- 1Research Center of Complex Systems Science, University of Shanghai for Science and Technology, Shanghai, China
- 2College of Information Engineering, Yangzhou University, Yangzhou, China
- 3Institute of Accounting and Finance, Shanghai University of Finance and Economics, Shanghai, China
- 4Shanghai Engineering Research Center of Finance Intelligence, Shanghai University of Finance and Economics, Shanghai, China
In this paper, we investigate the reconstruction of networks based on priori structure information by the Element Elimination Method (EEM). We firstly generate four types of synthetic networks as small-world networks, random networks, regular networks and Apollonian networks. Then, we randomly delete a fraction of links in the original networks. Finally, we employ EEM, the resource allocation (RA) and the structural perturbation method (SPM) to reconstruct four types of synthetic networks with 90% priori structure information. The experimental results show that, comparing with RA and SPM, EEM has higher indices of reconstruction accuracy on four types of synthetic networks. We also compare the reconstruction performance of EEM with RA and SPM on four empirical networks. Higher reconstruction accuracy, measured by local indices of success rates, could be achieved by EEM, which are improved by 64.11 and 47.81%, respectively.
1 Introduction
Reconstructing a network based on priori structure information has attracted lots of attention for the network science [1]. Prior information about the connectivity patterns or potential interactions of the networks are accessible via public database [2, 3], high-throughput experiments [4], or data mining of interaction knowledge [5–7]. A wide diversity of methods based on priori structure information have been developed for the problem of network reconstruction [1, 8, 9]. Among various models, a few reconstruction models would provide a reliable estimate of a network’s structure with priori structure information. Link prediction is a typical method which uses accessible structure to estimate the likelihood of existence of unobserved links or identifies spurious links in a network [10, 11]. The unknown structure of a network is then reconstructed by link prediction. A few link prediction models are validated in both synthetic networks and empirical networks, which are local similarity indices [12–14], maximum likelihood methods [11, 15] and methods based on predictability [16, 17].
The other method uses accessible structure information to reconstruct a class of networks with evolutionary games [18, 19]. Such model, known as compressive sensing reconstruction model (CSR), is initially proposed to solve the problems of global network reconstruction [20–22]. The CSR method provides theoretical framework to dealing with networks purely from measured time-series information. To reconstruct a network with N nodes, the CSR method reconstructs the adjacent matrix column by column and each column is a vector with N elements [23, 24]. Contrary to the CSR method, the adjacent matrix is reconstructed by the Element Elimination Method (EEM) in a similar fashion, but the number of elements in different column might be Ni(Ni ≤ N, i = 1, 2, … , N) because EEM initially eliminates coupling nodes based on priori structure information. Exploiting the natural sparsity of the vectors, the pioneering work has applied EEM to achieve a successful reconstruction in scale-free networks with a small fraction of hubs [25]. However, in many cases, examples of real-world networks are not characterized by scale-free [26], i.e., the collaboration network of film actors [27, 28], the neural network of the worm Caenorhabditis elegans [26], the power grid of the western United States [29, 30], and drug trafficking network [31], et al. In addition, unique structure could be observed in world airline networks [32, 33] and Apollonian networks [34–36], which are characterized by scale-free and also satisfies basic features of small-world. EEM for reconstructing networks characterized by other features has not been fully explored. We are interested in, to achieve a successful reconstruction, the detailed amount of time-series information required for EEM in spite of the priori structure information. This motivates us to investigate the application of EEM to other networks characterized by different features.
In this paper, we investigate the reconstruction of general networks, which are characterized by four types of synthetic networks as small-world networks, random networks, regular networks and Apollonian networks. Typically, the reconstruction accuracy of EEM is evaluated on four types of networks. We will show the performance of EEM, characterized by low information requirements and high reconstruction accuracy. Experiments on four synthetic networks demonstrate that comparing with the resource allocation (RA) [12] and the structural perturbation method (SPM) [16], EEM can effectively enhance the reconstruction accuracy. Further, three local indices of success rates demonstrate that the reconstruction accuracy obtained by EEM when reconstructing three separately local structure in a network is close. In addition, experiments on four empirical networks demonstrate that EEM outperforms RA and SPM. Compared with RA and SPM, EEM has higher reconstruction accuracy, measured by local indices of success rates, which are improved by 64.11 and 47.81%, respectively.
2 Methods and Models
2.1 The Procedure of the Network Reconstruction
Uncovering a network’s structure has many potential applications so that we can assess the system’s resilience [37–39], understand the dynamical mechanisms [40], identify significant nodes in a network [41, 42], detect community structure [43], locate diffusion sources Hu et al. [44, 45], and analyze the networks’ properties [46–48]. In this paper, an Element Elimination Method (EEM) [25] is employed to reconstruct the structure of networks. We then give the illustration of the procedures of employing EEM to reconstruct synthetic networks: 1) Generate synthetic networks. 2) Extract time-series information from observed data. 3) Reconstruct the networks with EEM. Noting that the adjacent relationships between nodes in the network are sparse and would not change over time, we could explore the casual relationships between nodes’ time-series information. Consequently, we could uncover the unknown link set EP of the networks by EEM based on priori link set ET.
