- 1Institute for Translational Medicine, The Affiliated Hospital, College of Medicine, Qingdao University, Qingdao, China
- 2College of Chinese Medicinal Materials, Jilin Agricultural University, Changchun, China
Type 2 diabetes (T2D) is a metabolic disease with complex etiology and mechanisms. Long non-coding ribonucleic acid (LncRNA) is a novel class of functional long RNA molecules that regulate multiple biological functions through various mechanisms. Studies in the past decade have shown that lncRNAs may play an important role in regulating insulin resistance and the progression of T2D. As a widely used biguanide drug, metformin has been used for glucose lowering effects in clinical practice for more than 60 years. For diabetic therapy, metformin reduces glucose absorption from the intestines, lowers hepatic gluconeogenesis, reduces inflammation, and improves insulin sensitivity. However, despite being widely used as the first-line oral antidiabetic drug, its mechanism of action remains largely elusive. Currently, an increasing number of studies have demonstrated that the anti-diabetic effects of metformin were mediated by the regulation of lncRNAs. Metformin-regulated lncRNAs have been shown to participate in the inhibition of gluconeogenesis, regulation of lipid metabolism, and be anti-inflammatory. Thus, this review focuses on the mechanisms of action of metformin in regulating lncRNAs in diabetes, including pathways altered by metformin via targeting lncRNAs, and the potential targets of metformin through modulation of lncRNAs. Knowledge of the mechanisms of lncRNA modulation by metformin in diabetes will aid the development of new therapeutic drugs for T2D in the future.
Introduction
Type 2 diabetes (T2DM) is a disease that is characterized by chronic high blood glucose and/or insulin resistance in multiple organs. If lifestyle changes and exercise in patients of T2DM cannot achieve satisfactory results, oral hypoglycemic drugs are introduced (Ganesan et al., 2023). Current treatment guidelines recommend the use of metformin as first-line therapy for T2DM (Lv and Guo, 2020). Metformin has been shown to lower fasting plasma glucose and HbA1c in a dose-related manner in patients with T2DM (Garber et al., 1997) and is associated with reduced body weight and waist circumference (Diabetes Prevention Program Research, 2012). Furthermore, although controversial, studies from the United Kingdom Prospective Diabetes Study and the Hyperinsulinaemia the Outcome of its Metabolic Effects (HOME) trial have shown that metformin has favorable effects on cardiovascular outcomes in both new-onset and advanced type 2 diabetes (Effect of intensive blood, 1998; Jager et al., 2005), as well as in kidney disease in patients with T2DM (Charytan et al., 2019the other hand, metformin inhibits mitochondrial). However, the molecular mechanisms of metformin remain largely elusive. In this paper, we highlight the mechanisms of metformin in current studies and focus on the action of metformin in regulating lncRNAs in T2DM. Exploring the action mechanisms of metformin will help to better guide clinical medication and find new drug targets for T2DM therapy.
Molecular pathway for the anti-diabetic effects of metformin
AMPK-dependent pathway
The molecular targets of metformin have been expanded upon in the recent decades (Foretz et al., 2019; Lu et al., 2021; Wu et al., 2023) but the primary mechanism of this biguanide remains unclear. The activation of adenosine 5’-monophosphate activated protein kinase (AMPK) is considered a core mechanism of metformin; AMPK phosphorylates several key enzymes that regulate the activity of transcription factors, coactivators, and corepressors, thus regulating lipid metabolism, inflammation, autophagy, and gluconeogenesis (Wang et al., 2019a; Yang et al., 2019; Zhang et al., 2022a; Li et al., 2022; Yang et al., 2022) (Figure 1). For example, AMPK activation suppresses the expression of a key lipogenic transcription factor, SREBP-1 in the liver, and activates AMPK phosphorylated acetyl-CoA carboxylase (ACC), which is an inhibitory phenotype and suppresses malonyl-CoA biosynthesis, thus inhibiting fatty acid synthesis and increasing fatty acid oxidation (Zhou et al., 2001). Furthermore, activated AMPK inhibits the activity of the mammalian target of rapamycin complex (mTORC) to induce cellular autophagy (Wang et al., 2021a) and decrease protein synthesis (Knudsen et al., 2020). The inhibition of fatty acids and protein synthesis can promote cellular glucose uptake and metabolism to lower glucose levels (Lundsgaard et al., 2020). On the other hand, metformin-induced activation of AMPK has been shown to phosphorylate CREB-regulated transcription coactivator 2 (CRTC2, also known as TORC2), which prevents its translocation to the nucleus, thus inhibiting the transcriptional activation of gluconeogenic genes (Koo et al., 2005). Moreover, metformin inhibits Th1 and Th17 cells differentiation while promoting Tregs development through activation of AMPK (Duan et al., 2019), which may contribute to its anti-inflammatory effects.
