- Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, Qingdao Medical College, Qingdao University, Qingdao, China
The DNA damage repair (DDR) pathway is a complex signaling cascade that can sense DNA damage and trigger cellular responses to DNA damage to maintain genome stability and integrity. A typical hallmark of cancer is genomic instability or nonintegrity, which is closely related to the accumulation of DNA damage within cancer cells. The treatment principles of radiotherapy and chemotherapy for cancer are based on their cytotoxic effects on DNA damage, which are accompanied by severe and unnecessary side effects on normal tissues, including dysregulation of the DDR and induced therapeutic tolerance. As a driving factor for oncogenes or tumor suppressor genes, noncoding RNA (ncRNA) have been shown to play an important role in cancer cell resistance to radiotherapy and chemotherapy. Recently, it has been found that ncRNA can regulate tumor treatment tolerance by altering the DDR induced by radiotherapy or chemotherapy in cancer cells, indicating that ncRNA are potential regulatory factors targeting the DDR to reverse tumor treatment tolerance. This review provides an overview of the basic information and functions of the DDR and ncRNAs in the tolerance or sensitivity of tumors to chemotherapy and radiation therapy. We focused on the impact of ncRNA (mainly microRNA [miRNA], long noncoding RNA [lncRNA], and circular RNA [circRNA]) on cancer treatment by regulating the DDR and the underlying molecular mechanisms of their effects. These findings provide a theoretical basis and new insights for tumor-targeted therapy and the development of novel drugs targeting the DDR or ncRNAs.
1 Introduction
Cells constantly respond to DNA damage caused by both endogenous and exogenous stimuli, such as replication pressure, telomere shortening, reactive oxygen species (ROS), ultraviolet radiation, ionizing radiation, and chemical toxins (Goldstein and Kastan, 2015). This process is known as DNA damage repair or the DNA damage response (DDR) and is defined as the cell’s response to DNA damage. DDR is a complex repair mechanism network composed of signaling pathways that maintain the stability and integrity of mammalian genomes (Li et al., 2024; Zuo et al., 2024). Based on the different types of DNA damage, DDR initiates repair processes through different pathways, such as mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), homologous recombination repair (HRR), and nonhomologous terminal junction (NHEJ) pathways (Liu et al., 2024) (Thote et al., 2024). Activation of the DDR often requires close coordination of these different DNA repair pathways, as well as signal transduction associated with cell cycle arrest, to allow for DNA damage repair and to prevent DNA damage from being replicated or transmitted to the next-generation (Ashwell and Zabludoff, 2008; Jackson and Bartek, 2009). Dysregulation of the DNA damage response (DDR) has been proven to be associated with susceptibility to the development of various diseases, including cancer, and can also lead to hypersensitivity or tolerance to treatment, which can be used to improve clinical treatment (Jurkovicova et al., 2022).
Choosing appropriate therapies and medication regimens remains a major challenge in cancer treatment. Chemotherapy and radiotherapy, which rely on cytotoxic DNA damage effects, are still the first-line treatment methods for many unresectable or metastatic malignant tumors. However, due to the unique strong adaptability of cancer cells, the DDR can be dysregulated, leading to hypersensitivity or drug resistance reactions against cancer agents in cancer cells (Jurkovicova et al., 2022). Resistance is believed to arise through genetic alterations and non genetic mechanisms, without altering the DNA sequence to reduce sensitivity to different cancer cells (Brauner et al., 2016; Rao and Ayres, 2017). A defective DDR drives cancer progression and recurrence by preselecting subclones with intrinsic or acquired drug resistance, leading to the development of tumor heterogeneity (Jurkovicova et al., 2022; Lytle et al., 2018; Bakhoum and Landau, 2017). At least 330 genes are involved in the imbalance of the DDR in tumor cells and are valuable targets for new cancer treatment methods (Vogelstein et al., 2013). These DDR-related genes have laid the foundation for a large number of new DDR inhibitors (DDRis) and DDR-based strategies. Recently, several DDRis, such as inhibitors of poly (ADP-ribose) polymerase (PARP), ataxia telangiectasia mutated (ATM) and ataxia–telangiectasia-mutated and Rad3-related kinase (ATR), have been identified as potential alternative strategies for overcoming cancer resistance, and their combination with radiation, cytotoxicity, or targeted drugs can maximize the benefits of DDR-targeted therapy (Cheng et al., 2022; Qiu et al., 2022). More importantly, recent studies have shown that DDR-related genes include not only protein-coding genes, such as kinases and pathway regulatory proteins but also noncoding genes, such as noncoding RNAs (ncRNAs).
ncRNAs have become important regulatory factors for gene expression by influencing chromatin structure, DNA/RNA/protein modifications, molecular stability, and RNA splicing (Tian et al., 2019).They are not only involved in physiological processes such as cell development and differentiation but also participate in the pathological processes of various diseases, especially tumors (Esteller, 2011; Rinn and Chang, 2012; Liu et al., 2023; Ao et al., 2023). Increasing evidence suggests that ncRNAs, especially microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs), are abnormally expressed in various cancers and are associated with the occurrence, development, and metastasis of tumors, as well as the acquisition of tumor resistance to treatment (Liu et al., 2022a; Zhou et al., 2022). They play important roles in tumor treatment resistance and tolerance by controlling multiple signaling pathways in cells, including the cell cycle, proliferation, apoptosis, DDR, and other key cellular signaling pathways, as well as regulating the expression of therapeutic targets or genes involved in therapeutic metabolism or transport (Fanale et al., 2016; Wei et al., 2020; Matsui and Corey, 2017; Xu et al., 2021). Moreover, the inhibition or overexpression of these ncRNAs related to tumor treatment has been demonstrated to regulate the activation or inactivation of their targeted oncogenes, tumor suppressor genes, or certain regulatory genes, possibly reversing the treatment tolerance of tumor cells and improving their sensitivity to therapeutic agents (Chen B. et al., 2022; Zhang et al., 2020; Liu et al., 2022b). Therefore, identifying and understanding ncRNAs and their mechanisms of action related to tumor treatment response can not only identify potential candidate molecules for predicting treatment response, but also develop personalized and more targeted treatment plans for cancer patients (Kutashev et al., 2024; Mamontova et al., 2024).
In this review, we first provide an overview of the basic information on the DDR (including major checkpoint kinases and repair pathways), as well as the regulatory roles of ncRNAs in the tolerance or sensitivity to tumor chemotherapy and radiation therapy. We focused on the impact of ncRNAs (mainly miRNAs, lncRNAs, and circRNAs) on cancer treatment through the regulation of the DDR. These findings provide both a theoretical basis for research on molecular markers or targeted therapies related to cancer treatment based on the DDR or ncRNAs and a scientific basis for rational planning of clinical medication and the development of innovative drugs.
2 The DDR in cancer therapy resistance
Tumor therapy resistance is mainly related to the intrinsic sensitivity of tumor cells and their microenvironment (Hu and Jiang, 2019). Therefore, different tumors exhibit different sensitivities or tolerances to different treatment methods, resulting in different treatment outcomes. At present, the factors related to therapeutic resistance mainly include cell hypoxia, the cell cycle, cell apoptosis, and the DDR (Bacová et al., 2000). In recent years, an increasing number of studies have focused on the role of the DDR in cancer therapy resistance. Many studies have shown that the DDR is a key inducer of drug resistance in cancer cell therapy, and alterations in the DDR of tumor cells commonly contribute to emerging resistance to tumor therapy (Eich et al., 2013; McMullen et al., 2020; Wang S. et al., 2022; Lim and Ma, 2019). Alterations in the DDR lead to genomic instability and the production of new antigens, upregulating the expression of programmed death ligand 1 (PD-L1), also known as differentiation cluster (CD) 279, which is an important immunosuppressive molecule (Liu et al., 2022b). PD-L1 interacts with cGAS-STING (cGAS-STING) signals, like interferon genes, to suppress immune responses and promote cancer cell tolerance by inhibiting T cell inflammation (Jiang et al., 2021). Moreover, many related genes or pathways are involved in the molecular mechanisms of tumor treatment tolerance mediated by the DDR and may vary depending on the treatment method.
2.1 The main checkpoint kinases involved in the DDR
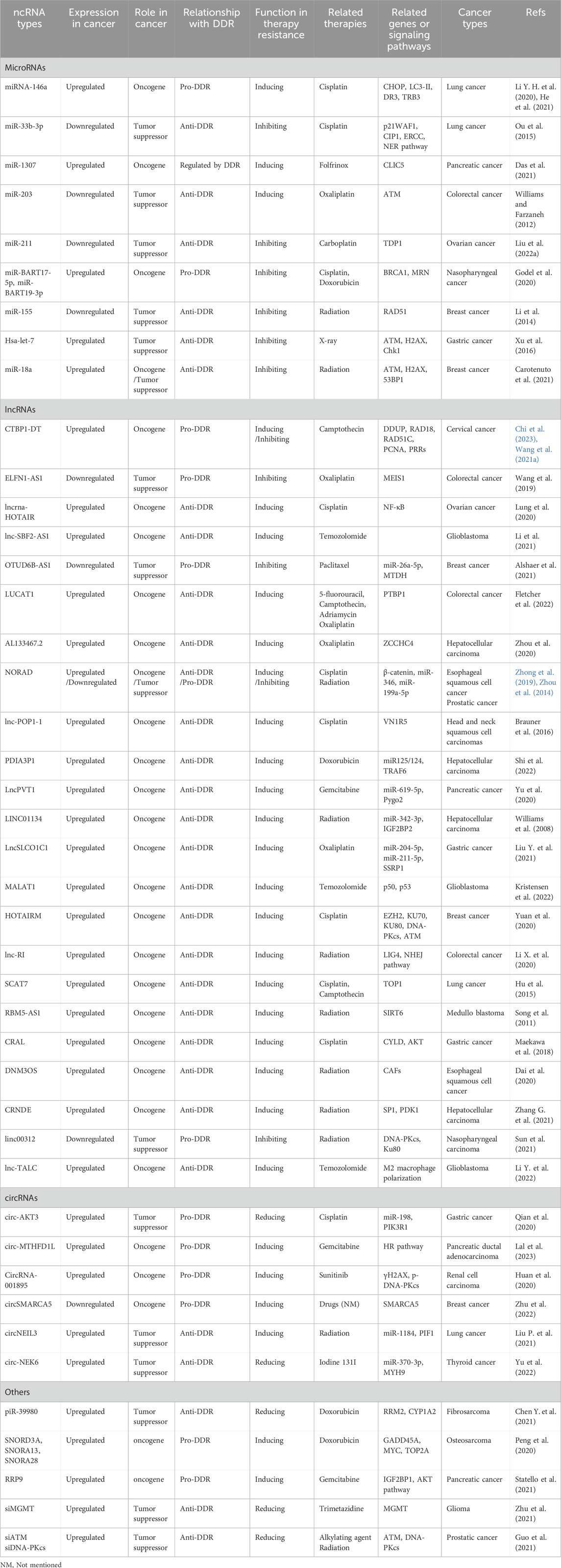
In response to DNA damage, the rapid activation of checkpoint kinases is the first signal transduction wave to be transmitted by cells, as shown in Figure 1. In vertebrate cells, at least three major protein kinases, namely, ATM, ATR and DNA-dependent protein kinase (DNA-PK), are associated with DDR (Blackford and Jackson, 2017). These kinases share similar upstream frap–atm–trap (FAT) domains and downstream phosphatidylinositol 3-kinase-like protein kinase (PIKK) regulatory domains (PRD). These proteins usually phosphorylate a serine or threonine residue followed by glutamine but can also be autophosphorylated (Blackford and Jackson, 2017). Moreover, to prevent the harmful effects of DNA single-strand break repair (SSBR) damage, which is one of the most common DNA lesions, cells are equipped with specialized enzymes, including apurinic/apyrimidinic (AP) endonuclease 1 (APE1), polynucleotide kinase-phosphatase (PNKP), tyrosine DNA phosphodiesterase 1 (TDP1), apatasin (APTX), and DNA ligases (Iyama and Wilson, 2013). These enzymes can produce appropriate 3′and 5′termini to cause chain breaks and interruptions (Iyama and Wilson, 2013). In addition, several other kinases related to cell vulnerability, such as breast-cancer susceptibility gene 1 (BRCA1), radiation sensitive 51 (RAD51), RAD52, xeroderma pigmentosum group C (XPC), and flap endonuclease 1 (FEN1), are involved in the DDR (Li E. et al., 2022). Among these modifications, ATM plays an important role in the response to DNA double-strand break (DSB) damage induced by exogenous sources such as radiation or anticancer chemotherapeutic agents (Shiloh, 2001). ATM is a product of a gene mutated in human hereditary ataxia degenerative disorder that belongs to the phosphatidylinositol 3-kinase superfamily. The activation of ATM can lead to the induction of p21/WAF1, the inhibition of cyclin-dependent kinase activity, and the failure to phosphorylate key substrates such as checkpoint kinase 2 (CHK2), p53, and BRCA1, ultimately leading to G1 arrest (Lavin, 1999; Jin and Oh, 2019). Correspondingly, abnormal ATM activation is often one of the key factors through which tumor cells develop tolerance to treatment (Jin and Oh, 2019; Choi et al., 2016). However, the inhibition of ATM increases the sensitivity of cancer cells to radiotherapy or chemotherapy (Lavin and Yeo, 2020). Therefore, several ATM inhibitors, such as KU-559933, CP-466722, AZ32, and AZD1390, have been developed and evaluated as anticancer agents in clinical applications (Jin and Oh, 2019).
