- 1The Second Clinical College, Guangzhou University of Chinese Medicine, Guangzhou, China
- 2College of Life Science and Technology, Mudanjiang Normal University, Mudanjiang, China
Artemisia annua L. is the main source of artemisinin, an antimalarial drug. High diversity of morphological characteristics and artemisinin contents of A. annua has affected the stable production of artemisinin while efficient discrimination method of A. annua strains is not available. The complete chloroplast (cp) genomes of 38 A. annua strains were assembled and analyzed in this study. Phylogenetic analysis of Artemisia species showed that distinct intraspecific divergence occurred in A. annua strains. A total of 38 A. annua strains were divided into two distinct lineages, one lineage containing widely-distributed strains and the other lineage only containing strains from northern China. The A. annua cp genomes ranged from 150, 953 to 150, 974 bp and contained 131 genes, and no presence or absence variation of genes was observed. The IRs and SC junctions were located in rps19 and ycf1, respectively, without IR contraction observed. Rich sequence polymorphisms were observed among A. annua strains, and a total of 60 polymorphic sites representing 14 haplotypes were identified which unfolding the cpDNA heteroplasmy of A. annua. In conclusion, this study provided valuable resource for A. annua strains identification and provided new insights into the evolutionary characteristics of A. annua.
1 Introduction
Artemisia annua L. is the only natural source for artemisinin which is used for the treatment of malaria (Klayman, 1985; Cheong et al., 2020; Septembre-Malaterre et al., 2020; Al-Khayri et al., 2022). The selection and identification of elite germplasm of A. annua are critical for the high-quality, stable and low-cost production of artemisinin (Ma et al., 2014; Chen et al., 2017; Li et al., 2017; Shen et al., 2017; Ding et al., 2023). The wide range of natural distribution and high genetic heterozygosity due to self-incompatibility of pollination brought great challenges to the breeding of A. annua (Ma et al., 2014; Li et al., 2017; Amiryousefi et al., 2018; Ma et al., 2018). Although the genetic background of A. annua is relatively complex, the traits, especially artemisinin content, is correlated with their geographical distribution (Li et al., 2017). Moreover, strains of A. annua from southern China still had higher artemisinin content than those of northern strains under the same cultivation environment (He et al., 2022), which indicated a relative stable correlation between genetic background and geographical distribution.
Previously, a number of identification and taxonomy studies were conducted at species level of A. annua and its closely related species (Li et al., 2017; Shi et al., 2018; Jiao et al., 2023). A. annua can be efficiently identified from other Artemisia L. species by ITS regions (Li et al., 2017). The nuclear single nucleotide polymorphisms of 205 Artemisia species were used to reconstruct the phylogenetic relationships of Artemisia (Jiao et al., 2023). Besides, different types of molecular markers were also applied to identify different A. annua germplasms. He et al. (2022) applied SSR molecular markers to distinguish A. annua strains from different habitats. Ding et al. (2023) found that the ITS2 haplotype analysis is an ideal tool for A. annua strains identification based on the polymorphism of ribosomal DNA (rDNA). The cp genome is also an ideal tool for variants/strains identification and the intra-species genetic variations of cp genomes have been reported in many species (Sabir et al., 2014; Lei et al., 2016; Sun et al., 2019; Kim et al., 2020; Song et al., 2020; Xu et al., 2022). Cells of flowering plants possess high copies of their cp genome, and the copy numbers were estimated to range from 1, 900 to 50, 000 copies per cell (Bendich, 1987; Johnson and Palmer, 1989; Morley and Nielsen, 2016). Notably, heteroplasmy of organellar genome that more than one types among multiple copies of organellar genomes, was observed in individuals or even within single cells (Johnson and Palmer, 1989; Sabir et al., 2014; Lei et al., 2016; Ramsey and Mandel, 2019). Compared to DNA fragments, the whole cp genome, with relative long size, may contain sufficient variation and exhibit uniparental unisexual inheritance (Jansen and Ruhlman, 2012; Jensen and Leister, 2014; Ruhlman and Jansen, 2014), which makes it an ideal source for germplasm discrimination and genetic characteristic analysis.
In this study, a total of 38 complete cp genomes of A. annua were assembled and annotated. The structures, sequence polymorphisms and phylogenetic relationships of cp genomes were first in-depth analyzed and compared among A. annua strains. The cp genome heteroplasmy of A. annua was first reported in this study. Intra- and inter-individual genetic diversity of cp genomes of A. annua revealed in this study provided important information for the identification and evolution of A. annua strains.
2 Materials and methods
2.1 Materials collection, DNA extraction and PCR amplification
Thirty-five individuals of A. annua were cultivated in Huairou District, Beijing, from seeds collected from four countries to test and verify the polymorphic loci (Supplementary Table 1). Fresh leaves were snap-frozen with liquid nitrogen and stored at −80°C (Eppendorf, Hamburg, Germany). The total DNA of A. annua individuals were extracted by the modified cetyltrimethylammonium bromide (3 × CTAB) method (Allen et al., 2006), and the DNA quality and concentration were measured by electrophoresis in 1.0% agarose gel and the NanoDrop2000 ultra-micro ultraviolet spectrophotometer (Thermo Scientific, MIT, United States). Primers were designed by Primer Premier 5 (Supplementary Table 2). PCR amplification was performed on 25 μL reaction mixtures containing 2X Pro Taq Master Mix (dye plus) 12.5 μL, nuclease-free water 8.5 μL, 1.0 μL each of 10 μM forward and reverse primers, and genomic DNA 1.0 μL. The PCR reaction conditions were listed in Supplementary Table 1. The amplified products were detected by electrophoresis in 1.0% agarose gel. The synthesis of primers, and sequencing of amplification products were conducted by Sangon Biotech Guangzhou branch office.
2.2 WGS data collection, cp genome assembly, and annotation
WGS (whole genome sequencing) datasets of LQ-9 and HAN1 strains, and whole genome resequencing datasets of 36 individuals were obtained from previous study (Liao et al., 2022a) (Supplementary Table 3). The quality of raw sequencing data was evaluated by FastQC 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and low quality bases and reads were trimmed and removed by Skewer (Jiang et al., 2014). The LQ-9 cp genome was assembled and as the reference genome for later analysis. WGS reads of LQ-9 were mapped to A. annua cp genome (GenBank Accession Number: MF623173) and mapped reads were extracted as cp-like reads. The extracted reads were then assembled into contigs by ABySS 2.0.0 (https://github.com/bcgsc/abyss) (Jackman et al., 2017). Finally, the cp sequence contigs were ordered and concatenated based on the collinearity with reference cp genome sequences. The initial gene annotation was conducted with plann 1.1.2 (Huang and Cronk, 2015) and then validated by BLAST and manually correction. The transport RNA (tRNA) genes were identified with tRNAscan-SE software (Lowe and Chan, 2016). Circular gene maps of the A. annua cp genomes were generated using Chloroplot software (Zheng et al., 2020). The cp genomes assembled in this study have been deposited in the Global Pharmacopoeia Genome Database (Liao et al., 2022b) at http://www.gpgenome.com/species/92 under the “SuperBarcodes” section.
