
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 30 October 2024
Sec. Pharmacogenetics and Pharmacogenomics
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1483723
Background: Antiseizure medications (ASM) exhibit considerable interindividual variability in terms of efficacy and adverse events. Genetic variation is thought to contribute to these differences in clinical outcomes. Specifically, the response to valproic acid (VPA), a widely used ASM, is influenced by multiple pharmacogenetic factors. However, and in contrast to other ASMs such as phenytoin and carbamazepine, there is a paucity of data on the association between VPA and various gene variants. The aim of this study was hence to evaluate the influence of candidate pharmacogenetic variants on VPA efficacy, toxicity and serum concentrations in a homogeneous cohort of patients newly diagnosed with genetic generalized epilepsies (GGE).
Methods: In this prospective cohort study, demographic, clinical and treatment outcomes of GGE patients were retrieved from their medical records. Whole exome sequencing was performed in collaboration with Epi25. Gene variants associated with VPA efficacy, metabolism and toxicities were retrieved from PharmGKB. An analysis was then conducted to explore potential associations between these gene variants and VPA clinical outcomes.
Results: Of the 166 patients included, 60 (36.1%) experienced treatment failure while 106 (63.9%) achieved treatment success. After adjusting for VPA maintenance dose, carriers of the rs3892097 (CYP2D6) variant were 2.5 times more likely to experience treatment failure compared to wildtype (p = 0.026). The rs1057910 variant (CYP2C9*3) was associated with increased serum VPA concentrations (p = 0.034). Moreover, the rs1137101 variant (LEPR gene, a metabolism regulator) was significantly associated with a higher risk of weight gain (regression coefficient of 3.430 [0.674; 6.186], p = 0.015) and a higher frequency of hair loss (OR = 3.394 [1.157; 9.956], p = 0.026), while the rs4480 variant (SOD2 gene, encoding for a mitochondrial scavenging enzyme) was correlated with a lower frequency of hair loss (OR = 0.276 [0.089; 0.858], p = 0.026).
Conclusion: These findings highlight the role of genetic factors in VPA treatment and underscore the potential for developing therapeutic strategies to enhance patient outcomes and minimize adverse effects.
Epilepsy, one of the most common neurological disorders, is characterized by recurrent, unprovoked seizures (Stafstrom and Carmant, 2015). Although its incidence is highest in infants and the elderly, epilepsy can manifest at any age, with diverse etiologies and clinical manifestations (Duncan et al., 2006).
Antiseizure medications (ASMs) control seizures in approximately 70% of newly diagnosed patients, thereby reducing morbidity and improving quality of life (Stafstrom and Carmant, 2015; Duncan et al., 2006). However, there is substantial interindividual variability in therapeutic response, dose-related adverse drug effects (ADEs), and drug plasma concentrations, even among patients with identical seizure types and treatment regimens (Loscher et al., 2009). Emerging evidence indicates that genetic variability substantially contributes to these differences in clinical outcomes (Loscher et al., 2009). This highlights the importance of pharmacogenetics, the study of genetic influences on drug response, in optimizing epilepsy treatment by elucidating the genetic factors influencing individual responses to ASMs.
Valproic acid (VPA), known for its broad spectrum of efficacy, is a widely used ASM, particularly in the management of genetic generalized epilepsies (GGE) (Marson et al., 2007; Marson et al., 2021; Epi25 collaborative, 2024). The pharmacogenetic landscape implicates numerous candidate genes in shaping inter-individual variability in VPA response based on its mechanisms of action and pharmacokinetic profile (PharmGKB, 2024c). For example, VPA enhances GABAergic activity by inhibiting ABAT and ALDH5A, enzymes crucial in gamma-aminobutyric acid (GABA) degradation (PharmGKB, 2024b; Johannessen and Johannessen, 2003). Additionally, VPA’s pharmacodynamic activity includes blocking voltage-gated sodium, potassium and calcium channels, including CACNA1C, CACNA1D, CACNA1N, and CACNA1F and the SCN family, thereby reducing high-frequency neuronal firing (Johannessen and Johannessen, 2003; Van den Berg et al., 1993). VPA undergoes extensive hepatic metabolism via enzymes such as UGT1A3, UGT1A4, UGT1A6, UGT1A8, UGT1A9, UGT1A10, UGT2B7 and UGT2B15, transforming VPA to valproate-glucuronide, its major urinary metabolite (Ito et al., 1990; Argikar and Remmel, 2009; Tan et al., 2010). Moreover, variants of the ABCB1 and ABCC1 gene encode efflux transporters at the blood-brain barrier that can expel VPA from the brain, lowering its concentration and potentially leading to VPA-resistant epilepsy (Kwan et al., 2009; Gibbs et al., 2004).
Despite its clinical significance, the pharmacogenetic data on VPA is limited compared to other ASMs such as carbamazepine and phenytoin, and the existing evidence is less robust (Ghodke-Puranik et al., 2013; Balestrini and Sisodiya, 2018). For instance, a multi-center study involving 142 adult Caucasian patients with epilepsy evaluated the association between the T1405 polymorphism in the carbamoyl phosphate synthetase 1 (CPS1) gene and VPA-induced hyperammonemia, demonstrating an increased risk in patients with the T1405 variant (Janicki et al., 2013). Similarly, a study on 52 pediatric patients with epilepsy showed that tailoring treatment according to CYP2C9 status reduced the incidence of hyperammonemia (Bűdi et al., 2015). In another study, several single nucleotide polymorphisms (SNPs) in the ABCB1 gene were analyzed in 170 Chinese children, revealing that homozygous carriers of rs1128503 were at higher risk of persistent seizures, while homozygous carriers of rs3789243 had a lower risk of gastrointestinal ADE (Zhu et al., 2023). However, the association between ABCB1 polymorphisms and ASM resistance remains inconclusive, and routine testing to predict response is not currently recommended (Balestrini and Sisodiya, 2018). Furthermore, a cohort study involving 212 Han Chinese patients with epilepsy treated with VPA identified associations between weight gain and polymorphisms in the leptin receptor (LEPR) and ankyrin repeat kinase domain containing 1 (ANKK1) genes (Mould and Upton, 2013). However, all of these studies require further validation because the evidence remains ambiguous (Balestrini and Sisodiya, 2018).
To address current gaps in the literature, we conducted a prospective cohort study aiming to evaluate the influence of candidate pharmacogenetic variants on VPA efficacy, toxicity and serum concentrations in a homogeneous cohort of patients newly diagnosed with GGE.
