
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 04 June 2024
Sec. Pharmacology of Infectious Diseases
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1339858
 Nicholas M. Smith1
Nicholas M. Smith1 Harpreet Kaur2
Harpreet Kaur2 Ravneet Kaur2
Ravneet Kaur2 Trisha Minoza2
Trisha Minoza2 Michael Kent2
Michael Kent2 Ayeh Barekat2
Ayeh Barekat2 Justin R. Lenhard2*
Justin R. Lenhard2*Objectives: We sought to evaluate the pharmacodynamics of β-lactam antibacterials against polymicrobial communities of clinically relevant gram-positive and gram-negative pathogens.
Methods: Two Enterococcus faecalis isolates, two Staphylococcus aureus isolates, and three Escherichia coli isolates with varying β-lactamase production were evaluated in static time-killing experiments. Each gram-positive isolate was exposed to a concentration array of ampicillin (E. faecalis) or cefazolin (S. aureus) alone and during co-culture with an E. coli isolate that was β-lactamase-deficient, produced TEM-1, or produced KPC-3/TEM-1B. The results of the time-killing experiments were summarized using an integrated pharmacokinetic/pharmacodynamics analysis as well as mathematical modelling to fully characterize the antibacterial pharmacodynamics.
Results: In the integrated analysis, the maximum killing of ampicillin (Emax) against both E. faecalis isolates was ≥ 4.11 during monoculture experiments or co-culture with β-lactamase-deficient E. coli, whereas the Emax was reduced to ≤ 1.54 during co-culture with β-lactamase-producing E. coli. In comparison to monoculture experiments, culturing S. aureus with KPC-producing E. coli resulted in reductions of the cefazolin Emax from 3.25 and 3.71 down to 2.02 and 2.98, respectively. Two mathematical models were created to describe the interactions between E. coli and either E. faecalis or S. aureus. When in co-culture with E. coli, S. aureus experienced a reduction in its cefazolin Kmax by 24.8% (23.1%RSE). Similarly, β-lactamase-producing E. coli preferentially protected the ampicillin-resistant E. faecalis subpopulation, reducing Kmax,r by 90.1% (14%RSE).
Discussion: β-lactamase-producing E. coli were capable of protecting S. aureus and E. faecalis from exposure to β-lactam antibacterials.
Combating the proliferation of antimicrobial resistance is an international goal that requires the cooperation of the global healthcare system for success (McEwen and Collignon, 2018; Septimus, 2018; Christaki et al., 2020; Morrison and Zembower, 2020). One of the main culprits of the spread of drug-resistant bacteria is the inappropriate use of antibacterial drugs. In recognition of the importance of judicious antibacterial use, many institutions have developed antimicrobial stewardship programs that focus on ensuring that patients receive appropriate anti-infective drug regimens with optimal doses, frequencies of administration, routes of administration, and durations of therapy (Marcelin et al., 2020; Pierce et al., 2020; Lanckohr and Bracht, 2022). Looking to the future, many experts are calling for individualized antimicrobial regimens that are tailored specifically to the patient and infective pathogens (Moser et al., 2019; Bulman et al., 2022; Ko and Tsalik, 2022); however, such a paradigm will not be possible unless the medical community gains a firm understanding of how antibacterial selection and dosing are impacted by the presence of multiple pathogens at the same site of infection.
At the moment, there is a critical lack of guidance for clinicians regarding how anti-infective regimens should be modified to target a specific polymicrobial community. For example, the Infectious Diseases Society of America guidelines on skin and soft tissue infections, community-acquired pneumonia, and hospital-acquired pneumonia do not provide recommendations for antibacterial selection or dosing based on the results of polymicrobial cultures (Stevens et al., 2014; Kalil et al., 2016; Metlay et al., 2019). Although investigators have started to evaluate the pharmacokinetics and pharmacodynamics (PK/PD) of antibacterials used against multiple pathogenic organisms, the focus in the literature has largely centered on the interactions of Staphylococcus aureus and Pseudomonas aeruginosa (DeLeon et al., 2014; Hoffman et al., 2006; Orazi and O'Toole, 2017; Orazi et al., 2019; Orazi et al., 2020; Lebrun et al., 1978; Michelsen et al., 2014; Dehbashi et al., 2021; Briaud et al., 2019; Tognon et al., 2017; Beaudoin et al., 2017; Cendra et al., 2019; Radlinski et al., 2017; Lenhard et al., 2019).
Enterobacterales, an order of gram-negative enteric bacteria, are becoming increasingly notorious for the spread of drug-resistant strains (Jean et al., 2022; Lepe and Martínez-Martínez, 2022). One of the most notorious resistance mechanisms utilized by pathogenic Enterobacterales is the production of β-lactamase enzymes that inactivate many of the most clinically relevant antibacterial agents (Rabaan et al., 2022; Caliskan-Aydogan and Alocilja, 2023). In the United States, Klebsiella pneumoniae carbapenemase (KPC) enzymes have become the most commonly encountered carbapenemase enzyme among carbapenem-resistant Enterobacterales (CRE), and the production of the enzyme has plagued many other countries as well (Hansen, 2021). The spread of CRE was identified as an “urgent” threat to public health by the Centers for Disease Control and Prevention and a “critical” priority for drug development by the World Health Organization (WHO, 2017; Centers for Disease Control, 2023).
Not only are Enterobacterales consistently encountered during polymicrobial intra-abdominal infections (Labricciosa et al., 2018; Cornely et al., 2020), but the organisms have been co-cultured along with gram-positive pathogens such as S. aureus and Enterococcus faecalis from skin and soft tissue infections (Shettigar et al., 2016), urinary tract infections (Siegman-Igra et al., 1994; Macleod and Stickler, 2007), pneumonia (Combes et al., 2002; Ferrer et al., 2015; Patil and Patil, 2017), and bacteremia as well (Pavlaki et al., 2013; Pammi et al., 2014; Yo et al., 2019). A prior investigation confirmed that carbapenemase-producing Acinetobacter baumannii are capable of sheltering adjacent gram-positive organisms from β-lactam exposure, but the ability of KPC-producing Enterobacterales to protect neighboring pathogens has not been defined (Liao et al., 2014). Given the spread of CRE, the PK/PD of β-lactam antibacterials against polymicrobial infections composed of Enterobacterales and gram-positive pathogens are needed to achieve truly individualized anti-infective regimens in the future.
Models of antibiotic pharmacodynamics often implement a subpopulation model structure to describe the bacterial cell population, which includes descriptions of a ‘susceptible’ subpopulation that constitutes the majority of cells (often >99%) and a ‘resistant’ subpopulation that constitutes the minority of cells (Jacobs et al., 2016; Minichmayr et al., 2022). Ultimately, these are data-driven strategies for characterizing antibiotic effects and rely on stepwise model building. Typically, the number of subpopulations are selected empirically based on likelihood ratio testing of the model objective function, however most models in the literature only require two to three subpopulations (Bulitta et al., 2015; Landersdorfer et al., 2018; Mi et al., 2022; Minichmayr et al., 2022; Smith et al., 2022). When modeling polymicrobial infections, each bacterial specie of interest would be modeled in a similar manner. An alternative model design can be implemented that models the full distribution of susceptibility among the bacteria but can be computationally expensive (Krzyzanski and Rao, 2017). Additionally, subpopulation models can be leveraged to facilitate tracking of resistance over time if the model is fit using both total bacterial counts along with population analysis profiles, but this process is labor-intensive (Landersdorfer et al., 2013).