2.3 Phylogenetic analysis
A total of 117 complete cp genome sequences from 38 Artemisia species, and one complete cp genome from Chrysanthemum were downloaded from the NCBI GenBank (Supplementary Table 4) for phylogenetic analysis. Among which, 115 cp genomes were reassembled since the direction of SSC regions were opposite to A. annua cp genome in this study. Multiple sequence alignments were performed with 156 cp genomes (Supplementary Tables 3, 4) using MAFFT 7.313 (Katoh and Standley, 2013). Maximum likelihood (ML) phylogenetic tree was performed by RAxML 8.2.11 (Stamatakis, 2014) with 1,000 bootstrap replicates under the GTR + G model. Neighbor-joining (NJ) tree was constructed using MEGA 7 (Kumar et al., 2016) under the Kimura two-parameter model with 1,000 bootstrap replicates.
2.4 Sequence comparison and nucleotide variation analyses
Whole cp genomes of 38 A. annua individuals were aligned using MAFFT 7.313 (Katoh and Standley, 2013). For rearrangement analysis, all aligned sequences were constructed with Geneious Prime program (Kearse et al., 2012). The IRscope program was used to evaluate the expansion and contraction of inverted repeat region (IR) (Amiryousefi et al., 2018). Comparative analysis of cp genomes was performed among Artemisia species and visualized by R. Furthermore, nucleotide diversity (Pi) was estimated by sliding window analysis using DnaSP 6.0 with a 600 bp window length and 200 bp step size (Rozas et al., 2017). High throughput sequencing (HTS) data of 38 A. annua individuals were aligned to the LQ-9 reference cp genome with Bowtie2 2.4.4 (Langmead and Salzberg, 2012). The variants were called and filtered using BCFtools (http://samtools.github.io/bcftools/) with a minor allele count higher than 1. False positives caused by sequencing errors were excluded by visualizing the sequencing data of mutation sites by Integrative Genomics Viewer (IGV) (Robinson et al., 2011).
3 Results
3.1 Interspecific divergence in Artemisia species based on cp genomes
A total of 38 complete A. annua cp genomes were assembled with WGS data of 38 distinct individuals released by previous study (Supplementary Table 3) (Liao et al., 2022a). To explore the phylogenetic and evolutionary relationships of A. annua populations, a phylogenetic tree was constructed using ML and NJ methods based on 156 complete cp genomes of 38 Artemisia species, and C. morifolium was included as the outgroup (Supplementary Table 4). The topological congruence between the ML and NJ phylogenetic trees was observed, with the majority of nodes exhibiting robust support values (>99%) (Figure 1; Supplementary Figure 1). Thirty-eight species were divided into two main branches, the first branch only comprising A. keiskeana (Figure 1, clade 1), while the second branch was further divided into seven well-supported clades (Figure 1, clades 2–8). Seriphidium was considered to be an independent genus (Lin et al., 2011; Haghighi et al., 2014), while this was not supported by the phylogenetic relationship of cp genomes as Seriphidium species were close related with A. schrenkiana and A. scopiformis. The phylogenetic tree also showed that all cp genomes from the same species clustered together, except A. argyi and A. selengensis. All A. annua cp genomes clustered into a same clade and shared the most recent common ancestor with A. fukudo and A. nakaii, which was consistent with the previous results of Shen et al. (2017) and Jin et al. (2023). Intriguingly, all A. annua strains were divided into two subgroups, forming two distinct lineages. Geographically, the first lineage (subgroup 1) containing a mixture of southern and northern strains was defined as a broad lineage, and the second (subgroup 2) was defined as a northern lineage containing only northern strains.
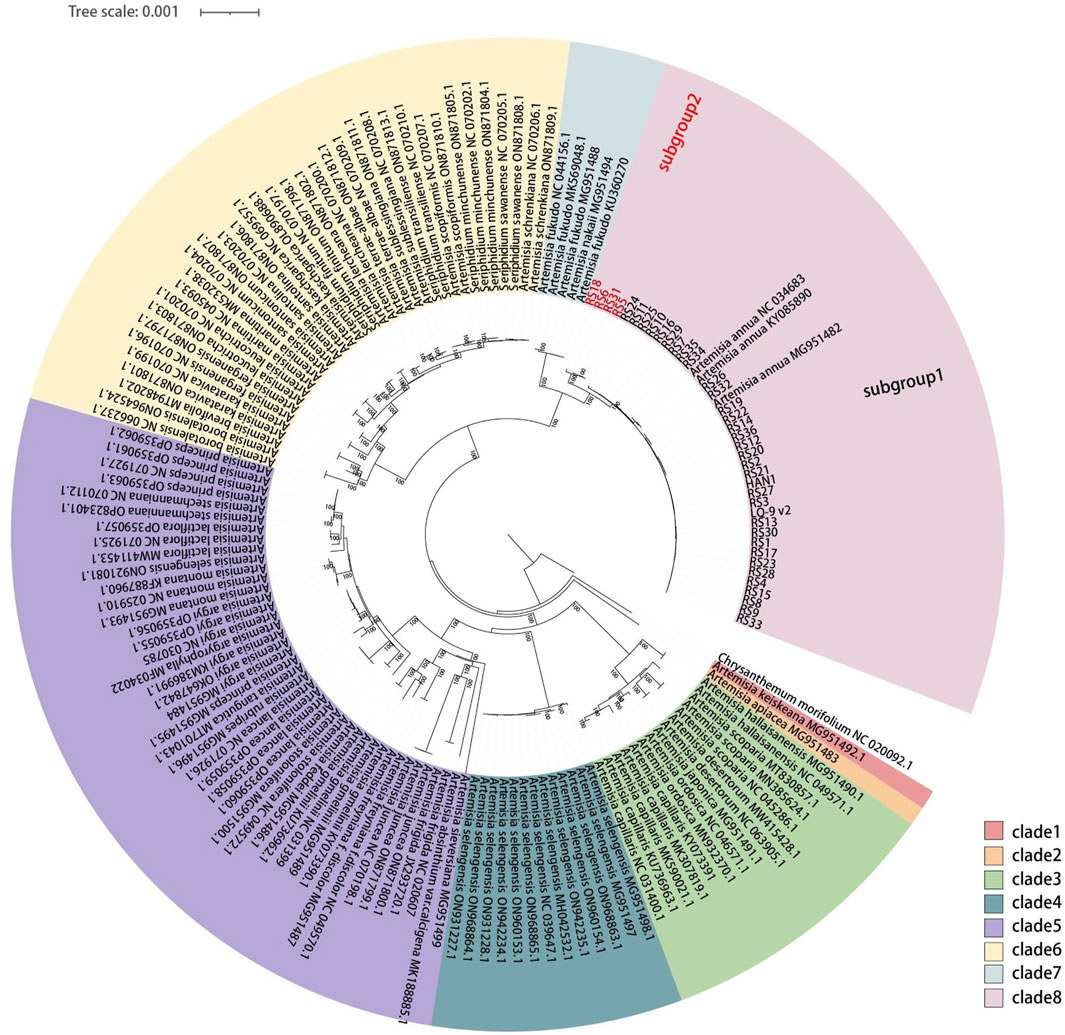
Figure 1. Phylogenetic tree obtained using the ML method for Artemisia species based on 156 complete cp genomes. Numbers above the branches indicate the ML bootstrap values.
