
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 06 March 2024
Sec. Predictive Toxicology
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1335374
 Nan Zhang1
Nan Zhang1 Gui-Yuan Song1,2
Gui-Yuan Song1,2 Qing-Hua Yu1,3
Qing-Hua Yu1,3 Xin-Ming Fan4
Xin-Ming Fan4 Wen-Shuo Zhang4
Wen-Shuo Zhang4 Yong-Jian Hu5
Yong-Jian Hu5 Tian-Zhu Chao1
Tian-Zhu Chao1 Yao-Yao Wu1
Yao-Yao Wu1 Shu-Yan Duan1
Shu-Yan Duan1 Fei Wang1
Fei Wang1 Rui-Peng Du1
Rui-Peng Du1 Ping Xu1*
Ping Xu1*Background: Previous studies have documented important roles for microRNA-147 (miR-147) in inflammation, radiation-induced injury, cancer, and a range of other diseases. Murine lungs exhibit high levels of miRNA, mRNA, and lncRNA expression. However, very little research to date has focused on the lncRNA-miRNA-mRNA competing endogenous RNA (ceRNA) networks associated with miR-147, and the regulation of lncRNAs and miRNAs in this setting remains poorly understood.
Methods: After establishing a miR-147−/− model mouse, samples of lung tissue were harvested for RNA-sequencing, and differentially expressed lncRNAs, miRNAs, and mRNAs were identified. The miRNA targets of these lncRNAs and the identified miRNAs were first overlapped to facilitate the prediction of target mRNAs, with analyses then examining the overlap between these targets and mRNAs that were differentially expressed. Then, these target mRNAs were subjected to pathway enrichment analyses. These results were ultimately used to establish a miR-147-related ceRNA network.
Results: Relative to wild-type mice, the lungs of miR-147−/− mice exhibited 91, 43, and 71 significantly upregulated lncRNAs, miRNAs, and mRNAs, respectively, together with 114, 31, and 156 that were significantly downregulated. The lncRNA-miRNA-mRNA network established based on these results led to the identification of Kcnh6 as a differentially expressed hub gene candidate and enabled the identification of a range of regulatory relationships. KEGG pathway enrichment showed that the mRNA targets of differentially expressed lncRNAs and miRNAs in the mice were associated with tumor-related signaling, endometrial cancer, bladder cancer, and ErbB signaling.
Conclusion: These results suggest that the identified ceRNA network in miR-147−/− mice shapes tumor-associated signaling activity, with miR-147 potentially regulating various lncRNAs and miRNAs through Kcnh6, ultimately influencing tumorigenesis. Future studies of the lncRNA, miRNA, and mRNA regulatory targets shown to be associated with miR-147 in the present study may ultimately lead to the identification of novel clinically relevant targets through which miR-147 shapes the pathogenesis of cancer and other diseases.
Non-coding RNAs (ncRNAs) have attracted much interest in recent years owing to advances in sequencing and bioinformatics techniques, contributing to a growing understanding of the biological importance and functions of microRNAs (miRNAs) and long ncRNAs (lncRNAs). As 21-24 nucleotide ncRNAs, miRNAs interact with the 3′-untranslated region (UTR) of specific mRNA targets and promoting degradation or reduced translation (Sandquist and Wong, 2014; Afonso-Grunz and Muller, 2015). Conversely, lncRNAs are over 200 nucleotides long and are capable of functioning as miRNA sponges that sequester these ncRNAs to interfere with their function, in addition to interacting with certain mRNAs(Mattick and Rinn, 2015). Dysregulated miRNA and lncRNA expression has increasingly been tied to the pathogenesis of inflammation, Alzheimer’s disease, cancer, and a range of other diseases (Cheng, 2015). The competing endogenous RNA (ceRNA) concept was first suggested by Salmena et al., in 2011, and posits that miRNA response elements (MREs) are present within mRNAs, lncRNAS, and other types of ncRNAs, thereby enabling ncRNAS to interfere with miRNA functionality by outcompeting mRNAs for sequence-specific mRNA binding (Salmena et al., 2011). While biologically interesting, research focused on validating this ceRNA hypothesis has been relatively limited to date.
First identified as a tumor suppressor candidate expressed in murine macrophages and splenic tissue, microRNA-147 (miR-147) has more recently been shown to regulate inflammation and macrophage functionality by targeting Toll-like receptor 4 (TLR4) in addition to shaping the incidence of inflammatory bowel disease and coronary atherosclerosis (Lagos-Quintana et al., 2002; Liu et al., 2009). The upregulation of this miRNA has been reported in squamous cell carcinomas of the tongue and esophagus, as well as gastric cancer, whereas its downregulation has been observed in colon cancer (Wong et al., 2008; Yao et al., 2009; Shen et al., 2018; Tang et al., 2018; Ning et al., 2019). At a functional level, miR-147 can reportedly target and suppress brain-derived neurotrophic factor (BDNF) expression to suppress non-small cell lung cancer (NSCLC) cell migratory, proliferative, and invasive activity (Li et al., 2020). In contrast, the upregulation of miR-147 in small-cell lung cancer has been tied to enhanced chemoresistance (Zhang et al., 2019). Prior research conducted by our group suggests that miR-147a is capable of regulating AKT and 3-phosphoinositide-dependent protein kinase 1 (PDPK1) transcription in the context of radiotherapy through its ability to bind the PDPK1 3′-UTR. By inhibiting miR-147 and stimulating nuclear paraspeckle assembly transcript 1 (NEAT1) to promote PDPK1 upregulation, trixrutin was further found to enhance radioprotection. Overall, these past data highlight the varied and complex roles that miR-147 played in various biological contexts. However, there has been little research to date focused on establishing a miR-147-related ceRNA network composed of lncRNAs, miRNAs, and mRNAs. As such, lung samples from miR-147−/− mice were herein used for transcriptomic sequencing and bioinformatics analyses aimed at clarifying the ceRNA networks associated with this miRNA. Together, these data will provide a foundation for research centered on clarifying the role that miR-147 plays in the pathogenesis of cancer and other diseases.