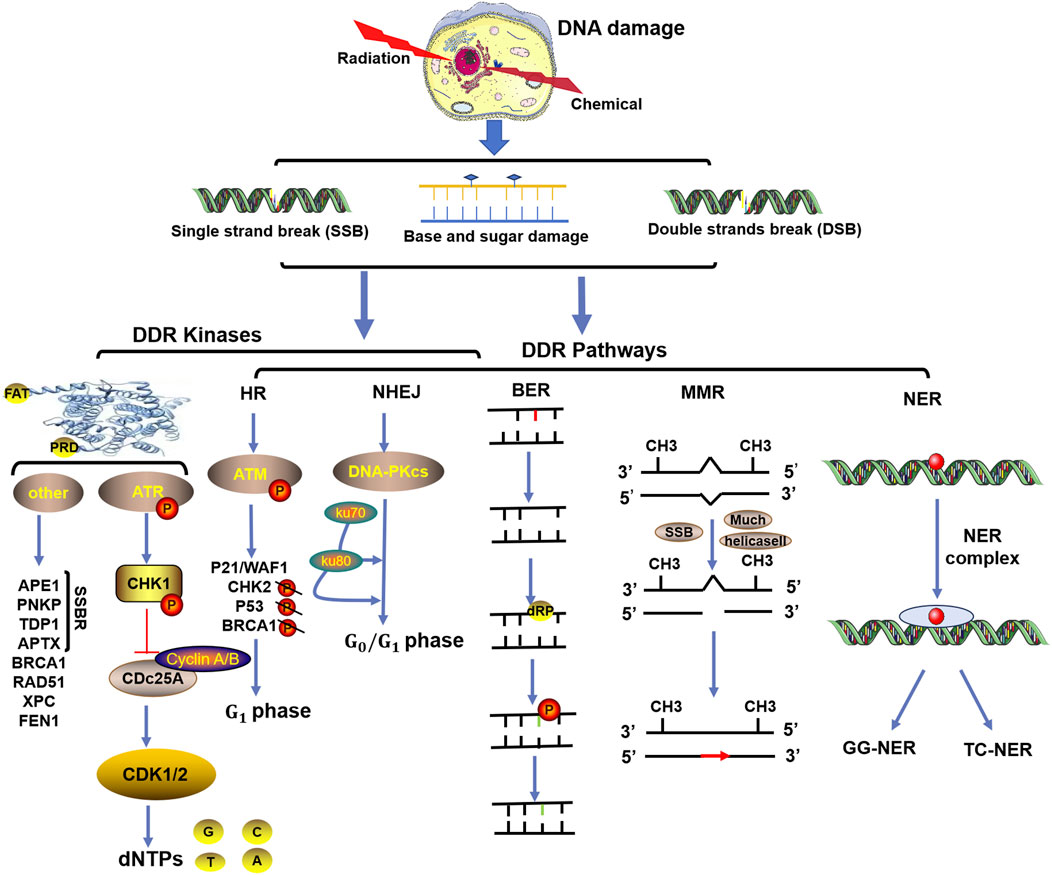
Figure 1. Functional complexes of DNA damage response (DDR) kinases and repair signaling pathways activated in response to radiation- or chemical-induced DNA damage. The three major types of DNA damage induced by radiation or chemical drugs include base and sugar damage, single-strand breaks (SSBs), and double-strand breaks (DSBs). In response to DNA damage, the rapid activation of checkpoint kinases (mainly ATM, ATR and DNA-PK) is the first signal transduction wave to be transmitted by cells. Based on the different types of DNA damage, the DNA damage response (DDR) initiates repair processes through different pathways, such as homologous recombination repair (HR), nonhomologous terminal junction (NHEJ), base excision repair (BER), mismatch repair (MMR), and nucleotide excision repair (NER).
In contrast to ATM, which is activated by DSBs, ATR proteins respond to SSBs. ATR is essential for cell proliferation, and one of its important functions is to activate checkpoint kinase 1 (CHK1). CHK1 degrades the phosphatase cell division cycle 25 A (Cdc25A) and inhibits cyclin-dependent kinases (CDKs), thereby arresting cell cycle progression and providing time for DNA repair (Blackford and Jackson, 2017). When challenged by chemotherapeutic drugs and/or radiation, the cell cycle checkpoint protein ATR and its major downstream effector CHK1 prevent the entry of cells with damaged or imperfectly replicated DNA into mitosis (Jin and Oh, 2019). This regulation is particularly noticeable in cells with a compromised G1 checkpoint, a characteristic frequently present in cancer cells (Jin and Oh, 2019). Additionally, ATR and/or CHK1, especially in cells with active oncogenes, limit excessive origin firing to minimize replication stress (RS), making them excellent therapeutic targets (Jin and Oh, 2019; Qiu et al., 2018; Lecona and Fernandez-Capetillo, 2018). The inhibition of ATR/CHK1 can be achieved by blocking cell cycle checkpoints, enhancing the killing effect of cytotoxic drugs or radiotherapy on tumor cells (Ma et al., 2011). At present, ATR/CHK1 inhibitors, such as UCN-01 (7-hydroxystaurosporine), AZD7762, LY2603618, and PF-00477736, have been developed for p53-deficient cells and have been used as single drugs or in combination with radiotherapy or multiple genotoxic chemotherapies in preclinical and clinical studies (Qiu et al., 2018).
Along with ATM and ATR, DNA-PK plays a key role in the DDR and is a multiprotein complex consisting of a catalytic subunit (DNA/PK) and the regulatory heterodimer Ku (Ku70/Ku80) (Yue et al., 2020). Like ATM, DNA-PKcs is activated by DSBs but stabilizes and aligns DNA ends to play an important role in maintaining genomic integrity (Damia, 2020). The interaction between two DNA-PKs located at the end of each DSB activates their intrinsic protein kinase activity, resulting in self-phosphorylation and dissociation of DNA-PK to activate the endonuclease and recruit different processing factors involved in the G0/G1 phases of the cell cycle (Medová et al., 2020). Abnormal expression of DNA-PK was shown to be beneficial for tumor development, progression, and treatment tolerance-related metastasis by regulating cell survival signaling pathways (Goodwin and Knudsen, 2014; George et al., 2019). Mutations and deletions in DNA PKcs are associated with enhanced chromosomal exchange and increased sensitivity to reagents that cause DNA DSBs (van der Burg et al., 2009; Woodbine et al., 2013). Therefore, many types of DNA-PK inhibitors, such as small-molecule substances, nucleotides, antisense oligonucleotides, and small interfering RNAs, have been developed and combined with chemical drugs to enhance the drug sensitivity of tumors (Damia, 2020). For example, the DNA-PK inhibitor AZD7648, developed by Jacqueline H.L. Fok, has been shown to be powerful and selective and to increase the effectiveness of both doxorubicin and irradiation. Additionally, the DNA-PK inhibitor olaparib has been approved for a number of applications in the treatment of breast and ovarian cancers (Fok et al., 2019).
2.2 The main repair pathways involved in the DDR
Generally, DNA repair systems are divided into direct and indirect repair systems. The direct repair system is involved in the repair of DNA during replication and is mediated by direct proteins catalyzed by the main DNA polymerase. O6 methylguanine-DNA methyltransferase (MGMT) repairs endogenous and alkylating agent-induced damage to the guanine O6 site and ultimately repairs pyrimidine dimers (Iyama and Wilson, 2013). The indirect DNA repair system includes at least five pathways, such as the MMR, NER, BER, HR, and NHEJ pathways (Iyama and Wilson, 2013), as shown in Figure 1. Among them, the HR and NHEJ pathways are involved in the response of double-strand break repair (DSBR) to SBRs. The HR pathway includes ATM as the core component and plays roles in cell and S phase division because it requires homologous sister chromatids to execute. NHEJ, with DNA-PK as its core component, can play roles in both dividing and nondividing cells and is independent of the cell cycle (Iyama and Wilson, 2013). Defects in HR or NHEJ can lead to the occurrence and development of many human diseases, including sensitivity to radiation and drug therapy for cancer (Difilippantonio et al., 2002).
BER is the most important repair pathway in the DNA repair system and has been proven to be related to the incidence of various malignant tumors. In the BER pathway, damaged bases are recognized and removed by DNA glycosidases, after which DNA damage is repaired by polymerase and nuclease (Grundy and Parsons, 2020). A significant portion of the DNA damage caused by external (photons and particle beams) and internal radiation technologies is the repair target of the BER pathway (Grundy and Parsons, 2020). For cancer treatment to be effective, the amount of DNA damage caused by radiotherapy must be greater than the repair ability of cancer cells. Recent studies have shown that PARP inhibitors (PARPis) promote the effectiveness of combination therapy with anti-PD-1 combined with ionizing radiation in colorectal cancer models, while PD-L1 expression is inversely proportional to BER gene expression, including the core genes eight-oxoguanine-DNA glycosylase (OGG1) and APE1 of the BER pathway (Permata et al., 2019).
The MMR pathway plays a crucial role in maintaining DNA replication fidelity and genomic stability; this pathway mainly involves detecting mismatches and inserting or deleting rings, inducing single-stranded incisions, and subsequently facilitating DNA repair through polymerase, nuclease, and ligase (Jiang et al., 2021). The two core protein complexes of the MMR pathway are MutSα (heterodimer of the MutS homolog 2 [MSH2] and MutS homolog 6 [MSH6] proteins) and MutS β (heterodimer of the MSH2 and MSH3 proteins), which can bind to mismatches in an adenosine triphosphate-dependent manner and subsequently recruit MutL α (heterodimer of the mutL homolog 1 [MLH1] and postmeiotic segregation increased 2 [PMS2] proteins) (Liu et al., 2017). Research has confirmed that MMR deficiency is a molecular characteristic that leads to microsatellite instability (MSI) and induces cancer (Koessler et al., 2008). Temozolomide (TMZ) and radiation therapy have antitumor effects on glioblastoma (GBM) mainly through ineffective MMR pathway activity and apoptosis induction (Ao et al., 2023). Research has shown that the MGMT pathway can repair DNA damage caused by TMZ, but GBM patients treated with TMZ may benefit from tumor cells that do not express the MGMT protein. Patients with low or missing MGMT expression levels still exhibit drug resistance. After MGMT deletion, O6 guanine remains in DNA and undergoes mismatch base pairing with thymine during DNA replication, ultimately leading to a lack of MMR (Li et al., 2023).
The NER pathway can handle spiral distortion lesions through two NER subpathways, which typically cleave 22 to 30 bases of oligonucleotides to eliminate damage, triggering the production of single-stranded DNA (ssDNA) (Li et al., 2023). One of the subpathways is the global genome NER (GG-NER), which is initiated by the HR23B/XPC complex, and distorted lesions are identified throughout the entire genome. Another pathway is transcriptional coupled repair (TCR), which plays a role in preventing damage to extended RNA polymerase (Hoeijmakers, 2001; Curtin, 2013). Genetic defects in the NER gene are the foundation of three serious genetic diseases, xeroderma pigmentosum (XP), Cockayne syndrome (CS), and lignosulfia malnutrition (TTD), which are associated with a high risk of skin cancer (Butkiewicz et al., 2002). Research has shown that the upregulation of NER genes may be an important reason for drug (such as cisplatin) resistance in some clinical cancer chemotherapeutic agents, such as ovarian and bladder cancers (Saldivar et al., 2007; Mouw, 2017).
Besides all these pathways and specific checkpoint kinase, nrecent studies have shown that ncRNAs can also participate in DDR by regulating the expression of oncogenes or suppressor genes during tumorigenesis. Moreover, different types of ncRNAs can target specific factors or pathways of different DDR, promoting or inhibiting tumor drug resistance, as described below.
3 Noncoding RNA regulation in cancer therapy resistance
ncRNAs, especially miRNAs, lncRNAs and circRNAs, have long been widely recognized as universal regulatory factors for various cancer characteristics, such as proliferation, apoptosis, invasion, and metastasis. However, for tumors, resistance or tolerance to chemotherapy, radiation therapy, targeted therapy, and immunotherapy remains a major setback. Recent studies have shown that ncRNAs play important roles as drivers of oncogenes or tumor suppressor genes to regulate cell proliferation or death signals in patients resistant to different cancer therapies, such as radiotherapy and chemotherapy, as shown in Figure 2.
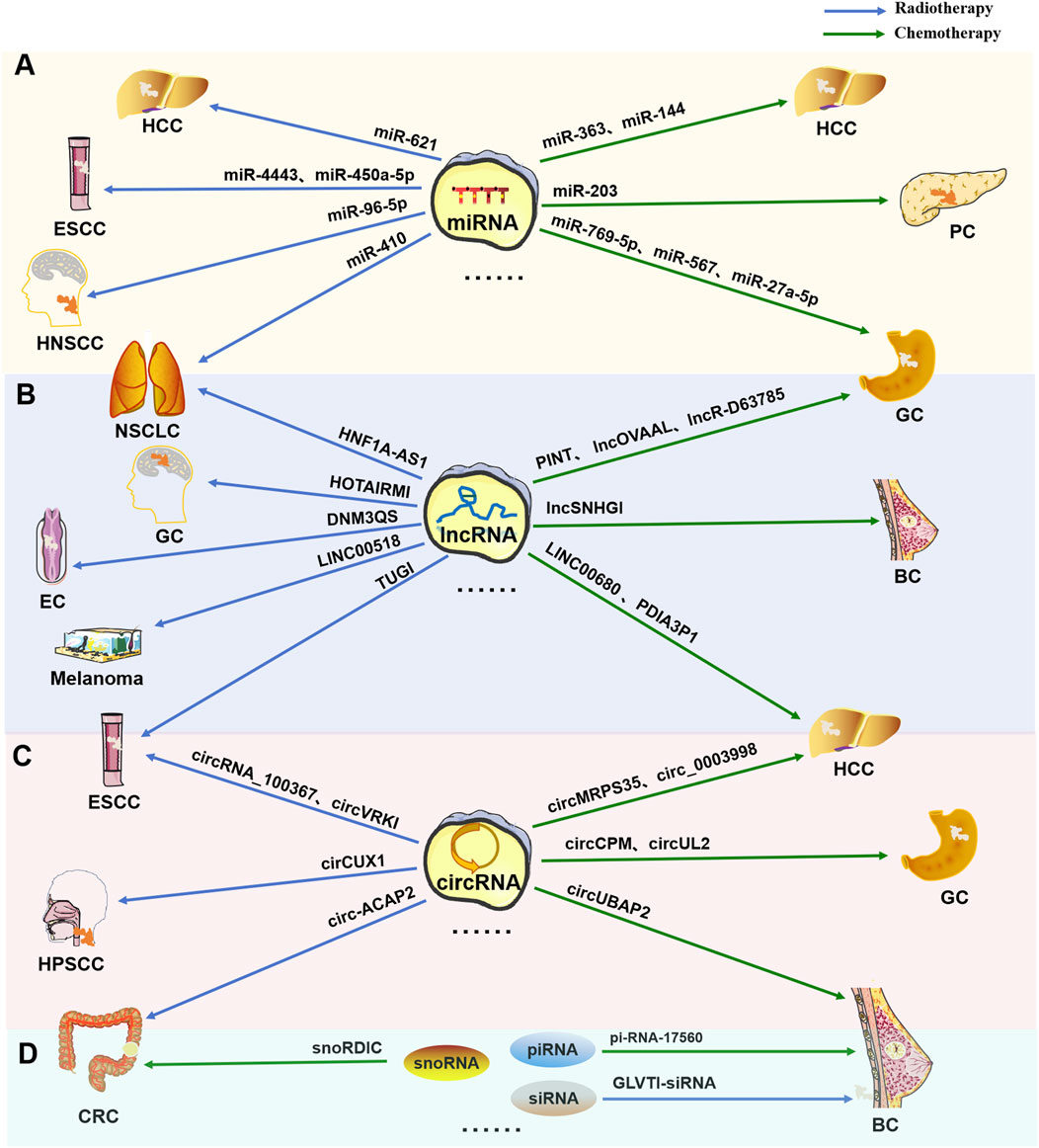
Figure 2. Multiple types of ncRNAs are related to tumor radiotherapy or chemotherapy tolerance. Typical tumor radiotherapy (blue arrow) or chemotherapy (green arrow) tolerance-related miRNAs (A), lncRNAs (B), circRNAs (C), and other types of RNAs (including snoRNAs, piRNAs, and siRNAs) (D).
