- 1College of Computer and Information Science, Chongqing Normal University, Chongqing, China
- 2Department of Dermatology, First Affiliated Hospital of Army Medical University, Chongqing, China
- 3School of Computer and Software Engineering, Xihua University, Chengdu, Sichuan, China
- 4Key Laboratory of Advanced Manufacturing Technology, Ministry of Education Guizhou University, Guiyang, Guizhou, China
- 5School of Science, Qingdao University of Technology, Qingdao, Shandong, China
Melanoma is a high-grade malignant tumor. Melanoma and mole lesions are highly similar and have a very high mortality rate. Early diagnosis and treatment have an important impact on the patient’s condition. The results of dermoscopy are usually judged visually by doctors through long-term clinical experience, and the diagnostic results may be different under different visual conditions. Computer-aided examinations can help doctors improve efficiency and diagnostic accuracy. The purpose of this paper is to use an improved quantum Inception-ResNet-V1 model to classify multiple types of skin lesion images and improve the accuracy of melanoma identification. In this study, the FC layer of Inception-ResNet-V1 is removed, the average pooling layer is the last, SVM is used as the classifier, and the convolutional layer is quantized. The performance of the model was tested experimentally on the ISIC 2019 dataset. To prevent the imbalance of the sample data set from affecting the experiment, the sample data is sampled with weight. Experiments show that the method used shows excellent performance, and the classification accuracy rate reaches 98%, which provides effective help for the clinical diagnosis of melanoma.
Introduction
Melanoma is one of the most harmful skin cancers, and it is a deadly malignant tumor [1–3]. There are many risk factors leading to the formation of melanoma, such as ultraviolet radiation, drug treatment, gene, family history, skin color, race, age, gender, etc. Although melanoma is not common, it is more lethal, and the incidence rate is still rising in the world. And the average diagnosis and treatment cost of melanoma is 10 times that of non-melanoma skin cancer [4–6]. Melanoma is cancer with the highest mortality among skin cancers. If melanoma is diagnosed at an early stage, a small operation can increase the chance of recovery and reduce the mortality rate of cancer. But without early detection and treatment, it can spread to other parts of the body [7]. Early and correct diagnosis is the key to ensuring the best prognosis for patients [8]. However, melanoma is misdiagnosed more than any cancer except breast cancer. Dermatoscopy is one of the most commonly used imaging techniques for dermatologists. It enlarges the surface of skin lesions, and its structure becomes more obvious to dermatologists [9,10]. The diagnosis of melanoma is usually carried out by using the vision of experienced doctors, first visually inspecting the skin lesions (usually using the ABCD rule and the seven-point inspection method) [11,12], analyzing the results of dermatoscopy and matching them with medical science [13]. The weakness of manual detection is greatly affected by human subjectivity, which makes it inconsistent under certain conditions because it is completely based on doctors’ vision and experience. Although the accuracy of suspicious cases can be further improved by using special high-resolution cameras and magnifying glasses to capture dermoscopic images for visual examination [14]. However, recent studies have shown that the classification method based on CNN has become the best choice for melanoma detection. The high accuracy of CNN based classifier for skin cancer image classification is equivalent to an experienced dermatologist [15].