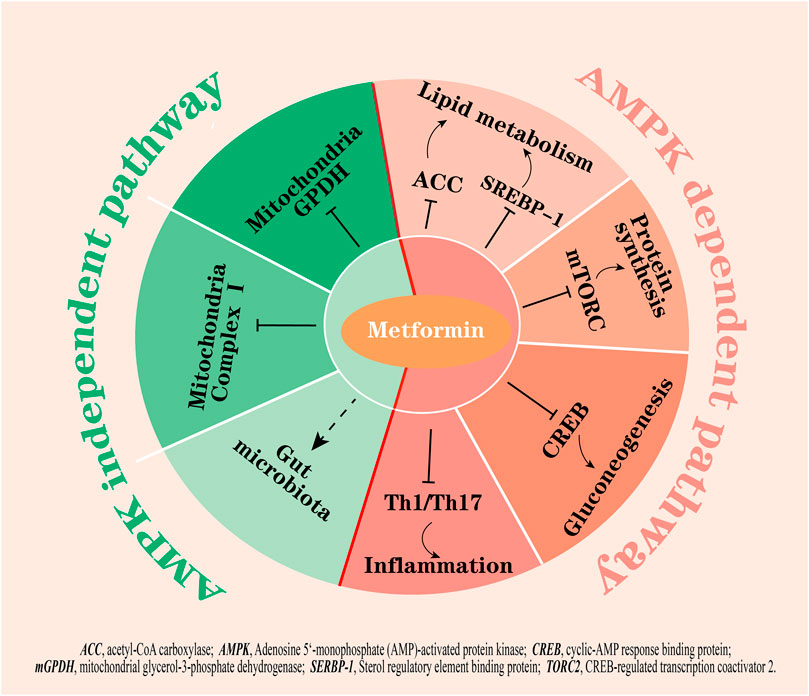
FIGURE 1. The hypoglycemic mechanisms of metformin by AMPK dependent and AMPK independent pathways. Activation of AMPK by metformin: 1) inhibits fatty acid synthesis by suppressing ACC and SREBP-1; 2) inhibits mTORC and CREB to inhibit protein synthesis and gluconeogenesis; and 3) inhibits the differentiation of Th1 and Th17 cells to exert anti-inflammation effects. On the other hand, metformin inhibits mitochondrial complex I and mGPDH, potentially modulates gut microbiota, and is not dependent on AMPK activation. Right part of red line, AMPK dependent pathways; left part of red line, AMPK independent pathways.
AMPK-independent pathway
However, many studies showed that the beneficial effects of metformin remained in a transgenic mouse model with lacked AMPK or its upstream activator LKB1 (Foretz et al., 2010), indicating that there are other critical targets other than AMPK for metformin action. In fact, increasing evidence points to other actions of metformin, for example, metformin has been shown to inhibit mitochondrial complex Ⅰ, which is independent of AMPK activity in cells (El-Mir et al., 2000; Owen et al., 2000); the inhibition results in a decrease in ATP production, thus suppressing gluconeogenesis in the liver. Additionally, metformin at low doses (<130 µM in plasma) was reported to inhibit mitochondrial glycerol-3-phosphate dehydrogenase (mGPDH) activity, resulting in an increased hepatic cytosolic NADH to NAD+ ratio (lactate to pyruvate ratio) independently of changes in intracellular levels of ATP and inhibition of glucose production from reduced gluconeogenic substrates (lactate and glycerol) (Madiraju et al., 2014; Madiraju et al., 2018). In addition to these, a study found that glucose tolerance improved in germ-free mice that received metformin-altered microbiota via fecal samples transfer, indicating that the altered gut microbiota mediates some of the antidiabetic effects of metformin (Wu et al., 2017) (Figure 1).
LncRNA biogenesis and expressions in T2DM
Long non-coding RNAs (LncRNAs) are non-coding RNA transcripts that are more than 200 nucleotides (nt) in length. According to their location in the gene relative to nearby protein-coding genes, lncRNAs are classified as intronic lncRNAs, antisense lncRNAs, and long “intergenic” non-coding RNAs (lincRNAs). The biogenesis of lncRNAs is similar to that of mRNAs, but lncRNAs have some unique features that can distinguish them from mRNAs, for example, lncRNAs have relatively low expression and the presence of fewer and longer exons than mRNAs (Liu et al., 2021; Liu et al., 2023). Although they cannot produce proteins, they can act as pathway switches to participate in the regulation of signaling pathways. LncRNAs can function as scaffolds (bringing two distant genes closer), decoys (binding to transcription factors to inhibit gene transcription), guiders (guiding regulatory proteins to gene sequences), and genomic targeting (directly binding to gene sequences to affect gene expression), thus participating in molecular and genomic modulation (Panni et al., 2020). Meanwhile, a single lncRNA may have multiple targets, so its function may be diverse; additionally, lncRNAs are less species conserved compared to miRNAs, which makes the investigation of lncRNAs difficult.
Various pathological processes of diseases are mediated by lncRNAs (Liu et al., 2022a; Zhou et al., 2022; Ao et al., 2023; Mattick et al., 2023). Several lncRNAs function as biomarkers or intervention targets that aid in the diagnosis and treatment of T2DM. For example, abnormal expressions of lncRNA, including CASC2, GAS5, ENST00000550337.1, HOTAIR, MEG3, H19, MALAT1, MIAT, ANRIL, XIST, PANDA, Linc-p21, PLUTO, and NBR2 in blood samples are closely associated with diagnostic outcomes of T2DM (Sathishkumar et al., 2018). Furthermore, some lncRNAs participate in the progression of diabetic complications, such as plasma taurine upregulated gene 1 (TUG1) which is associated with the progression of diabetic nephropathy in diabetic rats and Kcnq1ot1 which is involved in the progression of diabetic cardiomyopathy (Yang et al., 2018a). In addition, β cell replacement is the other way to treat diabetes; in recent years, 2D and 3D models based on induced pluripotent stem cells (iPSCs) have attracted much attention to provide a new approach to β-like cell replacement therapies, and lncRNAs play an essential role in the regulatory mechanisms of cell reprogramming and differentiation (Ilieva and Uchida, 2022). For example, the lncRNA Gm10451 is a potential regulator for the β-like cell differentiation of iPSCs (Huang et al., 2019).
