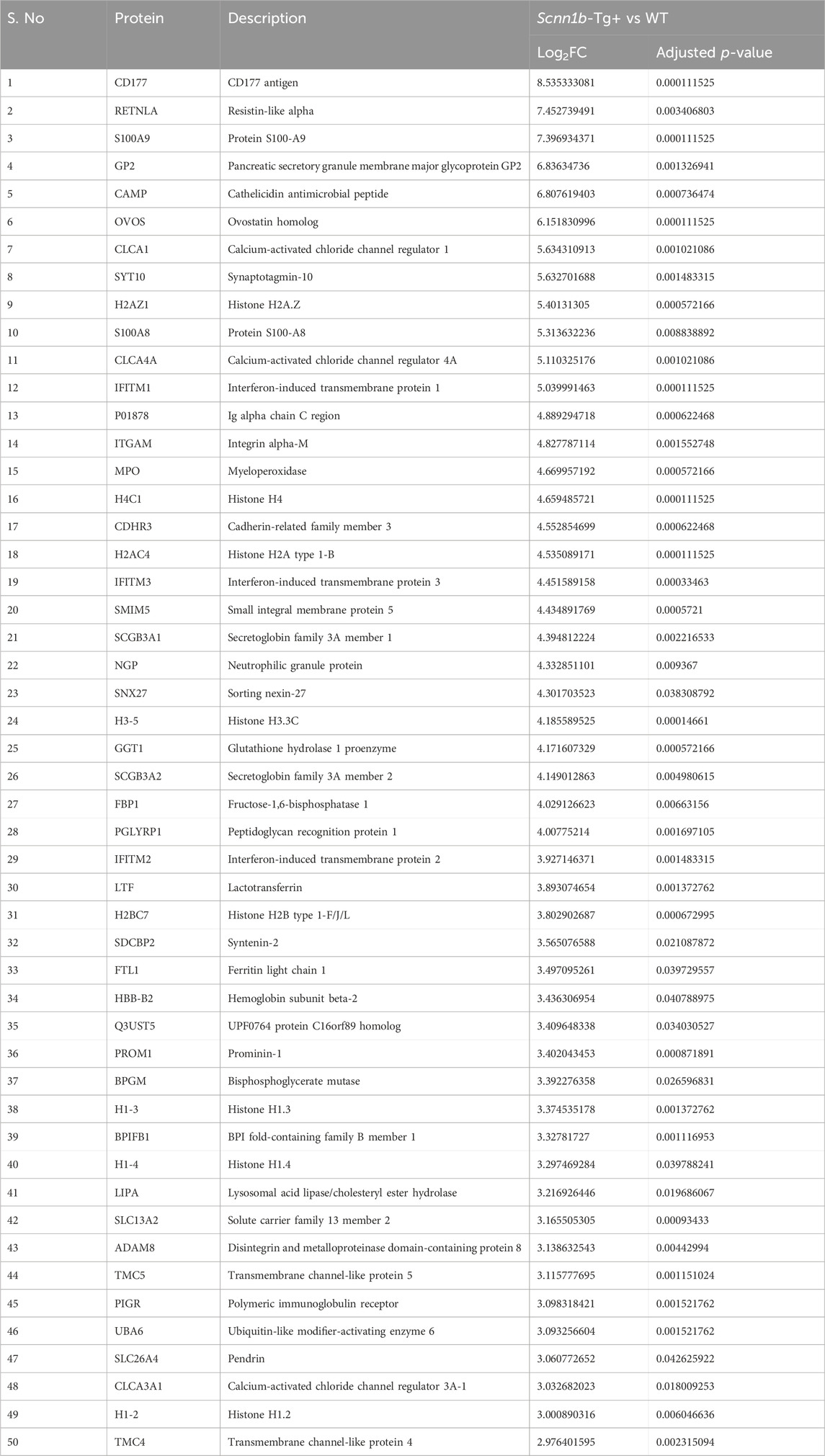
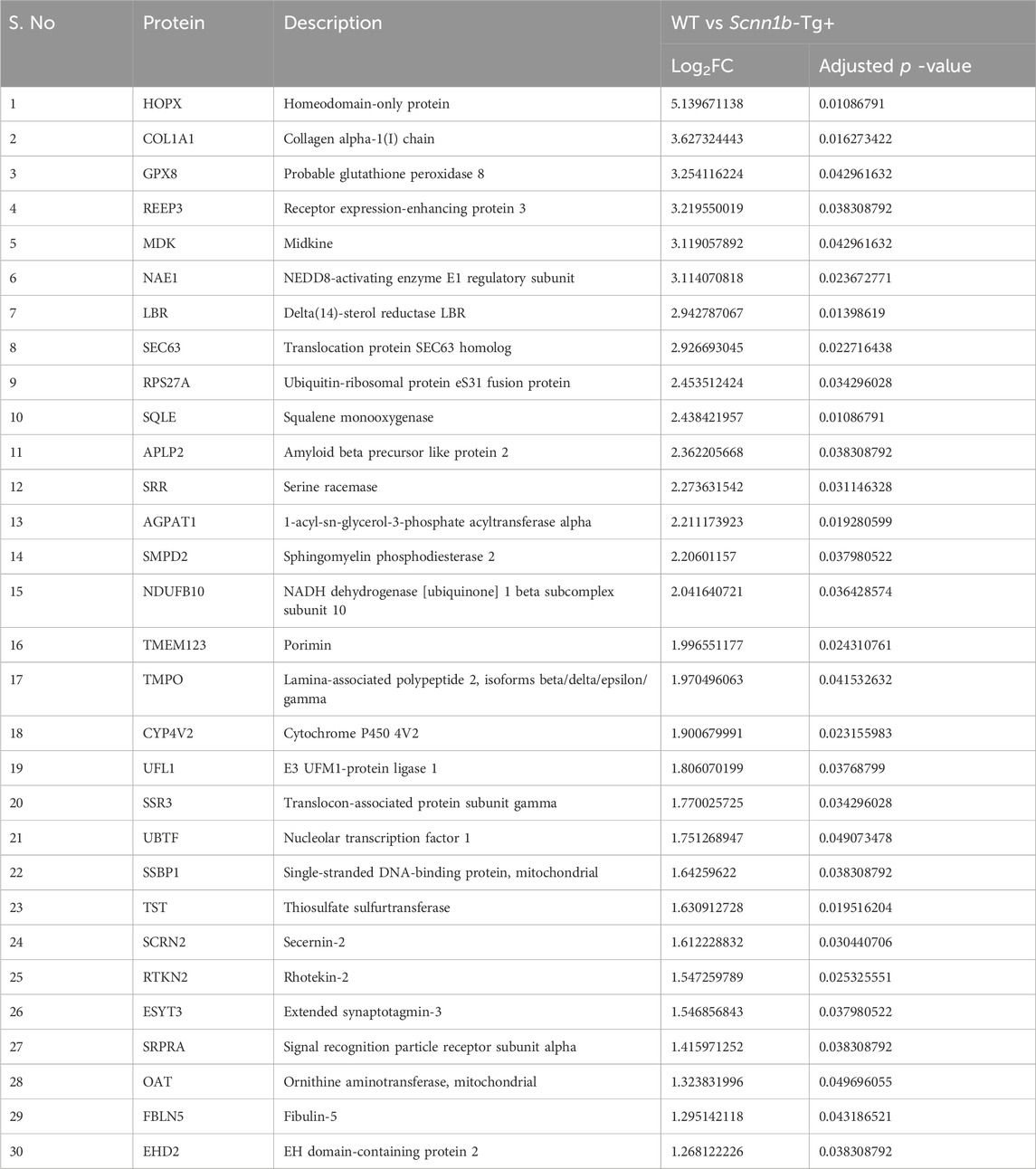
- 1Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
- 2Comparative and Molecular Pathogenesis Branch, Division of Translational Toxicology, National Institute of Environmental Health Sciences, Research Triangle Park, Durham, NC, United States
Exosomes, membrane-bound extracellular vesicles, ranging from approximately 30–200 nm in diameter, are released by almost all cell types and play critical roles in intercellular communication. In response to the prevailing stress, the exosome-bound protein signatures vary in abundance and composition. To identify the bronchoalveolar lavage fluid (BALF) exosome-bound proteins associated with mucoinflammatory lung disease and to gain insights into their functional implications, we compared BALF exosomes-derived proteins from adult Scnn1b transgenic (Scnn1b-Tg+) and wild type (WT) mice. A total of 3,144 and 3,119 proteins were identified in BALF exosomes from Scnn1b-Tg+ and WT mice, respectively. Using cutoff criteria (Log2 fold-change > 1 and adjusted p-value < 0.05), the comparison of identified proteins revealed 127 and 30 proteins that were significantly upregulated and downregulated, respectively, in Scnn1b-Tg+ versus WT mice. In addition, 52 and 27 proteins were exclusively enriched in Scnn1b-Tg+ and WT mice, respectively. The identified exosome-bound proteins from the homeostatic airspaces of WT mice were mostly relevant to the normal physiological processes. The protein signatures enriched in the BALF exosomes of Scnn1b-Tg+ mice were relevant to macrophage activation and mucoinflammatory processes. Ingenuity pathway analyses revealed that the enriched proteins in Scnn1b-Tg+ mice contributed to the inflammatory responses and antimicrobial defense pathways. Selective proteins including, RETNLA/FIZZ1, LGALS3/Galectin-3, S100A8/MRP8, and CHIL3/YM1 were immunolocalized to specific cell types. The comparative analysis between enriched BALF exosome proteins and previously identified differentially upregulated genes in Scnn1b-Tg+ versus WT mice suggested that the compartment-/cell-specific upregulation in gene expression dictates the enrichment of their respective proteins in the lung airspaces. Taken together, this study demonstrates that the BALF exosome-bound protein signatures reflect disease-relevant disturbances. Our findings suggest that the exosomes carry disease-relevant protein signatures that can be used as a diagnostic as well as predictive biomarkers for mucoinflammatory lung disease.
1 Introduction
Mucoinflammatory lung diseases including, cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), primary ciliary dyskinesia (PCD), and non-cystic fibrosis bronchiectasis, are characterized by mucus plugging, impaired mucociliary clearance (MCC), airway inflammation, and recurrent infections (Mall et al., 2018; Boucher, 2019; Burgel et al., 2009). Most of these pathological features of mucoinflammatory lung diseases are displayed by Scnn1b transgenic (Scnn1b-Tg+) mice (Mall et al., 2004; Mall et al., 2008). The Scnn1b-Tg+ mouse overexpresses sodium channel non-voltage-gated 1, beta subunit (Scnn1b) transgene in club cell secretory protein (CCSP) expressing airway epithelial cells. This overexpression of Scnn1b transgene causes hyperabsorption of sodium ions and, as a consequence, establishes an osmotic gradient that draws water into the airway epithelial cells leading to airway surface liquid dehydration, mucin hyperconcentration, and impaired MCC (Mall et al., 2004; Mall et al., 2008). In our previous report, transcriptomic analyses on whole lung and alveolar macrophages revealed differential gene signatures in the Scnn1b-Tg+ versus wild-type (WT) mice (Saini et al., 2014). However, apart from selective cytokine analyses (Mall et al., 2004; Mall et al., 2008; Lewis et al., 2020a; Choudhary et al., 2021a; Saini et al., 2016; Mao et al., 2023; Saini et al., 2018; Lewis et al., 2020b; Lewis et al., 2017), comprehensive analyses of airspace protein contents between Scnn1b-Tg+ versus WT mice have not yet been conducted.