[16] proposed a deep learning system for detecting melanoma lesions. They first performed illumination correction on the input image, and cut, scaled, and rotate the image. Then, they fed back the enhanced image to the pre-trained CNN for a large number of sample training and obtained an accuracy rate of 81%. [17] also preprocessed the skin image data set, segmented the region of interest (ROI) of the lesion area, extracted the features of the segmented image using the gray level co-occurrence matrix, and combined with the ABCD rule to identify and classify malignant tumors, achieving an accuracy rate of 92.1%. [18] used GoogLeNet to train the ISIC 2016 dataset and processed the samples of the ISIC 2016 dataset through the traditional data enhancement method to reduce the impact of the unbalanced training dataset on the CNN performance, with the maximum accuracy of 83.6%. [19] used the ResNet-152 model to classify clinical images of 12 skin diseases, fine-tuned the model using the training part of the Asan dataset, the MED-NODE dataset, and atlas site images, and the trained model passed Asan, Hallym and Edinburgh datasets for validation. Experiments demonstrate that the algorithm performance, tested with 480 Asan and Edinburgh images, is comparable to the results of 16 dermatologists. [20] used CNN to extract features from images, used SVM, RF, and NN to train and classify features, and processed the datasets ISIC 2017 and PH2 using data augmentation to avoid overfitting in accuracy. Due to the influence of the integration problem, the experiment obtained an accuracy of 89.2%. [21] used the improved Inception V4 model to classify skin cancer diseases, pre-trained the model on the ImageNet dataset, fused the low-level and high-level features of the image, and used the ISIC 2018 dataset to achieve 94.7% classification accuracy. [22] proposed an enhanced encoder-decoder network to overcome the limitations of uneven skin image features and blurred boundaries, which made the semantic level of the encoder feature map closer to the decoder feature map, and The model was tested on the ISIC 2017 and PH2 datasets for melanoma recognition and achieved 95% accuracy. [23] pre-trained Alex Net with the Softmax layer as the classification layer, and the model achieved 97% accuracy for skin cancer classification on three datasets: MED-NODE, Derm (IS & Quest), and ISIC. [24] analyzed ISIC images using a fully convolutional residual network (FCRN) and CNN to check for anomalies in the skin, a residual network was used to segment the images in the dataset, and a neural network was used for classification. [25] firstly preprocess the input color skin image to segment the region of interest (ROI); secondly, use traditional transformation to enhance the segmented ROI image. They evaluated the performance of the proposed method on three different datasets (MED-NODE, DermIS & DermQuest, and ISIC 2017) using the improved AlexNet, ResNet101, and GoogLeNet network structures, and achieved 99% accuracy on the MED-NODE dataset. [26] used a hybrid quantum mechanical system to encode and process image information to classify cancerous and noncancerous pigmented skin lesions in the HAM10000 dataset. [27] proposed a high-precision skin lesion classification model, using transfer learning and GoogLeNet pre-training model, to classify eight different categories of skin lesions in the dataset ISIC 2019, with an accuracy rate of 94%.
In this study, the trained model is applied to images of skin lesions, the classification layer is replaced with SVM, and the convolutional layer is quantized to improve the classification process. One of the difficulties in image classification is that the amount of computation in the classification process is very large, resulting in a relatively slow classification speed and consuming a lot of computing resources. [28, 29] Due to the characteristics of quantum parallel computing, the quantum image classification algorithm can still quickly complete the classification task in the case of a large amount of image data. [28] The contributions of the proposed method are as follows:
• This study quantizes the convolutional layers of Inception-ResNet-V1 to enhance the performance of the network.
• In this study, the FC layer of the network was removed, and SVM was used as the classifier because SVM also showed excellent performance in melanoma classification, which can be compared with the original model.
• This study adopts data augmentation and weighted sampling methods to alleviate the impact of data imbalance.
• The model exhibits a high accuracy classification rate.
Proposed method
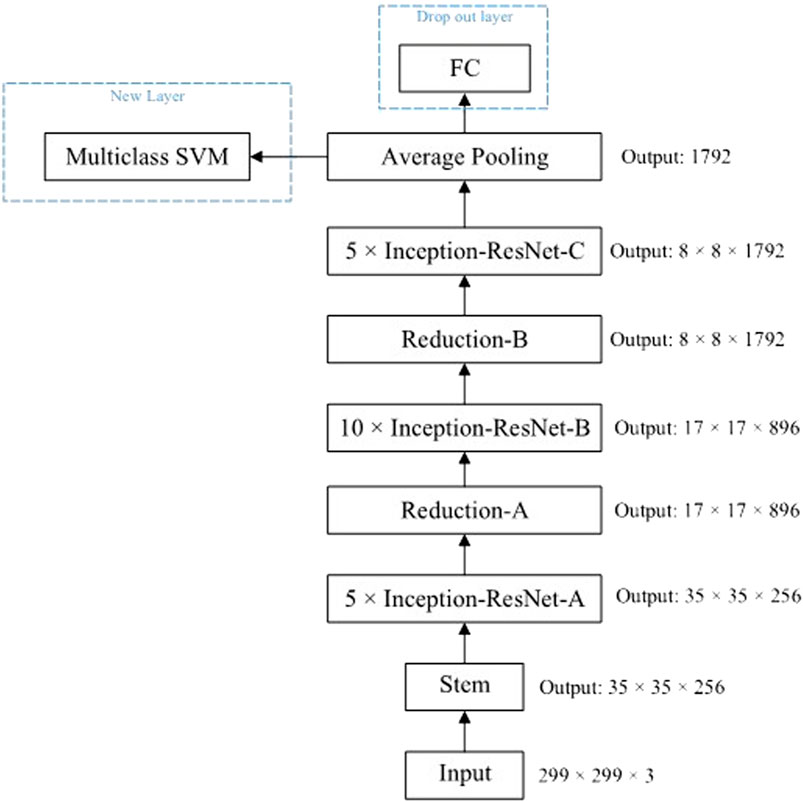
The proposed method is elaborated in this section, and the method structure is shown in Figure 1. The proposed method first augments and normalizes the image data, and then feeds the proposed model for training. The following is a detailed description of the proposed method.