Metformin mediates lncRNA expressions
Metformin treatment can alter the expressions of lncRNAs. How metformin acts on lncRNAs is not clear; a bioinformatics study showed that metformin promotes different expressions of lncRNA isoforms among lung cells, kidney cells, embryonic cells, hepatocytes, and pancreatic cells. For example, NEAT1-202, the canonical isoform of the NEAT1 gene, is upregulated in five out of six cell lines by metformin treatment, but other isoforms showed different fold change directions (Conceicao et al., 2022). This may explain different results for the same lncRNA in different studies. In addition, the presence of upregulated DNA methylation was found during metformin treatment. In these studies, DNMT1, the principal DNA methyltransferase in mammalian cells, was found to be increased by metformin. Hypermethylation can occur in lncRNAs; metformin has been shown to decrease H19 levels by inducing H19 methylation, which has been reported in cancer cells (Yan et al., 2015) and in animal models of pre-eclampsia (Shu et al., 2019).
Furthermore, the effects of metformin on lncRNAs depend on cell types and organs. For example, Dreh has been shown to be downregulated by a high-fat diet (HFD), and metformin has been shown to upregulate Dreh and improve lipid accumulation in mice livers (Guo et al., 2018). However, in C2C12 skeletal muscle cells, metformin-repressed Dreh expression induces an increase in glucose uptake (Takahashi et al., 2019). Furthermore, metformin was shown to decrease H19 in rodent models of HFD-induced liver disease (Guo et al., 2018), gastric cancer (Li et al., 2019), endometrial cancer (Aminimoghaddam et al., 2021), and breast cancer (Chen et al., 2022). However, metformin co-treatment with sitagliptin was shown to increase H19 expression to attenuate apoptosis and insulin resistance in rats with polycystic ovary syndrome (POCS) (Wang et al., 2019b). Similarly, the metformin-regulated expression of MALAT1, HOTAIR, and TUG1 was reported to be upregulated in breast cancer cells (Huang et al., 2021). However, other studies reported a decrease in the expression of TUG1 by metformin in vascular wall cells (You et al., 2020), a decrease in the expression of HOTAIR by metformin in MDA-MB-231 breast cancer cells (Golshan et al., 2021), and a decrease in the expression of MALAT1 by metformin in cervical cancer cells (Xia et al., 2018). The differences in metformin regulation suggest a necessity to differentiate the effects according to the state of the disease being treated. In diabetes conditions, metformin-induced alterations in lncRNA expression involve changes in gene transcription for glucose and lipid metabolism, the regulation of inflammatory cytokines, and the modulation of mitochondrial function.
The anti-diabetic action of metformin mediated by lncRNAs
Gluconeogenesis and glucose uptake
Inhibiting hepatic gluconeogenesis is the main mechanism of the blood glucose lowering action of metformin. However, the mechanisms underlying these effects are unclear. Studies have showed that the overexpression of the peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) preserved metformin’s ability to reduce glucose output regardless of AMPK expression (Foretz et al., 2010), suggesting that PGC-1α is a key factor for metformin’s gluconeogenic inhibitory effects. Concomitantly, another study found that cyclic adenosine monophosphate (cAMP), a gluconeogenic stimulus, induces changes in the expression of several lncRNAs in primary mouse hepatocytes. Furthermore, metformin treatment preserved 189 of the 456 upregulated lncRNAs and 167 of the 409 downregulated lncRNAs. Among those changed lncRNAs, NR_027710 and ENSMUST00000138573 were identified as having an association with the gene expression of PGC-1α (Wang et al., 2018), suggesting that metformin inhibiting gluconeogenesis could be mediated by the regulation of NR_027710 and ENSMUST00000138573. Consistent with these results, another study showed that 3,076 lncRNAs (1,823 upregulated and 1,253 downregulated) are altered in HFD-induced insulin resistant mice compared with the normal control group, and metformin treatment reversed 588 lncRNAs. Among those lncRNAs downregulated by metformin, NONMMUT031874.2 was highly expressed and found to target miR-7054-5p. Downregulating the expression of NONMMUT031874.2 by metformin could increase the expression of miR-7054-5p and then downregulate the expression of Socs3, which inhibits PEPCK to inhibit gluconeogenesis (Zhang et al., 2022b).
In addition, hepatic gluconeogenesis is catalyzed by multiple enzymes, including G6Pase, PEPCK, and FBP1 (Mues et al., 2009). Suppressing hepatic gluconeogenesis is usually accompanied by the activation of glycolysis enzymes, including GCK, PFKL, and PK (Figure 2) (Al-Oanzi et al., 2017). Metformin treatment decreased the expression of lncRNA-NONMMUT034936.2 in liver tissue from HFD mice and exhibited similar expression patterns with G6P target genes, indicating that metformin may affect gluconeogenesis by regulating NONMMUT034936.2 (Shu et al., 2021). Furthermore, transcription factor hepatocyte nuclear factor 4α (HNF-4α) binding to promoters of G6Pase or GCK is required for GCK gene activation and insulin-mediated G6Pase gene repression (Hirota et al., 2008). Metformin-exposed mouse fetuses were shown to elevate H19 expression (Deng et al., 2017), a lncRNA reported to be related to T2DM and influence DNA methylation (Ding et al., 2012), which induces hypomethylation and the overexpression of HNF-4α, thus efficiently repressing hepatic gluconeogenesis (Figure 2).
