- 1Laboratory of Biomedical Genomics and Oncogenetics, Institut Pasteur de Tunis, Tunis, Tunisia
- 2Genetic Typing Service, Institut Pasteur de Tunis, Tunis, Tunisia
- 3University of Tunis El Manar, Tunis, Tunisia
- 4Faculty of Medicine of Tunis, Tunis, Tunisia
- 5MitoLab Team, Unité MitoVasc, Unité Mixte de Recherche Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale U1083, SFR ICAT, University of Angers, Angers, France
Background: Chronic pain is a major socioeconomic burden in the Mediterranean region. However, we noticed an under-representation of these populations in the pharmacogenetics of pain management studies. In this context, we aimed 1) to decipher the pharmacogenetic variant landscape among Mediterranean populations compared to worldwide populations in order to identify therapeutic biomarkers for personalized pain management and 2) to better understand the biological process of pain management through in silico investigation of pharmacogenes pathways.
Materials and Methods: We collected genes and variants implicated in pain response using the Prisma guidelines from literature and PharmGK database. Next, we extracted these genes from genotyping data of 829 individuals. Then, we determined the variant distribution among the studied populations using multivariate (MDS) and admixture analysis with R and STRUCTURE software. We conducted a Chi2 test to compare the interethnic frequencies of the identified variants. We used SNPinfo web server, miRdSNP database to identify miRNA-binding sites. In addition, we investigated the functions of the identified genes and variants using pathway enrichment analysis and annotation tools. Finally, we performed docking analysis to assess the impact of variations on drug interactions.
Results: We identified 63 variants implicated in pain management. MDS analysis revealed that Mediterranean populations are genetically similar to Mexican populations and divergent from other populations. STRUCTURE analysis showed that Mediterranean populations are mainly composed of European ancestry. We highlighted differences in the minor allele frequencies of three variants (rs633, rs4680, and rs165728) located in the COMT gene. Moreover, variant annotation revealed ten variants with potential miRNA-binding sites. Finally, protein structure and docking analysis revealed that two missense variants (rs4680 and rs6267) induced a decrease in COMT protein activity and affinity for dopamine.
Conclusion: Our findings revealed that Mediterranean populations diverge from other ethnic groups. Furthermore, we emphasize the importance of pain-related pathways and miRNAs to better implement these markers as predictors of analgesic responses in the Mediterranean region.
Introduction
According to the International Association for the Study of Pain (IASP), pain is defined as an unpleasant sensory and emotional experience connected with actual or potential tissue damage, or described in terms of such damage (Raja et al., 2020). Excessive pain can significantly increase psychological health problems and affects the quality of life (Schuh et al., 2020). Pain includes several types such as neuropathic, nociceptive and nociplastic depending on its neuropsychological cause and duration (Isagulyan et al., 2022). It could be divided into two groups: acute and chronic. Chronic pain represents the most frequent condition (Raffaeli et al., 2021). More than 50% of hospitalized patients suffer from chronic pain in Spain and United Kingdom (Fayaz et al., 2016; Muñoz-Alvaredo et al., 2020) and 14% have moderate-to-severe disabling chronic pain (Dueñas et al., 2015; Yoshida et al., 2018; Mills et al., 2019). Therefore, an appropriate pain management by analgesics should be made. The most widely used analgesic class includes opioids, which are derived from natural compounds contained in opium poppy plants (Pathan and Williams, 2012). Opioids exert several effects in the central nervous system, including pain alleviation. They are commonly employed in a variety of therapeutic contexts. However, opioids could be responsible for sever adverse drug reactions (ADR) such as respiratory depression and tolerance. Therefore, it is important to conduct a tailored opioids medication to enhance patient’s quality of life and reduce the risk of induced ADR. There are various opium derivatives such as codeine, tramadol, fentanyl, sufentanil, alfentanil, and remifentanil (Kumar et al., 2019; Lambert, 2020). Inappropriate opioids use can induce moderate-to-severe ADR. For instance, opioid therapy can cause moderate to severe side effects in 20%–30% of patients such as respiratory depression and hypotension, and bradycardia in 11% of patients. These ADR are mainly due to the high interindividual opioid response variability. Therefore, individualized pain management for each patient is important for alleviating patient suffering and morbidity. Furthermore, tailoring the individual’s treatment can improve analgesic outcomes by maximizing efficacy and minimizing toxicities (Mahajan, 2014; Schuh et al., 2020). Interindividual diversity in pain management therapies is mainly driven by behavior, expectations, gender, renal function, psychosocial distress, ethnic origin and genetic variability (Molanaei et al., 2010). Hence, understanding how a patient’s background affects his response to painkillers is essential for developing personalized pain management therapies (Yoshida et al., 2018; Angus and Chang, 2021). The Mediterranean populations comprise regions around the Mediterranean basin including Southern European, Middle Eastern, and North African regions (de Larramendi et al., 2020). Currently clinical practices used for care pain management in these regions include painkiller drug administration, physical therapy and supplementary treatments (including massage and acupuncture). Painkiller prescription in Mediterranean region is based on governmental Survey and standardized guidelines (Polo-Santos et al., 2021). Nonetheless, there are a number of shortcomings in pain management strategies across the Mediterranean region. Current clinical guidelines often prescribe a straightforward drug regimen for pain management of different categories of patients. This could lead to serious pain persistence. Therefore, it is important to develop more specialized guidelines for chronic pain conditions taking into consideration patients’ intrinsic factor (age, sex, disease stage and the presence of other comorbidities) as well as its environmental factors (socioeconomic status and dietary habits). Moreover, primary care physicians (PCPs) often prescribe opioids as first line therapy which could lead to serious adverse drug reactions and drug addiction in some cases. This could be due to the absence of proper pain treatment policy and trainings for PCPs. Therefore, the lack of specialized pain clinics or trained healthcare providers could lead to serious socioeconomic burden in the region (Louriz et al., 2016; Hamdan and Mosleh, 2024). In the other hand, there are considerable evidences highlighting the role of genetic, epigenetic and ethnicity as pivotal factors on pain differences and management. Previous studies have demonstrated that genetic factors influence the individual response to opioids by 24%–60%. In addition, polymorphisms in transporters, receptors, drug-metabolizing enzymes, and other drug targets influences drug efficacy and toxicity. Thus, Pharmacogenomics studies suggest that an effective pain management strategy should take into account the patient’s genetic background and environmental adaptations, in order to maximize effectiveness and help mitigate impairment (Nahin, 2015). Despite the high prevalence of chronic pain in the Mediterranean populations and its socio-economic burden, only few studies have covered the pharmacogenetic of pain management in these populations (Dueñas et al., 2015). Indeed, several studies have reported a high incidence and prevalence of pain in European and North African countries of the Mediterranean region (Miró et al., 2007; Langley, 2011; Ahangar-Sirous et al., 2023). In addition, the incidence of chronic pain in the Mediterranean region was higher than those described in East Asia and American countries (Zimmer et al., 2022). Moreover, these regions are characterized by a high prevalence of drug dependence, especially opioids, compared to other regions. This suggests the presence of an ethnic disparity linked to the pharmacogenetic component in these populations (Amirkafi et al., 2023).
Taking into account all these elements, more pharmacogenetic studies and testing could revolutionize the field of pain management in the Mediterranean region through personalizing the disease treatment depending on the patient’s DNA profile (Kaye et al., 2019). Pharmacogenes that influence treatment outcomes can generally be split into two categories. On the one hand, pharmacodynamics-influencing genes, based on differences in drug target receptors and downstream signal transduction such as the opioid receptor; OPRM1, the enzyme catecholamine methyltransferase, COMT, etc. Pharmacokinetics genes, on the other hand, impact the cytochrome P450 family, enzymes involved in glucuronidation, and drug transporter proteins. Therefore, a better understanding of the biological pathways involved in the pain management process could help develop new therapeutic strategies. Furthermore, a number of genetic variations implicated in analgesics response variability can influence microRNA (miRNA) binding sites. MiRNAs have a crucial role as epigenetic markers in pain physiopathology, since they influence gene expression regulation and RNA silencing. They have the ability to control many target genes and metabolic activities. Indeed, several studies have documented alterations in miRNA expression in migraines, musculoskeletal diseases, and other pain conditions (Dayer et al., 2019; Polli et al., 2020). Furthermore, a single miRNA can regulate several target genes and metabolic processes that are involved in pain processing (Sabina et al., 2022). Determining the regulatory functions of miRNAs in intricate networks that control biological processes requires an understanding of the interactions between miRNAs and their targets. Since there are a large number of potential targets and a vast and increasing number of miRNA species, it is not possible to manually anticipate how a miRNA will interact with its target. Therefore, prediction databases are commonly used in silico analysis to determine potential target genes of miRNAs (Liu, Li, and Cairns 2014). These miRNAs can be used as biomarkers of therapeutic efficacy (Dai et al., 2018; Tramullas et al., 2018; Sabina et al., 2022).
In this purpose, our study aims 1) to decipher the pharmacogenetic landscape of chronic pain among Mediterranean populations compared to worldwide populations in order to identify therapeutic biomarkers for personalized pain management and 2) to better understand the biological process of pain management through in silico investigation of pharmacogenes pathways.
Materials and methods
To attend our objectives, we developed the present workflow (Figure 1). First, we conducted a general review of the literature to collect pharmacogenes and variants implicated in pain management. Second, we conducted a multidimensional scaling plot (MDS) and Structure analysis of these variants using publicly available genotyping data in order to explore the genetic landscape of pain management among Mediterranean populations compared to other populations. In the third step, we performed in silico pathway enrichment, miRNA analysis and simulation docking analysis.
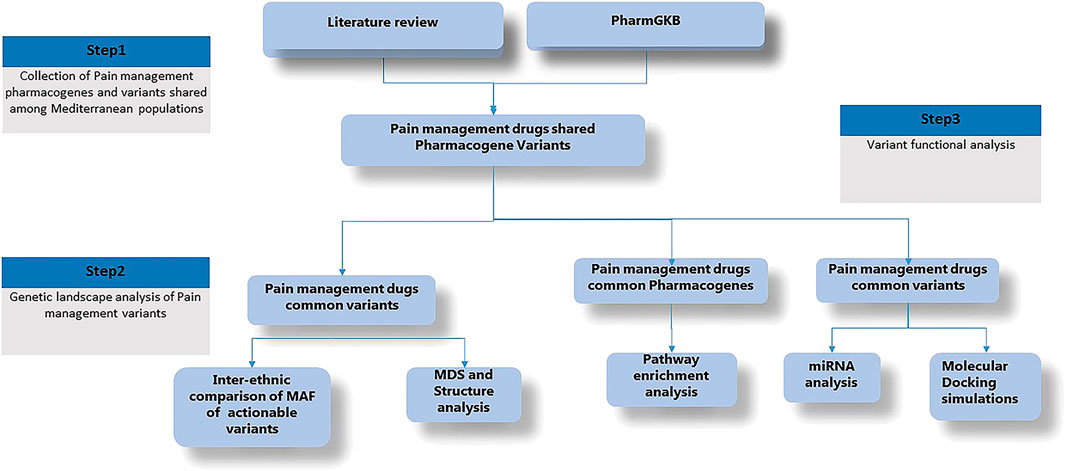
Figure 1. Detailled research workflow. Minor Allele Frequency (MAF), Multidimensional Scaling (MDS), microRNA (miRNA).
Step 1: Collection of Pain management pharmacogenes and variants shared among Mediterranean populations
Pain management pharmacogene selection
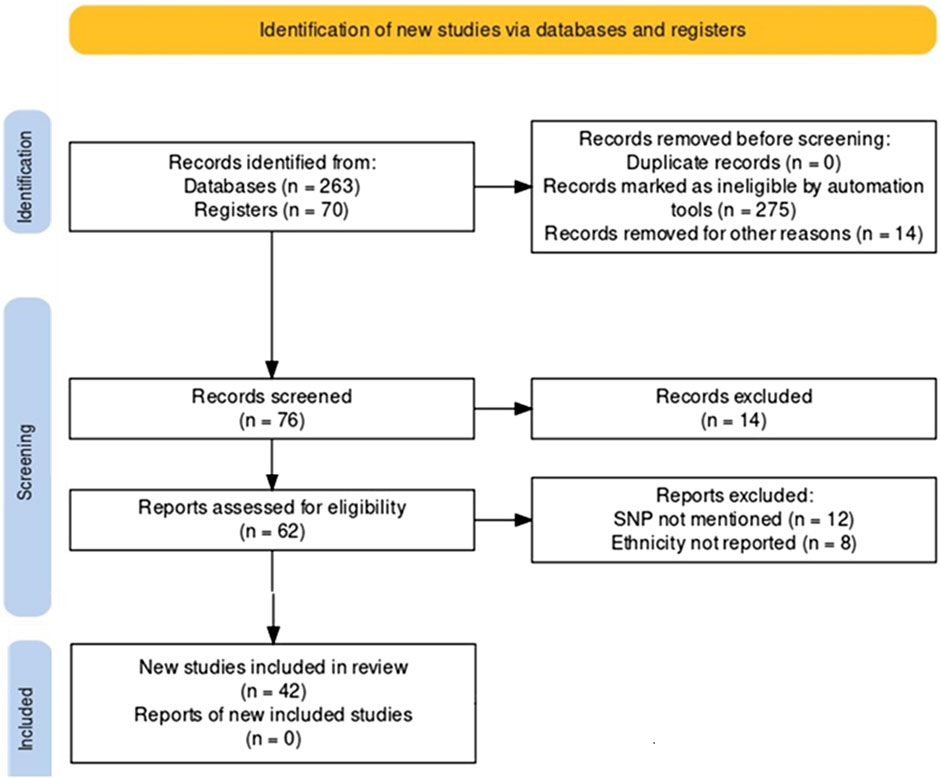
We conducted a systematic search of accessible, peer-reviewed, and complete articles published between January 2000 and April 2023 using the PRISMA guidelines (Page et al., 2021). Articles for review were selected from the MEDLINE (PubMed), Web of Science, and Google Scholar databases. Search terms included a combination of ‘pain management, pain pharmacogenomics of opioids, predictive genetic testing’. This search concerns chronic pain management. The full text version of the included article has been retrieved. In addition, reference lists of relevant studies were evaluated and cheeked by two assessors. Inclusion and exclusion criteria were documented according to the PRISMA rules and presented as a PRISMA flowchart (Figure 2). First, duplicate citations were removed from endnotes, then inappropriate citations linked to missing genotyping data, no ethnic reports, were removed after reading the full articles. Inclusion criteria were papers reporting polymorphisms associated with variation in analgesic response and studies presenting original data with clear and concise endpoint results. Exclusion criteria were non-English studies, abstract-only entries, papers without relevant topics, and studies containing no useful or overlapping data with previously published studies. Several genetic variants were identified according to the Prisma performance criteria. Clinical annotation of these variants was examined using the Pharmacogenomics Knowledge Base, « PharmGKB » http://www.pharmgkb.org, an interactive tool to investigate how genetic variation affects drug response. It displays genotype, molecular, and clinical knowledge integrated into pathway representations and Very Important Pharmacogene (VIP) summaries with links to additional external resources.
