
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 08 January 2024
Sec. Pharmacogenetics and Pharmacogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1334440
 Nathan D. Seligson1,2,3
Nathan D. Seligson1,2,3 Xunjie Zhang1
Xunjie Zhang1 Mark C. Zemanek1
Mark C. Zemanek1 Jasmine A. Johnson1
Jasmine A. Johnson1 Zachary VanGundy1
Zachary VanGundy1 Danxin Wang4
Danxin Wang4 Mitch A. Phelps3,5
Mitch A. Phelps3,5 Julianna Roddy1,2
Julianna Roddy1,2 Craig C. Hofmeister6
Craig C. Hofmeister6 Junan Li3,7*
Junan Li3,7* Ming J. Poi1,2,3*
Ming J. Poi1,2,3*Introduction: Polymorphisms in genes responsible for the metabolism and transport of tacrolimus have been demonstrated to influence clinical outcomes for patients following allogeneic hematologic stem cell transplant (allo-HSCT). However, the clinical impact of germline polymorphisms specifically for oral formulations of tacrolimus is not fully described.
Methods: To investigate the clinical impact of genetic polymorphisms in CYP3A4, CYP3A5, and ABCB1 on oral tacrolimus pharmacokinetics and clinical outcomes, we prospectively enrolled 103 adult patients receiving oral tacrolimus for the prevention of graft-versus-host disease (GVHD) following allo-HSCT. Patients were followed in the inpatient and outpatient phase of care for the first 100 days of tacrolimus therapy. Patients were genotyped for CYP3A5 *3 (rs776746), CYP3A4 *1B (rs2740574), ABCB1 exon 12 (rs1128503), ABCB1 exon 21 (rs2032582), ABCB1 exon 26 (rs1045642).
Results: Expression of CYP3A5 *1 was highly correlated with tacrolimus pharmacokinetics in the inpatient phase of care (p < 0.001) and throughout the entirety of the study period (p < 0.001). Additionally, Expression of CYP3A5 *1 was associated with decreased risk of developing AKI as an inpatient (p = 0.06). Variants in ABCB1 were not associated with tacrolimus pharmacokinetics in this study. We were unable to discern an independent effect of CYP3A4 *1B or *22 in this population.
Conclusion: Expression of CYP3A5 *1 is highly influential on the pharmacokinetics and clinical outcomes for patients receiving oral tacrolimus as GVHD prophylaxis following allo-HSCT.
Allogeneic hematopoietic stem cell transplantation (allo-HSCT), a mainstay treatment for a variety of hematologic malignancies (Sureda et al., 2015), is associated with a significant risk of graft-versus-host disease (GVHD) (Ziakas et al., 2014). Therapeutic immune suppression, often utilizing tacrolimus, is necessary to reduce the risk of GVHD (Hiraoka et al., 2001). Significant inter-patient variability in the pharmacokinetics of tacrolimus, along with a narrow therapeutic window, makes dosing of tacrolimus difficult. Sub-therapeutic levels of tacrolimus are associated with an increased risk of GVHD, while supra-therapeutic levels are associated with severe toxicity (Prezepiorka et al., 1999; Jacobson et al., 2001; Lee et al., 2002; Musuamba et al., 2009; Levine et al., 2010). Clinically, therapeutic drug monitoring of tacrolimus has been employed to personalize tacrolimus doses for patients and account for the known inter-patient pharmacokinetic and pharmacodynamic variability of tacrolimus (Elens et al., 2007; Thervet et al., 2008; Brunet et al., 2019).
Out of many factors associated with inter-patient pharmacokinetic and pharmacodynamics variabilities of tacrolimus, single nucleotide polymorphisms (SNPs) in cytochrome (CYP) 3A4 and 3A5 enzymes have been associated with clinical outcomes associated with tacrolimus (Kuehl et al., 2001; Thervet et al., 2010; Elens et al., 2011; Wang et al., 2011). The CYP3A5 *3 allele, the most frequently occurring CYP3A5 allele in populations of European ancestry, references an intronic SNP rs776746 that creates a cryptic splice site and an aberrant splice isoform with partial intron insertion, resulting in a premature stop codon and eliminating the expression of the functional CYP3A5 enzyme (Kuehl et al., 2001; Thervet et al., 2010). While the CYP3A5 *3 allele is the most prevalent allele, individuals with the wild type allele (designated *1) are considered CYP3A5 *1 expressers (*1/*1 or *1/*3). CYP3A5 *1 expressers required increased tacrolimus dose to maintain therapeutic levels of the drug compared to patients with the CYP3A5 *3/*3 genotype (Thervet et al., 2010; Khaled et al., 2016; Hamadeh et al., 2019). CYP3A4 also plays a critical role in tacrolimus metabolism (Elens et al., 2011; Wang et al., 2011). Two specific CYP3A4 alleles have been linked to variable tacrolimus metabolism: CYP3A4 *22, which refers to a polymorphism in intron 6 of CYP3A4 that reduces expression of the CYP3A4 mRNA and protein (Wang et al., 2011; Wang and Sadee, 2016), and CYP3A4 *1B which refers to a SNP on the promoter region of CYP3A4 whose impact remains to be further elucidated (Garcia-Martin et al., 2002; Spurdle et al., 2002; Gervasini et al., 2012; Zochowska et al., 2012). Adenosine triphosphate-binding cassette B1 (ABCB1) is a cell membrane transporter responsible for the active transport of tacrolimus (Op den Buijish et al., 2007). It has been proposed that ABCB1 plays a role in the bioavailability of tacrolimus in the setting of oral administration; however, little is known about the specific impacts of ABCB1 genomic variants on the pharmacokinetic and pharmacodynamic properties of tacrolimus in transplant patients (Mourad et al., 2006; Hawwa et al., 2009; Herrlinger et al., 2011; Kim et al., 2012; Tapiradamaz et al., 2014; Li et al., 2015; Debette-Gratien et al., 2016).
