- 1Key Laboratory of Neuroregeneration of Jiangsu and Ministry of Education, Co-Innovation Center of Neuroregeneration, Nantong University, Nantong, China
- 2Department of Orthopedic, Affiliated Hospital of Nantong University, Nantong, China
- 3Department of Orthopedic, Nantong Third People’s Hospital, Nantong University, Nantong, China
Injury to the peripheral nervous system disconnects targets to the central nervous system, disrupts signal transmission, and results in functional disability. Although surgical and therapeutic treatments improve nerve regeneration, it is generally hard to achieve fully functional recovery after severe peripheral nerve injury. A better understanding of pathological changes after peripheral nerve injury helps the development of promising treatments for nerve regeneration. Single-cell analyses of the peripheral nervous system under physiological and injury conditions define the diversity of cells in peripheral nerves and reveal cell-specific injury responses. Herein, we review recent findings on the single-cell transcriptome status in the dorsal root ganglia and peripheral nerves following peripheral nerve injury, identify the cell heterogeneity of peripheral nerves, and delineate changes in injured peripheral nerves, especially molecular changes in neurons, glial cells, and immune cells. Cell-cell interactions in peripheral nerves are also characterized based on ligand-receptor pairs from coordinated gene expressions. The understanding of cellular changes following peripheral nerve injury at a single-cell resolution offers a comprehensive and insightful view for the peripheral nerve repair process, provides an important basis for the exploration of the key regulators of neuronal growth and microenvironment reconstruction, and benefits the development of novel therapeutic drugs for the treatment of peripheral nerve injury.
Introduction
The mammalian peripheral nervous system is an essential nerve system component that transmits bioelectrical signals throughout the body’s neural network. The peripheral nervous system, according to its anatomical, structural, and functional characteristics, is generally subdivided into sensory, motor (nerve fibers of motor neurons), enteric, and autonomic domains (Murtazina and Adameyko, 2023). The sensory nerves, together with axons of motor neurons, form the somatic peripheral nervous system (Catala and Kubis, 2013). The enteric nerves comprise myenteric and submucosal plexuses (Sasselli et al., 2012). The autonomic nerves are composed of the sympathetic system that dominates “fight or flight” emergency responses and the parasympathetic system that mediates physiology balance (Zahalka and Frenette, 2020). The cranial peripheral nervous system (cranial nerves III to XIII) have both somatic and autonomous fibers innervation. The cranial nerves III have both motor and parasympathetic fibers while the cranial nerves VII, IX, and X have motor, sensory, and parasympathetic fibers (Catala and Kubis, 2013).
The somatic peripheral nervous system comprises eight pairs of cervical spinal nerves, twelve pairs of thoracic spinal nerves, five pairs of lumbar spinal nerves, five pairs of sacral spinal nerves, and a few pairs of coccygeal nerves (Catala and Kubis, 2013). The somatic peripheral nerves are delicate and commonly affected by traumatic events such as wars, natural disasters, construction accidents, and traffic accidents. Peripheral nerves can also be affected by iatrogenic injuries such as surgical injuries and drug injection injuries. In addition, numerous diseases, such as diabetes, neurological disorders (for example, Guillain-Barre syndrome and carpal tunnel syndrome), and autoimmune diseases (for example, Sjogren’s syndrome, lupus, and rheumatoid arthritis), may result in injuries to peripheral nerve trunks. The characterization and functional investigations of somatic sensory nerves are more comprehensive as compared with those of enteric and autonomic nerves (Drokhlyansky et al., 2020).
In peripheral nerve trunks, axons are wrapped with Schwann cells. Following peripheral nerve injury, especially those injuries with long nerve gaps, axon-associated Schwann cells undergo partial dedifferentiation and neuronal axons are transected. Afferent and efferent signals transmitted through neuronal axons are interrupted, inducing functional impairments, neuropathic pain, and physical disabilities. Peripheral nerve injury initiates acute immune responses within in nerve stumps and triggers inflammatory responses in neuronal somas (Kalinski et al., 2020). In both the proximal and the distal nerve stumps, numerous pro-inflammatory genes, such as Il6, Cxcl3, Ccl20, S100a8/a9, and Mmp9, are elevated and early immune responses are elicited (Chernov et al., 2015). Within 24–72 h after peripheral nerve injury, the axons distal to the injured sites and within a small zone distal to the proximal nerve stumps experience Wallerian degeneration, generating massive axonal- and myelin-derived debris. Schwann cells and immune cells are recruited into the injured sites to engulf and remove debris (Chen et al., 2015; Wong et al., 2017). Schwann cells sense injury signals, experience cellular reprogramming and are switched to a repair phenotype that benefits tissue homeostasis and regeneration via promoting the attraction of macrophages, the survival of neurons, and the elongation of axons (Jessen and Arthur-Farraj, 2019). Macrophages are polarized to an anti-inflammatory M2 phenotype to create a permissive microenvironment for nerve repair (Figure 1). Moreover, different from central nerves, in the peripheral nervous system, neurons retain regenerative ability following nerve injury (He and Jin, 2016). While distal axons undergo degeneration, axons at the proximal nerve stump are prepared for growth providing important basic for nerve repair and regeneration (Scheib and Höke, 2013). However, it is worth noting that although the peripheral nervous system obtains certain self-regeneration capacity, after long-distance peripheral nerve defect, peripheral nerve regeneration generally results in limited functional recovery without surgical and/or pharmacological treatments (Zou et al., 2023).
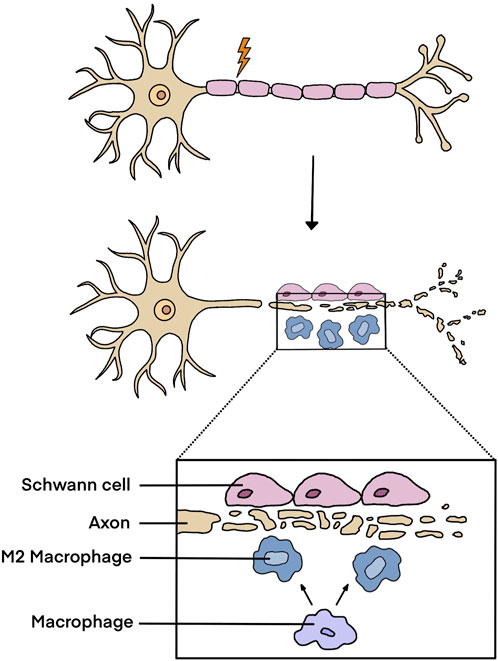
Figure 1. Injury responses in peripheral nerve stumps after peripheral nerve injury. Following peripheral nerve injury, Schwann cells and macrophages accumulate at the injured sites, clean axonal- and myelin-derived debris, and experience phenotype switch to generate a permissive microenvironment for nerve regeneration.
Pharmacological treatments for peripheral nerve injury
Surgical suture is a typical method for the treatment of peripheral nerve injury (Meng et al., 2023). The joint application of therapeutic drugs helps to accelerate the regeneration process and boost the functional repair of injured nerves. Subcutaneous application of cyclic guanosine monophosphate-dependent phospodiesterase-5 inhibitor sildenafil to rats that undergo peripheral nerve transection and epineurial sutures reveals that sildenafil slightly improves rat sciatic functional index, although the changes of sciatic function index are not significant, the numbers of myelinated axons are not different, and the motor nerve conduction velocity are not affected (Fang et al., 2013). After rat peripheral nerve transection and subsequent incision site suture, the administration of serine protease inhibitor ulinastatin increases the expression of nerve growth factor (NGF), elevates the production of many myelin associated proteins, stimulates axon regeneration and nerve remyelination, and advances sciatic functional index as well as the amplitude of compound muscle action potential (Zhang et al., 2020).