As illustrated in Figure 1, a procedure of network reconstruction is presented. Supposing the relationships between node 2 and other 5 nodes should be reconstructed, and only one adjacent relationship (a blue line in Figure 1A) is known. However, we are confused about which one is the original network from vastly different networks with possible connective relationships. Simultaneously, the network is evolving over the time, and a few time-series information of nodes’ strategies and payoffs could be obtained. We then build a model to bridge node 2’s strategies and its payoffs, as Figure 1B illustrated. Consequently, we can use EEM to reconstruct the network’s structure and obtain the adjacent relationships as shown in Figure 1C.
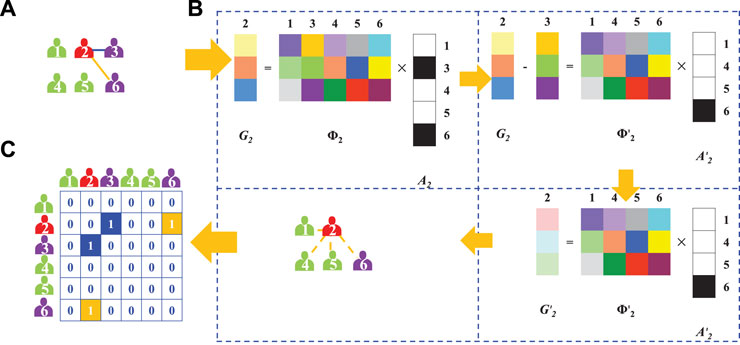
FIGURE 1. (Color Online) An illustration of reconstructing the hidden structure of a node based on priori structure information. (A) Original adjacent relationships of a node. For a node 2 in red with two neighbors, node 3 and node 6 in purple, we can observe a priori relationship, represented with a blue line, between node 2 and node 3. (B) EEM. We establish vector G2 and matrix Φ2 in the reconstruction form G2 = Φ2 ⋅A2 from time-series information, where vector A2 captures the adjacent relationships between node 2 and the other nodes. After subtracting time-series information determined by node 2 and priori neighbor, node 3, on the both sides of the equation, the unknown connections of node 2 can be reconstructed by optimizing the solution of the following equation
2.2 Generation of Synthetic Network
In order to evaluate the reconstruction performance of EEM in small-world networks and networks characterized by other features, we generate four types of synthetic networks. Noting that small-world network is a model of network that can be tuned between random network and regular network [26], we also consider the networks when their connection topology is assumed to be completely regular or completely random. Besides, the performance on the Apollonian networks by EEM has seldom been evaluated. Then, we generate four types of synthetic networks which are small-world networks, random networks, regular networks and Apollonian networks. The precedent findings indicate that the assortative coefficient has a direct influence on the accuracy of network reconstruction [49]. Therefore, some statistical properties have to be tuned when the networks are generated.
Supposing a network is composed of N nodes and |E| links. To minimize the influence from different network structure, we fix a default mean assortative coefficient
Due to privacy or confidentiality issues, the complete structure of a network is not accessible. In addition, it is an impossible mission for us to record nodes’ complete time-series information. In spite of the difficulties, some priori information about the adjacent relationships between a few nodes, and discrete records of nodes’ time-series information might be available. Despite the limited information, the connective relationships between nodes has a direct effect on the individual node, which contributes to node’s attitude or selection in the next time. The dependence from the network’s structure on nodes’ interactions provide information for us to utilize the time-series information of nodes to describe the adjacent relationships behind them [24, 50].
2.3 The Model of the Evolutionary Game
The main challenge lies in that the structure of the network is inaccessible, also in that merely limited nodes’ time-series information is available. Since the time-series information is closely related to the connective relationships between nodes, we can reconstruct the unknown structure from the limited time-series information.
We use an evolutionary game model, the Prisoner Dilemma Game (PDG) model, to describe the nodes’ dynamics [51–53]. In each round of the game, the nodes usually weigh the benefits against the risks and selects a strategy. Here, we use SYi(t) to define the strategy of node i. We denote vector SYi(t) = (1,0)T to represent a cooperation strategy, while we denote SYi(t) = (0,1)T to represent a defection strategy. Here, T stands for ‘transpose’.
When node i and node j trigger a game, the payoff of node i is dependent on both two nodes’ strategies and a uniform payoff matrix P, which is defined as:
where b (1 < b < 2) is a parameter characterizing the volume of payoff when node i select a defection strategy. In the t round, node i would play with all its different neighbors with the same strategy. When node i encounters a neighbor j, node i would gain payoff from node j as:
In the same round, node i’s total payoffs Gi would be calculated, and it is the sum of the payoffs from all node i’s neighbors.