Here, we report the results of a PK/PD analysis of β-lactam antibacterials against mixed cultures of Escherichia coli and either S. aureus or E. faecalis. Duos consisting of an E. coli isolate and a gram-positive organism were investigated in 24-h time-killing experiments to define the time course of bacterial killing in polymicrobial conditions. To determine the relevance of β-lactamase production, several E. coli isolates with different β-lactamase statuses were included in the investigation. Finally, a comprehensive analysis of the data was performed using several PK/PD and pharmacometric approaches.
The results of the ampicillin time-killing experiments against the E. faecalis isolates are summarized in Figure 1. When both E. faecalis isolates were cultured alone, ampicillin concentrations ≥ 6 mg/L achieved ≥ 3 log10 CFU/mL reductions by 24 h against both organisms. Similarly, ampicillin concentrations ≥ 6 mg/L achieved ≥ 2.5 log10 CFU/mL reductions by 24 h when both E. faecalis isolates were cultured with E. coli that do not produce a β-lactamase enzyme. In contrast, ampicillin concentrations up to 96 mg/L resulted in > 2 log10CFU/mL of E. faecalis growth above the starting inocula during co-culture with either of the β-lactamase-producing E. coli isolates. Despite having similar performance against both E. faecalis isolates in monoculture experiments, ampicillin was unable to achieve any killing against E. faecalis AR Bank # 0671 during co-culture with the KPC-producing E. coli, whereas ampicillin concentrations ≥ 6 mg/L reduced E. faecalis AR Bank #0573 counts by ≥ 1.8 log10 CFU/mL in the first 8 h before regrowth occurred by 24 h.
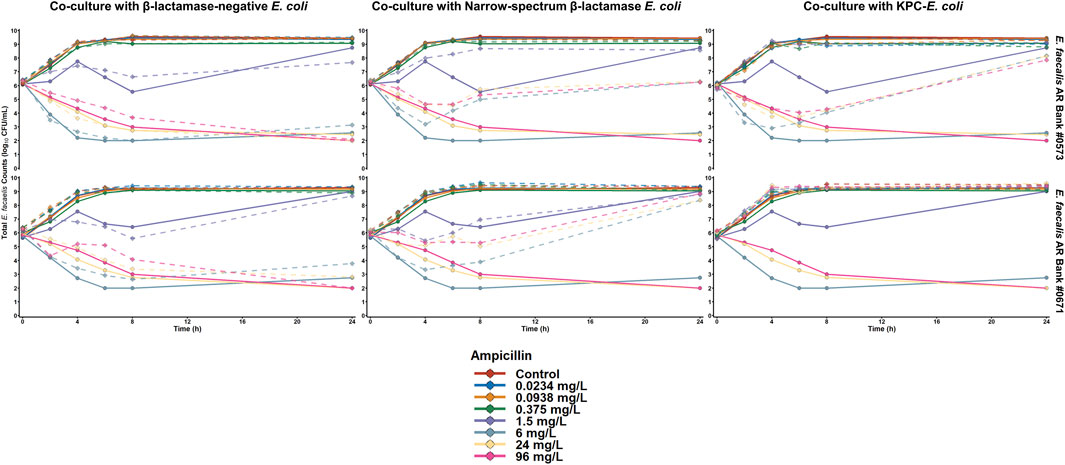
Figure 1. Time-killing plots depicting the activity of ampicillin against E. faecalis cultured alone or with E. coli that were either β-lactamase-deficient, produced a narrow spectrum TEM-1 enzyme, or produced TEM-1B and KPC-3. The quantity of E. faecalis is depicted for both monoculture (solid lines) and co-culture (dashed line) experiments.
The activity of cefazolin against both S. aureus isolates is depicted in Figure 2. When S. aureus ATCC 25923 was exposed to cefazolin alone or during co-culture with β-lactamase-deficient E. coli, cefazolin concentrations ≥ 0.25 mg/L achieved > 2 log10CFU/mL reductions by 24 h. In contrast, the maximum reductions of cefazolin concentrations between 0.25 and 4 mg/L were < 1 log10 CFU/mL during co-culture with either β-lactamase-producing E. coli isolate, and 16 mg/L achieved maximum reductions of 1.7 and 1.4 log10CFU/mL during c-culture with E. coli that produce TEM-1 and KPC-3/TEM-1B, respectively. Against S. aureus AR Bank # 0484 grown alone or cultured with E. coli with no β-lactamase production, cefazolin concentrations ≥ 1 mg/L achieved > 2.4 log10CFU/mL reductions by 24 h. During co-culture with TEM-1-producing E. coli, cefazolin concentrations of 1, 4, and 16 mg/L achieved maximum reductions of 1.2, 1.5, and 2.9 log10CFU/mL, respectively, whereas co-culture with KPC-3/TEM-1B-producing E. coli resulted in maximum reductions of 0.7, 0.6, and 1.1 log10CFU/mL, respectively.
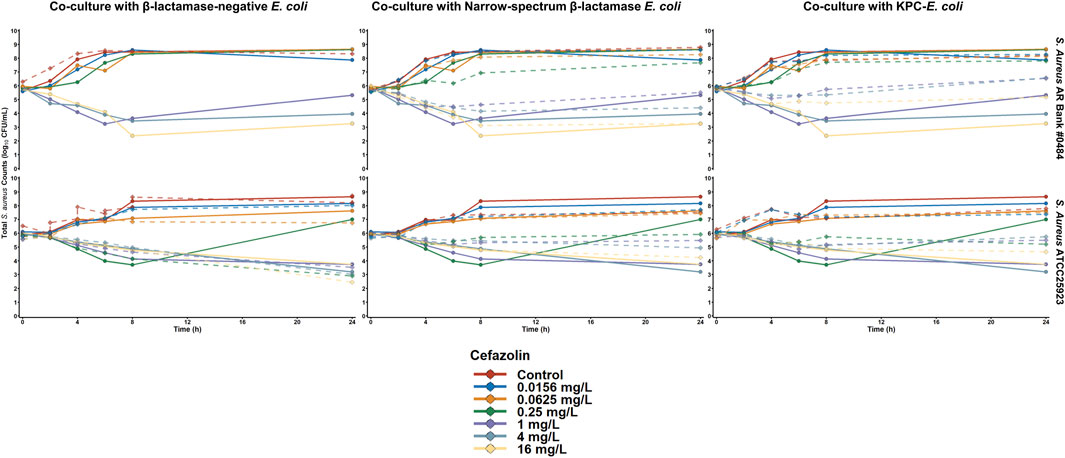
Figure 2. Time-killing plots depicting the activity of cefazolin against S. aureus cultured alone or with E. coli that were either β-lactamase-deficient, produced a narrow spectrum TEM-1 enzyme, or produced TEM-1B and KPC-3. The quantity of S. aureus is depicted for both monoculture (solid lines) and co-culture (dashed line) experiments.