Treatment with therapeutic drugs alone also benefits peripheral nerve regeneration, especially after relatively mild peripheral nerve injury such as nerve crush. Daily sildenafil treatment of sciatic nerve injured rats improves rat performance in the rotarod test (Korkmaz et al., 2016). Intraperitoneal injection of vitamin B complex containing B1, B2, B3, B6, and B12 to rats immediately after femoral nerve motor branch injury and every 24 h after surgery reduces the number/percentage of cells expressing pro-inflammatory cytokines and increases the number/percentage of cells expressing anti-inflammatory cytokines. The functional roles of vitamin B complex in local inflammation attenuation indicate that vitamin B can be used to treat peripheral nerve injury by reducing neuroinflammation (Ehmedah et al., 2019). The administration of immunosuppressant FK506 to rats undergo sciatic nerve crush injury directly reveals the promoting roles of immunosuppressive drugs in treating peripheral nerve injury. FK506 treatment accelerates the appearances of initial sign of return of toe movement and the initial return of ability to walk. Although serious quantification of myelin sheaths is not performed, morphological observations from light micrograph and electron microscopy show that daily subcutaneous injection of FK506 enhances nerve fiber myelination (Gold et al., 1994). Erythropoietin also exhibits immunomodulatory activity after peripheral nerve injury. Intraperitoneal injection of erythropoietin increases macrophage M2 activity, reduces in site apoptosis, bolsters debris phagocytosis, and benefits the myelination of nerve fibers as well as the recovery of sciatic functional index (Govindappa and Elfar, 2022). Co-treatment of erythropoietin with another anti-inflammatory drug dexamethasone further promotes the remyelination of injured peripheral nerves, stimulates functional recovery, and protects target muscle fibers from atrophy (Lee et al., 2020).
Current pharmacological treatments for peripheral nerve injury mainly take advantage of the anti-inflammatory effects of these drugs. Chronic inflammation may alter the behavior of cells in the nerve stumps, hamper the injury responses of Schwann cells, and diminish the regenerative capacity of injured nerves (Büttner et al., 2018). Hence, the application of drugs with anti-inflammatory and immunosuppressive properties may trigger the regeneration process. To identify novel therapeutic targets for the treatment of peripheral nerve injury, a recent study performs genome-wide CpG methylation profiling and identifies hypermethylation of formin-2 (fmn2) promoter as a key factor that mediates injury responses, microtubule dynamics at the growth cone, and axon regeneration. Next, by sequencing and in silico drug screen of fmn2 knockdown, small-molecule metaxalone has been discovered to be a regulator of microtubule dynamics and a novel therapeutic drug for the treatment of peripheral nerve injury (Au et al., 2023). Therefore, for the discovery of new therapeutic drugs, it is of great importance to determine genetic changes following peripheral nerve injury.
Attempts to identify molecular changes in the peripheral nervous system after nerve injury
High-throughput analysis provides an unprecedented ability to explore molecular signatures under physiological and pathological conditions. Sciatic nerve is the thickest peripheral nerve truck in mammals. Hence, injury to sciatic nerves is commonly used for the investigation of peripheral nerve injury and regeneration (Savastano et al., 2014). Using the sciatic nerve experimental model, emerging high-throughput studies have examined large-scale molecular expressions following peripheral nerve injury and enumerated a large number of essential factors for nerve regeneration. A large number of microarray assays have determined the expression changes of both coding and non-coding genes in the dorsal root ganglia (DRGs), the proximal nerve stumps, and distal nerve stumps of injured sciatic nerves (Jia et al., 2020; Kubo et al., 2002; Li et al., 2015; Lu et al., 2014; Sohn and Park, 2020; Wang et al., 2018). Microarray has certain disadvantages, especially the limited detections of transcripts (Murakami et al., 2014). RNA sequencing approach has no such limitation and can even discover previously unknown transcripts and isoforms (Marioni et al., 2008; Soneson and Delorenzi, 2013). Actually, the application of RNA sequencing in the DRGs and nerve stumps after peripheral nerve injury has enabled the characterization of novel microRNAs and alternative splicing events (Li et al., 2012; Li et al., 2011; Zhao and Yi, 2019). The functional involvement of these transcripts, such as the promoting role of alternative splicing of neuronal cell adhesion molecule Nrcam in axonal growth and elongation, have been elucidated on the basis of RNA sequencing (Mao et al., 2018).
Bulk sequencing generates transcriptome profiling by measuring averaged expressions of diverse constituent cells and thus masks specialized properties of single cells. Recently developed single-cell sequencing detects cell population heterogeneity and allows the identification of the states and functions of single cells (Tang et al., 2009). A series of single-cell sequencing technologies, ranging from targeted single-cell profiling to unbiased single-cell profiling, have been leveraged to study cellular population heterogeneity (Papalexi and Satija, 2018). Advances in droplet-based microfluidics push the development of massively parallel unbiased single-cell sequencing methods, such as indexing droplet sequencing (inDrop), Drop-seq, and 10X Genomics Chromium. The application of droplet-based microfluidics largely increases the number of cells in a single sequencing and decreases the cost for each cell (Zhang et al., 2019). These massively parallel unbiased sequencing technologies, especially the commercially available platform 10 × Genomics, have been widely used to decipher cellular populations in a variety of tissues and organs, including the peripheral nervous system (Nguyen et al., 2021; Xie et al., 2023; Zhang et al., 2022). For the 10 × Genomics analysis of the peripheral nervous system, tissues, such as the DRGs and peripheral nerve stumps, are processed to single-cell suspensions and functional gel-bead-in-emulsion (GEM) for dissecting cellular heterogeneity and detecting genetic expressions in specific cell types (Figure 2).
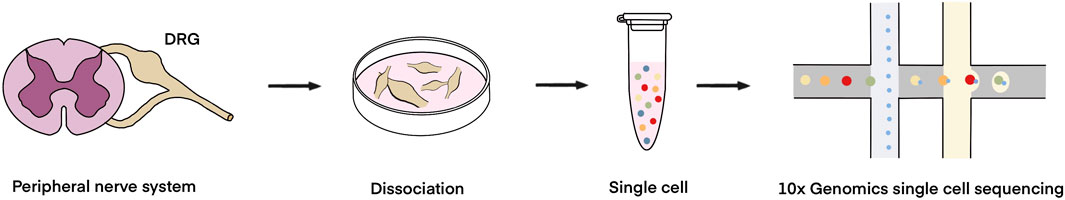
Figure 2. 10 × Genomics single-cell sequencing workflow for the investigation of peripheral nerves. The DRGs and/or peripheral nerve stumps are dissociated into small pieces and then enzymatic treated to collect cell populations. Single-cell suspensions are dispensed through microfluidic partitioning and mixed with barcode-containing gel beads, reverse transcription reagents, and oil to obtain GEM for sequencing analysis.
The single-nucleus sequencing method is also commonly used where a single nucleus is extracted, and nuclear transcriptomes are detected (Ding et al., 2020). Single-nuclei sequencing measures nuclear gene expressions and has been used as an alternative technology for optimal sample dissociation and cell storage (Denisenko et al., 2020). Compared with intact cells, a smaller amount of transcript is captured using single-nuclei sequencing because cytoplasmic information is lost. Still, single-nuclei sequencing has its own advantages and is extremely suitable for isolating cells from frozen tissues and formalin-fixed paraffin-embedded tissues as the freezing and fixation procedures interrupt cellular integrity and increase cytoplasmic transcript degradation (Cuevas-Diaz Duran et al., 2022). Single-nuclei sequencing is also more suitable to examine genetic features in cells with unusual morphologies and large sizes (Selewa et al., 2020). Some subtypes of neurons in the DRGs have large diameters and may not be able to successfully separated using microfluidic partitioning. For instance, the mean cell size of Ptgfr-expressing neuronal sub-type is reported to be 1,179 μm2 (Wang et al., 2023). Hence, it is appropriate to apply single-nuclei sequencing technology in the transcriptomic analysis of the DRGs as well as other neuron-containing tissues. Actually, single-nuclei sequencing has been frequently devoted to examine cellular heterogeneity in the peripheral nervous system (Avraham et al., 2022a; Yim et al., 2022). The application of these single-cell level analyses of the peripheral nervous system offers a wealth of knowledge of cellular and molecular processes under physiological and injury conditions.
