- 1WHO Collaborating Centre for Infectious Disease Epidemiology and Control, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 2Laboratory of Data Discovery for Health, Hong Kong Science and Technology Park, Hong Kong, Hong Kong SAR, China
- 3College of Information and Communication Engineering, Dalian Minzu University, Dalian, China
Given the worldwide pandemic of the novel coronavirus disease 2019 (COVID-19) and its continuing threat brought by the emergence of virus variants, there are great demands for accurate surveillance and monitoring of outbreaks. A valuable metric for assessing the current risk posed by an outbreak is the time-varying reproduction number (
Introduction
The novel coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to 257 million confirmed cases and 5.15 million deaths worldwide by November 22, 2021 [1]. The COVID-19 pandemic continues to pose substantial risks to public health, and the situation is worsened by the emergence of SARS-CoV-2 variants with potentially higher transmissibility [2].
Quantification of the transmissibility during epidemics is fundamental for designing and adjusting public health responses. The time-varying reproduction number
A large number of methods have been proposed to estimate
Recently, a new method was proposed by Hay et al. [15] using information inherent in cycle threshold (Ct) values from reverse transcription quantitative polymerase chain reaction (RT-qPCR) tests to estimate the time-varying reproduction number from positive samples. Ct values are semiquantitative results provided by RT-qPCR tests. It is common when testing for infectious diseases to use this quantification of sample viral load. Lower Ct values indicate higher viral loads, and a Ct value below 40 gives a positive result. Based on cross-sectional virologic surveys (observed viral loads), this method overcomes the biases in traditional approaches resulting from testing constraint, unrepresentative sampling, and reporting delays. They also developed the R package virosolver to infer epidemic dynamics including estimation of
In this study, we chose EpiEstim and virosolver as the representatives of traditional and new methods, respectively. Although the accuracies of the two approaches have been separately demonstrated, there is still a lack of comparison between the two methods to the best of our knowledge. Therefore, we quantify the accuracies of EpiEstim and virosolver in different transmission scenarios by individual-based simulations and develop a ready-to-use R package for researchers to compare different
Methods
SEIR-based simulation
To assess the performance of the methods, we simulate outbreaks in three scenarios with different basic reproduction numbers (
EpiEstim and virosolver methods were run separately on the same simulations for comparison. For EpiEstim, it relies on two inputs: incidence time series and the serial interval distribution. Incidence data by days since the start of outbreak were generated from the simulated SEIR epidemic. We used an empirical serial interval distribution informed by a previous outbreak of COVID-19 in Hong Kong in early 2020 [18], and we also used the simulated serial interval distribution for comparison, denoted by EpiEstim (empirical SI) and EpiEstim (simulated SI) in Figure 2. We assumed that the simulated serial interval distribution has the same standard variation as that of the empirical serial interval distribution and inferred the mean of the simulated serial interval distribution by conducting numerical experiments on a range of means from 1 to 10 with a step of 0.1 and chose the one yielding the least root mean square error (RMSE). For virosolver, the input data include population-level Ct values over days since the start of outbreak, and individual-level viral kinetics model over days since the infection. The Ct values were generated for all exposed, infectious, and recovered individuals when they were samples based on the Ct value model proposed by Hay et al. [15], and the viral kinetics parameters were also given in their study. We assumed that the Ct values were observed from randomized samples of the population at selected testing days, and Figure 2 shows the simulated Ct values of the sampled people every 14 days. Each panel presents the distribution of observed Ct values among sampled infected individuals on that testing day. Day 14 and Day 28 had no data because there was no infection among the samples at the early stage of the epidemic.
