
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 13 February 2025
Sec. Predictive Toxicology
Volume 16 - 2025 | https://doi.org/10.3389/fphar.2025.1489210
This article is part of the Research Topic Advances and Applications of Predictive Toxicology in Knowledge Discovery, Risk Assessment, and Drug Development View all 11 articles
 Qingshan Tang1†
Qingshan Tang1† Jiachen Dong1†
Jiachen Dong1† Feng Zhang1
Feng Zhang1 Dan Zhao1
Dan Zhao1 Qi Yang1
Qi Yang1 Jiayu Wen2
Jiayu Wen2 Yuhao Sun2
Yuhao Sun2 Jifu Wei2*†
Jifu Wei2*† Zhixian Liu2*†
Zhixian Liu2*†Background: The tyrosine receptor kinase inhibitor (TRKi) entrectinib is used to treat neurotrophic tyrosine receptor kinase (NTRK) fusion-positive solid tumors and ROS1-positive patients. Despite its impressive efficacy against cancer, the clinical application is still limited by the central nervous system (CNS)-related toxicities. However, the precise mechanism of such CNS-related toxicities remains elusive.
Methods: The effect of entrectinib-induced nerve cell damage was evaluated by the nerve cells (PC12, HT22 and SK-N-SH) based in vitro models. Various assays, including CCK-8, colony formation and EdU incorporation assays were utilized to estimate the cellular viability and proliferation ability. Cell apoptosis was measured by flow cytometry. Next, transcriptome sequencing technology was performed to identify differentially expressed genes (DEGs). Gene ontology (GO), kyoto encyclopedia of genes and genomes (KEGG) analysis and gene set enrichment analysis (GSEA) were applied to predict the potential functions of DEGs. Quantitative real time polymerase chain reaction (qRT-PCR) and Western blotting assays were performed to measure the expressions of thrombospondin-1 (THBS1), TGF-β1, PI3K, AKT and phosphorylated AKT (p-AKT) in the entrectinib-treated nerve cells. Additionally, we Preliminary observed and validated whether THBS1 overexpression could rescue nerve cell damage and the abnormalities in PI3K-AKT and TGF-β signaling pathways.
Results: Entrectinib significantly inhibited the nerve cells proliferation and colony formation, and induced nerve cells apoptosis. Transcriptome sequencing analysis and qRT-PCR revealed that THBS1 was downregulated within entrectinib treatment. KEGG and GSEA analysis also suggested that entrectinib directly caused the abnormalities in proliferation-related signaling pathway like PI3K-AKT pathway, and apoptosis-related signaling pathway including TGF-β pathway. We further demonstrated that THBS1, TGF-β1, PI3K, AKT and p-AKT were downregulated by entrectinib. Meanwhile, pretreatment with THBS1 overexpression plasmids significantly rescued nerve cells (PC12, HT22 and SK-N-SH) from cell death and the abnormalities in PI3K-AKT and TGF-β signaling pathways.
Conclusion: These results identified a critical role of entrectinib in promoting nerve cell damage by downregulating the expression of THBS1 while also inhibiting PI3K-AKT and TGF-β signaling pathways. Our findings will provide potential therapeutic targets for CNS-related toxicities.
In recent years, cancer has become the leading cause of mortality worldwide. The increasing burden of cancer has become a challenging major public health problem (Tu et al., 2016; Jassim et al., 2023). Conventionally, there are three therapeutic modalities of cancer treatment such as surgical treatment, radiotherapy, and chemical drug therapy (Zheng et al., 2018; Yang et al., 2022). Nevertheless, the systemic toxicity associated with above therapies pose a significant challenge to patient tolerance and compliance (Mun et al., 2018). With the development and application of genetic testing in clinical treatment, molecular-targeting therapy has drawn widespread concern, bringing further options for clinical application (Bhamidipati and Subbiah, 2023; Li et al., 2023). Currently, targeted therapy has been an indispensable alternation to manage the disease for numerous cancers with oncogene addiction (Yuan et al., 2019; Benitez et al., 2021).
Entrectinib is an orally active small-molecule, which targets tyrosine kinase inhibitor of neurotrophic tyrosine receptor kinase (NTRK), ROS1 and anaplastic lymphoma kinase (ALK) genes. As the first-generation tyrosine receptor kinase inhibitor (TRKi), it is approved for the treatment of patients with NTRK fusion-positive solid tumors and adults with ROS1 fusion-positive non-small cell lung cancer (Frampton, 2021; Desai et al., 2022). Despite its impressive efficacy against cancer, the serious toxicities on the central nervous system (CNS) have also been popularly concerned by the public during the course of cancer therapy (Marcus et al., 2021). For example, the overall incidence of fatigue was 45%, taste disturbance was 42.3%, dysesthesia was 29.0%, cognitive impairment was 24.2% and peripheral sensory neuropathies was 18%. These symptoms not only significantly reduce the patient’s quality of life but also pose substantial challenges in the management of treatment, and even result in treatment interruptions (Trendowski et al., 2019; Doebele et al., 2020; Frampton, 2021; Martineau et al., 2022). Nowadays, these clinical CNS-related toxicities were only addressed through dose modification and interruptions, even withdrawal of entrectinib (Marcus et al., 2021). Unfortunately, there has been no effective protocols available for relieving or treating these symptoms until now. Thus, it is crucial to illuminate the molecular mechanism underlying CNS-related toxicities.
Research indicated that the tropomyosin receptor kinases (TRK) protein, a member of the tyrosine kinase family, plays a critical role in modulating neuronal activity and axonal growth in both the central and peripheral nervous systems (Aydin-Abidin and Abidin, 2019; Jiang et al., 2021). Acting as a tyrosine receptor kinase inhibitor (TRKi), entrectinib can inhibit TRK, thus leading to on-target neurotoxicity (Giustini et al., 2022). Some chemotherapy drugs can lead to chemotherapy-induced peripheral neuropathy which is involved in nerve damage and axon loss (Fumagalli et al., 2020; Chen et al., 2024). Several factors, such as mitochondrial dysfunction, oxidative stress, microcirculation disturbance and neuroinflammation have been proposed as determinants of neurotoxicity (Staff et al., 2017; Wang et al., 2023; Chen et al., 2024). Evidence indicated that isoflurane could induce hippocampal cells apoptosis by inhibiting PI3K-AKT expression (Wang et al., 2016). However, due to the complexity of CNS-related toxicities, the mechanisms underlying entrectinib-induced neurotoxicity are not fully understood.
In this study, we aim to evaluate whether entrectinib could induce nerve cell damage in vitro model. The potential mechanism responsible for entrectinib-induced nerve cell damage will be investigated as well. Furthermore, this finding will offer important insights and potential therapeutic targets for the entrectinib-induced nerve cell damage.
The rat adrenal pheochromocytoma cells PC12 (FH0415), mouse hippocampal neuron cells HT22 (FH1027) and human neuroblastoma cells SK-N-SH (FH0164) were purchased from Shanghai Fuheng Biotechnology Co., Ltd. (Shanghai, China). PC12 cells were cultured in RPMI-1640 medium (KGM31800, KeyGEN, Nanjing, China) supplemented with 10% fetal bovine serum (FBS, A6901FBS, Invigentech, United States) and 1% penicillin and streptomycin (C0222, Beyotime, Shanghai, China). HT22 and SK-N-SH cells were cultured in DMEM medium (KGM12800, KeyGEN, Nanjing, China) with 10% FBS and 1% penicillin and streptomycin. All cells were incubated in a 37°C with 5% CO2 atmosphere.
Entrectinib (HY-12678) was obtained from MedChemExpress (MCE, Shanghai, China) with the purity of 99.87%. THBS1 overexpression plasmids were synthesized by Corues Biotchnology (Nanjing, China). The relevant materials used in this study were provided as following: EnoGeneCell Counting Kit-8 (CCK-8, E1CK-000208, EnoGene, Nanjing, China), BeyoClick™ EdU-555 cell proliferation detection kit (C0075S, Beyotime, Shanghai, China), Annexin V-FITC/PI apoptosis detection kit (KGA107, KeyGEN, Nanjing, China), 4% paraformaldehyde (PFA, BL539A, Biosharp, Hefei, China), crystal violet (C0121, Beyotime, Shanghai, China), Trizol (R0016, Beyotime, Shanghai, China), Freezol reagent (R711-01, Vazyme, Nanjing, China), Evo M-MLV RT mix kit (AG11728, Accurate, Changsha, China), AceQ Universal SYBR qPCR master mix (Q511-02, Vazyme, Nanjing, China), BCA protein assay kit (P0010, Beyotime, Shanghai, China), RIPA (P00138, Beyotime, Shanghai, China), ECL detection reagent (180-501, Tanon, Shanghai, China), lipofectamine 3000 transfection reagent (L3000015, Invitrogen, United States).