The activity of ampicillin was evaluated against the β-lactamase-deficient E. coli (E. coli AR Bank #0017) alone and during co-culture with each of the four gram-positive isolates (Supplementary Figure S1, 2). When the E. coli was alone, cultured with either E. faecalis isolate, or cultured with the β-lactamase-deficient S. aureus (ATCC 25923), 96 mg/L of ampicillin reduced the E. coli counts below the limit of detection by 24 h. In contrast, when the E. coli was cultured with the penicillin-resistant S. aureus AR Bank # 0484, a maximum reduction of 2.2 log10 CFU/mL of E. coli was achieved at 6 h followed by regrowth to 2.6 log10 CFU/mL above the starting inoculum by 24 h. When the activity of cefazolin was evaluated against E. coli alone or cultured with each S. aureus isolate, 16 mg/L of cefazolin achieved a > 4.6 log10 CFU/mL reduction against the E. coli regardless of the presence of S. aureus (Supplementary Figure S3).
The maximum β-lactam killing (Emax) of E. faecalis and S. aureus is depicted in Figure 3. Against both E. faecalis isolates, ampicillin achieved an Emax ≥ 4.11 during monoculture experiments and during co-culture with E. coli that do not produce a β-lactamase. When E. faecalis AR Bank # 0573 and E. faecalis AR Bank # 0671 were separately cultured with TEM-1-producing E. coli, the Emax was reduced to 1.21 and 1.25, respectively. The Emax of ampicillin was 1.54 when E. faecalis AR Bank # 0573 was cultured with KPC-3/TEM-1B-producing E. coli, whereas no apparent killing was achieved against the second E faecalis isolate (Emax could not be defined). Against S. aureus ATCC 25923 alone, the Emax of cefazolin was 3.25, but the Emax declined down to 2.81 and 2.02 during co-culture with β-lactamase-deficient and β-lactamase-producing E. coli, respectively. The Emax of cefazolin ranged from 3.47–3.71 against S. aureus AR Bank # 0484 in each experiment with the exception that the Emax was reduced to 2.98 during co-culture with the KPC-3/TEM-1B-producing E. coli isolate.
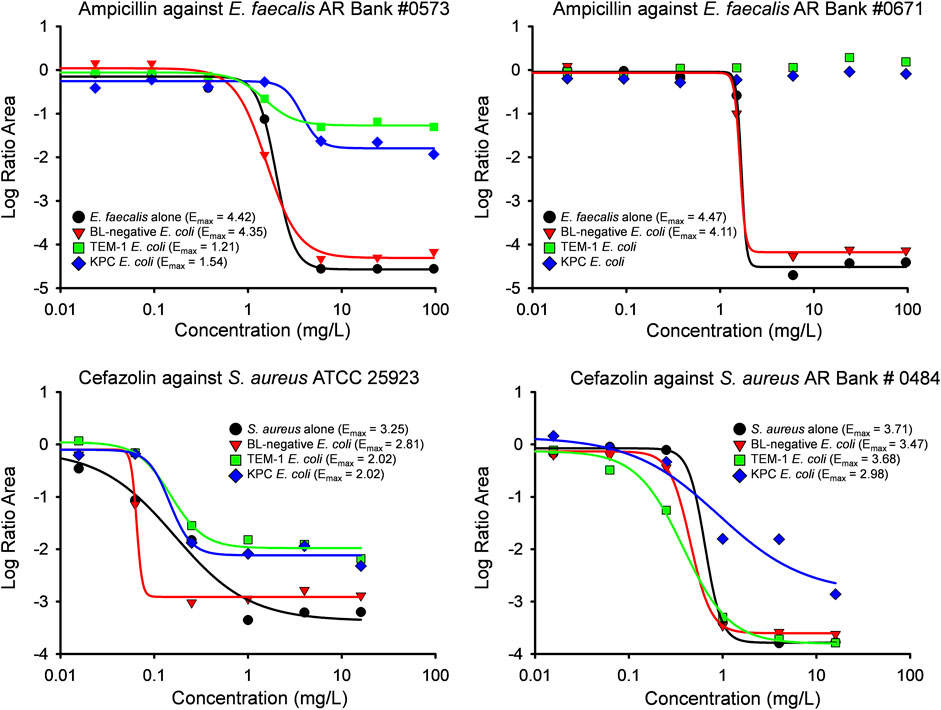
Figure 3. Results of an integrated PK/PD analysis are shown for both E. faecalis (top) and S. aureus (bottom) isolates investigated in the study. The Log Ratio Areas obtained from time-killing experiments were described by a mathematical Hill-type model in which Emax represents maximum antibacterial activity. The Log Ratio Areas of E. faecalis and S. aureus are shown for monoculture experiments (black circles) and co-culture with β-lactamase deficient E. coli (red triangles), E. coli producing TEM-1 (green squares), and E. coli producing KPC-3 and TEM-1B enzymes (blue diamonds). A Hill-type model was not able to describe some of the data for E. faecalis AR Bank #0671 due to lack of bacterial killing.
S. aureus in co-culture with E. coli was best described by a subpopulation-based model with susceptible and resistant sub-populations, which were characterized by unique mean generation times (MGT) and maximum rates of antibiotic-induced bacterial killing (Kmax) (Figure 4). Diagnostic plots showed that both S. aureus and E. coli total counts were well described by the model (Figure 5). Observed initial inocula for S. aureus and E. coli were 5.95 and 6.24 log10 CFU/mL with relative standard errors (RSEs) of 0.574 and 0.638, respectively, as compared to the target of six log10 CFU/mL (Table 1). For both S. aureus isolates, the “susceptible” subpopulations of the mechanism-based model had an estimated sensitivity (i.e., KC50) of 0.416 mg/L (15.9%) and 1.01 mg/L (36.6% RSE) for cefazolin and ampicillin, respectively. A strain-effect was observed with one isolate (S. aureus AR Bank # 0484), where the maximum rate of S. aureus killing by cefazolin was lowered by 0.357-fold (27.4%RSE) from 3.15 h-1 (22.7%RSE) to 1.12 h-1, as compared to S. aureus ATCC 25923. When in co-culture with E. coli, S. aureus experienced a reduction in its cefazolin Kmax by 24.8% (23.1%RSE), indicating a reduction in drug effect.
