- 1Hunan Provincial Maternal and Child Health Care Hospital, Changsha, China
- 2Department of Pharmacy, Xiangya Hospital, Central South University, Changsha, China
- 3Hunan Institute for Tuberculosis Control and Hunan Chest Hospital, Changsha, China
- 4Hunan Chest Hospital, Changsha, China
Background: Platinum-based chemotherapy bring severe hematological toxicity that can lead to dose reduction or discontinuation of therapy. Genetic variations have been reported to influence the risk and extent of hematological toxicity; however, the results are controversial and a comprehensive overview is lacking. This systematic review aimed to identify genetic biomarkers of platinum-induced hematological toxicity.
Method: Pubmed, Embase and Web of science database were systematically reviewed for studies that evaluated the association of genetic variants and platinum-related hematological toxicity in tumor patients with no prior history of chemotherapy or radiation, published from inception to the 28th of January 2022. The studies should have specific toxicity scoring system as well as defined toxicity end-point. The quality of reporting was assessed using the Strengthening the Reporting of Genetic Association Studies (STREGA) checklist. Results were summarized using narrative synthesis.
Results: 83 studies were eligible with over 682 single-nucleotide polymorphisms across 110 genes. The results are inconsistent and diverse with methodological issues including insufficient sample size, population stratification, various treatment schedule and toxicity end-point, and inappropriate statistics. 11 SNPs from 10 genes (ABCB1 rs1128503, GSTP1 rs1695, GSTM1 gene deletion, ERCC1 rs11615, ERCC1 rs3212986, ERCC2 rs238406, XPC rs2228001, XPCC1 rs25487, MTHFR rs1801133, MDM2 rs2279744, TP53 rs1042522) had consistent results in more than two independent populations. Among them, GSTP1 rs1695, ERCC1 rs11615, ERCC1 rs3212986, and XRCC1 rs25487 present the most promising results.
Conclusion: Even though the results are inconsistent and several methodological concerns exist, this systematic review identified several genetic variations that deserve validation in well-defined studies with larger sample size and robust methodology.
Systematic Review Registration: https://www.crd.york.ac.uk/, identifier CRD42021234164.
1 Introduction
Platinum agents, including cisplatin, carboplatin and oxaliplatin, are used effectively against various tumor diseases either as monotherapy or in combination with other chemotherapeutics, radiation therapy and/or surgery. However, they display a range of severe side effects due to their poor selectivity for cancerous tissue over normal tissue. Hematological toxicity caused by platinum drugs are those that affect bone marrow function and blood cell production, characterized by leukopenia, neutropenia, thrombocytopenia, and anemia (Oun et al., 2018). Leukopenia or neutropenia, can leave patients susceptible to infections. Platinum-induced anemia is persisting erythropoietin deficiency state correlated with renal tubular dysfunction (Wood and Hrushesky, 1995). Acute myelosuppression occurs shortly after chemotherapy, while residual bone marrow injury manifested by a decrease in hematopoietic stem cell reserves or a myelodysplastic syndrome (Wang et al., 2006).
All three platinum drugs can cause some form of hematological toxicity, and myelosuppression is the dose-limiting toxicity of carboplatin. In the majority of cases, neither cisplatin nor oxaliplatin is associated with severe myelosuppression (Rabik and Dolan, 2007). Carboplatin induced myelosuppression resulted in neutropenia and thrombocytopenia. Severe (grade 3 or 4) neutropenia occurs in approximately 18% of carboplatin-treated patients, whereas severe thrombocytopenia occurs in approximately 25% of cases (Go and Adjei, 1999). Cisplatin-induced hematological toxicity is usually mild at intermittent doses of 50–60 mg/m2 and myelosuppression presents in 25%–30% of patients (Prestayko et al., 1979). Myelosuppression caused by oxaliplatin is generally mild. Grade 3/4 anemia, neutropenia and thrombocytopenia are observed in only 2%–3% of patients (Hartmann and Lipp, 2003). Hematological toxicity aggravates when platinum agents were combined with other cytotoxic drugs. The degree of hematological toxicity varies upon different chemotherapy regimen. For example, hematological toxicity was more profound in lung cancer patients treated with platinum agents plus gemcitabine (Fisher and D’Orazio, 2000; Schiller et al., 2002).
Inhibition of cell proliferation is one of the major causes of platinum-induced myelosuppression and related complications. The cytotoxicity of platinum on hematopoietic stem cells is attributed to its highly reactive hydrated platinum complex that binds to DNA and form intra- and inter-strand crosslinks; thereby produce subsequent interference with DNA transcription and/or DNA replication (Das et al., 2008). The generation in oxidative stress is also responsible for platinum-induced bone marrow toxicity (Basu et al., 2015). Increased platinum influx, decreased platinum efflux, impaired cell detoxification, low or absent DNA damage repair and activated cell death signaling may be the reasons of platinum-induced hematological toxicity (Shaloam and Paul, 2014) (Figure 1).
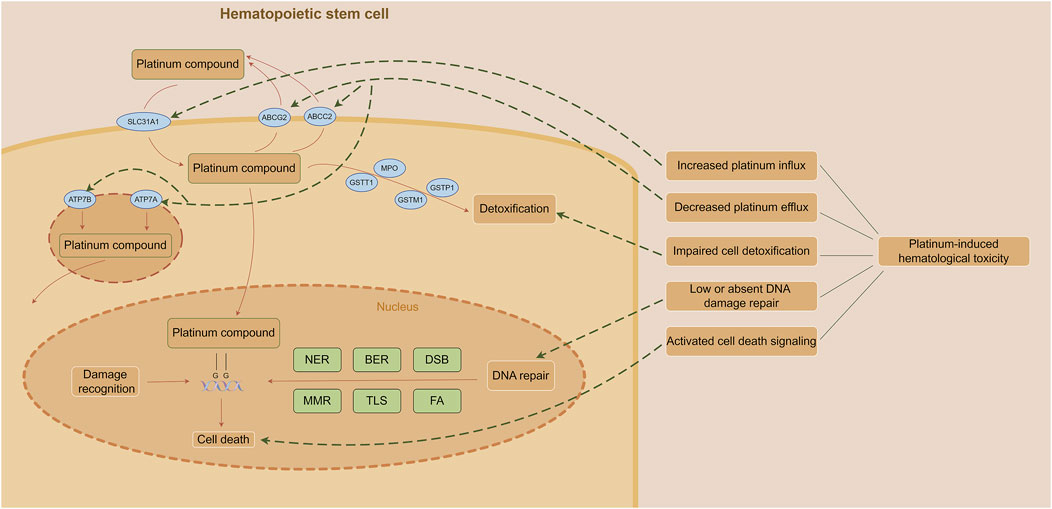
Figure 1. Mechanism of platinum-induced hematological toxicity. (The figure was made by Figdraw) Abbreviations: BER, base excision repair; DSB, double strand break repair; FA, fanconi anemia pathway; MMR, mismatch repair; NER, nucleotide excision repair; TLS, translesion DNA synthesis.
Identifying patients at greatest risk for these complications would be clinically useful for selecting patients for chemotherapy and planning the frequency of monitor and clinical treatment with colony-stimulating factor. Risk factors for hematological toxicities include kidney function, age, drug doses, combination chemotherapy, a poor performance status and prior chemotherapy exposure (Hartmann and Lipp, 2003; Ouyang et al., 2013). Furthermore, genetic variations in genes encoding proteins involved in pharmacokinetic and pharmacodynamic processes influence the occurrence and extent of adverse reactions (Zheng et al., 2020). Although several genetic polymorphisms have been identified to influence platinum-induced hematological toxicity, a comprehensive overview is lacking. We here provide an overview to identify which genetic variants consistently associated with hematological toxicity and discuss limitations of current pharmacogenetic analyses and formulate directions for further research.
2 Methods
2.1 Study eligibility
The systematic review was reported according to the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) checklist (Page et al., 2021) (Supplementary Table S1). The protocol was registered in the international prospective register of systematic reviews (PROSPERO; Registered number: CRD42021234164). The inclusion criteria were (Oun et al., 2018): studies that focus on the association between hematological toxicity and genetic polymorphisms (Wood and Hrushesky, 1995); studies including cancer patients using platinum-containing chemotherapy (Wang et al., 2006); studies that have specific toxicity scoring system and defined toxicity end-point. The exclusion criteria include the followings (Oun et al., 2018): preclinical studies (animal experiment or in vitro studies) (Wood and Hrushesky, 1995); studies in which patients were treated with concurrent radiotherapy (Wang et al., 2006); studies that were non-English, case report, review or meta-analysis (Rabik and Dolan, 2007); studies in which patients have prior history of chemotherapy and/or radiation.
2.2 Search strategy
PubMed/MEDLINE, EMBASE and Web of Science were searched for publications from inception to the 28th of January 2022. The literature search was conducted using Medical Subject Headings and combinations of relevant keywords. The detailed search strategy can be found in Supplementary Table S2. Additional research papers were identified by screening the reference sections of included articles. Two authors (YZ and MT) independently performed the data screening. Disagreements were consulted with a third arbiter (ZD).
2.3 Quality assessment
The quality of the studies will be assessed using a scoring system modified from a previously published study (Leusink et al., 2016) based on STREGA recommendations (Little et al., 2009) Supplementary Table S3. The scoring system contains ten items on five domains: clinical information, genotyping, study population origin, sample size and statistical correction for multiple testing, and study analysis. Each study included in this review was assessed for quality as good (overall quality score:7-10), moderate (overall quality score:4-6), or poor (overall quality score≤3) based on scores. Two reviewers (YZ and MT) will assess the quality independently and a third reviewer (PC) will be consulted in case of disagreement.
2.4 Data collection and analysis
The following data were extracted from each publication by two authors (YZ and MT): author, year, source of study (reference), sample size, ethnicity, type of cancer, number of treatment cycles, treatment schedule, toxicity scoring system, defined toxicity end-point, genetic polymorphisms involved and main study results.
Due to the heterogeneity in the patient population, treatment schedule, outcome definitions, meta-analysis was not appropriate. Studies were analyzed using a narrative synthesis approach.
3 Results
3.1 Study selection
The initial search delivered 2057 articles; after removal of duplicates, 1156 abstracts were primarily screened of which 207 full-text articles remained. After reading the full-text, 83 studies were eventually included in the present systematic review. The article selection process is shown in Figure 2.
3.2 Study characteristics and quality assessment
The study characteristics of the 83 included articles are shown in Table 1, Supplementary Tables S4, S5. 75 studies were candidate gene studies. Three studies were genome-wide association studies (GWAS) (Low et al., 2013; Huang et al., 2015; Cao et al., 2016), three studies were whole-exome sequencing (Gréen et al., 2016; Svedberg et al., 2020; Björn et al., 2020a). 1 study was whole-genome sequencing (WGS) (Björn et al., 2020b). 1 study used targeted resequencing of 100 pharmacokinetics-related genes (Yoshihama et al., 2018).
59 studies involved a single ethnic group [42 Chinese or Han Chinese, 5 Caucasian (Marsh et al., 2007; Tibaldi et al., 2008; Giovannetti et al., 2011; Ludovini et al., 2011; Lambrechts et al., 2015), 4 Japanese (Iwata et al., 2012; Low et al., 2013; Yoshihama et al., 2018; Nomura et al., 2020), 3 Indian (Ruzzo et al., 2014; Walia et al., 2021; Walia et al., 2022), 2 Korean (Kim et al., 2009; Lee et al., 2013), 1 Thai (Kumpiro et al., 2016), 1 Bangladeshi (Nairuz et al., 2021) and 1 eastern Slavonic origin (Khrunin et al., 2010)]. Three studies are mixed ancestry (Corrigan et al., 2014; Lambrechts et al., 2015; Ferracini et al., 2021). 21 studies did not explain how ancestry was determined.
Individual study sizes ranged from 32 to 1171 patients. 12 studies had fewer than 100 subjects. 10 studies involved discovery and replication sets (Marsh et al., 2007; Qian et al., 2012; Cao et al., 2016; Gréen et al., 2016; Jia et al., 2016; Yin et al., 2016; Zheng et al., 2017).
The studied cancer types were: lung cancer (n = 63), ovarian cancer (n = 6) (Marsh et al., 2007; Kim et al., 2009; Khrunin et al., 2010; Lambrechts et al., 2015; Liblab et al., 2020; Ferracini et al., 2021), malignant mesothelioma (n = 3) (Erčulj et al., 2012; Goričar et al., 2014; Senk et al., 2019), colorectal cancer (n = 3) (Cortejoso et al., 2013; Ruzzo et al., 2014), one study each evaluated colon cancer (Lee et al., 2013), gastric cancer (Seo et al., 2009), testicular cancer (Lee et al., 2013), esophageal cancer (Nomura et al., 2020), cervical cancer (Huang et al., 2015), pancreatic cancer (Giovannetti et al., 2011), four studies included various cancer types (Corrigan et al., 2014; Yoshihama et al., 2018) or did not mention the cancer type (Low et al., 2013).
64 studies used mixed combination chemotherapy: cisplatin/carboplatin-based chemotherapy (n = 51), cisplatin-based chemotherapy (n = 10), carboplatin-based chemotherapy (n = 3), 19 study uses single platinum-based chemotherapy. Platinum dosage and cycles varied according the tumor type: mostly cisplatin 75 mg/m2 or carboplatin AUC 5, both administered on day 1 every 3 weeks. The dose of oxaliplatin was 85 mg/m2 or 130 mg/m2 29 studies did not mention the dose of platinum agents.