The study was approved by the American University of Beirut Medical Center (AUBMC) Institutional Review Board (IRB) (IM.AB1.01). All subjects or parents signed a genetic informed consent, and children signed an assent form as applicable.
The patients included in this nested pharmacogenetic study were recruited from an ongoing prospective study, initiated in 2010 and focusing on children and adults newly diagnosed with epilepsy. As of August 2024, a total of 3010 subjects have been enrolled (Jomaa et al., 2023). This multicenter study is centralized at the AUBMC and conducted in association with the Lebanese Chapter of the International League against Epilepsy (ILAE). Adult and pediatric neurologists from across Lebanon refer patients with suspected newly diagnosed seizures to the AUBMC where a comprehensive evaluation is conducted. The work-up on each patient includes a 3-h sleep deprived video-EEG recording interpreted by two experienced electroencephalographers along with an epilepsy protocol brain MRI interpreted by a neuroradiologist blinded to the patient’s history. The results of the work-up are communicated to the referring physician who decides whether to initiate treatment and selects the appropriate ASM. If treatment with an ASM is initiated, the dose is adjusted until achieving seizure freedom or the occurrence of intolerable adverse events. Patients are subsequently evaluated every 3 months during the first year, and every 6 months thereafter, with additional visits as needed based on seizure recurrence or adverse events. Patients who experienced absence, myoclonic or primary generalized tonic-clonic seizures with evidence of generalized spike wave discharges on EEG were diagnosed with a GGE (Hirsch et al., 2022).
In 2017, the ongoing AUBMC cohort joined the Epi25 Collaboration (Epi25) consortium, which includes collaborators from all over the world. The consortium aims to perform Whole Exome Sequencing (WES) on a large population of patients with epilepsy to elucidate the genetic influences on both rare and common forms of epilepsy and to investigate whether genetic risk originates from de novo mutations or inherited variants (Epi25 collaborative, 2024). The samples are sequenced at the Broad Institute using the Illumina Infinium Global Screening Array, GSA-MD v1.0 (Illumina, San Diego, United States) (Epi25 Collaborative. Electronic, 2021; Niestroj et al., 2020; Montanucci et al., 2023).
Peripheral blood was collected into EDTA tubes, aliquoted and stored at −80°C until DNA isolation using the Flexigene DNA isolation kit (Qiagen, Ca, United States) as per manufacturer’s guidelines. The isolated DNA was stored at −20°C.
By 2020, a total of 1238 peripheral DNA samples were shipped from AUBMC to the Board Institute for WES. Following sequencing, BAM and VCF files were returned for storage and analysis by the institutional Bioinformatics core lab. Raw reads in unaligned BAM format (uBAM) were aligned to the human reference genome build 38 (hg38) using Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2010). The aligned reads were then sorted by chromosome coordinates, and duplicate reads were marked using the Genome Analysis Toolkit (GATK) MarkDuplicates tool (Van der Auwera et al., 2013). Base quality score recalibration was applied using the GATK ApplyBQSR tool. Single nucleotide variant (SNVs) and insertions/deletions (INDELs) were identified using the GATK HaplotypeCaller. The GATK GenomicsDBImport tool was used to merge GVCFs from multiple samples, followed by joint genotyping using the GenotypeGVCFs tool from GATK. Variant filtration and variant calling quality score recalibrations were performed using GATK VariantFiltration and Variant Quality Score Recalibration (applyVQSR) tools, respectively. The final variant call VCF file (format v4.2) was generated by the Broad variant calling pipeline with GATK version 4.1.1.0. Variants were annotated using Ensembl Variant Effect Predictor version 104 (Ensembl-VEP) (McLaren et al., 2016). Variants with a “PASS” filter tag were selected for downstream analysis. Variants and genotypes data extraction from the VCF file was performed using BCFtools (Danecek et al., 2021). The frequencies of the genetic polymorphisms were checked for conformity to the Hardy Weinberg Equilibrium (HWE) using the Chi-square test.
Patients included in this study had their samples sequenced up until 2020, were diagnosed with a GGE, initiated on VPA monotherapy, and had available clinical data. The following variables were collected from the patients’ charts, starting from the first visit to the last follow-up: gender, weight (kg), height (cm), age at diagnosis, electroclinical syndrome (ECS), VPA maintenance dose (mg/kg), VPA serum concentrations (mg/L) and corresponding doses, compliance, length of follow-up, and treatment discontinuation. Trough total VPA concentrations were determined from analysis of blood samples collected either eight to 12 h after the last VPA dose, or prior to the first dose of the day. Serum VPA concentrations were analyzed by a chemiluminescent micro-particle immunoassay (CMIA) using Architect from Abbott Laboratories (Abbott Park, Illinois, United States). The minimum detectable assay limit of the apparatus was 2 mg/L. At each follow-up visit or phone call, adherence to treatment was monitored through inquiries made to the caregiver/patient regarding the administration of ASM as prescribed. In addition, the duration of seizure remission was measured to evaluate treatment efficacy. Patients who were non-compliant and those with incomplete follow-up data were excluded from the study.
To identify target variants with genotype-phenotype associations involving VPA, a systematic search was conducted on 14 December 2023 using the “clinical annotations” and “variant annotations” feature of the PharmGKB curated database (PharmGKB, 2024a). This search focused exclusively on statistically significant (p-value <0.05) single nucleotide polymorphisms (SNPs), while excluding SNPs with unsupported evidence (level of evidence = 4). This search identified 31 variants of interest in 25 genes across four main phenotype categories: dosage, metabolism/pharmacokinetics, efficacy and toxicity. Further details on these variants, including classification based on VPA pharmacogenetic associations, mutations types and nucleotides changes, Genome Aggregation Database (gnomAD) (Genome Aggregation, 2024), and allele frequencies for all populations compiled and Europeans were extracted from the dbSNP database (Home - SNP - NCBI, 2024) (Supplementary Table S1).
Data were entered into SPSS (Version 25.0. Armonk, NY: IBM Corp.) to evaluate the potential associations between the various genotypes and treatment success, toxicity, and adjusted drug concentration of VPA for dose and weight (ADC) (Daci et al., 2015). Data are presented as mean ± standard deviation (SD) for continuous variables, and as counts and frequencies for categorical variables.