C57BL/6N and miR-147−/− mice (4 weeks old) were acquired from Beijing Wei-tong-li-Hua Experimental Animal Technology Co. And the Chinese-French Immunoregulatory Genes Laboratory of Xinxiang Medical University, respectively. The deletion of miR-147 in these mice and associated genotypic changes were reported previously (Song et al., 2023). Mice were individually housed under controlled conditions (22°C, 55% humidity, 12 h light/dark cycle). Animals were euthanized prior to lung tissue harvesting. The Science and Technology Ethics Committee of Zaozhuang University approved all animal protocols.
TRIzol (Invitrogen, MA, United States of America) was for RNA extraction from lung tissue samples. To construct a lncRNA library, Caliper Labchip GX and a DNA 1K Reagent Kit were used to assess RNA quality, using 500 ng of murine RNA. After rRNA removal with an RNase H kit, RNA fragmentation was performed, and RNAs were reverse transcribed for first-strand cDNA synthesis, followed by dUTP-based second-strand cDNA synthesis. Following end repair, 1 A nucleotide was ligated to the 3′ end of the cDNA segment, followed by PCR amplification with the following settings: 95°C for 3 min; 14 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 1 min; 72°C for 5 min. For miRNA library construction, an Agilent 2,100 Bioanalyzer and an Agilent DNA 1000 Kit were used for assessing RNA quality. Then, 5,000 ng samples of RNA were used to purify RNAs 18-30 nucleotides in length via polyacrylamide gel electrophoresis. These transcripts were then connected at the 3′ and 5’ ends, and reverse transcription and PCR amplification were performed, collecting PCR products via polyacrylamide gel electrophoresis and storing them in EB buffer. PCR products from these libraries were denatured into single-stranded forms and cyclized to yield single-stranded circular DNAs, which were then used for rolling cycle amplification, generating a DNA nanoball (DNB) containing multiple DNA copies. DNBs of sufficient quality were then loaded into patterned nanoarrays with a high-intensity DNA nanochip technique, followed by sequencing performed through combinatorial probe-anchor synthesis.
Sequencing data were initially filtered using SOAPnuke, and clean reads were stored in the FASTQ format, after which the Dr. Tom Multi-omics Data Mining System (https://biosys.bgi.com) was used for data analyses and data mining. Filtered clean reads were mapped with Bowtie2 to reference genomes and sRNA databases, after which RNAhybrid, miRanda, and TargetScan were used for predicting gene targets of lncRNAs and miRNAs, enabling the visualization of lncRNA-miRNA-mRNA ceRNA networks. RSEM (v1.3.1) was used to evaluate relative gene expression, and differentially expressed (DE) genes were identified with DESeq2 (v1.4.5) based on Q-values ≤0.05. GO and KEGG enrichments of DE mRNAs were analyzed with Phyper based upon hypergeometric distributions. Strong Q values (Q ≤ 0.05) were used to correct networks and significantly enriched terms.
BeNa Culture Collection of China (BNCC364345) and Cellverse Bioscience Technology Co., Ltd. (iCell-h017) were the sources of MLG2908 and HFL-1 cells, which were cultured with DMEM (Hyclone) or Ham’s F-12K(PM150910) medium that had been supplemented using 10% fetal bovine serum (FBS; Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL, Hyclone), with all culture being performed at 37°C in a 5% CO2 incubator.
Cells in confocal dishes were fixed using 4% paraformaldehyde, permeabilized for 10 min using 0.2% Triton X-100, rinsed thrice with PBS, blocked for 30 min with 1% BSA, and probed overnight with anti-Kcnh6 (Santa Cruz, sc-135959, 1:200) at 4°C. Cells were again washed, probed for 2 h with a secondary antibody (Beyotime, China), subjected to nuclear counterstaining for 10 min using DAPI (Beyotime, China), washed with PBS, and imaged via fluorescence microscopy.
Data are means ± standard deviation (SD) and were compared with GraphPad Prism 8. p < 0.05 was selected as the cut-off to define significance.
To begin exploring changes in transcriptional activity following the loss of miR-147, the lncRNA, miRNA, and mRNA expression profiles of miR-147−/− and wild-type mice were compared. In total, significant up- and downregulation of 71 and 156 mRNAs, respectively, were observed in miR-147−/− mice. Moreover, these mice exhibited 91 and 114 lncRNAs as well as 43 and 31 miRNAs that were significantly upregulated and downregulated, respectively (Q < 0.05, | log2FC | > 1). The most significant up- and downregulation, respectively, was observed in the lncRNAs BGIG10090_53735and Hoxb3os, while the respective miRNAs were mmu-miR-466b-3p (12.75-fold) and mmu-miR-466c-3p (10.65-fold), and mRNAs were Plac9a (8.50-fold) and Ifi30 (12.68-fold). These results were presented using volcano plots (Figure 1), and the top 5 differentially expressed (DE) transcripts in each category are presented in Tables 1–3.