47 studies analyzed the whole hematological toxicities. 56 studies analyzed detailed hematologic toxicity (leukopenia, neutropenia, thrombocytopenia or anemia). 20 studies analyzed both of them. Most studies (n = 80) use National Cancer Institute-Common Terminology Criteria for Adverse Events (NCI-CTCAE), three studies use WHO criteria (Isla et al., 2004; Chen et al., 2010; Huang et al., 2015).
The most used end-point of toxicity was the occurrence of grade ≥3 toxicity (n = 66). Other studies used a cut-off value of grade ≥1 (n = 12), grade ≥2 (n = 11), or grade 4 (n = 2) (Han et al., 2007; Marsh et al., 2007). One study described severe hematological toxicity including neutropenia G4, thrombocytopenia ≥ G3, and anemia ≥ G3 (Yoshihama et al., 2018). Eight studies analyzed more than one end-point of grading (Khrunin et al., 2010; Giovannetti et al., 2011; Erčulj et al., 2012; Lambrechts et al., 2015; Senk et al., 2019; Ferracini et al., 2021; Walia et al., 2021; Walia et al., 2022).
Among the 83 included studies, 28 (44.4%) were considered high quality, 46 (55.4%) moderate quality, and 9 (10.8%) were of low quality (Supplementary Table S6), the main reason for low study quality was the absence of calculation of sample size (n = 82), no consideration of population stratification (n = 80) and lack of statistical correction for multiple testing (n = 53).
3.3 Genetic associations investigated in platinum-induced hematological toxicity
51 variants were analyzed in more than one study (Tables 2, 3; Supplementary Table S7).
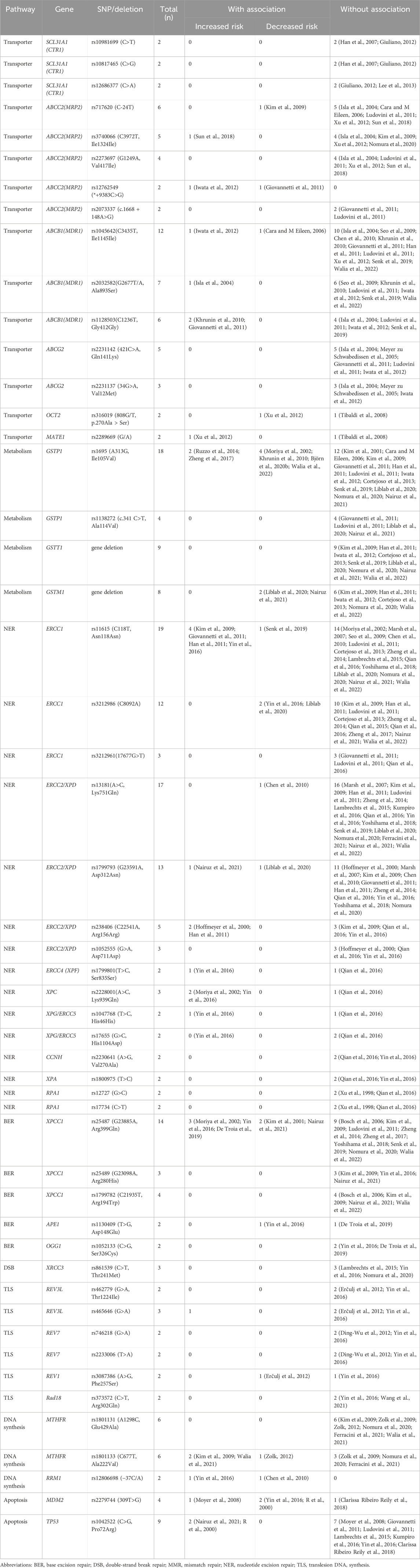
Table 2. Genetic polymorphisms investigated more than twice for association with platinum-induced hematological toxicity.
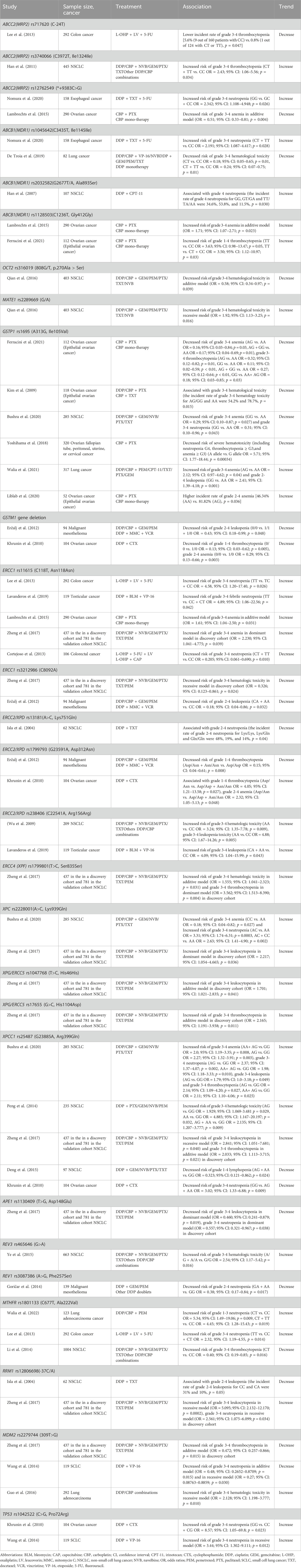
Table 3. Summary of positive associations in genetic polymorphisms that investigated more than twice.
3.3.1 Transportation for influx
3.3.1.1 SLC31A1 (CTR1)
The main route of platinum agents entering the cell is through the copper transporter CTR1 (SLC31A1) (Giuliano, 2012). Because of the lack of candidate SNPs, studies were limited. SLC31A1(CTR1) rs10981699 (Xu et al., 2012; Sun et al., 2018), rs10817465 (Xu et al., 2012; Sun et al., 2018), rs12686377 (Xu et al., 2012; Kumpiro et al., 2016) were analyzed in two studies respectively. No association has been found.
3.3.2 Transportation for efflux
3.3.2.1 ABCC2 (MRP2)
The multi-drug resistance protein (MRP2), encoded for by ABCC2, involved in pumping cisplatin out of the cell (Cara and M Eileen, 2006). The frequently investigated genetic polymorphisms in ABCC2 were rs717620 (C-24T) (n = 6), rs3740066 (C3972T, Ile1324Ile) (n = 5), rs2273697 (G1249A) (n = 4), rs12762549(*+9383C>G) (n = 2), and rs2073337 (c.1668 + 148A>G) (n = 2). ABCC2 rs717620 (C-24 T) and ABCC2 rs3740066 (C3972T, Ile1324Ile) may not affect the expression of MRP2 mRNA or protein (Moriya et al., 2002; Meyer zu Schwabedissen et al., 2005), while ABCC2 rs2273697 (G1249A) resulted in a significantly reduced expression of mRNA in human preterm placenta (Meyer zu Schwabedissen et al., 2005).
Positive association were reported in only few studies. ABCC2 rs717620 (C-24T) CC genotype present increased rate of grade 3-4 thrombocytopenia in contrast to CT and TT genotypes in 292 colon cancer patients treated with FOLFOX chemotherapy (5.6% vs. 0.8%, p = 0.047) (Lee et al., 2013). ABCC2 rs3740066(C3972T, Ile1324Ile) T allele was suggested as a risk factor for grade 3-4 thrombocytopenia in 445 NSCLC patients receiving platinum-based chemotherapy (OR = 2.43; 95% CI: 1.06–5.56; p = 0.034) (Han et al., 2011). As for ABCC2 rs12762549(G1249A), two studies show inconsistent results. Nomura et al. found a higher risk of grade 3-4 neutropenia in GG genotype in 158 esophageal cancer patients treated with docetaxel, cisplatin, and 5-fluorouracil chemotherapy (OR = 2.342; 95% CI: 1.108–4.948; p = 0.026) (Nomura et al., 2020). In contrast, ABCC2 rs12762549 is associated with decreased risk of grade 3-4 anemia in additive model (OR = 0.51; 95% CI: 0.33–0.81; p = 0.004) among 290 ovarian cancer patients upon treatment with paclitaxel and carboplatin (Lambrechts et al., 2015). No association has been found with the ABCC2 rs2073337 (Marsh et al., 2007; Lambrechts et al., 2015) and rs2273697 (Han et al., 2007; Marsh et al., 2007; Han et al., 2011; Qian et al., 2016).
3.3.2.2 ABCB1 (MDR1)
ABCB1 (MDR1 or p-glycoprotein) is thought to play a role in platinum efflux, although to a much lesser extent than the copper transporters or MRP2 (Cara and M Eileen, 2006). As to ABCB1 rs1045642(C3435T), the homozygous T-allele is associated with significantly lower duodenal MDR-1 expression and the highest digoxin plasma levels (Hoffmeyer et al., 2000). 12 studies were available for ABCB1 rs1045642(C3435T) but the results have not been consistent. De Troia B et al. reported that patients carrying 3435T allele had a lower risk of grade 3-4 neutropenia in 82 lung cancer patients treated with platinum-based chemotherapy (OR = 0.24; 95% CI: 0.07–0.75; p = 0.01) (De Troia et al., 2019). In contrast to these results, Nomura, H. et al. found T allele carriers were more likely to develop grade 3-4 neutropenia (OR = 2.191; 95% CI: 1.087–4.417; p = 0.028) in 158 esophageal cancer patients receiving docetaxel, cisplatin, and 5-fluorouracil therapy (Nomura et al., 2020).
Studies have also focused on ABCB1 rs2032582 (G2677TA, Ala893Ser) (n = 7) and ABCB1 rs1128503(C1236T) (n = 6). The two SNPs are associated with altered P-glycoprotein function, in which the Ser893 variant transporter resulted in a 47% lower intracellular digoxin concentration (p < .002) than did the Ala893 variant in vitro (Kim et al., 2001) and homozygous carriers of ABCB1 rs1128503(C1236T) polymorphism was significantly correlated with a decreased docetaxel clearance (Bosch et al., 2006). ABCB1 rs2032582 (G2677TA, Ala893Ser) displayed differential genotypic distribution between groups with grade 4 neutropenia and grade 1-3 neutropenia in 107 NSCLC patients (p = 0.030) (Han et al., 2007). ABCB1 rs1128503 (C1236T) were significantly associated with grade 3-4 anemia in additive model (OR = 1.71; 95% CI: 1.07–2.71; p = 0.023) in 290 ovarian cancer patients (Lambrechts et al., 2015). Similarly, another study in 112 ovarian cancer patients found that carriers of TT genotype were more frequently experienced grade 1-4 thrombocytopenia compared to those CC or CT genotype (OR = 3.50; 95% CI: 1.12–10.97; p = 0.03) (Ferracini et al., 2021).
3.3.2.3 ABCG2 (BCRP)
ABCG2 (breast cancer resistance protein or BCRP) is efflux transporter proteins that play a role in the development of chemoresistance to platinum agents (Ding-Wu et al., 2012). No association has been shown with the ABCG2 rs2231142 (Han et al., 2007; Marsh et al., 2007; Lambrechts et al., 2015; Nomura et al., 2020; Wang et al., 2021) and ABCG2 rs2231137 (Han et al., 2007; Nomura et al., 2020; Wang et al., 2021).
3.3.2.4 OCT2 (SLC22A2) and MATE1 (SLC47A1)
OCT2 is considered the predominant transporter mediating active accumulation of cisplatin in the kidney. MATE1 is thought to mediate the final step of renal tubular secretion of cisplatin (Giuliano, 2012). The OCT2 rs316019 (808G/T, p.270Ala > Ser) variant significantly impaired uptake kinetics of endogenous compounds and drugs (Zolk et al., 2009; Zolk, 2012). OCT2 rs316019 (808G/T, p.270Ala > Ser) and MATE1 rs2289669 (G/A) were analyzed in two studies (Iwata et al., 2012; Qian et al., 2016). Qian et al. found significant correlations with grade 3-4 hematological toxicity in these two SNPs among 403 NSCLC patients (Qian et al., 2016).
3.3.3 Metabolism
Platinum compounds can be detoxified by conjugation with glutathione through the aid of glutathione S-transferases (GSTs) (R et al., 2000). GSTP1, GSTM1, and GSTT1 belongs to Human GSTs and were mostly analyzed for the functional polymorphisms in gene regions.
3.3.3.1 GSTP1
Two common nonsynonymous polymorphisms in GSTP1, rs1695 (Ile105Val, A313G) and rs1138272 (Ala114Val), were shown to decrease GSTP1 enzyme activity (Moyer et al., 2008). GSTP1 rs1695 (Ile105Val, A313G) was the most common studied variant with 18 studies available. Four studies show a protective effect for hematological toxicity. One study in Brazil reported AG or GG genotype was associated with decreased risk of grade 3-4 anemia and grade 3-4 thrombocytopenia in 112 epithelial ovarian cancer patients (Ferracini et al., 2021). Another study in Korean showed that patients with A/G or G/G genotype showed lower rate of hematological toxicity (13/24, 54.2%) than those with A/A genotype (74/94, 78.7%) among 118 epithelial ovarian cancer patients (p = 0.015) (Kim et al., 2009). Bushra et al. found that GG genotype is associated with decreased risk of grade 3-4 anemia (OR = 0.29; 95% CI: 0.10–0.87; p = 0.027) and grade 3-4 neutropenia (OR = 0.31; 95% CI: 0.10–0.96; p = 0.043) in 285 NSCLC patients in Bangladesh (Bushra et al., 2020). Perhaps worth to mention is a comprehensive pharmacogenomic analysis in 320 gynecological cancers patients, of which GSTP1 rs1695 showed the lowest p-value against severe hematological toxicity (p = 0.00034) (Yoshihama et al., 2018). In contrast, Walia et al. found a significantly increased risk of grade 3-4 anemia with the AG genotype (OR = 2.12; 95% CI: 0.97–4.62; p = 0.04) and increased risk of grade 2-4 leukopenia with the GG genotype (OR = 2.41; 95% CI: 1.39–4.18; p = 0.001) in 317 North Indian lung cancer patients (Walia et al., 2021). Liblab et al. (2020) found that carriers of AG genotype were more frequently experienced grade 2-4 anemia than AA genotype in 52 ovarian cancer patients (81.82% vs. 46.34%, p = 0.036). Five studies demonstrated no significant association of GSTP1 rs1138272 (Ala114Val) polymorphism.
