
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 26 March 2024
Sec. Pharmacology of Anti-Cancer Drugs
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1273993
This article is part of the Research TopicNew Mechanisms for Anti-Cancer DrugsView all 22 articles
 Aqsa Khan1‡
Aqsa Khan1‡ Asifa Khan1†‡
Asifa Khan1†‡ Mohammad Aasif Khan1,2
Mohammad Aasif Khan1,2 Zoya Malik1
Zoya Malik1 Sheersh Massey1
Sheersh Massey1 Rabea Parveen1
Rabea Parveen1 Saad Mustafa1
Saad Mustafa1 Anas Shamsi3*
Anas Shamsi3* Syed A. Husain1*
Syed A. Husain1*For centuries, plants have been serving as sources of potential therapeutic agents. In recent years, there has been a growing interest in investigating the effects of plant-derived compounds on epigenetic processes, a novel and captivating Frontier in the field of epigenetics research. Epigenetic changes encompass modifications to DNA, histones, and microRNAs that can influence gene expression. Aberrant epigenetic changes can perturb key cellular processes, including cell cycle control, intercellular communication, DNA repair, inflammation, stress response, and apoptosis. Such disruptions can contribute to cancer development by altering the expression of genes involved in tumorigenesis. However, these modifications are reversible, offering a unique avenue for therapeutic intervention. Plant secondary compounds, including terpenes, phenolics, terpenoids, and sulfur-containing compounds are widely found in grains, vegetables, spices, fruits, and medicinal plants. Numerous plant-derived compounds have demonstrated the potential to target these abnormal epigenetic modifications, including apigenin (histone acetylation), berberine (DNA methylation), curcumin (histone acetylation and epi-miRs), genistein (histone acetylation and DNA methylation), lycopene (epi-miRs), quercetin (DNA methylation and epi-miRs), etc. This comprehensive review highlights these abnormal epigenetic alterations and discusses the promising efficacy of plant-derived compounds in mitigating these deleterious epigenetic signatures in human cancer. Furthermore, it addresses ongoing clinical investigations to evaluate the therapeutic potential of these phytocompounds in cancer treatment, along with their limitations and challenges.
Despite modern advances and therapeutics, cancer remains one of the most dreaded diseases globally. According to the GLOBOCAN cancer statistics 2020, female breast cancer incidence superseded lung cancer globally. Around 19.3 million new cancer cases have been reported worldwide, with 10 million cancer deaths. With a record of 2.3 million new diagnoses, female breast cancer has surpassed lung cancer as the most frequently diagnosed cancer, followed by colorectal (10.0%), prostate (7.3%), and abdominal (5.6%) cancers respectively. In 2040, the global cancer incidence is predicted to increase to 28.4 million cases, increasing 47% from 2020 (Sung et al., 2021). This creates an alarming situation towards the increasing number of cancer cases and calls for better and safer treatment options.
Cancer is a multifaceted disease with genetic mutations and abnormal epigenetic modifications (Baylin et al., 2001). According to Conrad Hal Waddington, epigenetics refers to heritable and reversible modifications in gene expression or changes in a chromosome without any alterations in DNA sequence (Henikoff and Matzke, 1997). The epigenetic regulation includes DNA methylation, posttranslational histone protein modifications such as acetylation, methylation, and phosphorylation, and microRNAs (noncoding RNA) that can inhibit translation or degrade mRNAs by modulating gene expression (Dehan et al., 2009). Epigenetic control plays a crucial role during early embryonic development, such as X-inactivation in females, genomic imprinting, development, and differentiation like the formation of long-term memory and behavior (Payer and Lee, 2008; Lubin et al., 2011). In addition to this, research studies suggest that epigenetic DNA modifications on uniparental disomy also serves as an underlying mechanism for developmental disorders, neurological diseases and tumorigenesis (Zoghbi and Beaudet, 2016; Tuna et al., 2009). A study on colorectal cancer showed several tumor suppressor genes (VCAN, FLT4, SFRP1 and GAS7) in the uniparental disomy and polysomy regions displaying elevated levels of DNA methylation (Torabi et al., 2015).
Deregulation of epigenetic modifications has critical implications on human health, such as cancer, metabolic syndrome, and neurodegenerative disorders like Alzheimer’s disease, Huntington’s disease, and amyotrophic lateral sclerosis (Berson et al., 2018; Smith and Ryckman, 2015). Recent studies highlighted that maternal behavioral patterns, diet choice, and exposure to other intrinsic or extrinsic factors have dramatically altered gene expression patterns and are implicated with epigenetic mechanisms (Chung and Herceg, 2020; Safi-Stibler and Gabory, 2020).
Since epigenetic modifications are reversible, a significant number of studies are now focused on the identification and development of pharmaceuticals that target these modulations (Singh et al., 2016; Liu Z. et al., 2019). In recent years, Food and Drug Administration, United States, approved several small synthetic molecule inhibitors that target distinct epigenetic modulations to treat several solid tumors and hematological malignancies. These clinically approved conventional drugs include Azacytidine and Decitabine (DNA methyltransferases (DNMT) inhibitors). Four histone deacetylases inhibitors (HDACi) are approved for the treatment of lymphomas, including vorinostat (SAHA), romidepsin (FK-228), belinostat (PXD-101), and tucidinostat (chidamide) (previous conditional approvals for panobinostat [LBH-589] for multiple myeloma [in combination with bortezomib] and romidepsin for peripheral T-cell lymphoma [PTCL] were recently withdrawn by the FDA) (Liu Z. et al., 2019; Xiao et al., 2023). On the contrary, several studies have found phytocompounds as potential regulators that can reverse these aberrant epigenetic modifications that induce tumor progression and eventually lead to cancer (Aggarwal et al., 2015; Montgomery and Srinivasan, 2019).
Drug development using naturally derived bioactive compounds has drawn a lot of interest in recent years as they are safe and economical. In contrast, present-day chemotherapeutic drugs are not only expensive but also toxic. One significant side effect of conventional medications is their ability to target normal cells in the body undergoing rapid proliferation, such as bone marrow cells, along with proliferating tumor cells. However, phytocompounds are nontoxic to normal cells and hence better tolerated (Singh et al., 2016). Phytocompounds are bioactive compounds, obtained from a wide variety of herbs, spices, vegetables, and fruits (Table 1), that possess anti-inflammatory, antioxidant, antimicrobial, anticancer, and anti-diabetic properties (Verma, 2016; Altaf et al., 2018; Gonelimali et al., 2018; Salehi et al., 2019). Furthermore, phytocompounds can target multiple cell cycle proteins, transcription factors, cell adhesion molecules, protein kinases, and anti-apoptotic factors (Aggarwal and Shishodia, 2006; Kundu and Surh, 2009; Bailon-Moscoso et al., 2017; Malik et al., 2021). Besides this, plant-based compounds possess the ability to target deregulated metabolic proteins and pathways in cancer, such as glycolysis, pentose phosphate pathway, lipid metabolism, amino acid metabolism, etc. (Pani et al., 2020; Khan et al., 2021a; Khan et al., 2021b). These multipronged functions of phytocompounds, targeted at various cellular pathways implicated epigenetically in cancer, can be a promising solution in the treatment of this deadliest disease.
This review highlights the prospective use of distinct phytocompounds to counteract epigenetic abnormalities that promote tumor development and progression in humans as well as their potential in anticancer therapeutics as summarized in Figure 1. Therefore, a comprehensive explanation of the most studied dietary compounds that critically modulate the epigenetic landscape in human cancer along with the latest developments, is provided extensively. In addition, list of all the phytocompounds that are discussed in this review and that are known to control epigenetic alterations are listed below in Table 1, along with their classification and sources.
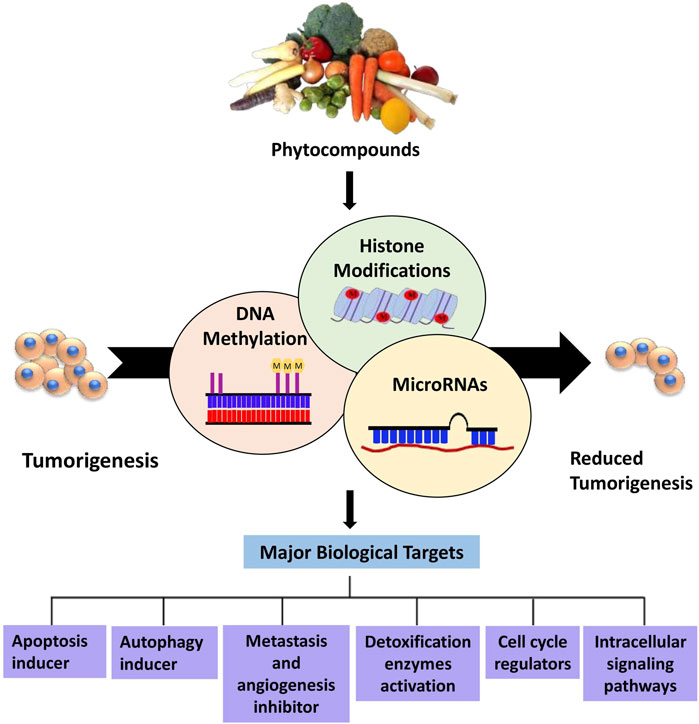
Figure 1. Overview of phytocompounds regulating major epigenetic modifications that involve regulation of key biological targets to reduce tumorigenesis.
Research articles, reviews, and abstracts were pulled for a literature survey from a variety of databases, including PubMed, Google Scholar, Springer, Wiley Online Library, ScienceDirect, etc., From the year 1998 and to 2022. Information from research articles, review articles, abstracts were used for this review. The literature search was conducted against the terminology “Bioactive compounds in cancer epigenetics” while other terminologies such as phytocompounds targeting DNA methylation, histone modification and epi-miRNAs were also included in this search as illustrated in Figure 2. Data was processed to find out general information about phytocompounds, with a strong focus on how they affect epigenetics modifications. Inclusion criteria was based on the topics that include phytocompounds targeting epigenetic modifications (DNA methylation, Histone modifications and miRNAs) in cancer. While the exclusion criteria, was based on studies demonstrating information duplication, such as reviews, material available in languages other than English, information unrelated to the subjects included in this analysis, insufficient information, and data older than 1998.
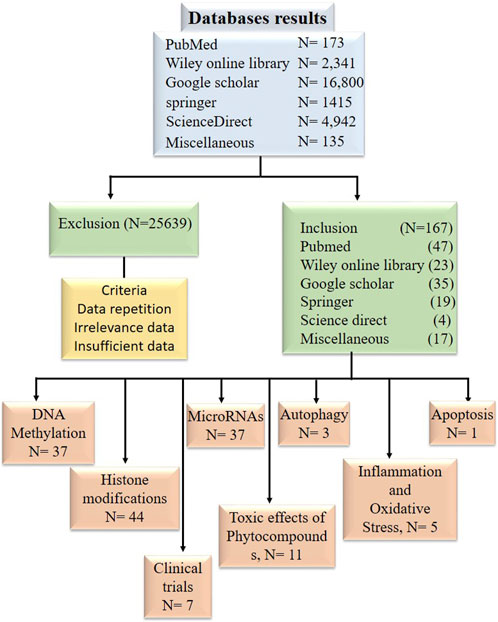
Figure 2. Diagram in block form illustrating the inclusion and exclusion of research publications in data tabulation.
DNA methylation is responsible for controlling gene expression and interacting with the nucleosomes that control DNA packaging, and it can influence entire DNA domains. It is a chemical, biological modification where cytosine residues are methylated at a 5′position. This modification mainly occurs at cytosine residues present in GC dinucleotide-rich regions clustered together to form the CpG islands spanned the 5′end region of several genes (Issa and Kantarjian, 2009).
The human genome contains approximately 28 million CpG sites. Under normal circumstances, most CpG islands, especially within the promoter region, are unmethylated except in the case of X-inactivation, genome imprinting, and repression of transposons. DNA methylation occurs during the early embryonic stages by a set of distinct enzymes called DNA methyltransferases (DNMTs) (Bird, 2002). DNA methyltransferases catalyze the formation of 5-methyl cytosine that involves the transfer of a methyl group from S-adenosyl-L- methionine (SAM) to the 5′position of the cytosine residue in CG dinucleotide (Santi et al., 1983). DNMT1, DNMT3A, and DNMT3B are the three major DNMTs involved in DNA methylation in mammals. Maintenance of methylation patterns during replication is one of the critical functions of DNMT1. During replication, DNMT1 adds a methyl group to hemi methylated CpG dinucleotides in the daughter strand as it shows 5–30 times greater affinity for hemi methylated substrates. In contrast, DNMT3A and DNMT3B are de novo methyltransferases exclusively involved in the methylation of previously unmethylated DNA sequences (Okano et al., 1999; Bestor, 2000). To ensure methylation, DNMTs must have access to the DNA that can be gained via perturbation of chromatin structure by specific chromatin remodeling proteins (As shown in Figure 3) (Baylin et al., 2001).