Patients who achieved at least 1 year of seizure freedom while on a maintenance dose of VPA monotherapy were categorized as having a successful treatment outcome. Conversely, patients who failed to achieve a 1-year remission, required treatment with an add-on ASM, switched medications after initiating VPA monotherapy, or discontinued VPA due to intolerable ADEs, were classified as treatment failures. Considered drug related toxicities included weight gain (calculated as the difference between the patient’s weight prior to VPA monotherapy from their last recorded weight while on VPA monotherapy), hair loss, nausea and/or vomiting and tremor.
For ADC analysis, all serum VPA concentrations were standardized by accounting for both the VPA dose and the patient weight using the following formula: serum VPA concentration (ug/mL)/[(patient weight (kg) * VPA dose (mg/day)], which yields the ADC variable in [(ug/mL/day)/(mg/kg)]. Weight values were recorded either at the time of VPA concentration measurement or within 6 months. If such data were unavailable, the average of the two closest weight measurements before and after the VPA concentration measurement was used. For patients with multiple VPA concentration readings at various time-points, the average ADC was computed.
Initially, a univariate association analysis was performed to assess the correlation of demographic variables and genetic polymorphisms with treatment success, toxicity, and ADC. Categorical variables were analyzed using the chi-square test. Continuous variables such as age at first visit, length of follow up, maintenance dose, VPA concentrations, and weight gain were assessed with the Mann–Whitney U test, after none were found to be normally distributed with the Shapiro-Wilk test for normality. The rest of the continuous variables were analyzed with Student t-test. Subsequently, a multivariate analysis was conducted to adjust for confounding variables. A binary logistic regression with Odds Ratios (OR) and 95% Confidence Intervals (CI) was performed to evaluate for the association with treatment success and toxicity (both categorized as yes/no), while a linear regression reporting Beta (β) and 95% CI was performed for ADC and weight gain. Confounding variables considered included gender, age at diagnosis, follow-up period and VPA dose per kilogram per day (mg/kg/d). Statistically significant (p < 0.05) confounding variables at the univariate level were adjusted for in the multivariate analysis. Significant differences in VPA maintenance doses were adjusted for in the treatment success and hair loss analyses. Significant differences in gender distribution were adjusted for in the nausea/vomiting and hair loss association analyses. Significant differences in age at initial diagnosis were adjusted for in the tremor and adjusted VPA concentration analyses. To account for children growth, we adjusted for age at first visit and length of follow-up in the multivariate analysis of the association of the gene variants with weight gain, even though age at first visit was not statically significant at the univariate level. Age at diagnosis was recorded as the age of the patient at the first visit. For VPA dose, the maintenance dose was used for cases of treatment success, the maximum dose for cases of inefficacy, and the dose at withdrawal for drug intolerability. The recessive genetic model was employed to evaluate the relationship between genes and outcome variables, comparing wildtype homozygous genotypes to the combined allele carriers and homozygous mutants.
Out of the 1238 sequenced samples, clinical data were reviewed for 218 consecutive patients who met the inclusion criteria. Fifty-two patients were excluded due to poor data quality, incomplete follow up, or non-compliance with medication. Thus, the final sample for the association analysis with treatment efficacy comprised 166 patients with an age range of 6 months–40 years (mean = 12.85 ± 7.28), and an initial weight range from 7.5 to 116 kg (mean = 47.32 ± 22.26, (N = 163)). For the toxicity analysis, four patients were excluded from the categories of hair loss (Supplementary Table S6) and nausea/vomiting (Supplementary Table S3) due to inadequate reporting of symptoms, thereby leaving 162 patients (N = 162) for the corresponding analyses. Three patients were excluded from the tremor analysis (Supplementary Table S4) for the same reason, thereby leaving 163 patients (N = 163) for the corresponding analysis. Weight gain analysis (Supplementary Table S5) was performed on 163 out of the initial 166 participants (N = 163), for which weight measurements were available. Finally, for the ADC analysis, VPA concentrations were available for only 150 out of the initial 166 participants (N = 150), on whom the corresponding analysis was conducted.
Out of the 31 listed genetic variants, three were not detected as all patients were wildtype homozygous, one was excluded due to the absence of wildtype alleles in our population sample, six lacked sufficient depth of coverage, and four did not satisfy HWE. This resulted in a total of 17 genetic variants for the association analyses, with minor allele frequencies comparable to those observed in Europeans populations. Five variants (rs2279020, rs9332120, rs3892097, rs7438284, rs2269577) from five genes (GABRA1, CYP2C9, CYP2D6, UGT2B7, XBP1) were analyzed for treatment outcome (success versus failure), four variants (rs1137101, rs1800497, rs4880, rs3087374) from four genes (LEPR, ANKK1, SOD2, POLG) were assessed for toxicity, and eight variants (rs6759892, rs1105879, rs1105880, rs7592281, rs2070959, rs1057910, rs1799853, rs7668258) from three genes (UGT1A, CYP2C9, UGT2B7) were examined for adjusted VPA drug concentration based on the phenotype categories from the PharmGKB (Supplementary Table S1).
Among the 166 patients included in the study, 60 (36.1%) experienced treatment failure while 106 (63.9%) achieved treatment success (Table 1). The treatment failure group had a mean maintenance VPA dose of 19.01 ± 9.03 mg/kg/d, significantly higher than the mean dose of 15.88 ± 6.7 mg/kg/d in the treatment success group (p = 0.036). This difference is expected, as patients resistant to treatment are likely to be prescribed higher doses of VPA. Other potential confounding factors did not show significant associations with treatment outcome.
Carriers of rs3892097 (CYP2D6) constituted 26.67% of the treatment failure group compared to 14.15% of the treatment success group (p = 0.047). The multivariate OR adjusted for maintenance VPA dose was 0.389 [0.169; 0.894] (p = 0.026), indicating that patients experiencing treatment failure were approximately 2.5 times more likely to carry this polymorphism. This underscores the detrimental effect of rs3892097 (CYP2D6) on the success rate of VPA monotherapy. No other SNPs showed significant associations with treatment outcome (Figure 1; Supplementary Table S2).