3.1 Regulation of miRNAs in tumor therapy resistance
miRNA is a type of short noncoding RNA with a length of approximately 19–25 nucleotides that can interact with the 3′ untranslated region (3′-UTR) of its target gene to regulate post transcriptional mechanisms of gene expression (Rykova et al., 2022). In recent years, an increasing number of miRNAs have been shown to be closely related to the resistance of tumors to radiotherapy and chemotherapy, as shown in Supplementary Table S1 and Supplementary Table S2.
3.1.1 Regulation of cancer chemotherapy resistance
There are numerous drugs that can treat multiple cancers, such as cisplatin (CDDP), 5-fluorouracil (5-FU), and doxorubicin (DOX). Correspondingly, many tumors have developed resistance to these spectral anticancer drugs. Moreover, some small RNAs have been reported to be associated with tolerance to these drugs. For example, researchers have demonstrated a significant decrease in miR-363, which leads to the release of its target gene myeloid cell leukemia 1 (Mcl-1) and enhances tumor resistance to CDDP (Ou et al., 2015). MiR-769-5p is upregulated in drug-resistant cancer (GC) cells and can target caspase-9 (CASP9) inactivation through exosomes, mediate the direct degradation of the p53 protein via the E3 ubiquitin ligase 4-like protein (NEDD4L), and ultimately induce resistance in CDDP-sensitive receptor cells (Jing et al., 2022). High expression of miR-203 targets DJ-1 mRNA and inhibit its expression, thereby negatively regulating the expression of chromosome ten (PTEN) and affecting the activity of the PTEN-phosphatidylinositol-3 kinase (PI3K)/protein kinase (AKT) pathway to influence proliferation, apoptosis, and drug resistance in pancreatic cancer cells (Du et al., 2019). In 5-FU-resistant tumors, miR-144 is significantly downregulated, and its upregulation may reverse the acquisition of 5-FU resistance by inhibiting the nuclear factor-erythroid 2-related factor 2 (Nrf2)-dependent pathway (Zhou et al., 2016). miR-567 is also significantly downregulated in cancer cells, and its upregulation can reduce drug resistance through the phosphoinositol-3-kinase adaptor protein 1 (PIK3AP1)–PI3K/AKT–c-MYC (MYC) feedback loop (Zhang F. et al., 2019). For Dox-resistant tumors, the overexpression of miR-27a-5p reportedly enhances the drug sensitivity of tumor cells through sarcoma (RAS)/mitogen-activated protein kinase (MEK)/c-FOS (FOS) and PTEN/AKT/SMAD family member 1 (SMAD1) pathway dependence (He et al., 2021).
3.1.2 Regulation in cancer radiotherapy resistance
Some miRNAs have been shown to be associated with tumor radiation resistance in various cancers and can affect treatment efficacy through different molecular pathways. miR-96-5p was found to promote the resistance of head and neck squamous cell carcinoma (HNSCC) to radiation therapy by inhibiting the expression of PTEN (Vahabi et al., 2019). miR-4443 is significantly upregulated in radiation-resistant esophageal squamous cell carcinoma (ESCC) cells and can promote cell radioresistance by targeting the protein tyrosine phosphatase receptor type PTPRJ (Shi et al., 2022). In hepatocellular carcinoma (HCC), the downregulation of miR-621 leads to an upsurge in SET Domain, Bifurcated 1 (SETDB1) activity, thereby augmenting gene silencing that impacts growth and DNA damage responses (Shao et al., 2019). Conversely, overexpressing miR-621 targets SETDB1, diminishing its functionality and resulting in increased expression of genes pivotal for DNA repair, apoptosis, and cell cycle regulation (Shao et al., 2019). Yue Yuan et al. reported that overexpression of miR-410 can enhance radioresistance in non-small cell lung cancer (NSCLC) by targeting the PTEN/PI3K/mTOR axis, suggesting that miR-410 is a therapeutic target for cancer (NSCLC) radiotherapy (Yuan et al., 2020). It was reported that overexpression of miR-450a-5p enhanced radiosensitivity by targeting bispecific phosphatase 10 (DUSP10) in ESCC (Dai et al., 2020). DUSP10 is associated with negative regulation of the mitogen activated protein kinase (MAPK) signaling pathway which plays a crucial role in cellular processes such as proliferation, differentiation, and response to DNA damage (Dai et al., 2020). That is to say, miR-450a-5p may enhance the activation of the MAPK pathway by downregulating DUSP10, thereby increasing sensitivity to radiotherapy by promoting apoptosis and inhibiting cell proliferation to cope with DNA damage, suggesting that miR-450a-5p is a potential radiosensitizer.
3.2 Regulation of lncRNAs in tumor therapy resistance
Unlike miRNAs, lncRNAs are a class of ncRNAs with lengths exceeding 200 bp that interact with DNA, RNA, and proteins to regulate gene expression (Xing et al., 2021; Tan et al., 2021). They also play important roles in tumor chemotherapy and radiation tolerance (McCabe and Rasmussen, 2021), as shown in Supplementary Table S1 and Supplementary Table S2.
3.2.1 Regulation in cancer chemotherapy resistance
Like miRNAs, lncRNAs have also been reported to be involved in the resistance of various tumors to spectral antitumor drugs, such as the cisplatin lncRNA SNHG1, which is significantly upregulated in breast cancer (BC) cells, and its silencing enhances the sensitivity of tumor cells to CDDP (Zhang M. et al., 2021). The lncRNA LINC-PINT is downregulated in CDDP-resistant GC cells, and its overexpression can enhance cell drug sensitivity by regulating the enhancer of zeste homolog 2 (EZH2)/autophagy-related gene 5 (ATG5) axis (Zhang C. et al., 2022). LINC00680 can activate AKT3 by absorbing miR-568 to reduce tumor cell resistance to 5-FU, which may lead to potential therapeutic targets (Shu et al., 2021). The lncRNA OVAAL could promote 5-FU resistance in GC cells by promoting pyrimidine biosynthesis (Tan et al., 2022). The lncRNA PDIA3P1, which is a component of the homolog of mRNA transport mutant 4 (hMTR4)-protein disulfide isomerase family A member three pseudogene 1 (PDIA3P1)/miR125/124–tumor necrosis factor receptor-associated factor 6 (TRAF6) axis, was upregulated in HCC cells treated with Dox (Xie et al., 2020). In addition, our previous research revealed that the lncRNA D63785 was highly expressed in GC tissues and cells and that its inhibition could increase the sensitivity of GC cells to Dox by upregulating the expression of miR-422a and inhibiting myocyte enhancer factor 2D (MEF2D) (Zhou et al., 2018).
3.2.2 Regulation in cancer radiotherapy resistance
Ulvi Ahmadov et al. reported that the lncRNA HOTAIRM1 was highly expressed in glioblastoma cells. HOTAIRM1 can reduce radiosensitivity by regulating mitochondrial function and ROS levels in glioblastoma cells (Ahmadov et al., 2021). Hongfang Zhang et al. reported that the lncRNA DNM3OS was upregulated in esophageal cancer cells and could confer radioresistance by regulating the DNA damage response (Zhang H. et al., 2019). In ESCC cells, lncTUG1 expression was upregulated and enhanced cell radiation tolerance by reducing miR-144-3p levels and regulating the mesenchymal–epithelial transition factor (MET)/epidermal growth factor receptor (EGFR)/AKT axis (Wang et al., 2020). Wang Zhiyu et al. indicated that the lncRNA HNF1A–AS1 was upregulated in NSCLC cells. The overexpression of HNF1A–AS1 could significantly enhance radioresistance by regulating the expression of miR-92a-3p (Wang et al., 2021a). In addition, LINC00518 was reported to be significantly upregulated in melanoma and to target the miR-33a-3p/hypoxia-inducible factor 1 (HIF-1α) negative feedback loop to enhance the radioresistance of cells (Liu Y. et al., 2021).
3.3 Regulation of circRNAs in tumor therapy resistance
CircRNAs are also a large class of ncRNAs approximately 100 nt in length. These RNAs have a covalent closed loop structure but do not have a 5′cap or 3′poly-A tail (Kristensen et al., 2022; Kristensen et al., 2019). They also play important roles in gene expression at the transcriptional and posttranscriptional levels by mainly acting as miRNA sponges and protein scaffolds (Zhou et al., 2021). CircRNAs are the third largest class of endogenous ncRNAs and, like miRNAs and lncRNAs, are associated with tumor treatment tolerance.
3.3.1 Regulation in cancer chemotherapy resistance
CircMRPS35 was highly expressed in HCC cells, and its encoded peptide (circMRPS35-168aa) promoted the resistance of tumor cells to CDDP (Li P. et al., 2022). CircUBAP2 was also significantly upregulated in triple-negative breast cancer (TNBC) cells, but it could reduce the resistance of tumor cells to CDDP by regulating the miR-300/anti-silencing function 1B (ASF1B) axis (Wang L. et al., 2022). In GC cells and tissues resistant to 5-FU, the expression of circCPM was significantly upregulated, which can play a role in 5-FU resistance by targeting the protein kinase AMP-activated α two catalytic subunit (PRKAA2) (Fang et al., 2022). For Dox resistance, circle-0003998 was highly expressed in HCC cells, but this resistance could be reduced by regulating the miR-218-5p/eukaryotic initiation factor-5A2 (EIF5A2) axis (Li X. et al., 2020). CircCUL2 was significantly downregulated in GC cells and tissues, and its overexpression could inhibit Dox resistance in tumor cells through miR-142-3p/Rho-associated coiled-coil-containing protein kinase (ROCK)-mediated autophagy activation (Peng et al., 2020).
3.3.2 Regulation in cancer radiotherapy resistance
It has been reported that circRNA_100367 is downregulated in ESCC cells and can reduce radioresistance via the miR-217/Wnt3 pathway (Liu J. et al., 2021). Wu Ping et al. indicated that the expression of the circRNA circux1 was prominently enhanced in hypopharyngeal squamous cell carcinoma (HPSCC) cells and that this circRNA could confer radioresistance to HPSCC through the caspase-1 pathway (Wu et al., 2021). Zhang Guifeng et al. reported that circ-ACAP2 was upregulated in colorectal cancer (CRC) cells and promoted radioresistance via the miR-143-3p/frizzled-4 (FZD4) axis (Zhang G. et al., 2021). CircMTDH.4 was highly expressed in NSCLC cells and can enhance radioresistance through the miR-630/astrocyte elevated gene-1 (AEG-1) axis (Li Y. H. et al., 2020). Moreover, He Yunlong et al. reported that the expression of circVRK1 decreased in ESCC cells and that its overexpression improved tumor radiosensitivity by regulating the miR-624-3p/PTEN/PI3K/AKT signaling pathway (He et al., 2019).
3.4 Regulation of other ncRNAs in tumor therapy resistance
In addition to the three classic ncRNAs mentioned above, several other ncRNAs are involved in tumor treatment tolerance, including Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and small interfering RNAs (siRNAs), as shown in Supplementary Table S1 and Supplementary Table S2. PiRNAs are a class of ncRNAs with lengths of 24–30 nt that are involved in maintaining the stability of the germline cell genome (Liu et al., 2019). According to previous reports, pi-R17560 is highly expressed in BC cells and may lead to chemotherapy resistance through fat mass- and obesity-associated protein (FTO)-mediated N6-methyladenosine (m6A) demethylation (Ou et al., 2022). Basudeb Das et al. reported that piR-39980 was downregulated in fibrosarcoma cells, where it induced Dox resistance by regulating drug accumulation and DNA repair (Das et al., 2021). SnoRNAs are small RNAs with lengths of 60–300 nucleotides that are present mainly in the nucleolus. They are considered important components of the cellular protein synthesis mechanism (Williams and Farzaneh, 2012). Liu Yonghui et al. indicated that SNORD1C was upregulated in colorectal cancer cells and maintained resistance to 5-FU by activating the Wnt signaling pathway (Liu et al., 2022c). Martina Godel et al. reported that SNORD3A was highly expressed in BC cells and could enhance 5-FU sensitivity in cells by negatively regulating miR-185-5p (Godel et al., 2020). SiRNAs are long double-stranded RNAs 21–23 nucleotides in length that play a role at the posttranscriptional level, especially in RNA interference (RNAi) (Alshaer et al., 2021). It was reported that siRNA targeting glucose transporter protein 1 (GLUT-1) could reduce the radioresistance of laryngeal cancer cells by helping to redistribute the cell cycle phases, reducing DNA repair ability and enhancing apoptosis (Zhong et al., 2019). Moreover, Li Qingjian et al. developed an intracellular pH-responsive nanoparticle (NP) that can transport siRNA and cisplatin targeting Rac-1 into cells to reduce cancer resistance to neoadjuvant chemotherapy (NAC) (Yu et al., 2020).
4 Regulatory roles of DDR-related ncRNAs in cancer therapy resistance
As mentioned above, numerous quantities and types of ncRNAs have been found to mediate or regulate the tolerance of cancer cells to radiotherapy and chemotherapy by targeting multiple genes or signaling pathways. Interestingly, several ncRNAs have recently been found to regulate tumor treatment tolerance by altering the DDR pathway, indicating that changes in DDR are crucial for the occurrence and development of tumor resistance, as shown in Table 1. These findings suggest that ncRNAs can also serve as regulatory factors for the DDR to reverse the tolerance of tumor cells, further supporting their potential value as tumor treatment targets.