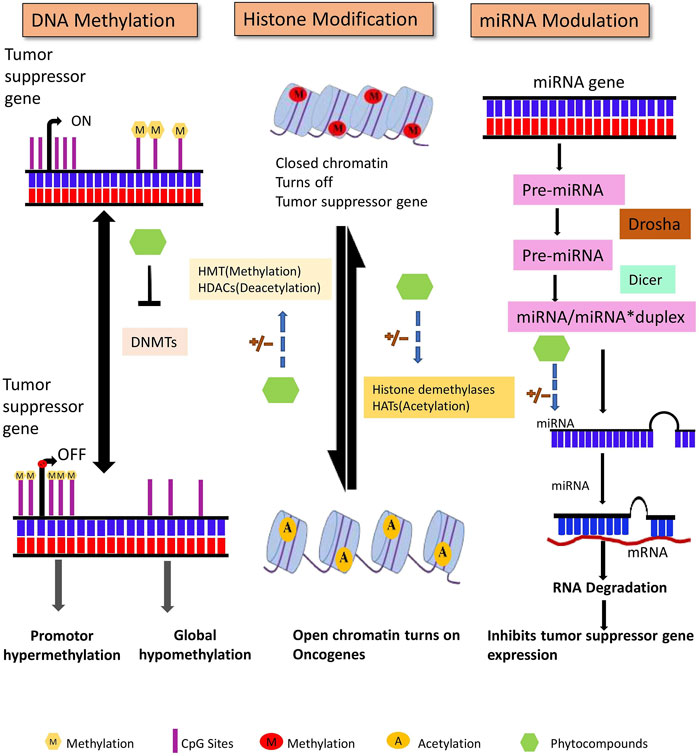
Figure 3. DNA methylation, posttranslational histone modifications, and microRNAs are all major epigenetic processes that regulate gene expression. These pathways are found to be dysregulated in cancer. Phytocompounds have shown to modulate abnormal epigenetic modifications.
In cancer, hypermethylation of CpG islands in the promoter sequence of some genes results in gene silencing by suppressing the transcriptional activity of tumor suppressor genes (Dehan et al., 2009). For instance, promoter hypermethylation of CpG sites of tumor suppressor genes like RARβ and RASSF1A can induce breast cancer development (Van Hoesel et al., 2013). Certain studies of malignant and tumor cells have shown hypomethylation of DNA sequences at CpG sites that can cause conformational and functional alterations in chromosomes (Gama-Sosa et al., 1983). In cancer there are several natural compounds known to influence the DNA methylation as listed in Table 2 in most of the cases by a single publication.
An increasing list of different phytocompounds targeting DNA methylation at distinct genes in various cancer cell lines has been discovered in the last two decades. This section summarizes the phytocompounds that have the potential to act as DNA methylation inhibitors and activators altering the tumor suppressor gene expression.
Apigenin, a polyphenol, has been shown to reduce the methylation of CpG sites in the Nrf2 promoter region, which contributed to increased mRNA levels of downstream genes. Nrf2 codes a transcription factor that regulates antioxidant enzymes. Furthermore, apigenin also decreased DNMT expression in mouse skin cancer cell line (Paredes-Gonzalez et al., 2014).
Neurog-1 gene plays a critical role in neuronal differentiation. Curcumin-induced treatment reported demethylation of 14 CpG sites of Neurog-1 promoter and downstream gene reactivation in prostate cancer cell line (Shu et al., 2011). BRCA1, a DNA repair gene, critically regulates cell cycle checkpoints and transcription. Triple-negative breast cancer (TNBC) cell lines were treated with curcumin, which caused the upregulation of the TET1 gene that promoted the BRCA1 promoter’s hypomethylation. Furthermore, it induced upregulation of DNMT3 that resulted in suppressed expression of proto-oncogene SNCG (Al-Yousef et al., 2020). Curcumin treatment reactivated RARβ by causing promoter hypomethylation in lung cancer cells. The treatment further resulted in increased RARβ protein and mRNA expression. Curcumin treatment also induced downregulation of DNMT3b, resulting in decreased expression of DNMT3b mRNA levels (Jiang et al., 2015).
Epigallocatechin 3-gallate (EGCG), a polyphenol obtained from green tea, is one of the most studied chemo preventive agents. Besides EGCG, epigallocatechin and epicatechin are also among the major constituents of green tea. Several studies have documented numerous medical benefits of EGCG; regulation of cancer cell growth is one of them (Shankar et al., 2007). According to an in vitro study, catechol inhibits DNMT activity by directly inhibiting DNMTs or by causing o-methylation of SAM by methyltransferases that result in increased SAM levels. Treatment of esophageal cancer cell lines with EGCG exhibited unmethylation specific bands for tumor suppressor p16INK4a, retinoic acid receptor β (RARβ), MGMT, and DNA repair hMLH1 genes. Furthermore, it demonstrated that EGCG reactivated the RARβ gene in prostate cancer and esophageal cancer cell lines (Fang et al., 2007). In another study, breast cancer cell lines reported downregulation of DNMT expression and restoration of the tumor suppressor gene SCUBE 2 expression, when treated with EGCG, that resulted in increased E-cadherin expression and suppression of cell migration and invasion (Sheng et al., 2019).
Genistein-induced treatment of HeLa cells reduced promoter 5′CpG methylation levels of various tumor suppressor genes involved in PI3K and MAPK signaling. The restoration of transcription levels of earlier hypermethylated genes (mentioned in Table 2) can be correlated to a decrease in methylation levels (Sundaram et al., 2019). In a triple-negative breast cancer cell, silencing of BRCA1 is often a result of overexpression of the aryl hydrocarbon receptor. In vivo study of the mammary gland of adult mice demonstrated that lifelong treatment with genistein reduced BRCA1 CpG methylation in the offspring’s mammary tissue (Donovan et al., 2019). Treatment with genistein and daidzein resulted in re-expression of BRCA1, EPHB2, and Glutathione S-transferase pi 1 (GSTP1) in prostate cancer cells, suggesting a preventive effect of soy phytoestrogens against prostate cancer (Adjakly et al., 2011).
GSTP1 encodes for a detoxifying enzyme that protects cells from genome-damaging stresses caused by reactive chemical species. Lycopene induced promoter demethylation of GSTP1 in prostate cancer cells, increasing the mRNA and protein levels. Further treatment with lycopene also decreased the protein levels of DNMT3a. Fu et al. (2014).
The hypermethylated RASSF1A promoter in prostate cancer cell line, when treated with phenethyl isothiocyanate (PEITC), reported a decrease in methylation of CpG sites by an average of 90% compared to untreated cells with 98% of methylation at 16 CpG sites. Promoter demethylation of the tumor suppressor gene RASSF1A correlated with decreased mRNA expression of DNMT1 and DNMT3A when treated with PEITC (Boyanapalli et al., 2016). PEITC induced CpG demethylation in GSTP1 promoter of LNCAP cells in a concentration-dependent manner and was almost similar or higher than the commonly used DNMT inhibitor 5-Aza-2′-deoxycytidine. On further analysis by pyrosequencing tool, it was revealed that PEITC treatment significantly reduced CpG methylation at positions 1 and 3 from 89.5% to 73.2% and 61.8%–6.5%, respectively, that was higher than the demethylating activity of 5′-Aza (Wang et al., 2007).
Quercetin inhibited the DNMT activity and resulted in the restoration of various tumor suppressor genes that were earlier hypermethylated at 5′CpG promoter sites in cervical cancer cells (mentioned in Table 2). Furthermore, in silico studies revealed that quercetin competitively inhibit DNMTs by binding to its catalytic active sites (Kedhari Sundaram et al., 2019). Quercetin treatment decreases the methylation levels of P16INK4a, Er-beta, and RASSF1A genes in bladder cancer cell lines. Furthermore, quercetin treatment inhibited the expression of mutant p53 and survivin proteins. P53 maintains cell cycle regulation, DNA repair, and apoptosis, and any alteration in p53 happens to decrease genomic stability and DNA repair. While Survivin acts as antagonists by inhibiting antiapoptotic pathways and promoting mitotic progression (Ma et al., 2006). In vitro and in vivo studies revealed that quercetin induced demethylation of highly methylated promoter sites of apoptosis-related genes BCL2L11 and DAPKlin, leukemia cell lines (Alvarez et al., 2018).
A study on rodent’s mammary tumor treated with resveratrol indicated a decrease in DNMT 3b expression. The study further reported that 26% of rats developed tumors when treated with a low dose of resveratrol, while only 18% developed tumors with high dose compared to 33% in the control group (Qin et al., 2014). Resveratrol upregulated the expression of cellular retinoic acid binding protein 2 (CRABP2) that mediates retinoic acid anticancer pathways in thyroid cancer cell lines by partial demethylation of CpG promoter sites. In addition, resveratrol significantly decreased DNMT1 and DNMT3A expression (Liu X. et al., 2019).
Sulforaphane treatment downregulated DNMT expression and can mediate promoter demethylation in prostate cancer cells (Wong et al., 2014). In colon cancer cells, sulforaphane inhibited the expression of DNMT1 and increased Nrf2 protein expression by decreasing the methylation of Nrf2 promoter region (Zhou J. W. et al., 2019).
In TNBC cells, withaferin A induced hypermethylation of tumor-promoting genes ADAM8, PLAU, TNFSF12, GSTM1, ME3, and hypomethylation of GLRX2, GFPT2, STX11, and VGF (vel Szic et al., 2017).
The reversible epigenetic process of DNA methylation regulates chromosomal integrity, tissue differentiation, and gene expression throughout embryogenesis. However, any aberrant epigenetic modifications during these processes can lead to tumorigenesis. As extensively discussed above, numerous studies provide evidence that these abnormal modifications can be undone by a plethora of phytocompound treatment. Phytocompounds targeting DNA methylation are listed below in Table 2.
The eukaryotic DNA is condensed in the form of chromatin. A nucleosome, the basic unit of chromatin, comprises of three parts, i.e., a core nucleosome, linker DNA, and histone H1. Each nucleosome core contains 2 copies of each histone H2A, H2B, H3, and H4, and approximately ∼147bp of DNA wrapped around it, in the form of a histone octamer. Core histone proteins comprise of three structural motifs, i.e., the histone fold regions, their diverse extensions, and histone tails. Histone tails are sites of posttranslational modifications and are extremely basic, which consists mainly of lysine and arginine amino acids (Luger and Richmond, 1998; Kornberg and Lorch, 1999). Histone modifications influence the regulation of chromatin dynamics in processes such as gene regulation, DNA repair, cell proliferation, and apoptosis. In cancer, deregulation of genes involved in these pathways may lead to unwarranted activation of oncogenes or inactivation of tumor suppressor genes (Audia and Campbell, 2016).
Histone acetyltransferases (HATs) catalyze the transfer of an acetyl group to the ε-amino group of the lysine side chains utilizing the cofactor acetyl CoA, thereby weakening the DNA and histone interactions. HATs have been classified into two- Type-A and Type-B. Further, HATs can be divided into three major categories- GNAT, MYST, and p300/CBP (Bannister and Kouzarides, 2011). Acetylation at K5 and K12 of newly synthesized H4 histones is catalyzed by type-B HAT (HAT1), along with specific sites at H3 histone (Kouzarides, 2007). Predominant transcriptional repressors, histone deacetylases (HDACs), catalyze the reverse lysine acetylation, thus restoring the positive charge of lysine and stabilizing the chromatin structure. The HDACs have been categorized into four major classes: classes I, II, III, and IV. HDACs 1, 2, 3 and belongs to class I HDACs, while class II comprises of HDACs 4, 5, 6, 7, 9, and 10; and only HDAC11 belongs to class IV HDACs. Sirtuins, another name for class III HDACs, are structurally distinct from the other classes and require a cofactor (NAD+) for its activity (Mottet and Castronovo, 2008).
Lysine and arginine residues are mostly favored for histone methylation. Lysine methyltransferases catalyze the transfer of a methyl group from SAM to a ε-amino group of lysine, whereas arginine methyltransferases catalyze the transfer of a methyl group from SAM to arginine’s ω-amino group. Histone demethylases have the opposite effect to histone methylases, both can activate or repress the transcriptional activity (Bannister et al., 2002). For example, methylation of histone H3 at K9 and K36 may negatively affect the promoter region while a positive one in the coding region (Kouzarides, 2007). Additionally, decreased levels of acetylation of histones H3 and H4 and elevated levels of DNA methyltransferases are usually found in prostate cancer cells (Baumgart and Haendler, 2017).
Histone phosphorylation is highly versatile and occurs predominantly at serine, threonine, and tyrosine residues. Phosphorylation results from the transfer of a phosphate group from ATP to the hydroxyl group of the amino acid side chain that is catalyzed by histone kinases (Xhemalce et al., 2011).
A recent study also revealed that lactate, produced by the incomplete oxidation of glucose through the Warburg effect in cancer cells, can regulate gene expression in macrophages by functioning as a new histone modification, i.e., lactylation. Excessive lactate production, i.e., Warburg effect benefits cancer in number of ways-promotes metastasis, angiogenesis, activation of T cells, polarization of macrophages. Now it is well documented that lactic acid also contributed in epigenetic modifications by adding the lactyl group on the ε-amino group of a lysine residue (Zhang et al., 2019).
In addition, there are diverse sets of posttranslational histone modifications that include deamination, ADP ribosylation, ubiquitylation, and sumoylation but are beyond the scope of this review.
In the last two decades, several dietary compounds have been confirmed to play a substantial role in the reversal of histone onco-modification and a few of the most important of them are discussed in detail below.
In prostate cancer cells, apigenin induced a decrease in HDAC activity, downregulated HDAC1 and 3 expression, and increased acetylation of histones H3 and H4. In vivo studies reveled that apigenin treatment reduced tumor growth and a significant decrease in HDAC activity that correlated with increased levels of p21/waf1and Bax protein along with a reduction in protein levels of bcl2 that favored apoptosis in tumor cells of the mice (Pandey et al., 2012). Another in vivo study on the breast cancer cell line of athymic nude mice revealed a decrease in HDAC activity on treatment with apigenin in a dose-dependent manner. Acetylation of histone H3 was shown to increase after treatment with apigenin correlated with transcriptional activation of p21WAF1/CIP1 gene (Tseng et al., 2017). Apigenin treatment lowered the protein expression levels of HDAC1, 3, 4, 5, 6, and 8 in the mouse skin cancer cell line in a dose-dependent manner (Paredes-Gonzalez et al., 2014).
