
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Phys. , 07 August 2015
Sec. Biophysics
Volume 3 - 2015 | https://doi.org/10.3389/fphy.2015.00055
A commentary has been posted on this article:
Chief Editor's Commentary on “Light Based Cellular Interactions: hypotheses and perspectives”
This work investigates the theoretical possibility of interactions between cells via light. We first take a brief look at the previous research done in the past to have a better understanding of the field and the origins of the concept of cellular interactions. Then we identify the different elements essential for interactions between two parties. We then compare the required elements with the known and studied elements and characteristics which are well defined in biology, chemistry and physics. This way we are able to set up four postulates required for cell interactions: I. A signal is present and subject to secondary modulation by the emitter cells. II. There is a plastic information medium that reacts directly to the metabolic state of the emitter and therefore carries information about the emitter. III. An optical signal can be perceived by cells on a molecular level by a multitude of different receptors. IV. The information can in theory be processed by cells and metabolic changes in reaction to the signals can be observed. We demonstrate that all required elements have been observed. Most of them have important and well-known roles in cells. Therefore, we suggest that our hypothetical model is a good explanation for light based cellular interactions.
The concept of cellular interactions via photons was proposed a long time ago [1] and there are several publications about light based interactions [1–15].
Gurwitsch was the first to claim the existence of a biological “radiation” which could affect the rate of mitosis. In many experiments he tried to figure out a good way to produce reproducible results [8].
From this moment on, almost all the experiments performed in this research field use a similar setup: Two living systems, chemically separated, but optically connected, show a reaction to each other's presence and/or activity.
As later described by Bateman [2] it was still unclear after several years if the “mitogenetic radiation” did really exist. Other groups tried to reproduce Gurwitsch's work, but many of them failed as described by Bateman.
However, others were able to observe interesting effects. For example, fertilized eggs of Parcentrotus lividus were incubated under the exposure of the radiation of B. tumefaciens cultures [7]. Choucron found that there was a causal effect between the existence of an action and the presence of the cultures, after observing deformations and abnormalities.
For a long time the concept of mitogenetic radiation was forgotten. But in the last four decades more and more researchers have become interested again in the topic. These are some more recent examples:
Quickenden [15] also reviewed the work done on mitogenetic radiation from Gurwitsch and other researchers, and asked interesting questions such as: Does UPE stimulate cell division? And: Can a cell culture affect the growth rate of another? Those questions are partially answered nowadays as we will see below.
By using Pseundomonas putida and spores of B. subtilis, a Russian group was able to influence germination efficiency, by using two different methods. It was concluded that growth-stimulating signals were transmittable and they couldn't be in the UV range, so the visible or near IR range could be responsible in this case. It also showed that the interaction could take place between different species [3].
In another publication, where human blood was used to irradiate radish seeds, an increase of the growth index could be observed. This system was also using a quartz cuvette. The effect was enhanced when fresh blood was used instead of 2 days old blood [6]. This suggests that the efficiency of the signaling is related to quality and viability of the samples.
Recently, new experiments were performed in Switzerland [12] with Paramecium caudatum with a “flask in flask” system, demonstrating the reproducibility of the phenomenon. Also several recent publications and reviews [10, 16–18] summarize the progress in the field in a very accurate way.
It was only with the development of photomultiplier tubes (PMTs) and the first measurements of photons coming from inside the cells that the research on ultra-weak photon emission (UPE) could begin. But it also showed that light was indeed produced by living systems and therefore cells could at least in theory use the photons to transfer information. We will not review the large number of publications that can be found on the UPE topic [15, 19–22] and which can be found in other reviews [17, 18, 23]. We will only look at a brief selection of publications which investigated cellular interactions.
In another publication [4] two E. coli cultures, separated by a glass absorbing light under 350 nm, were used. The growth rates in M9 and Luria Broth (LB) medium were greater than in controls and the generation times shorter than in the controls. Also different UPE spectra could be detected during lag, exponential and stationary phase of E. coli growth, in LB and M9 medium. It was therefore interpreted that E. coli could somehow react to their neighbors behind the glass barrier. Also, from another perspective, Australian researchers analyzed the spectra of the UPE from yeast [15, 24]. A decade later, yeast was used again to analyze the UPE during the mitotic cycle [25]. Yeast was chosen since there are possibilities to synchronize its mitotic cycle. It was observed that big changes occurred in the whole UPE spectra, those changes were with the very likely related to the mitotic phases.
We will see that these kinds of observations are critical to understand how the mitotic cycle can generate specific optical signals, which could be received and interpreted by other cells.
Finally we want to point to recent publications which explain the possible mechanisms for the emission of UPE [26] and set the appropriate definitions for the observed phenomena [18].
The improved molecular methods developed over the decades have helped researchers to better understand this phenomenon. Here are some examples: When the cells on one side of the separator were exposed to H2O2, they showed fascinating results. The NF-κB activation was measured after 10, 30 and 60 min in both compartments [5]. Also with different distances between emitter and receiver cells, the protein concentration was significantly decreased compared to the controls.
Only recently a work from Italy showed that mouse fribroblasts and adult human microvascular endothelial cells would react to each other's optical presence. The interaction between the immortalized mouse fibroblasts (NIH3T3) and adult human microvascular endothelial cells (HMVECad) could be observed. As a negative control a black filter was placed between the cultures [9, 10]. In prior work on fribroblasts (HS27), it was observed that the cells reacted to the presence of weak red light, by regulating 111 genes, which are related to proliferation and suppression of apoptosis [27].
Finally, in neuroscience, some advances have been made. Experiments on rat nerves showed that neurons could act as light guiding elements [28]. We should not forget that a lot of work is still required to really understand the exact meaning and purpose of such findings, but a “Dish in Dish” system from Chaban [14] could alter the calcium fluxes by using apoptotic dorsal root ganglion neurons or human neuroblastoma cells as emitter cells. Such a phenomenon has never been observed in the past. Those examples were chosen because they illustrate well how wide and diversified the topic of non-chemical distant cellular interactions is.