Single-cell-based analysis of the DRGs
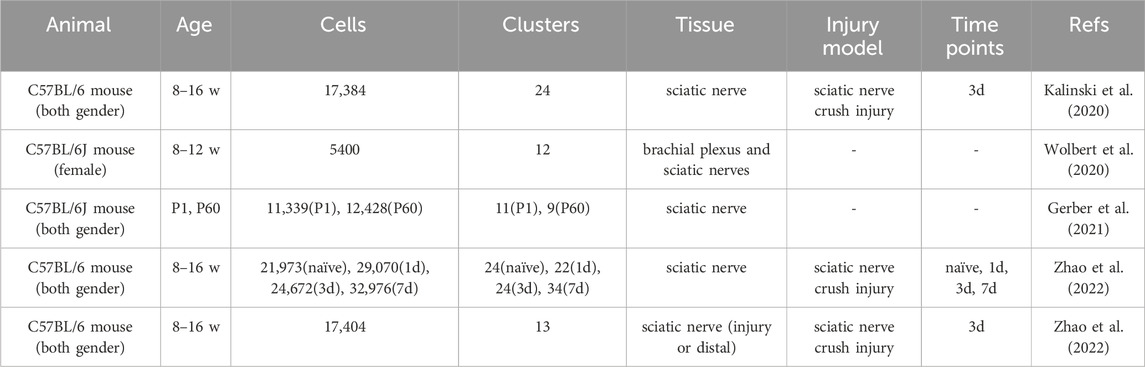
DRG neurons possess certain intrinsic regeneration capacity and their regeneration capacity may be influenced by surrounding cells via cell-cell interaction and secreted factors. Single-cell sequencing, especially 10X Genomics, has been widely used to examine cellular populations in the DRGs under the naïve and injured states (Table 1).
Cellular population in the DRGs
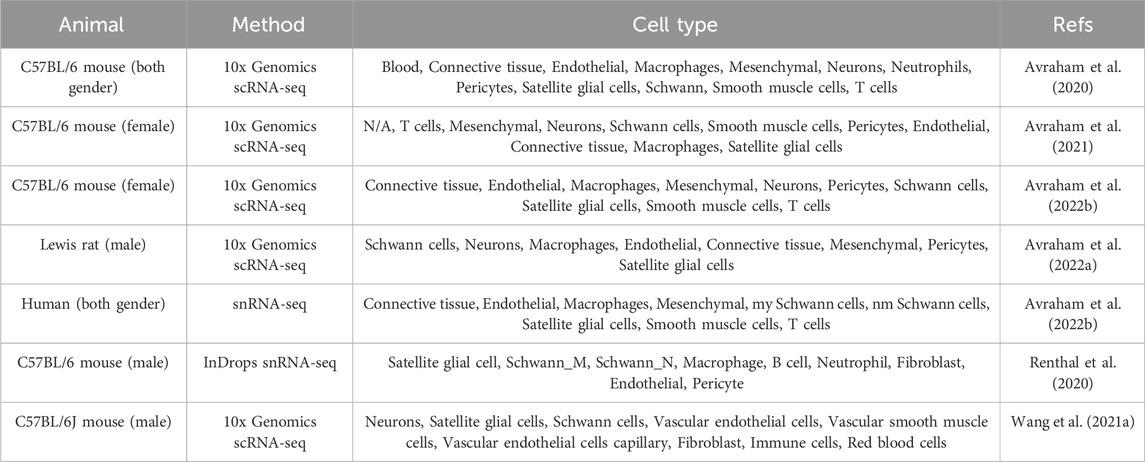
Neuronal somas are surrounded by peripheral glial and other non-neuronal cells in the DRGs. Diverse types of cells have been identified in the DRGs (Table 2). Single-cell profiling of mouse L4 and L5 DRGs collects a total of 6541 cells in the naïve DRGs and the DRGs at 3 days after sciatic nerve crush injury, detects 1500 genes per cell, and categorizes cell populations into 13 cell clusters, i.e., blood, connective tissue, endothelial cells, macrophages, mesenchymal cells, neurons, neutrophils, pericytes, satellite glial cells, Schwann cells, vasculature associated smooth muscle cells, T cells, and an unidentified group (Avraham et al., 2020). These major cellular subtypes, including connective tissue, endothelial cells, macrophages, mesenchymal cells, neurons, pericytes, satellite glial cells, Schwann cells, smooth muscle cells, T cells, are also identified in a later study that sequences a total of 25,154 mouse DRG cells under normal condition and following peripheral and central branch injury (Avraham et al., 2021). These cell types are further detected in a single-cell sequencing in the L4 and L5 DRGs from rats and in a single-nucleus sequencing from the human DRG samples, showing high similarities between rodents and humans and indicating the translational applicability of rodent models in peripheral neuropathies (Avraham et al., 2022b). Some other cell types, including fibroblasts and distinct types of immune cells, such as B cells, are also captured (Renthal et al., 2020).
Besides the recognition of cell type composition, single-cell sequencing also contributes to the categorization of neurons (Zeng and Sanes, 2017). Somatosensory neurons are traditionally classified to large diameter neurons (neurofilament-containing (NF) neurons) and small-medium diameter neurons (isolectin B4 (IB4)-positive non-peptide-containing (NP) neurons, peptidergic nociceptor-containing (PEP) neurons, and tyrosine hydroxylase-containing (TH) neurons) based on their morphological and molecular characteristics. These DRG neuron clusters have been discovered in a total of 622 neuronal cells in the mouse lumbar using single-cell tagged reverse transcription with the TH cluster occupies the largest population and the PEP cluster occupies the smallest population (Usoskin et al., 2015). Transcriptional profiling and functional assignment of single cells further allow the separation of low threshold mechanoreceptor (LTMR)-containing NF1-NF3 subpopulations and limb proprioceptive NF4 and NF5 subpopulations in the NF cluster, pruritus participating and itch sensing NP1-NP3 subpopulations in the NP cluster, thermosensitive PEP1 subpopulation and lightly myelinated Aδ nociceptor PEP1 subpopulation in the PEP cluster, and type C low-threshold mechanoreceptor TH neurons (Usoskin et al., 2015). A more detailed classification of DRG neurons has been obtained with high-coverage sequencing (Li et al., 2016; Wang K. et al., 2021). Comprehensive analyses of classification of DRG neurons have divided somatosensory neurons into a Oprk1-expressing sub-type, a SStr2-expressing sub-type, and a Gabrg3-expressing sub-type of small neurons for heat sensation, a Trpm8-expressing sub-type of small neurons for cold sensation, a Rxfp1-expressing sub-type of small neurons for cold sensation, a Nppb-expressing sub-type of small neurons, a Mrgpra3-positive and Mrgpra4-negative sub-type of small neurons, and two Mrgprd-expressing subtypes (Lpar3-positive or Lpar3-negative) of small neurons for itch sensation, two Fam19a4-expressing subtypes (Th-positive or Th-negative) of small neurons for c-LTMR sensation, a Mrgpra3-positive and Mrgpra4-positive sub-type of small neurons for message-like-stroking sensation, a Wnt7a-expressing sub-type of large neurons and a Prokr2-expressing sub-type of large neurons for proprioceptor sensation, a Ptgfr-expressing sub-type of large neurons for Aβ-LTMR sensation, a Cox6a2-expressing sub-type of large neurons for Aδ-LTMR sensation, as well as a Smr2-expressing sub-type of large neurons with undetermined function (Wang et al., 2023).
Non-neuronal cells, especially satellite glial cells, have also been grouped to diverse sub-types. Clustering of satellite glial cells from naïve DRG separates satellite glial cells to a Pou3f1-expressing sub-type that is associated with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways glycan biosynthesis and MAPK signaling, a Gm13889-expressing sub-type that is associated with cytokine and IL-17 signaling, a Aldh1l1-expressing sub-type that is associated with steroid biosynthesis and terpenoid backbone biosynthesis, and a Scn7a-expressing sub-type that is associated with ECM and cell adhesion pathways (Avraham et al., 2021).
