- 1Department of Critical Care Medicine, West China Hospital of Sichuan University, Chengdu, Sichuan, China
- 2Institute of Respiratory Health, West China Hospital of Sichuan University, Chengdu, Sichuan, China
Antibiotic resistance is a pressing global health challenge, and polymyxins have emerged as the last line of defense against multidrug-resistant Gram-negative (MDR-GRN) bacterial infections. Despite the longstanding utility of colistin, the complexities surrounding polymyxins in terms of resistance mechanisms and pharmacological properties warrant critical attention. This review consolidates current literature, focusing on polymyxins antibacterial mechanisms, resistance pathways, and innovative strategies to mitigate resistance. We are also investigating the pharmacokinetics of polymyxins to elucidate factors that influence their in vivo behavior. A comprehensive understanding of these aspects is pivotal for developing next-generation antimicrobials and optimizing therapeutic regimens. We underscore the urgent need for advancing research on polymyxins to ensure their continued efficacy against formidable bacterial challenges.
1 Introduction
Since the introduction of antibiotics in the last century, they have saved countless lives of patients with serious bacterial infections. In the past 50–60 years, doctors have come to expect that antibiotics would cure almost all patients with bacterial infections. However, due to the lack of early identification of the causative organisms and their antimicrobial susceptibility patterns in patients with bacteremia and severe infections in many healthcare facilities, broad-spectrum antibiotics have been heavily and mostly unnecessarily used since the 1990s (Akova, 2016). Consequently, this has led to the emergence of numerous drug-resistant bacteria and unregulated management of nosocomial infections, resulting in increased chances of transmission of drug-resistant bacteria, longer hospital stays, and higher mortality rates for patients (Akova, 2016). In 2017, the World Health Organization (WHO) added Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species to the list of key pathogens in urgent need of new antibiotics (Mancuso et al., 2021). While medical institutions have conducted certain control and preliminary systematic evaluations of these pathogens (Tomczyk et al., 2019), the overuse and misuse of antibiotics in healthcare, agriculture, and livestock have contributed to a significant increase in antimicrobial resistance (Nadimpalli et al., 2020; Schrader et al., 2020). WHO and the U.S. Centers for Disease Control and Prevention (CDC) have recognized antimicrobial resistance as a worldwide threat. Without effective management and scaling up the supply of antibiotics, nearly 10 million people worldwide are expected to die from drug-resistant infections by 2050 (Pulingam et al., 2022). Although the U.S. Food and Drug Administration (FDA) has approved several new antibiotics in recent years, the emergence of resistance has been reported (Abdallah et al., 2015; Giddins et al., 2018; Morrissey et al., 2020) (Figure 1). Therefore, there is an urgent global need for antimicrobials with innovative pharmacological activities and modes of action to combat the public health threat of antimicrobial resistance (Miethke et al., 2021).
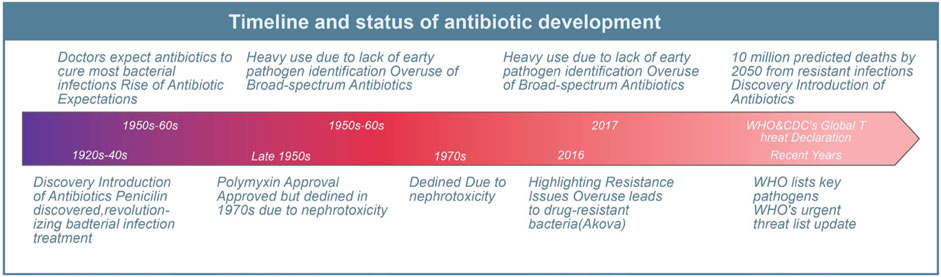
Figure 1. Antibiotic classification and current status of clinical resistance. This diagram illustrates the evolution and challenges of antibiotic use since its inception.
Polymyxins have received increasing attention in recent years, as they are considered a potential weapon against Gram-negative drug-resistant bacteria. Polymyxin was approved in the late 1950s (Li et al., 2006). However, its usage rapidly declined in the 1970s due to its nephrotoxicity. Nevertheless, with carbapenem-resistant Acinetobacter baumannii (CRAB), Pseudomonas aeruginosa, and Klebsiella pneumoniae resistance rates increasing each year (Nang et al., 2021), polymyxins have been reintroduced into the clinic as a last-resort salvage treatment option for these resistant organisms (Cai et al., 2012; Doi et al., 2015). Over the past two decades, significant progress has been made in the study of polymyxins, including their chemical structure, activity/toxicity relationship, antimicrobial activity, and polymyxin resistance mechanisms. This review summarizes the history of polymyxin development and provides an overview of the mechanisms of drug resistance. Additionally, it focuses on the research conducted to overcome colistin resistance and highlights the development of new antimicrobials that have entered clinical trials. Furthermore, the review presents the latest research progress in overcoming polymyxin resistance and sheds light on the pharmacokinetic behavior of polymyxins to improve the standardization and safety of their global clinical application.
2 Polymyxins: from discovery to re-emergence in the era of superbugs
Polymyxins, discovered in 1947, are antimicrobial cationic polypeptides produced by Bacillus polymyxins (Benedict and Langlykke, 1947). Figure 2 provides a timeline highlighting the major milestones in the discovery, use, and resurgence of polymyxins. Brownlee and Bushby (1948) isolated an antibiotic from Bacillus aerospore that exhibited antibacterial activity against Gram-negative bacteria. In 1949, White et al. conducted a comparative study on the antibacterial activities of polymyxins and “Neosporin” and found no significant difference between them, suggesting that both substances belong to the polymyxins class of antibiotics. Consequently, a nomenclature system was established for the polymyxins family (Stansly and Brownlee, 1949). To date, polymyxin A (also known as Neosporin), B, C, D (polymyxin), E (also known as colistin), F, M, P, S, and T have been identified from P. Polymysa strains (Shoji et al., 1977; Niu et al., 2013). After their discovery, many polymyxins were found to have reversible nephrotoxicity, leading to the clinical use of colistin and polymyxin B due to their relatively low nephrotoxicity. The main market products are polymyxin B and polymyxin E, which exhibit similar antimicrobial activity (Storm et al., 1977). There are currently three polymyxin analogues for injection that have been marketed both domestically and internationally: colistin methanesulfonate (CMS) for injection, colistin sulfate and polymyxin B sulfate for injection. The blood-brain barrier passage rate of polymyxin is low, and it is difficult to achieve effective drug concentration by intravenous administration. Local applications such as intracerebroventricular or intrathecal administration have been increasingly adopted by the clinic in recent years, but there is a lack of relevant standardized operational guidelines (Yang et al., 2023). In addition, the nebulized inhalation method can significantly increase the lung tissue concentration of polymyxin while decreasing the systemic exposure level of the drug, thus achieving the goal of improving the efficacy and reducing systemic adverse effects (Tang et al., 2023). Currently, several domestic and international guidelines and consensus recommend nebulized inhalation of polymyxin as one of the important therapeutic methods for multidrug-resistant gram-negative (MDR-GRN) bacterial induced pneumonia (Lin et al., 2022).
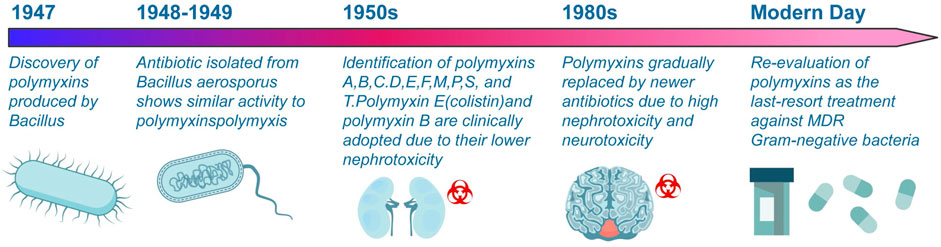
Figure 2. Evolution of polymyxin antibiotics. This illustration delineates the significant milestones in developing polymyxin antibiotics, from their discovery in 1947 to their contemporary relevance.
Polymyxin is a cyclic lipopeptide compound composed of 10 amino acids, characterized by a cationic polypeptide consisting of a cyclic heptapeptide and a tripeptide side chain (Rutten et al., 1990; Falagas et al., 2010). The primary difference between PMB and colistin lies in the amino acid variation at position 6 (R2), where PMB contains phenylalanine and colistin contains leucine (Figure 3) (Velkov et al., 2010; Nation et al., 2014). They have similar antimicrobial effects. Compared to the parent antibiotics, sulfomethyl derivatives of polymyxins exhibit lower toxicity and similar in vivo antibacterial activity. Consequently, sulfonated derivative polymyxin E has been commercially available for clinical use in Japan, Europe, and the United States since the 1950s. Conversely, colistin methanesulfonate sodium, an antibacterial active component covered by sulfate, lacks inherent antibacterial activity and serves as a precursor drug that exhibits bactericidal effects. However, CMS was gradually replaced by newer antibacterial drugs in the 1980s due to its high nephrotoxicity and neurotoxicity. Importantly, commercial polymyxin B, CMS, and colistin products are mixtures (Govaerts et al., 2002), leading to batch-to-batch differences in the abundance of individual ingredients.
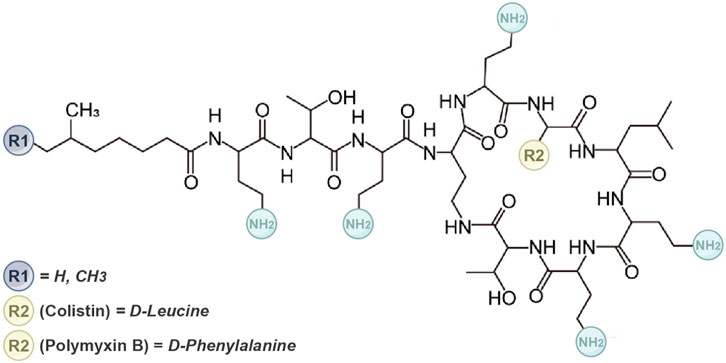
Figure 3. The molecular structure of the polymyxin. It outlines the structural distinctions between polymyxin B and colistin. Polymyxin B and colistin comprise five conserved L-α-γ-diaminobutyric acid (Dab) residues. These residues confer a net positive charge to polymyxin compounds at physiological pH. The cationic hydrophilicity of this macrocycle is pivotal for their antibacterial properties.
With limited antibiotic options available, the increasing bacterial resistance in clinical settings has necessitated the re-evaluation of “old” antibiotics, particularly polymyxin, which has shown effectiveness against many MDR Gram-negative bacteria. Polymyxins have been used in clinical practice for approximately 60 years, with polymyxin B and colistin now considered last-resort treatment options for infections caused by “superbugs."
