- 1QIMP Team, Center for Medical Physics and Biomedical Engineering, Medical University of Vienna, Vienna, Austria
- 2Division of Nuclear Medicine, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria
Medical imaging has evolved from a pure visualization tool to representing a primary source of analytic approaches toward in vivo disease characterization. Hybrid imaging is an integral part of this approach, as it provides complementary visual and quantitative information in the form of morphological and functional insights into the living body. As such, non-invasive imaging modalities no longer provide images only, but data, as stated recently by pioneers in the field. Today, such information, together with other, non-imaging medical data creates highly heterogeneous data sets that underpin the concept of medical big data. While the exponential growth of medical big data challenges their processing, they inherently contain information that benefits a patient-centric personalized healthcare. Novel machine learning approaches combined with high-performance distributed cloud computing technologies help explore medical big data. Such exploration and subsequent generation of knowledge require a profound understanding of the technical challenges. These challenges increase in complexity when employing hybrid, aka dual- or even multi-modality image data as input to big data repositories. This paper provides a general insight into medical big data analysis in light of the use of hybrid imaging information. First, hybrid imaging is introduced (see further contributions to this special Research Topic), also in the context of medical big data, then the technological background of machine learning as well as state-of-the-art distributed cloud computing technologies are presented, followed by the discussion of data preservation and data sharing trends. Joint data exploration endeavors in the context of in vivo radiomics and hybrid imaging will be presented. Standardization challenges of imaging protocol, delineation, feature engineering, and machine learning evaluation will be detailed. Last, the paper will provide an outlook into the future role of hybrid imaging in view of personalized medicine, whereby a focus will be given to the derivation of prediction models as part of clinical decision support systems, to which machine learning approaches and hybrid imaging can be anchored.
Introduction
Hybrid Imaging
Patient management today entails the use of non-invasive imaging methods. These fall into two categories: anatomical or morphological imaging and molecular or functional imaging. The first category includes imaging methods, such as X-ray, Computed Tomography (CT), or Ultrasound imaging (US) while the second category is the domain of nuclear medicine imaging, employing techniques such as Single Photon Emission Computer Tomography (SPECT) and Positron Emission Tomography (PET). Magnetic Resonance Imaging (MRI) is somewhat in between both categories for it provides anatomical details with high visual contrast while probing functional as well as insights into metabolic pathways [1].
Standalone imaging methods have been used for decades in patient diagnosis. Patients suffering from cancer, cardiovascular diseases or neurodegenerative disease, however, have been shown to benefit from the use of so-called combined, or hybrid imaging methods. Hybrid imaging describes the physical combination of complementary imaging systems, such as SPECT/CT [2], PET/CT [3], and PET/MRI [4], all of which provide “anato-metabolic” image information [5], that is based on intrinsically aligned morphological and functional data.
Routine diagnosis based on hybrid imaging employs mainly visual data interpretation [6, 7]. There is, however, much more information in these data that can be turned into knowledge [8]. Extraction and analysis of simple to higher-order radiomic features comprises a revolution in the field of in vivo disease characterization [7]. This concept appears to be particularly promising in the field of cancer research [8, 9], where diagnosis of tumors is typically performed with ex vivo approaches, such as biopsy. The drawback of biopsies—beyond being invasive—is that they provide only information about a small region of tumors from where the sample is taken (Figure 1). Furthermore, the majority of tumors are heterogeneous across all scales [10]. Hybrid imaging can help describe the overall heterogeneity of tumors on both morphological and metabolic levels [10, 11]. Therefore, hybrid imaging appears to be a key technology to build accurate, in vivo tumor characterization models [7].
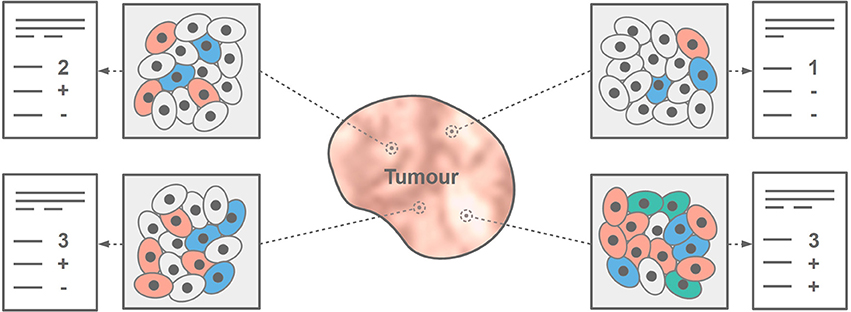
Figure 1. Tumor heterogeneity cannot be fully assessed with a core needle biopsy. Depending on the sampled region, histopathology results may differ, thus, pointing to different tumor biomarkers that subsequently affect the choice of therapy.
Hybrid Imaging and Medical Big Data
Our civilization has been dealing with finding ways to handle large-scale data sets for thousands of years [12, 13] (Figure 2). Thanks to multiple technological advances, our approach toward handling such data has continuously advanced, thus, resulting in the birth of the term “Big Data” [14–16]. The so-called 4 V model is one of the simplest ways to characterize Big Data [17] through four major features: volume, velocity, variety, and veracity (Figure 3), all of which help describe key observations of big data during their evaluation (Table 1).
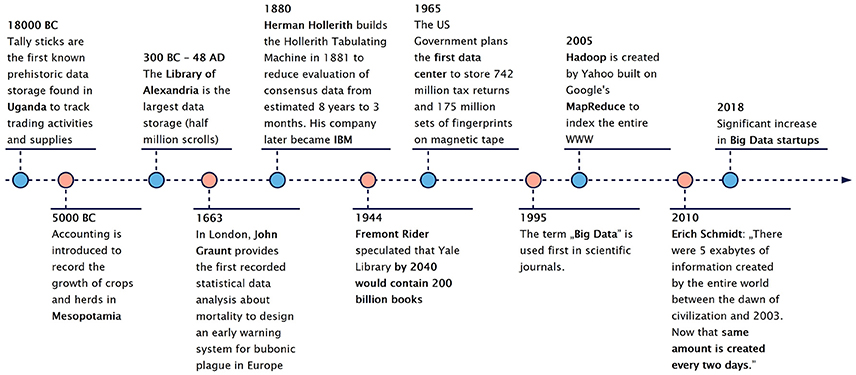
Figure 2. Data and Big data during the past eras of mankind [12, 13]. People were faced with the challenges of data storage from their beginning. In the modern history of mankind big data is bound to digital data storage, distributed supercomputing as well as novel evaluation endeavors.
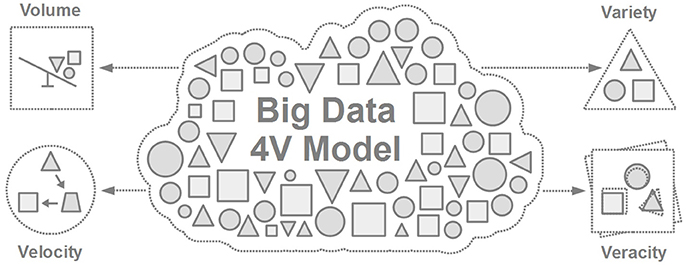
Figure 3. The 4V model of Big Data [17] referring to Volume (data size), Velocity (speed of change), Variety (different sources and formats of data), and Veracity (uncertainty in data).
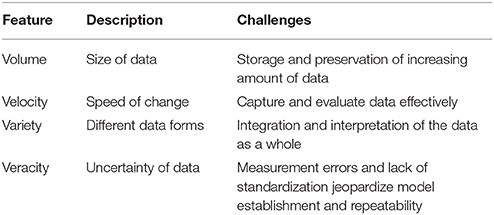
Table 1. Four major features of Big Data according to the 4V model [17].
For the past years, the volume of Big Data has grown exponentially. Based on a 2013 estimation, 90% of the world's data was generated in the two prior years (2011–2013) [18]. The Ponemon Institute estimated that 30% of all electronic data in 2012 was generated by healthcare alone [19]. According to a 2016 estimation from IBM researchers, 90% of all Medical Big Data is imaging data [20].
Modern PET/CT and PET/MRI systems provide gigabytes of datasets per study [21, 22]. Furthermore, hybrid imaging combines different modalities representing different image sizes and resolution levels, thus, it inherently results in datasets with a high variety. Both dynamic and gated acquisitions are subject to low sensitivity and longer acquisition times due to velocity of events in the living body [23]. The veracity in imaging patterns originating from varieties in imaging hardware, acquisition protocols and image reconstructions across vendors and system generations is understood, but remains a challenge for a better understanding of disease and multi-center data pooling alike [24]. All of these features eventually define hybrid imaging data a major component of Medical Big Data.
Machine Learning for Medical Big Data Analysis
Medical Big Data cannot be dealt with by traditional data processing applications [25], thus, novel data handling and evaluation approaches are required that extend beyond conventional software processing capabilities. Machine learning is a promising approach to deal with large-scale medical data [26]. In light of hybrid imaging, several groups have reported promising results for disease characterization by applying robust machine learning methods for combined in vivo analysis [27–29].
Furthermore, holistic approaches that combine clinical and imaging information become more popular [30–32]. Current technological advances support the collection of large-scale, heterogeneous information from living organisms, not only by hybrid imaging, but also by genomics [31], proteomics [33], or histopathology [34]. Such highly-heterogeneous datasets are very challenging to match, given the different time points and frequencies they have been acquired with. Nevertheless, the exploration of Medical Big Data is linked to a fully-personalized patient care [14, 35]. With the emergence of Medical Internet of Things (MIoT), real-time remote monitoring becomes feasible [36–38]. By collecting and combining MIoT information with hybrid imaging data, the accuracy of predictive analytics can be increased significantly [38] and can result in automated, Clinical Decision Support systems (CDSS) [39] (Figure 4). This leads to healthcare approaches that help improve patient comfort and reduce healthcare costs as part of a fully patient-centric, personalized medicine [40] (Figure 5). Hybrid imaging data, as a major constituent of this process, plays a central role in personalized medicine [41].

Figure 4. Representation of Medical Big Data as the result of various information capturing systems and examinations. The variety of Medical Big Data is manifested not only in the different structural nature of the collected data, but also in the various frequencies the given data can be collected from living beings. Machine Learning approaches can help in automatically exploring and analyzing this highly heterogeneous dataset, resulting in predictive models. These technological approaches can result in a Clinical Decision Support system (CDSS) that can help physicians to shift diagnosis and treatment toward precision medicine.
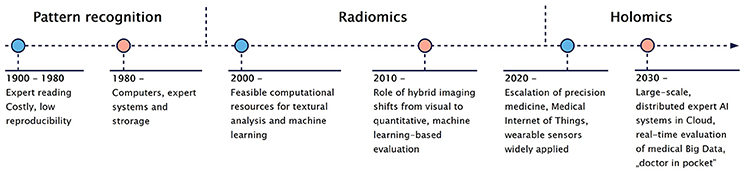
Figure 5. Brief introduction to the evolution of medical image interpretation and its incorporation to other Medical Big Data. From the early 1900s medical imaging data was interpreted manually by human based pattern recognition approaches. According to current trends, Medical Internet of Things [37] together with wearable sensors will support a real-time health status tracking. This trend will help to combine and analyse heterogeneous Medical Big Data (Holomics), which will be driven by expert AI systems operating in the cloud.
Technology
Machine Learning
Machine Learning refers to approaches that are able to identify and learn from patterns of datasets [7, 42, 43]. Even though ML algorithms can differ significantly, each of them can be characterized by the same logical structure composed of a model, a fitness measurement, and an optimizer (Figure 6).
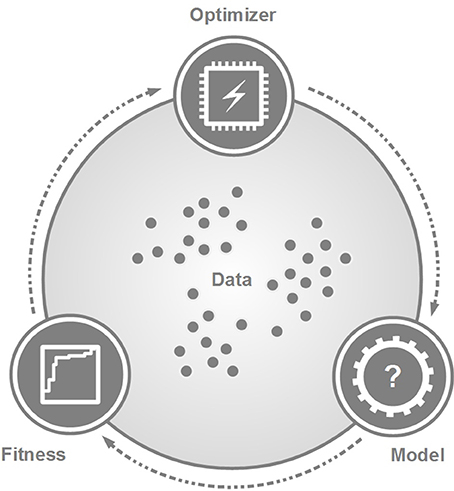
Figure 6. The general scheme of machine learning approaches. The data serve as an input to explore and to build a decision-making model by the optimizer. A fitness measurement characterizes the performance of the model. This scheme can symbolize a one-step or iterative process, depending on the given ML approach. Of note, the three modules may have different relationships with regards to the actual ML approach.