Phenotypic changes of DRG neurons in the DRGs after injury
Compared with the normal status, post injury changes are observed in neurons and other non-neuronal cells in DRG. Using 10X Genomics sequencing, in mouse L4 and L5 DRGs ipsilateral to the unilateral spared nerve injury, a surgical model that cuts the tibial and common peroneal nerve branches and induces alternations of neuronal firing activity and neuropathic pain, three new sub-types of neurons with high expressions of Atf3, including a Atf3/Gfra3/Gal sub-type, a Atf3/Mrgprd sub-type, and a Atf3/S100b/Gal sub-type, have been discovered from 24 h to 14 days after spared nerve injury. The Atf3/Mrgprd-labelled neurons peaks at 24 h post injury and rapidly disappears. This transiently existed neuron sub-type has high abundance of many regeneration-associated genes such as Sox11, Sprr1a, Cacna2d1, Gadd45a, and Cckbr and may switch to the Atf3/Gfra3/Gal neuron sub-type since 2 days post injury. The Atf3/Gfra3/Gal sub-type, as well as the Atf3/S100b/Gal sub-type, two neuron sub-types that are marked by axonal regeneration promoting neuropeptide galanin, are kept expressed at high levels in mouse DRGs from 2 days to 14 days after nerve injury and related with essential biological processes for nerve regeneration, such as inflammatory responses (Wang S. et al., 2021). Notably, the usage of spared nerve injury model to investigate DRG neuron changes after nerve injury may have some limitations as the spared nerve injury model induces partial denervation of one sciatic nerve branch and may affect other cell populations in the DRGs, including other connected neurons.
A recent single-cell sequencing study uses three different types of injury models, including L3 and L4 spinal nerve transection injury, sciatic nerve transection plus ligation injury, and sciatic nerve crush injury. A novel neuronal transcriptional state with high expressions of injury-induced genes Atf3, Sprr1a, Sox11, and Flrt3, is observed in all these injury models within 24 h after nerve injury (Renthal et al., 2020). A larger number of injured state neurons emerges after a more severe injury model. As high as 92.6% of neurons are recognized as the injured neuronal sub-type at 3 days after spinal nerve transection while only 41.4% of neurons switch to the injured state at the same time point after sciatic nerve transection plus ligation. These neurons at the injured state are capable to return to the native state at later time points after nerve injury and restore their original cell identify. Transcription factor ATF3, among high-expressed molecules in the injured neuronal sub-types, is found to be an indispensable driving factor for the transcriptional reprogramming of neurons (Renthal et al., 2020). Using an in vitro DRG neuron culture model, it is demonstrated that ATF3 facilitates DRG axon growth (Avraham et al., 2022a; Chandran et al., 2016). These findings validate previously observed injury-induced elevation of ATF3 in DRG neurons (Tsujino et al., 2000) and demonstrate that ATF3 is an important marker for neurons at the injured state. Besides Atf3, some other genes that are associated with neuronal repair process have been identified, such as Adcyap1, have been identified through exploring genetic changes in DRG neurons after nerve injury using experimental design single-cell RNA sequencing processing (Chen et al., 2023).
Phenotypic changes of glial cells in the DRGs after injury
Satellite glial cells, glial cells that surround the soma of DRG neurons, exhibit diverse expression patterns with unique biological implications, especially fatty acid metabolism, at 3 days after sciatic nerve crush injury (Avraham et al., 2020). Conditional knockout of Fasn, a regulator gene of endogenous fatty acid synthesis, in satellite glial cells hinders axon regeneration while the inhibitory role of Fasn deletion in axon growth can be rescued by PPARα agonist fenofibrate. These genetic and functional findings fully illuminate that satellite glial cells respond to peripheral nerve injury and regulate neuronal actions (Avraham et al., 2020). A unique sub-type of satellite glial cells that is associated with Gene Ontology (GO) Molecular Function nervous system development and axon regeneration emerges in the DRGs at 3 days after sciatic nerve crush injury. Differentially expressed genes in this injury-specific satellite glial cell sub-type are enriched with transcription factor Zeb1, a regulator of epithelial to mesenchymal transition, as well as transcription factor Rest, a regulator of pluripotency. These findings indicate that peripheral nerve injury initiates the transition of satellite glial cells to a more plastic state (Avraham et al., 2021).
Compared with the well-elucidated genetic changes of Schwann cells at the injured site, transcriptional changes of Schwann cells in the DRGs has not been clarified until recently. Gene profiling demonstrates that Schwann cells localized in the DRGs also experience transcriptional changes at 3 days after sciatic nerve crush injury. Specifically, after peripheral nerve injury, myelination-promoting gene Ngf is downregulated and regeneration-regulating gene Yap is upregulated in Schwann cells localized in the DRGs (Avraham et al., 2021).
Phenotypic changes of immune cells in the DRGs after injury
Injury-induced changes of immune cells in the DRGs, especially macrophages, have been explored as macrophages are implicated in neurite outgrowth and nerve regeneration (Niemi et al., 2013). Emerging studies demonstrate that macrophages not only infiltrate the injured peripheral nerve stumps but also accumulate at the DRGs (Zigmond and Echevarria, 2019). Besides the quantified enlarged cell number of macrophages in the DRGs, at 3 days post sciatic nerve injury, proliferation marker Mki67 is highly expressed in macrophages and proliferation-associated pathways such as cell cycle and DNA replication are significantly enriched (Avraham et al., 2021). Moreover, in the DRGs, a rare sub-type of macrophages that non only express macrophage markers but also express satellite glial cell markers is found to be increased after nerve injury (Avraham et al., 2021). Similarly, another study also reports that macrophages with a stellate morphology and enveloping extensions exist in the axotomized DRGs instead of the DRGs in the naïve state from 24 h after sciatic nerve crush injury (Kalinski et al., 2020).
Many other non-neuronal cells in the DRG, besides glial cells and immune cells, also exhibit different biological features after peripheral nerve injury. For instance, the expressions of tight junction genes Tjp1 and Tjp2 in DRG endothelial cells are reduced at 3 days after sciatic nerve injury. Decreased expressions of barrier components in DRG endothelial cells in the injured state may impair blood-nerve barrier and increase permeability (Avraham et al., 2021).
Single-cell-based analysis of peripheral nerves
As previously reported, sciatic nerve injury model is commonly used for the identification of cellular injury responses in the DRGs. Cellular and molecular changes in the injured peripheral nerves, especially injured sciatic nerves, are also commonly investigated to examine the wound environment after peripheral nerve injury (Table 3).
Cellular population in peripheral nerves
Peripheral nerve trunks are chiefly composed of fibroblasts, Schwann cells, and immune cells. A collection of 5400 viable cells from mouse brachial plexus and sciatic nerves is annotated to a variety of cell clusters using single-cell transcriptomes, including fibroblasts, Schwann cells, endothelial cells, T cells/natural killer cells, vascular smooth muscle cells, pericytes, macrophages, myeloid lineage cells, and endothelial cells, with Schwann cells and fibroblasts occupy 65% of the cell population (Wolbert et al., 2020). Schwann cells, vascular smooth muscle cells, pericytes, endothelial cells, and immune cells are also captured in another study that analyzes 4000 cells from mouse sciatic nerves. Fibroblasts and fibroblast-like cells or mesenchymal cells are alternatively recognized as Pcolce2 and Ly6c1-expressing epineurial cells, Itgb4 and Slc2a1-expressing perineurial cells, as well as Col2a1 and Sox9-expressing endoneurial cells in this study (Gerber et al., 2021).
Schwann cells, especially Schwann cells surrounding peripheral axons, are commonly separated to myelinating and non-myelinating Schwann cells based on their morphological features. Myelinating Schwann cells form myelin sheathes wrapping neuronal axons and facilitate the rapid conduction of action potentials. The functional roles of non-myelinating Schwann cells are relatively less understood. The application of single-cell sequencing to peripheral nerves now allows the splitting of Schwann cells that express pan markers Erbb3 and S100b to a myelinating sub-type that co-expresses myelin protein genes Mbp, Mpz, Ncmap, and Plp1 as well as a non-myelinating sub-type that expresses Ncam1 and Ngfr/p75. It is demonstrated that non-myelinating Schwann cells exist in a larger number than myelinating Schwann cells (Gerber et al., 2021; Wolbert et al., 2020). Notably, Ncam1 and Ngfr/p75 are typical markers of immature Schwann cells, Schwann cells in late embryonic period and early postnatal period. Immature Schwann cells develop to myelinating and non-myelinating Schwann cells postnatally (Jessen and Mirsky, 2019). However, as far as we know, there is no direct transcriptional comparison between immature Schwann cells in embryos and non-myelinating Schwann cells in adult animals. The advanced genetic based separation of Schwann cell sub-types thus provides important molecular basis for the comparison between immature Schwann cells in embryos and non-myelinating Schwann cells as well as the investigation of the biological functions of non-myelinating Schwann cells.