4.1 Regulation of DDR-related miRNAs in cancer therapy resistance
4.1.1 DDR-related miRNAs in cancer chemotherapy resistance
As shown in Figure 3, DDR-related miRNAs can strongly influence cancer chemotherapy tolerance. Tan W et al. reported that the upregulation of miRNA-146a increased resistance to chemotherapy (CDDP) by targeting the degradation of DNA damage-inducible transcript 3 (DDIT3) in lung cancer cells (Maekawa et al., 2018). It has long been known that the upregulation of miRNA-146a is closely related to the occurrence of lung cancer (Williams et al., 2008). Recently, it was further discovered that miRNA-146a can alter the chemotherapy sensitivity of cancer cells by downregulating C/EBP homologous protein (CHOP), leading to a decreased expression levels of light chain 3-II (LC3-II), death receptor 5 (DR5), and tribble homolog 3 (TRB3), further affecting the progression of lung cancer (Li et al., 2014). Xu S et al. reported that miR-33b-3p facilitates the DDR against cisplatin treatment in lung cancer cells by targeting p21WAF1/CIP1. Furthermore, they reported that ectopic expression of miR-33b-3p increased the expression of excision repair cross-complementation group 1 (ERCC1), which is crucial to the NER pathway for cisplatin-DDR (Xu et al., 2016). Carotenuto P et al. reported that an increase in the expression of MIR1307 can induce a reduction in chloride intracellular channel 5 (CLIC5) expression, resulting in protection against DNA damage caused by FOLFIRINOX (a combination of the chemical drugs fluorouracil, oxaliplatin, and irinotecan-FOI), ultimately leading to FOLFIRINOX resistance (Carotenuto et al., 2021). Zhou Y et al. reported that through inhibiting ATM kinase, miR-203 causes oxaliplatin resistance in CRC. During this process, cancer cells gain an improved capacity to repair damaged DNA or to decrease the activation of the DNA damage response system to prevent apoptosis (Zhou et al., 2014). Wang T et al. noted that miR-211 overexpression can promote platinum sensitivity in ovarian cancer by inhibiting the expression of the TDP1 gene and certain DDR genes, thereby affecting the prognosis of the disease (Wang et al., 2019). Lung RW et al. reported that Epstein–Barr virus (EBV)-encoded miRNAs (miR-BARTs, especially miR-BART17-5p and miR-BART19-3p) can reduce DDR ability by targeting BRCA1, increasing the sensitivity of nasopharyngeal carcinoma cells to cisplatin and doxorubicin (Lung et al., 2020).
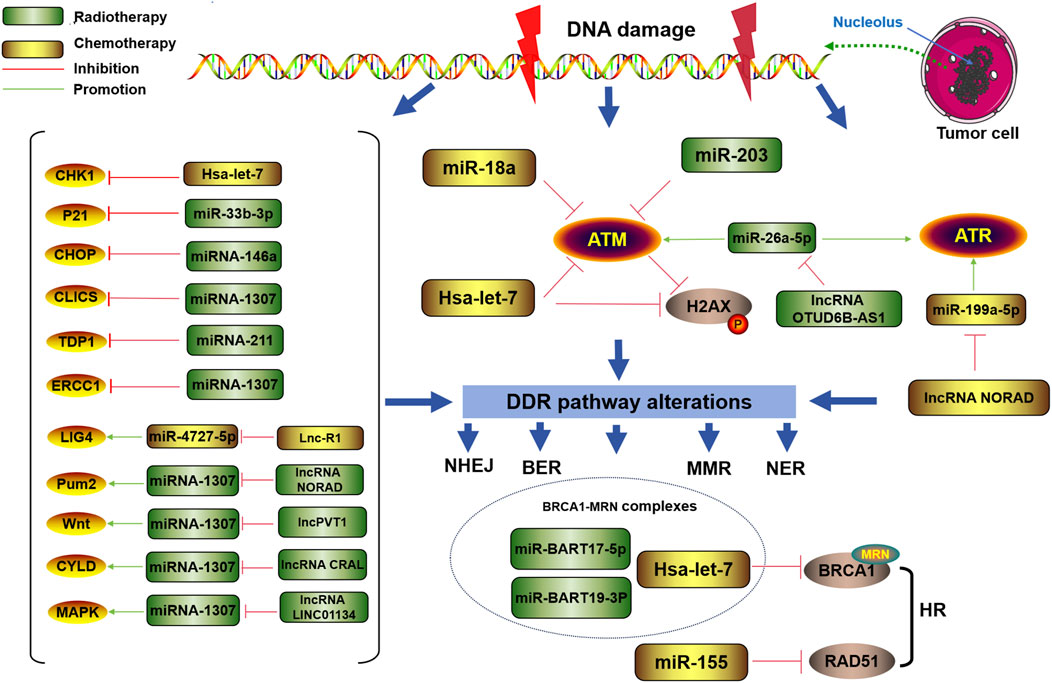
Figure 3. Regulatory role of DDR-related miRNAs in tumor chemotherapy or radiotherapy resistance DDR-related miRNAs can mediate changes in the DDR pathway to regulate radiation or chemotherapy resistance in tumor cells.
In addition, other types of ncRNAs can target miRNAs and regulate the chemotherapeutic tolerance of tumor cells via the DDR. Li PP et al. reported that overexpression of the lncRNA OTUD6B-AS1 increased paclitaxel resistance in triple-negative breast cancer patients through the downregulation of miR-26a-5p and the overexpression of metadherin (MTDH). The OTUD6B-AS1/miR-26a-5p/MTDH axis can inhibit DDR by blocking the phosphorylation and activation of RAD51, ATR, and ATM (Li et al., 2021). Fletcher CE et al. reported that the genomic protective lncRNA NORAD interacts with miR-346, blocking the association between pumilio RNA-binding family member 2 (Pum2) and miR-346 and increasing the turnover of DDR transcripts, thereby inducing the tolerance of prostate cancer to PARPis (Fletcher et al., 2022). The competitive binding of lncPVT1 with miR-619-5p regulates the target genes Pygopus 2 (PYGO2) and autophagy-related gene 14 (ATG14) of miR-619-5p to activate the Wnt/β-catenin pathway, increasing the chemoresistance of pancreatic cancer to gemcitabine (Zhou et al., 2020). The lncRNA CRAL has also been reported to enhance cisplatin resistance in GC cells by absorbing miR-505 and upregulating CYLD lysine 63 deubiquitinase (CYLD) expression (Chi et al., 2023). Interestingly, the lncRNA LINC01134 can promote the activation of miR-342-3p and insulin-like growth factor 2 (IGF2) mRNA-binding protein 2 (IGF2BP2), thereby activating the MAPK pathway to promote the repair of DNA damage and increase chemotherapy resistance in HCC (Wang et al., 2021b).
4.1.2 Regulation of DDR-related miRNAs in cancer radioresistance
DDR-related miRNAs play crucial roles in modulating cancer cell response to radiotherapy, influencing therapeutic efficacy through various molecular mechanisms (Figure 1). According to Gasparini et al., in triple-negative breast cancer cells with high expression of miR-155, homologous recombination and subsequent DNA damage repair after radiation therapy are reduced, increasing sensitivity to radiation. Furthermore, miR-155 can inhibit the recombinant enzyme RAD51 in cancer to control DNA repair activity and γ-radiation (IR) sensitivity (Gasparini et al., 2014). Hu H et al. reported that the miRNA Hsa let-7g indirectly inhibited DDR-related genes and increased the sensitivity of GC to treatment-induced oxidative stress (Hu et al., 2015). Song L et al. reported that ectopic expression of miR-18a inhibited IR-induced DDR and reduced the frequency of HRR, thereby increasing the radiosensitivity of breast cancer cells (Song et al., 2011).
In addition, miRNAs can also be targeted and regulated by lncRNAs to alter the radioresistance of tumor cells via the DDR. For example, the lncRNA NORAD is involved in inhibiting the expression of miR-199a-5p and the ATR/Chk1 pathway, ultimately weakening the effect of anti-PD-1 inhibitors and increasing radiation resistance in ESCC (Sun et al., 2021). Lnc-RI can competitively bind miR-4727-5p to promote the expression of DNA ligase IV (LIG4), thereby enhancing the NHEJ pathway or inducing decreased tumor cell apoptosis, leading to reduced radiation sensitivity in CRC cells (Li X. et al., 2020).
4.2 Regulation of DDR-related lncRNAs in cancer therapy resistance
Research has shown that abnormal expression of lncRNAs in cancer cells can reduce sensitivity to chemotherapy or radiotherapy by activating or inhibiting the DDR pathway. Chemotherapy drugs and radiation-mediated DNA damage can also induce changes in the expression of lncRNAs in tumor cells, thus regulating the DDR pathway and affecting cell tolerance, as shown in Figure 4. These data indicate that the overexpression or inhibition of these DDR-related lncRNAs can be an effective means of cancer treatment.
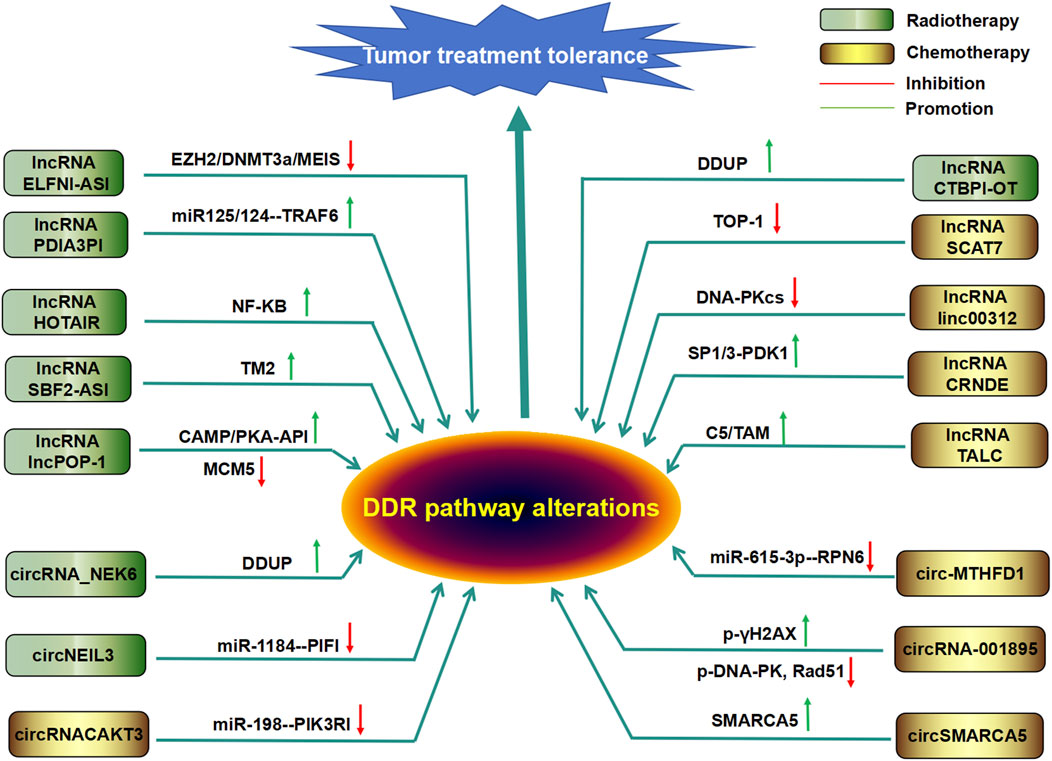
Figure 4. Regulatory roles of DDR-related lncRNAs and circRNAs in tumor chemotherapy or radiotherapy resistance Many DDR-related lncRNAs and circRNAs mediate or enhance tumor radiotherapy or chemotherapy.
4.2.1 DDR-related lncRNAs in cancer chemotherapy tolerance
According to reports, the integration site 1 (MEIS1) of neutrophil eosinophilic leukemia can damage the vitality of mouse CRC cells and tumor growth and enhance the sensitivity of CRC cells to oxaliplatin by preventing DNA damage repair (Zhang M. et al., 2021). However, in CRC, MEIS1 expression is significantly reduced and inversely proportional to patient survival. Further research has shown that lncRNA ELFN1–AS1 is localized in the promoter of MEIS1 by interacting with EZH2–DNA methyltransferase 3A (DNMT3A) to inhibit MEIS1 translation (Li Y. et al., 2022), suggesting that ELFN1 antisense RNA 1 (ELFN1-AS1) induces sensitivity to oxaliplatin in colon cancer cells through the EZH2/DNMT3a/MEIS1 axis. Upregulation of the lncRNA HOTAIR can induce nuclear factor (NF)-κB in ovarian cancer. Its prolonged activation or expression causes cells to enter a sustained state of maintaining the DDR to increase tumor cell resistance to CDDP chemotherapy (Qian et al., 2020). Similarly, lnc-SBF2-AS1 is highly expressed in glioblastoma and can enhance cell damage repair to DSBs, leading to tumor cells being chemically resistant to TMZ (Lal et al., 2023). In addition, some lncRNAs can bind to DDR-related molecules to regulate DDR in cancer. For example, the lncRNA LUCAT1 can bind to polypyrimidine tract-binding protein 1 (PTBP1), which is associated with DNA damage genes and contributes to chemotherapy tolerance in colorectal cancer cells (Huan et al., 2020). lncRNA-AL133467.2 inhibits DNA damage induced HCC cell apoptosis by interacting with zinc finger CCHC domain-containing protein 4 (ZCCHC4), thereby mediating tumor cell resistance to DNA damage agents (DDA) (Zhu et al., 2022). LncSLCO1C1 can interact with scaffold-based structure-specific recognition protein 1 (SSRP1)/H2A/H2B complexes and competitively bind with miR-211-5p and miR-204-5p to increase SSRP1 expression, enhance cell growth, prevent DNA damage, and induce chemotherapy drug tolerance in gastric cancer cells (Liu Y. et al., 2021). Of course, in addition to lncSLCO1C1, as mentioned above, there are also some lncRNAs that can mediate tumor cell tolerance to chemical drugs by regulating miRNAs and their target genes, including lncRNAOTUD6B-AS1, NORAD, lncPVT1, CRAL, and LINC01134 (Jackson and Bartek, 2009; Shao et al., 2019; Wang et al., 2020; Wang et al., 2021a; Wu et al., 2021).
