- Department of Physics and Biophysics, University of San Diego, San Diego, CA, United States
Volumetric microscopic imaging data acquired at high speeds is often needed in studies of soft matter. Several microscopy techniques exist for this purpose, but a relative newcomer is light sheet fluorescence microscopy (LSFM). This microscopy method has seen spectacular growth in the biological sciences over the past two decades. In this perspective, we highlight how LSFM may also apply to the field of soft matter. We review the principles and recent advances of LSFM and discuss how it has been used in prior soft matter studies. We demonstrate how a recent implementation of LSFM can be used to study capillary wave fluctuations and droplet coalescence in a colloidal fluid system.
Introduction
Light sheet fluorescence microscopy (LSFM) has attracted a significant and growing user base, chiefly among life scientists. This is due to LSFM’s ability to acquire optically sectioned images with economical use of excitation light. Though the development of light sheet microscopy can be traced back to the early 20th century, work in the early 21st century brought about a remarkable rise in the popularity of LSFM. Notably, in 2004, Jan Huisken and others in the lab of Ernst H. K. Stelzer demonstrated LSFM’s utility in imaging developing embryos over days without apparent photodamage [1]. In the years since, numerous LSFM designs have been published which push the technique’s capabilities in a range of directions from increased spatiotemporal resolution to aberration correction. Applications for these various LSFM designs have primarily been in the biological sciences. However, in this article we highlight work in the field of soft matter that has employed LSFM, describe recent advances in LSFM that may draw more scientists investigating nonliving materials into LSFM’s user base, and describe results using LSFM on colloidal samples in our own lab.
There are a multitude of LSFM configurations, but the demonstrations of the technique that initiated its renaissance over the last two decades [1–3] and a configuration which remains common today employ two orthogonally placed objective lenses with the sample at the common focal point. One objective lens, termed the excitation or illumination objective, focuses laser light to a thin sheet on the sample. The other objective lens, termed the imaging or detection objective, is aligned with its focal plane coincident with this sheet of excitation light. Optically sectioned images can then be captured at the frame rate of the camera, just like widefield methods and in contrast to the typically slower point scanning methods. For three-dimensional (3D) imaging, image stacks can be acquired by either translating the sample through the light sheet or by sweeping the light sheet in sync with translation of the imaging objective’s focal plane with, e.g., a scanning mirror and a piezoelectric stage. A key feature of this technique which fuels its appeal particularly among developmental biologists is its “gentleness.” [4] By confining the excitation light to the focal plane, photons are not wasted on (and not harming) regions of the sample not being imaged.
Over the last few years innovative LSFM designs have expanded the range of applications where the technique excels far beyond just the imaging of living whole organisms. This has been achieved by allowing for more flexibility in sample mounting procedures, increasing the 3D image stack acquisition speed, and improving the spatial resolution. While some of these advances in LSFM are difficult to replicate without considerable optical engineering knowhow, an increasing number of LSFM configurations are commercially available. Furthermore, multiple open access platforms and guides exist for researchers interested in constructing their own LSFM out of mostly off-the-shelf components [5–8].
While the vast majority of work in the field of LSFM focuses on the life sciences, we believe that the technique holds great potential for soft matter researchers and material scientists. Of course, the low phototoxicity and photobleaching that LSFM offers is less important to those studying nonliving materials. When dealing with colloids and emulsions rather than living cells and embryos, one does not have to be so frugal with photons. Nevertheless, the spatiotemporal resolution and data throughput rates achievable with LSFM make it competitive with the workhouse optical sectioning technique in many soft matter labs, the laser scanning confocal microscope. In fact, looking at the history of confocal microscopy’s usage in materials science and soft matter suggests that we may be coming upon an echo in time from a quarter of a century ago. In 1997, M. H. Chestnut wrote that confocal microscopy “has taken the world of cell biology by storm in the past decade, yet it remains infrequently applied to colloidal systems.” [9] The same could be said of LSFM today. We believe LSFM may follow a parallel trajectory to that of confocal microscopy, with increasing usage in soft matter labs after impressive demonstrations in biology labs.
In this Perspective article, we briefly describe recent developments in LSFM with attention paid to those applicable to soft matter research. We discuss how LSFM has been applied to soft matter research and highlight recent work from our own lab. In this short Perspective, we do not attempt to comprehensively review the field of LSFM nor document the history of its development, but we note several excellent review articles for the interested reader. [10–16]
Recent Developments in Light Sheet Microscopy
The number of LSFM configurations has grown remarkably over the last few years. Many recent designs take the latest concepts and methods from the larger optical microscopy field and apply them to the light-sheet modality. For example, adaptive optics for aberration correction [17, 18], fast movement of the focal plane with electrically tunable lenses [19], single-molecule localization microscopy [20], structured illumination [21], and multiphoton excitation [22] have all been used to improve the speed and/or spatial resolution of LSFM. Since many of these advances can apply to multiple microscopy modalities, not uniquely LSFM, we will not go into the details of their implementation and use. Instead, we will highlight the more LSFM-specific advances. These tend to focus on one of two major concerns: the shape of the excitation light sheet and the way samples must be mounted for imaging.