3.3.3.2 GSTT1 and GSTM1
Gene deletion in GSTT1 and GSTM1 can lead to an absence of enzymatic activity (Pemble et al., 1994; Xu et al., 1998). Nine studies showed no effect of GSTT1 gene deletion on the risk of hematological toxicity. Eight studies analyzed GSTM1 gene deletion, in which two studies showed a protective effect (Khrunin et al., 2010; Erčulj et al., 2012). In 94 malignant mesothelioma patients treated with platinum-based chemotherapy, Patients with homozygous GSTM1 gene deletion have a significantly decreased risk of grade 2-4 leukopenia compared with those of at least one functional allele (OR = 0.43; 95% CI: 0.18–0.99; p = 0.048) (Erčulj et al., 2012). Another study in 104 ovarian cancer patients that received paclitaxel and carboplatin found carriers of a homozygous GSTM1 gene deletion were less likely to develop grade 1-4 thrombocytopenia (OR = 0.13; 95% CI: 0.03–0.62; p = 0.005) and grade 2-4 anemia (OR = 0.29; 95% CI: 0.13–0.66; p = 0.003) than those of functional GSTM1 variants (Khrunin et al., 2010).
3.3.4 NER pathway
The bulky DNA intra-strand adducts generated by platinum are mainly repaired by the nucleotide-excision repair (NER) pathway (Clarissa Ribeiro Reily et al., 2018).
3.3.4.1 ERCC1
ERCC1 interact with XPF to make an incision at the damaged DNA lesion in NER pathway. ERCC1 rs11615 (C118T, Asn118Asn) (n = 19) and ERCC1 rs3212986 (C8092A) (n = 12) have been wildly studied. ERCC1 rs11615 reduces the transcription and mRNA levels of ERCC1, resulting in lower ERCC1 expression (Yu et al., 2000). ERCC1 rs3212986 resides in the 30-untranslated region that might affect mRNA stability (Chen et al., 2000). Four studies reported a significantly increased risk with ERCC1 rs11615 (C118T, Asn118Asn). In a study of 292 colon cancer patients receiving adjuvant oxaliplatin plus leucovorin plus 5-fluorouracil (FOLFOX) chemotherapy, TT genotype was more prone to develop grade 3-4 neutropenia (OR = 4.58, 95% CI: 1.20–17.40, p = 0.026) (Lee et al., 2013). Another study in 119 testicular cancer patients also reported an increased risk of grade 3-4 febrile neutropenia caused by bleomycin, etoposide, and cisplatin (BEP) chemotherapy in TT genotype (OR = 4.89; 95% CI: 1.06–22.56; p = 0.042) (Lavanderos et al., 2019). ERCC1 rs11615 was associated with an increased risk of grade 3-4 anemia in 290 ovarian cancer patients (OR = 1.61; 95% CI: 1.04–2.50; p = 0.031) (Lambrechts et al., 2015) and 437 NSCLC patients (OR = 2.230; 95% CI: 1.041–4.775; p = 0.039) (Zheng et al., 2017). On the contrary, Cortejoso et al. (2013) found individuals carrying T allele had a reduced risk of grade 3-4 neutropenia in 106 colon cancer patients (OR = 0.205; 95% CI: 0.061–0.690; p = 0.010). As for ERCC1 rs3212986(C8092A), two studies reported a decreased risk. Zheng et al. found that ERCC1 rs3212986 was significantly associated with a reduced risk of grade 3-4 hematologic toxicity in recessive model in 437 NSCLC patients (OR = 0.326; 95% CI: 0.123–0.861; p = 0.024) (Zheng et al., 2017). Erčulj et al. (2012) reported that patients carrying A allele had a lower risk of grade 2-4 leukopenia in 94 malignant mesothelioma patients (OR = 0.18; 95% CI: 0.04–0.86; p = 0.032). No association has been established between ERCC1 rs3212961 (17677G>T) and the development of hematologic toxicity (Marsh et al., 2007; Lambrechts et al., 2015; Song et al., 2017).
3.3.4.2 ERCC2/XPD
ERCC2 (XPD) is a helicase that can unwind the DNA strands to facilitate the binding of other NER proteins. ERCC2 rs13181 (Lys751Gln), rs1799793(Asp312Asn), rs238406 (C22541A, Arg156Arg), rs1052555 (Asp711Asp) have been evaluated in 17 studies, 13 studies, five studies and three studies respectively. ERCC2 rs13181(Lys751Gln) can alter mRNA transcription levels, leading to reduced levels of ERCC2 mRNA(82). For ERCC2 rs13181 (Lys751Gln), grade 2-4 neutropenia was more frequent among individual with 751 Lys/Lys genotype in 62 NSCLC patients treated with cisplatin and docetaxel (p = 0.04) (Isla et al., 2004).
ERCC2 rs1799793 (Asp312Asn) is associated with significantly decreased constitutive XPD mRNA levels in lymphocytes of healthy subjects (Wolfe et al., 2007). Results on rs1799793 (Asp312Asn) have not been consistent. Erčulj et al. found patients carrying 312Asp/Asn and 312Asn/Asn genotype had a lower risk of grade 1-4 thrombocytopenia in 94 malignant mesothelioma patients (OR = 0.15; 95% CI: 0.04–0.61; p = 0.008) (Erčulj et al., 2012). Whereas, Khrunin et al. (2010) found patients carrying Asp/Asn genotype were more prone to grade 1-4 thrombocytopenia (OR = 4.05; 95% CI: 1.21–13.58; p = 0.027) and grade 2-4 anemia (OR = 2.32; 95% CI: 1.05–5.13; p = 0.048) in 104 ovarian cancer patients. ERCC2 rs238406(C22541A, Arg156Arg) can reduce the levels of mRNA and ultimately affect XPD activity and function (Wolfe et al., 2007). The incidence of grade 3-4 hematologic toxicity (OR = 3.24; 95% CI: 1.35–7.78; p = 0.009) and grade 3-4 leukopenia (OR = 4.88; 95% CI: 1.67–14.26; p = 0.005) was significantly higher in variant homozygotes AA genotype, when compared with CC genotype in 209 NSCLC patients (Wu et al., 2009). Likewise, another study in 119 testicular cancer patients showed the increased risk of grade 3-4 leukopenia with the AA or CA genotype (OR = 4.09; 95% CI: 1.04–15.99; p = 0.043) (Lavanderos et al., 2019). Studies on ERCC2 rs1052555 (Asp711Asp) (n = 3) showed no significant difference (Wu et al., 2009; Song et al., 2017; Zheng et al., 2017).
3.3.4.3 ERCC4/XPF
ERCC4/XPF participates in the removal of damaged DNA strands by acting as an endonuclease with ERCC1. Of two studies in ERCC4 rs1799801 (T>C, Ser835Ser) (Song et al., 2017; Zheng et al., 2017), one study in 437 NSCLC patients demonstrated an association of the ERCC4 rs1799801 with grade 3-4 hematologic toxicity in additive model (OR = 1.555; 95% CI: 1.041–2.323; p = 0.031) and grade 3-4 thrombocytopenia in dominant model (OR = 3.562; 95% CI: 1.513–8.390; p = 0.004) (Zheng et al., 2017).
3.3.4.4 XPC
XPC cooperates with RAD23B to recognize DNA lesions and start the initial step of damage recognition. Three studies concentrate on XPC rs2228001(A>C, Lys939Gln) (Song et al., 2017; Zheng et al., 2017; Bushra et al., 2020). XPC rs2228001(A>C, Lys939Gln) was associated with lower DNA repair capacity (Zhu et al., 2008). In 285 Bangladesh NSCLC patients, carriers of XPC rs2228001 AC or CC genotypes showed significant suffering from grade 3-4 neutropenia (OR = 2.63; 95% CI: 1.41–4.90; p = 0.002), while those harboring CC genotypes are less likely to develop grade 3-4 anemia, compared with AA genotype carriers (OR = 0.18; 95% CI: 0.04–0.82; p = 0.027) (Bushra et al., 2020). Another study conducted in 437 Chinese NSCLC patients show XPC rs2228001 is associated with an increased risk of grade 3-4 leukocytopenia in dominant model (OR = 2.217; 95% CI: 1.054–4.665; p = 0.036) (Zheng et al., 2017).
3.3.4.5 ERCC5/XPG
ERCC5 (XPG) encodes a structure-specific endonuclease that can cleave the 3ʹ-end of damaged DNA lesions. Two studies focus on XPG rs1047768(A>C, Lys939Gln) and rs17655(A>C, Lys939Gln). Zheng et al. found a significant association of XPG rs1047768 with grade 3-4 leukocytopenia in additive model (OR = 1.701; 95% CI: 1.021–2.835; p = 0.041) and XPG rs17655 with grade 3-4 thrombocytopenia in additive model (OR = 2.165; 95% CI: 1.191–3.938; p = 0.011) (Zheng et al., 2017). The other study reported no association with these two investigated SNPs (Song et al., 2017).
3.3.4.6 Other NER genes
XPA rs1800975, RPA1 rs17734, RPA1 rs12727 and CCNH rs2230641 (Song et al., 2017; Zheng et al., 2017) were examined twice and showed no significant associations.
3.3.5 BER pathway
The base excision repair (BER) pathway is mainly responsible for removing the oxidative DNA lesions generated by platinum drug exposure (Jana et al., 2018).
3.3.5.1 XRCC1
XRCC1 acts as a central scaffolding protein to finish the final steps of BER. XPCC1 rs25487 (G>A, Arg399Gln) can cause missense mutation in the coding region, leading to decreased damage DNA repair activity (Wang et al., 2003; Vodicka et al., 2004). 14 studies investigated a link of XPCC1 rs25487 (G>A, Arg399Gln) with hematological toxicity and results have not been consistent. In a study of 285 NSCLC patients, XRCC1 rs25487 polymorphism showed significant associations with increased risk of grade 3-4 anemia, grade 3-4 neutropenia, grade 3-4 leukopenia and grade 3-4 thrombocytopenia (Bushra et al., 2020). A higher risk of grade 3-4 hematologic toxicity was also observed in AG and GG carriers in a cohort of 235 NSCLC patients (OR = 2.135; 95% CI: 1.207–3.777; p = 0.009) (Peng et al., 2014). In keeping with this, XRCC1 rs25487 was identified as risk factors for grade 3-4 leukocytopenia in recessive model (OR = 2.841; 95% CI: 1.051–7.681; p = 0.040), grade 3-4 thrombocytopenia in additive model (OR = 2.033; 95% CI: 1.113–3.715; p = 0.021) in discovery cohort of 437 NSCLC patients (Zheng et al., 2017). In contrast, AA or AG carriers had a decreased risk of grade 1-4 lymphopenia as observed in 94 NSCLC patients (OR = 0.323; 95% CI: 0.121–0.862; p = 0.024) (Deng et al., 2015). In 104 ovarian cancer that received cisplatin and cyclophosphamide, an increased risk of grade 3-4 neutropenia was found in GG wildtype carriers (OR = 3.02; 95% CI: 1.33–6.88; p = 0.009) (Khrunin et al., 2010). Other studies failed to reveal a positive association.
XPCC1 rs1799782 (Arg194Trp) can leading to decreased DNA repair activity (Wang et al., 2003). XPCC1 rs1799782 (Arg194Trp) (n = 4) (Wang et al., 2008; Kim et al., 2009; Khrunin et al., 2010; Lee et al., 2013) and XPCC1 rs25489 (Arg280His) (n = 3) (Khrunin et al., 2010; Lee et al., 2013; Zheng et al., 2017) showed no significant difference with hematological toxicity.
3.3.5.2 APE1 and OGG1
APE1 and OGG1 are key component in the BER pathway. Two studies examined APE1 rs1130409 (Asp148Glu) and OGG1 rs1052133 (Ser326Cys) (Peng et al., 2014; Zheng et al., 2017). APE1 rs1130409(Asp148Glu) variant exhibiting normal in vitro nuclease capacity (Daviet et al., 2007; Wilson et al., 2011) and OGG1 rs1052133(Ser326Cys) polymorphism negatively impacts OGG1 function (Wilson et al., 2011). APE1 rs1130409 was associated with a decreased risk of grade 3-4 leukocytopenia (OR = 0.460; 95% CI: 0.241–0.879; p = 0.019) and grade 3-4 neutropenia (OR = 0.557; 95% CI: 0.321–0.967; p = 0.038) in dominant model (Zheng et al., 2017). OGG1 rs1052133 failed to show significant association with hematological toxicity (Peng et al., 2014; Zheng et al., 2017).