3.2 Conserved cp genome structure in A. annua strains
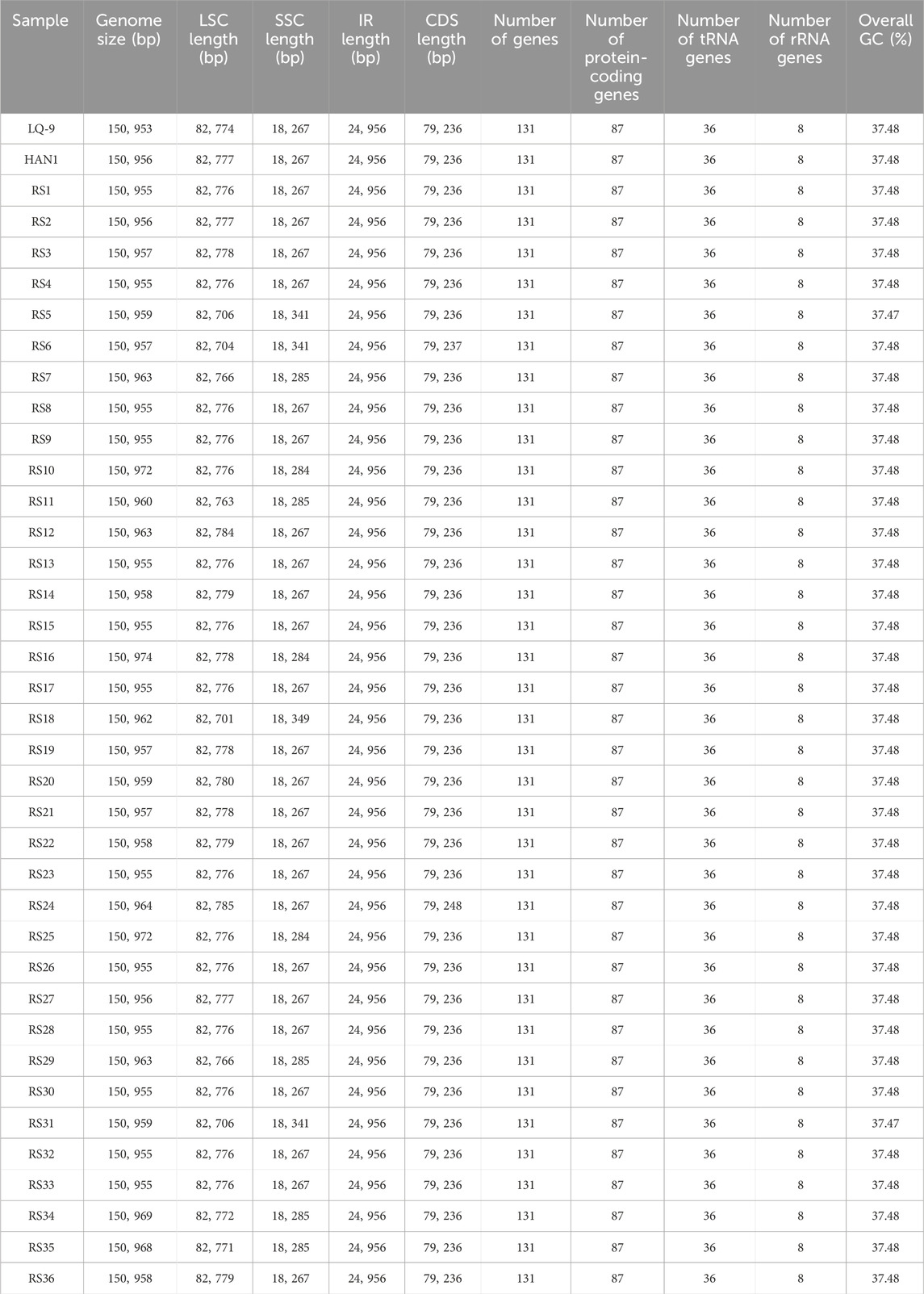
Thirty-eight assembled A. annua cp genomes were further annotated and analyzed. All cp genomes possessed the typical quadripartite structure which consisted of a large single-copy (LSC) region, a small single-copy (SSC) region, and two copies of IR regions (Figure 2). All cp genomes had lengths ranging from 150, 953 bp (LQ-9) to 150, 974 bp (RS16) with GC contents ranging from 37.47% to 37.48%. The LSC regions had lengths ranging from 82, 701 bp (RS18) to 82, 785 bp (RS24), and the SSC regions ranged from 18, 267 bp (RS24 et al.) to 18, 349 bp (RS18), while the lengths of IR regions were 24, 956 bp in all A. annua strains (Table 1), similar to the previously reported Artemisia cp genomes (Shen et al., 2017; Kim et al., 2020; Jin et al., 2023). Gene annotation showed that all cp genomes contained 131 genes, including 87 protein-coding genes, 36 tRNA genes and eight ribosomal RNA (rRNA) genes (Table 1). Of these, 17 genes (seven protein-coding genes, six tRNA genes and four rRNA genes) were duplicated in the IR regions in all A. annua strains (Supplementary Table 5). In addition, 17 intron-containing genes were identified, of which 15 contained one intron and two contained two introns (Supplementary Table 5).