Step 2: Genetic landscape of pain management variants
Genotyping data and quality control analysis
We extracted variants located in the selected pharmacogenes using the PLINK v2 toolkit (Chang et al., 2015). Genotypic data of 829 individuals from 16 populations were downloaded from the International 1,000 Genome Project phase III (www.internationalgenome.org) and published data (Li et al., 2008; Henn et al., 2012). The studied populations included those of Mediterranean ancestry: Toscani people of Italy (TSI), South Spain (Spain_S), North Spain (Spain_N), North West of Spain (Spain_NW) and Spain Basic populations (Spain_BASC), Northwestern and Western European ancestry populations of Utah from the CEPH collection (CEU), Algeria (Algeria), Egypt (Egypt), Libya (Libya), Tunisia Douiret (TN_Ber), South Morocco (Morocco_S), North Morocco (Morocco_N), American ancestry: African ancestry in the South Western United States of America (ASW) and people of Mexican ancestry living in Los Angeles, California, United States of America (MEX), individuals from East Asian ancestry: Han Chinese in Beijing, China (CHB), the Chinese population of metropolitan Denver, Colorado, United States of America (CHD) and Japanese in Tokyo, Japan; (JPT) (Supplementary Table S1).
PLINK v2 software was used to manage genomic data. We excluded variants deviating from the Hardy-Weinberg equilibrium (HWE) with a p-value of 10–4 and had missed genotyping rate ≤95% for each studied populations.
Statistical analysis
Merged genotyping data were pruned on the basis of physical distances and linkage disequilibrium (LD) between variants. High-density markers that do not provide additional information were excluded. Next, we used pruning data to study the landscape of the pain management pharmacogene by creating a multidimensional scaling plot (MDS).
To this end, a symmetric matrix of the identity-by-state (IBS) distances for all pairs of individuals was performed based on the proportion of shared common alleles. This analysis was performed using Plink (Chang et al., 2015) and R software (https://www.R-project.org/).
In addition, allele frequencies of druggable pharmacogene variants selected from common and rare shared variants were calculated. At this stage, the study populations were clustered according to their geographical origin. Three groups were formed; Mediterranean (MED), East Asian (EAS), and American (AMR) populations. A chi-square test was used to compare the allele frequencies of the selected variants between the MED populations and the other populations. Bonferroni’s adjustment was applied to the significance level and set to a p-value threshold of 5% divided by the number of tested variants. All analyzes were performed using R software.
Analysis of population genetic structures
The STRUCTURE ver. 2.3.4 software (Pritchard, Stephens, and Donnelly 2000; Falush, Stephens, and Pritchard 2007) was used to assess the population structure. This program implements a model-based clustering method to infer population structure using genotype data of unlinked markers. We used the admixture model and correlated allele frequency version of STRUCTURE. The Markov Chain Monte Carlo iteration for each structure analysis was run for 10,000 after an initial burn-in period of 10,000 steps. We calculated delta K as proposed by Evanno et al. (Evanno et al., 2005) to assess the most likely numbers of clusters. Next, we evaluate the similarity of the runs at each K level with the CLUMPAK software (Kopelman et al., 2015). We used the Distruct software in order to illustrate the optimal alignment of subpopulations inferring population substructure and individual assignment across the best runs at each k level (Rosenberg, 2004).
Step 3: Variant functional analysis
Pathway enrichment analysis
We have conducted a pathway enrichment analysis (PEA) to get a deeper insight on the biological functions and pathways involved in pain management. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), WikiPathway (UP_KW), InterPro pathway, SMART pathway, BioCarta Pathways and the Biological Biochemical Image Database (BBID) were investigated for functional annotation and enrichment analysis. A total of 381 pathways with minimum gene set size ≥2 were analyzed using Database for Annotation, Visualization and Integrated Discovery (DAVID), a web-based gene-enrichment analysis tool (https://david.ncifcrf.gov/). We have set a p-value = 0.05 as a significant threshold after multiple corrections. GO terms were then visualized using the Monash Gene Ontology (MonaGO) (https://monago.erc.monash.edu/) (Xin et al., 2022). It provides an intuitive, interactive and responsive interface for performing GO enrichment analysis and result visualization (Xin et al., 2022).
MiRNAs analysis
MiRNAs play a crucial role as pain efficacy indicators. To this end, we explored the SNPinfo web server to detect variants with potential miRNA-binding sites (Xu and Taylor, 2009). Next, we used miRdSNP database (Bruno et al., 2012) to identify the miRNAs of interest. Finally, the obtained list of miRNAs was explored to generate a heat map of GO pathways affected by this miRNA using miRPathD.
Structural effect of the selected variants
To investigate the effects of the identified Single Nucleotide Polymorphisms (SNP) on protein structure, we initially retrieved the 3D crystallized proteins from the RCSB database (https://www.rcsb.org/). Mutant proteins were created using UCSF Chimera 1.17.1 software, followed by an energy minimization step. We employed 1,000 steepest descent steps with a root mean square gradient of 0.02, using the Amber ff12SB force field. The energy minimization involved an update interval of 10 (Yousuf et al., 2017).
To compare the wild-type and mutant structures, we calculated the Root Mean Square Deviation (RMSD) using Chimera software. To assess the potential impact of the identified SNPs on protein stability, we used the mCSM (http://biosig.unimelb.edu.au/mcsm/stability), SDM (http://marid.bioc.cam.ac.uk/sdm2/), DUET (http://biosig.unimelb.edu.au/duet/stability) and INPS-3D (https://inpsmd.biocomp.unibo.it/inpsSuite/default/index3) servers for stability prediction.
Molecular docking simulations
To evaluate the impact of different SNPs on ligand interaction, we conducted molecular docking by comparing the wild-type and mutant proteins. The 3D structure of the ligand was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and subsequently optimized using Avogadro software version 1.95.1 (https://avogadro.cc/). Protein-substrate docking was performed using the AutoDock 4.2 program package. During the preparation of the receptor input file, we eliminated water molecules and heteroatoms, and then added missing hydrogens and Gasteiger charges. The conformation space search was conducted within a grid box, with a grid-point spacing of 0.375 Å and dimensions of (40 × 40 × 40) points. Finally, we performed molecular visualization using Discovery Studio 2017 R2 (https://www.3dsbiovia.com/products/collaborative-science/biovia-discoverystudio/).
Results
Step1: Collection of Pain management pharmacogenes and variants shared among Mediterranean populations
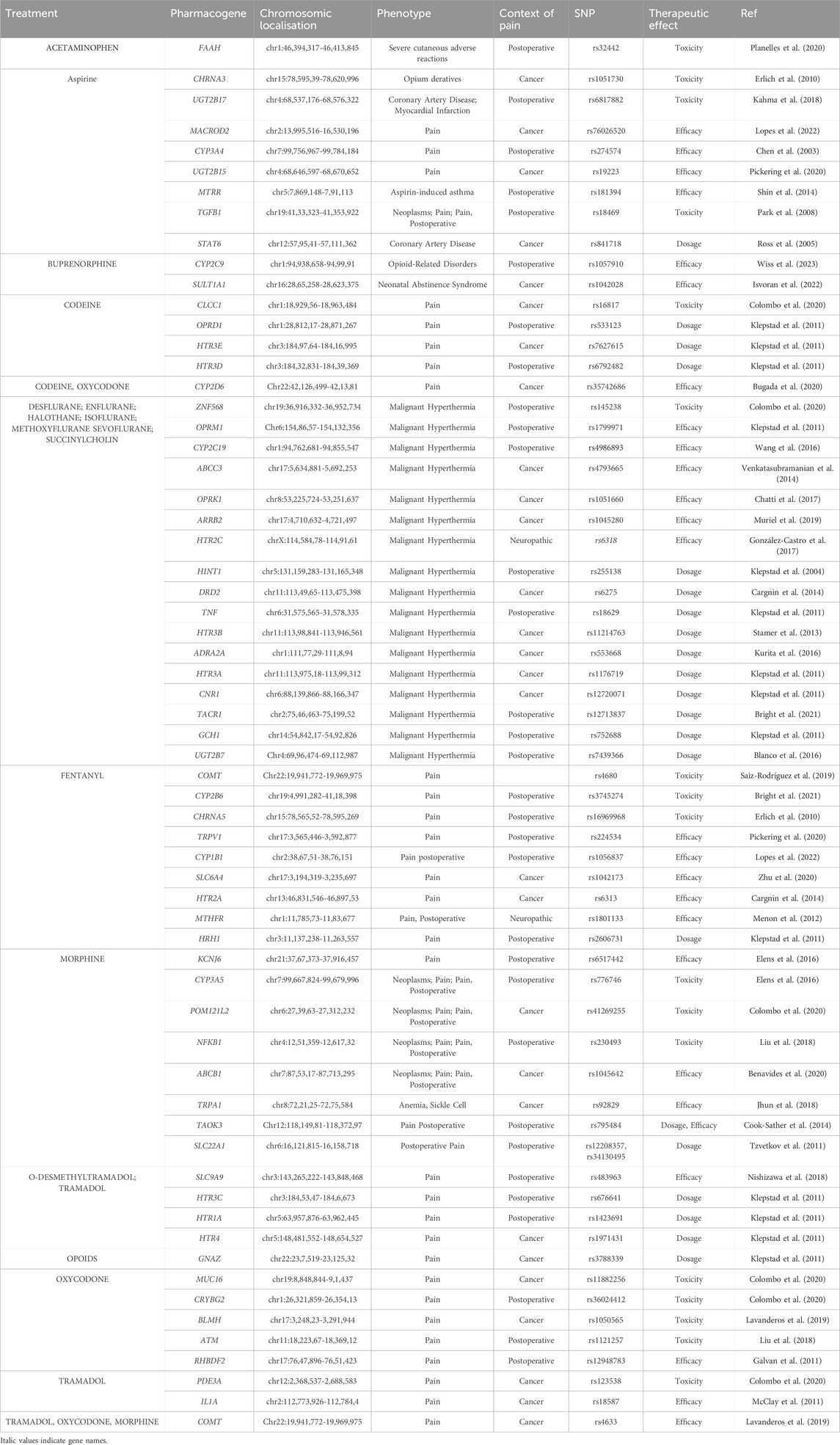
According to the PRISMA analysis we identified 263 papers investigating SNP involved in pharmacogenomic of pain management through PubMed, Web of Science and Google Scholar databases and an additional 70 papers through pharmGKB database. After removing duplicates, a total of 76 papers were screened for their relevance. Abstracts and titles screening identified 62 studies that met the inclusion criteria. After the full-text analysis, we excluded 20 studies not including genotyping and ethnic data. Hence, 42 studies were included in this study (Figure 2, Supplementary Table S2). Prisma reviewing and PharmGKB database screening have shown that 63 pharmacogenes are involved in the pharmacogenomics of pain management drugs. The results are shown in Table 1, which illustrates the pharmacogene’s name, chromosomal region, related drug, correlated phenotype, context, variant, and therapeutic effect.
Step 2: Genetic landscape of pain management variants
Statistical analysis
Variants located in pain management 63 pharmacogenes were extracted from the genotyping data of 829 studied individuals. A total of 1,450 common (p.value >5 × 10−2) and rare variants (p.value <5 × 10−2) were retained after quality control and genotypic data pruning. MDS analysis describing the genetic landscape of these genetic variants showed that North African populations (Algeria, Egypt, Libya, Morocco-N, Morocco-S, Tunisia) were clustered within the European populations (CEU, Spain-S, Spain-Basic, Spain-NW, and TSI) and have slight proximity to American population from Mexico. We showed that North African and European populations were distinguished from the American (ASW) and Asian (CHB, CHD, JPT) populations (Figure 3). Better individualization was observed in the MDS performed across continents. We observed a great genetic divergence between the Mediterranean (MED), American (AMR), and East Asian (EAS) groups. However, slight proximity was found between the MED and MEX populations.
