
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 31 January 2024
Sec. Pharmacology of Anti-Cancer Drugs
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1329409
This article is part of the Research Topic Advances in Drug-induced Diseases Volume II View all 43 articles
 Zhiwei Cui1†
Zhiwei Cui1† Zhen Zhai2†
Zhen Zhai2† De Xie3†
De Xie3† Lihui Wang1
Lihui Wang1 Feiyan Cheng1
Feiyan Cheng1 Siyu Lou4
Siyu Lou4 Fan Zou4
Fan Zou4 Rumeng Pan1
Rumeng Pan1 Shixue Chang5
Shixue Chang5 Haoyan Yao1
Haoyan Yao1 Jing She1
Jing She1 Yidan Zhang1
Yidan Zhang1 Xinyuan Yang1*
Xinyuan Yang1*Introduction: The discovery of neurotrophic tyrosine receptor kinase (NTRK) gene fusions has facilitated the development of precision oncology. Two first-generation NTRK inhibitors (larotrectinib and entrectinib) are currently approved for the treatment of patients with solid tumors harboring NTRK gene fusions. Nevertheless, comprehensive NTRK profiling at the pan-cancer genomic level and real-world studies pertaining to the adverse events of NTRK inhibitors are lacking.
Methods: We characterize the genome of NTRK at the pan-cancer level through multi-omics databases such as The Cancer Genome Atlas (TCGA). Through the FDA Adverse Event Reporting System (FAERS) database, we collect reports of entrectinib and larotrectinib-induced adverse events and perform a pharmacovigilance analysis using various disproportionality methods.
Results: NTRK1/2/3 expression is lower in most tumor tissues, while they have higher methylation levels. NTRK gene expression has prognostic value in some cancer types, such as breast invasive carcinoma (BRCA). The cancer type with highest NTRK alteration frequency is skin cutaneous melanoma (SKCM) (31.98%). Thyroid carcinoma (THCA) has the largest number of NTRK fusion cases, and the most common fusion pair is ETV6-NTRK3. Adverse drug events (ADEs) obtained from the FAERS database for larotrectinib and entrectinib are 524 and 563, respectively. At the System Organ Class (SOC) level, both drugs have positive signal value for “nervous system disorder”. Other positive signals for entrectinib include “cardiac disorders”, “metabolism and nutrition disorders”, while for larotrectinib, it is “hepatobiliary disorders”. The unexpected signals are also listed in detail. ADEs of the two NTRK inhibitors mainly occur in the first month. The median onset time of ADEs for entrectinib and larotrectinib was 16 days (interquartile range [IQR] 6–86.5) and 44 days ([IQR] 7–136), respectively.
Conclusion: Our analysis provides a broad molecular view of the NTRK family. The real-world adverse drug event analysis of entrectinib and larotrectinib contributes to more refined medication management.
Members of the tropomyosin receptor kinase (TRK) family include the TRKA, TRKB, and TRKC proteins, encoded by the neurotrophic tyrosine kinase receptor 1 (NTRK1), NTRK2, and NTRK3 genes, respectively (Khotskaya et al., 2017). Activation of neurotrophic factors, which are specific ligands for TRK receptors, triggers the activation of various downstream signaling cascades. These cascades include the mitogen-activated protein kinase (MAPK), phosphatidylinositol-3-kinase (PI3K), and phospholipase C-γ (PLC-γ) pathways. These pathways have significant effects on essential biological processes such as neuronal cell differentiation, survival, and proliferation (Bhangoo and Sigal, 2019; Jiang et al., 2022). Alterations in NTRK genes are closely associated with both tumor initiation and progression. Among these alterations, NTRK gene fusions are the most well-characterized aberrations. These gene fusions possess oncogenic properties as they promote tumorigenesis by constitutively activating downstream cell growth and proliferation pathways (Khotskaya et al., 2017; Cocco et al., 2018; Okamura et al., 2018). In addition to gene fusions, several studies have also characterized the genomic profile of NTRK. Light et al. reported that NTRK1/3 were highly expressed in neuroblastoma patients with a better prognosis, whereas NTRK2 was highly expressed in neuroblastoma patients with a poor prognosis (Light et al., 2012). Additionally, in 83 clinical samples of triple-negative breast cancers, NTRK1/2/3 were found to exhibit varying degrees of copy number gain and amplification (Zito Marino et al., 2023). However, these aforementioned studies are limited by focusing on specific cancer types and small sample sizes, highlighting the critical need for comprehensive analyses across multiple tumor types to investigate their functions.
The US Food and Drug Administration (FDA) has approved the first generation of NTRK inhibitors, which have shown favorable clinical outcomes. One such inhibitor is larotrectinib, a potent and highly selective small molecule inhibitor targeting three TRK proteins. Larotrectinib is prescribed for the treatment of patients with solid tumors who have NTRK gene fusions and have experienced disease progression after alternative therapies (Scott, 2019). Its recommended phase 2 dose is 100 mg twice daily (Laetsch et al., 2018). Whereas entrectinib is mainly used for the treatment of locally advanced or metastatic solid tumors harboring fusion mutations in the NTRK1/2/3, C-ros oncogene 1 (ROS1) and anaplastic lymphoma kinase (ALK) genes. The recommended adult dose of entrectinib is 600 mg/d, while the pediatric dose is based on body surface area (Frampton, 2021). The main difference between the two drugs is that entrectinib is indicated for adult and pediatric patients aged ≥12 years, while larotrectinib is indicated for adult and pediatric patients of all ages (Frampton, 2021). Both drugs prolong metastasis-free survival and overall survival and maintain health-related quality of life in patients with solid tumors (Drilon et al., 2022; Jiang et al., 2022). Adverse events were predominantly reported during clinical trials for both drugs, primarily in grades 1–2, such as fatigue and dizziness. Additionally, a variable proportion of patients (ranging from 2% to 40%) also experienced grade 3–4 adverse events, including anaemia and elevated aminotransferases (Doebele et al., 2020; Hong et al., 2020). However, the majority of clinical trials were unable to identify new signals of adverse drug reactions due to the limited number of patients included and the short duration of follow-up (Doz et al., 2022). Liguori et al. recently collected and descriptively analyzed reports of adverse events following the introduction of two first-generation NTRK inhibitors (Liguori et al., 2023). Nevertheless, there is still a lack of disproportionality analysis of the adverse reaction signals of the two first-generation NTRK inhibitors, identification of new adverse reaction signals, and detailed comparisons of the safety of the two drugs.
Evidence at the genomic level plays a crucial role in the foundation of drug design. Drugs with confirmed targets from human genetic studies are more likely to be successfully marketed than those lacking such evidence (Roden et al., 2019). Furthermore, mutations and epigenetic modifications of drug target genes can significantly and consistently alter cellular gene expression patterns and are strongly associated with drug efficacy and adverse drug reactions (Brockmöller and Tzvetkov, 2008; Borges et al., 2021). For instance, fibroblast growth factor receptor 2 (FGFR2) rearrangements in cholangiocarcinoma predict tumor sensitivity to FGFR2 inhibitors (Vogel et al., 2023). Clinical trials serve as a vital pre-market assessment of drugs and represent an important step in translating genomic evidence into clinical practice (Joffe et al., 2017). However, due to the relatively low exposure of clinical trials with clear inclusion criteria and drug treatment conditions, rare but serious adverse drug reactions are usually detected after marketing authorization and increased population exposure (Ehmann et al., 2014). Therefore, post-market real-world vigilance data on drugs are of paramount importance, complementing evidence from pharmacoepidemiology (Sabaté and Montané, 2023). By integrating data from three sources-genomics, clinical trials, and real-world pharmacovigilance, a comprehensive view is provided, which is not available from any independent data source (Figure 1). This approach also effectively overcomes the limitations of analyzing each data type separately (Jing et al., 2021).
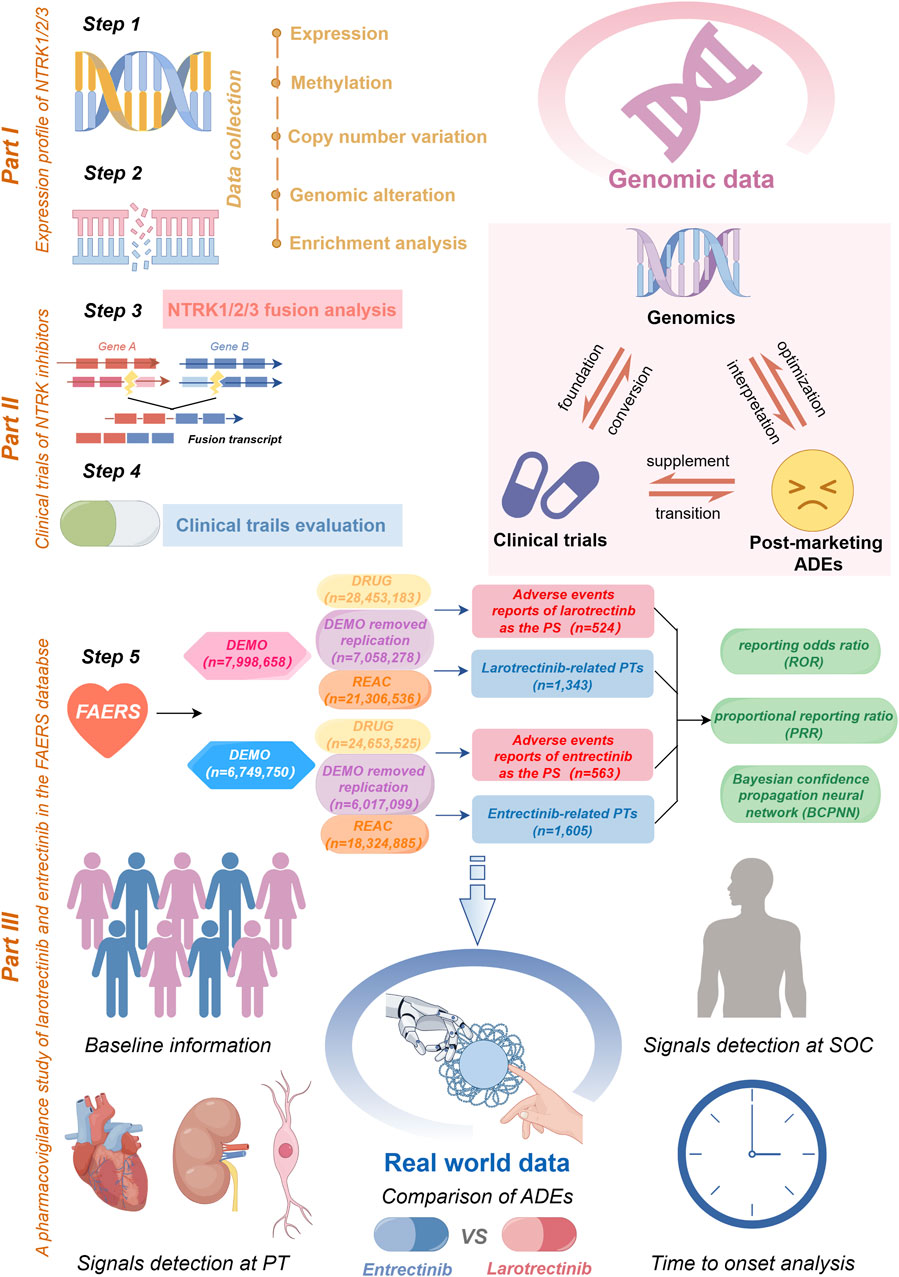
FIGURE 1. A flow chart of the whole study. In part I, we acquired NTRK expression profiling data and conducted an analysis of its mRNA expression, methylation, and CNV (step 1). Subsequently, we delved into the genomic alterations of NTRK, co-mutation pathways (step 2), and emphasized NTRK fusions (step 3). The part II involved a comprehensive review of the clinical trials of NTRK inhibitors (step 4). In part III, a safety assessment of the first-generation NTRK inhibitors was carried out utilizing real-world ADE reports sourced from the FAERS database (step 5). This encompassed data cleaning, baseline information description, signal detection, and time to onset analysis. Furthermore, we provided a summarization of the interrelationships between the three different data sources (top right of figure). CNV, copy number variation; FAERS, FDA Adverse Event Reporting System; SOC, System Organ Class; PT, preferred term; ADE, adverse drug event; PS, primary suspect.
This study aimed to characterize the molecular signature of NTRK using a large genomic dataset covering diverse tumor types, and to conduct a comprehensive analysis of the safety of first-generation NTRK inhibitors based on real-world adverse drug event (ADE) data. Firstly, we conducted a systematic analysis of NTRK expression, methylation, and genomic alterations across various cancer types. Additionally, we investigated NTRK fusions and examined the current status of clinical trials for the first-generation NTRK inhibitors, including larotrectinib and entrectinib. Lastly, we compared the differences in ADEs between the two drugs and assessed the time of onset for these ADEs. The integration of data from various sources in our study offers a comprehensive perspective on the NTRK gene.
The RNA sequencing data from a pan-cancer microarray (n = 15,776) was obtained from the UCSC Xena browser (https://xenabrowser.net/datapages/) on 1 August 2023. This dataset integrates various cancerous and normal tissues obtained from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) (Goldman et al., 2020). Secondly, these data were filtered and normalized before subsequent analyses were performed (Vivian et al., 2017; Zhao et al., 2020). For detailed procedures, please refer to a previously published article of us (Cui et al., 2023).
To assess the correlation between NTRK1/2/3 gene expression (with the median value as the cut-off) and patient prognosis, we utilized univariate Cox proportional hazards regression analysis to calculate hazard ratios (HR) and corresponding p-values. Patient survival indicators examined in this study comprised overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI). OS was defined as death from any cause, with surviving patients censored at the last follow-up. DSS was determined based on deaths attributed to a specific disease, and PFI represented the duration between the date of initial treatment in the randomized group and disease recurrence (Ekmekcioglu et al., 2016; Liu et al., 2018). During this process, key R packages was utilized, including “survival (version 3.3.1)” and “ggplot (version 3.3.6)”.
Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn/GSCA/#/) is an integrative platform for genomic cancer analysis (Liu et al., 2023). We employed the “Mutation” module to investigate the association between NTRK1/2/3 mRNA expression and copy number variation (CNV) as well as methylation across various tumor types. Additionally, we utilized the “Expression” module to explore the correlation between gene expression and pathway activity. All analyses were conducted on 1 August 2023.
