- 1Department of Medical Oncology, Universitair Ziekenhuis Brussel (UZBrussel), Brussels, Belgium
- 2Translational Oncology Research Center (TORC), Vrije Universiteit Brussel (VUB), Brussels, Belgium
- 3Pharmacy Department, Universitair Ziekenhuis Brussel (UZBrussel), Brussels, Belgium
- 4Faculty of Medicine & Pharmacy, Vrije Universiteit Brussel (VUB), Brussels, Belgium
- 5Centre for Medical Genetics, Research Group Genetics, Reproduction and Development, Clinical Sciences, Vrije Universiteit Brussel (VUB), Universitair Ziekenhuis Brussel (UZ Brussel), Brussels, Belgium
- 6Louvain Center for Toxicology and Applied Pharmacology, Institut de Recherche Expérimentale et Clinique (IREC), Université Catholique de Louvain (UCLouvain), Brussels, Belgium
- 7Department of Clinical Chemistry, Cliniques Universitaires Saint-Luc, Brussels, Belgium
Variations in the activity of the enzyme dihydropyrimidine dehydrogenase (DPD) are associated with toxicity to fluoropyrimidine-containing chemotherapy. Testing of DPD deficiency either by targeted genotyping of the corresponding DPYD gene or by quantification of plasma concentration of uracil and dihydrouracil (phenotyping approach) are the two main methods capable of predicting reduced enzymatic activity in order to reduce adverse reactions after fluoropyrimidine treatment. In this paper, we describe a patient with locally advanced colon carcinoma with severe toxicity following capecitabine therapy. Whereas targeted genotyping for the 4 most common DPYD variants analysis revealed heterozygous presence of the c.2846A>T variant, which is a relatively common variant associated with a partial deficiency, additional phenotyping was compatible with a complete DPD deficiency. Subsequent sequencing of the whole DPYD gene revealed the additional presence of the rare c.2872A>G variant, which is associated with a total loss of DPD activity. A clinical case of in trans compound heterozygosity of a common and a rare DPYD variant (c.2846A>T and c.2872A>G) has, to the best of our knowledge, not been previously described. Our case report shows the importance of performing either preemptive phenotyping or preemptive complete genetic analysis of the DPYD gene for patients planned for systemic fluoropyrimidines to identify rare and low frequency variants responsible for potentially life-threatening toxic reactions.
Introduction
Fluoropyrimidines belong to the class of antimetabolite drugs that form an integral backbone in the treatment of patients with cancers arising from the gastrointestinal tract, breast, head and neck. This includes 5-fluorouracil (5-FU) and the oral prodrugs capecitabine and tegafur. Conversion of 5-FU to dihydrofluorouracil is the first and rate-limiting step of the 5-FU degradation pathway and is regulated by the dihydropyrimidine dehydrogenase (DPD) enzyme, encoded by the DPYD gene (Etienne-Grimaldi et al., 2023). This enzyme converts up to 85% of 5-FU to inactive metabolites. Additionally, approximately 5% is excreted in urine. Deficiencies in DPD enzyme activity leads to increased intracellular concentrations of the active metabolites of 5-FU. This can lead to severe toxicity (neutropenia, mucositis and diarrhea), which is fatal in approximately 1% of patients. Because DPD deficiency affects 5%–7% of the Caucasian population, the European Medicines Agency (EMA) recommended in 2020 that prior to commencing fluoropyrimidine-containing therapy, patients should be tested either by genotyping the corresponding DPYD gene, or by phenotyping which is done by measuring plasma uracil concentrations or the dihydrouracil:uracil (UH2:U) ratio (Etienne-Grimaldi et al., 2023; European Medicines Agency, 2020). Both tests have their strengths and limitations, and no current recommendations are available regarding the preferred type of testing.
In this case report, we present a patient with severe 5-FU related toxicity, which was caused by a combination of a common DPYD variant with decreased DPD activity, and an initially undetected and uncommon DPYD variant associated with absent DPD activity. This case illustrates the added value of conducting a DPD phenotyping test and the potential danger of restricted testing of the locally most predominant DPYD variants. The CARE checklist was used when writing this report (Gagnier et al., 2013).