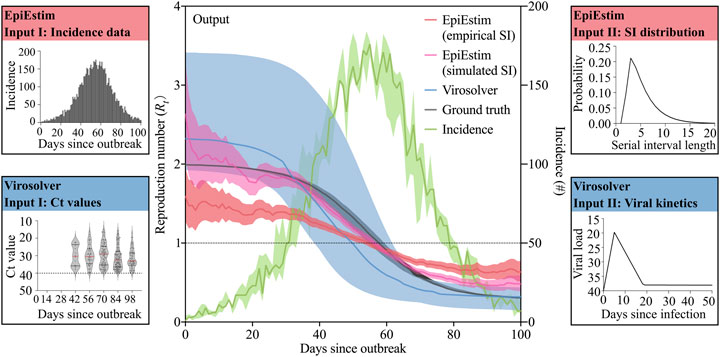
FIGURE 2. A schematic illustrating how our simulation platform generates a comparison of the estimated
EpiEstim
The framework of EpiEstim is based on statistical assumptions and Bayesian estimation. Transmission is modeled by a Poisson process so that the rate, at which individuals infected between infection and symptom onset generate new infections, is equal to
where
Using a Bayesian framework with a Gamma distributed prior with parameters of shape a and scale b for
Virosovler
The R package virosolver was developed by Hay et al. [15] using virological data and Ct values, to infer epidemic dynamics. Ct values are inversely correlated with
where
Results
We assessed the performance of EpiEstim and virosovler in three scenarios where
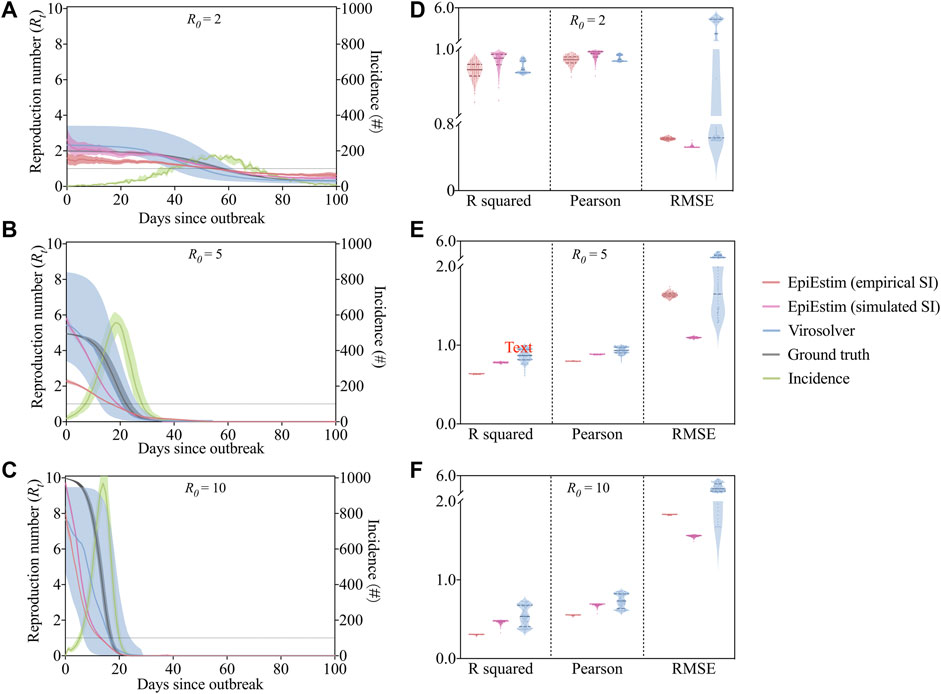
FIGURE 3. The output of
To quantify and compare the accuracies of the methods, we used multiple metrics including coefficient of determination (
Discussion
Quantifying disease transmissibility during outbreaks is crucial for designing effective control measures and assessing their effectiveness once implemented. In the situation where the incidence is still increasing while the time-varying reproduction number is actually dropping, there might be a very different outlook compared to if the incidence and the reproduction number are both increasing. The platform for estimating
The estimation tools we used here have several limitations and thus may result in potential bias. For EpiEstim, a preexisting estimate of serial interval distribution is required as the input data, which may account for the underestimation of reproduction number in our simulation study (Figure 3). If data on pairs of infector-infected individuals are available, the serial interval distribution can be estimated jointly, which leads to more precise estimates of transmissibility [21]. In addition, the inevitable delay between infection and case reporting (the incubation period) could also result in biased estimation of
The virologic data-based method, virosolver, as mentioned above, exhibits greater uncertainty of estimated than EpiEstim. This is probably caused by insufficient information on Ct value distribution and viral kinetics model. The viral load kinetics model used in virosolver was generated on the basis of observed properties of measured viral loads in the literature, and these results were applied to inform priors on key parameters when estimating reproduction numbers. The estimates can therefore be improved by choosing more precise, accurate priors relevant to the observations used during model fitting. For example, the model should be adjusted by specifying different distributions if results come from multiple testing platforms. Results may also be improved if individual-level features such as symptom status, age, antiviral treatment, and vaccination record are available and incorporated into the Ct value model.