The cBioPortal (https://www.cbioportal.org/) provides an open access to the interactive exploration of multidimensional cancer genomics data (Cerami et al., 2012; Gao et al., 2013). To systematically analyze the genomic alterations of NTRK1/2/3 genes across 32 different cancer types, we selected the “TCGA Pan Cancer Atlas Studies” cohort, which consisted of a total of 10,953 patients (including 10,967 samples) for further investigation. This cohort was chosen from the “Cancer Types Summary” and “Mutations” modules. To identify the most frequently altered genes, we utilized the “Genomic alterations” option under the “Comparison/Survival” module. Specifically, we focused on genes with alteration cases surpassing 100 in the altered group (totaling 2,799 genes) that were found to have q-values<0.01. The top 20 genes with the highest alteration frequencies in the altered group were subsequently visualized using a bar graph. To gain insights into the potential functions of these genes, a functional enrichment analysis was conducted. This analysis was performed using the R packages “clusterProfiler” (version 4.4.4) for enrichment analysis and “org.Hs.e.g.,.db” (version 3.1.0) for ID conversion (Yu et al., 2012). The p-values were adjusted using BH correction. All operational processes were conducted on 4 August 2023.
The fusion gene data of NTRK1/2/3 were retrieved from the TCGA Fusion Gene Database (https://www.tumorfusions.org/) on 5 August 2023. The database’s pipeline for RNA sequencing Data Analysis (PRADA) allowed us to identify fusion transcripts comprehensively and with high confidence, and provided a list of authentic fusion genes across 33 TCGA cancer types (Hu et al., 2018). The database defines four tiers to rank the screened fusion transcripts according to the abundance of supporting evidence. From tier 1 to tier 4 indicates strong to weak confidence levels.
The My Cancer Genome (MCG) database were launched in 2011 to guide clinicians in applying genomic test results to the treatment of cancer patients (Jain et al., 2020; Jain et al., 2021). Under the “Drugs” module, we selected two first-generation NTRK inhibitors (including larotrectinib and entrectinib) and systematically surveyed the top biomarker inclusion criteria for open clinical trials investigating these two drugs (5 August 2023).
Adverse drug events (ADEs) are unintended and harmful effects of medication use, and they commonly result in unplanned hospitalizations and fatalities (Bailey et al., 2016; Visacri et al., 2022). To assess the post-marketing safety of entrectinib and larotrectinib, we conducted a retrospective pharmacovigilance study with ADEs data extracted from the FAERS database. The FAERS data consists of seven datasets: demographic and administrative information (DEMO), drug information (DRUG), adverse drug reaction information (REAC), patient outcomes information (OUCT), reported sources (RPSR), drug therapy start dates and end dates (THER), and indications for drug administration (INDI). Considering the different timing of FDA approval for marketing of the two drugs, we collected information on all ADEs from the FAERS database from 2018 Q4 to 2023 Q1 (for larotrectinib, approved for marketing in 2018 Q4), and from 2019 Q3 to 2023 Q1 (for entrectinib, approved for marketing in 2019 Q3), respectively. As the database is updated on a quarterly basis, it is inevitable that there will be duplicates of previous published reports. In accordance with FDA recommendations, we performed deduplication prior to statistical analyses based on the following criteria: if CASEIDs were the same, the most recent FDA_DT was selected; if CASEIDs and FDA_DTs were the same, the higher PRIMARYIDs were selected (Shu et al., 2022). In this study, “larotrectinib” (brand name: VITRAKVI), and “entrectinib” (brand name: ROZLYTREK) were used to recognize records related to the two NTRK inhibitors. To analyze the role of drugs in ADEs, the drugs involved in ADE reports were classified as: primary suspect (PS), which denotes the primary drug considered to have possibly caused the ADE; second suspect (SS), which denotes the secondary drug considered to have possibly caused the ADE; concomitant (C), indicating the concurrent use of other drugs associated with the ADE; and interacting (I), representing possible drug interactions associated with the ADE. To improve accuracy, role codes for ADEs are reserved for entrectinib and larotrectinib as the PS drugs only, which resulted in ADEs associated with entrectinib and larotrectinib dosing being 563 and 524, respectively (Wu et al., 2023). System Organ Class (SOC) was the highest level of terminology in the Medical Dictionary for Regulatory Activities (MedDRA, version 26.0). All ADEs in the report were coded according to the preferred terms (PTs). Consequently, 1,343 larotrectinib-related PTs and 1,605 entrectinib-related PTs were screened out.
In pharmacovigilance studies, the term “signal” denotes a statistical association between a drug and an adverse event or drug-related adverse event (Hauben and Aronson, 2009; Sakaeda et al., 2013). Disproportionality analysis is a vital analytical method used to identify signals of adverse reactions associated with drugs, as well as to compare the occurrence of adverse reactions between a specific drug and all other drugs. This analysis helps uncover drug-related adverse reactions by evaluating the proportion of these reactions in relation to the overall adverse reaction pool (Montastruc et al., 2011). Therefore, in this study, we employed disproportionality analysis to identify potential correlations between the use of two NTRK inhibitors and all ADEs. We utilized two non-Bayesian algorithms, namely, the reporting odds ratio (ROR) and the proportional reporting ratio (PRR), along with a Bayesian algorithm known as the Bayesian confidence propagation neural network (BCPNN) (Montastruc et al., 2011). The Bayesian method (BCPNN) have sufficient power to detect unique signals even when few ADEs are reported for a drug (Bate, 2007). Generally, a higher parameter value indicates a stronger signal value. The specific formulas and criteria for detecting positive safety signals using the three algorithms are presented in Table 1. To enhance the reliability of our findings, we considered only adverse drug events (ADEs) with positive signal values that satisfied all three algorithms simultaneously. Furthermore, we excluded ADEs related to drug indications to ensure clarity in our presentation. Unexpected signals were determined as positive ADEs that were detected but not listed in the drug instruction. The primary analyses of this study are depicted in Figure 1. All data processing and statistical analyses were performed using SAS 9.4, Microsoft Excel 2019, and R software (version 4.2.1).
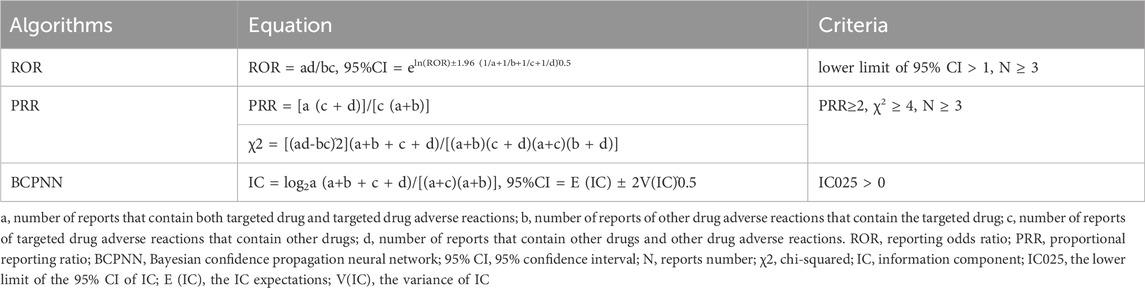
TABLE 1. Three disproportionality algorithms for assessing potential associations between NTRK inhibitors and ADEs.
The time to onset (TTO) for ADEs associated with NTRK inhibitor dosing was calculated as the interval between the ADEs onset date (EVENT_DT) within the DEMO file, and the start date of NTRK inhibitor dosing (START_DT) within the THER file. The exclusion criteria involved removing inaccurate or missing dates and cases where the ADEs onset date (EVENT_DT) preceded the start date of NTRK inhibitor dosing (START_DT). By utilizing the Weibull distribution, we could identify and estimate the increase or decrease in the incidence of ADE risk over time. The Weibull distribution employs two parameters, scale (α) and shape (β), to determine its shape (Kazi et al., 2020).
To gain a comprehensive understanding of NTRK genomic mRNA expression, we initially employed data from the TCGA and GTEx databases. NTRK mRNA expression was generally downregulated in tumor tissues compared to corresponding normal tissues. For NTRK1, NTRK2 and NTRK3, the number of tumors with significantly reduced expression was 20, 17, and 22, respectively (Figure 2A). Notably, there was a consistent trend towards significant downregulation of three NTRK genes expression in 15 tumor types (p < 0.05). However, in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) and thymoma (THYM), they were significantly upregulated. The full names of the 33 cancer abbreviations were shown in Supplementary Table S1.
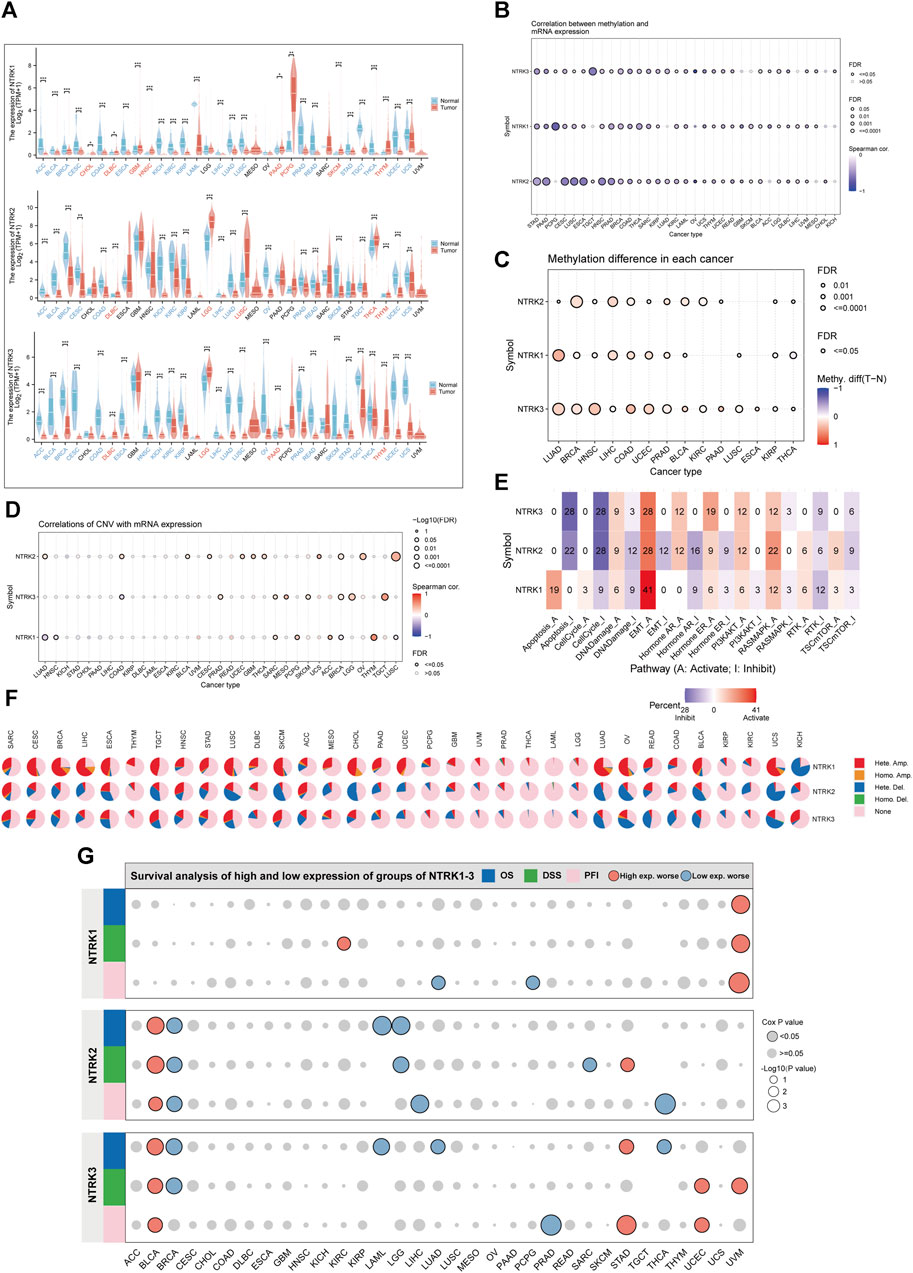
FIGURE 2. Genomic characterization of NTRK genes at the pan-cancer level. (A). Differential mRNA expression of NTRK1/2/3 in tumor and corresponding normal tissues. Gene expression is compared after Log2 (TPM+1) transformed. (B). Bubble plots demonstrate the correlation between NTRK genes expression and methylation levels. The bubble size positively correlates with the significance of FDR. The black outline border indicates FDR≤0.05. (C). Bubble plots depict the methylation differences of NTRK genes between tumor and normal samples. Blue dots and red color indicate methylation downregulation and upregulation in tumors, respectively. (D). Figure summarizes the correlations between CNV and NTRK genes expression in pan-cancer. (E). Percentage of cancers in which NTRK1/2/3 expression has a potential impact on pathway activity is shown. Pathway A and I represent activation or inhibition of this pathway. (F). Pie chart showing the percentage of different types of CNV in NTRK1/2/3 in a given cancer, with different colors representing different CNV types. (G). The relationship between NTRK genes expression and patient prognosis (OS, DSS, PFI) in pan-cancer. TPM, transcripts per million; FDR, false discovery rate; CNV, copy number variation; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval. *p < 0.05. **p < 0.01. ***p < 0.001. ns, no significance.
There is accumulating evidence indicating that both CNV and DNA methylation reshapes gene expression repertoire in cancers (Valsesia et al., 2012; Arechederra et al., 2018; Hammam et al., 2020; Ji et al., 2020). We therefore explored the relationship between NTRK genes expression and CNV, DNA methylation in different tumor types. Figure 2B indicated that in most tumor types, methylation levels and NTRK1/2/3 gene expression values were significantly negatively correlated (Spearman’s correlation<0, false discovery rate [FDR]<0.05). Moreover, we compared the methylation difference between tumor and normal and found that methylation of the NTRK genes was upregulated in various types, such as lung adenocarcinoma (LUAD), breast invasive carcinoma (BRCA), and head and neck squamous cell carcinoma (HNSC) (Figure 2C). Additionally, we evaluated the relationship between CNV and NTRK genes expression and observed a significant correlation in some tumors such as sarcoma (SARC), BRCA, and testicular germ cell tumors (TCGT) (Figure 2D). The comprehensive constitution of the heterozygous/homozygous CNV of NTRK1/2/3 in each cancer was displayed in Figure 2F.
We also analyzed the roles of NTRK genes in cancer pathways. The results suggested that NTRK1 had a relatively strong activation of apoptosis and a strong activation of epithelial-mesenchymal transition (EMT). NTRK2 and NTRK3 shared similar pathway activity. They exhibited strong inhibitory effect on apoptosis and cell cycle, and strong activation of EMT. Overall, NTRK genes had strong activating effect on EMT and potent inhibitory effect on the cell cycle (Figure 2E). The relationship between NTRK genes expression and patient prognosis was evaluated, as well. Increased NTRK1 expression was a risk factor for uveal melanoma (UVM), and high NTRK2 and NTRK3 expression was associated deceased OS, DSS, and PFI in bladder urothelial carcinoma (BLCA). However, elevated NTRK2 expression was protective in BRCA (Figure 2G).