In a new round, node i would attempt to maximize its payoffs by updating its strategy. According to Fermi rule [54], node i randomly select a node j from its neighbors after t round. In t + 1 round, node i would then adopt node j’s strategy with the probability
where TGi(t) is node i’s cumulative payoffs from 1 to t round. TGj(t) is similarly defined. Parameter κ characterizes node’s rationality when it update strategies. Parameter κ = 0 corresponds to rational selection behavior of nodes.
Since game occurs among connected nodes, the information of the adjacent relationships between nodes are hidden in their dynamical records of strategies or payoffs in the game. Then we can utilize the information to uncover a networks’ structure when we collect the time-series information about the strategies and payoffs of nodes. When we reconstruct a certain network, the limited time-series information is usually presented in a random sample of sufficient time-series information.
2.4 Element Elimination Method
Given limited time-series information of nodes, an EEM could be applied to reconstruct a network based on priori structure information. EEM is a variant of the CSR method, which utilizes priori structure information to exclude the priori connective relationships before reconstruction. Suppose that the relationships between nodes in a certain network can be represented by an adjacency matrix A with dimensions N × N, where N is the number of nodes in the network. EEM decomposes the process of reconstructing the entire network into many subnetwork recovery problems, and the network structure, namely, the adjacency matrix A, is reconstructed column by column [55,56]. An adjacency vector Ai of a node is used to describe the adjacent relationships between node i (i = 1, 2, … , N) and the other N − 1 nodes in the network, which contains no loop. The adjacency vector
The training set ET sheds light on the priori neighbor set
Most real-world networks are characterized by natural sparsity and the adjacency vector Ai of node i is sparse, which refers to vector Ai has only a few nonzero elements (i.e. aij = 1). Noting that the value of each element in node i’s priori adjacency vector
where
The elements in matrix
3 Experimental Results
3.1 Datasets
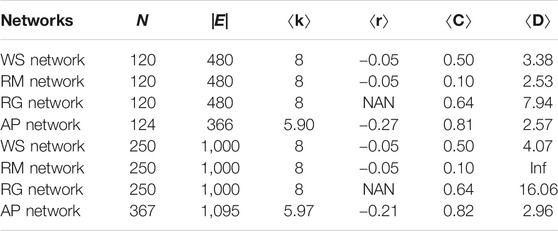
In order to understanding the performance of EEM in reconstructing the synthetic networks, the experiments are conducted in four types of networks. The basic statistical properties of the synthetic networks are presented in Table 1. N and |E| are the number of nodes and links.
We assume that the strategies and payoffs of each node in a certain round t is one piece of time-series information. In the experiments, we use M pieces of accessible time-series information obtained from discrete round t1 to round tM to reconstruct different networks. In this paper, we set N, namely the number of nodes in the network, as the maximum value of M. Then we use an index of information sufficiency η(η ≡ M/N) to represent the size of the time-series information used in the network reconstruction. Intuitively, the time-series information is sufficient when the pieces of the accessible time-series information M = N, while the time-series information is insufficient when 0 < M < N. Correspondingly, the accessible time-series information is sufficient when the index of information sufficiency η = 1 and the accessible time-series information is insufficient when 0 < η < 1. The reconstruction models are also applied to reconstruct networks with different priori information of the structure, measured by a probability Ps(0 ≤ Ps ≤ 1).
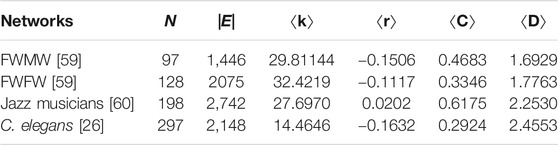
In addition, the performance of EEM is also evaluated in reconstructing the empirical networks. Table 2 shows the basic statistical properties of all four networks. These networks are chosen because they are characterized by large clustering coefficient and short distance.
3.2 Metrics
To test the EEM’s accuracy, the original existent link set, E, are randomly divided into two parts: the priori set ET, and the probe set EP. Clearly, E = ET ∪EP and ET ∩EP = ∅. In this paper, the priori set always contains Ps of links, and the remaining 1 − Ps of links constitute the probe set. We apply four standard indices to quantify the reconstruction accuracy: the success rates of existent links SR, the success rates of nonexistent links SN [24], precision PRE [61, 62] and the area under the receiver operating characteristic curve AUC [63] are applied. In addition, we apply local indices of success rates in the experiments.
Both the success rates of existent links SR and the success rates of nonexistent links SN estimate the similarity of the reconstructed networks and the original networks. The success rates of existent links SR denotes the ratio of the number of links reconstructed by the reconstruction models to the number of real existent links in the network. The success rates of nonexistent links SN denotes the ratio of the number of nonexistent links distinguished by the reconstruction models to the number of real nonexistent links in the network. We obtain
where Γio and Γir denote real neighbor set of node i and neighbor set of node i reconstructed by the reconstruction models, respectively. |⋅| denotes the number of elements in a set ⋅.