In the 2015 guidelines for the use of pharmacogenomic data to guide tacrolimus dosing, the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommended that patients expressing CYP3A5 *1 should receive doses 1.5-2-fold higher compared to standard dosing (Birdwell et al., 2015). While this recommendation is made for kidney, heart, lung, and allo-HSCT patients, it is notable that such recommendation in the setting of allo-HSCT is made based on evidence provided for intravenous tacrolimus (Woodahl et al., 2008; Yanagimachi et al., 2010; Onizuka et al., 2011; Yanagisawa et al., 2011; Iwamoto et al., 2015; Khaked et al., 2016; Hamadeh et al., 2019; Suztsugu et al., 2019). Very little data is available for patient receiving allo-HSCT alongside oral tacrolimus (Zhu et al., 2020). As such, clinicians have to extrapolate the findings of intravenous tacrolimus studies when considering this clinical problem. In addition to hepatic expression, CYPs in the 3A family are highly expressed in the intestine and are detectable in some brain regions (McFadyen et al., 1998; Paine et al., 2006). Given the potential for intestinal metabolism, as well as first-pass hepatic metabolism prior to reaching systemic circulation, the effect of genomic variants on tacrolimus administered orally poses an additional level of complexity in comparison to intravenous tacrolimus. While significant data exists demonstrating the role of pharmacogenomic variants on oral tacrolimus outside of allo-HSCT (Goto et al., 2004; Kato et al., 2018; Pasternak et al., 2019; Pasternak et al., 2022), this data may not fully apply in the setting of allo-HSCT given the differences in tacrolimus dosing and a desire for less aggressive early tacrolimus levels to confer a graft-versus-leukemia effect (Imado et al., 2004). It is therefore possible that existing findings using intravenous formulations of tacrolimus in the setting of allo-HSCT, and those of oral tacrolimus outside of allo-HSCT, may not fully reflect the pharmacogenomic implications of tacrolimus metabolism specific to the oral route in an allo-HSCT population. In this study, we present a prospective analysis of 103 patients who received oral tacrolimus for the prevention of GVHD following allo-HSCT.
A cohort of 103 adult patients receiving oral tacrolimus for the prevention of GVHD following allo-HSCT at The Ohio State University Wexner Medical Center (OSUMC) were prospectively enrolled into this study following approval from the Cancer Institutional Research Board’s guidance (IRB# 2012C0021) from 06/2012 until 07/2014. Inclusion criteria included age great than 18 years, patient to receive their first allo-HSCT during the study period, and absence of renal or hepatic dysfunction at baseline. Exclusion criteria included HIV positivity, presence of any medical conditions that would interfere with the patient’s ability to sign the informed consent, hypersensitivity to any component of tacrolimus, clinical history of any solid organ transplantation, previous chronic use of tacrolimus, or pregnancy.
Following patient consent, but prior to transplantation, whole blood samples were obtained to determine the patient’s CYP3A4, CYP3A5, ABCB1 genotypes. Clinical data was collected for patients for up to the first 100 days post-transplant or until the patient was lost to follow-up. As a standard of care in HSCT patients treated at OSUMC, tacrolimus therapeutic drug monitoring was employed using liquid chromatography/tandem mass spectrometry at least three times per week to measure trough drug concentration until goal steady state concentrations were reached. Standard of care at our institution includes starting immediate release oral tacrolimus 2 days prior to transplantation at a dose of 0.03 mg/kg. Twice daily dosing was considered standard; however, clinicians could elect to use daily dosing if necessary for the patient. Tacrolimus levels were normalized by both tacrolimus doses received as well as patient’s actual body weight as described in previous studies as follows: Normalized Tacrolimus level (ng/dL/mg/kg) = Tacrolimus level (ng/dL)/Total daily dose of Tacrolimus (mg)/Actual body weight (kg) (Sy et al., 2013; Cusinato et al., 2014; Ito et al., 2017).
To assess the proportion of patients who were within therapeutic range for each group during this study, we binned tacrolimus levels every 5 days starting from the day of transplant. We collected records of held tacrolimus doses due to supratherapeutic levels. We also collected the number of tacrolimus dose was changes during the inpatient stay. Concerning the number of dose changes a patient experienced, we considered held doses as a change in the dose. Due to the imbalance in the numbers of patients with different genotypes, we normalized both total dose changes and total doses held by the number of patients with a specific genotype. We also assessed the percentage of total days of therapy each patient had to hold at least one dose of tacrolimus. In addition, dose changes for each patient during the inpatient stay were also assessed as normalized by each inpatient length of stay (LOS).
Genomic DNA was extracted from patients’ blood samples using DNAeasy Blood Mini kit (QIAGEN, Germantown, MD, United States) following the manufacturer’s instructions. RNase A was included in the purification to remove potential RNA contamination in DNA products. The genotypes of CYP3A5*3 (A- > G, rs776746), ABCB1 exon 12 (C1246- > T, rs1128503), ABCB1 exon 21 (G2677- > A/T, rs2032582), and ABCB1 exon 26 (C3435- > T, rs1045642) were determined using polymerase chain reaction (PCR) - restriction fragment length polymorphism (RFLP) assays as previously described (Supplementary Table S1) (Fukuen et al., 2002; Owen et al., 2005; Dong et al., 2011). Patients were considered expressers of CYP3A5 *1 as long as they were not homozygous for the *3 allele (*1/*1 or *1/*3). Genotyping of CYP3A4*1B (A- > G, rs2740574) was conducted on a QuantStudioTM 7 Flex system (Thermo Fisher Scientific, Wakthan, MA, United States) using a Taqman® pre-validated genotyping kit, C__1837671_50 (Thermo Fisher Scientific) (Wang et al., 2011). The genotypes of CYP3A4*22 (C- > T, rs35599367) were determined using an allele-specific SYBR® green real-time PCR assay as previously described (Papp et al., 2003). All genotyping experiments were conducted in duplicate.
Pharmacokinetic endpoints were based on tacrolimus levels normalized to both the total daily dose of tacrolimus and the patient’s actual body weight. The goal range of serum tacrolimus levels for this study was defined as between 8 ng/mL and 12 ng/mL for the first 60 days after transplant as per our institution’s standard. After 60 days post-transplant, goal trough levels were determined by the preference of the treating physician. Analysis of time to therapeutic range was a simple comparison between the day number when the first therapeutic trough level was achieved as a continuous variable. Adverse event rates, including both acute kidney injury (AKI), tacrolimus-related neurotoxicity, and GVHD were collected. AKI was defined using KDIGO criteria (Khwaja, 2012). Neurotoxicity was defined by documentation of tremor or confusion attributed by the clinical team to be related to tacrolimus. GVHD was defined by documentation by the clinical team. For this study, only acute GVHD was included in our analysis. Patients without documentation of neurotoxicity or GVHD were assumed to not have experienced these adverse events.
All data analyses were conducted using R3.3.2 (The R Foundation; https://www.r-project.org). Patients’ characteristics were analyzed using descriptive statistics. Briefly, categorical data were presented as count and frequency; continuous data were summarized as mean and standard deviation (SD) except for age and The European Group for Blood and Marrow Transplantation (EBMT) risk score, which were summarized as median and range. Differences in categorical variables were analyzed using χ2 tests or Fisher’s exact tests. For continuous variables, Student’s t tests and one-way ANOVA were used to compare differences between two groups and among multiple groups where appropriate. For each SNP, the consistency between its distribution and the hyphenated Hardy-Weinberg principle was analyzed using χ2 tests. Potential statistical associations between different SNPs were also analyzed using χ2 tests using the R package genetics. Statistical tests were two-sided, and p values < 0.05 were considered statistically significant.
