- Department of Chemistry, The University of Hong Kong, Hong Kong, China
Introducing the concept of programmability paves the way for designing complex and intelligent materials, where the materials’ structural information is pre-encoded in the components that build the system. With highly tunable interactions, DNA-coated particles are promising building elements to program materials at the colloidal scale, but several grand challenges have prevented them from assembling into the desired structures and phases. In recent years, the field has seen significant progress in tackling these challenges, which has led to the realization of numerous colloidal structures and dynamics previously inaccessible, including the desirable colloidal diamond structure, that are useful for photonic and various other applications. We review this exciting progress, focusing in detail on how DNA-coated colloids can be designed to have a sophisticatedly tailored surface, shape, patches, as well as controlled kinetics, which are key factors that allow one to program in principle a limitless number of structures. We also share our view on how the field may be directed in future.
Introduction
DNA exists in almost all living creatures. It carries the genetic information of life through sequences of nucleotides, which program the diverse and sophisticated structures and functions of biomolecules. Yet, to materials scientists, who are concerned with how to arrange synthetic building blocks of various forms (molecules, polymers, and particles) into well-defined, complex structures, DNA means something more [1–4]. DNA strands with complementary sequences can form duplexes, a process driven by Watson-Crick base pairing, which is specific, accurate, reversible, tunable in strength, and most importantly, programmable by the DNA sequences [5, 6]. DNA is thus an ideal candidate that, once introduced in synthetic materials, can precisely regulate the interactions between the material’s building components and dictate their structures [7–11]. This philosophy has been the cornerstone for DNA nanotechnology, which utilizes DNA and its special properties for advancing materials design and engineering [2, 12–21].
While DNA has been used alone or combined with a number of synthetic materials, its integration with colloids to promote colloidal assembly has been of great interest for the soft matter community [4, 22–26]. Colloids are particles made from almost all compositions (metals, semiconductors, polymers, and oxides), with sizes ranging from several nanometers to several micrometers. The self-assembly of colloidal particles into superstructures holds much potential in applications including plasmonic [27], photonics [28], catalysis [11], and sensing [29]. Conventionally, the word “colloids” specifically refers to particles that are micrometers or sub-micrometers in size, whereas “nanoparticles” are those generally below 100 nm. The size difference has distinguished colloids with nanoparticles when it comes to the interparticle interactions and its range. For example, the range of DNA-mediated interaction between particles depends on the length of DNA, ranging from several nanometers to a few tens of nanometers. It is a short-ranged interaction for 1-µm particles but not so for 10-nm particles. In this context, while DNA was introduced in the nanoparticle systems as early as 1996 by tethering oligonucleotide strands on the nanoparticle surface to cause particle assembly [30, 31], the utilization of DNA on colloids and for a similar purpose have been much delayed [32–35].
Ideally, colloids coated with complementary DNA strands on the surface can assemble when the DNA form bridges. Although the interaction can be finely tuned by adjusting the DNA sequence, the self-assembly of spherical colloids (e.g., silica or polymer latex) only leads to structures analogous to those of metals (e.g., the face-centered cubic structure, or FCC) or ionic solids (e.g., cesium chloride). These structures can already be produced when capillary forces or Coulombic forces are utilized [36, 37]. To fully program the assembled structures to mimic the vast spectrum of atomic and molecular crystals, additional information is necessary [38]. In 2012, Wang and Pine et al. [39] introduced the concept of “colloids with valence”, or DNA-coated patchy particles, which laid down the grounds for realizing diverse colloidal structures found in atomic and molecular systems. In this case, colloids with localized surface patches are fabricated, to which DNA is site-specifically grafted. The anisotropic patch arrangement together with short-ranged DNA interaction provide essential information that endows colloids with directional and specific interactions, akin to the covalent bonds of atoms and molecules. For example, these particles have shown to form “colloidal molecules” and “polymers”. In particular, the particles with four patches in a tetrahedral geometry are the colloidal analogue of sp3 carbon atoms, which opened the possibility of making a diamond structure of colloids and injected enormous excitement into the field. Compared to the close-packed FCC structure, a diamond structure is of a four-fold coordination, having a packing fraction as low as 34%. When colloids several hundred nanometers in size are arranged this way, the assemblies possess complete photonic band gaps in the visible wavelength and are potentially useful for photonic materials that many have dreamed of [40–43].
However, diamond and other interesting structures do not just appear when DNA-patchy particles are mixed in a jar and maintained at a comfortable temperature. It was not until very recently that He and Pine et al. [44] reported its successful formation (via the patchy particle approach). To arrive at this point, the community faced two grand challenges. First, the DNA-coated colloids had failed to crystallize, meaning that they generally formed kinetically arrested structures rather than the equilibrium assemblies [25, 35, 45–50]. Second, the directing information coded in the particle surface/shape is still insufficient to program the desired structures. This paper describes these challenges and reviews the important progresses in tackling them, which eventually allowed the engineering of programmable materials by the self-assembly of DNA-coated colloids. Reviews that discuss other aspects of the field are available elsewhere [4, 22, 23, 26].
Crystallization of DNA-Coated Colloids
Although functionalizing nanoparticles with DNA can program a wide spectrum of three-dimensional crystal structures [30, 31, 51, 52], assembling micrometer-sized DNA-coated colloids into equilibrium crystalline phases has been far more challenging. DNA-colloids normally collide and attach like Velcro, forming random aggregates that are unable to further anneal towards the preprogrammed structures, which is known as the “hit and stick” problem [45–47, 53]. Although the bound particles can disassemble at an elevated temperature, they have difficulty in rolling over one another while they are bound, which limits the relative diffusion and leads to a kinetically trapped amorphous state. Several factors have been blamed for contributing to the particles’ inability to diffuse and rearrange, including the narrow temperature window for annealing and the inhomogeneity of the DNA coatings, among others [50, 54]. This had remained a serious issue for more than ten years since DNA was first introduced to colloids in around 2003 [32–35].
DNA-coated colloids condense at low temperatures by forming DNA bridges and disperse at above the melting temperature [47]. The transition occurs within a sharp temperature window [55]. The effective interaction between DNA-coated colloids has been modeled by several groups [56–60]. Assuming F to be the interparticle attractive potential, p to be the probability of a DNA sticky strand forming a bridge, N to be the total number of DNA strands on a colloid, k to be the Boltzmann constant, r to be the particle separation distance and T to be the temperature, then
In addition, the diffusion of bound particles can be limited by the inhomogeneity of the DNA strands grafted on the colloid surface. This imperfection may be introduced during the particle synthesis and DNA grafting. Consequently, it creates unwanted “patches” on the surface with high/low DNA areal density where the bound particle prefers to stay/escape. The problem is aggravated given the fact that the size of the DNA strands is much smaller than the colloid. The thin DNA coating is more sensitive to the roughness and inhomogeneity of the particles.
In addressing the mentioned issues, Leunissen and co-workers [50] hypothesized that the bound particles would diffuse and anneal if the DNA strands could move freely on the colloid surface. They introduced a strategy to fabricate colloids with mobile DNA linkers, whereby DNA strands are inserted into a lipid bilayer coated over solid colloids through a cholesterol anchor. The DNA strands are mobile because the hydrophobic anchors can diffuse within the lipid layer (Figures 1A–C). It was demonstrated that the mobility of DNA strands enabled particle diffusion (Figure 1D) and broadened the temperature-induced colloidal association/dissociation transition to a span of 10° (Figure 1E). Small two-dimensional superlattices were observed after annealing, as shown in Figures 1F,G. The feasibility of this idea was later validated by Frenkel et al. via a combination of theory and simulation [61]. They also introduced the concept of valency by using particles with mobile linkers.
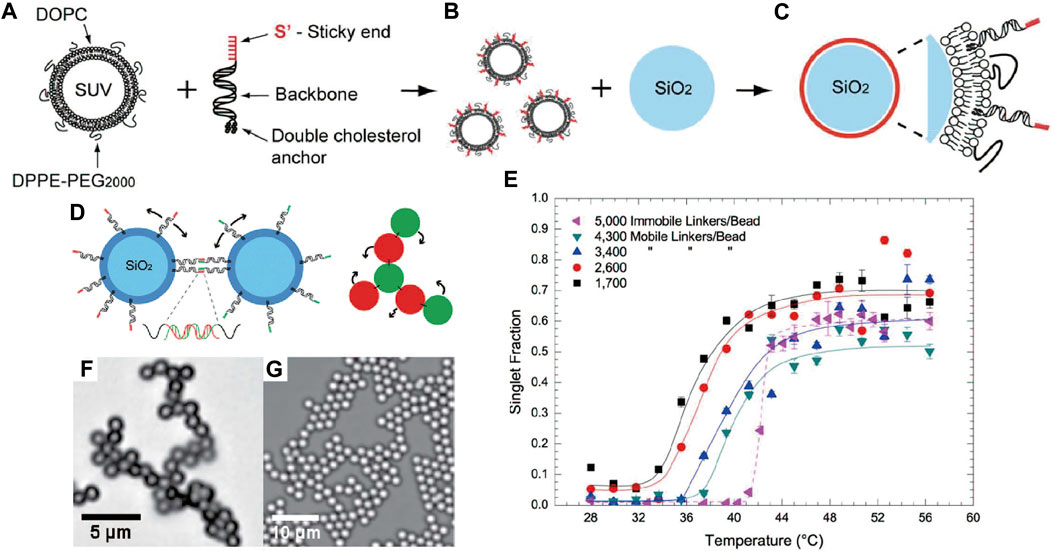
FIGURE 1. (A) Structure of lipid bilayer coated with DNA. (B) Coating SiO2 colloids with the DNA-functionalized lipid bilayer. (C) Colloids with mobile DNA strands. (D) Illustration of interaction between colloids. The mobile DNA bridges enable particles to roll while bound. (E) Singlet fraction of mobile DNA-coated colloids with different densities of DNA linker as a function of temperature, showing the broadened association/dissociation transition. Singlet fraction is defined as the fraction of unbound particles. Bright-field optical micrographs of colloidal assemblies with (F) 1.0-μm colloids, and (G) 2.0-μm colloids. Reproduced from ref. [50] with permission from the American Chemical Society, copyright 2013.
Rogers and Manoharan et al. [54] also believed that the sharp melting transition of DNA-coated colloids was the problem for obtaining equilibrium crystal phases. They tuned the phase behavior of particles by introducing extra free DNA strands that undergo the so-called toehold-mediated strand displacement with the strands grafted on the colloid surface (Figure 2A). Without the displacing strands, the free energy change of the DNA hybridization depends solely on the temperature. When the displacing strands are added, the enthalpy and the entropy changes of the displacement reactions depend on the sequence and the concentration of the displacing strands, respectively. In other words, the magnitude of the hybridization between DNA-coated colloids can be programmed by the displacing strands. If the displacing strands are chosen to make the enthalpy change of hybridization and displacement reaction the same, the temperature dependence of DNA-coated colloids can be eliminated. Thus, the colloidal crystallization via DNA interactions can be achieved over a broader range of temperature (Figure 2B). It was shown that the melting transition of DNA-coated colloids is roughly as wide as 10°C (Figure 2C). In addition, adding two types of displacing reactions allows the creation of a reentrant fluid phases (Figures 2D,E).