Curcumin treatment resulted in inhibition of cell proliferation in Raji cells of B-NHL cancer. The protein expression levels of HDAC1, 3, and 8 were also found to be downregulated and acetylation of H4 histone increased after treatment with curcumin in a dose- and time-dependent manner (Liu et al., 2005). In HeLa nuclear extracts, curcumin decreases the HDAC activity. Docking studies suggested curcumin as a potent inhibitor of HDAC8 than its carboxylic acid-derived pharmacological counterparts (Bora-Tatar et al., 2009). In human hepatoma Hep3B cells, curcumin induced a decrease in H3 and H4 histone acetylation. Further in vitro studies showed curcumin treatment decreased core histone acetylation catalyzed by HAT extracted from Hep3B cancer cell line that suggested the role of HAT in curcumin-induced histone hypoacetylation (Kang et al., 2005).
In a recent study, demethylzeylasteral treatment promoted decrease in tumor progression in liver stem cells by inhibiting H3 lactylation (H3K9la and H3K56la) (Pan et al., 2022).
Green tea polyphenol (GTP) treatment decreased a maximum of 43% HDAC activity in a time-dependent manner in prostate cancer cells, which was similar to Trichostatin A (TSA) that caused 45% inhibition in 24 h. Furthermore, a decrease in protein expression of HDAC1 and 3 was also observed. GTP treatment decreased the mRNA levels of HDAC1, 2, and 3 in a gradual time course, whereas no such changes were observed with TSA. Exposure to GTP further resulted in a 22-fold and 2.2-fold increase in H3 and H4 acetylation, respectively, in a gradual-time course (Pandey et al., 2010). EGCG treatment significantly reduced HDAC activity in skin cancer cell line A431. Acetylation and methylation levels of H3K9 were found to be increased and decreased, respectively, when treated with EGCG. Furthermore, acetylation of H3K4 and H4K5, 12, and 16 were shown to be increased after EGCG treatment, thus reactivating tumor suppressor genes (Nandakumar et al., 2011). In prostate cancer cells, GTP and its major constituent EGCG induced treatment caused a substantial decrease in the expression and activity of HDAC 1, 2, 3, and 8. On further investigation, EGCG acetylated the p53 gene at K373 and K382, which was found to be diminished when the EGCG treatment was withdrawn after a certain time period (Thakur et al., 2012).
Genistein treatment increased acetylation at H3, H4, and H3 dimethylated at K4 near the transcription start sites of tumor suppressor genes p21 and p16 in prostate cancer cell lines. ChIP analysis revealed an elevation in HAT activity, suggesting an increase in transcription level and gene activation after treatment with genistein (Majid et al., 2008). Genistein, eqoul, and AglyMax induced ER-mediated core histone acetylation via modulating the activity of HATs, and daidzein stimulated Erβ mediated histone acetylation (Hong et al., 2004).
Triple-negative breast cancer is considered the most aggressive subtype of breast cancer. An in vitro study on the effect of indole-3-carbinol on HCC70 triple-negative breast cancer cell lines has shown to inhibit the overall HDAC activity (Nouriemamzaden et al., 2020).
PEITC treatment of LNCaP prostate cancer cell line significantly reduced the protein levels of HDAC1, 2, 4, and 6 that correlated with promoter demethylation and activation of a tumor suppressor gene, RASSF1A (Boyanapalli et al., 2016). Histone hypoacetylation due to excessive HDAC activity is one of the hallmarks of leukemia. Mononuclear extracts from the bone marrow of acute myeloid leukemia patients showed limited or no H3 and H4 histone acetylation. However, after treatment with phenylhexyl isothiocyanate (PHI), there was a significant elevation in H3 and H4 histone acetylation compared to the control cultures (Xiao et al., 2010).
In HL-60 leukemia cancer cell lines, quercetin treatment induced FasL expression that triggered extrinsic apoptotic pathway, protein activation and conformational changes, and activation of ERK and JNK signaling pathways. Furthermore, H3 acetylation was found to be increased in quercetin treated HL-60 cells, and upregulated HAT activity and downregulated HDAC activity together resulted in stimulation of FasL expression (Lee et al., 2011). Leukemia cancer cell lines when treated with quercetin showed an increase in global histone acetylation of H3 and H4 histones. Promoter regions of proapoptotic genes experienced a three-to ten-fold increase in H3 and H4 acetylation in HL-60 and U937 cancer cells. In vitro and in vivo studies of two human xenograft myeloid leukemia models exhibited a decrease in HDAC1 and 2 activity after treatment with quercetin (Alvarez et al., 2018). Quercetin treatment significantly reduced the HDAC activity in a dose-dependent manner and HMT activity at H3 histone, which may methylate and trimethylate the ninth lysine residue in HeLa cancer cell lines. Molecular docking results of quercetin reported a decline in HDAC activity, suggesting that quercetin can competitively inhibit HDAC2, HDAC4, HDAC7, and HDAC8 by binding to the catalytic residue sites (Kedhari Sundaram et al., 2019).
In breast cancer cell lines, resveratrol decreases the protein levels of arginine methyltransferase PRMT5, lysine methyltransferase EZH2, and lysine deacetylase KDAC in a dose- and time-dependent manner that correlated with an increase in protein levels of BRAC1, p21, and p53 genes. After treatment with resveratrol, near to the proximity of the transcription start site of the mentioned genes, the levels of H4R3me2s and H3K27me3 were found to be decreased while that of H3K9ac and H3K27ac increased (Chatterjee et al., 2019). In human carcinoma cell line, resveratrol treatment increase the protein levels of acH3K9, acH3K14, acH3K12, acH4K5, and acH4K16, suggesting anti-tumor effects of resveratrol (Dai et al., 2020).
In human malignant melanoma cells, sulforaphane, decreases the protein expression levels of HDAC1, 2, 4, and 6. Sulforaphane also decreases the total HDAC activity and protein expression levels of CBP, CBP/p300, and PCAF. Furthermore, the protein expression levels of acH3K9, 14, and 27, and acH3K8 and 12 were significantly reduced after treatment with sulforaphane. The study also revealed a decrease HMT activity of SET7/9, further affecting the methylation at K9, 36, and 79 when treated with sulforaphane (Mitsiogianni et al., 2020). TERT (Telomerase Reverse Transcriptase) is associated with processes such as cell proliferation, senescence, cell differentiation, etc., and any alteration in it contributes to immortality and carcinogenesis. Sulforaphane treatment suppressed HDAC activity in prostate cancer cells and induced an increase in pan-acetylation of H3 and H4 histones of the hTERT promoter (Abbas et al., 2015).
Furthermore, the histone modulation activity of various phytocompounds like diallyl disulfide, garcinol, ginsenoside Rh2, phenyl hexyl isothiocyanate, rosmarinic acid, etc., in distinct cancer types is mentioned in Table 3.
Based on the studies mentioned above, it is evident that a wide range of phytocompounds (apigenin, curcumin, sulforaphane, resveratrol, genistein, quercetin, etc.) possess the ability to target major histone modifications that regulate gene expression of apoptosis, cell proliferation and inflammatory pathways (extensively discussed in section 6), all of which, when dysregulated can lead to carcinogenesis.
Non-coding RNAs without protein or peptide-coding potential are classified into two major categories: long ncRNA (200 nucleotides long) and short ncRNA that consists of miRNAs, siRNA, piwiRNA, etc. Non-coding RNAs, initially assumed to be junk in the transcriptome, have now been discovered to play a crucial role in cellular signaling pathways, including those that regulate cancer initiation and progression (Setoyama et al., 2011). miRNAs are single-stranded and 18–20 nucleotides in length that are formed after undergoing complex maturation steps (Figure 3). With the assistance of Drosha (RNase III enzyme) and Pasha (ds-RNA binding endonuclease), the primary miRNA stem-loop structure undergoes numerous modifications, which leaves a 70 nucleotide long pre-miRNA. This pre-miRNA is then transported to the cytoplasm from the nucleus and is subjected to further processing with DICER and TRBP (transactivating response RNA-binding protein) that generates a 22-nucleotide extended miRNA duplex. The duplex associates with the RISC (RNA inducing silencing complex) complex that targets the mRNA for gene regulation (Zhang et al., 2007; Shruti et al., 2011). Several studies have suggested that miRNA can influence cell signaling, regulation, proliferation, and apoptosis by controlling oncogenes and tumor suppressor gene expression (Mishra et al., 2016).
Any alterations during the biogenesis of miRNA or mutation of the factors can have profound implications. A category of miRNAs, termed as epi-miRNAs (epi-miRs), has recently been discovered to modulate the expression of genes encoding epigenetic reader proteins (Dai et al., 2014). Aberrant modulation of epi-miRs can induce epigenetic silencing of tumor suppressor genes or activation of oncogenes, resulting in carcinogenesis. MicroRNAs have been classically categorized as two distinct epi-miRs, namely, OncomiRs and tumor-suppressor miRs that play distinctive roles in tumorigenesis. OncomiRs are generally upregulated, resulting in enhanced cancer cell proliferation and metastasis, whereas tumor-suppressor miRs are downregulated leading to carcinogenesis (Svoronos et al., 2016; Sadakierska-Chudy, 2020). Phytocompounds that are known to alter the epi-miRNAs is the entire effect of the each phytocomponents is supported by a single publication are listed in Table 4.
A significant proportion of studies have demonstrated the potential role of phytocompounds in the regulation of epi-miRs in carcinogenesis. For instance, the apigenin-treated glioma cancer cell line U87 exhibited increased miR-16 expression, decreased BCL2 protein expression, and decreased expression of the MMP-9 gene. Anti-miR-16 transfection inhibited apigenin-induced miR-16 gene expression. Furthermore, anti-miR-16 transfection inhibited apigenin-induced miR-16 gene expression, increased BCL2 protein expression and NF-κB/MMP-9 levels (Chen et al., 2016).
Curcumin induced upregulation of miR-99a in retinoblastoma cancer cell line, SO-Rb50, and Y-79. When the cells were transfected with miR-99a inhibitor, the miR-99a expression decreased that correlated with anti-tumor activity of curcumin through enhancement of miR-99a expression. Phosphorylation levels of JAK1, STAT1, and STAT3 were significantly reduced when treated with curcumin, although no such effect was observed when miR-99a was knocked down (Li et al., 2018). Curcumin-treated breast cancer cell line MCF-10F resulted in decreased gene transcript and protein levels of Axl, Slug, CD24, and Rho-A, which are associated with epithelial-mesenchymal transition (EMT). An increased expression of miR-200a, let-7b, and miR-34a was found in the MCF-10F cancer cell line, while only miR-34a expression increased in the MDA-MB-231 cancer cell line after exposure to curcumin. An increase in expression of the examined genes occurred after the knockdown of miR-34a, while the genes were found downregulated when treated with curcumin and transfected with anti-miR-34a. The invasive and migrating capabilities of MCF-10F decreased when cells were transfected with anti-miR-34a and treated with curcumin (Gallardo et al., 2020).
Ellagitannin treated hepatocarcinoma cells showed a decrease in cell proliferation. The treatment further downregulated and upregulated the expression of various mi-RNAs (Table 4) (Wen et al., 2009).
Hepatocarcinoma cell line HepG2 exhibited decreased growth when treated with EGCG in a dose-dependent manner. miR-18a, miR-34b, miR-193b, miR-222, and miR-342 were found to be downregulated, while miR-16, miR-221, and let-7b were found to be upregulated after treatment with EGCG. Bcl-2 expression was suppressed after transfection with miR-16. Furthermore, transfection with miR-16 enhanced the activity of EGCG in Bcl-2 suppression and induction of apoptosis (Tsang and Kwok, 2010). Nasopharyngeal cancer cell line CNE2 was treated with EGCG where 32 miRNAs exhibited > 2-fold changes that have been shown to modulate cancer development, out of which 29 miRNAs were found to be upregulated and 1 miRNA was downregulated in a dose-dependent manner (Li et al., 2017).
Genistein-treated prostate cancer cell lines, exhibited an increase in miR-574-3p expression compared to the control. Transfection with pre-miR-574-3p miRNA precursors into PCa cell lines led to a significant increase in miR-574-3p expression and decreased cell invasion. In an in vivo study, the transfection of DU145 cells with miR-574-3p subcutaneously into nude mice resulted in tumor suppression due to overexpression of miR-574-3p (Chiyomaru et al., 2013). A significant increase in miR-145 expression was observed in genistein-treated retinoblastoma cancer cell line (Y79). Genistein treatment also reduced cancer cell proliferation and induced apoptosis. Y79 cells transfected with miR-145 specific siRNA resulted in the restoration of colony formation capacity and suppression of cell apoptosis that was induced due to genistein treatment. In silico studies suggested ABCE1 gene to be a potential target of miR-145. Furthermore, an in vivo study on the xenograft nude mice model revealed the suppression of tumor growth in Y79 cells administered with genistein (Wei et al., 2017).
Treatment with lycopene induced upregulation of miR-let-7f-1 in a dose and time-dependent manner in prostate cancer cells. Furthermore, transfection with miR mimics led to inhibition of cell proliferation and apoptosis induction (Li et al., 2016).
PEITC treated prostate cancer cell lines exhibited an increase in the expression of miR-194. Furthermore, the expression of the two matrix metalloproteinase, MMP2 and MMP9, which significantly contributes to tumor progression in terms of migration, invasion, and metastasis, decreased when treated with PEITC. An in silico study revealed BMP1 is a potential target of miR-194 and its inhibition tends to downregulate MMP2 and MMP9 levels (Zhang et al., 2016).
Quercetin-treated oral cancer cells exhibited an increase in miR-16 expression. Cell viability, migration, and invasive capabilities of oral cancer cells were found to be repressed when transfected with miR-16. HomeboxA10 (HOXA10) was found to be targeted by miR-16 after the bioinformatics analysis of the binding sites of HOXA10 and miR-16. HOXA10 is generally involved in the proliferation, invasion, and migration of cancer cells and is one of the potential biomarkers in oral cancer. Overexpressed miR-16 downregulated the protein levels of HOXA10, while the opposite was observed in miR-16 knockdown cells (Zhao et al., 2019). In breast cancer cells, miR-146a expression increased after treatment with quercetin in a dose-dependent manner. Furthermore, growth of the cells was inhibited after transfection with miR-146a mimic and treatment with quercetin. A substantial elevation in the expression of Bax and cleaved caspases was observed when transfected with miR-146a. Quercetin treatment for 8 weeks increased miR-146a expression and reduced tumor growth in a nude mouse orthotopic xenograft model (Tao et al., 2015).
