- 1Department of Hematology, Institute of Hematology, West China Hospital, Sichuan University, Chengdu, China
- 2West China School of Medicine and West China Hospital, Sichuan University, Chengdu, China
Background: Chidamide is subtype-selective histone deacetylase (HDAC) inhibitor that showed promising result in clinical trials to improve prognosis of angioimmunoblastic T-cell lymphoma (AITL) patients. However, in real world settings, contradictory reports existed as to whether chidamide improve overall survival (OS). Therefore, we aimed to develop an interpretable machine learning (Machine learning)–based model to predict the 2-year overall survival of AITL patients based on chidamide usage and baseline features.
Methods: A total of 183 patients with AITL were randomly divided into training set and testing set. We used 5 ML algorithms to build predictive models. Recursive feature elimination (RFE) method was used to filter for the most important features. The ML models were interpreted and the relevance of the selected features was determined using the Shapley additive explanations (SHAP) method and the local interpretable model–agnostic explanationalgorithm.
Results: A total of 183 patients with newly diagnosed AITL from 2012 to 2022 from 3 centers in China were enrolled in our study. Seventy-one patients were dead within 2 years after diagnosis. Five ML algorithms were built based on chidamide usage and 16 baseline features to predict 2-year OS. Catboost model presented to be the best predictive model. After RFE screening, 12 variables demonstrated the best performance (AUC = 0.8651). Using chidamide ranked third among all the variables that correlated with 2-year OS.
Conclusion: This study demonstrated that the Catboost model with 12 variables could effectively predict the 2-year OS of AITL patients. Combining chidamide in the treatment therapy was positively correlated with longer OS of AITL patients.
Introduction
Angioimmunoblastic T-cell lymphoma (AITL) is a distinct kind of peripheral T-cell lymphoma (PTCL) that has a poor prognosis (Swerdlow et al., 2016). For AITL patients, the 5-year overall survival (OS) rate was 44% and the progression-free survival (PFS) rate was 32% (Advani et al., 2021). Anthracycline-based chemotherapy regimens are frequently utilized, yet their effectiveness is constrained. Based on the unsatisfactory outcome of traditional treatment, the NCCN Clinical Practice Guidelines in Oncology recommended engaging in clinical trials as preferred management strategy (Horwitz et al., 2022). Notably, although some patients had the identical staging or prognostic scores that commonly used to evaluate T cell lymphoma, their clinical outcomes varied considerably. The differences in prognosis may be due to the heterogeneity of AITL (Zhang et al., 2023). Therefore, novel models that can better stratify patients are required.
Chidamide is a benzamide type of subtype-selective histone deacetylase (HDAC) inhibitor (Gong et al., 2012). In recent years, chidamide has appeared as a promising treatment in PTCL, especially in AITL. In phase II study of chidamide in relapsed or refractory (r/r) AITL, the overall response rate (ORR) was 50% (Shi et al., 2015). In a multicenter phase II clinical trial combining chidamide with prednisone, etoposide, and thalidomide in untreated AITL, the ORR was 90.2%. The 2-year progression-free survival (PFS) rate and overall survival (OS) rate were 66.5% and 82.2%, respectively (Wang et al., 2022b). However, in real world analysis, contradictory results exist as for whether combining chidamide with chemotherapy improves OS compared with chemotherapy alone (Shi et al., 2017; Liu et al., 2021; Wang et al., 2022a). Further evidence is required to clarify the efficiency of chidamide in real-world setting.
Machine learning (ML) algorithms is a key area of artificial intelligence, which may learn from complicated data by utilizing computational methods to identify possible features for prediction (Haug and Drazen, 2023). Compared with conventional generalized linear model, machine learning based on the advanced algorithm are more acceptable in terms of data distribution and integrity, as well as more flexible in terms of mining data value (Elemento et al., 2021). Therefore, machine learning has been widely used in the medical field in recent years and has developed into a potent tool for physicians to use when making clinical decisions (Radakovich et al., 2020; Haug and Drazen, 2023; Swanson et al., 2023). Hence, the purposes of the present study were to establish ML models to predict prognosis of AITL and to evaluate the benefit of chidamide in real-world setting.
Materials and methods
Patients
This retrospective multicenter study included patients with newly diagnosed AITL between 2012 and 2022 from three centers in China, and this study was approved by the Ethics Committees of three hospitals.