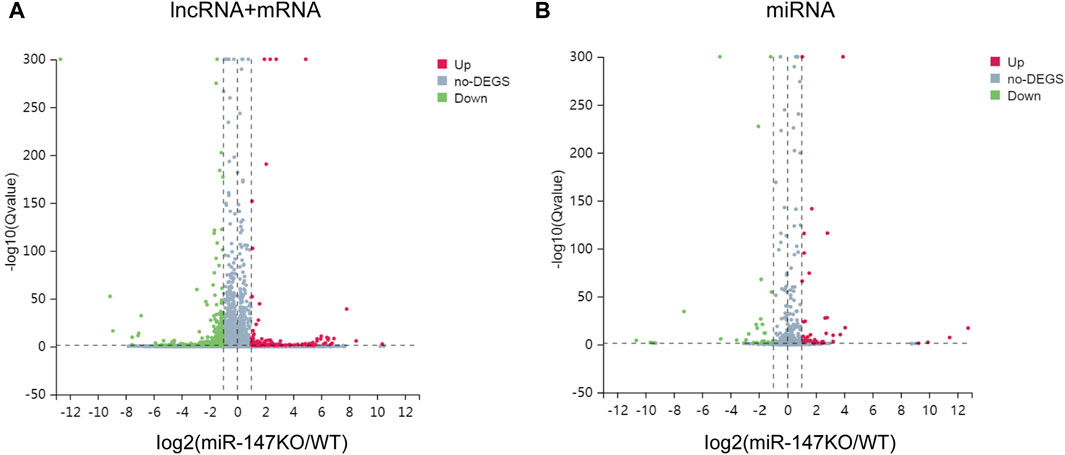
FIGURE 1. lncRNA, miRNA, and mRNA expression profiles in WT and miR-147−/− mice. (A)Volcano plots were generated to identify 91 upregulated and 114 downregulated lncRNAs as well as 71 upregulated and 156 downregulated mRNAs, or (B) 43 upregulated and 31 downregulated miRNAs. Upregulated and downregulated transcripts are respectively shown in red and green.
To explore the possible ceRNA-like regulatory activities of the DE lncRNAs and miRNAs identified in miR-147−/− mice, the putative miRNA targets of DE lncRNAs were identified, and the overlap between this list and DE miRNAs was assessed (Figure 2). In total, 46 overlapping DE miRNAs were identified through this approach, and their putative mRNA targets were then identified and compared with the list of DE mRNAs identified in the above transcriptomic analyses. This approach revealed 12 overlapping DE mRNA target genes. The overlapping target mRNAs of 128 DE lncRNAs and DE miRNAs were also identified (S-Figure 1). GO analyses of 986 putative miRNA target mRNAs were additionally performed, revealing that the most enriched molecular function terms associated with these DE lncRNA and miRNA targets included PDZ domain binding, voltage-gated ion channel activity, and ion channel activity (Figure 3A). GO enrichment analyses were also performed for the cellular component and biological process GO term categories when assessing these mRNA targets (S-Figure 2).
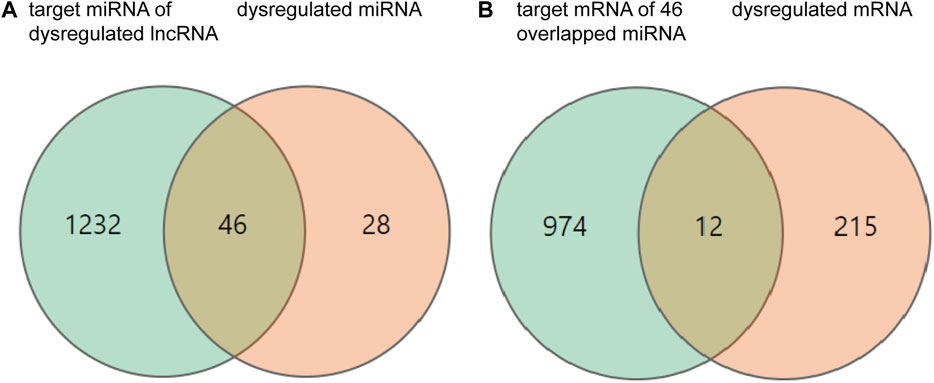
FIGURE 2. Differentially expressed (DE) mRNAs, miRNAs, and lncRNAs, and their relationships. (A) The orange and green circles respectively correspond to 74 DE miRNAs and 1,278 target miRNAs of DE lncRNAs. (B) The orange and green circles respectively correspond to 227 DE mRNAs and 986 mRNA targets of DE miRNAs.