The antibodies used in Western blotting assay were obtained as following: THBS1 (1:1,000, 18304-1-AP, Proteintech, Wuhan, China), PI3K (1:600, 20584-1-AP, Proteintech, Wuhan, China), AKT (1:5,000, 60203-2-AP, Proteintech, Wuhan, China), p-AKT (1:2,000, 4060T, Cell signaling technology, United States), TGF-β (1:3,000, 21898-1-AP, Proteintech, Wuhan, China), GAPDH (1:60,000, 60004-1-Ig, Proteintech, Wuhan, China), HRP-conjugated affinipure goat anti-rabbit IgG (1:5,000, SA00001-2, Proteintech, Wuhan, China) and HRP-conjugated affinipure goat anti-mouse IgG (1:4,000, SA00001-1, Proteintech, Wuhan, China).
To establish entrectinib activated cells, nerve cells (PC12, HT22 and SK-N-SH) were treated with entrectinib for 48 h at the concentrations of 0, 0.5, 1, 2, 5, 10, and 20 μmol/L, respectively. To explore whether entrectinib influences the cellular proliferation ability and apoptosis, we employed entrectinib (2.3, 4.2 and 4.3 μmol/L, respectively) to the PC12, HT22 and SK-N-SH cells for 48 h. To observe the effect of THBS1 on entrectinib activated cells, nerve cells were divided into five groups: the normal control (NC), the overexpression (OE) group transfected with THBS1 overexpression plasmids, the entrectinib group exposed to entrectinib (2.3, 4.2 and 4.3 μmol/L, respectively), the NC + entrectinib group and the THBS1 overexpression group cultured in medium containing entrectinib (2.3, 4.2 and 4.3 μmol/L, respectively) for 48 h.
PC12, HT22 and SK-N-SH cells were plated in 96-well plates at a density of 7 × 103 cells per well and incubated for 24 h. Subsequently, PC12, HT22 and SK-N-SH cells were treated with entrectinib and THBS1 overexpression plasmids for 48 h described above. After incubation, 10 μL CCK-8 solution was added into each well and incubated at 37°C for 2 h in the dark. Then, a microplate reader (Molecular Devices, Shanghai, China) was used to measure the absorbance at a wavelength of 450 nm.
PC12, HT22 and SK-N-SH cells were seeded into 24-well plates at a density of 3 × 104 cells per well and treated with entrectinib for 48 h. BeyoClick™ EdU-555 cell proliferation detection kit was used for the subsequent experiments. In brief, 20 μmol/L EdU was added into per well for 4 h. Hoechst 33342 was used to stain the nuclei for 10 min in the dark. Next, images were randomly captured by a fluorescence inverted microscope (Zeiss, Germany). Finally, ImageJ was utilized to count the total cell and proliferating cell numbers.
In six-well plates, PC12, HT22, SK-N-SH cells were cultured at a density of about 7 × 102 cells per well and then treated with entrectinib. The cultures were maintained for 14 days or until the number of single-cell clones exceeded 50. After rinsing with phosphate buffered saline (PBS), cells were fixed with 4% paraformaldehyde solution for 30 min. Next, 0.5% crystal violet solution was stained for 15 min. Finally, cells were washed several times with PBS and captured by a camera.
PC12, HT22 and SK-N-SH cells were seeded into 6-well plates and treated with entrectinib and THBS1 overexpression plasmids for 48 h described above. Then, the treated cells were collected and stained with Annexin V-FITC and PI staining solution for 10 min. Following the recommendations of the manufacturer, the cells were analyzed by flow cytometry (BD Biosciences, United States) within 1 h. Data were analyzed by Flowjo software (v.10.8.1).
PC12, HT22 and SK-N-SH cells were treated with entrectinib, respectively. After 48 h incubation, the total RNA was extracted by Trizol reagent and sequenced in Beijing Biomarker Technology Co., LTD. (Beijing, China). Briefly, according to the manufacturer’s instructions, the sequencing library was generated by Hieff NGS ultima dual-mode mRNA library prep kit (Yeasen, Shanghai, China). The libraries were quantified preliminarily with Qubit 3.0 fluorescence quantitative analyzer and sequenced on an Illumina novaseq platform to generate 150 bp paired-end reads. Clean data were obtained by removing reads containing adapter, ploy-N and low-quality from raw data. The analysis of differentially expressed genes (DEGs) between PC12 and HT22 cells was performed by DESeq2 software (Love et al., 2014), while SK-N-SH cells were analyzed by edgeR software (Robinson et al., 2010). Genes with a statistical threshold of false discovery rate (FDR) < 0.05 and |fold change| > 2 were considered to be significantly differentially expressed. Volcano plots, a visualized plots to illustrate DEGs, were generated by bioinformatics software available at https://www.bioinformatics.com.cn/.
Functional enrichment analyses of GO (Ashburner et al., 2000) and KEGG (Kanehisa et al., 2014) in DEGs from PC12 and HT22 cells were performed by the R package ClusterProfiler (v.4.6.2) (Yu et al., 2012). The R package ggplot2 (v.3.4.1) was utilized to visualize and screen biological functions and pathways associated with these DEGs. Enriched GO terms and KEGG pathways were presented to illustrate the analytical outcomes.
GSEA software (v.4.3.2) (Subramanian et al., 2005) was performed to investigate functional enrichment pathways and landmark gene sets in gene expression datasets. Signature gene sets were extracted from the Molecular Signatures Database (MSigDB, available at https://www.gsea-msigdb.org). Enrichment results were considered significant if the normalized enrichment score (NES) > 1 and the nominal P < 0.05 in the entrectinib treatment.
According to the manufacturer’s recommendations, total cellular RNA was extracted by Freezol reagent and reverse-transcribed into cDNA by the Evo M-MLV RT mix kit. The AceQ universal SYBR qPCR master mix kit was used for quantitative real-time PCR. The β-actin was served as the internal control. The expression of the target gene was calculated by the 2−ΔΔCT method. Primers used in this study were synthesized by Invitrogen Biotechnology Co., Ltd. (Invitrogen, United States), and their sequences were provided in Supplementary Table S1.
Total protein from cells was extracted through RIPA lysate with a mixture of phosphatase inhibitor and protease inhibitor. The concentration of extracted proteins was quantified through BCA protein assay kit. The protein samples were separated through 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. After blocking with 5% skim milk for 2 h, the PVDF membranes were incubated with the primary antibody overnight at 4°C. Subsequently, membranes were incubated with the corresponding secondary antibody and visualized by an enhanced chemiluminescence (ECL) reagent. Protein band intensities were analyzed by ImageJ software.
All experiments were conducted at least in triplicate to ensure reliability. Statistical analysis and graphical representation were carried out by GraphPad Prism (v.9.0). Differences in quantitative variables between two groups were assessed by the t-test, while differences among three or more groups were evaluated by one-way analysis of variance (ANOVA). P < 0.05 was considered as statistical significance.
Initially, the nerve cells (PC12, HT22 and SK-N-SH) were subjected to various concentrations of entrectinib (0, 0.5, 1, 2, 5, 10, and 20 μmol/L) for 48 h, and the impact of entrectinib on nerve cells viability was assessed by the CCK-8 assay. The results indicated that entrectinib significantly inhibited nerve cells viability compared with the control group, displaying a clear dose-dependent reduction trend (Figures 1A, C, E). Subsequently, the IC50 of entrectinib in PC12, HT22 and SK-N-SH cells was calculated as 2.3 μmol/L, 4.2 μmol/L, and 4.3 μmol/L, respectively (Figures 1B, D, F). Thus, the IC50 entrectinib incubation was selected for subsequent experiments. Collectively, these results preliminarily indicated that entrectinib showed an inhibitory effect on the nerve cells viability.