The ideal shape of the sheet of excitation light is thin, providing a high degree of optical sectioning and improved axial resolution, over a wide lateral extent, providing a large field of view. However, thinness of the sheet and its uniformity over a large field of view are conflicting demands. With a high numerical aperture (NA) illumination objective, one can achieve a thin light sheet. But there will be a limited lateral distance (i.e., distance along the excitation light’s propagation direction) over which the light sheet thickness can be approximated as uniform. This is often characterized with the Rayleigh length which defines the distance over which the excitation sheet’s thickness changes by a factor of
A common and straightforward approach to generating an illumination sheet is to use a cylindrical lens in the excitation path. To achieve uniform light sheet illumination even in the presence of scattering or absorbing sample features, this light sheet can be pivoted back and forth within the detection objective’s focal plane or two light sheets impinging on the sample from opposite directions can be used [24]. Another approach is to scan a thin pencil beam which averages out over time to a sheet, a method termed digital scanned laser light sheet fluorescence microscopy (DSLM) [25]. A further means of creating a very thin uniform light sheet over an extended field of view is to sweep the sheet in the same direction as that of the illumination light propagation, a technique that’s been called axially swept light sheet microscopy. [26–29]
While Gaussian beams are the most common in LSFM, the use of propagation-invariant beams like Bessel or Airy beams have been used to achieve a high degree of optical sectioning over an extended field of view. Scanning Bessel beams to create light sheets 0.5 µm thick, Planchon et al. in the lab of Eric Betzig achieved isotropic resolution of ∼0.3 µm [30]. Spatial and temporal resolutions were pushed further in work from the same lab using the coherent superposition of Bessel beams to create an optical lattice [31]. In keeping with the open access nature of much of the LSFM method development, Aakhte et al. released an open-source toolbox for shaping the excitation light sheet, allowing one to set the excitation pattern for optimizing, e.g., resolution, field of view, or penetration depth. [32]
A hurdle to more widespread LSFM adoption has been the requirements for sample mounting. For common LSFM configurations, like that shown in Figure 1A, the sample is often loaded in a thin capillary tube (by using tubing of fluorinated ethylene-propylene, the walls may be close to the refractive index of an aqueous sample) or, especially for live whole organism imaging, embedded in a cylinder of agarose or other gel. The sample can then be placed in the common focus of both the imaging and excitation objective lenses and moved with a motorized stage. While this mounting approach does provide exceptional optical access to the sample since rotating and translating the mounted sample can provide views from multiple perspectives, it prevents the use of popular sample preparations involving glass slides, petri dishes, multi-well plates or planar microfluidic devices.
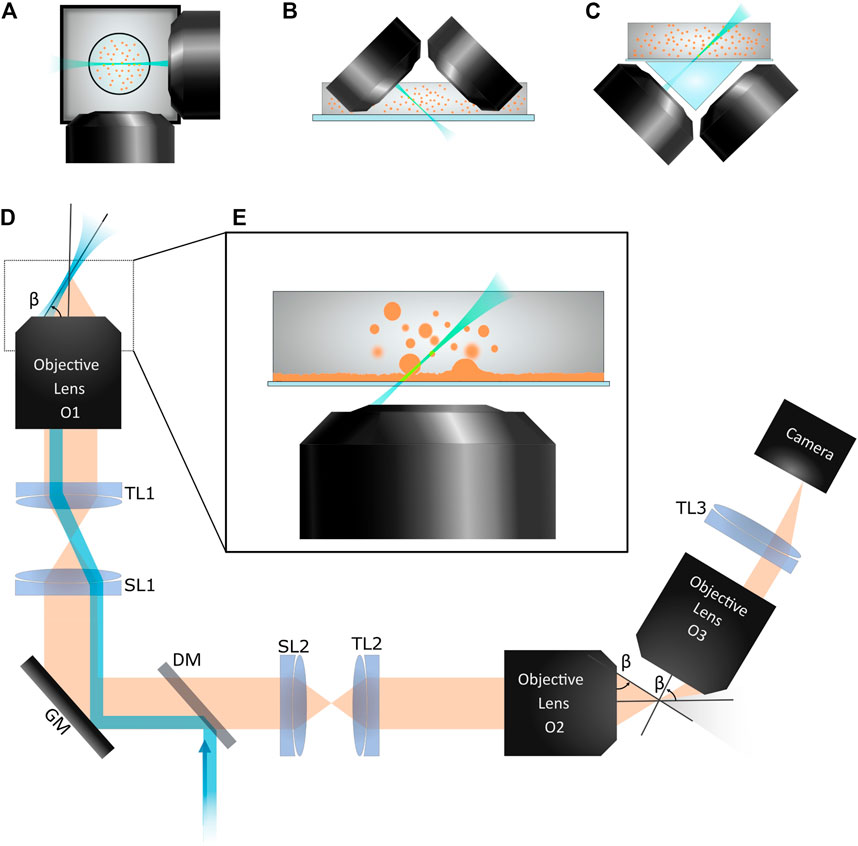
FIGURE 1. Various LSFM configurations are shown. (A) Two orthogonal objective lenses surround the sample contained in either a capillary tube or cylinder of gel. (B) An iSPIM system uses two objective lens that meet the flat-bottomed sample holder at 45°. This configuration is ideal for imaging into an open petri dish. (C) Open-top LSFM configurations image across a planar sample holder from below, similar to an inverted microscope. Since imaging is done across a titled interface, a water prism is used to minimize aberrations. (D) In oblique plane microscopy, the sheet of excitation light leaves the primary objective lens, O1, at an angle β relative to the front of the lens. Fluorescence emission is collected by O1. The second objective lens, O2, demagnifies the image and the third objective lens, O3, images the titled plane on the camera. TL1, TL2, and TL3: tube lenses. SL1 and SL2: scan lenses. GM: galvo mirror. DM: dichroic mirror. In our setup, O1 is a 40 × 1.15NA water immersion lens (Olympus); O2, 40 × 0.95NA lens (Olympus); O3, 50 × 0.75NA lens (Mitutoyo). (E) A close-up of O1 with a sample is shown. We use this OPM system to image a colloid-polymer system which phase separates into coexisting colloid-rich (liquid) and colloid-poor (gas) phases. We observe droplets coalesce with the bulk liquid phase as well as fluctuations of the planar liquid-gas interface.