One of the main problems that models in this field encounter is the following: How could such a weak signal as the UPE transfer information in presence of a much stronger noise, like the ambient daylight? To find the answer to this question is one of the greatest challenges in the field of non-chemical distant cellular interaction [29]. We will attempt to answer this later in Section The Processing.
Nevertheless, the problems and controversy of the question some hypothetical models have already been published [11, 30–32]. We will now present and discuss some of them briefly.
Austrian scientists proposed that UPE could form a sort of “holographic computer” within brains [11]. In our opinion even if the concept is interesting we have to conclude that this model is purely hypothetical and no experiments have been done to prove it. Nevertheless, the idea has been developed further by others [33, 34].
Another interesting hypothesis was published several years ago, claiming that mitochondria and microtubules could be seen as a biological version of optic fibers, guiding the photons generated by the respiratory chain to the right place [31].
With another approach Fels hypothesized in his work, that from two cell populations, only one should be sensitive to the repetition of the experiment. The other population, however, should establish a relation with the first one. This hypothesis considers a cell-to-cell relation as the analog to the relationship between quantum particles [30].
Also, a concept of interaction through vibration has been proposed in two hypothetical models [35, 36].
Despite the interesting ideas and aspects, all these theories are lacking an exact bio molecular explanation of the process of signal transfer.
Every form of interaction requires well defined elements, in a way that the information transfer can take place and function as required. To make the elements easier to grasp we will compare our model to the spoken language. For the physical elements there are the emitters, the medium and the receivers. For the spoken language the emitters would be the mouth, the tongue and the vocal cords, which can modulate the medium, which in this case is the sound or vibrations of the air. And the receivers would be the human inner and outer ears.
From the semantic point of view there must be “compliance.” The emitter and receiver have to be “adjusted” to understand each other. In our example we could say that the two people must speak the same language in order to understand each other. And finally there must be “processing.” The received signals must have an effect on the receiver: We could say that the information of the spoken message has to be processed by the brain and the person reacts to the information in some way. If there would be no reaction to the information then the information content can be seen as null from the point of view of the receiver and the term “interaction” becomes hardly applicable. We therefore retain the five important elements necessary for cellular interaction: Emitters, medium, receivers, compliance and processing. At last following characteristics of the required elements should be given:
- The emitters must be able to modulate the signal.
- The medium must be plastic and be able to travel from emitter to receiver.
- The receivers should be able to perceive the signals and to process the information.
- And finally a reproducible reaction to a defined signal has to be observable.
We will attempt to explain cellular interactions via light. Roughly speaking, our model can be described as following: The photons are first produced by excited species in the emitter cells. The signal is then transformed through absorption or/and spectrally modified by autofluorescence, by the emitter cell's contents, to then escape the emitter cells. Finally they are absorbed by different chromophores in the receiver cells to transmit the information to the nucleus or other biochemical pathways.
Such a model is purely hypothetical and may never be proven fully. But as we will see, many required elements are present in cells and their functionalities are well understood. Let us now take a closer look at the interaction model by understanding each element one by one.
For the existence of a working interactive system, first off an emitter is required. In the case of our model, the emitters are triplet excited carbonyl, singlet and triplet excited pigments and singlet oxygen. There are several publications showing the modulation of UPE, relative to the free radicals and excited molecules [such as reactive oxygen species (ROS)] [20, 21, 37–40]. The exact biochemical mechanisms were explained recently by Pospíšil [26]. Depending on what type of reactions are the sources and what kind of components are inside the cells, we will have different types of wavelength ranges and the intensities [15, 41, 42] of UPE. Also we should not omit that ROS are themselves part of multiple signaling pathways [43] and therefore already contain an interesting amount of information. We know that we have a measurable signal leaving the cells. We now can set our fist postulate: The signal is present and can in a second time be modulated by the cells. As for example: higher ATP production produces higher ROS concentration, which creates more photons [20, 44]. We will look at more ways to modulate the signal in the next paragraph.
The photons are the medium in our model. Light can transmit information in different ways. Following parameters can be modulated or may be different from the moment of their existence: Intensity, wavelength and statistical distribution. (We leave polarization and spin for now, since no biological research has been done on the subject as far as the author knows). The hypothesis that UPE could be a medium for biological information and that the information may lie inside the intensity, spectral distribution and statistical photon count distribution was already presented in 1992 [45].
As explained above, the intensity of the UPE is part of the information, because it may tell how many free radical and what kind of free radicals are in the cells. However, this is far away from being the most important part of the retrievable information.
The modulation of the signal can be achieved by two well-known effects:
The first one is attenuation, also known as absorption. In this case we can only talk about “absorption” when the photons are really absorbed according to the Beer–Lambert law [46], by cellular components and not just scattered. In the case of scattering the photon would still exist and would just change its direction. If we assume that the photons are not propagating in any specific direction and we consider that we have a statically equal amount of photons in every direction, we should omit all scattering effects. Therefore, only the attenuation matters, when the photon's energy is transformed into some other type of energy and the photon as such disappears. Only then it would have an impact on the signal, because it would specifically change the amount of photons relatively to the amount of present proteins absorbing at the specific wavelengths. The idea of information modulation by attenuation has been proposed before [11]. The fact that cells in different metabolic states have different cellular components and different absorption spectra is well known and described in the literature [47–49]. We will not discuss this work further.
The second phenomenon which is also well known and could modulate the signal is autofluorescence. Here the photons would be transformed into other photons with a longer wavelength, and therefore lower energy, by the proteins and cellular components. Also here the modulation would directly depend on the concentration of autofluorescent elements. The literature is full of lists containing autofluorescent proteins and biochemical components [50–52, 54]. We will therefore also not discuss this topic more deeply in this work.