Apart from the two methods presented in our study, many other approaches are still available, which we will include in this platform in the future to track disease transmissibility by using other data sources (e.g., hospitalization and death). Additionally, genomic data are also of great importance in the inference of transmissibility of COVID-19 considering recent emergence of virus variants [22]. We only provide incidence and
Our tool can also be applied to the new variants of SARS-CoV-2 as long as incidence data and Ct values of the infected people are available. Users can obtain a more accurate estimation by adopting the updated parameters of serial interval distribution and viral kinetics model for objective variants informed by recent studies [23–26]. In conclusion, we have established a platform for simulation and inference of time-varying reproduction numbers by incorporating two commonly used approaches. We would ensure our tool to epidemiologists and public health organizations in a wide range of future outbreak response scenarios.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
CL, LX, ZD, and BC: conceived the study, designed statistical and modeling methods, conducted analyses, interpreted results, and wrote and revised the manuscript; YB, XX, and EL: interpreted the results and revised the manuscript.
Conflict of Interest
BC reports honoraria from AstraZeneca, GSK, Moderna, Pfizer, Roche, and Sanofi Pasteur.
CL, LX, YB, EL, BC, and ZD were employed by the company Laboratory of Data Discovery for Health, Hong Kong Science and Technology Park.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the financial support from the Collaborative Research Fund (Project No. C7123-20G) of the Research Grants Council of the Hong Kong SAR Government.
References
1.Home - Johns Hopkins Coronavirus Resource center. Available from: https://coronavirus.jhu.edu/. Accessed 22 November 2021.
2. Sars- CDC. CoV-2 Variant Classifications and Definitions (2021). Available from: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-updates%2Fvariant-surveillance%2Fvariant-info.html (Accessed November 3, 2021).
3. Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford: OUP Oxford (1992).
4. Ferguson NM, Cummings DAT, Fraser C, Cajka JC, Cooley PC, Burke DS. Strategies for Mitigating an Influenza Pandemic. Nature (2006) 442:448–52. doi:10.1038/nature04795
5. Riley S, Fraser C, Donnelly CA, Ghani AC, Abu-Raddad LJ, Hedley AJ, et al. Transmission Dynamics of the Etiological Agent of SARS in Hong Kong: Impact of Public Health Interventions. Science (2003) 300:1961–6. doi:10.1126/science.1086478
6. Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, et al. Pandemic Potential of a Strain of Influenza A (H1N1): Early Findings. Science (2009) 324:1557–61. doi:10.1126/science.1176062
7. Ferguson NM, Donnelly CA, Anderson RM. Transmission Intensity and Impact of Control Policies on the Foot and Mouth Epidemic in Great Britain. Nature (2001) 413:542–8. doi:10.1038/35097116
8. Amundsen EJ, Stigum H, Røttingen J-A, Aalen OO. Definition and Estimation of an Actual Reproduction Number Describing Past Infectious Disease Transmission: Application to HIV Epidemics Among Homosexual Men in Denmark, Norway and Sweden. Epidemiol Infect (2004) 132:1139–49. doi:10.1017/s0950268804002997
9. Bettencourt LMA, Ribeiro RM. Real Time Bayesian Estimation of the Epidemic Potential of Emerging Infectious Diseases. PLoS One (2008) 3:e2185. doi:10.1371/journal.pone.0002185
10. Cintrón-Arias A, Castillo-Chávez C, Bettencourt LM, Lloyd AL, Banks HT. The Estimation of the Effective Reproductive Number from Disease Outbreak Data. Math Biosci Eng (2009) 6:261–82. doi:10.3934/mbe.2009.6.261
11. Howard SC, Donnelly CA. Estimation of a Time-Varying Force of Infection and Basic Reproduction Number with Application to an Outbreak of Classical Swine Fever. J Epidemiol Biostat (2000) 5:161–8.