Data collection
Data on the clinical and laboratory metrics of patients were obtained from three hospital databases. The inclusion criteria were strictly adhered to, ensuring that all participants: (1) had a confirmed pathological diagnosis of AITL, (2) newly diagnosed cases, (3) were aged over 18 years, and (4) were capable of independently signing informed consent. Baseline information includes a total of 16 variables: (1) basic condition: age, gender; (2) the condition of lymphoma: Ann Arbor staging, B symptom, extranodal involvement, Eastern Cooperative Oncology Group (ECOG) Performance Status, International Prognostic Index (IPI); (3) clinical symptoms: rash, edema/serous effusion; (4) laboratory parameters: hemoglobin (Hb), platelet (PLT), absolute lymphocyte count (ALC), absolute eosinophil count (AEC), serum albumin (ALB), serum globulin (GLB), lactate dehydrogenase (LDH).
Patients were treated with chemotherapy, mostly anthracycline-based regimens, with or without chidamide. We conducted patient follow-up monthly and recorded the detailed treatment strategy and clinical outcomes. Participants with significant data omissions were excluded to maintain the integrity and reliability of our research.
Study design and machine learning algorithms
A total of 17 variables were included to identify the prognostic value of these features, including 16 baseline features and “chidamide usage”. Among 183 patients, 71 patients died within 2 years after diagnosis, while 112 patients were still alive. About 75% (137) of patients were randomly selected into the training set, while the rest 46 patients fell into the validation set. Five models were built through logistics regression (LR), random forest (RF), light gradient boosting machine (LGBM), extreme gradient boosting (XGBoost), and categorical boosting (CatBoost) to predict 2-year OS. The selection of models was based on a comprehensive consideration of performance, interpretability, computational efficiency, generalizability, as well as support by literature documenting similar research contexts, ensuring the relevance and robustness of our approach.
Model validation
We explored five different machine learning algorithms and conducted grid search techniques to identify the best hyperparameter combinations, setting possible value ranges for different parameters and evaluating each parameter combination with 5-fold cross-validation. The receiver-operating characteristic (ROC) curve was used as the assessment metric to validate the performances of the different models. The performance of each model was evaluated by calculating the area under the curve (AUC), accuracy, precision, recall, specificity, and F1 score. After comparing the AUC values of the five algorithms, we chose CatBoost as our final model due to its superior performance and high AUC value on the test set. The parameters for five models were provided in Supplementary Table 1.
We implemented Recursive Feature Elimination (RFE) with the CatBoost algorithm to reduce the number of features and enhance model efficiency. RFE is a model-based feature selection method that methodically reduces the feature set by recursively removing the least significant feature during each iteration. By meticulously comparing the performance across different feature subsets, we selected 12 features that contribute most significantly to the model’s predictive power. The model demonstrated good performance and robustness on the test set, highlighting its generalizability.
Model interpretation
The interpretation of the predictions produced by the models was conducted using the Shapley additive explanations (SHAP) value. The SHAP technique is capable of providing a more comprehensive explanation of the significance of each variable in all component sequences by doing a marginal calculation of their contributions. The predictions were also explained using the local interpretable model–agnostic explanation (LIME). The trustworthiness of a model’s explanation for predicting a single sample using a local linear approximation of the model’s behavior can be enhanced.
Statistical analysis
Patients were categorized into two groups based on their 2-year status: alive and dead. Binary variables were subjected to either a Fisher exact test or a γ2 test. For continuous variables that adhere to the normal distribution, a Student’s t-test was employed, and the data were displayed as medians along with standard deviations. For variables that do not follow the normal distribution, a Mann-Whitney U test was utilized, and the data were presented as medians along with interquartile ranges (IQRs). The development and validation of the five machine learning algorithms were conducted using Python software. Statistical significance was set at p ≤ 0.05. Python software (version 3.9.13) was utilized to perform statistical analysis.