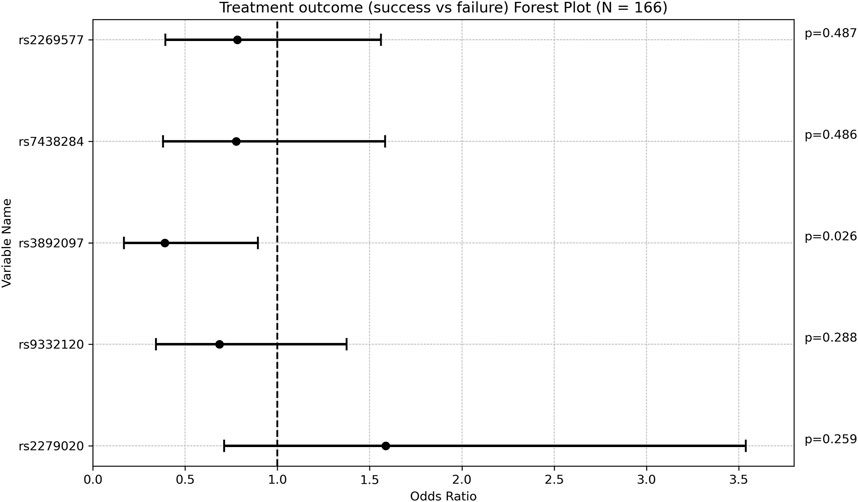
Figure 1. Forest plot representing the multivariate odds ratios (95%CI) of treatment success relative to treatment failure in each SNP.
In our study sample, ADEs from VPA monotherapy were spontaneously reported as follows: 4.93% of patients experienced nausea and/or vomiting, 12.26% had tremor and 11.72% reported hair loss.
All patients who experienced nausea and vomiting were females, whereas 42.86% of those without these symptoms were females (p = 0.002%). After adjusting for confounding variables, no SNP was found to be significantly associated with nausea and/or vomiting (Supplementary Table S3).
No statistically significant associations were identified between tremor and the genetic variants under investigation (Supplementary Table S4).
One SNP was found to be associated with increased weight gain after adjusting for VPA maintenance dose, age at diagnosis, and length of follow up. Specifically, rs1137101 (LEPR) had a multivariate regression coefficient of 3.430 [0.674; 6.186] with a p-value of 0.015. (Figure 2; Supplementary Table S5).
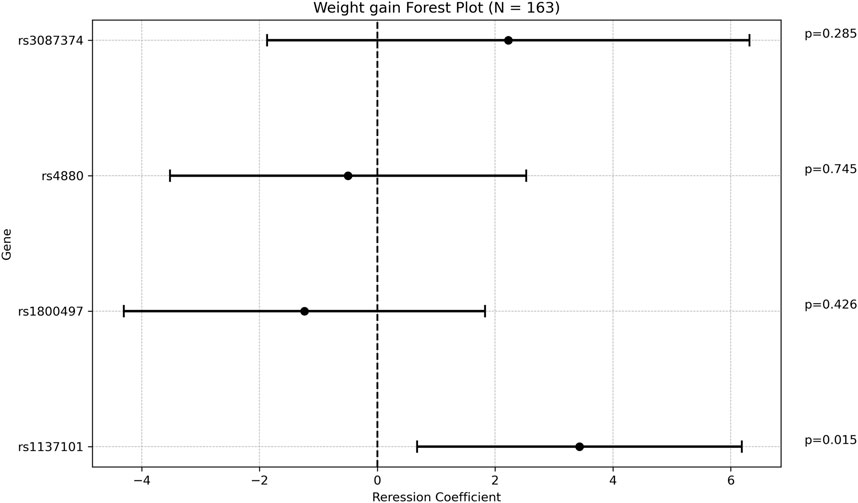
Figure 2. Forest plot representing the multivariate regression coefficients (95%CI) for weight gain in each SNP.
Regarding hair loss, patients who experienced this side effect had a significantly lower mean maintenance VPA dose compared to those who did not (13.38 ± 4.00 mg/kg/d vs 17.38 ± 8.04 mg/kg/d, respectively; p = 0.001. This finding is however inconclusive due to small percentage of patients who reported hair loss (Supplementary Table S6). A majority of patients (78.95%) who reported hair loss were females, compared to 41.26% in the group who did not report this side effect (p = 0.002). This finding is expected as females are more likely to notice and report hair loss. No other potential confounding factor was significantly associated with hair loss. The rs1137101 (LEPR) allele was present in 68.42% of patients with hair loss, compared to 40.56% of those without hair loss (p = 0.021). After adjusting for gender and VPA maintenance dose, the OR was 3.394 [1.157; 9.956], indicating that patients with this genetic variant had approximately three times the risk of developing hair loss. Conversely, rs4880 (SOD2) was found in 73.43% of patients without hair loss compared to 52.63% of those with hair loss (p = 0.061). This association became statistically significant when adjusting for gender and VPA maintenance dose, with a multivariate OR of 0.276 [0.089; 0.858], suggesting a protective effect of this SNP against hair loss. None of the remaining SNPs analyzed were significantly associated with hair loss (Figure 3).
There was a weak positive correlation between age at diagnosis and ADC, with a regression coefficient of 0.048 [0.011; 0.084], p = 0.0104 (Supplementary Table S7). The rs1057910 variant (CYP2C9*3) was associated with increased ADC with a mean of 4.9 ± 1.75 [(ug/mL/day)/(mg/kg)] in carriers vs 4.15 ± 1.61 in wildtype individuals (p = 0.028). In multivariate analysis, the regression coefficient was 0.722 [0.053; 1.391], p = 0.034. Of note that the variant rs7668258 (UGT2B7) was significantly associated with ADC only in the univariate analysis. None of the remaining SNPs were found to be significantly associated with ADC (Figure 4).
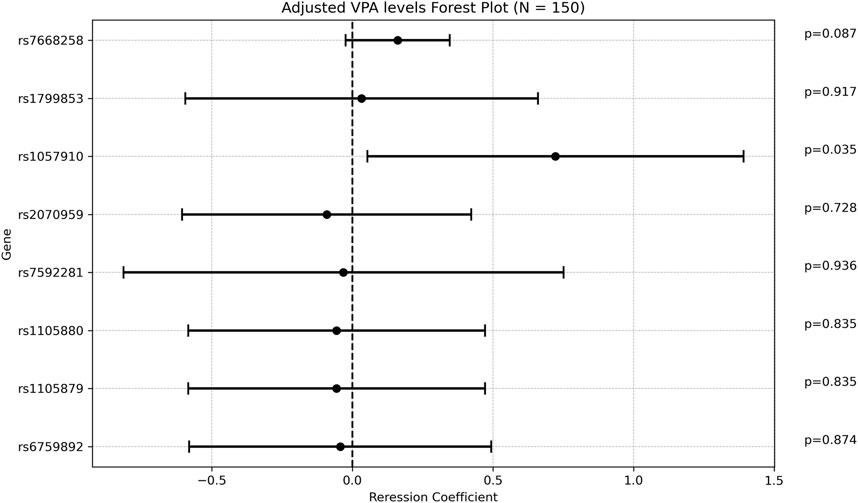
Figure 4. Forest plot representing the multivariate regression coefficients (95%CI) for ADC in each SNP.