The components which could modulate optical signals through absorption or autofluorescence could be:
Membranes, cytoplasmic proteins, organelles or vesicles and their contents, the nucleus and its contents, membrane proteins, nutrients which were assimilated by the cell, the cytoskeleton and others would fit to the requirements. Also the information could be modulated by extra cellular molecules, for example the surrounding medium or the extra cellular matrix in tissues.
Since cells in different metabolic states will have different concentrations of absorbing and autofluorescent components, these will also differ in their patterns of attenuation and autofluorescent reemission. Therefore, statistically speaking, only a specific photon spectral distribution will be able to escape the cells, specific to the metabolic activity of the cells. So in a manner of speaking we could say that the information lies in the photons which are “not present anymore” or “have another wavelength.” We can now set our second postulate: There is a plastic information medium that reacts directly to the metabolic state of the emitter (and maybe even its direct environment) and therefore carries information about the emitter.
To have effective receivers, we need biochemical compounds which can absorb light (also known as photoreceptors containing a chromophore) and have one or more aromatic components [53, 55]. We will now see that these required components are present in cells.
For example, recently a set of chromophores was discovered in non-phototrophic bacteria [56], which are able to act as gene transcription factors. Those proteins fulfill all the requisitions for receiver molecules. Another example can be found in rhodobacter. The blue light photoreceptor (BLUF) induces gene repression [57]. The next typical example of light sensing is the fungus Neurospora crassa where the gene expression for differentiation and metabolic activities is also photo-dependent. In this case flavins act as the main receptors, but also light dependent enzymes and regulators do exist [58]. In Drosophila melanogaster the photoreceptor CRY can even mediate neuronal firing. The same protein was found in humans but its function remains unclear [59]. In microorganisms the sensing and responding to light is well researched [60]. In cells various chromophores are known [61]. The chromophores mainly respond to photons by a cis/trans isomerization. The effect is always very wavelength specific. They will therefore only respond to photons with a specific wavelength.
Chromophores, like rhodopsin, are widely present in maybe all organisms and could be much more important than assumed until now. Flavin based photoreceptors are known but their photochemistry is still a partially unresolved question. E. coli does insert all known occurring flavins in the protein domain of a chromophore under growth conditions [62]. Orthologs of blue light receptors have also been found in, drosophila, arabidopsis, synechocystis, and humans [63, 64], and can also activate genes via dynamic conformational changes.
Also photosensitizers can absorb light and transfer the energy to other molecules which can produce singlet oxygen. Those can induce gene expression or even apoptosis [65]. Another receptor mechanism could be through photon induced cross-linking of transcription factors or other proteins [66].
We should not forget to mention that also the autofluorescent and absorbing elements in and on the receiver cells might modulate the signal as explained in Section The Medium. This might, on the one hand, block information that is not necessary if the two cells are already in the same state. On the other hand, it might allow a modulation of the information by the effect of autofluorescence so that the photons have the correct wavelengths to hit the right photoreceptors. But such statements are purely speculative.
Still, we can see through these examples that a large amount of photoreactive components exist in living systems and are well known. Definitely more will be discovered in the coming years. We can therefore set our third postulate: An optical signal could be perceived by cells on a molecular level by a multitude of different receptors.
The question of compliance is in this case quite difficult and we will not be able to answer it completely. How can we be sure that cells are talking the same language? As for today we cannot and we may never be able to. But if we take into account the fact that such an interaction would have been developed over the generations by natural selection and evolution. In this case a system without compliance would not make any sense, because it would not enhance the fitness of an optically interacting group. Therefore, we could allow ourselves to leave this question aside for the moment and simply assume that if there is an interaction then there must be compliance between the emitters and the receivers.
On the other hand, we can still ask ourselves: What would be the topic of such cellular conversation? What would cells or tissues talk about? Looking back at the introduction of this work, we can observe a recurring pattern: cell division. In an evolutionary context the topic makes absolute sense. The mitotic process costs lots of energy. Therefore, cells should have the right metabolic conditions and access to a profitable environment with enough nutrients to perform mitosis. Also the absence of pathogens or other disturbing elements can be beneficial to undertake a successful mitosis. If other organisms or cells surrounding cell are also dividing that could be interpreted as: “All prerequisites are set for mitosis.” But if for some reason the cells in the neighborhood are not dividing, it would be preferable to also wait for more favorable conditions. Since in our model such information would be generated anyway as a byproduct of the metabolic activity, why shouldn't other cells profit from this information?
Of course such assumptions are purely hypothetical and experimental evidence is still required to confirm the concept.
Finally in term of physical compliance we can take a look at the wavelength spectra of emitters and receivers and see if they are adjusted to each other.
If we take a look at the work from Pospíšil [26] where the different pathways for the electronically excited species productions are shown and the wavelengths of the emitted photons are described, we see that we have following emission spectra:
- 350–550 nm
- 550–750 nm
- 634 nm
- 1274 nm
We see that most parts of the visible spectra are covered with such emission patterns.
We now look at the possible receptors, such as the BLUF, LOV, CRY and PYP domains, the flavins and at the sensory rhodopsin. We see that these are just a few examples of possible absorption spectra and many more can be found in the literature:
- The BLUF domain responds to blue light: the peaks are at 375 and 450 nm [56] and other wavelengths [49].
- The PYP domain can respond to 446, 358, 434, and 465 nm depending on the cellular state [56, 67].
- Bacterial rhodopsin mostly absorbs green light ~500 nm [68].
- The LOV domain generally responds to blue-light ~450 nm [69].
- The Cryptochrome CRY has two chromophores: pterin which absorbs at a wavelength of 380 nm and flavin at 450 nm [70, 71].