Results
Patient characteristics
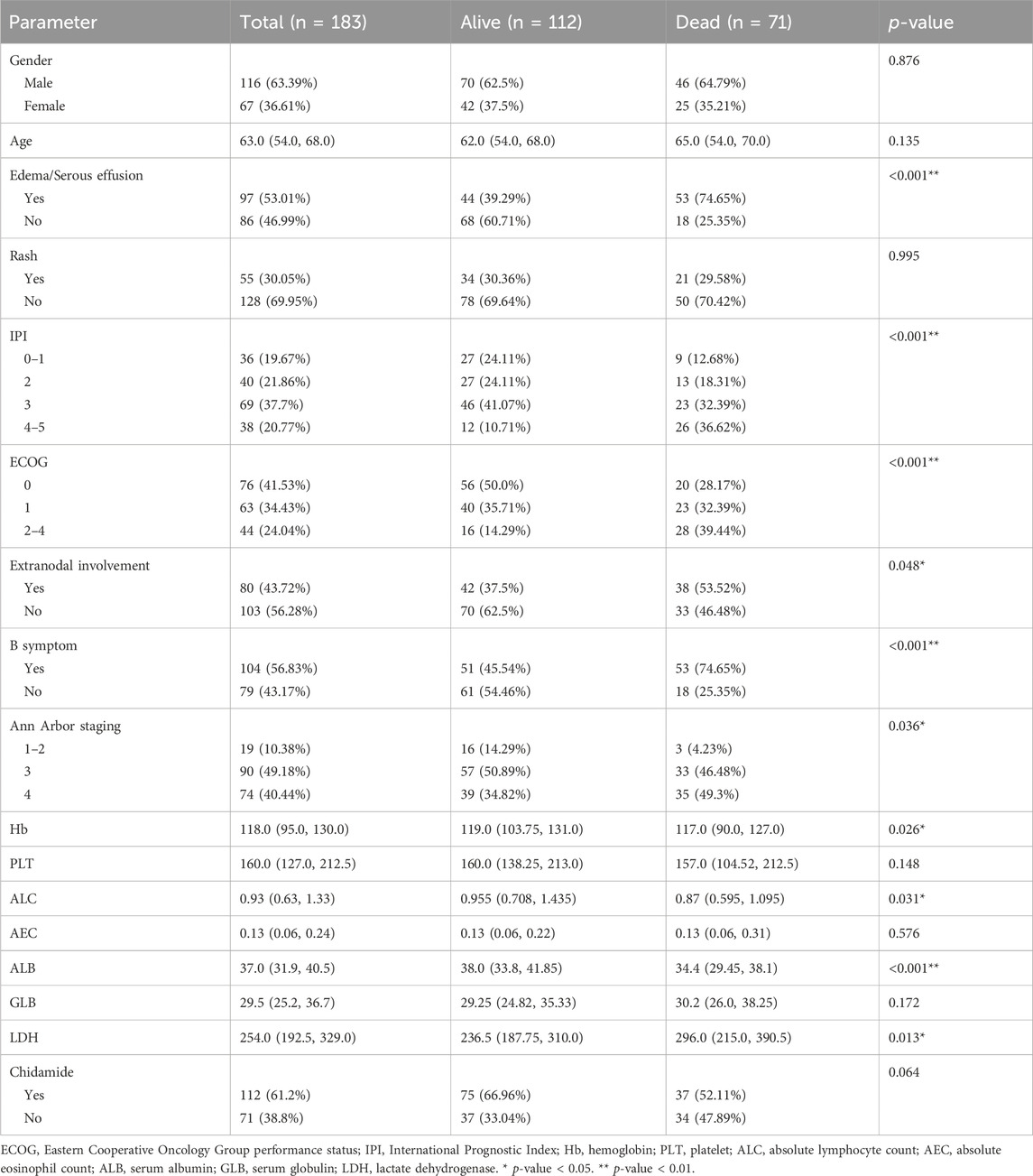
A total of 538 patients were newly diagnosed with AITL between 2012 and 2022 from three centers in China. Following the screening process, we included 183 patients who had a comprehensive baseline examination and long-term follow-up data of at least 24 months in our study (Figure 1). Cohort characteristics are presented in Table 1. The median age was 63 (IQR = 54–68) years and 63.39% were males. AITL patients were categorized into two groups based on 2-year OS. Among 183 patients, 112 patients had an OS longer than 2 years.
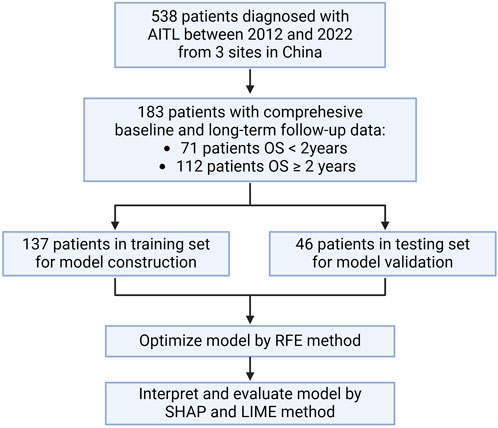
Figure 1. Flowchart. OS: overall survival; RFE: recursive feature elimination; SHAP: Shapley additive explanations; LIME, local interpretable model–agnostic explanation.