Our study identified a significant association between the efficacy of VPA and the rs3892097 variant (CYP2D6). Furthermore, we found that adjusted serum VPA concentrations were impacted by the presence of the rs1057910 variant (CYP2C9*3). Additionally, certain adverse effects of VPA were found to be influenced by the individual’s genetic profile. Specifically, weight gain was associated with the rs1137101 (LEPR), while hair loss was affected by the rs1137101 (LEPR) and rs4480 variants (SOD2).
Our findings revealed that rs3892097 variant in the CYP2D6 gene was associated with an increased rate of treatment failure. The CYP2D6 enzyme (debrisoquine hydroxylase) is involved in the metabolism of several antidepressants and neuroleptics (Kirchheiner et al., 2001; Wedlund and de Leon, 2004; Rau et al., 2004). Genetic variations in this enzyme significantly affect drug metabolism rates, leading to the categorization of different metabolizer groups: ultrarapid, extensive, intermediate, and poor metabolizers, which necessitates dosage adjustments (Xie et al., 2001; Shastry, 2005). While data on the interaction between the rs3892097 variant (CYP2D6) and VPA is limited, existing literature supports our findings. One study has previously linked the presence of rs3892097 (CYP2D6) to refractory epilepsy in pediatric patients (López-García et al., 2017). Our results are consistent with these observations, and a possible explanation could be the impaired VPA oxidation and high toxic metabolites levels associated with the rs3892097 variant (Shnayder et al., 2023).
Conversely, the rs1057910 variant of the CYP2C9 gene (also known as CYP2C9*3) was associated with significantly higher VPA serum concentrations compared to wild type cases. The CYP2C9 enzyme participates in the metabolism of different classes of drugs (CYP2C9 cytochrome, 2024) and is the primary enzyme responsible for the hydroxylation and desaturation of VPA (Kiang et al., 2006). Our findings are corroborated by a meta-analysis, which demonstrated that individuals heterozygous for the CYP2C9*3 polymorphism had higher VPA serum concentrations compared to homozygous wild type individuals (Yoon et al., 2020). Additionally, one study highlighted an inverse relationship between VPA serum concentrations and the seizure frequency (Gram et al., 1979).
Hair loss is a common side effect of VPA, with reported incidences ranging from 3.5% to 12% (Kakunje et al., 2018). Proposed mechanisms for VPA-induced hair loss include deficiencies in biotin, vitamin D, and other trace minerals, such as zinc, iron, copper and magnesium (Kakunje et al., 2018; Wolff et al., 2008; Plonka et al., 2005; Kil et al., 2013; Praharaj et al., 2022). Another suggested mechanism is VPA induced telogen effluvium, which may be linked to its aromatase-inhibiting effects, potentially leading to male pattern baldness through subsequent hyperandrogenism (Praharaj et al., 2022). In our study, the rs1137101 variant (LEPR) was associated with a significant increased risk of hair loss, a novel finding that has not been previously reported. Given that the leptin receptor plays an important role in inducing the anagen (growth) phase of the hair follicle (Sumikawa et al., 2014), it is plausible that the rs1137101 variant (LEPR) is associated with a defective leptin receptor. This defect could impair the initiation of the anagen phase, thereby increasing susceptibility to VPA-induced hair loss. Furthermore, our results indicated that patients with the rs4480 variant of the SOD2 gene was associated with a lower incidence of hair loss, suggesting a protective effect. The SOD2 encodes superoxide dismutase 2, a mitochondrial scavenging enzyme that converts superoxide byproducts of oxidative phosphorylation to the less reactive hydrogen peroxide (H2O2) and oxygen (O2) (Superoxide dismutase, 2024b). The rs4480 (SOD2) SNP (Val16Ala) yields the Ala variant, which is more efficiently imported into the mitochondrial matrix compared to the Val variant, thereby enhancing mitochondrial SOD2 activity (Sutton et al., 2003). Consistent with this, previous research has linked the Val to Ala polymorphism with a reduced risk of hepatotoxicity compared to the Val/Val genotype (Saruwatari et al., 2012). Our study is the first to relate rs4480 (SOD2) to VPA-induced hair loss. There is however circumstantial evidence suggesting that increased oxidative stress contributes to hair follicle damage and hair loss (Trüeb, 2009; Trüeb, 2021). VPA has been shown to increase oxidative stress in a study involving pediatric patients (Verrotti et al., 2008). It is therefore possible that the protective effect of increased SOD2 activity counteracts the oxidative stress effects induced by VPA, thereby mitigating hair loss. Additionally, VPA-induced deficiencies in trace elements like zinc and copper, which are cofactors for SOD1 (Superoxide dismutase, 2024a), may exacerbate oxidative stress and hair loss. Therefore, the enhanced SOD2 activity conferred by the rs4480 variant (SOD2) could help offset these adverse effects.
Our findings further reveal that the presence of rs1137101 variant (LEPR) was associated with an increased risk of weight gain while on VPA monotherapy. This SNP variant is located in the LEPR gene which encodes the leptin receptor, a key player in regulating metabolism and energy balance. The leptin receptor is responsible for signaling the brain about the body’s energy stores, influencing appetite and energy expenditure. Variants in the LEPR gene, such as rs1137101 (LEPR), can disrupt leptin signaling, potentially leading to leptin resistance, where the brain becomes less responsive to leptin’s regulatory effects. This disruption can interfere with metabolic pathways involved in glucose homeostasis and lipid metabolism, contributing to weight gain. Mutations in the LEPR gene have been associated with obesity and type II diabetes mellitus (Leptin receptor, 2024; Supti et al., 2024; Veerabathiran et al., 2023). Supporting our findings, a positive association between weight gain and the LEPR rs1137101 SNP was also demonstrated in a study on 212 epilepsy patients receiving VPA (Li et al., 2015).