Phenotypic changes of Schwann cells in peripheral nerves after injury
Schwann cells in peripheral nerve stumps undergo remarkable cellular reprogramming. Both myelinating and non-myelinating Schwann cells switch to a repair phenotype with enhanced proliferation and migration capacity following nerve injury (Jessen and Mirsky, 2019). Schwann cells collected from sciatic nerve trunks that contains the injury site and the distal nerve stumps at 3 days after sciatic nerve crush injury are classified to three sub-types, including a Mki67-marked proliferating sub-type that expresses of Ncam1, Cdl1, Erbb3, Epha5, Thbs2, Tnc, Hbegf, and Sostdc1, a pro-myelinating-associated sub-type that expresses Ngfr/p75, Nrcam, Gfra1, Btc, Gjb1, Cryab, Tnfrsf12a, and Gadd45b, as well as a flanked sub-type with high expressions of Nes, Cryab, and genes coding for extracellular matrix such as Bgn, Dcn, and Fn1 (Kalinski et al., 2020). However, due to a lack of tracing the trajectories of individual Schwann cells, the origins of these three Schwann cell sub-types are undetermined. It may be interesting to distinguish myelinating and non-myelinating Schwann cells and to explore the destination of myelinating and non-myelinating Schwann cells following nerve injury. In addition, the presences of immature Schwann cell marker Ncam1 in Mki67-marked proliferating Schwann cell sub-type in the injured sciatic nerve stumps as well as the expression of immature Schwann cell marker Ngfr/p75 in pro-myelinating-associated Schwann cell sub-type imply that there may exist certain similarity between immature Schwann cells and repair Schwann cells. Another study investigates the expressions of Schwann cell markers in peripheral nerves at multiple stages of embryonic and postnatal development (E13.5, E17.5, P1, P5, P14, P24, and P60) and detects the high abundance of proliferating Schwann cell markers Mki67 and Top2a as well as pro-myelinating Schwann cell marker Ncam1 in developing sciatic nerves (E17.5) (Gerber et al., 2021). The identification of proliferating and pro-myelinating Schwann cell markers in embryonic mice further reveals certain transcriptional similarity between the development and the regeneration process.
Phenotypic changes of immune cells in peripheral nerves after injury
The recruitment and infiltration of immune cells to the injured site are more vigorous as compared with the remote DRG tissues (Kalinski et al., 2020). The presence of immune cells, such as macrophages and neutrophils, contributes to debris removal during Wallerian degeneration, an essential preparatory stage for success regeneration (Lindborg et al., 2017; Zigmond and Echevarria, 2019). Single-cell sequencing of mouse sciatic nerves at the injured site and the distal nerve stumps at 3 days after sciatic nerve crush injury reveals that innate immune cells occupy approximately 44% of cell population and demonstrates the presence of T cells/natural killer cells, granulocytes, conventional dendritic cells, monocyte-derived dendritic cells, macrophages, and monocytes in sciatic nerves (Kalinski et al., 2020). Macrophages that express canonical markers Adgre1, Aif1, Cd68, and Cx3cr1 are further classified to distinct sub-populations with diverse distributions. For instance, Arg1+ macrophages are chiefly localized at the injured sites while F4/80+ macrophages are largely localized at the distal nerve stumps (Kalinski et al., 2020).
Steady-state sciatic nerve macrophages also demonstrate distribution-specific sub-populations. In the intact mouse sciatic nerves, a larger number of macrophages are Relmα− Mgl1- Iba-1+ macrophages and are localized inside the endoneurium of the sciatic nerve. A relative smaller number of macrophages are Relmα+ Mgl1+ Iba-1+ macrophages and are localized in the epineurium and the connective tissues surrounding nerve fascicles (Ydens et al., 2020). These two anatomically different sub-populations of macrophages exhibit dissimilar responses to peripheral nerve injury. Following peripheral nerve injury, epineurial Relmα+ Mgl1+ resident macrophages do not sense injury signals while endoneurial Relmα− Mgl1- macrophages are temporarily activated and can further be clustered to two sub-groups with differentially expressed genes enriched in canonical pathway granulocyte adhesion and diapedesis. Additionally, two novel clusters of macrophages, including a cluster of S100a4-expressing macrophages and a cluster of H2-Aa-expressing macrophages are found in the injured sciatic nerves at 1 day and 5 days after nerve injury, respectively. These emerged new macrophage sub-populations are considered to be newly recruited monocyte-derived macrophages and recruited macrophages that are further adapted to the wound microenvironment (Ydens et al., 2020). Single-cell sequencing of dissected the nerve injury site and distal nerve at 1 day, 3 days, and 7 days post sciatic nerve crush injury not only reveals the rapid accumulation of monocytes and macrophages in the injured nerve, but also demonstrates the metabolic reprogramming of immune cells, with monocytes and macrophages switch from a high glycolytic flux toward oxidative phosphorylation (Zhao et al., 2022).
Besides immune cells, structural cells in the peripheral nerve stumps also participate in shaping peripheral nerve inflammation. Mesenchymal progenitor cells are another main population in injured sciatic nerve stumps. Following sciatic nerve injury, mesenchymal progenitor cells express chemokines and display immune activity (Kalinski et al., 2020).
Cell-cell interactions in peripheral nerves
Separating heterogenetic cell populations at the single-cell level fosters the understanding of intercellular communications. Script and software, such as CellChat (Jin et al., 2021), and CellPhoneDB (Efremova et al., 2020), quantify associations among ligands and receptors and thus infer interactions between different cell types and even sub-types. By using CellPhoneDB to interpret single-cell sequencing atlas of rat DRG, active ligand-receptor bindings among endothelial cells, fibroblasts, and vascular smooth muscle cells are found in neonatal rat DRG (Zhang et al., 2021). In neonatal rat sciatic nerves, close interactions between endothelial cells and fibroblasts as well as receptor-ligand pairs a2b1 complex-Col1a1/Col3a1 between endothelial cells are revealed, providing important genetic foundation for the discovery of innovative signal transductions (Zhang et al., 2021). Interestingly, although Schwann cells occupy a large cell population in peripheral nerve stumps, there are relatively few receptor-ligand pairs between Schwann cells and other cell types in intact sciatic nerves. Analyses of Drop-seq data of injured mouse sciatic nerves with Python script Cellcellinteractnet, combined with cell-surface mass spectrometry, predict considerable interactions of Schwann cells and endoneurial cells with axons (Toma et al., 2020). Paracrine interactions between surrounding cells with axons may thus support the growth and elongation of axons and accelerate peripheral nerve regeneration. An advanced bioinformatic tool Scriabin has been recently developed to analysis cell-cell interaction matrix at single-cell resolution without cell aggregation and therefore more single-cell based interaction features are expected (Wilk et al., 2023).
Conclusion and perspective
The development of single-cell sequencing technologies and bioinformatic tools has enabled us to quickly obtain gene expression profiling of large numbers of individual cells and reveal the state and function of single cells. Herein, we summarize single-cell sequencing investigations of peripheral nerves, identify cellular and molecular hallmarks in the DRGs and peripheral nerve stumps in the native and injured states, and illuminate remote responses in axotomized DRGs and local response in injured peripheral nerve stumps. Still, it is worth mentioning that peripheral nerve injury also elicits changes in motor neurons whose neuronal somata are located in the spinal cord. The investigation of transcriptomic changes in motor neurons after peripheral nerve injury may contribute to the identification of potential strategies for the promotion of peripheral nerve regeneration, especially the recovery of motor functions.