In the present study, we assessed the impacts of various SNPs of CYP3A4, CYP3A5, and ABCB1 on clinical outcomes of a cohort of 103 patients who were given oral tacrolimus for the prevention of GVHD after allo-HSCT. Full demographic features of these patients listed in Table 1. Median follow up was 98 days (interquartile range 92.5–100 days). In this cohort, tacrolimus levels were not associated with the use of voriconazole (p = 0.59) or steroids (p = 0.36).
Each SNP tested in this study was assessed for Hardy Weinberg Equilibrium. While the distribution of CYP3A5 *3, CYP3A4 *22, ABCB1 exons 21 and 26 followed the Hardy Weinberg Equilibrium, CYP3A4 *1B and ABCB1 exon 12 deviated significantly (Table 2). In testing for linkage disequilibrium (Supplementary Table S2), an association was found between CYP3A5 *1 and the CYP3A4 *1B allele (p < 0.001; D’ = 0.48; R2 = 0.18), ABCB1 exon 12 and ABCB1 exon 21 (p < 0.001; D’ = 0.66; R2 = 0.22), ABCB1 exon 12 and ABCB1 exon 26 (p < 0.001; D’ = 0.45; R2 = 0.20), and ABCB1 exon 21 and ABCB1 exon 26 (p < 0.001; D’ = 0.71; R2 = 0.25). This is consistent with previous findings that CYP3A4 *1B may be in linkage disequilibrium with CYP3A5 *1 (Zeigler-Johnson et al., 2004; Miao et al., 2009). Additionally, three ABCB1 SNPs were associated with each other in this cohort of patients (ABCB1: exon 12 and exon 21, p < 0.001; exon 12 and exon 26, p < 0.001; exon 21 and exon 26, p < 0.001), while ABCB1 exon 21 was also associated with CYP3A5 *1 and CYP3A4 *1B (p = 0.001 and p = 0.045, respectively). We did not exclude variants from this study simply due to its deviation from Hardy Weinberg Equilibrium. Potential reasons for deviation from Hardy Weinberg Equilibrium include the relatively small sample size of this study, small variant allele frequency, and demographic homogeneity of this cohort.
To assess the effect of our tested genomic variants with tacrolimus levels across the full study period, we calculated the median normalized tacrolimus level for each patient for the first 100 days of therapy. Univariate analyses demonstrated that median normalized tacrolimus level for each individual patient over the entire study period was statistically associated with each of CYP3A4 *1B, CYP3A4 *22, and CYP3A5 *1 expression (p = 0.004, p = 0.03, p = 0.0009, respectively; Table 3). Multivariate analyses confirmed the association between normalized tacrolimus levels and CYP3A5 *1 expression alone (Table 3; Figure 1). Based on these results, we focused further study on CYP3A5 *1 expression.
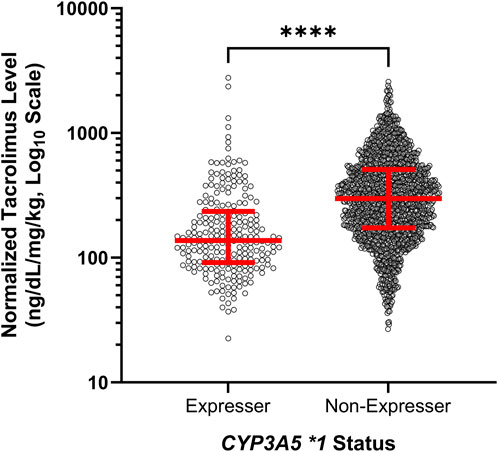
FIGURE 1. Expressers of CYP3A5 *1 exhibited lower normalized tacrolimus concentration across the first 100 days of therapy. Red lines represent the median and interquartile range. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
We then analyzed both weight normalized tacrolimus doses and tacrolimus levels at different time points during the inpatient stay of patients, including the first drawn level, day 6, day 10, and at discharge (±2 days). This analysis revealed a clinically important trend in dose requirements for CYP3A5 *1 expressers. While the weight normalized tacrolimus dose at the time of the first level did not differ significantly based on CYP3A5 genotype (p = 0.26; Figure 2A), the tacrolimus level at first assessment varied significantly (p < 0.001; Figure 2B). Over the course of the inpatient stay, CYP3A5 *1 expressers received increasing tacrolimus doses compared to non-expressers (day 6: p = 0.17; day 10: p = 0.008; Discharge: p < 0.001) while the discrepancy in tacrolimus levels slowly diminished (day 6: p < 0.001; day 10: p = 0.32. Discharge: p = 0.37). CYP3A5 genotype was clearly associated with changes in total daily dose of tacrolimus between the start of therapy and discharge, with the CYP3A5 *3/*3 genotype associated with a decreased dose from drug initiation to discharge (p = 0.0002) (Figure 2C). The CPIC guidelines for CYP3A5 genotype and tacrolimus dosing suggest a 1.5-2 times higher dose for patients who express CYP3A5 *1 (Birdwell et al., 2015). At time of discharge, with statistically equivalent tacrolimus drug concentrations, patients who express CYP3A5 *1 were stabilized on a median normalized tacrolimus dose 1.98 times higher than those who did not express CYP3A5 *1.