Predicting 2-year OS by baseline features
Our modeling utilized a comprehensive set of 17 variables, encompassing age, gender, Ann Arbor staging, B symptom, extranidal involvement, ECOG, IPI, rash, edema/serous effusion, Hb, PLT, ALC, AEC, ALB, GLB, LDH, and chidamide. The patients were divided into two groups, namely, the training set for model development and the testing set for model performance evaluation, using a random stratification method with a ratio of 3:1. The clinical characteristics of the two data sets are presented in Table 2. The p-values of all features in the training set and the test set were greater than 0.05, indicating that the division of the training and test sets is reasonable.
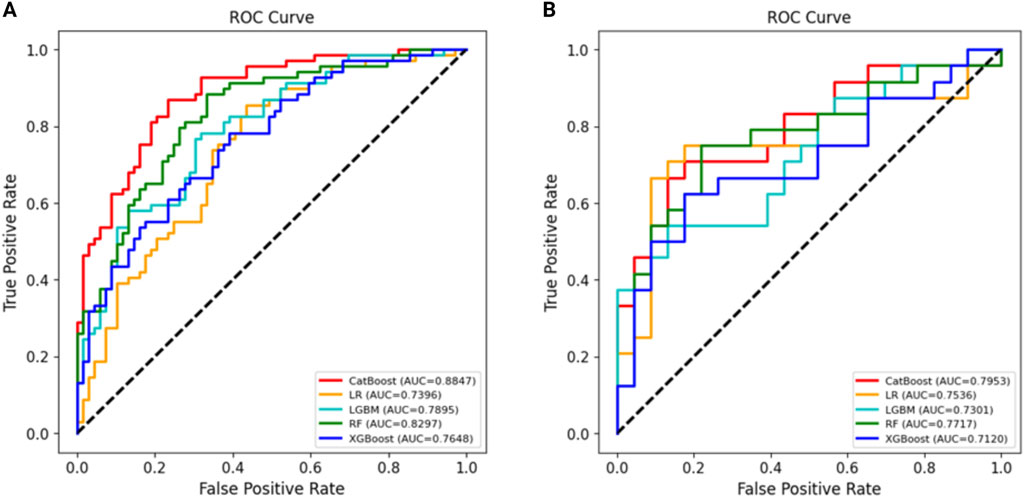
Using all 17 variables, predictive models for 2-year survival were developed based on five algorithms, including LR, RF, LGBM, XGBoost, and CatBoost. The best predictive performance was observed in CatBoost (training set AUC = 0.8949, testing set AUC = 0.8571). The AUC, accuracy, precision, recall, specificity, and F1 score of five models in the training and testing data sets were shown in Table 3. The ROC curves of five models were presented in Figure 2.
The Catboost model was further optimized by employing the recursive feature elimination (RFE) method to selectively filter for the most significant characteristics. The feature importance ranking by RFE was presented in Supplementary Figure 1. In the ideal Catboost algorithm, a total of 12 factors were utilized. These variables encompassed Age, B symptom, Extranodal involvement, ECOG, IPI, Edema/Serous effusion, ALC, AEC, ALB, GLB, LDH, and Chidamide (Figure 3). Table 4 showed the AUC, accuracy, precision, recall, specificity, and F1 score of the optimized Catboost model.

Figure 3. Using RFE method to screen the optimal variables on Catboost model (A). The ROC curves of the optimized Catboost model (B).
Interpretation and evaluation of machine learning model
We employed the SHAP approach to quantitatively assess the impact of each feature on the prediction outcomes of the model (Figure 4). The analysis of feature ranking revealed that the five most significant features were B symptom, Edema/Serous effusion, Chidamide, Extranodal involvement, and ECOG. Chidamide usage was negatively correlated with the outcome (OS shorter than 2 years), indicating the efficiency of chidamide on improving OS in AITL patients.