Exosomes are nano-sized extracellular vesicles (30–200 nm) released by almost all cell types and play critical roles in intercellular communications (Gurung et al., 2021; McVey et al., 2019; Théry et al., 2002). Exosomes are known to regulate homeostatic as well as pathological processes in various organ systems (Théry et al., 2002; Kalluri and LeBleu, 2020; Pathan et al., 2018; Holtzman and Lee, 2020). Multi-omics approaches including transcriptomics, metabolomics, and proteomics, have significantly enhanced our understanding of the exosome contents and their biological relevance during pathophysiological processes (Shaba et al., 2022; Chitoiu et al., 2020). Increasing evidence suggests that bronchoalveolar lavage fluid (BALF) exosomes contribute to inflammatory outcomes in various lung diseases, including asthma, COPD, CF, and PCD (Torregrosa Paredes et al., 2012; Rollet-Cohen et al., 2018; Kaur et al., 2021). Additionally, BALF-derived exosomal miRNAs from COPD patients showed unique miRNA signatures as compared to the healthy subjects, indicating their potential use as biomarkers to track the disease progression (Kaur et al., 2021). Recent studies indicate that exosomal proteomic analyses can be used to gain insights into lung diseases (Rollet-Cohen et al., 2018; Choudhary et al., 2021b; Velázquez-Enríquez et al., 2021). Our recent study revealed that exosome-bound protein signatures from ozone-stressed airspaces reflect the molecular processes involved in mucoinflammatory lung disease (Choudhary et al., 2021b).
In this study, to gain insights into the proteomic differences and their association with the pathological endpoints, we compared exosome-bound protein signatures between Scnn1b-Tg+ and WT adult mice. We hypothesized that alterations in the composition of exosome-bound protein signatures in Scnn1b-Tg+ mice are suggestive of mucoinflammatory lung disease. To test this hypothesis, exosomes harvested from the BALF of adult WT and Scnn1b-Tg+ mice were subjected to liquid chromatography-mass spectrometry (LC-MS/MS) followed by bioinformatic analyses. The results from this study provide critical insights into the relevance of exosome-bound protein signatures in the pathological manifestation of mucoinflammatory lung disease in this transgenic mouse model and suggest their potential application as diagnostic and predictive biomarkers for mucoinflammatory diseases.
2 Materials and methods
2.1 Animal husbandry
Male Scnn1b-Tg+ mice strain was procured from the Jackson Laboratory [Stock Number, 006438; Strain name, B6.Cg-Tg(Scgb1a1-Scnn1b)6608Bouc/J, Bar Harbor, ME]. All mice used in this study have a background of C57BL/6. The age of the mice were 6–7 weeks old. Mice were housed in individually ventilated, hot-washed cages on a 12-h light/12-h dark cycle at the Laboratory Animal Resources (LAR) facility at the College of Veterinary Medicine at North Carolina State University (NCSU). Animals were provided food and water ad libitum. All animal experiments were performed per NCSU Institutional Animal Care and Use Committee (IACUC) approval.
2.2 Exosome collection from bronchoalveolar lavage fluid (BALF)
BALF exosomes were collected by differential ultracentrifugation following a previously reported procedure (Choudhary et al., 2021b). Each exosome sample represents exosomes collected from BALF harvested from a total of three adult male mice. All the centrifugation steps were carried out at 4°C to prevent protein degradation (Figure 1A). The particle size distribution of exosomes was evaluated by a Nanoparticle tracking analyzer (ZetaView-version 8.05.16 SP3 software, Particle Metrix GmbH, Germany).
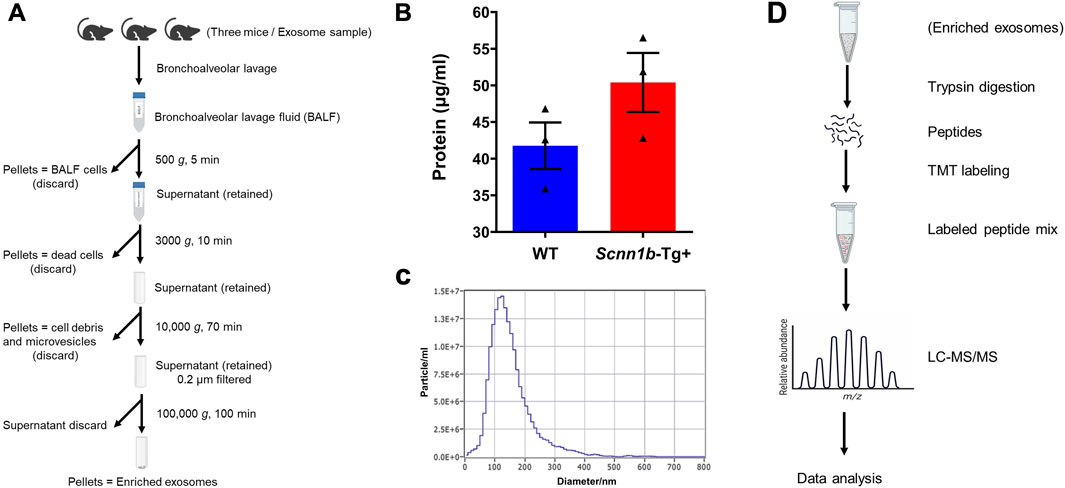
Figure 1. Outline of exosome isolation, characterization, and analysis approach to elucidate exosomal protein enrichment. (A) Schematic diagram of exosome isolation using differential ultracentrifugation. The diagram was created via www.biorender.com. (B) Total protein contents (μg/ml) in the BALF exosomes harvested from Scnn1b-Tg+ and WT mice (N = 3 for each genotype). Statistical analysis was performed by unpaired t-test. (C) The representative particle size distribution of exosomes was evaluated by a Nanoparticle tracking analyzer. The exosome samples collected in this study had an average diameter of 160.30 ± 4.85 nm. Error bars represent Mean ± Standard Error of the Mean (SEM). N = 3 for each genotype. (D) Flow diagram involved in exosome processing for proteomics data analyses. The diagram was created via www.biorender.com.
2.3 Sample preparation and data acquisition
Proteomic and data analyses were carried out by mass spectrometry (MS)-based quantitative proteomics as described previously (Choudhary et al., 2021b). Briefly, total protein from each sample was subjected to reduction, alkylation, and digestion using single-pot, solid-phase-enhanced sample preparation (SP3) (Hughes et al., 2019) with sequencing grade modified porcine trypsin (Promega, Madison, WI). Tryptic peptides were subsequently separated on an in-line 150 × 0.075 mm column filled with reverse phase XSelect CSH C18 2.5 µm resin (Waters Corporation, Milford, MA) using a Thermo UltiMate 3,000 RSLCnano system (Thermo Fisher Scientific, Waltham, MA). Peptides were eluted using a 60 min gradient from 98:2 to 65:35 buffer A (0.1% formic acid, 0.5% acetonitrile): B (0.1% formic acid, 99.9% acetonitrile) ratio. Eluted peptides were ionized by electrospray at 2.2 kV and subsequently analyzed using an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific, Waltham, MA). To construct a chromatogram library (Searle et al., 2018), we collected six gas-phase fractions on the Orbitrap Exploris using 4 m/z data-independent acquisition (DIA) spectra. Each spectrum employed the 4 m/z precursor isolation windows at 30,000 resolutions (normalized AGC target of 100% and maximum inject time of 66 m). After each DIA duty cycle, precursor spectra were obtained across the entire m/z range of the gas-phase fraction. For wide-window acquisitions, Orbitrap Exploris was set up to capture a precursor scan followed by 50 × 12 m/z DIA spectra.
2.4 Data analysis
Proteins were searched against the UniProt Mus musculus database (February 2024) using Spectronaut (Biognosys version 18.6) and employing the directDIA method. The data was acquired with quantity at the MS2 level, cross-run normalization to false, and protein grouping quantification based on median peptide and precursor abundance. Protein MS2 intensity values were evaluated for quality using ProteiNorm (Graw et al., 2020). The data was normalized using Cyclic Loess (Ritchie et al., 2015) and subjected to analysis using proteoDA (Thurman et al., 2023). Statistical analysis was performed by applying Linear Models for Microarray Data (limma) with empirical Bayes (eBayes) smoothing to the standard errors (Ritchie et al., 2015). Proteins meeting the criteria of an adjusted p-value < 0.05 and a Log2FC > 1 were considered statistically significant.
2.5 Biological pathway and protein interaction network analyses
Canonical pathways and biological networks were analyzed by Ingenuity Pathway Analysis (IPA) software (Qiagen, Redwood City, CA). The cutoff criteria (Log2FC > 1; adjusted p-value < 0.05) was used to screen proteins for analyses. Accordingly, differentially enriched proteins were uploaded into IPA software to analyze the enrichment of canonical pathways. The proteins with high abundance and low abundance were uploaded into IPA software to analyze their contribution to the enrichment of diseases and functions. The protein-protein interaction (PPI) network was analyzed by STRING (https://string-db.org), which is a database containing previously known and predicted interactions between proteins based on the provided input information (Szklarczyk et al., 2019). We used it to construct the PPI networks for differently enriched exosomal proteins with an interaction score greater than 0.4 or 0.7 as the cutoff value. The Cytoscape is a software for the network visualization (Kohl et al., 2011). Hub proteins in the network were identified by using the CytoHubba plugin of Cytoscape (Chin et al., 2014). Node degrees were ranked by score value.
2.6 Immunohistochemical localization of selected exosomal proteins
Left lung lobes were harvested from a separate cohort of mice and fixed in 10% neutral-buffered formalin. The formalin-fixed lung lobes were paraffin-embedded, cut into 5-μm sections, and placed on glass slides. Sections were subjected to immunohistochemical staining, as previously described (Choudhary et al., 2021b; Choudhary et al., 2021c; Lewis et al., 2020c). Rabbit polyclonal resistin-like alpha (RETNLA/FIZZ1) primary antibody (ab39626; 1:1,000 dilution; Abcam, Cambridge, MA), rabbit monoclonal S100 calcium-binding protein A8 (S100A8/MRP8) primary antibody (ab92331; 1:1,000 dilution; Abcam), rabbit monoclonal chitinase-3-like protein 3 (CHIL3/YM1) primary antibody (ab230610; 1:50,000 dilution; Abcam), and rabbit monoclonal galectin-3 [lectin, galactoside-binding, soluble, 3 (LGALS3)] primary antibody (ab76245; 1:1,000 dilution; Abcam) were used to assess the immunohistochemical localization of the corresponding antigens, respectively. The stained proteins were visualized using the ImmPACT NovaRED Substrate Kit (SK-4805; Vector Laboratories, Burlingame, CA). The immunostained slides were examined and photographed under the 4Χ or 40Χ objective of a light microscope with a DS-L3 digital camera (Eclipse Ci-L, Nikon Corporation, Tokyo, Japan).
2.7 Statistical analyses
Statistical analyses were performed by unpaired t-tests between groups. All data were presented as Mean ± Standard Error of the Mean (SEM). A p-value of less than 0.05 was considered statistically significant.