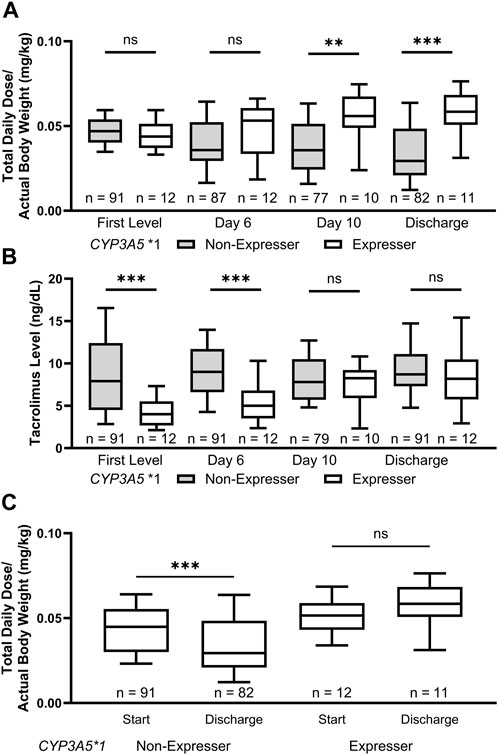
FIGURE 2. CYP3A5 *1 expressers experienced lower initial tacrolimus levels and required increased doses of tacrolimus at discharge. (A) The weight adjusted tacrolimus dose increased during the inpatient stay for CYP3A5 *1 expressers while the opposite was true for CYP3A5 *3/*3 patients. (B) There was a significant difference in the tacrolimus levels achieved in patients grouped by CYP3A5 genotype up until day 10 of therapy. (C) Patients with a CYP3A5 *3/*3 genotype were discharged on lower weight adjusted doses of tacrolimus to their starting dose. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
In addition, CYP3A5 *1 expressers spent significantly more time at subtherapeutic levels of tacrolimus compared to non-expressers throughout tacrolimus treatment (Figure 3). In the first 5 days after transplant, no CYP3A5 *1 expressers (0/12, 0%) were at or above the therapeutic range. In comparison, 41.8% of CYP3A5 *1 non-expressers were at or above the therapeutic range (38/91). Due to the significant impact of CYP3A5 *1 expression on the pharmacokinetics of tacrolimus, expressers of CYP3A5 *1 required more time to reach therapeutic range compared to non-expressers (13.9 ± 4.7 days vs. 9.9 ± 8.2 days; p = 0.003). Given this therapeutic complication, it is unsurprising that CYP3A5 *1 expressers experienced longer inpatient stays (27.7 ± 24.1 vs. 20.0 ± 5.7 days, p = 0.02). CYP3A5 *1 expressers also required fewer doses to be held (percentage of inpatient days held: 8.6% ± 12.6% vs. 30.8% ± 30.3%; p < 0.001) and, after normalization with the inpatient length of stay, required a lower number of dose changes during the inpatient stay compared to non-expressers (1.4 ± 0. 6 vs. 2.1 ± 0.8 changes/week; p = 0.008).
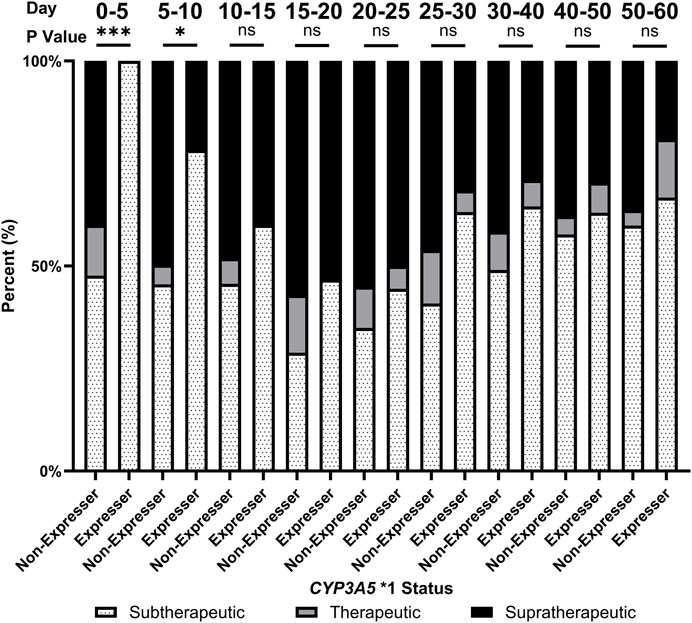
FIGURE 3. Proportion of patients achieving therapeutic levels of tacrolimus over the first 60 days of tacrolimus therapy by CYP3A5 status. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
To assess the effect of the tested genomic variants on clinical outcomes, we focused on the development of AKI and neurotoxicity in patients during the inpatient and outpatient stays. Expressers of CYP3A5 *1 showed a clinical trend towards lower rates of AKI compared to non-expressers during the inpatient stay (7.7% vs. 25%; p = 0.06); however, rates of AKI over the entire period of this study does not differ by CYP3A5 genotype (100% vs. 91.2%; p = 0.28). CYP3A5 *1 expressers exhibited statistically similar rates of neurotoxicity (33.3% vs. 20.1%; p = 0.33) and GVHD (50.0% vs. 49.5%; p = 0.97) as non-expressers.
Allo-HSCT is an integral therapy modality for the treatment of a number of hematologic malignancies requiring accurate and precise use of immune suppressing agents to prevent graft mediated harm to the patient. Multiple recent studies have confirmed the effect of CYP3A5, CYP3A4, and ABCB1 in patients receiving tacrolimus via intravenous infusion in the setting of allo-HSCT (Hamadeh et al., 2019; Yoshikawa et al., 2021). However, very little data is available in the setting of oral tacrolimus. In this study, we prospectively enrolled 103 patients who received oral tacrolimus for GVHD prophylaxis following an allo-HSCT, as is standard of care at our institution.
Previous studies have demonstrated that CYP3A5 *1 expressers and CYP3A4* 1B SNPs are associated with the pharmacokinetics and pharmacodynamic properties of tacrolimus (Semiz et al., 2011; Hamadeh et al., 2019; Zhu et al., 2020; Yoshikawa et al., 2021; Liu et al., 2022). Due to the potential linkage between the CYP3A5 *1 expressers and CYP3A4* 1B SNP, it is difficult to fully characterize the effect of each individual allele (Sinues et al., 2007; Estrela et al., 2008). In our study, we determined that CYP3A5 *1 expression was associated with an increased metabolism of tacrolimus leading to an increased dose requirement and time to reach therapeutic goals, while we were not able to identify an independent role for CYP3A4* 1B in tacrolimus metabolism (Musuamba et al., 2009; Liu et al., 2022). While most CYP3A5 *1 expressers required doses increases following the first tacrolimus level to reach therapeutic goals, overall, these patients received less dose adjustments than those with the CYP3A5 *3/*3 genotype. This observation could at least partially be attributed to clinicians’ reaction to high versus low tacrolimus levels. The clinician’s reaction to subtherapeutic tacrolimus levels, to an extent, dictates how much time is required to bring CYP3A5 *1 expressers to therapeutic tacrolimus levels. Proactive genotyping of CYP3A5 may allow clinicians to feel more confident in increasing the tacrolimus dose for carriers.
Genomic variations in ABCB1 as well as the CYP3A4 *22 SNP were not associated with tacrolimus pharmacokinetics. While ABCB1 is not directly implicated in the metabolism of tacrolimus, it plays a vital role in the active transport of tacrolimus (Brunet et al., 2019; Hamadeh et al., 2019). Since our study included only patients who were initiated on oral tacrolimus, this gene was anticipated to be of great importance in tacrolimus absorption. However, there is no evidence from our study in support of the association between the selected three ABCB1 SNPs and any clinically important endpoint as were found in previous studies (Hamadeh et al., 2019). We did observe a linkage association between SNPs in ABCB1 exon 12 and both exon 21 and exon 26, which has been previously reported, therefore complicating our analysis (Allabi et al., 2005; Qiu et al., 2012). Further study is needed to better understand the clinical actionability of ABCB1 in this setting.