Precision PRE is defined as the ratio of existent links reconstructed by models to the number of the whole unknown existent links. In our case, to calculate precision we need to rank all the unknown links in decreasing order according to existent possibilities computed by reconstruction models. Then we focus on the top-L (here L = |EP|) links. If there are H links successfully reconstructed, then
The area under the receiver operating characteristic curve AUC evaluates the reconstruction models’ performance according to the whole unknown link list. Provided the existent possibility of all unknown links, AUC can be interpreted as the probability that a randomly chosen unknown existent link is given a higher existent possibility than a randomly chosen nonexistent link. In the implementation, the value of AUC is calculated with a function perfcurve by Matlab.
Clearly, a higher value of the success rates of existent links SR, the success rates of nonexistent links SN, precision PRE or the area under the receiver operating characteristic curve AUC means a higher reconstruction accuracy. We conduct 50 times independent simulation for averaging the indices of reconstruction accuracy as the mean success rates of existent links
To understand the reconstruction performance of EEM when reconstructing local structure of the network divide the structure of each type of network into separately local structure. Supposing that the roles of nodes in the network are leaders, brokers and peripheral executors. We denote leaders are nodes with small degrees and the number of leaders in each type of network is 6. In addition, the subnetwork composed of leaders is a connected subgraph. Then brokers are nodes which are connected with leaders, and the residual nodes are peripheral executors. The sets of leaders, brokers and peripheral executors are not overlapped. We use letters L, B and P to represent the adjacent relationships between leaders, the adjacent relationships between leaders and brokers, and the adjacent relationships among peripheral executors and brokers, respectively. Then, we could obtain the success rates of existent links of each local structure normalized by the number of real existent links |Γio| of the network.
The sum of three local success rates of existent links is equal the global success rates of existent links.
Correspondingly, the maximum of three local success rates of existent links would be
when the original network is successfully reconstructed. To quantify the success rates of three different local structure, we define local indices of success rates as follows:
Similarly, a higher value of local index of success rates APPSRL, APPSRB, or APPSRP means a higher reconstruction accuracy. We conduct 50 times independent simulation for averaging the indices of success rates
3.3 Experimental Results on Synthetic and Empirical Networks
In order to understand the performance of EEM, four types of synthetic networks hosting a PDG dynamical process are considered in our paper. Figure 2A depicts the index of reconstruction accuracy for a synthetic small-world network, measured by the mean success rates of existent links
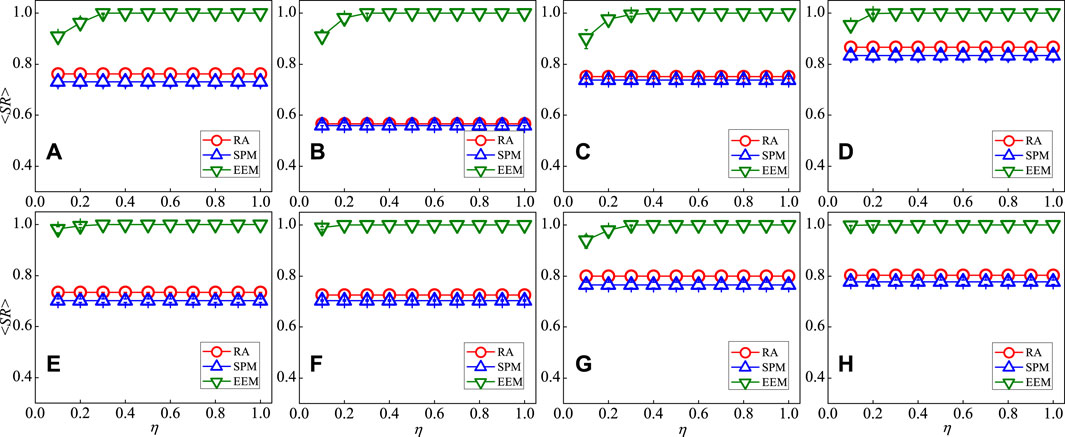
FIGURE 2. (Color Online) The mean success rates of existent links
Moreover, we compare the experimental results between EEM and two link prediction models which are the resource allocation (RA) and the structural perturbation method (SPM). Figures 2A–H show that when the index of information sufficiency η is low (i.e., η = 0.1), the mean success rates of existent links
Intuitively, a network’s structure would be accurately reconstructed when more priori information about the structure of the network are presented. Figure 3 shows the dependence of the values of
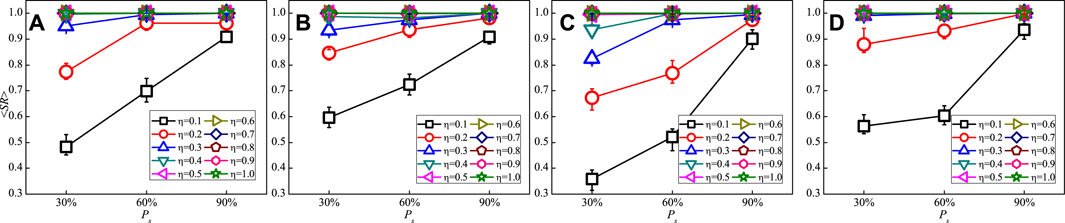
FIGURE 3. (Color Online) The mean success rates of existent links
In the following, we verify the performance of EEM in local structure of the networks. We divide the structure of each type of network into three separately local structure with subscript L, B, P for them. Figure 4A depicts reconstruction success rate of a small-world network, measured by the mean local index of success rates