However, over the last several years, multiple innovative designs have allowed LSFM to be used with more standard sample preparation protocols. In 2011, Wu et al. described an inverted selective plane illumination microscope (iSPIM), [33] so called since a module containing two perpendicular objective lenses could be mounted onto the illumination pillar of a standard inverted microscope frame (see Figure 1B). This system allows samples in petri dishes or similar geometries to be imaged. Taking the two objective lenses and moving them underneath the sample holder has resulted in multiple configurations referred to as open-top SPIM (see Figure 1C). Since imaging across a glass plate at a 45° angle will result in considerable aberrations, these open top setups use triangular prisms [34, 35], solid immersion lens [36, 37], and/or adaptive optics. [38, 39] These configurations allow the imaging of samples mounted between a glass slide and coverslip, in multi-well plates, or in microfluidic devices.
While the open-top LSFM configurations allow more standard sample preparations to be used, the fact that two objective lenses must surround the sample prohibits bulkier, high NA lenses from being used. However, recent LSFM designs have emerged which enable the use of high NA lenses with standard sample mountings. Instrumental to the development of many of these designs was the 2008 work of Chris Dunsby. [40] Dunsby described a way to illuminate the sample with a thin sheet of light and capture the emitted fluorescence with the same high NA objective lens (see Figures 1D,E). This class of LSFM is known as oblique plane microscopy (OPM). OPM relies on sending a beam of excitation light through the objective lens such that it exits the front surface of the lens at a steep angle, reminiscent of total internal reflection microscopy and similar to an approach known as highly inclined and laminated optical sheet (HILO) microscopy. [41] One then must image this tilted plane on a camera. Tilting the camera itself would result in large aberrations, but by using the principles of remote refocusing developed by Botcherby et al. [42], Dunsby used downstream optics to de-magnify the image and then re-magnify a tilted plane. In early implementations of OPM, 3D image stacks were obtained by either translating the sample stage or the second objective lens (the one which de-magnifies the image and labeled as O2 in Figure 1D). [43–45]
A stunning advance came in 2015 with the work of M. B. Bouchard and others in the lab of Elizabeth Hillman [46]. Their OPM implementation, termed swept confocally-aligned planar excitation (SCAPE) microscopy, used, similar to Dunsby’s design, a set of three objective lenses to collect light from the sample, de-magnify the image plane, and then re-magnify a tilted plane conjugate to the tilted sheet of excitation light. However, to achieve fast volumetric imaging rates, Bouchard et al. used a scanning mirror to sweep the excitation light across the sample and to de-scan the emitted light such that the image plane captured by the camera moved synchronously with the light sheet. A following paper by the same group described an improved SCAPE 2.0 which was used to image at high speeds and/or large fields of view (e.g., images at 100 volumes per second were recorded with 640 × 148 pixel frames and 127 slices per volume with an optical resolution of approximately 1.2, 0.6, and 1.6 µm in the three spatial dimensions) [47]. Subsequent microscopy designs which follow up on the work of Dunsby and Bouchard et al. have either demonstrated large field of view imaging or pushed the spatial resolution with high NA objectives. [7, 47–52] Of particular note are a few implementations which help to image samples with refractive index variations or absorbing regions. Designs by Yang et al. [53] and Sparks et al. [51] provide multiple views of the sample by illuminating and imaging a pair of nearly orthogonal planes (i.e., sending the excitation light sheet (and imaging plane) not just out from the left side of the objective lens at an angle of β as shown in Figure 1D, but also out from the right side of the objective at an angle of -β). Other implementations employ two-photon imaging for optically penetrating samples deeper than single-photon methods allow. [54, 55]
These latest LSFM designs cited in the previous paragraph are collectively referred to as single-objective light sheet (SOLS) methods. Although they often use three objective lenses, only one objective lens is positioned near the sample, similar to more conventional microscopy methods and distinct from the two objectives needed to surround the sample in most previous LSFMs. As these SOLS systems retain advantages of earlier LSFM designs, such as gentle optical sectioning and image acquisition rates limited by the camera, but allow for samples mounted in glass slides or similar planar configurations and are compatible with high NA lenses, it is likely they will find a wider range of applications. Importantly, we think these SOLS systems will generate new discoveries in soft matter and materials science.