As shown in Figure 3A, among the 32 cancer types observed, the frequency of mutations in the NTRK genes was not low overall. The frequency of mutations in the NTRK gene exceeded 15% in 19 cancer types. The mutation types were mainly composed of mRNA high, mutation, and amplification. The most frequent cancer types with NTRK genes alteration were skin cutaneous melanoma (SKCM) (31.98%), cholangiocarcinoma (CHOL) (30.56%), uterine carcinosarcoma (UCS) (29.82%), DLBC (27.08%), and SARC (26.67%). Instead, the mutation frequency of the NTRK genes in kidney chromophobe (KICH) (9.23%), glioblastoma multiforme (GBM) (7.77%), adrenocortical carcinoma (ACC) (6.5%), and UVM (5%) was relatively low, not exceeding 10%. Strikingly, all mutations in the NTRK gene in TCGT and UVM were mRNA high. In Figure 3B, the type, location and number of NTRK1/2/3 genetic alterations could further be observed. The hot spot mutation sites G169R (located in the linker) of NTRK1, A268V (located in the I-set) of NTRK2, and R153L/Q (located in the LRR-8) of NTRK3 were identified in 6, 3, and 5 patients. In general, the types of mutations at different amino acid sites of the NTRK1/2/3 gene were primarily missense mutations (Figure 3B). Further analysis of the mutations (Supplementary Figure S1A) revealed that most of the NTRK1/2/3 mutations were located in the structural domains of the Pkinase-Tyr and LRR-8. Moreover, the I-set domain in NTRK2/3 also contained close to a quarter of the number of mutations. The Ig-2 domain was specific to NTRK1 and contained about 5% of the mutations. In the final, we counted the genomic alterations of NTRK1/2/3 in the TCGA pan-cancer cohort in each patient. Of these, 645, 451, and 584 patients contained alterations in only one of the NTRK1, NTRK2, and NTRK3 genes, respectively. There were still 21 individuals with genomic alteration for NTRK1/2/3 simultaneously (Supplementary Figure S1B).
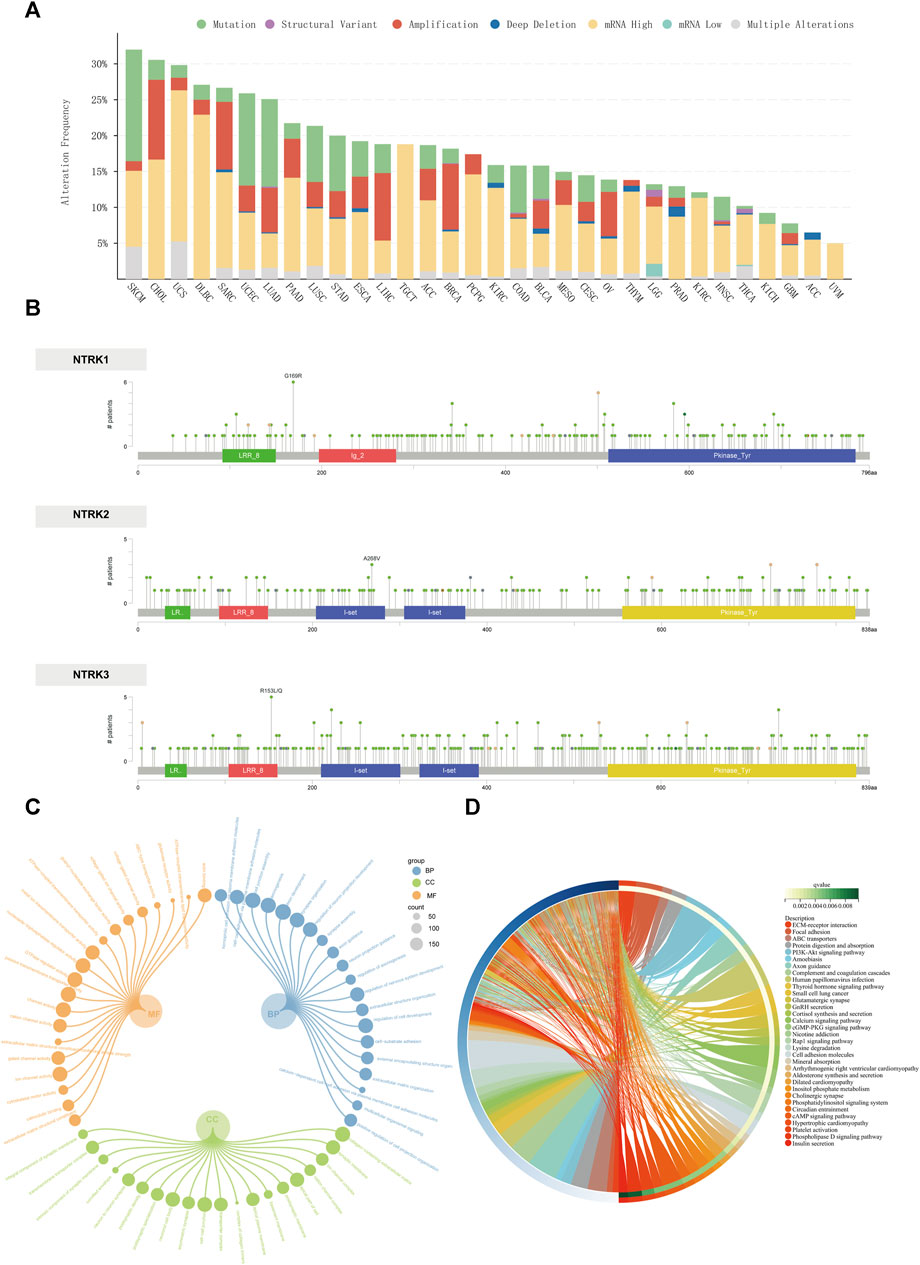
FIGURE 3. Genomic alterations of the NTRK genes at the pan-cancer level. (A). The alteration frequency of NTRK genes across 32 TCGA tumor types. (B). Amino acid mutation of in TCGA cancers. The GO and KEGG enrichment analyses of differentially altered genes between NTRK-gene-altered and unaltered groups are visualized using circular subgroup plot (C) and chord plot (D). GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
We also screened for altered genes that were significantly enriched in the NTRK-gene-altered group (compared to the unaltered group). The top 20 genes with the highest mutation frequencies in the NTRK-gene-altered group were displayed in Supplementary Figure S1C, and the full list of genes was provided in Supplementary Table S2. Genes with significant mutation differences were involved in biological processes including cell junctions, cell adhesion; involved in cellular components including extracellular matrix, synapses and ion channels; involved in molecular functions including cytoskeletal motility and ion channel activity (Figure 3C). Similarly, the signaling pathways and diseases in which these genes may be engaged included hormone synthesis and delivery (thyroid hormones, aldosterone hormone, insulin), some traditional signaling pathways (PI3K/Akt, cGMP-PKG, cAMP, etc.), as well as small-cell lung cancer, and human papillomavirus infection (Figure 3D). All enrichment analysis results were presented in Supplementary Table S3.
We detected fusion transcripts of the NTRK1/2/3 genes in pan-cancer through the TCGA Fusion Gene Database with high credibility (Supplementary Figure S2). In total, transcripts of NTRK fusion genes were detected in 11 tumor types, especially in thyroid carcinoma (THCA, n = 15), and brain lower grade glioma (LGG, n = 5). The NTRK3 gene had the highest number of fusion transcripts (n = 18). NTRK3-ETV6 (n = 10) had the highest percentage of all fusion pairs, especially in THCA (n = 6). Chromosomes 12 and 15 were the major chromosomes for 5′gene junction and 3′gene junction, respectively. The great majority of these NTRK fusion transcripts were classified as in-frame transcripts, while two fusion transcripts (NTRK2-RASEF, and FAT1-NTRK3) were classified as out-of-frame and one (PAN3-NTRK2) was classified as 5UTR-CDS. The complete data was available in Supplementary Table S4.
First-generation NTRK inhibitors (larotrectinib and entrectinib) treat patients with NTRK fusion-positive cancers with high remission rates, irrespective of tumor histology (Cocco et al., 2018). We have subsequently employed the MCG database to explore the investigation of these two drugs in clinical trials. The most frequent biomarkers in clinical trials investigating entrectinib were ROS1 fusion, NTRK1 fusion, and NTRK2 fusion (Figure 4A). For larotrectinib, they were NTRK1 fusion, NTRK2 fusion, and NTRK3 fusion (Figure 4B). Malignant solid tumors (n = 29), non-small cell lung carcinoma (n = 10), and colorectal carcinoma (n = 5) were the most common diseases studied in entrectinib clinical trials (Figure 4A), while malignant solid tumor (n = 21), non-Hodgkin lymphoma (n = 6), and congenital mesoblastic nephroma (n = 6) were the most prevalent diseases studied in larotrectinib clinical trials (Figure 4B).
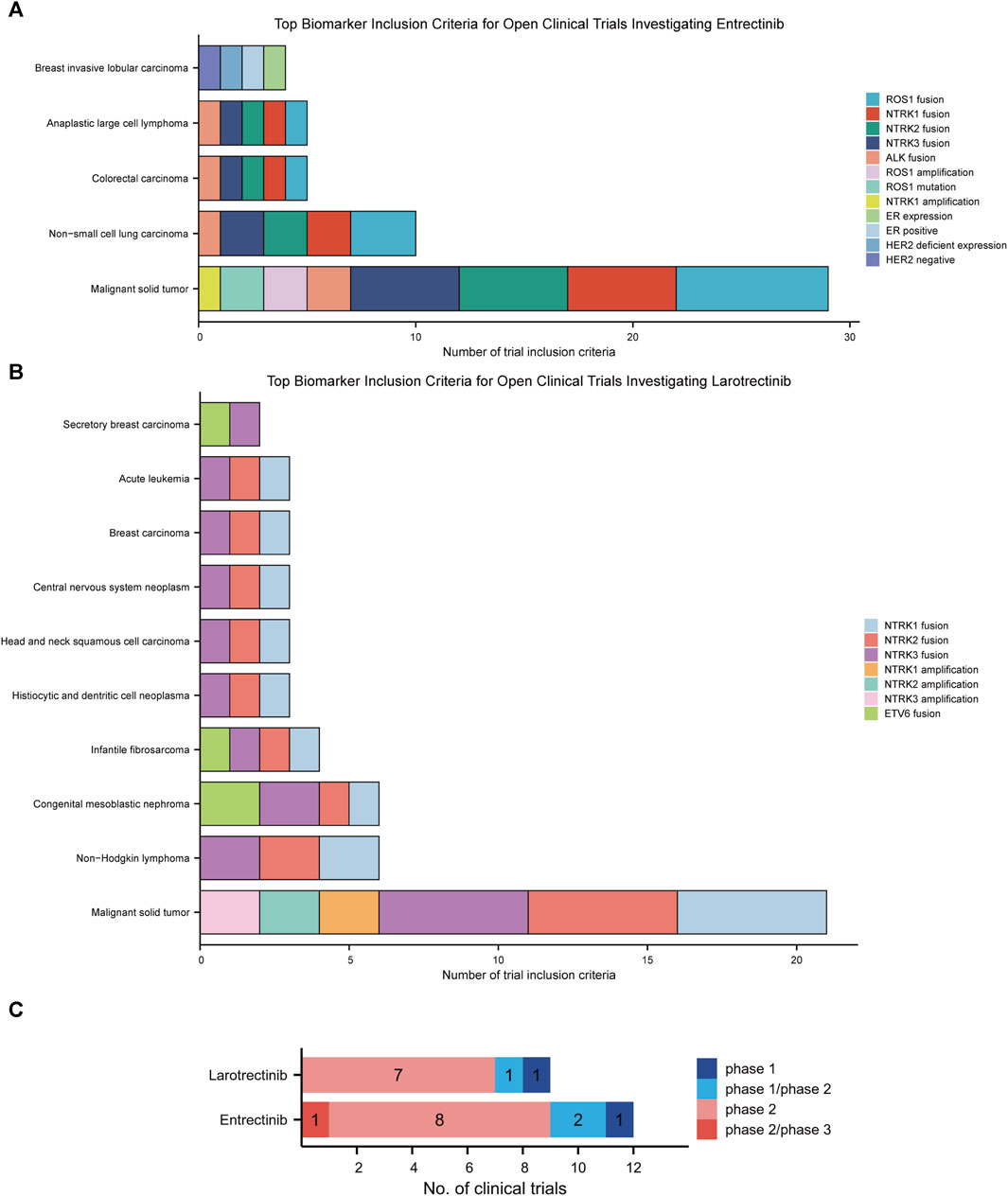
FIGURE 4. This chart shows the top biomarkers most frequently included in clinical trials investigating larotrectinib (A) and entrectinib (B), and the types of cancers associated with those biomarkers. (C). Current clinical trial status of larotrectinib and entrectinib.
Nine clinical trials studied larotrectinib, of which 9 were open and 0 were terminated. Of the trials that studied larotrectinib, 1 was early phase 1, 1 was phase 1/phase 2, and 7 were phase 2. Twelve clinical trials have studied entrectinib, 12 of which were open trials and 0 of which were closed trials. Of these trails, 1 was phase 1, 2 were phase 1/phase 2, 8 were phase 2, and 1 was phase 2/phase 3 (Figure 4C).
Subsequently, we conducted pharmacovigilance analyses in the FAERS database to investigate the signals of ADEs closely associated with the use of two first-generation NTRK inhibitors. In terms of the annual ADEs reported, entrectinib exhibited a steeper increase compared to larotrectinib, with both reaching their peaks in 2021. From that point, entrectinib continued to receive more ADE reports than larotrectinib (Figure 5A). Analysis of the baseline information for these ADEs revealed that the age group with the highest proportion of entrectinib users was below 18 years (5.7%), while for larotrectinib, it was the age group of 18–64 years (4.2%) (Table 2). However, it is important to note that approximately 90% of the reports had unknown age group information for both drugs. Regarding gender distribution, entrectinib had a higher proportion of female reports for ADEs (50.8%), whereas larotrectinib had a similar percentage of reports from males (42.0%) and females (42.4%). The top three countries contributing the highest number of reports were the United States (49.4%), Japan (24.7%), and Germany (2.5%) for entrectinib, and the United States (58.8%), France (7.3%), and Mexico (4.8%) for larotrectinib. In terms of report sources, health professionals accounted for the majority of reports for both drugs (65.5% for entrectinib and 73.3% for larotrectinib). While the primary indication for entrectinib was non-small cell lung carcinoma (29.7%), thyroid cancer (5.3%) was the main indication for larotrectinib. It is noteworthy that entrectinib (32.0%) had a higher proportion of reports of hospitalization compared to larotrectinib (22.6%) in cases of serious outcomes. However, both drugs exhibited similar proportions of reports for death, life-threatening events, and disability (Table 2).