3 Deciphering the multimodal antibacterial strategies of polymyxins
Understanding the mode of action of polymyxins is crucial for optimizing their use and developing new antibiotics. Polymyxin B and colistin, having similar chemical structures, exhibit comparable antibacterial mechanisms primarily against common Gram-negative bacteria (Kwa et al., 2007). Colistin demonstrates a narrow spectrum of antibiotics but shows activity against several clinically significant MDR Gram-negative bacteria, such as Pseudomonas aeruginosa, Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Enterobacter, and other enterobacteriaceae (Bialvaei and Samadi Kafil, 2015; Poirel et al., 2017; Doymaz and Karaaslan, 2019).
Both colistin and polymyxin B exert their effects on the bacterial membrane, causing damage. The outer membrane of Gram-negative bacteria serves as protection against various harmful substances, including antimicrobials. Extensive studies have demonstrated that polymyxins exerts its antibacterial effect by directly interacting with the lipid components of lipopolysaccharide (LPS), disrupting bacterial membranes (Nikaido, 2003). Electrostatic interactions occur between the positively charged α, γ-aminobutyric acid (Dab) residues of polymyxin and the phosphoric acid groups on the bacterial membrane, competitively displacing divalent cations (Ca2+ and Mg2+) through the negatively charged phosphoric acid groups in the lipid membrane (Dixon et al., 1986). In addition, polymyxins can neutralize endotoxins and inhibit the expression of cytokines such as TNF-α and IL-8, preventing tissue damage caused by excessive activation of inflammation (Schromm et al., 2021). Polymyxins binds to LPS through electrostatic and hydrophobic interactions (Velkov et al., 2010). Consequently, LPS becomes unstable, increasing bacterial membrane permeability, leading to the leakage of cytoplasmic contents and, ultimately, bacterial death (Schindler and Osborn, 1979; Falagas and Kasiakou, 2005). While the primary model for polymyxin’s antimicrobial activity involves the destruction of the bacterial adventitia and intima, additional mechanisms have also been proposed (Figure 4).
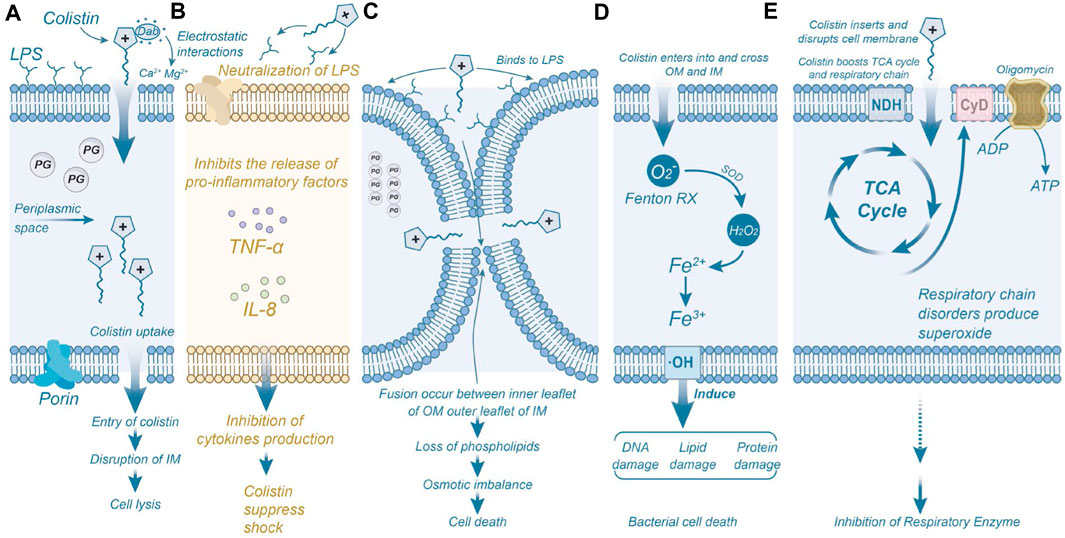
Figure 4. Mechanisms of Polymyxins-Induced Bacterial Inhibition and Death. (A) The Dab residue of colistin interacts electrostatically with the anionic phosphate group in the outer membrane of bacteria, resulting in structural disruption of the bacterial membrane. (B) Colistin neutralizes the activity of LPS molecules, inhibiting the induction of shock and consequent release of cytokines by immune cells, such as tumor necrosis factor-α and interleukin 8. (C) Colistin binds to phospholipids on the outer membrane of bacteria, causing depletion of phospholipids and leading to bacterial death. (D) Colistin triggers the production of reactive oxygen species (ROS) that damage DNA, lipids, and proteins, ultimately resulting in bacterial death. (E) Colistin inhibits the activity of respiratory enzymes. Permission to reproduce adapted from reference (El-Sayed Ahmed et al., 2020) has been obtained.
Another mechanism of polymyxin action is the inhibition of type II NADH-quinone oxidoreductase (NDH-2) activity, a significant respiratory enzyme in the bacterial inner membrane (Deris et al., 2014). After entering the bacteria, polymyxin inhibits the respiratory enzymes of the tricarboxylic acid (TCA) cycle and consumes ATP, leading to bacterial death. Studies have demonstrated that polymyxins inhibits NDH-2 activity in a concentration-dependent manner in various Gram-negative bacteria, including Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii (Mogi et al., 2009). These events are accompanied by activation of repair pathways and adventitial remodeling (Moffatt et al., 2019). Additionally, some researchers propose that most antibiotics induce bacterial death by perturbing bacterial metabolism, leading to increased production of reactive oxygen species (ROS), including superoxide (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (-OH), ultimately causing bacterial demise (Kohanski et al., 2007). Leveraging this property, many researchers have designed antimicrobial agents that promote bacterial death by stimulating the production of hydroxyl radicals through the Fenton reaction (Yeom et al., 2010). Elevated levels of hydroxyl radicals within bacteria can damage bacterial DNA, lipids, and protein synthesis, resulting in bacterial death (Dwyer et al., 2014). These findings indicate the importance of hydrophobic interaction in the antibacterial mechanism of polymyxin (Hancock, 1997).
4 Unraveling the complex web of polymyxin resistance: mechanisms, challenges, and countermeasures
The escalating issue of antibiotic resistance poses a significant threat to human health, particularly with the emergence of resistance in Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa against almost all available antibiotics. Such MDR bacteria are commonly found in intensive care units and among long-term hospitalized patients. Despite their potent bactericidal activity against many Gram-negative bacteria, the extensive use of polymyxins has led to the emergence of polymyxin-resistant strains, including Neisseria meningitidis, Proteus mirabilis, and Burkholderia spp. (Tzeng et al., 2019). Notably, nosocomial infections caused by MDR Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae are closely associated with increased morbidity and mortality, and their resistance to polymyxins has attracted significant attention (Barbier et al., 2013; Bassetti et al., 2018).
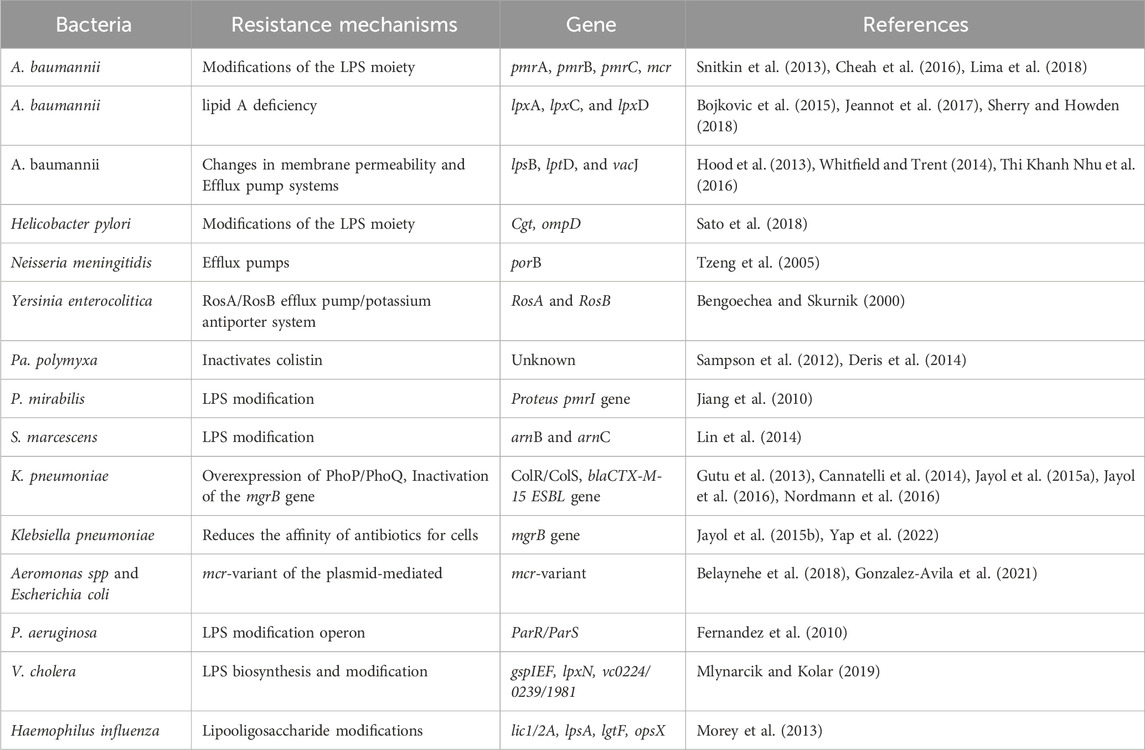
In this section, we present a systematic review of recent discoveries on the mechanisms of polymyxin resistance. In recent years, antibiotic resistance genes have become one of the greatest threats to human health in the 21st century. mcr-1, the first plasmid-mediated polymyxin resistance gene, was discovered in China in 2015, indicating that the last line of defense has also been breached, further exacerbating the threat of bacterial resistance to public health. Table 1 summarized the resistant bacteria and resistance genes to polymyxins in recent years. Some of the more attention-grabbing, classical mechanisms of polymyxin resistance are summarized in Figure 5. The phosphoethanolamine (pEtN) transferase encoded by mcr-1 delivers pEtN from the cell membrane through modification of lipid A, leading to colistin resistance (Figure 5A). This modification reduces the affinity of polymyxin for lipopolysaccharides, as depicted in Figure 5B (Needham and Trent, 2013; Liu et al., 2016). In addition to colistin resistance, mcr mediates bacterial resistance to antimicrobial peptides (AMPs). The amphiphilic structure of AMPs enables them to penetrate bacterial cell membranes, forming pores and disrupting the integrity of the cell membrane, leading to lysis and death of the bacteria (Nang et al., 2021; Rodriguez-Santiago et al., 2021). The emergence of the plasmid-mediated colistin resistance gene mcr-1 has attracted global attention and prompted several countries to adjust their policies on the use of colistin in food and animals. Currently, researchers have conducted extensive studies on the distribution, function, mechanism of action, transmission vectors, and origin of mcr, as well as prevention and control strategies for mcr-positive bacteria (Liu et al., 2024). Some researchers have recently identified the problem of polymyxin resistance mediated by multiple mcr gene plasmids (Liu et al., 2016; Schwarz and Johnson, 2016). Certain environments, such as hospitals with a high potential for transmission of resistant bacteria through food or surfaces, as well as heavily contaminated surface water, can serve as reservoirs for various infections.