- But flavins can have different oxidation states (the fully oxidized state, the semiquinone, a one electron reduced form and the hydroquinone, a two-electron reduced form). The fully oxidized absorption spectra are around 446, 370, and 265 nm. The semiquinone state absorbs around 650 nm and the hydroquinone species can absorb light in the UVB range [70].
As we see, the emission spectra and the receivers are therefore in compatible wavelength ranges. As for the modulation of the signal through absorption and autofluorescence, the spectra of the modulators in the UV-VIS-NIR regions of many molecules and proteins are well known [50–52]. We will therefore avoid losing ourselves in such extensive topics. But we can now assume that emitted photons could have an impact on the receiver cells. But we should not forget that, as for now, we don't know if UPE could provide enough energy to activate a photoreceptor.
To come back to our problem of signal-to-noise ratio, we will now take a look at the processing of the signal. Here again we have no proof for our theory, but we simply take a look at well-known mechanisms, described in molecular biology and observe if they would resolve the problem.
The problem of the signal-to-noise ratio is that the signals are very small and the weak changes are in the range of only few photons per second. While the noise, the surrounding daylight produces very large changes and relatively rapid fluctuations. But we should differ between two types of noises, rapid fluctuations of a light source (Poisson noise, see Figure 1C) and large jumps of intensity (when a light bulb is turned on or a cloud passes in front of the sun, see Figure 1B). While the fluctuations are much faster than biochemical reactions, such changes would be automatically average out, since the time scales are different. Or in other words: The biochemical pathways are in no way fast enough to react to every rapid fluctuation. Therefore, we estimate that such a problem can be left by side.
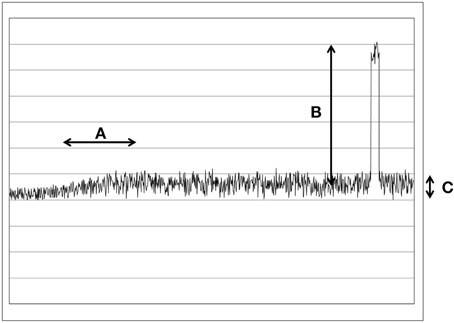
Figure 1. This figure illustrates the different types of changes and noises in a schematic way. (A) A slow and small change in the UPE. Typical for UPE changes. (B) A large and rapid change. Typical for daylight noises. (C) Poisson noise.
Obviously, a large number of averaging molecules need to be involved in the photon signal processing in order to decrease averaging time. Based on the known volumes and molar concentrations (if we estimate a few thousands of each signaling protein per cell), there actually could be a sufficient number of such averaging molecules and pathways to reduce the required averaging time. Also intercellular biochemical interactions within a population could also increase the number of such averaging entities.
On the other hand the large jumps in intensity would require a different mechanism so that these changes are not perceived as signals.
It would be a very simple mechanism to integrate a negative feedback loop [72] (as just one recent example of a negative feedback loop) that stops overregulation. When a certain level of signal is overstepped, then the negative feedback would become active and block the overregulation. This way the cells could filter out the small changes in large amounts of noise. The light from surrounding daylight noise source will always generate large changes (see Figure 1B), in the range of 1e14 photons/s/cm2. Those changes will be simply ignored because they would overregulate the pathway. Therefore, only small and slow changes (in the range of few hundred photons/s/cm2) would have an effect on the biochemical pathway (see Figure 1A).
Let us look at a theoretical example to better illustrate the signal filtering. Two groups of cells (an emitter and a receiver) are communicating at ambient light. They use two wavelength ranges (A and B). The signal has a total 150 photons/s/cm2 (100 photons/s/cm2 in range A and 50 photons/s/cm2 in range B). The ambient light has 1e14 photons/s/cm2 (5e13 photons/s/cm2in range A and 5e13 photons/s/cm2 in range B). In the next two steps two changes will happen. First a new light source is switched on in the room (as in Figure 1B), now the noise has 1.5e14 photons/s/cm2 (1e14 photons/s/cm2in range A and 5e13 photons/s/cm2 in range B). Therefore, the change of intensity in range A has a magnitude of 5e13 photons/s/cm2. Every change bigger than 1e6 photons/s/cm2 (This number is hypothetical and only serves as an example, still it has to be bigger than the fluctuation of the noise, see Figure 1C) still overregulates the pathway and is therefore ignored. In a second time the emitters send a new signal (as in Figure 1A) and the UPE in range A is decreased to 70 photons/s/cm2. The change is now small enough (30 photons/s/cm2) to have a slight impact on the biochemical pathway. The important concept here is that the cells are reacting to changes of light intensity and not actual intensity. Therefore, they must be able to differ between the amount of light they received before and the amount they receive now. The exact time scale between “before” and “now” cannot be defined since this parameter would depend on the reaction times, life time of the proteins and the concentration of the different yet unknown molecules of the pathways.
Such a mechanism is easy to understand and makes the concept that weak photon emission could still be a good medium for information quite plausible. But since for the moment there is no evidence for such a mechanism, the question if very few photons could transmit information, will remain unanswered [29] as long an experimental proof is inexistent.
Next we want to propose a second mechanism to show how easily information could be hidden and decoded in a little amount of light. Let us consider a set of different chromophores, each reacting only on one specific wavelength. Also each chromophore is at the beginning of at least one biochemical pathway. If now those pathways are interacting with each other, the information could be hidden in a relative amount of photons and not in an absolute number of photons. By using a relative signal of two (or more) wavelengths, information could reliably be transferred with very little light, independently of the amount of emitting cells. By coupling two or more detectors cells could also better differ between noise and signals. Therefore, we consider that interacting pathways as mandatory for our hypothesis. Also we should consider the concept that the different receptors could be distributed on more than one cell.
If the amount of photons in one wavelength range changes relatively to the photons amount in another wavelength range, then the ratio is modified. If the two pathways are interacting with each other it could end up a more complex regulation. Since interacting pathways are something absolutely common in biology, the assumption that the pathways of two or more photoreceptors are interacting, is not unreasonable. We should not omit the fact that interacting pathways of photoreceptors have already been observed in plants [73].