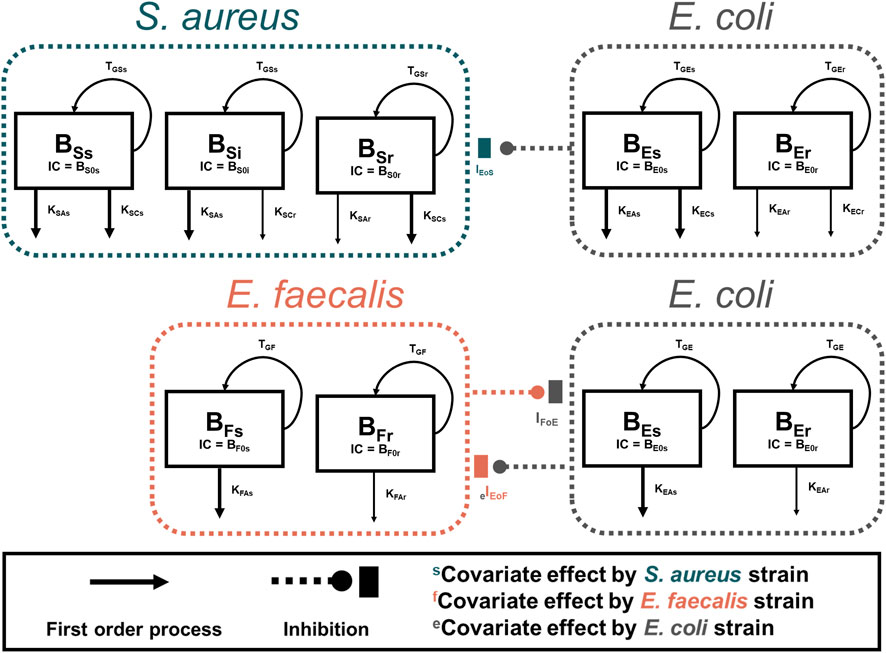
Figure 4. Model Diagrams. Two models were developed to describe each co-culture condition (S. aureus-E. coli and E. faecalis-E. coli). Experimental results of S. aureus in co-culture with E. coli were best described by using a subpopulation-based model where S. aureus was described by three sub-populations, principally differentiated by the sensitivity to either ampicillin or cefazolin. E. faecalis data were best characterized by two subpopulations with different rates of killing by ampicillin. For both models, E. coli total counts were best characterized by a two-subpopulation structure.
E. faecalis in co-culture with E. coli was best described with a subpopulation model that implemented susceptible- and resistant-subpopulations with unique maximum rates of ampicillin-induced killing (Figure 4). Diagnostics of the model predictions showed that both E. faecalis and E. coli data were well described (Figure 6). The observed starting inocula for E. faecalis and E. coli were 6.17 (0.472%RSE) and 6.03 (0.635%RSE) log10 CFU/mL, as compared to the target of six log10 CFU/mL (Table 2). β-lactamase-production by E. coli was found to reduce the maximum rate of ampicillin-induced killing of E. faecalis by 90.4% (0.978%RSE) versus a reduction of 70.6% (21.7%) for the β-lactamase-deficient E. coli. Maximum ampicillin-induced killing on E. coli (Kmax) was estimated to be 0.484 h-1, and was strongly influenced by isolate, with β-lactamase-deficient E. coli and TEM-1-producing E. coli having an estimated 8.33- and 2.75-fold increased rate of killing in comparison to the KPC-3/TEM-1B-producing E. coli, respectively.
Polymicrobial infections consisting of Enterobacterales and gram-positive pathogens are difficult to manage clinically, with optimal antibiotic strategies being unclear, especially in cases where one or more bacteria express β-lactamases or other antibiotic-modifying enzymes (Lenhard et al., 2019; Smith et al., 2021a). The situation is further complicated by the global spread of KPC-producing Enterobacterales, which are capable of inactivating the majority of β-lactam antibacterials (Hansen, 2021). Given the complexity of polymicrobial interactions, the present study sought to utilize novel mathematical modelling approaches to facilitate improved assessment of antibiotic action and potential methods to optimize therapy. The co-culture of E. coli with both S. aureus and E. faecalis resulted in significant protective effects that reduced β-lactam killing by 24.8%–90.4%. Furthermore, the extent of protection provided by E. coli was dependent on the type of β-lactamase harboured by the E. coli, with KPC-producing E. coli conferring the most substantial protection from β-lactams for two of the four gram-positive isolates. After simultaneous fitting of all data, residual variability was estimated as <0.35 log10 CFU/mL for bacteria studied, which is a typical finding for time kill experiments.
Although it is intuitive that the expression of β-lactamases may protect neighbouring pathogens from β-lactam exposure, the relationship between the production of drug-modifying enzymes and the impact on the pharmacodynamics of antibacterials against surrounding organisms is nuanced. A previous study observed that β-lactamase-producing S. aureus and Branhamella catarrhalis were capable of protecting Streptococcus pneumoniae from ampicillin in vivo, whereas Haemophilus influenzae did not augment the survival of the S. pneumoniae despite the production of β-lactamase enzymes (Renneberg and Walder, 1989). A subsequent in vivo investigation was able to confirm the inability of β-lactamase-producing H. influenzae to protect S. pneumoniae from an aminopenicillin (Westman et al., 2004). One variable that may impact the magnitude of a protective effect conferred by the production of drug-altering enzymes may be whether the β-lactamases are released into the extracellular space (Liao et al., 2014; Liao et al., 2015). In a prior in vitro investigation, aminoglycoside modifying enzyme-producing E. faecalis was not capable of appreciably protecting neighbouring gram-negative pathogens from gentamicin despite exposure of E. faecalis to lethal concentrations of ampicillin in an attempt to liberate intracellular enzymes (McMurtry et al., 2021). The relationship between resistance mechanisms and the pharmacodynamics of antibacterials during polymicrobial infections is therefore complex, and investigations of specific pathogen relationships and resistance mechanisms are likely needed to optimize antibacterial selection during polymicrobial infections. In the current study, not only were β-lactamase-producing E. coli capable of protecting S. aureus and E. faecalis from β-lactams, but penicillin-resistant S. aureus also demonstrated the ability to protect β-lactamase-deficient E. coli from drug exposure as well.
The management of polymicrobial infections has become further complicated by the increased prevalence of carbapenem resistance among drug-resistant gram-negative pathogens. Carbapenem-resistant nonfermenting organisms have already demonstrated the ability to protect neighboring pathogens from β-lactam exposure. A. baumannii, for example, shielded E. coli and S. aureus from carbapenems in two separate investigations (Liao et al., 2014; Smith et al., 2021b). Similarly, Stenotrophomonas maltophilia protected Serratia marcscens from imipenem and ceftazidime using inducible β-lactamase production (Kataoka et al., 2003). An equally frightening scenario, however, is the involvement of CRE in a polymicrobial infection. The transmissible spread of carbapenem resistance among Enterobacterales strains internationally has generated considerable concern for the global healthcare system, with KPC-producing Enterobacterales now representing some of the most relevant CRE internationally (Anonymous. WHO, 2017; Hansen, 2021; Centers for Disease Control, 2023). In the current investigation, the KPC-producing E. coli generated the most pronounced protective effect for both S. aureus and E. faecalis, indicating that CRE are capable of shielding neighboring pathogens from β-lactam exposure in a manner analogous to nonfermenting pathogens.