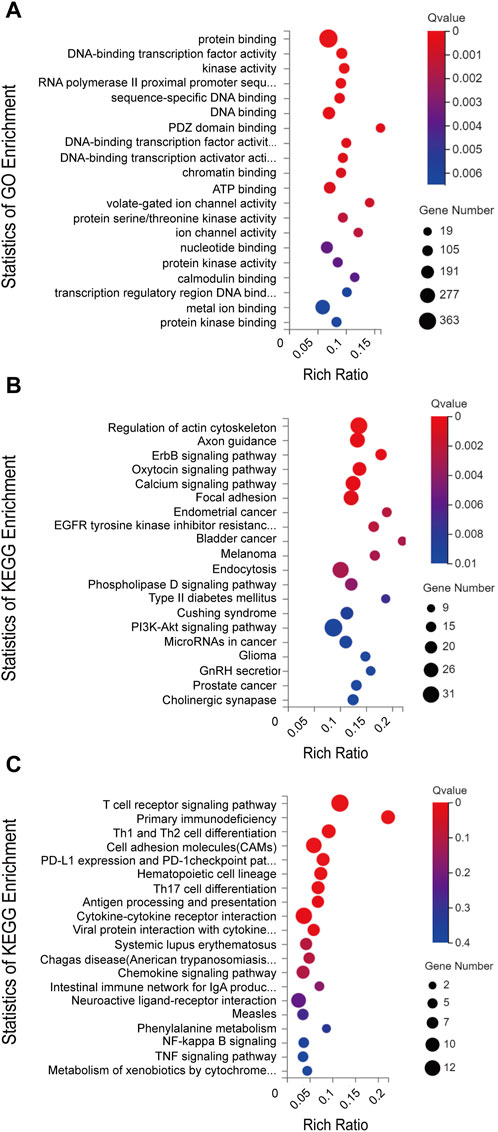
FIGURE 3. (A) GO annotations and (B) KEGG pathway analysis results for mRNAs regulated by the established ceRNA network. (C) KEGG pathway analysis results for differentially expressed mRNAs. The top 20 results are presented for each analysis based upon Q values.
Next, KEGG pathway analyses of mRNA targets of DE miRNAs were conducted, revealing that these genes were enriched in the bladder cancer (Cdh1, Cdkn1a, Dapk2, E2f1, Egfr, Erbb2, Fgfr3, Nras, Mapk1), endometrial cancer (Axin1, Cdh1, Cdkn1a, Egfr, Erbb2, Nras, Pik3cd, Pik3r1, Tcf7l1, Mapk1, Gsk3b), and ErbB signaling (Nrg2, Camk2g, Cdkn1a, Cdkn1b, Egfr, Eif4ebp1, Erbb2, Nras, Pik3cd, Pik3r1, Stat5b, Pak6, Shc2, Mapk1, Gsk3b) pathways (Figure 3B). KEGG enrichment analyses were also performed for DE mRNAs (Figure 3C).
To better clarify the significance of the DE lncRNAs and miRNAs identified in miR-147−/− mice, GO analyses were next used to screen for DE mRNAs associated with voltage-gated ion channel activity. Using Kcnh6 as targets, a ceRNA network was then constructed that included 41, 50, and 10 lncRNAs, miRNAs, and mRNAs, respectively (Figure 4). The visualized network constructed based on the selected filtered mRNAs may provide new insight into the mechanistic roles played by miR-147 in cancer. To further clarify the relationships among these different transcripts, the overlapping mRNAs between targets of DE lncRNAs and DE miRNAs were identified and used to establish a ceRNA network (S-Figure 3).
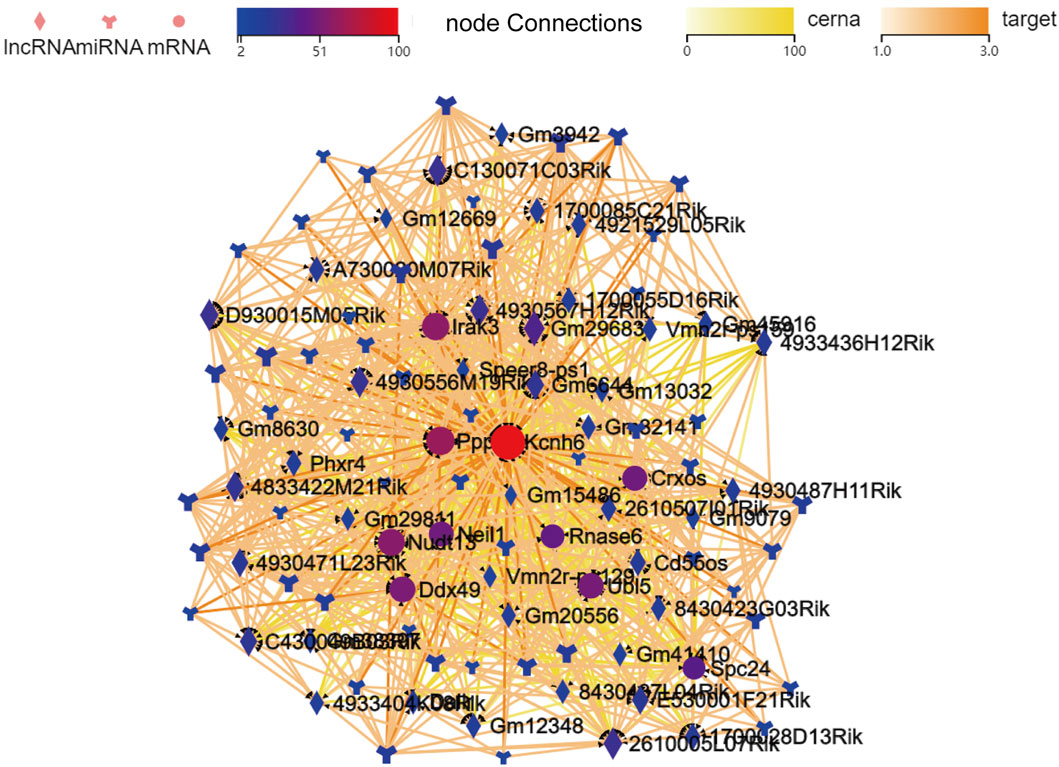
FIGURE 4. The established lncRNA-miRNA-mRNA network when filtered to focus on voltage-gated ion channel activity-related mRNAs identified in GO analyses. mRNAs, lncRNAs, and branching shapes respectively represent mRNAs, lncRNAs, and miRNAs, with associations between these transcripts being represented by connecting lines between nodes.