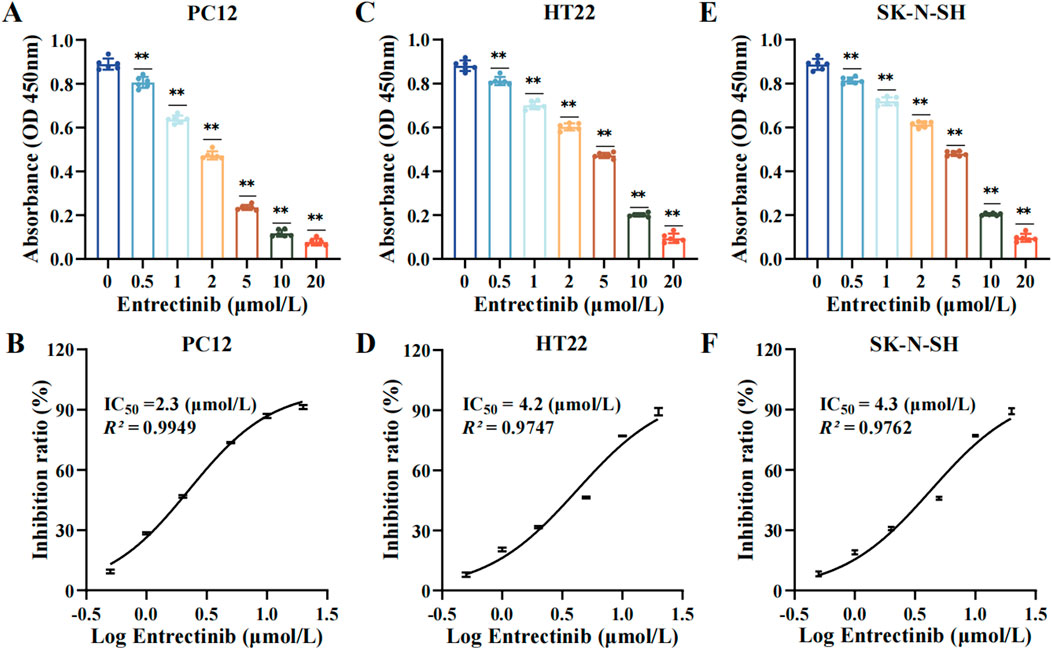
Figure 1. Entrectinib significantly inhibited nerve cells viability. (A, C, E) The absorbance of PC12, HT22 and SK-N-SH cells was subjected to the CCK-8 assay after treatment with entrectinib (0, 0.5, 1, 2, 5, 10, and 20 μmol/L). Data are shown as mean ± SD, n = 6 (**P < 0.01). (B, D, F) The IC50 value of PC12, HT22 and SK-N-SH cells was calculated by GraphPad Prism (v.9.0). Data are shown as mean ± SD, n = 6.
To further investigate entrectinib’s impact on nerve cell proliferation, we performed colony formation and EdU incorporation assays. As shown in Figures 2A–C, the relative colony number of nerve cells (PC12, HT22, SK-N-SH) was significantly decreased to 49.65%, 54.34%, and 50.21% after entrectinib treatment. In addition, EdU incorporation assay demonstrated that the red fluorescence had a notable reduction in the treatment of entrectinib, indicating that entrectinib could inhibit the replicative capacity of nerve cells (Figures 2D–F). These findings confirmed that entrectinib significantly inhibited the proliferation ability of nerve cells.
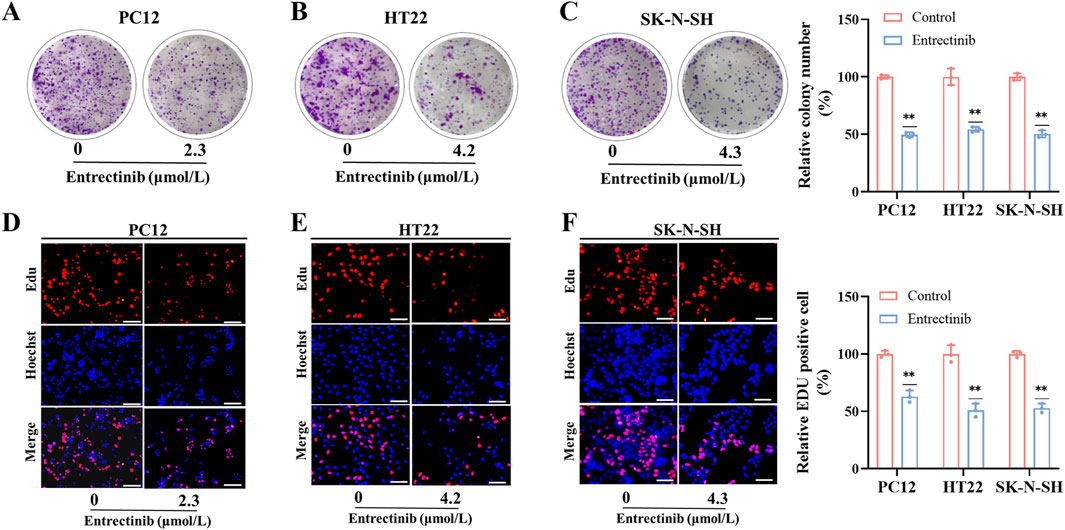
Figure 2. Entrectinib inhibited nerve cells proliferation. (A–C) PC12, HT22 and SK-N-SH cells treated with entrectinib (2.3, 4.2, and 4.3 μmol/L, respectively) were seeded into 6-plate wells, and the number of colonies was counted on the 14 days. The results are also shown in the bar chart. Data are shown as mean ± SD, n = 3 (**P < 0.01). (D–F) EdU incorporation assay was performed to determine the nerve cells proliferation ability of PC12, HT22 and SK-N-SH cells treated with entrectinib (2.3, 4.2, and 4.3 μmol/L, respectively), and the results are also shown in the bar chart. Scale bar = 40 μm. Data are shown as mean ± SD, n = 3 (**P < 0.01).
Moreover, Annexin V-FITC/PI apoptosis detection kit was employed to further explore the impact of entrectinib on nerve cells apoptosis. PC12, HT22 and SK-N-SH cells were treated with entrectinib of 2.3 μmol/L, 4.2 μmol/L and 4.3 μmol/L, respectively, and the apoptosis rate was significantly increased to 12.52%, 14.83% and 15.84% (Figures 3A–D). Taken together, these results indicated that entrectinib could significantly inhibit the viability and induce apoptosis within the nerve cells.
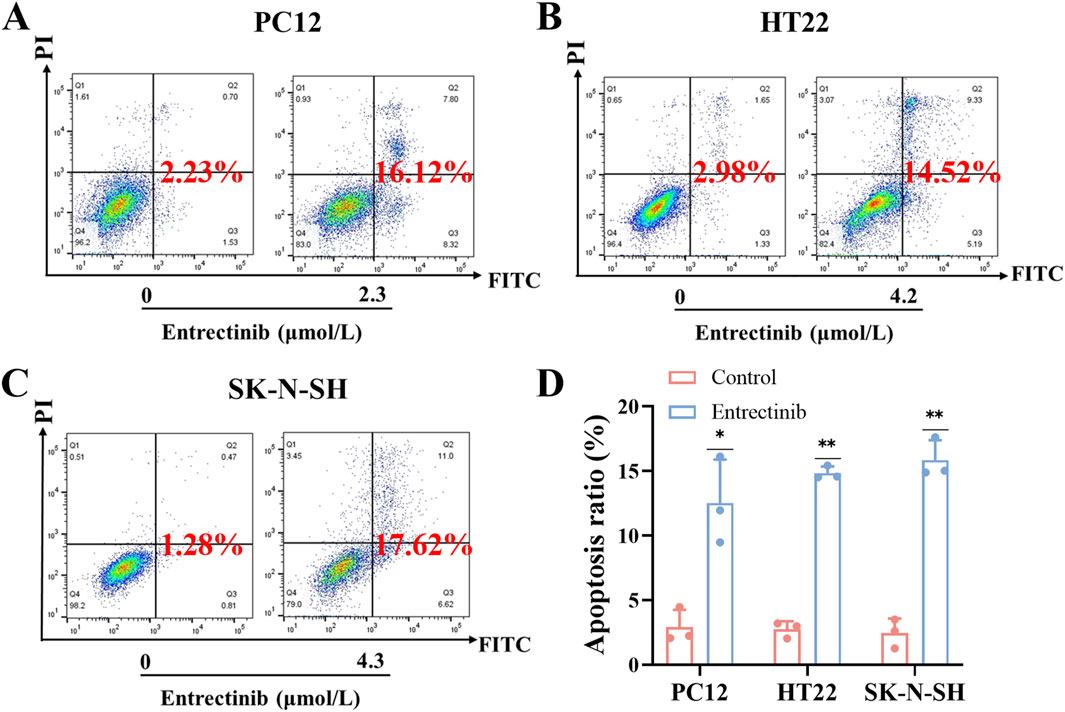
Figure 3. Entrectinib induced nerve cells apoptosis. (A–C) The effect of entrectinib on nerve cells (PC12, HT22 and SK-N-SH) apoptosis was measured by flow cytometry analysis. (D) Quantitative mean apoptosis ratio of (A–C). Data are shown as mean ± SD, n = 3 (**P < 0.01).