AKI is one of the most common adverse events for patients prescribed tacrolimus and is associated with high drug concentrations. Here, CY3A5 *1 expressers showed a clinical trend toward deceased rates of AKI during the inpatient stay, which dissipated over the course of the study. Expressers of CYP3A5 *1 spend significantly more time at subtherapeutic tacrolimus levels during the inpatient stay; however, once these patients reached therapeutic goals, the difference in AKI rates among patients with different genotypes disappeared. This finding is of significance clinically as proactive genotyping may help to identify patients who are at higher risk of early AKI development (Seligson et al., 2023). Patients who are not expressers of CY3A5 *1 may require closer renal monitoring during the inpatient stay while expressers of CY3A5 *1 should be followed closely in the outpatient setting as they reach therapeutic goals.
Finally, it is important to note key limitations to this study. First, this was a single center study that, despite strong enrollment, resulted in a relatively small number of patients with variant SNPs. For example, our study cohort did not have many patients who expressed CYP3A5 *1 or the CYP3A4 *1B allele, which compromised our effort to separate the potential impacts of CYP3A4 *1B. Our study also did not include SNPs in CYP3A5, notably *6 and *7, among other relevant SNPs in CYP3A4 and ABCB1 noted by the AMP and CPIC guidelines (Birdwell et al., 2015; Pratt et al., 2023). These SNPs were not well associated with tacrolimus metabolism when this study was designed. Further study should be sure to include these relevant SNPs in their analysis. From a clinical perspective, our study did not include any exclusions or measures of adherence to tacrolimus dosing after discharge. Data following discharge should then be considered as real-world data without a guarantee of adherence. Lastly, our study was reliant on chart review for many of the clinical variables assessed in this study, thus this study was not immune from errors in charting of the clinic course of a patient.
In conclusion, we provide evidence to support the role of CYP3A5 *1 expression in the pharmacokinetics of oral tacrolimus in a large cohort of adult patients with allo-HSCT and reflect other recent findings in similar patient populations (Yamashita et al., 2016; Zhu et al., 2020). Patients expressing CYP3A5 *1 required higher doses of tacrolimus to maintain therapeutic concentrations, suggesting that pharmacogenomics testing of CYP3A5 may be used to assist in the dosing of oral tacrolimus in this clinical setting. Prospective studies in large cohorts of patients at multiple clinical sites are warranted to verify the findings presented here.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
The studies involving humans were approved by The Ohio State University Cancer Institutional Research Board’s guidance (IRB# 2012C0021). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
NS: Investigation, Methodology, Writing–review and editing, Writing–original draft. XZ: Investigation, Writing–review and editing. MZ: Investigation, Writing–review and editing. JJ: Investigation, Writing–review and editing. ZV: Investigation, Writing–review and editing. DW: Methodology, Validation, Writing–review and editing. MP: Methodology, Resources, Writing–review and editing. JR: Methodology, Resources, Writing–review and editing. CH: Methodology, Resources, Writing–review and editing. JL: Formal Analysis, Investigation, Methodology, Supervision, Validation, Writing–review and editing. MP: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The startup fund to MP from The Ohio State University College of Pharmacy. This study was partly supported by a research grant from NIH, NIGMS/R35140845 (DW) and a startup fund from The Ohio State University College of Pharmacy (MP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1334440/full#supplementary-material
Allabi, A. C., Horsmans, Y., Issaoui, B., and Gala, J. L. (2005). Single nucleotide polymorphisms of ABCB1 (MDR1) gene and distinct haplotype profile in a West Black African population. Eur. J. Clin. Pharmacol. 61 (2), 97–102. doi:10.1007/s00228-004-0879-0
Birdwell, K. A., Decker, B., Barbarino, J. M., Peterson, J. F., Stein, C. M., Sadee, W., et al. (2015). Clinical pharmacogenetics implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharmacol. Ther. 98 (1), 19–24. doi:10.1002/cpt.113
Brunet, M., van Gelder, T., Åsberg, A., Haufroid, V., Hesselink, D. A., Langman, L., et al. (2019). Therapeutic drug monitoring of tacrolimus-personalized Therapy,Second consensus report. Ther. Drug Monit. 41 (3), 261–307. doi:10.1097/FTD.0000000000000640
Cusinato, D. A., Lacchini, R., Romao, E. A., Moysés-Neto, M., and Coelho, E. B. (2014). Relationship of CYP3A5 genotype and ABCB1 diplotype to tacrolimus disposition in Brazilian kidney transplant patients. Br. J. Clin. Pharmacol. 78 (2), 364–372. doi:10.1111/bcp.12345
Debette-Gratien, M., Woillard, J. B., Picard, N., Sebagh, M., Loustaud-Ratti, V., Sautereau, D., et al. (2016). Influence of donor and recipient CYP3A4, CYP3A5, and ABCB1 genotypes on clinical outcomes and nephrotoxicity in liver transplant recipients. Transplantation 100 (10), 2129–2137. doi:10.1097/TP.0000000000001394
Dong, L., Mao, M., Luo, R., Tong, Y., and Yu, D. (2011). Common ABCB1 polymorphisms associated with susceptibility to infantile spasms in the Chinese Han population. Genet. Mol. Res. 10 (4), 2569–2577. doi:10.4238/2001.October.19.3