Case description
A 56-year-old Caucasian male with no comorbidity was diagnosed in another hospital with a well-differentiated adenocarcinoma of the distal sigmoid after a positive fecal occult blood test (FOBT). Imaging showed no evidence of distant metastases. He underwent a laparoscopic rectosigmoidectomy plus a partial mesorectal excision (PME) in October 2023 with the tumor being staged as pT4aN1M0. He decided to transfer to our hospital for adjuvant chemotherapy. As per international guidelines, he started adjuvant therapy with CAPOX (oxaliplatin 130 mg/mg2 day 1, capecitabine 1,000 mg/mg2 for 14 days, every 3 weeks). Upon diagnosis, targeted DPYD genotyping had already been performed in the referring hospital for the most common variants, i.e., DPYD*2A, DPYD*13, HapB3 and c.2846A>T, and showed the patient was heterozygous for the c.2846A>T variant (p.D949V, rs67376798). This variant is well known and has been functionally characterized as conferring a decreased function to the DPYD enzyme (Offer et al., 2014) and is included in the 2017 update of the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline, the 2019 Dutch Pharmacogenetics Working Group (DPWG) guideline for fluoropyrimidine dosing and the PharmGKB database as a tier I variant (Amstutz et al., 2018; Lunenburg et al., 2020). As genotyping results were already available and further delay of adjuvant therapy was deemed undesirable, an additional phenotyping test that is normal practice in our hospital was not performed in this case. After consultation with the clinical pharmacist and in accordance with the guidelines, the first cycle was started with 50% dose reduction of capecitabine, with the caveat to monitor for possible additional toxicity in case of rare undetected variants. A treatment timeline is presented in Figure 1. After 5 days of capecitabine treatment, he presented to the emergency department with vomiting and diarrhea. Clinical examination was remarkable for facial erythema and stomatitis. He was admitted for rehydration and capecitabine was discontinued. Despite the use of supportive therapy, his symptoms progressively aggravated.
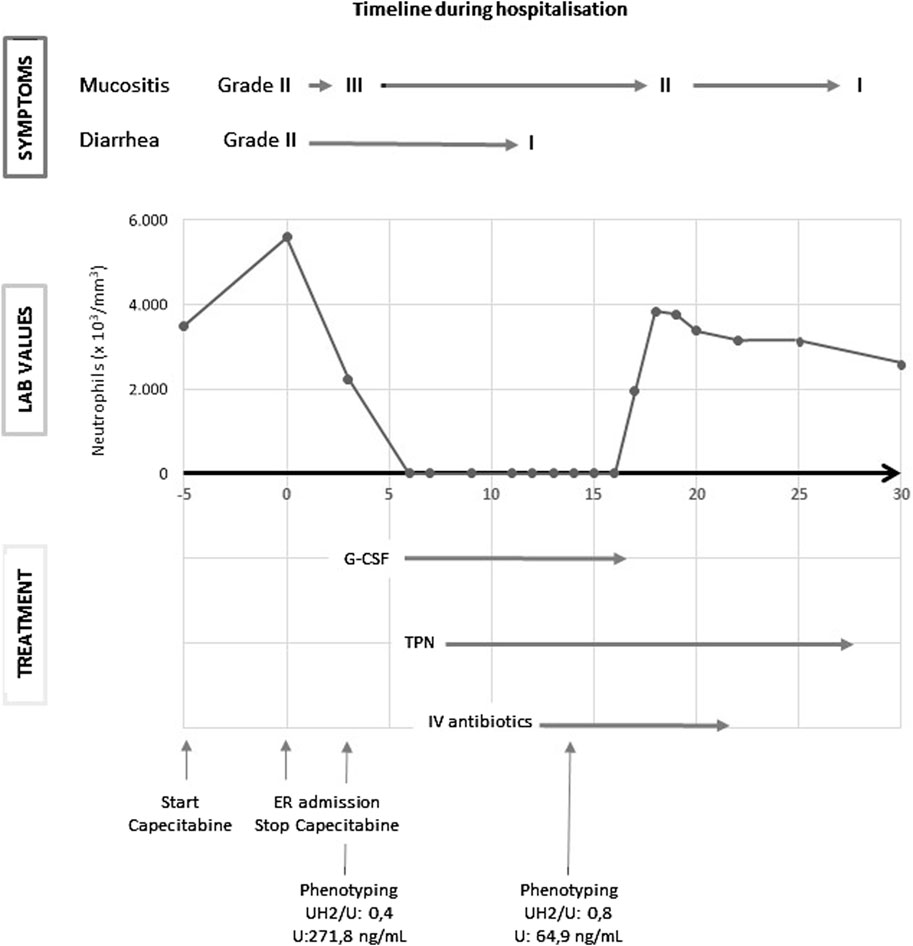
Figure 1. G-CSF: granulocyte colony-stimulating factor; IV: intravenous; TPN: total parenteral nutrition; U: uracil; UH2: dihydrouracil.