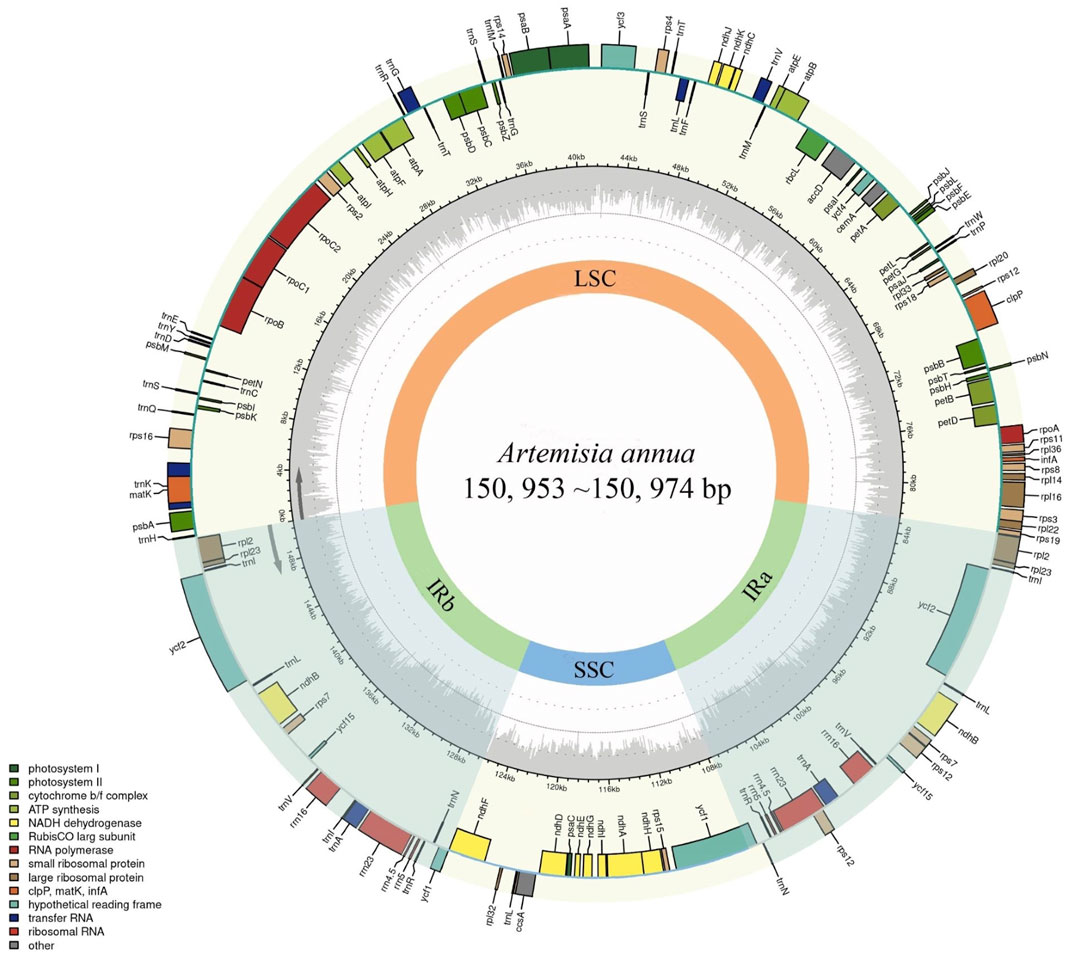
Figure 2. Circos plot of the A. annua cp genomes. Genes drawn inside the circle are transcribed in the clockwise direction, and those on the outside are transcribed in the counterclockwise direction. The genes are color-coded based on their function. The dark gray plot in the inner circle corresponds to GC content.
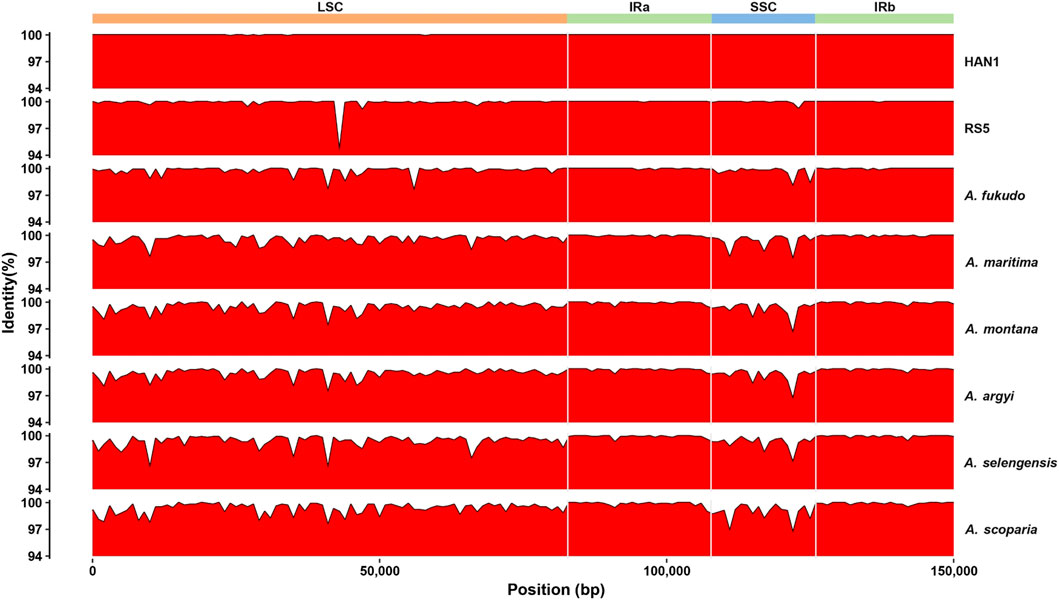
The IR border structure was conserved in A. annua that no IR contraction was observed. In each A. annua individual, the junctions between IRs and LSC and SSC were flanked by rps19 and ycf1, respectively (Supplementary Figure 2), in concordance with other Artemisia species (Kim et al., 2020; Jin et al., 2023). Comparison of genome structure showed that A. annua cp genomes were highly conserved, and no significant gene rearrangements were observed (Supplementary Figure 3). On the whole, the A. annua cp genomes were structural conserved.
3.3 Sequence polymorphisms in A. annua cp genomes
In this study, the intraspecific polymorphisms of A. annua cp genomes were assessed in inter-individual levels. Complete cp genomes of 38 A. annua individuals were aligned using the LQ-9 as the reference genome. In total, 60 polymorphic sites were identified after excluding false positives caused by sequencing errors, including 19 singleton variable sites and 41 parsimony informative sites, among which, 48 SNPs were identified in LSC, 11 in SSC and one in IR regions, respectively (Figure 3A; Table 2). The SSC regions were found to have the highest polymorphisms with a SNP site density of 8.8/10 kb, followed by LSC regions (5.3/10 kb), and the SNP density of IRs regions was only 0.4/10 kb (Table 2), revealing that the IR regions were more conserved than the single-copy regions, which is consistent with the comparative analysis of the whole cp genome of Artemisia species (Figure 4). Furthermore, the Pi values of 38 A. annua cp genomes were calculated using DnaSP v6.0, and the Pi values ranged from 0 to 0.0013 with an average of 0.00007. Based on DNA polymorphisms, seven highly diverged regions (Pi > 0.0007) were identified, including rps16-trnQ-UUG, petN-psbM, psbM-trnD-GUC, rpl20-clpP, clpP, trnL-UAG-rpl32 and rpl32 (Figure 3B), similar results have been observed in other Artemisia species (Shen et al., 2017; Kim et al., 2020; Jin et al., 2023). Fifteen protein-coding genes were found to containing variations, of which the ycf4 gene had the highest Pi value (0.0007) and the ndhF gene had the lowest Pi value (0.000002) (Figure 3C; Supplementary Table 6).