In the present study, the variability of the β-lactam protective effect conferred by the different E. coli isolates suggests that the management of polymicrobial infections may potentially be improved if therapy can be individualized for a specific polymicrobial community. Clinicians are beginning to recognize that polymicrobial communities have likely been underdiagnosed in the past, and new molecular techniques may allow for more rapid identification of a polymicrobial infectious process (Athamanolap et al., 2018; Zhang et al., 2019). An Israeli study found that about one in three cases of urosepsis at the authors’ institution were polymicrobial, with polymicrobial urosepsis being associated with a higher mortality rate than monomicrobial infections (28% versus 15%, p < 0.05) (Siegman-Igra et al., 1994). The most common gram-negative pathogens in polymicrobial urosepsis were Enterobacterales, whereas Enterococcus species were the most common gram-positive organisms. Similarly, another group observed that neonatal patients with polymicrobial bacteremia experienced a higher mortality rate in comparison to monomicrobial bacteremia (adjusted odds ratio 4.3, 95% CI 1.8–10.2) (Pammi et al., 2014). Enterococcus species and Klebsiella species were the most commonly encountered pathogens in polymicrobial bacteremia, and both organisms were isolated from polymicrobial infections over twice as frequently than from monomicrobial infections. Further translational and clinical investigations into the optimal antibacterial selection and dosing during polymicrobial infections may allow for clinical practice guidelines that address situations such as polymicrobial urinary tract infections, bacteremia, and other sites of infection that are traditionally viewed as monomicrobial.
The current study has multiple limitations that should be considered when interpreting the results of the investigation. Firstly, all of the experiments were completed in vitro using static concentrations of β-lactam antibacterials. Secondly, a limited number of isolates that were selected based on their production of β-lactamase enzymes were evaluated in the study. Lastly, there are many β-lactamase enzymes produced by Enterobacterales that were not included in the investigation. Future in vivo studies that utilize a diverse collection of pathogens isolated from polymicrobial infections will likely be able to expand upon the results of the present investigation.
In closing, the current investigation affirms the ability of β-lactamase-producing Enterobacterales to protect neighbouring gram-positive pathogens from β-lactam exposure. The degree of such a shielding effect likely depends on the enzymes produced by a given isolate, with KPC enzymes demonstrating a marked ability to augment the survival of adjacent organisms. Penicillin-resistant S. aureus also demonstrated the ability to protect β-lactamase-deficient E. coli from ampicillin exposure, highlighting the complexity of polymicrobial interactions. Further investigations that evaluate the optimal antibacterial regimens to use against specific groups of pathogens may assist with clinical decision making during polymicrobial infections.
Three E. coli isolates, two S. aureus isolates, and 2 E. faecalis isolates were included in the investigation. Each E. coli isolate possessed a different β-lactamase status, such that one organism did not produce a β-lactamase (E. coli AR Bank #0017, MICampicillin < 1 mg/L and MICcefazolin = 2 mg/L), one isolate produced TEM-1 (E. coli AR Bank #0019, MICampicillin > 32 mg/L and MICcefazolin = 8 mg/L), and the final isolate produced KPC-3 and TEM-1B (E. coli AR Bank #0114, MICampicillin > 32 mg/L and MICcefazolin > 32 mg/L). Both S. aureus isolates were susceptible to cefazolin (ATCC 25923 and S. aureus AR Bank #0484) and both E. faecalis isolates were susceptible to ampicillin (E. faecalis AR Bank #0573 and E. faecalis AR Bank #0671).
Time-killing experiments were conducted over 24 h as described previously (Smith et al., 2021b). Generally, isolates were originally studied in mono-culture, then, to evaluate possible bacteria-bacteria interactions, studied in combination as either S. aureus-E. coli or E. faecalis-E. coli combinations (Figure 7). In brief, overnight cultures were used to create a ∼106 CFU/mL inoculum of bacteria suspended in cation-adjusted Brain Heart Infusion broth. Each S. aureus and E. faecalis isolate was evaluated alone and during co-culture experiments in which the gram-positive organism was grown with one of the 3 E. coli isolates. In co-culture experiments, a 1:1 ratio of two organisms was used such that a total inoculum of 2 × 106 CFU/mL was achieved consisting of 1 × 106 CFU/mL of each pathogen. A concentration array of cefazolin ranging from 0.016–16 mg/L was used for experiments involving S. aureus, whereas ampicillin concentrations ranging from 0.023 to 96 mg/L were used for E. faecalis experiments. The cefazolin and ampicillin drug arrays were also evaluated against each E. coli isolate grown alone. A total suspension of 20 mL was contained in 50 mL conical tubes that were incubated at 37°C with constant shaking. At 0, 2, 4, 6, 8, and 24 h, 100 mcl samples were collected from the conical tubes, serially diluted in saline, and plated onto Brain Heart Infusion agar imbued with 8 mg/L of polymyxin B. During co-culture experiments, diluted samples were also plated onto Mueller-Hinton Agar impregnated with 4 mg/L of vancomycin to quantify the amount of E. coli. Plates were incubated at 37°C for 24 h and then used for viable cell counting.
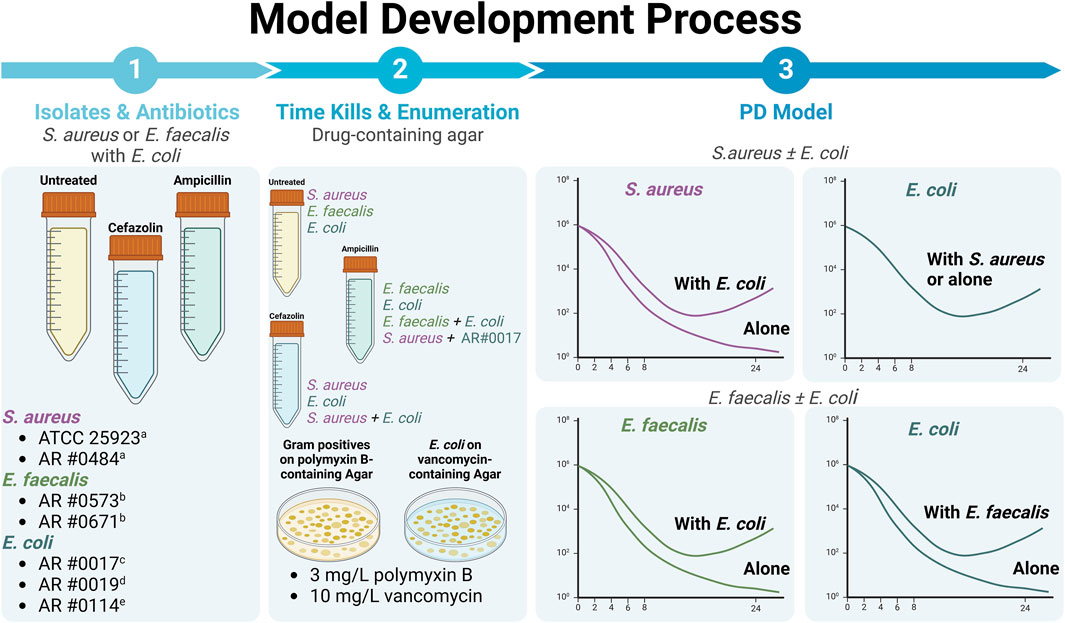
Figure 7. Our overall methodology involved detailed study of two different gram-positive bacteria, S. aureus and E. faecalis, alone and in co-culture with three different E. coli expressing different beta-lactamases. All experiments were performed using all possible combinations of S. aureus and E. coli strains along with all possible combinations of E. faecalis and E. coli strains, unless otherwise noted. Drug exposure were based on the typical first-line agent for each gram-positive. (i.e., primarily focused on use of cefazolin against S. aureus and ampicillin against E. faecalis). Please note that graphs of different bacterial killing effects are representative, only. aCefazolin susceptible. bAmpicillin susceptible. cNon-beta-lactamase producer. dTEM-1-producing. eKPC-3- and TEM-1B-co-producing (Created with BioRender.com).