3 Results
3.1 Identification of exosome-bound proteins enriched in Scnn1b-Tg+ airspaces
Exosomes were isolated from cell-free BALF using a differential centrifugation approach (Figure 1A). As shown in Figure 1B, the total protein concentrations in the exosomes from Scnn1b-Tg+ male mice (50.40 ± 4.03 μg/ml) trended higher as compared to WT male mice (41.77 ± 3.17 μg/ml), but the differences were not statistically significant. The average exosome size and concentration from male mice were 160.30 ± 4.85 nm and 4.0 × 1010 particle/ml, respectively (Figure 1C; Supplementary Figure S1). The differences of size and concentration were not statistically significant between Scnn1b-Tg+ and WT male mice (Supplementary Figure S1). The isolated exosomes were subjected to proteomics data generation and bioinformatic analysis (Figure 1D). Next, we retrieved a list of 1,222 exosome-specific protein signatures from the Exocarta Vesiclepedia database (Keerthikumar et al., 2016; Simpson et al., 2012). The comparative analysis between the Exocarta Vesiclepedia database and identified proteins from this study revealed that 742 out of 1,222 exosome-specific protein signatures were detected in BALF exosomes obtained in this study (Figure 2A; Supplementary Table S1). Out of the 742 exosome-specific protein signatures, 732 protein signatures were detected in BALF from both Scnn1b-Tg+ and WT mice, and 10 protein signatures were exclusively enriched in BALF from either Scnn1b-Tg+ or WT mice (Supplementary Table S1). Collectively, our data indicate that the exosome-specific protein signatures differ between Scnn1b-Tg+ and WT mice BALF, indicating the potential for the identification of novel exosome-based biomarkers for mucoinflammatory diseases.
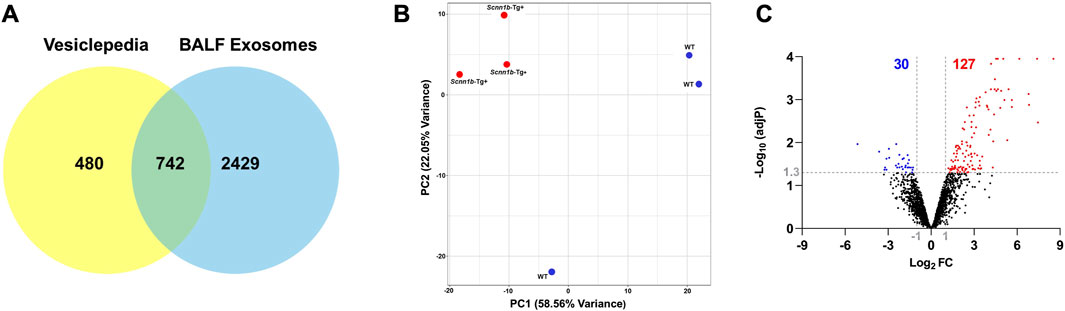
Figure 2. Differential enrichment of BALF exosome-bound proteins in Scnn1b-Tg+ versus WT mice. (A) Venn diagram depicts that 742 out of 1,222 exosome-specific markers obtained from the Vesiclepedia database were identified in exosomes harvested in this study. (B) Two-dimensional principal component (PC) analysis plot using PC1 and PC2 on differentially enriched proteins (after normalization) in exosomes from the Scnn1b-Tg+ and WT mice. (C) Volcano plots representing differentially abundant proteins in Scnn1b-Tg+ versus WT mice (Log2FC > 1; adjusted p-value < 0.05).
3.2 Differential enrichment of BALF exosome proteins in Scnn1b-Tg+ versus WT mice
The top two principal components (PC1 and PC2), which contribute to ∼80% variance, revealed that genotypic difference is the primary driver of differential protein enrichment in the Scnn1b-Tg+ versus WT mice (Figure 2B). Next, using cutoff criteria, i.e., Log2FC > 1 and adjusted p-value < 0.05, we identified differentially enriched protein signatures in BALF exosomes harvested from Scnn1b-Tg+ and WT mice. A total of 127 and 30 proteins were upregulated and downregulated, respectively, in Scnn1b-Tg+ versus WT mice (Figure 2C). The protein signatures enriched in Scnn1b-Tg+ mice BALF are listed in Table 1 (top 50 proteins) and Supplementary Table S2 (entire list). A list of 30 proteins with low abundance in exosomes from Scnn1b-Tg+ mice BALF is included in Table 2. Additionally, 52 and 27 proteins were exclusively expressed, respectively, in BALF from Scnn1b-Tg+ and WT mice (Supplementary Table S3).
3.3 Macrophage activation-associated proteins are differently enriched in BALF exosome of Scnn1b-Tg+ versus WT mice
Through a manual literature search, we prepared a list of protein signatures associated with macrophage activation and compared their levels in the exosomes from WT and Scnn1b-Tg+ mice BALF. The classically-activated macrophages CAM; M1-associated signatures, i.e., myristoylated alanine-rich C-kinase substrate (MARCKS), toll-like receptor 2 (TLR2), low-affinity immunoglobulin gamma Fc region receptor II (FCGR2), lipocalin 2 (LCN2), platelet-activating factor acetylhydrolase (PLA2G7), and FCGR3, were upregulated in BALF of Scnn1b-Tg+ mice as compared to WT mice (Figure 3A). The alternatively-activated macrophages AAM; M2 associated signatures, i.e., RETNLA/FIZZ1, TREM2, CHIL3/YM1, CHIL4, and LGALS3/Galectin-3 were upregulated in BALF from Scnn1b-Tg+ mice compared to WT mice (Figure 3A). Macrophage activation protein signatures, i.e., C-type mannose receptor 2 (MRC2) and prostaglandin G/H synthase 1 (PTGS1), were downregulated in BALF from Scnn1b-Tg+ mice compared to WT mice (Figure 3A).
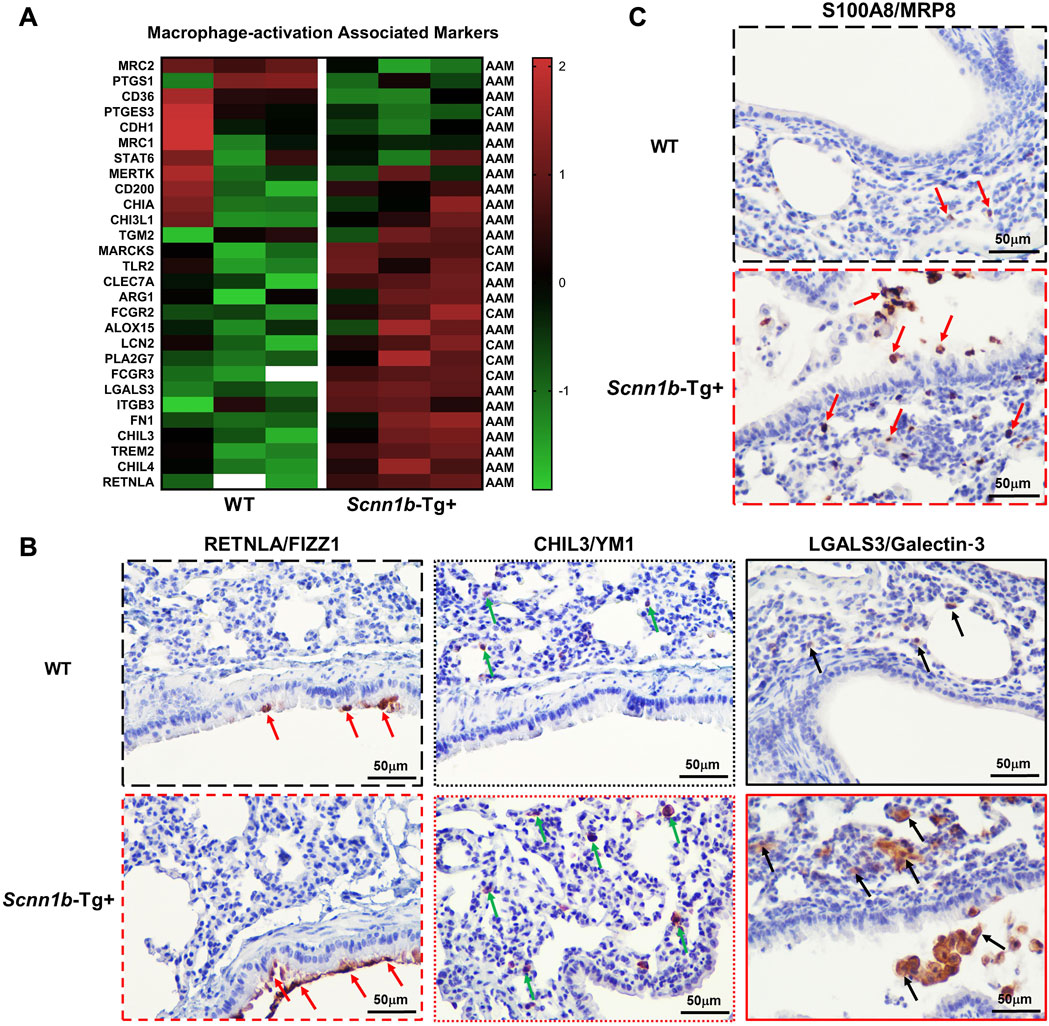
Figure 3. Exosome-bound proteins associated with activated macrophages and neutrophils in Scnn1b-Tg+ mice. Heat maps for normalized protein abundance values (Z-scores) of exosome-bound proteins associated with activated macrophages. (A) Classically activated macrophages (M1; CAM)- and alternatively activated macrophages (M2; AAM)-specific protein signatures were differently enriched in BALF exosome of Scnn1b-Tg+ and WT mice. Higher and lower expressions of each protein are denoted by red and green colors, respectively. The white blanks indicate the levels below the limit of detection. (B) Immunohistochemical staining of lung sections for cell-specific expression of AAM activation-associated signatures, FIZZ1 (RETNLA), YM1 (CHIL3), and Galectin-3 (LGALS3). Red arrows depict the FIZZ1-stained epithelial cells. Green arrows depict the YM1 staining in alveolar macrophages. Black arrows depict the Galectin-3-stained macrophages. (C) Immunohistochemical staining of lung sections for neutrophil-specific expression of MRP8 (S100A8). Red arrows point to the MRP8-stained neutrophils.
Next, to identify the cellular source of RETNLA/FIZZ1, CHIL3/YM1, and Galectin-3/LGALS3 proteins in exosomes, we conducted immunohistochemical staining on lung sections from Scnn1b-Tg+ and WT mice. RETNLA/FIZZ1 immunolocalization was restricted to the airway epithelial cells of Scnn1b-Tg+ mice (Figure 3B; Supplementary Figure S2). The intensity of CHIL3/YM1 and LGALS3/Galectin-3 were greater in the alveolar macrophages of Scnn1b-Tg+ mice versus WT mice (Figure 3B; Supplementary Figure S2). Myeloid-related protein 8 (MRP8/S100A8), is a neutrophil- and monocyte-specific protein (Frosch et al., 2000; Ryckman et al., 2003). To determine the cellular source of MRP8 in airspaces-derived exosomes, we performed immunohistochemical staining on lungs from Scnn1b-Tg+ and WT mice (Figure 3C; Supplementary Figure S2). The MRP8-stained cells in Scnn1b-Tg+ mice were exclusively neutrophils (Figure 3C; Supplementary Figure S2).