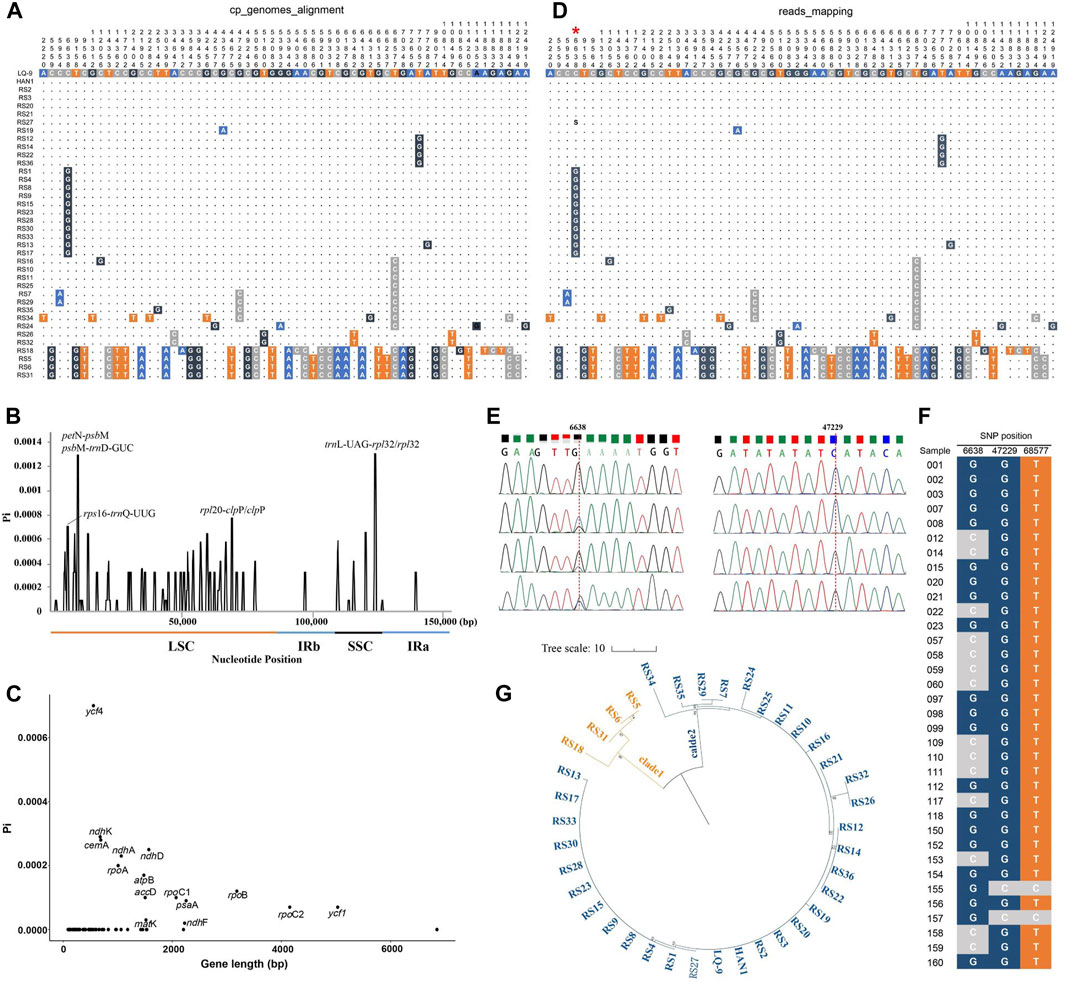
Figure 3. Variation of A. annua cp genomes. (A) Polymorphism analysis based on 38 A. annua global sequence alignment; (B) Nucleotide diversity of 38 A. annua cp genomes; (C) Nucleotide diversity of protein-coding genes; (D) Polymorphism analysis based on WGS reads mapping, red asterisk represents the pSNP site; (E) Partial peak diagram of Sanger sequencing of polymorphic sites 6,638 and 47,229, respectively; (F) Polymorphic sites confirmed by Sanger sequencing; (G) NJ tree constructed based on 60 polymorphic sites.
In addition, the WGS data of 38 individuals were aligned to LQ-9 reference genome with Bowtie2 for polymorphic sites validation. All 60 polymorphic sites were supported by mapped reads (Figures 3A, D). Notably, most of the polymorphic sites were single nucleotide polymorphisms (SNPs) between A. annua individuals, but partial single nucleotide polymorphism (pSNP) which contained more than one alternative bases in one site defined by James et al. (2009) were also found within individuals (Figure 3D). The polymorphic loci were confirmed by Sanger sequencing using the total DNA of 35 A. annua individuals (Figure 3F; Supplementary Table 1). In the Sanger sequencing results, the pSNP site generally contained multiple nested-peak which were identified as degenerate bases, and the SNP site showed clean single-peak representing complete base mutations (Figure 3E).
A total of 14 haplotypes were found in 38 A. annua individuals, which can be further divided into two distinct lineages as shown above (Figure 1; Figure 3G). Germplasm resources collected from the same location clustered together excluding Gansu, Hubei, and Xizang populations, but strains from different populations were also observed to have the same haplotype. Besides, polymorphism analysis showed that the subgroup 2 (the northern lineage) exhibited more unique polymorphic sites than the subgroup 1 (the broad lineage) and they shared only two polymorphic sites (Table 3). Other Artemisia species shared fifteen and four polymorphic sites with the northern lineage and the broad lineage, respectively (Table 3; Supplementary Figure 4). We proposed that the broad lineage may be the stable and specific evolutionary lineage during the speciation of A. annua, and the northern lineage may be the post-forming lineage. The above results showed that A. annua cp genomes were highly polymorphic, and the cp variants dataset can be used as the reference dataset for the identification and evolutionary analysis of A. annua strains.