FIGURE 4. (Color Online) The mean local indices of success rates
As illustrated in the main graph in Figure 4A, the mean local index of success rates
The underlying reason that EEM could obtain higher reconstruction accuracy than RA or SPM might be twofold. Firstly, EEM is applicable to reconstruct networks with sparse connective relationships because Wang et al. developed a paradigm [19, 24, 25] to address the network reconstruction problems and Candès et al. provided the theoretical framework for this paradigm [57, 58]. Both EEM and two link prediction models utilize the identical priori structure information of the network to obtain direct information of the unknown structure. In addition, EEM bridges the relationships between the nodes’ payoffs and strategies by virtue of time-series information because the payoffs can merely be obtained from each node’s neighbors. Then EEM could extract indirect information of the unknown structure from the above relationships which strengthens the reliability of the experimental results. RA and SPM could also extract valuable indirect information of the unknown structure, but the valuable information still originates from the priori structure information of the network due to lack of a universal theoretical framework.
Secondly, both the reconstruction accuracy of the local structure and the reconstruction accuracy of the global structure obtained by EEM highly consist. As illustrated in Figure 4, the absolute error between three mean local index of success rates
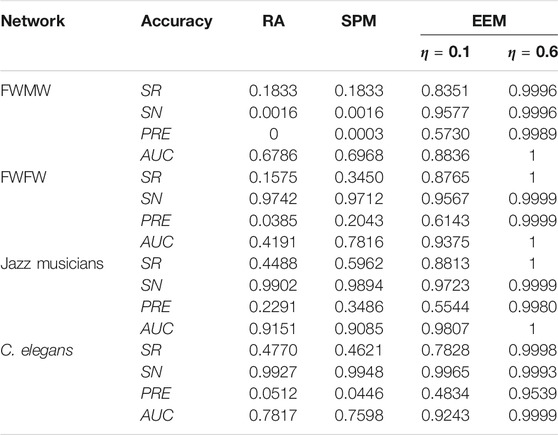
Finally, we test the results for four empirical networks. As shown in Table 3, we reconstruct the network structure by EEM, RA and SPM with 90% priori structure information. The empirical results indicate that four indices of reconstruction accuracy obtained by EEM are higher than RA and SPM for four empirical networks when the index of information sufficiency rate η = 0.1. Four indices of reconstruction accuracy obtained by EEM are higher than RA and SPM. Compared with RA, EEM’s reconstruction accuracy, measured by the mean success rates of existent links
3.4 Conclusion
In summary, we have investigated the performance of EEM for reconstructing synthetic networks, which are characterized by four types of networks as small-world networks, random networks, regular networks and Apollonian networks, based on priori structure information. The mean success rates of existent links
Although the efficiency of EEM has been measured in reconstructing network’s structure with both synthetic networks and empirical networks, there are still a lot of questions to be considered further. For example, the results show that EEM can give remarkably higher reconstruction accuracy on a network hosing a PDG dynamical process, but the performance of EEM has not been validated under another dynamical process. Although EEM could also be extended to cases with large-scale network, the computing time might increase exponentially. In addition, EEM’s capability to identify spurious links has not been explored. Noting that EEM can well capture the adjacent relationships from limited information and thus give more accurate reconstruction, such features make EEM appealing to reconstructing general networks with extremely low data requirement. Despite underlying challenges, we will make attempt to continue our research referring to the problems of network reconstruction.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
J-QF provided this topic and wrote the paper. QG, KY and J-GL guided, discussed and modified the manuscript. All authors contributed to manuscript and approved the submission version.
Funding
This work is supported by the National Natural Science Foundation of China (Grant Nos. 71771152 and 61773248), the National Social Science Fund of China (No.16BJY158), the Major Program of National Fund of Philosophy and Social Science of China (Nos. 20ZDA060 and 18ZDA088), and the Scientific Research Project of Shanghai Science and Technology Committee (Grant No. 19511102202).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the valuable discussion with Huan-Mei Qin, Guang Liang, Ren-De Li, Hong-Yi Ding, Shao-Yong Han.