Data augmentation and weighted sampling
ISIC 2019 dataset is an imbalanced dataset, which may make the model biased toward classes with a large number of samples during training. For example, the number of the most NV class is 50 times more than that of the least VASC class, which is likely to lead to the model being biased toward NV during training, thus affecting the accuracy of the model. To reduce the imbalance in the given dataset, this study uses the data augmentation method to expand the images of minority classes through rotation, cutting, flipping, and other ways [30,31], to reduce the image quantity gap between the majority class and the model, to reduce the influence of unbalanced data sets on the model. However, using traditional data augmentation alone has defects, because repeated samples lead to over-sampling, which will easily lead to overfitting of the learning algorithm. To solve this problem, this study applies the weighted random sampling method to the overlapping of repeated samples, that is, the weight of each instance is defined by the number of instances in the class [32], and this weight represents the probability of the instance being randomly sampled [32], which can offset the oversampling effect of the class with a small number of samples. The weighted sampling method is based on weight sampling, which can reserve more labels and meet the diversity. Although there are changes, they are still constrained by the relative weight size of labels [33].
Image normalization
For image data, the pixel value of the image is an integer between 0 and 255. When training a deep neural network for fitting, a small weight value is generally used. If the value of the training data is large, the model training process may be slowed down. Therefore, pixel normalization of image data is necessary. In this paper, Min-Max normalization is used to remove the pixel unit of image data and convert the data into dimensionless pure values. Specifically, after pixel normalization, the image pixel value is scaled to [0,1] [34].
Improved quantum Inception-ResNet-V1 model
The research uses residual connections to join the filter connection stage in the Inception architecture, which will allow the Inception architecture to retain its computational efficiency while gaining the benefits of the residual connection method. The residual version of the Inception network uses a more simplified Inception module than the source Inception uses. Each Inception module is followed by a filter expansion layer (i.e., a 1 × 1 convolutional layer without an activation function) that enlarges the dimension of the filter bank before adding it to match the input. This compensates for the dimensionality reduction in the Inception block. The research removes the FC of the Inception-ResNet-V1 model and uses the SVM as a classifier to test the performance. The architecture is shown in Figure 2.
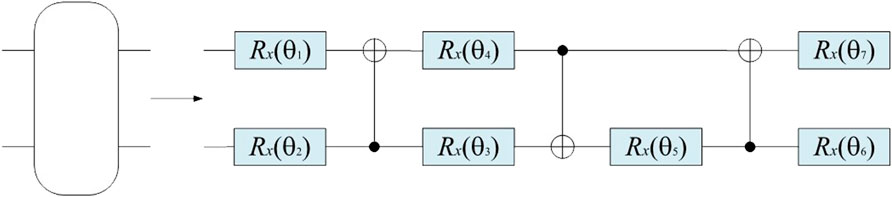
The backbone network Stem in Figure 2 uses a quantum convolution layer for feature extraction, which is composed of multiple parameterized quantum filters. Similar to the convolution kernel in the classical convolution layer, it uses a parameterized quantum filter to extract the characteristic information of all quantum bits in the local space of the data. The quantum filter includes many types of quantum bit gates, including single-bit gate and double-bit gate, which can perform unitary conversion of corresponding quantum bit, and impose a double-bit gate on adjacent quantum bits, thus causing quantum entanglement of adjacent quantum bits. In this paper, the quantum rotation gate R(θ) is used to transform the pixel value information of the image into quantum state information by quantum state encoding. On this basis, the obtained image feature information is converted into the angle of the quantum rotary gate. Each pixel value provides the corresponding parameters for the quantum rotary gate. Different quantum rotary gates act on the corresponding initial state |0⟩ of the quantum bit, and the feature information is stored in the quantum state, which can be used as the model input to the quantum convolution neural network [35]. For example, for n ×n, the quantum feature extraction function first encodes it into a quantum state through quantum bit coding, then evolves the quantum state through the parameterized quantum circuit, and finally outputs a real number through the expected value measurement. This method not only has the unique properties of quantum mechanics but also can keep the weight sharing of the convolutional kernel. In this study, we introduce a quantum circuit with parameters to enhance the performance of the network. The quantum convolution layer is shown in Figure 3.
The quantum filter used in this study consists of CNOT gate and rotary gate Rx(θ). The quantum circuit diagram is shown in Figure 4.