Our study has several significant limitations. Notably, the relatively small sample size may have contributed to wide confidence intervals in our findings. Additionally, despite being a prospective cohort study, we relied on data extracted from patient medical records, which inherently includes potential biases and inaccuracies. Another limitation is the subjective nature of some reported outcomes, particularly side effects, which are prone to recall bias. Finally, in a population with varied ages and weights like the one in this study, one would ideally use a population pharmacokinetic model to estimate VPA clearance for each individual (Mould and Upton, 2013). Such estimates would be more accurate and allow for more robust correlations with genetic factors. Despite these limitations, our study has notable strengths. It is among the first to explore the genetic variants rs1057910 (CYP2C9*3), rs1137101 (LEPR), rs4480(SOD2), and rs3892097 (CYP2D6) in the context of VPA treatment effectiveness and side effects. Our use of multivariate analysis adjusted for confounding factors, such as maintenance VPA dose, strengthens the validity of our conclusions.
In conclusion, our findings underscore the significant role of genetic variations in influencing the efficacy and adverse effects of VPA treatment for epilepsy. We identified a strong association between the rs3892097 (CYP2D6) variant and treatment failure, aligning with previous studies that suggest its role in drug metabolism. Additionally, we showed that carriers of the rs1057910 variants had higher adjusted VPA serum concentrations. Furthermore, the observed associations between rs1137101 (LEPR) and rs4480 variants (SOD2) with weight gain and hair loss further highlight the importance of personalized medicine in managing epilepsy. These insights into genetic influences on VPA treatment can guide more tailored therapeutic approaches, improving patient outcomes and minimizing adverse effects. Our study emphasizes the need for further research to elucidate the mechanistic pathways of these genetic variants and validate our findings in larger, prospective cohorts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by American University of Beirut Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
HY: Funding acquisition, Writing–review and editing, Data curation, Formal Analysis, Writing–original draft. JY: Data curation, Formal Analysis, Writing–original draft, Writing–review and editing. WN: Data curation, Writing–review and editing. AE: Writing–review and editing, Formal Analysis. NZ: Writing–review and editing, Conceptualization, Funding acquisition, Supervision. AB: Conceptualization, Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by an AUBFM Dean’s award for medical students’ research to Hady Yazbeck, and a grant from the American University of Beirut (Grant number: 1131613 52480 115372099990000).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1483723/full#supplementary-material
Argikar, U. A., and Remmel, R. P. (2009). Effect of aging on glucuronidation of valproic acid in human liver microsomes and the role of UDP-glucuronosyltransferase UGT1A4, UGT1A8, and UGT1A10. Drug Metab. Dispos. 37 (1), 229–236. doi:10.1124/dmd.108.022426
Balestrini, S., and Sisodiya, S. M. (2018). Pharmacogenomics in epilepsy. Neurosci. Lett. 667, 27–39. doi:10.1016/j.neulet.2017.01.014
Bűdi, T., Tóth, K., Nagy, A., Szever, Z., Kiss, Á., Temesvári, M., et al. (2015). Clinical significance of CYP2C9-status guided valproic acid therapy in children. Epilepsia 56 (6), 849–855. doi:10.1111/epi.13011
CYP2C9 cytochrome (2024). CYP2C9 cytochrome P450 family 2 subfamily C member 9 [Homo sapiens (human)] - gene - NCBI. Available at: https://www.ncbi.nlm.nih.gov/gene/1559.
Daci, A., Beretta, G., Vllasaliu, D., Shala, A., Govori, V., Norata, G. D., et al. (2015). Polymorphic variants of SCN1A and EPHX1 influence plasma carbamazepine concentration, metabolism and pharmacoresistance in a population of Kosovar Albanian epileptic patients. PLoS One 10 (11), e0142408. doi:10.1371/journal.pone.0142408
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10 (2), giab008. doi:10.1093/gigascience/giab008
Duncan, J. S., Sander, J. W., Sisodiya, S. M., and Walker, M. C. (2006). Adult epilepsy. Lancet 367 (9516), 1087–1100. doi:10.1016/S0140-6736(06)68477-8
Epi25 collaborative (2024). Epi25 collaborative. Available at: http://epi-25.org.
Epi25 Collaborative. Electronic (2021). Sub-genic intolerance, ClinVar, and the epilepsies: a whole-exome sequencing study of 29,165 individuals. Am. J. Hum. Genet. 108 (6), 965–982. doi:10.1016/j.ajhg.2021.04.009
Genome Aggregation (2024). Genome aggregation database (gnomAD) - registry of open data on AWS. Available at: https://registry.opendata.aws/broad-gnomad/.
Ghodke-Puranik, Y., Thorn, C. F., Lamba, J. K., Leeder, J. S., Song, W., Birnbaum, A. K., et al. (2013). Valproic acid pathway: pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics. 23 (4), 236–241. doi:10.1097/FPC.0b013e32835ea0b2
Gibbs, J. P., Adeyeye, M. C., Yang, Z., and Shen, D. D. (2004). Valproic acid uptake by bovine brain microvessel endothelial cells: role of active efflux transport. Epilepsy Res. 58 (1), 53–66. doi:10.1016/j.eplepsyres.2003.12.008
Gram, L., Flachs, H., Würtz-Jørgensen, A., Parnas, J., and Andersen, B. (1979). Sodium valproate, serum level and clinical effect in epilepsy: a controlled study. Epilepsia 20 (3), 303–311. doi:10.1111/j.1528-1157.1979.tb04808.x
Hirsch, E., French, J., Scheffer, I. E., Bogacz, A., Alsaadi, T., Sperling, M. R., et al. (2022). ILAE definition of the idiopathic generalized epilepsy syndromes: position statement by the ILAE task force on nosology and definitions. Epilepsia 63 (6), 1475–1499. doi:10.1111/epi.17236
Home - SNP - NCBI (2024). Available at: https://www.ncbi.nlm.nih.gov/snp/.