3.3.6 DSB pathway
Homologous recombination (HR) and non-homologous and joining pathways are responsible for repairing the DSBs generated by platinum-induced ICLs which are the most hazardous type of DNA damage. XRCC3 are one of the crucial proteins involved in mediating the HR pathway (Giovanna and Massimo, 2019). XRCC3 rs861539(C>T, Thr241Met) was show to reduce DNA damage repair capacity (Matullo et al., 2001). However, no association was detected in three analyses on XRCC3 rs861539 (Thr241Met) (Ludovini et al., 2011; Ruzzo et al., 2014; Zheng et al., 2017).
3.3.7 TLS
TLS is performed by a series of low-fidelity polymerases to tolerate platinum-induced DNA lesions (The Vinh and Orlando, 2010). REV3L rs462779 (Goričar et al., 2014; Zheng et al., 2017), REV3L rs465646 (Goričar et al., 2014; Ye et al., 2015; Zheng et al., 2017), REV7 rs746218 (Ye et al., 2015; Zheng et al., 2017), REV7 rs2233006 (Ye et al., 2015; Zheng et al., 2017), REV1 rs3087386 (Goričar et al., 2014; Zheng et al., 2017) and Rad18 rs373572 (Chu et al., 2016; Zheng et al., 2017) were analyzed in more than one study. REV3 rs465646 were associated with grade 3-4 hematologic toxicity in 663 NSCLC patients (OR = 2.54; 95% CI: 1.17–5.42; p = 0.016) (Ye et al., 2015). In a study of 139 malignant mesothelioma patients receiving cisplatin-based chemotherapy, carriers of the REV1 rs3087386 A allele were less prone to develop grade 2-4 neutropenia (OR = 0.38; 95% CI: 0.17–0.84; p = 0.017) (Goričar et al., 2014). Other studies show no significant associations.
3.3.8 DNA synthesis
3.3.8.1 MTHFR
MTHFR is involved in folate metabolism, essential for the synthesis of nucleic acids and amino acids. Two common nonsynonymous polymorphisms in MTHFR, rs1801133 (C677T, Ala222Val) and rs1801131 (A1298C, Glu429Ala) were shown to decrease MTHFR enzyme activity in vitro (van der Put et al., 1998). These two SNPs were analyzed in six studies (Lee et al., 2013; Corrigan et al., 2014; Kanazawa et al., 2014; Li et al., 2014; Ruzzo et al., 2014; Walia et al., 2022). The results for MTHFR rs1801133 (C677T, Ala222Val) were inconsistent. An increased risk of grade 1-3 neutropenia was detected in carriers of the T allele (OR = 4.45; 95% CI: 1.28–15.43; p = 0.019) among 123 lung adenocarcinoma patients treated with platinum and pemetrexed chemotherapy (Walia et al., 2022). Likewise, carriers of the TT genotype were more likely to develop grade 3-4 neutropenia compared with those carrying CC and CT genotype in 292 colon cancer patients (OR = 2.32, 95% CI: 1.19–4.55, p = 0.014) (Lee et al., 2013). While, the heterozygote CT genotype show a protective effect for grade 3-4 thrombocytopenia in Li et al.’s study of 1004 NSCLC patients (OR = 0.40; 95% CI: 0.19–0.85; p = 0.016) (Li et al., 2014). MTHFR rs1801131 (A1298C, Glu429Ala) showed no effect on risk of hematological toxicity.
3.3.8.2 RRM1
RRM1 encodes the regulatory subunit of ribonucleotide reductase and is involved in the production of deoxyribonucleotides during DNA synthesis. RRM1 – 37A allele impacted promoter activity with increased RRM1 mRNA expression in vitro (Bepler et al., 2005; Rodriguez et al., 2011). RRM1 rs12806698 (−37C/A) was suggested as a candidate risk factor for grade 3-4 leukocytopenia (OR = 5.095; 95% CI: 2.132–12.170; p = 0.0002) and grade 3-4 neutropenia (OR = 2.561; 95% CI: 1.075–6.099; p = 0.034) in recessive model in 437 NSCLC patients (Zheng et al., 2017), while in another study of 63 NSCLC patients, grade 2-4 leukopenia was more frequent among patients with wild-type RRM1 rs12806698 CC genotype than carriers of CA genotype (37% versus 10%, p = 0.05) (Isla et al., 2004).
3.3.9 Apoptosis
3.3.9.1 MDM2
MDM2 rs2279744 (309T>G) resulted in higher levels of MDM2 RNA and protein and the subsequent attenuation of the p53 DNA damage response (Bond et al., 2004). For MDM2 rs2279744 (309T>G) (n = 4), two studies in lung cancer patients observed a protective effect for grade 3-4 thrombocytopenia (OR = 0.472; 95% CI: 0.257–0.866; p = 0.015) (Zheng et al., 2017) and grade 3-4 neutropenia (OR = 0.27; 95% CI: 0.08763–0.8859; p = 0.030) respectively (Wang et al., 2014). Another study in 292 lung adenocarcinoma patients show opposite results that MDM2 rs2279744 was associated with an increased risk of grade 3-4 hematologic toxicity in recessive model (OR = 2.128; 95% CI: 1.198–3.777; p = 0.010) (Guo et al., 2016).
3.3.9.2 TP 53
TP53 (p53) is a key regulator of cell cycle control and DNA damage response as well as apoptosis initiation. TP53 rs1042522 (Arg72Pro) Arg72 variant is more efficient than the Pro72 variant in inducing apoptosis (Dumont et al., 2003). TP53 rs1042522 (Arg72Pro) was associated with an increased risk of grade 3-4 neutropenia in recessive model (OR = 3.44; 95% CI: 1.302–9.111; p = 0.012) in 119 SCLC patients treated with cisplatin and etoposide (Wang et al., 2014). Another study in 104 ovarian cancer patients showed similar result that patients carrying GG genotype were prone to experience grade 3-4 neutropenia, compared with CC + CG genotype (OR = 8.57; 95% CI: 1.05–69.8; p = 0.023) (Khrunin et al., 2010). The remaining studies (n = 7) showed no significant difference.
3.4 Significant associations in other less commonly investigated variants
The array of SNPs that investigated only once are listed in Supplementary Table S8. Lots of studies analyzed those less commonly investigated variants in main candidate genes such as SLC31A1 (CTR1) (Xu et al., 2012; Sun et al., 2018), ATP7A (Deng et al., 2015; Li et al., 2018), ATP7B (Li et al., 2018), ABCC2 (Ruzzo et al., 2014), ABCB1 (Chen et al., 2010; Chen et al., 2016), ABCG2 (Chen et al., 2016; Wang et al., 2021), OCT2 (Chen et al., 2016; Qian et al., 2016), MDM2 (Zheng et al., 2014; Qian et al., 2015), TP53 (Khrunin et al., 2010) or tagSNPs on genes that are not directly affect the influx, efflux and metabolism of platinum agent or play as mediators or modulators of center cellular activities. Those genes and the functions were listed in Supplementary Table S8, including the transport genes (SLCs, AQP2, AQP9, TMEM205, SIRT1, ABCC1, MVP, AQP1) (Chen et al., 2016; Senk et al., 2019), Metabolism gene (CYP2E1, GSTA1, GSTM3, AGXT, MAPT, MPO) (Marsh et al., 2007; Khrunin et al., 2010; Lee et al., 2013), MicroRNAs (Zhan et al., 2012; Fang et al., 2017), Long Non-Coding RNA (Hu et al., 2016; Gong et al., 2017). Apoptosis-related genes (BCL2, BAX, CASP3, CASP8, CASP10, TNFα and MIF) (Gu et al., 2012; Qian et al., 2012; Liu D. et al., 2017a), EPO (Zheng et al., 2021), VCP (Peng et al., 2013), STAT3 (Gong et al., 2019), WISP1 (Chen et al., 2014), TERT (Zhao et al., 2015), CHEK2 (Xu et al., 2016), MMP-2 (Zhao et al., 2012), RICTOR (Wang et al., 2016), eIF3 a (Yin et al., 2015), HSP genes and Rho family genes (Zou et al., 2016), CDC25 family genes (Cai et al., 2014), The JNK and P38 MAPK pathways genes (Jia et al., 2016), MIF signaling pathway (Tan et al., 2014). As to DNA repair pathway, there were studies focus on tagSNPs in less investigated NER genes (Chen et al., 2016; Song et al., 2017; Zheng et al., 2017) and BER genes (Peng et al., 2014; Zheng et al., 2017). DNA repair pathway that not directly involved in repairing platinum-induced DNA lesions like MMR (Liu JY. et al., 2017b; Zheng et al., 2017), DSB (Zheng et al., 2017), TLS (Goričar et al., 2014; Shao et al., 2014; Ye et al., 2015; Chu et al., 2016; Zheng et al., 2017), FA pathway (Zheng et al., 2017), have received more attention recently. Some significant association was found, while these SNPs were not previously analyzed and should be validated in future studies.
3.5 High throughput researches
Four studies are derived from the same cohort in Sweden population of 215 NSCLC patients treated with gemcitabine/carboplatin chemotherapy (Gréen et al., 2016; Svedberg et al., 2020; Björn et al., 2020a; Björn et al., 2020b). Three of them were whole-exome sequence studies concentrating of gemcitabine/carboplatin-induced grade 3-4 thrombocytopenia (Gréen et al., 2016; Björn et al., 2020a), leukopenia (Svedberg et al., 2020) and neutropenia (Gréen et al., 2016; Svedberg et al., 2020). The fourth study performed a GWAS in a subset of 96 patients (Björn et al., 2020b). These studies identified and validated several genetic variations, genes and hematopoiesis-related pathways to be potential significance and created weighted genetic risk score (wGRS) prediction models for predicting the risk of chemotherapy-induced hematological toxicity. There were GWAS concentrating on NSCLC (Cao et al., 2016), cervical cancer (Huang et al., 2015), and unclassified carcinomas (Low et al., 2013). A list of novel genetic variants such as rs13014982 at 2q24.3 and rs9909179 at 17p12 were identified (Cao et al., 2016) and the artificial neural networks model based on the multiple risk factors were constructed (Huang et al., 2015). Yin et al. developed a strategy to establish a predicted model of toxicity integrating both genetic and clinical factors using DM techniques (Yin et al., 2016).
4 Discussion
4.1 Main findings
This systematic review has reported and evaluated the findings of published studies that investigated the genetic associations with hematological toxicities in the cancer patients receiving platinum-based chemotherapy. We found that ABCC2 rs12762549, ABCB1 rs1045642, ABCB1 rs1128503, GSTP1 rs1695, GSTM1 gene deletion, ERCC1 rs11615, ERCC1 rs3212986, ERCC2 rs238406, ERCC2 rs1799793, XPC rs2228001, XPCC1 rs25487, MTHFR rs1801133, MDM2 rs2279744, RRM1 rs12806698, TP53 rs1042522 show positive associations in more than two studies, but most associations were not consistently replicated across the reviewed studies. Among them, ABCB1 rs1128503, GSTP1 rs1695, GSTM1 gene deletion, ERCC1 rs11615, ERCC1 rs3212986, ERCC2 rs238406, XPC rs2228001, XPCC1 rs25487, MTHFR rs1801133, MDM2 rs2279744, TP53 rs1042522 had consistent results across at least two independent populations. These genetic variants may provide insights into the molecular mechanisms towards platinum-induced hematological toxicities (Qihai et al., 2023).
It's also worth mentioning that GSTP1 rs1695 (A313G, Ile105Val) show significant association in six studies. Among them, four studies show protective role of GSTP1 rs1695 against platinum-induced hematological toxicities, including a comprehensive analysis that targeted resequencing of 100 notable pharmacokinetics-related genes in which GSTP1 rs1695 showed the smallest p-value (p = 0.00034) (Yoshihama et al., 2018). Positive associations of GSTP1 rs1695 with increased risk of platinum-induced hematological toxicity was found in two meta-analysis (Lv et al., 2018; Kim et al., 2022), while these two meta-analysis studies face the limitations of insufficient data availability. Alteration of DNA repair ability might play an important role in the development of platinum-induced hematological toxicity. Genetic variants in the candidate NER genes may affect the repair function and are most promising in predicting platinum-related hematological toxicity, since it is the main pathway responsible of repairing the bulky DNA intra-strand adducts generated by platinum agents (Hilary et al., 2024). ERCC1 rs11615 (C118T, Asn118Asn) and ERCC1 rs3212986 (C8092A) are two common variant that affect ERCC1 (key enzyme in NER pathway) mRNA expression or mRNA stability (Chen et al., 2000; Yu et al., 2000). ERCC1 rs11615 present consistent results with increased risk of grade 3-4 neutropenia or anemia in four studies, and ERCC1 rs3212986 show decreased risk of grade 3-4 hematologic toxicity in two studies. These two SNPs may be important molecular biomarkers for predicting platinum-induced hematological toxicities. XRCC1 rs25487 (G23885A, Arg399Gln) show positive association in six studies, emphasizing the potential contribution of BER pathway in oxidative stress in platinum-induced hematological toxicity, since cisplatin can exert cytotoxic effects through the generation of ROS (Zheng et al., 2020).
Apart from that, lots of genetic variants located in genes not directly linked to drug exposure are investigated. The GWAS have identified a handful of candidate genetic variants associated with platinum-based hematological toxicities and novel biologic pathways of potential impact. But these studies still face the challenge of statistically underpowered and stringent threshold of multiple testing. Additional validation in multiple independent sample sets or functional analyses are required to further elucidate the gene-phenotype relationship (Low et al., 2014).