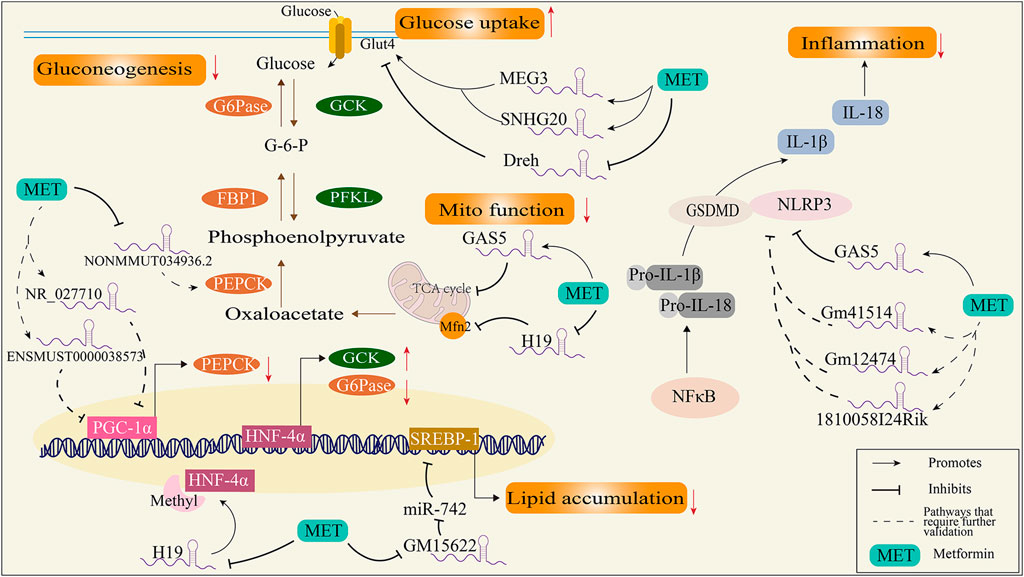
FIGURE 2. Schematic diagram of the mechanism of metformin regulating diabetes through lncRNAs. FBP1, fructose-1,6-bisphosphatase; G6Pase, glucose-6-phosphatase; GCK, glucokinase; GSDMD, gasdermin-D; HNF-4α, transcription factor hepatocyte nuclear factor 4α; IL-18, interleukin 18; IL-1β, interleukin 1β; Mfn2, mitofusion protein 2; NLRP-3, NOD-like receptor protein; NF-κB, nuclear factor-κB; PEPCK, phosphoenolpyruvate carboxylase kinase; PFKL, phosphofructokinase; PGC-1α, peroxisome proliferator-activated receptor-gamma coactivator; SERBP-1, sterol regulatory element binding protein.
On the other hand, the increased glucose uptake effects by metformin are mediated by lncRNAs. LncRNA-Dreh is reduced by metformin in C2C12 myotubes. Furthermore, knockdown of Dreh resulted in reduced glucose concentrations in the culture medium and increased levels of GLUT4 protein in the plasma membrane. The overexpression of Dreh attenuated the glucose-lowering effect of metformin in myotubes (Takahashi et al., 2019), suggesting the blood glucose lowering actions of metformin are mediated by Dreh in the skeletal muscle cells. Moreover, metformin treatment is reported to increase GLUT4 expression by inducing expression of MEG3 and SNHG20, thus improving insulin resistance in the endometrium of PCOS rats (Liu et al., 2022b), indicating the metformin regulation of lncRNAs in glucose uptake is not specific to a type of lncRNA.
Lipid metabolism
Dyslipidemia is common among patients with T2DM (prevalence > 75%) (Athyros et al., 2018). Aberrations in lipid metabolism contribute to the progression of T2DM and the risk of diabetic complications. For example, excessive dietary lipid intake and enhanced lipogenesis can cause ectopic lipids to accumulate in adipose, pancreatic, vascular, and cardiac cells (Yang et al., 2018b), resulting in atherosclerosis and cardiomyopathy due to insulin resistance (Wolf et al., 2014). Metformin has been shown to inhibit lipogenesis by suppressing sterol regulatory element binding protein 1 (SREBP-1) (Madsen et al., 2015), which is a transcriptional factor for lipogenesis. In particular, the inhibition of SREBP-1 by metformin is reported to be mediated by lncRNA; a study found that lncRNA-Gm15622 was upregulated in the liver of obese mice fed a HFD and in liver cells treated with saturated fatty acid. Metformin suppresses lncRNA-Gm15622 both in vivo and in vitro, thus decreasing the expression of SREBP-1c and inhibiting lipid accumulation by sponge for SREBP-1 targeted microRNA (Ma et al., 2020). Additionally, MEG3 and SNHG20 have been shown to be downregulated in HFD-induced fatty liver mice, and metformin improves lipid accumulation by reversing the expressions of MEG3 and SNHG20 (Guo et al., 2018). As stated above, MEG3 and SNHG20 were shown to regulate glucose uptake, indicating that these two lncRNAs may mediate the cross-talk of lipid and glucose metabolism via metformin.