Experiments and results
Dataset description
The data in this article comes from the ISIC 2019 [36] challenge (Skin Lesion Analysis Towards Melanoma Detection). The ISIC 2019 dataset contains 25,331 dermoscopy images in 8 categories, namely actinic keratosis (AKIEC): 867, basal cell carcinoma (BCC): 3,323, benign keratosis (BKL): 2,624, skin fibers Tumor (DF): 239, Melanoma (MEL): 4,522, Melanocytic nevus (NV): 12,875, Angiosarcoma (VASC): 253, Squamous cell carcinoma (SCC): 628, as shown in Table 1. In the experiments, we randomly 80% of images (about 20,231 images) of the dataset for training, 10% images (about 2,550 images) for testing, and 10% images (about 2,550 images) for validation.
Experiment
The study conducted three experiments using the ISIC 2019 dataset. The first is to evaluate the proposed method using the original dataset without image augmentation. The second approach is to augment the dataset and re-evaluate the proposed method. The third is to use the processed dataset to evaluate the proposed method after processing the dataset using image augmentation and weighted sampling. All experiments are performed with fixed values, i.e. batch size 10, number of training 32, and initial learning rate 0.001. The proposed model was evaluated using four performance metrics [37]: accuracy, precision, sensitivity, and specificity.
where tp, fp, tn, and fn refer to true positives, false positives, true negatives, and false negatives, respectively.
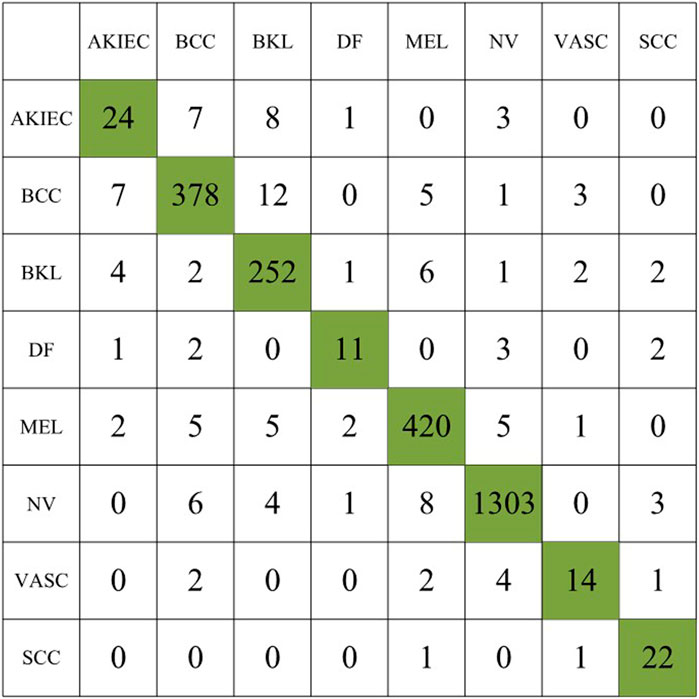
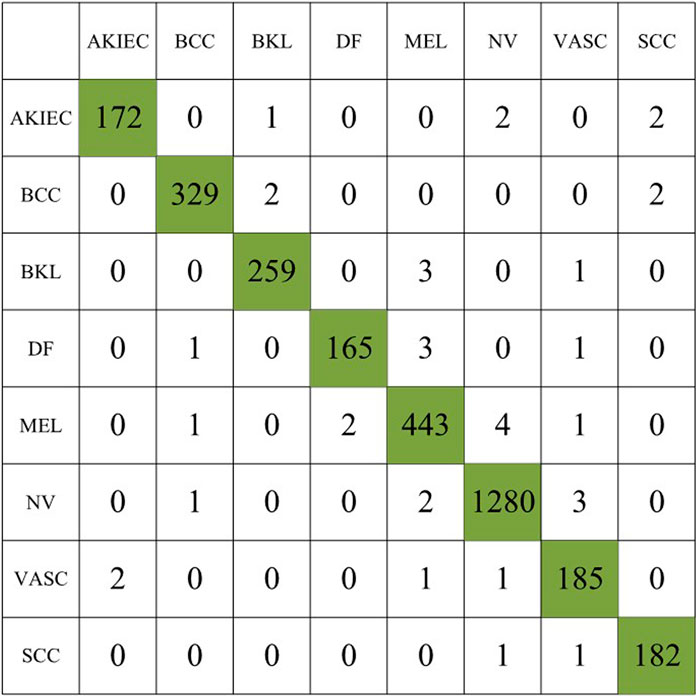
In the first experiment, we use the original dataset to evaluate the proposed method, the experimental results are summarized in Table 2, and the confusion matrix of the first experiment is shown in Figure 5.