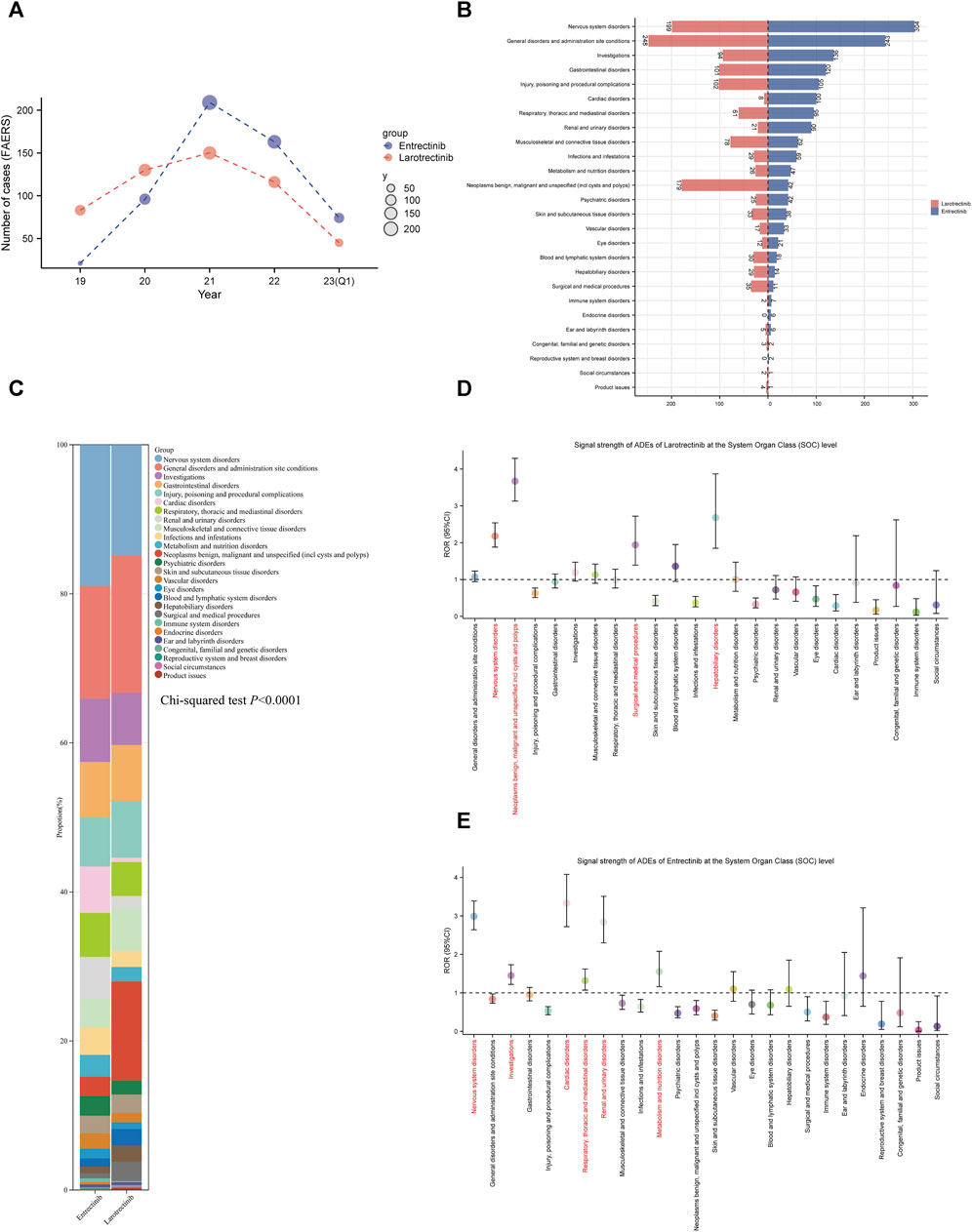
FIGURE 5. Signal detection at the SOC level. (A). Annual distribution of ADE reports of the two first-generation NTRK inhibitors (entrectinib, blue; larotrectinib, red), from 2019 Q1 to 2023 Q1. (B). Number of ADE reports for entrectinib and larotrectinib at the SOC level. (C). The bar scale graph shows the percentage of ADEs at the SOC level. The ROR values and their corresponding 95% confidence intervals for larotrectinib (D) and entrectinib (E) are displayed for different levels of SOC. ADEs, adverse drug events; SOC, System Organ Class; Q1, first quarter; ROR, reporting odds ratio.
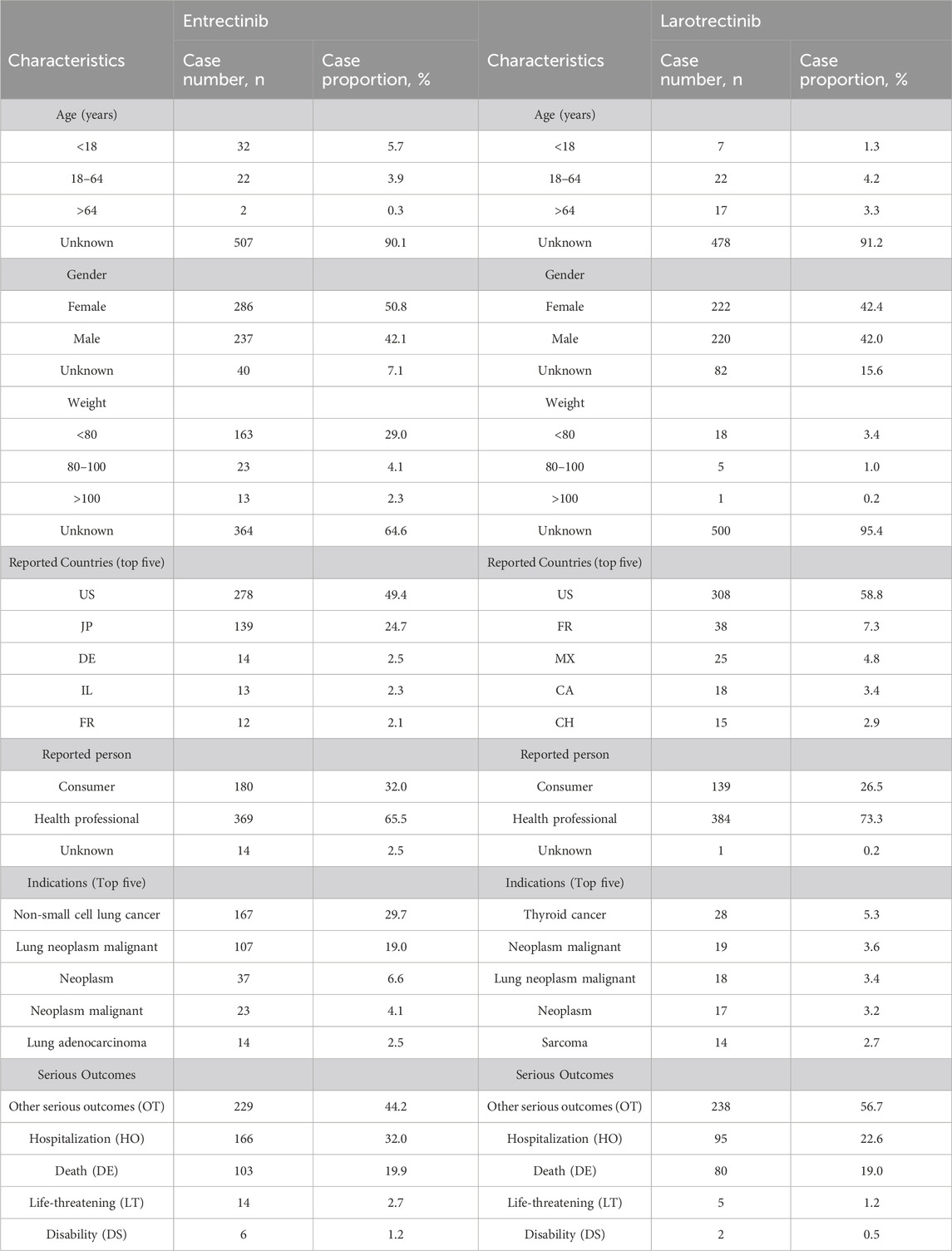
TABLE 2. The demographic baseline data for adverse drug event (ADE) reports involving entrectinib and larotrectinib as the primary suspect (PS) drugs.
Figure 5B presented the reported numbers of the two NTRK inhibitors at the SOC level. For entrectinib and larotrectinib, ADEs were reported at 26 and 24 organ systems, respectively. Interestingly, no ADEs were reported for larotrectinib at the “endocrine disorder” and “reproductive system and breast disorders” SOCs (n = 0). In larotrectinib, the top three SOCs in terms of ADEs numbers were nervous system disorders (n = 304), general disorders and administration site conditions (n = 243), and investigations (n = 136). In entrectinib, they were, general disorders and administration site conditions (n = 248), nervous system disorders (n = 199), and neoplasms benign, malignant and unspecified (n = 179) (Figure 5B). Comparing the composition of each SOC between the two drugs, we found that entrectinib had a higher percentage of “nervous system disorders” (18.94% versus [vs.] 14.82%), “cardiac disorders” (6.23% vs. 0.60%), and “renal and urinary disorders” (5.61% vs. 1.56%) than larotrectinib, while “general disorders and administration site conditions” (18.47% vs. 15.14%) and “neoplasms benign, malignant and unspecified” (13.33% vs. 2.62%) were higher for larotrectinib. Statistical analyses also suggested that there was a significant difference in the proportion of SOCs between the two drugs (p < 0.0001, Figure 5C).
The disproportionality results of the three different algorithms at various SOC levels were shown in Table 3. Wherein, the number of SOCs meeting at least one algorithm signal value threshold was 4 and 6 for larotrectinib and entrectinib, respectively. For larotrectinib, they were nervous system disorders (SOC code:10029205, ROR 2.18 [1.88–2.54]), neoplasms benign, malignant and unspecified (SOC code:10029104, ROR 3.67 [3.13–4.29]), hepatobiliary disorders (SOC code:10019805, ROR 2.68 [1.85–3.87]), and surgical and medical procedures (SOC code:10042613, ROR 1.94 [1.39–2.72]). For entrectinib, they were nervous system disorders (SOC code:10029205, ROR 2.99 [2.64–3.39]), investigations (SOC code:10022891, ROR 1.45 [1.22–1.73]), cardiac disorders (SOC code:10007541, ROR 3.33 [2.72–4.08]), respiratory, thoracic and mediastinal disorders (SOC code:10038738, ROR 1.32 [1.07–1.62]), renal and urinary disorders (SOC code:10038359, ROR 2.84 [2.30–3.51]), and metabolism and nutrition disorders (SOC code:10027433, ROR 1.55 [1.16–2.08]). To improve visualization, Figures 5D,E depicted the signal strength (ROR, with 95% confidence interval [CI]) at the SOC level for larotrectinib and entrectinib, respectively.
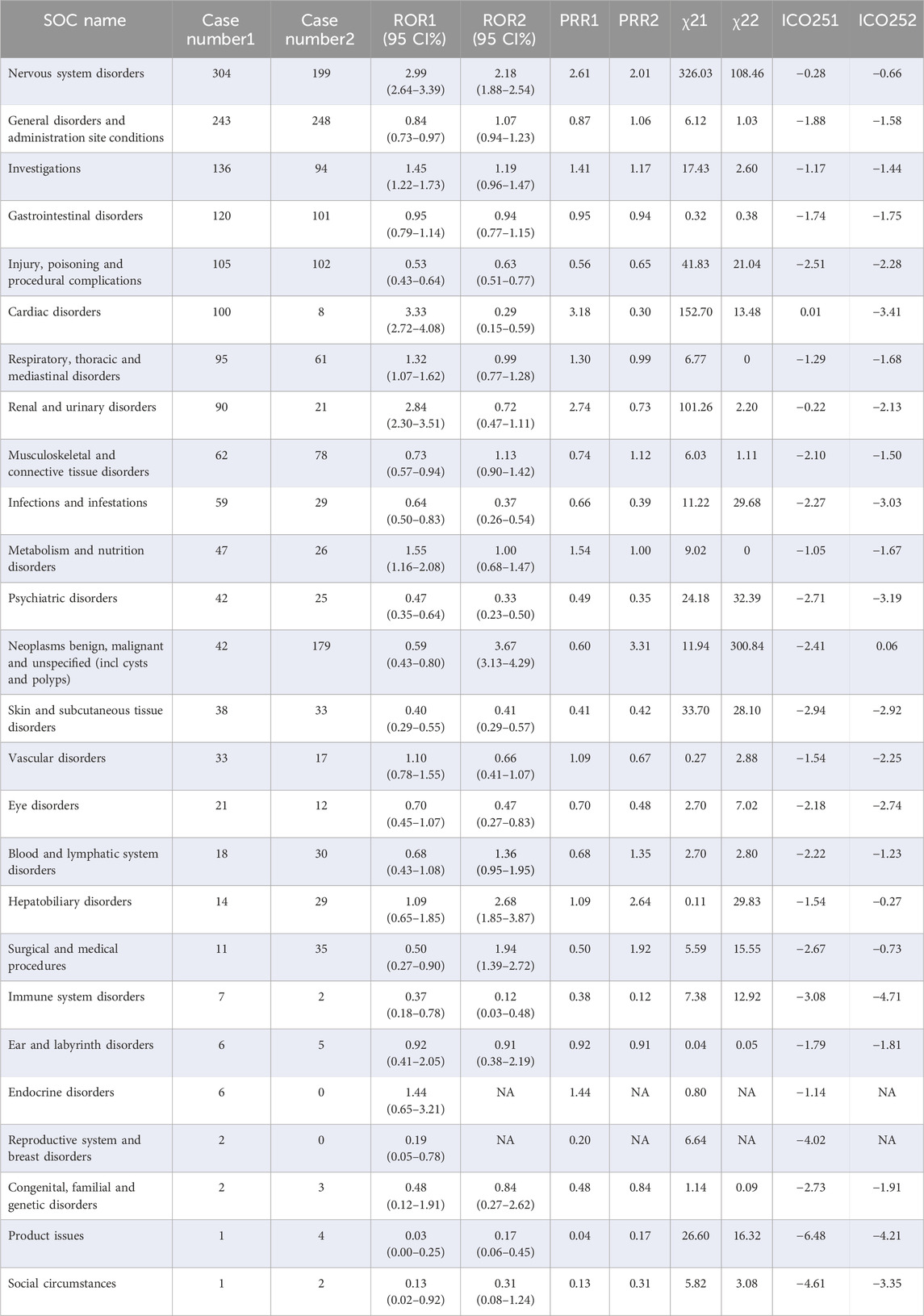
TABLE 3. Signal detection at the SOC level. Lower right marker 1, entrectinib vs all other drugs; Lower right marker 2, larotrectinib vs all other drugs. ROR, reporting odds ratio; CI, confidence interval; PRR, proportional reporting ratio; χ2, chi-squared; IC, information component; IC025, the lower limit of the 95% CI of IC; SOC, System Organ Class.