Suspecting 5-FU-related toxicity, DPD phenotyping was performed. Quantification of uracil (U) and dihydrouracil (UH2) was performed by liquid chromatography with tandem mass spectrometry after protein precipitation followed by liquid-liquid extraction using a fully validated method certified according to ISO-15189 standards, with additional external quality control organized by Asqualab, France (ISO-9001 certified EQC programme) to ensure accuracy of the results. According to literature, in particular Belgian recommendations, a reference value of >14 ng/mL for pre-treatment uracilemia was used to detect DPD deficiency, provided that pre-analytical conditions have been respected (plasma freezing time less than 1h30) and the patient’s renal function has been taken into account (eGFR >60 mL/min) (Casneuf et al., 2022). Thresholds for differentiating between partial and total DPD deficiency are less clear, although a value of 100 ng/mL has been proposed in the Belgian guidelines. A UH2/U ratio value of <1 is usually proposed to establish a diagnosis of total DPD deficiency, whereas the value of this ratio is usually >10 in patients with normal DPD activity. A first phenotyping test was sent on day 2 of hospitalization but was reported as non-interpretable (uracil (U): 271.8 ng/mL, dihydrouracil (UH2): 116.8 ng/mL; UH2/U: 0.4) due to the detected presence of 5-FU artificially increasing uracilemia (Thomas et al., 2021). On day 2 of hospitalization, he developed transfusion-dependent pancytopenia, which deepened over the next days. Despite the administration of granulocyte colony-stimulating factor (G-CSF), the nadir for neutrophil count was at 0,00 ×103/µL on day 10 of hospitalization, and for thrombocytes 10 × 103/µL on day 7. On day 5 of hospitalization total parental nutrition (TPN) was initiated due to severe mucositis. On day 5 of hospitalization, he developed neutropenic fever, which was empirically treated with piperacillin/tazobactam and vancomycin. Staphylococcus aureus was cultured from his sputum, upon which vancomycin was switched to flucloxacillin. Due to clinical improvement and recovery of cytopenia, flucloxacillin was discontinued on day 15 of hospitalization and piperacillin/tazobactam on day 21. From day 31 of hospitalization his TPN was gradually diminished due to increased oral intake. On day 28 of hospitalization a second phenotyping test (at a distance from capecitabine administration and interference by competitive inhibition of the DPD enzyme) was reported to be compatible with a complete DPD deficiency, which was in line with the clinical situation of our patient (U: 64.9 ng/mL; UH2: 54.4 ng/mL; UH2/U: 0.8). Our patient was finally discharged on day 34. After extensive counseling he decided not to restart further adjuvant treatment and is now closely clinically monitored with regular scans. At the time of writing, he is still in remission.
Considering his extreme toxicity despite a 50% dose reduction of capecitabine and a complete DPD deficiency according the second phenotyping test, DPYD full sequencing was carried out (using SOPHiA DDM® for Pharmacogenomics on a MiSeq Illumina Inc., United States) confirming the additional heterozygous presence of a NM_000110.4:c.2872A>G (p.K958E, rs141044036) variant, known to be associated with a total loss of DPD activity (Offer et al., 2014). The identification of two DPYD variants combined with the observed severe toxicity suggested the patient was an in trans compound heterozygous carrier. To confirm this hypothesis, next-generation sequencing using a pan-cancer capture based 380 gene panel, was performed on a NovaSeq 6000 (Illumina Inc., United States). Sequencing of the entire DPYD gene confirmed the presence of both, already identified, variants. Read visualisation at the level of their genomic locations (c.2846A>T: g.97547947; c.2872A>G: g.97547921) showed they were present in separate reads (variant allele frequencies: 50% and 48% respectively), confirming the in trans compound heterozygous state of the patient (Figure 2). This in trans compound heterozygous state of two not fully functional DPYD alleles (c.2846A>T/c.2872A>G) corresponds to a DPD activity score of 0.5 and therefore a poor metabolizer matched phenotype as per CPIC guideline (Amstutz et al., 2018).