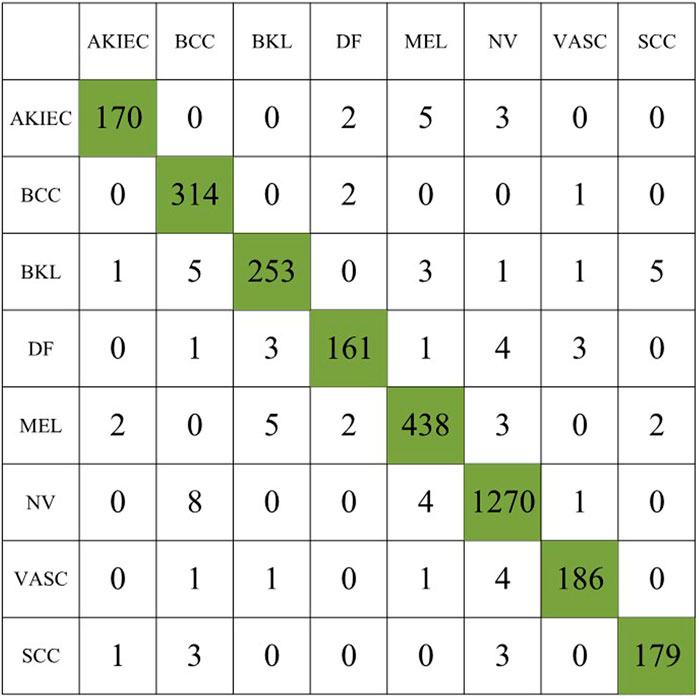
In the second experiment, we augment the number of images for the minority classes AKIEC, DF, VASC, and SCC to 1743, 1,667, 1920, and 1856, respectively, resulting in a total of 3,053 images. Table 3 summarizes the experimental results, and the confusion matrix for the second experiment is shown in Figure 6.
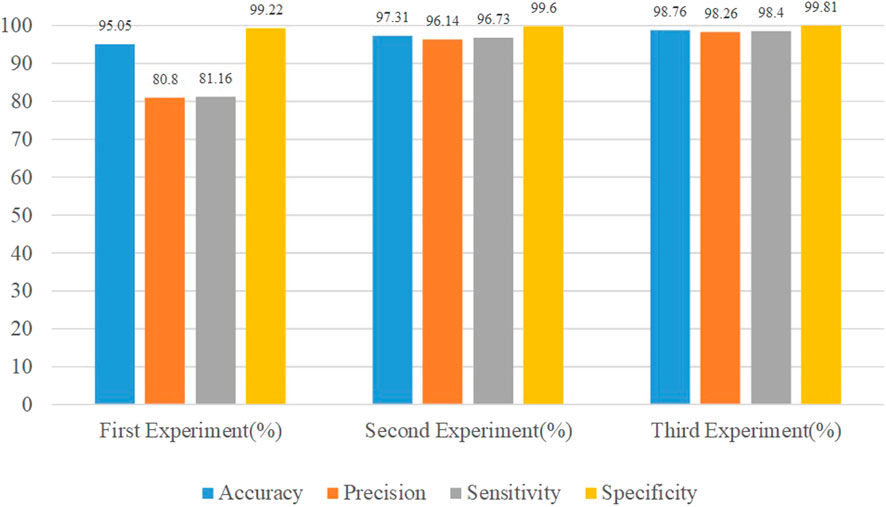
Compared to the first experiment, we observed a significant increase in sensitivity and precision. The imbalance gap in the number of minority class images is reduced. The accuracy of the model also increases to 97.31%, indicating that data augmentation plays an important role in model performance.
In the third experiment, we augmented the images of the minority classes AKIEC, DF, VASC, and SCC to 1743, 1,667, 1920, and 1856, respectively, and used the weighted sampling method for the augmented class images to prevent duplicate samples affect the experimental accuracy. Table 4 summarizes the experimental results, and the confusion matrix for the second experiment is shown inFigure 7.
The accuracy of the model after using the weighted sampling method is increased to 98.76%. Compared with the first two experiments, each index has been improved to varying degrees. The performance comparison of the three experiments is shown in Figure 8.
Experimental results show the lowest performance when using the original dataset in the first experiment. In the second experiment, the obtained results were improved. The third experiment shows the best value for the performance metric. The accuracy of the model increased from 95.05% to 98.765%, the Precision increased from 80.8% to 98.26%, the Sensitivity increased from 81.16% to 98.45%, and the Specificity increased from 99.22% to 99.81%.
The sampling process can be combined with the memorylessness of the Markovian effect, that is, the system does not remember the previous state of the current state, and only decides what state to transition to at the next moment based on the current state. Markov Decision Processes (MDPs) maximize returns by using methods such as dynamic programming, random sampling, etc.