The lncRNA CTBP1-DT is highly expressed in cancer cells, acts as an oncogene, and is further regulated by the cellular environment (Liu P. et al., 2021). Interestingly, camptothecin (CPT) was reported to not affect the expression of the lncRNA CTBP1-DT but can significantly induce the expression of the lncRNA CTBP1-DT, which encodes the microprotein DNA damage-upregulated protein (DDUP) (Yu et al., 2022). Furthermore, CPT-induced DNA damage-mediated phosphorylation of DDUP results in a drastic “dense to loose” conformational change in DDUP structure, maintaining the retention of radiation-sensitive 18 (RAD18) at the site of DNA damage, thereby promoting DNA damage repair through dependent RAD51C-mediated HRR and single ubiquitination proliferating cell nuclear antigen (PCNA)-mediated pattern recognition receptor (PRR) mechanisms (Yu et al., 2022). Interestingly, treatment with the ATR inhibitor berzoserib can significantly inhibit the formation of DDUP lesions, leading to the rapid elimination of RAD18-and PCNA-expressing lesions and ultimately to the hypersensitivity of ovarian cancer cells to DNA destructive chemotherapy drugs (Yu et al., 2022). The lncRNA lnc-POP1-1 is induced by vomeronal type 1 receptor 5 (VN1R5), a protein whose expression is significantly increased in cisplatin-resistant HNSCC cells and tissues, through the activation of specific protein 1 (Sp1) transcription factors via the cyclic AMP (cAMP)/protein kinase A (PKA) pathway. Additionally, lnc-POP1-1 directly binds to the microsomal maintenance deficient type 5 (MCM5) protein and slows the degradation of MCM5 by inhibiting its ubiquitination, thereby promoting the repair of DNA damage caused by cisplatin (Brauner et al., 2016). The lncRNA PDIA3P1 can be upregulated by silencing hMTR4, which assists tumors in preventing apoptosis caused by Dox (Xie et al., 2020). Mechanistically, PDIA3P1 combines with the miR-125/124 pathway to promote the expression of TRAF6, which can increase chemoresistance (Xie et al., 2020). In addition, the expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was increased in a manner dependent on nuclear factor-κB (NF-κB) and p53 in GBM cells treated with temozolomide (Kristensen et al., 2022). The depletion of MALAT1 significantly increased the cytotoxicity of GBM cells to temozolomide, while the in vivo application of anti-MALAT1 siRNA increased the efficacy of temozolomide in glioma xenograft mice (Kristensen et al., 2022).
4.2.2 DDR-related lncRNAs in cancer radiotherapy resistance
The lncRNA HOTAIR is highly expressed in cancer cells and promotes the expression of DDR factors, leading to radioresistance in breast cancer (Chen Y. et al., 2021). The DNA damage-induced lncRNA SCAT7 targets the degradation of topoisomerase I (TOP1), induces the activation and maintenance of DDR, and leads to chemotherapy resistance of lung adenocarcinoma to cisplatin and camptothecin (Statello et al., 2021). Cancer-associated fibroblasts (CAF) can enhance the synthesis of lncRNA dynamin three opposite strand (DNM3OS) in a platelet-derived growth factor-β (PDGFβ/platelet-derived growth factor receptor β(PDGFRβ)/Forkhead box transcription factor O1 (FOXO1) signaling pathway-dependent manner, leading to the DDR and increased radiosensitivity in ESCC (Zhang H. et al., 2019). The interaction between lncRNA RBM5-AS1 and SIRT6 can also induce the DDR to enhance radiation resistance in medulloblastoma (Zhu et al., 2021). lncRNA CRNDE has also been reported to participate in radiation resistance by modulating the SP1/3-phosphoinositide-dependent protein kinase 1 (PDK1) axis in hepatocellular carcinoma (Zhang G. et al., 2021). In contrast, overexpression of lncRNA linc00312 in nasopharyngeal carcinoma can inhibit the expression of DNA-dependent protein kinase catalytic subunit (DNA-PKcs), thereby reducing their binding to the Ku80 protein, inhibiting DNA damage repair, and reducing the radiation resistance of tumor cells (Guo et al., 2021).
In addition, the lncRNA TMZ-related lncRNA (lnc-TALC) in cases of GBM recurrence can be secreted into extracellular vesicles and transmitted to tumor-associated macrophages to promote the M2 polarization of microglia (Li Y. et al., 2022). The M2 polarization of microglia can induce the secretion of complement component C5/C5a, promote the phosphorylation of p38 MAPK and the repair of DNA damage induced by TMZ, and lead to chemotherapy resistance (Li Y. et al., 2022). This suggests that lnc-TALC transported by extracellular vesicles can reshape the GBM microenvironment and reduce the sensitivity of tumors to TMZ chemotherapy.
4.3 Regulation of cancer therapy resistance by DDR-related circRNAs
Compared to those of the other two types of ncRNAs, there are currently fewer circRNAs found in tumors related to chemotherapy or radiotherapy tolerance.
4.3.1 DDR-related circRNAs in cancer chemotherapy resistance
CircAKT3 (hsa_circ_0000199, a circRNA originating from exons 8, 9, 10, and 11 of the AKT3 gene) was upregulated in CDDP-resistant GC tissues and cells compared with CDDP-sensitive samples (Huang et al., 2019). Further in vivo and in vitro experiments have confirmed that circAKT3 promotes DNA damage repair and inhibits apoptosis by absorbing miR-198 to promote the expression of PI3 kinase p85 alpha (PIK3R1) in GC cells (Huang et al., 2019). Hsa_circ_0078297 (circ-MTHFD1L) was also significantly increased in pancreatic ductal adenocarcinoma (PDAC) tissues and cells (Chen Z. et al., 2022). Circ-MTHFD1L can also upregulate the expression of the ribophorin family 6 (RPN6), an endogenous miR-615-3p molecular sponge, thereby promoting the HR pathway for DNA damage repair and enhancing the chemotherapeutic resistance of tumor cells to gemcitabine. However, the combination of silent circ-MTHFD1L and olaparib can increase the sensitivity of cancer to gemcitabine (Chen Z. et al., 2022). Similarly, circRNA-001895 was significantly upregulated in sunitinib-resistant renal cell carcinoma (RCC) tissues and cells (Tan et al., 2023). Knockout of circRNA_001895 in sunitinib-resistant RCC cells inhibited cell proliferation and promoted cell apoptosis by inducing the phosphorylation of the histone H2AX (γH2AX) and reducing phosphorylation of DNA-dependent protein kinase (p-DNA-PK) and Rad51 (Tan et al., 2023). In addition, in contrast to that in the host gene SMARCA5 (an ATPase of the ISWI class of chromatin remodelers), the expression level of circSMARCA5 in breast cancer tissue is significantly reduced, and circSMARCA5 can bind to its parent locus to form an R ring, which leads to the suspension of the transcription of exon 15 of SMARCA5 (Dogan et al., 2023). Overexpression of circSMARCA5 can induce drug sensitivity in breast cancer cell lines by inhibiting the expression of SMARCA5 and the production of truncated nonfunctional proteins (Dogan et al., 2023). These data suggest that circRNAs may serve as therapeutic targets for treating drug resistance.
4.3.2 DDR-related circRNAs in cancer radioresistance
CircNEIL3 is a significantly downregulated circRNA in lung adenocarcinoma (LUAD) cells treated with 0, 2, or 4 Gy of radiation (Zhang T. et al., 2022). Overexpression of circNEIL3 can significantly inhibit radiation-induced cell ptosis by regulating the miR-1184/phytochrome-interacting factor 1 (PIF1) axis and triggering the activation of absent in melanoma 2 (AIM2) inflammasomes, inducing tumor cell tolerance to radiation therapy (Zhang T. et al., 2022). The expression of circ-NEK6 was significantly increased in iodine-131 (131I)-resistant differentiated thyroid cancer (DTC) tissues and cell lines (Chen F. et al., 2021). Silencing of circ-NEK6 in iodine 131I-resistant cells inhibited cell proliferation, migration, and invasion while inducing cell apoptosis and DNA damage by regulating the miR-370-3p/nonmuscle myosin heavy chain IIA (MYH9) axis (Yu et al., 2022). These results indicate that CircNEIL3 and circ_NIMA-related kinase-6 (NEK6) may be potential biomarkers and therapeutic targets for cancer patients with radiation resistance.
4.4 Other DDR-related ncRNAs regulate cancer therapy resistance
There are also some other types of DDR-related ncRNAs involved in tumor chemotherapy drug tolerance or radiotherapy tolerance, including piRNAs, snoRNAs, and siRNAs (Table 1). For example, the overexpression of piR-39980 not only significantly inhibits the proliferation, migration, ROS generation, and colony formation of tongue squamous cell carcinoma (TSCC) cells but also inhibits the expression of farnesyl-diphosphate farnesyltransferase 1 (FDFT1). Moreover, the inhibition of FDFT1 induces hypoxia, which slows DNA repair and the accumulation of damaged DNA, leading to tumor cell death (Chattopadhyay and Mallick, 2023). piR-39980 can decrease fibrosarcoma chemoresistance to DOX by regulating ribonucleotide reductase subunit M2 (RRM2) and cytochrome P450 enzyme 1A2 (CYP1A2), indicating that piRNAs can bind to DOX for cancer treatment (Das et al., 2021). A group of upregulated snoRNAs, particularly SNORD3A, SNORA13, and SNORA28, have been detected in human anti-Dox osteosarcoma cells (Godel et al., 2020). Overexpression of these three snoRNAs can significantly reduce the cytotoxicity of Dox, induce upregulation of growth arrest and DNA damage protein 45A (GADD45A) and MYC, and downregulate type II topoisomerase A (TOP2A). These findings suggest that snoRNAs induce tumor cell resistance to Dox by regulating the expression of genes involved in DNA damage perception, DNA repair, ribosomal biogenesis, and proliferation (Godel et al., 2020). Ribosomal RNA processing 9 (RRP9) is an evolutionarily conserved U3-snoRNP protein that is crucial for early preribosomal RNA (rRNA) processing and cleavage and is expressed at higher levels in pancreatic cancer tissues than in normal tissues (Zhang Z. et al., 2022). RRP9 has been shown to activate the AKT signaling pathway by interacting with the deoxyribonucleic acid-binding region of IGF2 mRNA-binding protein 1 (IGF2BP1) in vivo and in vitro, thereby promoting the resistance of prostate cancer cells to gemcitabine (Zhang Z. et al., 2022). Hypoxia radiation-sensitive nanoparticles (RDPP (Met)/TMZ/siMGMT) that can effectively release TMZ and small interfering O6 methylguanine DNA methyltransferase RNA (siMGMT) have been reported to effectively penetrate the blood‒brain barrier, accurately target glioma cells, and inhibit cell proliferation (Xie et al., 2022). These findings suggest that RDPP (Met)/TMZ/siMGMT can effectively improve the chemotherapeutic efficacy and radiation sensitivity of temozolomide in glioma (Xie et al., 2022). In addition, siRNA-mediated ATR silencing promoted the sensitivity of cancer cells to methyl methanesulfonate (MMS) (Collis et al., 2003), further supporting the enormous potential of using siRNAs in tumor treatment.
5 Conclusions, perspectives and discussions
In summary, many recent studies have shown that ncRNAs (mainly miRNAs, lncRNAs and circRNAs) play important regulatory roles in tumor chemotherapy and radiation tolerance by targeting changes in the DDR. These ncRNAs either activate or maintain the DDR to induce therapeutic tolerance in tumor cells or block the DDR to induce cell apoptosis and enhance the cytotoxicity of chemotherapy drugs or radiotherapy. Cancer has become the second leading cause of death worldwide due to its rapid metastasis, spread, recurrence, and other factors (Xu et al., 2016). Therefore, cancer is the main focus of ongoing worldwide research in biomedicine. Radiotherapy and chemotherapy are important methods for treating malignant tumors, and tumor tolerance is a key factor influencing the effectiveness of tumor treatment. Recent observations based on increasing evidence suggest that ncRNAs may affect DNA damage repair and play important regulatory roles in tumor radiotherapy and chemotherapy tolerance (Volpato et al., 2007). Moreover, DDR-related ncRNAs may be targets for estimating cancer treatment responses, thus contributing to the development of treatment designs targeting unique individuals. Many ncRNAs have been proven to be important molecules that affect the occurrence and development of tumors. Indeed, with the deepening of clinical research, a new ncRNA scoring system has emerged that combines PD-L1 expression, tumor change load, and cytotoxic T lymphocyte (CTL) infiltration for precise tumor immunotherapy (Zhong et al., 2019). In addition, a new biomarker, extracellular ncRNAs, has emerged for clinical diagnosis. As a liquid biopsy method, these extracellular ncRNAs can be used to predict and diagnose the recovery of physiological and pathological states after treatment with platinum-containing neoadjuvant chemotherapy (Thomsen and Vitetta, 2018). This is partly due to their stability in human body fluids and peripheral blood, as well as their specific expression in diseases. For example, certain miRNAs (such as miR-136 and upregulated miR-27b) exhibit differential expression between healthy subjects and oral cancer patients (Williams et al., 2008; Baraniskin and Schroers, 2021). Similarly, the high expression levels of many lncRNAs, such as LINC00665, can distinguish between diseased breast cells/cancer tissues and normal tissues, indicating the potential diagnostic value of lncRNAs in tumors (Godel et al., 2020). In addition, due to the positive correlations between the expression of certain ncRNAs and clinical stage, metastasis, and patient survival, ncRNAs can be considered prognostic molecular markers for oral cancer and other cancers (Crooke et al., 2018). Therefore, the DDR-related ncRNAs mentioned in this article may be ideal biomarkers for cancer prognosis and recurrence diagnosis.
Translating miRNA-based strategies into clinical applications for cancer treatment presents significant challenges that need to be addressed. These challenges include effectively delivering miRNAs to target cells, minimizing off-target effects, managing immune responses, optimizing dosing, and conducting rigorous clinical validation. However, along with these challenges come promising opportunities. miRNA-based therapies have the potential for precision medicine by allowing tailored treatments based on individual tumor profiles. They can also be integrated with existing therapies to enhance efficacy and overcome resistance. Additionally, certain miRNAs can serve as biomarkers for diagnosing and predicting prognosis in cancer patients. The diverse regulatory roles of miRNAs in oncogenic pathways provide numerous therapeutic targets. Advancements in delivery technologies such as nanoparticles and viral vectors are improving the specificity and efficiency of miRNA delivery. Furthermore, evolving regulatory frameworks support the translation of ncRNA-based therapeutics into clinical practice. Addressing these challenges while capitalizing on these opportunities is crucial for realizing the transformative potential of miRNA-based strategies in improving cancer treatment outcomes.