The real information would therefore not lie in the absolute number of photons, but in the relative numbers of photons in the different wavelength ranges. Thus, we could express the photo-information matrix Θ with, for example four wavelength ranges (as in the example in Figure 2) we would have six ratio values in the
Θ matrix= (a; b; c; d; e; f)
which would be the 6 ratios (a; b; c; d; e; f) of wavelengths ranges of the 4 photoreceptors (Pr):
Pr1, Pr2, Pr3 and Pr4 as
a = Pr1/Pr2; b = Pr1/Pr3; c = Pr1/Pr4; d = Pr2/Pr3; e = Pr2/Pr4; f = Pr3/Pr4.
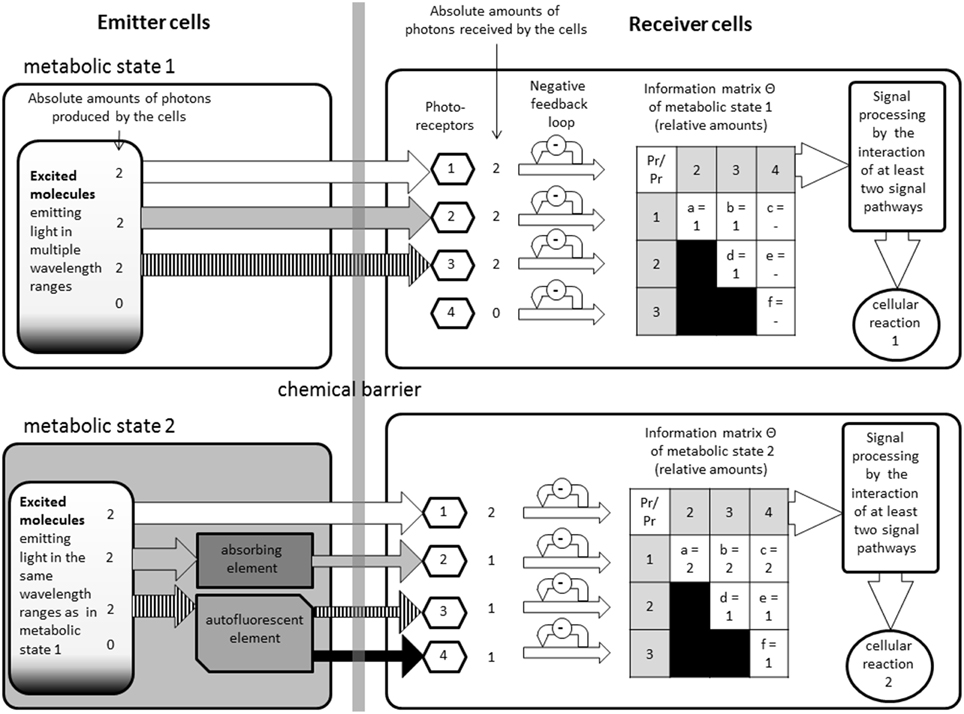
Figure 2. This figure illustrates how the model could function, but it does not reflect any specific proteins or pathways. On the left side are the emitter cells in two different metabolic states. Depending on the state, different amounts of light will be able to escape the cells in different wavelengths. On the right are the receiver cells. In between there is a chemical barrier which restrains chemical interactions but makes an optical interaction possible. The information matrixes show the different ratios that could be found if certain wavelengths modulators (autofluorescent or absorbing elements) are present or not, without changing the original emission spectra.
Since the photo-information Θ is expressed by a number of photons per second divided by another number of photons per second, it has no unit. This kind of matrixes combined with the information about the absorption spectra and the autofluorescence spectra of the samples could help researchers to analyze the UPE data. By identifying the exact chromophores and their wavelength ranges, we should be able to reproduce the signals. We could then verify this hypothesis by observing the cellular effects.
If we take a look at the list of chromophores [56] present in different organisms we can see that around a third of all listed organisms have more than one known chromophore, and could therefore be candidates for more complex signal integration. A list for further hypothetical receiver pathway like metabolic processes can be found in another work [11].
Since the research about photosensory proteins is only at its beginning, many other photoreceptors may be discovered in the next years. Some pathways are already known [49, 55, 74]. We hope that this model inspires researchers to look at and for integrated pathways of photoreceptors.
Taking into account the modulation through absorption, the negative feedback loop and the relative amount of photons, it is understandable that not a high amount of photons will have a high amount of information, but a low amount of photons will. We have also understood that the ratios and therefore the shape of wavelength spectra of the emission patterns are the key factors and not the total intensity of UPE.
We can now see that the processing of the information could take place in the cells and the information could be “understood” and a cellular reaction could take place. As mentioned before such phenomenon have already been observed in several experiments [4, 5, 12]. Knowing that cell cultures can react on a genetic level to optical signals [27] we can set our last postulate: The information could in theory be processed by cells and metabolic changes in reaction to the signals can be observed.
We have demonstrated that all elements for non-chemical distant cellular interactions are or could be present in cells and an information exchange could theoretically occur via light. Since no proven biological model exists at this moment to explain this phenomenon we consider our model as a good preliminary solution. But we must also consider that for such a model, following mechanisms must exist:
- Cells must emit photons.
- Photons must be modulated (partially absorbed or transformed by autofluorescence) by the cell contents or other extra-cellular molecules.
- Different photoreceptors must be present in the cells.
- The biochemical pathways of the photoreceptors must interact with each other and have a negative feedback loop to filter out the noise.