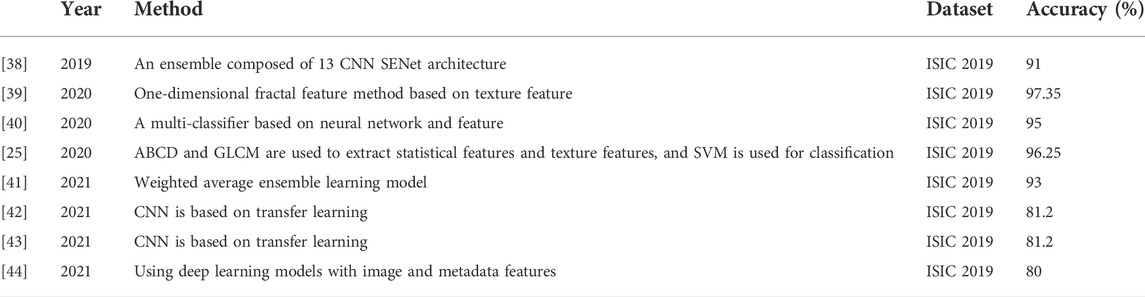
The performance of this method is compared with that of existing skin cancer classification methods, and Table 5 summarizes the data image types, used models, and accuracy rates of existing methods. Table 5 clearly shows that this method outperforms the literature methods listed in the table.
Conclusion
In this study, the improved quantum Inception-ResNet-V1 network was used, and after data augmentation and weighted sampling of the ISIC 2019 dataset, the skin damage images were classified, and the classification accuracy was as high as 98.76%. Inception with residuals makes the network need to learn less knowledge and the data distribution of each layer is close, making it easy to learn. The feature extraction function of quantum convolution can extract features in a larger space and achieve higher learning accuracy. In the quantum convolution layer, a single quantum gate applies operations to adjacent qubits, and the same quantum convolution is performed. Within the layers, all quantum gates have tunable parameters, preserving the properties of local connections and weight sharing in convolutional neural networks. These two characteristics enable the quantum convolutional neural network to effectively extract image features, reduce the complexity of the network model, and significantly improve the computational efficiency of the model.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
ZL proposed the content and methods of the research and organized the database to conduct experiments, and wrote the first draft. XC and ZC performed statistical analysis. YW and HM provides direction for dissertation medical content. DH and YD participated in the revision and reading of the manuscript.
Funding
The National Natural Science Foundation of China (No. 61772295, 61572270). The Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJZD-M202000501). Chongqing Technology Innovation and application development special general project (cstc2020jscxlyjsAX0002). Chongqing Technology Foresight and Institutional Innovation Project (cstc2021jsyj-yzysbAX0011).
Acknowledgments
Thanks for the important technical help given by colleagues in the laboratory, and the fund support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Vuković P, Lugović-Mihić L, Ćesić D, Novak-Bilić G, Šitum M, Spoljar S. Melanoma development: Current knowledge on melanoma pathogenesis. Acta Dermatovenerologica Croatica (2020) 28:163.
2. Amaria RN, Menzies AM, Burton EM, Scolyer RA, Tetzlaff MT, Antdbacka R, et al. Neoadjuvant systemic therapy in melanoma: Recommendations of the international neoadjuvant melanoma consortium. Lancet Oncol (2019) 20:e378–89. doi:10.1016/S1470-2045(19)30332-8
3. Saginala K, Barsouk A, Aluru JS, Rawla P, Barsouk A. Epidemiology of melanoma. Med Sci (2021) 9:63. doi:10.3390/medsci9040063
4. Carr S, Smith C, Wernberg J. Epidemiology and risk factors of melanoma. Surg Clin North Am (2020) 100:1–12. doi:10.1016/j.suc.2019.09.005
5. Hartman RI, Lin JY. Cutaneous melanoma—A review in detection, staging, and management. Hematol Oncol Clin North Am (2019) 33:25–38. doi:10.1016/j.hoc.2018.09.005
6. Pérez E, Reyes O, Ventura S. Convolutional neural networks for the automatic diagnosis of melanoma: An extensive experimental study. Med image Anal (2021) 67:101858. doi:10.1016/j.media.2020.101858
7. Naeem A, Farooq MS, Khelifi A, Abid A. Malignant melanoma classification using deep learning: Datasets, performance measurements, challenges and opportunities. IEEE Access (2020) 8:110575–97. doi:10.1109/access.2020.3001507
8. Marconcini R, Spagnolo F, Stucci LS, Ribero S, Marra E, De Rosa F, et al. Current status and perspectives in immunotherapy for metastatic melanoma. Oncotarget (2018) 9:12452–70. doi:10.18632/oncotarget.23746
9. Jenkins RW, Fisher DE. Treatment of advanced melanoma in 2020 and beyond. J Invest Dermatol (2021) 141:23–31. doi:10.1016/j.jid.2020.03.943