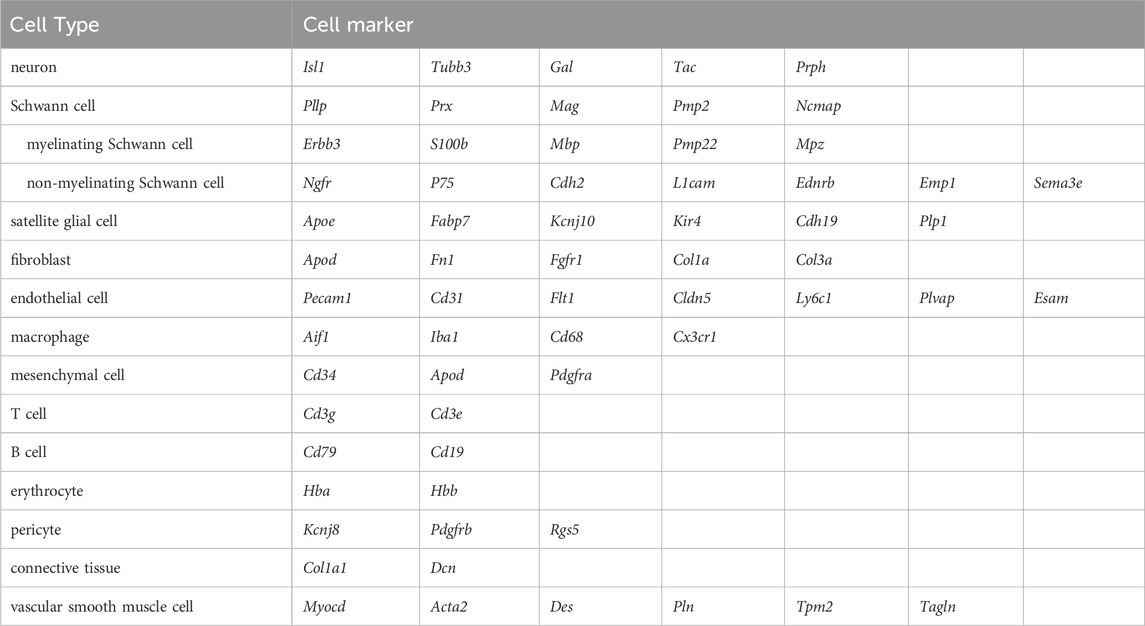
Additionally, numerous cellular markers are either validated or newly-identified using single-cell sequencing. For instance, fatty acid binding protein gene Fabp7, a gene that is top enriched in the satellite glial cell cluster in the DRGs, clearly labels satellite glial cells enveloping neuronal soma, but does not label Schwann cells, and consequently is recognized as a novel specific marker for satellite glial cells (Avraham et al., 2020). Lipoprotein gene Apod, a gene that has been previously demonstrated to be present in Schwann cells, is expressed in a robustly higher abundance in non-myelinating Schwann cells as compared with myelinating Schwann cells and accordingly can be used for the identification of non-myelinating Schwann cells (Wolbert et al., 2020). Markers of peripheral cells are summarized and listed in Table 4. Improved algorithm software, such as OnClass, helps the categorization of cell populations and the identification of marker genes on a data-centric basis, regardless of the training data, and therefore may help paving the way for a more unseen cell types and sub-types (Wang K. et al., 2021).
As a recently developed technology, single-cell sequencing technology still has some limitations (Garcia-Flores et al., 2023; Mutisheva et al., 2022). Complete separation of cell populations and acquisition of single cells are initial steps that fundamentally affect the precision of sequencing data. Take rodent sciatic nerve tissues, for example, it generally takes hours from tissue collection to cell suspension loading. Extended time may impact the quality of isolated cells and induce inaccurate outcomes. Proper enzyme digestion methods and single-cell separation procedures are thus critical for gene expression profiling. Another weakness is that it is hard to obtain a consistent and comparable cellular types and sub-types among different single-cell sequencing data due to inaccurate measurement, differently employed marker genes, and diverse clustering. Improved sequencing technologies and bioinformatic tools as well as enlarged recognition of marker genes, especially marker genes for rare cells, may help to solve the existing problem and offer matched data for molecular characterization and biological implication (Pullin and McCarthy, 2024).
Taken together, a better understanding of the peripheral nerve heterogeneity during the peripheral nerve injury and regeneration process benefit the cognition of essential molecules and novel cell sub-populations that regulate peripheral nerve regeneration and hence may contribute to the discovery new therapeutic drugs targeting these molecules and cell sub-populations.
Author contributions
LZ: Data curation, Visualization, Writing–review and editing. CJ: Data curation, Visualization, Writing–review and editing. BY: Conceptualization, Writing–original draft. JZ: Data curation, Writing–review and editing. YS: Conceptualization, Writing–original draft. SY: Conceptualization, Writing–original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Key R&D Program of China (2021YFA1201404), Jiangsu Provincial Research (YJXYY202204), and Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX23_3382).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Au, N. P. B., Wu, T., Chen, X., Gao, F., Li, Y. T. Y., Tam, W. Y., et al. (2023). Genome-wide study reveals novel roles for formin-2 in axon regeneration as a microtubule dynamics regulator and therapeutic target for nerve repair. Neuron 111, 3970–3987.e8. doi:10.1016/j.neuron.2023.11.011
Avraham, O., Chamessian, A., Feng, R., Yang, L., Halevi, A. E., Moore, A. M., et al. (2022a). Profiling the molecular signature of satellite glial cells at the single cell level reveals high similarities between rodents and humans. Pain 163, 2348–2364. doi:10.1097/j.pain.0000000000002628
Avraham, O., Deng, P. Y., Jones, S., Kuruvilla, R., Semenkovich, C. F., Klyachko, V. A., et al. (2020). Satellite glial cells promote regenerative growth in sensory neurons. Nat. Commun. 11, 4891. doi:10.1038/s41467-020-18642-y
Avraham, O., Feng, R., Ewan, E. E., Rustenhoven, J., Zhao, G., and Cavalli, V. (2021). Profiling sensory neuron microenvironment after peripheral and central axon injury reveals key pathways for neural repair. eLife 10, e68457. doi:10.7554/eLife.68457
Avraham, O., Le, J., Leahy, K., Li, T., Zhao, G., and Cavalli, V. (2022b). Analysis of neuronal injury transcriptional response identifies CTCF and YY1 as co-operating factors regulating axon regeneration. Front. Mol. Neurosci. 15, 967472. doi:10.3389/fnmol.2022.967472
Büttner, R., Schulz, A., Reuter, M., Akula, A. K., Mindos, T., Carlstedt, A., et al. (2018). Inflammaging impairs peripheral nerve maintenance and regeneration. Aging Cell. 17, e12833. doi:10.1111/acel.12833
Catala, M., and Kubis, N. (2013). Gross anatomy and development of the peripheral nervous system. Handb. Clin. Neurol. 115, 29–41. doi:10.1016/b978-0-444-52902-2.00003-5
Chandran, V., Coppola, G., Nawabi, H., Omura, T., Versano, R., Huebner, E. A., et al. (2016). A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89, 956–970. doi:10.1016/j.neuron.2016.01.034
Chen, P., Piao, X., and Bonaldo, P. (2015). Role of macrophages in Wallerian degeneration and axonal regeneration after peripheral nerve injury. Acta Neuropathol. 130, 605–618. doi:10.1007/s00401-015-1482-4
Chen, Q., Zhang, X. Y., Wang, Y. P., Fu, Y. J., Cao, F., Xu, Y. N., et al. (2023). Unveiling adcyap1 as a protective factor linking pain and nerve regeneration through single-cell RNA sequencing of rat dorsal root ganglion neurons. BMC Biol. 21, 235. doi:10.1186/s12915-023-01742-8
Chernov, A. V., Dolkas, J., Hoang, K., Angert, M., Srikrishna, G., Vogl, T., et al. (2015). The calcium-binding proteins S100A8 and S100A9 initiate the early inflammatory program in injured peripheral nerves. J. Biol. Chem. 290, 11771–11784. doi:10.1074/jbc.M114.622316
Cuevas-Diaz Duran, R., González-Orozco, J. C., Velasco, I., and Wu, J. Q. (2022). Single-cell and single-nuclei RNA sequencing as powerful tools to decipher cellular heterogeneity and dysregulation in neurodegenerative diseases. Front. Cell. Dev. Biol. 10, 884748. doi:10.3389/fcell.2022.884748
Denisenko, E., Guo, B. B., Jones, M., Hou, R., de Kock, L., Lassmann, T., et al. (2020). Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21, 130. doi:10.1186/s13059-020-02048-6
Ding, J., Adiconis, X., Simmons, S. K., Kowalczyk, M. S., Hession, C. C., Marjanovic, N. D., et al. (2020). Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746. doi:10.1038/s41587-020-0465-8
Drokhlyansky, E., Smillie, C. S., Van Wittenberghe, N., Ericsson, M., Griffin, G. K., Eraslan, G., et al. (2020). The human and mouse enteric nervous system at single-cell resolution. Cell. 182, 1606–1622. doi:10.1016/j.cell.2020.08.003