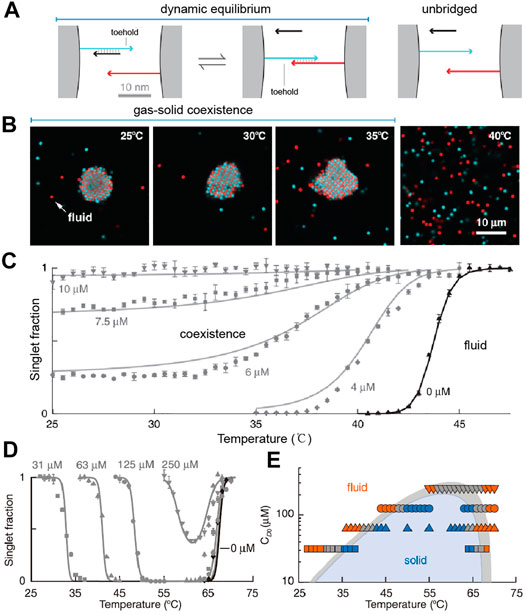
FIGURE 2. (A) illustration of toehold-mediated strands’ displacement between colloids. (B) Confocal fluorescence micrographs of colloidal phases at various temperatures. (C) The singlet fraction of DNA-coated colloids when they are mixed with different concentrations of free displacing strands. (D) Singlet fraction of colloids when two types of displacing strands are introduced. (E) A phase diagram by varying the temperature and the displacing strand concentrations. Reproduced from ref. [54] with permission from the American Association for the Advancement of Science, copyright 2015.
Rogers et al. [62–66] further investigated the physical behavior of the assembly of DNA-coated colloids mediated by free DNA strands. They argued that encoding information into the sequence and concentration of the free strands instead of the grafted strands could overcome the problems conventional colloidal self-assembly suffers, including uncollaborative mutual interactions and a limited number of interactions. For example, they found that the phase behavior of DNA-coated colloids can be altered by adding free DNA strands for displacement reaction, as mentioned above [62]. Then, using free DNA strands as the linkers to bridge grafted DNA on colloids’ surface, they reported a reentrant melting transition upon increasing concentration of linker as well as the possibility to encode multiple interactions spontaneously [63]. In addition, colloidal crystals are constructed using free linker DNA, depending on the concentration and the symmetry of the linker. It was interesting to find that the phase behavior becomes less dependent on the linker concentration when the asymmetry of linker grows [64]. Xia and Ni et al. [67] formulated a mean-field theory to simulate the self-assembly of colloids with mobile DNA coating and free DNA linkers. They showed that the interaction between colloids mediated by DNA linker changes nonmonotonically as the concentration of linker increases, as the result of the competition between the entropies of various species in the system.
While the narrow temperature window was a big hurdle to crystallize DNA-coated colloids, Wang, Weck, and Pine et al. [48] considered the problems from another perspective: surface inhomogeneity. One hypothesis was that a high DNA coverage on colloids is needed, which can alleviate the possibly uneven DNA distribution and eliminate heterogeneity. Another hypothesis was that, because the DNA coating is very thin, the surface of the colloids should be smooth. A rough colloid surface may cause some DNA to be inaccessible, thus creating areas that other particles cannot explore during the crystallization. Based on the two assumptions, the authors developed a strategy to synthesize azide-functionalized TPM colloids [TPM: 3-(Trimethoxysilyl)propyl methacrylate], to which DNA strands can be grafted using the strain-prompted azide-alkyne cycloaddition reaction (SPAAC) (Figure 3A). The resulting colloids have a smooth surface, and more importantly, a DNA coating 5–25 times higher in the areal density than particles made by previous methods [33, 45]. Due to the high DNA coverage, it was also possible to shorten the DNA sticky ends to only four bases and still keep the melting temperature of particles at a moderate temperature, between 44–46°C (Figure 3B). It is generally agreed that high DNA coatings and short sticky ends can facilitate the kinetics of the system to reach equilibrium [23]. For a given sticky end, a high coating increases the melting temperature, which increases the dissociation rate of the DNA bridges and the diffusivity of particles. Rogers et al. believed that for particles with fewer strands that bind strongly, the lifetime of the DNA bonds may be large to slow down the rotational diffusion of the particles [23]. Theoretical works have been recently developed to rationalize the experiment findings [64, 68, 69]. For example, by using a simulation method that tracks the single binding and unbinding between complementary linkers, Mognetti et al. proved that particles could move relative to each other while being cross-linked. Moreover, they clarified that the dissociation rate of the DNA bridges determines the colloid’s diffusivity [68].
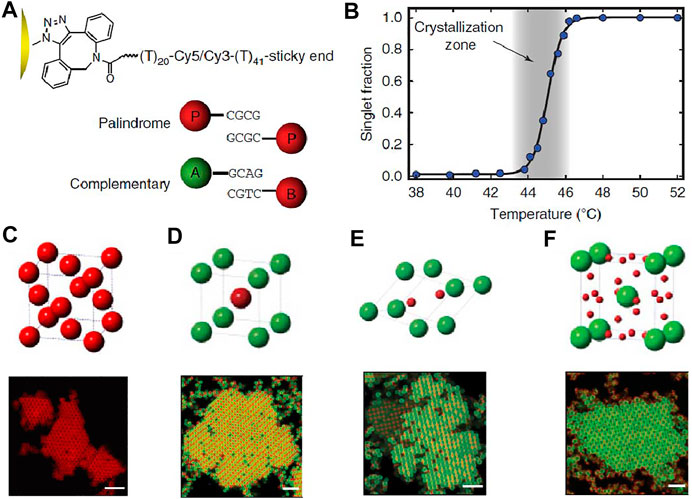
FIGURE 3. Schematic illustration of DNA-coated colloids and their crystals. (A) DNA is modified onto the colloids’ surface via strain-promoted azide-alkyne cycloaddition, SPAAC. A 40-60 base long polyT sequence serves as a flexible linker between the particle surface and the sticky ends. (B) The melting transition of DNA-coated colloids, indicated by the singlet fraction as a function of temperature. Cartoon and confocal fluorescence micrographs of (C) FCC lattice from 1.0-μm colloids functionalized with self-complementary sticky ends, (D) AB lattice (isostructural to CsCl) from 1.0-μm A and 1.0-μm B, (E) AB2 lattice (isostructural to AlB2) from 1.0-μm A and 0.54-μm B colloids, and (F) AB6 lattice (isostructural to Cs6C60) from 1.5-μm A and 0.54-μm B colloids with hetero-complementary sticky ends. Scale bars, 5 μm. Reproduced from ref. [48] with permission from the Springer Nature, copyright 2015.
With the capability to anneal, the crystal structures of DNA-coated colloids can be further programmed by the particle types and sizes. For example, FCC crystal lattice was obtained from the assembly of colloids with a palindromic, self-complementary DNA sticky end (Figure 3C). Binary AB lattice (isostructural to CsCl) was obtained from the co-assembly of 1.0-µm A colloids and 1.0-µm B colloids with hetero-complementary DNA sticky ends (Figure 3D). AB2 lattice (isostructural to AlB2) was obtained from 1.0-µm A colloids and 0.54-µm B colloids (Figure 3E). AB6 lattice (isostructural to Cs6C60) was obtained from 1.5-µm A colloids and 0.54-µm B colloids (Figure 3F). Moreover, the crystallization process is completed in minutes to hours (not days), so the real time nucleation, growth, restructuring, and defect formation of bulk crystals of DNA-coated colloids could be studied.
The significance of this work is that it unveiled the myth of the crystallization of DNA-coated colloids and allowed the verification of various related hypotheses. For example, a smooth particle surface is important; particles with a rough surface were tested and they failed to crystallize [70]. It was also indicated that a flexible backbone for the DNA brush on the surface is important, as it allows the DNA sticky ends more freedom to explore appropriate conformations to bind [35]. This is in stark contrast with the crystallization of DNA-coated nanoparticles, where double-stranded DNA is normally used [7].
Subsequently, the crystallization was extended to other commonly used particles, including those made from titanium oxide, silica, polystyrene (PS), and poly (methyl methacrylate) (PMMA) [70]. The synthetic procedures are revisited and modified to achieve smooth particle surfaces and a dense layer of DNA coating (Figure 4A). Although the maximum number of DNA conjugated to different particles may vary, all particles crystallized. Similarly, FCC lattice, AB lattice, AB2 lattice, and AB6 lattice can be produced (Figures 4B–E). Instead of using particles of the same type, particles with different compositions were mixed to adopt predetermined spatial arrangements. For example, PS colloids, which have a low index of refraction, are co-assembled with titania colloids, which have a higher index of refraction. In other words, one can program crystals from a mixture of arbitrary colloids.
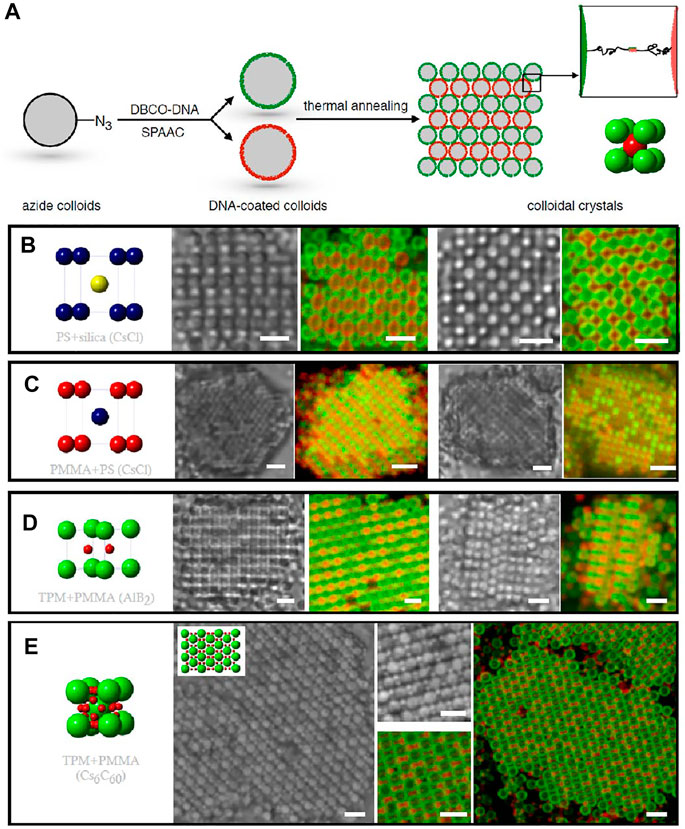
FIGURE 4. (A) Illustration of the crystallization of DNA-coated colloids. Cartoon, bright-field and confocal fluorescence micrographs of AB lattice (isostructural to CsCl) from (B) 1.0-μm PS and 1.0-μm silica colloids and (C) 1.0-μm PMMA and 1.0-μm PS colloids. (D) AB2 lattice (isostructural to AlB2) from 1.0-μm TPM and 500-nm PMMA colloids. (E) AB6 lattice (isostructural to Cs6C60) from 1.5-μm TPM and 500-nm PMMA colloids. Scale bars in (B–C), 2 μm. Scale bars in (D–E), 3 μm. Reproduced from ref. [70] with permission from the American Chemical Society, copyright 2015.
The demand for high DNA coverage has later stimulated the development of new methods for functionalizing DNA on colloids. For example, Oh et al. [71] reported a swell-deswelling method, which introduced azide-modified polystyrene-b-polyethylene oxide (PS-PEO-N3) di-block copolymers onto PS colloids (Figures 5A,B). The method was inspired by previous works by Crocker et al. [35, 72] When the PS particles are plasticized with organic solvents, the PS blocks of the copolymer can insert into the particles. As the particles de-swelled by solvent evaporation, the block copolymers are firmly trapped in the particles by anchoring the PS blocks in the particles’ polymer matrix. A high grafting density was achieved because they become more concentrated when the particles shrink during the deswelling. When DNA is attached to the azide groups at the end of the PEO blocks by SPAAC reaction, a high DNA areal density, 115,000 strands on a 1.0-μm colloid, is realized (Figure 5B). Using these particles, the authors obtained various single and binary crystals (Figures 5C–F). The method has also been employed to coat DNA on PS particles with different surface charges; it works when the PS is slightly crosslinked (up to 10% crosslinking).