In order to integrate the data obtained from time-killing experiments into a single quantifiable analysis, a mathematical approach was used to calculate maximum antibacterial activity during monoculture and co-culture conditions as described previously (McMurtry et al., 2021). In brief, the area under the CFU curve was calculated for each ampicillin or cefazolin concentration in each time-killing experiment using Systat Software version 14.0. The Log Ratio Area was then calculated by normalizing the area under the CFU curve of each drug concentration using the corresponding growth control (Eq. 1). Lastly, a Hill-type mathematical model was then used to describe the data, where Emax represents the maximum killing of either ampicillin or cefazolin (Eq. 2).
Data were modelled using Monolix (2022R1, Lixoft, Antony, France) by the stochastic approximation expectation maximization algorithm. Standard errors and likelihood were calculated using the linearization method, and observations below the limit of quantification (<102 CFU/mL) were modelled as censored data. In general, the model development process for the subpopulation models was performed stepwise. First, resistant subpopulations were characterized by a mutation frequency parameter which identifies the starting concentration of the resistant bacterial subpopulation. Resistant subpopulations were then tested for differences in maximum killing rate (kmax), sensitivity to the antibiotic of interest (KC50), or both, as done previously. After identifying parameters of antibiotic action, we tested for differences in growth rate between the subpopulations. To ensure the most parsimonious model is selected, likelihood ratio testing (for nested models) or comparison of the Schwartz Criterion was used. The mechanism-based model for co-culture conditions were performed as described above by first developing a base model for each bacterium in monoculture assuming a susceptible and resistant subpopulation with identical growth rates. Hill-type functions were utilized to describe β-lactam killing effects, based on previous studies that demonstrated saturable killing and underlying mechanism of (Regoes et al., 2004). Unique mechanistic interactions between bacteria were tested as either unidirectional or bidirectional effects on either the bacterial growth rate or antibiotic killing rate. Interactions were included based on reduction in model AIC. Given known differences in β-lactamase production, enzyme status was tested for statistical significance as a pharmacodynamic covariate on drug sensitivity (i.e., KC50) or maximum effect (i.e., Kmax). Because most experiments were performed in singlicate, we characterized experimental variability using constant, residual variability parameters that are informed by all datapoints collected.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
NS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. HK: Data curation, Investigation, Writing–review and editing. RK: Data curation, Investigation, Writing–review and editing. TM: Data curation, Investigation, Writing–review and editing. MK: Data curation, Investigation, Writing–review and editing. AB: Data curation, Investigation, Writing–review and editing. JL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The current work was supported by California Northstate University College of Pharmacy. NS is currently supported by NIH/NIAID under award L30AI164450.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1339858/full#supplementary-material
Centers for Disease Control (2023). Biggest threats and data. Available at: https://www.cdc.gov/drugresistance/biggest-threats.html (Accessed on July 26, 2023).
WHO (2017). WHO Publishes list of bacteria for which new antibiotics are urgently needed. WHO. Media Centre. News Release. Available at: https://www.who.int/en/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed July 25, 2023).
Athamanolap, P., Hsieh, K., and Wang, A. T. (2018). Integrated bacterial identification and antimicrobial susceptibility testing for polymicrobial infections using digital PCR and digital high-resolution melt in a microfluidic array platform. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 5346–5349. doi:10.1109/EMBC.2018.8513472
Beaudoin, T., Yau, Y. C. W., Stapleton, P. J., Gong, Y., Wang, P. W., Guttman, D. S., et al. (2017). Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 3, 25. doi:10.1038/s41522-017-0035-0
Briaud, P., Camus, L., Bastien, S., Doléans-Jordheim, A., Vandenesch, F., and Moreau, K. (2019). Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci. Rep. 9, 16564. doi:10.1038/s41598-019-52975-z
Bulitta, J. B., Ly, N. S., Landersdorfer, C. B., Wanigaratne, N. A., Velkov, T., Yadav, R., et al. (2015). Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob. Agents Chemother. 59, 2315–2327. doi:10.1128/AAC.04099-14
Bulman, Z. P., Wicha, S. G., Nielsen, E. I., Lenhard, J. R., Nation, R. L., Theuretzbacher, U., et al. (2022). Research priorities towards precision antibiotic therapy to improve patient care. Lancet Microbe 3, e795–e802. doi:10.1016/S2666-5247(22)00121-5
Caliskan-Aydogan, O., and Alocilja, E. C. (2023). A review of carbapenem resistance in Enterobacterales and its detection techniques. Microorganisms 11, 1491. doi:10.3390/microorganisms11061491
Cendra, M. D. M., Blanco-Cabra, N., Pedraz, L., and Torrents, E. (2019). Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms. Sci. Rep. 9, 16284. doi:10.1038/s41598-019-52726-0
Christaki, E., Marcou, M., and Tofarides, A. (2020). Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J. Mol. Evol. 88, 26–40. doi:10.1007/s00239-019-09914-3
Combes, A., Figliolini, C., Trouillet, J. L., Kassis, N., Wolff, M., Gibert, C., et al. (2002). Incidence and outcome of polymicrobial ventilator-associated pneumonia. Chest 121, 1618–1623. doi:10.1378/chest.121.5.1618
Cornely, O. A., Cisneros, J. M., Torre-Cisneros, J., Rodríguez-Hernández, M. J., Tallón-Aguilar, L., Calbo, E., et al. (2020). Pharmacokinetics and safety of aztreonam/avibactam for the treatment of complicated intra-abdominal infections in hospitalized adults: results from the REJUVENATE study. J. Antimicrob. Chemother. 75, 618–627. doi:10.1093/jac/dkz497