Inflammation
Chronic inflammation is found in adipose tissues, pancreatic islets, and the liver (Lontchi-Yimagou et al., 2013), which contributes to insulin resistance and progression to T2DM and the long-term complications of diabetes (Berbudi et al., 2020; Gasmi et al., 2021). The nucleotide-binding oligomerization domain leucine-rich repeat and pyrin domain-containing protein 3 (NLRP3) inflammasome has emerged as a key mediator of pathological inflammation in diabetes (Esser et al., 2014). Firstly, inflammatory stimuli (such as TNFα, SFA, or LPS) induce NF-κB expression and result in the expression of pro-IL-1β and pro-IL-18; secondly, NLRP3 and GSDMD mediated the maturation and secretion of IL-1β and IL-18 after activation. Subsequently, cells secrete a large number of pro-inflammatory cytokines (e.g., IL-1β and IL-18) and induce a type of cell death called pyroptosis (Ding et al., 2019). The specific regulatory mechanisms of NLRP3 inflammasome activation remain unclear, and lncRNAs were shown to modulate NLRP3 activation in diabetes. For example, growth arrest-specific 5 (GAS5) overexpression suppressed NLRP3 inflammasome activation-mediated pyroptosis by regulating the miR-34b-3p/AHR axis in diabetic cardiomyocytes (Xu et al., 2020a). Although there is no direct evidence showing that metformin could regulate NLRP3 by regulating GAS5, clinical trials showed that taking metformin for 2 months was shown to increase GAS5 in patients with T2D (Parvar et al., 2023), indicating its anti-inflammatory role in the diabetic heart. Furthermore, metformin significantly alleviated NLRP3 and GSDMD expressions in diabetic periodontitis mice with a severe inflammation state, and in this study, lncRNA_1810058I24Rik, lncRNA_Gm12474, and lncRNA_Gm41514 were found to be co-expressed with NLRP3 and GSDMD (Zhou et al., 2020), which may potentially be regulated by metformin. However, a study conducted on the mouse glomerular membrane epithelial cell line SV40-MES-13 showed that metformin treatment efficiently inhibits elevation of H19 via high glucose in cells, with decreased levels of TNF-a, IL-6, and TGFβ-1, and overexpressed H19 abolishes the effects of metformin in SV40-MES-13 cells (Xu et al., 2020b), suggesting that the anti-inflammation effects of metformin are related to the regulation of lncRNAs.
Mitochondria
Mitochondrial dysfunction is a key pathological factor for diabetes. Reduced oxidative phosphorylation and excessive ROS production have been found in patients with obesity and type 2 diabetes (Wada and Nakatsuka, 2016; Luc et al., 2019). In addition, mitochondrial dynamic changes, such as mitochondrial fusion and fission, have been considered to be closely related to insulin sensitivity (Watanabe et al., 2014; Rovira-Llopis et al., 2017). For example, mice with knockdown of liver-specific mitofusin 2 (Mfn-2, a key protein for mitochondrial fusion) exhibit hepatic insulin resistance and glucose intolerant (Sebastian et al., 2012). For a decade, mitochondria have been considered a classic target for the blood glucose lowering actions of metformin. A most recent study identified the possible binding sites for biguanides on mitochondrial complex I protein subunits (Bridges et al., 2023). Additionally, mGPDH and complex IV have also been considered targets of metformin in animal experiments (Madiraju et al., 2014; LaMoia et al., 2022), and more targets have been found in the research into the actions of metformin in mitochondria (Pecinova et al., 2019; MacDonald et al., 2021). LncRNAs regulated by metformin potentially affect mitochondrial function; clinical research on DNA methylation in plasma from metformin-treated patients with T2DM showed that 57 lncRNA-associated DNA methylation regions were found, which included the mitochondrial ATP synthase-coupling factor 6 (ATP5J) (Solomon et al., 2020), suggesting that metformin may regulate ATP5J through lncRNAs. In addition, metformin increases GAS5 expression in HepG2 cells and the plasma of patients with T2DM (Parvar et al., 2023), and elevated GAS5 decreases mitochondrial tricarboxylic acid flux by disrupting the metabolic enzyme tandem of the tricarboxylic acid cycle (Sang et al., 2021), providing another explanation for the mitochondrial inhibition effects of metformin. Moreover, metformin decreases the expression of H19, while H19 has been shown to downregulate the expression of Mfn-2, a key protein for mitochondrial fusion, in type 1 diabetic renal tissues (Sultan et al., 2021), and a decline in Mfn2 was observed in both type 1 and type 2 diabetic rodents (Gao et al., 2012; Veeranki et al., 2016), suggesting that metformin may regulate Mfn2 by decreasing H19.
Limitations and future directions
On one hand, the doses of metformin used in experiments are critical for its molecular actions on tissues or organs. It is reported that exposing the liver to metformin concentrations of >1 mM is reported to be supra-pharmacologic and will likely result in non-clinically relevant effects of metformin, and the inhibition effects of complex I and activation of AMPK by metformin are reported to be the results of these supra-pharmacologic effects. Indeed, studies have shown that micromolar metformin concentrations suppress glucose production in primary mouse hepatocytes that may not be observed in clinical conditions. In our paper, we found the lncRNA studies related to the anti-diabetic effects of metformin were mostly conducted using an oral administration of 100–300 mg/kg/d in mice (Table 1), which achieves approximate plasma concentrations of 50–150 μM according to other animal studies (LaMoia and Shulman, 2021), similar to humans taking 1 g of metformin twice daily (35 μM), but in cell experiments, the dosage range of metformin was wide, ranging from in 10 μM to 2 mM mostly achieving maximum effectiveness at 1 mM (Table 1). However, the mechanism studies have mostly been proved in cell experiments; therefore, further investigations are needed to confirm the results. Furthermore, the understanding of the molecular mechanisms of metformin at low doses in the treatment of diabetes is expanding a lot, such as its impact on the gut microbiota, its targeting of the lysosomal PEN2–ATP6AP1 axis (Shin et al., 2020; Sugawara and Ogawa, 2023), and its immunomodulatory effects (Alrifai et al., 2019; Saito et al., 2020), but the evidence for lncRNA participation in these effects is lacking.