References
1. Liao JC, Boscolo R, Yang Y-L, Tran LM, Sabatti C, Roychowdhury VP. Network Component Analysis: Reconstruction of Regulatory Signals in Biological Systems. Proc Natl Acad Sci (2003) 100:15522–7. doi:10.1073/pnas.2136632100
2. Matys V, Fricke E, Geffers R, Göβling E, Haubrock M, Hehl R. TRANSFAC(R): Transcriptional Regulation, from Patterns to Profiles. Nucleic Acids Res (2003) 31:374–8. doi:10.1093/nar/gkg108
3. Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT. Ecocyc: A Comprehensive Database Resource for Escherichia Coli. Nucleic Acids Res (2004) 33:D334–D337. doi:10.1093/nar/gki108
4. Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK. Transcriptional Regulatory Networks in Saccharomyces Cerevisiae. Science (2002) 298:799–804. doi:10.1126/science.1075090
5. Dong GG, Wang F, Shekhtmane LM, Danziger MM, Fan JF, Du RJ. Optimal Resilience of Modular Interacting Networks. Proc Natl Acad Sci USA. (2021) 118:e1922831118. doi:10.1073/pnas.1922831118
6. Bussemaker HJ, Li H, Siggia ED. Building a Dictionary for Genomes: Identification of Presumptive Regulatory Sites by Statistical Analysis. Proc Natl Acad Sci (2000) 97:10096–100. doi:10.1073/pnas.180265397
7. Bussemaker HJ, Li H, Siggia ED. Regulatory Element Detection Using Correlation With Expression. Nat Genet (2001) 27:167–71. doi:10.1038/84792
8. Chang C, Ding Z, Hung YS, Fung PCW. Fast Network Component Analysis (Fastnca) for Gene Regulatory Network Reconstruction From Microarray Data. Bioinformatics (2008) 24:1349–58. doi:10.1093/bioinformatics/btn131
9. Cugueró-Escofet MÀ, Quevedo J, Alippi C, Roveri M, Puig V, García D. Model- vs. Data-Based Approaches Applied to Fault Diagnosis in Potable Water Supply Networks. J Hydroinformatics (2016) 18:831–50. doi:10.2166/hydro.2016.218
10. Lü L, Zhou T. Link Prediction in Complex Networks: A Survey. Physica A: Stat Mech Its Appl (2011) 390:1150–70. doi:10.1016/j.physa.2010.11.027
11. Clauset A, Moore C, Newman MEJ. Hierarchical Structure and the Prediction of Missing Links in Networks. Nature (2008) 453:98–101. doi:10.1038/nature06830
12. Zhou T, Lü L, Zhang Y-C. Predicting Missing Links via Local Information. Eur Phys J B (2009) 71:623–30. doi:10.1140/epjb/e2009-00335-8
13. Lü L, Jin C-H, Zhou T. Similarity Index Based on Local Paths for Link Prediction of Complex Networks. Phys Rev E (2009) 80:046122. doi:10.1103/PhysRevE.80.046122
14. Liben-Nowell D, Kleinberg J. The Link-Prediction Problem for Social Networks. J Am Soc Inf Sci (2007) 58:1019–31. doi:10.1002/asi.20591
15. Guimerà R, Sales-Pardo M. Missing and Spurious Interactions and the Reconstruction of Complex Networks. Proc Natl Acad Sci (2009) 106:22073–8. doi:10.1073/pnas.0908366106
16. Lü L, Pan L, Zhou T, Zhang Y-C, Stanley HE. Toward Link Predictability of Complex Networks. Proc Natl Acad Sci USA (2015) 112:2325–30. doi:10.1073/pnas.1424644112
17. Sun J, Feng L, Xie J, Ma X, Wang D, Hu Y. Revealing the Predictability of Intrinsic Structure in Complex Networks. Nat Commun (2020) 11:1–10. doi:10.1038/s41467-020-14418-6
18. Zhang H-F, Wang W-X. Complex System Reconstruction. Acta Physica Sinica (2020) 69:088906. doi:10.7498/aps.69.20200001
19. Wang W-X, Lai Y-C, Grebogi C. Data Based Identification and Prediction of Nonlinear and Complex Dynamical Systems. Phys Rep (2016) 644:1–76. doi:10.1016/j.physrep.2016.06.004
20. Xu M, Xu C-Y, Wang H, Li Y-K, Hu J-B, Cao K-F. Global and Partitioned Reconstructions of Undirected Complex Networks. Eur Phys J B (2016) 89:1–6. doi:10.1140/epjb/e2016-60956-2
21. Barranca VJ, Zhou D. Compressive Sensing Inference of Neuronal Network Connectivity in Balanced Neuronal Dynamics. Front Neurosci (2019) 13:1101. doi:10.3389/fnins.2019.01101
22. Li R-D, Guo Q, Ma H-T, Liu J-G. Network Reconstruction of Social Networks Based on the Public Information. Chaos (2021) 31:033123. doi:10.1063/5.0038816