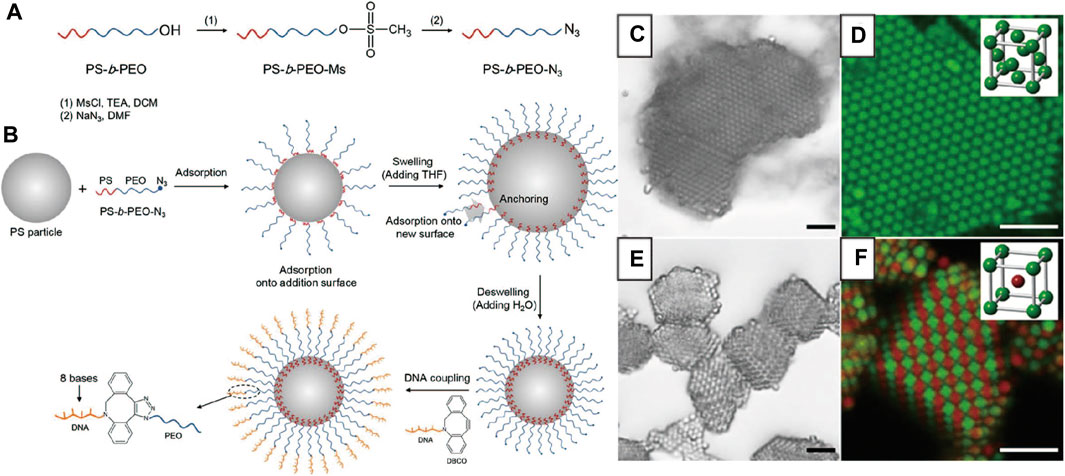
FIGURE 5. (A) The preparation of PS-PEO-N3. (B) Illustration of modification of PS-PEO-N3 and DNA on PS colloids. Bright-field and confocal fluorescence micrographs of (C, D) FCC lattice from PS colloids with self-complementary DNA (E, F) BCC lattice (or AB lattice) from A and B colloids with hetero-complementary DNA. Scale bars, 5 μm. Reproduced from ref. [71] with permission from the American Chemical Society, copyright 2015.
Oh et al. [73] later reported another effective strategy to incorporate homogeneous and high-density DNA coatings onto colloids. As shown in Figure 6A, PS colloids with carboxylate groups are activated with 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMTMM). Then, heterobifunctional polyethylene glycol (NH2-PEG-N3) is added to react with DMTMM-activated colloids through nucleophilic substitution. The azide terminal group on the PEG can undergo SPAAC reaction with DBCO-modified DNA strands. The obtained DNA-coated colloids have DNA coatings up to 1,100,000 DNA strands per 1-µm particle, equivalent with 1 DNA/2.9 nm2, four times higher than the previous record [35]. Regular colloidal crystallization is constructed from particles fabricated by this method. The melting profile and the obtained crystals are shown (Figures 6B,C).
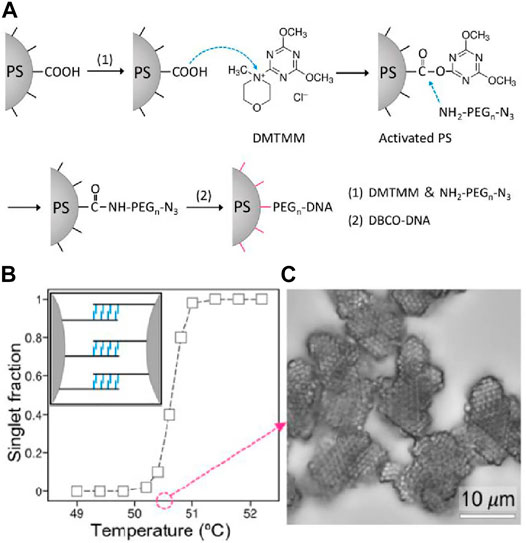
FIGURE 6. (A) Scheme of preparing high-density DNA-coated colloids. (B)The singlet fraction of particle as a function of temperature. (C) Colloidal crystals of FCC lattice obtained and observed under bright-field microscope. Reproduced from ref. [73] with permission from the American Chemical Society, copyright 2020.
The above reports focused on binary colloidal systems where only two types of DNA strands are employed. Oh et al. [74] demonstrated multiple phase transitions and responsive multistep reorganization of DNA-coated colloids by using three different DNA strands that vary on the binding strength. Free DNA strands are added to regulate the binding selectivity between DNA-coated colloids via toehold-mediated strand displacement [54]. For convenience, the DNA strands modified on the colloid surface are noted as A, B, and P (Palindrome, self-complementary), while the free DNA strands with complementary sequence to A and B strands are noted as DA and DB respectively (Figure 7A). As shown in Figures 7B,C, the binding strength of each duplex are designed as follows: A-B > A-DA, B-DB > P-P. At 35°C, P-P, A-DA, B-DB duplexes coexist, leading to amorphous aggregation. Increasing the temperature to between 36 and 44°C, the P-P duplex will break, while the A-DA and B-DB duplexes remained intact; dispersed singlet colloids were observed. As the temperature was increased to a range of 45 and 63°C, the A-DA and B-DB duplexes will break, leaving the A strands hybridizing with B strands on the colloids; aggregation will again form. When the temperature was raised up to 65°C, the aggregation melts again as the A-B duplexes destabilize. Crystallization was observed at around 36 and 45°C. It is demonstrated that the colloidal superlattice can be programmed by both the size ratio of colloids and the temperature. For example, when the size ratio is 1:1, the FCC lattice will be favored at low temperature, while the CsCl lattice is formed at high temperature (Figure 7D). Changing the size ratio to 1:0.6, the lattice at the high temperature changed from CsCl to AlB2 (Figure 7E). When the third particle is introduced and their size ratio is set to be 1:1:0.6, the CsCl lattice and FCC lattice co-exist at low temperature and convert to AlB2 lattice at a high temperature (Figure 7F).
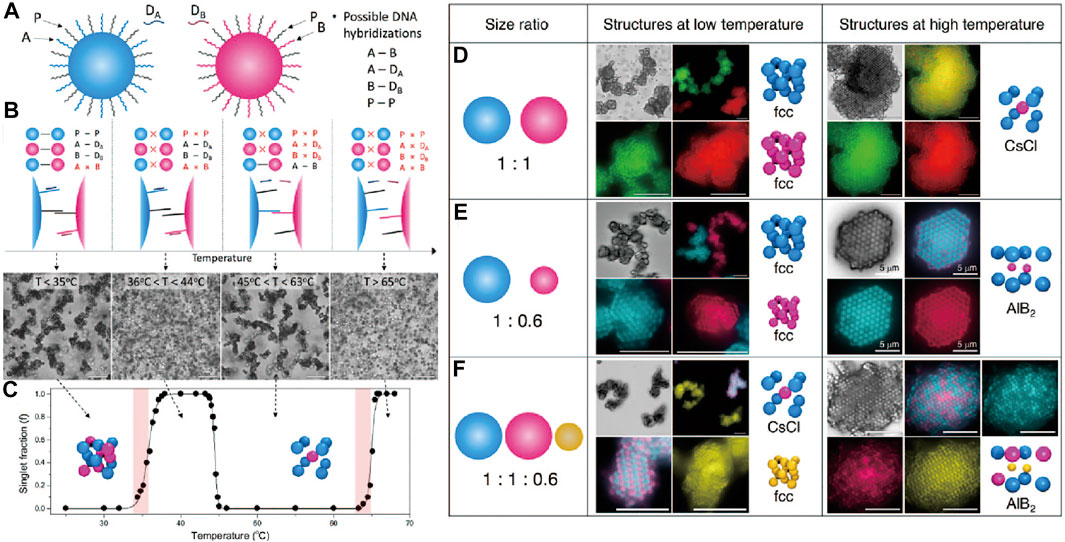
FIGURE 7. (A) Colloids with three different types of DNA strands, labeled as A, B, and P; free strands for strands displacements are labeled as DA and DB. (B) Illustration of how colloids undergo several phase transitions when temperature is increased. (C) Bright-field micrographs and the singlet fraction show the phases of DNA-coated colloids as a function of temperature. (D–F) Cartoon, bright-field and confocal micrographs of crystals obtained. (D) Two types of colloids with size ratio 1:1, which form two separate FCC crystal at lower temperature and a CsCl crystal at higher temperature. (E) Two types of colloids with the size ratio of 1:0.6 form two separate FCC crystals at lower temperature and a AlB2 crystal at higher temperature. (F) Three types of colloids with the size ratio of 1:1:0.6 will form a CsCl crystal and FCC crystal at lower temperature and a AlB2 crystal at higher temperature. Scale bars, 10 μm. Reproduced from ref. [74] with permission from the American Chemical Society, copyright 2020.
Introducing Anisotropic Information to DNA-Coated Colloids
In parallel to the deepened understanding of DNA-mediated colloidal interactions, the past decade has witnessed an explosion in the type of colloids that can be synthesized, featuring anisotropic particles with non-spherical shapes [75] (e.g., cubes, rods, and triangular prisms), surface patches [76], or a combination of both [52, 77]. Compared to the simple microspheres, these new particles possess extra information to guide the packing and assembly of particles, allowing access to structures with reduced symmetries and directionality. While DNA-coated spheres have already allowed access to numerous crystals, the integration of anisotropic particles with DNA can ultimately program a much wider spectrum of structures, including those found in atomic and molecular crystals. In this section, we focus on how the anisotropic information is introduced and how they play a vital role in diversifying the assembled structures.
As mentioned, Wang, Weck, and Pine et al. [39] introduced the DNA patchy colloids into the field, which are particles with localized surface regions that are functionalized with DNA. The patchy particles are synthesized by a cluster-encapsulation method, in which small clusters consisting of several microspheres in symmetric configurations are encapsulated (Figures 8A,B). The encapsulation condition is controlled so that the vertices of the clusters are left out; they bear a different type of chemical group—in this case amidine—from the encapsulating materials and serve as the chemical patches. The DNA with sticky ends is site-specifically coated on the patches via a biotin-streptavidin-biotin linkage, where biotin molecules are conjugated to the amidine patch in advance (Figures 8A,C,D). The patches, with controlled numbers, adopted highly symmetric positions around the particles, such as linear, triangular, or tetrahedral, imitating the hybridized atomic orbitals, such as sp, sp2, or sp3 (Figure 8D). They have the ability to form colloidal bonds with directionality (according to the patch position) and specificity (due to the properties of DNA). A number of colloidal structures, such as colloidal molecules and polymers, are observed when DNA patchy particles are assembled (Figures 8E–I).