4.2 Quality and inconsistency among studies
Genetic association studies require a large number of patients to provide adequate power, as a rare variant with large effect, or common variant with modest effect is more probable in genetic epidemiology (Campbell et al., 1995). While the majority of the studies in our systematic review did not indicate the sample size calculation in their statistical analysis, and the sample size in most studies in our systematic review are much smaller than that would be implied, which may be underpowered to detect a statistically significant relationship (Jorgensen and Williamson, 2008).
One of the challenges in pharmacogenomics is the ethnic background of the study population. The prevalence of toxicity varies according to the ethnic background (O’Donnell and Dolan, 2009; Li and Meyre, 2013). For example, higher rates of toxicities have been observed in east Asian populations compared to European and North American populations (Watanabe et al., 2003). In addition, allele frequency of genetic variants vary depending on the ethnic background or even the geographical location. When cases and controls are drawn from multiple ethnic or geographic groups, population stratification exists, which may put the study at risk of confounding and can lead to false positive associations (Jorgensen and Williamson, 2008). Population stratification was seldom assessed in most genetic association studies, which has been cited as a major reason for lack of replication.
Different treatment protocols are recommended according to cancer types. The composition, proportion and cycles of chemotherapy regimen can influence the incidence and degree of hematological toxicity. Furthermore, treatment protocols may also differ with regard to the use of concomitant supportive treatments. The time and dose of granulocyte colony-stimulating factor administered may be various across institutions, which are seldom mentioned in the method part and may bring confounds in genetic association studies. Moreover, there are overlap in metabolic pathway between platinum and other antineoplastic drugs, which can alter the pharmacogenetic effects of polymorphisms. For example, The ABC transporter ABCB1 and ABCC2 is responsible for the efflux of many commonly used antineoplastic drugs that usually used in combination with platinum agents, including taxanes (Marsh, 2006; Lambrechts et al., 2015). If Pt-DNA lesions are not repaired, the DNA lesion triggers activation of the apoptosis pathway, an essential step for the effectiveness of platinum-based chemotherapeutics for killing tumor cells, which is also the apoptosis pathway of many other cytotoxic drugs (Fulda and Debatin, 2006). Apart from the role of participating in DNA synthesis, MTHFR encode key enzymes for the metabolism of 5-FU (Lee et al., 2013) and RRM1 was also the primary target for gemcitabine (Yuan et al., 2015).
Genetic association studies are heavily reliant on the phenotype, but it may be difficult to establish the true phenotype. Although myelosuppression is quantitative, the degree of myelosuppression could be missed based on the frequency of measurement. Apart from that, differences in the toxicity assessment criterion and endpoints may hamper reproducibility of previous findings. Some studies analyzed the total hematological toxicity (Wang et al., 2008; Kim et al., 2009; Gu et al., 2012), while other studies analyzed the detail hematological toxicity (leukopenia, neutropenia, thrombocytopenia or anemia) (Isla et al., 2004; Tibaldi et al., 2008; Xu et al., 2012). The detail hematological toxicity may be more accurate and clinically relevant, but its low incidence requires cohort study with large sample size, which are unattainable in the current researches. Furthermore, methodological flaws were observed in the imprecise dichotomization of patients with mild toxicity and severe toxicity. The majority use the occurrence of grade 3-4 as endpoints, other studies use grade 2-4 (Isla et al., 2004; Erčulj et al., 2012; Huang et al., 2015; Jia et al., 2016; Liblab et al., 2020) or grade 1-4 (Chen et al., 2010; Erčulj et al., 2012; Deng et al., 2015; Walia et al., 2022). Medical interventions of dose reduction and/or treatment discontinuation were taken when grade 3-4 hematological toxicity arises. Therefore, the toxicity endpoints of grade 2-4 or grade 1-4 may not be clinically relevant (Supplementary Table S4).
There is statistical heterogeneity in the data analysis (Supplementary Table S5). Some studies just used chi-square test or fisher’s exact test to estimate the difference of genotype distribution in cases and controls (Kimcurran et al., 2011; Kalikaki et al., 2015). Others use logistic regression to make comparisons between groups and generate odds ratios and 95% confidence intervals. Logistic regression is more appropriate as it can provide a quantitative measure of the relationship between the groups, allow adjustment for confounding factors, and detect gene-gene or gene-environment interactions. Some studies made a clear statement about mode of inheritance assumed for analysis, and used more than one assumption (Peng et al., 2014; Bushra et al., 2020; Ferracini et al., 2021; Walia et al., 2022), while other studies only compared the three categories of genotype frequencies (homozygous wild type, heterozygous, homozygous variant) between cases and controls (Isla et al., 2004; Han et al., 2007; Kim et al., 2009; Lee et al., 2013; Liblab et al., 2020). 30 of the included studies performed the correction for multiple comparisons and only 10 studies performed validation of the results or by splitting the cohort for a primary and an exploratory analysis (Marsh et al., 2007; Qian et al., 2012; Cao et al., 2016; Gréen et al., 2016; Jia et al., 2016; Yin et al., 2016; Zheng et al., 2017; Björn et al., 2020a; Björn et al., 2020b; Svedberg et al., 2020).
4.3 Limitations
This review has several limitations, which mainly reflects the status of current genetic association studies. In the search process, many studies had to be excluded because hematological toxicity was not clearly described or there is no toxicity grading criterion. What’s more, we observed that the most studies did not provide toxicity statistics data (no p-value) or just provide insufficient toxicity data (for example, some studies only mentioned correlation in the results section without providing the original statistical data), which may raise some doubts of data authenticity. Also, some studies did not control exclusion criteria of prior history of chemotherapy and/or radiation or a considerable proportion of the patients receive chemotherapy combined with radiation or other non-platinum chemotherapy regimens, which may increase the likelihood of cumulative hematological toxicity. There is much heterogeneity between incorporated studies, thus we were unable to perform a quantitative comparison and meta-analysis.
4.4 Implications for clinical practice and research
Future studies should focus on the following aspects. Firstly, genetic association study inevitably faces the concerns of bias and confounding, and are susceptible to inappropriate conclusions, therefore it calls for careful planning of study design to improving quality of methodology (Saito et al., 2006). Secondly, the majority genetic polymorphisms identified in the eligible publications were repeated in only one or two studies (Hilary et al., 2024). SNPs screened out as potential factors in susceptibility to hematological toxicity in our systematic review require well-planned, methodologically robust studies to validate them. Thirdly, functional predictions of significant genetic variants needed to be confirmed and validated in vitro or in vivo work (Felipe Antonio de Oliveira et al., 2022). Fourthly, more studies should emphasize on the hematopoiesis-related pathways identified in a whole-exome sequenced study of 215 NSCLC patients treated with a single treatment (Björn et al., 2020a). Moreover, clinical trial with large sample size to perform subgroup analysis should be conducted to allow proper stratified analysis. Finally, owing to the complexity of mechanisms of platinum action, a single SNP alone may have low effect to platinum response, thus supporting a polygenetic effect in platinum-induced hematological toxicity (Daniel et al., 2023). Future direction should be establishing appropriate statistical methods with capacity to integrate multiple genetic, phenotypic, epidemiological and clinical variables effects.
5 Conclusion
To summarize, this systematic review has successfully reported and evaluated studies on genetic associations of platinum-based hematological toxicities. Review of these studies identified several genetic variants that potentially affect the risk of platinum-induced hematological toxicity. Many methodological issues exist that may affect reproducibility of results and lead to inconsistency, including insufficient sample size, population stratification, various treatment schedule, heterogeneity in the assessment of hematological toxicity and statistics. Well-designed studies with sufficient samples sizes and standardization of phenotypes are warranted to address the limitations of the current studies and to ensure the robust findings that can be more effective to be used in personalized therapeutics.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
YZ: Conceptualization, Formal Analysis, Methodology, Supervision, Writing–original draft. MT: Conceptualization, Formal Analysis, Methodology, Writing–review and editing. ZD: Conceptualization, Formal Analysis, Methodology, Writing–review and editing. PC: Conceptualization, Formal Analysis, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Health Research Project of Hunan Provincial Health Commission (D202302046032, W20243057), National Key Clinical Specialty Scientific Research Project (Z2023110), as well as National Natural Science Foundation of China (81903111).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1445328/full#supplementary-material
Abbreviations
AUC, area under curve; BER, base excision repair; CI, confidence interval; DSB, double-strand break repair; FA, fanconi anemia; FOLFOX, oxaliplatin, leucovorin and 5-fluorouracil; FR, flanking region; GWAS, genome-wide association studies; HR, homologous recombination; ICL, interstrand crosslink; MMR, mismatch repair; NCI-CTCAE, National Cancer Institute-Common Terminology Criteria for Adverse Events; NSCLC, non-small cell lung cancer; NER, nucleotide excision repair; OR, odds ratios; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses; ROS, reactive oxygen species; SNP, single nucleotide polymorphism; TLS, translesion DNA synthesis; UTR, untranslated region; WHO, World Health Organization.
References
Basu, A., Ghosh, P., Bhattacharjee, A., Patra, A. R., and Bhattacharya, S. (2015). Prevention of myelosuppression and genotoxicity induced by cisplatin in murine bone marrow cells: effect of an organovanadium compound vanadium(III)-l-cysteine. Mutagenesis 30 (4), 509–517. doi:10.1093/mutage/gev011
Bepler, G., Zheng, Z., Gautam, A., Sharma, S., Cantor, A., Sharma, A., et al. (2005). Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer 47 (2), 183–192. doi:10.1016/j.lungcan.2004.07.043
Björn, N., Badam, T. V. S., Spalinskas, R., Brandén, E., Koyi, H., Lewensohn, R., et al. (2020b). Whole-genome sequencing and gene network modules predict gemcitabine/carboplatin-induced myelosuppression in non-small cell lung cancer patients. NPJ Syst. Biol. Appl. 6 (1), 25. doi:10.1038/s41540-020-00146-6
Björn, N., Sigurgeirsson, B., Svedberg, A., Pradhananga, S., Brandén, E., Koyi, H., et al. (2020a). Genes and variants in hematopoiesis-related pathways are associated with gemcitabine/carboplatin-induced thrombocytopenia. Pharmacogenomics J. 20 (2), 179–191. doi:10.1038/s41397-019-0099-8
Bond, G. L., Hu, W., Bond, E. E., Robins, H., Lutzker, S. G., Arva, N. C., et al. (2004). A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119 (5), 591–602. doi:10.1016/j.cell.2004.11.022
Bosch, T. M., Huitema, A. D., Doodeman, V. D., Jansen, R., Witteveen, E., Smit, W. M., et al. (2006). Pharmacogenetic screening of CYP3A and ABCB1 in relation to population pharmacokinetics of docetaxel. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 12 (19), 5786–5793. doi:10.1158/1078-0432.CCR-05-2649
Bushra, M. U., Rivu, S. F., Sifat, A. E., Nahid, N. A., Ahmed, M. U., Al-Mamun, M. M. A., et al. (2020). Genetic polymorphisms of GSTP1, XRCC1, XPC and ERCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer patients of Bangladesh. Mol. Biol. Rep. 47 (9), 7073–7082. doi:10.1007/s11033-020-05771-2
Cai, W., Chen, C., Li, X., Shi, J., Sun, Q., Liu, D., et al. (2014). Association of CDC25 phosphatase family polymorphisms with the efficacy/toxicity of platinum-based chemotherapy in Chinese advanced NSCLC patients. Future Oncol. 10 (7), 1175–1185. doi:10.2217/fon.14.25
Campbell, M. J., Julious, S. A., and Altman, D. G. (1995). Estimating sample sizes for binary, ordered categorical, and continuous outcomes in two group comparisons. BMJ 311 (7013), 1145–1148. doi:10.1136/bmj.311.7013.1145
Cao, S., Wang, S., Ma, H., Tang, S., Sun, C., Dai, J., et al. (2016). Genome-wide association study of myelosuppression in non-small-cell lung cancer patients with platinum-based chemotherapy. Pharmacogenomics J. 16 (1), 41–46. doi:10.1038/tpj.2015.22
Cara, A. R., and M Eileen, D. (2006). Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 33 (1), 9–23. doi:10.1016/j.ctrv.2006.09.006
Chen, J., Wu, L., Wang, Y., Yin, J., Li, X., Wang, Z., et al. (2016). Effect of transporter and DNA repair gene polymorphisms to lung cancer chemotherapy toxicity. Tumor Biol. 37 (2), 2275–2284. doi:10.1007/s13277-015-4048-0
Chen, J., Yin, J., Li, X., Wang, Y., Zheng, Y., Qian, C., et al. (2014). WISP1 polymorphisms contribute to platinum-based chemotherapy toxicity in lung cancer patients. Int. J. Mol. Sci. 15 (11), 21011–21027. doi:10.3390/ijms151121011
Chen, P., Wiencke, J., Aldape, K., Kesler-Diaz, A., Miike, R., Kelsey, K., et al. (2000). Association of an ERCC1 polymorphism with adult-onset glioma. Cancer Epidemiol. biomarkers and Prev. a Publ. Am. Assoc. Cancer Res. cosponsored by Am. Soc. Prev. Oncol. 9 (8), 843–847.