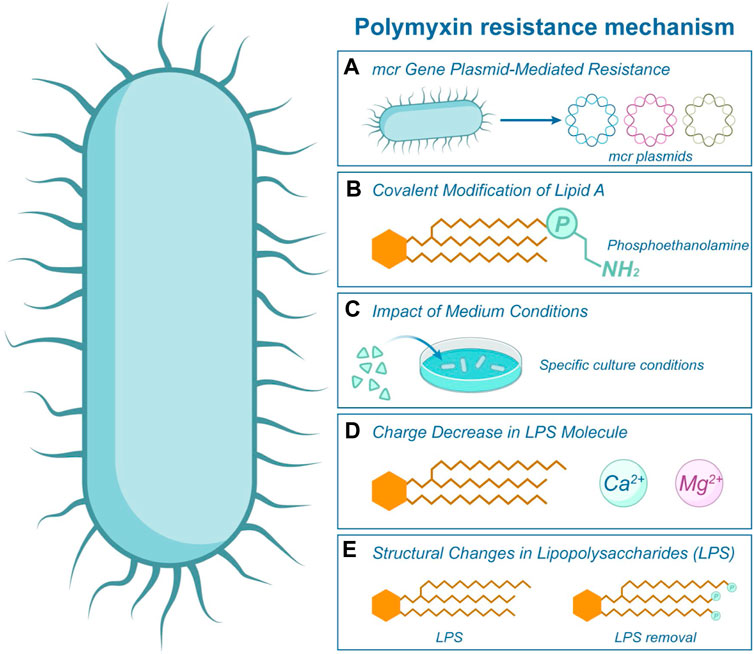
Figure 5. Polymyxin Resistance Mechanism. This illustration showcases the various mechanisms by which bacteria develop resistance to polymyxin. Central to this resistance is the structure of Lipopolysaccharide (LPS) present in the bacterial cell, as highlighted in figure. (A) The mcr gene on bacterial plasmids facilitates the primary resistance mechanism, as depicted by the label mcr in the image. Furthermore, resistance can arise from covalent modifications to Lipid A (B) or structural alterations in the LPS (E). The efficacy of polymyxin is further diminished by the reduced charge on LPS (D), especially when it interacts with cations such as Mg2+. (C) The growth medium conditions in which bacteria reside can also influence their susceptibility to polymyxin.
It has also been observed that the conditions of the medium influence the sensitivity of some bacteria to polymyxins. Salmonella enterica cells lacking carbon, nitrogen or phosphate ions, and serum and quiescence cells exhibit reduced sensitivity to polymyxin (Figure 5C) (McLeod and Spector, 1996). An alternative mechanism involves the activation of the pmrA genes in Salmonella typhimurium, triggered by low levels of Mg2+ ions. It decreases the lipopolysaccharide molecule’s negative charge and subsequently reduces polymyxin’s binding affinity (Groisman et al., 1997), as depicted in Figure 5D.
Previous studies have demonstrated that the resistance mechanism in most Gram-negative bacteria is associated with structural changes in lipopolysaccharides (Moore et al., 1984; Pelletier et al., 2013). Bacteria can develop resistance to polymyxin either by modifying the phosphate group of lipid A or by directly removing LPS (Moffatt et al., 2010; Olaitan et al., 2014), as depicted in Figure 5E. For instance, in the presence of high concentrations of polymyxin in Pseudomonas aeruginosa, the bacterial acidic phospholipids undergo conversion to neutral lipids, along with changes in proteins and carbohydrates, ultimately leading to the development of resistance (Gilleland and Lyle, 1979; Gilleland and Farley, 1982). Zhao et al. found that the increase of polymyxin concentration would affect the dynamics of genetic variants in the flora and lead to different degrees of evolution of resistance, and emphasized that during the use of polymyxin, the evolutionary findings were integrated into pharmacokinetics/pharmacodynamics to improve the antibacterial efficacy of patients (Zhao et al., 2022).
Antibiotic resistance has significantly compromised the effectiveness of antibiotics, leading to a substantial burden on medical care improvement and cost control. To address this issue, healthcare professionals must use antibiotics judiciously, preventing misuse and overuse, which can delay the emergence of resistance and reduce healthcare expenses for patients. Governments and medical institutions should also manage and optimize antibiotic use patterns, selecting the most suitable treatment plan based on recommended dosages and durations to achieve optimal clinical outcomes while minimizing toxicity and the risk of subsequent resistance. Furthermore, establishing an integrated and specialized antibiotic use monitoring system can help detect and prevent the emergence of antibiotic resistance in advance, addressing the problem at its source. Pharmaceutical researchers, in particular, should intensify their efforts in developing polymyxin antibiotics to counter emerging or potentially resistant bacteria in the future.
5 Innovative strategies in combating antibiotic resistance
Antibiotic resistance poses a significant challenge to global health, necessitating the exploration of diverse strategies to counteract its proliferation. Yan Zhu et al. proposed the combination of genome-scale metabolic modeling with multi-omics data elucidated the mechanisms by which A. baumannii cells respond to colistin treatment, including (i) upregulation of gluconeogenesis, pentose phosphate pathway, amino acid, and nucleotide biosynthesis fluxes; (ii) downregulation of TCA cycling, peptidoglycan, and lipopolysaccharide biogenesis; and (iii) alterations in respiratory chain fluxes. The findings elucidate the interaction of multiple metabolic pathways in A. baumannii when treated with colistin and provide key mechanistic insights for optimizing polymyxin combination therapy (Zhu et al., 2019). Li J et al. revealed the key pathways associated with the synergistic activity of polymyxin B and rifampicin in combination against multidrug-resistant Acinetobacter baumannii by comparative metabolomics. They found that polymyxin B monotherapy significantly disrupted glycerophospholipid and fatty acid metabolism within 1 h, reflecting its activity against the bacterial outer membrane. Rifampicin monotherapy significantly disrupted glycerophospholipid, nucleotide, and amino acid metabolism, which was associated with inhibition of RNA synthesis, and with the combination, polymyxin B initially affected pathways associated with outer membrane biogenesis, whereas rifampicin affected them through inhibition of RNA synthesis, and the findings provide new mechanistic insights into optimizing this synergistic combination in patients (Zhao et al., 2021). As bacteria continue to evolve and resist conventional treatments, the scientific community has responded with vigor, delving deep into combination therapies, chemobiological innovations, and the potential of nanotechnology.
5.1 Polymyxin combination therapies: overcoming bacterial resistance
Combining polymyxin with sensitizing drugs represents a promising strategy to overcome polymyxin resistance and restore its sensitivity. Srisakul et al. proposed the combination of polymyxin and sulbactam as a means to overcome lipid A-mediated colistin resistance (Srisakul et al., 2022). Moreover, Chen et al. reported that Anthranilyl-CoA Synthetase PqsA effectively enhanced the activity of polymyxin B against MDR Pseudomonas aeruginosa-associated infections (Chen J. et al., 2022). In a study by Li et al., it was demonstrated that the combined use of the guanidine derivative isopropoxy benzene guanidine with low-level colistin enhanced the permeability of the bacterial outer membrane and increased the accumulation of reactive oxygen species, thereby combating MDR Escherichia coli (Li et al., 2023). Shein et al. suggested combining colistin and EDTA could overcome mgrB-mediated colistin resistance in carbapenem-resistant Klebsiella pneumoniae (Shein et al., 2022). Wang et al. showed that combination therapy using colistin and resveratrol improved the membrane permeability of bacteria and increased the sensitivity of Pseudomonas aeruginosa to colistin (Wang et al., 2023). The plasmid-mediated resistance gene mcr-1, a homolog of eptA, confers resistance by modifying lipid A through cationic phosphoethanolamine (Liu et al., 2016). MacNair et al. (2018) demonstrated that combining polymyxin with antibiotics targeting Gram-positive bacteria effectively treated infections caused by drug-resistant Gram-negative bacteria expressing mcr-1 (MacNair et al., 2018).
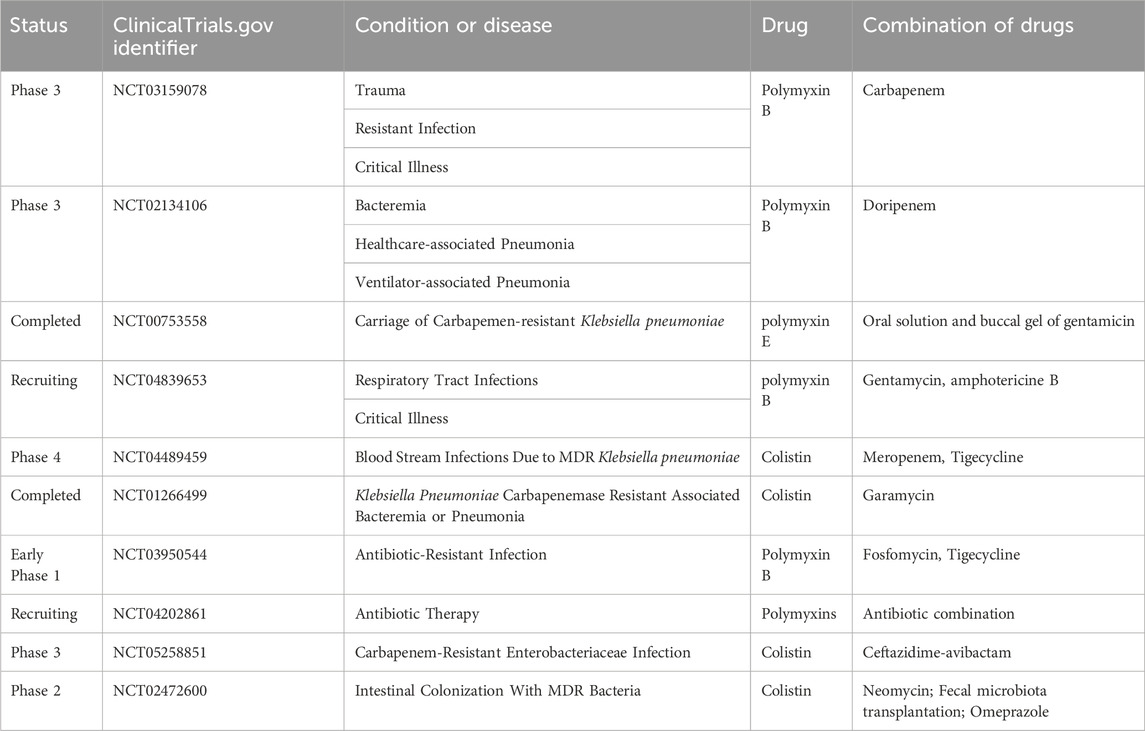
Additionally, combination therapy using melatonin and colistin has shown efficacy in eradicating mcr-positive pathogens and exhibits a favorable biosafety profile. The combined antibacterial mechanism of polymyxin and melatonin involves enhancing bacterial outer membrane permeability, promoting oxidative damage, and inhibiting the expression of bacterial efflux proteins. Liu et al. demonstrated that the combination of polymyxin and melatonin significantly restored the efficacy of colistin in three animal models of E. coli infection carrying mcr-1 (Liu et al., 2020). Table 2 presents an overview of clinical trials investigating polymyxin combination therapy to overcome drug resistance. Lindsey A Carfrae et al. proposed that the biotin biosynthesis inhibitor MAC13772 acted synergistically with colchicine, indirectly disrupting fatty acid synthesis (FAS) via MAC13772, leading to changes in phospholipid composition and restoring susceptibility to the antibiotic colchicine. In addition, the investigators propose that combination therapy using colchicine and the clinically relevant FabI inhibitor Debio1452-NH3 is efficacious against systemic infections in mice with mcr-1-positive Klebsiella pneumoniae and colchicine-resistant Escherichia coli. We explored the mechanism of this interaction using chemogenomics, lipidomics, and transcriptomics (Carfrae et al., 2023).