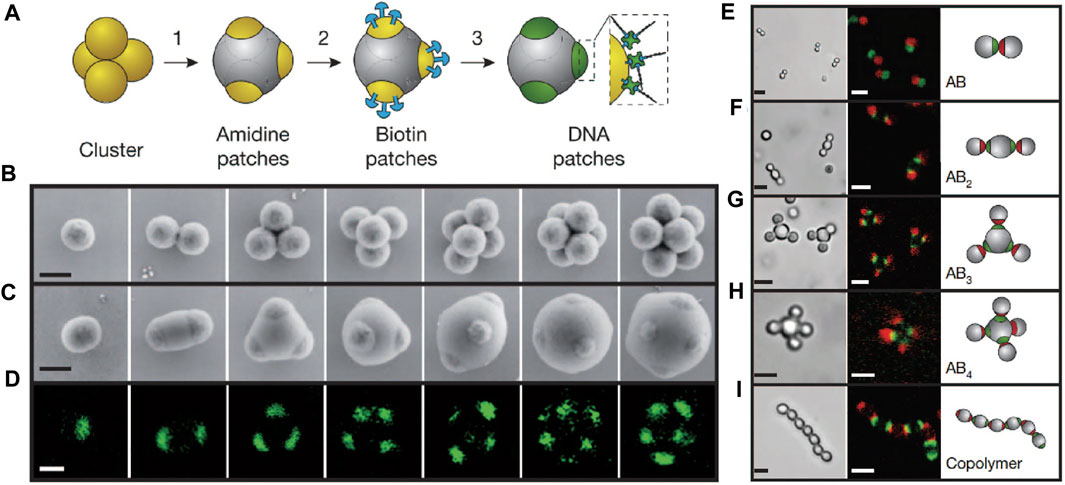
FIGURE 8. (A) Preparation of DNA patchy colloids. (B–D) Electron and confocal fluorescence micrographs of clusters and patchy colloids. (E–I) “Colloidal molecules” assembled from patchy colloids. Bright-field and confocal micrographs of colloidal molecules are shown. The fluorescence comes from the dye-labelled streptavidin linked with DNA, showing that only the patches are modified. Scale bars in (B–D), 500 nm. Scale bars in (E–I), 2 μm. Reproduced from ref. [39] with permission from the Springer Nature, copyright 2012.
The milestone work has stimulated much effort in developing various anisotropic particles coated with DNA. For example, Feng and Chaikin et al. [78] reported the fabrication of DNA Janus colloid—essentially one-patch particles. DNA-coated microspheres were allowed to bind to a DNA-functionalized substrate. The binding is achieved via free DNA strands in the solution which simultaneously hybridize to the strands on the particles and the substrate. A patch was created where the sphere touches the substrate and when the free DNA strands are permanently crosslinked to the DNA on the particles.
Crocker and co-workers [79] used colloidal crystals as the template to make DNA-functionalized clusters. A mixture of particles with two different sizes and complementary DNA strands were subject to sedimentation, which induced the formation of colloidal crystals (FCC) (Figure 9A). The small particles occupy the interstitial sites of the big particles, while they also bind to the big particles by DNA hybridization (Figures 9B,C). At this stage, T4 DNA ligase is employed to form covalent phosphodiester bonds to crosslink the hybridized DNA strands (Figure 9D). Further purification steps can break the sedimentation and clusters can be recovered (Figure 9E). The clusters obtained have DNA functionalized on the surface capable of directional bonding (Figure 9F), from which an icosahedral cluster (Figures 9G–I) can be fabricated by mixing the clusters with spherical particles bearing complementary DNA. The symmetries of such ordered clusters can be altered by adjusting the size ratio of spherical colloids. For example, tetrahedral and octahedral clusters were obtained when the size ratio is approximately 4:1 and 2:1, respectively.
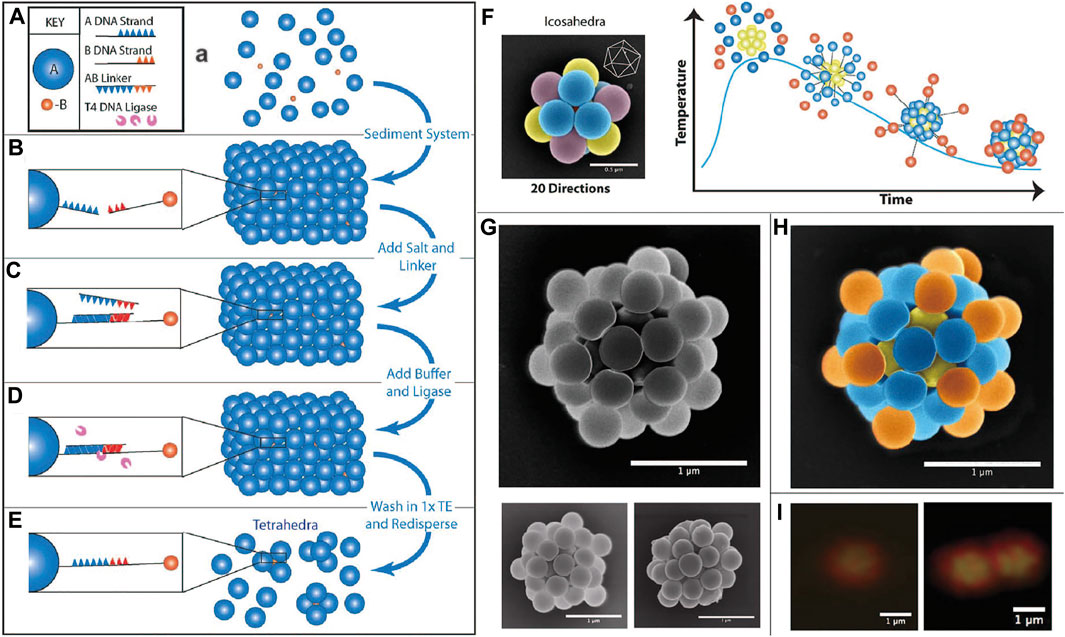
FIGURE 9. (A–E) Schematic procedure of making tetrahedra colloids via sedimentation and forming DNA bridges. (F) Directional bonding of icosahedral colloids with other colloids. (G) SEM images of decorated icosahedral colloids. (H) Colored SEM images of decorated icosahedral colloids. (I) Confocal images of a decorated icosahedral colloid (left) and two icosahedral colloids interacting with each other (right). Scale bars in (G–I), 1 µm. Scale bar in (F), 500 nm. Reproduced from ref. [79] with permission from the American Chemical Society, copyright 2015.
Also by using colloidal crystals as a template, Pine et al. reported the synthesis of faceted DNA patchy particles (Figure 10A) [80]. Deformable emulsion droplets were coated with DNA strands and formed binary crystals. The DNA strands are mobile on the droplet surface and were recruited to the adhesion zones where the droplets touch and flatten, as shown in Figure 10B. DNA-patches are formed after solidification of droplets by rapid photo-polymerization under UV light (Figure 10C). The obtained faceted DNA-patchy colloids show directional bonding with other DNA-coated spheres with complementary DNA strands (Figures 10D,E).
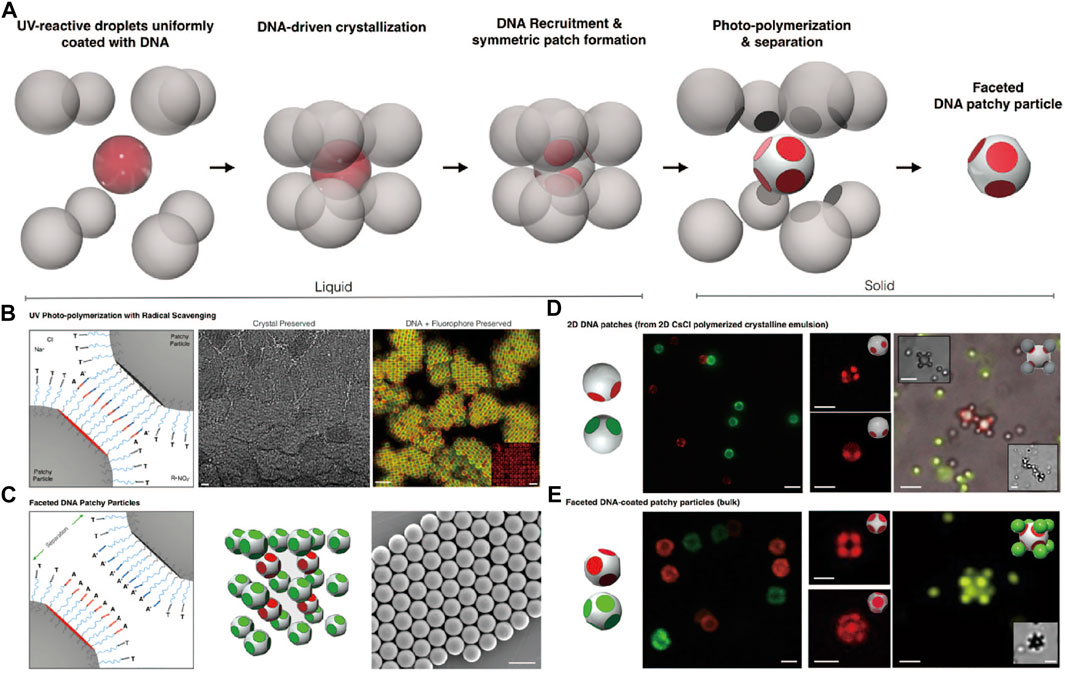
FIGURE 10. (A) The schematic procedure of producing faceted DNA patchy colloids. (B) Photo-polymerization of crystalline emulsion under UV lights (left). Bright-field micrograph of CsCl lattice of colloidal crystal after photo-polymerization (middle, scale bars, 10 μm). Confocal fluorescence micrograph of colloidal crystal shows that DNA fluorescence is preserved (right, scale bars, 5 μm). (C) Patches on colloids. SEM images of patchy colloids after photo-polymerization (right, scale bars, 2 μm). (D, E) Colloidal assemblies of patchy colloids with PS colloids via DNA interactions. Scale bars in (D), 2 μm. Scale bars in (E), 1 μm. Reproduced from ref. [80] with permission from the National Academy of Sciences, copyright 2020.
To build complex structures, a prerequisite is to introduce multiple interactions, which calls for orthogonal functionalization of patchy particles. Zheng et al. [81] designed bifunctional patchy particles with two different DNAs, one modified on the patches and the other on the matrix. To achieve this, chlorine-modified microspheres and clusters were first fabricated, which was partially encapsulated with the TPM oil. Methacrylate acid was introduced to the TPM oil before it solidified, resulting in bifunctional colloids with chloride on the patches and carboxylic acid on the matrix (Figure 11A). Then, two different DNA strands were conjugated separately on bifunctional colloids via orthogonal coupling reactions, SPAAC for the patches and carbodiimide-mediated reaction for the matrix (Figure 11B). Microspheres coated with complementary DNA were then allowed to co-assemble with the bi-functional patchy particles. By tuning the DNA sequences and the temperature, the patchy particles assemble with one or two types of spheres (Figure 11C).
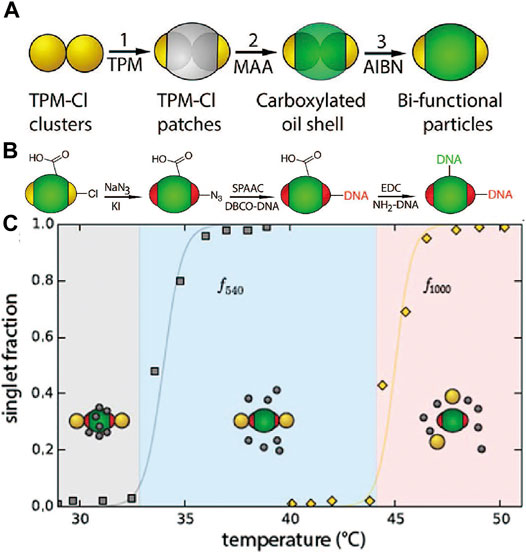
FIGURE 11. (A) Scheme for the preparation of the bifunctional colloids. (B) Scheme of modification of two different DNA onto the patch and shell of the bifunctional colloids. (C) Melting curve of bifunctional colloids, represented with two types of TPM and their singlet fraction. Reproduced from ref. [81] with permission from the American Chemical Society, copyright 2016.