The model predicts new information from the data and it is the result of any ML training process. The optimizer generates the model in a way that its fitness value is maximized. Specific algorithms may interpret the relationship of the model, the fitness measure and the optimizer in different ways. In case of unsupervised machine learning, for example, the fitness measure can be an internal step of the optimizer by measuring, e.g., within-cluster variances [44]. In contrast, supervised ML approaches may consider the fitness measure as an independent component from the optimizer [45]. Furthermore, the optimizer and model may not be independent but represent an integrative structure [46, 47].
There are numerous ways to categorize ML algorithms. One way of categorization relies on the nature of the data to be analyzed, thus, leading to the categories of supervised, unsupervised, and reinforcement methods (Figure 7). Supervised machine learning is a classification or regression approach that builds a predictive model based on labeled reference data [45]. Several research groups focusing on imaging analysis apply supervised machine learning to retrospective datasets [7, 10, 43].
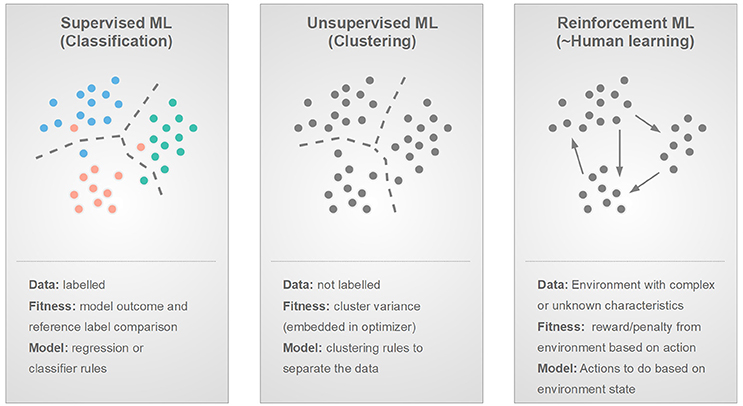
Figure 7. Three main types of machine learning based on the nature of the input data or environment: supervised, unsupervised and reinforcement learning and their main properties by the means of data, fitness, and model.
Unsupervised machine learning operates with unlabeled data, hence, it can be characterized as a clustering approach [44]. Research groups focusing on exploratory analysis of their imaging dataset without a ground truth, utilize unsupervised machine learning methodologies [46, 48–50].
Reinforcement learning mimics human learning, thus, it considers that there is an environment with a certain state that can be changed by an ML “agent” through certain actions [46, 51]. Whenever, an action is taken and the environment state changes, a reward or punishment is issued back to the agent. The goal is to build a set of actions that maximize the reward and minimize the punishment regardless of the current environment stage. Reinforcement learning can be utilized in case a given environment is very complex, prone to changes, or—generally speaking—is of unknown nature [51]. To date, reinforcement learning is underrepresented in the field of medical imaging with only limited applications [46, 52].
Another type of categorizing ML methods follows the nature of the feature extraction and analysis of the data. In this case two main groups can be identified, such as shallow learning methods building on engineered (or handcrafted) features [8, 53] and deep learning (DL) methods building on automatically comprehended, multi-layer representation of the data [54]. Several related works utilize machine learning built on engineered features [55–59]. These approaches typically employ feature selection [60] in the form of feature redundancy reduction [61] or feature ranking [8].
Deep learning is reported to generally outperform shallow learning ML approaches, as it is able to decompose and analyse the data on different levels of information complexity [54, 62]. Nevertheless, the true potential of DL is seen only in view of data having a complex, hierarchical structure. This is a challenging requirement to fulfill, and, thus, DL to date is underrepresented in the context of tumor characterization and hybrid imaging [62, 63]. On the other hand, DL appears to be a promising approach to synthetize artificial CT from MRI for the attenuation correction of PET images acquired by hybrid PET/MRI imaging systems [64]. In addition, DL appears helpful for dealing with brain disease characterization, such as AD/MCI based on PET/MRI data [65, 66].
Despite the wide range of ML approaches available today, there is no unique ML method which generally outperforms all others [67]. Therefore, testing of multiple ML approaches is encouraged to identify the most suitable for a particular evaluation [68]. Table 2 provides an overview of the most common ML algorithms applied in medical science.
High-Performance and Cloud Computing
Machine learning evaluation of Medical Big Data requires high-performance computational resources [50]. As the amount of data increases exponentially, progressively complex computational architectures are needed for the storage, processing, and analysis of the data [98].
Distributed systems, such as the Hadoop ecosystem are potential solutions to deal with Medical Big Data [99, 100]. The foundation of the Hadoop ecosystem is Apache Hadoop with two major functional components: the MapReduce model for data processing and the Hadoop Distributed File System (HDFS) for storage. MapReduce splits the input data set into independent pieces processed in parallel by map tasks, while the “reduce” component combines the outputs of the map tasks afterwards [101]. The HDFS is a fault-tolerant distributed file system designed to run on low-cost hardware, which is suitable for medical image data applications [102]. Hadoop has been applied to address numerous tasks in medical imaging, such as parameter optimization for lung texture segmentation, content-based medical image indexing, and 3D directional wavelet analysis for solid texture classification [99, 103, 104].
Next to Apache Hadoop, several distributed high-performance computing platforms are available as well [99, 100]. One example is the open-source Apache Spark [105, 106], which has a better ability of computing compared to Hadoop [107, 108]. It has been shown that Spark is up to 20 times faster than Hadoop for iterative applications, it accelerates a real-world data analytics report by a factor of 40 and it can be used interactively to scan a 1 TB dataset within a few seconds [109]. These characteristics enable Spark to serve as an efficient tool for medical imaging data analysis tasks, such as the computation of voxel-wise local clustering coefficients of fMRI data [107].
To date, major industry leaders provide cloud storage with the combination of cloud ML engines to address the need of Big Data evaluation [15, 110]. These systems are potentially ideal frameworks for large-scale hybrid imaging data evaluation [111, 112]. An example service is the Google Cloud Platform used by both academic research institutions and by a variety of healthcare companies. Here, the Google Cloud Machine Learning Engine can be utilized to submit anonymized MRI scans to an ML-enabled AI platform to help diagnose prostate cancer [113].
Similar to the Google Cloud Platform, Amazon provides a suite of services called the Amazon Web Services including cloud computing and machine learning tools. The Amazon Elastic Compute Cloud, for example, is an Infrastructure as a Service which offers the possibility to rent virtual computers. It is being used to develop a technology for supporting radiologists to identify abnormalities in medical images across different modalities as well as for providing a blood flow imaging solution that enables doctors to render MRI scans in multi-dimensional models and better diagnose patients for cardiovascular diseases [114, 115]1.
Another package for cloud-based services is Azure provided by Microsoft. Azure has been employed for medical image classification using algorithms, such as support vector machines and k-nearest neighbors [116, 117]. A few machine learning examples for medical imaging analysis utilizing Azure are covered in Criminisi [118].
Data Handling
Data Preservation and Reproduction
Beyond technical considerations of evaluating large-scale medical data, persistent storage is a challenge for various medical institutions. To date, hospitals that generate and collect medical data are also responsible for archiving the data [119]. The process is further defined in the triangle of legal obligations, practical considerations as well as financial resources. The time period a hospital needs to archive patient data varies with the country, however, mandatory preservation periods are generally between 10 and 100 years [120]. However, these storage periods are much longer than the estimated storage durability of memory technologies available today (Table 3). Medical imaging related research, particularly research involving longitudinal and/or large-scale population analysis, may mandate data sets that have been acquired over decades. In the context of hybrid imaging, state-of-the-art PET/CT and PET/MRI systems provide large datasets, as a raw PET list mode data may grow over several gigabytes of storage space, while a multi-slice CT may correspond to ~2 gigabytes of data [21]. Furthermore, a wide range of MRI sequences can be acquired as part of a PET/MRI study [22], that further add to the issue of dealing with large data. Since PET raw data (aka list mode files) is considerably large, it is frequently not archived at all [122]. This prevents scientists from retrospectively optimizing image quality by new image reconstruction approaches and eventually to standardize protocols for accurate, population-wide evaluations. Since the required data preservation for both routine and research purposes is not feasible by conventional tools, there is an urgent need to shift focus toward more persistent solutions. As an example, cloud storage and evaluation approaches [111, 112] can support convenient data sharing and repeatability of published results across various research groups.

Table 3. Estimated lifespan of some media storage technologies according to Morgan [121].
Data Sharing
Clinical research is an essential building block for the concept of efficient patient management. Research studies are generally complex and the resulting data are valuable, not only to the principal investigator but to society as a whole [123, 124]. Nonetheless, many researchers remain reluctant to share their data with an expert audience [125, 126] beyond describing them as part of peer-reviewed publications. In contrast, sharing research data in a structured and tangible way has been shown to yield benefits for both the principal investigators and other experts in the field who may re-use the data with alternative evaluation approaches to extract new information that may subsequently benefit patient management [124]. Journals, like “Science” or “Nature” expect data to be made public and, therefore, provide the necessary means [124]. However, the quality of public supplementary material collections of published studies is variable, and frequently the re-use of these data is not possible [127]. The same holds true for the quality of alternative public data archives that were shown to contain incomplete data and data archived only partially in over 56% cases that prevented re-use [126].
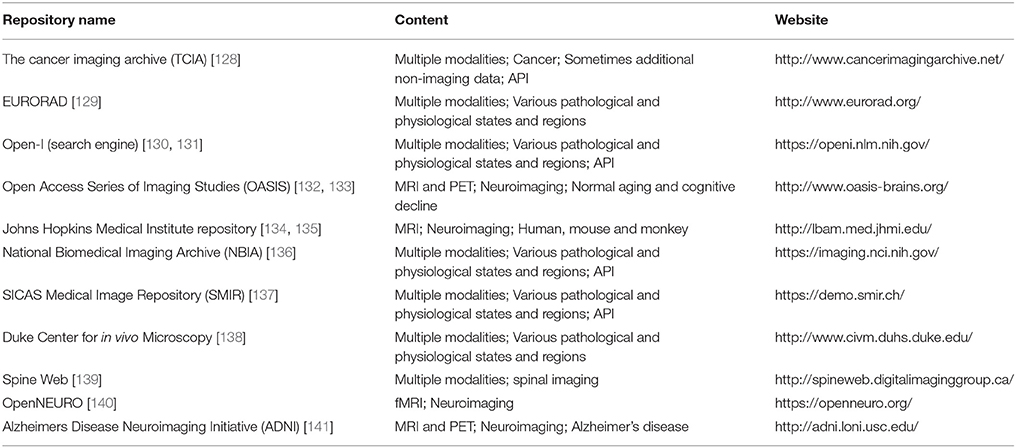
In the light of medical imaging and hybrid imaging in particular, a wide range of imaging data from different fields is already available for researchers worldwide (Table 4). Some of these databases are dedicated to the collection and sharing of very heterogeneous data from different modalities, various diseases, and different body regions.
One such data source is The Cancer Imaging Archive (TCIA). TCIA is an open-access repository, funded by the National Cancer Institute, containing several million medical images from various cancer types2. The data is partitioned into collections based on characteristics, such as cancer type or affected region in common. Apart from supplying users with massive amounts of high quality data, it also provides an application programming interface (API) for automated data access. EURORAD, which is operated by the European Society of Radiology also includes but is not limited to cancer images. Nevertheless, it mainly focuses on the training of radiologists and provides no automated data access3 Open-i is a service hosted by the National Library of Medicine (NLM) [130]. It provides a search engine and a download API for accessing images from PubMed Central articles, NLM History of Medicine collection and other sources. Nevertheless, not all images which can be retrieved using open-i are free to use. Another major source for in vivo medical images is the National Biomedical Imaging Archive (NBIA) which includes clinical and genetic data associated to the images [136].
In addition to these more general data repositories, there are many specialized data sources for medical images. The Open Access Series of Imaging Studies (OASIS) for example, includes comparative data for patients with Alzheimer's disease and normal physiological conditions [132]. In addition to neuroimaging data, it also includes clinical and biomarker information. Another specialized data base is the medical image repository of the Johns Hopkins Medical Institute [134]. It includes MRI images of human, mouse and monkey brain images. Further data sources for medical images can be found in Table 4.
Joint Data Exploration
To date, in vivo disease characterization with hybrid imaging data—especially in the light of oncological applications—is performed mainly by analyzing engineered features [63, 129]. This process is widely referred to as “Radiomics,” even though, this kind of approach was originally applied to morphological images only [8]. In an early publication “Radiomics” was defined as “the high-throughput extraction of large amounts of image features from radiographic images” [128]. For the sake of consistency, we employ the term “radiomics” in the context of in-vivo feature analysis, including those derived from functional and hybrid imaging. However, we introduce a different term, “Holomics,” to address combinations of imaging and non-imaging data, and that we consider more appropriate than the term “imiomics,” as suggested in Strand et al. [142] for the combined analysis of imaging and—omics data.