To further explore the impact of entrectinib on nerve cells transcription levels, we employed high-throughput sequencing technology on the entrectinib-treated nerve cells (PC12, HT22 and SK-N-SH) samples. Differential expression analysis revealed that 413, 338, and 481 DEGs were in PC12, HT22 and SK-N-SH cells, respectively (Figures 4A–C; Supplementary Table S2). We then determined the intersection of DEGs present in the nerve cells, where 4 DEGs (FTL1, THBS1, FTL1-PS1, COL3A1) were intersected (Figure 4D). Notably, thrombospondin-1 (THBS1) was significantly downregulated in entrectinib treatment, implying that it might be involved in nerve cell damage (Figure 4E). These findings confirmed that entrectinib could induce extensive transcriptomic changes, and THBS1 was the most significant.
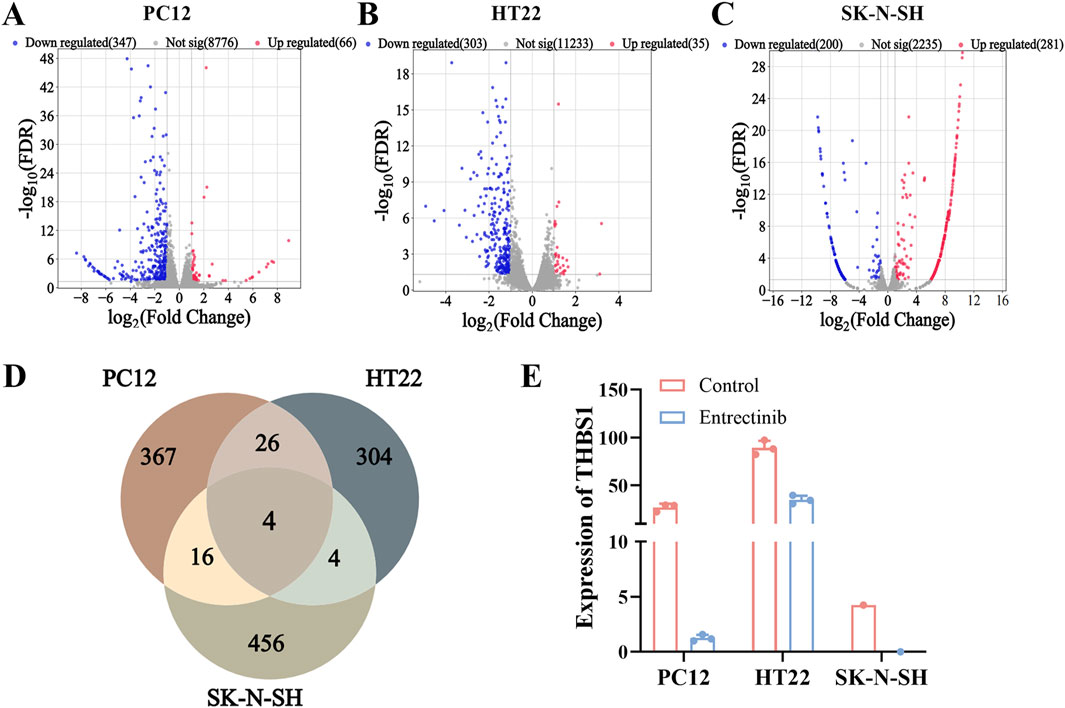
Figure 4. Transcriptome sequencing analyses identified THBS1 target involved in nerve cell damage. (A–C) The DEGs of PC12, HT22 and SK-N-SH cells were presented in the form of volcano plot, where red represents upregulated genes, blue represents downregulated genes, and gray represents genes with insignificant differences (Set threshold FDR < 0.05 and |fold change| > 2). (D) Venn diagram was performed to illustrate overlap between DEGs by PC12, HT22 and SK-N-SH cells. (E) The expression of THBS1 was identified by transcriptome sequencing analyses.
Next, we conducted GO and KEGG pathway enrichment analyses to uncover the biological functions and pathways associated with DEGs. GO results revealed that DEGs were involved in biological processes including peptide cross-linking and epithelial cell proliferation regulation. Meanwhile, it also showed the relationship with cellular components and molecular functions, such as extracellular matrix, transcription repressor complex, growth factor activity and extracellular matrix binding (Figure 5A; Supplementary Table S3). Futhermore, KEGG analysis implicated that DEGs were enriched in pathways such as PI3K-AKT signaling pathway, TGF-β signaling pathway and p53 signaling pathway, et al. (Figure 5B; Supplementary Table S4). Similarly, GSEA was also performed across hallmark gene sets to identify potential signatures of response. The results showed that PI3K-AKT-mTOR signaling pathway, TGF-β signaling pathway and p53 signaling pathway (Figure 5C; Supplementary Table S5) were enriched in entrectinib treatment. The above results suggested that the entrectinib-induced nerve cell damage may be related to the PI3K-AKT-mTOR signaling pathway, p53 signaling pathway and TGF-β signaling pathway.
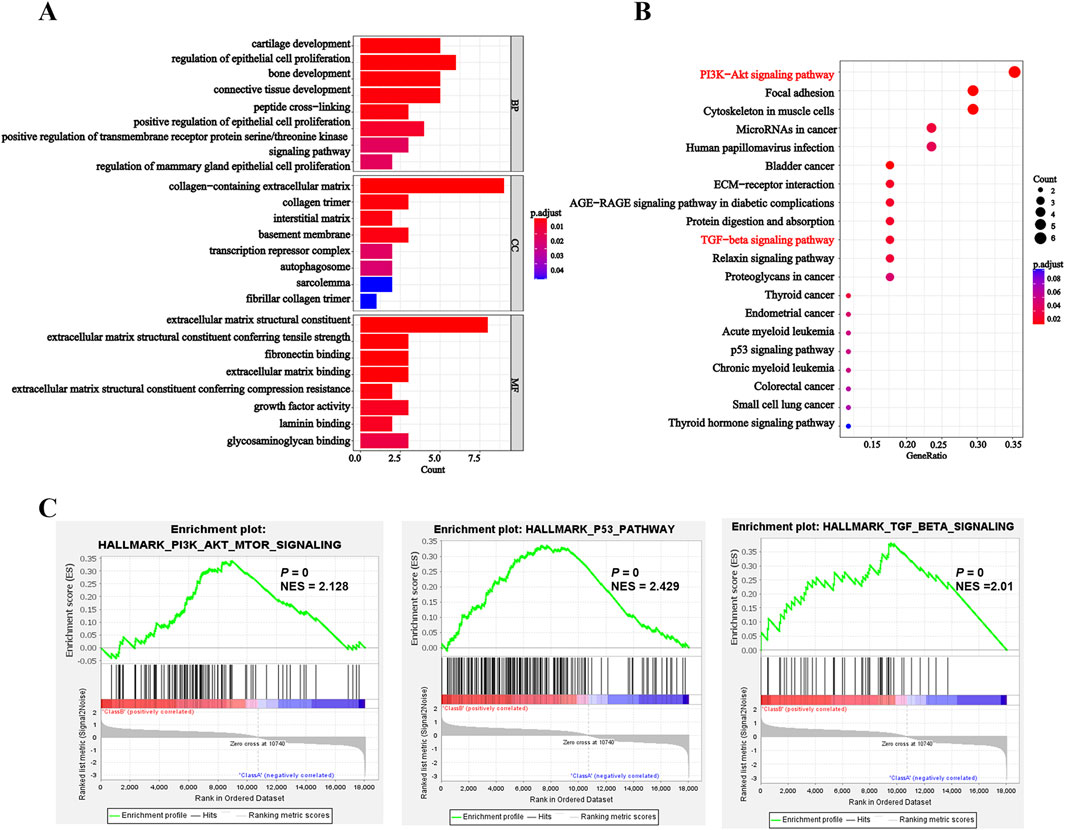
Figure 5. Entrectinib induced the nerve cell damage by affecting the PI3K-AKT-mTOR signaling pathway, p53 signaling pathway and TGF-β signaling pathway. (A, B) The R package ClusterProfiler (v.4.6.2) was used for clustering analysis of potential targets and pathways of entrectinib-induced nerve cell damage. Histogram and bubble plots showed the results of GO and KEGG enrichment analysis. (C) GSEA of hallmark gene sets between control groups and entrectinib-treated groups.