Colorectal cancer cell line HCT-116 exhibited a decrease in cell viability in a dose-and-time-dependent manner when treated with resveratrol. HCT-116 cells transfected with LNA miR inhibitor showed a dramatic decline in miR-200c expression compared with the un-transfected and scrambled groups. After treatment with resveratrol, miR-200c expression increased significantly in both transfected and un-transfected cells. Resveratrol treatment reduced the mRNA and protein expression of vimentin and ZEB1, while that of E-cadherin increased in both groups that correlated with EMT induction (Karimi Dermani et al., 2017).
Rosmarinic acid decreased cell viability, cell proliferation, invasion, and migration and suppressed EMT while promoting apoptosis in pancreatic cancer cell lines. Further treatment resulted in increased miR-506 levels that correlated with suppression of MMP2 and MMP16 proteins. The xenograft mouse model also exhibited a reduction in tumor growth after treatment with rosmarinic acid (Han et al., 2019).
Sulforaphane-treated pancreatic cancer cell lines showed a substantial increase in miR135b-5p. Cells transfected with liposomes of miR-135b-5p mimics exhibited overexpression of miR-135b-3p that resulted in reduced cell viability, migration, and colony-forming capacity. In silico analysis revealed RASAL2 to be a potential target of miR-135b-3p. Furthermore, an in vivo studies on tumor xenografts, where BxPC-3 cells were transfected with miR-135b-3p mimics, resulted in decreased tumor size that correlated with overexpression of miR-135b-3p and RASAL2 (Yin et al., 2019). In colorectal cancer cell, sulforaphane treatment inhibited oncogenic miR-21, decreased cell viability, induced apoptosis, and downregulated the expression of hTERT (Martin et al., 2017).
In human glioblastoma cells, miR-21 levels were increased that has been shown to target a positive regulator of apoptosis, the PDCD4 gene. Ursolic acid treatment decreased cell proliferation and induced apoptosis while suppressing the levels of miR-21 that eventually led to enhanced expression of PDCD4 (Wang et al., 2012).
All these studies discuss the therapeutic effect of phytocompounds targeting epi-miRs that either decreases or increases miRNA level to eventually reduce tumorigenesis. An elaborate list of phytocompounds modulating miRNA in specific cancer cell types is provided in Table 4.
As discussed above, aberrant epigenetic modulation can alter the function and activity of typical genes and transcriptional factors involved in various essential pathways like autophagy, apoptosis inflammation, etc., that can lead to cancer.
Autophagy plays a dual role in cancer by promoting tumorigenesis or suppressing tumor progression. To satisfy the high metabolic requirements of proliferating tumor cells, autophagy recycles the cells’ intracellular constituents to supply nutrients, while suppression or inhibition of autophagy genes may result in cancer cell death. Autophagy also promotes inflammation in tumor cells, which eventually results in tumor progression (Yun and Lee, 2018). Post-translational modifications like acetylation, phosphorylation, ubiquitination, nitrosylation can influence autophagy by regulating the activity of ATG proteins and the expression of genes involved in autophagy (Botti-Millet et al., 2016). As described above, many studies have shown that deregulation of ATG genes through different epigenetic modulations can cause tumor progression. For instance, curcumin treatment inhibited DNMT1 and DNMT3B expression in prostate cancer cells that resulted in promoter hypomethylation of miR-143 and miR-145. This restoration of miRNAs further downregulated ATG2B expression, thus inhibiting autophagy (Liu et al., 2017).
Dysregulation of apoptosis is one of the hallmarks of cancer. Cancer cells tend to survive longer and accumulate mutations due to deregulation of apoptosis over a period of time. Furthermore, deviations from normal apoptotic pathways can enhance the invasiveness of cancer cells and promote angiogenesis. In most cancer cells, BCL-2 is generally overexpressed while the function of caspases is found to be disabled (Pfeffer and Singh, 2018). As mentioned above, emerging studies have shown that phytocompounds can correct epigenetic modulations that interfere with apoptotic pathways that result in cancer progression.
Excessive aggregation of reactive oxygen species (ROS) has been observed in cancer. ROS is involved in inflammation, cell transformation, tumor cell survival, proliferation, invasion, angiogenesis, and metastasis, mediated through transcription factors such as NF-κB, AP-1, STAT3, etc. (Gupta et al., 2012; Prasad et al., 2017). Furthermore, the overexpression of MMPs can be correlated with enhanced invasiveness and angiogenesis in distinct cancer types (Reunanen and Kähäri, 2013). Besides this, ROS can also regulate the expression of multiple tumor suppressor genes like p53, Nrf2, PTEN, and Rb (Gupta et al., 2012). Nrf2 plays a crucial role in homeostasis maintenance and regulation of genes that produce anti-inflammatory and anti-cancer effects (Wu et al., 2019). Additionally, it is evident from the studies mentioned that abnormal epigenetic modulation can alter the function and activity of transcription factors and genes involved in the production of ROS in cancer. However, distinct phytocompounds (curcumin, resveratrol, berberine, luteolin, etc.) as mentioned above, possess the ability to reverse the activity of all these transcription factors and genes involved in the accumulation of ROS back to normal, which have been implicated in abnormal epigenetic modulation.
Therefore, it is evident from all these studies and observations that autophagy, apoptosis, inflammation, and oxidative stress play a crucial role in cancer. This warrants additional studies to understand the underlying mechanisms of genes, transcription factors, and pathways implicated in cancer. Furthermore, these implications can also be used as biomarkers in the diagnosis of distinct cancer types.
Most in vitro and in vivo studies suggested the use of high concentrations of phytocompounds for tumor growth suppression and chemoprevention. However, in humans, certain elevated levels of these phytocompounds may not be reached due to their poor bioavailability and may lack therapeutic efficacy. Studies also show that such high doses of phytocompounds over a prolonged duration may exhibit high toxicity. For instance, resveratrol was administered to rats for 4 weeks in a dose-dependent manner. Serious side effects such as increased kidney weight, increased plasma BUN, and creatinine levels, which gradually contributed to nephrotoxicity, were observed at a concentration of 3,000 mg resveratrol per kg body weight of rats (Crowell et al., 2004). In another study on rat thymocytes, resveratrol at 10 µM concentration raised the shrunken cell population and exerted cytotoxic effects on normal cells by inducing apoptosis (Fujimoto et al., 2009). Despite evidence indicating the antioxidant activity of curcumin, several studies also demonstrated the pro-oxidant activity of curcumin, increasing ROS levels in cells (Yoshino et al., 2004; Su et al., 2006). The pro-oxidant nature of curcumin would increase cellular ROS levels at higher doses, potentially contributing to carcinogenesis (López-Lázaro, 2008). Furthermore, excessive quercetin intake exacerbates tumorigenicity induced by a chemical carcinogen N-ethyl-N′-nitro-N-nitrosoguanidine (ENNG), in the duodenum of mice (Matsukawa et al., 2002). While the immense health-promoting benefits of epigallocatechin-3-gallate, a study demonstrated that high dose administration of EGCG in mice resulted in hepatotoxicity correlated with inhibition of antioxidant enzymes and Nrf2 targeted genes (Wang et al., 2015). Treatment of female CD-1 mice with genistein at environmentally appropriate doses resulted in irregular estrous cycles, early reproductive senescence, impaired ovarian activity, diminished fertility, while at higher doses, the number of stillbirths increased (Jefferson et al., 2005). In an in vivo study on Swiss mice, administration of higher doses (100 and 200 mg/kg) of apigenin resulted in elevated serum levels of alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, and reactive oxygen species that eventually contributed to liver damage (Singh et al., 2012).
In addition to this, a variety of phytocompounds that humans have consumed for decades have been shown to possess carcinogenic properties. For example, capsaicin, cycasin, phytoestrogens, safrole, amygdalin, phorbol esters, pyrrolizidine alkaloids, obtained from different dietary sources, may serve as potential carcinogens or promoters of tumors (Bode and Dong, 2015; Guldiken et al., 2018).
Several experiments have been performed with numerous phytocompounds to examine their possible positive and detrimental biological effects. However, all natural substances should not be considered healthy and attention should also be given to their toxic dose-related effects. Furthermore, all these observations warrant additional studies and humanized clinical trials regarding the adverse and chronic effects of high toxic doses of distinct phytocompounds.
Phytocompounds disrupt the process of epigenetic transformation in cancer by directly inhibiting epigenetic modulations and also by modulating epigenetic regulators. As more evidence emerges highlighting the therapeutic importance of epigenetic modifications of cancer and the ability of phytocompounds to target these modifications further endorses their clinical relevance. The potential of phytocompounds already known to have anticancer effects to target epigenetics should be thoroughly assessed and may be used as a criterion for inclusion of compounds for further clinical evaluation. For this context, we review a few plant-based compounds known to suppress epigenetic modifications that are currently in various phases of clinical studies below.
Several preclinical studies have demonstrated the efficacy of phytocompounds in regulating epigenetic changes for chemotherapeutic purposes, there have been insufficient clinical trials to back this up. For instance, curcumin regulated the activity of MMP-2, Bcl-2, Nrf-2, Bax, and PIK3/AkT signaling in a randomized clinical trial to study the effects of paclitaxel and curcumin combined in breast cancer to mitigate multidrug resistance. In patients with advanced breast cancer, curcumin is under investigation as monotherapy (NCT03980509) or in combination with paclitaxel in phase II clinical trial (NCT03072992). The main object of these clinical studies is to determine the effect of curcumin on the development of advanced breast cancer and to estimate the risk of adverse effects. In a phase II study of the effect of sulforaphane-rich extracts in men with recurrent cancer, 20 subjects were treated with 200 µmoles/day of sulforaphane extract for 20 weeks. Out of the 20 subjects, six showed an increase in histone acetylation following sulforaphane treatment (Alumkal et al., 2015). In an ongoing randomized pilot study in phase I clinical trial investigating the impact of quercetin on EGCG uptake in prostate cancer, the downregulating effects of quercetin on enzyme function and protein and gene expression of COMT (catechol-O-methyltransferase) and DNMT1 are being assessed (NCT01912820). Treatment with 3,3′-Diindolylmethane enhanced the expression of let-7, miR-27b, miR-34a, miR-124, miR-200, and miR-320, which led to downregulation of androgen receptor activity, EMT, and stem cell markers, all of which were associated with enzalutamide resistance in Castration-resistant prostate cancer (Li and Sarkar, 2016).
Additionally, these natural compounds can be categorized in to 3 different phases as per their success in treating cancer are compounds under preclinical trials, clinical trials and those are used in current cancer therapy. Phytocompounds those are in pre-clinical trials are ursolic acid (Prasad et al., 2012; Zhang et al., 2018), withaferin A (Choi and Kim, 2015; Suman et al., 2016; Kuppusamy et al., 2017), curcumin (Kunnumakkara et al., 2017), baicalein (Dou et al., 2018; Tao et al., 2018), EGCG (Thangapazham et al., 2007), apigenin (Chang et al., 2018; Yan et al., 2018), genistein (Zhang et al., 2013; Hsiao et al., 2019), resveratrol (Banerjee et al., 2002), sulphorane (Qazi et al., 2010), thymoquinone (Zhu et al., 2016), etc. Phytocompounds that go for clinical trial focus on three major aspects of cancer research: 1) improving the response of cancer cells towards standard chemo- and radiotherapy, 2) reducing the severe adverse effects of standard cancer therapy, and 3) looking for unwanted interactions with standard therapy. Preclinical studies have shown the effectiveness of various phytochemicals as mentioned above. The phytochemicals which are currently under clinical trials against various cancers are Berberine (NCT03281096), curcumin (NCT03072992), EGCG (NCT02891538), lycopene (NCT03167268), quercetin (NCT01912820), resveratrol (NCT01476592) and sulphorane (NCT03232138). Several compounds are in clinical use are vincristine, vinblastine, paclitaxel, etoposide to name a few. This underscores the significance of natural chemicals and the imperative to further investigate their potential in developing the most efficacious and secure pharmaceutical interventions for cancer therapy.
Research on cancer-fighting phytocompounds is still in their infancy since only a small number of phytocompounds (such as paclitaxel, docetaxel, and vinblastine) have been granted clinical use licenses. With an increasing understanding of the range of anticancer effects of plant-based compounds, such as inhibition of cancer epigenetics, there is an urgent need to screen more therapeutically effective phytocompounds. In addition, no clinical investigation has been conducted to date to assess how phytocompounds modify micro RNAs in the context of cancer epigenetics therapy. In the majority of related clinical studies, methodological flaws such as the absence of a control or placebo group, small sample sizes, and brief trial duration are observed. Therefore, for many phytochemicals, it is too early to conclude their anticancer actions and hence large-scale and well-controlled clinical trials are required to validate their efficacies, adverse effects, and safeties before their use for the treatment of cancer. To achieve the international standard, promising phytochemicals require extensive standardization in terms of methods for evaluating their bioavailability, efficacy, safety, quality, composition, manufacturing processes, regulatory and approval practices. In order to enhance the clinical assessment of phytocompounds, we must establish an evaluation pipeline. A methodology that takes into account drug optimization, effectiveness assessment, tissue toxicity and distribution, chemical accessibility, pharmacokinetics, absorption, and most importantly bioavailability should be created for an enhanced evaluation of phytocompounds. The stability and availability of phytocompounds in blood can be improved by using stable synthetic analogues, chemically modified derivatives, micelle-coated medications, liposomal conjugates, phospholipid complexes, adjuvants, and nanoparticles. The effectiveness of plant-based medicines can also be augmented by using other techniques including structure-activity relationship, directed optimization, and pharmacophore-oriented molecular design (Xiao et al., 2016; Atanasov et al., 2021; Mohammadi et al., 2022).