Moreover, with the development of biotechnology, such as high-throughput sequencing, functional research on ncRNAs related to the DDR could provide new prospects for cancer treatment. Numerous studies have elucidated the relationship between DDR-related ncRNAs and tumor treatment tolerance. Therefore, DDR-related ncRNAs are currently among the most interesting topics for researchers in anticancer therapy, with the goal of thoroughly understanding the pathways that regulate and control their molecular mechanisms. A large amount of evidence suggests that tumor cells typically exhibit altered DNA damage repair ability, and the underlying mechanisms are being increasingly studied. However, the mechanisms of action of these DDR-related ncRNAs involve multiple complex factors, and the current review clarifies only the tip of the iceberg by combining previous literature and research results. However, further studies are needed to determine the optimal detection methods for ncRNAs related to the DDR in patients, their diagnostic value for cancer recurrence, and the development of personalized therapies. In addition, further research is needed to evaluate the effectiveness of DDR-related ncRNA drugs in clinical settings. Overall, a comprehensive understanding of the important role of ncRNAs in tumor treatment tolerance will be beneficial for improving the rational use of drugs and the development of new drugs.
Author contributions
ZG: Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis. XLu: Conceptualization, Data curation, Formal Analysis, Writing–original draft, Writing–review and editing. XW: Data curation, Formal Analysis, Writing–original draft. TH: Data curation, Formal Analysis, Writing–original draft. XLi: Data curation, Formal Analysis, Writing–original draft. ZL: Data curation, Formal Analysis, Writing–original draft. PL: Funding acquisition, Investigation, Supervision, Validation, Visualization, Writing–review and editing. ZZ: Funding acquisition, Investigation, Supervision, Validation, Visualization, Writing–review and editing, Writing–original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Natural Science Foundation of Shandong Province of China (ZR2020MH250), and the National Natural Science Foundation of China (92249303 and 81502063).
Acknowledgments
This study was supported by the Natural Science Foundation of Shandong Province of China (ZR2020MH250), and the National Natural Science Foundation of China (92249303 and 81502063).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1390300/full#supplementary-material
References
Ahmadov, U., Picard, D., Bartl, J., Silginer, M., Trajkovic-Arsic, M., Qin, N., et al. (2021). The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 12, 885. doi:10.1038/s41419-021-04146-0
Alshaer, W., Zureigat, H., Al Karaki, A., Al-Kadash, A., Gharaibeh, L., Hatmal, M. M., et al. (2021). siRNA: mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 905, 174178. doi:10.1016/j.ejphar.2021.174178
Ao, X., Ding, W., Li, X., Xu, Q., Chen, X., Zhou, X., et al. (2023). Non-coding RNAs regulating mitochondrial function in cardiovascular diseases. J. Mol. Med. Berlin, Ger. 101, 501–526. doi:10.1007/s00109-023-02305-8
Ashwell, S., and Zabludoff, S. (2008). DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin. Cancer Res. 14, 4032–4037. doi:10.1158/1078-0432.ccr-07-5138
Bacová, G., Hunáková, L. E., Chorváth, M., Boljesíková, E., Chorváth, B., Sedlák, J., et al. (2000). Radiation-induced DNA damage and repair evaluated with 'comet assay' in human ovarian carcinoma cell lines with different radiosensitivities. Neoplasma 47, 367–374.
Bakhoum, S. F., and Landau, D. A. (2017). Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 7, a029611. doi:10.1101/cshperspect.a029611
Baraniskin, A., and Schroers, R. (2021). Liquid biopsy and other non-invasive diagnostic measures in PCNSL. Cancers (Basel) 13, 2665. doi:10.3390/cancers13112665
Blackford, A. N., and Jackson, S. P. (2017). ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817. doi:10.1016/j.molcel.2017.05.015
Brauner, A., Fridman, O., Gefen, O., and Balaban, N. Q. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330. doi:10.1038/nrmicro.2016.34
Butkiewicz, D., Rusin, M., Pawlas, M., Czarny, M., and Chorazy, M. (2002). Repair of DNA damage using nucleotide excision repair (NER)--relationship with cancer risk. Postepy Hig. i Med. doswiadczalnej 56, 485–498.
Carotenuto, P., Amato, F., Lampis, A., Rae, C., Hedayat, S., Previdi, M. C., et al. (2021). Modulation of pancreatic cancer cell sensitivity to FOLFIRINOX through microRNA-mediated regulation of DNA damage. Nat. Commun. 12, 6738. doi:10.1038/s41467-021-27099-6
Chattopadhyay, T., and Mallick, B. (2023). FDFT1 repression by piR-39980 prevents oncogenesis by regulating proliferation and apoptosis through hypoxia in tongue squamous cell carcinoma. Life Sci. 329, 121954. doi:10.1016/j.lfs.2023.121954
Chen, B., Dragomir, M. P., Yang, C., Li, Q., Horst, D., and Calin, G. A. (2022a). Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target Ther. 7, 121. doi:10.1038/s41392-022-00975-3
Chen, F., Yin, S., Feng, Z., Liu, C., Lv, J., Chen, Y., et al. (2021b). Knockdown of circ_NEK6 decreased 131I resistance of differentiated thyroid carcinoma via regulating miR-370-3p/MYH9 Axis. Technol. cancer Res. and Treat. 20, 15330338211004950. doi:10.1177/15330338211004950
Chen, Y., Li, Z., Chen, X., and Zhang, S. (2021a). Long non-coding RNAs: from disease code to drug role. Acta Pharm. Sin. B 11, 340–354. doi:10.1016/j.apsb.2020.10.001
Chen, Z., Hu, J. F., Wang, Z. W., Liao, C. Y., Kang, F. P., Lin, C. F., et al. (2022b). Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma. J. Exp. and Clin. cancer Res. CR 41, 153. doi:10.1186/s13046-022-02343-z
Cheng, B., Pan, W., Xing, Y., Xiao, Y., Chen, J., and Xu, Z. (2022). Recent advances in DDR (DNA damage response) inhibitors for cancer therapy. Eur. J. Med. Chem. 230, 114109. doi:10.1016/j.ejmech.2022.114109
Chi, X., Chen, Z., Chen, Y., Hong, H., Yu, J., and Lv, L. (2023). Upregulation of lncRNA PTOV1-AS1 in hepatocellular carcinoma contributes to disease progression and sorafenib resistance through regulating miR-505. J. Biochem. Mol. Toxicol. 37, e23437. doi:10.1002/jbt.23437
Choi, M., Kipps, T., and Kurzrock, R. A. T. M. (2016). ATM mutations in cancer: therapeutic implications. Mol. cancer Ther. 15, 1781–1791. doi:10.1158/1535-7163.mct-15-0945
Collis, S., Swartz, M., Nelson, W., and DeWeese, T. (2003). Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res. 63, 1550–1554.
Crooke, S. T., Witztum, J. L., Bennett, C. F., and Baker, B. F. (2018). RNA-targeted therapeutics. Cell metab. 27, 714–739. doi:10.1016/j.cmet.2018.03.004
Curtin, N. J. (2013). Inhibiting the DNA damage response as a therapeutic manoeuvre in cancer. Br. J. Pharmacol. 169, 1745–1765. doi:10.1111/bph.12244
Dai, J., Su, Y., Zhong, S., Cong, L., Liu, B., Yang, J., et al. (2020). Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 5, 145. doi:10.1038/s41392-020-00261-0
Damia, G. (2020). Targeting DNA-PK in cancer. Mutat. Res. 821, 111692. doi:10.1016/j.mrfmmm.2020.111692
Das, B., Jain, N., and Mallick, B. (2021). piR-39980 mediates doxorubicin resistance in fibrosarcoma by regulating drug accumulation and DNA repair. Commun. Biol. 4, 1312. doi:10.1038/s42003-021-02844-1
Difilippantonio, M. J., Petersen, S., Chen, H. T., Johnson, R., Jasin, M., Kanaar, R., et al. (2002). Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196, 469–480. doi:10.1084/jem.20020851
Dogan, I., Aydin, E., Khanmammadov, N., Paksoy, N., Ferhatoğlu, F., Ak, N., et al. (2023). Long-term outcomes and predictors of recurrence in node-negative early stage breast cancer patients. J. cancer Res. Clin. Oncol. 149, 14833–14841. doi:10.1007/s00432-023-05276-y
Du, S., Xu, L. Y., Gao, P., Liu, Q. S., Lu, F. F., Mo, Z. H., et al. (2019). MiR-203 regulates DJ-1 expression and affects proliferation, apoptosis and DDP resistance of pancreatic cancer cells. Eur. Rev. Med. Pharmacol. Sci. 23, 8833–8840. doi:10.26355/eurrev_201910_19278
Eich, M., Roos, W. P., Nikolova, T., and Kaina, B. (2013). Contribution of ATM and ATR to the resistance of glioblastoma and malignant melanoma cells to the methylating anticancer drug temozolomide. Mol. cancer Ther. 12, 2529–2540. doi:10.1158/1535-7163.Mct-13-0136
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi:10.1038/nrg3074
Fanale, D., Castiglia, M., Bazan, V., and Russo, A. (2016). Involvement of non-coding RNAs in chemo- and radioresistance of colorectal cancer. Adv. Exp. Med. Biol. 937, 207–228. doi:10.1007/978-3-319-42059-2_11
Fang, L., Lv, J., Xuan, Z., Li, B., Li, Z., He, Z., et al. (2022). Circular CPM promotes chemoresistance of gastric cancer via activating PRKAA2-mediated autophagy. Clin. Transl. Med. 12, e708. doi:10.1002/ctm2.708
Fletcher, C. E., Deng, L., Orafidiya, F., Yuan, W., Lorentzen, M. P. G. S., Cyran, O. W., et al. (2022). A non-coding RNA balancing act: miR-346-induced DNA damage is limited by the long non-coding RNA NORAD in prostate cancer. Mol. cancer 21, 82. doi:10.1186/s12943-022-01540-w
Fok, J. H. L., Ramos-Montoya, A., Vazquez-Chantada, M., Wijnhoven, P. W. G., Follia, V., James, N., et al. (2019). AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 10, 5065. doi:10.1038/s41467-019-12836-9
Gasparini, P., Lovat, F., Fassan, M., Casadei, L., Cascione, L., Jacob, N. K., et al. (2014). Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. U. S. A. 111, 4536–4541. doi:10.1073/pnas.1402604111
George, V. C., Ansari, S. A., Chelakkot, V. S., Chelakkot, A. L., Chelakkot, C., Menon, V., et al. (2019). DNA-dependent protein kinase: epigenetic alterations and the role in genomic stability of cancer. Mutat. Res. Rev. Mutat. Res. 780, 92–105. doi:10.1016/j.mrrev.2018.06.001
Godel, M., Morena, D., Ananthanarayanan, P., Buondonno, I., Ferrero, G., Hattinger, C. M., et al. (2020). Small nucleolar RNAs determine resistance to doxorubicin in human osteosarcoma. Int. J. Mol. Sci. 21, 4500. doi:10.3390/ijms21124500
Goldstein, M., and Kastan, M. B. (2015). The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 66, 129–143. doi:10.1146/annurev-med-081313-121208
Goodwin, J. F., and Knudsen, K. E. (2014). Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 4, 1126–1139. doi:10.1158/2159-8290.cd-14-0358
Grundy, G. J., and Parsons, J. L. (2020). Base excision repair and its implications to cancer therapy. Essays Biochem. 64, 831–843. doi:10.1042/ebc20200013
Guo, Z., Wang, Y. H., Xu, H., Yuan, C. S., Zhou, H. H., Huang, W. H., et al. (2021). LncRNA linc00312 suppresses radiotherapy resistance by targeting DNA-PKcs and impairing DNA damage repair in nasopharyngeal carcinoma. Cell Death Dis. 12, 69. doi:10.1038/s41419-020-03302-2
He, H., Song, F., Gao, Q., Lu, Z., Yuan, Y., Li, X., et al. (2021). The APEX1/miRNA-27a-5p axis plays key roles in progression, metastasis and targeted chemotherapy of gastric cancer. Int. J. Pharm. 599, 120446. doi:10.1016/j.ijpharm.2021.120446
He, Y., Mingyan, E., Wang, C., Liu, G., Shi, M., and Liu, S. (2019). CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int. J. Biol. Macromol. 125, 116–123. doi:10.1016/j.ijbiomac.2018.11.273
Hoeijmakers, J. H. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374. doi:10.1038/35077232
Hu, C., and Jiang, X. (2019). The SUMO-specific protease family regulates cancer cell radiosensitivity. Biomed. Pharmacother. 109, 66–70. doi:10.1016/j.biopha.2018.10.071
Hu, H., Zhao, X., Jin, Z., and Hou, M. (2015). Hsa-let-7g miRNA regulates the anti-tumor effects of gastric cancer cells under oxidative stress through the expression of DDR genes. J. Toxicol. Sci. 40, 329–338. doi:10.2131/jts.40.329
Huan, L., Guo, T., Wu, Y., Xu, L., Huang, S., Xu, Y., et al. (2020). Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol. cancer 19, 11. doi:10.1186/s12943-019-1122-z
Huang, X., Li, Z., Zhang, Q., Wang, W., Li, B., Wang, L., et al. (2019). Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. cancer 18, 71. doi:10.1186/s12943-019-0969-3