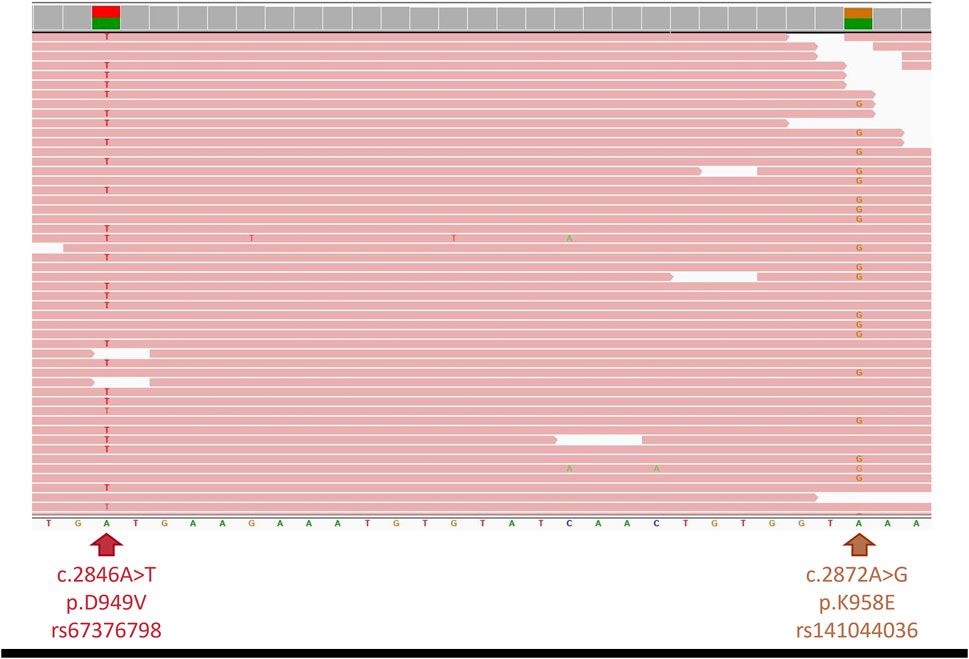
Figure 2. Integrative Genomics Viewer screenshot of genomic region. Next-generation sequencing showing both DPYD variants (c.2846A>T (left) and c.2872A>G (right)) present in different sequence reads, confirming in trans phasing.
Discussion
This case report highlights the clinical importance of screening for DPD deficiency prior to initiating fluoropyrimidine therapy using either targeted genotyping or phenotyping by measuring pre-treatment uracilemia. Although not yet recommended in the United States by the Food and Drug Administration (FDA), this screening strategy is currently strongly recommended in Europe by EMA since 2020 (European Medicines Agency, 2020; de With et al., 2023) and without it, our patient would have received a standard dose of 5-FU and would most likely not have survived his treatment.
The question arises which method forms the optimal strategy for preemptive DPD screening. The phenotyping test would have been able to predict a major risk of toxicity in our patient whereas the used targeted genotyping test failed to do, as the additional rare variant was not represented. The phenotyping test, however, has drawbacks too. Its main constraint is that it requires plasma to have been separated and frozen within 90 min of blood collection (Maillard et al., 2023; Thomas et al., 2023). Furthermore, knowledge of the patient’s glomerular filtration rate (eGFR) is necessary, as an increase in U and UH2 is observed in case of renal failure, without being linked to DPD deficiency. In this case UH2/U ratio remains in the normal range as both components are increased, still allowing a reliable interpretation of results (Narjoz et al., 2023). In addition, there is a between-subject variability and possible circadian rhythm in DPD enzyme activity (Jacobs et al., 2016). Finally, large between-center differences in uracil levels have been observed although the situation has considerably improved recently due to the introduction of external quality control programs (e.g., Asqualab, France) (de With et al., 2022). As was demonstrated in our patient, this test cannot be correctly interpreted when fluoropyrimidines are taken simultaneously or were only recently stopped, due to competitive inhibition of the DPD enzyme (Thomas et al., 2021). It is therefore very important to keep in mind that this phenotyping test should ideally be carried out before any fluoropyrimidine is taken. In case of unexplained fluoropyrimidine toxicity, clinicians should not forget to respect this delay when considering the phenotyping test.