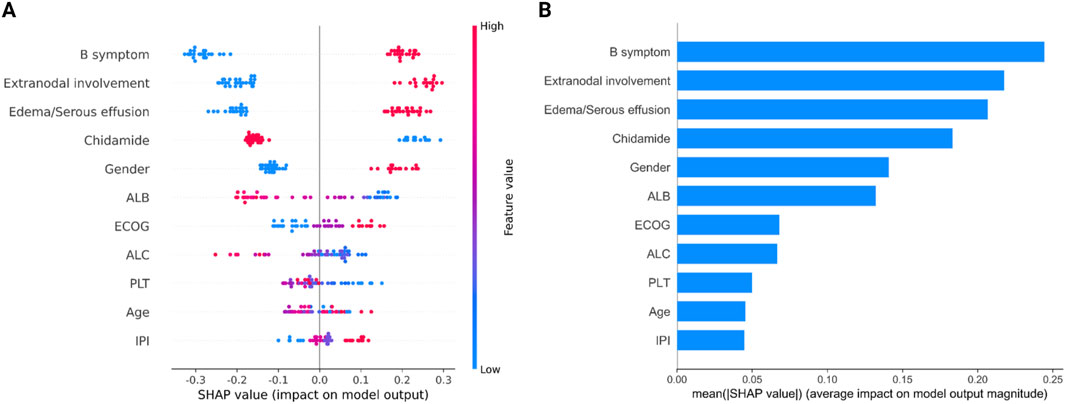
Figure 4. Attribution of 12 features in the optimized Catboost model based on the SHAP algorithm. (A) Summary of SHAP analysis on the data set. One dot represents a case in the data set, and the color of a dot indicates the value of the feature. Blue indicates the lowest range and red the highest range. (B) Ranking of feature importance indicated by SHAP. SHAP values provide a clear depiction of how each feature influences the model’s prediction, indicating whether the impact is positive or negative. For example, from the SHAP plot, we can observe that B symptoms have the most significant contribution. A higher value of B symptoms corresponds to a positive SHAP value, suggesting a positive impact on the prediction, supporting a likelihood of death. Conversely, a lower value of B symptoms results in a negative SHAP value, indicating a negative impact on the prediction, supporting survival.
Moreover, LIME algorithm was applied to explain the influence of different variables of the Catboost model on the prediction results. Two cases were randomly selected to interpret the visualized prediction results (Figure 5).
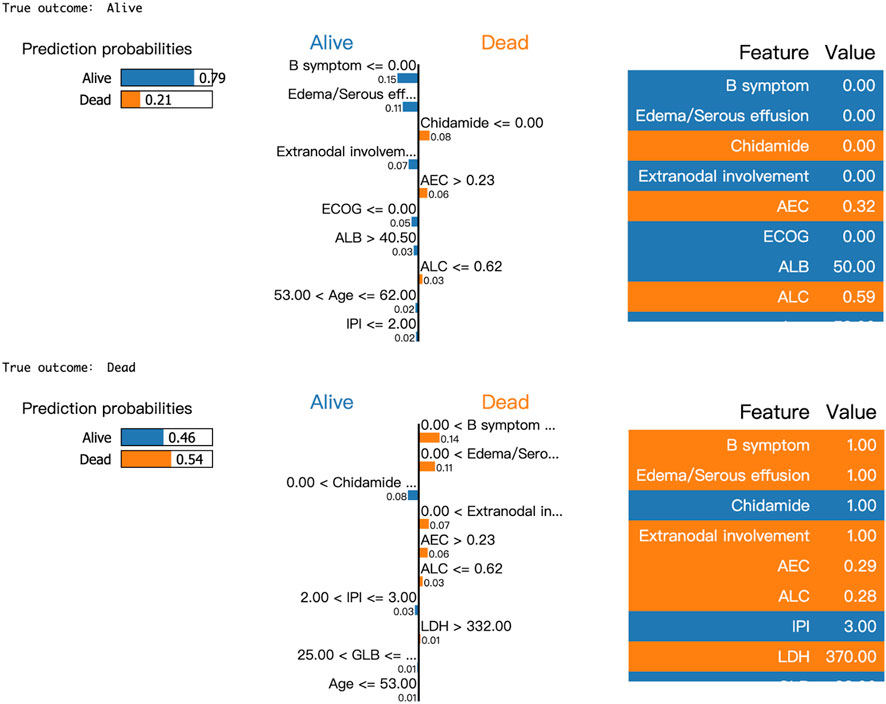
Figure 5. Results of LIME with Catboost applied to two randomly selected patients. Orange attributes support death, while blue attributes support survival. Taking the first patient as an example, the upper left corner of the LIME plot shows a survival prediction probability of 0.79 and a death prediction probability of 0.21, indicating a higher likelihood of survival for the patient within 2 years. The middle section lists the individual features along with their corresponding coefficients, which reflect their impact on the prediction. For instance, the coefficient for B symptoms is 0.14, shown in blue, suggesting that the absence of B symptoms increases the model’s predicted probability of survival by 0.14. Similarly, the absence of Edema/Serous effusion increases the survival probability by 0.11, while the absence of Chidamide use increases the probability of death by 0.08.