After satisfying the thresholds of the three algorithms simultaneously, we detected 67 entrectinib-related ADEs and 57 larotrectinib-related ADEs at the PT level, respectively. We sorted these screened ADEs by case number and IC025 value, respectively. For entrectinib, signals including dizziness (n = 58), renal impairment (n = 35), and taste disorder (n = 26) had the highest reported cases, which suggested that they were more common in reports of ADEs following entrectinib dosing (Figure 6A). Dysplasia (6.13), ataxia (4.57), and troponin I increased (4.14) had the highest IC025 values, indicating that they were more associated with entrectinib dosing than the other drugs (Figure 6D). For larotrectinib, dizziness (n = 35), neuropathy peripheral (n = 26), and paraesthesia (n = 17) had the largest number of reports (Figure 6B), while adenocarcinoma of salivary gland (9.43), astrocytoma (7.51), and glioblastoma multiforme (6.98) had the highest IC025 values (Figure 6E).
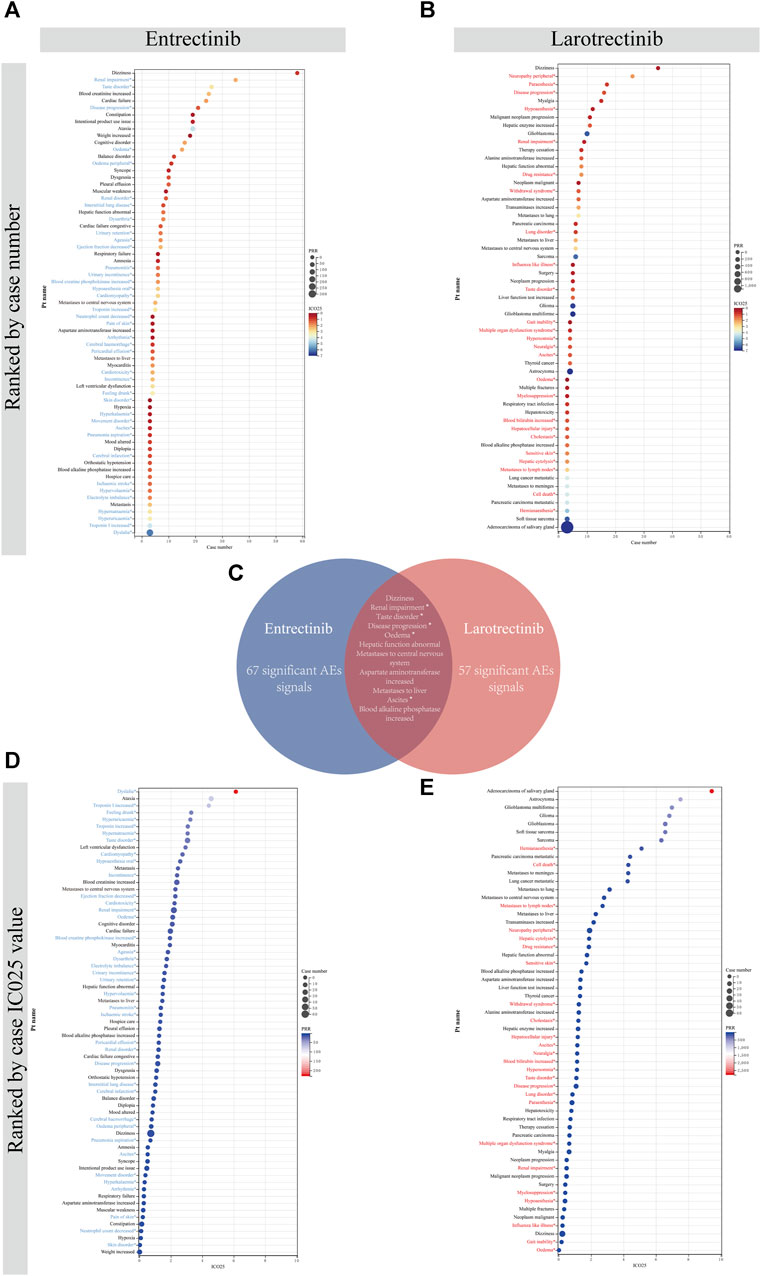
FIGURE 6. Comparison of signal values of the two first-generation NTRK inhibitors for ADEs at the PT level. All positive signals for entrectinib (A) and larotrectinib (B) are ranked by case number. (C). Intersection of positive signals of two drugs. All positive signals for entrectinib (D) and larotrectinib (E) are ranked by IC025 value. An asterisk indicates that the signal is not indicated in the drug instruction. PRR, proportional reporting ratio; PT, preferred term; IC, information component; IC025, the lower limit of the 95% confidence interval of IC.
Besides, a large number of unexpected signals were identified and highlighted in prominent colors. These signals need to be refined in subsequent updates of the drug instruction. After taking the intersection of ADEs for both drugs, we found a total of 11 overlapping signals (Figure 6C). Of them, renal impairment, taste disorder, disease progression, oedema, and ascites were intersected unexpected signals, which needed to be given sufficient attention in subsequent clinical studies. All signals and calculations that satisfied the thresholds of the three algorithms were listed in Supplementary Table S5.
Recognizing the onset time of these ADEs can enable healthcare professionals better target their post-medication monitoring. After data filtering (removing missing and incorrect reporting times), 243 and 113 TTO reports were collected related to entrectinib and larotrectinib, respectively. The median TTO for entrectinib and larotrectinib was 16 days (Figure 7E) and 44 days (Figure 7F). Observing these TTO at the SOC level, we noticed that the SOCs with longer onset after entrectinib administration included “blood and lymphatic system disorders” (median TTO: 23 days), and “neoplasms benign, malignant and unspecified” (median TTO: 17 days), while “psychiatric disorders” (median TTO: 1 day), “eye disorders” (median TTO: 4 days), and “gastrointestinal disorders” (median TTO: 4 days) had shorter onset (Figure 7A). For larotrectinib, “infections and infestations” (median TTO: 242 days) and “neoplasms benign, malignant and unspecified” (median TTO: 101 days) had longer onset time, while “psychiatric disorders” (median TTO: 4 days), “skin and subcutaneous tissue disorders” (median TTO: 8 days), and “nervous system disorders” (median TTO: 9 days) had shorter onset time (Figure 7B). The specific statistical description of TTO was in Supplementary Table S6.
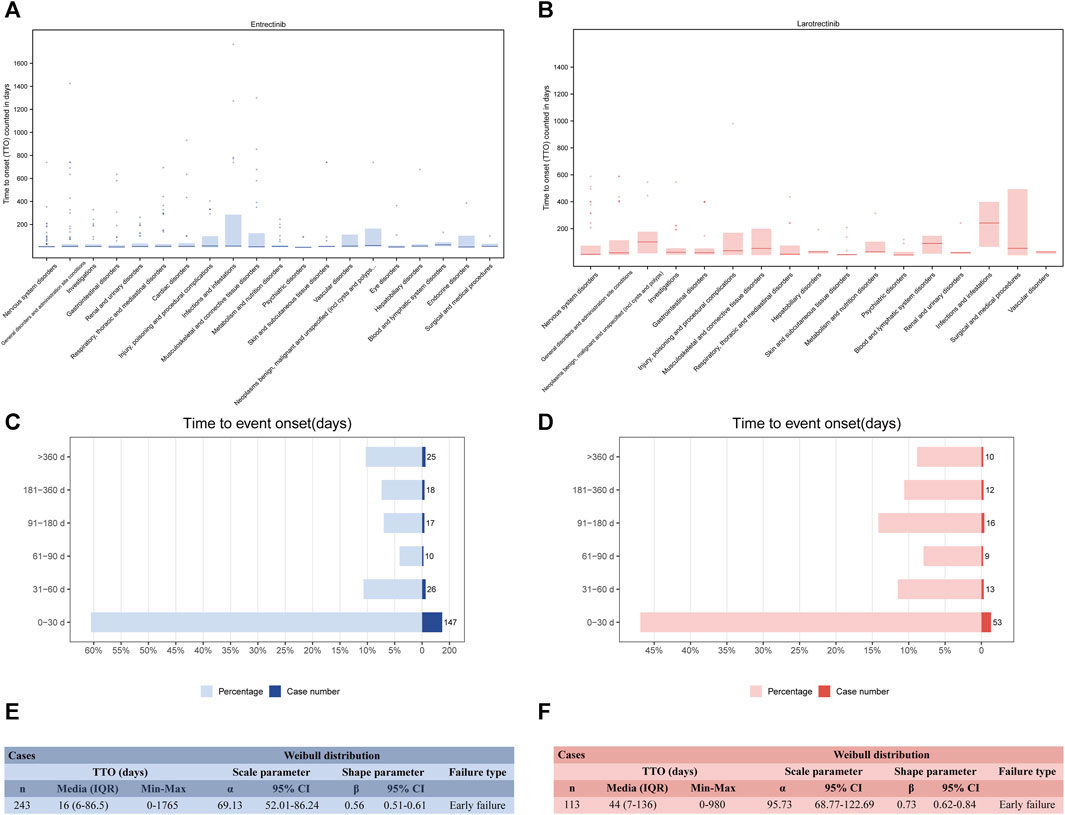
FIGURE 7. Time to onset (TTO) analysis (counted in days). Box plot of the TTO at the SOC level for entrectinib (A) and larotrectinib (B). Bold bar within the stick: median TTO; Lower end of the stick: 1/4 quantile of the TTO; Upper end of the stick: 3/4 quantile of the TTO. Number and proportion of all TTO reports in different time periods for entrectinib (C) and larotrectinib (D). Weibull distribution test of TTO for entrectinib (E) and larotrectinib (F). A TTO of 0 days signifies that the adverse event occurred on the same day as the start of treatment. IQR, interquartile range.
Concerning the distribution of ADEs over time, Figures 7C,D indicated that the majority of ADEs occurred within the first month after NTRK inhibitors use (n = 147, 60.5% for entrectinib; n = 53, 46.9% for larotrectinib). As time was delayed, the number of ADEs decreased but leveled off. Of note, our data showed that ADEs could still occur after 1 year of drug administration (n = 25, 10.3% for entrectinib; n = 10, 8.9% for larotrectinib) (Supplementary Table S6). The Weibull shape parameter analysis revealed calculated shape parameters β) of 0.56 (0.51–0.61) for entrectinib (Figure 7E) and 0.73 (0.62–0.84) for larotrectinib (Figure 7F), both with β values <1, indicating an early failure type. This suggested that for both NTRK inhibitors, the incidence of ADE decreased over time.
Precision medicine endeavors to develop medical interventions personalized to an individual’s genetic, environmental, and lifestyle factors (Wahida et al., 2023). In recent years, with the rapid advancement of precision medicine, there has been a shift in clinical trials from being focused on specific tumor types to being oriented towards the genome and agnostic to histology (Fountzilas et al., 2022). In this broader context, genomics offers valuable insights into identifying dysregulated genes, associated pathways, and histological knowledge within complex disease pathology. Clinical trials play a crucial role in translating genomic evidence into effective drug treatments, bridging the gap between bench research and bedside application (Knoepfler, 2015). Pharmacovigilance, as a critical component of drug safety regulation, is essential in identifying ADEs that occur during clinical trials and post-marketing, ensuring medication safety. Conversely, analyzing ADEs in isolation without understanding the underlying molecular etiology of the observed signals can lead to “fragmentation” in the relationship between ADEs and genomic alterations. By integrating genomic-level evidence and participant target data from clinical trials, this fragmentation can be partially mitigated, aiding in the identification of high-risk individuals and populations for ADEs, thus enhancing drug safety, efficacy, and overall public health (Zhou et al., 2023). Recently, studies have emerged linking genomics and pharmacovigilance. For instance, Jing et al. assessed the relationship between multiple genomic factors and immune-associated adverse events in various cancer types by combining genomic data with pharmacovigilance data (Jing et al., 2020). In our study, we conducted a comprehensive mapping of the NTRK genome, utilizing multi-omics data from TCGA, followed by a review of clinical trials involving NTRK inhibitors and an extensive pharmacovigilance analysis using the FAERS database. Our genomic evidence provides valuable insights into mitigating adverse effects of first-generation NTRK inhibitors, such as drug resistance, while pharmacovigilance analyses offer crucial additions by uncovering undetected adverse effects in clinical trials and aiding in optimal drug design at the genomic level.
After analyzing the expression profiles of NTRK genes, we observed that the mRNA expression of NTRK1/2/3 was generally downregulated in tumor tissues compared to normal tissues across most cancers. Additionally, we identified a significant negative correlation between the hypermethylation state of the NTRK gene promoters and their mRNA expression in multiple cancer types. Previous studies have also reported aberrant methylation patterns in the NTRK genes in various cancers. For instance, Yamada et al. demonstrated elevated methylation levels in PRAD cell lines for NTRK2, which resulted in decreased mRNA expression (Yamada et al., 2004). Similarly, hypermethylation status of NTRK3 was reported in cervical cancer through analyzing sequencing data and further validated in clinical samples (Ji et al., 2021). Considering that epigenetic changes are an essential mechanism for regulating gene expression, aberrant DNA methylation might be a plausible explanation for the dysregulation of NTRK gene expression (Arechederra et al., 2018). Moreover, CNV of DNA sequences can remodel gene structure, regulate gene expression, and facilitate significant phenotypic variation, including NTRK genes (Zhou et al., 2011; Tsai et al., 2016; Yu et al., 2021). Our study also uncovered the presence of CNVs in NTRK genes in many cancers, particularly in UCS and LUAD. This observation suggests that CNVs might influence the mRNA expression and function of these genes. However, further experiments are needed to validate this hypothesis, as there is limited evidence available from previous studies. Additionally, we observed a protective role of NTRK3 in BRCA and PRAD, as well as a protective role of NTRK2 in LGG. These findings are consistent with previous research and suggest that these genes may exert a protective effect through mechanisms such as immune cell recruitment, angiogenesis inhibition, and apoptosis induction (Bouzas-Rodriguez et al., 2010; Luo et al., 2013; Bagherabadi et al., 2022; Fang et al., 2023). However, we also noticed that high NTRK expression was hazardous for patients with BLCA and UVM. The varying prognoses in different types of tumors may be attributed to the fact that the function of NTRK genes is receptor-dependent. In the presence of a ligand, these genes transmit positive, proliferative signals, whereas they induce apoptosis in the absence of a ligand (Tauszig-Delamasure et al., 2007; Luo et al., 2013). Also, the effect of tumor heterogeneity and different splicing patterns on its function cannot be overlooked (Bagherabadi et al., 2022).