Ito, M., Ikeda, Y., Arnez, J. G., Finocchiaro, G., and Tanaka, K. (1990). The enzymatic basis for the metabolism and inhibitory effects of valproic acid: dehydrogenation of valproyl-CoA by 2-methyl-branched-chain acyl-CoA dehydrogenase. Biochim. Biophys. Acta 1034 (2), 213–218. doi:10.1016/0304-4165(90)90079-c
Janicki, P. K., Bezinover, D., Postula, M., Thompson, R. S., Acharya, J., Acharya, V., et al. (2013). Increased occurrence of valproic acid-induced hyperammonemia in carriers of T1405N polymorphism in carbamoyl phosphate synthetase 1 gene. ISRN Neurol, 261497. doi:10.1155/2013/261497
Johannessen, C. U., and Johannessen, S. I. (2003). Valproate: past, present, and future. CNS Drug Rev. 9 (2), 199–216. doi:10.1111/j.1527-3458.2003.tb00249.x
Jomaa, N., Nasreddine, W., Hmeimess, G., Beaini, S., Beydoun, A., Hotait, M., et al. (2023). Risk of recurrence in patients with an unprovoked tonic-clonic seizure and generalized epileptiform discharges on EEG. Epilepsia 64 (8), 2153–2161. doi:10.1111/epi.17671
Kakunje, A., Prabhu, A., Sindhu Priya, E., Karkal, R., Kumar, P., Gupta, N., et al. (2018). Valproate: it’s effects on hair. Int. J. Trichology 10 (4), 150–153. doi:10.4103/ijt.ijt_10_18
Kiang, T. K. L., Ho, P. C., Anari, M. R., Tong, V., Abbott, F. S., and Chang, T. K. H. (2006). Contribution of CYP2C9, CYP2A6, and CYP2B6 to valproic acid metabolism in hepatic microsomes from individuals with the CYP2C9*1/*1 genotype. Toxicol. Sci. 94 (2), 261–271. doi:10.1093/toxsci/kfl096
Kil, M. S., Kim, C. W., and Kim, S. S. (2013). Analysis of serum zinc and copper concentrations in hair loss. Ann. Dermatol. 25 (4), 405–409. doi:10.5021/ad.2013.25.4.405
Kirchheiner, J., Brøsen, K., Dahl, M. L., Gram, L. F., Kasper, S., Roots, I., et al. (2001). CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: a first step towards subpopulation-specific dosages. Acta Psychiatr. Scand. 104 (3), 173–192. doi:10.1034/j.1600-0447.2001.00299.x
Kumar, R., Arora, R., Sarangi, S. C., Ganeshan, N. S., Agarwal, A., Kaleekal, T., et al. (2021). Pharmacodynamic and pharmacokinetic interactions of hydroalcoholic leaf extract of Centella asiatica with valproate and phenytoin in experimental models of epilepsy in rats. J. Ethnopharmacol. 270, 113784. doi:10.1016/j.jep.2021.113784
Kwan, P., Wong, V., Ng, P. W., Lui, C. H. T., Sin, N. C., Poon, W. S., et al. (2009). Gene-wide tagging study of association between ABCB1 polymorphisms and multidrug resistance in epilepsy in Han Chinese. Pharmacogenomics 10 (5), 723–732. doi:10.2217/pgs.09.32
Leptin receptor (2024). LEPR leptin receptor Homo sapiens human - gene - NCBI. Available at: https://www.ncbi.nlm.nih.gov/gene/3953.
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26 (5), 589–595. doi:10.1093/bioinformatics/btp698
Li, H., Wang, X., Zhou, Y., Ni, G., Su, Q., Chen, Z., et al. (2015). Association of LEPR and ANKK1 gene polymorphisms with weight gain in epilepsy patients receiving valproic acid. Int. J. Neuropsychopharmacol. 18 (7), pyv021. doi:10.1093/ijnp/pyv021
López-García, M. A., Feria-Romero, I. A., Serrano, H., Rayo-Mares, D., Fagiolino, P., Vázquez, M., et al. (2017). Influence of genetic variants of CYP2D6, CYP2C9, CYP2C19 and CYP3A4 on antiepileptic drug metabolism in pediatric patients with refractory epilepsy. Pharmacol. Rep. 69 (3), 504–511. doi:10.1016/j.pharep.2017.01.007
Loscher, W., Klotz, U., Zimprich, F., and Schmidt, D. (2009). The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia 50 (1), 1–23. doi:10.1111/j.1528-1167.2008.01716.x
Marson, A., Burnside, G., Appleton, R., Smith, D., Leach, J. P., Sills, G., et al. (2021). The SANAD II study of the effectiveness and cost-effectiveness of valproate versus levetiracetam for newly diagnosed generalised and unclassifiable epilepsy: an open-label, non-inferiority, multicentre, phase 4, randomised controlled trial. Lancet 397 (10282), 1375–1386. doi:10.1016/S0140-6736(21)00246-4
Marson, A. G., Al-Kharusi, A. M., Alwaidh, M., Appleton, R., Baker, G. A., Chadwick, D. W., et al. (2007). The SANAD study of effectiveness of valproate, lamotrigine, or topiramate for generalised and unclassifiable epilepsy: an unblinded randomised controlled trial. Lancet 369 (9566), 1016–1026. doi:10.1016/S0140-6736(07)60461-9
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R. S., Thormann, A., et al. (2016). The Ensembl variant effect predictor. Genome Biol. 17 (1), 122. doi:10.1186/s13059-016-0974-4
Montanucci, L., Lewis-Smith, D., Collins, R. L., Niestroj, L. M., Parthasarathy, S., Xian, J., et al. (2023). Genome-wide identification and phenotypic characterization of seizure-associated copy number variations in 741,075 individuals. Nat. Commun. 14 (1), 4392. doi:10.1038/s41467-023-39539-6
Mould, D. R., and Upton, R. N. (2013). Basic concepts in population modeling, simulation, and model-based drug development-part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacometrics Syst. Pharmacol. 2 (4), e38. doi:10.1038/psp.2013.14
Niestroj, L. M., Perez-Palma, E., Howrigan, D. P., Zhou, Y., Cheng, F., Saarentaus, E., et al. (2020). Epilepsy subtype-specific copy number burden observed in a genome-wide study of 17 458 subjects. Brain 143 (7), 2106–2118. doi:10.1093/brain/awaa171
PharmGKB (2024a). Available at: https://www.pharmgkb.org/.
PharmGKB (2024b). Valproic acid pathway. Pharmacodynamics. Available at: https://www.pharmgkb.org/pathway/PA165959313.
PharmGKB (2024c). Valproic acid pathway, pharmacokinetics. Available at: https://www.pharmgkb.org/pathway/PA165964265.