3.4 Mucoinflammation and mucous cell metaplasia (MCM) signatures are differently enriched in BALF exosome of Scnn1b-Tg+ versus WT mice
We prepared a list of signatures known to be associated with mucoinflammatory lung diseases in mice and humans (Choudhary et al., 2021b; Choudhary et al., 2021d; Saini et al., 2014) and compared their levels in the airspace-derived exosomes from WT and Scnn1b-Tg+ mice. Our analyses revealed significant enrichment of the mucoinflammatory signatures, i.e., S100A9, cathelicidin antimicrobial peptide (CAMP), S100A8, peptidoglycan recognition protein 1 (PGLYRP1), lactotransferrin (LTF), BPI fold-containing family B member 1 (BPIFB1), lysosomal acid lipase/cholesteryl ester hydrolase (LIPA), apolipoprotein B-100 (APOB), apolipoprotein C-III (APOC3), cluster of differentiation 14 (CD14), CD5 antigen-like (CD5L), cathepsin D (CTSD), and LGALS3, in BALF exosome of Scnn1b-Tg+ versus WT mice (Figures 4A, B; Supplementary Figure S3; Supplementary Table S4). Although non-significantly, some mucoinflammatory protein signatures, i.e., high mobility group protein B1 (HMGB1), complement factor D (CFD), plasminogen, orosomucoid-1 (ORM1), alpha-1-microglobulin/bikunin precursor (AMBP), and glutathione peroxidase 3 (GPX3), were relatively less abundant, in BALF exosome of Scnn1b-Tg+ mice versus WT mice (Figures 4A, B; Supplementary Figure S1; Supplementary Table S4). MCM is a consistent feature of Scnn1b-Tg+ lung disease (Mall et al., 2004; Mall et al., 2008; Lewis et al., 2020c). The MCM-associated protein signatures including, secretoglobin family 1A member 1 (SCGB1A1) and calcium-activated chloride channel regulator 1 (CLCA1), were significantly enriched in BALF exosome of Scnn1b-Tg+ mice as compared to WT mice (Figure 4C).
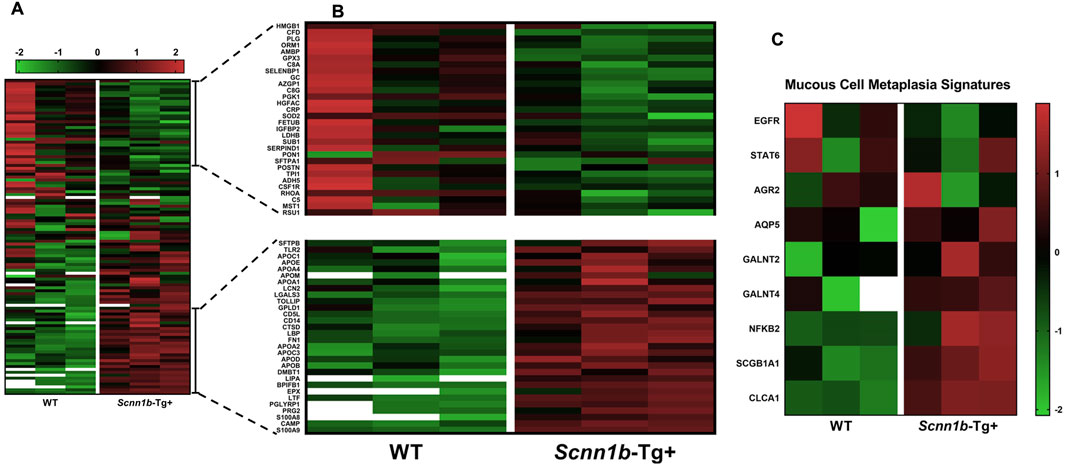
Figure 4. Heat maps for normalized protein abundance values (Z-scores) of exosome-bound proteins relevant to the mucoinflammatory disturbances in mice and humans. (A) Low-resolution heat map of exosome-bound protein signatures associated with mucoinflammatory disease in mice and humans (A high-resolution heat map with protein names is provided as Supplementary Figure S3). The heat map representing low abundance [(B); top panel] or enriched [(B); bottom panel] protein signatures in BALF exosomes from Scnn1b-Tg+ mice. (C) Heat map for exosome-bound protein signatures associated with mucous cell metaplasia. The white blanks indicate the levels below the limit of detection.
3.5 Pathway analysis on BALF exosome-bound proteins
To identify canonical pathways that are enriched due to the abundance of exosome-bound proteins harvested in this study, we employed ingenuity pathways analysis (IPA) approach (cutoff criteria-Log2FC > 1 and adjusted p-value < 0.05). Our analyses revealed that enriched proteins in BALF exosomes of Scnn1b-Tg+ were associated with the upregulation of pathways including neutrophil degranulation, antimicrobial peptides, endogenous ligand-mediated TLR regulation, neutrophil extracellular trap signaling pathway, and Fc-gamma receptor (FCGR) dependent phagocytosis (Figure 5A).
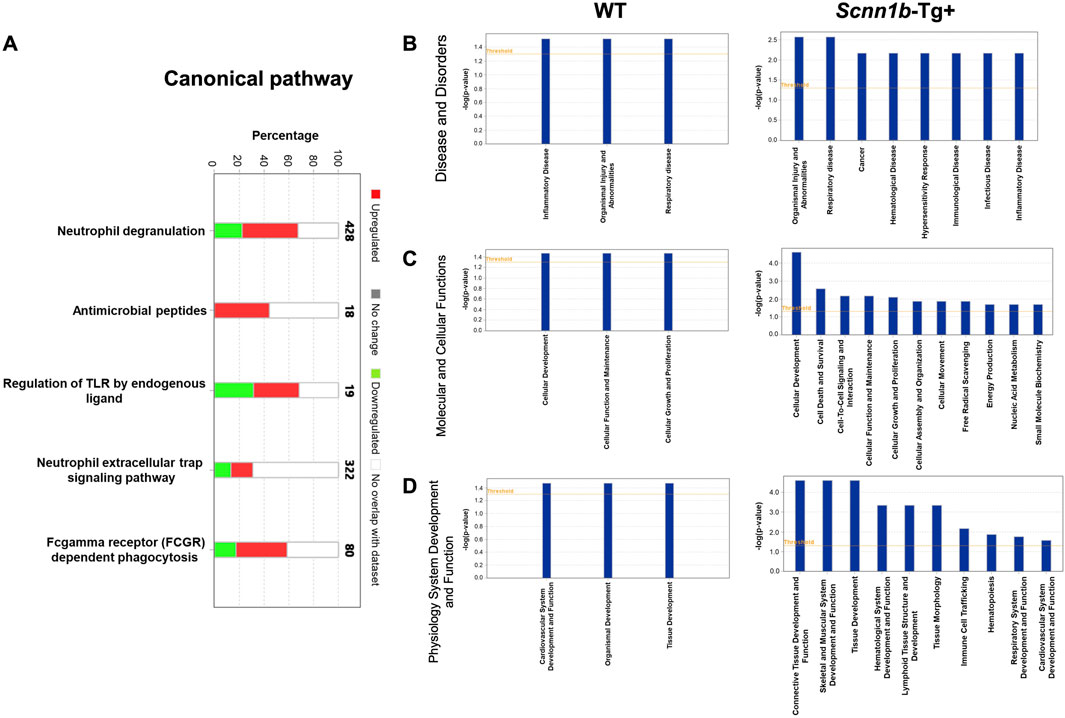
Figure 5. Ingenuity Pathway (IP) analyses of BALF exosome-bound proteins enriched in Scnn1b-Tg+ and WT mice. (A) IP analysis for the enrichment of canonical pathways influenced by the significantly enriched proteins in Scnn1b-Tg+ versus WT mice (Log2FC > 1; adjusted p-value < 0.05). Stacked bar graph indicates the proportions of total enriched (red) and low abundance (green) proteins associated with a particular pathway. IP analysis for the enrichment of disease and disorders (B) Molecular and cellular functions (C), and physiology system development and function (D) influenced by the abundant proteins in BALF exosomes from Scnn1b-Tg+ and WT mice.
The top diseases and biological functions influenced by the enriched proteins in BALF exosome of Scnn1b-Tg+ mice included organismal injury and abnormalities, inflammatory disease, cancer, hematological disease, hypersensitivity response, immunological disease, infectious disease, and respiratory disease (Figure 5B). The top molecular and cellular functions influenced by the enriched proteins in BALF exosome of Scnn1b-Tg+ mice included cellular development, cell death and survival, cell-to-cell signaling and interaction, cellular function and maintenance, cellular growth and proliferation, cellular assembly and organization, cellular movement, free radical scavenging, energy production, nucleic acid metabolism, and small molecule biochemistry. In WT mice, the enriched proteins were relevant to cellular development, function, maintenance, growth, and proliferation (Figure 5C). The top physiology systems development and functions associated with the enriched exosomal proteins in Scnn1b-Tg+ mice included connective tissue development and function, skeletal and muscular system development and function, tissue development, hematological system development and function, lymphoid tissue structure and development, tissue morphology, immune cell trafficking, hematopoiesis, respiratory system development and function, and cardiovascular system development and function (Figure 5D).
3.6 Protein-protein interaction network analysis on BALF exosome-bound proteins
To identify the protein-protein interaction network, we performed STRING analyses on 179 proteins that were abundant in BALF exosomes from Scnn1b-Tg+ mice. These proteins included the 127 significantly enriched proteins in BALF from Scnn1b-Tg+ versus WT mice (Log2FC > 1; adjusted p-value < 0.05), and 52 proteins that were exclusively found in BALF from Scnn1b-Tg+ mice. The significantly influenced protein-protein interaction networks due to the enriched protein signatures in the BALF exosomes of Scnn1b-Tg+ versus WT mice included neutrophil degranulation, immune system, innate immune system, and antimicrobial peptides (Figure 6A). Furthermore, the top 10 proteins, i.e., LTF, myeloperoxidase (MPO), TLR4, integrin subunit alpha M (ITGAM), CAMP, matrix metalloproteinase 9 (MMP9), proteinase 3 (PRTN3), neutrophil elastase (ELANE), cathepsin G (CTSG), and S100 calcium-binding protein A9 (S100A9), were involved in network ranked by degree method using Cytoscape-CytoHubba plugin (Figure 6B). To further explore the functional relevance of 30 protein signatures, which were more abundant in BALF from WT mice, and 27 protein signatures, which were exclusively present in BALF from WT mice, STRING database protein-protein interaction network analyses were conducted. The output of STRING analysis indicated that the top networks were involved in the endoplasmic reticulum, i.e., endomembrane system, endoplasmic reticulum, and endoplasmic reticulum membrane (Figure 6C).