Radiomics
In general, any radiomics-based exploration of imaging data requires object delineation, followed by feature extraction and evaluation [128, 143, 144]. Extracted features cover the range of first to higher order, wavelet, Laplacian, and fractal features [6, 8, 145, 146]. Some of these features—referred to as textural features—characterize certain spatial patterns in images. The concept of textural evaluation in medical images has been first introduced by Haralick et al. [147] in the 1970s. At the time neither the image quality nor the computational capacity was sufficient to operate with textural features. Thanks to recent advances in imaging and computational fields, several groups have investigated the potential of textural features in light of in vivo disease characterization [8, 28, 56, 148–150].
Routine clinical evaluation of PET images relies on the statistical analysis of Standardized Uptake Values (SUV) [151]. In case of oncology imaging, semi-quantitative variants, such as SUV max, SUV peak [152–154], total lesion glycolysis (TLG) [155, 156], and metabolic tumor volume (MTV) [157, 158] are used to differentiate non-/malignant lesions. However, these values are insufficient to describe the stage of a tumors or to account for tumor heterogeneity [6]. Instead, textural features by their nature, appear to be ideal candidates for metabolic tumor heterogeneity analysis [159, 160]. Accordingly, promising results in the field of treatment outcome [161], therapy response [156, 162], survival [163–166] as well prognostic stratification [167] have been proposed. Several studies perform conventional correlation analysis [156, 160, 165, 168, 169], as well as robust machine learning evaluation [10, 170, 171] of textural features to characterize tumors in vivo. An oncological review of PET-based radiomic approaches concluded that it is a promising method for personalized medicine as it can enhance cancer management [172].
State-of-the-art ML approaches and hybrid imaging appear to be synergistic partners [7, 63, 173]. There is increasing evidence for in vivo tissue characterization with both PET/CT and PET/MRI hybrid imaging [11, 146, 174, 175]. In one study, PET, CT, and PET/CT features were used to predicting local tumor control in head and neck cancer [129] by multivariate cox regression with a confidence interval (CI) CICT and CIPET/CT of 0.73, however, CT-based radiomics overestimated the probability of tumor control in the poor prognostic groups. Another study found a correlation of PET and CT features using lymph node density [176] and concluded that CT density measurements together with PET uptake analysis increases the differentiation between malignant and benign LN. Disease-free survival prediction in non-small cell lung cancer patients can be performed in PET/CT images with an area under the receiver operator characteristic curve (AUC) of 0.68 when employing combined PET/CT features [177]. Another group combined PET uptake measures with CT textural features for radiation pneumonitis diagnosis [178]. They reported an AUC increase of 0.04–0.08 in the combined model compared to single-modality classifiers (AUC 0.71–0.81).
Outcome prediction of locally advanced cervical cancer based on PET/CT and MRI is a topic of ongoing research [174]. The combined analysis of PET and ADC features resulted in an accuracy of 94% for predicting recurrence and 100% for predicting lack of loco-regional control compared to clinical parameters (51–60% accuracy). Combined PET and CT analysis was used to predict FMISO uptake in head-and-neck [179]. The group identified that the combined PET and CT features provide the highest AUC (0.79) for the prediction of tumor hypoxia as evaluated by FMISO PET. Joint fusion features can also be used, for example, to predict lung metastasis in soft-tissue sarcomas [57]. Here, the best performance was achieved with a combined PET/T1 and PET/T2FS textural analysis resulting AUC 0.984, which was significantly higher than that for single modality approaches. A systematic review focusing on oncological applications of radiomics approaches is presented in Avanzo et al. [175].
Holomics
There are several studies that go beyond the utilization of hybrid imaging and incorporate additional non-imaging data for increased predictive accuracy [180–182]. This kind of approach successfully increased risk assessment of head-and-neck cancer built on in vivo and clinical variables with utilizing random forest ML approaches [29]. Independent cross-cohort validation revealed an AUC of 0.69 and a CI of 0.67 for predicting loco-regional recurrences, while distant metastases were predicted with an AUC of 0.86 and a CI of 0.88.
Associations between tumor vascularity, VEGF expression and PET/MRI features in primary clear-cell-renal-cell-carcinoma have been discussed in Yin et al. [32]. The authors reported the highest correlation of tumor microvascular density and PET/MRI features compared to PET or MRI features alone. Correlation of [18F]FDG PET textural features with gene expression in pharyngeal cancer was performed in Chen et al. [183]. The study demonstrated that the overexpression status of vascular endothelial growth factor (VEGF) together with PET features was prognostic, thus, allowing to better stratify treatment response compared to PET-only parameters. Late life depression classification and response prediction with ML based on clinical and imaging features was presented in Patel et al. [30]. The study revealed an accuracy (ACC) of 87% for the classification of late-life depression and ACC of 89% to predict treatment response. Combined ML analysis of in vivo, ex vivo, and patient demographics features to predict 36-months glioma survival was presented in Papp et al. [184]. Comparison of the combined model (M36IEP) with the ex vivo and patient (M36EP), imaging and patient (M36IP), and imaging-only (M36I) models revealed an AUC of 0.9, 0.87, 0.77, and 0.72, respectively in a Monte Carlo cross-validation scheme.
Holomics also introduces several technological challenges. The “curse of dimensionality” refers to the phenomenon that by increasing the dimension of a data, the volume of feature space increases, hence the data becomes sparse [185]. Therefore, it is suggested that the number of data points shall increase exponentially in order to derive accurate models from high dimensional data. To overcome this issue, several dimensionality reduction methods are applied to the combined analysis of imaging and non-imaging data [186]. Similarly, feature selection methods, such as pre-filtering [8, 61] or wrapper and embedded approaches [187–189] can be utilized.
Standardization
Machine learning approaches operating over medical big data require a large amount of standardized data to generate accurate predictive models [10, 63]. Nevertheless, access to standardized multi-center data in the field of hybrid imaging is a challenge [54, 63] which necessitates multi-center standardization efforts [7]. Standardization of hybrid imaging techniques through patient preparation, imaging protocols as well as data evaluation is already of general interest in the field of medical imaging [22, 28, 55, 190–193].
Imaging Protocol
Functional imaging through SPECT and PET aim at the assessment of physiological parameters, as metabolic activity or perfusion. However, these parameters depend on various factors, and, thus, are unstable. For example, the uptake of glucose—and concomitantly—[18F]FDG in brown fat is dependent on its activation, which seems to be triggered by the surrounding temperature [194]. The uptake of glucose in the myocardium depends on the current metabolic pathway of the heart. The heart gains its energy almost exclusively from carbohydrates (primarily glucose) or from metabolizing fatty acids, whereas the pathway used depends on the availability of these substances, and, therefore, on the diet the patient followed prior to the examination [195]. As a consequence of these variabilities, standardized procedures in functional imaging demand the standardization of the entire workflow, including appropriate patient preparation [196].
International organizations, such as the IAEA, EANM, SNM, or ACR have proposed guidelines for patient preparation, imaging and evaluation approaches [197–199]. Accreditation programmes have been set up, such as EARL or the accreditation programs of the ACR, to reach at least a minimum of comparability of imaging data between different centers. However, despite such standardization efforts, site and system specific configurations still result in highly heterogeneous imaging patterns [24]. The reasons are manifold; as explained above, the patient preparation affects the outcome of a functional study. The physiological mechanisms behind this are in general understood and can be handled using appropriate protocols [196]. However, in clinical practice it is often difficult to adhere to these protocols in all details. For example, for outpatients it is almost impossible to check to what extent a required diet was followed.
Another source of variability are differences in the imaging systems and image processing chains. Different imaging system come with different detectors, detector arrangements, and electronics leading to differences in sensitivity and resolution [200]. Further, differences in image reconstruction algorithms, data correction techniques as scatter- and attenuation correction, used image matrices and voxel size as well as applied post filtering processing steps can substantially influence image appearance, quantitative readings, and noise properties of the image data [201]. All of these issues broaden the variability of data quality between different systems and imaging centers, and, therefore, contribute to a limited comparability of image-based ML studies [10, 28, 202, 203].
Delineation
Feature engineering requires the object of interest to be delineated first [8, 145, 204–206] with reproducibility [28]. Lesion segmentation can be performed manually, or semi-/automatically [151, 207]. Manual delineation employs slice-by-slice contouring tools to delineate objects in medical images, which is subject to inter-observer variability depending on the level of expertise of the operator [158, 208].
Semi-automated delineation with fixed thresholds is a popular approach when delineating objects in functional images [151]. These approaches either determine the threshold level by a certain SUV level [209] or by a percentage of the maximum SUV value in a given lesion [210]. Unfortunately, fixed thresholds are prone to the presence of different noise patterns originated from differences in the acquisition and reconstruction protocols [207, 211]. Therefore, different research groups that dichotomize PET tracer avid lesions by fixed thresholds reported contradicting results [212–214]. Inter-observer variabilities can be reduced by training programmes [215] or by collecting and pre-selecting many observer's contouring about the same lesion to achieve an average or consensus contour [151].
Automated delineation methods, such as the Fuzzy Local Adaptive Bayesian (FLAB) were reported to be robust for various, even heterogeneous object delineation tasks in PET [151, 205, 211]. This approach has been also successfully applied to hybrid imaging data, such as PET/CT and PET/MRI [216]. Similarly, random walk approaches have been proven to be effective delineation tools especially over noisy images [216–218]. A comparative study of 13 PET segmentation methods over 157 simulated, phantom and clinical PET images was presented in Hatt et al. [207]; here, a method built on a convolutional neural network (CNN) was found to be the most accurate.
Despite the known drawbacks of manual and semi-automated approaches and the emerging success of automated contouring, to date, the latter solution is still underrepresented in clinical routine [151, 219–221]. This indicates the necessity to extend the evaluation and cross-validation of popular delineation approaches in a large-scale multi-center environment.
Feature Engineering
Given the popularity of textural features in functional and hybrid imaging, their variability with respect to noise, acquisition protocols, and sample size is reasonably well understood [222, 223]. However, technical parameters, such as textural matrix bin size as well as value range intervals appear to greatly affect textural feature repeatability as well [55, 224]. Some in vivo features are not yet unified with regards to a common naming convention and the underlying equation itself [10]. Discussions are ongoing as to the impact of variations in imaging protocols, reconstruction parameters and choice of delineation on textural parameters [10, 160, 225–227]. As an example, while numerous studies utilized a fixed number of bins for textural analysis, recent studies suggested that a fixed bin size with variable number of bins per lesion provides better comparability and reproducibility of textural features [10, 55, 184]. Furthermore, image resolution normalization [228], or normalization of already extracted radiomic features in the feature space [229] have been proposed. In addition, guidelines are available focusing on imaging, feature extraction, analysis and cross-validation standardization of radiomic studies [230–232]. Even though these initiatives point toward a repeatable radiomic research, to date, there are still no standardized, widely accepted and followed radiomics protocols established in the field [232].
Machine Learning Performance Evaluation
Machine learning methods can establish highly accurate predictive models [42, 184, 233]. Nevertheless, inaccurate representation of performance values may lead to misinterpretation of results. Even though this issue is not exclusive to ML approaches [234], predictive models established by ML are prone to such misrepresentations. The training phase of each ML approach optimizes a predictive model over a training dataset. This implicates that the established model may become over-fitted to the training data resulting in a poor performance with independent data. Striking a balance between the training and validation errors is a challenge and referred to as bias-variance trade-off [235].
To estimate the performance of the model in single-center studies, cross-validation approaches shall be utilized [236], such as the leave-one-out method [188, 237], k-fold and stratified k-fold cross-validation techniques [233, 238] as well as Monte Carlo approaches [184, 239]. Likewise, multi-center validation schemes [69, 128, 170] shall be preferred over single-center schemes when estimating the reproducibility of reported results.
Robust machine learning approaches that intend to properly estimate the performance of its predictive models generally split the data into three subsets [240]. Initially, a part of the whole data set is taken out and categorized as test set, the remaining samples are categorized as training set. Test and training set are supposed to follow the same distribution, so they correctly represent the same underlying sample population [241, 242] Splitting is usually conducted to obtain approximate set sizes of 70 and 30% of the original data set for training and test set, respectively [243, 244]. The training set is partitioned using different techniques listed in Table 5. Using these methods, the training set is further divided into an actual training set and a validation set. Training and validation set are again supposed to follow the same distribution. The resulting training-validation pairs can be used to train and tune the ML-established models, respectively, while the remaining test set can be utilized to estimate the performance of the models over an independent dataset. For most of the listed techniques, this procedure occurs several times on each of these training-validation set pairs. A common approach when partitioning the data into training and validation set is the use of stratification [246]. In stratified validation, the sets have the same fraction of labels as the data of origin. This is particularly important when dealing with data sets where the number of samples corresponding to the different labels are imbalanced.