Elens, L., Bouamar, R., Hesselink, D. A., Haufroid, V., van der Heiden, I. P., van Gelder, T., et al. (2011). A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin. Chem. 57 (11), 1574–1583. doi:10.1373/clinchem.2011.165613
Elens, L., Capron, A., van Kerckhove, V., Lerut, J., Mourad, M., Lison, D., et al. (2007). 1199G>A and 2677G>T/A polymorphisms of ABCB1 independently affect tacrolimus concentration in hepatic tissue after liver transplantation. Pharmacogenet Genomics 17 (10), 873–883. doi:10.1097/FPC.0b013e3282e9a533
Estrela, R. C., Ribeiro, F. S., Carvalho, R. S., Gregório, S. P., Dias-Neto, E., Struchiner, C. J., et al. (2008). Distribution of ABCB1 polymorphisms among Brazilians: impact of population admixture. Pharmacogenomics 9 (3), 267–276. doi:10.2217/14622416.9.3.267
Fukuen, S., Fukuda, T., Maune, H., Ikenaga, Y., Yamamoto, I., Inaba, T., et al. (2002). Novel detection assay by PCR-RFLP and frequency of the CYP3A5 SNPs, CYP3A5*3 and *6, in a Japanese population. Pharmacogenetics 12 (4), 331–334. doi:10.1097/00008571-200206000-00009
García-Martín, E., Martínez, C., Pizarro, R. M., García-Gamito, F. J., Gullsten, H., Raunio, H., et al. (2002). CYP3A4 variant alleles in white individuals with low CYP3A4 enzyme activity. Clin. Pharmacol. Ther. 71 (3), 196–204. doi:10.1067/mcp.2002.121371
Gervasini, G., Garcia, M., Macias, R. M., Cubero, J. J., Caravaca, F., and Benitez, J. (2012). Impact of genetic polymorphisms on tacrolimus pharmacokinetics and the clinical outcome of renal transplantation. Transpl. Int. 25 (4), 471–480. doi:10.1111/j.1432-2277.2012.01446.x
Goto, M., Masuda, S., Kiuchi, T., Ogura, Y., Oike, F., Okuda, M., et al. (2004). CYP3A5*1-carrying graft liver reduces the concentration/oral dose ratio of tacrolimus in recipients of living-donor liver transplantation. Pharmacogenetics 14 (7), 471–478. doi:10.1097/01.fpc.0000114747.08559.49
Hamadeh, I. S., Zhang, Q., Steuerwald, N., Hamilton, A., Druhan, L. J., McSwain, M., et al. (2019). Effect of CYP3A4, CYP3A5, and ABCB1 polymorphisms on intravenous tacrolimus exposure and adverse events in adult allogeneic stem cell transplant patients. Biol. Blood Marrow Transpl. 25 (4), 656–663. doi:10.1016/j.bbmt.2018.12.766
Hawwa, A. F., McKiernan, P. J., Shields, M., Millership, J. S., Collier, P. S., and McElnay, J. C. (2009). Influence of ABCB1 polymorphisms and haplotypes on tacrolimus nephrotoxicity and dosage requirements in children with liver transplant. Br. J. Clin. Pharmacol. 68 (3), 413–421. doi:10.1111/j.1365-2125.2009.03461.x
Herrlinger, K. R., Koc, H., Winter, S., Teml, A., Stange, E. F., Fellermann, K., et al. (2011). ABCB1 single-nucleotide polymorphisms determine tacrolimus response in patients with ulcerative colitis. Clin. Pharmacol. Ther. 89 (3), 422–428. doi:10.1038/clpt.2010.348
Hiraoka, A., Ohashi, Y., Okamoto, S., Moriyama, Y., Nagao, T., Kodera, Y., et al. (2001). Phase III study comparing tacrolimus (FK506) with cyclosporine for graft-versus-host disease prophylaxis after allogeneic bone marrow transplantation. Bone Marrow Transpl. 8 (2), 181–185. doi:10.1038/sj.bmt.1703097
Imado, T., Iwasaki, T., Kuroiwa, T., Sano, H., and Hara, H. (2004). Effect of FK506 on donor T-cell functions that are responsible for graft-versus-host disease and graft-versus-leukemia effect. Transplantation 77 (3), 391–398. doi:10.1097/01.TP.0000111759.48240.F5
Ito, A., Okada, Y., Hashita, T., Aomori, T., Hiromura, K., Nojima, Y., et al. (2017). Sex differences in the blood concentration of tacrolimus in systemic lupus erythematosus and rheumatoid arthritis patients with CYP3A5*3/*3. Biochem. Genet. 55 (3), 268–277. doi:10.1007/s10528-017-9795-8
Iwamoto, T., Monma, F., Fujieda, A., Nakatani, K., Gayle, A. A., Nobori, T., et al. (2015). Effect of genetic polymorphism of CYP3A5 and CYP2C19 and concomitant use of voriconazole on blood tacrolimus concentration in patients receiving hematopoietic stem cell transplantation. Ther. Drug Monit. 37 (5), 581–588. doi:10.1097/FTD.0000000000000182
Jacobson, P., Ng, J., Ratanatharathorn, V., Uberti, J., and Brundage, R. C. (2001). Factors affecting the pharmacokinetics of tacrolimus (FK506) in hematopoietic cell transplant (HCT) patients. Bone Marrow Transpl. 28 (8), 753–758. doi:10.1038/sj.bmt.1703224
Kato, H., Usui, M., Muraki, Y., Okuda, M., Nakatani, K., Hayasaki, A., et al. (2018). Intravenous administration of tacrolimus stabilizes control of blood concentration regardless of CYP3A5 polymorphism in living donor liver transplantation: comparison of intravenous infusion and oral administration in early postoperative period. Transpl. Proc. 50 (9), 2684–2689. doi:10.1016/j.transproceed.2018.03.049
Khaled, S. K., Palmer, J. M., Herzog, J., Stiller, T., Tsai, N. C., Senitzer, D., et al. (2016). Influence of absorption, distribution, metabolism, and excretion genomic variants on tacrolimus/sirolimus blood levels and graft-versus-host disease after allogeneic hematopoietic cell transplantation. Biol. Blood Marrow Transpl. 22 (2), 268–276. doi:10.1016/j.bbmt.2015.08.027
Khwaja, A. (2012). KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin. Pract. 120 (4), c179–c184. doi:10.1159/000339789
Kim, I. W., Moon, Y. J., Ji, E., Kim, K. I., Han, N., Kim, S. J., et al. (2012). Clinical and genetic factors affecting tacrolimus trough levels and drug-related outcomes in Korean kidney transplant recipients. Eur. J. Clin. Pharmacol. 68 (5), 657–669. doi:10.1007/s00228-011-1182-5
Kuehl, P., Zhang, J., Lin, Y., Lamba, J., Assem, M., Schuetz, J., et al. (2001). Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27 (4), 383–391. doi:10.1038/86882