On the other hand, even in the same organ/tissue, metformin acts on multiple lncRNAs, and further research is needed on the lncRNAs that play the main role. Furthermore, the mechanism of actions of lncRNAs are not fully understood, and whether the regulatory effect of metformin on lncRNAs is the result of metabolic regulation or the reason for metabolic changes still needs further research. Moreover, current research on the mechanisms of lncRNAs focuses mainly on their function as molecular sponges for miRNA, but metformin itself also plays a role in miRNA expression (Wang et al., 2021b; Ao et al., 2023). Therefore, the relationship between the beneficial effects of metformin on metabolism and non-coding RNAs needs further clarification. Additionally, many lncRNAs, such as H19 and GAS5, have been shown to mediate the anti-diabetic effects of metformin, so the development of drugs targeting these lncRNAs may be a future direction for new drug research. However, the specificity of drug regulation of lncRNAs and the avoidance of possible side effects are problems that need to be solved.
Conclusion
In this review, we found that metformin regulates lncRNAs to inhibit gluconeogenesis, increase glucose uptake, decrease lipid deposition, and alleviate inflammation, as well as potentially target mitochondria, thus demonstrating that metformin exerts global anti-diabetic effects by targeting lncRNAs and that these effects involve multiple signal pathways. In addition, we found that metformin doses may cause differences in experimental and clinical effects. A unified dose or finding a reasonable explanation is a direction of further research into the mechanisms of action of metformin. Overall, regulation of lncRNAs is emerging as a supplementary mechanism for the anti-diabetic action of metformin; the understanding of the diversity of lncRNA modification by metformin will deepen our knowledge of its pharmaceutical effects and provide new insights for diabetic therapy.
Author contributions
WC: Conceptualization, Writing–original draft. WL: Writing–review and editing. PL: Funding acquisition, Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
(1998). Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). UK Prospective Diabetes Study (UKPDS) Group. Lancet 352 (9131), 854–865.
Al-Oanzi, Z. H., Fountana, S., Moonira, T., Tudhope, S. J., Petrie, J. L., Alshawi, A., et al. (2017). Opposite effects of a glucokinase activator and metformin on glucose-regulated gene expression in hepatocytes. Diabetes Obes. Metab. 19 (8), 1078–1087. doi:10.1111/dom.12910
Alrifai, T., Ali, F. S., Saleem, S., Ruiz, D. C. M., Rifai, D., Younas, S., et al. (2019). Immune checkpoint inhibitor induced diabetes mellitus treated with insulin and metformin: evolution of diabetes management in the era of immunotherapy. Case Rep. Oncol. Med. 2019, 8781347. doi:10.1155/2019/8781347
Aminimoghaddam, S., Fooladi, B., Noori, M., Nickhah Klashami, Z., Kakavand Hamidi, A., and M Amoli, M. (2021). The effect of metformin on expression of long non-coding RNA H19 in endometrial cancer. Med. J. Islam Repub. Iran. 35, 155. doi:10.47176/mjiri.35.155
Ao, X., Ding, W., Li, X., Xu, Q., Chen, X., Zhou, X., et al. (2023). Non-coding RNAs regulating mitochondrial function in cardiovascular diseases. J. Mol. Med. Berl. 101 (5), 501–526. doi:10.1007/s00109-023-02305-8
Athyros, V. G., Doumas, M., Imprialos, K. P., Stavropoulos, K., Georgianou, E., Katsimardou, A., et al. (2018). Diabetes and lipid metabolism. Horm. (Athens) 17 (1), 61–67. doi:10.1007/s42000-018-0014-8
Berbudi, A., Rahmadika, N., Tjahjadi, A. I., and Ruslami, R. (2020). Type 2 diabetes and its impact on the immune system. Curr. Diabetes Rev. 16 (5), 442–449. doi:10.2174/1573399815666191024085838
Bridges, H. R., Blaza, J. N., Yin, Z., Chung, I., Pollak, M. N., and Hirst, J. (2023). Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides. Science 379 (6630), 351–357. doi:10.1126/science.ade3332
Charytan, D. M., Solomon, S. D., Ivanovich, P., Remuzzi, G., Cooper, M. E., McGill, J. B., et al. (2019). Metformin use and cardiovascular events in patients with type 2 diabetes and chronic kidney disease. Diabetes Obes. Metab. 21 (5), 1199–1208. doi:10.1111/dom.13642
Chen, J., Qin, C., Zhou, Y., Chen, Y., Mao, M., and Yang, J. (2022). Metformin may induce ferroptosis by inhibiting autophagy via lncRNA H19 in breast cancer. FEBS Open Bio 12 (1), 146–153. doi:10.1002/2211-5463.13314
Conceicao, I., Luscher-Dias, T., Queiroz, L. R., de Melo, A. G. B., Machado, C. R., Gomes, K. B., et al. (2022). Metformin treatment modulates long non-coding RNA isoforms expression in human cells. Noncoding RNA 8 (5), 68. doi:10.3390/ncrna8050068
Deng, J., Mueller, M., Geng, T., Shen, Y., Liu, Y., Hou, P., et al. (2017). H19 lncRNA alters methylation and expression of Hnf4α in the liver of metformin-exposed fetuses. Cell Death Dis. 8 (12), e3175. doi:10.1038/cddis.2017.392
Diabetes Prevention Program Research, G. (2012). Long-term safety, tolerability, and weight loss associated with metformin in the Diabetes Prevention Program Outcomes Study. Diabetes Care 35 (4), 731–737. doi:10.2337/dc11-1299
Ding, G. L., Wang, F. F., Shu, J., Tian, S., Jiang, Y., Zhang, D., et al. (2012). Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 61 (5), 1133–1142. doi:10.2337/db11-1314
Ding, S., Xu, S., Ma, Y., Liu, G., Jang, H., and Fang, J. (2019). Modulatory mechanisms of the NLRP3 inflammasomes in diabetes. Biomolecules 9 (12), 850. doi:10.3390/biom9120850
Duan, W., Ding, Y., Yu, X., Ma, D., Yang, B., Li, Y., et al. (2019). Metformin mitigates autoimmune insulitis by inhibiting Th1 and Th17 responses while promoting Treg production. Am. J. Transl. Res. 11 (4), 2393–2402.