In our further exploration of the genomic alterations of NTRK genes, we discovered that in pan-cancer, alterations in these genes were predominantly characterized by high mRNA expression, amplifications, and mutations. The most frequently mutated amino acid sites in NTRK1/2/3 were G169R, A268V, and R153L/Q, respectively. Prior research has indicated that non-fusion NTRK alterations, such as mutations and amplifications, are associated with the response to NTRK inhibitors and their therapeutic efficacy (Okamura et al., 2018; Amatu et al., 2019; Zito Marino et al., 2023). In particular, mutations at the G595R and G667C sites of NTRK1, and at G696A and G623R of NTRK3, have been associated with the acquisition of resistance to first-generation NTRK inhibitors, possibly due to the fact that these point mutations alter the three-dimensional conformation of the kinase’s structural domains, thereby reducing or abolishing binding to the inhibitors (Russo et al., 2016; Drilon et al., 2017; Somwar et al., 2020). In contrast, in a case report by Hempel et al., a patient with ESCA with NTRK1 gene amplification experienced shrinkage of primary and metastatic tumors within 6 weeks after treatment with the NTRK inhibitor larotrectinib, accompanied by a decrease in tumor markers (Hempel et al., 2020). Finally, we also explored potential signaling pathways for genes co-altered with NTRK genes, which included PI3K/Akt, cGMP-PKG, and cAMP, etc. Mutational pathways co-activated with target genes are frequently observed in targeted therapies (Moore et al., 2020). For example, resistance to BRAF inhibitors is linked to the activation of the MAPK pathway, necessitating the co-administration of MEK inhibitors under certain circumstances (Yi et al., 2022). Likewise, combining NTRK inhibitors with co-mutational pathway inhibitors (such as PI3K inhibitors) could represent a potential therapeutic strategy for patients resistant to first-generation NTRK inhibitors. In conclusion, our study offers a comprehensive understanding of NTRK genomic alterations. While non-fusion alterations have not yet demonstrated consistent and long-lasting response to targeted therapies, they may still yield valuable insights into mechanisms of drug resistance.
Structural rearrangements of the genome can lead to the formation of fusion genes, which possess oncogenic properties and subsequently result in the overexpression of aberrant proteins. These abnormal proteins are known to drive tumorigenesis and present potential targets for therapeutic intervention (Mitelman et al., 2007; Bastus et al., 2010; Best et al., 2018). While NTRK gene fusions occur at a low frequency in common cancer types like lung and colorectal cancers (<25%), they are observed at a high frequency in specific rare tumor types (>80%). These fusions activate downstream cell growth and proliferative pathways, thereby promoting tumorigenesis (Vaishnavi et al., 2015; Amatu et al., 2016; Amatu et al., 2019). Moreover, constitutively active TRK kinase generated by gene fusion becomes a target for the action of NTRK inhibitors (Wang et al., 2015; Khotskaya et al., 2017). Our study provided a comprehensive analysis of fusion mutations involving NTRK genes across various cancer types. We observed that the majority of these fusion mutations occurred in NTRK3, with THCA having the highest reported incidence. Among the fusion pairs, ETV6-NTRK3 was found to have the highest percentage. Furthermore, most of these fusion mutations were in-frame mutations. Consistent with our findings, research studies have increasingly demonstrated the presence of the ETV6-NTRK3 fusion in various tumor types, including glioblastoma, ductal carcinoma, fibrosarcoma, and THCA (Tognon et al., 2002; Bastos et al., 2018; Biswas et al., 2022; Jiang, 2022). Interestingly, the study by Kinnunen et al. employed proximity-labeled mass spectrometry to establish a stable, reliable association of the ETV6-NTRK3 fusion with several key signaling pathways, including ERBB, IRS-1, and JAK/STAT (Kinnunen et al., 2023). As another of the more common and first reported NTRK fusion mutations, TPM3-NTRK1 was initially identified in colorectal cancer (CRC) samples, and subsequent studies have confirmed that chromosomal rearrangements in this manner make patients with CRC highly sensitive to TRKA inhibitors (Martin-Zanca et al., 1986; Ardini et al., 2014; Créancier et al., 2015). Other fusion types, such as SQSTM1-NTRK1, although carried by only one patient with THCA in our study, a recent case report demonstrated that patients carrying SQSTM1-NTRK1 had a 51% reduction in tumor burden after 18 months of treatment with larotrectinib, and this impressive efficacy continued (Bargas et al., 2022). Of course, we have identified other novel fusion types including SQSTM1-NTRK2, FAT1-NTRK3, and AKAP13-NTRK3, however the relevant reports are very limited so far. Whether they could become new targets for NTRK inhibitors needs to be validated in subsequent clinical trials (Bargas et al., 2022).
Genomic evidence indicates that both larotrectinib and entrectinib are pan-TRK inhibitors, targeting TRKA, TRKB, and TRKC. However, it is important to note that these TRK receptors also play essential roles in neurodevelopment. Consequently, the use of these pan-TRK inhibitors may lead to treatment-related side effects due to the inhibition of TRK signaling in normal tissues (Bhangoo and Sigal, 2019). Our pharmacovigilance study revealed that entrectinib had positive signaling values at six SOC levels compared to four for larotrectinib. Of particular note, both entrectinib (ROR: 2.99 [2.64–3.39]) and larotrectinib (ROR: 2.18 [1.88–2.54]) had positive signal values at the “nervous system disorders”. Generally, neurological ADEs are primarily thought to be related to the on-target effects of TRK inhibitors (Kummar et al., 2023). Considering the critical role of NTRK1 and NTRK3 in the normal functioning of sensory neurons, their loss-of-function mutations may lead to dizziness, sensory abnormalities, headaches and gait disturbances (Cocco et al., 2018; Amatu et al., 2019). Another anticipated ADE is weight gain, due to the vital role of NTRK2 in controlling energy balance and appetite (An et al., 2020; Houtz et al., 2021) Consistently, at the PT level, we also identified several common, drug-related on-target adverse reactions, including dizziness, constipation, ataxia, weight increased, balance disorder, and dysgeusia. These ADEs we identified are generally in line with previously reported adverse events in clinical trials of entrectinib. Although the majority of these treatment-related adverse events (TRAEs) were grade 1/2, weight gain (5%–10%) was considered the most common grade 3/4 event, and central nervous system toxicity was reported as the most severe TRAE (3%–4%) (Al-Salama and Keam, 2019; Doebele et al., 2020; Drilon et al., 2020; Dziadziuszko et al., 2021). Therefore, it is crucial to implement timely monitoring and provide appropriate guidance to patients. However, it is interesting to note that entrectinib has shown a potential protective effect in patients with tumors that may have a higher risk of central nervous system metastasis. This could be attributed to its ability to more easily penetrate the blood-brain barrier (Dziadziuszko et al., 2021; Frampton, 2021). How to balance adverse effects and potential benefits to optimize drug selection in the future also needs to be explored in depth.
Although previous clinical trials have shown similar safety profiles for both drugs, in our study we demonstrated that entrectinib had stronger signal values for “cardiac disorders”, “respiratory, thoracic and mediastinal disorders”, “renal and urinary disorders” and “metabolism and nutrition disorders”, while larotrectinib had stronger signal values for “neoplasms benign, malignant and unspecified”, and “hepatobiliary disorders”. For entrectinib, cardiac problem is the other most common type of severe TRAE (Drilon et al., 2020; Marcus et al., 2021). In a case of NSCLC with ROS1 rearrangement, the patient developed drug-induced heart failure after treatment with entrectinib, and the symptoms improved after drug discontinuation (Otsu et al., 2022). Results from another previous pharmacovigilance analysis also revealed that ALK and ROS1 inhibitors induced higher odds of cardiac conduction disease than other targeted therapies (Waliany et al., 2021). Moreover, at the PT level, we also detected positive signal values including “interstitial lung disease”, “respiratory failure”, and “pneumonitis” with a relatively high number of cases at the respiratory level, which may be due to the rich blood supply of the lungs, causing accumulation of the drug, or the release of toxic substances (Long and Suresh, 2020; Spagnolo et al., 2022). Elevated aminotransferase occurred in half of the adverse event reports during the clinical trials of larotrectinib and was also a primary cause of drug dose reductions (Drilon et al., 2018; Laetsch et al., 2018; Hong et al., 2020; Le et al., 2022). In light of these findings, it is important to conduct regular and dynamic monitoring of the patient’s liver function and to administer suitable hepatoprotective agents. Additionally, we observed several ADEs related to neoplasm progression for larotrectinib, including “malignant neoplasm progression,” “neoplasm malignant,” and “metastases to lung”. These adverse signals may be more associated with disease progression rather than being solely attributed to adverse reactions caused by larotrectinib. This is supported by the fact that a significant majority of patients experienced objective remission following treatment with the larotrectinib (Doebele et al., 2020; Doz et al., 2022; Drilon et al., 2022). In conclusion, our analysis has provided a comparative evaluation of the signal strength in different organs for both larotrectinib and entrectinib. This comprehensive assessment can contribute to a more detailed and proactive clinical medication monitoring approach, enhancing patient safety and optimizing the effectiveness of treatment.
Furthermore, our study revealed some unexpected signals. For entrectinib, we observed unexpected signals with a high number of cases, such as taste disorder and renal impairment. Furthermore, we identified unexpected signals with a low number of cases but a high signal intensity, including dyslalia and hyperuricemia. It is worth noting that taste disorder and dyslalia have been hypothesized to be a result of the potential on-target effects of NTRK inhibitors (Naik et al., 2010; Ma et al., 2011; Szobota et al., 2019). Hyperuricemia has been recognized in previous clinical trials as a serious toxicity induced by entrectinib treatment that required intervention, but the exact cause is unknown (Dziadziuszko et al., 2021; Marcus et al., 2021). Similarly, regarding larotrectinib, there were more case reports of neuropathy peripheral, paresthesia, and stronger signal strength for hemianesthesia, and drug-resistance. Regarding the mechanism of resistance to larotrectinib, in addition to being associated with mutations or altered pathway activation as described above, it may also relate to the mode of administration (e.g., intermittent dosing), which may require a combination of other drugs to overcome (Amatu et al., 2016; Cocco et al., 2019; Kummar et al., 2023). Remarkably, unexpected signals found in both drugs included, renal impairment, taste disorder, disease progression, edema, and ascites. Some previous clinical trials failed to detect the new signals, whereas our results complement the previous studies and are indicative of subsequent updates of drug instruction (Doebele et al., 2020; Hong et al., 2020; Doz et al., 2022). Furthermore, as both NTRK inhibitors are predominantly metabolized by the enzyme cytochrome P450 (CYP450), which has been strongly linked to hepatic and renal drug toxicity, investigating CYP450 site variation at the pharmacogenomic level could yield distinctive insights into the toxicity of NTRK inhibitors (Quintanilha et al., 2017; Wang et al., 2020).
It is important to note that while we have made certain genomic insights regarding the adverse effects (e.g., drug resistance) of first-generation NTRK inhibitors, more reliable conclusions about causality may require higher-level genetic evidence in the future. This could involve identifying single nucleotide polymorphism (SNP) loci significantly associated with ADEs through genome-wide association studies (Ghouse et al., 2022; Giles et al., 2022). Such an approach could enable the screening and identification of patients susceptible to adverse effects, thereby improving efficacy and concurrently mitigating adverse effects.
Some ADEs can occur shortly after starting treatment, ranging from minutes to hours. However, other ADEs may manifest days, weeks, months, or even years after exposure. The timing of these events can vary depending on various factors, including the drug’s pharmacokinetics and its metabolites, as well as the underlying pathophysiological mechanism of action (Leroy et al., 2014). Previous studies have provided information on various ADEs, but the precise timing of these events remains largely unknown. In our study, we found that both drugs exhibited earlier onset of ADEs in the SOC categories of “psychiatric disorders” and “nervous system disorders,” while ADEs related to “infections and infestations” and “neoplasms benign, malignant, and unspecified” occurred later. However, there were notable differences between the two drugs. For instance, ADEs in the category of “musculoskeletal and connective tissue disorders” had a median time to onset of 6 days in entrectinib and 53 days in larotrectinib. In both drugs, these ADEs predominantly occurred within the first month of administration, with a progressive decrease in their probability of occurrence over time. This highlights the significance of early medication surveillance. Overall, our analysis provided valuable insights into the onset timing of ADEs for two NTRK inhibitors, facilitating a more meticulous approach to preventing or diagnosing the occurrence of ADEs.
Our study exhibits several strengths. Firstly, the comprehensive analysis of data from multiple sources contributes to a more thorough understanding of the NTRK genes. Secondly, the precise application of multiple analysis methods and the depth of the results further underscore the strength of this study. However, it is crucial to acknowledge certain limitations. Firstly, the small sample sizes for specific cancer types may introduce bias into the analyses concerning these types of cancer. Moreover, the FAERS database does not establish a causal relationship between drug use and ADEs, and limited drug dosage information hindered our analysis of the correlation between ADEs and drug dosage. Lastly, due to the recent introduction of the two NTRK inhibitors to the market, reported ADEs were limited, necessitating further validation of our results with larger sample sizes in future studies.
This study delineated the genomic features of NTRK, encompassing expression, methylation, and gene fusion, through the analysis of multi-omics data. Subsequently, comprehensive analyses were conducted to compare the safety profiles of two first-generation NTRK inhibitors using the FAERS database. These analyses offer valuable insights for healthcare professionals, aiding their understanding of the mechanisms of resistance to NTRK inhibitors and facilitating the monitoring of adverse effects associated with entrectinib and larotrectinib.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
ZC: Conceptualization, Writing–original draft, Writing–review and editing. ZZ: Conceptualization, Writing–original draft, Writing–review and editing. DX: Formal Analysis, Writing–original draft. LW: Formal Analysis, Writing–original draft. FC: Formal Analysis, Writing–original draft. SL: Visualization, Writing–original draft. FZ: Visualization, Writing–original draft. RP: Visualization, Writing–original draft. SC: Visualization, Writing–original draft. HY: Visualization, Writing–original draft. JS: Writing–review and editing. YZ: Writing–review and editing. XY: Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the fund of “Project A of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU-2021-01).”