Lee, S. J., Klein, J. P., Barrett, A. J., Ringden, O., Antin, J. H., Cahn, J. Y., et al. (2002). Severity of chronic graft-versus-host disease: association with treatment-related mortality and relapse. Blood 100 (2), 406–414. doi:10.1182/blood.v100.2.406
Levine, J. E., Logan, B., Wu, J., Alousi, A. M., Ho, V., Bolaños-Meade, J., et al. (2010). Graft-versus-host disease treatment: predictors of survival. Biol. Blood Marrow Transpl. 16 (12), 1693–1699. doi:10.1016/j.bbmt.2010.05.019
Li, Y., Yan, L., Shi, Y., Bai, Y., Tang, J., and Wang, L. (2015). CYP3A5 and ABCB1 genotype influence tacrolimus and sirolimus pharmacokinetics in renal transplant recipients. Springerplus 4, 637. doi:10.1186/s40064-015-1425-5
Liu, X. L., Guan, Y. P., Wang, Y., Huang, K., Jiang, F. L., Wang, J., et al. (2022). Population pharmacokinetics and initial dosage optimization of tacrolimus in pediatric hematopoietic stem cell transplant patients. Front. Pharmacol. 13, 891648. doi:10.3389/fphar.2022.891648
McFadyen, M. C. E., Melvin, W. T., and Murray, G. I. (1998). Regional distribution of individual forms of cytochrome P450 mRNA in normal adult human brain. Biochem. Pharmacol. 55 (6), 825–830. doi:10.1016/s0006-2952(97)00516-9
Miao, J., Jin, Y., Marunde, R. L., Gorski, C. J., Kim, S., Quinney, S., et al. (2009). Association of genotypes of the CYP3A cluster with midazolam disposition in vivo. Pharmacogenomics J. 9 (5), 319–326. doi:10.1038/tpj.2009.21
Mourad, M., Wallemacq, P., De Meyer, M., Brandt, D., Van Kerkhove, V., Malaise, J., et al. (2006). The influence of genetic polymorphisms of cytochrome P450 3A5 and ABCB1 on starting dose- and weight-standardized tacrolimus trough concentrations after kidney transplantation in relation to renal function. Clin. Chem. Lab. Med. 44 (10), 1192–1198. doi:10.1515/CCLM.2006.229
Musuamba, F. T., Mourad, M., Haufroid, V., Delattre, I. K., Verbeeck, R. K., and Wallemacq, P. (2009). Time of drug administration, CYP3A5 and ABCB1 genotypes, and analytical method influence tacrolimus pharmacokinetics: a population pharmacokinetic study. Ther. Drug Monit. 31 (6), 734–742. doi:10.1097/FTD.0b013e3181bf8623
Onizuka, M., Kunii, N., Toyosaki, M., Machida, S., Ohgiya, D., Ogawa, Y., et al. (2011). Cytochrome P450 genetic polymorphisms influence the serum concentration of calcineurin inhibitors in allogeneic hematopoietic SCT recipients. Bone Marrow Transpl. 46 (8), 1113–1117. doi:10.1038/bmt.2010.273
Op den Buijsch, R. A., Christians, M. H., Stolk, L. M., de Vries, J. E., Cheung, C. Y., Undre, N. A., et al. (2007). Tacrolimus pharmacokinetics and pharmacogenetics: influence of adenosine triphosphate-binding cassette B1 (ABCB1) and cytochrome (CYP) 3A polymorphisms. Fundam. Clin. Pharmacol. 21 (4), 427–435. doi:10.1111/j.1472-8206.2007.00504.x
Owen, A., Goldring, C., Morgan, P., Chadwick, D., Park, B. K., and Pirmohamed, M. (2005). Relationship between the C3435T and G2677T(A) polymorphisms in the ABCB1 gene and P-glycoprotein expression in human liver. Br. J. Clin. Pharmacol. 59 (3), 365–370. doi:10.1111/j.1365-2125.2005.02229.x
Paine, M. F., Hart, H. L., Ludington, S. S., Haining, R. L., Rettie, A. E., and Zeldin, D. C. (2006). The human intestinal cytochrome P450 "pie. Drug Metab. Dispos. 34 (5), 880–886. doi:10.1124/dmd.105.008672
Papp, A. C., Pinsonneault, J. K., Cooke, G., and Sadée, W. (2003). Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques 34 (5), 1068–1072. doi:10.2144/03345dd03
Pasternak, A. L., Kidwell, K. M., Dempsey, J. M., Gersch, C. L., Pesch, A., Sun, Y., et al. (2019). Impact of CYP3A5 phenotype on tacrolimus concentrations after sublingual and oral administration in lung transplant. Pharmacogenomics 20 (6), 421–432. doi:10.2217/pgs-2019-0002
Pasternak, A. L., Marcath, L. A., Li, Y., Nguyen, V., Gersch, C. L., Rae, J. M., et al. (2022). Impact of pharmacogenetics on intravenous tacrolimus exposure and conversions to oral therapy. Transpl. Cell Ther. 28 (1), 19.e1. doi:10.1016/j.jtct.2021.09.011
Pratt, V. M., Cavallari, L. H., Fulmer, M. L., Gaedigk, A., Hachad, H., Ji, Y., et al. (2023). CYP3A4 and CYP3A5 genotyping recommendations: a joint consensus recommendation of the association for molecular pathology, clinical pharmacogenetics implementation Consortium, College of American pathologists, Dutch pharmacogenetics working group of the royal Dutch pharmacists association, European society for pharmacogenomics and personalized therapy, and pharmacogenomics knowledgebase. J. Mol. Diagn 25 (9), 619–629. doi:10.1016/j.jmoldx.2023.06.008
Przepiorka, D., Nash, R. A., Wingard, J. R., Zhu, J., Maher, R. M., Fitzsimmons, W. E., et al. (1999). Relationship of tacrolimus whole blood levels to efficacy and safety outcomes after unrelated donor marrow transplantation. Biol. Blood Marrow Transpl. 5 (2), 94–97. doi:10.1053/bbmt.1999.v5.pm10371361
Qiu, H., Dong, H., Pan, S., and Miao, K. (2012). The single nucleotide polymorphism and haplotype analysis of MDR1 in Jiangsu Han population of China. Biomed. Pharmacother. 66 (6), 459–463. doi:10.1016/j.biopha.2012.03.007
Seligson, N. D., Kolesar, J. M., Alam, B., Baker, L., Lamba, J. K., Fridley, B. L., et al. (2023). Integrating pharmacogenomic testing into paired germline and somatic genomic testing in patients with cancer. Pharmacogenomics 24 (13), 731–738. doi:10.2217/pgs-2023-0125
Semiz, S., Dujić, T., Ostanek, B., Prnjavorac, B., Bego, T., Malenica, M., et al. (2011). Analysis of CYP3A4*1B and CYP3A5*3 polymorphisms in population of Bosnia and Herzegovina. Med. Glas. (Zenica) 8 (1), 84–89.