To validate the transcriptome sequencing analysis results, we further performed qRT-PCR and Western blotting assays. Compared with the control group, the expression of THBS1 was significantly decreased after entrectinib treatment within the nerve cells (Figures 6A, B), which was in parallel with sequencing results. Additionally, the levels of relative proteins involved in PI3K-AKT and TGF-β signaling pathways were examined as well. Obviously, entrectinib could downregulate the expressions of PI3K, AKT, phosphorylated AKT (p-AKT) and TGF-β1 proteins within the nerve cells (PC12, HT22 and SK-N-SH) (Figures 6B–C). Therefore, these results demonstrated that entrectinib could downregulate THBS1 expression while also inhibiting PI3K-AKT and TGF-β signaling pathways.
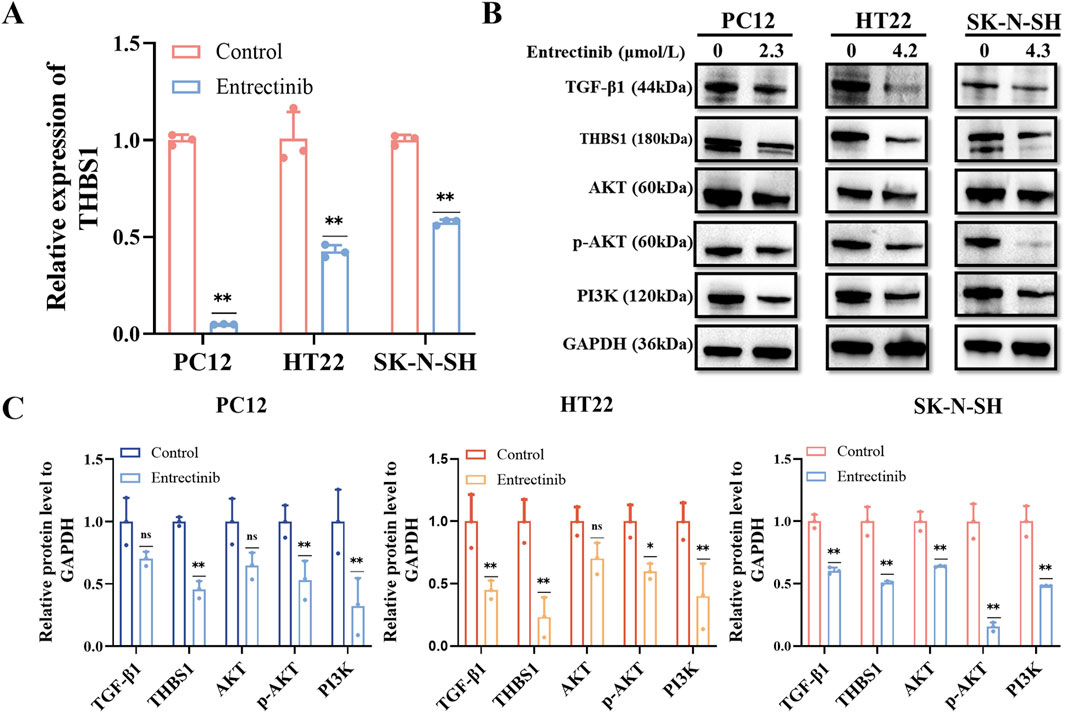
Figure 6. Entrectinib downregulated THBS1 expression while also inhibiting PI3K-AKT and TGF-β signaling pathways. (A) The expression of THBS1 in PC12, HT22 and SK-N-SH cells treated with entrectinib was detected by qRT-PCR. Data are shown as mean ± SD, n = 3 (**P < 0.01). (B) The expressions of PI3K, AKT, p-AKT, TGF-β and THBS1 proteins in PC12, HT22, SK-N-SH cells treated with entrectinib were detected by Western blotting assay. (C) The quantitative analyses of TGF-β1, THBS1, AKT, p-AKT and PI3K proteins in (B) are shown. Data are shown as mean ± SD, n = 3 (*P < 0.05, **P < 0.01).
To preliminary observe and validate whether THBS1 overexpression could rescue nerve cell damage and the abnormalities in PI3K-AKT and TGF-β signaling pathways, nerve cells were transfected with THBS1 overexpression plasmids. As shown in Figures 7A–E, THBS1 overexpression can rescue nerve cell damage induced by entrectinib. Additionally, the expressions of PI3K, AKT, phosphorylated AKT (p-AKT) and TGF-β1 proteins were also upregulated by THBS1 overexpression in the entrectinib-treated cells (Figures 7F–H). These above results suggested that THBS1 plays an important functional role in rescuing nerve cell damage and the abnormalities in the PI3K-AKT and TGF-β signaling pathways induced by entrectinib.
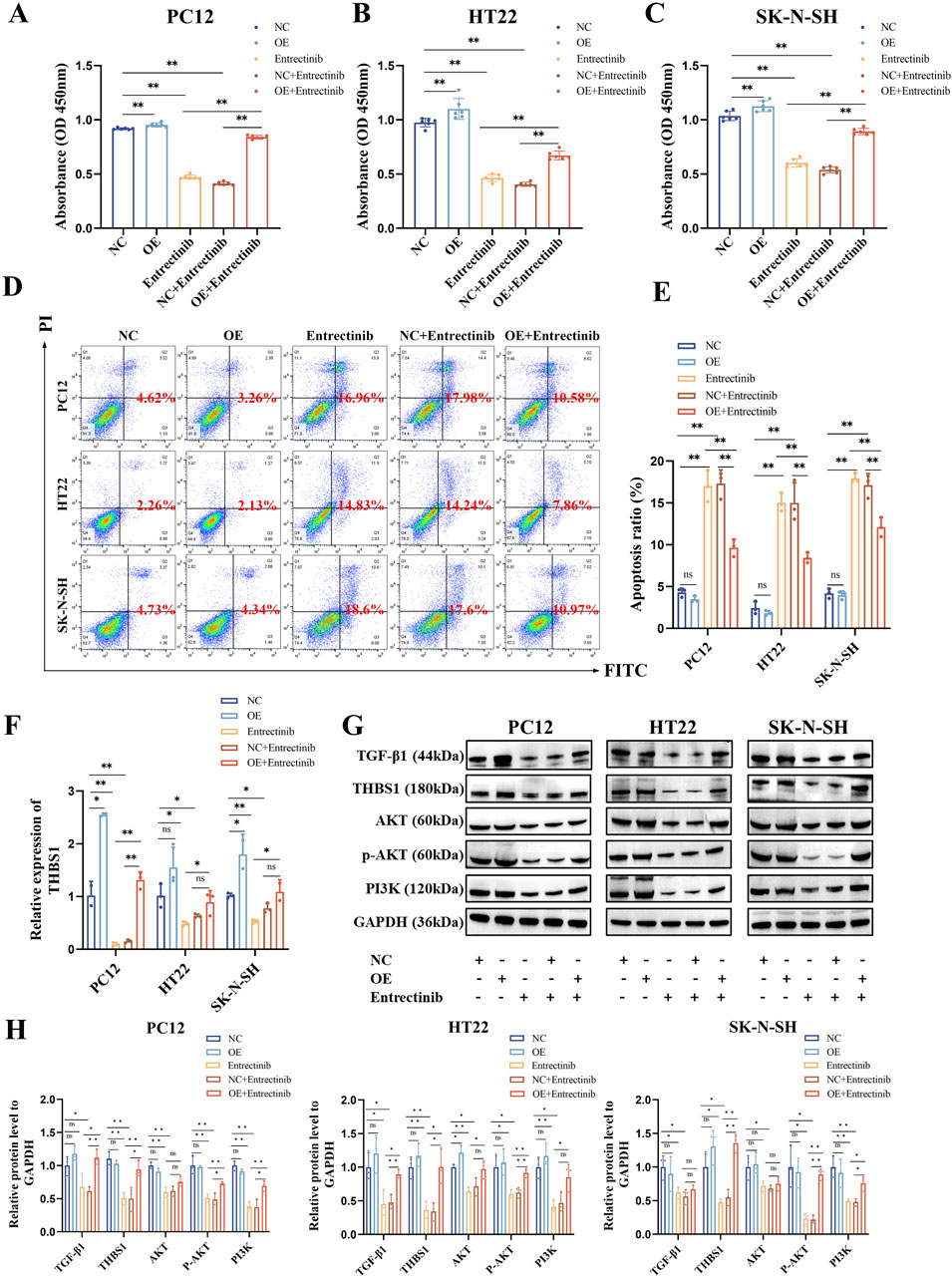
Figure 7. THBS1 overexpression rescue nerve cell damage and the abnormalities in the PI3K-AKT and TGF-β signaling pathways induced by entrectinib. (A–C) Overexpression of THBS1 in entrectinib-treated cells rescued cell viability. Data are shown as mean ± SD, n = 6 (**P < 0.01). (D, E) Overexpression of THBS1 in entrectinib-treated cells rescued cell apoptosis. Data are shown as mean ± SD, n = 3 (**P < 0.01). (F) qRT-PCR measured the efficiency of THBS1 overexpression. Data are shown as mean ± SD, n = 3 (*P < 0.05, **P < 0.01). (G) Western blotting assay examined the levels of PI3K, AKT, p-AKT, TGF-β and THBS1 proteins in PC12, HT22, SK-N-SH cells after treatement with entrectinib or THBS1 overexpression plasmids or the combination for 48 h. (H) Representative quantification were shown. Data are shown as mean ± SD, n = 3 (*P < 0.05, **P < 0.01).