All these elements have been summarized in our graphical model (see Figure 2). This figure illustrates how the model could function, but it does not reflect any specific components:
The emitters are chemically, but not optically, separated from the receivers through a barrier. On the upper left side we see an emitter cell with is emitting light in three different wavelength ranges. As an example the absolute amounts of photons were defined as “2” (This number is purely theoretical and does not reflect any real values, it was only chosen to simplify the calculations) on all ranges in this example. Once the photons are received by the photoreceptors, the signals are filtered by a negative feedback loop to avoid over regulation by stronger noise. In the next step the biochemical pathways are then combined generating the information matrix with the relative amount of photons integrated.
On the lower side we added in this example, one absorbing and one autofluorescent elements, emerging from the altered metabolic state. The two metabolic states could be, for example: the cells are undergoing mitosis or not. This causes that one of the ranges is partially absorbed, which is also setting the absolute amount of received photons to “1” (This number is purely theoretical and does not reflect any real values, it was only chosen to simplify the calculations). Another one is partially transformed by autofluorescence into a fourth wavelength range; therefore those two ranges have now a relative amount of photons of “1.” Depending on the concentrations of the autofluorescent molecules and absorbing molecules the amount of light would relatively change. We do not take into consideration the probability that the photons actually reach the receivers. We consider that the amount of cells on both side have to be large enough to neglect such calculations. We can see how easily the smallest changes of the concentration of a specific molecule can alter the relative information matrix perceived by another biological system. Such changes could then alter gene activities or have other metabolic effects.
We hope that this model will be confirmed by future experiments and research projects. We also prospect that in the future more chromophores and biochemical pathways, connecting the receptors between them, will be identified to close the remaining gaps.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
3. Nikolaev YA, El'-Registan GI, Desu SB. Distant interaction during germination of bacillus subtillis spores. In: Beloussov LV, Voeikov VL, Martynyuk VS, editors. Biophotonics and Coherent Systems in Biology. Moscow: Springer (2007). p. 159–66
4. Trushin MV. Studies on distant regulation of bacterial regulation of growth and light emission. Microbiology (2003) 149:363–8. doi: 10.1099/mic.0.25825-0
5. Farhadi A, Forsyth C, Banan A, Shaik M, Engen P, Fields JZ, et al. Evidence for non-chemical, non electrical intercellular signaling in intestinal cells. Bioelectrochemistry (2007) 71:142–8. doi: 10.1016/j.bioelechem.2007.03.001
6. Budagovsky A, Budagovska O, Budagovsky I. Biological structure as a converter of coherent radiation. In: Beloussov LV, Voeikov VL, Martynyuk, VS, editors. Biophotonics and Coherent Systems in Biology. Moscow: Springer (2007). p. 47–63.
7. Choucroun N. On the hypothesis of mitogenetic radiation. J Mar Biolo Assoc UK. (1930) 17:65–74. doi: 10.1017/S0025315400051791
8. Gurwitsch A. Die Mitogenetische Strahlung. Monographien aus dem Gesamtgebiet der Physiologie der Pflanzen und der Tiere. Berlin: Springer (1932).
9. Rossi C, Foletti A, Magnani A, Lamponi S. New perspectives in cell communication: bioelectromagneticinteractions. Semin Cancer Biol. (2011) 21:207–14. doi: 10.1016/j.semcancer.2011.04.003
10. Prasad A, Rossi C, Lamponi S, Pospíšil P, Foletti A. New perspective in cell communication: potential role of ultra-weak photon emission. J Photochem Photobiol B Biol. (2014). 139:47–53 doi: 10.1016/j.jphotobiol.2014.03.004
11. Grass F, Klima H, Kasper S. Biophotons, microtubules and CNS, is our brain a “Holographic computer”? Med Hypotheses (2004) 62:169–72. doi: 10.1016/S0306-9877(03)00308-6
12. Fels D. Cellular communication through light. PLoS ONE (2009) 4:e5086. doi: 10.1371/journal.pone.0005086
14. Chaban VV, Cho T, Reid CB, Norris KC. Physically disconnected non-diffusible cell-to-cell communication between neuroblastoma SH-SY5Y and DRG primary sensory neurons. Am J Transl Res. (2013) 5:69–79.
15. Quickenden TI, Que Hee SS. On the existence of mitogenetic radiation. Speculation Sci Technol. (1981) 4:453–64
16. Scholkmann F, Fels D, Cifra M. Non-chemical and non-contact cell-to-cell communication: a short review. Am J Transl Res. (2013) 5:586–93.
17. Cifra M, Fields JZ, Farhadi A. Electromagnetic cellular interactions. Prog Biophys Mol Biol. (2011) 105:223–46. doi: 10.1016/j.pbiomolbio.2010.07.003
18. Cifra M, Pospíšil P. Ultra-weak photon emission from biological samples: definition, mechanisms, properties, detection and applications. J Photochem Photobiol B Biol. (2014) 139:2–10. doi: 10.1016/j.jphotobiol.2014.02.009
19. Kobayashi M, Tadeka M, Sato T, Yamazaki Y, Kaneko K, Ito KI, et al. In vivo imaging of utlraweak photon emission from a rats brain correlated with cerebral energy metabolism and oxidative stress. Neurosci Res. (1999) 34:103–13. doi: 10.1016/S0168-0102(99)00040-1
20. Laager F, Park S-H, Yang J-M, Song W, Soh K-S. Effects of exercises on biophoton emission of the wrist. Eur J Appl Physiol. (2007) 102:463–9. doi: 10.1007/s00421-007-0607-4
21. Vogel R, Süssmuth RE. A model for the generation of low level chemiluminescence from microbiological growth media and its depletion by bacterial cells, Bioelectrochemistry and Bioenergetics (1999) 48:375–82. doi: 10.1016/S0302-4598(99)00006-9
22. Musumeci F, Guiseppe P, Scordino A, Tudisco S, Lo Presti C, Applegate LA, et al. Discrimintation between normal an cancer cells by using sprectral analysis of delayed luminescence. Appl Phys Lett. (2005) 86:153902. doi: 10.1063/1.1900317
23. Ives JA, van Wijk EPA, Bat N, Crawford C, Walter A, Jonas WB, et al. Ultraweak photon emission as a non-invasive health assessment: a systematic review. PLoS ONE (2014) 9:e87401. doi: 10.1371/journal.pone.0087401
24. Quickenden TI. Weak luminescence from the yeast Saccharomyces cerevisiae and the existence of mitogenetic radiation. Biochem Biophys Res Commun. (1974) 60:764–70.