Chen, S., Huo, X., Lin, Y., Ban, H., Lin, Y., Li, W., et al. (2010). Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients. Int. J. Hyg. Environ. Health 213 (2), 140–145. doi:10.1016/j.ijheh.2010.01.004
Chu, T. Q., Li, R., Shao, M. H., Ye, J. Y., and Han, B. H. (2016). RAD18 polymorphisms are associated with platinum-based chemotherapy toxicity in Chinese patients with non-small cell lung cancer. Acta Pharmacol. Sin. 37 (11), 1490–1498. doi:10.1038/aps.2016.100
Clarissa Ribeiro Reily, R., Matheus Molina, S., Annabel, Q., Januario Bispo, C.-N., and Carlos Frederico Martins, M. (2018). DNA repair pathways and cisplatin resistance: an intimate relationship. Clin. (Sao Paulo) 73 (Suppl. 1), e478s. doi:10.6061/clinics/2018/e478s
Corrigan, A., Walker, J. L., Wickramasinghe, S., Hernandez, M. A., Newhouse, S. J., Folarin, A. A., et al. (2014). Pharmacogenetics of pemetrexed combination therapy in lung cancer: pathway analysis reveals novel toxicity associations. pharmacogenomics J. 14 (5), 411–417. doi:10.1038/tpj.2014.13
Cortejoso, L., García, M. I., García-Alfonso, P., González-Haba, E., Escolar, F., Sanjurjo, M., et al. (2013). Differential toxicity biomarkers for irinotecan- and oxaliplatin-containing chemotherapy in colorectal cancer. Cancer Chemother. Pharmacol. 71 (6), 1463–1472. doi:10.1007/s00280-013-2145-6
Daniel, Z. H., Thaned, C. C. O., Dhayan, P. T., Hui, T. A. T., Eunice, D. K., Wan, Q. C., et al. (2023). Systematic review and meta-analysis of the influence of genetic variation on ototoxicity in platinum-based chemotherapy. Otolaryngol. Head. Neck Surg. 168 (6), 1324–1337. doi:10.1002/ohn.222
Das, B., Antoon, R., Tsuchida, R., Lotfi, S., Morozova, O., Farhat, W., et al. (2008). Squalene selectively protects mouse bone marrow progenitors against cisplatin and carboplatin-induced cytotoxicity in vivo without protecting tumor growth. Neoplasia (New York, NY) 10 (10), 1105–1119. doi:10.1593/neo.08466
Daviet, S., Couvé-Privat, S., Gros, L., Shinozuka, K., Ide, H., Saparbaev, M., et al. (2007). Major oxidative products of cytosine are substrates for the nucleotide incision repair pathway. DNA repair 6 (1), 8–18. doi:10.1016/j.dnarep.2006.08.001
Deng, J. H., Deng, J., Shi, D. H., Ouyang, X. N., and Niu, P. G. (2015). Clinical outcome of cisplatin-based chemotherapy is associated with the polymorphisms of GSTP1 and XRCC1 in advanced non-small cell lung cancer patients. Clin. Transl. Oncol. 17 (9), 720–726. doi:10.1007/s12094-015-1299-6
De Troia, B., Dalu, D., Filipazzi, V., Isabella, L., Tosca, N., Ferrario, S., et al. (2019). ABCB1 c.3435C>T polymorphism is associated with platinum toxicity: a preliminary study. Cancer Chemother. Pharmacol. 83 (4), 803–808. doi:10.1007/s00280-019-03794-6
Ding-Wu, S., Lynn, M. P., Matthew, D. H., and Michael, M. G. (2012). Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 64 (3), 706–721. doi:10.1124/pr.111.005637
Dumont, P., Leu, J. I., Della Pietra, A. C., George, D. L., and Murphy, M. (2003). The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33 (3), 357–365. doi:10.1038/ng1093
Erčulj, N., Kovač, V., Hmeljak, J., and Dolžan, V. (2012). The influence of platinum pathway polymorphisms on the outcome in patients with malignant mesothelioma. Ann. Oncol. 23 (4), 961–967. doi:10.1093/annonc/mdr324
Fang, C., Li, X. P., Gong, W. J., Wu, N. Y., Tang, J., Yin, J. Y., et al. (2017). Age-related common miRNA polymorphism associated with severe toxicity in lung cancer patients treated with platinum-based chemotherapy. Clin. Exp. Pharmacol. Physiology 44, 21–29. doi:10.1111/1440-1681.12704
Felipe Antonio de Oliveira, G., Edilene Santos, dA., and Edenir Inez, P. (2022). Insights on variant analysis in silico tools for pathogenicity prediction. Front. Genet. 13, 1010327. doi:10.3389/fgene.2022.1010327
Ferracini, A. C., Lopes-Aguiar, L., Lourenço, G. J., Yoshida, A., Lima, C. S. P., Sarian, L. O., et al. (2021). GSTP1 and ABCB1 polymorphisms predicting toxicities and clinical management on carboplatin and paclitaxel-based chemotherapy in ovarian cancer. Clin. Transl. Sci. 14 (2), 720–728. doi:10.1111/cts.12937
Fisher, M. D., and D’Orazio, A. (2000). Phase II and III trials: comparison of four chemotherapy regimens in advanced non small-cell lung cancer (ECOG 1594). Clin. Lung Cancer 2 (1), 21–22. doi:10.1016/s1525-7304(11)70620-9
Fulda, S., and Debatin, K. M. (2006). Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 25 (34), 4798–4811. doi:10.1038/sj.onc.1209608
Giovanna, D., and Massimo, B. (2019). Platinum resistance in ovarian cancer: role of DNA repair. Cancers (Basel) 11 (1), 119. doi:10.3390/cancers11010119
Giovannetti, E., Pacetti, P., Reni, M., Leon, L. G., Mambrini, A., Vasile, E., et al. (2011). Association between DNA-repair polymorphisms and survival in pancreatic cancer patients treated with combination chemotherapy. Pharmacogenomics 12 (12), 1641–1652. doi:10.2217/pgs.11.109
Giuliano, C. (2012). Membrane transporters as mediators of Cisplatin effects and side effects. Sci. (Cairo). 2012, 473829. doi:10.6064/2012/473829
Go, R. S., and Adjei, A. A. (1999). Review of the comparative pharmacology and clinical activity of cisplatin and carboplatin. J. Clin. Oncol. 17 (1), 409–422. doi:10.1200/JCO.1999.17.1.409
Gong, W. J., Ma, L. Y., Hu, L., Lv, Y. N., Huang, H., Xu, J. Q., et al. (2019). STAT3 rs4796793 contributes to lung cancer risk and clinical outcomes of platinum-based chemotherapy. Int. J. Clin. Oncol. 24 (5), 476–484. doi:10.1007/s10147-018-01386-7
Gong, W. J., Peng, J. B., Yin, J. Y., Li, X. P., Zheng, W., Xiao, L., et al. (2017). Association between well-characterized lung cancer lncRNA polymorphisms and platinum-based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacol. Sin. 38 (4), 581–590. doi:10.1038/aps.2016.164
Goričar, K., Kovač, V., and Dolžan, V. (2014). Polymorphisms in translesion polymerase genes influence treatment outcome in malignant mesothelioma. Pharmacogenomics 15 (7), 941–950. doi:10.2217/pgs.14.14
Gréen, H., Hasmats, J., Kupershmidt, I., Edsgärd, D., De Petris, L., Lewensohn, R., et al. (2016). Using whole-exome sequencing to identify genetic markers for carboplatin and gemcitabine-induced toxicities. Clin. Cancer Res. 22 (2), 366–373. doi:10.1158/1078-0432.CCR-15-0964
Gu, S., Wu, Q., Zhao, X., Wu, W., Gao, Z., Tan, X., et al. (2012). Association of CASP3 polymorphism with hematologic toxicity in patients with advanced non-small-cell lung carcinoma treated with platinum-based chemotherapy. Cancer Sci. 103 (8), 1451–1459. doi:10.1111/j.1349-7006.2012.02323.x
Guo, D., Zhou, Y., Guo, Z., Gong, J., Wei, Y., and Zhu, W. (2016). Association of common polymorphisms in p53 and mdm2 with platinum-related grade iii/iv toxicities in Chinese advanced lung adenocarcinoma patients. Int. J. Clin. Exp. Med. 9 (9), 18410–18418.
Han, B., Gao, G., Wu, W., Gao, Z., Zhao, X., Li, L., et al. (2011). Association of ABCC2 polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients. Lung Cancer 72 (2), 238–243. doi:10.1016/j.lungcan.2010.09.001
Han, J. Y., Lim, H. S., Yoo, Y. K., Shin, E. S., Park, Y. H., Lee, S. Y., et al. (2007). Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer 110 (1), 138–147. doi:10.1002/cncr.22760
Hartmann, J. T., and Lipp, H. P. (2003). Toxicity of platinum compounds. Expert Opin. Pharmacother. 4 (6), 889–901. doi:10.1517/14656566.4.6.889
Hilary, S., Mohamad Ayub Khan, S., Md Asiful, I., and Shing, C. T. (2024). Genetic variants associated with response to platinum-based chemotherapy in non-small cell lung cancer patients: a field synopsis and meta-analysis. Br. J. Biomed. Sci. 81, 11835. doi:10.3389/bjbs.2024.11835
Hoffmeyer, S., Burk, O., von Richter, O., Arnold, H. P., Brockmöller, J., Johne, A., et al. (2000). Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. U. S. A. 97 (7), 3473–3478. doi:10.1073/pnas.050585397
Hu, L., Chen, S. H., Lv, Q. L., Sun, B., Qu, Q., Qin, C. Z., et al. (2016). Clinical significance of long non-coding RNA CASC8 rs10505477 polymorphism in lung cancer susceptibility, platinum-based chemotherapy response, and toxicity. Int. J. Environ. Res. Public Health 13 (6), 545. doi:10.3390/ijerph13060545
Huang, K., Luo, A., Li, X., Li, S., and Wang, S. (2015). Chemotherapy-induced neutropenia during adjuvant treatment for cervical cancer patients: development and validation of a prediction model. Int. J. Clin. Exp. Med. 8 (7), 10835–10844.
Isla, D., Sarries, C., Rosell, R., Alonso, G., Domine, M., Taron, M., et al. (2004). Single nucleotide polymorphisms and outcome in docetaxel–cisplatin-treated advanced non-small-cell lung cancer. Ann. Oncol. 15 (8), 1194–1203. doi:10.1093/annonc/mdh319
Iwata, K., Aizawa, K., Kamitsu, S., Jingami, S., Fukunaga, E., Yoshida, M., et al. (2012). Effects of genetic variants in SLC22A2 organic cation transporter 2 and SLC47A1 multidrug and toxin extrusion 1 transporter on cisplatin-induced adverse events. Clin. Exp. Nephrol. 16 (6), 843–851. doi:10.1007/s10157-012-0638-y
Jana, S., Mariangela, S., Cristina, R.-S., Colin, S., Arjan, F. T., Wim, V., et al. (2018). Base and nucleotide excision repair facilitate resolution of platinum drugs-induced transcription blockage. Nucleic Acids Res. 46 (18), 9537–9549. doi:10.1093/nar/gky764
Jia, M., Zhu, M., Wang, M., Sun, M., Qian, J., Ding, F., et al. (2016). Genetic variants of GADD45A, GADD45B and MAPK14 predict platinum-based chemotherapy-induced toxicities in Chinese patients with non-small cell lung cancer. Oncotarget 7 (18), 25291–25303. doi:10.18632/oncotarget.8052
Jorgensen, A. L., and Williamson, P. R. (2008). Methodological quality of pharmacogenetic studies: issues of concern. Statistics Med. 27 (30), 6547–6569. doi:10.1002/sim.3420
Kalikaki, A., Voutsina, A., Koutsopoulos, A., Papadaki, C., Sfakianaki, M., Yachnakis, E., et al. (2015). ERCC1 SNPs as potential predictive biomarkers in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Investig. 33 (4), 107–113. doi:10.3109/07357907.2014.1001897
Kanazawa, K., Yokouchi, H., Wang, X., Ishida, T., Fujita, Y., Fujiuchi, S., et al. (2014). Phase II trial of carboplatin and pemetrexed as first-line chemotherapy for non-squamous non-small cell lung cancer, and correlation between the efficacy/toxicity and genetic polymorphisms associated with pemetrexed metabolism: hokkaido Lung Cancer Clinical Study Group Trial (HOT) 0902. Cancer Chemother. Pharmacol. 74 (6), 1149–1157. doi:10.1007/s00280-014-2589-3
Khrunin, A. V., Moisseev, A., Gorbunova, V., and Limborska, S. (2010). Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharmacogenomics J. 10 (1), 54–61. doi:10.1038/tpj.2009.45
Kim, H. S., Kim, M. K., Chung, H. H., Kim, J. W., Park, N. H., Song, Y. S., et al. (2009). Genetic polymorphisms affecting clinical outcomes in epithelial ovarian cancer patients treated with taxanes and platinum compounds: a Korean population-based study. Gynecol. Oncol. 113 (2), 264–269. doi:10.1016/j.ygyno.2009.01.002
Kim, R. B., Leake, B. F., Choo, E. F., Dresser, G. K., Kubba, S. V., Schwarz, U. I., et al. (2001). Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin. Pharmacol. Ther. 70 (2), 189–199. doi:10.1067/mcp.2001.117412
Kim, W., Cho, Y. A., Kim, D. C., and Lee, K. E. (2022). Association between genetic polymorphism of GSTP1 and toxicities in patients receiving platinum-based chemotherapy: a systematic review and meta-analysis. Pharm. (Basel) 15 (4), 439. doi:10.3390/ph15040439
Kimcurran, V., Zhou, C., Schmid-Bindert, G., Shengxiang, R., Zhou, S., Zhang, L., et al. (2011). Lack of correlation between ERCC1 (C8092A) single nucleotide polymorphism and efficacy/toxicity of platinum based chemotherapy in Chinese patients with advanced non-small cell lung cancer. Adv. Med. Sci. 56 (1), 30–38. doi:10.2478/v10039-011-0013-3
Kumpiro, S., Sriuranpong, V., and Areepium, N. (2016). Impact of the copper transporter protein 1 (CTR1) polymorphism on adverse events among advanced NonSmall cell lung cancer patients treated with a carboplatin/gemcitabine regimen. Asian Pac. J. cancer Prev. APJCP 17 (9), 4391–4394.