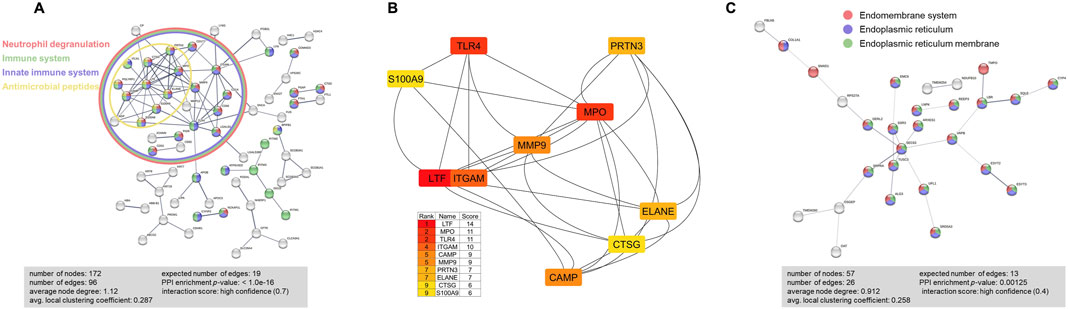
Figure 6. STRING database protein-protein interaction network analyses were performed on the abundant BALF exosome proteins from Scnn1b-Tg+ versus WT mice. (A) A total of 179 (127 enriched proteins and 52 exclusively abundant in Scnn1b-Tg+ mice) protein signatures were used for this analysis. The protein-protein interactions were determined based on evidence, using the high confidence level (0.7) setting. 172 nodes and 96 edges were identified. Disconnected nodes were selected to be hidden. PPI enrichment p-value < 1.0e-16. Proteins involved in Neutrophil degranulation (Counts = 36/526; FDR adjusted p-value < 1.57e-19), Immune system (Counts = 49/1,615; FDR adjusted p-value < 9.34e-14), Innate immune system (Counts = 44/945; FDR adjusted p-value < 1.44e-18), and antimicrobial peptides (Counts = 10/84; FDR adjusted p-value < 1.41e-06) were enriched. (B) Top 10 hub proteins in the network ranked by score value using the Cytoscape-CytoHubba plugin. These proteins included lactotransferrin (LTF), myeloperoxidase (MPO), toll-like receptor 4 (TLR4), integrin subunit alpha M (ITGAM), cathelicidin antimicrobial peptide (CAMP), matrix metalloproteinase 9 (MMP9), proteinase 3 (PRTN3), neutrophil elastase (ELANE), cathepsin G (CTSG), and S100 calcium binding protein A9 (S100A9). (C) STRING database protein-protein interaction network analyses on differently expressed proteins that had high abundance and exclusively expressed (total of 57) in WT mice. The protein-protein interactions were determined based on evidence, using the high confidence level (0.4) setting. 57 nodes and 26 edges were identified. Disconnected nodes were selected to be hidden. PPI enrichment p-value < 0.00125. Proteins involved in the Endomembrane system (Counts = 32/4360; FDR adjusted p-value < 5.15e-07), Endoplasmic reticulum (Counts = 27/1926; FDR adjusted p-value < 1.66e-11), and endoplasmic reticulum membrane (Counts = 24/1,100; FDR adjusted p-value < 2.67e-13) were enriched.
3.7 Comparative analysis between enriched BALF exosome proteins and transcriptomes from Scnn1b-Tg+ versus WT mice
To investigate the cellular origin of proteins and ascertain whether the enriched exosomal proteins mirror their upregulation at the mRNA levels, we cross-referenced significantly enriched proteins from Scnn1b-Tg+ airspace with the differentially upregulated transcripts from the whole lung and alveolar macrophages (purified BALF macrophages) of Scnn1b-Tg+ versus WT mice (Saini et al., 2014). The 127 enriched exosomal proteins (Log2FC > 1 and adjusted p-value < 0.05) and the 52 exosomal proteins exclusively expressed in the airspace of Scnn1b-Tg+ mice were used for this comparative analysis. Our analyses revealed that ∼46% (31 out of 67) and ∼26% (94 out of 362) of the upregulated transcripts (FC > 2 and adjusted p-value < 0.05) from whole lung and alveolar macrophages of Scnn1b-Tg+ mice, respectively, were identified as protein signatures in the BALF exosomes of Scnn1b-Tg+ mice (Table 3). Among these, 77% (24 out of 31) and 61% (57 out of 94) of the whole lung- and alveolar macrophage-relevant proteins, respectively, were significantly enriched in BALF exosome of Scnn1b-Tg+ versus WT mice. Additionally, 9 out of the 24 and 18 out of the 57 of the whole lung- and alveolar macrophage-relevant proteins, respectively, trended higher in BALF exosome of Scnn1b-Tg+ versus WT mice (Table 3). For example, pendrin, an anion exchanger associated with the airway surface liquid homeostasis, and its mRNA, i.e., Slc26a4, were significantly abundant in BALF exosomes and overexpressed in whole lung, respectively, from the Scnn1b-Tg+ versus WT mice. Similarly, proteins including ITGAM (CD11B), CD300A, S100A8/9, and their respective mRNAs were significantly elevated in BALF exosomes from the Scnn1b-Tg+ versus WT mice. The enrichment status of 13 proteins, i.e., CD177, GP2, LIPA, PIGR, SLC26A4, CD68, CTSD, RETNLA, FBP1, GPNMB, CCL9, GLA, MMP12, was consistent with their upregulated gene expression in the whole lung (Figure 7A; Table 3). The enrichment of 25 proteins, including S100A9, S100A8, ITGAM, IFITM3, SCGB3A1, SCGB3A2, BPIFB1, ADAM8, SCGB1A1, BASP1, VNN3, MFGE8, ABCG2, MLPH, CEACAM1, CYFIP2, RETNLA, FBP1, GPNMB, CD300A, STEAP3, PHLDA3, CCL9, GLA, MMP12, mirrored their upregulated gene expression in alveolar macrophages from Scnn1b-Tg+ versus WT adult mice. (Figure 7B; Table 3). These analyses highlight the impact of the cell/compartment-specific gene expression changes on the protein composition of the BALF exosomes.
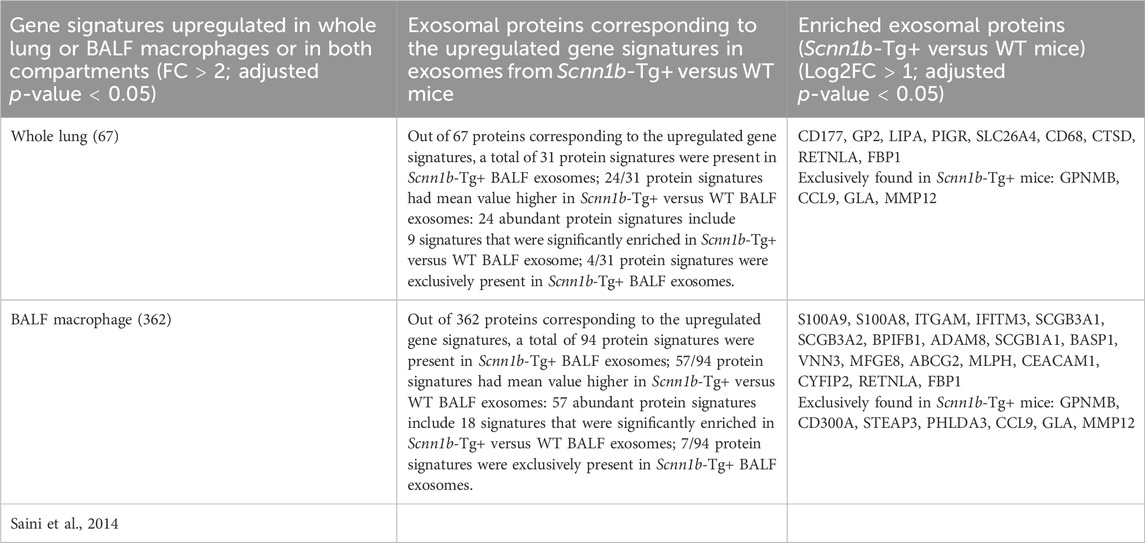
Table 3. Comparative analysis of enriched protein signatures from Scnn1b-Tg+ mice (current study) and transcriptome from whole lung and purified BALF macrophage (Saini et al., 2014).
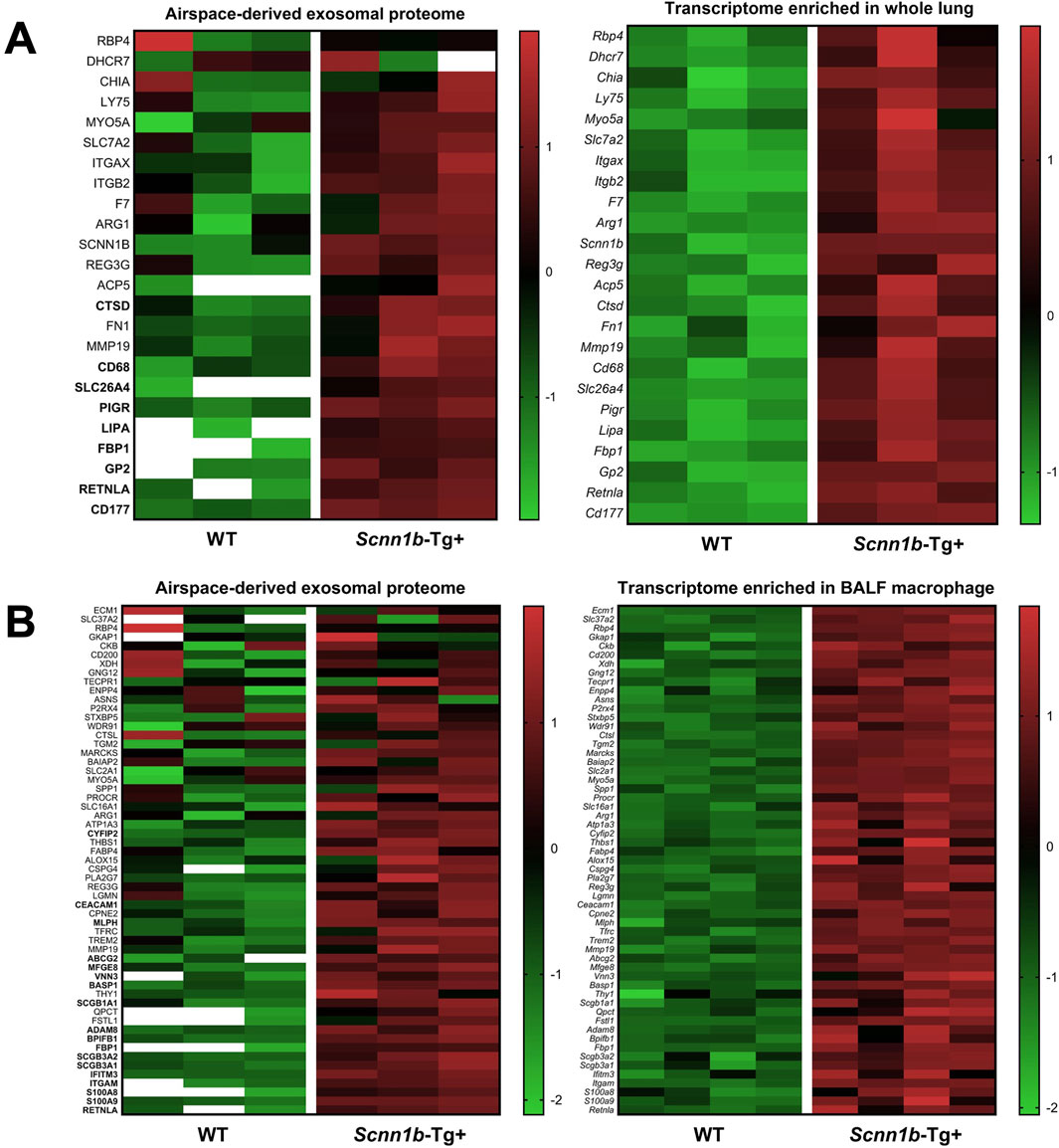
Figure 7. Comparative proteins and transcriptome analysis. Comparative analysis of protein signatures (Log2FC > 1 and adjusted p-value < 0.05) enriched in Scnn1b-Tg+ BALF exosomes and differentially upregulated genes (FC > 2 and adjusted p-value < 0.05) in whole lung (A) and purified BALF macrophage (B) of Scnn1b-Tg+ mice. Heat maps for normalized values (Z-scores) of upregulated differentially expressed genes (right panels) from the whole lung and BALF macrophage of Scnn1b-Tg+ versus WT mice and normalized values (Z-scores) of corresponding exosome proteins identified in the airspace of Scnn1b-Tg+ versus WT mice (left panels). The data with white blank in the heat maps means it is below limit of detection.