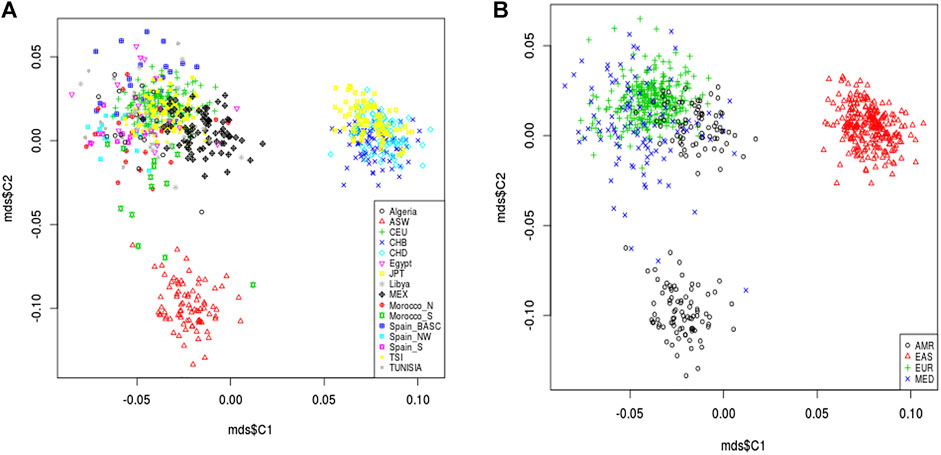
Figure 3. Multidimensional scaling plot of pain management pharmacogene shared variants. The plot shows the proximity of North African populations (Algeria, Egypt, Libya, Morocco-N, Morocco-S, Tunisia) and the European populations (CEU, Sapin-S, Spain-Basic, Spain-NW, and TSI) in the Mediterranean region which are clustered together and distinguished from the American (ASW, MEX) and Asian (CHB, CHD, JPT) populations (A). Better individualization was observed in MDS performed across continents. In addition, there is a great genetic divergence among the Mediterranean (MED), American (AMR) and East Asian (EAS) groups. However, slight proximity was observed between the MED cluster and AMR from Mexico (B).
Inferring admixture events between Mediterranean populations
The inference of admixture events to the overall pharmacogene variation in pain management was estimated using STRUCTURE software. Ancestry proportions for the studied individuals were estimated from hypothetical ancestors K = 2 to K = 16. The best K value estimated using the Evanno’s ΔK method was 3 (Supplementary Figures S1, S2). We observed the emergence of specific population clusters at this ancestry (K = 3). The ancestry assignment was mainly differentiated between African (blue), European (green), and Asian (red). The Mediterranean population (MED) was clearly differentiated from the American (AMR) and Asiatic populations (EAS) (Figure 4, Supplementary Figure S3). The number of clusters are confirmed by the estimated membership coefficients using the Q statistic (Figure 4B). The Triangle plot shows that the MED are distinct from ASW, EAS and AMR.
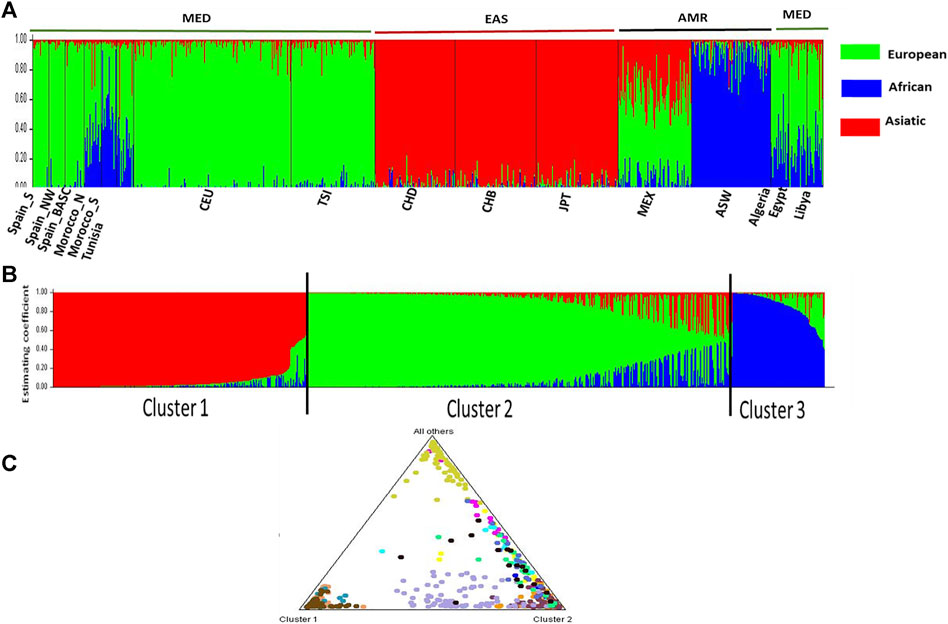
Figure 4. STRUCTURE analysis Bar plot (K = 3). (A) describes the genetic relationship between 16 populations at the best estimated k level. K presents the possible numbers of parental population clusters. One color represents one parental population into different color segments. The proportion of each ancestral component in a single individual is represented by a vertical bar divided into 3 colors red, green and blue. In this analysis, 1,450 markers study display results for runs with highest likelihood out of 16 runs in each cluster K3 to 16. Black vertical lines identify the population boundaries. The height extent of each color within an individual’s color bar corresponds to the estimated membership of the individual in one of the clusters; each cluster is assigned a separate color. The bars with multiple colors can be interpreted as genetic admixture or as relative probabilities of belonging to the different clusters. (B) Three estimated clusters of the 829 individuals presented with different colors inferred by STRUCTURE analysis. Clusters 1–3 were presented by red, green, blue, and yellow, respectively. Each bar represents an individual, in which a different color represents the estimated membership coefficients using the Q statistic. (C) The triangle result shows that the North African populations are close to the European, forming a cluster 2 which is distinct from the EAS cluster and AMR cluster.
Allelic frequency of variants with relevant therapeutic effect
Our statistical analysis revealed significant differences in MAF of three variants with high therapeutic effects (rs4633, rs4680, and rs165728) located in the Catechol-O-methyltransferase protein (COMT) gene among the MED, AMR, and EAS populations (Table 2). The frequency of the minor allele “T” of rs4633 in the MED populations was higher than the AMR and EAS populations by 36% and 79%, respectively (p = 0.0087). On the other hand, the frequency of “A” allele of rs4680 is associated with low frequency in MED compared to AMR and EAS (42% and 57%, respectively) (p.value = 0.019). Moreover, the “C” allele of rs1655728 in the MED population was significantly lower than AMR (p = 0.006) and EAS (−75% and −87%, respectively) (p = 8.856e-08). The “G” allele of rs13070715 (reindexed within dbSNP as rs2227931) represented higher frequencies in the MED than AMR and EAS studied populations (43% and 12%, respectively).
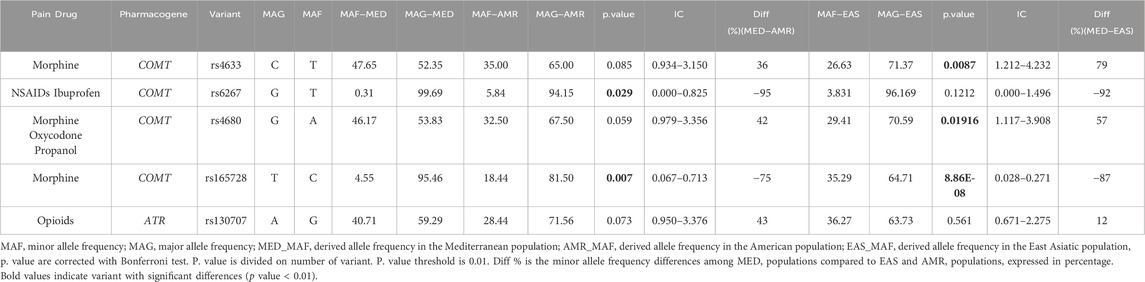
Table 2. Allelic frequencies comparison of pain therapeutic high impact variants among studied populations.
Step 3: Variant functional analysis
Pathway enrichment analysis
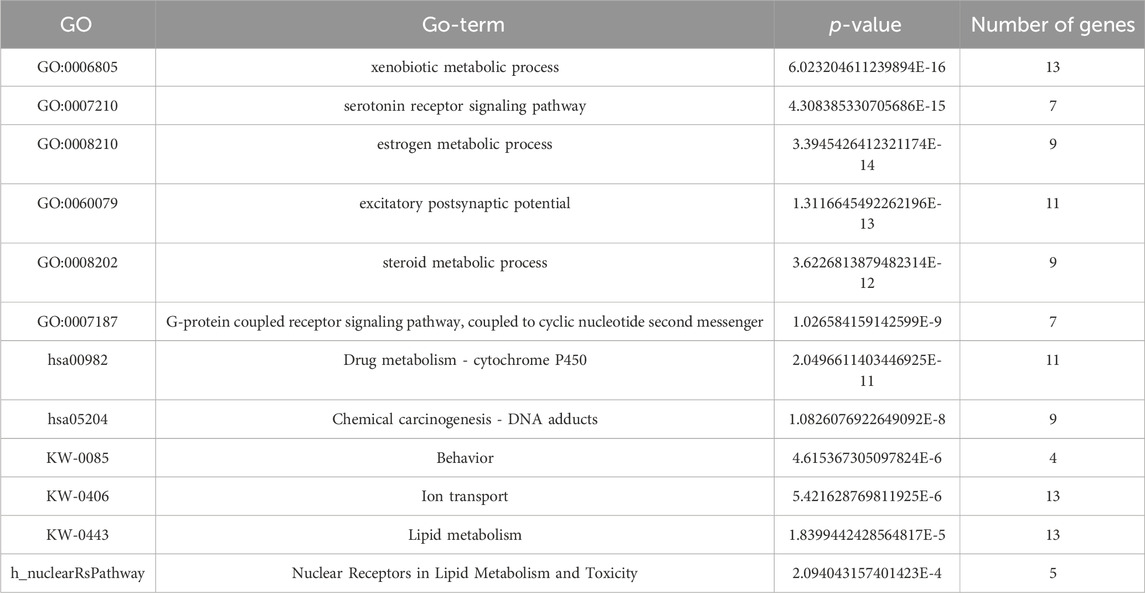
We performed a pathway enrichment analysis of the obtained gene list using the DAVID online tool. A total of 47 biological pathways were enriched in pain management genes with p-value <0.05. Results are shown in Table 3. The MonaGO chord diagram showed that pain management genes were enriched in 30 GO biological functions with cut-off p-value<2 × 10−4 (Figure 5). The most GO enriched terms were: xenobiotic metabolic process, serotonin receptor signaling pathway, estrogen metabolic process, excitatory postsynaptic potential, and steroid metabolic process. The most enriched KEGG pathways were: Serotonergic synapse, Drug metabolism—cytochrome P450, Chemical carcinogenesis—DNA adducts, and Neuroactive ligand-receptor interaction. Wikipathway revealed four significant pathways enriched in pain management: Steroid metabolism, Behavior, Ion transport, and Lipid metabolism. Whereas BIOCARTA showed one significant pathway; Nuclear Receptors in Lipid Metabolism and Toxicity (p-value = 2.09 × 10−4) (Supplementary Table S3).
MiRNAs analysis
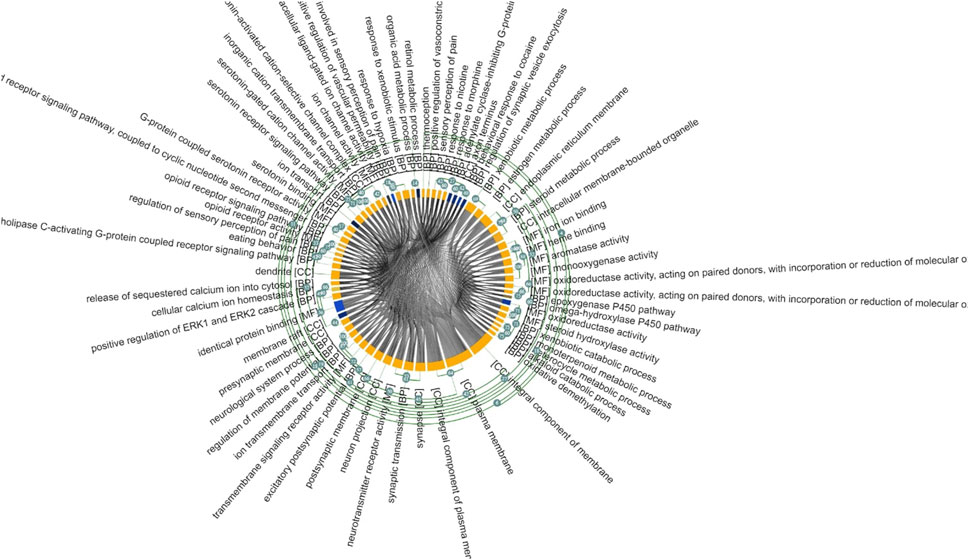
The SNPinfo shows that ten variants have potential miRNA-binding sites (Table 4). After the investigation of miRdSNP database, we noticed that only eight out of ten variants dispose of potential miRNA-binding sites. Pathway analysis revealed that detected miRNA was enriched in multiple GO pathways. A number of these pathways was also identified by our GO pathway analysis of pain management associated genes (Figure 6). These pathways are: regulation of acute inflammatory response, cellular calcium ion homeostasis, response to drug, cellular response to DNA damage, cellular response to toxic substance, regulation of chronic inflammation response, synaptic transmission GABAergic, response to estrogen, and positive regulation of metabolic process. The main enriched miRNAs associated with these pathways were: hsa-miR-130a-3p, hsa-miR-223-3p, hsa-miR-15a-5p, hsa-miR-128-3p, hsa-miR-30c-5p, hsa-miR-19a-3p, andhsa-miR-24-3p. Results are shown in Table 4.