Targeted DPYD genotyping for the four most common variants (DPYD*2A, DPYD*13, HapB3 and c.2846A>T (median allele frequency (MAF): 0.28457%; gnomAD v2.1.1, exome data)) is a well-standardized method with a good level of evidence supporting clinical effect and is more cost-effective than more advanced sequencing techniques (Lunenburg et al., 2020). However, as demonstrated in our case, it missed the concurring and rarer variant c.2872A>G (Offer et al., 2014). Moreover, while these variants are the most common causes of 5-FU toxicity in a Caucasian population, this is not the case in other ethnic groups that have been under-represented in large case-control clinical association studies of 5-FU toxicity (White et al., 2021). Because of this, several international expert working groups (Pratt et al., 2024; Garcia-Alfonso et al., 2022), suggest expanding testing to include additional clinically relevant DPYD variants, especially in non-European populations and even recommend considering testing for rare variants. Furthermore, with general populations becoming more diverse, testing for only a limited number of variants should be either avoided or accompanied by phenotyping. To date, a combined genotype-phenotype approach prior to initiation of fluoropyrimidine therapy does not appear to necessarily improve toxicity prediction (Etienne-Grimaldi et al., 2017). However, as Next-Generation Sequencing (NGS) technology becomes increasingly cheaper and accessible, more advanced genotyping such as whole genome sequencing and long-read sequencing could potentially offer a more robust solution to detect common as well as rare variants, including compound heterozygosity (Caspar et al., 2021). Although its use in a clinical setting is still limited and requires more research, including the involvement of microRNA, it may substantially improve the prediction of DPYD activity (De Luca et al., 2022; Deac et al., 2021). Nevertheless, DPYD phenotyping will persist in its role and may contribute to elucidating the DPYD functionalities associated with newly identified unknown variants of which the number will rise with performing whole DPYD gene sequencing. Administering therapy based on these unknown variants without understanding their DPYD functionality could result in fatal outcomes or reduced treatment efficacy. In retrospect, our patient should have been classified according DPWG as having an activity score of 0.5, prompting the need for additional phenotyping before making any decisions on the final dose (Lunenburg et al., 2020).
Co-existence of two loss-of-function DPYD variants has previously been described in oncological patients with confirmed reduced DPD activity and/or observed fluoropyrimidine toxicity (De Falco et al., 2019; Henricks et al., 2017; Baiardi et al., 2023; Johnson et al., 2002; Lau et al., 2023; Shrestha et al., 2018; Detailleur et al., 2021). However, in these cases, the co-existing variants were (moderately) common whereas in our case report the c.2872A>G variant was very rare and occurred only in 0.00199% (5/251,256) of alleles captured in the gnomAD database (v2.1.1, exome data). Furthermore, in previous reports compound heterozygosity was suspected, but in trans phasing of the variants could not be confirmed due to the large genomic distance between variants. Confirming that the two inactivating variants occur on the same DPYD allele (in cis) or on separate DPYD alleles (in trans) is vital and a particular strength in our case report, as only the latter will lead to non-production of functional DPD and subsequently a poor metabolizer status. Previously, only one patient captured in the 1,000 Genomes database was found to be in trans compound heterozygous for two common DPYD variants (c.1236G>A and c.2846A>T) (Lunenburg et al., 2018). To our knowledge, the in trans compound heterozygosity with the common c.2846A>T variant and the rare c.2872A>G variant identified in our patient has not previously been described. Calculated frequencies for compound heterozygosity for four common DPYD variants (DPYD*2A, DPYD*13, c.1236G>A and c.2846A>T) range from 0.0001% to 0.008% (Lunenburg et al., 2018). The calculated frequency of the c.2846A>T/c.2872A>G combination occurring in our patient was 0.000006%. However, despite its rarity the presence of this combination had profound clinical impact on our patient. This highlights the importance of keeping in mind that there is always a small, but by no means ignorable, chance a patient might harbour two DPYD variants, resulting in a poor metabolizer status. Furthermore, a recent study of 3,000 patients who underwent both DPD phenotyping and DPYD genotyping showed that a considerable amount of interindividual DPD activity could be attributed to rare DPYD variants (Larrue et al., 2024). In another study, multivariate analysis showed an increased risk of developing severe fluoropyrimidine toxicity in patients harbouring at least one very rare variant (MAF <0.1%) (De Mattia et al., 2022). Furthermore, inclusion of DPYD variants with higher incidence rates in populations of non-European ancestry compared to the European population might improve patient safety and reduce severe fluoropyrimidine-related toxicity (Chan et al., 2024). This issue of rare variants is broader than the DPYD gene as it was shown that 10.8% of putatively functional pharmacogenetic variants was considered rare (Ingelman-Sundberg et al., 2018). These results combined with this case report again underscore the importance of integrating rare variants in pharmacogenetic testing, despite the challenges it poses (De Mattia et al., 2024).