Lambrechts, S., Lambrechts, D., Despierre, E., Van Nieuwenhuysen, E., Smeets, D., Debruyne, P. R., et al. (2015). Genetic variability in drug transport, metabolism or DNA repair affecting toxicity of chemotherapy in ovarian cancer. Bmc Pharmacol. and Toxicol. 16, 2. doi:10.1186/s40360-015-0001-5
Lavanderos, M. A., Cayún, J. P., Roco, Á., Sandoval, C., Cerpa, L., Rubilar, J. C., et al. (2019). Association study among candidate genetic polymorphisms and chemotherapy-related severe toxicity in testicular cancer patients. Front. Pharmacol. 10, 206. doi:10.3389/fphar.2019.00206
Lee, K.-H., Chang, H. J., Han, S.-W., Oh, D.-Y., Im, S.-A., Bang, Y.-J., et al. (2013). Pharmacogenetic analysis of adjuvant FOLFOX for Korean patients with colon cancer. Cancer Chemother. Pharmacol. 71 (4), 843–851. doi:10.1007/s00280-013-2075-3
Leusink, M., Onland-Moret, N. C., de Bakker, P. I., de Boer, A., and Maitland-van der Zee, A. H. (2016). Seventeen years of statin pharmacogenetics: a systematic review. Pharmacogenomics 17 (2), 163–180. doi:10.2217/pgs.15.158
Li, A., and Meyre, D. (2013). Challenges in reproducibility of genetic association studies: lessons learned from the obesity field. Int. J. Obes. 37 (4), 559–567. doi:10.1038/ijo.2012.82
Li, X., Shao, M., Wang, S., Zhao, X., Chen, H., Qian, J., et al. (2014). Heterozygote advantage of methylenetetrahydrofolate reductase polymorphisms on clinical outcomes in advanced non-small cell lung cancer (NSCLC) patients treated with platinum-based chemotherapy. Tumor Biol. 35 (11), 11159–11170. doi:10.1007/s13277-014-2427-6
Li, Y. Q., Zhang, X. Y., Chen, J., Yin, J. Y., and Li, X. P. (2018). ATP7B rs9535826 is associated with gastrointestinal toxicity of platinum-based chemotherapy in nonsmall cell lung cancer patients. J. Cancer Res. Ther. 14 (4), 881–886. doi:10.4103/jcrt.JCRT_890_17
Liblab, S., Vusuratana, A., and Areepium, N. (2020). ERCC1, XRCC1, and GSTP1 polymorphisms and treatment outcomes of advanced epithelial ovarian cancer patients treated with platinum-based chemotherapy. Asian Pac. J. cancer Prev. APJCP 21 (7), 1925–1929. doi:10.31557/APJCP.2020.21.7.1925
Little, J., Higgins, J. P., Ioannidis, J. P., Moher, D., Gagnon, F., von Elm, E., et al. (2009). STrengthening the REporting of genetic association studies (STREGA): an extension of the STROBE statement. PLoS Med. 6 (2), e22. doi:10.1371/journal.pmed.1000022
Liu, D., Xu, W., Ding, X., Yang, Y., Lu, Y., Fei, K., et al. (2017a). Caspase 8 polymorphisms contribute to the prognosis of advanced lung adenocarcinoma patients after platinum-based chemotherapy. Cancer Biol. Ther. 18 (12), 948–957. doi:10.1080/15384047.2016.1276128
Liu, J. Y., Qian, C. Y., Gao, Y. F., Chen, J., Zhou, H. H., and Yin, J. Y. (2017b). Association between DNA mismatch repair gene polymorphisms and platinum-based chemotherapy toxicity in non-small cell lung cancer patients. Chin. J. Cancer 36 (1), 12. doi:10.1186/s40880-016-0175-2
Low, S. K., Chung, S., Takahashi, A., Zembutsu, H., Mushiroda, T., Kubo, M., et al. (2013). Genome-wide association study of chemotherapeutic agent-induced severe neutropenia/leucopenia for patients in Biobank Japan. Cancer Sci. 104 (8), 1074–1082. doi:10.1111/cas.12186
Low, S. K., Takahashi, A., Mushiroda, T., and Kubo, M. (2014). Genome-wide association study: a useful tool to identify common genetic variants associated with drug toxicity and efficacy in cancer pharmacogenomics. Clin. cancer Res. 20 (10), 2541–2552. doi:10.1158/1078-0432.CCR-13-2755
Ludovini, V., Floriani, I., Pistola, L., Minotti, V., Meacci, M., Chiari, R., et al. (2011). Association of cytidine deaminase and xeroderma pigmentosum group D polymorphisms with response, toxicity, and survival in cisplatin/gemcitabine-treated advanced non-small cell lung cancer patients. J. Thorac. Oncol. 6 (12), 2018–2026. doi:10.1097/JTO.0b013e3182307e1f
Lv, F., Ma, Y., Zhang, Y., and Li, Z. (2018). Relationship between GSTP1 rs1695 gene polymorphism and myelosuppression induced by platinum-based drugs: a meta-analysis. Int. J. Biol. markers 33 (4), 364–371. doi:10.1177/1724600818792897
Marsh, S., Paul, J., King, C. R., Gifford, G., McLeod, H. L., and Brown, R. (2007). Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: the scottish randomised trial in ovarian cancer. J. Clin. Oncol. 25 (29), 4528–4535. doi:10.1200/JCO.2006.10.4752
Matullo, G., Palli, D., Peluso, M., Guarrera, S., Carturan, S., Celentano, E., et al. (2001). XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis 22 (9), 1437–1445. doi:10.1093/carcin/22.9.1437
Meyer zu Schwabedissen, H. E., Jedlitschky, G., Gratz, M., Haenisch, S., Linnemann, K., Fusch, C., et al. (2005). Variable expression of MRP2 (ABCC2) in human placenta: influence of gestational age and cellular differentiation. Drug metabolism Dispos. Biol. fate Chem. 33 (7), 896–904. doi:10.1124/dmd.104.003335
Moriya, Y., Nakamura, T., Horinouchi, M., Sakaeda, T., Tamura, T., Aoyama, N., et al. (2002). Effects of polymorphisms of MDR1, MRP1, and MRP2 genes on their mRNA expression levels in duodenal enterocytes of healthy Japanese subjects. Biol. and Pharm. Bull. 25 (10), 1356–1359. doi:10.1248/bpb.25.1356
Moyer, A. M., Salavaggione, O. E., Wu, T. Y., Moon, I., Eckloff, B. W., Hildebrandt, M. A., et al. (2008). Glutathione s-transferase p1: gene sequence variation and functional genomic studies. Cancer Res. 68 (12), 4791–4801. doi:10.1158/0008-5472.CAN-07-6724
Nairuz, T., Bushra, Y. U., and Kabir, Y. (2021). Effect of XPD and TP53 gene polymorphisms on the risk of platinum-based chemotherapy induced toxicity in Bangladeshi lung cancer patients. Asian Pac. J. cancer Prev. APJCP 22 (12), 3809–3815. doi:10.31557/APJCP.2021.22.12.3809
Nomura, H., Tsuji, D., Demachi, K., Mochizuki, N., Matsuzawa, H., Yano, T., et al. (2020). ABCB1 and ABCC2 genetic polymorphism as risk factors for neutropenia in esophageal cancer patients treated with docetaxel, cisplatin, and 5-fluorouracil chemotherapy. Cancer Chemother. Pharmacol. 86 (2), 315–324. doi:10.1007/s00280-020-04118-9
O’Donnell, P. H., and Dolan, M. E. (2009). Cancer pharmacoethnicity: ethnic differences in susceptibility to the effects of chemotherapy. Clin. cancer Res. 15 (15), 4806–4814. doi:10.1158/1078-0432.CCR-09-0344
Oun, R., Moussa, Y. E., and Wheate, N. J. (2018). The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans. Camb. Engl. 2003 47 (19), 6645–6653. doi:10.1039/c8dt00838h
Ouyang, Z., Peng, D., and Dhakal, D. P. (2013). Risk factors for hematological toxicity of chemotherapy for bone and soft tissue sarcoma. Oncol. Lett. 5 (5), 1736–1740. doi:10.3892/ol.2013.1234
Page, M. J., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ Clin. Res. ed 372, n71. doi:10.1136/bmj.n71
Pemble, S., Schroeder, K. R., Spencer, S. R., Meyer, D. J., Hallier, E., Bolt, H. M., et al. (1994). Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem. J. 300 (Pt 1), 271–276. doi:10.1042/bj3000271
Peng, J., Yang, L. X., Zhao, X. Y., Gao, Z. Q., Yang, J., Wu, W. T., et al. (2013). VCP gene variation predicts outcome of advanced non-small-cell lung cancer platinum-based chemotherapy. Tumor Biol. 34 (2), 953–961. doi:10.1007/s13277-012-0631-9
Peng, Y., Li, Z., Zhang, S., Xiong, Y., Cun, Y., Qian, C., et al. (2014). Association of DNA base excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with outcome to platinum-based chemotherapy in advanced nonsmall-cell lung cancer patients. Int. J. Cancer 135 (11), 2687–2696. doi:10.1002/ijc.28892
Prestayko, A. W., D'Aoust, J. C., Issell, B. F., and Crooke, S. T. (1979). Cisplatin (cis-diamminedichloroplatinum II). Cancer Treat. Rev. 6 (1), 17–39. doi:10.1016/s0305-7372(79)80057-2
Qian, C. Y., Zheng, Y., Wang, Y., Chen, J., Liu, J. Y., Zhou, H. H., et al. (2016). Associations of genetic polymorphisms of the transporters organic cation transporter 2 (OCT2), multidrug and toxin extrusion 1 (MATE1), and ATP-binding cassette subfamily C member 2 (ABCC2) with platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Chin. J. cancer 35 (1), 85. doi:10.1186/s40880-016-0145-8
Qian, J., Liu, H., Gu, S., Wu, Q., Zhao, X., Wu, W., et al. (2015). Genetic variants of the MDM2 gene are predictive of treatment-related toxicities and overall survival in patients with advanced NSCLC. Clin. Lung Cancer 16 (5), e37–e53. doi:10.1016/j.cllc.2015.02.001
Qian, J., Qu, H. Q., Yang, L., Yin, M., Wang, Q., Gu, S., et al. (2012). Association between CASP8 and CASP10 polymorphisms and toxicity outcomes with platinum-based chemotherapy in Chinese patients with non-small cell lung cancer. Oncologist 17 (12), 1551–1561. doi:10.1634/theoncologist.2011-0419
Qihai, S., Zhengyang, H., Xing, J., Yunyi, B., Jiaqi, L., Huan, Z., et al. (2023). The genomic signature of resistance to platinum-containing neoadjuvant therapy based on single-cell data. Cell Biosci. 13 (1), 103. doi:10.1186/s13578-023-01061-z
R, C. S., P, W. J., and A, A. F. (2000). Glutathione S-transferase: genetics and role in toxicology. Toxicol. Lett. 112-113, 357–363. doi:10.1016/s0378-4274(99)00230-1
Rabik, C. A., and Dolan, M. E. (2007). Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 33 (1), 9–23. doi:10.1016/j.ctrv.2006.09.006
Rodriguez, J., Boni, V., Hernández, A., Bitarte, N., Zarate, R., Ponz-Sarvisé, M., et al. (2011). Association of RRM1 -37A>C polymorphism with clinical outcome in colorectal cancer patients treated with gemcitabine-based chemotherapy. Eur. J. cancer 47 (6), 839–847. doi:10.1016/j.ejca.2010.11.032
Ruzzo, A., Graziano, F., Galli, F., Giacomini, E., Floriani, I., Galli, F., et al. (2014). Genetic markers for toxicity of adjuvant oxaliplatin and fluoropyrimidines in the phase III TOSCA trial in high-risk colon cancer patients. Sci. Rep. 4, 6828. doi:10.1038/srep06828
Saito, Y. A., Talley, N. J., de Andrade, M., and Petersen, G. M. (2006). Case-control genetic association studies in gastrointestinal disease: review and recommendations. Am. J. gastroenterology 101 (6), 1379–1389. doi:10.1111/j.1572-0241.2006.00587.x
Schiller, J. H., Harrington, D., Belani, C. P., Langer, C., Sandler, A., Krook, J., et al. (2002). Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl. J. Med. 346 (2), 92–98. doi:10.1056/NEJMoa011954
Senk, B., Goricar, K., Kovac, V., Dolzan, V., and Franko, A. (2019). Genetic polymorphisms in aquaporin 1 as risk factors for malignant mesothelioma and biomarkers of response to cisplatin treatment. Radiology Oncol. 53 (1), 96–104. doi:10.2478/raon-2019-0009
Seo, B. G., Kwon, H. C., Oh, S. Y., Lee, S., Kim, S. G., Kim, S. H., et al. (2009). Comprehensive analysis of excision repair complementation group 1, glutathione S-transferase, thymidylate synthase and uridine diphosphate glucuronosyl transferase 1A1 polymorphisms predictive for treatment outcome in patients with advanced gastric cancer treated with FOLFOX or FOLFIRI. Oncol. Rep. 22 (1), 127–136. doi:10.3892/or_00000415
Shaloam, D., and Paul, B. T. (2014). Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 740, 364–378. doi:10.1016/j.ejphar.2014.07.025