Dehbashi, S., Alikhani, M. Y., Tahmasebi, H., and Arabestani, M. R. (2021). The inhibitory effects of Staphylococcus aureus on the antibiotic susceptibility and virulence factors of Pseudomonas aeruginosa: a549 cell line model. Amb. Express 11, 50. doi:10.1186/s13568-021-01210-y
DeLeon, S., Clinton, A., Fowler, H., Everett, J., Horswill, A. R., and Rumbaugh, K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728. doi:10.1128/IAI.02198-14
Ferrer, M., Difrancesco, L. F., Liapikou, A., Rinaudo, M., Carbonara, M., Li Bassi, G., et al. (2015). Polymicrobial intensive care unit-acquired pneumonia: prevalence, microbiology and outcome. Crit. Care 19, 450. doi:10.1186/s13054-015-1165-5
Hansen, G. T. (2021). Continuous evolution: perspective on the epidemiology of carbapenemase resistance among Enterobacterales and other gram-negative bacteria. Infect. Dis. Ther. 10, 75–92. doi:10.1007/s40121-020-00395-2
Hoffman, L. R., Deziel, E., D'Argenio, D. A., Lepine, F., Emerson, J., McNamara, S., et al. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103, 19890–19895. doi:10.1073/pnas.0606756104
Jacobs, M., Gregoire, N., Couet, W., and Bulitta, J. B. (2016). Distinguishing antimicrobial models with different resistance mechanisms via population pharmacodynamic modeling. PLoS Comput. Biol. 12, e1004782. doi:10.1371/journal.pcbi.1004782
Jean, S. S., Harnod, D., and Hsueh, P. R. (2022). Global threat of carbapenem-resistant gram-negative bacteria. Front. Cell Infect. Microbiol. 12, 823684. doi:10.3389/fcimb.2022.823684
Kalil, A. C., Metersky, M. L., Klompas, M., Muscedere, J., Sweeney, D. A., Palmer, L. B., et al. (2016). Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious Diseases society of America and the American thoracic society. Clin. Infect. Dis. 63, e61–e111. doi:10.1093/cid/ciw353
Kataoka, D., Fujiwara, H., Kawakami, T., Tanaka, Y., Tanimoto, A., Ikawa, S., et al. (2003). The indirect pathogenicity of Stenotrophomonas maltophilia. Int. J. Antimicrob. Agents 22, 601–606. doi:10.1016/s0924-8579(03)00244-9
Ko, E. R., and Tsalik, E. L. (2022). A new era in host response biomarkers to guide precision medicine for infectious Diseases. J. Pediatr. Infect. Dis. Soc. 11, 477–479. doi:10.1093/jpids/piac081
Krzyzanski, W., and Rao, G. G. (2017). Multi-scale model of drug induced adaptive resistance of Gram-negative bacteria to polymyxin B. PLoS One 12, e0171834. doi:10.1371/journal.pone.0171834
Labricciosa, F. M., Sartelli, M., Abbo, L. M., Barbadoro, P., Ansaloni, L., Coccolini, F., et al. (2018). Epidemiology and risk factors for isolation of multi-drug-resistant organisms in patients with complicated intra-abdominal infections. Surg. Infect. (Larchmt) 19, 264–272. doi:10.1089/sur.2017.217
Lanckohr, C., and Bracht, H. (2022). Antimicrobial stewardship. Curr. Opin. Crit. Care 28, 551–556. doi:10.1097/MCC.0000000000000967
Landersdorfer, C. B., Ly, N. S., Xu, H., Tsuji, B. T., and Bulitta, J. B. (2013). Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob. Agents Chemother. 57, 2343–2351. doi:10.1128/AAC.00092-13
Landersdorfer, C. B., Rees, V. E., Yadav, R., Rogers, K. E., Kim, T. H., Bergen, P. J., et al. (2018). Optimization of a meropenem-tobramycin combination dosage regimen against hypermutable and nonhypermutable Pseudomonas aeruginosa via mechanism-based modeling and the hollow-fiber infection model. Antimicrob. Agents Chemother. 62, e02055. doi:10.1128/AAC.02055-17
Lebrun, M., de Repentigny, J., and Mathieu, L. G. (1978). Diminution de l'activité antibactérienne d'antibiotiques dans des cultures et des infections expérimentales mixtes. Can. J. Microbiol. 24, 154–161. doi:10.1139/m78-028
Lenhard, J. R., Smith, N. M., Quach, C. D., Nguyen, T. Q., Doan, L. H., and Chau, J. (2019). Bacterial brothers in arms: cooperation of Staphylococcus aureus and Pseudomonas aeruginosa during antimicrobial exposure. J. Antimicrob. Chemother. 74, 2657–2665. doi:10.1093/jac/dkz247
Lepe, J. A., and Martínez-Martínez, L. (2022). Resistance mechanisms in Gram-negative bacteria. Med. Intensiva Engl. Ed. 46, 392–402. doi:10.1016/j.medine.2022.05.004
Liao, Y. T., Kuo, S. C., Chiang, M. H., Lee, Y. T., Sung, W. C., Chen, Y. H., et al. (2015). Acinetobacter baumannii extracellular OXA-58 is primarily and selectively released via outer membrane vesicles after sec-dependent periplasmic translocation. Antimicrob. Agents Chemother. 59, 7346–7354. doi:10.1128/AAC.01343-15
Liao, Y. T., Kuo, S. C., Lee, Y. T., Chen, C. P., Lin, S. W., Shen, L. J., et al. (2014). Sheltering effect and indirect pathogenesis of carbapenem-resistant Acinetobacter baumannii in polymicrobial infection. Antimicrob. Agents Chemother. 58, 3983–3990. doi:10.1128/AAC.02636-13
Macleod, S. M., and Stickler, D. J. (2007). Species interactions in mixed-community crystalline biofilms on urinary catheters. J. Med. Microbiol. 56, 1549–1557. doi:10.1099/jmm.0.47395-0
Marcelin, J. R., Chung, P., and Van Schooneveld, T. C. (2020). Antimicrobial stewardship in the outpatient setting: a review and proposed framework. Infect. Control Hosp. Epidemiol. 41, 833–840. doi:10.1017/ice.2020.94
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 6. doi:10.1128/microbiolspec.ARBA-0009-2017
McMurtry, T. A., Barekat, A., Rodriguez, F., Purewal, P., Bulman, Z. P., and Lenhard, J. R. (2021). Capability of Enterococcus faecalis to shield Gram-negative pathogens from aminoglycoside exposure. J. Antimicrob. Chemother. 76, 2610–2614. doi:10.1093/jac/dkab211
Metlay, J. P., Waterer, G. W., Long, A. C., Anzueto, A., Brozek, J., Crothers, K., et al. (2019). Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American thoracic society and infectious Diseases society of America. Am. J. Respir. Crit. Care Med. 200, e45–e67. doi:10.1164/rccm.201908-1581ST
Mi, K., Zhou, K., Sun, L., Hou, Y., Ma, W., Xu, X., et al. (2022). Application of semi-mechanistic pharmacokinetic and pharmacodynamic model in antimicrobial resistance. Pharmaceutics 14, 246. doi:10.3390/pharmaceutics14020246
Michelsen, C. F., Christensen, A. M., Bojer, M. S., Høiby, N., Ingmer, H., and Jelsbak, L. (2014). Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J. Bacteriol. 196, 3903–3911. doi:10.1128/JB.02006-14