25. Mei WP. Dissertation, Ultraschwache Photoemission bei Synchroniserten Hefezellen in Abhängigkeit vom Zellteilungszyklus. Fachereich Biologie Universität Hannover (1991).
26. Pospíšil P, Prasad A, Rác M. Role of reactive oxygen species in ultra-weak photon emission in biological systems. J Photochem Photobiol B. (2014) 39:11–23. doi: 10.1016/j.jphotobiol.2014.02.008
27. Zhang Y, Song S, Fong C-C, Tsang C-H, Yang Z, Yang M. cDNA microarray analysis of gene expression profiles in human fibroblast cells irradiated with red light. J Invest Dermatol. (2003) 120:849–57. doi: 10.1046/j.1523-1747.2003.12133.x
28. Sun Y, Wang C, Dai J. Biophotons as neural communication signals demonstrated by in situ biophoton autography. Photochem Photobiol Sci. (2010) 9:315–22. doi: 10.1039/b9pp00125e
29. Kucera O, Cifra M. Cell-to-cell signaling through light: just a ghost of chance? Cell Commun. Signal. (2013) 11:87. doi: 10.1186/1478-811X-11-87
30. Fels D. Analogy between quantum and cell relations. Axiomathes (2012) 22:509–20. doi: 10.1007/s10516-011-9156-x
31. Thar R, Kühl M. Propagation of electromagnetic in mitochondria? J Theor Biol. (2004) 230:261–70. doi: 10.1016/j.jtbi.2004.05.021
32. Reguera G. When microbial conversations get physical. Trends Microbiol. (2011) 19:105–13. doi: 10.1016/j.tim.2010.12.007
33. Bókkon I, Salari V. Information storing by biomagnetites. J Biol Phys. (2009) 36:109–20. doi: 10.1007/s10867-009-9173-9
34. Dotta BT, Saroka KS, Persinger MA. Increased photon emission from the head while imagining white light is correlated with changes in electroencephalographic power: support for Bokkon's biophoton hypothesis. Neurosci. Lett. (2012) 513:151–4. doi: 10.1016/j.neulet.2012.02.021
35. Dent L. A new model of intracellular communication based on coherent, high-frequency vibrations in biomolecules. Biol Inf. (2013) 435–49. doi: 10.1142/9789814508728_0019
36. Havelka D, Kucera O, Deriu MA, Cifra M. Electro-acoustic behavior of the mitotic spindle: a semi-classical coarse-grained model. PLoS ONE (2014) 9:e86501. doi: 10.1371/journal.pone.0086501
37. Laager F, Soh K-S. Effects of carbonyl cyanide 3-chlorophenylhydrazone on yeast biophoton emission. J Korean Jugshin Sci (2007) 11, 1–8.
38. Rastogi A, Pospíšil P. Spontaneous ultraweak photon emission imaging of oxidative metabolic processes in human skin: effect of molecular oxygen and antioxidant defense system. J Biomed Opt. (2011) 16. doi: 10.1117/1.3616135
39. Hideg É, Björn LO. Ultraweak light emission, free radicals, chilling and light sensitivity. Physiol. Plant. (2008) 98:223–8. doi: 10.1034/j.1399-3054.1996.980201.x
40. Birtic S, Ksas B, Genty B, Mueller MJ, Triantaphylide C, Havaux M. Using spontaneous photon emission to image lipid oxidation patterns in plant tissues. Plant J. (2011) 67:1103–15. doi: 10.1111/j.1365-313X.2011.04646.x
41. Makino T. Biophoton emission and defense systems in plants. In: Shen X, Wijk RV editors. Biophotonic Optical Science and Engineering for the 21st Century New York, NY: Springer Science (2005). p. 205–218.
42. Gallep CDM. Ultraweak, spontaneous photon emission in seedlings: toxicological and chronobiological applications. Luminescence (2014) 29:963–68. doi: 10.1002/bio.2658
43. Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. (2000) 279:L1005–28.
44. Laager FM, Becker NM, Park S-H, Soh K-S. Effects of Lac operon activation, deletion of the Yhha gene, and the removal of oxygen on the ultra-weak photon emission of Escherichia coli. Electromag Biol Med. (2009) 28:240–9. doi: 10.1080/15368370903065820
45. Slawinski J, Ezzahir A, Godlewski M, Kwiecinska T, Rajfur Z, Sitko D, et al. Stress-induced photon emission from perturbed organisms. Experiementia (1992) 48:1041–58. doi: 10.1007/BF01947992
46. Beer, A. Bestimmung der Absorption des rothen Lichts in farbigen Flüssigkeiten (Determination of the absorption of red light in colored liquids). Ann Phys Chem. (1852) 86:78–88.
47. Brown JQ, Vishwanath K, Palmer GM, Ramanujam N. Advances in quantitative UV-visible spectroscopy for clinical and pre-clinical application in cancer. Curr Opin Biotechnol. (2009) 20:119–31. doi: 10.1016/j.copbio.2009.02.004
48. Nonoyama A. Using Multiwavelength UV-Visible Spectroscopy for the Characterization of Red Blood Cells: An Investigation of Hypochromism. Dissertation, College of Arts and Sciences University of South Florida (2004).