LSFM for Soft Matter Research
While recent microscopy designs make a transition from widefield or confocal fluorescence microscopy to LSFM more straightforward, there have already been multiple studies employing earlier LSFM designs for soft matter systems. It is important to note that the use of laser light sheet imaging has long been used to study the packing and flow of granular media and non-Brownian particles. These studies typically investigate millimeter-scale particles in refractive index matched solutions. [56] While our focus in this article is on microscopic imaging, prior work on granular matter has much to offer those employing light sheet illumination on any scale, micro or macro. For example, Salili et al. have explored methods for combating striping artifacts, demonstrated with a sample of millimeter sized PMMA spheres in an index matched solution. [57] Such striping artifacts are problematic in light sheet imaging at both the microscopic [58] and macroscopic scales. Huang et al. used a clever light-sheet method to image a granular packing of 7 mm spheres across a flat interface at 45° using a liquid-filled prism [59], a technique also applied to microscopic light sheet imaging. [34]
Soft matter research using light sheet imaging on the microscopic scale include the work of Loftus et al. in the lab of R. Parthasarathy. In that work, a LSFM of the variety depicted in Figure 1A was used to measure the contours of a thermally fluctuating vesicle. [60] Optically sectioned images acquired at 60 to 100 frames per second allowed the authors to track edges of vesicles with a precision of approximately 10 nm and to determine vesicle bending rigidities. Again studying vesicles, Jahl and Parthasarathy measured the hydrodynamic drag of phospholipid vesicles by tracking their motion using LSFM. [61]
Other works have used LSFM to measure the diffusive dynamics of colloidal particles. Ritter et al. demonstrated how LSFM provides improved contrast over wide-field microscopy and fast imaging rates for single-particle tracking applications. [62] They tracked quantum dots diffusing in a 3D environment using a 2 µm thick light sheet and 243 frames per second imaging. Their system’s lateral and axial optical resolutions were approximately 350 nm and 1.1 µm, respectively. More recently, LSFM has been used to measure the subdiffusive dynamics of micron-sized particles in crowded polymer networks. [63]
These soft matter studies cited in the previous two paragraphs which have utilized LSFM[60–63] have all done so using Gaussian excitation beams. We are not aware of work in the field of soft matter which has used non-Gaussian beams or structured illumination with LSFM as mentioned in the previous section. However, the use of such light-sheet-shaping methods may be of interest to those looking to push the limits of their optical resolution.
We are not aware of any published work using SOLS microscopes for soft matter research. However, in our own lab we have recently constructed such a setup roughly following the designs presented by Yang et al. [49] and Millet-Sikking and York. [7] We used three objective lenses with a 40× water immersion 1.15 NA as our primary (O1) lens (see Figure 1D). With our downstream optics imaging a tilted plane onto our camera, the effective NA of our system is about 0.8. The laser light sheet is generated with a series of cylindrical lenses before focusing at a galvanometer scanning mirror (Thorlabs, GVS011) placed conjugate to the back focal planes of both O1 and O2. By scanning this galvo mirror, the excitation light sheet will sweep across the sample and the plane imaged onto the camera (Thorlabs, CS2100M-USB) by O3 will move in sync with the light sheet. This is one of most notable advantages of such SOLS systems. Volumetric imaging is achieved solely by scanning a mirror; the sample stage and all lenses remain static. As the sample stage and objective lenses all have much greater inertia than the scanning mirror, one can quickly acquire image stacks. Furthermore, there is no mechanical perturbation of the sample during image stack acquisition, a concern when using an immersion objective lens which physically contacts the sample.
We use our SOLS system to image a colloid-polymer mixture and measure capillary wave dynamics and droplet coalescence (Figure 2). Similar studies have been conducted by Aarts et al. using confocal microscopy. [64] The benefit of using LSFM is that we can acquire optically sectioned images of the liquid-gas interface at rates limited only by the camera speed. Such an image is shown in Figure 2A, where we observe the interface between colloid-rich and colloid-poor fluid phases. These coexisting fluids phases emerge after mixing fluorescently labeled poly(N-isopropylacrylamide) (pNIPAM) microgel colloidal particles (∼330 nm in diameter) with xanthan which acts as a depletant. This colloid-polymer mixture is loaded into a chamber made of a glass slide and coverslip and allowed to sit undisturbed for several hours so that a colloid-rich layer develops on the coverslip surface. It is important to note that this image shows a plane aligned with the excitation light sheet as depicted in Figures 1D,E where, in our case, the angle β is 35°. After recording a time series of such images, we find the location of the interface (marked by the cyan line in Figure 2A) to determine the height of the interface as a function of position and time,
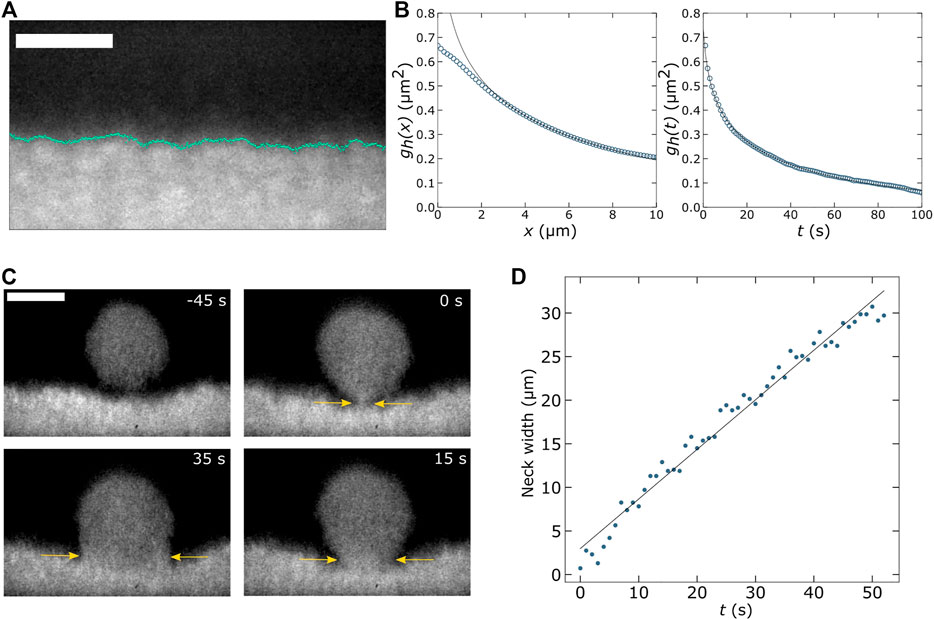
FIGURE 2. (A) Using the OPM setup shown in Figures 1D,E we measure thermally fluctuating capillary waves in a phase separated colloid-polymer mixture. This mixture consists of ∼330-nm-diameter fluorescent poly(N-isopropylacrylamide) microgel particles and xanthan, as described in our early work [73]. Scale bar is 20 µm. (B) After tracking the liquid-gas interface over a lateral distance of 78 µm for 700 s, we compute the height-height correlation function,
Furthermore, we can acquire volumetric image stacks quickly by sweeping the light sheet and image plane across the sample using the galvo mirror. We can observe the process of colloid-rich droplets coalescing with the bulk colloid-rich phase by recording volumes of about 70 × 78 × 71 μm3 at a rate of 1 Hz. For each volume, we record 30 slices. From this four-dimensional data set, we find the width of the neck that forms between the falling droplet and the bulk phase (yellow arrows indicate this neck in Figure 2C). We find an approximately linear relationship between this neck width and time which yields a capillary velocity of ∼0.3 μm/s (Figure 2D). These capillary dynamics are slow in comparison to many systems of interest to soft matter researchers. However, with a high-speed camera or more limited field of view, faster dynamics could easily be studied. SOLS systems are well suited for fast image acquisition, as previously mentioned, given the fact that images are acquired at the camera frame rate and that the image plane is swept across the sample using a galvo mirror, rather than moving a bulkier optical component.