4 Discussion
Exosome-mediated intercellular communications play important roles in normal and pathological conditions through the transfer of various bioactive compounds including proteins, lipids, DNA, and RNA molecules (Kalluri and LeBleu, 2020; De Toro et al., 2015; Stoorvogel et al., 2002). Our recent proteomic-oriented report revealed interesting findings from airspaces-derived exosomes from ozone-stressed airspaces in mice (Choudhary et al., 2021b). In this study, to gain insights into the proteomic differences and their association with the pathological endpoints, we hypothesized that alterations in the composition of exosome-bound protein signatures in Scnn1b-Tg+ mice are suggestive of mucoinflammatory lung disease. To investigate this, we addressed a series of questions: (1) Do Scnn1b-Tg+ mice exhibit differential composition of proteins in the airspace exosomes as compared to WT mice? (2) What proteins are enriched in exosomes derived from Scnn1b-Tg+ airspaces? (3) Do the exosomal proteins reflect macrophage activation patterns and mucoinflammatory disturbances in the Scnn1b-Tg+ airspace? (4) Which biological pathways are influenced by exosome-bound proteins in Scnn1b-Tg+ versus WT mice?
Protein signatures found enriched in the BALF exosomes from WT mice were suggestive of normal tissue functions. For example, homeodomain-only protein homeobox (HOPX), which is required for developing mouse neocortex (Vaid et al., 2018), was enriched in WT mice. Glutathione peroxidase 8 (GPX8), which protects cytosol from H2O2 leakage in the endoplasmic reticulum (Ramming et al., 2014), and the growth factor midkine (MDK) which regulates the renin-angiotensin system and mediates the interaction between the kidneys and lungs in mice (Hobo et al., 2009), were enriched in WT mice BALF exosomes. Interestingly, several exosomal proteins that were exclusively expressed in WT mice BALF exosomes were also involved in normal biosynthesis, i.e., keratin, type I cuticular Ha2 (KRT32), a type I hair keratin, involved in exoskeleton formation (Adav et al., 2023), pleckstrin homology domain-containing, family G member 5 (PLEKHG5), a guanine-nucleotide exchange factors to activate ras homolog family member A (RhoA) (De Toledo et al., 2001; Qian et al., 2019), and antigen peptide transporter 1 (TAP1), the subunit of transporter associated with antigen processing (Gaudet and Wiley, 2001a; Gaudet and Wiley, 2001b). These data indicate that under homeostatic conditions, exosome-bound proteins in bronchoalveolar airspaces are relevant to normal physiological processes.
A large number of protein signatures relevant to inflammation, macrophage activation, and mucous cell metaplasia were found enriched in the BALF exosomes from Scnn1b-Tg+ mice. CD177, a specific marker of neutrophil activation (Lévy et al., 2021; Bayat et al., 2010), was the most abundant protein present within the exosomes from Scnn1b-Tg+ mice. Extracellular vesicle-derived ITGAM, which is involved in acute lung injury (Hu et al., 2023), was highly expressed in the exosome of Scnn1b-Tg+ versus WT mice. Neutrophil elastase (NE), a key inflammatory protease (Voynow and Shinbashi, 2021; Pham, 2006), and a known promoter of mucous cell metaplasia in the Scnn1b-Tg+ mice (Gehrig et al., 2014), Lymphocyte antigen 6 family member G (Ly6G) and MPO, two distinct neutrophil markers (Lee et al., 2013; Aratani, 2018), and CTSG, a serine protease, acting as a chemoattractant for mononuclear cells and neutrophils (Chertov et al., 1997), were exclusively expressed in BALF exosomes of Scnn1b-Tg+ mice. Moreover, MMP-9 and MMP-12, the proteolytic enzymes, were also exclusively expressed in the BALF exosome of Scnn1b-Tg+ mice. Of note, the roles of MMP-9 and MMP-12 are well-established in the pathogenesis of COPD (Molet et al., 2005; Papakonstantinou et al., 2015; Liu et al., 2020).
Our previous report indicated that an admixture of activated macrophages was present in the BALF of Scnn1b-Tg+ mice, with predominantly classically-activated (M1) macrophages in neonates and alternatively-activated (M2) macrophages in adulthood (Saini et al., 2014). Consistent with this report, M2 activation-relevant protein signatures showed a significant enrichment within the BALF exosomes of Scnn1b-Tg+ mice. CHIL3/YM1, an M2 alternative activation marker in mice (Raes et al., 2005; Raes et al., 2002), forms eosinophilic crystals in the cytoplasm of alveolar macrophages and multinucleate giant cells, and within the alveolar spaces (Guo et al., 2000). YM1/2 contributes to Th2 cytokine production and allergic airway inflammation (Cai et al., 2009). Chil3, an mRNA for CHIL3/YM1, showed significantly upregulated expression in the alveolar macrophage of Scnn1b-Tg+ mice (Saini et al., 2014). Consistent with this report, CHIL3/YM1 was enriched in the BALF exosomes from Scnn1b-Tg+ mice and the macrophage-specific immunolocalization of this protein suggests its origination from macrophages. Galectin-3 (LGALS3), a β-galactoside-binding lectin, is also expressed by alternatively activated macrophages (MacKinnon et al., 2008; Nishi et al., 2007). Galectin-3 was enriched in BALF exosomes from ozone-exposed mice (Choudhary et al., 2021b). Galectin-3 was exclusively enriched in airspace-derived exosomes from Scnn1b-Tg+ mice and had macrophage-specific immunolocalization in Scnn1b-Tg+ lungs. FIZZ1 (Found in Inflammatory Zone 1/RETNLA), another well-known alternative macrophage activation marker in mice (Raes et al., 2002), was found significantly upregulated in the alveolar macrophages of Scnn1b-Tg+ lungs (Saini et al., 2014). The airway epithelial-specific immunolocalization of FIZZ1 has been reported in hypoxia-exposed (Teng et al., 2003) and ozone-exposed mice (Choudhary et al., 2021b). Consistent with these reports, FIZZ1 immunostaining was localized to the airway epithelium of Scnn1b-Tg+ mice. These reports suggest that exosome-bound FIZZ1 originates from airway epithelial cells in Scnn1b-Tg+ airspace. While we were able to identify the cellular sources of exosomes-bound protein signatures, it remains unclear whether the exosomes continue to exchange their cargo after their extracellular release.
Mucoinflammatory lung diseases are characterized by intrapulmonary accumulations of hyper-concentrated mucus, which contributes to the vulnerability to recurrent infections (Boucher, 2019; Livraghi-Butrico et al., 2017). To investigate the BALF exosome-bound protein signatures potentially relevant to the development of lung mucoinflammation, we compared the presence of protein signatures associated with mucoinflammatory lung diseases in the BALF exosomes of Scnn1b-Tg+ mice versus WT mice. Numerous proteins previously shown to be involved in mucoinflammatory lung diseases were enriched in the exosomes from Scnn1b-Tg+ mice, including S100A9, CAMP, S100A8, PGLYRP1, LTF, BPIFB1, LIPA, APOB, APOC3, CD5L, CD14, CTSD, and LGALS3. Scnn1b-Tg+ mice present with neutrophilic inflammation in airspace (Mall et al., 2004) and enriched S100A8 was found in airspaces-derived exosomes of ozone-challenged mice (Choudhary et al., 2021b). These findings were consistent with the increased S100A8 expression in BALF exosome and neutrophils of Scnn1b-Tg+ mice (Figure 3C; Figures 4A, B). Similarly, the upregulation of protein signatures relevant to MCM, i.e., SCGB1A1 and CLCA1, was consistent with the previous study that Scnn1b-Tg+ mice have increased numbers of mucous cells in airways versus their WT littermates (Mall et al., 2008).
BALF exosome-bound proteins from stressed airways might contribute to the functional and disease-associated pathways (Choudhary et al., 2021b). Ingenuity pathways analysis (IPA) on the differentially expressed proteins within the exosomes gathered in this study revealed activation of several canonical pathways including neutrophil degranulation, antimicrobial peptides, regulation of TLR by endogenous ligand, neutrophil extracellular trap signaling pathway, and Fc-gamma receptor-dependent phagocytosis. Additionally, enriched protein-protein interaction (PPI) networks from airspace-derived exosomes from BALF of Scnn1b-Tg+ mice revealed the major subsets including neutrophil degranulation, immune system, innate immune system, and antimicrobial peptides. However, the PPI analyses from protein signatures upregulated and exclusively expressed in WT mice presented endoplasmic reticulum-associated networks including the endomembrane system, endoplasmic reticulum, and endoplasmic reticulum membrane, which might potentially help maintain the homeostatic conditions of biosynthesis.
Increasing research on exosomes have showed its potential in translational medicine and have provided novel insights for the clinical diagnostics and therapeutic strategies as biomarkers, vaccines, and drug carriers (Khan et al., 2015; Alvarez-Erviti et al., 2011; Tan et al., 2014; Li et al., 2013). This is the first study to provide a detailed proteomics analysis of exosomes from Scnn1b-Tg+ airspaces and our findings revealed that BALF exosomes-bound protein signatures reflect the muco-obstructive lung disease-relevant disturbances. These protein signatures could be used for diagnostic or prognostic monitoring of patients with muco-obstructive lung disease, as well as to design novel exosome-based therapies. There are also limitations to this study. First, this study characterized proteomic signatures in male mice only. Although the size and concentration of the BALF exosome in Scnn1b-Tg+ male and Scnn1b-Tg+ female animals were comparable, the sex-specific differential enrichment of exosomal proteins cannot be ruled out. Second, we were unable to perform functional studies to determine the biological influence of exosome-derived proteins in cell-based or animal models.