As the challenges for crystallization of DNA-coated spheres are tackled, Pine and coworkers aimed to explore equilibrium assemblies using DNA patchy particles. As smooth surfaces and high DNA areal density are needed, new methods for making patchy particles are developed. The crystallization of DNA Janus particles was first studied. A new scheme was designed so the Janus particles obtained had smooth surfaces and high DNA coverage, needed for crystallization [82]. As Figure 12A shows, the synthesis begins with the partial encapsulation of PS microspheres with TPM oil droplet. Toluene is then added to plasticize the PS particle, which becomes a liquid and phase separates with the TPM oil phase. Removing the toluene followed by solidification of the TPM oil creates patchy particles. The patch size can be finely controlled by adjusting the ratio of TPM to PS. To functionalize DNA on the PS patches, the swelling-deswelling method previously mentioned was used. TPM is highly crosslinked and thus not functionalized. The Janus particles show cooperative assembly resulting in dimer chains, trimer chains, and bilayers depending on the Janus balance (patch/matrix ratio) as shown in Figures 12B–H. For example, when the ratio was in the range between 0.3 and 0.41, dimer chains were favored, when the ratio is between 0.41 and 0.48, trimer chains were formed, and when the ratio is between 0.6 and 0.68, bilayers were observed. Here, the patchiness contains important information that programs the self-assembly behavior; the DNA serves as a glue that allows the formation of equilibrium structures.
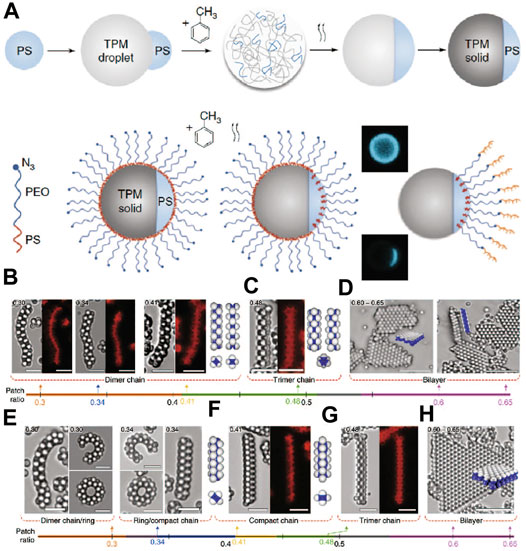
FIGURE 12. (A) The preparation of DNA-coated Janus colloids. (B–H) The patchy particles assemble to form various colloidal superstructures (chains and layers) depending on the size ratios between the patch and the matrix. Cartoons, bright-field and confocal fluorescence micrographs show these structures. The grey-scale pictures are obtained from bright-field microscopy and the colored pictures are from confocal fluorescent microscopy. Scale bars: 5 µm. Reproduced from ref. [82] with permission from the Springer Nature, copyright 2019.
Later, the above system has been modified to host two types of DNA [83]. A 60-bases DNA (PH) is introduced onto the TPM matrix, and a 20-bases DNA (PL) is coated on the PS patch (Figures 13A–C). Free DNA strands (D) are also introduced, which can undergo strand displacement with the PH strands to alter the binding behavior of the particles. Specifically, the PH-PH duplexes are stronger than the PH-D duplexes, which are stronger than PL-PL duplexes. This means that the self-assembly behavior of the patchy particles can be controlled by temperature, which determines the preferred DNA duplexes that form, as well as by the anisotropic distribution of the DNA on the particle surface (Figure 13D). For example, at a low temperature (below 44°C), patchy colloids form amorphous aggregates via PL-PL hybridization; the PH is blocked by the D strands. Bilayer structures are formed when the temperature is kept between 44.0 and 46.0°C. As temperature is increased, between 46.5 and 57.5°C, where PL-PL hybridization is broken while the PH-D hybridization remains intact, the patchy colloids disperse. Further temperature increases could free PH strands from the PH-D duplexes, and the PH-PH duplexes form to make patchy colloids aggregate again. Small clusters are observed between 57.5 and 58.5°C, while chain-like structures with or without branches have formed between 58.5 and 70°C. Small clusters form again between 70 and 71.5°C, and everything finally disperses when the temperature is above 71.5°C. In this work, the information derived from the DNA sequences (both specificity and dynamic) and from the particle anisotropy is simultaneously coded into the particle system, programming the temperature dependent, reconfigurable self-assembly.
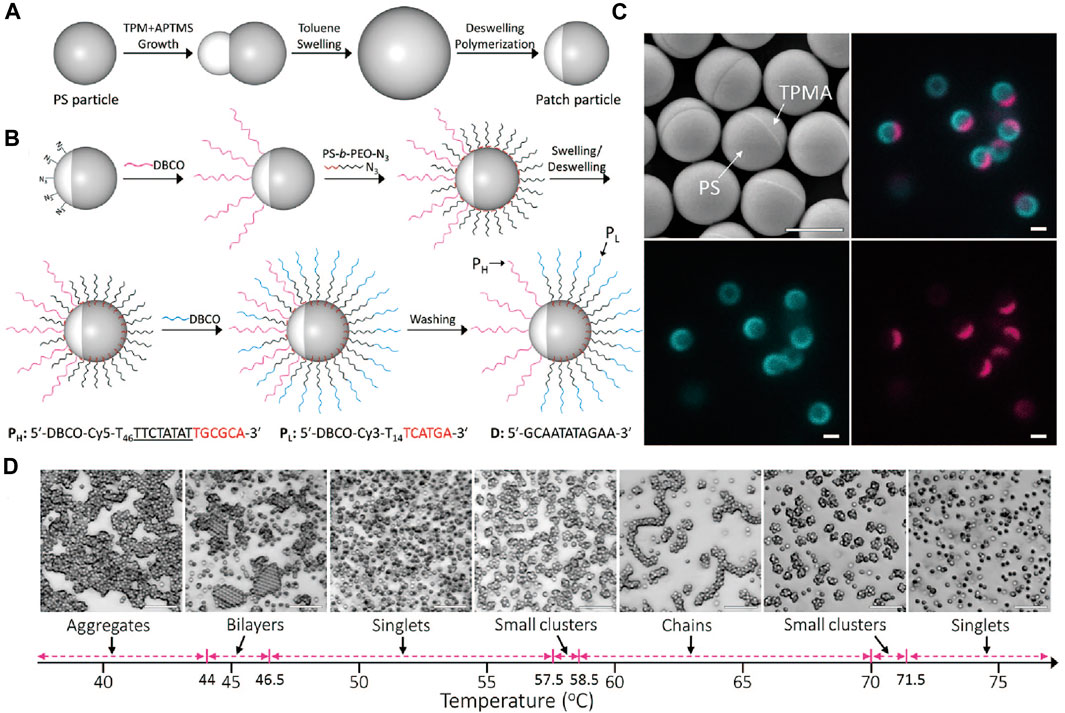
FIGURE 13. (A) Preparation of Janus colloids composed of TPM and PS. (B) Illustration of modification of two different types of DNA on Janus colloids. (C) SEM and confocal images of DNA-grafted patchy colloids. Different DNAs are labeled with different colors. Scale bars, 1 μm. (D) Assembly of DNA-coated patchy colloids show different structures (aggregates, bilayers, chains and clusters) at different temperatures. Scale bars, 10 μm. Reproduced from ref. [83] with permission from the American Chemical Society, copyright 2020.
Next, two-dimensional (2D) superlattices can be programmed by using patchy particles with two patches. Liu and Weck et al. [84] demonstrated that quasi-2D colloidal superlattices were constructed by ellipsoidal di-patch colloids via both depletion and DNA-mediated interactions. The di-patch colloids were fabricated from PS-N3 dimer clusters via partial encapsulation with TPM, followed by DNA modification via SPAAC. The assembled structures were determined by several factors, including the patch size, the annealing temperature, and the strength of the depletion interaction. For di-patch colloids with θ = 86° (θ is defined as the half of the opening angle and characterized for patch size), flower-like Kagome structures were formed when the annealing temperature was at 41–46°C and brick-wall like structures were formed when they were annealed at 57–62°C; the two structures co-existed at 47–53°C. When the patch size θ was changed to 70° and with depletion interaction, the quasi-2D superlattices were different. Orthogonal packed single or double-layer structures were mainly observed at 42–46°C with 1.0 wt% of Pluronic F127, whereas wrinkled monolayer structures were the case at 46–50°C with 3.0 wt% of Synperonic F108; the co-existence of both structures was found at 44–48°C with 1.8 wt% Pluronic F127. An empirical phase diagram of the self-assembly of di-patch colloids was constructed to show how superlattices were influenced by various conditions.
As demonstrated, DNA patchy particles show unique, directional self-assembly behavior enabling complex structures programmed by the patch number, size, and arrangement. Using particles with four patches in a tetrahedral geometry, it is possible to achieve the colloidal diamond, one of the most desirable colloidal structures to build photonic crystals with an optical band gap [40, 43]. Although the directional interaction is available, it is still difficult to build colloidal diamonds from DNA patchy particles for two reasons. First, the formation of the diamond structure is entropically unfavored. The packing fraction of cubic diamond is 34% [85], much lower than close packing of spheres, which is 74% [86]. Second, amorphous liquid or gel with tetrahedral topology is more likely to occur during the assembly of tetrahedrally patched colloids.
To overcome the mentioned difficulties, there have been two proposals. One is to incorporate the lattice of diamond crystals into a denser or more coordinated structure to avoid the problem of low filling fraction. Crocker et al. [87] explored this route by using DNA-coated microspheres, which forms a “double diamond” structure. Two slightly differently sized PS colloids (named “A” and “B”) are modified with two different DNA strands that can both hybridize to a linker strand (Figures 14A–C). Both “A” and “B” can assemble into diamond crystals that interpenetrate each other, isomorphic to the NaTl Zintl phase in atomic crystals (Figures 14D–H). There are several empirical requirements for the formation of the “double diamond” superlattice. For example, the size ratio of the “A” and “B” particles can be 0.96, 0.88, or 0.85 but cannot be 1. Also, strong unlike interactions, UAB >> UBB, and weak interactions between the larger spheres, UBB > 0, should both exist in the system. It is important to note that the attempt to simulate colloidal “double diamond” from a fluid phase has failed, indicating that the nucleate mechanism behind is unclear.
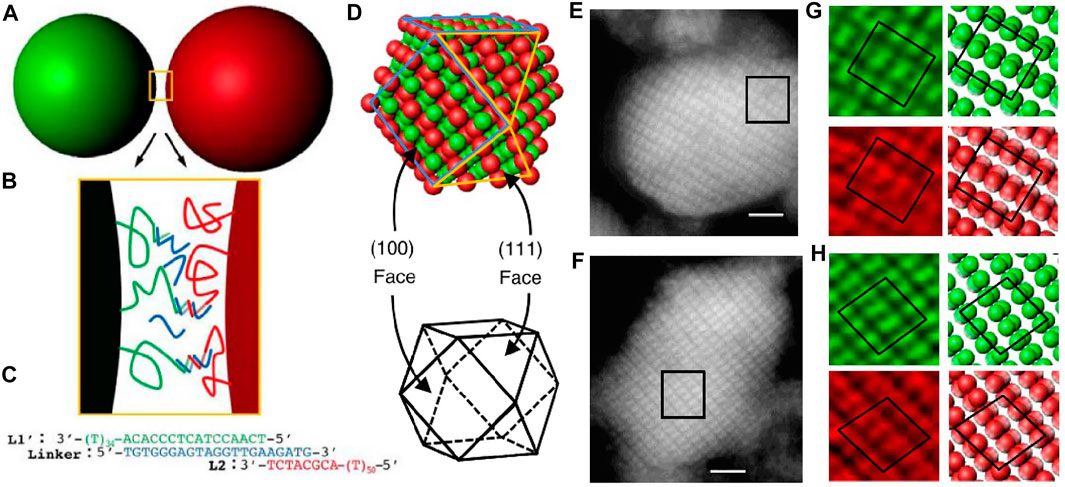
FIGURE 14. (A) Illustration of DNA-coated colloids. (B) The DNA interactions between colloids. (C) The bridge formed between colloids. (D) The double diamond structure after self-assembly. (E-H) The confocal section of double diamond in different views. Scale bars, 2 μm. Reproduced from ref. [87] with permission from the Springer Nature, copyright 2017.