In summary, the case described here highlights the importance of preemptive screening for DPD deficiency prior to initiating fluoropyrimidine therapy to avoid severe adverse drug reactions. In addition, it underlines the importance of conducting a DPD phenotyping test, provided it is carried out correctly and in compliance with pre-analytical conditions, as such a test can be more sensitive than a targeted genotyping test, due to the possible presence of rare DPYD variants that may affect DPD enzyme activity. The advent of more advanced NGS tests is promising and may circumvent some limitations of both tests. Still, phenotyping will maintain its clinical role in final determination of DPD activity, especially in the case of rare variants.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
AH-H: Writing–original draft, Writing–review and editing, Conceptualization, Investigation. P-JC: Writing–original draft, Writing–review and editing, Formal Analysis. JV: Investigation, Writing–review and editing. PV: Visualization, Writing–review and editing, Investigation. SS: Writing–review and editing. FV: Formal Analysis, Validation, Writing–review and editing. VH: Conceptualization, Formal Analysis, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Additional genetic analysis for this case report was supported by the UZ Brussel Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amstutz, U., Henricks, L. M., Offer, S. M., Barbarino, J., Schellens, J. H. M., Swen, J. J., et al. (2018). Clinical pharmacogenetics implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin. Pharmacol. Ther. 103 (2), 210–216. doi:10.1002/cpt.911
Baiardi, G., Clavarezza, M., Stella, M., Casazza, S., De Censi, A., and Mattioli, F. (2023). Precision fluoropyrimidines dosing in a compound heterozygous variant carrier of the DPYD gene: a case report. Cancer Chemother. Pharmacol. 91 (5), 435–439. doi:10.1007/s00280-023-04515-w
Casneuf, V., Borbath, I., Van den Eynde, M., Verheezen, Y., Demey, W., Verstraete, A. G., et al. (2022). Joint Belgian recommendation on screening for DPD-deficiency in patients treated with 5-FU, capecitabine (and tegafur). Acta Clin. Belg 77 (2), 346–352. doi:10.1080/17843286.2020.1870855
Caspar, S. M., Schneider, T., Stoll, P., Meienberg, J., and Matyas, G. (2021). Potential of whole-genome sequencing-based pharmacogenetic profiling. Pharmacogenomics 22 (3), 177–190. doi:10.2217/pgs-2020-0155
Chan, T. H., Zhang, J. E., and Pirmohamed, M. (2024). DPYD genetic polymorphisms in non-European patients with severe fluoropyrimidine-related toxicity: a systematic review. Br. J. Cancer 131 (3), 498–514. doi:10.1038/s41416-024-02754-z
Deac, A. L., Burz, C. C., Militaru, C., Bocsan, I. C., Pop, R. M., Achimas-Cadariu, P., et al. (2021). Role of microRNAs in fluoropyrimidine-related toxicity: what we know. Eur. Rev. Med. Pharmacol. Sci. 25 (8), 3306–3315. doi:10.26355/eurrev_202104_25742
De Falco, V., Natalicchio, M. I., Napolitano, S., Coppola, N., Conzo, G., Martinelli, E., et al. (2019). A case report of a severe fluoropyrimidine-related toxicity due to an uncommon DPYD variant. Med. Baltim. 98 (21), e15759. doi:10.1097/MD.0000000000015759
De Luca, O., Salerno, G., De Bernardini, D., Torre, M. S., Simmaco, M., Lionetto, L., et al. (2022). Predicting dihydropyrimidine dehydrogenase deficiency and related 5-fluorouracil toxicity: opportunities and challenges of DPYD exon sequencing and the role of phenotyping assays. Int. J. Mol. Sci. 23 (22), 13923. doi:10.3390/ijms232213923
De Mattia, E., Milan, N., Assaraf, Y. G., Toffoli, G., and Cecchin, E. (2024). Clinical implementation of rare and novel DPYD variants for personalizing fluoropyrimidine treatment: challenges and opportunities. Int. J. Biol. Sci. 20 (10), 3742–3759. doi:10.7150/ijbs.97686
De Mattia, E., Silvestri, M., Polesel, J., Ecca, F., Mezzalira, S., Scarabel, L., et al. (2022). Rare genetic variant burden in DPYD predicts severe fluoropyrimidine-related toxicity risk. Biomed. Pharmacother. 154, 113644. doi:10.1016/j.biopha.2022.113644
Detailleur, S., Segelov, E., Re, M. D., and Prenen, H. (2021). Dihydropyrimidine dehydrogenase deficiency in patients with severe toxicity after 5-fluorouracil: a retrospective single-center study. Ann. Gastroenterol. 34 (1), 68–72. doi:10.20524/aog.2020.0551
de With, M., Knikman, J., de Man, F. M., Lunenburg, C., Henricks, L. M., van Kuilenburg, A. B. P., et al. (2022). Dihydropyrimidine dehydrogenase phenotyping using pretreatment uracil: a note of caution based on a large prospective clinical study. Clin. Pharmacol. Ther. 112 (1), 62–68. doi:10.1002/cpt.2608
de With, M., Sadlon, A., Cecchin, E., Haufroid, V., Thomas, F., Joerger, M., et al. (2023). Implementation of dihydropyrimidine dehydrogenase deficiency testing in Europe. ESMO Open 8 (2), 101197. doi:10.1016/j.esmoop.2023.101197
European Medicines Agency, (2020). EMA recommendations on DPD testing prior to treatment with fluorouracil, capecitabine, tegafur and flucytosine 2020. Available at: https://www.ema.europa.eu/en/medicines/human/referrals/fluorouracil-and-fluorouracil-related-substances-capecitabine-tegafur-and-flucytosine-containing-medicinal-products (Accessed March 06 2024).