Kcnh6 was identified as a differentially expressed mRNA that was also a core gene within the established lncRNA-miRNA-mRNA network, emphasizing it as being closely related to miR-147. To validate the results of these sequencing analyses, miR-147−/− mice were analyzed, with genotyping confirming that these knockout mice had been successfully established (Figure 5A). WB and qPCR demonstrated significant Kcnh6 upregulation in these mice lacking miR-147 expression (Figures 5B, C), with IHC staining providing additional confirmation of a significant rise in Kcnh6 levels within the lungs of these miR-147−/− mice (Figure 5D). Kcnh6 was also strongly expressed in the livers of the miR-147−/− mice (S-Figure 4).
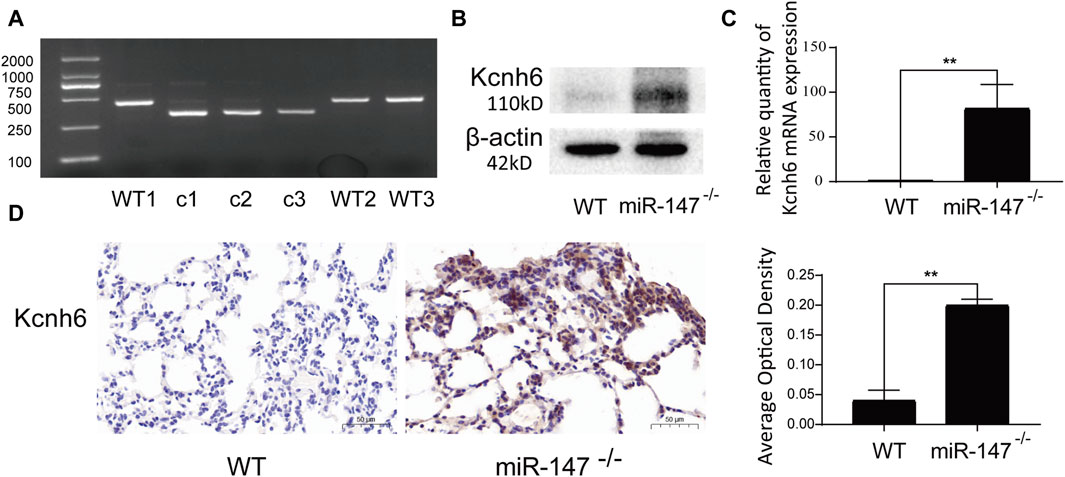
FIGURE 5. Kcnh6 expression in miR147−/− mice. PCR analyses were used for gene identification. (A) In the WT and miR-147−/− mice, the sequences were 435 bp and 324 bp, respectively. Kcnh6 upregulation in miR-147−/− mice was confirmed via (B) Western immunoblotting, (C) qPCR, and (D) immunohistochemistry, with GraphPad Prism 8.0 being used for these analyses. **p < 0.01.
To clarify possible modulatory associations between miR-147 and Kcnh6, miR-147 was knocked down in both murine MLG2908 lung fibroblasts and human HFL-1 embryonic lung fibroblasts, after which qPCR, immunofluorescence, and Western immunoblotting were employed to assess Kcnh6 expression. These analyses demonstrated that Kcnh6 protein and mRNA levels rose upon miR-147 knockdown, in line with the results shown above (Figure 6).
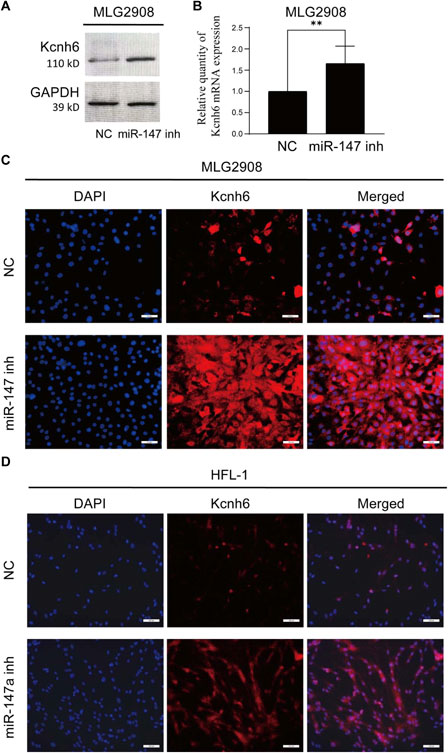
FIGURE 6. Assessment of the expression of Kcnh6 in miR-147 knockdown cells. After the knockdown of miR-147 within MLG2908 cells, the upregulation of Kcnh6 was detected via (A) Western immunoblotting, (B) qPCR, and (C) immunofluorescence. (D) Following miR-147a knockdown within HFL-1 cells, the upregulation of Kcnh6 was detected by immunofluorescence. GraphPad Prism 8.0 was used to analyze the results. **p < 0.01.