10. Chin R, Chen K, Abraham C, Robinson C, Perkins S, Johanns T, et al. Brain metastases in metastatic cutaneous melanoma: Patterns of care and clinical outcomes in the era of immunotherapy and targeted therapy. Int J Radiat Oncology*Biology*Physics (2021) 111:e565–6. doi:10.1016/j.ijrobp.2021.07.1528
11. Nachbar F, Stolz W, Merkle T, Cognetta AB, Vogt T, Landthaler M, et al. The abcd rule of dermatoscopy: High prospective value in the diagnosis of doubtful melanocytic skin lesions. J Am Acad Dermatol (1994) 30:551–9. doi:10.1016/s0190-9622(94)70061-3
12. Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: Comparison of the abcd rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol (1998) 134:1563–70. doi:10.1001/archderm.134.12.1563
13. Vuković P, Lugović-Mihić L, Ćesić D, Novak-Bilić G, Šitum M, Spoljar S. Melanoma development: Current knowledge on melanoma pathogenesis. Acta Dermatovenerologica Croatica (2020) 28:163.
14. Atkins MB, Curiel-Lewandrowski C, Fisher DE, Swetter SM, Tsao H, Aguirre-Ghiso JA, et al. The state of melanoma: Emergent challenges and opportunities. Clin Cancer Res (2021) 27:2678–97. doi:10.1158/1078-0432.ccr-20-4092
15. Wouters J, Kalender-Atak Z, Minnoye L, Spanier KI, De Waegeneer M, Bravo González-Blas C, et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat Cel Biol (2020) 22:986–98. doi:10.1038/s41556-020-0547-3
16. Nasr-Esfahani E, Samavi S, Karimi N, Soroushmehr SMR, Jafari MH, Ward K, et al. (2016). Melanoma detection by analysis of clinical images using convolutional neural network. In 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC IEEE), 1373–6.
17. Alquran H, Qasmieh IA, Alqudah AM, Alhammouri S, Alawneh E, Abughazaleh A, et al. (2017). The melanoma skin cancer detection and classification using support vector machine. In 2017 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies (AEECT). 1–5. doi:10.1109/AEECT.2017.8257738
18. Vasconcelos CN, Vasconcelos BN. Experiments using deep learning for dermoscopy image analysis. Pattern Recognition Lett (2020) 139:95–103. doi:10.1016/j.patrec.2017.11.005
19. Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol (2018) 138:1529–38. doi:10.1016/j.jid.2018.01.028
20. Pham T-C, Luong C-M, Visani M, Hoang V-D (2018). Deep cnn and data augmentation for skin lesion classification. In Asian Conference on Intelligent Information and Database Systems (Springer), 573–82.
21. Emara T, Afify HM, Ismail FH, Hassanien AE (2019). A modified inception-v4 for imbalanced skin cancer classification dataset. In 2019 14th International Conference on Computer Engineering and Systems (ICCES) (IEEE), 28–33.
22. Adegun AA, Viriri S. Deep learning-based system for automatic melanoma detection. IEEE Access (2019) 8:7160–72. doi:10.1109/access.2019.2962812
23. Hosny KM, Kassem MA, Foaud MM. Classification of skin lesions using transfer learning and augmentation with alex-net. PloS one (2019) 14:e0217293. doi:10.1371/journal.pone.0217293
24. Yu L, Chen H, Dou Q, Qin J, Heng P-A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging (2017) 36 (4):994–1004. doi:10.1109/TMI.2016.2642839
25. Hosny KM, Kassem MA, Foaud MM. Skin melanoma classification using roi and data augmentation with deep convolutional neural networks. Multimed Tools Appl (2020) 79:24029–55. doi:10.1007/s11042-020-09067-2
26. Iyer V, Ganti B, Hima Vyshnavi A, Krishnan Namboori P, Iyer S. Hybrid quantum computing based early detection of skin cancer. J Interdiscip Math (2020) 23:347–55. doi:10.1080/09720502.2020.1731948
27. Kassem MA, Hosny KM, Fouad MM. Skin lesions classification into eight classes for isic 2019 using deep convolutional neural network and transfer learning. IEEE Access (2020) 8:114822–32. doi:10.1109/access.2020.3003890
28. Zhou N-R, Liu X-X, Chen Y-L, Du N-S. Quantum K-nearest-neighbor image classification algorithm based on KL transform. Int J Theor Phys. Springer (2021) 60 (3):1209–1224.