While such studies of capillary wave dynamics have been conducted previously using confocal microscopy, we hope the results in Figure 2 demonstrate that similar work can be done with LSFM, which has a number of strengths as we have described in this Perspective. We also note that the LSFM we have used (Figure 1D) was assembled from the off-the-shelf parts at a fraction of what a commercial confocal microscope would have cost.
Conclusion and Outlook
The rise of LSFM in biology has been dramatic this century. Due to its ability to gently optically section specimen at high speeds and its growing popularity in biology research, it was declared Method of the Year by Nature Methods in 2014. [65] Developments in LSFM technology since that time have pushed the spatiotemporal resolution and allowed for a broader range of sample preparations to be used. The advent of SOLS microscopes is likely to broaden light sheet’s reach even more and fuel future discoveries. In Nature Methods in 2021, Senior Editor Rita Strack wrote that SOLS will “further propel light sheet microscopy into the mainstream.” [66]
While granular matter physicists have long used light sheet illumination to image into thick 3D samples, those studying particles and droplets that are orders of magnitude smaller have been relatively less disposed to use LSFM. In this article, we have noted a few of the instances where LSFM was used to investigate soft matter and we end by echoing a sentiment expressed in one of those works. In their article on measuring membrane bending moduli with LSFM, Loftus et al. wrote that LSFM’s “utility for nonliving systems has been remarkably unrealized, however, and we look forward to its further applications to dynamic interfaces and soft materials.”[60]
We believe that LSFM is still underutilized in the study of nonliving systems. But the recent innovations we have covered here are likely to change that. LSFM gives one the ability to optically section samples with the speed of widefield image acquisition. While confocal microscopes using multipoint or resonant scanning can rival LSFM’s speed and spatial resolution and while confocal microscopy’s less economical use of photons is not such a grave issue for nonliving systems as it is for living ones, there are still multiple reasons that soft matter researchers might pursue LSFM. LSFM can be the more affordable option in a lab with sufficient optical engineering knowhow. And multiple do-it-yourself (DIY) guides to LSFM construction exist for those with less optics experience. [5–8] Additionally, if one takes the DIY approach, as so many in the LSFM community do, then one can construct a microscope specifically suited to a given experiment. This is often done in the granular matter community where light sheet setups are built around containers where samples are sheared or otherwise perturbed. [67–70] Finally, since many LSFM designs leave the sample quite accessible, one may have an easier time coupling this type of microscopy to other instruments such as rheometers, optical or magnetic tweezers, or microfluidics. As examples, Taormina et al. combine a light sheet of the geometry shown in Figure 1A with magnetic tweezers to measure the viscoelasticity of mucus [71] and Jiang et al. built a custom LSFM for work with droplet-based microfluidics. [72]
We look forward to future work using LSFM to probe nonliving materials and other systems of interest to soft matter researchers. We note that far more LSFM configurations exist than we had space to describe here and encourage interested readers to discover those in the more thorough review articles we have cited.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
RY constructed the LSFM and collected data with supervision from RM. Both authors wrote the manuscript.
Funding
This work was supported by a Cottrell Scholars Award to RM from the Research Corporation for Science Advancement. Funds for publication were provided by a grant from the College of Arts and Sciences, University of San Diego.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Huisken J, Swoger J, Bene FD, Wittbrodt J, Stelzer EHK. Optical Sectioning Deep inside Live Embryos by Selective Plane Illumination Microscopy. Science (2004) 305(5686):1007–9. doi:10.1126/science.1100035
2. Voie AH, Burns DH, Spelman FA. Orthogonal-plane Fluorescence Optical Sectioning: Three-Dimensional Imaging of Macroscopic Biological Specimens. J Microsc (1993) 170(Pt 3):229–36. doi:10.1111/j.1365-2818.1993.tb03346.x
3. Fuchs E, Jaffe J, Long R, Azam F. Thin Laser Light Sheet Microscope for Microbial Oceanography. Opt Express (2002) 10(2):145–54. doi:10.1364/oe.10.000145
4. Huisken J. Slicing Embryos Gently with Laser Light Sheets. BioEssays (2012) 34(5):406–11. doi:10.1002/bies.201100120
5. Pitrone PG, Schindelin J, Stuyvenberg L, Preibisch S, Weber M, Eliceiri KW, et al. . OpenSPIM: an Open-Access Light-Sheet Microscopy Platform. Nat Methods (2013) 10(7):598–9. doi:10.1038/nmeth.2507
6. Gualda EJ, Vale T, Almada P, Feijó JA, Martins GG, Moreno N. OpenSpinMicroscopy: an Open-Source Integrated Microscopy Platform. Nat Methods (2013) 10(7):599–600. doi:10.1038/nmeth.2508
7. Millett-Sikking A, York AG. High NA Single-Objective Lightsheet. Githubio (2019). doi:10.5281/zenodo.3244420
8. Kumar M, Kishore S, McLean D, Kozorovitskiy Y. Crossbill: an Open Access Single Objective Light-Sheet Microscopy Platform. bioRxiv (2021).