Discussion
AITL is a life-threatening lymphoma with heterogeneous nature. Current staging systems are far from satisfactory to stratify AITL patients. Novel models that can better stratify patients are required. By establishing Catboost model, our study demonstrated that 12 features, including Age, B symptom, Extranodal involvement, ECOG, IPI, Edema/Serous effusion, ALC, AEC, ALB, GLB, LDH, and Chidamide, were significantly related with 2-year OS of AITL patients.
Chidamide, a HDAC inhibitor, has presented as a promising target therapy in recent years in PTCL patients. In clinical trials, chidamide remarkably prolonged the OS of AITL patients. However, in real world analysis, contradictory results exist as for whether combining chidamide with chemotherapy improves OS compared with chemotherapy alone (Shi et al., 2017; Liu et al., 2021; Wang et al., 2022a). Further evidence is required to clarify the efficiency of chidamide in real-world setting. Using ML algorithm, we demonstrated that using chidamide was among the most important features that influent 2-year OS. Specially, in the optimized Catboost model, chidamide ranked third among all the variables that correlated with OS. In general, our study supported that in real-world setting, combining chidamide in treatment strategy could prolong OS of AITL patients.
Chidamide was administered as the initial treatment for 68 patients, as the second line of treatment for 22 patients, and as the maintenance treatment for 43 patients. For patients treated with chemotherapy without Chidamide, no other substitute was added. Nevertheless, the subset analysis revealed that the disparity in response rates between chemotherapy combined with chidamide and without chidamide was not statistically significant in both first line and second line treatment, owing to the small sample size. Out of the 43 patients who received chidamide as a form of maintenance therapy, 24 patients had available treatment responses both before and after the maintenance period. The treatment responses in the majority of patients (22 cases) were consistent both before and after maintenance. It is worth mentioning that two patients demonstrated improved treatment response following the administration of chidamide. There was no evidence of any progressive disease. Previously, the benefit of chidamide as maintenance therapy in real world setting was also reported by Guo et al. (2022).
Similar to the commonly used prognostic scores in T-cell lymphoma, including International Prognostic Index (IPI), Prognostic Index for T-cell lymphoma (PIT), International peripheral T-cell lymphoma Project score (IPTCLP) and modified Prognostic Index for T-cell lymphoma (mPIT), age, B symptom, Extranodal involvement, ECOG, PLT, and LDH were correlated with OS in our study (Gutiérrez-García et al., 2011). Moreover, ALC was also significantly related with 2-year OS, which was in agreement with the AITL score established based on 282 patients with AITL enrolled between 2006 and 2018 in the international prospective T-cell Project (Advani et al., 2021). Interestingly, we also showed that AEC was associated with prognosis, which has not been reported before.
Remarkably, unlike aforementioned prognostic scores, we showed that Edema/Serous effusion and ALB level were also significantly correlated with prognosis. In the optimized Catboost model, Edema/Serous effusion was found to be the second most significant variable. Previously, Sun et al. reported the correlation of serous effusion with OS based on a cohort of 55 AITL patients (Sun et al., 2021), while Huang et al. reported that ALB <30 g/L was significantly associated with poor prognosis base on a cohort of 64 AITL patients (Huang et al., 2020). From a clinical perspective, there was a correlation between Edema/Serous effusion and a reduction in ALB levels. This pair of features warrant more attention in further studies of AITL.