4 Discussion
In this study, the phylogenetic relationships, structures, sequence polymorphisms of A. annua cp genomes were analyzed. All A. annua cp genomes showed highly conserved in structures, gene numbers and gene order. Minor differences were observed in genome sizes, but the lengths of IR regions were identical in all strains without contraction, similar to the observation in other Artemisia species (Shen et al., 2017; Kim et al., 2020; Jin et al., 2023). CpDNA is multicopy and abundant in plant (Bendich, 1987; McPherson et al., 2013; Malé et al., 2014; Morley and Nielsen, 2016), and were thought to be very conserved, as each species is typically characterized by a single cpDNA type. Recently, some genome-based studies have focused on population-level resolution and a single individual per species, and intraspecific genetic variations were detected in many species (Cui et al., 2020; Chen et al., 2022; Huang et al., 2022). Similarly, high sequence polymorphisms were found among A. annua populations. Distinct intraspecific divergence was observed, in which a more complex population structure was assumed in broad lineage due to their multiple maternal lineages and relatively higher number of haplotypes.
Although multiple copies and heteroplasmy of organelle genomes had been discovered over a century, the phenomenon of heteroplasmy was often overlooked in previous studies (Bendich, 1987; Ramsey and Mandel, 2019). The mitochondrial heteroplasmy in humans has been studied extensively (Li et al., 2010; Jiang et al., 2023) as its occurrence is strongly related with mitochondrial diseases (Wallace, 1992; Wallace, 1994; Chinnery and Turnbull, 1999; Chinnery et al., 2002). Besides, the heteroplasmy characteristic of mitochondria have also been used as genetic markers in molecular identification (Salas et al., 2001; Salas et al., 2005; Jiang et al., 2023). Plants possess high copy numbers of cp genomes, and polymorphisms of cp genomes are common at the level of population or species (Johnson and Palmer, 1989; Kim et al., 2020; Song et al., 2020; Xu et al., 2022). However, intra and inter-individual heteroplasmy of cp genomes are rarely reported (Fitter et al., 1996; García et al., 2004; Frey et al., 2005). In this study, the heteroplasmy of A. annua cp genomes were first discovered and verified, which may provide an important genetic basis for the accurate identification and evolutionary analysis of A. annua strains in the future.
This study represents the first in-depth understanding of A. annua cp genomes, and the rich maternal haplotypes composition can be used as the reference dataset for strains identification and breeding of A. annua. As cp genome is inherited matrilineally and relatively conserved, the combination of cp genome haplotypes and trait-related loci from nuclear genome should be further applied to A. annua strains screening in the future.
5 Conclusion
In the present study, the phylogenetic relationships, structures, sequence polymorphisms of A. annua cp genomes were characterized. Phylogenetic analysis of Artemisia species showed that intraspecific divergence occurred during the speciation of A. annua, forming two distinct lineages, the broad lineage and the northern lineage. The structures of A. annua cp genomes were extremely conserved without IR contraction and gene rearrangements observed. Rich sequence polymorphisms were observed among A. annua strains, and cpDNA heteroplasmy has been first reported and verified in A. annua. A total of 60 polymorphic sites were identified by global sequence alignment and WGS reads mapping, which can be further divided into 14 haplotypes, representing the cp variants dataset of A. annua. Our work provides important information for the identification and evolution of A. annua strains.
Data availability statement
The WGS data and assembled cp genome data were deposited in the Global Pharmacopoeia Genome Database at http://www.gpgenome.com/species/92 under the “SuperBarcodes” section. Data supporting the findings of this work are available within the paper and its supplemental information files. The datasets generated and analyzed during the study are available from the corresponding author upon reasonable request.
Author contributions
XD: Writing–review and editing, Writing–original draft, Visualization, Validation, Methodology, Data curation, Conceptualization. HP: Writing–original draft, Validation, Software, Investigation, Data curation, Conceptualization. PS: Writing–original draft, Formal Analysis, Data curation, Conceptualization. SiZ: Writing–original draft, Validation, Investigation, Data curation. SB: Writing–original draft, Validation, Methodology, Data curation, Conceptualization. ShZ: Writing–original draft, Validation, Software. CD: Writing–original draft, Validation, Investigation. JC: Writing–original draft, Visualization, Software. LG: Writing–review and editing, Formal Analysis, Data curation. DZ: Writing–review and editing, Methodology. XQ: Writing–review and editing, Writing–original draft, Supervision, Resources, Methodology, Investigation. BL: Writing–review and editing, Writing–original draft, Supervision, Software, Methodology, Funding acquisition, Conceptualization. ZH: Writing–review and editing, Writing–original draft, Supervision, Resources, Methodology, Investigation, Formal Analysis, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (grant number 82204548), Science and Technology Projects in Guangzhou (grant number 2023A04J0466), the Young Elite Scientists Sponsorship Program from China Association of Chinese Medicine (grant number CACM-2022-QNRC2-B30), Forestry Bureau of Guangdong Province (grant number 1247246), and Traditional Chinese Medicine Bureau of Guangdong Province (grant number 20232032).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1466578/full#supplementary-material
Abbreviations
cp, chloroplast; rDNA, ribosomal DNA; WGS, whole genome sequencing; tRNA, transport RNA; ML, maximum likelihood; NJ, neighbor-joining; IR, inverted repeat region; Pi, nucleotide diversity; HTS, high-throughput sequencing; IGV, Integrative Genomics Viewer; LSC, large single-copy region; SSC, small single-copy region; rRNA, ribosomal RNA; CDS, coding sequence; SNPs, single nucleotide polymorphisms; pSNP, partial single nucleotide polymorphism.