Figure 6. Heatmap of pain pharmacogene miRNAs enriched pathways. Our result showed that miRNAs were mapped to several pathways involved in regulation of acute inflammatory response, cellular calcium ion homeostasis, response to drug, cellular response to DNA damage, cellular response to toxic substance, regulation of chronic inflammation response, synaptic transmission GABAergic, response to estrogen, and positive regulation of metabolic process.
Predicting the structure impact of nsSNP
We focused our structural analysis primarily on three non-synonymous single nucleotide polymorphisms “nsSNPs” (rs4680, rs6267, and rs2227931). The first two variants (rs4680, and rs6267) are located in the COMT gene. We utilized the crystal structure of COMT (PDB ID: 3IBW) with a resolution of 1.93 Å to create the corresponding mutated proteins. The RMSD analysis revealed slight deviations of 0.179 Å and 0.187 Å, respectively. Moreover, the tools: mCSM, SDM, DUET and INPS-3D predicted that these mutations would destabilize the protein, as indicated by negative ΔΔG values of −0.697, −1.250, −0.857 and −0.381 kcal/mol for rs6267, and -1.372, −2.450, −1.507 and −0.74 kcal/mol for rs4680. The rs2227931 SNP is located within the ATR serine/threonine kinase ATR gene and causes a premature stop codon at position 1736. As a consequence, this mutation leads to the loss of 34% (908 amino acids) of the protein sequence (Figure 7). Specifically, it results in the deletion of the Kinase domain, PIKK regulatory domain (PRD), FAT C-terminus domain (FATC), and a significant portion of the FAT domain. These alterations in the protein structure may disrupt the function of the ATR protein.
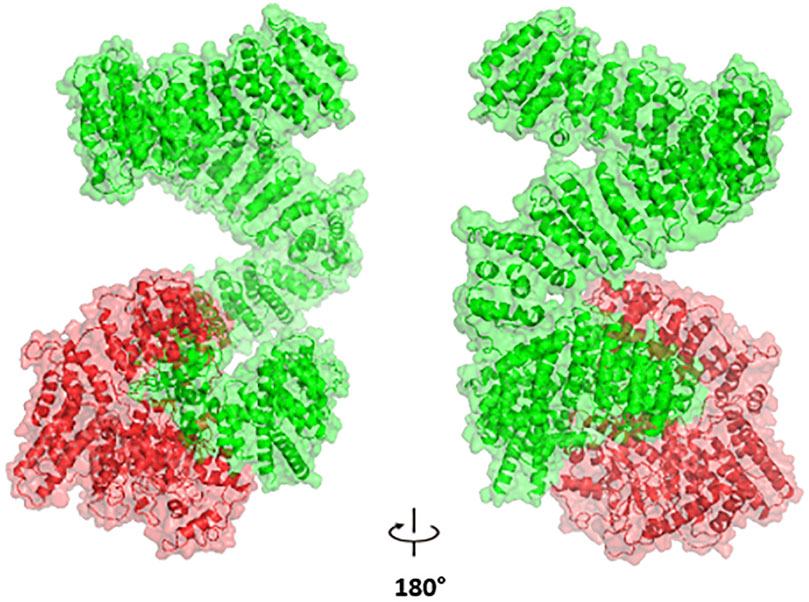
Figure 7. The Impact of p. Tyr1736Ter Nonsense Variant (rs2227931) on the ATR Protein: depiction of the truncated protein segment highlighted in red.
Molecular docking and binding energy calculation
In our study, we employed a molecular docking approach to investigate the interaction between wild-type and mutant COMT proteins, specifically rs4680 and rs6267 variants, with dopamine. The calculation of binding energy plays a pivotal role in discerning the affinity between the protein and ligand. By conducting a thorough analysis of the docking complex, we were able to identify noteworthy characteristics. Notably, we observed a decrease in binding energy for the dopamine complex in comparison to the wild-type protein complex (Table 5). This decrease suggests a decline in stability, which may potentially lead to lower enzyme activity. Furthermore, we noticed a decrease in the number of interactions in these complexes (Figure 8). In the wild-type protein complex, three hydrogen bonds were observed with Met40, Asn170, and Glu199, which are specific substrate binding residues. However, in the mutant proteins rs4680 and rs6267, only one hydrogen bond was present with Glu199 and Asn170, respectively.
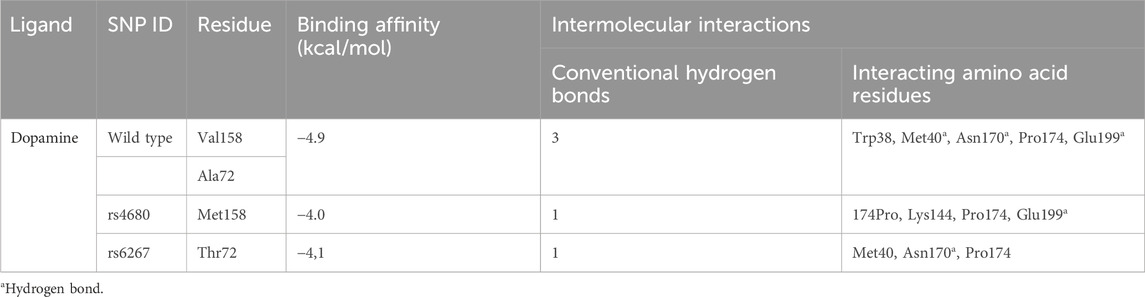
Table 5. Dopamine interactions with the active site of COMT wild type and mutant protein: binding affinity, number of conventional hydrogen bonds, and interacting amino acid residues.
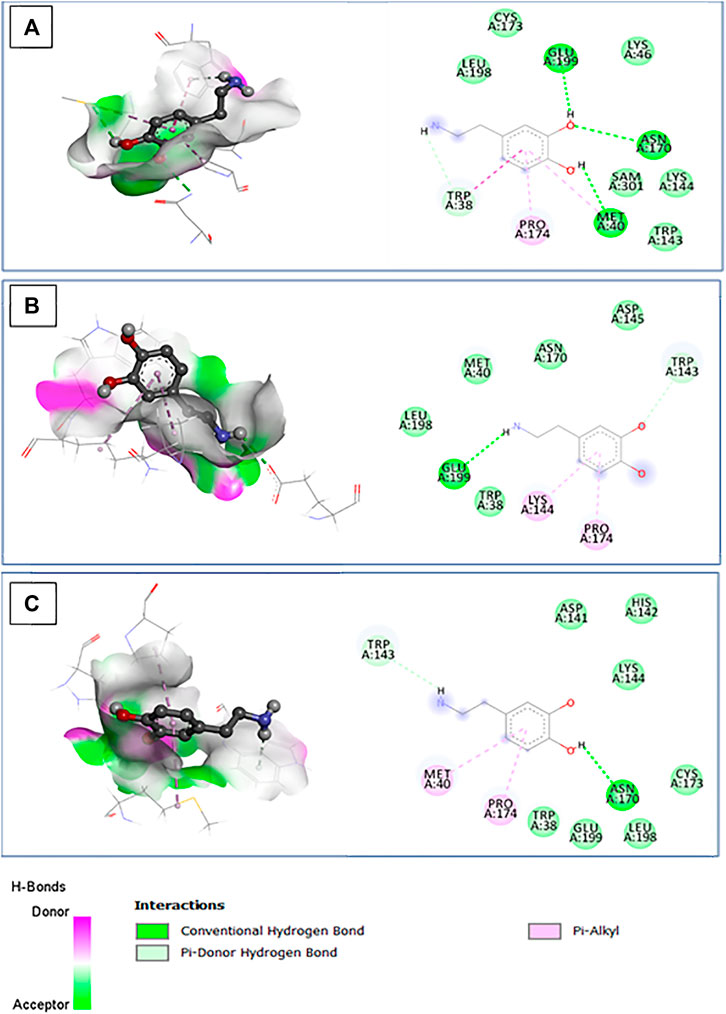
Figure 8. The 3D structure of the docked dopamine bound to the pocket region of the wild type (A) and mutant COMT protein (B): rs4680; (C): rs6267, along with hydrogen bonds and the corresponding 2D diagram of the interactions.
Discussion
Differences in allele frequencies may introduce genetic diversity among different ethnic groups which has the potential to alter the therapeutic efficacy of commonly used treatments (Goodman and Brett, 2021) such as analgesics. Therefore, incorporation of pharmacogenomic assessments prior to treatment would be extremely beneficial in terms of cost, quality of life, and therapeutic resource optimization.
Indeed, pharmacogenetics relies on genotype-based prescribing decisions to maximize efficacy and mitigate side effect risks. Due to the limited access to genotyping tests, ancestral markers are often used to estimate the likely genotype of a patient. This is based on the frequency of genetic variation in a patient’s ethnic group, as some variations are specific to certain ethnic groups. Thus, a deeper understanding of ancestral pharmacogenetic markers may lead to a better understanding of pharmacogenetic mechanisms in Mediterranean populations, which could enhance the development of tailored pain therapies for these populations (Shah and Gaedigk, 2018). Moreover, the ancestral marker data may be very helpful in developing pharmacogenetic tests that can be easily translated into clinical care practices. As a result, it can reduce the socio-economic and psychological burdens of pain conditions. Furthermore, reducing the harmful toxicities and risk of addiction associated with pain care requires the discovery of these ancestral markers. Hence, a wide representation of different ethnic groups is essential for the development of tailored pain management strategies. Pharmacogenetics studies are mainly conducted on white-non hispanic population (Ortega and Meyers, 2014), and little is still known about the pharmacogenetics of the Mediterranean basin, especially the southern countries of this region. In the present study, we investigated the pharmacogenetic landscape of variants implicated in pain management across the Mediterranean region, compared to other groups of populations. Next, we explored their role in different biological processes to identify potential therapeutic targets. Finally, we studied the protein-drug reaction to better understand the role of these variants in the therapeutic efficacy and toxicity.
Genetic landscape of pain management
Our MDS analysis revealed a high genetic similarity between North African and European populations of the MED region. However, we observed a great genetic divergence between the MED populations group and the two other groups (AMR and EAS). These results were further validated when conducting admixture analysis. Indeed, we noticed that the MED group is composed mainly of European and African components with low genetic contribution of the EAS ancestries. These findings are consistent with previous results demonstrating the impact of ethnicity on genetic variability of pain pharmacogenes (Perry et al., 2019). In fact, a similar genetic positioning was also observed in a previous study conducted on pharmacogenes implicated in metabolic syndrome drug response (Jmel et al., 2018). Furthermore, Mezzi et al., demonstrated a high genetic homogeneity between European populations (Mizzi et al., 2014). These results could be explained by the ancestral invasion and migration events especially between the North African populations and the South-west European populations. On the other hand, the high genetic heterogeneity between MED group and EAS populations could be due to an ancient divergence between the two groups (Boukhalfa et al., 2023). Interestingly, we notice a genetic proximity between MED populations and Mexican populations. This could be explained by the colonial history of Mexico, especially from circum-Mediterranean regions of Spain (Gómez et al., 2021).
The high genetic admixture of the MED populations in comparison to other ethnic groups could explain their high genetic variability regarding pain management pharmacogenes. Indeed, the minor allele frequency (MAF) comparison has shown the difference in MAF of three variants (rs4680, rs4633, and rs165728) located in COMT gene. The COMT gene encodes the catechol-O-methyltransferase, a key enzyme that regulates cognitive functions, mood and pain perception through regulating the catecholamine concentrations (Sadhasivam et al., 2014). The first variant, rs4680 (G/A) induces an amino acid change from Valine to Methionine. It was predicted to induce COMT protein instability. Our docking analysis revealed a decrease in binding energy and a reduction in the number of interactions. These findings align with previous studies in the literature, which have indicated that the presence of the A allele is associated with lower enzymatic activity, consequently leading to elevated levels of prefrontal dopamine (Mir et al., 2018). Our results showed a lower frequency of the “A” allele of rs4680 in MED compared to AMR and EAS population. This variant is associated with increased analgesic response to morphine (Elens et al., 2016). According to a study conducted by Rakvåg et al., Indian individuals with cancer from various sources (breast, lung, abdominal cavity, and urogenital system) who had the GG genotype of the COMT rs4680 received an average of 50% higher daily doses of morphine compared to those with the AA genotype (Rakvåg et al., 2005). Similarly, a previous study conducted on American patients after nephrectomy showed that carriers of the GG genotype had a higher morphine consumption in comparison to the AA genotype carriers (Candiotti et al., 2014). In contrast, the AA genotype was significantly associated with moderate to higher-pain in South African breast cancer survivors (Firfirey et al., 2022). However, no association between this variant and morphine clinical efficacy in Tunisian cancer patients (Chatti et al., 2017).