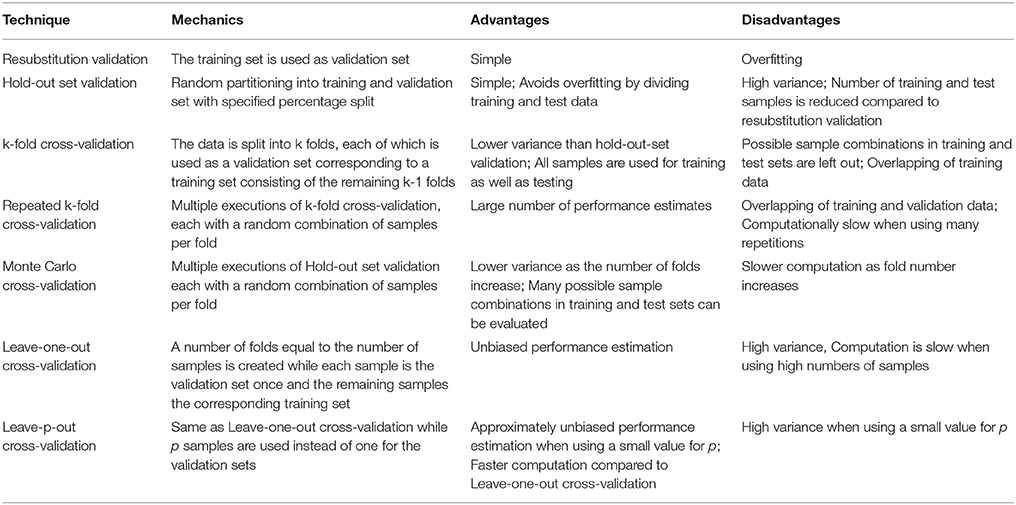
Table 5. List of cross-validation techniques as discussed in Upadhaya et al. [164], Mi et al. [188], Beleites et al. [238], Xu and Liang [239], and Ross et al. [245].
In summary, ML performance shall never be reported over training sets, as the performance values over this set are overestimated, especially in case of overfitting. If a validation set is used to guide model selection or optimization, its performance shall not be reported either, as it becomes the part of the ML optimization process. To properly estimate the performance of the models, independent test sets shall be utilized, that had not been part of any ML decision making process in the given cross-validation fold.
Outlook
Images are data [8] and data is knowledge; this statement applies to all types of data, not only in medicine. It is our task to turn this knowledge into a patient benefit. One option is to build clinical decision support systems that are trained and validated on these data, and, therefore, embrace non-invasive imaging and non-imaging data potentially linked through machine learning as described here.
Nonetheless, restricted data access and variable data formats challenge the build-up of knowledge databases and the adoption of CDSS in modern healthcare. Across all disciplines and specialties data come in different formats. In medicine alone, available data come in the form of 2D and 3D images, they may entail serial information, additional raw (measured) data may be attached, data further include clinical tests, blood samples, genomic analysis, and so on. From a patient's perspective, these information is scattered across multiple systems, including the electronic medical record (EMR) system, laboratories, picture archiving systems (PACS), and alike. These data must be made available, accessible and tangible in order to apply any type of knowledge generation.
The sourcing of knowledge, to help an individual patient now, or to derive new therapeutics for more patients in the future, is inherently linked to the concept of “big data.” Big data, in combination with ML, can help uncover associations between various types of data (assuming that data silos can be torn down and data can be accessed) and it can help build prediction models for diagnosis and disease progression as well as therapy response assessment [247]. The use of big data requires a so called end-to-end strategy in which “IT departments or groups are the technical enablers; but key executives, business groups, and other stakeholders help set objectives, identify critical success factors, and make relevant decisions” [247]. Such strategy entails multiple milestones, including the validation of ML algorithms, the standardization of features and the general accessibility (and willingness to share) of data.
Novel Heterogeneity Phantoms
In view of the rapid growth of ML and hybrid imaging, the importance of image quality naturally shifts from visual interpretation toward quantitative, automated evaluation. This trend requires a change of focus toward standardization efforts across all scales of hybrid imaging. To date, there are no standardized, commercially available physical “heterogeneity” phantoms that allow us to validate and optimize hybrid imaging procedures for ML evaluation on-site or across multiple sites. We believe, such efforts are required to support the adoption of ML in the context of hybrid imaging.
Open Data, Open Cloud
The combination of open science and cloud computing is in the focus of several public initiatives. In 2016, the European Commission announced the creation of the European Open Science Cloud in order to promote scientific data access and evaluation with high-performance cloud computing technologies [248]. According to their report, all scientific data created under the umbrella of the Horizon 2020 research and innovation programme will be open data to support the scientific community. In addition, acceleration of quantum computing technology will be initiated by 2018 to support the construction of the next generation of supercomputers. By 2020 a large scale European high performance computing data storage and network infrastructure will be deployed to establish the base of future research and innovation in Europe [248]. Such efforts, in combination with existing open source date can help synchronize hypothesis-driven data cohorts for an efficient application of ML approaches with the purpose to generate knowledge from image data. To date, hybrid image data are not yet widely dispersed in such data initiatives, but increased awareness and ease-of-use of data repositories may facilitate a growth in accessible hybrid image data.
Doctor in Pocket
Machine Learning together with widely-accessible medical Big Data promises an era, where computer-aided diagnosis (CAD) and clinical decision support systems (CDSS) will contribute to routine decision making [39]. Artificial Intelligence (AI) assistants [249] will be able to process and provide personalized, real-time feedback to individuals over their Medical Big Data through their smartphones. These AI assistants could follow our physiological and mental wellness. While they could have access to massive population-wide medical information to learn from, they could dynamically change their model of ours to end up with fully-personalized predictive models (Figure 5).
Conclusions
Medical imaging originated from technological progress and innovation proposed by cross-specialists, including physicists, engineers, medical doctors, biologists, mathematicians, chemists, and alike. Medical imaging research has always been a data-driven science. Lately, medical practice, and healthcare in general, has moved into big data, as a modernist's view on data-driven science.
Medical Big data offers the ability to source unique knowledge from the available data, which, however, are spread across various formats and information contents and which may not be equally well accessible and assessable. Joint efforts are needed to turn medical big data into useful medical big data, for example by harmonizing data access and by moving from single site to multi-centric data cohorts and repositories.
While we have access to computer algorithms that can deduce higher-order information from available data, their validation hinges on the availability of large scale, high-quality, and standardized reference data. Only recently we have seen a growing awareness for the need for standardized imaging and data collection procedures, as pre-requisites for the use of machine learning and the construction of clinical decision support systems that can be employed in routine practice.
In this context, hybrid imaging contains a multitude of valuable information that, if combined with complementary non-imaging data, has been shown to yield surprisingly accurate insights into the causes of disease. If adopted carefully in the context of CDSS, hybrid imaging may contribute to an improved diagnosis of patients, and, in turn, to a more efficient therapy planning.
Author Contributions
All authors contributed to drafting and establishing the main skeleton of the paper. LP contributed to the Technology, Data Handling, Joint Data Exploration and Standardization sections. CS contributed to the Technology section. IR contributed to the Standardization section. MH contributed to the Data Handling section. TB was the primary reviewer of the paper and contributed to the Outlook and Conclusions sections. All authors reviewed and approved the paper for submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1. ^AWS Case Study: Arterys. Available online at: https://aws.amazon.com/de/solutions/case-studies/arterys/
2. ^Cancer Imaging Archive. Available online at: http://www.cancerimagingarchive.net/.
3. ^Eurorad: Radiological Case Database. Available online at: http://www.eurorad.org/.
References
1. Rinck PA. Magnetic Resonance in Medicine. 11th ed. The Round Table Foundation: BoD (2017). Available online at: http://www.magnetic-resonance.org/
2. Beyer T, Freudenberg LS, Townsend DW, Czernin J. The future of hybrid imaging—part 1: hybrid imaging technologies and SPECT/CT. Insights Imaging (2011) 2:161–9. doi: 10.1007/s13244-010-0063-2
3. Beyer T, Townsend DW, Czernin J, Freudenberg LS. The future of hybrid imaging—part 2: PET/CT. Insights Imaging (2011) 2:225–34. doi: 10.1007/s13244-011-0069-4
4. Beyer T, Freundenberg LS, Czernin J, Townsend DW. The future of hybrid imaging-part 3: Pet/mr, small-animal imaging and beyond. Insights Imaging (2012) 3:189. doi: 10.1007/s13244-011-0136-x
5. Wahl RL, Quint LE, Cieslak RD, Aisen AM, Koeppe RA, Meyer CR. “Anatometabolic” tumor imaging: fusion of FDG PET with CT or MRI to localize foci of increased activity. J Nucl Med. (1993) 34:1190–7.
6. Visvikis D, Hatt M, Tixier F, Rest CC, Le. The age of reason for FDG PET image-derived indices. Eur J Nucl Med Mol Imaging (2012) 39:1670–2. doi: 10.1007/s00259-012-2239-0
7. Gillies RJ, Beyer T. PET and MRI: is the whole greater than the sum of its parts? Cancer Res. (2016) 76:6163–6. doi: 10.1158/0008-5472.CAN-16-2121
8. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology (2015) 278:563–77. doi: 10.1148/radiol.2015151169
9. Court LE, Fave X, Mackin D, Lee J, Yang J, Zhang L. Computational resources for radiomics. Transl Cancer Res. (2016) 5:340–8. doi: 10.21037/tcr.2016.06.17
10. Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D. Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging (2017) 44:151–65. doi: 10.1007/s00259-016-3427-0
11. Cook GJR, Siddique M, Taylor BP, Yip C, Chicklore S, Goh V. Radiomics in PET: principles and applications. Clin Transl Imaging (2014) 2:269–76. doi: 10.1007/s40336-014-0064-0
12. Press G,. A Very Short History of Data Science. Forbes (2013). Available online at: https://www.forbes.com/sites/gilpress/2013/05/28/a-very-short-history-of-data-science/
13. Marr B,. A Brief History of Big Data Everyone Should Read. World Econ Forum (2015) Available online at: https://www.weforum.org/agenda/2015/02/a-brief-history-of-big-data-everyone-should-read/
14. Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Heal Inf Sci Syst. (2014) 2:3. doi: 10.1186/2047-2501-2-3
15. Qiu J, Wu Q, Ding G, Xu Y, Feng S. A survey of machine learning for big data processing. EURASIP J Adv Signal Process. (2016) 2016:67. doi: 10.1186/s13634-016-0355-x
16. Kotrotsou A, Zinn PO, Colen RR. Radiomics in brain tumors: an emerging technique for characterization of tumor environment. Magn Reson Imaging Clin N Am. (2016) 24:719–29 doi: 10.1016/j.mric.2016.06.006
17. Guo H, Wang L, Chen F, Liang D. Scientific big data and Digital Earth. Chin Sci Bull. (2014) 59:5066–73. doi: 10.1007/s11434-014-0645-3