5.2 Chemobiological advancements: enhancing antimicrobial efficacy against drug-resistant pathogens
Jonathan M. Stokes et al. found that pentamidine and its structural analogues sensitize Gram-negative pathogens to antibiotics and overcome acquired resistance to polymyxins (Stokes et al., 2017). Velkov and Li et al. optimized the structure of polymyxin by chemical biology and successfully developed a new lipopeptide, which has significant antibacterial activity against a variety of drug-resistant pathogens. The antimicrobial peptide has entered phase I clinical trials (Roberts et al., 2022). Zsolt Szűcs et al. prepared a vancomycin polycationic glycogen derivative with an n-decane side chain and 5 aminoethyl groups, which has a structure similar to that of polymyxin and can act synergistically against Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii by combining with teicoplanin (Szucs et al., 2022). Lindsey A. Carfrae et al. have found that the sensitivity of colistin therapy can be restored by inhibiting the synthesis of fatty acids in bacteria and finally changing the lipid composition of bacterial membranes (Carfrae et al., 2023).
5.3 Harnessing nanotechnology: a frontier in optimizing antibiotic delivery and performance
Nanomaterial-based therapy is a promising tool against refractory bacterial infections, characterized by the ability to evade the existing mechanisms related to acquired drug resistance and increase the activity of antimicrobials (Makabenta et al., 2021). Figure 6 summarizes some nanotechnologies to overcome antibiotic resistance. Nano-form metals, metal oxides, and other nano-drugs also have direct antibacterial effects (Lee et al., 2019; Elbourne et al., 2020; Zhang et al., 2022).
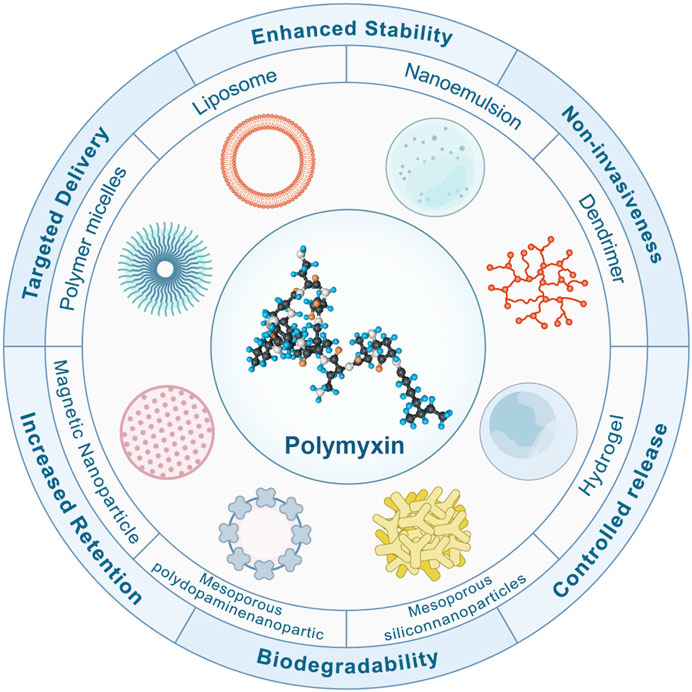
Figure 6. Features of Nanotechnology in Improving Polymyxin Resistance. Nanotechnology has emerged as a cornerstone in the battle against antibiotic resistance. Nano-carriers bolster the resilience of antibiotics amidst the intricacies of physiopathological landscapes. Harnessing the potential of gels and polymer nanofibers, the longevity of antibiotic release is notably augmented. Moreover, these carriers adeptly prolong the drug’s residence at injury locales. Their biodegradable and non-invasive nature ensures a harmonious interaction with the human physiology. These nano-carriers deftly modulate drug release within the gastrointestinal milieu, optimizing absorption and maximizing bioavailability. Beyond that, the strategic design of nanoparticles and microparticles ensures a precision-guided delivery to infection epicentres, adeptly curbing side effects and magnifying therapeutic efficacy.
Nanotechnology represents a promising platform for drug delivery, particularly in the fight against antibiotic resistance. The key advantages of these nanocarriers lie in their ability to optimize and enhance the therapeutic effects of drugs (Figure 6). Nanocarriers greatly improve the stability of antibiotics in complex physiological and pathological environments. For example, nano-metals and metal oxides offer a protective barrier against threats such as hydrolysis, oxidation, pH fluctuations, and enzymatic attacks, thereby ensuring the preservation of their activity until they reach the intended target site (Elbourne et al., 2020). Moreover, nanocarriers, including gels and polymer nanofibers, facilitate the sustained release of antibiotics due to their unique design (Thapa et al., 2020). This feature ensures consistent drug concentrations and helps reduce dosing frequencies, thereby enhancing patient compliance. Furthermore, their small size allows nanocarriers to reside longer at damaged or infected sites, enabling effective infection control precisely where it is most needed (Mofazzal Jahromi et al., 2018; Liang et al., 2022). Significantly, these nanocarriers possess biodegradable and non-invasive properties, allowing for their safe breakdown and elimination from the body after fulfilling their purpose without causing any further harm (Ohnstedt et al., 2019). Moreover, nanocarriers provide a protective environment for drugs in the gastrointestinal tract, regulating their release. For instance, combining or encapsulating polymyxins with polymers and liposomes can protect drugs in the gastrointestinal tract, leading to enhanced drug absorption (Maher et al., 2016; Faustino and Pinheiro, 2020). Furthermore, the design of nanos and microparticles ensures targeted drug delivery, enabling drugs to be directed straight to the infection site. This precision allows for more accurate treatments, reduced drug dosages, and diminished risks of systemic side effects (Lee et al., 2019; Zigrayova et al., 2023). Additionally, when combined with the most effective antibiotics, nano preparations have exhibited synergistic effects and the potential to address the emerging global crisis of bacterial resistance (Lee et al., 2019; Zigrayova et al., 2023). Chengyuan Qin et al. Overcoming colistin resistance in bacterial infections by negatively charged polyethylene glycol functionalized liposomal co-delivery of curcumin and colistin. Liposomes restored the affinity of mucilage to the bacterial membrane and increased the uptake of curcumin, thereby decreasing efflux pump activity and realizing the synergistic effect of mucilage and curcumin. The liposome-loaded group did not exhibit any toxicity at effective antibacterial doses (Qin et al., 2023).
6 The critical role and complexities of polymyxins in modern medicine
Polymyxins, specifically Polymyxin B and Colistin (Polymyxin E), have emerged as pivotal agents in the fight against antibiotic-resistant bacterial infections. Polymyxin B is often available as a sulphate for injection, while colistin is available as polymyxin E sulphate for injection and colistin methane sulfonate for injection. CMS is a prodrug that is converted to colistin in the body after administration in order to exert antimicrobial activity. As the global medical community grapples with the challenges posed by multi-drug resistant Gram-negative bacteria (MDR-GNB), the significance and intricacies of these drugs have gained paramount importance. This article delves into the types, pharmacokinetic properties, differences, and the potential of personalized treatments with polymyxins. Additionally, it sheds light on the future research directions, emphasizing the need to further elucidate their behavior and interactions, especially in conjunction with contemporary medical treatments (Figure 7). CMS is a precursor drug that needs to be converted in the kidneys, and its conversion rate is affected by a number of factors, resulting in large individual variations in PK parameters, and blood concentrations are affected by renal function. PMB is mainly eliminated by non-renal routes and the total clearance has little correlation with renal function. Dosage does not need to be adjusted according to renal function, and effective blood mass concentrations can be achieved rapidly and are less affected by renal function. Therefore, PMB is more suitable for bloodstream infections, whereas CMS is more suitable for urinary tract infections. The antimicrobial activity of different polymyxin E formulations varies, with CMS being less active than polymyxin E sulphate and less toxic, while polymyxin E sulphate is less commonly used due to toxicity and is mainly used for drug sensitivity testing (Infectious Diseases Society of China et al., 2021). Currently, there are fewer data from studies related to polymyxin E sulphate, and it is expected that more relevant studies will enrich the clinical options in the future.
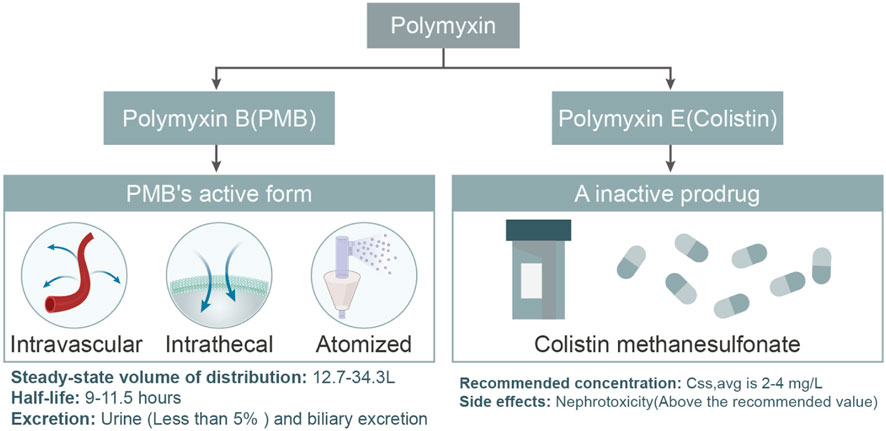
Figure 7. Comparative Pharmacokinetic Profiles of PMB and Colistin. This flowchart elucidates the distinct pharmacokinetic properties of PMB and Colistin. For PMB, the diagram underscores its distribution, half-life, and excretion pathways, notably the minimal renal excretion and a potential route through biliary excretion. In contrast, colistin is emphasized for its intravenous administration as the inactive prodrug, colistin methanesulphonate. The recommended concentrations for both drugs are depicted, with an accent on potential side effects, especially the heightened risk of nephrotoxicity when exceeding the advised levels.