In addition, a previous study reported reduced morphine needs related to the rs4680 minor allele (A) in people of European Caucasian and Asian origin. The effects of rs4680 are not consistently supported in pediatricians but only in adults. The rs4680 minor allele A is associated with increased pain during mobilization following surgery and decreased postoperative analgesic administration (Li et al., 2019). In contrast, the minor A-allele of this polymorphism decreases catabolic enzyme activity by 25%, resulting in more dopamine in the prefrontal cortex (Tchivileva et al., 2010). However, the significance of opioids’ stimulating effects to their addiction potential is unknown. Some studies believe that dopamine-mediated reward deficit is an incentive for drug abuse (Blum et al., 2000). This energizing effect might be an endophenotype (Gottesman and Gould, 2003), indicating a desire for non-medical opioid usage. Furthermore, earlier research has linked rs4680 to variations in opioid usage in chronic pain patients after surgery (Yennurajalingam et al., 2021). Previous study has shown that rs4680 decreases COMT enzyme activity, resulting in poor noradrenaline metabolism and increased noradrenaline concentrations in the bloodstream, as well as impacts on the brain’s arousal regions, which explains the decrease in somnolence. The increasing noradrenaline concentration may have also an effect on the analgesic effects (Valomon et al., 2014). A clinical study of cancer pain in Asian patients found that GG carriers require a greater intravenous morphine dosage than AA carriers (Tanaka et al., 2021), with the lack of drug preventing or lowering opioid-induced somnolence (Cargnin et al., 2014). We showed significant MAF differences of rs4633 (C>T) between The MED, AMR and EAS populations. This variant has previously been linked to fibromyalgia in Korean patients (Knezevic et al., 2018b) and postoperative pain in American children (Sadhasivam et al., 2014), where researchers discovered that children with the (CC) genotype require more morphine and analgesic intervention postoperatively than children with the (TT) genotype (De Gregori et al., 2016). The rs4633 variant allele “T” is more common in the MED population than in the AMR and EAS populations. Morphine appeared to be more effective in populations with MED. This finding may influence the morphine dose in MED patients suffering from extreme pain as well as the development of treatment algorithms.
However, allele “C” of rs4633 is less frequent in the MED population than AMR and EAS. This allele is associated with increased severity of Pain when treated with propranolol in Caucasian women with temporomandibular joint disorders and pain (Tchivileva et al., 2010).
The variant rs2227931 located in the overlapping region of the ATR-ATM genes is highly prevalent in MED and AMR populations than EAS population. This variant is known to be implicated in DNA damage leading to drug resistance. Our modeling result showed that the rs2227931 caused the deletion of the Kinase domain, PIKK regulatory domain (PRD), FATC, and a significant portion of the FAT domain. These alterations in the ATR protein structure disrupt the function of the ATR and lead to opioids resistance (Karran, 2001). Finally, rs165728 is located in the 3′UTR region of the COMT gene. A negative association between this variant and opioid daily dose in advanced cancer patients have been recently detected (Yennurajalingam et al., 2021). The study included American patients from three ethnic groups (black non-hispanic, hispanic, and white non-hispanic). Our results pinpointed that the “C” allele of rs165728 is rare in the MED population compared to AMR and EAS studied populations. Moreover, previous studies showed that Allele “C” is associated with increased dose of morphine in Caucasian individuals with Neoplasms as compared to allele T (Rakvåg et al., 2008). We suggest that MED populations require a lower dose of morphine to accomplish the target therapeutic effect.
Pathway enrichment analysis
We identified 30 enriched GO pathways for pain management. The most enriched GO pathways identified by MonaGO were: xenobiotic metabolic process, serotonin receptor signaling pathway, estrogen metabolic process, excitatory postsynaptic potential, and steroid metabolic process. Xenobiotic metabolic process involves the different pathways implicated in the metabolism of exogenous components like drugs. Serotonin receptor signaling pathway has been widely implicated in stress response as well as in pain signaling (Haleem, 2019). A large body of evidence demonstrates the implication of serotonin1A receptor (5-HT1A receptor) in the control of pain. Thus, multiple therapeutic strategies suggest 5-HT1A receptor as potential pharmacological target for treating pain (Haleem, 2019). Interestingly, there are seven serotonin (5-HT) receptors (5-HT1 to 5-HT7) differentially distributed in the brain (Viguier et al., 2013). A recent study conducted on mild traumatic brain injury (mTBI) showed that 5-HT could enhance the nociception sensitization directly via 5-HT3 receptor signaling or indirectly through the upregulation of spinal chemokine production (Sahbaie et al., 2019). Estrogen receptors (ERs) include two nuclear types of receptors (ERα and ERβ), mainly enriched in the cytoplasm and can also be recruited into the cell membrane. Different studies have demonstrated the role of ERs in alleviating nociception (Chen et al., 2021). For example, a previous study has proven the therapeutic effect of Erβ in Inflammatory Bowel Disease (IBD) rat model through the suppression of the P2X purinoceptor 3 receptor (P2X3R) (Jiang et al., 2017). Similarly, Erα poses an anti-nociception action via decreasing ATP-P2X3-mediated Ca2+ influx (Cho and Chaban, 2012). Excitatory postsynaptic potential (EPSE) describes the process that leads to a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. A previous study conducted by Ren et al. revealed that monosynaptic EPSPE increased significantly in the arthritis pain rat model through the regulation of metabotropic glutamate receptor subtype 1 (mGluR1) signaling (Ren and Neugebauer, 2010). The steroid metabolic process appears as one of the most enriched pathways in our analysis. Similar results were found regarding therapeutic targets of low back pain (Wen et al., 2022). Steroids and corticosteroids have been widely used as pain relief medications. For example, corticosteroids could directly or indirectly decrease the production of pro-inflammatory cytokines by inhibiting Phospholipase A2 and the ensuing arachidonic acid metabolic pathway (Knezevic et al., 2018a). Genes associated with pain management drugs have been found enriched in multiple other pathways such as: Chemical carcinogenesis - DNA adducts, Neuroactive ligand-receptor interaction, Behavior, Ion transport, and Lipid metabolism. Morphine and related opioids are powerful yet addictive pain drugs. Opium poses several carcinogenic effects. For example, opium smoke could contain carcinogenic chemicals like matic hydrocarbons (PAHs), arsenic and lead. Furthermore, opium alkaloids could induce genotoxic effects such as: chromosome damage, micronuclei, DNA fragmentation related to morphine, and sister chromatid exchanges related to codeine. Finally, opioid receptor activation could enhance angiogenesis and neovascularization, impairment of immune function, and facilitation of tumor initiation, proliferation, and migration (Sheikh et al., 2023). However, contradictory results have been presented regarding their role as chemical carcinogenesis. This discrepancy could be due to the type of the used opium (mixed or pure), condition and intensity (Kosciuczuk et al., 2020; López-Lázaro, 2022). Neuroactive ligand-receptor interaction and Ion transport were significantly enriched in pain management. These findings are in line with those of Chidambaran et al. reporting that Neuroactive ligand-receptor interaction and ion channels were similarly enriched in chronic and acute postoperative pain (Chidambaran et al., 2020). It is widely known that ions, especially calcium (Ca2+) plays a critical role in neuronal excitability and synaptic transmission and thus it is inevitable in the pain perception process (Harding and Zamponi, 2022). Interestingly, behavior appears as one of the most enriched pathways in pain management genes. Alterations of brain reward-related behavior have been widely proposed in relation to painkillers use, especially opioids. They can enhance drug-seeking and drug-taking behaviors (Merlin et al., 2018; Grecco and Atwood, 2020; Maumus et al., 2020) specifically through μ opioid receptors (Sikora et al., 2019). Thus, medical professionals should develop a personalized management of patients using opioids. Finally, lipid metabolism plays an important role in chronic pain. For example, a systematic review revealed a significant association between serum HDL, LDL, triglyceride and musculoskeletal pain (Tilley et al., 2015). Furthermore, lipid bioactive mediators such as: endocannabinoids, endogenous PPAR-α activators, and oxidative products of PUFA metabolism appear to regulate the transmission of nociceptive information from peripheral sites of injury and inflammation to the central nervous system (Piomelli et al., 2014).
MiRNAs analysis
Our in silico analysis revealed ten variants were predicted to affect miRNA implicated in several pathways related to pain management such as: regulation of acute inflammatory response, cellular calcium ion homeostasis, response to drug, cellular response to DNA damage, synaptic transmission GABAergic, response to estrogen, and positive regulation of metabolic process. Differential expressions of miR-130a-3p and miR-128-3p have been observed in rat models of spinal cord injury (SCI) (Hu et al., 2021). Recent studies have shown the downregulation of miR-130a-3p in SCI rat models attenuated NP via inhibiting inflammatory markers (IL-1β, IL-6 and TNF-α), mitigated apoptosis, activated microglia and upregulated the IGF-1/IGF-1R signaling axis (Yao et al., 2021). However, that upregulation of miR-128-3p could alleviate the neuropathic pain (NP) in SCI rat models via regulating aquaporin-4 (AQP4) pathway (Xian et al., 2021). Other study suggests the potential role of miR-128-3p in alleviating NP through regulating neuroinflammation mediated by ZEB1 (zinc finger E-box binding homeobox 1) in chronic constriction injury (CCI) rat model (Zhang et al., 2020). ZEB1 can inhibit cell adhesion and has been identified as a potential target of several miRNAs (miR-28-5p, miR-200b, miR429, and miR-150) in neuropathic pain (Zhang et al., 2020). miR-15a is a tumor suppressor in chronic lymphocytic leukemia, pituitary adenomas, and prostate cancer. Recent study by Cai et al. has successfully demonstrated the therapeutic effect of this miRNA in alleviating CCI- induced NP through AKT3 mediated autophagy process (Cai et al., 2020). A previous study suggests that over-expression of miR-30c-5p in plasma and Cerebrospinal Fluid Markers (CSF) in combination with other clinical variables could predict NP in patients with chronic peripheral ischemia (Tramullas et al., 2018). The same study has shown that miR-30c-5p inhibitor delayed neuropathic pain development and reversed fully established allodynia in rodents through regulating mechanisms mediated by TGF-b and involved the endogenous opioid system (Tramullas et al., 2018). Moreover, downregulation of miR-19a-3p appears to mediate NP through MeCP2-mediated BDNF upregulation (Manners et al., 2015). BDNF is a main regulatory factor of sensory neurotransmission in nociceptive pathways and hyperalgesia (Manners et al., 2015). A very recent computational study by Yu et al. (Yu et al., 2023), identified a differential expression of miR-223-3p in NP. These results have been supported by previous findings. Significant downregulation of miR-223-3p has been previously reported in fibromyalgia patients (Bjersing et al., 2013). Furthermore, upregulation of miR-223-3p could alleviate trigeminal neuralgia NP through targeting MKNK2 and MAPK/ERK signals (Huang et al., 2022). In addition, over-expression of miR-223-3p induced by electroacupuncture significantly reduces postherpetic neuralgia pain by inhibiting neuron cell autophagy (Toyama et al., 2017). miR-24-3p is a master regulator of cancer development and occurrence (Wang et al., 2020). Previous study has demonstrated a significant upregulation of miR-24-3p upon opioid treatment suggesting its crucial role in pain management (Toyama et al., 2017). Together, these data demonstrate the potential role of miRNAs in regulating pain sensation through regulation of inflammatory and immune pathways. Finally, these miRNAs could serve as promising biomarkers for monitoring pain-killers activation in the body. Further clinical trials are needed to prove their role in pain management. The implementation of aforementioned biomarkers (SNP and miRNA) in pharmacogenetic testing through genetic and molecular screening of these biomarkers could be advantageous for patient’s tailored treatment (Roberts et al., 2023; Selvaraj et al., 2023). Indeed, genotyping data will be then communicated to the healthcare provider through established Clinical Decision Support Systems (CDSS) or innovative bioinformatic algorithms in order to assess clinical decision-making. This approach will assist physicians in prescribing the most adapted therapeutic molecule and dose for the patient ensuring a personalized and patient-centric approach.
This practices contribute to minimize the side effects and optimize the efficiency of analgesics and opioids. Thus, could be of great interest to better clinical decision-making and reduce socioeconomic disparities such as minimizing healing time and the risk of drug dependency.
Conclusion
Our findings show that Mediterranean populations are characterized by high genetic heterogeneity in terms of the investigated pharmacogenes involved in chronic pain management. Our results highlight the important role of the identification of ancestral markers, miRNAs and pain related pathways for better clinical decision making and pain management. We suggest that the implementation of these markers through effective pharmacogenetic strategies could improve pain response rate, reduce side effects and healing-time. However, further research is needed to close this gap and explore pain-related pharmacogenetic markers in under-represented population such as Mediterranean populations.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: www.internationalgenome.org, https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1002397, https://www.science.org/doi/10.1126/science.1153717?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed. Generated results are available in the Supplementary Material of the paper.
Ethics statement
Ethical approval was not required for the studies involving humans because the data relating to participants were obtained from public data. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements because the data used in our study was obtained from the International 1,000 genome project phase III database and published data from Li et al., 2008 and Henn et al., 2012.