18. SINTEF. Big Data, for Better or Worse: 90% of World's Data Generated Over Last Two Years. SINTEF (2013).
19. Brown N. Healthcare Data Growth: An Exponential Problem (2015). Available online at: http://www.nextech.com/blog/healthcare-data-growth-an-exponential-problem
20. Landi H. IBM Unveils Watson-Powered Imaging Solutions at RSNA (2016). Available online at: https://www.healthcare-informatics.com/news-item/analytics/ibm-unveils-watson-powered-imaging-solutions-rsna
21. Smith EM. Storage management: what radiologists need to know. Appl Radiol. (2009) 38:13–8. Available online at: https://appliedradiology.com/articles/storage-management-what-radiologists-need-to-know
22. Rosenkrantz AB, Friedman K, Chandarana H, Melsaether A, Moy L, Ding YS, et al. Current status of hybrid PET/MRI in oncologic imaging. Am J Roentgenol. (2016) 206:162–72. doi: 10.2214/AJR.15.14968
23. Karakatsanis NA, Lodge MA, Tahari AK, Zhou Y, Wahl RL, Rahmim A. Dynamic whole-body PET parametric imaging: I. Concept, acquisition protocol optimization and clinical application. Phys Med Biol. (2013) 58:7391. doi: 10.1088/0031-9155/58/20/7391
24. Rausch I, Bergmann H, Geist B, Schaffarich M, Hirtl A, Hacker M, et al. Variation of system performance, quality control standards and adherence to international FDG-PET/CT imaging guidelines. Nuklearmedizin (2014) 53:242–8. doi: 10.3413/Nukmed-0665-14-05
25. Mehta N, Pandit A. Concurrence of big data analytics and healthcare: a systematic review. Int J Med Inform. (2018) 114:57–65. doi: 10.1016/j.ijmedinf.2018.03.013
26. Erickson BJ. Machine learning: discovering the future of medical imaging. J Digit Imaging (2017) 30:391. doi: 10.1007/s10278-017-9994-1
27. Chiang S, Guindani M, Yeh HJ, Dewar S, Haneef Z, Stern JM, et al. A hierarchical Bayesian model for the identification of PET markers associated to the prediction of surgical outcome after anterior temporal lobe resection. Front Neurosci. (2017) 11:669. doi: 10.3389/fnins.2017.00669
28. Larue RTHM, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. (2017) 90:20160665. doi: 10.1259/bjr.20160665
29. Vallières M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts HJWL, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. (2017) 7:10117. doi: 10.1038/s41598-017-10371-5
30. Patel MJ, Andreescu C, Price JC, Edelman KL, Reynolds CF, Aizenstein HJ. Machine learning approaches for integrating clinical and imaging features in late-life depression classification and response prediction. Int J Geriatr Psychiatry (2015) 30:1056–67. doi: 10.1002/gps.4262
31. Gevaert O. SP-0596: machine learning and bioinformatics approaches to combine imaging with non-imaging data for outcome prediction. Radiother Oncol. (2017) 123:S313. doi: 10.1016/S0167-8140(17)31036-8
32. Yin Q, Hung S-C, Wang L, Lin W, Fielding JR, Rathmell WK, et al. Associations between tumor vascularity, vascular endothelial growth factor expression and PET/MRI radiomic signatures in primary clear-cell–renal-cell-carcinoma: proof-of-concept study. Sci Rep. (2017) 7:43356. doi: 10.1038/srep43356
33. May M. LIFE SCIENCE TECHNOLOGIES: big biological impacts from big data. Science (2014) 344:1298–300. doi: 10.1126/science.344.6189.1298
34. Gu J, Taylor CR. Practicing pathology in the era of big data and personalized medicine. Appl Immunohistochem Mol Morphol. (2014) 22:1–9. doi: 10.1097/PAI.0000000000000022
35. Murdoch T, Detsky A. The inevitable application of big data to health care. J Am Med Inform Assoc. (2013) 309:1351–2. doi: 10.1001/jama.2013.393
36. Luo J, Chen Y, Tang K, Luo J. “Remote monitoring information system and its applications based on the Internet of Things,” in 2009 International Conference on Future BioMedical Information Engineering (FBIE) (Sanya) (2009). p. 482–5.
37. Jara AJ, Zamora-Izquierdo MA, Skarmeta AF. Interconnection framework for mhealth and remote monitoring based on the internet of things. IEEE J Sel Areas Commun. (2013) 31:47–65. doi: 10.1109/JSAC.2013.SUP.0513005
38. Hu F, Xie D, Shen S. On the application of the internet of things in the field of medical and health care. In: 2013 IEEE Int Conf Green Comput Commun IEEE Internet Things IEEE Cyber, Phys Soc Comput. (Beijing) (2013). p. 2053–8.
39. Wagholikar KB, Sundararajan V, Deshpande AW. Modeling paradigms for medical diagnostic decision support: a survey and future directions. J Med Syst. (2012) 36:3029. doi: 10.1007/s10916-011-9780-4
40. Mathur S, Sutton J. Personalized medicine could transform healthcare (Review). Biomed Rep. (2017) 7:3–5. doi: 10.3892/br.2017.922
41. Aboagye EO, Kraeber-Bodéré F. Highlights lecture EANM 2016: “Embracing molecular imaging and multi-modal imaging: a smart move for nuclear medicine towards personalized medicine.” Eur J Nucl Med Mol Imaging (2017) 44:1559–74. doi: 10.1007/s00259-017-3704-6
42. Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. (2006) 2:59–77. doi: 10.1177/117693510600200030
43. Kourou K, Exarchos TP, Exarchos KP, Karamouzis M V., Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. (2015) 13:8–17. doi: 10.1016/j.csbj.2014.11.005
44. Tarca AL, Carey VJ, Chen X, Romero R, Drăghici S. Machine learning and its applications to biology. PLoS Comput Biol. (2007) 3:e30116. doi: 10.1371/journal.pcbi.0030116
45. Miles N. Wernick, Yang Y, Brankov JG, Yourganov G, Strother SC. Machine learning in medical imaging. IEEE Signal Process Mag. (2014) 27:25–38. doi: 10.1109/MSP.2010.936730
46. Schmidhuber J. Deep Learning in neural networks: an overview. Neural Netw. (2015) 61:85–117. doi: 10.1016/j.neunet.2014.09.003
47. Teramoto A, Fujita H, Yamamuro O, Tamaki T. Automated detection of pulmonary nodules in PET/CT images: ensemble false-positive reduction using a convolutional neural network technique. Med Phys. (2016) 43:2821–7. doi: 10.1118/1.4948498
48. Gatidis S, Scharpf M, Martirosian P, Bezrukov I, Küstner T, Hennenlotter J, et al. Combined unsupervised-supervised classification of multiparametric PET/MRI data: application to prostate cancer. NMR Biomed. (2015) 28:914–22. doi: 10.1002/nbm.3329
49. Wu J, Cui Y, Sun X, Cao G, Li B, Ikeda DM, et al. Unsupervised clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin Cancer Res. (2017) 23:3334–42. doi: 10.1158/1078-0432.CCR-16-2415
50. Najafabadi MM, Villanustre F, Khoshgoftaar TM, Seliya N, Wald R, Muharemagic E. Deep learning applications and challenges in big data analytics. J Big Data (2015) 2:1. doi: 10.1186/s40537-014-0007-7
51. Sutton RS, Bartoa G. Reinforcement learning: an introduction. IEEE Trans Neural Netw. (1998) 9:1054. doi: 10.1109/TNN.1998.712192
52. Tseng H-H, Luo Y, Cui S, Chien J-T, Ten Haken RK, Naqa I El. Deep reinforcement learning for automated radiation adaptation in lung cancer. Med Phys. (2017) 44:6690–705. doi: 10.1002/mp.12625
53. Buizza G. Classifying Patients' Response to Tumour Treatment From PET/CT Data: A Machine Learning Approach [Internet] [Dissertation]. (2017). TRITA-STH. Available online at: http://urn.kb.se/resolve?urn=urn:nbn:se:kth:diva-200916
54. Greenspan H, van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging (2016) 35:1153–9. doi: 10.1109/TMI.2016.2553401
55. Leijenaar RTH, Nalbantov G, Carvalho S, van Elmpt WJC, Troost EGC, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. (2015) 5:11075. doi: 10.1038/srep11075
56. Cameron A, Khalvati F, Haider MA, Wong A. MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng. (2016) 63:1145–56. doi: 10.1109/TBME.2015.2485779
57. Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. (2015) 60:5471–96. doi: 10.1088/0031-9155/60/14/5471
58. Sala E, Mema E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. (2017) 72:3–10. doi: 10.1016/j.crad.2016.09.013
59. Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol. (2016) 61:R150–66. doi: 10.1088/0031-9155/61/13/R150
60. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. (2003) 3:1157–82. Available online at: https://dl.acm.org/citation.cfm?id=944968
61. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. (2014) 5:4006. doi: 10.1038/ncomms5006
62. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
63. Hatt M, Tixier F, Visvikis D, Cheze Le Rest C. Radiomics in PET/CT: more than meets the eye? J Nucl Med. (2017) 58:365–66. doi: 10.2967/jnumed.116.184655
64. Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. (2017) 44:1408–19. doi: 10.1002/mp.12155
65. Li R, Zhang W, Suk H-I, Wang L, Li J, Shen D, et al. Deep learning based imaging data completion for improved brain disease diagnosis. Med Image Comput Comput Assist Interv. (2014) 17:305–12. doi: 10.1007/978-3-319-10443-0_39
66. Suk H-I, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv. (2013) 16:583–90. doi: 10.1007/978-3-642-40763-5_72
67. Ho YC, Pepyne DL. Simple explanation of the no free lunch theorem of optimization. In: 40th IEEE Conf Decis Control. Orlando, FL (2001). p. 4409–14.
68. Wolpert DH, Macready WG. No free lunch theorems for optimization. IEEE Trans Evol Comput. (1997) 1:67–82. doi: 10.1109/4235.585893
69. Wnuk P, Kowalewski M, Małkowski B, Bella M, Dancewicz M, Szczesny T, et al. PET-CT derived Artificial Neural Network can predict mediastinal lymph nodes metastases in Non-Small Cell Lung Cancer patients. Preliminary report and scoring model. Q J Nucl Med Mol Imaging (2014). [Epub ahead of print].
70. Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Direct PseudoCT generation for pelvis PET/MRI attenuation correction using deep convolutional neural networks with multi-parametric MRI: zero echo-time and dixon deep pseudoCT (ZeDD-CT). J Nucl Med. (2017) 59:852–8. doi: 10.2967/jnumed.117.198051
71. Zhang B, He X, Ouyang F, Gu D, Dong Y, Zhang L, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. (2017) 403:21–7. doi: 10.1016/j.canlet.2017.06.004
72. Wang H, Zhou Z, Li Y, Chen Z, Lu P, Wang W, et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. (2017) 7:11. doi: 10.1186/s13550-017-0260-9
73. Fournel AP, Reynaud E, Brammer MJ, Simmons A, Ginestet CE. Group analysis of self-organizing maps based on functional MRI using restricted Frechet means. Neuroimage (2013) 76:373–85. doi: 10.1016/j.neuroimage.2013.02.043
74. Hu J, Wu W, Zhu B, Wang H, Liu R, Zhang X, et al. Cerebral glioma grading using Bayesian network with features extracted from multiple modalities of magnetic resonance imaging. PLoS ONE (2016) 11:e0153369. doi: 10.1371/journal.pone.0157095
75. Li R, Yu J, Zhang S, Bao F, Wang P, Huang X, et al. Bayesian network analysis reveals alterations to default mode network connectivity in individuals at risk for Alzheimer's disease. PLoS ONE (2013) 8:e82104. doi: 10.1371/journal.pone.0082104
76. Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. (2017) 19:221–48. doi: 10.1146/annurev-bioeng-071516-044442
77. van Tulder G, de Bruijne M. Combining generative and discriminative representation learning for lung CT analysis with convolutional restricted boltzmann machines. IEEE Trans Med Imaging (2016) 35:1262–72. doi: 10.1109/TMI.2016.2526687
78. Hein E, Albrecht A, Melzer D, Steinhöfel K, Rogalla P, Hamm B, et al. Computer-assisted diagnosis of focal liver lesions on CT images. Acad Radiol. (2005) 12:1205–10. doi: 10.1016/j.acra.2005.05.009
79. Berthon B, Marshall C, Evans M, Spezi E. ATLAAS: an automatic decision tree-based learning algorithm for advanced image segmentation in positron emission tomography. Phys Med Biol. (2016) 61:4855–69. doi: 10.1088/0031-9155/61/13/4855
80. Markel D, Caldwell C, Alasti H, Soliman H, Ung Y, Lee J, et al. Automatic segmentation of lung carcinoma using 3D texture features in 18-FDG PET/CT. Int J Mol Imaging (2013) 2013:980769. doi: 10.1155/2013/980769
81. Khatami A, Khosravi A, Nguyen T, Lim CP, Nahavandi S. Medical image analysis using wavelet transform and deep belief networks. Expert Syst Appl. (2017) 86:190–8. doi: 10.1016/j.eswa.2017.05.073
82. Akhbardeh A, Jacobs MA. Comparative analysis of nonlinear dimensionality reduction techniques for breast MRI segmentation. Med Phys. (2012) 39:2275–89. doi: 10.1118/1.3682173
83. Ha S, Choi H, Cheon GJ, Kang KW, Chung J-K, Kim EE, et al. Autoclustering of non-small cell lung carcinoma subtypes on 18F-FDG PET Using texture analysis: a preliminary result. Nucl Med Mol Imaging (2014) 48:278–86. doi: 10.1007/s13139-014-0283-3
84. Baum KG, Schmidt E, Rafferty K, Krol A, Helguera M. Evaluation of novel genetic algorithm generated schemes for positron emission tomography (PET)/MAGNETIC Resonance Imaging (MRI) image fusion. J Digit Imaging (2011) 24:1031–43. doi: 10.1007/s10278-011-9382-1
85. Panda R, Agrawal S, Sahoo M, Nayak R. A novel evolutionary rigid body docking algorithm for medical image registration. Swarm Evol Comput. (2017) 33:108–18. doi: 10.1016/j.swevo.2016.11.002
86. Ortiz A, Gorriz JM, Ramirez J, Salas-Gonzalez D. Improving MR brain image segmentation using self-organising maps and entropy-gradient clustering. Inf Sci. (2014) 262:117–36. doi: 10.1016/j.ins.2013.10.002
87. Hanzouli-Ben Salah H, Lapuyade-Lahorgue J, Bert J, Benoit D, Lambin P, Van Baardwijk A, et al. A framework based on hidden Markov trees for multimodal PET/CT image co-segmentation. Med Phys. (2017) 44:5835–48. doi: 10.1002/mp.12531
88. Chen Y, Juttukonda M, Lee YZ, Su Y, Espinoza F, Lin W, et al. MRI based attenuation correction for PET/MRI via MRF segmentation and sparse regression estimated CT. In: 2014 IEEE 11th International Symposium on Biomedical Imaging, ISBI 2014 Inst Electr Electron Eng Inc. Beijing (2014) p. 1364–7.