El-Mir, M. Y., Nogueira, V., Fontaine, E., Avéret, N., Rigoulet, M., and Leverve, X. (2000). Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 275 (1), 223–228. doi:10.1074/jbc.275.1.223
Esser, N., Legrand-Poels, S., Piette, J., Scheen, A. J., and Paquot, N. (2014). Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 105 (2), 141–150. doi:10.1016/j.diabres.2014.04.006
Foretz, M., Guigas, B., and Viollet, B. (2019). Understanding the glucoregulatory mechanisms of metformin in type 2 diabetes mellitus. Nat. Rev. Endocrinol. 15 (10), 569–589. doi:10.1038/s41574-019-0242-2
Foretz, M., Hébrard, S., Leclerc, J., Zarrinpashneh, E., Soty, M., Mithieux, G., et al. (2010). Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Invest. 120 (7), 2355–2369. doi:10.1172/JCI40671
Ganesan, K., Rana, M. B. M., and Sultan, S. (2023). Oral Hypoglycemic Medications, in StatPearls. 2023: treasure Island (FL) ineligible companies. Disclosure: muhammad Burhan Majeed Rana declares no relevant financial relationships with ineligible companies. Disclosure: senan Sultan declares no relevant financial relationships with ineligible companies.
Gao, Q., Wang, X. M., Ye, H. W., Yu, Y., Kang, P. F., Wang, H. J., et al. (2012). Changes in the expression of cardiac mitofusin-2 in different stages of diabetes in rats. Mol. Med. Rep. 6 (4), 811–814. doi:10.3892/mmr.2012.1002
Garber, A. J., Duncan, T. G., Goodman, A. M., Mills, D. J., and Rohlf, J. L. (1997). Efficacy of metformin in type II diabetes: results of a double-blind, placebo-controlled, dose-response trial. Am. J. Med. 103 (6), 491–497. doi:10.1016/s0002-9343(97)00254-4
Gasmi, A., Noor, S., Menzel, A., Doşa, A., Pivina, L., and Bjørklund, G. (2021). Obesity and insulin resistance: associations with chronic inflammation, genetic and epigenetic factors. Curr. Med. Chem. 28 (4), 800–826. doi:10.2174/0929867327666200824112056
Golshan, M., Khaleghi, S., Shafiee, S. M., Valaee, S., Ghanei, Z., Jamshidizad, A., et al. (2021). Metformin modulates oncogenic expression of HOTAIR gene via promoter methylation and reverses epithelial-mesenchymal transition in MDA-MB-231 cells. J. Cell Biochem. 122 (3-4), 385–393. doi:10.1002/jcb.29867
Guo, J., Zhou, Y., Cheng, Y., Fang, W., Hu, G., Wei, J., et al. (2018). Metformin-induced changes of the coding transcriptome and non-coding RNAs in the livers of non-alcoholic fatty liver disease mice. Cell Physiol. Biochem. 45 (4), 1487–1505. doi:10.1159/000487575
Hirota, K., Sakamaki, J. i., Ishida, J., Shimamoto, Y., Nishihara, S., Kodama, N., et al. (2008). A combination of HNF-4 and Foxo1 is required for reciprocal transcriptional regulation of glucokinase and glucose-6-phosphatase genes in response to fasting and feeding. J. Biol. Chem. 283 (47), 32432–32441. doi:10.1074/jbc.M806179200
Huang, Y., Xu, Y., Lu, Y., Zhu, S., Guo, Y., Sun, C., et al. (2019). lncRNA Gm10451 regulates PTIP to facilitate iPSCs-derived beta-like cell differentiation by targeting miR-338-3p as a ceRNA. Biomaterials 216, 119266. doi:10.1016/j.biomaterials.2019.119266
Huang, Y., Zhou, Z., Zhang, J., Hao, Z., He, Y., Wu, Z., et al. (2021). lncRNA MALAT1 participates in metformin inhibiting the proliferation of breast cancer cell. J. Cell Mol. Med. 25 (15), 7135–7145. doi:10.1111/jcmm.16742
Ilieva, M., and Uchida, S. (2022). Long noncoding RNAs in induced pluripotent stem cells and their differentiation. Am. J. Physiol. Cell Physiol. 322 (4), C769–C774. doi:10.1152/ajpcell.00059.2022
Jager, J. De., Kooy, A., Lehert, Ph., Bets, D., Wulffelé, M. G., Teerlink, T., et al. (2005). Effects of short-term treatment with metformin on markers of endothelial function and inflammatory activity in type 2 diabetes mellitus: a randomized, placebo-controlled trial. J. Intern. Med. 257 (1), 100–109. doi:10.1111/j.1365-2796.2004.01420.x
Knudsen, J. R., Li, Z., Persson, K. W., Li, J., Henriquez-Olguin, C., and Jensen, T. E. (2020). Contraction-regulated mTORC1 and protein synthesis: influence of AMPK and glycogen. J. Physiol. 598 (13), 2637–2649. doi:10.1113/JP279780
Koo, S. H., Flechner, L., Qi, L., Zhang, X., Screaton, R. A., Jeffries, S., et al. (2005). The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437 (7062), 1109–1111. doi:10.1038/nature03967