Author contributions
HJ: Validation, Writing–original draft, Software, Methodology, Investigation, Formal Analysis, Data curation, Conceptualization. WB: Validation, Writing–review and editing, Software, Methodology, Formal Analysis, Data curation. IG: Validation, Software, Methodology, Formal Analysis, Data curation, Writing–review and editing. RS: Writing–review and editing. HD: Writing–review and editing. RK: Methodology, Writing–review and editing, Visualization, Validation, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. HJ and DH are supported by the MOBIDOC scheme, funded by the Tunisian Ministry of Higher Education and Scientific Research through the PromESsE project and managed by the ANPR. RK is supported by the Tunisian Ministry of Higher Education and Scientific Research and the Tunisian Ministry of Public Health.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1380613/full#supplementary-material
Abbreviations
IASP, International Association for the Study of Pain; ADR, Adverse Drug Reactions; COMT, Catéchol-O-Méthyl-Transférase; OPRM1, Opioid Receptor Mu 1; PharmGKB, Pharmacogenomics Knowledge Base; VIP, Very Important Pharmacogene; miRNA, microRNA; HWE, Hardy-Weinberg equilibrium; MDS, Multidimensional scaling plot; IBS, Identity-by-state; PEA, Pathway Enrichment Analysis; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; UP_KW, WikiPathway; BBID, BioCarta Pathways and the Biological Biochemical Image Database; MonaGO, Monash Gene Ontology; RMSD, Root Mean Square Deviation; SNP, Single Nucleotide Polymorphisms; MAF, Minor Allele Frequency; MED, Mediterranean; AMR, American; EAS, East Asiatic; ERs, Estrogen receptors; IBD, Inflammatory Bowel Disease; EPSE, Excitatory postsynaptic potential; mGluR1, Regulation of Metabotropic Glutamate Receptor subtype 1; HDL, Height, Density Lipoprotein; LDL, Low Density Lipoprotein; SCI, Spinal Cord Injury (SCI); FATC, FAT C-terminus domain; CSF, Cerebrospinal Fluid Markers.
References
Ahangar-Sirous, R., Alizadeh, M., Nejadghaderi, S. A., Noori, M., Khabbazi, A., Sullman, M. J. M., et al. (2023). The burden of neck pain in the Middle East and North Africa region, 1990-2019. Heliyon 9, e21296. doi:10.1016/j.heliyon.2023.e21296
Amirkafi, A., Mohammadi, F., Tehrani-Banihashemi, A., Abbasi-Kangevari, M., Abbasi-Kangevari, Z., Abdollahi, M., et al. (2023). Drug-use disorders in the eastern mediterranean region: a glance at GBD 2019 findings. Soc. Psychiatry Psychiatric Epidemiol. doi:10.1007/s00127-023-02587-w
Angus, D. C., and Chang, C. H. (2021). Heterogeneity of treatment effect: estimating how the effects of interventions vary across individuals. Jama 326, 2312–2313. doi:10.1001/jama.2021.20552
Benavides, R., Vsevolozhskaya, O., Cattaneo, S., Zaykin, D., Brenton, A., Parisien, M., et al. (2020). A functional polymorphism in the ATP-Binding Cassette B1 transporter predicts pharmacologic response to combination of nortriptyline and morphine in neuropathic pain patients. Pain 161, 619–629. doi:10.1097/j.pain.0000000000001750
Bjersing, J. L., Lundborg, C., Bokarewa, M. I., and Mannerkorpi, K. (2013). Profile of cerebrospinal microRNAs in fibromyalgia. PLoS One 8, e78762. doi:10.1371/journal.pone.0078762
Blanco, F., Muriel, C., Labrador, J., Gonzalez-Porras, J. R., Gonzalez-Sarmiento, R., and Lozano, F. S. (2016). Influence of UGT2B7, CYP3A4, and OPRM1 gene polymorphisms on transdermal buprenorphine pain control in patients with critical lower limb ischemia awaiting revascularization. Pain Pract. 16, 842–849. doi:10.1111/papr.12343
Blum, K., Braverman, E. R., Holder, J. M., Lubar, J. F., Monastra, V. J., Miller, D., et al. (2000). The reward deficiency syndrome: a biogenetic model for the diagnosis and treatment of impulsive, addictive and compulsive behaviors. J. Psychoact. Drugs 32 (i-iv), 1–112. doi:10.1080/02791072.2000.10736099
Boukhalfa, W., Jmel, H., Kheriji, N., Gouiza, I., Dallali, H., Hechmi, M., et al. (2023). Decoding the genetic relationship between Alzheimer’s disease and type 2 diabetes: potential risk variants and future direction for North Africa. Front. Aging Neurosci. 15, 1114810. doi:10.3389/fnagi.2023.1114810
Bright, D., Langerveld, A., Devuyst-Miller, S., Saadeh, C., Choker, A., Lehigh, E., et al. (2021). Identification of a sex-stratified genetic algorithm for opioid addiction risk. Pharmacogenomics J. 21, 326–335. doi:10.1038/s41397-021-00212-0
Bruno, A. E., Li, L., Kalabus, J. L., Pan, Y., Yu, A., and Hu, Z. (2012). miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3'UTRs of human genes. BMC Genomics 13, 44. doi:10.1186/1471-2164-13-44
Bugada, D., Lorini, L. F., Fumagalli, R., and Allegri, M. (2020). Genetics and opioids: towards more appropriate prescription in cancer pain. Cancers (Basel) 12, 1951. doi:10.3390/cancers12071951
Cai, L., Liu, X., Guo, Q., Huang, Q., Zhang, Q., and Cao, Z. (2020). MiR-15a attenuates peripheral nerve injury-induced neuropathic pain by targeting AKT3 to regulate autophagy. Genes Genomics 42, 77–85. doi:10.1007/s13258-019-00881-z
Candiotti, K. A., Yang, Z., Buric, D., Arheart, K., Zhang, Y., Rodriguez, Y., et al. (2014). Catechol-o-methyltransferase polymorphisms predict opioid consumption in postoperative pain. Anesth. Analg. 119, 1194–1200. doi:10.1213/ANE.0000000000000411
Cargnin, S., Viana, M., Sances, G., Bianchi, M., Ghiotto, N., Tassorelli, C., et al. (2014). Combined effect of common gene variants on response to drug withdrawal therapy in medication overuse headache. Eur. J. Clin. Pharmacol. 70, 1195–1202. doi:10.1007/s00228-014-1726-6
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. doi:10.1186/s13742-015-0047-8
Chatti, I., Woillard, J. B., Mili, A., Creveaux, I., Ben Charfeddine, I., Feki, J., et al. (2017). Genetic analysis of mu and kappa opioid receptor and COMT enzyme in cancer pain Tunisian patients under opioid treatment. Iran. J. Public Health 46, 1704–1711.
Chen, X. P., Tan, Z. R., Huang, S. L., Huang, Z., Ou-Yang, D. S., and Zhou, H. H. (2003). Isozyme-specific induction of low-dose aspirin on cytochrome P450 in healthy subjects. Clin. Pharmacol. Ther. 73, 264–271. doi:10.1067/mcp.2003.14
Chidambaran, V., Ashton, M., Martin, L. J., and Jegga, A. G. (2020). Systems biology-based approaches to summarize and identify novel genes and pathways associated with acute and chronic postsurgical pain. J. Clin. Anesth. 62, 109738. doi:10.1016/j.jclinane.2020.109738
Cho, T., and Chaban, V. V. (2012). Interaction between P2X3 and oestrogen receptor (ER)α/ERβ in ATP-mediated calcium signalling in mice sensory neurones. J. Neuroendocrinol. 24, 789–797. doi:10.1111/j.1365-2826.2011.02272.x
Colombo, F., Pintarelli, G., Galvan, A., Noci, S., Corli, O., Skorpen, F., et al. (2020). Identification of genetic polymorphisms modulating nausea and vomiting in two series of opioid-treated cancer patients. Sci. Rep. 10, 542. doi:10.1038/s41598-019-57358-y
Cook-Sather, S. D., Li, J., Goebel, T. K., Sussman, E. M., Rehman, M. A., and Hakonarson, H. (2014). TAOK3, a novel genome-wide association study locus associated with morphine requirement and postoperative pain in a retrospective pediatric day surgery population. Pain 155, 1773–1783. doi:10.1016/j.pain.2014.05.032
Dai, Z., Chu, H., Ma, J., Yan, Y., Zhang, X., and Liang, Y. (2018). The regulatory mechanisms and therapeutic potential of MicroRNAs: from chronic pain to morphine tolerance. Front. Mol. Neurosci. 11, 80. doi:10.3389/fnmol.2018.00080
Dayer, C. F., Luthi, F., Le Carré, J., Vuistiner, P., Terrier, P., Benaim, C., et al. (2019). Differences in the miRNA signatures of chronic musculoskeletal pain patients from neuropathic or nociceptive origins. PLoS One 14, e0219311. doi:10.1371/journal.pone.0219311
De Gregori, M., Diatchenko, L., Ingelmo, P. M., Napolioni, V., Klepstad, P., Belfer, I., et al. (2016). Human genetic variability contributes to postoperative morphine consumption. J. Pain 17, 628–636. doi:10.1016/j.jpain.2016.02.003
De Larramendi, H., Dunne, C. W., Lange, M. A., and Teevan, C. (2020). “IEMed mediterranean yearbook 2020,” in European institute of the mediterranean (IEMed) Barcelona, Spain.
Dueñas, M., Salazar, A., Ojeda, B., Fernández-Palacín, F., Micó, J. A., Torres, L. M., et al. (2015). A nationwide study of chronic pain prevalence in the general Spanish population: identifying clinical subgroups through cluster analysis. Pain Med. 16, 811–822. doi:10.1111/pme.12640
Elens, L., Norman, E., Matic, M., Rane, A., Fellman, V., and Van Schaik, R. H. (2016). Genetic predisposition to poor opioid response in preterm infants: impact of KCNJ6 and COMT polymorphisms on pain relief after endotracheal intubation. Ther. Drug Monit. 38, 525–533. doi:10.1097/FTD.0000000000000301
Erlich, P. M., Hoffman, S. N., Rukstalis, M., Han, J. J., Chu, X., Linda Kao, W. H., et al. (2010). Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum. Genet. 128, 491–499. doi:10.1007/s00439-010-0876-6
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Fayaz, A., Croft, P., Langford, R. M., Donaldson, L. J., and Jones, G. T. (2016). Prevalence of chronic pain in the UK: a systematic review and meta-analysis of population studies. BMJ Open 6, e010364. doi:10.1136/bmjopen-2015-010364
Firfirey, F., September, A. V., and Shamley, D. (2022). ABCB1 and OPRM1 single-nucleotide polymorphisms collectively modulate chronic shoulder pain and dysfunction in South African breast cancer survivors. Pharmacogenomics 23, 513–530. doi:10.2217/pgs-2022-0020
Galvan, A., Skorpen, F., Klepstad, P., Knudsen, A. K., Fladvad, T., Falvella, F. S., et al. (2011). Multiple Loci modulate opioid therapy response for cancer pain. Clin. Cancer Res. 17, 4581–4587. doi:10.1158/1078-0432.CCR-10-3028
Gómez, R., Schurr, T. G., and Meraz-Ríos, M. A. (2021). “139C12Diversity of Mexican paternal lineages reflects evidence of migration and 500 Years of admixture,” in Human migration: biocultural perspectives. Editors M. D. L. MUÑOZ-MORENO, and M. H. CRAWFORD (Oxford University Press).