Collectively, our data provides a detailed proteomics analysis of the exosomes from Scnn1b-Tg+ and WT mice airspaces. With the immunolocalization approach, we were able to identify cellular sources of some of the disease-relevant proteins. The comparative analysis between enriched BALF exosome proteins and upregulated gene transcriptomes in Scnn1b-Tg+ versus WT mice revealed the relevance of the mRNA level changes and their impact on the protein signatures in the airspaces. We believe this study will create a foundation for future mechanistic studies aiming at understanding the pathogenesis of mucoinflammatory lung diseases and facilitating the identification of novel molecular and cellular processes for the eventual therapeutic targeting during the progression of pulmonary diseases. This study also suggests that the exosomes from BALF carry disease-relevant protein signatures that can be used as diagnostic predictive biomarkers for mucoinflammatory diseases.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Our raw data is available in MassIVE database (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000095568). Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by NCSU Institutional Animal Care and Use Committee (IACUC). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing–original draft, Writing–review and editing. AS: Investigation, Methodology, Writing–review and editing. SP: Conceptualization, Formal Analysis, Investigation, Resources, Writing–review and editing. YS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Financial Support: The work was supported by NIH R01 (R01ES030125) and R21 (R21ES034509) grants.
Acknowledgments
We thank IDeA National Resource for Quantitative Proteomics (NIH/NIGMS grant R24GM137786) for proteomic and data analyses. We thank Thao Vo for assistance with bioinformatic analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1460692/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Concentration (A) and size (B) of isolated exosome. Each exosome sample represents exosomes collected from BALF harvested from a total of one, two, or three adult (6–7 weeks old) mice. Each mouse has the same volume in all the exosome samples. Error bars represent Mean ± Standard Error of the Mean (SEM).
SUPPLEMENTARY FIGURE S2 | Low resolution of immunohistochemical staining for panel B and C of Figure 3. The broad area of the lung section images for immunohistochemical staining of RETNLA/FIZZ1 (A), CHIL3/YM1 (B), LGALS3/Galectin-3 (C), and S100A8/MRP8 (D) were captured at 4× objective.
SUPPLEMENTARY FIGURE S3 | High resolution heat maps for panel A of Figure 4 depicting normalized protein abundance values (Z-scores) of exosome-bound proteins associated with mucoinflammatory disease (mice and humans) and mucous cell metaplasia. The white blanks indicate the levels below the limit of detection.
References
Adav, S. S., Leung, C. Y., and Ng, K. W. (2023). Profiling of hair proteome revealed individual demographics. Forensic Sci. Int. Genet. 66, 102914. doi:10.1016/j.fsigen.2023.102914
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. A. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29 (4), 341–345. doi:10.1038/nbt.1807
Aratani, Y. (2018). Myeloperoxidase: its role for host defense, inflammation, and neutrophil function. Archives Biochem. biophysics 640, 47–52. doi:10.1016/j.abb.2018.01.004
Bayat, B., Werth, S., Sachs, U. J. H., Newman, D. K., Newman, P. J., and Santoso, S. (2010). Neutrophil transmigration mediated by the neutrophil-specific antigen CD177 is influenced by the endothelial S536N dimorphism of platelet endothelial cell adhesion molecule-1. J. Immunol. 184 (7), 3889–3896. doi:10.4049/jimmunol.0903136
Boucher, R. C. (2019). Muco-obstructive lung diseases. N. Engl. J. Med. 380 (20), 1941–1953. doi:10.1056/NEJMra1813799
Burgel, P.-R., Nesme-Meyer, P., Chanez, P., Caillaud, D., Carré, P., Perez, T., et al. (2009). Cough and sputum production are associated with frequent exacerbations and hospitalizations in COPD subjects. Chest 135 (4), 975–982. doi:10.1378/chest.08-2062
Cai, Y., Kumar, R. K., Zhou, J., Foster, P. S., and Webb, D. C. (2009). Ym1/2 promotes Th2 cytokine expression by inhibiting 12/15 (S)-lipoxygenase: identification of a novel pathway for regulating allergic inflammation. J. Immunol. 182 (9), 5393–5399. doi:10.4049/jimmunol.0803874
Chertov, O., Ueda, H., Xu, L. L., Tani, K., Murphy, W. J., Wang, J. M., et al. (1997). Identification of human neutrophil-derived cathepsin G and azurocidin/CAP37 as chemoattractants for mononuclear cells and neutrophils. J. Exp. Med. 186 (5), 739–747. doi:10.1084/jem.186.5.739
Chin, C.-H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11–S17. doi:10.1186/1752-0509-8-S4-S11
Chitoiu, L., Dobranici, A., Gherghiceanu, M., Dinescu, S., and Costache, M. (2020). Multi-omics data integration in extracellular vesicle biology—utopia or future reality? Int. J. Mol. Sci. 21 (22), 8550. doi:10.3390/ijms21228550
Choudhary, I., Vo, T., Paudel, K., Patial, S., and Saini, Y. (2021d). Compartment-specific transcriptomics of ozone-exposed murine lungs reveals sex- and cell type-associated perturbations relevant to mucoinflammatory lung diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 320 (1), L99–l125. doi:10.1152/ajplung.00381.2020
Choudhary, I., Vo, T., Paudel, K., Wen, X., Gupta, R., Kesimer, M., et al. (2021b). Vesicular and extravesicular protein analyses from the airspaces of ozone-exposed mice revealed signatures associated with mucoinflammatory lung disease. Sci. Rep. 11 (1), 23203. doi:10.1038/s41598-021-02256-5
Choudhary, I., Vo, T., Paudel, K., Yadav, R., Mao, Y., Patial, S., et al. (2021a). Postnatal ozone exposure disrupts alveolar development, exaggerates mucoinflammatory responses, and suppresses bacterial clearance in developing Scnn1b-Tg+ mice lungs. J. Immunol. 207 (4), 1165–1179. doi:10.4049/jimmunol.2001286
Choudhary, I., Vo, T., Paudel, K., Yadav, R., Mao, Y., Patial, S., et al. (2021c). Postnatal ozone exposure disrupts alveolar development, exaggerates mucoinflammatory responses, and suppresses bacterial clearance in developing scnn1b-tg(+) mice lungs. J. Immunol. 207 (4), 1165–1179. doi:10.4049/jimmunol.2001286
De Toledo, M., Coulon, V., Schmidt, S., Fort, P., and Blangy, A. (2001). The gene for a new brain specific RhoA exchange factor maps to the highly unstable chromosomal region 1p36. 2–1p36. 3. Oncogene 20 (50), 7307–7317. doi:10.1038/sj.onc.1204921
De Toro, J., Herschlik, L., Waldner, C., and Mongini, C. (2015). Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front. Immunol. 6, 203. doi:10.3389/fimmu.2015.00203
Frosch, M., Strey, A., Vogl, T., Wulffraat, N. M., Kuis, W., Sunderkötter, C., et al. (2000). Myeloid-related proteins 8 and 14 are specifically secreted during interaction of phagocytes and activated endothelium and are useful markers for monitoring disease activity in pauciarticular-onset juvenile rheumatoid arthritis. Arthritis & Rheumatism Official J. Am. Coll. Rheumatology 43 (3), 628–637. doi:10.1002/1529-0131(200003)43:3<628::AID-ANR20>3.0.CO;2-X
Gaudet, R., and Wiley, D. C. (2001a). Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 20 (17), 4964–4972. doi:10.1093/emboj/20.17.4964
Gaudet, R., and Wiley, D. C. (2001b). Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 20, 4964–4972. doi:10.1093/emboj/20.17.4964
Gehrig, S., Duerr, J., Weitnauer, M., Wagner, C. J., Graeber, S. Y., Schatterny, J., et al. (2014). Lack of neutrophil elastase reduces inflammation, mucus hypersecretion, and emphysema, but not mucus obstruction, in mice with cystic fibrosis–like lung disease. Am. J. Respir. Crit. care Med. 189 (9), 1082–1092. doi:10.1164/rccm.201311-1932OC
Graw, S., Tang, J., Zafar, M. K., Byrd, A. K., Bolden, C., Peterson, E. C., et al. (2020). proteiNorm–A user-friendly tool for normalization and analysis of TMT and label-free protein quantification. ACS omega 5 (40), 25625–25633. doi:10.1021/acsomega.0c02564
Guo, L., Johnson, R. S., and Schuh, J. C. L. (2000). Biochemical characterization of endogenously formed eosinophilic crystals in the lungs of mice. J. Biol. Chem. 275 (11), 8032–8037. doi:10.1074/jbc.275.11.8032
Gurung, S., Perocheau, D., Touramanidou, L., and Baruteau, J. (2021). The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal 19 (1), 47. doi:10.1186/s12964-021-00730-1
Hobo, A., Yuzawa, Y., Kosugi, T., Kato, N., Asai, N., Sato, W., et al. (2009). The growth factor midkine regulates the renin-angiotensin system in mice. J. Clin. investigation 119 (6), 1616–1625. doi:10.1172/JCI37249
Holtzman, J., and Lee, H. (2020). Emerging role of extracellular vesicles in the respiratory system. Exp. Mol. Med. 52 (6), 887–895. doi:10.1038/s12276-020-0450-9
Hu, Q., Zhang, S., Yang, Y., Li, J., Kang, H., Tang, W., et al. (2023). Extracellular vesicle ITGAM and ITGB2 mediate severe acute pancreatitis-related acute lung injury. ACS nano 17 (8), 7562–7575. doi:10.1021/acsnano.2c12722
Hughes, C. S., Moggridge, S., Müller, T., Sorensen, P. H., Morin, G. B., and Krijgsveld, J. (2019). Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14 (1), 68–85. doi:10.1038/s41596-018-0082-x
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Kaur, G., Maremanda, K. P., Campos, M., Chand, H. S., Li, F., Hirani, N., et al. (2021). Distinct exosomal miRNA profiles from BALF and lung tissue of COPD and IPF patients. Int. J. Mol. Sci. 22 (21), 11830. doi:10.3390/ijms222111830
Keerthikumar, S., Chisanga, D., Ariyaratne, D., Al Saffar, H., Anand, S., Zhao, K., et al. (2016). ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 428 (4), 688–692. doi:10.1016/j.jmb.2015.09.019
Khan, M., Nickoloff, E., Abramova, T., Johnson, J., Verma, S. K., Krishnamurthy, P., et al. (2015). Embryonic stem cell–derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circulation Res. 117 (1), 52–64. doi:10.1161/CIRCRESAHA.117.305990
Kohl, M., Wiese, S., and Warscheid, B. (2011). Cytoscape: software for visualization and analysis of biological networks. Data Min. proteomics Stand. Appl. 696, 291–303. doi:10.1007/978-1-60761-987-1_18
Lee, P. Y., Wang, J. X., Parisini, E., Dascher, C. C., and Nigrovic, P. A. (2013). Ly6 family proteins in neutrophil biology. J. Leukoc. Biol. 94 (4), 585–594. doi:10.1189/jlb.0113014