Iyama, T., and Wilson, D. M. (2013). DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 12, 620–636. doi:10.1016/j.dnarep.2013.04.015
Jackson, S., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078. doi:10.1038/nature08467
Jiang, M., Jia, K., Wang, L., Li, W., Chen, B., Liu, Y., et al. (2021). Alterations of DNA damage response pathway: biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm. Sin. B 11, 2983–2994. doi:10.1016/j.apsb.2021.01.003
Jin, M. H., and Oh, D. Y. (2019). ATM in DNA repair in cancer. Pharmacol. and Ther. 203, 107391. doi:10.1016/j.pharmthera.2019.07.002
Jing, X., Xie, M., Ding, K., Xu, T., Fang, Y., Ma, P., et al. (2022). Exosome-transmitted miR-769-5p confers cisplatin resistance and progression in gastric cancer by targeting CASP9 and promoting the ubiquitination degradation of p53. Clin. Transl. Med. 12, e780. doi:10.1002/ctm2.780
Jurkovicova, D., Neophytou, C. M., Gašparović, A., and Gonçalves, A. C. (2022). DNA damage response in cancer therapy and resistance: challenges and opportunities. Int. J. Mol. Sci. 23, 14672. doi:10.3390/ijms232314672
Koessler, T., Oestergaard, M. Z., Song, H., Tyrer, J., Perkins, B., Dunning, A. M., et al. (2008). Common variants in mismatch repair genes and risk of colorectal cancer. Gut 57, 1097–1101. doi:10.1136/gut.2007.137265
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Kristensen, L. S., Jakobsen, T., Hager, H., and Kjems, J. (2022). The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19, 188–206. doi:10.1038/s41571-021-00585-y
Kutashev, K., Meschichi, A., Reeck, S., Fonseca, A., Sartori, K., White, C. I., et al. (2024). Differences in RAD51 transcriptional response and cell cycle dynamics reveal varying sensitivity to DNA damage among Arabidopsis thaliana root cell types. New phytologist 243, 966–980. doi:10.1111/nph.19875
Lal, B., Kulkarni, A., McDermott, J., Rais, R., Alt, J., Wu, Y., et al. (2023). Preclinical efficacy of LP-184, a tumor site activated synthetic lethal therapeutic, in glioblastoma. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 29, 4209–4218. doi:10.1158/1078-0432.ccr-23-0673
Lavin, M. F. (1999). ATM: the product of the gene mutated in ataxia-telangiectasia. Int. J. Biochem. and cell Biol. 31, 735–740. doi:10.1016/s1357-2725(99)00028-x
Lavin, M. F., and Yeo, A. J. (2020). Clinical potential of ATM inhibitors. Mutat. Res. 821, 111695. doi:10.1016/j.mrfmmm.2020.111695
Lecona, E., and Fernandez-Capetillo, O. (2018). Targeting ATR in cancer. Nat. Rev. Cancer 18, 586–595. doi:10.1038/s41568-018-0034-3
Li, E., Xia, M., Du, Y., Long, K., Ji, F., Pan, F., et al. (2022a). METTL3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the EGF/RAD51 axis. Elife 11, e75231. doi:10.7554/eLife.75231
Li, H., Shi, Y., Li, Y., Wu, S., Yang, R., Liu, Q., et al. (2024). DNA damage response-related signatures characterize the immune landscape and predict the prognosis of HCC via integrating single-cell and bulk RNA-sequencing. Int. Immunopharmacol. 137, 112475. doi:10.1016/j.intimp.2024.112475
Li, J., Song, C., Gu, J., Li, C., Zang, W., Shi, L., et al. (2023). RBBP4 regulates the expression of the Mre11-Rad50-NBS1 (MRN) complex and promotes DNA double-strand break repair to mediate glioblastoma chemoradiotherapy resistance. Cancer Lett. 557, 216078. doi:10.1016/j.canlet.2023.216078
Li, J., Yang, H., Li, Y., Liu, Y., Chen, S., Qi, C., et al. (2014). microRNA-146 up-regulation predicts the prognosis of non-small cell lung cancer by miRNA in situ hybridization. Exp. Mol. pathology 96, 195–199. doi:10.1016/j.yexmp.2013.11.004
Li, P., Song, R., Yin, F., Liu, M., Liu, H., Ma, S. Q., et al. (2022b). circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol. Ther. 30, 431–447. doi:10.1016/j.ymthe.2021.08.027
Li, P. P., Li, R. G., Huang, Y. Q., Lu, J. P., Zhang, W. J., and Wang, Z. Y. (2021). LncRNA OTUD6B-AS1 promotes paclitaxel resistance in triple negative breast cancer by regulation of miR-26a-5p/MTDH pathway-mediated autophagy and genomic instability. Aging 13, 24171–24191. doi:10.18632/aging.203672
Li, X., He, J., Ren, X., Zhao, H., and Zhao, H. (2020a). Circ_0003998 enhances doxorubicin resistance in hepatocellular carcinoma by regulating miR-218-5p/EIF5A2 pathway. Diagn. Pathol. 15, 141. doi:10.1186/s13000-020-01056-1
Li, Y., Gan, Y., Liu, J., Li, J., Zhou, Z., Tian, R., et al. (2022c). Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal Transduct. Target. Ther. 7, 87. doi:10.1038/s41392-022-00902-6
Li, Y. H., Xu, C. L., Pu, H. H., Liu, J. L., and Wang, Y. (2020b). circMTDH.4/miR-630/AEG-1 axis participates in the regulation of proliferation, migration, invasion, chemoresistance, and radioresistance of NSCLC. Mol. Carcinog. 59, 141–153. doi:10.1002/mc.23135
Lim, Z. F., and Ma, P. C. (2019). Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. and Oncol. 12, 134. doi:10.1186/s13045-019-0818-2
Liu, D., Keijzers, G., and Rasmussen, L. J. (2017). DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. Rev. Mutat. Res. 773, 174–187. doi:10.1016/j.mrrev.2017.07.001
Liu, H. L., Nan, H., Zhao, W. W., Wan, X. B., and Fan, X. J. (2024). Phase separation in DNA double-strand break response. Nucl. (Austin, Tex.) 15, 2296243. doi:10.1080/19491034.2023.2296243
Liu, J., Xue, N., Guo, Y., Niu, K., Gao, L., Zhang, S., et al. (2021b). Correction for: CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY) 13, 23868–23870. doi:10.18632/aging.203664
Liu, P., Fu, R., Chen, K., Zhang, L., Wang, S., Liang, W., et al. (2021c). ETV5-mediated upregulation of lncRNA CTBP1-DT as a ceRNA facilitates HGSOC progression by regulating miR-188-5p/MAP3K3 axis. Cell Death Dis. 12, 1146. doi:10.1038/s41419-021-04256-9
Liu, Y., Ao, X., Wang, Y., Li, X., and Wang, J. (2022a). Long non-coding RNA in gastric cancer: mechanisms and clinical implications for drug resistance. Front. Oncol. 12, 841411. doi:10.3389/fonc.2022.841411
Liu, Y., Ding, W., Wang, J., Ao, X., and Xue, J. (2023). Non-coding RNA-mediated modulation of ferroptosis in cardiovascular diseases. Biomed. Pharmacother. 164, 114993. doi:10.1016/j.biopha.2023.114993
Liu, Y., Dou, M., Song, X., Dong, Y., Liu, S., Liu, H., et al. (2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18, 123. doi:10.1186/s12943-019-1052-9
Liu, Y., He, D., Xiao, M., Zhu, Y., Zhou, J., and Cao, K. (2021a). Long noncoding RNA LINC00518 induces radioresistance by regulating glycolysis through an miR-33a-3p/HIF-1α negative feedback loop in melanoma. Cell Death Dis. 12, 245. doi:10.1038/s41419-021-03523-z
Liu, Y., Wang, Y., Li, X., Jia, Y., Wang, J., and Ao, X. (2022b). FOXO3a in cancer drug resistance. Cancer Lett. 540, 215724. doi:10.1016/j.canlet.2022.215724
Liu, Y., Zhao, C., Wang, G., Chen, J., Ju, S., Huang, J., et al. (2022c). SNORD1C maintains stemness and 5-FU resistance by activation of Wnt signaling pathway in colorectal cancer. Cell death Discov. 8, 200. doi:10.1038/s41420-022-00996-5
Lung, R. W., Tong, J. H. M., Ip, L. M., Lam, K. H., Chan, A. W. H., Chak, W. P., et al. (2020). EBV-encoded miRNAs can sensitize nasopharyngeal carcinoma to chemotherapeutic drugs by targeting BRCA1. J. Cell. Mol. Med. 24, 13523–13535. doi:10.1111/jcmm.16007
Lytle, N. K., Barber, A. G., and Reya, T. (2018). Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 18, 669–680. doi:10.1038/s41568-018-0056-x
Ma, C. X., Janetka, J. W., and Piwnica-Worms, H. (2011). Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol. Med. 17, 88–96. doi:10.1016/j.molmed.2010.10.009
Maekawa, T., Fukaya, R., Takamatsu, S., Itoyama, S., Fukuoka, T., Yamada, M., et al. (2018). Possible involvement of Enterococcus infection in the pathogenesis of chronic pancreatitis and cancer. Biochem. biophysical Res. Commun. 506, 962–969. doi:10.1016/j.bbrc.2018.10.169
Mamontova, V., Trifault, B., and Burger, K. (2024). Nono induces Gadd45b to mediate DNA repair. Life Sci. alliance 7, e202302555. doi:10.26508/lsa.202302555
Matsui, M., and Corey, D. R. (2017). Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16, 167–179. doi:10.1038/nrd.2016.117
McCabe, E. M., and Rasmussen, T. P. (2021). lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Seminars cancer Biol. 75, 38–48. doi:10.1016/j.semcancer.2020.12.012
McMullen, M., Karakasis, K., Madariaga, A., and Oza, A. M. (2020). Overcoming platinum and PARP-inhibitor resistance in ovarian cancer. Cancers 12, 1607. doi:10.3390/cancers12061607
Medová, M., Medo, M., Hovhannisyan, L., Muñoz-Maldonado, C., Aebersold, D. M., and Zimmer, Y. (2020). DNA-PK in human malignant disorders: mechanisms and implications for pharmacological interventions. Pharmacol. and Ther. 215, 107617. doi:10.1016/j.pharmthera.2020.107617
Mouw, K. W. (2017). DNA repair pathway alterations in bladder cancer. Cancers (Basel) 9, 28. doi:10.3390/cancers9040028
Ou, B., Liu, Y., Gao, Z., Xu, J., Yan, Y., Li, Y., et al. (2022). Senescent neutrophils-derived exosomal piRNA-17560 promotes chemoresistance and EMT of breast cancer via FTO-mediated m6A demethylation. Cell Death Dis. 13, 905. doi:10.1038/s41419-022-05317-3
Ou, Y., Zhai, D., Wu, N., and Li, X. (2015). Downregulation of miR-363 increases drug resistance in cisplatin-treated HepG2 by dysregulating Mcl-1. Gene 572, 116–122. doi:10.1016/j.gene.2015.07.002
Peng, L., Sang, H., Wei, S., Li, Y., Jin, D., Zhu, X., et al. (2020). circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol. Cancer 19, 156. doi:10.1186/s12943-020-01270-x
Permata, T. B. M., Hagiwara, Y., Sato, H., Yasuhara, T., Oike, T., Gondhowiardjo, S., et al. (2019). Base excision repair regulates PD-L1 expression in cancer cells. Oncogene 38, 4452–4466. doi:10.1038/s41388-019-0733-6
Qian, L., Fei, Q., Zhang, H., Qiu, M., Zhang, B., Wang, Q., et al. (2020). lncRNA HOTAIR promotes DNA repair and radioresistance of breast cancer via EZH2. DNA cell Biol. 39, 2166–2173. doi:10.1089/dna.2020.5771
Qiu, Y., Hu, X., Zeng, X., and Wang, H. (2022). Triple kill: DDR inhibitors, radiotherapy and immunotherapy leave cancer cells with no escape. Acta biochimica biophysica Sinica 54, 1569–1576. doi:10.3724/abbs.2022153
Qiu, Z., Oleinick, N. L., and Zhang, J. (2018). ATR/CHK1 inhibitors and cancer therapy. Radiotherapy Oncol. J. Eur. Soc. Ther. Radiology Oncol. 126, 450–464. doi:10.1016/j.radonc.2017.09.043
Rao, S., and Ayres, J. S. (2017). Resistance and tolerance defenses in cancer: lessons from infectious diseases. Seminars Immunol. 32, 54–61. doi:10.1016/j.smim.2017.08.004
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Rykova, E., Ershov, N., Damarov, I., and Merkulova, T. (2022). SNPs in 3'UTR miRNA target sequences associated with individual drug susceptibility. Int. J. Mol. Sci. 23, 13725. doi:10.3390/ijms232213725
Saldivar, J. S., Wu, X., Follen, M., and Gershenson, D. (2007). Nucleotide excision repair pathway review I: implications in ovarian cancer and platinum sensitivity. Gynecol. Oncol. 107, S56–S71. doi:10.1016/j.ygyno.2007.07.043
Shao, Y., Song, X., Jiang, W., Chen, Y., Ning, Z., Gu, W., et al. (2019). MicroRNA-621 acts as a tumor radiosensitizer by directly targeting SETDB1 in hepatocellular carcinoma. Mol. Ther. J. Am. Soc. Gene Ther. 27, 355–364. doi:10.1016/j.ymthe.2018.11.005
Shi, X., Liu, X., Huang, S., Hao, Y., Pan, S., Ke, Y., et al. (2022). miR-4443 promotes radiation resistance of esophageal squamous cell carcinoma via targeting PTPRJ. J. Transl. Med. 20, 626. doi:10.1186/s12967-022-03818-5
Shiloh, Y. (2001). ATM (ataxia telangiectasia mutated): expanding roles in the DNA damage response and cellular homeostasis. Biochem. Soc. Trans. 29, 661–666. doi:10.1042/0300-5127:0290661
Shu, G., Su, H., Wang, Z., Lai, S., Wang, Y., Liu, X., et al. (2021). LINC00680 enhances hepatocellular carcinoma stemness behavior and chemoresistance by sponging miR-568 to upregulate AKT3. J. Exp. Clin. Cancer Res. 40, 45. doi:10.1186/s13046-021-01854-5
Song, L., Lin, C., Wu, Z., Gong, H., Zeng, Y., Wu, J., et al. (2011). miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PloS one 6, e25454. doi:10.1371/journal.pone.0025454