89. Kebir S, Khurshid Z, Gaertner FC, Essler M, Hattingen E, Fimmers R, et al. Unsupervised consensus cluster analysis of [18F]-fluoroethyl-L-tyrosine positron emission tomography identified textural features for the diagnosis of pseudoprogression in high-grade glioma. Oncotarget (2017) 8:8294–304. doi: 10.18632/oncotarget.14166
90. Parmar C, Leijenaar RTH, Grossmann P, Rios Velazquez E, Bussink J, Rietveld D, et al. Radiomic feature clusters and Prognostic Signatures specific for Lung and Head & Neck cancer. Sci Rep. (2015) 5:11044. doi: 10.1038/srep11044
91. Sharma N, Ray A, Shukla K, Sharma S, Pradhan S, Srivastva A, et al. Automated medical image segmentation techniques. J Med Phys. (2010) 35:3–14. doi: 10.4103/0971-6203.58777
92. Pecori B, Lastoria S, Caracò C, Celentani M, Tatangelo F, Avallone A, et al. Sequential PET/CT with [18F]-FDG predicts pathological tumor response to preoperative short course radiotherapy with delayed surgery in patients with locally advanced rectal cancer using logistic regression analysis. PLoS ONE (2017) 12:e0169462. doi: 10.1371/journal.pone.0169462
93. Kim S-A, Roh J-L, Kim JS, Lee JH, Lee SH, Choi S-H, et al. 18F-FDG PET/CT surveillance for the detection of recurrence in patients with head and neck cancer. Eur J Cancer (2017) 72:62–70. doi: 10.1016/j.ejca.2016.11.009
94. Kim M, Ghate A, Phillips MH. A Markov decision process approach to temporal modulation of dose fractions in radiation therapy planning. Phys Med Biol. (2009) 54:4455–76. doi: 10.1088/0031-9155/54/14/007
95. Bousse A, Pedemonte S, Thomas BA, Erlandsson K, Ourselin S, Arridge S, et al. Markov random field and Gaussian mixture for segmented MRI-based partial volume correction in PET. Phys Med Biol. (2012) 57:6681–705. doi: 10.1088/0031-9155/57/20/6681
96. Held K, Kops ER, Krause BJ, Wells WM, Kikinis R, Muller-Gartner H-W. Markov random field segmentation of brain MR images. IEEE Trans Med Imaging (1997) 16:878–86.
97. Prasad MN, Brown MS, Ahmad S, Abtin F, Allen J, da Costa I, et al. Automatic segmentation of lung parenchyma in the presence of diseases based on curvature of ribs. Acad Radiol. (2008) 15:1173–80. doi: 10.1016/j.acra.2008.02.004
98. Landset S, Khoshgoftaar TM, Richter AN, Hasanin T. A survey of open source tools for machine learning with big data in the Hadoop ecosystem. J Big Data (2015) 2:24. doi: 10.1186/s40537-015-0032-1
99. Bao S, Weitendorf FD, Plassard AJ, Huo Y, Gokhale A, Landman BA. Theoretical and empirical comparison of big data image processing with Apache Hadoop and Sun Grid Engine. Proc SPIE Int Soc Opt Eng. (2017) 2017:10138. doi: 10.1117/12.2254712
100. Davies B. What Are Hadoop Alternatives and Should You Look for One? (2017) Available online at: http://bigdata-madesimple.com/what-are-hadoop-alternatives-and-should-you-look-for-one/
101. Mackey G, Sehrish S, Bent J, Lopez J, Habib S, Wang J. Introducing map-reduce to high end computing. In: 2008 3rd Petascale Data Storage Workshop (Austin, TX: IEEE). p. 1–6.
102. Borthakur D,. HDFS Architecture Guide. Hadoop Apache Project (2008). Available online at: http://doc.kevenking.cn/hadoop/hadoop-1.0.4/hdfs_design.pdf
103. Markonis D, Schaer R, Eggel I, Müller H, Depeursinge A. Using MapReduce for large-scale medical image analysis. arXiv:1510.06937 (2015), p. 1–10.
104. Yang C-T, Chen L-T, Chou W-L, Wang K-C. Implementation of a medical image file accessing system on cloud computing. In: 2010 13th IEEE International Conference on Computational Science and Engineering Hong Kong: IEEE (2010). p. 321–6.
105. Meng X, Bradley J, Yavuz B, Sparks E, Venkataraman S, Liu D, et al. MLlib: machine learning in apache spark. arXiv:1505.06807 (2015). p. 1–7.
106. Shanahan JG, Dai L. Large scale distributed data science using apache spark. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD '15. New York, NY: ACM Press (2015). p. 2323–4.
107. Zaharia M, Franklin MJ, Ghodsi A, Gonzalez J, Shenker S, Stoica I, et al. Apache spark. Commun ACM (2016) 59:56–65. doi: 10.1145/2934664
108. Fu J, Sun J, Wang K. SPARK—A Big Data Processing Platform for Machine Learning. In: 2016 International Conference on Industrial Informatics - Computing Technology, Intelligent Technology, Industrial Information Integration (ICIICII). Wuhan (2016). p. 48–51.
109. Zaharia M, Chowdhury M, Das T, Dave A, Ma J, McCauly M, et al. Resilient distributed datasets: a fault-tolerant abstraction for in-memory cluster computing. In: Presented as Part of the 9th {USENIX} Symposium on Networked Systems Design and Implementation ({NSDI} 12). San Jose, CA: USENIX (2012). p. 15–28.
110. Pop D. Machine Learning and Cloud Computing: Survey of Distributed and SaaS Solutions. Inst e-Austria Timisoara, Tech Rep 1 (2012).
111. Bao S, Plassard AJ, Landman BA, Gokhale A. Cloud engineering principles and technology enablers for medical image processing-as-a-service. In: 2017 IEEE International Conference on Cloud Engineering (IC2E). Vancouver, BC (2017). p. 127–37.
112. Kagadis GC, Kloukinas C, Moore KL, Philbin J, Papadimitroulas P, Alexakos C, et al. Cloud computing in medical imaging. Med Phys. (2013) 40:70901. doi: 10.1118/1.4811272
113. GCP. Google Cloud Platform: Maxwell MRI Revolutionises Healthcare Delivery with AI on GCP (2017). Available online at: https://cloud.google.com/customers/maxwell-mri/
114. Njenga PW. AWS Startups Blog: Adapting Deep Learning to Medicine With Behold.ai (2016). Available online at: https://aws.amazon.com/de/blogs/startups/adapting-deep-learning-to-medicine-with-behold-ai/
115. Dormer K, Patni H, Bidulock D, et al. Medical imaging and efficient sharing of medical imaging information. Pub. No.: US 2017/0076043 A1 (2017).
116. Chaithra R. Secure medical Image classification based on Azure Cloud. Int J Eng Sci Comput. (2017). 7:12185–8.
117. Roychowdhury S, Bihis M. AG-MIC: azure-based generalized flow for medical image classification. IEEE Access. (2016) 4:5243–57. doi: 10.1109/ACCESS.2016.2605641
118. Criminisi A. Machine learning for medical images analysis. Med Image Anal. (2016) 33:91–3. doi: 10.1016/j.media.2016.06.002
119. Milieu Time.lex. Overview of the National Laws on Electronic Health Records in the EU Member States and Their Interaction With the Provision of Cross-Border eHealth Services Final Report and Recommendations (2014). Available online at: https://ec.europa.eu/health//sites/health/files/ehealth/docs/laws_report_recommendations_en.pdf
120. Ruotsalainen P, Manning B. A notary archive model for secure preservation and distribution of electrically signed patient documents. Int J Med Inform. (2007) 76:449–53. doi: 10.1016/j.ijmedinf.2006.09.011
121. Morgan C. Data Storage Lifespans: How Long Will Media Really Last? Storagecraft Available online at: https://www.storagecraft.com/blog/data-storage-lifespan/ (2017).
122. Kesner A, Koo P. A consideration for changing our PET data saving practices: a cost/benefit analysis. J Nucl Med. (2016) 57:1912. Available online at: http://jnm.snmjournals.org/content/57/supplement_2/1912.abstract
123. Kohli MD, Summers RM, Geis JR. Medical image data and datasets in the era of machine learning—whitepaper from the 2016 C-MIMI meeting dataset session. J Digit Imaging (2017) 30:392–9. doi: 10.1007/s10278-017-9976-3
124. Costello MJ. Motivating online publication of data. Bioscience (2009) 59:418–27. doi: 10.1525/bio.2009.59.5.9
125. Wicherts JM, Bakker M, Molenaar D. Willingness to share research data is related to the strength of the evidence and the quality of reporting of statistical results. PLoS ONE (2011) 6:e26828. doi: 10.1371/journal.pone.0026828
126. Roche DG, Kruuk LEB, Lanfear R, Binning SA. Public data archiving in ecology and evolution: how well are we doing? PLoS Biol. (2015) 13:e1002295. doi: 10.1371/journal.pbio.1002295
127. Santos C, Blake J, States DJ. Supplementary data need to be kept in public repositories. Nature (2005) 438:738. doi: 10.1038/438738a
128. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer (2012) 48:441–6. doi: 10.1016/j.ejca.2011.11.036
129. Bogowicz M, Riesterer O, Stark LS, Studer G, Unkelbach J, Guckenberger M, et al. Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma. Acta Oncol. (2017) 56:1531–6. doi: 10.1080/0284186X.2017.1346382
130. Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, et al. Preparing a collection of radiology examinations for distribution and retrieval. J Am Med Informatics Assoc. (2016) 23:304–10. doi: 10.1093/jamia/ocv080
131. Jaeger S, Candemir S, Antani S, Wang Y-XJ, Lu P-X, Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. (2014) 4:475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20
132. Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. (2007) 19:1498–507. doi: 10.1162/jocn.2007.19.9.1498
133. Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies: longitudinal mri data in nondemented and demented older adults. J Cogn Neurosci. (2010) 22:2677–84. doi: 10.1162/jocn.2009.21407
134. Zhang J, Peng Q, Li Q, Jahanshad N, Hou Z, Jiang M, et al. Longitudinal characterization of brain atrophy of a Huntington's disease mouse model by automated morphological analyses of magnetic resonance images. Neuroimage (2010) 49:2340–51. doi: 10.1016/j.neuroimage.2009.10.027
135. Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, et al. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer's disease participants. Neuroimage (2009) 46:486–99.