6.1 Types and drug forms of polymyxins
In current clinical practice, Polymyxin B and Colistin are widely regarded as the two primary polymyxins, both boasting impressive antibacterial properties. Due to the rising antibiotic resistance of MDR-GNB globally, many previously effective antibiotics have become less effective against MDR Gram-negative pathogens (El-Sayed Ahmed et al., 2020). This resurgence of colistin in the mid-1990s emerged as a crucial weapon against MDR Gram-negative pathogens (El-Sayed Ahmed et al., 2020). Notably, Klebsiella pneumoniae, a common cause of healthcare-associated infections, often uses colistin as the treatment of choice (Uzairue et al., 2022). However, these two polymyxins differ in their modes and forms of administration. Polymyxin E is commonly administered intravenously as the prodrug colistin methanesulphonate, while PMB can be directly intravenously administered as its active form under physiological pH (Nation et al., 2015) (Figure 7). Alarmingly, with the clinical discovery of more and more carbapenem resistant pseudomonas, especially resistance to polymyxins has begun to appear (Qureshi et al., 2015).
6.2 Pharmacokinetic properties of polymyxin B
The administration of PMB is versatile, including intravenous, intrathecal, or nebulized inhalation routes. After intravenous administration, its steady-state distribution volume ranges from 12.7–34.3 L, with a half-life of about 9–11.5 h. This provides valuable guidance for clinicians on dose adjustments. Interestingly, in both animals and humans, less than 5% of PMB is excreted in the urine, indicating that the kidneys are not the primary route of elimination for PMB (Zavascki et al., 2008; Manchandani et al., 2016). Although biliary excretion might contribute to PMB’s clearance, further studies are needed to identify other potential clearance mechanisms (Manchandani et al., 2016). It is worth noting that the intensity of concentration imposed by polymyxins affects the dynamics of genetic variation within the bacterial population, leading to different evolutionary outcomes of resistance. Jinxin Zhao et al. demonstrated that polymyxin B recurs at a critical threshold concentration (1 mg/L; i.e., 4× MIC) with low levels of resistance, but without fixed mutations, and that this resistance reverses upon removal of the antibiotic. This contrasts with the evolution of polymyxin B at ultra-MIC concentrations (≥4 × MIC), which drives the evolution of irreversible resistance, with molecular evolution occurring more rapidly at higher antibiotic concentrations. This study highlights the important role of combining evolutionary findings with pharmacokinetics/pharmacodynamics to optimize antibiotic use in patients (Zhao et al., 2022). V Aranzana-Climent et al. investigated a semi-mechanistic PK/PD model for the combination of polymyxin B and minocycline against polymyxin-resistant Acinetobacter baumannii. The combination effect was driven by minocycline, with PMB as an adjunct; simulations at clinically achievable concentrations indicated that 1.5 mg/L minocycline +0.2 mg/L PMB was expected to produce sustained killing for more than 30 h, whereas 0.3 mg/L minocycline +1 mg/L PMB was sufficient for bacterial regeneration. Interaction equations indicated that synergistic effects were maximized at PMB concentrations ≥0.1 mg/L and minocycline concentrations ≥1 mg/L. The possible mechanism is that PMB opens the bacterial membrane and increases the entry of minocycline into the cell, and the intracellular concentration of minocycline increased in bacteria treated with 0.5 mg/L PMB and minocycline in combination (Aranzana-Climent et al., 2020). Further studies on the protein binding rates of polymyxins and their combined antimicrobials in humans are necessary before any definitive recommendations can be issued. And, caution should be used in interpreting these modeling results based on in vitro results.
To provide the best treatment for each patient more accurately, researchers have begun to explore population pharmacokinetics (PPK). This method considers various physiological, pathological, and genetic factors that might influence drug efficacy and safety. Combined with pharmacodynamic (PD) metrics, this allows physicians to better adjust treatment strategies, achieving personalized treatment and enhancing therapeutic outcomes (van der Leeuw et al., 2014; Chen N. et al., 2022). Recent studies have assessed the pharmacokinetics/pharmacodynamics of Polymyxin B in patients with carbapenem-resistant K. pneumoniae bloodstream infections (Yu et al., 2022). Furthermore, for other drugs like paroxetine, population pharmacokinetic models have been successfully applied to guide personalized treatments (Li et al., 2022). These studies offer a framework for understanding how to leverage pharmacokinetic/pharmacodynamic principles to optimize polymyxin treatment strategies.
6.3 Metabolism, excretion of polymyxin B, and future research directions
While we have a certain understanding of many aspects of PMB, its metabolism and excretion mechanisms in the body remain somewhat elusive. Future research should focus more on these areas, especially considering the increasing bacterial resistance to conventional antibiotics. Additionally, with the advancements in medical technology, more patients now require continuous renal replacement therapy (CRRT) or extracorporeal membrane oxygenation (ECMO) support. These treatments might influence the pharmacokinetics of polymyxins, making it crucial to understand their interactions. A recent study reported a patient with septic shock induced by severe acute pancreatitis who received life support through ECMO and CRRT and multiple anti-infective drug treatments. The study monitored the plasma concentration of Colistin sulfate during ECMO and CRRT, finding no significant difference before and after ECMO and CRRT (Peng et al., 2022), implying that ECMO and CRRT might not significantly influence the pharmacokinetics of Colistin sulfate. Moreover, different connection modes for ECMO and CRRT have shown that both modalities can achieve therapeutic goals without necessitating higher levels of anticoagulation therapy (Liu et al., 2021), providing important guidance for clinicians on the use of polymyxins in patients receiving ECMO and CRRT treatments. In conclusion, as our understanding of the behavior of polymyxins in the body deepens, future research should focus on their interactions with modern medical technologies, such as ECMO and CRRT, to optimize treatment strategies and improve therapeutic outcomes.
7 Conclusion
This review provides an overview of the latest discoveries and development history of polymyxins, as well as the mechanisms underlying multidrug resistance to polymyxins. Additionally, we discuss current research directions to overcome polymyxin resistance and highlight new antibiotics undergoing clinical research. Furthermore, we outline the future challenges and prospects of polymyxins in treating bacterial infectious diseases, particularly in relation to MDR bacterial infections. Over the past 5 years, numerous studies have explored the impact of the mcr gene on polymyxin resistance, revealing a much more complex mechanism of bacterial drug resistance than previously understood. Future research should focus on elucidating the causes of colistin resistance to enable the precise design and development of antibiotic drugs targeting drug-resistant bacteria and optimize drug administration strategies. These efforts are crucial to minimize the development of resistance and prolong the effectiveness of polymyxin as a last-line treatment.
Different forms of polymyxins exhibit significant differences in pharmacokinetics and toxicity, with some forms being more prone to induce nephrotoxicity or neurotoxicity. This necessitates a deep evaluation and careful consideration by physicians when selecting and using polymyxins (Nation et al., 2014). Specifically, Polymyxin B is seen as the last line of defense against carbapenem-resistant microbes, but its common side effects, such as neurotoxicity and nephrotoxicity, cannot be overlooked (Nation and Li, 2009; Yu et al., 2022). The toxicity observed in clinical settings stems from colistin’s antibacterial mechanism, which involves the interaction and damage inflicted on bacterial bilayer membranes. When administered at high doses, this same mechanism can cause severe damage to the cell membranes of human organs, including the liver and kidneys. Toxicity is typically reversible upon discontinuation, and its severity is dose-dependent (Li et al., 2006). To address these issues, developing new polymyxin preparations must focus on reducing dosage, achieving targeted drug delivery, and controlling drug release. Using polymyxin-based nanoparticles, liposomes, microneedles, and composite nanomaterials necessitates collaborative efforts across disciplines such as chemistry, nanomedicine, and materials science to address polymyxin delivery, drug resistance, and toxicity challenges. Conjugation of polymyxin is also a promising approach that can enhance intestinal permeability and absorption while preventing microbial resistance. However, it is crucial to investigate the impact of molecular modifications on drug stability, antimicrobial activity, and the ability to overcome microbial drug resistance. The emergence of drug resistance poses a significant threat to the treatment of MDR-GNB infections since polymyxin serves as the last line of defense. Therefore, the future of antibiotic drug development lies in optimizing existing polymyxin drugs to reduce dosage, increase efficiency, mitigate toxicity, and overcome the emergence of drug resistance.
Addressing antibiotic resistance demands a comprehensive approach that includes political agendas, legislation, treatment development, and educational initiatives. Regular surveillance, policies, and the implementation of new medical therapies targeting resistant bacteria are essential for combating antibiotic resistance in human and agricultural contexts. Given the varying rates of resistance development across different antibiotics, multifaceted measures are necessary to ensure the sustainable development of healthcare. The increasing reliance on colistin as a last resort antimicrobial necessitates urgent exploration of its antimicrobial potential and the development of new, more effective antimicrobial agents to safeguard public health in the future. Looking ahead, there is growing interest in using new technological approaches to overcome polymyxin resistance. Continued advancements in analytical techniques for the identification and structural interpretation of natural products will likely lead to the discovery of novel polymyxin groups and new lipopeptide components within existing polymyxin groups.
Author contributions
SY: Conceptualization, Formal Analysis, Validation, Visualization, Writing–original draft, Writing–review and editing. HW: Formal Analysis, Investigation, Validation, Visualization, Writing–review and editing. DZ: Investigation, Supervision, Validation, Visualization, Writing–review and editing. SZ: Formal Analysis, Investigation, Validation, Visualization, Writing–review and editing. CH: Conceptualization, Formal Analysis, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was supported by the Sichuan Science and Technology Program, China (Grant Nos. 2023YFS0322 and 2024NSFSC1735), 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYGD23012).