Alternatively, Ducrot and Pine et al. [88] coupled two low-coordinated architectures, the diamond and pyrochlore, into an MgCu2 superlattice by co-assembling spherical colloids and tetrahedral clusters with complementary DNA (Figures 15A–C). The morphology of tetrahedral cluster is characterized by the compression ratio rcc/2a, which indicates the degree of overlap between constituent spheres, as illustrated in Figure 15C. It was hypothesized that the co-nucleation of the two colloidal species with an attracting potential would lead to the formation of a MgCu2 structure. This idea is verified by Brownian dynamic simulation. Crystallites can be observed from experiments when annealing the system (Figures 15D,E). The compression ratio has a great impact on the colloidal MgCu2 superlattice. For example, when the compression ratio is lower than one, MgCu2 structure was observed after slowly annealing. However, no MgCu2 structure was observed when the compression ratio was close to one. This phenomenon can be explained by a geometric reason that a singlet sphere can form no more than six bonds with surrounding tetrahedral clusters that are uncompressed (compressed ratio equals one), while it can form up to twelve bonds with surrounding tetrahedral clusters that are compressed, providing more binding energy to stabilize the MgCu2 superlattice.
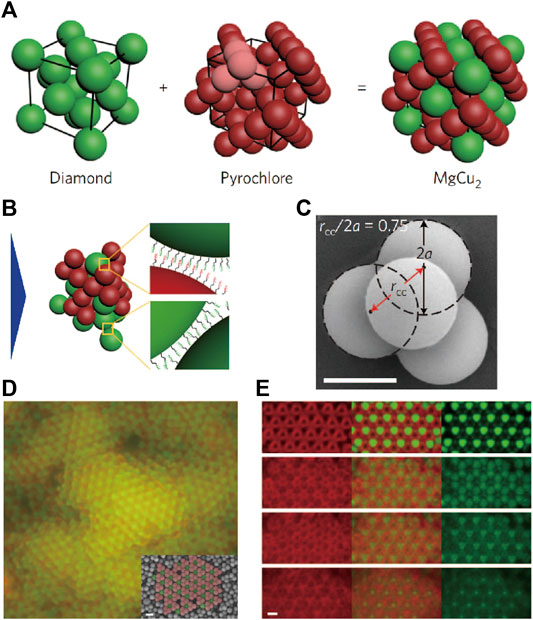
FIGURE 15. (A, B) Illustration of colloidal MgCu2 superlattice via DNA interactions. (C) Illustration of compression ratio rcc/2a (which indicates the degree of overlap between constituent spheres). (D, E) Confocal images of colloidal MgCu2 superlattice. The inset image in (D) is obtained from SEM. Scale bar, 1 μm. Reproduced from ref. [88] with permission from the Springer Nature, copyright 2017.
The idea of using tetrahedral patchy particles to build the diamond structure was recently realized by the He and Pine et al [44]. In the atomic diamond, the adjacent sp3 carbons adopt a staggered conformation due to steric effect. While in the previous design of patchy particles, no mechanism is available to control the colloidal conformation, although they are four-fold coordinated. To address this problem, He, Sacanna, and Pine designed patchy particles with retracted patches and a tetrahedral shape. In brief, a tetrahedral cluster is designed where one single spherical colloid partially overlapped with four tetrahedrally coordinated spherical lobes. The center of each tetrahedral cluster is modified with DNA, serving as retracted patches (Figure 16A). Such a design ensures that, when the DNA-modified patches reach each other and bind, the tetrahedral lobes are oriented to the staggered conformation (Figure 16B). In other words, the particle lobes function to alter the orientation during colloidal nucleation while the center part is the DNA-coated patch to stabilize the overall colloidal structure.
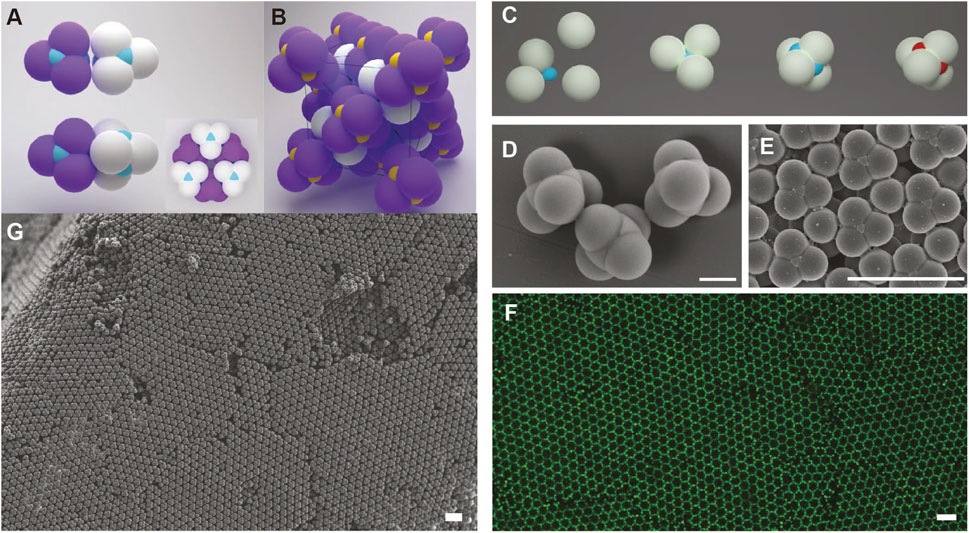
FIGURE 16. (A) Illustration of colloidal cubic diamond via DNA-mediated assembly, where DNA is modified on the surface of the center part (blue) of each patchy particle. The tetrahedrally coordinated spherical lobes (purple and white) serve to control the orientation of particle to adopt a staggered conformation. (B) Unit cell of a colloidal cubic diamond. Preparation (C) and SEM images (D) of patchy colloids. (F) Confocal microscope image of colloidal cubic diamond (E–G) SEM images of colloidal cubic diamond. Scale bar of (D), 1 µm. Scale bars of (E–G), 5 m. Reproduced from ref. [44] with permission from the Springer Nature, copyright 2020.
The patchy particles were fabricated using a strategy called colloidal fusion, developed earlier by Gong and Sacanna et al [89] (Figure 16C). First, solid non-crosslinked PS particles are mixed with smaller TPM oil droplets. The stoichiometric aggregation of PS particles around the TPM oil leads to the tetrahedral cluster. THF is added to change the compression ratio by deforming PS lobes. After evaporation of THF and solidification of the TPM oil, patchy particles are obtained (Figure 16D). Azide groups are modified on the TPM’s surface, followed by the introduction of DBCO-functionalized DNA (Figure 16C). After carefully controlled crystallization upon annealing, the colloidal diamond is obtained, as shown from the SEM and fluorescent images in Figures 16E–G.
The photonic bandgap of the obtained diamond structure was calculated using MIT Photonic Bands software [90]. By changing the compression ratio from 0.1 to 0.8, the colloidal cubic diamond shows bandgap for the inverse lattice, i.e., structure defined by the void space of the colloidal diamond structure. The widest bandgap is achieved around a compression ratio of 0.6. Therefore, the colloidal cubic diamond has promising optical properties and a highly constrained and mechanically stable structure, suitable as a template for making high-dielectric-contrast photonic crystal with cubic diamond symmetry.
The next steps for fabricating photonic materials based on the method described are as follows. First, template the inverse diamond structure with high refractive index materials. PS and TPM have relatively lower refractive indices, of 1.6 and 1.4, respectively, which is not enough to open up a photonic bandgap. Therefore, using the colloidal systems described as a template to make an inverse diamond structure by backfilling the template with a high-refractive-index material such as TiO2 will be promising in making future optic devices. Second, produce the colloidal diamond in a large scale. The already-obtained colloidal diamond is of 80 µm in lateral dimension and 40 µm in thickness. Strategies leading to large crystals should be applied for making optical waveguides, lasers, and other optical applications.
Summary and Future Perspectives
In summary, the structures of self-assembled materials by DNA-coated colloids can be programmed according to the information encoded in the sequences of various DNA strands and the morphology of the colloids. They are potentially useful for further applications, for example, the next generation of optical devices. Despite the great achievements, there are still many challenges. First, the role of DNA in colloidal self-assembly is not fully explored yet as it functions more like a “glue” to maintain the overall self-assembly structure. There is still space for the enhancement of DNA programmability in colloidal self-assembly. For example, most systems only exploit two or three types of DNA strands, while multiple types of DNA strands are engaged in the nanoparticle system to build intricate structures [8]. It will be promising to see more complex self-assembled structures by grafting multiple types of DNA strands onto the colloids and/or introducing free strands in the solution. Besides, the DNA strands of the current studies are one dimensional, while investigations of more complex DNA structures such as DNA tiles and DNA origami are rare. DNA origami, for example, carry more information by having prescribed shape and binding sites and thus have greater programmability in creating 3D structures. The Gang group [91, 92], Shih group [93], and Seeman group [94]had already reported interesting results in this direction, though mostly focusing on nanoparticles. A notable example is the use of tetrahedral origami cages to direct spherical gold nanoparticles to form a diamond lattice [95]. For colloids, the Chaikin and Seeman group have designed a cross-shaped DNA origami to facilitate the formation of chiral colloidal clusters with precision [9]. It can be expected that the combination of spherical or patchy colloids with DNA tiles or origamis will be a new interesting way to construct ordered assemblies with higher complexity. Second, whether the use of DNA can shed light on the use of other biomolecules or biomolecular analogues, such as peptide, deserves further attention. Only in saline buffer will the DNA-coated colloids self-assemble, which greatly limits its applications in other fields, such as low-salt solution environments. DNA alternatives, such as peptide nucleic acids (PNA) and nanocomposite tectons (NCTs), have been recently developed to overcome the weakness [96, 97]. Third, the self-assemblies obtained from DNA-coated colloids fall in the periodic crystal lattice, while other structures [98–100], such as quasicrystals, have not been reported yet. Previous research has demonstrated the possibility to produce 2D quasicrystal from nanoparticles [101]. It will be possible to build colloidal quasicrystal with DNA-coated colloids.
Author Contributions
TZ and YW wrote the manuscript. All authors discussed about the contents.