Oncogenic transformation occurs through a cumulative process in which multiple incremental changes in cellular characteristics ultimately lead to unrestrained growth. A range of systemic factors, including insulin-related signaling, however, can also induce carcinogenic signaling and shape cancer phenotypes (Dearth et al., 2007). Insulin is secreted into systemic circulation whereupon it binds to cell surface insulin receptors, activating intracellular PI3K signaling activity that may contribute to malignant tumor development (Ruderman et al., 1990; Gallagher and LeRoith, 2011). In line with this hypothesis, hyperinsulinemia is associated with more rapid tumor development in both mice and humans (Novosyadlyy et al., 2010; Orgel and Mittelman, 2013), whereas reducing insulin levels yields the opposite effect (Nencioni et al., 2018). Kcnh6 is a voltage-dependent K+ (Kv) channel that primarily influences the secretion of insulin (Vandenberg et al., 2012; Hylten-Cavallius et al., 2017). Indeed, Kcnh6 dysfunction has been reported to result in inappropriate short-term insulin secretion as well as β-cell failure over longer time periods (Yang et al., 2018). The Kv inhibitor BBR has shown promise as an approach to targeting Kcnh6 and thereby treating diabetes in humans (Zhao et al., 2021). Kcnh6 is also capable of reportedly protecting pancreatic β cells against ER-related stress and apoptotic death (Lu et al., 2020). The functional importance of Kcnh6 in tumors, however, remains to be studied in detail.
Several published articles have highlighted the biological functions of miR-147 and suggested that it may offer value as a biomarker for a range of diseases when assessed in combination with other miRNAs or validated biomarkers (Enes et al., 2016; Michalovicz et al., 2019). In one report, HPM was found to alleviate colon damage and suppress related inflammatory activity owing to its ability to promote miR-147 upregulation, thereby reducing inflammation-related mRNA expression and downstream inflammatory cytokine levels (Wei et al., 2015). Other studies have established miR-147 and a lncRNA MEG3 target that is closely tied to the proliferative activity and apoptotic death of leukemia cells in vitro (Li et al., 2018). The downregulation of miR-147 has been reported in NSCLC tumors as compared to paracancerous tissues, with corresponding miR-147 downregulation in NSCLC patient serum and further evidence that lower serum miR-147 is independently associated with poorer NSCLC patient prognosis (Chu et al., 2016; Li et al., 2020). In a previous study, our group determined that knocking down miR-147 was associated with significant changes in thymic lncRNA, miRNA, and mRNA expression in mice. In radiation-protected mouse lungs, the ability of troxerutin to upregulate PDPK1 was found to be mediated through miR-147 targeting and the consequent activation of AKT together with the inhibition of JNK activity (Song et al., 2023). To better clarify the role that knocking out miR-147 has on the murine lungs, lung tissues from miR-147−/−mice were thus used for sequencing. Here, Kcnh6 upregulation was observed in the lungs of miR-147−/− mice, and associated transcriptomic changes were enriched in tumor-associated signaling pathways. While these results supported the ability of miR-147 to serve as a regulator of Kcnh6, they did not demonstrate the status of Kcnh6 as a direct miR-147 target. Accordingly, the targeting relationship between the two was probed in greater detail. There is not a high degree of miR-147 conservation between mice and humans, with slight differences in the sequences of the mouse (miR-147) and human (miR-147a) versions of this miRNA. To validate the regulatory relationship between miR-147 and Kcnh6, this miRNA was inhibited in both murine and human embryonic lung fibroblasts, yielding comparable results in both cases. This suggests that miR-147 may suppress insulin secretion via the inhibition of Kcnh6, thereby suppressing PI3K signaling activity and influencing tumorigenesis.
Since the first proposal of the ceRNA network model, researchers have increasingly sought to clarify the regulatory links among lncRNAs, mRNAs, and miRNAs based on the hypothesis that MRE-containing ceRNAs can modulate miRNA functionality by competing with target mRNAs for miRNA binding (Salmena et al., 2011). In some reports, lncRNAs have been demonstrated to modulate tumor-associated gene expression through their ability to impact tumor-related mRNA expression (Wang et al., 2020). Here, the overlap between DE miRNAs and the targets of DE lncRNAs was used to identify miRNA-mRNA targeting relationships. This ultimately enabled the construction of a ceRNA network centered on the DEG Kcnh6, providing predictive insight into the lncRNAs, miRNAs, and mRNAs that may be associated with Kcnh6. To date, few articles have focused on tumor treatment via modulating the insulin-PI3K axis, and the present results thus highlight a promising new target for antitumor therapeutic strategies focused on this axis.