The current investigation possesses several limitations that should not be ignored. First, this study was conducted retrospectively and had a limited sample size. Additional validation in large cohorts is required to further substantiate the efficacy of the model. Second, present study identified chidamide as an effective treatment to AITL patients that prolonged their OS. Nevertheless, the treatment regimens employing chidamide exhibited variability among patients in this retrospective investigation, encompassing factors such as the timing and duration of therapy. Hence, it is imperative to conduct additional validation of the efficacy of chidamide in a substantial prospective cohort within a real-world context. Furthermore, the absence of complete data has resulted in the exclusion of certain potential influencing elements, such as C-reactive protein and Beta-2-microglobulin, from the establishment of machine learning models. Moreover, along with the development of genome and transcriptome analysis, molecular abnormalities which may impact the prognosis of lymphoma were gradually unveiled (Cortes et al., 2018; Lone et al., 2021; Leca et al., 2023; Zhang et al., 2024). In the age of precision medicine, as sequencing technology becomes increasingly prevalent, it is imperative to incorporate these molecular abnormalities into future prognostic models of AITL.
Conclusion
The Catboost model developed in our study shown a strong prediction capability for the 2-year OS of patients with AITL. Chidamide, in particular, was one of the most significant factors that influenced OS. The utilization of chidamide exhibited a strong correlation with an improved outcome. Further validation of the model’s predictive value should be conducted using larger cohorts.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Ethics Committee on Biomedical Research, West China Hospital of Sichuan University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
CZ: Writing–original draft, Conceptualization, Formal Analysis. JX: Conceptualization, Writing–review and editing. MG: Investigation, Writing–review and editing. TY: Data curation, Writing–review and editing. WT: Data curation, Writing–review and editing. JW: Methodology, Writing–review and editing. QL: Methodology, Writing–review and editing. YY: Methodology, Writing–review and editing. XZ: Validation, Writing–review and editing. CX: Funding acquisition, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Sichuan science and technology department for key research and development projects (Grant/Award Number: 2019YFS0027).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1435284/full#supplementary-material
References
Advani, R. H., Skrypets, T., Civallero, M., Spinner, M. A., Manni, M., Kim, W. S., et al. (2021). Outcomes and prognostic factors in angioimmunoblastic T-cell lymphoma: final report from the international T-cell Project. Blood 138 (3), 213–220. doi:10.1182/blood.2020010387
Cortes, J. R., Ambesi-Impiombato, A., Couronné, L., Quinn, S. A., Kim, C. S., da Silva Almeida, A. C., et al. (2018). RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell 33 (2), 259–273. doi:10.1016/j.ccell.2018.01.001
Elemento, O., Leslie, C., Lundin, J., and Tourassi, G. (2021). Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer 21 (12), 747–752. doi:10.1038/s41568-021-00399-1
Gong, K., Xie, J., Yi, H., and Li, W. (2012). CS055 (Chidamide/HBI-8000), a novel histone deacetylase inhibitor, induces G1 arrest, ROS-dependent apoptosis and differentiation in human leukaemia cells. Biochem. J. 443 (3), 735–746. doi:10.1042/bj20111685
Guo, W., Wang, X., Li, J., Yin, X., Zhao, Y., Tang, Y., et al. (2022). Chidamide maintenance therapy following induction therapy in patients with peripheral T-cell lymphoma who are ineligible for autologous stem cell transplantation: case series from China. Front. Oncol. 12, 875469. doi:10.3389/fonc.2022.875469
Gutiérrez-García, G., García-Herrera, A., Cardesa, T., Martínez, A., Villamor, N., Ghita, G., et al. (2011). Comparison of four prognostic scores in peripheral T-cell lymphoma. Ann. Oncol. 22 (2), 397–404. doi:10.1093/annonc/mdq359
Haug, C. J., and Drazen, J. M. (2023). Artificial intelligence and machine learning in clinical medicine, 2023. N. Engl. J. Med. 388 (13), 1201–1208. doi:10.1056/NEJMra2302038