Entrectinib, a novel multi-target TRKi, has demonstrated considerable therapeutic efficacy in tumors harboring NTRK, ROS1 or ALK gene fusions (Desai et al., 2022). It can penetrate the blood-brain barrier and exhibit CNS activity, underscoring its potential in treating brain tumors (Liu et al., 2018; Desai et al., 2022; Jiang et al., 2022). However, bulk evidences suggest that entrectinib can cause serious neurotoxicity following prolonged application, such as sensory neuropathy and peripheral neuropathy (Delgado et al., 2021; Giustini et al., 2022). However, the underlying mechanism of entrectinib-induced neurotoxicity remains elusive.
In this study, we investigated the effects of entrectinib on PC12, HT22 and SK-N-SH cells in vitro, focusing on its impact on the proliferation and apoptosis of nerve cells. Initially, a CCK-8 assay was carried out to determine cellular viability, we demonstrated that entrectinib inhibited the cell viability in a dose-dependent trend. Furthermore, EdU incorporation and colony formation assays were used to analyze the nerve cells proliferation. We also observed a significant inhibition of cell proliferation and clonogenic potential after entrectinib administration, confirming its anti-proliferative effects in vitro. Flow cytometry analysis further demonstrated that entrectinib effectively increased the apoptosis ratio of nerve cells. Taken together, these results indicated that entrectinib could significantly inhibit proliferation ability and induce apoptosis within the nerve cells.
Subsequently, transcriptome sequencing was performed to explore the gene expression changes of entrectinib-treated nerve cells. The transcriptome sequencing analysis identified THBS1 was intersected by PC12, HT22 and SK-N-SH cells, and it showed a downregulated trend. THBS1 belongs to adhesion glycoprotein, which plays a pivotal role in mediating cell-cell and cell-matrix interactions (Huang et al., 2017; Yao et al., 2023). THBS1, first discovered in platelets, but now many studies indicate that it plays a crucial role in the development of diseases (Firlej et al., 2011). Moreover, THBS1 is also involved in the physiological and pathological processes of the nervous system and indispensable for axon regeneration (Bray et al., 2019; Yao et al., 2023). Therefore, we further examined the expression of THBS1 within entrectinib treatment. The results revealed that the expression of THBS1 was significantly decreased by entrectinib, which was in parallel with sequencing results. In addition, THBS1 overexpression rescued entrectinib-induced nerve cell damage. Collectively, these results indicated that entrectinib-induced nerve cell damage may be related to the downregulation of THBS1.
KEGG and GSEA analysis results revealed that the PI3K-AKT and TGF-β signaling pathways were significantly enriched in entrectinib treatment. The PI3K-AKT signaling pathway plays an important role in intracellular signal transduction and regulates diverse cellular processes including cell cycle, adhesion, migration, inflammation, metabolism and survival (Jafari et al., 2019; Xiao et al., 2022). In addition, the PI3K-AKT signaling pathway also exerts profound influence on nervous system physiology, governing processes such as myelin formation (Gaesser and Fyffe-Maricich, 2016), axon regeneration (Huang et al., 2019), nerve cell regeneration (Luo et al., 2019) and apoptosis (Wang et al., 2022; Kilic et al., 2017; Lu et al., 2022). It was reported that PI3K-AKT signaling pathway could regulate neurotoxicity and mediate the survival of neurons (Goyal et al., 2023). Receptor tyrosine kinase (RTK), serving as principal upstream regulator of the PI3K-AKT pathway, modulated AKT activity through activating PI3K (Haddadi et al., 2018; Rai et al., 2019). It is worth noting that PI3K, as a crucial anti-apoptotic regulator, triggers the activation of its downstream target AKT upon its activation. Phosphorylation of transmembrane receptors such as RTK can lead to the activation of PI3K and p-AKT, thus activating neuroprotective effects (Griffin et al., 2005; Wang et al., 2022). Therefore, Western blotting assay was performed to examine the protein levels of PI3K, AKT and p-AKT within entrectinib treatment, we found that entrectinib could downregulate the levels of these proteins. Moreover, THBS1 overexpression can rescue the levels of relative proteins involved in PI3K-AKT signaling pathways in the entrectinib-treated nerve cells (PC12, HT22 and SK-N-SH). Thus, the reduction of the PI3K-AKT signaling pathway caused by THBS1 inhibition may be related to the entrectinib-induced nerve cell damage.
As we all known, THBS1 is a potential upstream target with TGF-β signaling pathway (Sun et al., 2022). TGF-β is a multifunctional peptide that governs the diverse cellular processes like cell proliferation, differentiation, death and migration (Jakowlew, 2006; Hata and Chen, 2016; Deng et al., 2024; Giarratana et al., 2024). TGF-β signaling pathway is also involved in neurotrophic signaling transmission and is closely related to the normal development and function of nerves (Meyers and Kessler, 2017; Ding et al., 2024). Brionne et al. (2003) research showed that the downregulation of TGF-β1 in primary neurons resulted in a strong reduction of survival. In addition, TGF-β1 heterozygous knockout mice displayed heightened sensitivity to toxic insults. THBS1 serves as a key activator of TGF-β, significantly activates TGF-β1 factor and TGF-β signaling pathway (Atanasova et al., 2019; Bedolla et al., 2024). Additionally, previous publications have reported that TGF-β signaling pathway induced renal injury may be related to the regulation of THBS1 (Sun et al., 2022). Taken together, we conducted qPCR and Western blotting assays to examine the expressions of THBS1 and TGF-β1 within entrectinib treatment. We found that entrectinib significantly downregulated the expressions of THBS1 and TGF-β1. Additionally, THBS1 overexpression can rescue TGF-β1 expression in the entrectinib-treated nerve cells, implicating that the reduction of the TGF-β signaling pathway caused by THBS1 inhibition may be a potential mechanism for the entrectinib-induced nerve cell damage.
In conclusion, our findings proposed a putative mechanism whereby entrectinib-induced nerve cell damage may downregulate THBS1 expression while also inhibiting PI3K-AKT and TGF-β signaling pathways (Figure 8). Although our current study offers valuable insights, there still exists some limitations. Due to the limited availability of appropriate animal models, we were unable to investigate the effects of entrectinib on the proteins and gene expression levels within the nerve cells in vivo. Thus, future studies should aim to comprehensively illustrate the intricate mechanism based on in vivo experiments. In a word, our study contributed important insights into the molecular mechanism underlying entrectinib’s neurotoxic effects and proposes potential therapeutic targets.
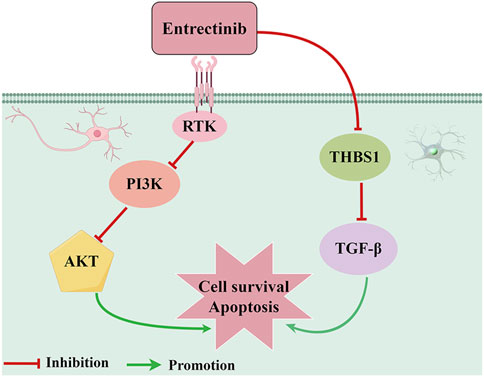
Figure 8. Proposed action pathway of entrectinib in inducing nerve cell damage. Entrectinib induced nerve cell damage by downregulating THBS1 expression while also inhibiting PI3K-AKT and TGF-β signaling pathways. The figure was drawn by Figdraw.