These analyses revealed that abnormal RNA regulation may contribute to the incidence of various malignancies including endometrial, bladder, and lung cancers, with the ErbB signaling playing a key role in the biological changes that develop following the knockout of miR-147 through shifts in the pulmonary lncRNA-miRNA-mRNA network. Further studies of the miR-147-related ceRNA network and efforts to target specific lncRNAs and miRNAs are thus expected to yield new insight into how miR-147 shapes the pathogenesis of cancers and other diseases. However, additional analyses will be essential to validate the lncRNA-miRNA-mRNA network relationships, providing a clear avenue for future research efforts.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was approved by The Science and Technology Ethics Committee of Zaozhuang University. The study was conducted in accordance with the local legislation and institutional requirements.
NZ: Data curation, Writing–original draft, Writing–review and editing, Validation. G-YS: Validation, Writing–original draft, Writing–review and editing, Software. Q-HY: Validation, Writing–original draft, Writing–review and editing. X-MF: Resources, Writing–original draft. W-SZ: Validation, Writing–original draft. Y-JH: Validation, Writing–original draft. T-ZC: Methodology, Writing–original draft. Y-YW: Methodology, Writing–original draft. S-YD: Validation, Writing–original draft. FW: Validation, Writing–original draft. R-PD: Software, Writing–original draft. PX: Funding acquisition, Methodology, Resources, Writing–original draft, Writing–review and editing, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (NSFC) (Nos. 81773358, 11705158, U1504824), the Natural Science Foundation of Shandong Province (No. ZR2023QH137) and the Doctoral Research Initiation Fund of Zaozhuang University (Nos. 745010210 and 1020727).
We thank Beijing Genomics institution (BGI) Gene Co., Ltd. for the sequencing work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1335374/full#supplementary-material
Afonso-Grunz, F., and Muller, S. (2015). Principles of miRNA-mRNA interactions: beyond sequence complementarity. Cell Mol. Life Sci. 72 (16), 3127–3141. doi:10.1007/s00018-015-1922-2
Cheng, G. (2015). Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 81, 8175–8193. doi:10.1016/j.addr.2014.09.001
Chu, G., Zhang, J., and Chen, X. (2016). Serum level of microRNA-147 as diagnostic biomarker in human non-small cell lung cancer. J. Drug Target 24 (7), 613–617. doi:10.3109/1061186X.2015.1116539
Dearth, R. K., Cui, X., Kim, H. J., Hadsell, D. L., and Lee, A. V. (2007). Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle 6 (6), 705–713. doi:10.4161/cc.6.6.4035
Enes, C. M., Kervancioglu, M., Oztuzcu, S., Yilmaz, C. F., Ergun, S., Baspinar, O., et al. (2016). Plasma microRNA profiling of children with idiopathic dilated cardiomyopathy. Biomarkers 21 (1), 56–61. doi:10.3109/1354750X.2015.1118533
Gallagher, E. J., and LeRoith, D. (2011). Minireview: IGF, insulin, and cancer. Endocrinology 152 (7), 2546–2551. doi:10.1210/en.2011-0231
Hylten-Cavallius, L., Iepsen, E. W., Wewer, A. N., Svendstrup, M., Lubberding, A. F., Hartmann, B., et al. (2017). Patients with long-QT syndrome caused by impaired hERG-encoded K(v)11.1 potassium channel have exaggerated endocrine pancreatic and incretin function associated with reactive hypoglycemia. Circulation 135 (18), 1705–1719. doi:10.1161/CIRCULATIONAHA.116.024279
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12 (9), 735–739. doi:10.1016/s0960-9822(02)00809-6
Li, F., Wang, X., and Yang, L. (2020). MicroRNA-147 targets BDNF to inhibit cell proliferation, migration and invasion in non-small cell lung cancer. Oncol. Lett. 20 (2), 1931–1937. doi:10.3892/ol.2020.11715
Li, Z. Y., Yang, L., Liu, X. J., Wang, X. Z., Pan, Y. X., and Luo, J. M. (2018). The long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia. EBioMedicine 34, 3461–3475. doi:10.1016/j.ebiom.2018.07.013
Liu, G., Friggeri, A., Yang, Y., Park, Y. J., Tsuruta, Y., and Abraham, E. (2009). miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 106 (37), 15819–15824. doi:10.1073/pnas.0901216106
Lu, J., Shen, H., Li, Q., Xiong, F., Xie, R., Yuan, M., et al. (2020). KCNH6 protects pancreatic β-cells from endoplasmic reticulum stress and apoptosis. FASEB J. 34 (11), 15015–15028. doi:10.1096/fj.202001218R
Mattick, J. S., and Rinn, J. L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22 (1), 5–7. doi:10.1038/nsmb.2942