Funding
YW acknowledges support from the start-up fund of The University of Hong Kong. This project is also partially supported by the Early Career Scheme (ECS) from the Research Grants Council (RGC) of Hong Kong (Project number: 27303817), General Research Fund (GRF) of the RGC of Hong Kong (Project number: 17302820), and Croucher Innovation Award 2019 (Croucher Foundation, Hong Kong).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Chen J, Seeman NC Synthesis from DNA of a Molecule with the Connectivity of a Cube. Nature (1991) 350(6319):631–3. doi:10.1038/350631a0
2. Rothemund PWK Folding DNA to Create Nanoscale Shapes and Patterns. Nature (2006) 440(7082):297–302. doi:10.1038/nature04586
3. Yin P, Hariadi RF, Sahu S, Choi HMT, Park SH, LaBean TH, et al. Programming DNA Tube Circumferences. Science (2008) 321(5890):824–6. doi:10.1126/science.1157312
4. Jones MR, Seeman NC, Mrkin CA Programmable Materials and the Nature of the DNA Bond. Science (2015) 347(6224):1260901. doi:10.1126/science.1260901
5. SantaLucia J A Unified View of Polymer, Dumbbell, and Oligonucleotide DNA Nearest-Neighbor Thermodynamics. Proc Natl Acad Sci (998) 95(4):1460–5. doi:10.1073/pnas.95.4.1460
6. Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, et al. Thermodynamic Parameters for an Expanded Nearest-Neighbor Model for Formation of RNA Duplexes with Watson−Crick Base Pairs†. Biochemistry (1998) 37(42):14719–35. doi:10.1021/bi9809425
7. Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA DNA-programmable Nanoparticle Crystallization. Nature (2008) 451(7178):553–6. doi:10.1038/nature06508
8. Macfarlane RJ, Jones MR, Lee B, Auyeung E, Mirkin CA Topotactic Interconversion of Nanoparticle Superlattices. Science (2013) 341(6151):1222–5. doi:10.1126/science.1241402
9. Ben Zion MY, He X, Maass CC, Sha R, Seeman NC, Chaikin PM Self-assembled Three-Dimensional Chiral Colloidal Architecture. Science (2017) 358(6363):633–6. doi:10.1126/science.aan5404
10. Wang S, Du JS, Diercks NJ, Zhou W, Roth EW, Dravid VP, et al. Colloidal Crystal "Alloys". J Am Chem Soc (2019) 141(51):20443–50. doi:10.1021/jacs.9b11109
11. Wang S, Park SS, Buru CT, Lin H, Chen PC, Roth EW, et al. Colloidal crystal Engineering with Metal-Organic Framework Nanoparticles and DNA. Nat Commun (2020) 11(1):2495. doi:10.1038/s41467-020-16339-w
12. He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, et al. Hierarchical Self-Assembly of DNA into Symmetric Supramolecular Polyhedra. Nature (2008) 452(7184):198–201. doi:10.1038/nature06597
13. Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, et al. Self-assembly of a Nanoscale DNA Box with a Controllable Lid. Nature (2009) 459(7243):73–6. doi:10.1038/nature07971
14. Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM Self-assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature (2009) 459(7245):414–8. doi:10.1038/nature08016
15. Woo S, Rothemund PWK Programmable Molecular Recognition Based on the Geometry of DNA Nanostructures. Nat Chem (2011) 3(8):620–7. doi:10.1038/nchem.1070
16. Udomprasert A, Bongiovanni MN, Sha R, Sherman WB, Wang T, Arora PS, et al. Amyloid Fibrils Nucleated and Organized by DNA Origami Constructions. Nat Nanotech (2014) 9(7):537–41. doi:10.1038/nnano.2014.102
17. Veneziano R, Ratanalert S, Zhang K, Zhang F, Yan H, Chiu W, et al. Designer Nanoscale DNA Assemblies Programmed from the Top Down. Science (2016) 352(6293):1534. doi:10.1126/science.aaf4388
18. Tikhomirov G, Petersen P, Qian L Fractal Assembly of Micrometre-Scale DNA Origami Arrays with Arbitrary Patterns. Nature (2017) 552(7683):67–71. doi:10.1038/nature24655
19. Yao G, Zhang F, Wang F, Peng T, Liu H, Poppleton E, et al. Meta-DNA Structures. Nat Chem (2020) 12(11):1067–75. doi:10.1038/s41557-020-0539-8
20. Pinheiro AV, Han D, Shih WM, Yan H Challenges and Opportunities for Structural DNA Nanotechnology. Nat Nanotech (2011) 6(12):763–72. doi:10.1038/nnano.2011.187
21. Zhang DY, Seelig G Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat Chem (2011) 3(2):103–13. doi:10.1038/nchem.957
22. Laramy CR, O’Brien MN, Mirkin CA Crystal Engineering with DNA. Nat Rev Mater (2019) 4(3):201–24. doi:10.1038/s41578-019-0087-2
23. Rogers WB, Shih WM, Manoharan VN Using DNA to Program the Self-Assembly of Colloidal Nanoparticles and Microparticles. Nat Rev Mater (2016) 1(3):16008. doi:10.1038/natrevmats.2016.8
24. Joshi D, Bargteil D, Caciagli A, Burelbach J, Xing Z, Nunes AS, et al. Kinetic Control of the Coverage of Oil Droplets by DNA-Functionalized Colloids. Sci Adv (2016) 2(8):e1600881. doi:10.1126/sciadv.1600881
25. Di Michele L, Varrato F, Kotar J, Nathan SH, Foffi G, Eiser E Multistep Kinetic Self-Assembly of DNA-Coated Colloids. Nat Commun (2013) 4:2007. doi:10.1038/ncomms3007
26. Moon J, Jo I-S, Ducrot E, Oh JS, Pine DJ, Yi G-R DNA-coated Microspheres and Their Colloidal Superstructures. Macromol Res (2018) 26(12):1085–94. doi:10.1007/s13233-018-6151-8
27. Mueller NS, Okamura Y, Vieira BGM, Juergensen S, Lange H, Barros EB, et al. Deep strong Light-Matter Coupling in Plasmonic Nanoparticle Crystals. Nature (2020) 583(7818):780–4. doi:10.1038/s41586-020-2508-1
28. Wang J, Sultan U, Goerlitzer ESA, Mbah CF, Engel M, Vogel N Structural Color of Colloidal Clusters as a Tool to Investigate Structure and Dynamics. Adv Funct Mater (2019) 30(26):1907730. doi:10.1002/adfm.201907730
29. Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA Selective Colorimetric Detection of Polynucleotides Based on the Distance-dependent Optical Properties of Gold Nanoparticles. Science (1997) 277(5329):1078–81. doi:10.1126/science.277.5329.1078
30. Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature (1996) 382(6592):607–9. doi:10.1038/382607a0
31. Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, et al. Organization of 'nanocrystal Molecules' Using DNA. Nature (1996) 382(6592):609–11. doi:10.1038/382609a0
32. Soto CM, Srinivasan A, Ratna BR Controlled Assembly of Mesoscale Structures Using DNA as Molecular Bridges. J Am Chem Soc (2002) 124(29):8508–9. doi:10.1021/ja017653f
33. Milam VT, Hiddessen AL, Crocker JC, Graves DJ, Hammer DA DNA-driven Assembly of Bidisperse, Micron-Sized Colloids. Langmuir (2003) 19(24):10317–23. doi:10.1021/la034376c
34. Rogers PH, Michel E, Bauer CA, Vanderet S, Hansen D, Roberts BK, et al. Selective, Controllable, and Reversible Aggregation of Polystyrene Latex Microspheres via DNA Hybridization. Langmuir (2005) 21(12):5562–9. doi:10.1021/la046790y
35. Kim AJ, Biancaniello PL, Crocker JC Engineering DNA-Mediated Colloidal Crystallization. Langmuir (2006) 22(5):1991–2001. doi:10.1021/la0528955
36. Leunissen ME, Christova CG, Hynninen A-P, Royall CP, Campbell AI, Imhof A, et al. Ionic Colloidal Crystals of Oppositely Charged Particles. Nature (2005) 437(7056):235–40. doi:10.1038/nature03946
37. Hueckel T, Hocky GM, Palacci J, Sacanna S Ionic Solids from Common Colloids. Nature (2020) 580(7804):487–90. doi:10.1038/s41586-020-2205-0
38. Glotzer SC, Solomon MJ Anisotropy of Building Blocks and Their Assembly into Complex Structures. Nat Mater (2007) 6(8):557–62. doi:10.1038/nmat1949
39. Wang Y, Wang Y, Breed DR, Manoharan VN, Feng L, Hollingsworth AD, et al. Colloids with Valence and Specific Directional Bonding. Nature (2012) 491(7422):51–5. doi:10.1038/nature11564
40. Ho KM, Chan CT, Soukoulis CM Existence of a Photonic gap in Periodic Dielectric Structures. Phys Rev Lett (1990) 65(25):3152–5. doi:10.1103/physrevlett.65.3152
41. John S Strong Localization of Photons in Certain Disordered Dielectric Superlattices. Phys Rev Lett (1987) 58(23):2486–9. doi:10.1103/physrevlett.58.2486
42. Yablonovitch E Inhibited Spontaneous Emission in Solid-State Physics and Electronics. Phys Rev Lett (1987) 58(20):2059–62. doi:10.1103/physrevlett.58.2059
43. Joannopoulos JD. Photonic Crystals : Molding the Flow of Light. 2nd ed.. Princeton: Princeton University Press (2008).