9. Chestnut MH. Confocal Microscopy of Colloids. Curr Opin Colloid Interf Sci (1997) 2(2):158–61. doi:10.1016/s1359-0294(97)80020-9
10. Huisken J, Stainier DYR. Selective Plane Illumination Microscopy Techniques in Developmental Biology. Development (2009) 136(12):1963–75. doi:10.1242/dev.022426
11. Ntziachristos V. Going Deeper Than Microscopy: the Optical Imaging Frontier in Biology. Nat Methods (2010) 7(8):603–14. doi:10.1038/nmeth.1483
12. Tomer R, Khairy K, Keller PJ. Shedding Light on the System: Studying Embryonic Development with Light Sheet Microscopy. Curr Opin Genet Dev (2011) 21(5):558–65. doi:10.1016/j.gde.2011.07.003
13. Reynaud EG, Peychl J, Huisken J, Tomancak P. Guide to Light-Sheet Microscopy for Adventurous Biologists. Nat Methods (2015) 12(1):30–4. doi:10.1038/nmeth.3222
14. Stelzer EHK. Light-sheet Fluorescence Microscopy for Quantitative Biology. Nat Methods (2015) 12(1):23–6. doi:10.1038/nmeth.3219
15. Olarte OE, Andilla J, Gualda EJ, Loza-Alvarez P. Light-sheet Microscopy: a Tutorial. Adv Opt Photon (2018) 10(1):111–79. doi:10.1364/aop.10.000111
16. Girkin JM, Carvalho MT. The Light-Sheet Microscopy Revolution. J Opt (2018) 20(5):053002. doi:10.1088/2040-8986/aab58a
17. Turaga D, Holy TE. Image-based Calibration of a Deformable Mirror in Wide-Field Microscopy. Appl Opt (2010) 49(11):2030–40. doi:10.1364/ao.49.002030
18. Bourgenot C, Saunter CD, Taylor JM, Girkin JM, Love GD. 3D Adaptive Optics in a Light Sheet Microscope. Opt Express (2012) 20(12):13252–61. doi:10.1364/oe.20.013252
19. Fahrbach FO, Voigt FF, Schmid B, Helmchen F, Huisken J. Rapid 3D Light-Sheet Microscopy with a Tunable Lens. Opt Express (2013) 21(18):21010–26. doi:10.1364/oe.21.021010
20. Cella Zanacchi F, Lavagnino Z, Perrone Donnorso M, Del Bue A, Furia L, Faretta M, et al. . Live-cell 3D Super-resolution Imaging in Thick Biological Samples. Nat Methods (2011) 8(12):1047–9. doi:10.1038/nmeth.1744
21. Breuninger T, Greger K, Stelzer EHK. Lateral Modulation Boosts Image Quality in Single Plane Illumination Fluorescence Microscopy. Opt Lett (2007) 32(13):1938–40. doi:10.1364/ol.32.001938
22. Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and Fast Live Imaging with Two-Photon Scanned Light-Sheet Microscopy. Nat Methods (2011) 8(9):757–60. doi:10.1038/nmeth.1652
23. Gebhardt JCM, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, et al. . Single-molecule Imaging of Transcription Factor Binding to DNA in Live Mammalian Cells. Nat Methods (2013) 10(5):421–6. doi:10.1038/nmeth.2411
24. Huisken J, Stainier DYR. Even Fluorescence Excitation by Multidirectional Selective Plane Illumination Microscopy (mSPIM). Opt Lett (2007) 32(17):2608–10. doi:10.1364/ol.32.002608
25. Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of Zebrafish Early Embryonic Development by Scanned Light Sheet Microscopy. Science (2008) 322(5904):1065–9. doi:10.1126/science.1162493
26. Dean KM, Fiolka R. Uniform and Scalable Light-Sheets Generated by Extended Focusing. Opt Express (2014) 22(21):26141–52. doi:10.1364/oe.22.026141
27. Dean KM, Roudot P, Welf ES, Danuser G, Fiolka R. Deconvolution-free Subcellular Imaging with Axially Swept Light Sheet Microscopy. Biophysical J (2015) 108(12):2807–15. doi:10.1016/j.bpj.2015.05.013
28. Duocastella M, Arnold CB, Puchalla J. Selectable Light-Sheet Uniformity Using Tuned Axial Scanning. Microsc Res Tech (2016).