Epigenetic aberrations remarkably contribute to cancer incidence. As mentioned above, distinct studies highlighted the potential of phytocompounds in preventing tumorigenesis through regulation of epigenetic modulation by targeting the activity and expression of DNMTs, HDACs, HMTs, epi-miRNAs. The dietary phytocompounds analyzed tend to modulate epigenetic modifications in vitro and in some in vivo cancer models, thus inhibiting or suppressing cancer cell viability, proliferation, and growth. However, the limited number of studies and insufficient preclinical and clinical data on the effect of phytocompounds on the epigenetic landscape still remains a challenge. Future research should focus on clinical studies regarding the optimal dose and duration of phytocompounds as epidrugs. One of the studies exhibited that a low dietary dose of resveratrol compared with a 200-fold higher dose tends to suppress colorectal cancer development in human and mice tissues (Cai et al., 2015). This result indicates that a low dose of phytocompounds can inhibit tumor progression, thus making it essential to analyze the optimal dose and toxicity of the phytocompounds. To develop anticancer therapeutics, the issue of poor bioavailability of phytocompounds needs to be addressed. Furthermore, significant epidemiological studies revealed that phytocompounds interact with other bioactive compounds that may interfere with their intestinal absorption (Phan et al., 2018). A promising approach to overcome these challenges is the utilization of modern drug delivery systems such as nanoparticles, micelles, liposomes, etc., to enhance bioavailability and overcome systemic toxicity (Aqil et al., 2013). Accumulating shreds of evidence have also demonstrated the synergistic effects of various phytochemicals with chemotherapeutic drugs to be more effective in chemoprevention and cure (Tan and Norhaizan, 2019). However, research regarding the mechanism and course of action of these combined phytocompounds and drugs is still in their infancy. More studies are essential to fully comprehend the mode of action of phytocompounds that have been shown to target a single cell type or are tissue/organ-specific. Besides in vivo and in vitro studies, mechanistic studies using bioinformatics and high-throughput sequencing methods can help us to better understand and target altered epigenetic modifications in cancer. Dietary phytocompounds (as listed in Tables 2–4) may offer a cost-effective method for chemoprevention, hence improving global health by decreasing the incidence of cancer. Numerous shreds of evidence point to the potential of phytocompounds to target aberrant epigenetic alterations in different forms of cancer. As was already established, genistein functions as a DNMT inhibitor, promotes histone acetylation, and increases levels of miRNAs, all of which contribute to therapeutic impact on prostate cancer. Such phytochemicals need to undergo a thorough evaluation for preclinical and clinical investigations taking into consideration their therapeutic potential in the treatment of cancer. Our study concludes by highlighting the potential of natural compounds in addressing the epigenetic vulnerabilities of cancer, as well as the possible therapeutic benefits that can be identified by advancing our knowledge in this field.
AqK: Writing–review and editing, Conceptualization, Investigation, Writing–original draft. AsK: Conceptualization, Investigation, Writing–original draft, Writing–review and editing. MK: Conceptualization, Investigation, Writing–review and editing, Formal Analysis. ZM: Conceptualization, Formal Analysis, Investigation, Writing–review and editing. SM: Conceptualization, Formal Analysis, Writing–review and editing. RP: Conceptualization, Formal Analysis, Writing–review and editing, Investigation. SM: Conceptualization, Formal Analysis, Investigation, Writing–review and editing. SH: Writing–review and editing, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AK acknowledges the support provided by Department of Bioscience, Jamia Millia Islamia. SH acknowledges the support provided by the Department of Bioscience, Faculty of Natural Sciences, Jamia Millia Islamia. AS is grateful to Ajman University, UAE for supporting this publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbas, A., Hall, J. A., Patterson, W. L., Ho, E., Hsu, A., Al-Mulla, F., et al. (2015). Sulforaphane modulates telomerase activity via epigenetic regulation in prostate cancer cell lines. Biochem. Cell Biol. 94 (1), 71–81. doi:10.1139/bcb-2015-0038
Adjakly, M., Bosviel, R., Rabiau, N., Boiteux, J. P., Bignon, Y. J., Guy, L., et al. (2011). DNA methylation and soy phytoestrogens: quantitative study in DU-145 and PC-3 human prostate cancer cell lines. Epigenomics 3, 795–803. doi:10.2217/epi.11.103
Aggarwal, B. B., and Shishodia, S. (2006). Molecular targets of dietary agents for prevention and therapy of cancer. Biochem. Pharmacol. 71, 1397–1421. doi:10.1016/j.bcp.2006.02.009
Aggarwal, R., Jha, M., Shrivastava, A., and Jha, A. K. (2015). Natural compounds: role in reversal of epigenetic changes. Biochem. 80 (8), 972–989. doi:10.1134/S0006297915080027
Altaf, M. M., Ahmad Khan, M. S., and Ahmad, I. (2018). “Diversity of bioactive compounds and their therapeutic potential,” in New look to phytomedicine: advancements in herbal products as novel drug leads (Elsevier), 15–34.
Alumkal, J. J., Slottke, R., Schwartzman, J., Cherala, G., Munar, M., Graff, J. N., et al. (2015). A phase II study of sulforaphane-rich broccoli sprout extracts in men with recurrent prostate cancer. Invest. New Drugs 33 (2), 480–489. doi:10.1007/s10637-014-0189-z
Alvarez, M. C., Maso, V., Torello, C. O., Ferro, K. P., and Saad, S. T. O. (2018). The polyphenol quercetin induces cell death in leukemia by targeting epigenetic regulators of pro-apoptotic genes. Clin. Epigenetics 10 (1), 139. doi:10.1186/s13148-018-0563-3
Al-Yousef, N., Shinwari, Z., Al-Shahrani, B., Al-Showimi, M., and Al-Moghrabi, N. (2020). Curcumin induces Re-expression of BRCA1 and suppression of γ synuclein by modulating DNA promoter methylation in breast cancer cell lines. Oncol. Rep. 43 (3), 827–838. doi:10.3892/or.2020.7473
Aqil, F., Munagala, R., Jeyabalan, J., and Vadhanam, M. V. (2013). Bioavailability of phytochemicals and its enhancement by drug delivery systems. Cancer Lett. 28, 133–141. doi:10.1016/j.canlet.2013.02.032
Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., Orhan, I. E., Banach, M., Rollinger, J. M., et al. (2021). Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 20, 200–216. doi:10.1038/s41573-020-00114-z
Audia, J. E., and Campbell, R. M. (2016). Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8 (4), a019521. doi:10.1101/cshperspect.a019521
Bailon-Moscoso, N., Cevallos-Solorzano, G., Romero-Benavides, J., and Ramirez Orellana, M. (2017). Natural compounds as modulators of cell cycle arrest: application for anticancer chemotherapies. Curr. Genomics 18, 106–131. doi:10.2174/1389202917666160808125645
Banerjee, S., Bueso-Ramos, C., and Aggarwal, B. B. (2002). Suppression of 7,12-dimethylbenz(a)anthracene-induced mammary carcinogenesis in rats by resveratrol: role of nuclear factor-kappaB, cyclooxygenase 2, and matrix metalloprotease 9. Cancer Res. 62 (17), 4945–4954.
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi:10.1038/cr.2011.22
Bannister, A. J., Schneider, R., and Kouzarides, T. (2002). Histone methylation: dynamic or static? Cell 109 (7), 801–806. doi:10.1016/s0092-8674(02)00798-5
Bashir, A. O., El-Mesery, M. E., Anwer, R., and Eissa, L. A. (2020). Thymoquinone potentiates MiR-16 and MiR-375 expressions in hepatocellular carcinoma. Life Sci., 254. doi:10.1016/j.lfs.2020.117794
Baumgart, S. J., and Haendler, B. (2017). Exploiting epigenetic alterations in prostate cancer. Int. J. Mol. Sci. 18, 1017. doi:10.3390/ijms18051017
Baylin, S. B., Esteller, M., Rountree, M. R., Bachman, K. E., Schuebel, K., and Herman, J. G. (2001). Abberant patterns of DNA methylation, chromatin formation and gene expression in cancer. Human molecular genetics. Hum. Mol. Genet. 10, 687–692. doi:10.1093/hmg/10.7.687
Berger, A., Venturelli, S., Kallnischkies, M., Böcker, A., Busch, C., Weiland, T., et al. (2013). Kaempferol, a new nutrition-derived pan-inhibitor of human histone deacetylases. J. Nutr. Biochem. 24 (6), 977–985. doi:10.1016/j.jnutbio.2012.07.001
Berson, A., Nativio, R., Berger, S. L., and Bonini, N. M. (2018). Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 41, 587–598. doi:10.1016/j.tins.2018.05.005
Bestor, T. H. (2000). The DNA methyltransferases of mammals. Hum. Mol. Genet. 9 (16), 2395–2402. doi:10.1093/hmg/9.16.2395
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16 (1), 6–21. doi:10.1101/gad.947102
Bode, A. M., and Dong, Z. (2015). Toxic phytochemicals and their potential risks for human cancer. Cancer Prev. Res. 1, 1–8. doi:10.1158/1940-6207.CAPR-14-0160
Bora-Tatar, G., Dayangaç-Erden, D., Demir, A. S., Dalkara, S., Yelekçi, K., and Erdem-Yurter, H. (2009). Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: activity and docking studies. Bioorg. Med. Chem. 17 (14), 5219–5228. doi:10.1016/j.bmc.2009.05.042
Botti-Millet, J., Nascimbeni, A. C., Dupont, N., Morel, E., and Codogno, P. (2016). Fine-tuning autophagy: from transcriptional to posttranslational regulation. Am. J. Physiol. Physiol. 311 (3), C351–C362. doi:10.1152/ajpcell.00129.2016
Boyanapalli, S. S. S., Li, W., Fuentes, F., Guo, Y., Ramirez, C. N., Gonzalez, X. P., et al. (2016). Epigenetic reactivation of RASSF1A by phenethyl isothiocyanate (PEITC) and promotion of apoptosis in LNCaP cells. Pharmacol. Res. 114, 175–184. doi:10.1016/j.phrs.2016.10.021
Cai, H., Scott, E., Kholghi, A., Andreadi, C., Rufini, A., Karmokar, A., et al. (2015). Cancer chemoprevention: evidence of a nonlinear dose response for the protective effects of resveratrol in humans and mice. Sci. Transl. Med. 7 (298), 298ra117. doi:10.1126/scitranslmed.aaa7619
Chang, J. H., Cheng, C. W., Yang, Y. C., Chen, W. S., Hung, W. Y., Chow, J. M., et al. (2018). Downregulating CD26/DPPIV by apigenin modulates the interplay between akt and snail/slug signaling to restrain metastasis of lung cancer with multiple EGFR statuses. J. Exp. Clin. Cancer Res. 37 (1), 199. doi:10.1186/s13046-018-0869-1
Chatterjee, B., Ghosh, K., and Kanade, S. R. (2019). Resveratrol modulates epigenetic regulators of promoter histone methylation and acetylation that restores BRCA1, P53, P21CIP1 in human breast cancer cell lines. BioFactors 45 (5), 818–829. doi:10.1002/biof.1544
Chen, X. J., Wu, M. Y., Li, D. H., and You, J. (2016). Apigenin inhibits glioma cell growth through promoting MicroRNA-16 and suppression of BCL-2 and nuclear factor-ΚB/MMP-9. Mol. Med. Rep. 14 (3), 2352–2358. doi:10.3892/mmr.2016.5460
Chiyomaru, T., Yamamura, S., Fukuhara, S., Hidaka, H., Majid, S., Saini, S., et al. (2013). Genistein up-regulates tumor suppressor MicroRNA-574-3p in prostate cancer. PLoS One 8 (3), e58929. doi:10.1371/journal.pone.0058929
Choi, B. Y., and Kim, B.-W. (2015). Withaferin-A inhibits colon cancer cell growth by blocking STAT3 transcriptional activity. J. Cancer Prev. 20 (3), 185–192. doi:10.15430/JCP.2015.20.3.185
Chung, F. F. L., and Herceg, Z. (2020). The promises and challenges of toxico-epigenomics: environmental chemicals and their impacts on the epigenome. Environ. Health Perspect. 128, 15001. doi:10.1289/EHP6104
Crowell, J. A., Korytko, P. J., Morrissey, R. L., Booth, T. D., and Levine, B. S. (2004). Resveratrol-associated renal toxicity. Toxicol. Sci. 82 (2), 614–619. doi:10.1093/toxsci/kfh263
