in DIC). Meanwhile, it is also based on epi-illumination, preserving an “open top” to the experimental platform. This means our method can be naturally combined with plastic microfluidic devices, which enable large scale screening but require a certain space to operate. More importantly, our label-free method enables the study of any strain of worm while the traditional fluorescence-based methods resort to specific strains which could influence the interpretation of the data. Our method also “saves” the fluorescent channel for other proteins or pathways that might be of interest.
In summary, we have developed a label-free method to analyze the lipid droplets in vivo and can naturally integrate with other technologies to conduct more comprehensive and in-depth study of C. elegans and other small model organisms.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
RS, ZS, and KC conceived the project. RS performed experiments including system construction, data acquisition and data preprocessing. YS assisted in system construction, data collection and deep learning segmentation. JF contributed the DL code for the input and test parts of the dataset. XC provided the C. elegans samples and helped to design the starvation experiments. ZS and KC supervised the project.
National Key R&D Program of China (2017YFA0505300); National Natural Science Foundation of China (NSFC) (31670803); University of Science and Technology of China Innovation Team (2017–2019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2022.894797/full#supplementary-material
1. Lemieux GA, Ashrafi K. Insights and Challenges in usingC. Elegansfor Investigation of Fat Metabolism. Crit Rev Biochem Mol Biol (2015) 50:69–84. doi:10.3109/10409238.2014.959890
2. Gao B, Sun Q. Programming Gene Expression in Multicellular Organisms for Physiology Modulation through Engineered Bacteria. Nat Commun (2021) 12:1–8. doi:10.1038/s41467-021-22894-7
3. Zhang Y, Zou X, Ding Y, Wang H, Wu X, Liang B. Comparative Genomics and Functional Study of Lipid Metabolic Genes in caenorhabditis Elegans. BMC genomics (2013) 14:164–13. doi:10.1186/1471-2164-14-164
4. Zhang SO, Trimble R, Guo F, Mak HY. Lipid Droplets as Ubiquitous Fat Storage Organelles in c. elegans. BMC Cel Biol (2010) 11:96–11. doi:10.1186/1471-2121-11-96
5. Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, et al. Genetic and Dietary Regulation of Lipid Droplet Expansion in caenorhabditis Elegans. Proc Natl Acad Sci U.S.A (2010) 107:4640–5. doi:10.1073/pnas.0912308107
6. Li S, Xu S, Ma Y, Wu S, Feng Y, Cui Q, et al. A Genetic Screen for Mutants with Supersized Lipid Droplets in caenorhabditis Elegans. G3: Genes, Genomes, Genet (2016) 6:2407–19. doi:10.1534/g3.116.030866
7. Mak HY. Visualization of Lipid Droplets in c. elegans by Light and Electron Microscopy. Methods Cel Biol (2013) 116:39–51. doi:10.1016/b978-0-12-408051-5.00003-6
8. Witting M, Schmitt-Kopplin P. The Caenorhabditis elegans Lipidome. Arch Biochem Biophys (2016) 589:27–37. doi:10.1016/j.abb.2015.06.003
9. Tipping WJ, Lee M, Serrels A, Brunton VG, Hulme AN. Imaging Drug Uptake by Bioorthogonal Stimulated Raman Scattering Microscopy. Chem Sci (2017) 8:5606–15. doi:10.1039/c7sc01837a
10. Cherkas A, Mondol AS, Rüger J, Urban N, Popp J, Klotz L-O, et al. Label-free Molecular Mapping and Assessment of Glycogen in c. elegans. Analyst (2019) 144:2367–74. doi:10.1039/c8an02351d
11. Chen W-W, Lemieux GA, Camp CH, Chang T-C, Ashrafi K, Cicerone MT. Spectroscopic Coherent Raman Imaging of caenorhabditis Elegans Reveals Lipid Particle Diversity. Nat Chem Biol (2020) 16:1087–95. doi:10.1038/s41589-020-0565-2
12. Chowdhury S, Chen M, Eckert R, Ren D, Wu F, Repina N, et al. High-resolution 3d Refractive index Microscopy of Multiple-Scattering Samples from Intensity Images. Optica (2019) 6:1211–9. doi:10.1364/optica.6.001211
13. Song W, Matlock A, Fu S, Qin X, Feng H, Gabel CV, et al. Led Array Reflectance Microscopy for Scattering-Based Multi-Contrast Imaging. Opt Lett (2020) 45:1647–50. doi:10.1364/ol.387434
14. Chen M, Ren D, Liu H-Y, Chowdhury S, Waller L. Multi-layer Born Multiple-Scattering Model for 3d Phase Microscopy. Optica (2020) 7:394–403. doi:10.1364/optica.383030
15. Nguyen TH, Kandel ME, Rubessa M, Wheeler MB, Popescu G. Gradient Light Interference Microscopy for 3d Imaging of Unlabeled Specimens. Nat Commun (2017) 8:210–9. doi:10.1038/s41467-017-00190-7
16. Li J, Matlock AC, Li Y, Chen Q, Zuo C, Tian L. High-speed In Vitro Intensity Diffraction Tomography. Adv Photon (2019) 1:066004. doi:10.1117/1.ap.1.6.066004
17. Shi R, Chen X, Huo J, Guo S, Smith ZJ, Chu K. Epi-illumination Dark-Field Microscopy Enables Direct Visualization of Unlabeled Small Organisms with High Spatial and Temporal Resolution. J Biophotonics (2022) 15:e202100185. doi:10.1002/jbio.202100185
18. Cáceres Mendieta Id. C. On-chip Phenotypic Screening and Characterization of C. elegans Enabled by Microfluidics and Image Analysis Methods. Ph.D. thesis. Atlanta, GA, USA: Georgia Institute of Technology (2013).