The raw data presented in the study are deposited in the Sequence Read Archive (SRA) repository, accession number is PRJNA1219491.
QT: Data curation, Investigation, Writing–original draft. JD: Conceptualization, Writing–original draft. FZ: Writing–original draft, Supervision. DZ: Formal Analysis, Writing–original draft. QY: Methodology, Writing–original draft. JYW: Visualization, Writing–original draft. YS: Software, Writing–original draft. JFW: Funding acquisition, Supervision, Writing–review and editing. ZL: Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by China International Medical Foundation (Z-2021-46-2101-2023 to ZL) and Yishan Research Project of Jiangsu Cancer Hospital (No. YSZD202406 to JW).
We acknowledge all authors participating in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1489210/full#supplementary-material
SUPPLEMENTARY TABLE S1 | The qPCR primers used in this study.
SUPPLEMENTARY TABLE S2 | The differentially expressed genes between control groups and entrectinib-treated groups in this study.
SUPPLEMENTARY TABLE S3 | The results of GO analysis in this study.
SUPPLEMENTARY TABLE S4 | The results of KEGG analysis in this study.
SUPPLEMENTARY TABLE S5 | The results of GSEA analysis in this study.
SUPPLEMENTARY TABLE S6 | The raw data of transcriptome sequencing in this study.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 (1), 25–29. doi:10.1038/75556
Atanasova, V. S., Russell, R. J., Webster, T. G., Cao, Q., Agarwal, P., Lim, Y. Z., et al. (2019). Thrombospondin-1 is a major activator of TGF-β signaling in recessive dystrophic epidermolysis bullosa fibroblasts. J. Invest. Dermatol. 139 (7), 1497–1505.e5. doi:10.1016/j.jid.2019.01.011
Aydin-Abidin, S., and Abidin, I. (2019). 7,8-Dihydroxyflavone potentiates ongoing epileptiform activity in mice brain slices. Neurosci. Lett. 703, 25–31. doi:10.1016/j.neulet.2019.03.013
Bedolla, A., Wegman, E., Weed, M., Stevens, M. K., Ware, K., Paranjpe, A., et al. (2024). Adult microglial TGFβ1 is required for microglia homeostasis via an autocrine mechanism to maintain cognitive function in mice. Nat. Commun. 15 (1), 5306. doi:10.1038/s41467-024-49596-0
Benitez, J. C., Geraud, A., Texier, M., Massard, C., Paci, A., Soria, J. C., et al. (2021). Late phase 1 studies: concepts and outcomes. Lancet Oncol. 22 (10), e446–e455. doi:10.1016/S1470-2045(21)00467-8
Bhamidipati, D., and Subbiah, V. (2023). Impact of tissue-agnostic approvals for patients with gastrointestinal malignancies. Trends Cancer 9 (3), 237–249. doi:10.1016/j.trecan.2022.11.003
Bray, E. R., Yungher, B. J., Levay, K., Ribeiro, M., Dvoryanchikov, G., Ayupe, A. C., et al. (2019). Thrombospondin-1 mediates axon regeneration in retinal ganglion cells. Neuron 103 (4), 642–657.e7. doi:10.1016/j.neuron.2019.05.044
Brionne, T. C., Tesseur, I., Masliah, E., and Wyss-Coray, T. (2003). Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron 40 (6), 1133–1145. doi:10.1016/s0896-6273(03)00766-9
Chen, X., Gan, Y., Au, N. P. B., and Ma, C. H. E. (2024). Current understanding of the molecular mechanisms of chemotherapy-induced peripheral neuropathy. Front. Molec. Neurosci. 17, 1345811. doi:10.3389/fnmol.2024.1345811
Delgado, J., Pean, E., Melchiorri, D., Migali, C., Josephson, F., Enzmann, H., et al. (2021). The European Medicines Agency review of entrectinib for the treatment of adult or paediatric patients with solid tumours who have a neurotrophic tyrosine receptor kinase gene fusions and adult patients with non-small-cell lung cancer harbouring ROS1 rearrangements. ESMO Open 6 (2), 100087. doi:10.1016/j.esmoop.2021.100087
Deng, Z., Fan, T., Xiao, C., Tian, H., Zheng, Y., Li, C., et al. (2024). TGF-β signaling in health, disease, and therapeutics. Signal Transduct. Target. Ther. 9 (1), 61. doi:10.1038/s41392-024-01764-w
Desai, A. V., Robinson, G. W., Gauvain, K., Basu, E. M., Macy, M. E., Maese, L., et al. (2022). Entrectinib in children and young adults with solid or primary CNS tumors harboring NTRK, ROS1, or ALK aberrations (STARTRK-NG). Neuro-Oncology 24 (10), 1776–1789. doi:10.1093/neuonc/noac087
Ding, Z., Jiang, M., Qian, J., Gu, D., Bai, H., Cai, M., et al. (2024). Role of transforming growth factor-β in peripheral nerve regeneration. Neural Regen. Res. 19 (2), 380–386. doi:10.4103/1673-5374.377588
Doebele, R. C., Drilon, A., Paz-Ares, L., Siena, S., Shaw, A. T., Farago, A. F., et al. (2020). Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 21 (2), 271–282. doi:10.1016/S1470-2045(19)30691-6
Firlej, V., Mathieu, J. R., Gilbert, C., Lemonnier, L., Nakhle, J., Gallou-Kabani, C., et al. (2011). Thrombospondin-1 triggers cell migration and development of advanced prostate tumors. Cancer Res. 71 (24), 7649–7658. doi:10.1158/0008-5472.CAN-11-0833
Frampton, J. E. (2021). Entrectinib: a review in NTRK+ solid tumours and ROS1+ nsclc. Drugs 81 (6), 697–708. doi:10.1007/s40265-021-01503-3
Fumagalli, G., Monza, L., Cavaletti, G., Rigolio, R., and Meregalli, C. (2020). Neuroinflammatory process involved in different preclinical models of chemotherapy-induced peripheral neuropathy. Front. Immunol. 11, 626687. doi:10.3389/fimmu.2020.626687
Gaesser, J. M., and Fyffe-Maricich, S. L. (2016). Intracellular signaling pathway regulation of myelination and remyelination in the CNS. Exp. Neurol. 283 (Pt B), 501–511. doi:10.1016/j.expneurol.2016.03.008
Giarratana, A. O., Prendergast, C. M., Salvatore, M. M., and Capaccione, K. M. (2024). TGF-β signaling: critical nexus of fibrogenesis and cancer. J. Transl. Med. 22 (1), 594. doi:10.1186/s12967-024-05411-4
Giustini, N. P., Oh, H., and Eaton, K. D. (2022). Development of neuropathic arthropathy with entrectinib: case report. JTO Clin. Res. Rep. 3 (11), 100419. doi:10.1016/j.jtocrr.2022.100419
Goyal, A., Agrawal, A., Verma, A., and Dubey, N. (2023). The PI3K-AKT pathway: a plausible therapeutic target in Parkinson's disease. Exp. Mol. Pathol. 129, 104846. doi:10.1016/j.yexmp.2022.104846
Griffin, R. J., Moloney, A., Kelliher, M., Johnston, J. A., Ravid, R., Dockery, P., et al. (2005). Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer's disease pathology. J. Neurochem. 93 (1), 105–117. doi:10.1111/j.1471-4159.2004.02949.x
Haddadi, N., Lin, Y., Travis, G., Simpson, A. M., Nassif, N. T., and McGowan, E. M. (2018). PTEN/PTENP1: 'Regulating the regulator of RTK-dependent PI3K/Akt signalling', new targets for cancer therapy. Mol. Cancer 17 (1), 37. doi:10.1186/s12943-018-0803-3
Hata, A., and Chen, Y. G. (2016). TGF-Β signaling from receptors to smads. Cold Spring Harb. Perspect. Biol. 8 (9), a022061. doi:10.1101/cshperspect.a022061
Huang, H., Kaur, S., and Hu, Y. (2019). Lab review: molecular dissection of the signal transduction pathways associated with PTEN deletion-induced optic nerve regeneration. Restor. Neurol. Neurosci. 37 (6), 545–552. doi:10.3233/RNN-190949
Huang, T., Wang, L., Liu, D., Li, P., Xiong, H., Zhuang, L., et al. (2017). FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin-1. Int. J. Oncol. 50 (5), 1501–1512. doi:10.3892/ijo.2017.3927