Efremova, M., Vento-Tormo, M., Teichmann, S. A., and Vento-Tormo, R. (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506. doi:10.1038/s41596-020-0292-x
Ehmedah, A., Nedeljkovic, P., Dacic, S., Repac, J., Draskovic Pavlovic, B., Vucevic, D., et al. (2019). Vitamin B complex treatment attenuates local inflammation after peripheral nerve injury. Molecules 24, 4615. doi:10.3390/molecules24244615
Fang, T., Shao, Y., Oswald, T., Lineaweaver, W. C., and Zhang, F. (2013). Effect of sildenafil on peripheral nerve regeneration. Ann. Plast. Surg. 70, 62–65. doi:10.1097/SAP.0b013e31826a1aff
Garcia-Flores, V., Xu, Y., Pusod, E., Romero, R., Pique-Regi, R., and Gomez-Lopez, N. (2023). Preparation of single-cell suspensions from the human placenta. Nat. Protoc. 18, 732–754. doi:10.1038/s41596-022-00772-w
Gerber, D., Pereira, J. A., Gerber, J., Tan, G., Dimitrieva, S., Yángüez, E., et al. (2021). Transcriptional profiling of mouse peripheral nerves to the single-cell level to build a sciatic nerve ATlas (SNAT). eLife 10, e58591. doi:10.7554/eLife.58591
Gold, B. G., Storm-Dickerson, T., and Austin, D. R. (1994). The immunosuppressant FK506 increases functional recovery and nerve regeneration following peripheral nerve injury. Restor. Neurol. Neurosci. 6, 287–296. doi:10.3233/rnn-1994-6404
Govindappa, P. K., and Elfar, J. C. (2022). Erythropoietin promotes M2 macrophage phagocytosis of Schwann cells in peripheral nerve injury. Cell. Death Dis. 13, 245. doi:10.1038/s41419-022-04671-6
He, Z., and Jin, Y. (2016). Intrinsic control of axon regeneration. Neuron 90, 437–451. doi:10.1016/j.neuron.2016.04.022
Jessen, K. R., and Arthur-Farraj, P. (2019). Repair Schwann cell update: adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia 67, 421–437. doi:10.1002/glia.23532
Jessen, K. R., and Mirsky, R. (2019). Schwann cell precursors; multipotent glial cells in embryonic nerves. Front. Mol. Neurosci. 12, 69. doi:10.3389/fnmol.2019.00069
Jia, Y., Zhang, M., Li, P., Tang, W., Liu, Y., Hu, Y., et al. (2020). Bioinformatics analysis of long non-coding RNAs involved in nerve regeneration following sciatic nerve injury. Mol. Pain 16, 1744806920971918. doi:10.1177/1744806920971918
Jin, S., Guerrero-Juarez, C. F., Zhang, L., Chang, I., Ramos, R., Kuan, C. H., et al. (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088. doi:10.1038/s41467-021-21246-9
Kalinski, A. L., Yoon, C., Huffman, L. D., Duncker, P. C., Kohen, R., Passino, R., et al. (2020). Analysis of the immune response to sciatic nerve injury identifies efferocytosis as a key mechanism of nerve debridement. Elife 9, e60223. doi:10.7554/eLife.60223
Korkmaz, M. F., Parlakpınar, H., Ceylan, M. F., Ediz, L., Şamdancı, E., Kekilli, E., et al. (2016). The effect of sildenafil on recuperation from sciatic nerve injury in rats. Balk. Med. J. 33, 204–211. doi:10.5152/balkanmedj.2016.14701
Kubo, T., Yamashita, T., Yamaguchi, A., Hosokawa, K., and Tohyama, M. (2002). Analysis of genes induced in peripheral nerve after axotomy using cDNA microarrays. J. Neurochem. 82, 1129–1136. doi:10.1046/j.1471-4159.2002.01060.x
Lee, J. I., Hur, J. M., You, J., and Lee, D. H. (2020). Functional recovery with histomorphometric analysis of nerves and muscles after combination treatment with erythropoietin and dexamethasone in acute peripheral nerve injury. PLoS One 15, e0238208. doi:10.1371/journal.pone.0238208
Li, C. L., Li, K. C., Wu, D., Chen, Y., Luo, H., Zhao, J. R., et al. (2016). Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell. Res. 26, 83–102. doi:10.1038/cr.2015.149
Li, S., Xue, C., Yuan, Y., Zhang, R., Wang, Y., Wang, Y., et al. (2015). The transcriptional landscape of dorsal root ganglia after sciatic nerve transection. Sci. Rep. 5, 16888. doi:10.1038/srep16888
Li, S., Yu, B., Wang, S., Gu, Y., Yao, D., Wang, Y., et al. (2012). Identification and functional analysis of novel micro-RNAs in rat dorsal root ganglia after sciatic nerve resection. J. Neurosci. Res. 90, 791–801. doi:10.1002/jnr.22814
Li, S., Yu, B., Wang, Y., Yao, D., Zhang, Z., and Gu, X. (2011). Identification and functional annotation of novel microRNAs in the proximal sciatic nerve after sciatic nerve transection. Sci. China Life Sci. 54, 806–812. doi:10.1007/s11427-011-4213-7
Lindborg, J. A., Mack, M., and Zigmond, R. E. (2017). Neutrophils are critical for myelin removal in a peripheral nerve injury model of wallerian degeneration. J. Neurosci. 37, 10258–10277. doi:10.1523/jneurosci.2085-17.2017
Lu, A., Huang, Z., Zhang, C., Zhang, X., Zhao, J., Zhang, H., et al. (2014). Differential expression of microRNAs in dorsal root ganglia after sciatic nerve injury. Neural Regen. Res. 9, 1031–1040. doi:10.4103/1673-5374.133164
Mao, S., Zhang, S., Zhou, Z., Shi, X., Huang, T., Feng, W., et al. (2018). Alternative RNA splicing associated with axon regeneration after rat peripheral nerve injury. Exp. Neurol. 308, 80–89. doi:10.1016/j.expneurol.2018.07.003
Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M., and Gilad, Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517. doi:10.1101/gr.079558.108
Meng, Q., Burrell, J. C., Zhang, Q., and Le, A. D. (2023). Potential application of orofacial MSCs in tissue engineering nerve guidance for peripheral nerve injury repair. Stem Cell. Rev. Rep. 19, 2612–2631. doi:10.1007/s12015-023-10609-y
Murakami, Y., Tanahashi, T., Okada, R., Toyoda, H., Kumada, T., Enomoto, M., et al. (2014). Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One 9, e106314. doi:10.1371/journal.pone.0106314
Murtazina, A., and Adameyko, I. (2023). The peripheral nervous system. Development 150, dev201164. doi:10.1242/dev.201164
Mutisheva, I., Robatel, S., Bäriswyl, L., and Schenk, M. (2022). An innovative approach to tissue processing and cell sorting of fixed cells for subsequent single-cell RNA sequencing. Int. J. Mol. Sci. 23, 10233. doi:10.3390/ijms231810233
Nguyen, M. Q., von Buchholtz, L. J., Reker, A. N., Ryba, N. J., and Davidson, S. (2021). Single-nucleus transcriptomic analysis of human dorsal root ganglion neurons. Elife 10, e71752. doi:10.7554/eLife.71752
Niemi, J. P., DeFrancesco-Lisowitz, A., Roldán-Hernández, L., Lindborg, J. A., Mandell, D., and Zigmond, R. E. (2013). A critical role for macrophages near axotomized neuronal cell bodies in stimulating nerve regeneration. J. Neurosci. 33, 16236–16248. doi:10.1523/jneurosci.3319-12.2013
Papalexi, E., and Satija, R. (2018). Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18, 35–45. doi:10.1038/nri.2017.76
Pullin, J. M., and McCarthy, D. J. (2024). A comparison of marker gene selection methods for single-cell RNA sequencing data. Genome Biol. 25, 56. doi:10.1186/s13059-024-03183-0
Renthal, W., Tochitsky, I., Yang, L., Cheng, Y. C., Li, E., Kawaguchi, R., et al. (2020). Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron 108, 128–144. doi:10.1016/j.neuron.2020.07.026