Dai, E., Yu, X., Zhang, Y., Meng, F., Wang, S., Liu, X., et al. (2014). EpimiR: a database of curated mutual regulation between MiRNAs and epigenetic modifications. Database 2014, bau023. doi:10.1093/database/bau023
Dai, L., Chen, L., Wang, W., and Lin, P. (2020). Resveratrol inhibits ACHN cells via regulation of histone acetylation. Pharm. Biol. 58 (1), 231–238. doi:10.1080/13880209.2020.1738503
Dawood, M., Ooko, E., and Efferth, T. (2019). Collateral sensitivity of parthenolide via NF-κB and HIF-α inhibition and epigenetic changes in drug-resistant cancer cell lines. Front. Pharmacol. 10, 542. doi:10.3389/fphar.2019.00542
Dehan, P., Kustermans, G., Guenin, S., Horion, J., Boniver, J., and Delvenne, P. (2009). DNA methylation and cancer diagnosis: new methods and applications. Expert Rev. Mol. Diagnostics 9, 651–657. doi:10.1586/erm.09.53
Dittharot, K., Dakeng, S., Suebsakwong, P., Suksamrarn, A., Patmasiriwat, P., and Promkan, M. (2019). Cucurbitacin B induces hypermethylation of oncogenes in breast cancer cells. Planta Med. 85 (5), 370–378. doi:10.1055/a-0791-1591
Donovan, M. G., Selmin, O. I., Doetschman, T. C., and Romagnolo, D. F. (2019). Epigenetic activation of BRCA1 by genistein in vivo and triple negative breast cancer cells linked to antagonism toward aryl hydrocarbon receptor. Nutrients 11 (11), 2559. doi:10.3390/nu11112559
Dou, J., Wang, Z., Ma, L., Peng, B., Mao, K., Li, C., et al. (2018). Baicalein and baicalin inhibit colon cancer using two distinct fashions of apoptosis and senescence. Oncotarget 9 (28), 20089–20102. doi:10.18632/oncotarget.24015
Druesne, N., Pagniez, A., Mayeur, C., Thomas, M., Cherbuy, C., Duée, P. H., et al. (2004). Diallyl disulfide (DADS) increases histone acetylation and P21waf1/cip1 expression in human colon tumor cell lines. Carcinogenesis 25, 1227–1236. doi:10.1093/carcin/bgh123
Fang, L., Xu, W., and Kong, D. (2019). Icariin inhibits cell proliferation, migration and invasion by down-regulation of MicroRNA-625-3p in thyroid cancer cells. Biomed. Pharmacother. 109, 2456–2463. doi:10.1016/j.biopha.2018.04.012
Fang, M., Chen, D., and Yang, C. S. (2007). Dietary polyphenols may affect DNA methylation. J. Nutr. 137 (1), 223S-228S–228S. doi:10.1093/jn/137.1.223S
Fu, L. J., Ding, Y. B., Wu, L. X., Wen, C. J., Qu, Q., Zhang, X., et al. (2014). The effects of lycopene on the methylation of the GSTP1 promoter and global methylation in prostatic cancer cell lines PC3 and LNCaP. Int. J. Endocrinol. 2014, 620165. doi:10.1155/2014/620165
Fujimoto, A., Sakanashi, Y., Matsui, H., Oyama, T., Nishimura, Y., Masuda, T., et al. (2009). Cytometric analysis of cytotoxicity of polyphenols and related phenolics to rat thymocytes: potent cytotoxicity of resveratrol to normal cells. Basic Clin. Pharmacol. Toxicol. 104 (6), 455–462. doi:10.1111/j.1742-7843.2009.00386.x
Gallardo, M., Kemmerling, U., Aguayo, F., Bleak, T. C., Muñoz, J. P., and Calaf, G. M. (2020). Curcumin rescues breast cells from epithelial-mesenchymal transition and invasion induced by anti-miR-34a. Int. J. Oncol. 56, 480–493. doi:10.3892/ijo.2019.4939
Gama-Sosa, M. A., Slagel, V. A., Trewyn, R. W., Oxenhandler, R., Kuo, K. C., Gehrke, C. W., et al. (1983). The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 11 (19), 6883–6894. doi:10.1093/nar/11.19.6883
Gonelimali, F. D., Lin, J., Miao, W., Xuan, J., Charles, F., Chen, M., et al. (2018). Antimicrobial properties and mechanism of action of some plant extracts against Food pathogens and spoilage microorganisms. Front. Microbiol. 9, 1639. doi:10.3389/fmicb.2018.01639
Guldiken, B., Ozkan, G., Catalkaya, G., Ceylan, F. D., Ekin Yalcinkaya, I., and Capanoglu, E. (2018). Phytochemicals of herbs and spices: health versus toxicological effects. Food Chem. Toxicol. 119, 37–49. doi:10.1016/j.fct.2018.05.050
Guo, J., Mei, Y., Li, K., Huang, X., and Yang, H. (2016). Downregulation of MiR-17-92a cluster promotes autophagy induction in response to celastrol treatment in prostate cancer cells. Biochem. Biophys. Res. Commun. 478 (2), 804–810. doi:10.1016/j.bbrc.2016.08.029
Gupta, S. C., Hevia, D., Patchva, S., Park, B., Koh, W., and Aggarwal, B. B. (2012). Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxidants Redox Signal. 1, 1295–1322. doi:10.1089/ars.2011.4414
Han, X., Liu, C. F., Gao, N., Zhao, J., and Xu, J. (2018). RETRACTED: kaempferol suppresses proliferation but increases apoptosis and autophagy by up-regulating microRNA-340 in human lung cancer cells. Biomed. Pharmacother. 108, 809–816. doi:10.1016/j.biopha.2018.09.087
Han, Y., Ma, L., Zhao, L., Feng, W., and Zheng, X. (2019). Rosmarinic inhibits cell proliferation, invasion and migration via up-regulating MiR-506 and suppressing MMP2/16 expression in pancreatic cancer. Biomed. Pharmacother. 115, 108878. doi:10.1016/j.biopha.2019.108878
Hang, S., Wang, X., and Li, H. (2019). Triptolide inhibits viability and migration while promotes apoptosis in nephroblastoma cells by regulation of MiR-193b-3p. Exp. Mol. Pathol. 108, 80–88. doi:10.1016/j.yexmp.2019.04.006
Henikoff, S., and Matzke, M. A. (1997). Exploring and explaining epigenetic effects. Trends Genet. 13 (8), 293–295. doi:10.1016/s0168-9525(97)01219-5
Hong, T., Nakagawa, T., Pan, W. J., Kim, M. Y., Kraus, W. L., Ikehara, T., et al. (2004). Isoflavones stimulate estrogen receptor-mediated core histone acetylation. Biochem. Biophys. Res. Commun. 317 (1), 259–264. doi:10.1016/j.bbrc.2004.03.041
Hsiao, Y. C., Peng, S. F., Lai, K. C., Liao, C. L., Huang, Y. P., Lin, C. C., et al. (2019). Genistein induces apoptosis in vitro and has antitumor activity against human leukemia HL-60 cancer cell xenograft growth in vivo. Environ. Toxicol. 34 (4), 443–456. doi:10.1002/tox.22698
Issa, J. P. J., and Kantarjian, H. M. (2009). Targeting DNA methylation. Clin. Cancer Res. 15 (12), 3938–3946. doi:10.1158/1078-0432.CCR-08-2783
Jang, S. Y., Hong, D., Jeong, S. Y., and Kim, J. H. (2015). Shikonin causes apoptosis by up-regulating P73 and down-regulating ICBP90 in human cancer cells. Biochem. Biophys. Res. Commun. 465 (1), 71–76. doi:10.1016/j.bbrc.2015.07.131
Jang, Y. G., Hwang, K. A., and Choi, K. C. (2018). Rosmarinic acid, a component of rosemary tea, induced the cell cycle arrest and apoptosis through modulation of HDAC2 expression in prostate cancer cell lines. Nutrients 10, 1784. doi:10.3390/nu10111784
Jefferson, W. N., Padilla-Banks, E., and Newbold, R. R. (2005). Adverse effects on female development and reproduction in CD-1 mice following neonatal exposure to the phytoestrogen genistein at environmentally relevant doses. Biol. Reprod. 73 (4), 798–806. doi:10.1095/biolreprod.105.041277
Jiang, A., Wang, X., Shan, X., Li, Y., Wang, P., Jiang, P., et al. (2015). Curcumin reactivates silenced tumor suppressor gene RARβ by reducing DNA methylation. Phyther. Res. 29, 1237–1245. doi:10.1002/ptr.5373
Kang, J., Chen, J., Shi, Y., Jia, J., and Zhang, Y. (2005). Curcumin-induced histone hypoacetylation: the role of reactive oxygen species. Biochem. Pharmacol. 69 (8), 1205–1213. doi:10.1016/j.bcp.2005.01.014
Kang, K. A., Piao, M. J., Hyun, Y. J., Zhen, A. X., Cho, S. J., Ahn, M. J., et al. (2019). Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with P53 in human colon cancer cells. Exp. Mol. Med. 51 (4), 1–14. doi:10.1038/s12276-019-0238-y
Karimi Dermani, F., Saidijam, M., Amini, R., Mahdavinezhad, A., Heydari, K., and Najafi, R. (2017). Resveratrol inhibits proliferation, invasion, and epithelial–mesenchymal transition by increasing MiR-200c expression in HCT-116 colorectal cancer cells. J. Cell. Biochem. 118 (6), 1547–1555. doi:10.1002/jcb.25816
Kedhari Sundaram, M., Hussain, A., Haque, S., Raina, R., and Afroze, N. (2019). Quercetin modifies 5′CpG promoter methylation and reactivates various tumor suppressor genes by modulating epigenetic marks in human cervical cancer cells. J. Cell. Biochem. 120 (10), 18357–18369. doi:10.1002/jcb.29147
Khan, A., Mohammad, T., Shamsi, A., Hussain, A., Alajmi, M. F., Husain, S. A., et al. (2021b). Identification of plant-based hexokinase 2 inhibitors: combined molecular docking and dynamics simulation studies. J. Biomol. Struct. Dyn. 40, 10319–10331. doi:10.1080/07391102.2021.1942217
Khan, A., Siddiqui, S., Husain, S. A., Mazurek, S., and Iqbal, M. A. (2021a). Phytocompounds targeting metabolic reprogramming in cancer: an assessment of role, mechanisms, pathways, and therapeutic relevance. J. Agric. Food Chem. 16, 6897–6928. doi:10.1021/acs.jafc.1c01173
Kim, D.-Y., Kim, S.-H., Cheong, H.-T., Ra, C.-S., Rhee, K.-J., and Jung, B. D. (2020). Berberine induces P53-dependent apoptosis through inhibition of DNA Methyltransferase3b in Hep3B cells. Korean J. Clin. Lab. Sci. 52 (1), 69–77. doi:10.15324/kjcls.2020.52.1.69
Kornberg, R. D., and Lorch, Y. (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294. doi:10.1016/s0092-8674(00)81958-3
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 23, 693–705. doi:10.1016/j.cell.2007.02.005
Krifa, M., Leloup, L., Ghedira, K., Mousli, M., and Chekir-Ghedira, L. (2014). Luteolin induces apoptosis in BE colorectal cancer cells by downregulating calpain, UHRF1, and DNMT1 expressions. Nutr. Cancer 66 (7), 1220–1227. doi:10.1080/01635581.2014.951729
Kundu, J. K., and Surh, Y. J. (2009). Molecular basis of chemoprevention with dietary phytochemicals: redox-regulated transcription factors as relevant targets. Phytochem. Rev. 8 (2), 333–347. doi:10.1007/s11101-009-9132-x
Kunnumakkara, A. B., Bordoloi, D., Harsha, C., Banik, K., Gupta, S. C., and Aggarwal, B. B. (2017). Curcumin mediates anticancer effects by modulating multiple cell signaling pathways. Clin. Sci. 1, 1781–1799. doi:10.1042/CS20160935
Kuppusamy, P., Nagalingam, A., Muniraj, N., Saxena, N. K., and Sharma, D. (2017). Concomitant activation of ETS-like transcription factor-1 and death receptor-5 via extracellular signal-regulated kinase in withaferin A-mediated inhibition of hepatocarcinogenesis in mice. Sci. Rep. 7 (1), 17943. doi:10.1038/s41598-017-18190-4
Lea, M. A., Randolph, V. M., Lee, J. E., and DesBordes, C. (2001). Induction of histone acetylation in mouse erythroleukemia cells by some organosulfur compounds including allyl isothiocyanate. Int. J. Cancer 92 (6), 784–789. doi:10.1002/ijc.1277
Lee, W. J., Chen, Y. R., and Tseng, T. H. (2011). Quercetin induces FasL-related apoptosis, in part, through promotion of histone H3 acetylation in human leukemia HL-60 cells. Oncol. Rep. 25 (2), 583–591. doi:10.3892/or.2010.1097
Li, B. B., Huang, G. L., Li, H. H., Kong, X., and He, Z. W. (2017). Epigallocatechin-3-Gallate modulates MicroRNA expression profiles in human nasopharyngeal carcinoma CNE2 cells. Chin. Med. J. Engl. 130 (1), 93–99. doi:10.4103/0366-6999.196586
Li, D., Chen, L., Zhao, W., Hao, J., and An, R. (2016). MicroRNA-Let-7f-1 is induced by lycopene and inhibits cell proliferation and triggers apoptosis in prostate cancer. Mol. Med. Rep. 13 (3), 2708–2714. doi:10.3892/mmr.2016.4841
Li, Y., and Sarkar, F. H. (2016). Role of BioResponse 3,3′-diindolylmethane in the treatment of human prostate cancer: clinical experience. Med. Princ. Pract. 25 (2), 11–17. doi:10.1159/000439307
Li, Y., Sun, W., Han, N., Zou, Y., and Yin, D. (2018). Curcumin inhibits proliferation, migration, invasion and promotes apoptosis of retinoblastoma cell lines through modulation of MiR-99a and JAK/STAT pathway. BMC Cancer 18 (1), 1230. doi:10.1186/s12885-018-5130-y
Link, A., Balaguer, F., Shen, Y., Lozano, J. J., Leung, H. C. E., Boland, C. R., et al. (2013). Curcumin modulates DNA methylation in colorectal cancer cells. PLoS One 8 (2), e57709. doi:10.1371/journal.pone.0057709
Liu, H. L., Chen, Y., Cui, G. H., and Zhou, J. F. (2005). Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-nhl cell line Raji proliferation. Acta Pharmacol. Sin. 26 (5), 603–609. doi:10.1111/j.1745-7254.2005.00081.x