Michalovicz, L. T., Kelly, K. A., Vashishtha, S., Ben-Hamo, R., Efroni, S., Miller, J. V., et al. (2019). Astrocyte-specific transcriptome analysis using the ALDH1L1 bacTRAP mouse reveals novel biomarkers of astrogliosis in response to neurotoxicity. J. Neurochem. 150 (4), 420–440. doi:10.1111/jnc.14800
Nencioni, A., Caffa, I., Cortellino, S., and Longo, V. D. (2018). Fasting and cancer: molecular mechanisms and clinical application. Nat. Rev. Cancer 18 (11), 707–719. doi:10.1038/s41568-018-0061-0
Ning, X., Wang, C., Zhang, M., and Wang, K. (2019). Ectopic expression of miR-147 inhibits stem cell marker and epithelial-mesenchymal transition (EMT)-Related protein expression in colon cancer cells. Oncol. Res. 27 (4), 399–406. doi:10.3727/096504018X15179675206495
Novosyadlyy, R., Lann, D. E., Vijayakumar, A., Rowzee, A., Lazzarino, D. A., Fierz, Y., et al. (2010). Insulin-mediated acceleration of breast cancer development and progression in a nonobese model of type 2 diabetes. Cancer Res. 70 (2), 741–751. doi:10.1158/0008-5472.CAN-09-2141
Orgel, E., and Mittelman, S. D. (2013). The links between insulin resistance, diabetes, and cancer. Curr. Diab Rep. 13 (2), 213–222. doi:10.1007/s11892-012-0356-6
Ruderman, N. B., Kapeller, R., White, M. F., and Cantley, L. C. (1990). Activation of phosphatidylinositol 3-kinase by insulin. Proc. Natl. Acad. Sci. U. S. A. 87 (4), 1411–1415. doi:10.1073/pnas.87.4.1411
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146 (3), 353–358. doi:10.1016/j.cell.2011.07.014
Sandquist, M., and Wong, H. R. (2014). Biomarkers of sepsis and their potential value in diagnosis, prognosis and treatment. Expert Rev. Clin. Immunol. 10 (10), 1349–1356. doi:10.1586/1744666X.2014.949675
Shen, J., Niu, W., Zhang, H., Jun, M., and Zhang, H. (2018). Downregulation of MicroRNA-147 inhibits cell proliferation and increases the chemosensitivity of gastric cancer cells to 5-fluorouracil by directly targeting PTEN. Oncol. Res. 26 (6), 901–911. doi:10.3727/096504017X15061902533715
Song, G. Y., Hu, Y. J., Zhang, N., Wang, F., Chao, T. Z., Wu, Y. Y., et al. (2023). Construction and validation of a competing endogenous RNA network in the thymus of miR-147(-/-) mice. Int. Immunopharmacol. 117, 117109896. doi:10.1016/j.intimp.2023.109896
Tang, Y., Li, Z., and Shi, Z. X. (2018). Mechanisms of the suppression of proliferation and invasion ability mediated by microRNA-147b in esophageal squamous cell carcinoma. Zhonghua Yi Xue Za Zhi 98 (26), 2092–2098. doi:10.3760/cma.j.issn.0376-2491.2018.26.007
Vandenberg, J. I., Perry, M. D., Perrin, M. J., Mann, S. A., Ke, Y., and Hill, A. P. (2012). hERG K(+) channels: structure, function, and clinical significance. Physiol. Rev. 92 (3), 1393–1478. doi:10.1152/physrev.00036.2011
Wang, J. Y., Yang, Y., Ma, Y., Wang, F., Xue, A., Zhu, J., et al. (2020). Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed. Pharmacother. 121, 121109627. doi:10.1016/j.biopha.2019.109627
Wei, K., Zhang, D., Hong, J., Zhang, C., Feng, X., Huang, Y., et al. (2015). Herb-partitioned moxibustion and the miRNAs related to crohn's disease: a study based on rat models. Evid. Based Complement. Altern. Med. 2015, 2015265238. doi:10.1155/2015/265238
Wong, T. S., Liu, X. B., Wong, B. Y., Ng, R. W., Yuen, A. P., and Wei, W. I. (2008). Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 14 (9), 2588–2592. doi:10.1158/1078-0432.CCR-07-0666
Yang, J. K., Lu, J., Yuan, S. S., Asan Cao, X., Qiu, H. Y., Shi, T-T., et al. (2018). From hyper-to hypoinsulinemia and diabetes: effect of KCNH6 on insulin secretion. Cell Rep. 25 (13), 3800–3810. doi:10.1016/j.celrep.2018.12.005
Yao, Y., Suo, A. L., Li, Z. F., Liu, L. Y., Tian, T., Ni, L., et al. (2009). MicroRNA profiling of human gastric cancer. Mol. Med. Rep. 2 (6), 963–970. doi:10.3892/mmr_00000199
Zhang, W. C., Wells, J. M., Chow, K. H., Huang, H., Yuan, M., Saxena, T., et al. (2019). miR-147b-mediated TCA cycle dysfunction and pseudohypoxia initiate drug tolerance to EGFR inhibitors in lung adenocarcinoma. Nat. Metab. 1 (4), 460–474. doi:10.1038/s42255-019-0052-9
Keywords: miRNA-147, KCNH6, ceRNA, lncRNA-miRNA-mRNA network, cancer
Citation: Zhang N, Song G-Y, Yu Q-H, Fan X-M, Zhang W-S, Hu Y-J, Chao T-Z, Wu Y-Y, Duan S-Y, Wang F, Du R-P and Xu P (2024) Evaluation of the lncRNA-miRNA-mRNA ceRNA network in lungs of miR-147 −/− mice. Front. Pharmacol. 15:1335374. doi: 10.3389/fphar.2024.1335374
Received: 08 November 2023; Accepted: 20 February 2024;
Published: 06 March 2024.
Edited by:
Gang Chen, University of Kentucky, United StatesCopyright © 2024 Zhang, Song, Yu, Fan, Zhang, Hu, Chao, Wu, Duan, Wang, Du and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Xu, MTMyNzM3MzAyNzFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.