Shao, M., Jin, B., Niu, Y., Ye, J., Lu, D., and Han, B. (2014). Association of POLK polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients. Cell Biochem. biophysics 70 (2), 1227–1237. doi:10.1007/s12013-014-0046-x
Song, X., Wang, S., Hong, X., Li, X., Zhao, X., Huai, C., et al. (2017). Single nucleotide polymorphisms of nucleotide excision repair pathway are significantly associated with outcomes of platinum-based chemotherapy in lung cancer. Sci. Rep. 7 (1), 11785. doi:10.1038/s41598-017-08257-7
Sun, C., Zhang, Z., Qie, J., Wang, Y., Qian, J., Wang, J., et al. (2018). Genetic polymorphism of SLC31A1 is associated with clinical outcomes of platinum-based chemotherapy in non-small-cell lung cancer patients through modulating microRNA-mediated regulation. Oncotarget 9 (35), 23860–23877. doi:10.18632/oncotarget.24794
Svedberg, A., Björn, N., Sigurgeirsson, B., Pradhananga, S., Brandén, E., Koyi, H., et al. (2020). Genetic association of gemcitabine/carboplatin-induced leukopenia and neutropenia in non-small cell lung cancer patients using whole-exome sequencing. Lung Cancer 147, 106–114. doi:10.1016/j.lungcan.2020.07.005
Tan, X., Wu, Q., Cai, Y., Zhao, X., Wang, S., Gao, Z., et al. (2014). Novel association between CD74 polymorphisms and hematologic toxicity in patients with nsclc after platinum-based chemotherapy. Clin. Lung Cancer 15 (1), 67–78. doi:10.1016/j.cllc.2013.08.006
The Vinh, H., and Orlando, D. S. (2010). Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ. Mol. Mutagen 51 (6), 552–566. doi:10.1002/em.20573
Tibaldi, C., Giovannetti, E., Vasile, E., Mey, V., Laan, A. C., Nannizzi, S., et al. (2008). Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin. Cancer Res. 14 (6), 1797–1803. doi:10.1158/1078-0432.CCR-07-1364
van der Put, N. M., Gabreëls, F., Stevens, E. M., Smeitink, J. A., Trijbels, F. J., Eskes, T. K., et al. (1998). A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am. J. Hum. Genet. 62 (5), 1044–1051. doi:10.1086/301825
Vodicka, P., Kumar, R., Stetina, R., Sanyal, S., Soucek, P., Haufroid, V., et al. (2004). Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis 25 (5), 757–763. doi:10.1093/carcin/bgh064
Walia, H. K., Singh, N., and Sharma, S. (2021). GSTP1 Ile<i>105</i>Val polymorphism among North Indian lung cancer patients treated using monotherapy and poly-pharmacy. Hum. Exp. Toxicol. 40 (12_Suppl. l), S739–S752. doi:10.1177/09603271211059496
Walia, H. K., Singh, N., and Sharma, S. (2022). MTHFR polymorphism as a predictive biomarker for gastrointestinal and hematological toxicity in North Indian adenocarcinoma patients. J. Chemother. 34 (5), 326–340. doi:10.1080/1120009X.2021.1997008
Wang, L., Sun, C., Li, X., Mao, C., Qian, J., Wang, J., et al. (2021). A pharmacogenetics study of platinum-based chemotherapy in lung cancer: ABCG2 polymorphism and its genetic interaction with SLC31A1 are associated with response and survival. J. Cancer 12 (5), 1270–1283. doi:10.7150/jca.51621
Wang, S., Song, X., Li, X., Zhao, X., Chen, H., Wang, J., et al. (2016). RICTOR polymorphisms affect efficiency of platinum-based chemotherapy in Chinese non-small-cell lung cancer patients. Pharmacogenomics 17 (15), 1637–1647. doi:10.2217/pgs-2016-0070
Wang, X., Wang, Y. Z., Ma, K. W., Chen, X., and Li, W. (2014). MDM2 rs2279744 and TP53 rs1042522 polymorphisms associated with etoposide- and cisplatin-induced grade III/IV neutropenia in Chinese extensive-stage small-cell lung cancer patients. Oncol. Res. Treat. 37 (4), 176–180. doi:10.1159/000360785
Wang, Y., Probin, V., and Zhou, D. (2006). Cancer therapy-induced residual bone marrow injury-Mechanisms of induction and implication for therapy. Curr. cancer Ther. Rev. 2 (3), 271–279. doi:10.2174/157339406777934717
Wang, Y., Spitz, M. R., Zhu, Y., Dong, Q., Shete, S., and Wu, X. (2003). From genotype to phenotype: correlating XRCC1 polymorphisms with mutagen sensitivity. DNA repair 2 (8), 901–908. doi:10.1016/s1568-7864(03)00085-5
Wang, Z., Xu, B., Lin, D., Tan, W., Leaw, S., Hong, X., et al. (2008). XRCC1 polymorphisms and severe toxicity in lung cancer patients treated with cisplatin-based chemotherapy in Chinese population. Lung Cancer 62 (1), 99–104. doi:10.1016/j.lungcan.2008.02.019
Watanabe, A., Taniguchi, M., and Sasaki, S. (2003). Induction chemotherapy with docetaxel, cisplatin, fluorouracil and l-leucovorin for locally advanced head and neck cancers: a modified regimen for Japanese patients. Anticancer Drugs 14 (10), 801–807. doi:10.1097/00001813-200311000-00005
Wilson, D. M., Kim, D., Berquist, B. R., and Sigurdson, A. J. (2011). Variation in base excision repair capacity. Mutat. Res. 711 (1-2), 100–112. doi:10.1016/j.mrfmmm.2010.12.004
Wolfe, K. J., Wickliffe, J. K., Hill, C. E., Paolini, M., Ammenheuser, M. M., and Abdel-Rahman, S. Z. (2007). Single nucleotide polymorphisms of the DNA repair gene XPD/ERCC2 alter mRNA expression. Pharmacogenet Genomics 17 (11), 897–905. doi:10.1097/FPC.0b013e3280115e63
Wood, P. A., and Hrushesky, W. J. (1995). Cisplatin-associated anemia: an erythropoietin deficiency syndrome. J. Clin. investigation 95 (4), 1650–1659. doi:10.1172/JCI117840
Wu, W., Zhang, W., Qiao, R., Chen, D., Wang, H., Wang, Y., et al. (2009). Association of XPD polymorphisms with severe toxicity in non-small cell lung cancer patients in a Chinese population. Clin. Cancer Res. 15 (11), 3889–3895. doi:10.1158/1078-0432.CCR-08-2715
Xu, S., Wang, Y., Roe, B., and Pearson, W. R. (1998). Characterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletion. J. Biol. Chem. 273 (6), 3517–3527. doi:10.1074/jbc.273.6.3517
Xu, W., Liu, D., Yang, Y., Ding, X., Sun, Y., Zhang, B., et al. (2016). Association of CHEK2 polymorphisms with the efficacy of platinum-based chemotherapy for advanced non-small-cell lung cancer in Chinese never-smoking women. J. Thorac. Dis. 8 (9), 2519–2529. doi:10.21037/jtd.2016.08.70
Xu, X., Ren, H., Zhou, B., Zhao, Y., Yuan, R., Ma, R., et al. (2012). Prediction of copper transport protein 1 (CTR1) genotype on severe cisplatin induced toxicity in non-small cell lung cancer (NSCLC) patients. Lung Cancer 77 (2), 438–442. doi:10.1016/j.lungcan.2012.03.023
Ye, J., Chu, T., Li, R., Niu, Y., Jin, B., Xia, J., et al. (2015). Pol zeta polymorphisms are associated with platinum based chemotherapy response and side effects among non-small cell lung cancer patients. Neoplasma 62 (5), 833–839. doi:10.4149/neo_2015_101
Yin, J. Y., Li, X., Li, X. P., Xiao, L., Zheng, W., Chen, J., et al. (2016). Prediction models for platinum-based chemotherapy response and toxicity in advanced NSCLC patients. Cancer Lett. 377 (1), 65–73. doi:10.1016/j.canlet.2016.04.029
Yin, J. Y., Meng, X. G., Qian, C. Y., Li, X. P., Chen, J., Zheng, Y., et al. (2015). Association of positively selected eIF3a polymorphisms with toxicity of platinum-based chemotherapy in NSCLC patients. Acta Pharmacol. Sin. 36 (3), 375–384. doi:10.1038/aps.2014.160
Yoshihama, T., Fukunaga, K., Hirasawa, A., Nomura, H., Akahane, T., Kataoka, F., et al. (2018). GSTP1 rs1695 is associated with both hematological toxicity and prognosis of ovarian cancer treated with paclitaxel plus carboplatin combination chemotherapy: a comprehensive analysis using targeted resequencing of 100 pharmacogenes. Oncotarget 9 (51), 29789–29800. doi:10.18632/oncotarget.25712
Yu, J. J., Lee, K. B., Mu, C., Li, Q., Abernathy, T. V., Bostick-Bruton, F., et al. (2000). Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int. J. Oncol. 16 (3), 555–560. doi:10.3892/ijo.16.3.555
Yuan, Z. J., Zhou, W. W., Liu, W., Wu, B. P., Zhao, J., Wu, W., et al. (2015). Association of GSTP1 and RRM1 polymorphisms with the response and toxicity of gemcitabine-cisplatin combination chemotherapy in Chinese patients with non-small cell lung cancer. Asian Pac J. Cancer Prev. 16 (10), 4347–4351. doi:10.7314/apjcp.2015.16.10.4347
Zhan, X., Wu, W., Han, B., Gao, G., Qiao, R., Lv, J., et al. (2012). Hsa-miR-196a2 functional SNP is associated with severe toxicity after platinum-based chemotherapy of advanced nonsmall cell lung cancer patients in a Chinese population. J. Clin. Laboratory Analysis 26 (6), 441–446. doi:10.1002/jcla.21544
Zhao, X., Wang, S., Wu, J., Li, X., Wang, X., Gao, Z., et al. (2015). Association of TERT polymorphisms with clinical outcome of non-small cell lung cancer patients. PLoS ONE 10 (5), e0129232. doi:10.1371/journal.pone.0129232
Zhao, X., Wang, X., Wu, W., Gao, Z., Wu, J., Garfield, D. H., et al. (2012). Matrix metalloproteinase-2 polymorphisms and clinical outcome of Chinese patients with nonsmall cell lung cancer treated with first-line, platinum-based chemotherapy. Cancer 118 (14), 3587–3598. doi:10.1002/cncr.26669
Zheng, D., Chen, Y., Gao, C., Wei, Y., Cao, G., Lu, N., et al. (2014). Polymorphisms of p53 and MDM2 genes are associated with severe toxicities in patients with non-small cell lung cancer. Cancer Biol. Ther. 15 (11), 1542–1551. doi:10.4161/15384047.2014.956599
Zheng, Y., Deng, Z., Tang, M., and Cai, P. (2021). Erythropoietin promoter polymorphism is associated with treatment efficacy and severe hematologic toxicity for platinum-based chemotherapy. Expert Opin. Drug Metabolism Toxicol. 17 (4), 495–502. doi:10.1080/17425255.2021.1879048
Zheng, Y., Deng, Z., Tang, M., Xiao, D., and Cai, P. (2020). Impact of genetic factors on platinum-induced gastrointestinal toxicity. Mutat. Res. Rev. Mutat. Res. 786, 108324. doi:10.1016/j.mrrev.2020.108324
Zheng, Y., Deng, Z., Yin, J., Wang, S., Lu, D., Wen, X., et al. (2017). The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. Int. J. Cancer 141 (11), 2336–2347. doi:10.1002/ijc.30921
Zhu, Y., Yang, H., Chen, Q., Lin, J., Grossman, H. B., Dinney, C. P., et al. (2008). Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA repair 7 (2), 141–148. doi:10.1016/j.dnarep.2007.08.006
Zolk, O. (2012). Disposition of metformin: variability due to polymorphisms of organic cation transporters. Ann. Med. 44 (2), 119–129. doi:10.3109/07853890.2010.549144
Zolk, O., Solbach, T. F., König, J., and Fromm, M. F. (2009). Functional characterization of the human organic cation transporter 2 variant p.270Ala>Ser. Drug metabolism Dispos. Biol. fate Chem. 37 (6), 1312–1318. doi:10.1124/dmd.108.023762
Keywords: platinum, chemotherapy, hematological toxicity, polymorphisms, pharmacogenomics
Citation: Zheng Y, Tang M, Deng Z and Cai P (2024) Genetic polymorphisms and platinum-induced hematological toxicity: a systematic review. Front. Pharmacol. 15:1445328. doi: 10.3389/fphar.2024.1445328
Received: 07 June 2024; Accepted: 05 August 2024;
Published: 21 August 2024.
Edited by:
Roberto Rodríguez-Labrada, Cuban Neuroscience Center, CubaReviewed by:
Md. Siddiqul Islam, Southeast University, BangladeshMukerrem Betul Yerer Aycan, Erciyes University, Türkiye
Copyright © 2024 Zheng, Tang, Deng and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei Cai, Y2FpcGVpMTk4NTAxMDRAMTI2LmNvbQ==; Zheng Deng, enJzdWl5dWVAc2luYS5jb20=
†These authors have contributed equally to this work
 Yi Zheng
Yi Zheng Mimi Tang
Mimi Tang Zheng Deng
Zheng Deng Pei Cai
Pei Cai