Liu, J., Li, M., Wang, Y., and Luo, J. (2017). Curcumin sensitizes prostate cancer cells to radiation partly via epigenetic activation of MiR-143 and MiR-143 mediated autophagy inhibition. J. Drug Target. 25 (7), 645–652. doi:10.1080/1061186X.2017.1315686
Liu, X., Li, H., Wu, M. L., Wu, J., Sun, Y., Zhang, K. L., et al. (2019b). Resveratrol reverses retinoic acid resistance of anaplastic thyroid cancer cells via demethylating CRABP2 gene. Front. Endocrinol. (Lausanne). 10, 734. doi:10.3389/fendo.2019.00734
Liu, Z., Gao, Y., and Li, X. (2019a). Cancer epigenetics and the potential of epigenetic drugs for treating solid tumors. Expert Rev. Anticancer Ther. 19, 139–149. doi:10.1080/14737140.2019.1552139
López-Lázaro, M. (2008). Anticancer and carcinogenic properties of curcumin: considerations for its clinical development as a cancer chemopreventive and chemotherapeutic agent. Mol. Nutr. Food Res. 52, S103–S127. doi:10.1002/mnfr.200700238
Lubin, F. D., Gupta, S., Parrish, R. R., Grissom, N. M., and Davis, R. L. (2011). Epigenetic mechanisms: critical contributors to long-term memory formation. Neurosci 17 (6), 616–632. doi:10.1177/1073858411386967
Luger, K., and Richmond, T. J. (1998). The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8, 140–146. doi:10.1016/s0959-437x(98)80134-2
Ma, L., Feugang, J. M., Konarski, P., Wang, J., Lu, J., Fu, S., et al. (2006). Growth inhibitory effects of quercetin on bladder cancer cell. Front. Biosci. 11 (Suppl. 1), 2275–2285. doi:10.2741/1970
Majid, S., Kikuno, N., Nelles, J., Noonan, E., Tanaka, Y., Kawamoto, K., et al. (2008). Genistein induces the P21WAF1/CIP1 and P16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 68 (8), 2736–2744. doi:10.1158/0008-5472.CAN-07-2290
Malik, Z., Parveen, R., Parveen, B., Zahiruddin, S., Aasif Khan, M., Khan, A., et al. (2021). Anticancer potential of andrographolide from andrographis paniculata (Burm.f.) nees and its mechanisms of action. J. Ethnopharmacol. 272, 113936. doi:10.1016/j.jep.2021.113936
Martin, S. L., Kala, R., and Tollefsbol, T. O. (2017). Mechanisms for the inhibition of colon cancer cells by sulforaphane through epigenetic modulation of MicroRNA-21 and human telomerase reverse Transcriptase (HTERT) down-regulation. Curr. Cancer Drug Targets 18 (1), 97–106. doi:10.2174/1568009617666170206104032
Matsukawa, Y., Nishino, H., Yoshida, M., Sugihara, H., Katsura, K., Takamatsu, T., et al. (2002). Quercetin enhances tumorigenicity induced by N-ethyl-N’-Nitro-N-nitrosoguanidine in the duodenum of mice. Environ. Health Prev. Med. 6 (4), 235–239. doi:10.1007/BF02897975
Mishra, S., Yadav, T., and Rani, V. (2016). Exploring MiRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncology/Hematology 98, 12–23. doi:10.1016/j.critrevonc.2015.10.003
Mitsiogianni, M., Trafalis, D. T., Franco, R., Zoumpourlis, V., Pappa, A., and Panayiotidis, M. I. (2020). Sulforaphane and iberin are potent epigenetic modulators of histone acetylation and methylation in malignant melanoma. Eur. J. Nutr. 60, 147–158. doi:10.1007/s00394-020-02227-y
Mohammadi, K., Sani, M. A., Azizi-Lalabadi, M., and McClements, D. J. (2022). Recent progress in the application of plant-based colloidal drug delivery systems in the pharmaceutical Sciences. Adv. Colloid Interface Sci. 307, 102734. doi:10.1016/j.cis.2022.102734
Montgomery, M., and Srinivasan, A. (2019). Epigenetic gene regulation by dietary compounds in cancer prevention. Adv. Nutr. 10, 1012–1028. doi:10.1093/advances/nmz046
Mottet, D., and Castronovo, V. (2008). Histone deacetylases: target enzymes for cancer therapy. Clin. Exp. Metastasis 25, 183–189. doi:10.1007/s10585-007-9131-5
Mu, J., Zhu, D., Shen, Z., Ning, S., Liu, Y., Chen, J., et al. (2017). The repressive effect of MiR-148a on wnt/β-catenin signaling involved in glabridin-induced anti-angiogenesis in human breast cancer cells. BMC Cancer 17 (1), 307. doi:10.1186/s12885-017-3298-1
Nandakumar, V., Vaid, M., and Katiyar, S. K. (2011). (-)-Epigallocatechin-3-Gallate reactivates silenced tumor suppressor genes, Cip1/P21 and P16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 32, 537–544. doi:10.1093/carcin/bgq285
Nardi, I., Reno, T., Yun, X., Sztain, T., Wang, J., Dai, H., et al. (2018). Triptolide inhibits wnt signaling in NSCLC through upregulation of multiple wnt inhibitory factors via epigenetic modifications to histone H3. Int. J. Cancer 143 (10), 2470–2478. doi:10.1002/ijc.31756
Nouriemamzaden, F., Word, B., Cotton, E., Hawkins, A., Littlejohn, K., Moore, R., et al. (2020). Modulation of estrogen α and progesterone receptors in triple negative breast cancer cell lines: the effects of vorinostat and indole-3-carbinol in vitro. Anticancer Res. 40 (7), 3669–3683. doi:10.21873/anticanres.14356
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 (3), 247–257. doi:10.1016/s0092-8674(00)81656-6
Paluszczak, J., Krajka-Kuźniak, V., and Baer-Dubowska, W. (2010). The effect of dietary polyphenols on the epigenetic regulation of gene expression in MCF7 breast cancer cells. Toxicol. Lett. 192, 119–125. doi:10.1016/j.toxlet.2009.10.010
Pan, L., Feng, F., Wu, J., Fan, S., Han, J., Wang, S., et al. (2022). Demethylzeylasteral targets lactate by inhibiting histone lactylation to suppress the tumorigenicity of liver cancer stem cells. Pharmacol. Res. 181, 106270. doi:10.1016/j.phrs.2022.106270
Pandey, M., Kaur, P., Shukla, S., Abbas, A., Fu, P., and Gupta, S. (2012). Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: in vitro and in vivo study. Mol. Carcinog. 51 (12), 952–962. doi:10.1002/mc.20866
Pandey, M., Shukla, S., and Gupta, S. (2010). Promoter demethylation and chromatin remodeling by green tea polyphenols leads to Re-expression of GSTP1 in human prostate cancer cells. Int. J. Cancer 126 (11), 2520–2533. doi:10.1002/ijc.24988
Pani, S., Sahoo, A., Patra, A., and Debata, P. R. (2020). Phytocompounds curcumin, quercetin, indole-3-carbinol, and resveratrol modulate lactate–pyruvate level along with cytotoxic activity in HeLa cervical cancer cells. Biotechnol. Appl. Biochem. 68, 1396–1402. doi:10.1002/bab.2061
Parbin, S., Shilpi, A., Kar, S., Pradhan, N., Sengupta, D., Deb, M., et al. (2016). Insights into the molecular interactions of thymoquinone with histone deacetylase: evaluation of the therapeutic intervention potential against breast cancer. Mol. Biosyst. 12 (1), 48–58. doi:10.1039/c5mb00412h
Paredes-Gonzalez, X., Fuentes, F., Su, Z. Y., and Kong, A. N. T. (2014). Apigenin reactivates Nrf2 anti-oxidative stress signaling in mouse skin epidermal JB6 P + cells through epigenetics modifications. AAPS J. 16 (4), 727–735. doi:10.1208/s12248-014-9613-8
Payer, B., and Lee, J. T. (2008). X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 42, 733–772. doi:10.1146/annurev.genet.42.110807.091711
Peng, Z., and Zhang, Y. (2017). Methyl jasmonate induces the apoptosis of human colorectal cancer cells via downregulation of EZH2 expression by MicroRNA-101. Mol. Med. Rep. 15 (2), 957–962. doi:10.3892/mmr.2016.6061
Pfeffer, C. M., and Singh, A. T. K. (2018). Apoptosis: a target for anticancer therapy. Int. J. Mol. Sci. 2, 448. doi:10.3390/ijms19020448
Phan, M. A. T., Paterson, J., Bucknall, M., and Arcot, J. (2018). Interactions between phytochemicals from fruits and vegetables: effects on bioactivities and bioavailability. Crit. Rev. Food Sci. Nutr. 58 (8), 1310–1329. doi:10.1080/10408398.2016.1254595
Prajapati, K. S., Shuaib, M., Gupta, S., and Kumar, S. (2022). Withaferin A mediated changes of MiRNA expression in breast cancer-derived mammospheres. Mol. Carcinog. 61 (9), 876–889. doi:10.1002/mc.23440
Prasad, S., Gupta, S. C., and Tyagi, A. K. (2017). Reactive oxygen species (ROS) and cancer: role of antioxidative nutraceuticals. Cancer Lett. 28, 95–105. doi:10.1016/j.canlet.2016.03.042
Prasad, S., Yadav, V. R., Sung, B., Reuter, S., Kannappan, R., Deorukhkar, A., et al. (2012). Ursolic acid inhibits growth and metastasis of human, colorectal cancer in an orthotopic nude mouse model by targeting multiple cell signaling pathways: chemosensitization with capecitabine. Clin. Cancer Res. 18 (18), 4942–4953. doi:10.1158/1078-0432.CCR-11-2805
Qazi, A., Pal, J., Maitah, M., Fulciniti, M., Pelluru, D., Nanjappa, P., et al. (2010). Anticancer activity of a broccoli derivative, sulforaphane, in barrett adenocarcinoma: potential use in chemoprevention and as adjuvant in chemotherapy. Transl. Oncol. 3 (6), 389–399. doi:10.1593/tlo.10235
Qin, W., Zhang, K., Clarke, K., Weiland, T., and Sauter, E. R. (2014). Methylation and MiRNA effects of resveratrol on mammary tumors vs. Normal tissue. Nutr. Cancer 66 (2), 270–277. doi:10.1080/01635581.2014.868910
Reunanen, N., and Kähäri, V. (2013). “Matrix metalloproteinases in cancer cell invasion,” in Madame curie bioscience database (Austin, TX: Landes Bioscience).
Sadakierska-Chudy, A. (2020). Micrornas: diverse mechanisms of action and their potential applications as cancer epi-therapeutics. Biomolecules 10, 1285. doi:10.3390/biom10091285
Safi-Stibler, S., and Gabory, A. (2020). Epigenetics and the developmental origins of health and disease: parental environment signalling to the epigenome, critical time windows and sculpting the adult phenotype. Seminars Cell Dev. Biol. 97, 172–180. doi:10.1016/j.semcdb.2019.09.008
Salehi, B., Ata, A., Kumar, N. V. A., Sharopov, F., Ramírez-Alarcón, K., Ruiz-Ortega, A., et al. (2019). Antidiabetic potential of medicinal plants and their active components. Biomolecules 9, 551. doi:10.3390/biom9100551
Santi, D. V., Garrett, C. E., and Barr, P. J. (1983). On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell 33, 9–10. doi:10.1016/0092-8674(83)90327-6
Setoyama, T., Ling, H., Natsugoe, S., and Calin, G. A. (2011). Non-coding RNAs for medical practice in oncology. Keio J. Med. 60 (4), 106–113. doi:10.2302/kjm.60.106
Shankar, S., Ganapathy, S., and Srivastava, R. K. (2007). Green tea polyphenols: biology and therapeutic implications in cancer. Front. Biosci. 12, 4881–4899. doi:10.2741/2435
Sheng, J., Shi, W., Guo, H., Long, W., Wang, Y., Qi, J., et al. (2019). The inhibitory effect of (-)-Epigallocatechin-3 -gallate on breast cancer progression via reducing SCUBE2 methylation and DNMT activity. Molecules 24 (16), 2899. doi:10.3390/molecules24162899
Shi, Q., Li, J., Feng, Z., Zhao, L., Luo, L., You, Z., et al. (2014). Effect of ginsenoside Rh2 on the migratory ability of HepG2 liver carcinoma cells: recruiting histone deacetylase and inhibiting activator protein 1 transcription factors. Mol. Med. Rep. 10 (4), 1779–1785. doi:10.3892/mmr.2014.2392
Shruti, K., Shrey, K., and Vibha, R. (2011). Micro RNAs: tiny sequences with enormous potential. Biochem. Biophys. Res. Commun. 15, 445–449. doi:10.1016/j.bbrc.2011.03.058
Shu, L., Khor, T. O., Lee, J. H., Boyanapalli, S. S. S., Huang, Y., Wu, T. Y., et al. (2011). Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 13 (4), 606–614. doi:10.1208/s12248-011-9300-y
Singh, P., Mishra, S. K., Noel, S., Sharma, S., and Rath, S. K. (2012). Acute exposure of apigenin induces hepatotoxicity in Swiss mice. PLoS One 7, e31964. doi:10.1371/journal.pone.0031964
Singh, S., Sharma, B., Kanwar, S. S., and Kumar, A. (2016). Lead phytochemicals for anticancer drug development. Front. Plant Sci. 7, 1667. doi:10.3389/fpls.2016.01667
Smith, C. J., and Ryckman, K. K. (2015). Epigenetic and developmental influences on the risk of obesity, diabetes, and metabolic syndrome. Diabetes, Metabolic Syndrome Obes. 8, 295–302. doi:10.2147/DMSO.S61296
Su, C. C., Lin, J. G., Li, T. M., Chung, J. G., Yang, J. S., Ip, S. W., et al. (2006). Curcumin-induced apoptosis of human colon cancer colo 205 cells through the production of ROS, Ca2+ and the activation of caspase-3. Anticancer Res. 26, 4379–4389.