Acknowledgments
The author extends the appreciation to the Sichuan Science and Technology Program, China (Grant Nos. 2023YFS0322 and 2024NSFSC1735), 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYGD23012).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdallah, M., Olafisoye, O., Cortes, C., Urban, C., Landman, D., and Quale, J. (2015). Activity of eravacycline against Enterobacteriaceae and Acinetobacter baumannii, including multidrug-resistant isolates, from New York City. Antimicrob. Agents Chemother. 59 (3), 1802–1805. doi:10.1128/AAC.04809-14
Akova, M. (2016). Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 7 (3), 252–266. doi:10.1080/21505594.2016.1159366
Aranzana-Climent, V., Buyck, J. M., Smani, Y., Pachon-Diaz, J., Marchand, S., Couet, W., et al. (2020). Semi-mechanistic PK/PD modelling of combined polymyxin B and minocycline against a polymyxin-resistant strain of Acinetobacter baumannii. Clin. Microbiol. Infect. 26 (9), 1254 e1259–e1254. doi:10.1016/j.cmi.2020.01.017
Barbier, F., Andremont, A., Wolff, M., and Bouadma, L. (2013). Hospital-acquired pneumonia and ventilator-associated pneumonia: recent advances in epidemiology and management. Curr. Opin. Pulm. Med. 19 (3), 216–228. doi:10.1097/MCP.0b013e32835f27be
Bassetti, M., Righi, E., Vena, A., Graziano, E., Russo, A., and Peghin, M. (2018). Risk stratification and treatment of ICU-acquired pneumonia caused by multidrug-resistant/extensively drug-resistant/pandrug-resistant bacteria. Curr. Opin. Crit. Care 24 (5), 385–393. doi:10.1097/MCC.0000000000000534
Belaynehe, K. M., Shin, S. W., Park, K. Y., Jang, J. Y., Won, H. G., Yoon, I. J., et al. (2018). Emergence of mcr-1 and mcr-3 variants coding for plasmid-mediated colistin resistance in Escherichia coli isolates from food-producing animals in South Korea. Int. J. Infect. Dis. 72, 22–24. doi:10.1016/j.ijid.2018.05.011
Benedict, R. G., and Langlykke, A. F. (1947). Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 54 (1), 24.
Bengoechea, J. A., and Skurnik, M. (2000). Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 37 (1), 67–80. doi:10.1046/j.1365-2958.2000.01956.x
Bialvaei, A. Z., and Samadi Kafil, H. (2015). Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 31 (4), 707–721. doi:10.1185/03007995.2015.1018989
Bojkovic, J., Richie, D. L., Six, D. A., Rath, C. M., Sawyer, W. S., Hu, Q., et al. (2015). Characterization of an acinetobacter baumannii lptD deletion strain: permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J. Bacteriol. 198 (4), 731–741. doi:10.1128/JB.00639-15
Brownlee, G., and Bushby, S. R. (1948). Chemotherapy and pharmacology of aerosporin; a selective gram-negative antibiotic. Lancet 1 (6491), 127–132. doi:10.1016/s0140-6736(48)90090-7
Cai, Y., Chai, D., Wang, R., Liang, B., and Bai, N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67 (7), 1607–1615. doi:10.1093/jac/dks084
Cannatelli, A., Giani, T., D'Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58 (10), 5696–5703. doi:10.1128/AAC.03110-14
Carfrae, L. A., Rachwalski, K., French, S., Gordzevich, R., Seidel, L., Tsai, C. N., et al. (2023). Inhibiting fatty acid synthesis overcomes colistin resistance. Nat. Microbiol. 8 (6), 1026–1038. doi:10.1038/s41564-023-01369-z
Cheah, S. E., Johnson, M. D., Zhu, Y., Tsuji, B. T., Forrest, A., Bulitta, J. B., et al. (2016). Polymyxin resistance in acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci. Rep. 6, 26233. doi:10.1038/srep26233
Chen, J., Lu, Y., Ye, F., Zhang, H., Zhou, Y., Li, J., et al. (2022a). A small-molecule inhibitor of the anthranilyl-CoA Synthetase PqsA for the treatment of multidrug-resistant Pseudomonas aeruginosa. Microbiol. Spectr. 10 (4), e0276421. doi:10.1128/spectrum.02764-21
Chen, N., Guo, J., Xie, J., Xu, M., Hao, X., Ma, K., et al. (2022b). Population pharmacokinetics of polymyxin B: a systematic review. Ann. Transl. Med. 10 (4), 231. doi:10.21037/atm-22-236
Deris, Z. Z., Akter, J., Sivanesan, S., Roberts, K. D., Thompson, P. E., Nation, R. L., et al. (2014). A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J. Antibiot. (Tokyo) 67 (2), 147–151. doi:10.1038/ja.2013.111
Dixon, R. A., and Chopra, I. (1986). Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob. Agents Chemother. 29 (5), 781–788. doi:10.1128/AAC.29.5.781
Doi, Y., Murray, G. L., and Peleg, A. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 36 (1), 85–98. doi:10.1055/s-0034-1398388
Doymaz, M. Z., and Karaaslan, E. (2019). Comparison of antibacterial activities of polymyxin B and colistin against multidrug resistant Gram negative bacteria. Infect. Dis. (Lond) 51 (9), 676–682. doi:10.1080/23744235.2019.1640386
Dwyer, D. J., Belenky, P. A., Yang, J. H., MacDonald, I. C., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U. S. A. 111 (20), E2100–E2109. doi:10.1073/pnas.1401876111
Elbourne, A., Cheeseman, S., Atkin, P., Truong, N. P., Syed, N., Zavabeti, A., et al. (2020). Antibacterial liquid metals: biofilm treatment via magnetic activation. ACS Nano 14 (1), 802–817. doi:10.1021/acsnano.9b07861
El-Sayed Ahmed, M. A. E., Zhong, L. L., Shen, C., Yang, Y., Doi, Y., and Tian, G. B. (2020). Colistin and its role in the Era of antibiotic resistance: an extended review (2000-2019). Emerg. Microbes Infect. 9 (1), 868–885. doi:10.1080/22221751.2020.1754133
Falagas, M. E., and Kasiakou, S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40 (9), 1333–1341. doi:10.1086/429323
Falagas, M. E., Rafailidis, P. I., and Matthaiou, D. K. (2010). Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat 13 (4-5), 132–138. doi:10.1016/j.drup.2010.05.002
Faustino, C., and Pinheiro, L. (2020). Lipid systems for the delivery of amphotericin B in antifungal therapy. Pharmaceutics 12 (1), 29. doi:10.3390/pharmaceutics12010029
Fernandez, L., Gooderham, W. J., Bains, M., McPhee, J. B., Wiegand, I., and Hancock, R. E. (2010). Adaptive resistance to the "last hope" antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54 (8), 3372–3382. doi:10.1128/AAC.00242-10
Giddins, M. J., Macesic, N., Annavajhala, M. K., Stump, S., Khan, S., McConville, T. H., et al. (2018). Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in bla(KPC-2)-Harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob. Agents Chemother. 62 (3), e02101. doi:10.1128/AAC.02101-17
Gilleland, H. E., and Farley, L. B. (1982). Adaptive resistance to polymyxin in Pseudomonas aeruginosa due to an outer membrane impermeability mechanism. Can. J. Microbiol. 28 (7), 830–840. doi:10.1139/m82-125
Gilleland, H. E., and Lyle, R. D. (1979). Chemical alterations in cell envelopes of polymyxin-resistant Pseudomonas aeruginosa isolates. J. Bacteriol. 138 (3), 839–845. doi:10.1128/jb.138.3.839-845.1979
Gonzalez-Avila, L. U., Loyola-Cruz, M. A., Hernandez-Cortez, C., Bello-Lopez, J. M., and Castro-Escarpulli, G. (2021). Colistin resistance in aeromonas spp. Int. J. Mol. Sci. 22 (11), 5974. doi:10.3390/ijms22115974
Govaerts, C., Orwa, J., Van Schepdael, A., Roets, E., and Hoogmartens, J. (2002). Characterization of polypeptide antibiotics of the polymyxin series by liquid chromatography electrospray ionization ion trap tandem mass spectrometry. J. Pept. Sci. 8 (2), 45–55. doi:10.1002/psc.367
Groisman, E. A., Kayser, J., and Soncini, F. C. (1997). Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179 (22), 7040–7045. doi:10.1128/jb.179.22.7040-7045.1997
Gutu, A. D., Sgambati, N., Strasbourger, P., Brannon, M. K., Jacobs, M. A., Haugen, E., et al. (2013). Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob. Agents Chemother. 57 (5), 2204–2215. doi:10.1128/AAC.02353-12
Hancock, R. E. (1997). The bacterial outer membrane as a drug barrier. Trends Microbiol. 5 (1), 37–42. doi:10.1016/S0966-842X(97)81773-8
Hood, M. I., Becker, K. W., Roux, C. M., Dunman, P. M., and Skaar, E. P. (2013). Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 81 (2), 542–551. doi:10.1128/IAI.00704-12
Infectious Diseases Society of China, , Chinese Thoracic Society, , Chinese Society of Critical Care Medicine, , Chinese Society of Hematology, , Chinese Society of Bacterial Infection and Resistance, , Committee of Drug Clinical Evaluate Research of Chinese Pharmaceutical Association, , et al. (2021). Multi-disciplinary expert consensus on the optimal clinical use of the polymyxins in China. Zhonghua Jie He He Hu Xi Za Zhi 44 (4), 292–310. doi:10.3760/cma.j.cn112147-20201109-01091
Jayol, A., Nordmann, P., Brink, A., and Poirel, L. (2015a). Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59 (5), 2780–2784. doi:10.1128/AAC.05055-14
Jayol, A., Nordmann, P., Desroches, M., Decousser, J. W., and Poirel, L. (2016). Acquisition of broad-spectrum cephalosporin resistance leading to colistin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 60 (5), 3199–3201. doi:10.1128/AAC.00237-16
Jayol, A., Poirel, L., Villegas, M. V., and Nordmann, P. (2015b). Modulation of mgrB gene expression as a source of colistin resistance in Klebsiella oxytoca. Int. J. Antimicrob. Agents 46 (1), 108–110. doi:10.1016/j.ijantimicag.2015.02.015
Jeannot, K., Bolard, A., and Plesiat, P. (2017). Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 49 (5), 526–535. doi:10.1016/j.ijantimicag.2016.11.029
Jiang, S. S., Liu, M. C., Teng, L. J., Wang, W. B., Hsueh, P. R., and Liaw, S. J. (2010). Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrob. Agents Chemother. 54 (4), 1564–1571. doi:10.1128/AAC.01219-09
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A., and Collins, J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130 (5), 797–810. doi:10.1016/j.cell.2007.06.049