49. Han Y, Braatsch S, Osterloh L, Klug G. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci USA. (2004) 101:12306–311. doi: 10.1073/pnas.0403547101
50. Schönenbrücher H, Adhikary R, Mukherjee P, Casey TA, Rasmussen MA, Maistrovich FD, et al. Fluorescence-based method, exploiting lipofuscin, for real-time detection of central nervous system tissues on bovine carcasses. J Agric Food Chem. (2008) 56:6220–6. doi: 10.1021/jf0734368
51. Zipfel WR, Williams RM, Christie R, Nikitin Y, Hyman BT, Webb WW. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci USA. (2003) 100:7075–80. doi: 10.1073/pnas.0832308100
52. Fritzsche M, Mandenius C-F. Fluorescent cell-based sensing approaches for toxicity testing. Anal Bioanal Chem. (2010) 398:181–91. doi: 10.1007/s00216-010-3651-6
53. Gauden M, Grinstead JS, Laan W, van Stokkum IHM, Avila-Perez M, Toh KC, et al. On the role of aromatic side chain in the photoactivation of BLUF domains. Biochemistry (2007) 46:7405–15. doi: 10.1021/bi7006433
54. Gallas JM, Eisner M. Fluorescne of melanin-dependence upon excitation wavelength and concentration. Photochem Photobiol. (1987) 45:595–600. doi: 10.1111/j.1751-1097.1987.tb07385.x
55. Kennis JTM, Mathes T. Molecular eyes: proteins that transform light into biological information. Interface Focus (2013) 3:20130005. doi: 10.1098/rsfs.2013.0005
56. Van der Horst M, Key J, Hellingwerf KJ. Photosensing in chemotrophic, non phototrophic bacteria, let there be light sensing too. Trends Microbiol. (2007) 15:554–62. doi: 10.1016/j.tim.2007.09.009
57. Grinstead JS, Avila-Perez M, Hellingwerf KJ, Boelens R, Kaptein R. Light-induced Flipping of a conserved Glutamine Sidechain and its orientation in AppA BLUF Domain. J Am Chem Soc. (2006) 128:15066–7. doi: 10.1021/ja0660103
58. Kritsky MS, Belozerskaya TA, Sokolovsky VY, Filippovich SY. Photoreceptor apparatus of Fungus Neurospora crassa. Mol Biol. (2005) 39:514–28. doi: 10.1007/s11008-005-0068-y
59. Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science (2011) 331:1409–13. doi: 10.1126/science.1199702
60. Hellingwerf KJ. The molecualr basis of sensing and responding to light in microorganisms. Antonie van Leewenhoek (2002) 81:51–9. doi: 10.1023/A:1020521424582
61. Karu T, Pyatibrat LV, Afanasyeva NI. A novel mitochondrial Signaling pathway activated by visible to near Infrared radiation. Photochem Photobiol. (2004) 80:366–72. doi: 10.1562/2004-03-25-RA-123.1
62. Laan W, Hellingwerf KJ. (2004). Structural charaterisation of flavin based photoreceptors. In: ESF LESC Exploratory Workshop. Parma (2004). p. 8–9
63. Liscum E, Hodgson DW, Campbell J. Blue light signaling through the crytochromes and phototropins. So thats what the blues is all about. Plant Physiol. (2003) 133:1429–36. doi: 10.1104/pp.103.030601
64. Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, Wu W, et al. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. (2008) 6:e160. doi: 10.1371/journal.pbio.0060160
65. Kochevar IE. Single oxygen signaling: from intimate to Global. Sci STKE (2004) 7:7. doi: 10.1126/stke.2212004pe7
66. Fancy DA, Kodadek T. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross linking triggered by long wavelength light. Proc Natl Acad Sci USA. (1999) 96:6020–4. doi: 10.1073/pnas.96.11.6020
67. Kyndt JA, Fitch JC, Meyer TE, Cusanovich MA. Thermochromatium tepidum photoactive yellow protein/bacteriophytochrome/diguanylate cyclase: characterization of the PYP domain. Biochemistry (2005) 44:4755–64. doi: 10.1021/bi047373n
68. Hayashi S, Tajkhorshid E, Schulten K. Molecular Dynamics Simulation of Bacteriorhodopsin's Photoisomerization Using Ab Initio Forces for the Excited Chromophore. Biophys J. (2003) 85:1440–9. doi: 10.1016/S0006-3495(03)74576-7
69. Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Plant Biol. (1999) 96:8779–83. doi: 10.1073/pnas.96.15.8779
70. Song SH, Dick B, Penzkofer A, Pokorny R, Batschauer A, Essen LO. Absorption and fluorescence spectroscopic characterization of cryptochrome 3 from Arabidopsis thaliana. J Photochem Photobiol B. (2006) 85:1–16. doi: 10.1109/OCEANS.2006.306784
71. Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell (2012) 24:2992–3008. doi: 10.1105/tpc.112.098947
72. Padovani D, Folly-Klan M, Labarde A, Boulakirba S, Campanacci V, Franco M, et al. EFA6 controls Arf1 and Arf6 activation through a negative feedback loop. Proc Natl Acad Sci USA. (2014) 111:12378–83. doi: 10.1073/pnas.1409832111
73. Casal JJ. Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Phtochem. Photobiol. (2000) 71:1–11. doi: 10.1562/0031-8655(2000)0710001PCPPII2.0.CO2
Keywords: ultra-weak photon emission (UPE), light based cellular interactions, absorption, non-chemical interactions, singlet oxygen
Citation: Laager F (2015) Light based cellular interactions: hypotheses and perspectives. Front. Phys. 3:55. doi: 10.3389/fphy.2015.00055
Received: 29 September 2014; Accepted: 15 July 2015;
Published: 07 August 2015.
Edited by:
Dieter W. Heermann, Heidelberg University, GermanyReviewed by:
Anna Carbone, Politecnico di Torino, ItalyCopyright © 2015 Laager. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frédéric Laager, SUPER Lab, Marie-Curie-Straße 1, 26129 Oldenburg, GermanyZi5sYWFnZXJAc3VwZXItbGFiLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.