Etienne-Grimaldi, M. C., Boyer, J. C., Beroud, C., Mbatchi, L., van Kuilenburg, A., Bobin-Dubigeon, C., et al. (2017). New advances in DPYD genotype and risk of severe toxicity under capecitabine. PLoS One 12 (5), e0175998. doi:10.1371/journal.pone.0175998
Etienne-Grimaldi, M. C., Pallet, N., Boige, V., Ciccolini, J., Chouchana, L., Barin-Le Guellec, C., et al. (2023). Current diagnostic and clinical issues of screening for dihydropyrimidine dehydrogenase deficiency. Eur. J. Cancer 181, 3–17. doi:10.1016/j.ejca.2022.11.028
Gagnier, J. J., Kienle, G., Altman, D. G., Moher, D., Sox, H., Riley, D., et al. (2013). The CARE Guidelines: Consensus-based Clinical Case Reporting Guideline Development. Glob. Adv. Health. Med. 2 (5), 38–43. doi:10.7453/gahmj.2013.008
Garcia-Alfonso, P., Saiz-Rodriguez, M., Mondejar, R., Salazar, J., Paez, D., Borobia, A. M., et al. (2022). Consensus of experts from the Spanish Pharmacogenetics and Pharmacogenomics Society and the Spanish Society of Medical Oncology for the genotyping of DPYD in cancer patients who are candidates for treatment with fluoropyrimidines. Clin. Transl. Oncol. 24 (3), 483–494. doi:10.1007/s12094-021-02708-4
Henricks, L. M., Kienhuis, E., de Man, F. M., van der Veldt, A. A. M., Hamberg, P., van Kuilenburg, A. B. P., et al. (2017). Treatment algorithm for homozygous or compound heterozygous DPYD variant allele carriers with low-dose capecitabine. JCO Precis. Oncol. 1, 1–10. doi:10.1200/po.17.00118
Ingelman-Sundberg, M., Mkrtchian, S., Zhou, Y., and Lauschke, V. M. (2018). Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genomics 12 (1), 26. doi:10.1186/s40246-018-0157-3
Jacobs, B. A., Deenen, M. J., Pluim, D., van Hasselt, J. G., Krahenbuhl, M. D., van Geel, R. M., et al. (2016). Pronounced between-subject and circadian variability in thymidylate synthase and dihydropyrimidine dehydrogenase enzyme activity in human volunteers. Br. J. Clin. Pharmacol. 82 (3), 706–716. doi:10.1111/bcp.13007
Johnson, M. R., Wang, K., and Diasio, R. B. (2002). Profound dihydropyrimidine dehydrogenase deficiency resulting from a novel compound heterozygote genotype. Clin. Cancer Res. 8 (3), 768–774.