González-Castro, T. B., Hernandez-Diaz, Y., Juárez-Rojop, I. E., López-Narváez, L., Tovilla-Zárate, C. A., Rodriguez-Perez, J. M., et al. (2017). The role of the Cys23Ser (rs6318) polymorphism of the HTR2C gene in suicidal behavior: systematic review and meta-analysis. Psychiatr. Genet. 27, 199–209. doi:10.1097/YPG.0000000000000184
Goodman, C. W., and Brett, A. S. (2021). Race and pharmacogenomics-personalized medicine or misguided practice? Jama 325, 625–626. doi:10.1001/jama.2020.25473
Gottesman, I. I., and Gould, T. D. (2003). The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry 160, 636–645. doi:10.1176/appi.ajp.160.4.636
Grecco, G. G., and Atwood, B. K. (2020). Prenatal opioid exposure enhances responsiveness to future drug reward and alters sensitivity to pain: a review of preclinical models and contributing mechanisms. eNeuro 7, 0393–2020. doi:10.1523/ENEURO.0393-20.2020
Haleem, D. J. (2019). Targeting Serotonin1A receptors for treating chronic pain and depression. Curr. Neuropharmacol. 17, 1098–1108. doi:10.2174/1570159X17666190811161807
Hamdan, A., and Mosleh, R. (2024). How does the general population approach their pain? A cross-sectional study in Palestine. SAGE Open Med. 12, 20503121231223442. doi:10.1177/20503121231223442
Harding, E. K., and Zamponi, G. W. (2022). Central and peripheral contributions of T-type calcium channels in pain. Mol. Brain 15, 39. doi:10.1186/s13041-022-00923-w
Henn, B. M., Botigué, L. R., Gravel, S., Wang, W., Brisbin, A., Byrnes, J. K., et al. (2012). Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 8, e1002397. doi:10.1371/journal.pgen.1002397
Huang, B., Guo, S., Zhang, Y., Lin, P., Lin, C., Chen, M., et al. (2022). MiR-223-3p alleviates trigeminal neuropathic pain in the male mouse by targeting MKNK2 and MAPK/ERK signaling. Brain Behav. 12, e2634. doi:10.1002/brb3.2634
Hu, C., He, M., Xu, Q., and Tian, W. (2021). Advances with non-coding RNAs in neuropathic pain. Front. Neurosci. 15, 760936. doi:10.3389/fnins.2021.760936
Isagulyan, E. D., Makashova, E. S., Myasnikova, L. K., Sergeenko, E. V., Aslakhanova, K. S., Tomskiy, A. A., et al. (2022). Psychogenic (nociplastic) pain: current state of diagnosis, treatment options, and potentials of neurosurgical management. Prog. Brain Res. 272, 105–123. doi:10.1016/bs.pbr.2022.03.008
Isvoran, A., Peng, Y., Ceauranu, S., Schmidt, L., Nicot, A. B., and Miteva, M. A. (2022). Pharmacogenetics of human sulfotransferases and impact of amino acid exchange on Phase II drug metabolism. Drug Discov. Today 27, 103349. doi:10.1016/j.drudis.2022.103349
Jhun, E. H., Hu, X., Sadhu, N., Yao, Y., He, Y., Wilkie, D. J., et al. (2018). Transient receptor potential polymorphism and haplotype associate with crisis pain in sickle cell disease. Pharmacogenomics 19, 401–411. doi:10.2217/pgs-2017-0198
Jiang, Q., Li, W. X., Sun, J. R., Zhu, T. T., Fan, J., Yu, L. H., et al. (2017). Inhibitory effect of estrogen receptor beta on P2X3 receptors during inflammation in rats. Purinergic Signal 13, 105–117. doi:10.1007/s11302-016-9540-5
Jmel, H., Romdhane, L., Ben Halima, Y., Hechmi, M., Naouali, C., Dallali, H., et al. (2018). Pharmacogenetic landscape of Metabolic Syndrome components drug response in Tunisia and comparison with worldwide populations. PLoS One 13, e0194842. doi:10.1371/journal.pone.0194842
Kahma, H., Filppula, A. M., Neuvonen, M., Tarkiainen, E. K., Tornio, A., Holmberg, M. T., et al. (2018). Clopidogrel carboxylic acid glucuronidation is mediated mainly by UGT2B7, UGT2B4, and UGT2B17: implications for pharmacogenetics and drug-drug interactions( ). Drug Metab. Dispos. 46, 141–150. doi:10.1124/dmd.117.078162
Karran, P. (2001). Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis 22, 1931–1937. doi:10.1093/carcin/22.12.1931
Kaye, A. D., Garcia, A. J., Hall, O. M., Jeha, G. M., Cramer, K. D., Granier, A. L., et al. (2019). Update on the pharmacogenomics of pain management. Pharmgenomics Pers. Med. 12, 125–143. doi:10.2147/PGPM.S179152
Klepstad, P., Fladvad, T., Skorpen, F., Bjordal, K., Caraceni, A., Dale, O., et al. (2011). Influence from genetic variability on opioid use for cancer pain: a European genetic association study of 2294 cancer pain patients. Pain 152, 1139–1145. doi:10.1016/j.pain.2011.01.040
Klepstad, P., Rakvåg, T. T., Kaasa, S., Holthe, M., Dale, O., Borchgrevink, P. C., et al. (2004). The 118 A > G polymorphism in the human mu-opioid receptor gene may increase morphine requirements in patients with pain caused by malignant disease. Acta Anaesthesiol. Scand. 48, 1232–1239. doi:10.1111/j.1399-6576.2004.00517.x
Knezevic, N. N., Jovanovic, F., Voronov, D., and Candido, K. D. (2018a). Do corticosteroids still have a place in the treatment of chronic pain? Front. Pharmacol. 9, 1229. doi:10.3389/fphar.2018.01229
Knezevic, N. N., Tverdohleb, T., Knezevic, I., and Candido, K. D. (2018b). The role of genetic polymorphisms in chronic pain patients. Int. J. Mol. Sci. 19, 1707. doi:10.3390/ijms19061707
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A., and Mayrose, I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191. doi:10.1111/1755-0998.12387
Kosciuczuk, U., Knapp, P., and Lotowska-Cwiklewska, A. M. (2020). Opioid-induced immunosuppression and carcinogenesis promotion theories create the newest trend in acute and chronic pain pharmacotherapy. Clin. (Sao Paulo) 75, e1554. doi:10.6061/clinics/2020/e1554
Kumar, S., Kundra, P., Ramsamy, K., and Surendiran, A. (2019). Pharmacogenetics of opioids: a narrative review. Anaesthesia 74, 1456–1470. doi:10.1111/anae.14813
Kurita, G. P., Ekholm, O., Kaasa, S., Klepstad, P., Skorpen, F., and Sjøgren, P. (2016). Genetic variation and cognitive dysfunction in opioid-treated patients with cancer. Brain Behav. 6, e00471. doi:10.1002/brb3.471
Lambert, D. G. (2020). Opioids and the COVID-19 pandemic: does chronic opioid use or misuse increase clinical vulnerability? Br. J. Anaesth. 125, e382–e383. doi:10.1016/j.bja.2020.07.004
Langley, P. C. (2011). The prevalence, correlates and treatment of pain in the European Union. Curr. Med. Res. Opin. 27, 463–480. doi:10.1185/03007995.2010.542136
Lavanderos, M. A., Cayún, J. P., Roco, Á., Sandoval, C., Cerpa, L., Rubilar, J. C., et al. (2019). Association study among candidate genetic polymorphisms and chemotherapy-related severe toxicity in testicular cancer patients. Front. Pharmacol. 10, 206. doi:10.3389/fphar.2019.00206
Li, J. Z., Absher, D. M., Tang, H., Southwick, A. M., Casto, A. M., Ramachandran, S., et al. (2008). Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104. doi:10.1126/science.1153717
Li, J., Wei, Z., Zhang, J., Hakonarson, H., and Cook-Sather, S. D. (2019). Candidate gene analyses for acute pain and morphine analgesia after pediatric day surgery: African American versus European Caucasian ancestry and dose prediction limits. Pharmacogenomics J. 19, 570–581. doi:10.1038/s41397-019-0074-4
Liu, J., Tang, X., Shi, F., Li, C., Zhang, K., Liu, J., et al. (2018). Genetic polymorphism contributes to (131)I radiotherapy-induced toxicities in patients with differentiated thyroid cancer. Pharmacogenomics 19, 1335–1344. doi:10.2217/pgs-2018-0070
Lopes, G. S., Lopes, J. L., Bielinski, S. J., Armasu, S. M., Zhu, Y., Cavanaugh, D. C., et al. (2022). Identification of sex-specific genetic associations in response to opioid analgesics in a White, non-Hispanic cohort from Southeast Minnesota. Pharmacogenomics J. 22, 117–123. doi:10.1038/s41397-022-00265-9
López-Lázaro, M. (2022). Opium, street opium, and cancer risk. Curr. Pharm. Des. 28, 2039–2042. doi:10.2174/1381612828666220607104805
Louriz, M., Belayachi, J., Madani, N., Abidi, K., Dendane, T., Belabes Benchekroun, A., et al. (2016). Practices and perceived barriers regarding pain management among Emergency Department physicians: a nationwide multicenter survey in Moroccan hospitals. Acute Med. Surg. 3, 360–363. doi:10.1002/ams2.201
Mahajan, P. B. (2014). Will pharmacogenomics take the pain out of pain medication. J. Pharmacogenomics Pharmacoproteomics 6, e142. doi:10.4172/2153-0645.1000e142
Manners, M. T., Tian, Y., Zhou, Z., and Ajit, S. K. (2015). MicroRNAs downregulated in neuropathic pain regulate MeCP2 and BDNF related to pain sensitivity. FEBS Open Bio 5, 733–740. doi:10.1016/j.fob.2015.08.010
Maumus, M., Mancini, R., Zumsteg, D. M., and Mandali, D. K. (2020). Aberrant drug-related behavior monitoring. Ochsner J. 20, 358–361. doi:10.31486/toj.20.0108
Mcclay, J. L., Adkins, D. E., Aberg, K., Bukszár, J., Khachane, A. N., Keefe, R. S., et al. (2011). Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology 36, 616–626. doi:10.1038/npp.2010.193
Menon, S., Lea, R. A., Roy, B., Hanna, M., Wee, S., Haupt, L. M., et al. (2012). Genotypes of the MTHFR C677T and MTRR A66G genes act independently to reduce migraine disability in response to vitamin supplementation. Pharmacogenet Genomics 22, 741–749. doi:10.1097/FPC.0b013e3283576b6b
Merlin, J. S., Young, S. R., Starrels, J. L., Azari, S., Edelman, E. J., Pomeranz, J., et al. (2018). Managing concerning behaviors in patients prescribed opioids for chronic pain: a delphi study. J. Gen. Intern Med. 33, 166–176. doi:10.1007/s11606-017-4211-y
Mills, S. E. E., Nicolson, K. P., and Smith, B. H. (2019). Chronic pain: a review of its epidemiology and associated factors in population-based studies. Br. J. Anaesth. 123, e273–e283. doi:10.1016/j.bja.2019.03.023
Miró, J., Paredes, S., Rull, M., Queral, R., Miralles, R., Nieto, R., et al. (2007). Pain in older adults: a prevalence study in the Mediterranean region of Catalonia. Eur. J. Pain 11, 83–92. doi:10.1016/j.ejpain.2006.01.001
Mir, R., Bhat, M., Javid, J., Jha, C., Saxena, A., and Banu, S. (2018). Potential impact of COMT-rs4680 G > A gene polymorphism in coronary artery disease. J. Cardiovasc Dev. Dis. 5, 38. doi:10.3390/jcdd5030038
Mizzi, C., Peters, B., Mitropoulou, C., Mitropoulos, K., Katsila, T., Agarwal, M. R., et al. (2014). Personalized pharmacogenomics profiling using whole-genome sequencing. Pharmacogenomics 15, 1223–1234. doi:10.2217/pgs.14.102
Molanaei, H., Carrero, J. J., Heimbürger, O., Nordfors, L., Lindholm, B., Stenvinkel, P., et al. (2010). Influence of the CYP2D6 polymorphism and hemodialysis on codeine disposition in patients with end-stage renal disease. Eur. J. Clin. Pharmacol. 66, 269–273. doi:10.1007/s00228-009-0759-8
Muñoz-Alvaredo, L., López Vallecillo, M., Jiménez Pérez, J. M., Martín-Gil, B., Muñoz Moreno, M. F., and Fernández-Castro, M. (2020). Prevalence, pain management and registration in Internal Medicine units. Enferm. Clin. Engl. Ed. 30, 275–281. doi:10.1016/j.enfcli.2018.11.004
Muriel, J., Margarit, C., Barrachina, J., Ballester, P., Flor, A., Morales, D., et al. (2019). Pharmacogenetics and prediction of adverse events in prescription opioid use disorder patients. Basic Clin. Pharmacol. Toxicol. 124, 439–448. doi:10.1111/bcpt.13155
Nahin, R. L. (2015). Estimates of pain prevalence and severity in adults: United States, 2012. J. Pain 16, 769–780. doi:10.1016/j.jpain.2015.05.002
Nishizawa, D., Mieda, T., Tsujita, M., Nakagawa, H., Yamaguchi, S., Kasai, S., et al. (2018). Genome-wide scan identifies candidate loci related to remifentanil requirements during laparoscopic-assisted colectomy. Pharmacogenomics 19, 113–127. doi:10.2217/pgs-2017-0109
Ortega, V. E., and Meyers, D. A. (2014). Implications of population structure and ancestry on asthma genetic studies. Curr. Opin. Allergy Clin. Immunol. 14, 381–389. doi:10.1097/ACI.0000000000000102
Page, M. J., Mckenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Bmj 372, n71. doi:10.1136/bmj.n71
Park, H. J., Ye, Y. M., Hur, G. Y., Kim, S. H., and Park, H. S. (2008). Association between a TGFbeta1 promoter polymorphism and the phenotype of aspirin-intolerant chronic urticaria in a Korean population. J. Clin. Pharm. Ther. 33, 691–697. doi:10.1111/j.1365-2710.2008.00957.x
Pathan, H., and Williams, J. (2012). Basic opioid pharmacology: an update. Br. J. Pain 6, 11–16. doi:10.1177/2049463712438493