19. Casas ME. Analysis of Lipid Storage in C. elegans Enabled by Image Processing and Microfluidics. Ph.D. thesis. Atlanta, GA, USA: Georgia Institute of Technology (2017).
20. Mehta SB, Sheppard CJR. Quantitative Phase-Gradient Imaging at High Resolution with Asymmetric Illumination-Based Differential Phase Contrast. Opt Lett (2009) 34:1924–6. doi:10.1364/ol.34.001924
21. Ford TN, Chu KK, Mertz J. Phase-gradient Microscopy in Thick Tissue with Oblique Back-Illumination. Nat Methods (2012) 9:1195–7. doi:10.1038/nmeth.2219
22. Ledwig P, Robles FE. Quantitative 3d Refractive index Tomography of Opaque Samples in Epi-Mode. Optica (2021) 8:6–14. doi:10.1364/optica.410135
23. Tian L, Waller L. Quantitative Differential Phase Contrast Imaging in an Led Array Microscope. Opt Express (2015) 23:11394–403. doi:10.1364/oe.23.011394
24. Chaphalkar AR, Jawale YK, Khatri D, Athale CA. Quantifying Intracellular Particle Flows by Dic Object Tracking. Biophysical J (2021) 120:393–401. doi:10.1016/j.bpj.2020.12.013
25. Cheng S, Fu S, Kim YM, Song W, Li Y, Xue Y, et al. Single-cell Cytometry via Multiplexed Fluorescence Prediction by Label-free Reflectance Microscopy. Sci Adv (2021) 7:eabe0431. doi:10.1126/sciadv.abe0431
26. Guo S, Ma Y, Pan Y, Smith ZJ, Chu K. Organelle-specific Phase Contrast Microscopy Enables Gentle Monitoring and Analysis of Mitochondrial Network Dynamics. Biomed Opt Express (2021) 12:4363–79. doi:10.1364/boe.425848
27. Ronneberger O, Fischer P, Brox T. U-net: Convolutional Networks for Biomedical Image Segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Berlin, Germany: Springer (2015). p. 234–41. doi:10.1007/978-3-319-24574-4_28
28. Ounkomol C, Seshamani S, Maleckar MM, Collman F, Johnson GR. Label-free Prediction of Three-Dimensional Fluorescence Images from Transmitted-Light Microscopy. Nat Methods (2018) 15:917–20. doi:10.1038/s41592-018-0111-2
29. Fouad AD, Pu SH, Teng S, Mark JR, Fu M, Zhang K, et al. Quantitative Assessment of Fat Levels in caenorhabditis Elegans Using Dark Field Microscopy. G3: Genes, Genomes, Genet (2017) 7:1811–8. doi:10.1534/g3.117.040840
30. Chen M, Tian L, Waller L. 3d Differential Phase Contrast Microscopy. Biomed Opt Express (2016) 7:3940–50. doi:10.1364/boe.7.003940
Keywords: asymmetrical illumination, epi-illumination, dark field microscopy, lipid droplets, C. elegans, segmentation
Citation: Shi R, Sun Y, Fang J, Chen X, Smith ZJ and Chu K (2022) Asymmetrical Illumination Enables Lipid Droplets Segmentation in Caenorhabditis elegans Using Epi-Illumination Dark Field Microscopy. Front. Phys. 10:894797. doi: 10.3389/fphy.2022.894797
Received: 12 March 2022; Accepted: 30 May 2022;
Published: 16 June 2022.
Edited by:
Jun Zhang, Tianjin University, ChinaReviewed by:
Ryan McGorty, University of San Diego, United StatesCopyright © 2022 Shi, Sun, Fang, Chen, Smith and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zachary J. Smith, enNtaXRoQHVzdGMuZWR1LmNu; Kaiqin Chu, a3FjaHVAdXN0Yy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.