Suman, S., Das, T. P., Sirimulla, S., Alatassi, H., Ankem, M. K., and Damodaran, C. (2016). Withaferin-A suppress AKT induced tumor growth in colorectal cancer cells. Oncotarget 7 (12), 13854–13864. doi:10.18632/oncotarget.7351
Sundaram, M. K., Unni, S., Somvanshi, P., Bhardwaj, T., Mandal, R. K., Hussain, A., et al. (2019). Genistein modulates signaling pathways and targets several epigenetic markers in hela cells. Genes (Basel). 10 (12), 955. doi:10.3390/genes10120955
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Svoronos, A. A., Engelman, D. M., and Slack, F. J. (2016). OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 1, 3666–3670. doi:10.1158/0008-5472.CAN-16-0359
Tan, B. L., and Norhaizan, M. E. (2019). Curcumin combination chemotherapy: the implication and efficacy in cancer. Molecules 24, 2527. doi:10.3390/molecules24142527
Tang, J., Wang, Z., Chen, L., Huang, G., and Hu, X. (2015). Gossypol acetate induced apoptosis of pituitary tumor cells by targeting the BCL-2 via the upregulated MicroRNA MiR-15a. Int. J. Clin. Exp. Med. 8 (6), 9079–9085.
Tao, S. F., He, H. F., and Chen, Q. (2015). Quercetin inhibits proliferation and invasion acts by up-regulating MiR-146a in human breast cancer cells. Mol. Cell. Biochem. 402 (1–2), 93–100. doi:10.1007/s11010-014-2317-7
Tao, Y., Zhan, S., Wang, Y., Zhou, G., Liang, H., Chen, X., et al. (2018). Baicalin, the major component of traditional Chinese medicine scutellaria baicalensis induces colon cancer cell apoptosis through inhibition of OncomiRNAs. Sci. Rep. 8 (1), 14477. doi:10.1038/s41598-018-32734-2
Thakur, V. S., Gupta, K., and Gupta, S. (2012). Green tea polyphenols increase P53 transcriptional activity and acetylation by suppressing class I histone deacetylases. Int. J. Oncol. 41 (1), 353–361. doi:10.3892/ijo.2012.1449
Thangapazham, R. L., Singh, A. K., Sharma, A., Warren, J., Gaddipati, J. P., and Maheshwari, R. K. (2007). Green tea polyphenols and its constituent epigallocatechin gallate inhibits proliferation of human breast cancer cells in vitro and in vivo. Cancer Lett. 245 (1–2), 232–241. doi:10.1016/j.canlet.2006.01.027
Torabi, K., Miró, R., Fernández-Jiménez, N., Quintanilla, I., Ramos, L., Prat, E., et al. (2015). Patterns of somatic uniparental disomy identify novel tumor suppressor genes in colorectal cancer. Carcinogenesis 36 (10), 1103–1110. doi:10.1093/carcin/bgv115
Tsang, W. P., and Kwok, T. T. (2010). Epigallocatechin gallate up-regulation of MiR-16 and induction of apoptosis in human cancer cells. J. Nutr. Biochem. 21 (2), 140–146. doi:10.1016/j.jnutbio.2008.12.003
Tseng, T. H., Chien, M. H., Lin, W. L., Wen, Y. C., Chow, J. M., Chen, C. K., et al. (2017). Inhibition of MDA-MB-231 breast cancer cell proliferation and tumor growth by apigenin through induction of G2/M arrest and histone H3 acetylation-mediated P21WAF1/CIP1 expression. Environ. Toxicol. 32 (2), 434–444. doi:10.1002/tox.22247
Tuna, M., Knuutila, S., and Mills, G. B. (2009). Uniparental disomy in cancer. Trends Mol. Med. 15, 120–128. doi:10.1016/j.molmed.2009.01.005
Van Hoesel, A. Q., Sato, Y., Elashoff, D. A., Turner, R. R., Giuliano, A. E., Shamonki, J. M., et al. (2013). Assessment of DNA methylation status in early stages of breast cancer development. Br. J. Cancer 108 (10), 2033–2038. doi:10.1038/bjc.2013.136
vel Szic, K. S., Declerck, K., Crans, R. A. J., Diddens, J., Scherf, D. B., Gerhäuser, C., et al. (2017). Epigenetic silencing of triple negative breast cancer hallmarks by withaferin A. Oncotarget 8, 40434–40453. doi:10.18632/oncotarget.17107
Verma, S. (2016). Medicinal plants with anti-inflammatory activity. J. Phytopharm. 5 (4), 157–159. doi:10.31254/phyto.2016.5407
Wang, D., Wang, Y., Wan, X., Yang, C. S., and Zhang, J. (2015). Green tea polyphenol (-)-Epigallocatechin-3-Gallate triggered hepatotoxicity in mice: responses of major antioxidant enzymes and the Nrf2 rescue pathway. Toxicol. Appl. Pharmacol. 283 (1), 65–74. doi:10.1016/j.taap.2014.12.018
Wang, J., Li, Y., Wang, X., and Jiang, C. (2012). Ursolic acid inhibits proliferation and induces apoptosis in human glioblastoma cell lines U251 by suppressing TGF-β1/MiR-21/PDCD4 pathway. Basic Clin. Pharmacol. Toxicol. 111, 106–112. doi:10.1111/j.1742-7843.2012.00870.x
Wang, J., Wu, M., Zheng, D., Zhang, H., Lv, Y., Zhang, L., et al. (2020). Garcinol inhibits esophageal cancer metastasis by suppressing the P300 and TGF-β1 signaling pathways. Acta Pharmacol. Sin. 41 (1), 82–92. doi:10.1038/s41401-019-0271-3
Wang, L. G., Beklemisheva, A., Liu, X. M., Ferrari, A. C., Feng, J., and Chiao, J. W. (2007). Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol. Carcinog. 46 (1), 24–31. doi:10.1002/mc.20258
Wei, D., Yang, L., Lv, B., and Chen, L. (2017). Genistein suppresses retinoblastoma cell viability and growth and induces apoptosis by upregulating MiR-145 and inhibiting its target ABCE1. Mol. Vis. 23, 385–394.
Wen, X. Y., Wu, S. Y., Li, Z. Q., Liu, Z. Q., Zhang, J. J., Wang, G. F., et al. (2009). Ellagitannin (BJA3121), an anti-proliferative natural polyphenol compound, can regulate the expression of MiRNAs in HepG2 cancer cells. Phyther. Res. 23, 778–784. doi:10.1002/ptr.2616
Wong, C. P., Hsu, A., Buchanan, A., Palomera-Sanchez, Z., Beaver, L. M., Houseman, E. A., et al. (2014). Effects of sulforaphane and 3,3′-diindolylmethane on genome-wide promoter methylation in normal prostate epithelial cells and prostate cancer cells. PLoS One 9 (1), e86787. doi:10.1371/journal.pone.0086787
Wu, S., Lu, H., and Bai, Y. (2019). Nrf2 in cancers: a double-edged sword. Cancer Med. 8, 2252–2267. doi:10.1002/cam4.2101
Xhemalce, B., Dawson, M. A., and Bannister, A. J. (2011). “Histone modifications,” in Encyclopedia of molecular cell biology and molecular medicine (American Cancer Society).
Xiao, L., Huang, Y., Zhen, R., Chiao, J. W., Liu, D., and Ma, X. (2010). Deficient histone acetylation in acute leukemia and the correction by an isothiocyanate. Acta Haematol. 123 (2), 71–76. doi:10.1159/000264628
Xiao, Y., Hale, S., Awasthee, N., Meng, C., Zhang, X., Liu, Y., et al. (2023). HDAC3 and HDAC8 PROTAC dual degrader reveals roles of histone acetylation in gene regulation. Cell Chem. Biol. doi:10.1016/j.chembiol.2023.07.010
Xiao, Z., Morris-Natschke, S. L., and Lee, K. H. (2016). Strategies for the optimization of natural leads to anticancer drugs or drug candidates. Med. Res. Rev. 36 (1), 32–91. doi:10.1002/med.21377
Yan, X., Xie, T., Wang, S., Wang, Z., Li, H. Y., and Ye, Z. M. (2018). Apigenin inhibits proliferation of human chondrosarcoma cells via cell cycle arrest and mitochondrial apoptosis induced by ROS generation-an in vitro and in vivo study. J. Clin. Exp. Med. 11 (3), 1615–1631.
Ye, Y., Fang, Y., Xu, W., Wang, Q., Zhou, J., and Lu, R. (2016). 3,3′-Diindolylmethane induces anti-human gastric cancer cells by the MiR-30e-ATG5 modulating autophagy. Biochem. Pharmacol. 115, 77–84. doi:10.1016/j.bcp.2016.06.018
Yin, L., Xiao, X., Georgikou, C., Luo, Y., Liu, L., Gladkich, J., et al. (2019). Sulforaphane induces MiR135b-5p and its target gene, RASAL2, thereby inhibiting the progression of pancreatic cancer. Mol. Ther. - Oncolytics 14, 74–81. doi:10.1016/j.omto.2019.03.011
Yoshino, M., Haneda, M., Naruse, M., Htay, H. H., Tsubouchi, R., Qiao, S. L., et al. (2004). Prooxidant activity of curcumin: copper-dependent formation of 8-hydroxy-2′-deoxyguanosine in DNA and induction of apoptotic cell death. Toxicol. Vitr. 18 (6), 783–789. doi:10.1016/j.tiv.2004.03.009
Yun, C. W., and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466. doi:10.3390/ijms19113466
Zhang, B., Wang, Q., and Pan, X. (2007). MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiology 210, 279–289. doi:10.1002/jcp.20869
Zhang, C., Shu, L., Kim, H., Khor, T. O., Wu, R., Li, W., et al. (2016). Phenethyl isothiocyanate (PEITC) suppresses prostate cancer cell invasion epigenetically through regulating MicroRNA-194. Mol. Nutr. Food Res. 60 (6), 1427–1436. doi:10.1002/mnfr.201500918
Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y., et al. (2019). Metabolic regulation of gene expression by histone lactylation. Nature 574 (7779), 575–580. doi:10.1038/s41586-019-1678-1
Zhang, H., Jia, R., Wang, C., Hu, T., and Wang, F. (2014). Piceatannol promotes apoptosis via up-regulation of MicroRNA-129 expression in colorectal cancer cell lines. Biochem. Biophys. Res. Commun. 452 (3), 775–781. doi:10.1016/j.bbrc.2014.08.150
Zhang, Y., Huang, L., Shi, H., Chen, H., Tao, J., Shen, R., et al. (2018). Ursolic acid enhances the therapeutic effects of oxaliplatin in colorectal cancer by inhibition of drug resistance. Cancer Sci. 109 (1), 94–102. doi:10.1111/cas.13425
Zhang, Y., Li, Q., Zhou, D., and Chen, H. (2013). Genistein, a soya isoflavone, prevents azoxymethane-induced up-regulation of WNT/β-Catenin signalling and reduces colon pre-neoplasia in rats. Br. J. Nutr. 109 (1), 33–42. doi:10.1017/S0007114512000876
Zhao, J., Fang, Z., Zha, Z., Sun, Q., Wang, H., Sun, M., et al. (2019). Quercetin inhibits cell viability, migration and invasion by regulating MiR-16/HOXA10 Axis in oral cancer. Eur. J. Pharmacol. 847, 11–18. doi:10.1016/j.ejphar.2019.01.006
Zheng, M., and Wu, Y. (2020). Piceatannol suppresses proliferation and induces apoptosis by regulation of the MicroRNA-21/phosphatase and tensin homolog/protein kinase B signaling pathway in osteosarcoma cells. Mol. Med. Rep. 22 (5), 3985–3993. doi:10.3892/mmr.2020.11484
Zhou, J., Liu, M., Chen, Y., Xu, S., Guo, Y., and Zhao, L. (2019b). Cucurbitacin B suppresses proliferation of pancreatic cancer cells by CeRNA: effect of MiR-146b-5p and LncRNA-AFAP1-AS1. J. Cell. Physiol. 234 (4), 4655–4667. doi:10.1002/jcp.27264
Zhou, J. W., Wang, M., Sun, N. X., Qing, Y., Yin, T. F., Li, C., et al. (2019a). Sulforaphane-induced epigenetic regulation of Nrf2 expression by DNA methyltransferase in human caco-2 cells. Oncol. Lett. 18 (3), 2639–2647. doi:10.3892/ol.2019.10569
Zhu, W. Q., Wang, J., Guo, X. F., Liu, Z., and Dong, W. G. (2016). Thymoquinone inhibits proliferation in gastric cancer via the STAT3 pathway in vivo and in vitro. World J. Gastroenterol. 22 (16), 4149–4159. doi:10.3748/wjg.v22.i16.4149
Zoghbi, H. Y., and Beaudet, A. L. (2016). Epigenetics and human disease. Cold Spring Harb. Perspect. Biol. 8 (2), a019497. doi:10.1101/cshperspect.a019497
Zuo, Q., Wu, R., Xiao, X., Yang, C., Yang, Y., Wang, C., et al. (2018). The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J. Cell. Biochem. 119 (11), 9573–9582. doi:10.1002/jcb.27275
Keywords: cancer, cancer epigenetics, DNMTs, HDACs, miRNAs, phytocompounds, bioactive compounds, cancer prevention
Citation: Khan A, Khan A, Khan MA, Malik Z, Massey S, Parveen R, Mustafa S, Shamsi A and Husain SA (2024) Phytocompounds targeting epigenetic modulations: an assessment in cancer. Front. Pharmacol. 14:1273993. doi: 10.3389/fphar.2023.1273993
Received: 07 August 2023; Accepted: 09 November 2023;
Published: 26 March 2024.
Edited by:
Daiqing Liao, University of Florida, United StatesReviewed by:
Lei Huang, University of Massachusetts Medical School, United StatesCopyright © 2024 Khan, Khan, Khan, Malik, Massey, Parveen, Mustafa, Shamsi and Husain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Syed A. Husain, c2h1c2FpbkBqbWkuYWMuaW4=; Anas Shamsi, YW5hcy5zaGFtc2kxOEBnbWFpbC5jb20=
†Present address: Asifa Khan, Department of Molecular, Cell and Cancer Biology, University of Massachusetts Chan Medical School, Worcester, MA, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.