12. Kelly HA, Mercer GN, Fielding JE, Dowse GK, Glass K, Carcione D, et al. Pandemic (H1N1) 2009 Influenza Community Transmission Was Established in One Australian State when the Virus Was First Identified in North America. PLoS One (2010) 5:e11341. doi:10.1371/journal.pone.0011341
13. Wallinga J, Teunis P. Different Epidemic Curves for Severe Acute Respiratory Syndrome Reveal Similar Impacts of Control Measures. Am J Epidemiol (2004) 160:509–16. doi:10.1093/aje/kwh255
14. Cori A, Ferguson NM, Fraser C, Cauchemez S. A New Framework and Software to Estimate Time-Varying Reproduction Numbers during Epidemics. Am J Epidemiol (2013) 178:1505–12. doi:10.1093/aje/kwt133
15. Hay JA, Kennedy-Shaffer L, Kanjilal S, Lennon NJ, Gabriel SB, Lipsitch M, et al. Estimating Epidemiologic Dynamics from Cross-Sectional Viral Load Distributions. Science (2021) 373:373. doi:10.1126/science.abh0635
16. Alimohamadi Y, Taghdir M, Sepandi M. Estimate of the Basic Reproduction Number for COVID-19: A Systematic Review and Meta-Analysis. J Prev Med Public Health (2020) 53:151–7. doi:10.3961/jpmph.20.076
17. Liu Y, Rocklöv J. The Reproductive Number of the Delta Variant of SARS-CoV-2 Is Far Higher Compared to the Ancestral SARS-CoV-2 Virus. J Trav Med (2021) 28. doi:10.1093/jtm/taab124
18. Zhao S, Gao D, Zhuang Z, Chong MKC, Cai Y, Ran J, et al. Estimating the Serial Interval of the Novel Coronavirus Disease (COVID-19): A Statistical Analysis Using the Public Data in Hong Kong from January 16 to February 15, 2020. Front Phys (2020) 8:347. doi:10.3389/fphy.2020.00347
19. Gostic KM, McGough L, Baskerville EB, Abbott S, Joshi K, Tedijanto C, et al. Practical Considerations for Measuring the Effective Reproductive Number, Rt. Plos Comput Biol (2020) 16:e1008409. doi:10.1371/journal.pcbi.1008409
20. Real-time Dashboard . Available from: https://covid19.sph.hku.hk/. Accessed 10 November 2021.
21. Thompson RN, Stockwin JE, van Gaalen RD, Polonsky JA, Kamvar ZN, Demarsh PA, et al. Improved Inference of Time-Varying Reproduction Numbers during Infectious Disease Outbreaks. Epidemics (2019) 29:100356. doi:10.1016/j.epidem.2019.100356
22. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early Transmissibility Assessment of the N501Y Mutant Strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance (2021) 26:2002106. doi:10.2807/1560-7917.es.2020.26.1.2002106
23. Ryu S, Kim D, Lim J-S, Ali ST, Cowling BJ. Serial Interval and Transmission Dynamics during SARS-CoV-2 Delta Variant Predominance, South Korea. Emerg Infect Dis (2022) 28:407–10. doi:10.3201/eid2802.211774
24. Pung R, Mak TM, Kucharski AJ, Lee VJCMMID COVID-19 working group. Serial Intervals in SARS-CoV-2 B.1.617.2 Variant Cases. The Lancet (2021) 398:837–8. doi:10.1016/s0140-6736(21)01697-4
25. Singanayagam A, Hakki S, Dunning J, Community Transmission and Viral Load Kinetics of the SARS-CoV-2 delta (B. 1.617. 2) Variant in Vaccinated and Unvaccinated Individuals in the UK: a Prospective, Longitudinal, Cohort Study. Lancet Infect Dis (2021) 22(2):183–195. doi:10.1016/s1473-3099(21)00648-4
Keywords: epidemics (covid 19), surveillance, infectious disease, package, modeling
Citation: Liu C, Xu L, Bai Y, Xu X, Lau EHY, Cowling BJ and Du Z (2022) Local Surveillance of the COVID-19 Outbreak. Front. Phys. 10:824369. doi: 10.3389/fphy.2022.824369
Received: 29 November 2021; Accepted: 13 January 2022;
Published: 03 March 2022.
Edited by:
Huijia Li, Central University of Finance and Economics, ChinaReviewed by:
Yongxing Li, Beijing University of Technology, ChinaXueyan Liu, Jilin University, China
Copyright © 2022 Liu, Xu, Bai, Xu, Lau, Cowling and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanwei Du, ZHV6aGFud2VpMEBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
 Caifen Liu
Caifen Liu Lingfeng Xu1,2,3†
Lingfeng Xu1,2,3† Eric H. Y. Lau
Eric H. Y. Lau Zhanwei Du
Zhanwei Du