References
Al-Khayri, J. M., Sudheer, W. N., Lakshmaiah, V. V., Mukherjee, E., Nizam, A., Thiruvengadam, M., et al. (2022). Biotechnological approaches for production of artemisinin, an anti-malarial drug from Artemisia annua L. Molecules 27 (9), 3040. doi:10.3390/molecules27093040
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1 (5), 2320–2325. doi:10.1038/nprot.2006.384
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34 (17), 3030–3031. doi:10.1093/bioinformatics/bty220
Bendich, A. J. (1987). Why do chloroplasts and mitochondria contain so many copies of their genome? Bioessays 6 (6), 279–282. doi:10.1002/bies.950060608
Chen, C., Miao, Y., Luo, D., Li, J., Wang, Z., Luo, M., et al. (2022). Sequence characteristics and phylogenetic analysis of the Artemisia argyi chloroplast genome. Front. Plant Sci. 13, 906725. doi:10.3389/fpls.2022.906725
Chen, S., Xiang, L., Li, L., Wu, L., Huang, L., Zhang, D., et al. (2017). Global strategy and raw material production on artemisinin resources regeneration. Chin. Sci. Bull. 62, 1982–1996. doi:10.1360/N972017-00286
Cheong, D. H. J., Tan, D. W. S., Wong, F. W. S., and Tran, T. (2020). Anti-malarial drug, artemisinin and its derivatives for the treatment of respiratory diseases. Pharmacol. Res. 158, 104901. doi:10.1016/j.phrs.2020.104901
Chinnery, P. F., Samuels, D. C., Elson, J., and Turnbull, D. M. (2002). Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet 360 (9342), 1323–1325. doi:10.1016/s0140-6736(02)11310-9
Chinnery, P. F., and Turnbull, D. M. (1999). Mitochondrial DNA and disease. Lancet 354 (Suppl. 1), Si17–21. doi:10.1016/s0140-6736(99)90244-1
Cui, H., Ding, Z., Zhu, Q., Wu, Y., and Gao, P. (2020). Population structure and genetic diversity of watermelon (Citrullus lanatus) based on SNP of chloroplast genome. 3 Biotech. 10 (8), 374. doi:10.1007/s13205-020-02372-5
Ding, X., Chen, J., Dai, C., Shi, P., Pan, H., Lin, Y., et al. (2023). Developing population identification tool based on polymorphism of rDNA for traditional chinese medicine: Artemisia annua L. Phytomedicine 116, 154882. doi:10.1016/j.phymed.2023.154882
Fitter, J. T., Thomas, M. R., Rose, R. J., and Steele-Scott, N. (1996). Heteroplasmy of the chloroplast genome of Medicago sativa L. cv ‘Regen S’ confirmed by sequence analysis. Theor. Appl. Genet. 93 (5), 685–690. doi:10.1007/BF00224063
Frey, J. E., Frey, B., and Forcioli, D. (2005). Quantitative assessment of heteroplasmy levels in Senecio vulgaris chloroplast DNA. Genetica 123 (3), 255–261. doi:10.1007/s10709-004-3711-y
García, M. A., Nicholson, E. H., and Nickrent, D. L. (2004). Extensive intraindividual variation in plastid rDNA sequences from the holoparasite Cynomorium coccineum (Cynomoriaceae). J. Mol. Evol. 58 (3), 322–332. doi:10.1007/s00239-003-2554-y
Haghighi, A. R., Belduz, A. O., Vahed, M. M., Coskuncelebi, K., and Terzioglu, S. (2014). Phylogenetic relationships among Artemisia species based on nuclear ITS and chloroplast psbA-trnH DNA markers. Biologia 69 (7), 834–839. doi:10.2478/s11756-014-0379-3
He, W., Liao, B., Shen, X., Guo, S., Liao, X., Liang, Y., et al. (2022). Development of genomic SSR molecular markers with low coverage and high precision in Artemisia annua linn. Mol. Plant Breed. 20 (18), 6087–6096. doi:10.13271/j.mpb.020.006087
Huang, D. I., and Cronk, Q. C. (2015). Plann: a command-line application for annotating plastome sequences. Appl. Plant Sci. 3 (8), apps1500026. doi:10.3732/apps.1500026
Huang, X., Coulibaly, D., Tan, W., Ni, Z., Shi, T., Li, H., et al. (2022). The analysis of genetic structure and characteristics of the chloroplast genome in different japanese apricot germplasm populations. BMC Plant Biol. 22 (1), 354. doi:10.1186/s12870-022-03731-5
Jackman, S. D., Vandervalk, B. P., Mohamadi, H., Chu, J., Yeo, S., Hammond, S. A., et al. (2017). ABySS 2.0: resource-efficient assembly of large genomes using a bloom filter. Genome Res. 27 (5), 768–777. doi:10.1101/gr.214346.116
James, S. A., O'kelly, M. J., Carter, D. M., Davey, R. P., van Oudenaarden, A., and Roberts, I. N. (2009). Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res. 19 (4), 626–635. doi:10.1101/gr.084517.108
Jensen, P. E., and Leister, D. (2014). Chloroplast evolution, structure and functions. F1000Prime Rep. 6, 40. doi:10.12703/p6-40
Jiang, H., Lei, R., Ding, S. W., and Zhu, S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf 15, 182. doi:10.1186/1471-2105-15-182
Jiang, L., Liu, J., Li, S., Wen, Y., Zheng, X., Qin, L., et al. (2023). CmVCall: an automated and adjustable nanopore analysis pipeline for heteroplasmy detection of the control region in human mitochondrial genome. Forensic Sci. Int. Genet. 67, 102930. doi:10.1016/j.fsigen.2023.102930
Jiao, B., Chen, C., Wei, M., Niu, G., Zheng, J., Zhang, G., et al. (2023). Phylogenomics and morphological evolution of the mega-diverse genus Artemisia (Asteraceae: Anthemideae): implications for its circumscription and infrageneric taxonomy. Ann. Bot. 131 (5), 867–883. doi:10.1093/aob/mcad051
Jin, G., Li, W., Song, F., Yang, L., Wen, Z., and Feng, Y. (2023). Comparative analysis of complete Artemisia subgenus Seriphidium (Asteraceae: Anthemideae) chloroplast genomes: insights into structural divergence and phylogenetic relationships. BMC Plant Biol. 23 (1), 136. doi:10.1186/s12870-023-04113-1
Johnson, L. B., and Palmer, J. D. (1989). Heteroplasmy of chloroplast DNA in medicago. Plant Mol. Biol. 12 (1), 3–11. doi:10.1007/bf00017442
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi:10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. doi:10.1093/bioinformatics/bts199
Kim, G. B., Lim, C. E., Kim, J. S., Kim, K., Lee, J. H., Yu, H. J., et al. (2020). Comparative chloroplast genome analysis of Artemisia (Asteraceae) in East Asia: insights into evolutionary divergence and phylogenomic implications. BMC Genomics 21 (1), 415. doi:10.1186/s12864-020-06812-7
Klayman, D. L. (1985). Qinghaosu (artemisinin): an antimalarial drug from China. Science 228 (4703), 1049–1055. doi:10.1126/science.3887571
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/nmeth.1923
Lei, W., Ni, D., Wang, Y., Shao, J., Wang, X., Yang, D., et al. (2016). Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 6, 21669. doi:10.1038/srep21669
Li, L., Josef, B. A., Liu, B., Zheng, S., Huang, L., and Chen, S. (2017). Three-dimensional evaluation on ecotypic diversity of traditional chinese medicine: a case study of Artemisia annua L. Front. Plant Sci. 8, 1225. doi:10.3389/fpls.2017.01225
Li, M., Schönberg, A., Schaefer, M., Schroeder, R., Nasidze, I., and Stoneking, M. (2010). Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 87 (2), 237–249. doi:10.1016/j.ajhg.2010.07.014
Liao, B., Hu, H., Xiao, S., Zhou, G., Sun, W., Chu, Y., et al. (2022b). Global pharmacopoeia genome database is an integrated and mineable genomic database for traditional medicines derived from eight international pharmacopoeias. Sci. China Life Sci. 65 (4), 809–817. doi:10.1007/s11427-021-1968-7
Liao, B., Shen, X., Xiang, L., Guo, S., Chen, S., Meng, Y., et al. (2022a). Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield. Mol. Plant 15 (8), 1310–1328. doi:10.1016/j.molp.2022.05.013
Lin, Y., Humphries, C. J., and Gilbert, M. G. (2011). Flora of china volume 20–21 (Seriphidium). Beijing: Missouri Botanical Garden Press.