We are very grateful to the developers and maintainers of the various databases mentioned in the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1329409/full#supplementary-material
Al-Salama, Z. T., and Keam, S. J. (2019). Entrectinib: first global approval. Drugs 79 (13), 1477–1483. doi:10.1007/s40265-019-01177-y
Amatu, A., Sartore-Bianchi, A., Bencardino, K., Pizzutilo, E. G., Tosi, F., and Siena, S. (2019). Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 30 (8), viii5–viii15. doi:10.1093/annonc/mdz383
Amatu, A., Sartore-Bianchi, A., and Siena, S. (2016). NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 1 (2), e000023. doi:10.1136/esmoopen-2015-000023
An, J. J., Kinney, C. E., Tan, J.-W., Liao, G.-Y., Kremer, E. J., and Xu, B. (2020). TrkB-expressing paraventricular hypothalamic neurons suppress appetite through multiple neurocircuits. Nat. Commun. 11 (1), 1729. doi:10.1038/s41467-020-15537-w
Ardini, E., Bosotti, R., Borgia, A. L., De Ponti, C., Somaschini, A., Cammarota, R., et al. (2014). The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol. Oncol. 8 (8), 1495–1507. doi:10.1016/j.molonc.2014.06.001
Arechederra, M., Daian, F., Yim, A., Bazai, S. K., Richelme, S., Dono, R., et al. (2018). Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat. Commun. 9 (1), 3164. doi:10.1038/s41467-018-05550-5
Bagherabadi, A., Hooshmand, A., Shekari, N., Singh, P., Zolghadri, S., Stanek, A., et al. (2022). Correlation of NTRK1 downregulation with low levels of tumor-infiltrating immune cells and poor prognosis of prostate cancer revealed by gene network analysis. Genes 13 (5), 840. doi:10.3390/genes13050840
Bailey, C., Peddie, D., Wickham, M. E., Badke, K., Small, S. S., Doyle-Waters, M. M., et al. (2016). Adverse drug event reporting systems: a systematic review. Br. J. Clin. Pharmacol. 82 (1), 17–29. doi:10.1111/bcp.12944
Bargas, S., Mc Leer, A., Mondet, J., Chabre, O., and Laramas, M. (2022). An impressive response with larotrectinib in a patient with a papillary thyroid carcinoma harboring an SQSTM1-NTRK1 fusion. Eur. J. Endocrinol. 186 (4), K5–K8. doi:10.1530/EJE-21-0509
Bastos, A. U., de Jesus, A. C., and Cerutti, J. M. (2018). ETV6-NTRK3 and STRN-ALK kinase fusions are recurrent events in papillary thyroid cancer of adult population. Eur. J. Endocrinol. 178 (1), 83–91. doi:10.1530/EJE-17-0499
Bastus, N. C., Boyd, L. K., Mao, X., Stankiewicz, E., Kudahetti, S. C., Oliver, R. T. D., et al. (2010). Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 70 (23), 9544–9548. doi:10.1158/0008-5472.CAN-10-1638
Bate, A. (2007). Bayesian confidence propagation neural network. Drug Saf. 30 (7), 623–625. doi:10.2165/00002018-200730070-00011
Best, S. A., Harapas, C. R., Kersbergen, A., Rathi, V., Asselin-Labat, M.-L., and Sutherland, K. D. (2018). FGFR3-TACC3 is an oncogenic fusion protein in respiratory epithelium. Oncogene 37 (46), 6096–6104. doi:10.1038/s41388-018-0399-5
Bhangoo, M. S., and Sigal, D. (2019). TRK inhibitors: clinical development of larotrectinib. Curr. Oncol. Rep. 21 (2), 14. doi:10.1007/s11912-019-0761-y
Biswas, A., Rajesh, Y., Das, S., Banerjee, I., Kapoor, N., Mitra, P., et al. (2022). Therapeutic targeting of RBPJ, an upstream regulator of ETV6 gene, abrogates ETV6-NTRK3 fusion gene transformations in glioblastoma. Cancer Lett. 544, 215811. doi:10.1016/j.canlet.2022.215811
Borges, J. B., Oliveira, V. F. d., Ferreira, G. M., Los, B., Barbosa, T. K. A. A., Marçal, E. d.S. R., et al. (2021). Genomics, epigenomics and pharmacogenomics of familial hypercholesterolemia (FHBGEP): a study protocol. Res. Soc. Adm. Pharm. RSAP 17 (7), 1347–1355. doi:10.1016/j.sapharm.2020.10.007
Bouzas-Rodriguez, J., Cabrera, J. R., Delloye-Bourgeois, C., Ichim, G., Delcros, J.-G., Raquin, M.-A., et al. (2010). Neurotrophin-3 production promotes human neuroblastoma cell survival by inhibiting TrkC-induced apoptosis. J. Clin. Investigation 120 (3), 850–858. doi:10.1172/JCI41013
Brockmöller, J., and Tzvetkov, M. V. (2008). Pharmacogenetics: data, concepts and tools to improve drug discovery and drug treatment. Eur. J. Clin. Pharmacol. 64 (2), 133–157. doi:10.1007/s00228-007-0424-z
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 (5), 401–404. doi:10.1158/2159-8290.CD-12-0095
Cocco, E., Scaltriti, M., and Drilon, A. (2018). NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 15 (12), 731–747. doi:10.1038/s41571-018-0113-0
Cocco, E., Schram, A. M., Kulick, A., Misale, S., Won, H. H., Yaeger, R., et al. (2019). Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat. Med. 25 (9), 1422–1427. doi:10.1038/s41591-019-0542-z
Créancier, L., Vandenberghe, I., Gomes, B., Dejean, C., Blanchet, J.-C., Meilleroux, J., et al. (2015). Chromosomal rearrangements involving the NTRK1 gene in colorectal carcinoma. Cancer Lett. 365 (1), 107–111. doi:10.1016/j.canlet.2015.05.013
Cui, Z., Zou, F., Wang, R., Wang, L., Cheng, F., Wang, L., et al. (2023). Integrative bioinformatics analysis of WDHD1: a potential biomarker for pan-cancer prognosis, diagnosis, and immunotherapy. World J. Surg. Oncol. 21 (1), 309. doi:10.1186/s12957-023-03187-3
Doebele, R. C., Drilon, A., Paz-Ares, L., Siena, S., Shaw, A. T., Farago, A. F., et al. (2020). Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet. Oncol. 21 (2), 271–282. doi:10.1016/S1470-2045(19)30691-6
Doz, F., van Tilburg, C. M., Geoerger, B., Højgaard, M., Øra, I., Boni, V., et al. (2022). Efficacy and safety of larotrectinib in TRK fusion-positive primary central nervous system tumors. Neuro-oncology 24 (6), 997–1007. doi:10.1093/neuonc/noab274
Drilon, A., Laetsch, T. W., Kummar, S., DuBois, S. G., Lassen, U. N., Demetri, G. D., et al. (2018). Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 378 (8), 731–739. doi:10.1056/NEJMoa1714448
Drilon, A., Nagasubramanian, R., Blake, J. F., Ku, N., Tuch, B. B., Ebata, K., et al. (2017). A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov. 7 (9), 963–972. doi:10.1158/2159-8290.CD-17-0507
Drilon, A., Siena, S., Dziadziuszko, R., Barlesi, F., Krebs, M. G., Shaw, A. T., et al. (2020). Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet. Oncol. 21 (2), 261–270. doi:10.1016/S1470-2045(19)30690-4
Drilon, A., Tan, D. S. W., Lassen, U. N., Leyvraz, S., Liu, Y., Patel, J. D., et al. (2022). Efficacy and safety of larotrectinib in patients with tropomyosin receptor kinase fusion-positive lung cancers. JCO Precis. Oncol. 6, e2100418. doi:10.1200/PO.21.00418
Dziadziuszko, R., Krebs, M. G., De Braud, F., Siena, S., Drilon, A., Doebele, R. C., et al. (2021). Updated integrated analysis of the efficacy and safety of entrectinib in locally advanced or metastatic ROS1 fusion-positive non-small-cell lung cancer. J. Clin. Oncol. 39 (11), 1253–1263. doi:10.1200/JCO.20.03025
Ehmann, F., Caneva, L., and Papaluca, M. (2014). European Medicines Agency initiatives and perspectives on pharmacogenomics. Br. J. Clin. Pharmacol. 77 (4), 612–617. doi:10.1111/bcp.12319
Ekmekcioglu, S., Davies, M. A., Tanese, K., Roszik, J., Shin-Sim, M., Bassett, R. L., et al. (2016). Inflammatory marker testing identifies CD74 expression in melanoma tumor cells, and its expression associates with favorable survival for stage III melanoma. Clin. Cancer Res. 22 (12), 3016–3024. doi:10.1158/1078-0432.CCR-15-2226
Fang, Y., Zhang, Q., Chen, C., Chen, Z., Zheng, R., She, C., et al. (2023). Identification and comprehensive analysis of epithelial-mesenchymal transition related target genes of miR-222-3p in breast cancer. Front. Oncol. 13, 1189635. doi:10.3389/fonc.2023.1189635
Fountzilas, E., Tsimberidou, A. M., Vo, H. H., and Kurzrock, R. (2022). Clinical trial design in the era of precision medicine. Genome Med. 14 (1), 101. doi:10.1186/s13073-022-01102-1
Frampton, J. E. (2021). Entrectinib: a review in NTRK+ solid tumours and ROS1+ NSCLC. Drugs 81 (6), 697–708. doi:10.1007/s40265-021-01503-3
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6 (269), pl1. doi:10.1126/scisignal.2004088
Ghouse, J., Tragante, V., Muhammad, A., Ahlberg, G., Skov, M. W., Roden, D. M., et al. (2022). Polygenic risk score for ACE-inhibitor-associated cough based on the discovery of new genetic loci. Eur. Heart J. 43 (45), 4707–4718. doi:10.1093/eurheartj/ehac322
Giles, J. B., Steiner, H. E., Rollin, J., Shaffer, C. M., Momozawa, Y., Mushiroda, T., et al. (2022). Genome-wide association study of platelet factor 4/heparin antibodies in heparin-induced thrombocytopenia. Blood Adv. 6 (14), 4137–4146. doi:10.1182/bloodadvances.2022007673
Goldman, M. J., Craft, B., Hastie, M., Repečka, K., McDade, F., Kamath, A., et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38 (6), 675–678. doi:10.1038/s41587-020-0546-8
Hammam, E., Ananda, G., Sinha, A., Scheidig-Benatar, C., Bohec, M., Preiser, P. R., et al. (2020). Discovery of a new predominant cytosine DNA modification that is linked to gene expression in malaria parasites. Nucleic Acids Res. 48 (1), 184–199. doi:10.1093/nar/gkz1093
Hauben, M., and Aronson, J. K. (2009). Defining 'signal' and its subtypes in pharmacovigilance based on a systematic review of previous definitions. Drug Saf. 32 (2), 99–110. doi:10.2165/00002018-200932020-00003
Hempel, D., Wieland, T., Solfrank, B., Grossmann, V., Steinhard, J., Frick, A., et al. (2020). Antitumor activity of larotrectinib in esophageal carcinoma with NTRK gene amplification. Oncol. 25 (6), e881–e886. doi:10.1634/theoncologist.2019-0641
Hong, D. S., DuBois, S. G., Kummar, S., Farago, A. F., Albert, C. M., Rohrberg, K. S., et al. (2020). Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet. Oncol. 21 (4), 531–540. doi:10.1016/S1470-2045(19)30856-3
Houtz, J., Liao, G.-Y., An, J. J., and Xu, B. (2021). Discrete TrkB-expressing neurons of the dorsomedial hypothalamus regulate feeding and thermogenesis. Proc. Natl. Acad. Sci. U. S. A. 118 (4), e2017218118. doi:10.1073/pnas.2017218118
Hu, X., Wang, Q., Tang, M., Barthel, F., Amin, S., Yoshihara, K., et al. (2018). TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 46 (D1), D1144–D1149. doi:10.1093/nar/gkx1018
Jain, N., Mittendorf, K. F., Holt, M., Lenoue-Newton, M., Maurer, I., Miller, C., et al. (2020). The My Cancer Genome clinical trial data model and trial curation workflow. J. Am. Med. Inf. Assoc. 27 (7), 1057–1066. doi:10.1093/jamia/ocaa066
Jain, N. M., Holt, M., Micheel, C., and Levy, M. (2021). Landscape analysis of breast cancer and acute myeloid leukemia trials using the my cancer genome clinical trial data model. JCO Clin. Cancer Inf. 5, 975–984. doi:10.1200/CCI.21.00082
Ji, A. L., Rubin, A. J., Thrane, K., Jiang, S., Reynolds, D. L., Meyers, R. M., et al. (2020). Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182 (2), 1661–1662. doi:10.1016/j.cell.2020.08.043
Ji, H., Li, K., Jiang, W., Li, J., Zhang, J.-A., and Zhu, X. (2021). MRVI1 and NTRK3 are potential tumor suppressor genes commonly inactivated by DNA methylation in cervical cancer. Front. Oncol. 11, 802068. doi:10.3389/fonc.2021.802068
Jiang, H. (2022). A novel ETV6-NTRK3 gene fusion in primary renal fibrosarcoma. Eur. Rev. Med. Pharmacol. Sci. 26 (13), 4705–4708. doi:10.26355/eurrev_202207_29195
Jiang, Q., Li, M., Li, H., and Chen, L. (2022). Entrectinib, a new multi-target inhibitor for cancer therapy. Biomed. Pharmacother. = Biomedecine Pharmacother. 150, 112974. doi:10.1016/j.biopha.2022.112974
Jing, Y., Liu, J., Ye, Y., Pan, L., Deng, H., Wang, Y., et al. (2020). Multi-omics prediction of immune-related adverse events during checkpoint immunotherapy. Nat. Commun. 11 (1), 4946. doi:10.1038/s41467-020-18742-9
Jing, Y., Zhang, Y., Wang, J., Li, K., Chen, X., Heng, J., et al. (2021). Association between sex and immune-related adverse events during immune checkpoint inhibitor therapy. J. Natl. Cancer Inst. 113 (10), 1396–1404. doi:10.1093/jnci/djab035
Joffe, E., Iasonos, A., and Younes, A. (2017). Clinical trials in the genomic era. J. Clin. Oncol. Official J. Am. Soc. Clin. Oncol. 35 (9), 1011–1017. doi:10.1200/JCO.2016.70.8891
Kazi, D. S., Bellows, B. K., Baron, S. J., Shen, C., Cohen, D. J., Spertus, J. A., et al. (2020). Cost-effectiveness of tafamidis therapy for transthyretin amyloid cardiomyopathy. Circulation 141 (15), 1214–1224. doi:10.1161/CIRCULATIONAHA.119.045093