Horwitz, S. M., Ansell, S., Ai, W. Z., Barnes, J., Barta, S. K., Brammer, J., et al. (2022). T-cell lymphomas, version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 20 (3), 285–308. doi:10.6004/jnccn.2022.0015
Huang, C., Zhang, H., Gao, Y., Diao, L., and Liu, L. (2020). Development of a novel clinical prognostic model for patients with angioimmunoblastic T-cell lymphoma. Technol. Cancer Res. Treat. 19, 1533033820964231. doi:10.1177/1533033820964231
Leca, J., Lemonnier, F., Meydan, C., Foox, J., El Ghamrasni, S., Mboumba, D. L., et al. (2023). IDH2 and TET2 mutations synergize to modulate T Follicular Helper cell functional interaction with the AITL microenvironment. Cancer Cell 41 (2), 323–339.e10. doi:10.1016/j.ccell.2023.01.003
Liu, W., Zhao, D., Liu, T., Niu, T., Song, Y., Xu, W., et al. (2021). A multi-center, real-world study of chidamide for patients with relapsed or refractory peripheral T-cell lymphomas in China. Front. Oncol. 11, 750323. doi:10.3389/fonc.2021.750323
Lone, W., Bouska, A., Sharma, S., Amador, C., Saumyaranjan, M., Herek, T. A., et al. (2021). Genome-wide miRNA expression profiling of molecular Subgroups of peripheral T-cell lymphoma. Clin. Cancer Res. 27 (21), 6039–6053. doi:10.1158/1078-0432.Ccr-21-0573
Radakovich, N., Nagy, M., and Nazha, A. (2020). Machine learning in haematological malignancies. Lancet Haematol. 7 (7), e541–e550. doi:10.1016/s2352-3026(20)30121-6
Shi, Y., Dong, M., Hong, X., Zhang, W., Feng, J., Zhu, J., et al. (2015). Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann. Oncol. 26 (8), 1766–1771. doi:10.1093/annonc/mdv237
Shi, Y., Jia, B., Xu, W., Li, W., Liu, T., Liu, P., et al. (2017). Chidamide in relapsed or refractory peripheral T cell lymphoma: a multicenter real-world study in China. J. Hematol. Oncol. 10 (1), 69. doi:10.1186/s13045-017-0439-6
Sun, J., He, S., Cen, H., Zhou, D., Li, Z., Wang, M. Y., et al. (2021). A novel prognostic model for angioimmunoblastic T-cell lymphoma: a retrospective study of 55 cases. J. Int. Med. Res. 49 (5), 3000605211013274. doi:10.1177/03000605211013274
Swanson, K., Wu, E., Zhang, A., Alizadeh, A. A., and Zou, J. (2023). From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 186 (8), 1772–1791. doi:10.1016/j.cell.2023.01.035
Swerdlow, S. H., Campo, E., Pileri, S. A., Harris, N. L., Stein, H., Siebert, R., et al. (2016). The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127 (20), 2375–2390. doi:10.1182/blood-2016-01-643569
Wang, J., Su, N., Fang, Y., Ma, S., Zhang, Y., Cai, J., et al. (2022a). Comparison of chemotherapy combined with chidamide versus chemotherapy in the frontline treatment for peripheral T-cell lymphoma. Front. Immunol. 13, 835103. doi:10.3389/fimmu.2022.835103
Wang, Y., Zhang, M., Song, W., Cai, Q., Zhang, L., Sun, X., et al. (2022b). Chidamide plus prednisone, etoposide, and thalidomide for untreated angioimmunoblastic T-cell lymphoma in a Chinese population: a multicenter phase II trial. Am. J. Hematol. 97 (5), 623–629. doi:10.1002/ajh.26499
Zhang, C., Mou, B., Xu, J., Wang, J., Liu, Q., Yang, Y., et al. (2023). Angioimmunoblastic T-cell lymphoma: novel recurrent mutations and prognostic biomarkers by cell-free DNA profiling. Br. J. Haematol. 203 (5), 807–819. doi:10.1111/bjh.19089
Keywords: machine learning, chidamide, prognosis, angioimmunoblastic T-cell lymphoma, biomarker
Citation: Zhang C, Xu J, Gu M, Tang Y, Tang W, Wang J, Liu Q, Yang Y, Zhong X and Xu C (2024) Predicting the efficiency of chidamide in patients with angioimmunoblastic T-cell lymphoma using machine learning algorithm. Front. Pharmacol. 15:1435284. doi: 10.3389/fphar.2024.1435284
Received: 20 May 2024; Accepted: 16 August 2024;
Published: 28 August 2024.
Edited by:
Sheema Khan, The University of Texas Rio Grande Valley, United StatesReviewed by:
Ambreen Hamadani, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, IndiaShabia Shabir, Islamic University of Science and Technology, India
Copyright © 2024 Zhang, Xu, Gu, Tang, Tang, Wang, Liu, Yang, Zhong and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caigang Xu, eHVjYWlnYW5nQHdjaHNjdS5jbg==
†These authors have contributed equally to this work
 Chunlan Zhang
Chunlan Zhang Juan Xu1†
Juan Xu1† Wenjiao Tang
Wenjiao Tang Jie Wang
Jie Wang