29. Chakraborty T, Driscoll MK, Jeffery E, Murphy MM, Roudot P, Chang B-J, et al. . Light-sheet Microscopy of Cleared Tissues with Isotropic, Subcellular Resolution. Nat Methods (2019) 16(11):1109–13. doi:10.1038/s41592-019-0615-4
30. Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, et al. . Rapid Three-Dimensional Isotropic Imaging of Living Cells Using Bessel Beam Plane Illumination. Nat Methods (2011) 8(5):417–23. doi:10.1038/nmeth.1586
31. Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. . Lattice Light-Sheet Microscopy: Imaging Molecules to Embryos at High Spatiotemporal Resolution. Science (2014) 346(6208):1257998. doi:10.1126/science.1257998
32. Aakhte M, Akhlaghi EA, Müller H-AJ. SSPIM: a Beam Shaping Toolbox for Structured Selective Plane Illumination Microscopy. Sci Rep (2018) 8(1):10067. doi:10.1038/s41598-018-28389-8
33. Wu Y, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, et al. . Inverted Selective Plane Illumination Microscopy (iSPIM) Enables Coupled Cell Identity Lineaging and Neurodevelopmental Imaging in Caenorhabditis elegans. Proc Natl Acad Sci (2011) 108(43):17708–13. doi:10.1073/pnas.1108494108
34. McGorty R, Liu H, Kamiyama D, Dong Z, Guo S, Huang B. Open-top Selective Plane Illumination Microscope for Conventionally Mounted Specimens. Opt Express (2015) 23(12):16142. doi:10.1364/oe.23.016142
35. Kim B, Na M, Park S, Kim K, Park J-H, Chung E, et al. . Open-top Axially Swept Light-Sheet Microscopy. Biomed Opt Express (2021) 12(4):2328–38. doi:10.1364/boe.419030
36. Barner LA, Glaser AK, True LD, Reder NP, Liu JTC. Solid Immersion Meniscus Lens (SIMlens) for Open-Top Light-Sheet Microscopy. Opt Lett (2019) 44(18):4451–4. doi:10.1364/ol.44.004451
37. Glaser AK, Reder NP, Chen Y, Yin C, Wei L, Kang S, et al. . Multi-immersion Open-Top Light-Sheet Microscope for High-Throughput Imaging of Cleared Tissues. Nat Commun (2019) 10(1):2781. doi:10.1038/s41467-019-10534-0
38. Mcgorty R, Xie D, Huang B. High-NA Open-Top Selective-Plane Illumination Microscopy for Biological Imaging. Opt Express (2017) 25(15):17798–810. doi:10.1364/oe.25.017798
39. Vladimirov N, Preusser F, Wisniewski J, Yaniv Z, Desai RA, Woehler A, et al. . Dual-view Light-Sheet Imaging through a Tilted Glass Interface Using a Deformable Mirror. Biomed Opt Express (2021) 12(4):2186–203. doi:10.1364/boe.416737
40. Dunsby C. Optically Sectioned Imaging by Oblique Plane Microscopy. Opt Express (2008) 16(25):20306–16. doi:10.1364/oe.16.020306
41. Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly Inclined Thin Illumination Enables clear Single-Molecule Imaging in Cells. Nat Methods (2008) 5(2):159–61. doi:10.1038/nmeth1171
42. Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. Aberration-free Optical Refocusing in High Numerical Aperture Microscopy. Opt Lett (2007) 32(14):2007–9. doi:10.1364/ol.32.002007
43. Kumar S, Wilding D, Sikkel MB, Lyon AR, MacLeod KT, Dunsby C. High-speed 2D and 3D Fluorescence Microscopy of Cardiac Myocytes. Opt Express (2011) 19(15):13839–47. doi:10.1364/oe.19.013839
44. Cutrale F, Gratton E. Inclined Selective Plane Illumination Microscopy Adaptor for Conventional Microscopes. Microsc Res Tech (2012) 75(11):1461–6. doi:10.1002/jemt.22089
45. Li T, Ota S, Kim J, Wong ZJ, Wang Y, Yin X, et al. . Axial Plane Optical Microscopy. Sci Rep (2014) 4(1):7253. doi:10.1038/srep07253
46. Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, et al. . Swept Confocally-Aligned Planar Excitation (SCAPE) Microscopy for High-Speed Volumetric Imaging of Behaving Organisms. Nat Photon (2015) 9(2):113–9. doi:10.1038/nphoton.2014.323
47. Voleti V, Patel KB, Li W, Perez Campos C, Bharadwaj S, Yu H, et al. . Real-time Volumetric Microscopy of In Vivo Dynamics and Large-Scale Samples with SCAPE 2.0. Nat Methods (2019) 16(10):1054–62. doi:10.1038/s41592-019-0579-4
48. Kim J, Wojcik M, Wang Y, Moon S, Zin EA, Marnani N, et al. . Oblique-plane Single-Molecule Localization Microscopy for Tissues and Small Intact Animals. Nat Methods (2019) 16(9):853–7. doi:10.1038/s41592-019-0510-z
49. Yang B, Chen X, Wang Y, Feng S, Pessino V, Stuurman N, et al. . Epi-illumination SPIM for Volumetric Imaging with High Spatial-Temporal Resolution. Nat Methods (2019) 16(6):501–4. doi:10.1038/s41592-019-0401-3
50. Sapoznik E, Chang BJ, Huh J, Ju RJ, Azarova EV, Pohlkamp T, et al. . A Versatile Oblique Plane Microscope for Large-Scale and High-Resolution Imaging of Subcellular Dynamics. eLife (2020) 9:e57681. doi:10.7554/eLife.57681
51. Sparks H, Dent L, Bakal C, Behrens A, Salbreux G, Dunsby C, et al. . Dual-view Oblique Plane Microscopy (dOPM). Biomed Opt Express (2020) 11(12):7204–20. doi:10.1364/boe.409781
52. Shao W, Kilic K, Yin W, Wirak G, Qin X, Feng H, et al. . Wide Field-Of-View Volumetric Imaging by a Mesoscopic Scanning Oblique Plane Microscopy with Switchable Objective Lenses. Quantitative Imaging Med Surg (2021) 11(3):98397–997. doi:10.21037/qims-20-806
53. Yang B, Lange M, Millett-Sikking A, Solak AC, Kumar SV, Wang W, et al. . High-Resolution, Large Imaging Volume, and Multi-View Single Objective Light-Sheet Microscopy. bioRxiv (2021).