Khotskaya, Y. B., Holla, V. R., Farago, A. F., Mills Shaw, K. R., Meric-Bernstam, F., and Hong, D. S. (2017). Targeting TRK family proteins in cancer. Pharmacol. Ther. 173, 58–66. doi:10.1016/j.pharmthera.2017.02.006
Kinnunen, M., Liu, X., Niemelä, E., Öhman, T., Gawriyski, L., Salokas, K., et al. (2023). The impact of ETV6-NTRK3 oncogenic gene fusions on molecular and signaling pathway alterations. Cancers 15 (17), 4246. doi:10.3390/cancers15174246
Knoepfler, P. S. (2015). From bench to FDA to bedside: US regulatory trends for new stem cell therapies. Adv. Drug Deliv. Rev. 82-83, 192–196. doi:10.1016/j.addr.2014.12.001
Kummar, S., Shen, L., Hong, D. S., McDermott, R., Keedy, V. L., Casanova, M., et al. (2023). Larotrectinib efficacy and safety in adult patients with tropomyosin receptor kinase fusion sarcomas. Cancer 129, 3772–3782. doi:10.1002/cncr.35036
Laetsch, T. W., DuBois, S. G., Mascarenhas, L., Turpin, B., Federman, N., Albert, C. M., et al. (2018). Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet. Oncol. 19 (5), 705–714. doi:10.1016/S1470-2045(18)30119-0
Le, X., Baik, C., Bauman, J., Gilbert, J., Brose, M. S., Grilley-Olson, J. E., et al. (2022). Larotrectinib treatment for patients with TRK fusion-positive salivary gland cancers. Oncol., oyac080. doi:10.1093/oncolo/oyac080
Leroy, F., Dauxois, J.-Y., Théophile, H., Haramburu, F., and Tubert-Bitter, P. (2014). Estimating time-to-onset of adverse drug reactions from spontaneous reporting databases. BMC Med. Res. Methodol. 14, 17. doi:10.1186/1471-2288-14-17
Light, J. E., Koyama, H., Minturn, J. E., Ho, R., Simpson, A. M., Iyer, R., et al. (2012). Clinical significance of NTRK family gene expression in neuroblastomas. Pediatr. Blood Cancer 59 (2), 226–232. doi:10.1002/pbc.23343
Liguori, V., Gaio, M., Zinzi, A., Cagnotta, C., Riccardi, C., Docimo, G., et al. (2023). The safety profiles of two first-generation NTRK inhibitors: analysis of individual case safety reports from the FDA adverse event reporting system (FAERS) database. Biomedicines 11 (9), 2538. doi:10.3390/biomedicines11092538
Liu, C.-J., Hu, F.-F., Xie, G.-Y., Miao, Y.-R., Li, X.-W., Zeng, Y., et al. (2023). GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Briefings Bioinforma. 24 (1), bbac558. doi:10.1093/bib/bbac558
Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., Lazar, A. J., Cherniack, A. D., et al. (2018). An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173 (2), 400–416.e11. doi:10.1016/j.cell.2018.02.052
Long, K., and Suresh, K. (2020). Pulmonary toxicity of systemic lung cancer therapy. Respirol. Carlt. Vic.) 25 (2), 72–79. doi:10.1111/resp.13915
Luo, Y., Kaz, A. M., Kanngurn, S., Welsch, P., Morris, S. M., Wang, J., et al. (2013). NTRK3 is a potential tumor suppressor gene commonly inactivated by epigenetic mechanisms in colorectal cancer. PLoS Genet. 9 (7), e1003552. doi:10.1371/journal.pgen.1003552
Ma, L., Wang, D.-D., Zhang, T.-Y., Yu, H., Wang, Y., Huang, S.-H., et al. (2011). Region-specific involvement of BDNF secretion and synthesis in conditioned taste aversion memory formation. J. Neurosci. 31 (6), 2079–2090. doi:10.1523/JNEUROSCI.5348-10.2011
Marcus, L., Donoghue, M., Aungst, S., Myers, C. E., Helms, W. S., Shen, G., et al. (2021). FDA approval summary: entrectinib for the treatment of NTRK gene fusion solid tumors. Clin. Cancer Res. 27 (4), 928–932. doi:10.1158/1078-0432.CCR-20-2771
Martin-Zanca, D., Hughes, S. H., and Barbacid, M. (1986). A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature 319 (6056), 743–748. doi:10.1038/319743a0
Mitelman, F., Johansson, B., and Mertens, F. (2007). The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7 (4), 233–245. doi:10.1038/nrc2091
Montastruc, J.-L., Sommet, A., Bagheri, H., and Lapeyre-Mestre, M. (2011). Benefits and strengths of the disproportionality analysis for identification of adverse drug reactions in a pharmacovigilance database. Br. J. Clin. Pharmacol. 72 (6), 905–908. doi:10.1111/j.1365-2125.2011.04037.x
Moore, A. R., Rosenberg, S. C., McCormick, F., and Malek, S. (2020). RAS-targeted therapies: is the undruggable drugged? Nat. Rev. Drug Discov. 19 (8), 533–552. doi:10.1038/s41573-020-0068-6
Naik, B. S., Shetty, N., and Maben, E. V. S. (2010). Drug-induced taste disorders. Eur. J. Intern. Med. 21 (3), 240–243. doi:10.1016/j.ejim.2010.01.017
Okamura, R., Boichard, A., Kato, S., Sicklick, J. K., Bazhenova, L., and Kurzrock, R. (2018). Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis. Oncol. 2018, 1–20. doi:10.1200/PO.18.00183
Otsu, Y., Kata, Y., Takayasu, H., Inoue, S., and Kaneko, T. (2022). Entrectinib-induced heart failure in a patient with metastatic lung adenocarcinoma: a case report. Cureus 14 (12), e32174. doi:10.7759/cureus.32174
Quintanilha, J. C. F., de Sousa, V. M., Visacri, M. B., Amaral, L. S., Santos, R. M. M., Zambrano, T., et al. (2017). Involvement of cytochrome P450 in cisplatin treatment: implications for toxicity. Cancer Chemother. Pharmacol. 80 (2), 223–233. doi:10.1007/s00280-017-3358-x
Roden, D. M., McLeod, H. L., Relling, M. V., Williams, M. S., Mensah, G. A., Peterson, J. F., et al. (2019). Pharmacogenomics. Lancet (London, Engl. 394 (10197), 521–532. doi:10.1016/S0140-6736(19)31276-0
Russo, M., Misale, S., Wei, G., Siravegna, G., Crisafulli, G., Lazzari, L., et al. (2016). Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 6 (1), 36–44. doi:10.1158/2159-8290.CD-15-0940
Sabaté, M., and Montané, E. (2023). Pharmacoepidemiology: an overview. J. Clin. Med. 12 (22), 7033. doi:10.3390/jcm12227033
Sakaeda, T., Tamon, A., Kadoyama, K., and Okuno, Y. (2013). Data mining of the public version of the FDA adverse event reporting system. Int. J. Med. Sci. 10 (7), 796–803. doi:10.7150/ijms.6048
Scott, L. J. (2019). Larotrectinib: first global approval. Drugs 79 (2), 201–206. doi:10.1007/s40265-018-1044-x
Shu, Y., Ding, Y., Liu, Y., Wu, P., He, X., and Zhang, Q. (2022). Post-marketing safety concerns with secukinumab: a disproportionality analysis of the FDA adverse event reporting system. Front. Pharmacol. 13, 862508. doi:10.3389/fphar.2022.862508
Somwar, R., Hofmann, N. E., Smith, B., Odintsov, I., Vojnic, M., Linkov, I., et al. (2020). NTRK kinase domain mutations in cancer variably impact sensitivity to type I and type II inhibitors. Commun. Biol. 3 (1), 776. doi:10.1038/s42003-020-01508-w
Spagnolo, P., Bonniaud, P., Rossi, G., Sverzellati, N., and Cottin, V. (2022). Drug-induced interstitial lung disease. Eur. Respir. J. 60 (4), 2102776. doi:10.1183/13993003.02776-2021
Szobota, S., Mathur, P. D., Siegel, S., Black, K., Saragovi, H. U., and Foster, A. C. (2019). BDNF, NT-3 and Trk receptor agonist monoclonal antibodies promote neuron survival, neurite extension, and synapse restoration in rat cochlea ex vivo models relevant for hidden hearing loss. PloS One 14 (10), e0224022. doi:10.1371/journal.pone.0224022
Tauszig-Delamasure, S., Yu, L.-Y., Cabrera, J. R., Bouzas-Rodriguez, J., Mermet-Bouvier, C., Guix, C., et al. (2007). The TrkC receptor induces apoptosis when the dependence receptor notion meets the neurotrophin paradigm. Proc. Natl. Acad. Sci. U. S. A. 104 (33), 13361–13366. doi:10.1073/pnas.0701243104
Tognon, C., Knezevich, S. R., Huntsman, D., Roskelley, C. D., Melnyk, N., Mathers, J. A., et al. (2002). Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2 (5), 367–376. doi:10.1016/s1535-6108(02)00180-0
Tsai, C.-T., Hsieh, C.-S., Chang, S.-N., Chuang, E. Y., Ueng, K.-C., Tsai, C.-F., et al. (2016). Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat. Commun. 7, 10190. doi:10.1038/ncomms10190
Vaishnavi, A., Le, A. T., and Doebele, R. C. (2015). TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 5 (1), 25–34. doi:10.1158/2159-8290.CD-14-0765
Valsesia, A., Stevenson, B. J., Waterworth, D., Mooser, V., Vollenweider, P., Waeber, G., et al. (2012). Identification and validation of copy number variants using SNP genotyping arrays from a large clinical cohort. BMC Genomics 13, 241. doi:10.1186/1471-2164-13-241
Visacri, M. B., Duarte, N. C., Lima, T. d.M., de Souza, R. N., Cobaxo, T. S., Teixeira, J. C., et al. (2022). Adverse reactions and adherence to capecitabine: a prospective study in patients with gastrointestinal cancer. J. Oncol. Pharm. Pract. 28 (2), 326–336. doi:10.1177/1078155221989420
Vivian, J., Rao, A. A., Nothaft, F. A., Ketchum, C., Armstrong, J., Novak, A., et al. (2017). Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35 (4), 314–316. doi:10.1038/nbt.3772
Vogel, A., Segatto, O., Stenzinger, A., and Saborowski, A. (2023). FGFR2 inhibition in cholangiocarcinoma. Annu. Rev. Med. 74, 293–306. doi:10.1146/annurev-med-042921-024707
Wahida, A., Buschhorn, L., Fröhling, S., Jost, P. J., Schneeweiss, A., Lichter, P., et al. (2023). The coming decade in precision oncology: six riddles. Nat. Rev. Cancer 23 (1), 43–54. doi:10.1038/s41568-022-00529-3
Waliany, S., Zhu, H., Wakelee, H., Padda, S. K., Das, M., Ramchandran, K., et al. (2021). Pharmacovigilance analysis of cardiac toxicities associated with targeted therapies for metastatic NSCLC. J. Thorac. Oncol. 16 (12), 2029–2039. doi:10.1016/j.jtho.2021.07.030
Wang, D., Li, D., Qin, G., Zhang, W., Ouyang, J., Zhang, M., et al. (2015). The structural characterization of tumor fusion genes and proteins. Comput. Math. Methods Med. 2015, 912742. doi:10.1155/2015/912742
Wang, Y., Sparidans, R. W., Li, W., Lebre, M. C., Beijnen, J. H., and Schinkel, A. H. (2020). OATP1A/1B, CYP3A, ABCB1, and ABCG2 limit oral availability of the NTRK inhibitor larotrectinib, while ABCB1 and ABCG2 also restrict its brain accumulation. Br. J. Pharmacol. 177 (13), 3060–3074. doi:10.1111/bph.15034
Wu, Y., Wei, M., and Zhang, J. (2023). A real-world pharmacovigilance analysis of FDA adverse event reporting system database for upadacitinib. Front. Pharmacol. 14, 1200254. doi:10.3389/fphar.2023.1200254
Yamada, Y., Toyota, M., Hirokawa, Y., Suzuki, H., Takagi, A., Matsuzaki, T., et al. (2004). Identification of differentially methylated CpG islands in prostate cancer. Int. J. Cancer 112 (5), 840–845. doi:10.1002/ijc.20335
Yi, Q., Peng, J., Xu, Z., Liang, Q., Cai, Y., Peng, B., et al. (2022). Spectrum of BRAF aberrations and its potential clinical implications: insights from integrative pan-cancer analysis. Front. Bioeng. Biotechnol. 10, 806851. doi:10.3389/fbioe.2022.806851
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics a J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Yu, Z., Wang, H., Song, Q., Huang, J., Xu, J., Su, J., et al. (2021). Prognostic value and characterization of NTRK1 variation by fluorescence in situ hybridization in esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 147 (10), 3113–3121. doi:10.1007/s00432-021-03578-7
Zhao, S., Ye, Z., and Stanton, R. (2020). Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA (New York, N.Y.) 26 (8), 903–909. doi:10.1261/rna.074922.120
Zhou, C., Peng, S., Lin, A., Jiang, A., Peng, Y., Gu, T., et al. (2023). Psychiatric disorders associated with immune checkpoint inhibitors: a pharmacovigilance analysis of the FDA Adverse Event Reporting System (FAERS) database. EClinicalMedicine 59, 101967. doi:10.1016/j.eclinm.2023.101967
Zhou, J., Lemos, B., Dopman, E. B., and Hartl, D. L. (2011). Copy-number variation: the balance between gene dosage and expression in Drosophila melanogaster. Genome Biol. Evol. 3, 1014–1024. doi:10.1093/gbe/evr023
Keywords: NTRK, gene fusion, entrectinib, larotrectinib, FAERS, adverse drug event, pharmacovigilance
Citation: Cui Z, Zhai Z, Xie D, Wang L, Cheng F, Lou S, Zou F, Pan R, Chang S, Yao H, She J, Zhang Y and Yang X (2024) From genomic spectrum of NTRK genes to adverse effects of its inhibitors, a comprehensive genome-based and real-world pharmacovigilance analysis. Front. Pharmacol. 15:1329409. doi: 10.3389/fphar.2024.1329409
Received: 30 October 2023; Accepted: 15 January 2024;
Published: 31 January 2024.
Edited by:
Patricia Moriel, State University of Campinas, BrazilReviewed by:
Carolina Dagli Hernandez, Independent Researcher, Sao Paulo, BrazilCopyright © 2024 Cui, Zhai, Xie, Wang, Cheng, Lou, Zou, Pan, Chang, Yao, She, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyuan Yang, eGlueXVhbnlhbmcyMDIyQDE2My5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.