29. Gong L-H, Xiang L-Z, Liu S-H, Zhou N-R. Born machine model based on matrix product state quantum circuit. Phys A: Stat Mech Appl. Elsevier (2022) 593:126907.
30. Abdelhalim ISA, Mohamed MF, Mahdy YB. Data augmentation for skin lesion using self-attention based progressive generative adversarial network. Expert Syst Appl (2021) 165:113922. doi:10.1016/j.eswa.2020.113922
31. Bissoto A, Perez F, Valle E, Avila S. Skin lesion synthesis with generative adversarial networks. In: OR 2.0 context-aware operating theaters, computer assisted robotic endoscopy, clinical image-based procedures, and skin image analysis. Springer (2018). p. 294–302.
32. Ren J, Yang Q, Zhu Y, Zhu X, Tian H, Wang W, et al. Complex social contagions on weighted networks considering adoption threshold heterogeneity. IEEE Access (2020) 8:61905–14. doi:10.1109/access.2020.2984615
33. Athey S, Imbens GW. Design-based analysis in difference-in-differences settings with staggered adoption. J Econom (2022) 226:62–79. doi:10.1016/j.jeconom.2020.10.012
34. Marsh-Armstrong B, Migacz J, Jonnal R, Werner JS. Automated quantification of choriocapillaris anatomical features in ultrahigh-speed optical coherence tomography angiograms. Biomed Opt Express (2019) 10:5337–50. doi:10.1364/boe.10.005337
35. Ye T-Y, Li H-K, Hu J-L. Semi-quantum key distribution with single photons in both polarization and spatial-mode degrees of freedom. Int J Theor Phys (Dordr) (2020) 59:2807–15. doi:10.1007/s10773-020-04540-y
36. Combalia M, Codella NC, Rotemberg V, Helba B, Vilaplana V, Reiter O, et al. Bcn20000: Dermoscopic lesions in the wild (2019). arXiv preprint arXiv:1908.02288.
37. Fawcett T. An introduction to roc analysis. Pattern recognition Lett (2006) 27:861–74. doi:10.1016/j.patrec.2005.10.010
38. Pacheco AG, Ali A-R, Trappenberg T. Skin cancer detection based on deep learning and entropy to detect outlier samples (2019). arXiv preprint arXiv:1909.04525.
39. Molina-Molina EO, Solorza-Calderón S, Álvarez-Borrego J. Classification of dermoscopy skin lesion color-images using fractal-deep learning features. Appl Sci (2020) 10:5954. doi:10.3390/app10175954
40. El-Khatib H, Popescu D, Ichim L. Deep learning–based methods for automatic diagnosis of skin lesions. Sensors (2020) 20:1753. doi:10.3390/s20061753
41. Monika MK, Vignesh NA, Kumari CU, Kumar M, Lydia EL. Skin cancer detection and classification using machine learning. Mater Today Proc (2020) 33:4266–70. doi:10.1016/j.matpr.2020.07.366
42. Rahman Z, Hossain MS, Islam MR, Hasan MM, Hridhee RA. An approach for multiclass skin lesion classification based on ensemble learning. Inform Med Unlocked (2021) 25:100659. doi:10.1016/j.imu.2021.100659
43. Cauvery K, Siddalingaswamy P, Pathan S, D’souza N (2021). A multiclass skin lesion classification approach using transfer learning based convolutional neural network. In 2021 Seventh International conference on Bio Signals, Images, and Instrumentation (ICBSII) (IEEE), 1–6.
Keywords: melanoma, deep learning, CNN, quantum computing, skin cancer, ISIC 2019
Citation: Li Z, Chen Z, Che X, Wu Y, Huang D, Ma H and Dong Y (2022) A classification method for multi-class skin damage images combining quantum computing and Inception-ResNet-V1. Front. Phys. 10:1046314. doi: 10.3389/fphy.2022.1046314
Received: 16 September 2022; Accepted: 19 October 2022;
Published: 03 November 2022.
Edited by:
Tianyu Ye, Zhejiang Gongshang University, ChinaReviewed by:
Suzhen Yuan, Chongqing University of Posts and Telecommunications, ChinaH. Z. Shen, Northeast Normal University, China
Copyright © 2022 Li, Chen , Che , Wu , Huang , Ma and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumin Dong, ZHltQGNxbnUuZWR1LmNu
 Ziyi Li1
Ziyi Li1 Yaguang Wu
Yaguang Wu  Yumin Dong
Yumin Dong