Kwa, A., Kasiakou, S. K., Tam, V. H., and Falagas, M. E. (2007). Polymyxin B: similarities to and differences from colistin (polymyxin E). Expert Rev. Anti Infect. Ther. 5 (5), 811–821. doi:10.1586/14787210.5.5.811
Lee, N. Y., Ko, W. C., and Hsueh, P. R. (2019). Nanoparticles in the treatment of infections caused by multidrug-resistant organisms. Front. Pharmacol. 10, 1153. doi:10.3389/fphar.2019.01153
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6 (9), 589–601. doi:10.1016/S1473-3099(06)70580-1
Li, J., Zhang, X., Han, N., Wan, P., Zhao, F., Xu, T., et al. (2023). Mechanism of action of isopropoxy benzene guanidine against multidrug-resistant pathogens. Microbiol. Spectr. 11 (1), e0346922. doi:10.1128/spectrum.03469-22
Li, X. L., Huang, S. Q., Xiao, T., Wang, X. P., Kong, W., Liu, S. J., et al. (2022). Pharmacokinetics of immediate and sustained-release formulations of paroxetine: population pharmacokinetic approach to guide paroxetine personalized therapy in Chinese psychotic patients. Front. Pharmacol. 13, 966622. doi:10.3389/fphar.2022.966622
Liang, Y., Liang, Y., Zhang, H., and Guo, B. (2022). Antibacterial biomaterials for skin wound dressing. Asian J. Pharm. Sci. 17 (3), 353–384. doi:10.1016/j.ajps.2022.01.001
Lima, W. G., Alves, M. C., Cruz, W. S., and Paiva, M. C. (2018). Chromosomally encoded and plasmid-mediated polymyxins resistance in Acinetobacter baumannii: a huge public health threat. Eur. J. Clin. Microbiol. Infect. Dis. 37 (6), 1009–1019. doi:10.1007/s10096-018-3223-9
Lin, H., Liu, X., and Sun, P. (2022). Effects of aerosol inhalation combined with intravenous drip of polymyxin B on bacterial clearance, symptoms improvement, and serum infection indexes in patients with pneumonia induced by multidrug-resistant gram-negative bacteria. Emerg. Med. Int. 2022, 5244538. doi:10.1155/2022/5244538
Lin, Q. Y., Tsai, Y. L., Liu, M. C., Lin, W. C., Hsueh, P. R., and Liaw, S. J. (2014). Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob. Agents Chemother. 58 (9), 5181–5190. doi:10.1128/AAC.00013-14
Liu, J. H., Liu, Y. Y., Shen, Y. B., Yang, J., Walsh, T. R., Wang, Y., et al. (2024). Plasmid-mediated colistin-resistance genes: mcr. Trends Microbiol. 32 (4), 365–378. doi:10.1016/j.tim.2023.10.006
Liu, M., Yan, Y., Li, G., Zhang, Y., and Guo, F. (2021). Comparison of clinical outcomes of different connection modes of extracorporeal membrane oxygenation combine with continuous renal replacement therapy. Heart Surg. Forum 24 (6), E1018–E1022. doi:10.1532/hsf.4335
Liu, Y., Jia, Y., Yang, K., Tong, Z., Shi, J., Li, R., et al. (2020). Melatonin overcomes MCR-mediated colistin resistance in Gram-negative pathogens. Theranostics 10 (23), 10697–10711. doi:10.7150/thno.45951
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 (2), 161–168. doi:10.1016/S1473-3099(15)00424-7
MacNair, C. R., Stokes, J. M., Carfrae, L. A., Fiebig-Comyn, A. A., Coombes, B. K., Mulvey, M. R., et al. (2018). Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 9 (1), 458. doi:10.1038/s41467-018-02875-z
Maher, S., Mrsny, R. J., and Brayden, D. J. (2016). Intestinal permeation enhancers for oral peptide delivery. Adv. Drug Deliv. Rev. 106 (Pt B), 277–319. doi:10.1016/j.addr.2016.06.005
Makabenta, J. M. V., Nabawy, A., Li, C. H., Schmidt-Malan, S., Patel, R., and Rotello, V. M. (2021). Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat. Rev. Microbiol. 19 (1), 23–36. doi:10.1038/s41579-020-0420-1
Manchandani, P., Zhou, J., Ledesma, K. R., Truong, L. D., Chow, D. S., Eriksen, J. L., et al. (2016). Characterization of polymyxin B biodistribution and disposition in an animal model. Antimicrob. Agents Chemother. 60 (2), 1029–1034. doi:10.1128/AAC.02445-15
Mancuso, G., Midiri, A., Gerace, E., and Biondo, C. (2021). Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10 (10), 1310. doi:10.3390/pathogens10101310
McLeod, G. I., and Spector, M. P. (1996). Starvation- and Stationary-phase-induced resistance to the antimicrobial peptide polymyxin B in Salmonella typhimurium is RpoS (sigma(S)) independent and occurs through both phoP-dependent and -independent pathways. J. Bacteriol. 178 (13), 3683–3688. doi:10.1128/jb.178.13.3683-3688.1996
Miethke, M., Pieroni, M., Weber, T., Bronstrup, M., Hammann, P., Halby, L., et al. (2021). Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 5 (10), 726–749. doi:10.1038/s41570-021-00313-1
Mlynarcik, P., and Kolar, M. (2019). Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc Czech Repub. 163 (1), 28–38. doi:10.5507/bp.2018.070
Mofazzal Jahromi, M. A., Sahandi Zangabad, P., Moosavi Basri, S. M., Sahandi Zangabad, K., Ghamarypour, A., Aref, A. R., et al. (2018). Nanomedicine and advanced technologies for burns: preventing infection and facilitating wound healing. Adv. Drug Deliv. Rev. 123, 33–64. doi:10.1016/j.addr.2017.08.001
Moffatt, J. H., Harper, M., and Boyce, J. D. (2019). Mechanisms of polymyxin resistance. Adv. Exp. Med. Biol. 1145, 55–71. doi:10.1007/978-3-030-16373-0_5
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54 (12), 4971–4977. doi:10.1128/AAC.00834-10
Mogi, T., Murase, Y., Mori, M., Shiomi, K., Omura, S., Paranagama, M. P., et al. (2009). Polymyxin B identified as an inhibitor of alternative NADH dehydrogenase and malate: quinone oxidoreductase from the Gram-positive bacterium Mycobacterium smegmatis. J. Biochem. 146 (4), 491–499. doi:10.1093/jb/mvp096
Moore, R. A., Chan, L., and Hancock, R. E. (1984). Evidence for two distinct mechanisms of resistance to polymyxin B in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26 (4), 539–545. doi:10.1128/AAC.26.4.539
Morey, P., Viadas, C., Euba, B., Hood, D. W., Barberan, M., Gil, C., et al. (2013). Relative contributions of lipooligosaccharide inner and outer core modifications to nontypeable Haemophilus influenzae pathogenesis. Infect. Immun. 81 (11), 4100–4111. doi:10.1128/IAI.00492-13
Morrissey, I., Olesky, M., Hawser, S., Lob, S. H., Karlowsky, J. A., Corey, G. R., et al. (2020). In vitro activity of eravacycline against gram-negative bacilli isolated in clinical Laboratories worldwide from 2013 to 2017. Antimicrob. Agents Chemother. 64 (3), e01699. doi:10.1128/AAC.01699-19
Nadimpalli, M. L., Marks, S. J., Montealegre, M. C., Gilman, R. H., Pajuelo, M. J., Saito, M., et al. (2020). Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat. Microbiol. 5 (6), 787–795. doi:10.1038/s41564-020-0722-0
Nang, S. C., Azad, M. A. K., Velkov, T., Zhou, Q. T., and Li, J. (2021). Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 73 (2), 679–728. doi:10.1124/pharmrev.120.000020
Nation, R. L., and Li, J. (2009). Colistin in the 21st century. Curr. Opin. Infect. Dis. 22 (6), 535–543. doi:10.1097/QCO.0b013e328332e672
Nation, R. L., Li, J., Cars, O., Couet, W., Dudley, M. N., Kaye, K. S., et al. (2015). Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect. Dis. 15 (2), 225–234. doi:10.1016/S1473-3099(14)70850-3
Nation, R. L., Velkov, T., and Li, J. (2014). Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin. Infect. Dis. 59 (1), 88–94. doi:10.1093/cid/ciu213
Needham, B. D., and Trent, M. S. (2013). Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11 (7), 467–481. doi:10.1038/nrmicro3047
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 (4), 593–656. doi:10.1128/MMBR.67.4.593-656.2003
Niu, B., Vater, J., Rueckert, C., Blom, J., Lehmann, M., Ru, J. J., et al. (2013). Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol. 13, 137. doi:10.1186/1471-2180-13-137
Nordmann, P., Jayol, A., and Poirel, L. (2016). Rapid detection of polymyxin resistance in enterobacteriaceae. Emerg. Infect. Dis. 22 (6), 1038–1043. doi:10.3201/eid2206.151840
Ohnstedt, E., Lofton Tomenius, H., Vagesjo, E., and Phillipson, M. (2019). The discovery and development of topical medicines for wound healing. Expert Opin. Drug Discov. 14 (5), 485–497. doi:10.1080/17460441.2019.1588879
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643. doi:10.3389/fmicb.2014.00643
Pelletier, M. R., Casella, L. G., Jones, J. W., Adams, M. D., Zurawski, D. V., Hazlett, K. R., et al. (2013). Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 57 (10), 4831–4840. doi:10.1128/AAC.00865-13
Peng, D., Zhang, F., Lv, P., Chen, Y., Yang, J., Zhu, W., et al. (2022). Plasma concentrations of Colistin sulfate in a patient with septic shock on extracorporeal membrane oxygenation and continuous renal replacement therapy: a case report. Ann. Transl. Med. 10 (10), 614. doi:10.21037/atm-22-2081
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30 (2), 557–596. doi:10.1128/CMR.00064-16
Pulingam, T., Parumasivam, T., Gazzali, A. M., Sulaiman, A. M., Chee, J. Y., Lakshmanan, M., et al. (2022). Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 170, 106103. doi:10.1016/j.ejps.2021.106103
Qin, C., Tang, N., Gan, Y., Zhao, H., Li, Y., Tian, G. B., et al. (2023). Liposomes Co-delivering curcumin and colistin to overcome colistin resistance in bacterial infections. Adv. Healthc. Mater 12 (24), e2202903. doi:10.1002/adhm.202202903
Qureshi, Z. A., Hittle, L. E., O'Hara, J. A., Rivera, J. I., Syed, A., Shields, R. K., et al. (2015). Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin. Infect. Dis. 60 (9), 1295–1303. doi:10.1093/cid/civ048
Roberts, K. D., Zhu, Y., Azad, M. A. K., Han, M. L., Wang, J., Wang, L., et al. (2022). A synthetic lipopeptide targeting top-priority multidrug-resistant Gram-negative pathogens. Nat. Commun. 13 (1), 1625. doi:10.1038/s41467-022-29234-3