Perry, M., Baumbauer, K., Young, E. E., Dorsey, S. G., Taylor, J. Y., and Starkweather, A. R. (2019). The influence of race, ethnicity and genetic variants on postoperative pain intensity: an integrative literature review. Pain Manag. Nurs. 20, 198–206. doi:10.1016/j.pmn.2018.11.002
Pickering, G., Creveaux, I., Macian, N., and Pereira, B. (2020). Paracetamol and pain modulation by TRPV1, UGT2B15, SULT1A1 genotypes: a randomized clinical trial in healthy volunteers. Pain Med. 21, 661–669. doi:10.1093/pm/pnz037
Piomelli, D., Hohmann, A. G., Seybold, V., and Hammock, B. D. (2014). A lipid gate for the peripheral control of pain. J. Neurosci. 34, 15184–15191. doi:10.1523/JNEUROSCI.3475-14.2014
Planelles, B., Margarit, C., Inda, M. D., Ballester, P., Muriel, J., Barrachina, J., et al. (2020). Gender based differences, pharmacogenetics and adverse events in chronic pain management. Pharmacogenomics J. 20, 320–328. doi:10.1038/s41397-019-0118-9
Polli, A., Godderis, L., Ghosh, M., Ickmans, K., and Nijs, J. (2020). Epigenetic and miRNA expression changes in people with pain: a systematic review. J. Pain 21, 763–780. doi:10.1016/j.jpain.2019.12.002
Polo-Santos, M., Videla-Cés, S., Pérez-Hernández, C., Mayoral-Rojals, V., Ribera-Canudas, M. V., and Sarría-Santamera, A. (2021). An update on resources, procedures and healthcare provision in pain units: a survey of Spanish practitioners. Int. J. Environ. Res. Public Health 18, 451. doi:10.3390/ijerph18020451
Raffaeli, W., Tenti, M., Corraro, A., Malafoglia, V., Ilari, S., Balzani, E., et al. (2021). Chronic pain: what does it mean? A review on the use of the term chronic pain in clinical practice. J. Pain Res. 14, 827–835. doi:10.2147/JPR.S303186
Raja, S. N., Carr, D. B., Cohen, M., Finnerup, N. B., Flor, H., Gibson, S., et al. (2020). The revised International Association for the Study of Pain definition of pain: concepts, challenges, and compromises. Pain 161, 1976–1982. doi:10.1097/j.pain.0000000000001939
Rakvåg, T. T., Klepstad, P., Baar, C., Kvam, T.-M., Dale, O., Kaasa, S., et al. (2005). The Val158Met polymorphism of the human catechol-O-methyltransferase (COMT) gene may influence morphine requirements in cancer pain patients. Pain 116, 73–78. doi:10.1016/j.pain.2005.03.032
Rakvåg, T. T., Ross, J. R., Sato, H., Skorpen, F., Kaasa, S., and Klepstad, P. (2008). Genetic variation in the catechol-O-methyltransferase (COMT) gene and morphine requirements in cancer patients with pain. Mol. Pain 4, 64. doi:10.1186/1744-8069-4-64
Ren, W., and Neugebauer, V. (2010). Pain-related increase of excitatory transmission and decrease of inhibitory transmission in the central nucleus of the amygdala are mediated by mGluR1. Mol. Pain 6, 93. doi:10.1186/1744-8069-6-93
Roberts, B., Cooper, Z., Lu, S., Stanley, S., Majda, B. T., Collins, K. R. L., et al. (2023). Utility of pharmacogenetic testing to optimise antidepressant pharmacotherapy in youth: a narrative literature review. Front. Pharmacol. 14, 1267294. doi:10.3389/fphar.2023.1267294
Rosenberg, N. A. (2004). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. notes 4, 137–138. doi:10.1046/j.1471-8286.2003.00566.x
Ross, J. R., Rutter, D., Welsh, K., Joel, S. P., Goller, K., Wells, A. U., et al. (2005). Clinical response to morphine in cancer patients and genetic variation in candidate genes. Pharmacogenomics J. 5, 324–336. doi:10.1038/sj.tpj.6500327
Sabina, S., Panico, A., Mincarone, P., Leo, C. G., Garbarino, S., Grassi, T., et al. (2022). Expression and biological functions of miRNAs in chronic pain: a review on human studies. Int. J. Mol. Sci. 23, 6016. doi:10.3390/ijms23116016
Sadhasivam, S., Chidambaran, V., Olbrecht, V. A., Esslinger, H. R., Zhang, K., Zhang, X., et al. (2014). Genetics of pain perception, COMT and postoperative pain management in children. Pharmacogenomics 15, 277–284. doi:10.2217/pgs.13.248
Sahbaie, P., Irvine, K. A., Liang, D. Y., Shi, X., and Clark, J. D. (2019). Mild traumatic brain injury causes nociceptive sensitization through spinal chemokine upregulation. Sci. Rep. 9, 19500. doi:10.1038/s41598-019-55739-x
Saiz-Rodríguez, M., Ochoa, D., Herrador, C., Belmonte, C., Román, M., Alday, E., et al. (2019). Polymorphisms associated with fentanyl pharmacokinetics, pharmacodynamics and adverse effects. Basic Clin. Pharmacol. Toxicol. 124, 321–329. doi:10.1111/bcpt.13141
Schuh, M. J., Randles, H., and Crosby, S. (2020). Improving pain management with pharmacogenomics: a general introduction. J. Pain Palliat. Care Pharmacother. 34, 114–119. doi:10.1080/15360288.2020.1734140
Selvaraj, V., Sekaran, S., and Rajamani Sekar, S. K. (2023). Advancing postoperative pain management in humans with miRNA-based therapeutic strategies: lacunae that need to be addressed. Int. J. Surg. 109, 2849–2850. doi:10.1097/JS9.0000000000000514
Shah, R. R., and Gaedigk, A. (2018). Precision medicine: does ethnicity information complement genotype-based prescribing decisions? Ther. Adv. Drug Saf. 9, 45–62. doi:10.1177/2042098617743393
Sheikh, M., Brennan, P., Mariosa, D., and Robbins, H. A. (2023). Opioid medications: an emerging cancer risk factor? Br. J. Anaesth. 130, e401–e403. doi:10.1016/j.bja.2022.12.007
Shin, S. W., Park, B. L., Chang, H., Park, J. S., Bae, D. J., Song, H. J., et al. (2014). Exonic variants associated with development of aspirin exacerbated respiratory diseases. PLoS One 9, e111887. doi:10.1371/journal.pone.0111887
Sikora, M., Skupio, U., Jastrzebska, K., Rodriguez Parkitna, J., and Przewlocki, R. (2019). Antagonism of μ-opioid receptors reduces sensation seeking-like behavior in mice. Behav. Brain Res. 359, 498–501. doi:10.1016/j.bbr.2018.11.039
Stamer, U. M., Zhang, L., Book, M., Lehmann, L. E., Stuber, F., and Musshoff, F. (2013). CYP2D6 genotype dependent oxycodone metabolism in postoperative patients. PLoS One 8, e60239. doi:10.1371/journal.pone.0060239
Tanaka, R., Sato, J., Ishikawa, H., Sato, T., Shino, M., Ohde, Y., et al. (2021). Influence of genetic variants of opioid-related genes on opioid-induced adverse effects in patients with lung cancer: a STROBE-compliant observational study. Med. Baltim. 100, e27565. doi:10.1097/MD.0000000000027565
Tchivileva, I. E., Lim, P. F., Smith, S. B., Slade, G. D., Diatchenko, L., Mclean, S. A., et al. (2010). Effect of catechol-O-methyltransferase polymorphism on response to propranolol therapy in chronic musculoskeletal pain: a randomized, double-blind, placebo-controlled, crossover pilot study. Pharmacogenet Genomics 20, 239–248. doi:10.1097/FPC.0b013e328337f9ab
Tilley, B. J., Cook, J. L., Docking, S. I., and Gaida, J. E. (2015). Is higher serum cholesterol associated with altered tendon structure or tendon pain? A systematic review. Br. J. Sports Med. 49, 1504–1509. doi:10.1136/bjsports-2015-095100
Toyama, K., Kiyosawa, N., Watanabe, K., and Ishizuka, H. (2017). Identification of circulating miRNAs differentially regulated by opioid treatment. Int. J. Mol. Sci. 18, 1991. doi:10.3390/ijms18091991
Tramullas, M., Francés, R., De La Fuente, R., Velategui, S., Carcelén, M., García, R., et al. (2018). MicroRNA-30c-5p modulates neuropathic pain in rodents. Sci. Transl. Med. 10, eaao6299. doi:10.1126/scitranslmed.aao6299
Tzvetkov, M. V., Saadatmand, A. R., Lötsch, J., Tegeder, I., Stingl, J. C., and Brockmöller, J. (2011). Genetically polymorphic OCT1: another piece in the puzzle of the variable pharmacokinetics and pharmacodynamics of the opioidergic drug tramadol. Clin. Pharmacol. Ther. 90, 143–150. doi:10.1038/clpt.2011.56
Valomon, A., Holst, S. C., Bachmann, V., Viola, A. U., Schmidt, C., Zürcher, J., et al. (2014). Genetic polymorphisms of DAT1 and COMT differentially associate with actigraphy-derived sleep-wake cycles in young adults. Chronobiol Int. 31, 705–714. doi:10.3109/07420528.2014.896376
Venkatasubramanian, R., Fukuda, T., Niu, J., Mizuno, T., Chidambaran, V., Vinks, A. A., et al. (2014). ABCC3 and OCT1 genotypes influence pharmacokinetics of morphine in children. Pharmacogenomics 15, 1297–1309. doi:10.2217/pgs.14.99
Viguier, F., Michot, B., Hamon, M., and Bourgoin, S. (2013). Multiple roles of serotonin in pain control mechanisms--implications of 5-HT₇ and other 5-HT receptor types. Eur. J. Pharmacol. 716, 8–16. doi:10.1016/j.ejphar.2013.01.074
Wang, H., Chen, C., Ding, K., Zhang, W., and Hou, J. (2020). MiR-24-3p as a prognostic indicator for multiple cancers: from a meta-analysis view. Biosci. Rep. 40. doi:10.1042/BSR20202938
Wang, X., Lai, Y., Luo, Y., Zhang, X., Zhou, H., Ye, Z., et al. (2016). Relationship between clopidogrel-related polymorphisms and variable platelet reactivity at 1 year: a cohort study from Han Chinese. J. Res. Med. Sci. 21, 111. doi:10.4103/1735-1995.193502
Wen, F., Yu, J., and Cheng, Y. (2022). Network pharmacology-based dissection of the mechanism of drynariae rhizoma for low back pain. Biomed. Res. Int. 2022, 6092424. doi:10.1155/2022/6092424
Wiss, F. M., Stäuble, C. K., Meyer Zu Schwabedissen, H. E., Allemann, S. S., and Lampert, M. L. (2023). Pharmacogenetic analysis enables optimization of pain therapy: a case report of ineffective oxycodone therapy. J. Pers. Med. 13, 829. doi:10.3390/jpm13050829
Xian, S., Ding, R., Li, M., and Chen, F. (2021). LncRNA NEAT1/miR-128-3p/AQP4 axis regulating spinal cord injury-induced neuropathic pain progression. J. Neuroimmunol. 351, 577457. doi:10.1016/j.jneuroim.2020.577457
Xin, Z., Cai, Y., Dang, L. T., Burke, H. M. S., Revote, J., Charitakis, N., et al. (2022). MonaGO: a novel gene ontology enrichment analysis visualisation system. BMC Bioinforma. 23, 69. doi:10.1186/s12859-022-04594-1
Xu, Z., and Taylor, J. A. (2009). SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 37, W600–W605. doi:10.1093/nar/gkp290
Yao, L., Guo, Y., Wang, L., Li, G., Qian, X., Zhang, J., et al. (2021). Knockdown of miR-130a-3p alleviates spinal cord injury induced neuropathic pain by activating IGF-1/IGF-1R pathway. J. Neuroimmunol. 351, 577458. doi:10.1016/j.jneuroim.2020.577458
Yennurajalingam, S., Astolfi, A., Indio, V., Beccaro, M., Schipani, A., Yu, R., et al. (2021). Genetic factors associated with pain severity, daily opioid dose requirement, and pain response among advanced cancer patients receiving supportive care. J. Pain Symptom Manage 62, 785–795. doi:10.1016/j.jpainsymman.2021.03.024
Yoshida, K., Nishizawa, D., Ide, S., Ichinohe, T., Fukuda, K. I., and Ikeda, K. (2018). A pharmacogenetics approach to pain management. Neuropsychopharmacol. Rep. 38, 2–8. doi:10.1002/npr2.12003
Yousuf, Z., Iman, K., Iftikhar, N., and Mirza, M. U. (2017). Structure-based virtual screening and molecular docking for the identification of potential multi-targeted inhibitors against breast cancer. Breast Cancer (Dove Med. Press) 9, 447–459. doi:10.2147/BCTT.S132074
Yu, Y., Xu, X., Lin, C., and Liu, R. (2023). Systematic identification of potential key microRNAs and circRNAs in the dorsal root ganglia of mice with sciatic nerve injury. Front. Mol. Neurosci. 16, 1119164. doi:10.3389/fnmol.2023.1119164
Zhang, X., Zhang, Y., Cai, W., Liu, Y., Liu, H., Zhang, Z., et al. (2020). MicroRNA-128-3p alleviates neuropathic pain through targeting ZEB1. Neurosci. Lett. 729, 134946. doi:10.1016/j.neulet.2020.134946
Zhu, X. L., Han, X., Xin, X. F., Song, H. J., and Guo, H. (2020). Correlations of analgesic dosage of morphine with SLC6A4 gene polymorphisms in patients with lung cancer. Eur. Rev. Med. Pharmacol. Sci. 24, 5046–5052. doi:10.26355/eurrev_202005_21197
Keywords: pharmacogenes, opioids, Mediterranean populations, adverse drug reaction, admixture
Citation: Jmel H, Boukhalfa W, Gouiza I, Seghaier RO, Dallali H and Kefi R (2024) Pharmacogenetic landscape of pain management variants among Mediterranean populations. Front. Pharmacol. 15:1380613. doi: 10.3389/fphar.2024.1380613
Received: 01 February 2024; Accepted: 05 April 2024;
Published: 15 May 2024.
Edited by:
Georgia Ragia, Democritus University of Thrace, GreeceCopyright © 2024 Jmel, Boukhalfa, Gouiza, Seghaier, Dallali and Kefi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rym Kefi, cnltLmtlZmlAcGFzdGV1ci51dG0udG4=
 Haifa Jmel
Haifa Jmel Wided Boukhalfa
Wided Boukhalfa Ismail Gouiza
Ismail Gouiza Roua Ouled Seghaier1
Roua Ouled Seghaier1 Hamza Dallali
Hamza Dallali Rym Kefi
Rym Kefi