Jafari, M., Ghadami, E., Dadkhah, T., and Akhavan-Niaki, H. (2019). PI3k/AKT signaling pathway: erythropoiesis and beyond. J. Cell. Physiol. 234 (3), 2373–2385. doi:10.1002/jcp.27262
Jakowlew, S. B. (2006). Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 25 (3), 435–457. doi:10.1007/s10555-006-9006-2
Jassim, A., Rahrmann, E. P., Simons, B. D., and Gilbertson, R. J. (2023). Cancers make their own luck: theories of cancer origins. Nat. Rev. Cancer 23 (10), 710–724. doi:10.1038/s41568-023-00602-5
Jiang, Q., Li, M., Li, H., and Chen, L. (2022). Entrectinib, a new multi-target inhibitor for cancer therapy. Biomed. Pharmacother. 150, 112974. doi:10.1016/j.biopha.2022.112974
Jiang, T., Wang, G., Liu, Y., Feng, L., Wang, M., Liu, J., et al. (2021). Development of small-molecule tropomyosin receptor kinase (TRK) inhibitors for NTRK fusion cancers. Acta Pharm. Sin. B 11 (2), 355–372. doi:10.1016/j.apsb.2020.05.004
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42 (Database issue), D199–D205. doi:10.1093/nar/gkt1076
Kilic, U., Caglayan, A. B., Beker, M. C., Gunal, M. Y., Caglayan, B., Yalcin, E., et al. (2017). Particular phosphorylation of PI3K/Akt on Thr308 via PDK-1 and PTEN mediates melatonin's neuroprotective activity after focal cerebral ischemia in mice. Redox Biol. 12, 657–665. doi:10.1016/j.redox.2017.04.006
Li, Z., Zou, J., and Chen, X. (2023). In response to precision medicine: current subcellular targeting strategies for cancer therapy. Adv. Mater. 35 (21), e2209529. doi:10.1002/adma.202209529
Liu, D., Offin, M., Harnicar, S., Li, B. T., and Drilon, A. (2018). Entrectinib: an orally available, selective tyrosine kinase inhibitor for the treatment of NTRK, ROS1, and ALK fusion-positive solid tumors. Ther. Clin. Risk Manag. 14, 1247–1252. doi:10.2147/TCRM.S147381
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lu, T., Li, H., Zhou, Y., Wei, W., Ding, L., Zhan, Z., et al. (2022). Neuroprotective effects of alisol A 24-acetate on cerebral ischaemia-reperfusion injury are mediated by regulating the PI3K/AKT pathway. J. Neuroinflamm. 19 (1), 37. doi:10.1186/s12974-022-02392-3
Luo, S., Li, H., Mo, Z., Lei, J., Zhu, L., Huang, Y., et al. (2019). Connectivity map identifies luteolin as a treatment option of ischemic stroke by inhibiting MMP9 and activation of the PI3K/Akt signaling pathway. Exp. Mol. Med. 51 (3), 1–11. doi:10.1038/s12276-019-0229-z
Marcus, L., Donoghue, M., Aungst, S., Myers, C. E., Helms, W. S., Shen, G., et al. (2021). FDA approval summary: entrectinib for the treatment of NTRK gene fusion solid tumors. Clin. Cancer Res. 27 (4), 928–932. doi:10.1158/1078-0432.CCR-20-2771
Martineau, C., Turcotte, M. K., Otis, N., Provost, F., Themens, L., Guay, M. P., et al. (2022). Management of adverse events related to first-generation tyrosine receptor kinase inhibitors in adults: a narrative review. Support. Care Cancer 30 (12), 10471–10482. doi:10.1007/s00520-022-07401-y
Meyers, E. A., and Kessler, J. A. (2017). TGF-Β family signaling in neural and neuronal differentiation, development, and function. Cold Spring Harb. Perspect. Biol. 9 (8), a022244. doi:10.1101/cshperspect.a022244
Mun, E. J., Babiker, H. M., Weinberg, U., Kirson, E. D., and Von Hoff, D. D. (2018). Tumor-treating fields: a fourth modality in cancer treatment. Clin. Cancer Res. 24 (2), 266–275. doi:10.1158/1078-0432.CCR-17-1117
Rai, S. N., Dilnashin, H., Birla, H., Singh, S. S., Zahra, W., Rathore, A. S., et al. (2019). The role of PI3K/akt and ERK in neurodegenerative disorders. Neurotox. Res. 35 (3), 775–795. doi:10.1007/s12640-019-0003-y
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi:10.1093/bioinformatics/btp616
Staff, N. P., Grisold, A., Grisold, W., and Windebank, A. J. (2017). Chemotherapy-induced peripheral neuropathy: a current review. Ann. Neurol. 81 (6), 772–781. doi:10.1002/ana.24951
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Sun, J., Ge, X., Wang, Y., Niu, L., Tang, L., and Pan, S. (2022). USF2 knockdown downregulates THBS1 to inhibit the TGF-β signaling pathway and reduce pyroptosis in sepsis-induced acute kidney injury. Pharmacol. Res. 176, 105962. doi:10.1016/j.phrs.2021.105962
Trendowski, M. R., El Charif, O., Dinh, P. C., Travis, L. B., and Dolan, M. E. (2019). Genetic and modifiable risk factors contributing to cisplatin-induced toxicities. Clin. Cancer Res. 25 (4), 1147–1155. doi:10.1158/1078-0432.CCR-18-2244
Tu, Z., Xiao, R., Xiong, J., Tembo, K. M., Deng, X., Xiong, M., et al. (2016). CCR9 in cancer: oncogenic role and therapeutic targeting. J. Hematol. Oncol. 9, 10. doi:10.1186/s13045-016-0236-7
Wang, C. M., Cai, X. L., and Wen, Q. P. (2016). Astaxanthin reduces isoflurane-induced neuroapoptosis via the PI3K/Akt pathway. Mol. Med. Rep. 13 (5), 4073–4078. doi:10.3892/mmr.2016.5035
Wang, C. Y., Lin, T. T., Hu, L., Xu, C. J., Hu, F., Wan, L., et al. (2023). Neutrophil extracellular traps as a unique target in the treatment of chemotherapy-induced peripheral neuropathy. EBioMedicine 90, 104499. doi:10.1016/j.ebiom.2023.104499
Wang, Q., Shen, Z. N., Zhang, S. J., Sun, Y., Zheng, F. J., and Li, Y. H. (2022). Protective effects and mechanism of puerarin targeting PI3K/Akt signal pathway on neurological diseases. Front. Pharmacol. 13, 1022053. doi:10.3389/fphar.2022.1022053
Xiao, C. L., Yin, W. C., Zhong, Y. C., Luo, J. Q., Liu, L. L., Liu, W. Y., et al. (2022). The role of PI3K/Akt signalling pathway in spinal cord injury. Biomed. Pharmacother. 156, 113881. doi:10.1016/j.biopha.2022.113881
Yang, J., Zhao, Y., Zhou, Y., Wei, X., Wang, H., Si, N., et al. (2022). Advanced nanomedicines for the regulation of cancer metabolism. Biomaterials 286, 121565. doi:10.1016/j.biomaterials.2022.121565
Yao, L., Lu, F., Koc, S., Zheng, Z., Wang, B., Zhang, S., et al. (2023). LRRK2 Gly2019Ser mutation promotes ER stress via interacting with THBS1/TGF-β1 in Parkinson's disease. Adv. Sci. (Weinh) 10 (30), e2303711. doi:10.1002/advs.202303711
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16 (5), 284–287. doi:10.1089/omi.2011.0118
Yuan, M., Huang, L. L., Chen, J. H., Wu, J., and Xu, Q. (2019). The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 4, 61. doi:10.1038/s41392-019-0099-9
Keywords: entrectinib, nerve cell damage, THBS1, Pi3k-akt, TGF-β
Citation: Tang Q, Dong J, Zhang F, Zhao D, Yang Q, Wen J, Sun Y, Wei J and Liu Z (2025) Entrectinib can induce nerve cell damage by inhibiting PI3K-AKT and TGF-β signaling pathways. Front. Pharmacol. 16:1489210. doi: 10.3389/fphar.2025.1489210
Received: 31 August 2024; Accepted: 17 January 2025;
Published: 13 February 2025.
Edited by:
Huaqiang Zhai, Beijing University of Chinese Medicine, ChinaReviewed by:
Yu Zhijian, Shenzhen University, ChinaCopyright © 2025 Tang, Dong, Zhang, Zhao, Yang, Wen, Sun, Wei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jifu Wei, d2VpamlmdUBuam11LmVkdS5jbg==; Zhixian Liu, bGl1emhpeGlhbkBuam11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.