54. Kumar M, Kishore S, Nasenbeny J, McLean DL, Kozorovitskiy Y. Integrated One- and Two-Photon Scanned Oblique Plane Illumination (SOPi) Microscopy for Rapid Volumetric Imaging. Opt Express (2018) 26(10):13027–41. doi:10.1364/oe.26.013027
55. Shin Y, Kim D, Kwon H-S. Oblique Scanning 2-photon Light-Sheet Fluorescence Microscopy for Rapid Volumetric Imaging. J Biophotonics (2018) 11(5):e201700270. doi:10.1002/jbio.201700270
56. Dijksman JA, Rietz F, Lőrincz KA, van Hecke M, Losert W. Invited Article: Refractive index Matched Scanning of Dense Granular Materials. Rev Scientific Instr (2012) 83(1):011301. doi:10.1063/1.3674173
57. Salili SM, Harrington M, Durian DJ. Note: Eliminating Stripe Artifacts in Light-Sheet Fluorescence Imaging. Rev Scientific Instr (2018) 89(3):036107. doi:10.1063/1.5016546
58. Ricci P, Gavryusev V, Müllenbroich C, Turrini L, de Vito G, Silvestri L, et al. . Removing Striping Artifacts in Light-Sheet Fluorescence Microscopy: a Review. Prog Biophys Mol Biol (2021). doi:10.1016/j.pbiomolbio.2021.07.003
59. Huang AYL, Huang MYF, Capart H, Chen R-H. Optical Measurements of Pore Geometry and Fluid Velocity in a Bed of Irregularly Packed Spheres. Exp Fluids (2008) 45(2):309–21. doi:10.1007/s00348-008-0480-x
60. Loftus AF, Noreng S, Hsieh VL, Parthasarathy R. Robust Measurement of Membrane Bending Moduli Using Light Sheet Fluorescence Imaging of Vesicle Fluctuations. Langmuir (2013) 29(47):14588–94. doi:10.1021/la403837d
61. Jahl PE, Parthasarathy R. Lipid Bilayer Hydrodynamic Drag. Phys Rev Res (2020) 2(1):013132. doi:10.1103/physrevresearch.2.013132
62. Ritter JG, Veith R, Siebrasse J-P, Kubitscheck U. High-contrast Single-Particle Tracking by Selective Focal Plane Illumination Microscopy. Opt Express (2008) 16(10):7142–52. doi:10.1364/oe.16.007142
63. Anderson SJ, Matsuda C, Garamella J, Peddireddy KR, Robertson-Anderson RM, McGorty R. Filament Rigidity Vies with Mesh Size in Determining Anomalous Diffusion in Cytoskeleton. Biomacromolecules (2019) 20:4380–4388. doi:10.1021/acs.biomac.9b01057
64. Aarts DGAL, Schmidt M, Lekkerkerker HNW. Direct Visual Observation of thermal Capillary Waves. Science (2004) 304:847–50. doi:10.1126/science.1097116
66. Strack R. Single-objective Light Sheet Microscopy. Nat Methods (2021) 18(1):28. doi:10.1038/s41592-020-01027-w
67. Rietz F, Radin C, Swinney HL, Schröter M. Nucleation in Sheared Granular Matter. Phys Rev Lett (2018) 120(5):055701. doi:10.1103/PhysRevLett.120.055701
68. Houssais M, Ortiz CP, Durian DJ, Jerolmack DJ. Onset of Sediment Transport Is a Continuous Transition Driven by Fluid Shear and Granular Creep. Nat Commun (2015) 6(1):6527. doi:10.1038/ncomms7527
69. Harrington M, Lin M, Nordstrom KN, Losert W. Experimental Measurements of Orientation and Rotation of Dense 3D Packings of Spheres. Granular Matter (2014) 16(2):185–91. doi:10.1007/s10035-013-0474-0
70. Philippe P, Badiane M. Localized Fluidization in a Granular Medium. Phys Rev E Stat Nonlin Soft Matter Phys (2013) 87(4):042206. doi:10.1103/PhysRevE.87.042206
71. Taormina MJ, Hay EA, Parthasarathy R. Passive and Active Microrheology of the Intestinal Fluid of the Larval Zebrafish. Biophysical J (2017) 113(4):957–65. doi:10.1016/j.bpj.2017.06.069
72. Jiang H, Zhu T, Zhang H, Nie J, Guan Z, Ho C-M, et al. . Droplet-based Light-Sheet Fluorescence Microscopy for High-Throughput Sample Preparation, 3-D Imaging and Quantitative Analysis on a Chip. Lab Chip (2017) 17(13):2193–7. doi:10.1039/c7lc00164a
Keywords: light sheet fluorescence microscopy, microscopy, 3D imaging, colloidal fluids, capillary waves
Citation: You R and McGorty R (2021) Light Sheet Fluorescence Microscopy Illuminating Soft Matter. Front. Phys. 9:760834. doi: 10.3389/fphy.2021.760834
Received: 18 August 2021; Accepted: 28 September 2021;
Published: 12 October 2021.
Edited by:
Marco Laurati, University of Florence, ItalyReviewed by:
Erick Sarmiento-Gomez, University of Guanajuato, MexicoBenoit Loppinet, Foundation for Research and Technology Hellas (FORTH), Greece
Copyright © 2021 You and McGorty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryan McGorty, cm1jZ29ydHlAc2FuZGllZ28uZWR1
 Ruilin You
Ruilin You Ryan McGorty
Ryan McGorty