Minichmayr, I. K., Aranzana-Climent, V., and Friberg, L. E. (2022). Pharmacokinetic/pharmacodynamic models for time courses of antibiotic effects. Int. J. Antimicrob. Agents 60, 106616. doi:10.1016/j.ijantimicag.2022.106616
Morrison, L., and Zembower, T. R. (2020). Antimicrobial resistance. Gastrointest. Endosc. Clin. N. Am. 30, 619–635. doi:10.1016/j.giec.2020.06.004
Moser, C., Lerche, C. J., Thomsen, K., Hartvig, T., Schierbeck, J., Jensen, P., et al. (2019). Antibiotic therapy as personalized medicine - general considerations and complicating factors. Apmis 127, 361–371. doi:10.1111/apm.12951
Orazi, G., Jean-Pierre, F., and O'Toole, G. A. (2020). Pseudomonas aeruginosa PA14 enhances the efficacy of norfloxacin against Staphylococcus aureus newman biofilms. J. Bacteriol. 202, e00159. doi:10.1128/JB.00159-20
Orazi, G., and O'Toole, G. A. (2017). Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio 8, e00873. doi:10.1128/mBio.00873-17
Orazi, G., Ruoff, K. L., and O'Toole, G. A. (2019). Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio 10, e01501. doi:10.1128/mBio.01501-19
Pammi, M., Zhong, D., Johnson, Y., Revell, P., and Versalovic, J. (2014). Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect. Dis. 14, 390. doi:10.1186/1471-2334-14-390
Patil, H. V., and Patil, V. C. (2017). Incidence, bacteriology, and clinical outcome of ventilator-associated pneumonia at tertiary care hospital. J. Nat. Sci. Biol. Med. 8, 46–55. doi:10.4103/0976-9668.198360
Pavlaki, M., Poulakou, G., Drimousis, P., Adamis, G., Apostolidou, E., Gatselis, N. K., et al. (2013). Polymicrobial bloodstream infections: epidemiology and impact on mortality. J. Glob. Antimicrob. Resist 1, 207–212. doi:10.1016/j.jgar.2013.06.005
Pierce, J., Apisarnthanarak, A., Schellack, N., Cornistein, W., Maani, A. A., Adnan, S., et al. (2020). Global antimicrobial stewardship with a focus on low- and middle-income countries. Int. J. Infect. Dis. 96, 621–629. doi:10.1016/j.ijid.2020.05.126
Rabaan, A. A., Eljaaly, K., Alhumaid, S., Albayat, H., Al-Adsani, W., Sabour, A. A., et al. (2022). An overview on phenotypic and genotypic characterisation of carbapenem-resistant Enterobacterales. Med. Kaunas. 58, 1675. doi:10.3390/medicina58111675
Radlinski, L., Rowe, S. E., Kartchner, L. B., Maile, R., Cairns, B. A., Vitko, N. P., et al. (2017). Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 15, e2003981. doi:10.1371/journal.pbio.2003981
Regoes, R. R., Wiuff, C., Zappala, R. M., Garner, K. N., Baquero, F., and Levin, B. R. (2004). Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob. Agents Chemother. 48, 3670–3676. doi:10.1128/AAC.48.10.3670-3676.2004
Renneberg, J., and Walder, M. (1989). The role of beta-lactamase in mixed infections in mice in relation to treatment with ampicillin. J. Infect. Dis. 160, 337–341. doi:10.1093/infdis/160.2.337
Septimus, E. J. (2018). Antimicrobial resistance: an antimicrobial/diagnostic stewardship and infection prevention approach. Med. Clin. North Am. 102, 819–829. doi:10.1016/j.mcna.2018.04.005
Shettigar, K., Jain, S., Bhat, D. V., Acharya, R., Ramachandra, L., Satyamoorthy, K., et al. (2016). Virulence determinants in clinical Staphylococcus aureus from monomicrobial and polymicrobial infections of diabetic foot ulcers. J. Med. Microbiol. 65, 1392–1404. doi:10.1099/jmm.0.000370
Siegman-Igra, Y., Kulka, T., Schwartz, D., and Konforti, N. (1994). Polymicrobial and monomicrobial bacteraemic urinary tract infection. J. Hosp. Infect. 28, 49–56. doi:10.1016/0195-6701(94)90152-x
Smith, N. M., Ang, A., Tan, F., Macias, K., James, S., Sidhu, J., et al. (2021a). Interaction of Staphylococcus aureus and acinetobacter baumannii during in vitro β-lactam exposure. Antimicrob. Agents Chemother. 65, e02414. doi:10.1128/AAC.02414-20
Smith, N. M., Ang, A., Tan, F., Macias, K., James, S., Sidhu, J., et al. (2021b). Interaction of Staphylococcus aureus and Acinetobacter baumannii during in vitro β-lactam exposure. Antimicrob. Agents Chemother. 65, e02414. doi:10.1128/aac.02414-20
Smith, N. M., Boissonneault, K. R., Chen, L., Petraitis, V., Petraitiene, R., Tao, X., et al. (2022). Mechanistic insights to combating NDM- and CTX-M-coproducing Klebsiella pneumoniae by targeting cell wall synthesis and outer membrane integrity. Antimicrob. Agents Chemother. 66, e0052722. doi:10.1128/aac.00527-22
Stevens, D. L., Bisno, A. L., Chambers, H. F., Dellinger, E. P., Goldstein, E. J., Gorbach, S. L., et al. (2014). Practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 59, e10–e52. doi:10.1093/cid/ciu444
Tognon, M., Köhler, T., Gdaniec, B. G., Hao, Y., Lam, J. S., Beaume, M., et al. (2017). Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. Isme J. 11, 2233–2243. doi:10.1038/ismej.2017.83
Westman, E., Lundin, S., Hermansson, A., and Melhus, A. (2004). Beta-lactamase-producing nontypeable Haemophilus influenzae fails to protect Streptococcus pneumoniae from amoxicillin during experimental acute otitis media. Antimicrob. Agents Chemother. 48, 3536–3542. doi:10.1128/AAC.48.9.3536-3542.2004
Yo, C. H., Hsein, Y. C., Wu, Y. L., Hsu, W. T., Ma, M. H., Tsai, C. H., et al. (2019). Clinical predictors and outcome impact of community-onset polymicrobial bloodstream infection. Int. J. Antimicrob. Agents 54, 716–722. doi:10.1016/j.ijantimicag.2019.09.015
Keywords: polymicrobial, pharmacodynamic, beta-lactam, mechanism-based modeling, Staphylococcus aureus, Escherichia coli, Enterococcus faecalis, systems microbiology
Citation: Smith NM, Kaur H, Kaur R, Minoza T, Kent M, Barekat A and Lenhard JR (2024) Influence of β-lactam pharmacodynamics on the systems microbiology of gram-positive and gram-negative polymicrobial communities. Front. Pharmacol. 15:1339858. doi: 10.3389/fphar.2024.1339858
Received: 16 November 2023; Accepted: 06 May 2024;
Published: 04 June 2024.
Edited by:
Adline Princy Solomon, SASTRA Deemed to be University, IndiaReviewed by:
Payam Behzadi, Islamic Azad University, ShahreQods, IranCopyright © 2024 Smith, Kaur, Kaur, Minoza, Kent, Barekat and Lenhard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin R. Lenhard, anVzdGluLmxlbmhhcmRAY25zdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.