136. Freymann JB, Kirby JS, Perry JH, Clunie DA, Jaffe CC. Image data sharing for biomedical research—meeting HIPAA requirements for de-identification. J Digit Imaging (2012) 25:14–24. doi: 10.1007/s10278-011-9422-x
137. Gerber N, Reyes M, Barazzetti L, Kjer HM, Vera S, Stauber M, et al. A multiscale imaging and modelling dataset of the human inner ear. Sci Data (2017) 4:170132. doi: 10.1038/sdata.2017.132
138. Petiet AE, Kaufman MH, Goddeeris MM, Brandenburg J, Elmore SA, Johnson GA. High-resolution magnetic resonance histology of the embryonic and neonatal mouse: a 4D atlas and morphologic database. Proc Natl Acad Sci USA. (2008) 105:12331–6. doi: 10.1073/pnas.0805747105
139. Yao J, Burns JE, Forsberg D, Seitel A, Rasoulian A, Abolmaesumi P, et al. A multi-center milestone study of clinical vertebral CT segmentation. Comput Med Imaging Graph. (2016) 49:16–28. doi: 10.1016/j.compmedimag.2015.12.006
140. Esteban O, Birman D, Schaer M, Koyejo OO, Poldrack RA, Gorgolewski KJ. MRIQC: advancing the automatic prediction of image quality in MRI from unseen sites. PLoS ONE (2017) 12:e0184661. doi: 10.1371/journal.pone.0184661
141. Weiner MW, Aisen PS, Jack CR, Jagust WJ, Trojanowski JQ, Shaw L, et al. The Alzheimer's disease neuroimaging initiative: progress report and future plans. Alzheimer's Dement. (2010) 6:202–11.e7. doi: 10.1016/j.jalz.2010.03.007
142. Strand R, Malmberg F, Johansson L, Lind L, Sundbom M, Ahlström H, et al. A concept for holistic whole body MRI data analysis, Imiomics. PLoS ONE (2017) 12:e0169966. doi: 10.1371/journal.pone.0169966
143. Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol. (2004) 59:1061–9. doi: 10.1016/j.crad.2004.07.008
144. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging (2012) 30:1234–48. doi: 10.1016/j.mri.2012.06.010
145. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE (2014) 9:e102107. doi: 10.1371/journal.pone.0102107
146. Scrivener M, de Jong EEC, van Timmeren JE, Pieters T, Ghaye B, Geets X. Radiomics applied to lung cancer: a review. Transl Cancer Res. (2016) 5:398–409. doi: 10.21037/tcr.2016.06.18
147. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. Syst Man Cybern. (1973) 3:610–21. doi: 10.1109/TSMC.1973.4309314
148. Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, et al. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep. (2017) 7:14331. doi: 10.1038/s41598-017-14753-7
149. Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res. (2017) 6:86–91. doi: 10.21037/tlcr.2017.01.04
150. Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging (2014) 27:805–23. doi: 10.1007/s10278-014-9716-x
151. Foster B, Bagci U, Mansoor A, Xu Z, Mollura DJ. A review on segmentation of positron emission tomography images. Comput Biol Med (2014) 50:76–96. doi: 10.1016/j.compbiomed.2014.04.014
152. Lopci E, Riva M, Olivari L, Raneri F, Soffietti R, Piccardo A, et al. Prognostic value of molecular and imaging biomarkers in patients with supratentorial glioma. Eur J Nucl Med Mol Imaging (2017) 44:1155–64. doi: 10.1007/s00259-017-3618-3
153. Van Velden FHP, Cheebsumon P, Yaqub M, Smit EF, Hoekstra OS, Lammertsma AA, et al. Evaluation of a cumulative SUV-volume histogram method for parameterizing heterogeneous intratumoural FDG uptake in non-small cell lung cancer PET studies. Eur J Nucl Med Mol Imaging (2011) 38:1636–47. doi: 10.1007/s00259-011-1845-6
154. Lodge MA, Chaudhry MA, Wahl RL. Standardized uptake value noise considerations for PET quantification using maximum and peak noise considerations for PET quantification using maximum and peak standardized uptake value. J Nucl Med J Nucl Med. (2012) 53:1041–7. doi: 10.2967/jnumed.111.101733
155. Hatt M, Visvikis D, Albarghach NM, Tixier F, Pradier O, Cheze-Le Rest C. Prognostic value of 18F-FDG PET image-based parameters in oesophageal cancer and impact of tumour delineation methodology. Eur J Nucl Med Mol Imaging (2011) 38:1191–202. doi: 10.1007/s00259-011-1755-7
156. Bundschuh RA, Dinges J, Neumann L, Seyfried M, Zsótér N, Papp L, et al. Textural parameters of tumor heterogeneity in 18F-FDG PET/CT for therapy response assessment and prognosis in patients with locally advanced rectal cancer. J Nucl Med. (2014) 55:891–7. doi: 10.2967/jnumed.113.127340
157. Yoo MY, Paeng JC, Cheon GJ, Lee DS, Chung JK, Kim EE, et al. Prognostic value of metabolic tumor volume on 11C-methionine PET in predicting progression-free survival in high-grade glioma. Nucl Med Mol Imaging (2015) 49:291–7. doi: 10.1007/s13139-015-0362-0
158. Shah B, Srivastava N, Hirsch AE, Mercier G, Subramaniam RM. Intra-reader reliability of FDG PET volumetric tumor parameters: effects of primary tumor size and segmentation methods. Ann Nucl Med. (2012) 26:707–14. doi: 10.1007/s12149-012-0630-3
159. Tixier F, Le Rest CC, Hatt M, Albarghach NM, Pradier O, Metges J-PJ-PP, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. (2011) 52:369–78. doi: 10.2967/jnumed.110.082404
160. Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D, et al. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. (2012) 53:693–700. doi: 10.2967/jnumed.111.099127
161. El Naqa I, Grigsby PW, Apte A, Kidd E, Donnelly E, Khullar D, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recogn (2009) 42:1162–71. doi: 10.1016/j.patcog.2008.08.011
162. George J, Claes P, Vunckx K, Tejpar S, Deroose CM, Nuyts J, et al. A textural feature based tumor therapy response prediction model for longitudinal evaluation with PET imaging. In: 2012 9th IEEE Int Symp Biomed Imaging. Barcelona (2012). p. 1048–51.
163. Pyka T, Bundschuh RA, Andratschke N, Mayer B, Specht HM, Papp L, et al. Textural features in pre-treatment [F18]-FDG-PET/CT are correlated with risk of local recurrence and disease-specific survival in early stage NSCLC patients receiving primary stereotactic radiation therapy. Radiat Oncol. (2015) 10:100. doi: 10.1186/s13014-015-0407-7
164. Upadhaya T, Morvan Y, Stindel E, Le Reste PJ, Hatt M. A framework for multimodal imaging-based prognostic model building: preliminary study on multimodal MRI in Glioblastoma Multiforme. IRBM (2015) 36:345–50. doi: 10.1016/j.irbm.2015.08.001
165. Win T, Miles KA, Janes SM, Ganeshan B, Shastry M, Endozo R, et al. Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer. Clin Cancer Res. (2013) 19:3591–9. doi: 10.1158/1078-0432.CCR-12-1307
166. Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: preliminary evidence of an association with tumour metabolism, stage, and survival. Clin Radiol. (2012) 67:157–64. doi: 10.1016/j.crad.2011.08.012
167. Desseroit MC, Visvikis D, Tixier F, Majdoub M, Perdrisot R, Guillevin R, et al. Development of a nomogram combining clinical staging with 18F-FDG PET/CT image features in non-small-cell lung cancer stage I-III. Eur J Nucl Med Mol Imaging (2016) 43:1477–85. doi: 10.1007/s00259-016-3325-5
168. Cook GJR, Yip C, Siddique M, Goh V, Chicklore S, Roy A, et al. Are pretreatment 18F-FDG PET tumor textural features in non–small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. (2013) 54:19–26. doi: 10.2967/jnumed.112.107375
169. Lovinfosse P, Polus M, Van Daele D, Martinive P, Daenen F, Hatt M, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging (2017) 45:365–75. doi: 10.1007/s00259-017-3855-5
170. Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. (2015) 5:13087. doi: 10.1038/srep13087
171. Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. (2016) 6:71. doi: 10.3389/fonc.2016.00071
172. Lee JW, Lee SM. Radiomics in oncological PET/CT: clinical applications. Nucl Med Mol Imaging (2010) (2017) doi: 10.1007/s13139-017-0500-y
173. Bailey DL, Pichler BJ, Gückel B, Barthel H, Beer AJ, Botnar R, et al. Combined PET/MRI: from status quo to status go. Summary report of the fifth international workshop on PET/MR imaging; February 15–19, 2016; Tübingen, Germany. Mol Imaging Biol. (2016) 18:637–50. doi: 10.1007/s11307-016-0993-2
174. Lucia F, Visvikis D, Desseroit M-C, Miranda O, Malhaire J-P, Robin P, et al. Prediction of outcome using pretreatment 18F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging (2017) 45:768–86. doi: 10.1007/s00259-017-3898-7
175. Avanzo M, Stancanello J, El Naqa I. Beyond imaging: the promise of radiomics. Phys Med. (2017) 38:122–39. doi: 10.1016/j.ejmp.2017.05.071
176. Giesel FL, Schneider F, Kratochwil C, Rath D, Moltz J, Holland-Letz T, et al. Correlation between SUV max and CT radiomic analysis using lymph node density in PET/CT-based lymph node staging. J Nucl Med. (2017) 58:282–7. doi: 10.2967/jnumed.116.179648
177. Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging (2017) 45:207–17. doi: 10.1007/s00259-017-3837-7
178. Anthony GJ, Cunliffe A, Castillo R, Pham N, Guerrero T, Armato SG, et al. Incorporation of pre-therapy 18 F-FDG uptake data with CT texture features into a radiomics model for radiation pneumonitis diagnosis. Med Phys. (2017) 44:3686–94. doi: 10.1002/mp.12282
179. Crispin-Ortuzar M, Apte A, Grkovski M, Oh JH, Lee N, Humm J, et al. Combined FDG and CT radiomics features predict FMISO uptake in head and neck cancer. Int J Radiat Oncol. (2017) 99:S49–50. doi: 10.1016/j.ijrobp.2017.06.126
180. Way GP, Allaway RJ, Bouley SJ, Fadul CE, Sanchez Y, Greene CS. A machine learning classifier trained on cancer transcriptomes detects NF1 inactivation signal in glioblastoma. BMC Genomics (2017) 18:127. doi: 10.1186/s12864-017-3519-7
181. Emblem KE, Pinho MC, Zöllner FG, Due-Tonnessen P, Hald JK, Schad LR, et al. A generic support vector machine model for preoperative glioma survival associations. Radiology (2015) 275:228–34. doi: 10.1148/radiol.14140770
182. Zhang H, Molitoris J, Tan S, Giacomelli I, Scartoni D, Gzell C, et al. SU-F-R-04: radiomics for survival prediction in glioblastoma (GBM). Med Phys. (2016) 43:3373. doi: 10.1118/1.4955776
183. Chen SW, Shen WC, Lin YC, Chen RY, Hsieh TC, Yen KY, et al. Correlation of pretreatment 18F-FDG PET tumor textural features with gene expression in pharyngeal cancer and implications for radiotherapy-based treatment outcomes. Eur J Nucl Med Mol Imaging (2017) 44:567–80. doi: 10.1007/s00259-016-3580-5
184. Papp L, Poetsch N, Grahovac M, Schmidbauer V, Woehrer A, Preusser M, et al. Glioma survival prediction with the combined analysis of in vivo 11C-MET-PET, ex vivo and patient features by supervised machine learning. J Nucl Med. (2017). 6:86–91. doi: 10.2967/jnumed.117.202267
185. Verleysen M, François D. The curse of dimensionality in data mining and time series prediction. In: Cabestany J, Prieto A, Sandoval F, editors. Computational Intelligence and Bioinspired Systems. IWANN 2005. Lecture Notes in Computer Science, Vol. 3512. Berlin; Heidelberg: Springer (2005).
186. Viswanath SE, Tiwari P, Lee G, Madabhushi A. Dimensionality reduction-based fusion approaches for imaging and non-imaging biomedical data: concepts, workflow, and use-cases. BMC Med Imaging (2017) 17:2. doi: 10.1186/s12880-016-0172-6
187. Jung M, Zscheischler J. A guided hybrid genetic algorithm for feature selection with expensive cost functions. Proc Comput Sci. (2013) 18:2337–46. doi: 10.1016/j.procs.2013.05.405
188. Mi H, Petitjean C, Dubray B, Vera P, Ruan S. Robust feature selection to predict tumor treatment outcome. Artif Intell Med. (2015) 64:195–204. doi: 10.1016/j.artmed.2015.07.002
189. Adegoke B., Ola BO, Omotayo M. Review of feature selection methods in medical image processing. IOSR J Eng. (2014) 4:1–5. doi: 10.9790/3021-04140105
190. Hricak H, Choi BI, Scott AM, Sugimura K, Muellner A, von Schulthess GK, et al. Global trends in hybrid imaging. Radiology (2010) 257:498–506. doi: 10.1148/radiol.10100579
191. Leijenaar RTH, Carvalho S, Velazquez ER, van Elmpt WJC, Parmar C, Hoekstra OS, et al. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. (2013) 52:1391–7. doi: 10.3109/0284186X.2013.812798
192. Soussan M, Orlhac F, Boubaya M, Zelek L, Ziol M, Eder V, et al. Relationship between tumor heterogeneity measured on FDG-PET/CT and pathological prognostic factors in invasive breast cancer. PLoS ONE (2014) 9:e94017. doi: 10.1371/journal.pone.0094017
193. Orlhac F, Nioche C, Soussan M, Buvat I. Understanding changes in tumor texture indices in PET: a comparison between visual assessment and index values in simulated and patient data. J Nucl Med. (2017) 58:387–92. doi: 10.2967/jnumed.116.181859
194. Martinez-Tellez B, Sanchez-Delgado G, Garcia-Rivero Y, Alcantara JMA, Martinez-Avila WD, Muñoz-Hernandez MV, et al. A new personalized cooling protocol to activate brown adipose tissue in young adults. Front Physiol (2017) 8:863. doi: 10.3389/fphys.2017.00863
195. Williams G, Kolodny GM. Suppression of myocardial 18 F-FDG uptake by preparing patients with a high-fat, low-carbohydrate diet. Am J Roentgenol. (2008) 190:W151–6. doi: 10.2214/AJR.07.2409
196. Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nucl Med. (2009) 50:11–20. doi: 10.2967/jnumed.108.057182
197. EANM. EANM Guidelines. www.eanm.org (2017). Available online at: http://www.eanm.org/publications/guidelines/
198. IAEA. Standard Operating Procedures for PET/CT: A Practical Approach for Use in Adult Oncology. 26th ed. IAEA Human Health Series (2013).