Plonka, P. M., Handjiski, B., Popik, M., Michalczyk, D., and Paus, R. (2005). Zinc as an ambivalent but potent modulator of murine hair growth in vivo-preliminary observations. Exp. Dermatol 14 (11), 844–853. doi:10.1111/j.1600-0625.2005.00365.x
Praharaj, S. K., Munoli, R. N., Udupa, S. T., and Vaidyanathan, S. (2022). Valproate-associated hair abnormalities: pathophysiology and management strategies. Hum. Psychopharmacol. 37 (1), e2814. doi:10.1002/hup.2814
Rau, T., Wohlleben, G., Wuttke, H., Thuerauf, N., Lunkenheimer, J., Lanczik, M., et al. (2004). CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants-a pilot study. Clin. Pharmacol. Ther. 75 (5), 386–393. doi:10.1016/j.clpt.2003.12.015
Saruwatari, J., Deguchi, M., Yoshimori, Y., Noai, M., Yoshida, S., Ogusu, N., et al. (2012). Superoxide dismutase 2 Val16Ala polymorphism is a risk factor for the valproic acid-related elevation of serum aminotransferases. Epilepsy Res. 99 (1), 183–186. doi:10.1016/j.eplepsyres.2011.10.033
Shastry, B. S. (2005). Genetic diversity and new therapeutic concepts. J. Hum. Genet. 50 (7), 321–328. doi:10.1007/s10038-005-0264-6
Shnayder, N. A., Grechkina, V. V., Khasanova, A. K., Bochanova, E. N., Dontceva, E. A., Petrova, M. M., et al. (2023). Therapeutic and toxic effects of valproic acid metabolites. Metabolites 13 (1), 134.
Stafstrom, C. E., and Carmant, L. (2015). Seizures and epilepsy: an overview for neuroscientists. Cold Spring Harb. Perspect. Med. 5 (6), a022426. doi:10.1101/cshperspect.a022426
Sumikawa, Y., Inui, S., Nakajima, T., and Itami, S. (2014). Hair cycle control by leptin as a new anagen inducer. Exp. Dermatol. 23 (1), 27–32. doi:10.1111/exd.12286
Superoxide dismutase (2024a). SOD1 superoxide dismutase 1 [Homo sapiens (human)] - gene - NCBI. Available at: https://www.ncbi.nlm.nih.gov/gene/6647.
Superoxide dismutase (2024b). SOD2 superoxide dismutase 2 [Homo sapiens (human)] - gene - NCBI. Available at: https://www.ncbi.nlm.nih.gov/gene/6648#bibliography.
Supti, D. A., Akter, F., Rahman, M. I., Munim, M. A., Tonmoy, M. I. Q., Tarin, R. J., et al. (2024). Meta-analysis investigating the impact of the LEPR rs1137101 (A>G) polymorphism on obesity risk in Asian and Caucasian ethnicities. Heliyon 10 (6), e27213. doi:10.1016/j.heliyon.2024.e27213
Sutton, A., Khoury, H., Prip-Buus, C., Cepanec, C., Pessayre, D., and Degoul, F. (2003). The Ala16Val genetic dimorphism modulates the import of human manganese superoxide dismutase into rat liver mitochondria. Pharmacogenetics 13 (3), 145–157. doi:10.1097/01.fpc.0000054067.64000.8f
Tan, L., Yu, J. T., Sun, Y. P., Ou, J. R., Song, J. H., and Yu, Y. (2010). The influence of cytochrome oxidase CYP2A6, CYP2B6, and CYP2C9 polymorphisms on the plasma concentrations of valproic acid in epileptic patients. Clin. Neurol. Neurosurg. 112 (4), 320–323. doi:10.1016/j.clineuro.2010.01.002
Trüeb, R. M. (2009). Oxidative stress in ageing of hair. Int. J. Trichology 1 (1), 6–14. doi:10.4103/0974-7753.51923
Trüeb, R. M. (2021). Oxidative stress and its impact on skin, scalp and hair. Int. J. Cosmet. Sci. 43 (S1), S9–S13. doi:10.1111/ics.12736
Van den Berg, R. J., Kok, P., and Voskuyl, R. A. (1993). Valproate and sodium currents in cultured hippocampal neurons. Exp. Brain Res. 93 (2), 279–287. doi:10.1007/BF00228395
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43 (1110), 11.10.1–11. doi:10.1002/0471250953.bi1110s43
Veerabathiran, R., P, A., Bk, I., D, R., and Rs, A. H. (2023). Genetic predisposition of LEPR (rs1137101) gene polymorphism related to type 2 diabetes mellitus – a meta-analysis. Ann. Med. 55 (2), 2302520. doi:10.1080/07853890.2024.2302520
Verrotti, A., Scardapane, A., Franzoni, E., Manco, R., and Chiarelli, F. (2008). Increased oxidative stress in epileptic children treated with valproic acid. Epilepsy Res. 78 (2), 171–177. doi:10.1016/j.eplepsyres.2007.11.005
Wedlund, P. J., and de Leon, J. (2004). Cytochrome P450 2D6 and antidepressant toxicity and response: what is the evidence? Clin. Pharmacol. Ther. 75 (5), 373–375. doi:10.1016/j.clpt.2004.01.002
Wolff, K., Goldsmith, L., Katz, S., Gilchrest, B., Paller, A., and Leffell, D. (2008). Fitzpatrick’s dermatology in general medicine. 7th Edition. New York: McGraw-Hill.
Xie, H. G., Kim, R. B., Wood, A. J., and Stein, C. M. (2001). Molecular basis of ethnic differences in drug disposition and response. Annu. Rev. Pharmacol. Toxicol. 41, 815–850. doi:10.1146/annurev.pharmtox.41.1.815
Yoon, H. Y., Ahn, M. H., Yee, J., Lee, N., Han, J. M., and Gwak, H. S. (2020). Influence of CYP2C9 and CYP2A6 on plasma concentrations of valproic acid: a meta-analysis. Eur. J. Clin. Pharmacol. 76 (8), 1053–1058. doi:10.1007/s00228-020-02872-6
Keywords: pharmacogenetics, VPA, epilepsy, efficacy, toxicity, concentrations
Citation: Yazbeck H, Youssef J, Nasreddine W, El Kurdi A, Zgheib N and Beydoun A (2024) The role of candidate pharmacogenetic variants in determining valproic acid efficacy, toxicity and concentrations in patients with epilepsy. Front. Pharmacol. 15:1483723. doi: 10.3389/fphar.2024.1483723
Received: 20 August 2024; Accepted: 18 October 2024;
Published: 30 October 2024.
Edited by:
Antonio Javier Carcas Sansuán, Universidad Autónoma de Madrid, SpainReviewed by:
Marisol López López, Universidad Autónoma Metropolitana, MexicoCopyright © 2024 Yazbeck, Youssef, Nasreddine, El Kurdi, Zgheib and Beydoun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathalie Zgheib, bmsxNkBhdWIuZWR1Lmxi; Ahmad Beydoun, YWIyOUBhdWIuZWR1Lmxi
†ORCID: Nathalie Zgheib, orcid.org/0000-0002-0571-768X; Ahmad Beydoun, orcid.org/0000-0002-9047-1185
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.