23. Shen Z, Wang W-X, Fan Y, Di Z, Lai Y-C. Reconstructing Propagation Networks With Natural Diversity and Identifying Hidden Sources. Nat Commun (2014) 5:1–10. doi:10.1038/ncomms5323
24. Wang W-X, Lai Y-C, Grebogi C, Ye J. Network Reconstruction Based on Evolutionary-Game Data via Compressive Sensing. Phys Rev X (2011) 1:021021. doi:10.1103/PhysRevX.1.021021
25. Ma L, Han X, Shen Z, Wang W-X, Di Z. Efficient Reconstruction of Heterogeneous Networks From Time Series via Compressed Sensing. PLoS One (2015) 10:e0142837. doi:10.1371/journal.pone.0142837
26. Watts DJ, Strogatz SH. Collective Dynamics of 'Small-World' Networks. Nature (1998) 393:440–2. doi:10.1038/30918
27. Amaral LAN, Scala A, Barthélémy M, Stanley HE. Classes of Small-World Networks. Proc Natl Acad Sci (2000) 97:11149–52. doi:10.1073/pnas.200327197
28. Ravasz E, Barabási A-L. Hierarchical Organization in Complex Networks. Phys Rev E (2003) 67. doi:10.1103/PhysRevE.67.026112
29. Albert R, Albert I, Nakarado GL. Structural Vulnerability of the North American Power Grid. Phys Rev E Stat Nonlin Soft Matter Phys (2004) 69:025103. doi:10.1103/PhysRevE.69.025103
30. Crucitti P, Latora V, Marchiori M. A Topological Analysis of the Italian Electric Power Grid. Physica A: Stat Mech Its Appl (2004) 338:92–7. doi:10.1016/j.physa.2004.02.029
31. Bright D, Koskinen J, Malm A. Illicit Network Dynamics: The Formation and Evolution of a Drug Trafficking Network. J Quant Criminol (2019) 35:237–58. doi:10.1007/s10940-018-9379-8
32. Barrat A, Barthélemy M, Pastor-Satorras R, Vespignani A. The Architecture of Complex Weighted Networks. Proc Natl Acad Sci (2004) 101:3747–52. doi:10.1073/pnas.0400087101
33. Verma T, Araújo NAM, Nagler J, Andrade JS, Herrmann HJ. Model for the Growth of the World Airline Network. Int J Mod Phys C (2016) 27:1650141. doi:10.1142/S0129183116501412
34. Soares DJB, Andrade JS, Herrmann HJ, da Silva LR. Three-Dimensional Apollonian Networks. Int J Mod Phys C (2006) 17:1219–26. doi:10.1142/S0129183106009175
35. Andrade RFS, Andrade JS, Herrmann HJ. Ising Model on the Apollonian Network With Node-Dependent Interactions. Phys Rev E (2009) 79:036105. doi:10.1103/PhysRevE.79.036105
36. Araújo NA, Andrade RFS, Herrmann HJ. Q-State Potts Model on the Apollonian Network. Phys Rev E (2010) 82:046109. doi:10.1103/physreve.82.046109
37. Dong G, Wang F, Shekhtman LM, Danziger MM, Fan J, Du R. Optimal Resilience of Modular Interacting Networks. Proc Natl Acad Sci USA (2021) 118:e1922831118. doi:10.1073/pnas.1922831118
38. Dong G, Fan J, Shekhtman LM, Shai S, Du R, Tian L. Resilience of Networks With Community Structure Behaves as if Under an External Field. Proc Natl Acad Sci USA (2018) 115:6911–5. doi:10.1073/pnas.1801588115
39. Gao J, Barzel B, Barabási A-L. Universal Resilience Patterns in Complex Networks. Nature (2016) 530:307–12. doi:10.1038/nature16948
40. Xu X, Zhang J, Small M. Superfamily Phenomena and Motifs of Networks Induced From Time Series. Proc Natl Acad Sci (2008) 105:19601–5. doi:10.1073/pnas.0806082105
41. Ren Z-M. Age Preference of Metrics for Identifying Significant Nodes in Growing Citation Networks. Physica A: Stat Mech its Appl (2019) 513:325–32. doi:10.1016/j.physa.2018.09.001
42. Liu J-G, Ren Z-M, Guo Q. Ranking the Spreading Influence in Complex Networks. Physica A: Stat Mech its Appl (2013) 392:4154–9. doi:10.1016/j.physa.2013.04.037
43. Pan Y, Li D-H, Liu J-G, Liang J-Z. Detecting Community Structure in Complex Networks via Node Similarity. Physica A: Stat Mech its Appl (2010) 389:2849–57. doi:10.1016/j.physa.2010.03.006
44. Hu Z-L, Shen Z, Tang C-B, Xie B-B, Lu J-F. Localization of Diffusion Sources in Complex Networks With Sparse Observations. Phys Lett A (2018) 382:931–7. doi:10.1016/j.physleta.2018.01.037