44. He M, Gales JP, Ducrot É, Gong Z, Yi G-R, Sacanna S, et al. Colloidal diamond. Nature (2020) 585(7826):524–9. doi:10.1038/s41586-020-2718-6
45. Dreyfus R, Leunissen ME, Sha R, Tkachenko A, Seeman NC, Pine DJ, et al. Aggregation-disaggregation Transition of DNA-Coated Colloids: Experiments and Theory. Phys Rev E Stat Nonlin Soft Matter Phys (2010) 81(4 Pt 1):041404. doi:10.1103/physreve.81.041404
46. Michele LD, Eiser E Developments in Understanding and Controlling Self Assembly of DNA-Functionalized Colloids. Phys Chem Chem Phys (2013) 15(9):3115. doi:10.1039/c3cp43841d
47. Angioletti-Uberti S, Mognetti BM, Frenkel D Re-entrant Melting as a Design Principle for DNA-Coated Colloids. Nat Mater (2012) 11(6):518–22. doi:10.1038/nmat3314
48. Wang Y, Wang Y, Zheng X, Ducrot E, Yodh JS, Weck M, et al. Crystallization of DNA-Coated Colloids. Nat Commun (2015) 6:7253. doi:10.1038/ncomms8253
49. Licata NA, Tkachenko AV Dynamics of Particles with "Key-Lock" Interactions. Europhys Lett (2008) 81(4):48009. doi:10.1209/0295-5075/81/48009
50. van der Meulen SAJ, Leunissen ME Solid Colloids with Surface-Mobile DNA Linkers. J Am Chem Soc (2013) 135(40):15129–34. doi:10.1021/ja406226b
51. Macfarlane RJ, Lee B, Jones MR, Harris N, Schatz GC, Mirkin CA Nanoparticle Superlattice Engineering with DNA. Science (2011) 334(6053):204–8. doi:10.1126/science.1210493
52. Lu F, Yager KG, Zhang Y, Xin H, Gang O Superlattices Assembled through Shape-Induced Directional Binding. Nat Commun (2015) 6:6912. doi:10.1038/ncomms7912
53. Schade NB, Holmes-Cerfon MC, Chen ER, Aronzon D, Collins JW, Fan JA, et al. Tetrahedral Colloidal Clusters from Random Parking of Bidisperse Spheres. Phys Rev Lett (2013) 110(14):148303. doi:10.1103/physrevlett.110.148303
54. Rogers WB, Manoharan VN Programming Colloidal Phase Transitions with DNA Strand Displacement. Science (2015) 347(6222):639–42. doi:10.1126/science.1259762
55. Mognetti BM, Leunissen ME, Frenkel D Controlling the Temperature Sensitivity of DNA-Mediated Colloidal Interactions through Competing Linkages. Soft Matter (2012) 8(7):2213. doi:10.1039/c2sm06635a
56. Biancaniello PL, Kim AJ, Crocker JC Colloidal Interactions and Self-Assembly Using DNA Hybridization. Phys Rev Lett (2005) 94(5):058302. doi:10.1103/physrevlett.94.058302
57. Rogers WB, Crocker JC Direct Measurements of DNA-Mediated Colloidal Interactions and Their Quantitative Modeling. Proc Natl Acad Sci (2011) 108(38):15687–92. doi:10.1073/pnas.1109853108
58. Rogers WB, Sinno T, Crocker JC Kinetics and Non-exponential Binding of DNA-Coated Colloids. Soft Matter (2013) 9(28):6412. doi:10.1039/c3sm50593f
59. Varilly P, Angioletti-Uberti S, Mognetti BM, Frenkel D A General Theory of DNA-Mediated and Other Valence-Limited Colloidal Interactions. J Chem Phys (2012) 137(9):094108. doi:10.1063/1.4748100
60. Angioletti-Uberti S, Varilly P, Mognetti BM, Tkachenko AV, Frenkel D Communication: a Simple Analytical Formula for the Free Energy of Ligand-Receptor-Mediated Interactions. J Chem Phys (2013) 138(2):021102. doi:10.1063/1.4775806
61. Angioletti-Uberti S, Varilly P, Mognetti BM, Frenkel D Mobile Linkers on DNA-Coated Colloids: Valency without Patches. Phys Rev Lett (2014) 113(12):128303. doi:10.1103/physrevlett.113.128303
62. Gehrels EW, Rogers WB, Manoharan VN Using DNA Strand Displacement to Control Interactions in DNA-Grafted Colloids. Soft Matter (2018) 14(6):969–84. doi:10.1039/c7sm01722g
63. Lowensohn J, Oyarzún B, Narváez Paliza G, Mognetti BM, Rogers WB Linker-Mediated Phase Behavior of DNA-Coated Colloids. Phys Rev X (2019) 9(4):041054. doi:10.1103/physrevx.9.041054
64. Lowensohn J, Hensley A, Perlow-Zelman M, Rogers WB Self-Assembly and Crystallization of DNA-Coated Colloids via Linker-Encoded Interactions. Langmuir (2020) 36(25):7100–8. doi:10.1021/acs.langmuir.9b03391
65. Rogers WB A Mean-Field Model of Linker-Mediated Colloidal Interactions. J Chem Phys (2020) 153(12):124901. doi:10.1063/5.0020578
66. Fang H, Hagan MF, Rogers WB Two-step Crystallization and Solid-Solid Transitions in Binary Colloidal Mixtures. Proc Natl Acad Sci USA (2020) 117(45):27927–33. doi:10.1073/pnas.2008561117
67. Xia X, Hu H, Ciamarra MP, Ni R Linker-mediated Self-Assembly of mobile DNA-Coated Colloids. Sci Adv (2020) 6(21):eaaz6921. doi:10.1126/sciadv.aaz6921
68. Jana PK, Mognetti BM Translational and Rotational Dynamics of Colloidal Particles Interacting through Reacting Linkers. Phys Rev E (2019) 100(6-1):060601. doi:10.1103/physreve.100.060601
69. Lee-Thorp JP, Holmes-Cerfon M Modeling the Relative Dynamics of DNA-Coated Colloids. Soft Matter (2018) 14(40):8147–59. doi:10.1039/c8sm01430b
70. Wang Y, Wang Y, Zheng X, Ducrot É, Lee M-G, Yi G-R, et al. Synthetic Strategies toward DNA-Coated Colloids that Crystallize. J Am Chem Soc (2015) 137(33):10760–6. doi:10.1021/jacs.5b06607
71. Oh JS, Wang Y, Pine DJ, Yi G-R High-Density PEO-B-DNA Brushes on Polymer Particles for Colloidal Superstructures. Chem Mater (2015) 27(24):8337–44. doi:10.1021/acs.chemmater.5b03683
72. Kim AJ, Manoharan VN, Crocker JC Swelling-Based Method for Preparing Stable, Functionalized Polymer Colloids. J Am Chem Soc (2005) 127(6):1592–3. doi:10.1021/ja0450051
73. Oh JS, He M, Yi G-R, Pine DJ High-Density DNA Coatings on Carboxylated Colloids by DMTMM- and Azide-Mediated Coupling Reactions. Langmuir (2020) 36(13):3583–9. doi:10.1021/acs.langmuir.9b03386
74. Oh JS, Yi G-R, Pine DJ Reconfigurable Self-Assembly and Kinetic Control of Multiprogrammed DNA-Coated Particles. ACS Nano (2020) 14(4):4595–600. doi:10.1021/acsnano.0c00164
75. Elechiguerra JL, Reyes-Gasga J, Yacaman MJ The Role of Twinning in Shape Evolution of Anisotropic noble Metal Nanostructures. J Mater Chem (2006) 16(40):3906. doi:10.1039/b607128g
76. Chen Q, Bae SC, Granick S Directed Self-Assembly of a Colloidal Kagome Lattice. Nature (2011) 469(7330):381–4. doi:10.1038/nature09713
77. O'Brien MN, Lin HX, Girard M, Olvera de la Cruz M, Mirkin CA Programming Colloidal Crystal Habit with Anisotropic Nanoparticle Building Blocks and DNA Bonds. J Am Chem Soc (2016) 138(44):14562–5. doi:10.1021/jacs.6b09704
78. Feng L, Dreyfus R, Sha R, Seeman NC, Chaikin PM DNA Patchy Particles. Adv Mater (2013) 25(20):2779–83. doi:10.1002/adma.201204864
79. McGinley JT, Wang Y, Jenkins IC, Sinno T, Crocker JC Crystal-templated Colloidal Clusters Exhibit Directional DNA Interactions. ACS Nano (2015) 9(11):10817–25. doi:10.1021/acsnano.5b03272
80. Diaz A JA, Oh JS, Yi GR, Pine DJ Photo-printing of Faceted DNA Patchy Particles. Proc Natl Acad Sci U S A (2020) 117(20):10645–53. doi:10.1073/pnas.1918504117
81. Zheng X, Wang Y, Wang Y, Pine DJ, Weck M Thermal Regulation of Colloidal Materials Architecture through Orthogonal Functionalizable Patchy Particles. Chem Mater (2016) 28(11):3984–9. doi:10.1021/acs.chemmater.6b01313
82. Oh JS, Lee S, Glotzer SC, Yi G-R, Pine DJ Colloidal Fibers and Rings by Cooperative Assembly. Nat Commun (2019) 10(1):3936. doi:10.1038/s41467-019-11915-1
83. Oh JS, Yi G-R, Pine DJ Reconfigurable Transitions between One- and Two-Dimensional Structures with Bifunctional DNA-Coated Janus Colloids. ACS Nano (2020) 14(11):15786–92. doi:10.1021/acsnano.0c06846
84. Liu M, Zheng X, Grebe V, He M, Pine DJ, Weck M Two-Dimensional (2D) or Quasi-2D Superstructures from DNA-Coated Colloidal Particles. Angew Chem Int Ed (2021) 60(11):5744–8. doi:10.1002/anie.202014045
85. Askeland DR, Fulay PP. The Science and Engineering of Materials. 5th ed.. Toronto, London: Nelson; Thomson (2006).
86. Pusey PN, van Megen W Phase Behaviour of Concentrated Suspensions of Nearly Hard Colloidal Spheres. Nature (1986) 320(6060):340–2. doi:10.1038/320340a0
87. Wang Y, Jenkins IC, McGinley JT, Sinno T, Crocker JC Colloidal Crystals with diamond Symmetry at Optical Lengthscales. Nat Commun (2017) 8:14173. doi:10.1038/ncomms14173
88. Ducrot É, He M, Yi G-R, Pine DJ Colloidal Alloys with Preassembled Clusters and Spheres. Nat Mater (2017) 16(6):652–7. doi:10.1038/nmat4869
89. Gong Z, Hueckel T, Yi G-R, Sacanna S Patchy Particles Made by Colloidal Fusion. Nature (2017) 550(7675):234–8. doi:10.1038/nature23901
90. Johnson S, Joannopoulos J Block-iterative Frequency-Domain Methods for Maxwell's Equations in a Planewave Basis. Opt Express (2001) 8(3):173. doi:10.1364/oe.8.000173
91. Majewski PW, Michelson A, Cordeiro MAL, Tian C, Ma C, Kisslinger K, et al. Resilient Three-Dimensional Ordered Architectures Assembled from Nanoparticles by DNA. Sci Adv (2021) 7(12):eabf0617. doi:10.1126/sciadv.abf0617
92. Shani L, Michelson AN, Minevich B, Fleger Y, Stern M, Shaulov A, et al. DNA-assembled Superconducting 3D Nanoscale Architectures. Nat Commun (2020) 11(1):5697. doi:10.1038/s41467-020-19439-9
93. Sun W, Shen J, Zhao Z, Arellano N, Rettner C, Tang J, et al. Precise Pitch-Scaling of Carbon Nanotube Arrays within Three-Dimensional DNA Nanotrenches. Science (2020) 368(6493):874–7. doi:10.1126/science.aaz7440
94. Pei H, Sha R, Wang X, Zheng M, Fan C, Canary JW, et al. Organizing End-Site-Specific SWCNTs in Specific Loci Using DNA. J Am Chem Soc (2019) 141(30):11923–8. doi:10.1021/jacs.9b03432
95. Liu W, Tagawa M, Xin HL, Wang T, Emamy H, Li H, et al. Diamond Family of Nanoparticle Superlattices. Science (2016) 351(6273):582–6. doi:10.1126/science.aad2080
96. Santos PJ, Gabrys PA, Zornberg LZ, Lee MS, Macfarlane RJ Macroscopic Materials Assembled from Nanoparticle Superlattices. Nature (2021) 591(7851):586–91. doi:10.1038/s41586-021-03355-z
97. Chakrabarti R, Klibanov AM Nanocrystals Modified with Peptide Nucleic Acids (PNAs) for Selective Self-Assembly and DNA Detection. J Am Chem Soc (2003) 125(41):12531–40. doi:10.1021/ja035399g
98. Zeravcic Z, Manoharan VN, Brenner MP Size Limits of Self-Assembled Colloidal Structures Made Using Specific Interactions. Proc Natl Acad Sci USA (2014) 111(45):15918–23. doi:10.1073/pnas.1411765111
99. Tkachenko AV Morphological Diversity of DNA-Colloidal Self-Assembly. Phys Rev Lett (2002) 89(14):148303. doi:10.1103/physrevlett.89.148303
100. Jacobs WM, Reinhardt A, Frenkel D Rational Design of Self-Assembly Pathways for Complex Multicomponent Structures. Proc Natl Acad Sci USA (2015) 112(20):6313–8. doi:10.1073/pnas.1502210112
Keywords: DNA colloidal crystals, patchy particle, anisotropic particle, programmable self-assembly, self-assembly
Citation: Zhang T, Lyu D, Xu W, Mu Y and Wang Y (2021) Programming Self-Assembled Materials With DNA-Coated Colloids. Front. Phys. 9:672375. doi: 10.3389/fphy.2021.672375
Received: 25 February 2021; Accepted: 25 May 2021;
Published: 19 July 2021.
Edited by:
Stefano Angioletti-Uberti, Imperial College London, United KingdomReviewed by:
Bortolo Matteo Mognetti, Université libre de Bruxelles, BelgiumW. Benjamin Rogers, Brandeis University, United States
Copyright © 2021 Zhang, Lyu, Xu, Mu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yufeng Wang, d2FuZ2xhYkBoa3UuaGs=
 Tianran Zhang
Tianran Zhang Dengping Lyu
Dengping Lyu Wei Xu
Wei Xu Yijiang Mu
Yijiang Mu Yufeng Wang
Yufeng Wang