Sasselli, V., Pachnis, V., and Burns, A. J. (2012). The enteric nervous system. Dev. Biol. 366, 64–73. doi:10.1016/j.ydbio.2012.01.012
Savastano, L. E., Laurito, S. R., Fitt, M. R., Rasmussen, J. A., Gonzalez Polo, V., and Patterson, S. I. (2014). Sciatic nerve injury: a simple and subtle model for investigating many aspects of nervous system damage and recovery. J. Neurosci. Methods 227, 166–180. doi:10.1016/j.jneumeth.2014.01.020
Scheib, J., and Höke, A. (2013). Advances in peripheral nerve regeneration. Nat. Rev. Neurol. 9, 668–676. doi:10.1038/nrneurol.2013.227
Selewa, A., Dohn, R., Eckart, H., Lozano, S., Xie, B., Gauchat, E., et al. (2020). Systematic comparison of high-throughput single-cell and single-nucleus transcriptomes during cardiomyocyte differentiation. Sci. Rep. 10, 1535. doi:10.1038/s41598-020-58327-6
Sohn, E. J., and Park, H. T. (2020). Differential expression of circular RNAs in the proximal and distal segments of the sciatic nerve after injury. Neuroreport 31, 76–84. doi:10.1097/wnr.0000000000001371
Soneson, C., and Delorenzi, M. (2013). A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinforma. 14, 91. doi:10.1186/1471-2105-14-91
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382. doi:10.1038/nmeth.1315
Toma, J. S., Karamboulas, K., Carr, M. J., Kolaj, A., Yuzwa, S. A., Mahmud, N., et al. (2020). Peripheral nerve single-cell analysis identifies mesenchymal ligands that promote axonal growth. eNeuro 7, ENEURO.0066–20.2020. doi:10.1523/eneuro.0066-20.2020
Tsujino, H., Kondo, E., Fukuoka, T., Dai, Y., Tokunaga, A., Miki, K., et al. (2000). Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol. Cell. Neurosci. 15, 170–182. doi:10.1006/mcne.1999.0814
Usoskin, D., Furlan, A., Islam, S., Abdo, H., Lönnerberg, P., Lou, D., et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153. doi:10.1038/nn.3881
Wang, H., Wu, J., Zhang, X., Ding, L., and Zeng, Q. (2018). Microarray analysis of the expression profile of lncRNAs reveals the key role of lncRNA BC088327 as an agonist to heregulin‑1β‑induced cell proliferation in peripheral nerve injury. Int. J. Mol. Med. 41, 3477–3484. doi:10.3892/ijmm.2018.3571
Wang, K., Cai, B., Song, Y., Chen, Y., and Zhang, X. (2023). Somatosensory neuron types and their neural networks as revealed via single-cell transcriptomics. Trends Neurosci. 46, 654–666. doi:10.1016/j.tins.2023.05.005
Wang, K., Wang, S., Chen, Y., Wu, D., Hu, X., Lu, Y., et al. (2021a). Single-cell transcriptomic analysis of somatosensory neurons uncovers temporal development of neuropathic pain. Cell. Res. 31, 904–918. doi:10.1038/s41422-021-00479-9
Wang, S., Pisco, A. O., McGeever, A., Brbic, M., Zitnik, M., Darmanis, S., et al. (2021b). Leveraging the Cell Ontology to classify unseen cell types. Nat. Commun. 12, 5556. doi:10.1038/s41467-021-25725-x
Wilk, A. J., Shalek, A. K., Holmes, S., and Blish, C. A. (2023). Comparative analysis of cell-cell communication at single-cell resolution. Nat. Biotechnol. 42, 470–483. doi:10.1038/s41587-023-01782-z
Wolbert, J., Li, X., Heming, M., Mausberg, A. K., Akkermann, D., Frydrychowicz, C., et al. (2020). Redefining the heterogeneity of peripheral nerve cells in health and autoimmunity. Proc. Natl. Acad. Sci. U. S. A. 117, 9466–9476. doi:10.1073/pnas.1912139117
Wong, K. M., Babetto, E., and Beirowski, B. (2017). Axon degeneration: make the Schwann cell great again. Neural Regen. Res. 12, 518–524. doi:10.4103/1673-5374.205000
Xie, K., Cheng, X., Zhu, T., and Zhang, D. (2023). Single-cell transcriptomic profiling of dorsal root ganglion: an overview. Front. Neuroanat. 17, 1162049. doi:10.3389/fnana.2023.1162049
Ydens, E., Amann, L., Asselbergh, B., Scott, C. L., Martens, L., Sichien, D., et al. (2020). Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat. Neurosci. 23, 676–689. doi:10.1038/s41593-020-0618-6
Yim, A. K. Y., Wang, P. L., Bermingham, J. R., Hackett, A., Strickland, A., Miller, T. M., et al. (2022). Disentangling glial diversity in peripheral nerves at single-nuclei resolution. Nat. Neurosci. 25, 238–251. doi:10.1038/s41593-021-01005-1
Zahalka, A. H., and Frenette, P. S. (2020). Nerves in cancer. Nat. Rev. Cancer 20, 143–157. doi:10.1038/s41568-019-0237-2
Zeng, H., and Sanes, J. R. (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546. doi:10.1038/nrn.2017.85
Zhang, C., Hu, M. W., Wang, X. W., Cui, X., Liu, J., Huang, Q., et al. (2022). scRNA-sequencing reveals subtype-specific transcriptomic perturbations in DRG neurons of EGFPf mice in neuropathic pain condition. Elife 11, e76063. doi:10.7554/eLife.76063
Zhang, J., Zhang, Y., Chen, L., Rao, Z., and Sun, Y. (2020). Ulinastatin promotes regeneration of peripheral nerves after sciatic nerve injury by targeting let-7 microRNAs and enhancing NGF expression. Drug Des. Devel Ther. 14, 2695–2705. doi:10.2147/dddt.S255158
Zhang, R., Chen, S., Wang, X., Gu, X., and Yi, S. (2021). Cell populations in neonatal rat peripheral nerves identified by single-cell transcriptomics. Glia 69, 765–778. doi:10.1002/glia.23928
Zhang, X., Li, T., Liu, F., Chen, Y., Yao, J., Li, Z., et al. (2019). Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell. 73, 130–142. doi:10.1016/j.molcel.2018.10.020
Zhao, L., and Yi, S. (2019). Transcriptional landscape of alternative splicing during peripheral nerve injury. J. Cell. Physiol. 234, 6876–6885. doi:10.1002/jcp.27446
Zhao, X. F., Huffman, L. D., Hafner, H., Athaiya, M., Finneran, M. C., Kalinski, A. L., et al. (2022). The injured sciatic nerve atlas (iSNAT), insights into the cellular and molecular basis of neural tissue degeneration and regeneration. Elife 11, e80881. doi:10.7554/eLife.80881
Zigmond, R. E., and Echevarria, F. D. (2019). Macrophage biology in the peripheral nervous system after injury. Prog. Neurobiol. 173, 102–121. doi:10.1016/j.pneurobio.2018.12.001
Keywords: peripheral nervous system, peripheral nerve injury, dorsal root ganglion, sciatic nerve, single-cell sequencing
Citation: Zhao L, Jiang C, Yu B, Zhu J, Sun Y and Yi S (2024) Single-cell profiling of cellular changes in the somatic peripheral nerves following nerve injury. Front. Pharmacol. 15:1448253. doi: 10.3389/fphar.2024.1448253
Received: 13 June 2024; Accepted: 20 September 2024;
Published: 02 October 2024.
Edited by:
Juana Gallar, Miguel Hernández University of Elche, SpainReviewed by:
Victor Guaiquil, University of Illinois Chicago, United StatesIgnacio Alcalde, Instituto Universitario Fernández-Vega, Spain
Copyright © 2024 Zhao, Jiang, Yu, Zhu, Sun and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwei Zhu, emh1amlhbndlaV9udEAxNjMuY29t; Yuyu Sun, c3VueXV5dW50QDEyNi5jb20=; Sheng Yi, c3lpQG50dS5lZHUuY24=
†These authors have contributed equally to this work
 Li Zhao1†
Li Zhao1† Bin Yu
Bin Yu Sheng Yi
Sheng Yi