45. Hu Z-L, Han X, Lai Y-C, Wang W-X. Optimal Localization of Diffusion Sources in Complex Networks. R Soc Open Sci (2017) 4:170091. doi:10.1098/rsos.170091
46. Wang X, Su J, Ma F, Yao B. Mean First-Passage Time on Scale-free Networks Based on Rectangle Operation. Front Phys (2021) 9:238. doi:10.3389/fphy.2021.675833
47. Ren Z-M, Zeng A, Zhang Y-C. Bridging Nestedness and Economic Complexity in Multilayer World Trade Networks. Humanit Soc Sci Commun (2020) 7:1–8. doi:10.1057/s41599-020-00651-3
48. Buldyrev SV, Parshani R, Paul G, Stanley HE, Havlin S. Catastrophic cascade of Failures in Interdependent Networks. Nature (2010) 464:1025–8. doi:10.1038/nature08932
49. Guo Q, Liang G, Fu JQ, Han JT, Liu JG. Roles of Mixing Patterns in the Network Reconstruction. Phys Rev E (2016) 94:052303. doi:10.1103/PhysRevE.94.052303
50. Han X, Shen Z, Wang W-X, Lai Y-C, Grebogi C. Reconstructing Direct and Indirect Interactions in Networked Public Goods Game. Sci Rep (2016) 6:1–12. doi:10.1038/srep30241
51. Nowak MA, May RM. Evolutionary Games and Spatial Chaos. Nature (1992) 359:826–9. doi:10.1038/359826a0
52. Rong Z, Yang H-X, Wang W-X. Feedback Reciprocity Mechanism Promotes the Cooperation of Highly Clustered Scale-free Networks. Phys Rev E (2010) 82:047101. doi:10.1103/PhysRevE.82.047101
53. Tang Y, Jing M, Yu Y. Conditional Neutral Reward Promotes Cooperation in the Spatial Prisoner's Dilemma Game. Front Phys (2021) 9:79. doi:10.3389/fphy.2021.639252
54. Szabó G, Tőke C. Evolutionary Prisoner's Dilemma Game on a Square Lattice. Phys Rev E (1998) 58:69–73. doi:10.1103/PhysRevE.58.69
55. Wang W-X, Yang R, Lai Y-C, Kovanis V, Harrison MAF. Time-Series-Based Prediction of Complex Oscillator Networks via Compressive Sensing. Epl (Europhysics Letters) (2011) 94:48006. doi:10.1209/0295-5075/94/48006
56. Han X, Shen Z, Wang WX, Di Z. Robust Reconstruction of Complex Networks From Sparse Data. Phys Rev Lett (2015) 114:028701. doi:10.1103/PhysRevLett.114.028701
57. Candès EJ, Romberg J, Tao T. Robust Uncertainty Principles: Exact Signal Reconstruction from Highly Incomplete Frequency Information. IEEE Trans Inform Theor (2006) 52:489–509. doi:10.1109/TIT.2005.862083
58. Candès EJ, Wakin MB. An Introduction to Compressive Sampling. IEEE Signal Process Mag (2008) 25:21–30. doi:10.1109/msp.2007.914731
59. Baird D, Luczkovich J, Christian RR. Assessment of Spatial and Temporal Variability in Ecosystem Attributes of the St marks National Wildlife Refuge, Apalachee bay, florida. Estuarine, Coastal Shelf Sci (1998) 47:329–49. doi:10.1006/ecss.1998.0360
60. Gleiser PM, Danon L. Community Structure in Jazz. Advs Complex Syst (2003) 06:565–73. doi:10.1142/S0219525903001067
61. Zhou T, Ren J, Medo M, Zhang YC. Bipartite Network Projection and Personal Recommendation. Phys Rev E Stat Nonlin Soft Matter Phys (2007) 76:046115. doi:10.1103/PhysRevE.76.046115
62. Liu JG, Shi K, Guo Q. Solving the Accuracy-Diversity Dilemma via Directed Random Walks. Phys Rev E (2012) 85:016118. doi:10.1103/PhysRevE.85.016118
Keywords: network reconstruction, element elimination method, priori structure information, time-series information, evolutionary game
Citation: Fu J-Q, Guo Q, Yang K and Liu J-G (2021) Network Reconstruction in Terms of the Priori Structure Information. Front. Phys. 9:732835. doi: 10.3389/fphy.2021.732835
Received: 29 June 2021; Accepted: 27 July 2021;
Published: 11 August 2021.
Edited by:
Mahdi Jalili, RMIT University, AustraliaReviewed by:
Francisco Welington Lima, Federal University of Piauí, BrazilKe Hu, Xiangtan University, China
Copyright © 2021 Fu, Guo, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Guo Liu, bGl1amcwMDRAdXN0Yy5lZHUuY24=
 Jia-Qi Fu
Jia-Qi Fu Qiang Guo1
Qiang Guo1 Kai Yang
Kai Yang Jian-Guo Liu
Jian-Guo Liu