Lévy, Y., Wiedemann, A., Hejblum, B. P., Durand, M., Lefebvre, C., Surénaud, M., et al. (2021). CD177, a specific marker of neutrophil activation, is associated with coronavirus disease 2019 severity and death. iScience 24 (7), 102711. doi:10.1016/j.isci.2021.102711
Lewis, B. W., Choudhary, I., Paudel, K., Mao, Y., Sharma, R., Wang, Y., et al. (2020b). The innate lymphoid system is a critical player in the manifestation of mucoinflammatory airway disease in mice. J. Immunol. 205 (6), 1695–1708. doi:10.4049/jimmunol.2000530
Lewis, B. W., Sultana, R., Sharma, R., Noël, A., Langohr, I., Patial, S., et al. (2017). Early postnatal secondhand smoke exposure disrupts bacterial clearance and abolishes immune responses in muco-obstructive lung disease. J. Immunol. 199 (3), 1170–1183. doi:10.4049/jimmunol.1700144
Lewis, B. W., Vo, T., Choudhary, I., Kidder, A., Bathula, C., Ehre, C., et al. (2020a). Ablation of IL-33 suppresses Th2 responses but is accompanied by sustained mucus obstruction in the Scnn1b transgenic mouse model. J. Immunol. 204 (6), 1650–1660. doi:10.4049/jimmunol.1900234
Lewis, B. W., Vo, T., Choudhary, I., Kidder, A., Bathula, C., Ehre, C., et al. (2020c). Ablation of IL-33 suppresses Th2 responses but is accompanied by sustained mucus obstruction in the Scnn1b transgenic mouse model. J. Immunol. 204 (6), 1650–1660. doi:10.4049/jimmunol.1900234
Li, J., Liu, K., Liu, Y., Xu, Y., Zhang, F., Yang, H., et al. (2013). Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 14 (8), 793–803. doi:10.1038/ni.2647
Liu, Y. N., Guan, Y., Shen, J., Jia, Y. L., Zhou, J. C., Sun, Y., et al. (2020). Shp2 positively regulates cigarette smoke-induced epithelial mesenchymal transition by mediating MMP-9 production. Respir. Res. 21 (1), 161. doi:10.1186/s12931-020-01426-9
Livraghi-Butrico, A., Grubb, B. R., Wilkinson, K. J., Volmer, A. S., Burns, K. A., Evans, C. M., et al. (2017). Contribution of mucus concentration and secreted mucins Muc5ac and Muc5b to the pathogenesis of muco-obstructive lung disease. Mucosal Immunol. 10 (2), 829–407. doi:10.1038/mi.2017.29
MacKinnon, A. C., Farnworth, S. L., Hodkinson, P. S., Henderson, N. C., Atkinson, K. M., Leffler, H., et al. (2008). Regulation of alternative macrophage activation by galectin-3. J. Immunol. 180 (4), 2650–2658. doi:10.4049/jimmunol.180.4.2650
Mall, M., Grubb, B. R., Harkema, J. R., O'Neal, W. K., and Boucher, R. C. (2004). Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat. Med. 10 (5), 487–493. doi:10.1038/nm1028
Mall, M. A., Danahay, H., and Boucher, R. C. (2018). Emerging concepts and therapies for mucoobstructive lung disease. Ann. Am. Thorac. Soc. 15 (Suppl. 3), S216-S226–S226. doi:10.1513/AnnalsATS.201806-368AW
Mall, M. A., Harkema, J. R., Trojanek, J. B., Treis, D., Livraghi, A., Schubert, S., et al. (2008). Development of chronic bronchitis and emphysema in beta-epithelial Na+ channel-overexpressing mice. Am. J. Respir. Crit. care Med. 177 (7), 730–742. doi:10.1164/rccm.200708-1233OC
Mao, Y., Patial, S., and Saini, Y. (2023). Airway epithelial cell-specific deletion of HMGB1 exaggerates inflammatory responses in mice with muco-obstructive airway disease. Front. Immunol. 13, 944772. doi:10.3389/fimmu.2022.944772
McVey, M. J., Maishan, M., Blokland, K. E. C., Bartlett, N., and Kuebler, W. M. (2019). Extracellular vesicles in lung health, disease, and therapy. Am. J. Physiology-Lung Cell. Mol. Physiology 316, L977-L989. doi:10.1152/ajplung.00546.2018
Molet, S., Belleguic, C., Lena, H., Germain, N., Bertrand, C. P., Shapiro, S. D., et al. (2005). Increase in macrophage elastase (MMP-12) in lungs from patients with chronic obstructive pulmonary disease. Inflamm. Res. 54 (1), 31–36. doi:10.1007/s00011-004-1319-4
Nishi, Y., Sano, H., Kawashima, T., Okada, T., Kuroda, T., Kikkawa, K., et al. (2007). Role of galectin-3 in human pulmonary fibrosis. Allergol. Int. 56 (1), 57–65. doi:10.2332/allergolint.O-06-449
Papakonstantinou, E., Karakiulakis, G., Batzios, S., Savic, S., Roth, M., Tamm, M., et al. (2015). Acute exacerbations of COPD are associated with significant activation of matrix metalloproteinase 9 irrespectively of airway obstruction, emphysema and infection. Respir. Res. 16 (1), 78. doi:10.1186/s12931-015-0240-4
Pathan, M., Fonseka, P., Chitti, S. V., Kang, T., Sanwlani, R., Van Deun, J., et al. (2018). Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 47 (D1), D516-D519–D519. doi:10.1093/nar/gky1029
Pham, C. T. (2006). Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 6 (7), 541–550. doi:10.1038/nri1841
Qian, M., Chen, Z., Wang, S., Guo, X., Zhang, Z., Qiu, W., et al. (2019). PLEKHG5 is a novel prognostic biomarker in glioma patients. Int. J. Clin. Oncol. 24 (11), 1350–1358. doi:10.1007/s10147-019-01503-0
Raes, G., De Baetselier, P., Noël, W., Beschin, A., Brombacher, F., and Hassanzadeh Gh, G. (2002). Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J. Leukoc. Biol. 71 (4), 597–602. doi:10.1189/jlb.71.4.597
Raes, G., Van den Bergh, R., De Baetselier, P., Ghassabeh, G. H., Scotton, C., Locati, M., et al. (2005). Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J. Immunol. 174 (11), 6561–6562. doi:10.4049/jimmunol.174.11.6561
Ramming, T., Hansen, H. G., Nagata, K., Ellgaard, L., and Appenzeller-Herzog, C. (2014). GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 70, 106–116. doi:10.1016/j.freeradbiomed.2014.01.018
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids Res. 43 (7), e47. doi:10.1093/nar/gkv007
Rollet-Cohen, V., Bourderioux, M., Lipecka, J., Chhuon, C., Jung, V. A., Mesbahi, M., et al. (2018). Comparative proteomics of respiratory exosomes in cystic fibrosis, primary ciliary dyskinesia and asthma. J. Proteomics 185, 1–7. doi:10.1016/j.jprot.2018.07.001
Ryckman, C., Vandal, K., Rouleau, P., Talbot, M., and Tessier, P. A. (2003). Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 170 (6), 3233–3242. doi:10.4049/jimmunol.170.6.3233
Saini, Y., Dang, H., Livraghi-Butrico, A., Kelly, E. J., Jones, L. C., O'Neal, W. K., et al. (2014). Gene expression in whole lung and pulmonary macrophages reflects the dynamic pathology associated with airway surface dehydration. BMC Genomics 15 (1), 726. doi:10.1186/1471-2164-15-726
Saini, Y., Lewis, B. W., Yu, D., Dang, H., Livraghi-Butrico, A., Del Piero, F., et al. (2018). Effect of LysM+ macrophage depletion on lung pathology in mice with chronic bronchitis. Physiol. Rep. 6 (8), e13677. doi:10.14814/phy2.13677
Saini, Y., Wilkinson, K. J., Terrell, K. A., Burns, K. A., Livraghi-Butrico, A., Doerschuk, C. M., et al. (2016). Neonatal pulmonary macrophage depletion coupled to defective mucus clearance increases susceptibility to pneumonia and alters pulmonary immune responses. Am. J. Respir. cell Mol. Biol. 54 (2), 210–221. doi:10.1165/rcmb.2014-0111OC
Searle, B. C., Pino, L. K., Egertson, J. D., Ting, Y. S., Lawrence, R. T., MacLean, B. X., et al. (2018). Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 9 (1), 5128. doi:10.1038/s41467-018-07454-w
Shaba, E., Vantaggiato, L., Governini, L., Haxhiu, A., Sebastiani, G., Fignani, D., et al. (2022). Multi-omics integrative approach of extracellular vesicles: a future challenging milestone. Proteomes 10 (2), 12. doi:10.3390/proteomes10020012
Simpson, R. J., Kalra, H., and Mathivanan, S. (2012). ExoCarta as a resource for exosomal research. J. Extracell. vesicles 1 (1), 18374. doi:10.3402/jev.v1i0.18374
Stoorvogel, W., Kleijmeer, M. J., Geuze, H. J., and Raposo, G. (2002). The biogenesis and functions of exosomes. Traffic 3 (5), 321–330. doi:10.1034/j.1600-0854.2002.30502.x
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids Res. 47 (D1), D607-D613–D613. doi:10.1093/nar/gky1131
Tan, C. Y., Lai, R. C., Wong, W., Dan, Y. Y., Lim, S. K., and Ho, H. K. (2014). Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res. Ther. 5 (3), 76. doi:10.1186/scrt465
Teng, X., Li, D., Champion, H. C., and Johns, R. A. (2003). FIZZ1/RELMalpha, a novel hypoxia-induced mitogenic factor in lung with vasoconstrictive and angiogenic properties. Circulation Res. 92 (10), 1065–1067. doi:10.1161/01.RES.0000073999.07698.33
Théry, C., Zitvogel, L., and Amigorena, S. (2002). Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2 (8), 569–579. doi:10.1038/nri855
Thurman, T. J., Washam, C. L., Alkam, D., Bird, J. T., Gies, A., Dhusia, K., et al. (2023). proteoDA: a package for quantitative proteomics. J. Open Source Softw. 8 (85), 5184. doi:10.21105/joss.05184
Torregrosa Paredes, P., Esser, J., Admyre, C., Nord, M., Rahman, Q. K., Lukic, A., et al. (2012). Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy 67 (7), 911–919. doi:10.1111/j.1398-9995.2012.02835.x
Vaid, S., Camp, J. G., Hersemann, L., Eugster Oegema, C., Heninger, A. K., Winkler, S., et al. (2018). A novel population of Hopx-dependent basal radial glial cells in the developing mouse neocortex. Development 145 (20), dev169276. doi:10.1242/dev.169276
Velázquez-Enríquez, J. M., Santos-Álvarez, J. C., Ramírez-Hernández, A. A., Reyes-Jiménez, E., López-Martínez, A., Pina-Canseco, S., et al. (2021). Proteomic analysis reveals key proteins in extracellular vesicles cargo associated with idiopathic pulmonary fibrosis in vitro. Biomedicines 9 (8), 1058. doi:10.3390/biomedicines9081058
Keywords: exosome, proteomics, airspace, Scnn1b-Tg+, lung, mucoinflammation
Citation: Mao Y, Suryawanshi A, Patial S and Saini Y (2024) Airspaces-derived exosomes contain disease-relevant protein signatures in a mouse model of cystic fibrosis (CF)-like mucoinflammatory lung disease. Front. Pharmacol. 15:1460692. doi: 10.3389/fphar.2024.1460692
Received: 06 July 2024; Accepted: 26 August 2024;
Published: 25 September 2024.
Edited by:
Ranu Surolia, University of Alabama at Birmingham, United StatesReviewed by:
Bhagavatula Moorthy, Baylor College of Medicine, United StatesGagandeep Kaur, University of Rochester Medical Center, United States
Pawan Sharma, Temple University, United States
Copyright © 2024 Mao, Suryawanshi, Patial and Saini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yogesh Saini, eXNhaW5pQG5jc3UuZWR1
 Yun Mao
Yun Mao Amol Suryawanshi
Amol Suryawanshi Sonika Patial
Sonika Patial Yogesh Saini
Yogesh Saini