Larrue, R., Fellah, S., Hennart, B., Sabaouni, N., Boukrout, N., Van der Hauwaert, C., et al. (2024). Integrating rare genetic variants into DPYD pharmacogenetic testing may help preventing fluoropyrimidine-induced toxicity. Pharmacogenomics J. 24 (1), 1. doi:10.1038/s41397-023-00322-x
Lau, D. K., Fong, C., Arouri, F., Cortez, L., Katifi, H., Gonzalez-Exposito, R., et al. (2023). Impact of pharmacogenomic DPYD variant guided dosing on toxicity in patients receiving fluoropyrimidines for gastrointestinal cancers in a high-volume tertiary centre. BMC Cancer 23 (1), 380. doi:10.1186/s12885-023-10857-8
Lunenburg, C., Henricks, L. M., van Kuilenburg, A. B. P., Mathijssen, R. H. J., Schellens, J. H. M., Gelderblom, H., et al. (2018). Diagnostic and therapeutic strategies for fluoropyrimidine treatment of patients carrying multiple DPYD variants. Genes (Basel) 9 (12), 585. doi:10.3390/genes9120585
Lunenburg, C., van der Wouden, C. H., Nijenhuis, M., Crommentuijn-van Rhenen, M. H., de Boer-Veger, N. J., Buunk, A. M., et al. (2020). Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 28 (4), 508–517. doi:10.1038/s41431-019-0540-0
Maillard, M., Launay, M., Royer, B., Guitton, J., Gautier-Veyret, E., Broutin, S., et al. (2023). Quantitative impact of pre-analytical process on plasma uracil when testing for dihydropyrimidine dehydrogenase deficiency. Br. J. Clin. Pharmacol. 89 (2), 762–772. doi:10.1111/bcp.15536
Narjoz, C., Nadour, Z., Zaanan, A., Taieb, J., Loriot, M. A., and Pallet, N. (2023). Screening for dihydropyrimidine dehydrogenase deficiency by measuring uracilemia in chronic kidney disease patients is associated with a high rate of false positives. Clin. Chim. Acta 543, 117326. doi:10.1016/j.cca.2023.117326
Offer, S. M., Fossum, C. C., Wegner, N. J., Stuflesser, A. J., Butterfield, G. L., and Diasio, R. B. (2014). Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity. Cancer Res. 74 (9), 2545–2554. doi:10.1158/0008-5472.Can-13-2482
Pratt, V. M., Cavallari, L. H., Fulmer, M. L., Gaedigk, A., Hachad, H., Ji, Y., et al. (2024). DPYD genotyping recommendations: a joint consensus recommendation of the association for molecular pathology, American college of medical genetics and genomics, clinical pharmacogenetics implementation Consortium, college of American pathologists, Dutch pharmacogenetics working group of the royal Dutch pharmacists association, European society for Pharmacogenomics and personalized therapy, Pharmacogenomics knowledgebase, and pharmacogene variation Consortium. J. Mol. Diagn. doi:10.1016/j.jmoldx.2024.05.015
Shrestha, S., Tapper, E. E., Trogstad-Isaacson, C. S., Hobday, T. J., Offer, S. M., and Diasio, R. B. (2018). Dose modification for safe treatment of a compound complex heterozygous DPYD variant carrier with fluorouracil. JCO Precis. Oncol. 2, 1–5. doi:10.1200/po.18.00179
Thomas, F., Launay, M., Guitton, J., Loriot, M. A., Boyer, J. C., Haufroid, V., et al. (2023). Plasma uracil as a DPD phenotyping test: pre-analytical handling matters. Clin. Pharmacol. Ther. 113 (3), 471–472. doi:10.1002/cpt.2772
Thomas, F., Maillard, M., Launay, M., Tron, C., Etienne-Grimaldi, M. C., Gautier-Veyret, E., et al. (2021). Artificial increase of uracilemia during fluoropyrimidine treatment can lead to DPD deficiency misinterpretation. Ann. Oncol. 32 (6), 810–811. doi:10.1016/j.annonc.2021.02.020
Keywords: case report, capecitabine, DPYD gene polymorphism, pheno- and genotyping, rare variant
Citation: de Haar-Holleman A, Cortoos P-J, Vlaeminck J, Van Landuyt P, Steurbaut S, Vaeyens F and Haufroid V (2024) Case report: A case of severe capecitabine toxicity due to confirmed in trans compound heterozygosity of a common and rare DPYD variant. Front. Pharmacol. 15:1459565. doi: 10.3389/fphar.2024.1459565
Received: 04 July 2024; Accepted: 09 September 2024;
Published: 23 September 2024.
Edited by:
Georgia Ragia, Democritus University of Thrace, GreeceReviewed by:
Francisco Abad-Santos, Universidad Autónoma de Madrid, SpainNicolas Pottier, Université de Lille, France
Copyright © 2024 de Haar-Holleman, Cortoos, Vlaeminck, Van Landuyt, Steurbaut, Vaeyens and Haufroid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pieter-Jan Cortoos, cGlldGVyLWphbi5jb3J0b29zQHV6YnJ1c3NlbC5iZQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Amy de Haar-Holleman
Amy de Haar-Holleman Pieter-Jan Cortoos
Pieter-Jan Cortoos Jelle Vlaeminck
Jelle Vlaeminck Paulien Van Landuyt3
Paulien Van Landuyt3 Stephane Steurbaut
Stephane Steurbaut Freya Vaeyens
Freya Vaeyens Vincent Haufroid
Vincent Haufroid