199. MacFarlane CR, American College of Radiologists. ACR accreditation of nuclear medicine and PET imaging departments. J Nucl Med Technol. (2006) 34:18–24.
200. Peng H S. Levin C. Recent developments in PET instrumentation. Curr Pharm Biotechnol. (2010) 11:555–71. doi: 10.2174/138920110792246555
201. Feliciani G, Bertolini M, Fioroni F, Iori M. Texture analysis in CT and PET: a phantom study for features variability assessment. Phys Med. (2016) 32:77. doi: 10.1016/j.ejmp.2016.01.263
202. Shiri I, Rahmim A, Ghaffarian P, Geramifar P, Abdollahi H, Bitarafan-Rajabi A. The impact of image reconstruction settings on 18F-FDG PET radiomic features: multi-scanner phantom and patient studies. Eur Radiol. (2017) 27:4498–509. doi: 10.1007/s00330-017-4859-z
203. Fried D, Meier J, Mawlawi O, Zhou S, Ibbott G, Liao Z, et al. MO-DE-207B-07: assessment of reproducibility of FDG-PET-based radiomics features across scanners using phantom imaging. Med Phys. (2016) 43:3705–6. doi: 10.1118/1.4957256
204. Schaefer A, Kim YJ, Kremp S, Mai S, Fleckenstein J, Bohnenberger H, et al. PET-based delineation of tumour volumes in lung cancer: comparison with pathological findings. Eur J Nucl Med Mol Imaging (2013) 40:1233–44. doi: 10.1007/s00259-013-2407-x
205. Hatt M, Cheze le Rest C, Descourt P, Dekker A, De Ruysscher D, Oellers M, et al. Accurate automatic delineation of heterogeneous functional volumes in positron emission tomography for oncology applications. Int J Radiat Oncol. (2010) 77:301–8. doi: 10.1016/j.ijrobp.2009.08.018
206. Carles M, Torres-Espallardo I, Alberich-Bayarri A, Olivas C, Bello P, Nestle U, et al. Evaluation of PET texture features with heterogeneous phantoms: complementarity and effect of motion and segmentation method. Phys Med Biol. (2017) 62:652–68. doi: 10.1088/1361-6560/62/2/652
207. Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. (2017) 44:177–95. doi: 10.1016/j.media.2017.12.007
208. Vorwerk H, Beckmann G, Bremer M, Degen M, Dietl B, Fietkau R, et al. The delineation of target volumes for radiotherapy of lung cancer patients. Radiother Oncol. (2009) 91:455–60. doi: 10.1016/j.radonc.2009.03.014
209. Nestle U, Kremp S, Schaefer-Schuler A, Sebastian-Welsch C, Hellwig D, Rübe C, et al. Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-Small cell lung cancer. J Nucl Med. (2005) 46:1342–8.
210. Schinagl DAX, Vogel W V., Hoffmann AL, van Dalen JA, Oyen WJ, Kaanders JHAM. Comparison of five segmentation tools for 18F-fluoro-deoxy-glucose-positron emission tomography-based target volume definition in head and neck cancer. Int J Radiat Oncol Biol Phys. (2007) 69:1282–9. doi: 10.1016/j.ijrobp.2007.07.2333
211. Hatt M, Visvikis D, Le Rest CC. Autocontouring versus manual contouring. J Nucl Med. (2011) 52:658–9. doi: 10.2967/jnumed.110.084897
212. Nariai T, Tanaka Y, Wakimoto H, Aoyagi M, Tamaki M, Ishiwata K, et al. Usefulness of L-[methyl-11C] methionine-positron emission tomography as a biological monitoring tool in the treatment of glioma. J Neurosurg. (2005) 103:498–507. doi: 10.3171/jns.2005.103.3.0498
213. Kobayashi K, Hirata K, Yamaguchi S, Manabe O, Terasaka S, Kobayashi H, et al. Prognostic value of volume-based measurements on 11C-methionine PET in glioma patients. Eur J Nucl Med Mol Imaging (2015) 42:1071–80. doi: 10.1007/s00259-015-3046-1
214. Takano K, Kinoshita M, Arita H, Okita Y, Chiba Y, Kagawa N, et al. Diagnostic and prognostic value of 11C-methionine pet for nonenhancing gliomas. Am J Neuroradiol. (2016) 37:44–50. doi: 10.3174/ajnr.A4460
215. Konert T, Vogel WV, Everitt S, MacManus MP, Thorwarth D, Fidarova E, et al. Multiple training interventions significantly improve reproducibility of PET/CT-based lung cancer radiotherapy target volume delineation using an IAEA study protocol. Radiother Oncol. (2016) 121:39–45. doi: 10.1016/j.radonc.2016.09.00
216. Bagci U, Udupa JK, Mendhiratta N, Foster B, Xu Z, Yao J, et al. Joint segmentation of anatomical and functional images: applications in quantification of lesions from PET, PET-CT, MRI-PET, and MRI-PET-CT images. Med Image Anal. (2013) 17:929–45. doi: 10.1016/j.media.2013.05.004
217. Mi H, Petitjean C, Dubray B, Vera P, Ruan S. Automatic lung tumor segmentation on {PET} images based on random walks and tumor growth model. In: 2014 IEEE 11th Int Symp Biomed Imaging. Bejing (2014). p. 1385–8.
218. Onoma DP, Ruan S, Thureau S, Nkhali L, Modzelewski R, Monnehan GA, et al. Segmentation of heterogeneous or small FDG PET positive tissue based on a 3D-locally adaptive random walk algorithm. Comput Med Imaging Graph. (2014) 38:753–63. doi: 10.1016/j.compmedimag.2014.09.007
219. Layer T, Blaickner M, Knäusl B, Georg D, Neuwirth J, Baum RP, et al. PET image segmentation using a Gaussian mixture model and Markov random fields. EJNMMI Phys. (2015) 2:9. doi: 10.1186/s40658-015-0110-7
220. Boellaard R, Delgado-Bolton R, Oyen WJG, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging (2015) 42:328–54. doi: 10.1007/s00259-014-2961-x
221. Dolz J, Kirişli HA, Fechter T, Karnitzki S, Oehlke O, Nestle U, et al. Interactive contour delineation of organs at risk in radiotherapy: clinical evaluation on NSCLC patients. Med Phys. (2016) 43:2569–80. doi: 10.1118/1.4947484
222. Brooks FJ, Grigsby PW. The effect of small tumor volumes on studies of intratumoral heterogeneity of tracer uptake. J Nucl Med. (2014) 55:37–42. doi: 10.2967/jnumed.112.116715
223. Hatt M, Majdoub M, Vallieres M, Tixier F, Le Rest CC, Groheux D, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med. (2015) 56:38–44. doi: 10.2967/jnumed.114.14405
224. Buvat I, Orlhac F, Soussan M. Tumor texture analysis in PET: where do we stand? J Nucl Med. (2015) 56:1642–4. doi: 10.2967/jnumed.115.163469
225. Zhao B, Tan Y, Tsai W-Y, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. (2016) 6:23428. doi: 10.1038/srep23428
226. Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging (2015) 2:41002. doi: 10.1117/1.JMI.2.4.041002
227. Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. (2015) 50:757–65. doi: 10.1097/RLI.0000000000000180
228. Mackin D, Fave X, Zhang L, Yang J, Jones AK, Ng CS, et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS ONE (2017) 12:e0178524. doi: 10.1371/journal.pone.0178524
229. Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, et al. A post-reconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. (2018). doi: 10.2967/jnumed.117.199935. [Epub ahead of print].
230. Zwanenburg A, Leger S, Vallières M, Löck S. Initiative for the IBS. Image biomarker standardisation initiative. arXiv: 1612.07003 (2016).
231. Maastro. Decoding the Tumor Phenotype with Non-Invasive Imaging (2017). Available online at: http://www.radiomics.world/
232. Vallieres M, Zwanenburg A, Badic B, Cheze-Le Rest C, Visvikis D, Hatt M. Responsible radiomics research for faster clinical translation. J Nucl Med. (2017) 59:189–93. doi: 10.2967/jnumed.117.200501
233. Ahmad LG, Eshlaghy AT, Poorebrahimi A, Ebrahimi M, Razavi AR. Using three machine learning techniques for predicting breast cancer recurrence. J Heal Med Inform (2013) 4:2–4. doi: 10.4172/2157-7420.1000124
234. Chalkidou A, O'Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS ONE (2015) 10:e0124165. doi: 10.1371/journal.pone.0124165
235. Birlutiu A, Groot P, Heskes T. Efficiently learning the preferences of people. Mach Learn. (2013) 90:1–28. doi: 10.1007/s10994-012-5297-4
236. Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. RadioGraph. (2017) 37:505–15. doi: 10.1148/rg.2017160130
237. Holman AG, Gabuzda D. A machine learning approach for identifying amino acid signatures in the HIV env gene predictive of dementia. PLoS ONE (2012) 7:e49538. doi: 10.1371/journal.pone.0049538
238. Beleites C, Baumgartner R, Bowman C, Somorjai R, Steiner G, Salzer R, et al. Variance reduction in estimating classification error using sparse datasets. Chemom Intell Lab Syst. (2005) 79:91–100. doi: 10.1016/j.chemolab.2005.04.008
239. Xu Q-S, Liang Y-Z. Monte Carlo cross validation. Chemom Intell Lab Syst. (2001) 56:1–11. doi: 10.1016/S0169-7439(00)00122-2
240. Mallick S. Bias-Variance Tradeoff in Machine Learning. Learn Open CV. Available online at: http://www.learnopencv.com/bias-variance-tradeoff-in-machine-learning/ (2017).
241. Dai W, Yang Q, Xue G-R, Yu Y. Boosting for transfer learning. In: Proceedings of the 24th International Conference on Machine Learning - ICML '07. New York, NY; Corvalis: ACM Press. (2007) p. 193–200.
242. González CR, Abu-Mostafa YS. Mismatched training and test distributions can outperform matched ones. Neural Comput. (2015) 27:365–87. doi: 10.1162/NECO_a_00697
243. Buntine W, Niblett T. A further comparison of splitting rules for decision-tree induction. Mach Learn. (1992) 8:75–85.
244. Holzreiter SH, Köhle ME. Assessment of gait patterns using neural networks. J Biomech. (1993) 26:645–51.
245. Ross KA, Jensen CS, Snodgrass R, Dyreson CE, Jensen CS, Snodgrass R, et al. Cross-Validation. In: Ling L, Tamer ÖM, editors. Encyclopedia of Database Systems. Boston, MA: Springer US (2009). p. 532–8.
246. Diamantidis NA, Karlis D, Giakoumakis EA. Unsupervised stratification of cross-validation for accuracy estimation. Artif Intell. (2000) 116:1–16. doi: 10.1016/S0004-3702(99)00094-6
248. EC-Brussels. European Cloud Initiative to give Europe a Global Lead in the Data-Driven Economy. Eur Comm. Available online at: http://europa.eu/rapid/press-release_IP-16-1408_en.htm (2016).
249. Charara S. The Future of Wearables: From Fitness Trackers to Always on Assistants. www.wearable.com. Available online at: https://www.wareable.com/headphones/the-future-of-hearables-from-fitness-trackers-to-always-on-assistants (2015).
Keywords: hybrid imaging, medical big data, machine learning, radiomics, holomics, personalized medicine
Citation: Papp L, Spielvogel CP, Rausch I, Hacker M and Beyer T (2018) Personalizing Medicine Through Hybrid Imaging and Medical Big Data Analysis. Analysis. Front. Phys. 6:51. doi: 10.3389/fphy.2018.00051
Received: 30 December 2017; Accepted: 09 May 2018;
Published: 07 June 2018.
Edited by:
Claudia Kuntner, Austrian Institute of Technology, AustriaReviewed by:
Issam El Naqa, University of Michigan, United StatesWolfgang Huf, Hietzing Hospital, Austria
Copyright © 2018 Papp, Spielvogel, Rausch, Hacker and Beyer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Beyer, dGhvbWFzLmJleWVyQG1lZHVuaXdpZW4uYWMuYXQ=
 Laszlo Papp
Laszlo Papp Clemens P. Spielvogel
Clemens P. Spielvogel Ivo Rausch
Ivo Rausch Marcus Hacker2
Marcus Hacker2 Thomas Beyer
Thomas Beyer