Statello, L., Ali, M. M., Reischl, S., Mahale, S., Kosalai, S. T., Huarte, M., et al. (2021). The DNA damage inducible lncRNA SCAT7 regulates genomic integrity and topoisomerase 1 turnover in lung adenocarcinoma. Nar. cancer 3, zcab002. doi:10.1093/narcan/zcab002
Sun, Y., Wang, J., Ma, Y., Li, J., Sun, X., Zhao, X., et al. (2021). Radiation induces NORAD expression to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1 signalling and by inhibiting pri-miR-199a1 processing and the exosomal transfer of miR-199a-5p. J. Exp. and Clin. cancer Res. CR 40, 306. doi:10.1186/s13046-021-02084-5
Tan, J. N., Zhou, S. N., Zhang, W., Yang, B., Zhong, G. Y., Huang, J., et al. (2022). Long noncoding RNA OVAAL enhances nucleotide synthesis through pyruvate carboxylase to promote 5-fluorouracil resistance in gastric cancer. Cancer Sci. 113, 3055–3070. doi:10.1111/cas.15453
Tan, L., Huang, Z., Chen, Z., Chen, S., Ye, Y., Chen, T., et al. (2023). CircRNA_001895 promotes sunitinib resistance of renal cell carcinoma through regulation of apoptosis and DNA damage repair. J. Chemother. (Florence, Italy) 35, 11–18. doi:10.1080/1120009x.2021.2009990
Tan, Y. T., Lin, J. F., Li, T., Li, J. J., Xu, R. H., and Ju, H. Q. (2021). LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. (Lond) 41, 109–120. doi:10.1002/cac2.12108
Thomsen, M., and Vitetta, L. (2018). Adjunctive treatments for the prevention of chemotherapy- and radiotherapy-induced mucositis. Integr. cancer Ther. 17, 1027–1047. doi:10.1177/1534735418794885
Thote, V., Dinesh, S., and Sharma, S. (2024). Prediction of deleterious non-synonymous SNPs of human MDC1 gene: an in silico approach. Syst. Biol. reproductive Med. 70, 101–112. doi:10.1080/19396368.2024.2325699
Tian, X. J., Ferro, M. V., and Goetz, H. (2019). Modeling ncRNA-mediated circuits in cell fate decision. Methods Mol. Biol. Clift. N.J. 1912, 411–426. doi:10.1007/978-1-4939-8982-9_16
Vahabi, M., Pulito, C., Sacconi, A., Donzelli, S., D'Andrea, M., Manciocco, V., et al. (2019). miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J. Exp. and Clin. cancer Res. CR 38, 141. doi:10.1186/s13046-019-1119-x
van der Burg, M., van Dongen, J. J., and van Gent, D. C. (2009). DNA-PKcs deficiency in human: long predicted, finally found. Curr. Opin. allergy Clin. Immunol. 9, 503–509. doi:10.1097/ACI.0b013e3283327e41
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A., and Kinzler, K. W. (2013). Cancer genome landscapes. Sci. (New York, N.Y.) 339, 1546–1558. doi:10.1126/science.1235122
Volpato, L. E., Silva, T. C., Oliveira, T. M., Sakai, V. T., and Machado, M. A. (2007). Radiation therapy and chemotherapy-induced oral mucositis. Braz. J. otorhinolaryngology 73, 562–568. doi:10.1016/s1808-8694(15)30110-5
Wang, L., Yang, X., Zhou, F., Sun, X., and Li, S. (2022b). Circular RNA UBAP2 facilitates the cisplatin resistance of triple-negative breast cancer via microRNA-300/anti-silencing function 1B histone chaperone/PI3K/AKT/mTOR axis. Bioengineered 13, 7197–7208. doi:10.1080/21655979.2022.2036894
Wang, P., Yang, Z., Ye, T., Shao, F., Li, J., Sun, N., et al. (2020). lncTUG1/miR-144-3p affect the radiosensitivity of esophageal squamous cell carcinoma by competitively regulating c-MET. J. Exp. Clin. Cancer Res. 39, 7. doi:10.1186/s13046-019-1519-y
Wang, S., Wang, Y., Li, Q., Li, X., and Feng, X. (2022a). A novel circular RNA confers trastuzumab resistance in human epidermal growth factor receptor 2-positive breast cancer through regulating ferroptosis. Environ. Toxicol. 37, 1597–1607. doi:10.1002/tox.23509
Wang, T., Hao, D., Yang, S., Ma, J., Yang, W., Zhu, Y., et al. (2019). miR-211 facilitates platinum chemosensitivity by blocking the DNA damage response (DDR) in ovarian cancer. Cell death and Dis. 10, 495. doi:10.1038/s41419-019-1715-x
Wang, Z., Liu, L., Du, Y., Mi, Y., and Wang, L. (2021a). The HNF1A-AS1/miR-92a-3p axis affects the radiosensitivity of non-small cell lung cancer by competitively regulating the JNK pathway. Cell Biol. Toxicol. 37, 715–729. doi:10.1007/s10565-021-09595-z
Wang, Z., Wang, X., Rong, Z., Dai, L., Qin, C., Wang, S., et al. (2021b). LncRNA LINC01134 contributes to radioresistance in hepatocellular carcinoma by regulating DNA damage response via MAPK signaling pathway. Front. Pharmacol. 12, 791889. doi:10.3389/fphar.2021.791889
Wei, L., Sun, J., Zhang, N., Zheng, Y., Wang, X., Lv, L., et al. (2020). Noncoding RNAs in gastric cancer: implications for drug resistance. Mol. Cancer 19, 62. doi:10.1186/s12943-020-01185-7
Williams, A. E., Perry, M. M., Moschos, S. A., Larner-Svensson, H. M., and Lindsay, M. A. (2008). Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem. Soc. Trans. 36, 1211–1215. doi:10.1042/bst0361211
Williams, G. T., and Farzaneh, F. (2012). Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 12, 84–88. doi:10.1038/nrc3195
Woodbine, L., Neal, J. A., Sasi, N. K., Shimada, M., Deem, K., Coleman, H., et al. (2013). PRKDC mutations in a SCID patient with profound neurological abnormalities. J. Clin. investigation 123, 2969–2980. doi:10.1172/jci67349
Wu, P., Fang, X., Liu, Y., Tang, Y., Wang, W., Li, X., et al. (2021). N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 12, 298. doi:10.1038/s41419-021-03558-2
Xie, C., Zhang, L. Z., Chen, Z. L., Zhong, W. J., Fang, J. H., Zhu, Y., et al. (2020). A hMTR4-PDIA3P1-miR-125/124-TRAF6 regulatory Axis and its function in NF kappa B signaling and chemoresistance. Hepatology 71, 1660–1677. doi:10.1002/hep.30931
Xie, Y., Lu, X., Wang, Z., Liu, M., Liu, L., Wang, R., et al. (2022). A hypoxia-dissociable siRNA nanoplatform for synergistically enhanced chemo-radiotherapy of glioblastoma. Biomaterials Sci. 10, 6791–6803. doi:10.1039/d2bm01145j
Xing, C., Sun, S. G., Yue, Z. Q., and Bai, F. (2021). Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother. 134, 111158. doi:10.1016/j.biopha.2020.111158
Xu, S., Huang, H., Chen, Y. N., Deng, Y. T., Zhang, B., Xiong, X. D., et al. (2016). DNA damage responsive miR-33b-3p promoted lung cancer cells survival and cisplatin resistance by targeting p21(WAF1/CIP1). Cell cycleGeorget. Tex. 15, 2920–2930. doi:10.1080/15384101.2016.1224043
Xu, Y., Qiu, M., Shen, M., Dong, S., Ye, G., Shi, X., et al. (2021). The emerging regulatory roles of long non-coding RNAs implicated in cancer metabolism. Mol. Ther. 29, 2209–2218. doi:10.1016/j.ymthe.2021.03.017
Yu, R., Hu, Y., Zhang, S., Li, X., Tang, M., Yang, M., et al. (2022). LncRNA CTBP1-DT-encoded microprotein DDUP sustains DNA damage response signalling to trigger dual DNA repair mechanisms. Nucleic acids Res. 50, 8060–8079. doi:10.1093/nar/gkac611
Yu, S., Li, Y., Liao, Z., Wang, Z., Wang, Z., Li, Y., et al. (2020). Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 69, 540–550. doi:10.1136/gutjnl-2019-318860
Yuan, Y., Liao, H., Pu, Q., Ke, X., Hu, X., Ma, Y. F., et al. (2020). miR-410 induces both epithelial-mesenchymal transition and radioresistance through activation of the PI3K/mTOR pathway in non-small cell lung cancer. Signal Transduct. Target. Ther. 5, 85. doi:10.1038/s41392-020-0182-2
Yue, X., Bai, C., Xie, D., Ma, T., and Zhou, P. K. (2020). DNA-PKcs: a multi-faceted player in DNA damage response. Front. Genet. 11, 607428. doi:10.3389/fgene.2020.607428
Zhang, C., Kang, T., Wang, X., Wang, J., Liu, L., Zhang, J., et al. (2022a). LINC-PINT suppresses cisplatin resistance in gastric cancer by inhibiting autophagy activation via epigenetic silencing of ATG5 by EZH2. Front. Pharmacol. 13, 968223. doi:10.3389/fphar.2022.968223
Zhang, F., Li, K., Yao, X., Wang, H., Li, W., Wu, J., et al. (2019a). A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine 44, 311–321. doi:10.1016/j.ebiom.2019.05.003
Zhang, G., Liu, Z., Zhong, J., and Lin, L. (2021b). Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis. Eur. J. Clin. investigation 51, e13607. doi:10.1111/eci.13607
Zhang, H., Hua, Y., Jiang, Z., Yue, J., Shi, M., Zhen, X., et al. (2019b). Cancer-associated fibroblast-promoted LncRNA DNM3OS confers radioresistance by regulating DNA damage response in esophageal squamous cell carcinoma. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 25, 1989–2000. doi:10.1158/1078-0432.ccr-18-0773
Zhang, M., Yang, L., Hou, L., and Tang, X. (2021a). LncRNA SNHG1 promotes tumor progression and cisplatin resistance through epigenetically silencing miR-381 in breast cancer. Bioengineered 12, 9239–9250. doi:10.1080/21655979.2021.1996305
Zhang, T., Wu, D. M., Luo, P. W., Liu, T., Han, R., Deng, S. H., et al. (2022b). CircNEIL3 mediates pyroptosis to influence lung adenocarcinoma radiotherapy by upregulating PIF1 through miR-1184 inhibition. Cell death and Dis. 13, 167. doi:10.1038/s41419-022-04561-x
Zhang, X., Xie, K., Zhou, H., Wu, Y., Li, C., Liu, Y., et al. (2020). Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 19, 47. doi:10.1186/s12943-020-01171-z
Zhang, Z., Yu, H., Yao, W., Zhu, N., Miao, R., Liu, Z., et al. (2022c). RRP9 promotes gemcitabine resistance in pancreatic cancer via activating AKT signaling pathway. Cell Commun. Signal 20, 188. doi:10.1186/s12964-022-00974-5
Zhong, J. T., Yu, Q., Zhou, S. H., Yu, E., Bao, Y. Y., Lu, Z. J., et al. (2019). GLUT-1 siRNA enhances radiosensitization of laryngeal cancer stem cells via enhanced DNA damage, cell cycle redistribution, and promotion of apoptosis in vitro and in vivo. OncoTargets Ther. 12, 9129–9142. doi:10.2147/ott.s221423
Zhou, C., Yi, C., Yi, Y., Qin, W., Yan, Y., Dong, X., et al. (2020). LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. cancer 19, 118. doi:10.1186/s12943-020-01237-y
Zhou, S., Ye, W., Zhang, Y., Yu, D., Shao, Q., Liang, J., et al. (2016). miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am. J. Transl. Res. 8, 2992–3002.
Zhou, X., Ao, X., Jia, Z., Li, Y., Kuang, S., Du, C., et al. (2022). Non-coding RNA in cancer drug resistance: underlying mechanisms and clinical applications. Front. Oncol. 12, 951864. doi:10.3389/fonc.2022.951864
Zhou, Y., Wan, G., Spizzo, R., Ivan, C., Mathur, R., Hu, X., et al. (2014). miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol. Oncol. 8, 83–92. doi:10.1016/j.molonc.2013.09.004
Zhou, Z., Lin, Z., He, Y., Pang, X., Wang, Y., Ponnusamy, M., et al. (2018). The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Mol. Ther. Nucleic acids 12, 405–419. doi:10.1016/j.omtn.2018.05.024
Zhou, Z., Zhang, Y., Gao, J., Hao, X., Shan, C., Li, J., et al. (2021). Circular RNAs act as regulators of autophagy in cancer. Mol. Ther. oncolytics 21, 242–254. doi:10.1016/j.omto.2021.04.007
Zhu, C., Li, K., Jiang, M., and Chen, S. (2021). RBM5-AS1 promotes radioresistance in medulloblastoma through stabilization of SIRT6 protein. Acta neuropathol. Commun. 9, 123. doi:10.1186/s40478-021-01218-2
Zhu, H., Chen, K., Chen, Y., Liu, J., Zhang, X., Zhou, Y., et al. (2022). RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduct. Target. Ther. 7, 240. doi:10.1038/s41392-022-01033-8
Zuo, Y., He, J., Zhou, Z., Sun, J., Ouyang, C., Huang, H., et al. (2024). Long non-coding RNA LIP interacts with PARP-1 influencing the efficiency of base excision repair. Non-coding RNA Res. 9, 649–658. doi:10.1016/j.ncrna.2024.03.010
Glossary
Keywords: DNA damage response (DDR), non coding RNA (ncRNA), resistance, radiotherapy, chemotherapy
Citation: Gao Z, Luan X, Wang X, Han T, Li X, Li Z, Li P and Zhou Z (2024) DNA damage response-related ncRNAs as regulators of therapy resistance in cancer. Front. Pharmacol. 15:1390300. doi: 10.3389/fphar.2024.1390300
Received: 23 February 2024; Accepted: 14 August 2024;
Published: 26 August 2024.
Edited by:
Chunbo He, University of Oklahoma Health Sciences Center, United StatesReviewed by:
Seema Kumari, Gandhi Institute of Technology and Management (GITAM), IndiaEduardo López-Urrutia, National Autonomous University of Mexico, Mexico
Copyright © 2024 Gao, Luan, Wang, Han, Li, Li, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peifeng Li, cGVpZmxpQHFkdS5lZHUuY24=; Zhixia Zhou, emhvdV96aGl4aWFAcWR1LmVkdS5jbg==
†These authors have contributedequally to this work
 Ziru Gao
Ziru Gao Xinchi Luan
Xinchi Luan Xuezhe Wang
Xuezhe Wang Tianyue Han
Tianyue Han Zeyang Li
Zeyang Li Peifeng Li
Peifeng Li Zhixia Zhou
Zhixia Zhou