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 (W1), W54–W57. doi:10.1093/nar/gkw413
Ma, J., Xiang, J., Yang, Y., Long, L., Li, H., Qin, D., et al. (2014). Current situation of breeding and progress of systematic breeding of new varieties of Artemisia annua. Hubei Agric. Sci. 53 (19), 4520–4524. doi:10.14088/j.cnki.issn0439-8114.2014.19.003
Ma, T., Xiang, L., Zhang, D., Shi, Y., Ding, D., Shen, X., et al. (2018). Statues and research strategy of molecular breeding in Artemisia annua. Zhongguo Zhong Yao Za Zhi 43 (15), 3041–3050. doi:10.19540/j.cnki.cjcmm.2018.0093
Malé, P. J., Bardon, L., Besnard, G., Coissac, E., Delsuc, F., Engel, J., et al. (2014). Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Mol. Ecol. Resour. 14 (5), 966–975. doi:10.1111/1755-0998.12246
Mcpherson, H., Van Der Merwe, M., Delaney, S. K., Edwards, M. A., Henry, R. J., McIntosh, E., et al. (2013). Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 13, 8. doi:10.1186/1472-6785-13-8
Morley, S. A., and Nielsen, B. L. (2016). Chloroplast DNA copy number changes during plant development in organelle DNA polymerase mutants. Front. Plant Sci. 7, 57. doi:10.3389/fpls.2016.00057
Ramsey, A. J., and Mandel, J. R. (2019). When one genome is not enough: organellar heteroplasmy in plants. Annu. Plant Rev. Online 2, 619–658. doi:10.1002/9781119312994.apr0616
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29 (1), 24–26. doi:10.1038/nbt.1754
Rozas, J., Ferrer-Mata, A., Sánchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302. doi:10.1093/molbev/msx248
Ruhlman, T. A., and Jansen, R. K. (2014). The plastid genomes of flowering plants. Methods Mol. Biol. 1132, 3–38. doi:10.1007/978-1-62703-995-6_1
Sabir, J. S., Arasappan, D., Bahieldin, A., Abo-Aba, S., Bafeel, S., Zari, T. A., et al. (2014). Whole mitochondrial and plastid genome SNP analysis of nine date palm cultivars reveals plastid heteroplasmy and close phylogenetic relationships among cultivars. PLoS One 9 (4), e94158. doi:10.1371/journal.pone.0094158
Salas, A., Carracedo, A., Macaulay, V., Richards, M., and Bandelt, H. J. (2005). A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem. Biophys. Res. Commun. 335 (3), 891–899. doi:10.1016/j.bbrc.2005.07.161
Salas, A., Lareu, M. V., and Carracedo, A. (2001). Heteroplasmy in mtDNA and the weight of evidence in forensic mtDNA analysis: a case report. Int. J. Leg. Med. 114 (3), 186–190. doi:10.1007/s004140000164
Septembre-Malaterre, A., Lalarizo Rakoto, M., Marodon, C., Bedoui, Y., Nakab, J., Simon, E., et al. (2020). Artemisia annua, a traditional plant brought to light. Int. J. Mol. Sci. 21 (14), 4986. doi:10.3390/ijms21144986
Shen, X., Wu, M., Liao, B., Liu, Z., Bai, R., Xiao, S., et al. (2017). Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 22 (8), 1330. doi:10.3390/molecules22081330
Shi, Y., Zou, R., Tang, J., Jiang, Y., Xiong, Z., and Wei, X. (2018). Combination of FTIR and DNA barcode for identification of Artemisia annual L. And its closely related species. Mol. Plant Breed. 16 (15), 5117–5125. doi:10.13271/j.mpb.016.005117
Song, F., Li, T., Burgess, K. S., Feng, Y., and Ge, X. J. (2020). Complete plastome sequencing resolves taxonomic relationships among species of Calligonum L. (Polygonaceae) in China. BMC Plant Biol. 20 (1), 261. doi:10.1186/s12870-020-02466-5
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. doi:10.1093/bioinformatics/btu033
Sun, S., Zhou, X., Li, Z., Song, H., Long, Z., and Fu, P. (2019). Intra-individual heteroplasmy in the gentiana tongolensis plastid genome (Gentianaceae). PeerJ 7, e8025. doi:10.7717/peerj.8025
Wallace, D. C. (1992). Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science 256 (5057), 628–632. doi:10.1126/science.1533953
Wallace, D. C. (1994). Mitochondrial DNA sequence variation in human evolution and disease. Proc. Natl. Acad. Sci. 91, 8739–8746. doi:10.1073/pnas.91.19.8739
Xu, K., Lin, C., Lee, S. Y., Mao, L., and Meng, K. (2022). Comparative analysis of complete Ilex (Aquifoliaceae) chloroplast genomes: insights into evolutionary dynamics and phylogenetic relationships. BMC Genomics 23 (1), 203. doi:10.1186/s12864-022-08397-9
Keywords: Artemisia annua, chloroplast genome, genetic diversity, strains identification, comparative analysis
Citation: Ding X, Pan H, Shi P, Zhao S, Bao S, Zhong S, Dai C, Chen J, Gong L, Zhang D, Qiu X, Liao B and Huang Z (2024) A comparative analysis of chloroplast genomes revealed the chloroplast heteroplasmy of Artemisia annua. Front. Pharmacol. 15:1466578. doi: 10.3389/fphar.2024.1466578
Received: 18 July 2024; Accepted: 31 July 2024;
Published: 14 August 2024.
Edited by:
Da-Cheng Hao, Dalian Jiaotong University, ChinaReviewed by:
Shuai Guo, China Academy of Chinese Medical Sciences, ChinaYue Liu, Chengdu University of Traditional Chinese Medicine, China
Jun Qian, Shanghai Biozeron Biotechnology Co., Ltd., China
Copyright © 2024 Ding, Pan, Shi, Zhao, Bao, Zhong, Dai, Chen, Gong, Zhang, Qiu, Liao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Qiu, cWl1eGlhb2h1aUBnenVjbS5lZHUuY24=; Baosheng Liao, bGlhb2Jhb3NoZW5nQGd6dWNtLmVkdS5jbg==; Zhihai Huang, emhodWFuZzczMDhAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xiaoxia Ding
Xiaoxia Ding Hengyu Pan1†
Hengyu Pan1† Lu Gong
Lu Gong Danchun Zhang
Danchun Zhang Xiaohui Qiu
Xiaohui Qiu Zhihai Huang
Zhihai Huang