Rodriguez-Santiago, J., Cornejo-Juarez, P., Silva-Sanchez, J., and Garza-Ramos, U. (2021). Polymyxin resistance in Enterobacterales: overview and epidemiology in the Americas. Int. J. Antimicrob. Agents 58 (5), 106426. doi:10.1016/j.ijantimicag.2021.106426
Rutten, A. A., Bequet-Passelecq, B. G., and Koeter, H. B. (1990). Two-compartment model for rabbit skin organ culture. Vitro Cell Dev. Biol. 26 (4), 353–360. doi:10.1007/BF02623826
Sampson, T. R., Liu, X., Schroeder, M. R., Kraft, C. S., Burd, E. M., and Weiss, D. S. (2012). Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 56 (11), 5642–5649. doi:10.1128/AAC.00756-12
Sato, T., Shiraishi, T., Hiyama, Y., Honda, H., Shinagawa, M., Usui, M., et al. (2018). Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b:H4-st131-H30Rx and non-x. Antimicrob. Agents Chemother. 62 (9), e00864. doi:10.1128/AAC.00864-18
Schindler, M., and Osborn, M. J. (1979). Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry 18 (20), 4425–4430. doi:10.1021/bi00587a024
Schrader, S. M., Vaubourgeix, J., and Nathan, C. (2020). Biology of antimicrobial resistance and approaches to combat it. Sci. Transl. Med. 12 (549), eaaz6992. doi:10.1126/scitranslmed.aaz6992
Schromm, A. B., Paulowski, L., Kaconis, Y., Kopp, F., Koistinen, M., Donoghue, A., et al. (2021). Cathelicidin and PMB neutralize endotoxins by multifactorial mechanisms including LPS interaction and targeting of host cell membranes. Proc. Natl. Acad. Sci. U. S. A. 118 (27), e2101721118. doi:10.1073/pnas.2101721118
Schwarz, S., and Johnson, A. P. (2016). Transferable resistance to colistin: a new but old threat. J. Antimicrob. Chemother. 71 (8), 2066–2070. doi:10.1093/jac/dkw274
Shein, A. M. S., Wannigama, D. L., Higgins, P. G., Hurst, C., Abe, S., Hongsing, P., et al. (2022). High prevalence of mgrB-mediated colistin resistance among carbapenem-resistant Klebsiella pneumoniae is associated with biofilm formation, and can be overcome by colistin-EDTA combination therapy. Sci. Rep. 12 (1), 12939. doi:10.1038/s41598-022-17083-5
Sherry, N., and Howden, B. (2018). Emerging Gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam - epidemiology, laboratory detection and treatment implications. Expert Rev. Anti Infect. Ther. 16 (4), 289–306. doi:10.1080/14787210.2018.1453807
Shoji, J., Kato, T., and Hinoo, H. (1977). The structures of two new polymyxin group antibiotics. J. Antibiot. (Tokyo) 30 (5), 427–429. doi:10.7164/antibiotics.30.427
Snitkin, E. S., Zelazny, A. M., Gupta, J., Program, N. C. S., Palmore, T. N., Murray, P. R., et al. (2013). Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res. 23 (7), 1155–1162. doi:10.1101/gr.154328.112
Srisakul, S., Wannigama, D. L., Higgins, P. G., Hurst, C., Abe, S., Hongsing, P., et al. (2022). Overcoming addition of phosphoethanolamine to lipid A mediated colistin resistance in Acinetobacter baumannii clinical isolates with colistin-sulbactam combination therapy. Sci. Rep. 12 (1), 11390. doi:10.1038/s41598-022-15386-1
Stansly, P. G., and Brownlee, G. (1949). Nomenclature of polymyxin antibiotics. Nature 163 (4146), 611. doi:10.1038/163611a0
Stokes, J. M., MacNair, C. R., Ilyas, B., French, S., Cote, J. P., Bouwman, C., et al. (2017). Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2, 17028. doi:10.1038/nmicrobiol.2017.28
Storm, D. R., Rosenthal, K. S., and Swanson, P. E. (1977). Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46, 723–763. doi:10.1146/annurev.bi.46.070177.003451
Szucs, Z., Bereczki, I., Fenyvesi, F., Herczegh, P., Ostorhazi, E., and Borbas, A. (2022). Synthesis of an amphiphilic vancomycin aglycone derivative inspired by polymyxins: overcoming glycopeptide resistance in Gram-positive and Gram-negative bacteria in synergy with teicoplanin in vitro. Sci. Rep. 12 (1), 20921. doi:10.1038/s41598-022-24807-0
Tang, T., Li, Y., Xu, P., Zhong, Y., Yang, M., Ma, W., et al. (2023). Optimization of polymyxin B regimens for the treatment of carbapenem-resistant organism nosocomial pneumonia: a real-world prospective study. Crit. Care 27 (1), 164. doi:10.1186/s13054-023-04448-z
Thapa, R. K., Diep, D. B., and Tonnesen, H. H. (2020). Topical antimicrobial peptide formulations for wound healing: current developments and future prospects. Acta Biomater. 103, 52–67. doi:10.1016/j.actbio.2019.12.025
Thi Khanh Nhu, N., Riordan, D. W., Do Hoang Nhu, T., Thanh, D. P., Thwaites, G., Huong Lan, N. P., et al. (2016). The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6, 28291. doi:10.1038/srep28291
Tomczyk, S., Zanichelli, V., Grayson, M. L., Twyman, A., Abbas, M., Pires, D., et al. (2019). Control of carbapenem-resistant enterobacteriaceae, acinetobacter baumannii, and Pseudomonas aeruginosa in healthcare facilities: a systematic review and reanalysis of quasi-experimental studies. Clin. Infect. Dis. 68 (5), 873–884. doi:10.1093/cid/ciy752
Tzeng, Y. L., Ambrose, K. D., Zughaier, S., Zhou, X., Miller, Y. K., Shafer, W. M., et al. (2005). Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187 (15), 5387–5396. doi:10.1128/JB.187.15.5387-5396.2005
Tzeng, Y. L., Berman, Z., Toh, E., Bazan, J. A., Turner, A. N., Retchless, A. C., et al. (2019). Heteroresistance to the model antimicrobial peptide polymyxin B in the emerging Neisseria meningitidis lineage 11.2 urethritis clade: mutations in the pilMNOPQ operon. Mol. Microbiol. 111 (1), 254–268. doi:10.1111/mmi.14153
Uzairue, L. I., Rabaan, A. A., Adewumi, F. A., Okolie, O. J., Folorunso, J. B., Bakhrebah, M. A., et al. (2022). Global prevalence of colistin resistance in Klebsiella pneumoniae from bloodstream infection: a systematic review and meta-analysis. Pathogens 11 (10), 1092. doi:10.3390/pathogens11101092
van der Leeuw, J., Ridker, P. M., van der Graaf, Y., and Visseren, F. L. (2014). Personalized cardiovascular disease prevention by applying individualized prediction of treatment effects. Eur. Heart J. 35 (13), 837–843. doi:10.1093/eurheartj/ehu004
Velkov, T., Thompson, P. E., Nation, R. L., and Li, J. (2010). Structure--activity relationships of polymyxin antibiotics. J. Med. Chem. 53 (5), 1898–1916. doi:10.1021/jm900999h
Wang, L., Zhang, Y., Lin, Y., Cao, J., Xu, C., Chen, L., et al. (2023). Resveratrol increases sensitivity of clinical colistin-resistant Pseudomonas aeruginosa to colistin in vitro and in vivo. Microbiol. Spectr. 11 (1), e0199222. doi:10.1128/spectrum.01992-22
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi:10.1146/annurev-biochem-060713-035600
Yang, X., Guo, C., Wu, G., Zhao, K., Xiang, D., Xu, D., et al. (2023). Treatment of central nervous system infection caused by multidrug-resistant acinetobacter baumannii with intravenous and intraventricular colistin sulfate: a case report and literature review. Infect. Drug Resist 16, 6029–6038. doi:10.2147/IDR.S425415
Yap, P. S., Cheng, W. H., Chang, S. K., Lim, S. E., and Lai, K. S. (2022). MgrB mutations and altered cell permeability in colistin resistance in Klebsiella pneumoniae. Cells 11 (19), 2995. doi:10.3390/cells11192995
Yeom, J., Imlay, J. A., and Park, W. (2010). Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J. Biol. Chem. 285 (29), 22689–22695. doi:10.1074/jbc.M110.127456
Yu, Z., Liu, X., Du, X., Chen, H., Zhao, F., Zhou, Z., et al. (2022). Pharmacokinetics/pharmacodynamics of polymyxin B in patients with bloodstream infection caused by carbapenem-resistant Klebsiella pneumoniae. Front. Pharmacol. 13, 975066. doi:10.3389/fphar.2022.975066
Zavascki, A. P., Goldani, L. Z., Cao, G., Superti, S. V., Lutz, L., Barth, A. L., et al. (2008). Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin. Infect. Dis. 47 (10), 1298–1304. doi:10.1086/592577
Zhang, P., Qiu, Y., Wang, Y., Xiao, L., Yu, S., Shi, M., et al. (2022). Nanoparticles promote bacterial antibiotic tolerance via inducing hyperosmotic stress response. Small 18 (19), e2105525. doi:10.1002/smll.202105525
Zhao, J., Han, M. L., Zhu, Y., Lin, Y. W., Wang, Y. W., Lu, J., et al. (2021). Comparative metabolomics reveals key pathways associated with the synergistic activity of polymyxin B and rifampicin combination against multidrug-resistant Acinetobacter baumannii. Biochem. Pharmacol. 184, 114400. doi:10.1016/j.bcp.2020.114400
Zhao, J., Zhu, Y., Lin, Y. W., Yu, H., Wickremasinghe, H., Han, J., et al. (2022). Polymyxin dose tunes the evolutionary dynamics of resistance in multidrug-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 28 (7), 1026.e1–1026.e5. doi:10.1016/j.cmi.2022.02.043
Zhu, Y., Zhao, J., Maifiah, M. H. M., Velkov, T., Schreiber, F., and Li, J. (2019). Metabolic responses to polymyxin treatment in acinetobacter baumannii ATCC 19606: integrating transcriptomics and metabolomics with genome-scale metabolic modeling. mSystems 4 (1), e00157. doi:10.1128/mSystems.00157-18
Keywords: antibacterial mechanism, drug resistance mechanisms, nanotechnology, polymyxins, pharmacokinetics
Citation: Yang S, Wang H, Zhao D, Zhang S and Hu C (2024) Polymyxins: recent advances and challenges. Front. Pharmacol. 15:1424765. doi: 10.3389/fphar.2024.1424765
Received: 28 April 2024; Accepted: 04 June 2024;
Published: 21 June 2024.
Edited by:
Xiangji Liu, Frontage Laboratories Inc., United StatesReviewed by:
Gabriela Cristina Fernandez, National Scientific and Technical Research Council (CONICET), ArgentinaSujeet Kumar, The Ohio State University, United States
Copyright © 2024 Yang, Wang, Zhao, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenggong Hu, aHVjaGVuZ2dvbmdAc2N1LmVkdS5jbg==
 Shan Yang1
Shan Yang1 Chenggong Hu
Chenggong Hu