Sinues, B., Vicente, J., Fanlo, A., Vasquez, P., Medina, J. C., Mayayo, E., et al. (2007). CYP3A5*3 and CYP3A4*1B allele distribution and genotype combinations: differences between Spaniards and Central Americans. Ther. Drug Monit. 29 (4), 412–416. doi:10.1097/FTD.0b013e31811f390a
Spurdle, A. B., Goodwin, B., Hodgson, E., Hopper, J. L., Chen, X., Purdie, D. M., et al. (2002). The CYP3A4*1B polymorphism has no functional significance and is not associated with risk of breast or ovarian cancer. Pharmacogenetics 12 (5), 355–366. doi:10.1097/00008571-200207000-00003
Suetsugu, K., Mori, Y., Yamamoto, N., Shigematsu, T., Miyamoto, T., Egashira, N., et al. (2019). Impact of CYP3A5, POR, and CYP2C19 polymorphisms on trough concentration to dose ratio of tacrolimus in allogeneic hematopoietic stem cell transplantation. Int. J. Mol. Sci. 20 (10), 2413. doi:10.3390/ijms20102413
Sureda, A., Bader, P., Cesaro, S., Dreger, P., Duarte, R. F., Dufour, C., et al. (2015). Indications for allo- and auto-SCT for haematological diseases, solid tumours and immune disorders: current practice in Europe, 2015. Bone Marrow Transpl. 50 (8), 1037–1056. doi:10.1038/bmt.2015.6
Sy, S. K., Heuberger, J., Shilbayeh, S., Conrado, D. J., and Derendorf, H. (2013). A Markov chain model to evaluate the effect of CYP3A5 and ABCB1 polymorphisms on adverse events associated with tacrolimus in pediatric renal transplantation. AAPS J. 15 (4), 1189–1199. doi:10.1208/s12248-013-9528-9
Tapirdamaz, Ö., Hesselink, D. A., el Bouazzaoui, S., Azimpour, M., Hansen, B., van der Laan, L. J., et al. (2014). Genetic variance in ABCB1 and CYP3A5 does not contribute toward the development of chronic kidney disease after liver transplantation. Pharmacogenet Genomics 24 (9), 427–435. doi:10.1097/FPC.0000000000000063
Thervet, E., Anglicheau, D., Legendre, C., and Beaune, P. (2008). Role of pharmacogenetics of immunosuppressive drugs in organ transplantation. Ther. Drug Monit. 30 (2), 143–150. doi:10.1097/FTD.0b013e31816babef
Thervet, E., Loriot, M. A., Barbier, S., Buchler, M., Ficheux, M., Choukroun, G., et al. (2010). Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin. Pharmacol. Ther. 87 (6), 721–726. doi:10.1038/clpt.2010.17
Wang, D., Guo, Y., Wrighton, S. A., Cooke, G. E., and Sadee, W. (2011). Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 11 (4), 274–286. doi:10.1038/tpj.2010.28
Wang, D., and Sadee, W. (2016). CYP3A4 intronic SNP rs35599367 (CYP3A4*22) alters RNA splicing. Pharmacogenet Genomics 26 (1), 40–43. doi:10.1097/FPC.0000000000000183
Woodahl, E. L., Hingorani, S. R., Wang, J., Guthrie, K. A., McDonald, G. B., Batchelder, A., et al. (2008). Pharmacogenomic associations in ABCB1 and CYP3A5 with acute kidney injury and chronic kidney disease after myeloablative hematopoietic cell transplantation. Pharmacogenomics J. 8 (4), 248–255. doi:10.1038/sj.tpj.6500472
Yamashita, T., Fujishima, N., Miura, M., Niioka, T., Abumiya, M., Shinohara, Y., et al. (2016). Effects of CYP3A5 polymorphism on the pharmacokinetics of a once-daily modified-release tacrolimus formulation and acute kidney injury in hematopoietic stem cell transplantation. Cancer Chemother. Pharmacol. 78 (1), 111–118. doi:10.1007/s00280-016-3060-4
Yanagimachi, M., Naruto, T., Tanoshima, R., Kato, H., Yokosuka, T., Kajiwara, R., et al. (2010). Influence of CYP3A5 and ABCB1 gene polymorphisms on calcineurin inhibitor-related neurotoxicity after hematopoietic stem cell transplantation. Clin. Transpl. 24 (6), 855–861. doi:10.1111/j.1399-0012.2009.01181.x
Yanagisawa, R., Katsuyama, Y., Shigemura, T., Saito, S., Tanaka, M., Nakazawa, Y., et al. (2011). Engraftment syndrome, but not acute GVHD, younger age, CYP3A5 or MDR1 polymorphisms, increases tacrolimus clearance in pediatric hematopoietic SCT. Bone Marrow Transpl. 46 (1), 90–97. doi:10.1038/bmt.2010.64
Yoshikawa, N., Takeshima, H., Sekine, M., Akizuki, K., Hidaka, T., Shimoda, K., et al. (2021). Relationship between CYP3A5 polymorphism and tacrolimus blood concentration changes in allogeneic hematopoietic stem cell transplant recipients during continuous infusion. Pharm. (Basel) 14 (4), 353. doi:10.3390/ph14040353
Zeigler-Johnson, C., Friebel, T., Walker, A. H., Wang, Y., Spangler, E., Panossian, S., et al. (2004). CYP3A4, CYP3A5, and CYP3A43 genotypes and haplotypes in the etiology and severity of prostate cancer. Cancer Res. 64 (22), 8461–8467. doi:10.1158/0008-5472.CAN-04-1651
Zhu, J., Patel, T., Miller, J. A., Torrice, C. D., Aggarwal, M., Sketch, M. R., et al. (2020). Influence of germline genetics on tacrolimus pharmacokinetics and pharmacodynamics in allogeneic hematopoietic stem cell transplant patients. Int. J. Mol. Sci. 21 (3), 858. doi:10.3390/ijms21030858
Ziakas, P. D., Zervou, F. N., Zacharioudakis, I. M., and Mylonakis, E. (2014). Graft-versus-host disease prophylaxis after transplantation: a network meta-analysis. PLoS One 9 (12), e114735. doi:10.1371/journal.pone.0114735
Keywords: tacrolimus, single nucleotide polymorphism, precision medicine, hematologic stem cell transplant, CYP3A4, CYP3A5
Citation: Seligson ND, Zhang X, Zemanek MC, Johnson JA, VanGundy Z, Wang D, Phelps MA, Roddy J, Hofmeister CC, Li J and Poi MJ (2024) CYP3A5 influences oral tacrolimus pharmacokinetics and timing of acute kidney injury following allogeneic hematopoietic stem cell transplantation. Front. Pharmacol. 14:1334440. doi: 10.3389/fphar.2023.1334440
Received: 07 November 2023; Accepted: 21 December 2023;
Published: 08 January 2024.
Edited by:
Luis Andrés López-Fernández, Gregorio Marañón Hospital, SpainReviewed by:
Simran D. S. Maggo, Children’s Hospital of Los Angeles, United StatesCopyright © 2024 Seligson, Zhang, Zemanek, Johnson, VanGundy, Wang, Phelps, Roddy, Hofmeister, Li and Poi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junan Li, bGkuMjI1QG9zdS5lZHU=; Ming J. Poi, cG9pLjJAb3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.