LaMoia, T. E., Butrico, G. M., Kalpage, H. A., Goedeke, L., Hubbard, B. T., Vatner, D. F., et al. (2022). Metformin, phenformin, and galegine inhibit complex IV activity and reduce glycerol-derived gluconeogenesis. Proc. Natl. Acad. Sci. U. S. A. 119 (10), e2122287119. doi:10.1073/pnas.2122287119
LaMoia, T. E., and Shulman, G. I. (2021). Cellular and molecular mechanisms of metformin action. Endocr. Rev. 42 (1), 77–96. doi:10.1210/endrev/bnaa023
Li, P., Tong, L., Song, Y., Sun, J., Shi, J., Wu, Z., et al. (2019). Long noncoding RNA H19 participates in metformin-mediated inhibition of gastric cancer cell invasion. J. Cell Physiol. 234 (4), 4515–4527. doi:10.1002/jcp.27269
Li, Z., Liu, L., Yang, Y., Zheng, H., Cai, Y., Ma, Y., et al. (2022). Metformin ameliorates senescence of adipose-derived mesenchymal stem cells and attenuates osteoarthritis progression via the AMPK-dependent autophagy pathway. Oxid. Med. Cell Longev. 2022, 4620254. doi:10.1155/2022/4620254
Liu, J., Zhao, Y., Chen, L., Li, R., Ning, Y., and Zhu, X. (2022b). Role of metformin in functional endometrial hyperplasia and polycystic ovary syndrome involves the regulation of MEG3/miR-223/GLUT4 and SNHG20/miR-4486/GLUT4 signaling. Mol. Med. Rep. 26 (1), 218. doi:10.3892/mmr.2022.12734
Liu, Y., Ao, X., Wang, Y., Li, X., and Wang, J. (2022a). Long non-coding RNA in gastric cancer: mechanisms and clinical implications for drug resistance. Front. Oncol. 12, 841411. doi:10.3389/fonc.2022.841411
Liu, Y., Ding, W., Wang, J., Ao, X., and Xue, J. (2023). Non-coding RNA-mediated modulation of ferroptosis in cardiovascular diseases. Biomed. Pharmacother. 164, 114993. doi:10.1016/j.biopha.2023.114993
Liu, Y., Ding, W., Yu, W., Zhang, Y., Ao, X., and Wang, J. (2021). Long non-coding RNAs: biogenesis, functions, and clinical significance in gastric cancer. Mol. Ther. Oncolytics 23, 458–476. doi:10.1016/j.omto.2021.11.005
Lontchi-Yimagou, E., Sobngwi, E., Matsha, T. E., and Kengne, A. P. (2013). Diabetes mellitus and inflammation. Curr. Diab Rep. 13 (3), 435–444. doi:10.1007/s11892-013-0375-y
Lu, J., Huang, Y., Zhang, X., Xu, Y., and Nie, S. (2021). Noncoding RNAs involved in DNA methylation and histone methylation, and acetylation in diabetic vascular complications. Pharmacol. Res. 170, 105520. doi:10.1016/j.phrs.2021.105520
Luc, K., Schramm-Luc, A., Guzik, T. J., and Mikolajczyk, T. P. (2019). Oxidative stress and inflammatory markers in prediabetes and diabetes. J. Physiol. Pharmacol. 70 (6). doi:10.26402/jpp.2019.6.01
Lundsgaard, A. M., Fritzen, A. M., Nicolaisen, T. S., Carl, C. S., Sjøberg, K. A., Raun, S. H., et al. (2020). Glucometabolic consequences of acute and prolonged inhibition of fatty acid oxidation. J. Lipid Res. 61 (1), 10–19. doi:10.1194/jlr.RA119000177
Lv, Z., and Guo, Y. (2020). Metformin and its benefits for various diseases. Front. Endocrinol. (Lausanne) 11, 191. doi:10.3389/fendo.2020.00191
Ma, M., Duan, R., Shen, L., Liu, M., Ji, Y., Zhou, H., et al. (2020). The lncRNA Gm15622 stimulates SREBP-1c expression and hepatic lipid accumulation by sponging the miR-742-3p in mice. J. Lipid Res. 61 (7), 1052–1064. doi:10.1194/jlr.RA120000664
MacDonald, M. J., Ansari, I. U. H., Longacre, M. J., and Stoker, S. W. (2021). Metformin's therapeutic efficacy in the treatment of diabetes does not involve inhibition of mitochondrial glycerol phosphate dehydrogenase. Diabetes 70 (7), 1575–1580. doi:10.2337/db20-1143
Madiraju, A. K., Erion, D. M., Rahimi, Y., Zhang, X. M., Braddock, D. T., Albright, R. A., et al. (2014). Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510 (7506), 542–546. doi:10.1038/nature13270
Madiraju, A. K., Qiu, Y., Perry, R. J., Rahimi, Y., Zhang, X. M., Zhang, D., et al. (2018). Metformin inhibits gluconeogenesis via a redox-dependent mechanism in vivo. Nat. Med. 24 (9), 1384–1394. doi:10.1038/s41591-018-0125-4
Madsen, A., Bozickovic, O., Bjune, J. I., Mellgren, G., and Sagen, J. V. (2015). Metformin inhibits hepatocellular glucose, lipid and cholesterol biosynthetic pathways by transcriptionally suppressing steroid receptor coactivator 2 (SRC-2). Sci. Rep. 5, 16430. doi:10.1038/srep16430
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24 (6), 430–447. doi:10.1038/s41580-022-00566-8
Mues, C., Zhou, J., Manolopoulos, K. N., Korsten, P., Schmoll, D., Klotz, L. O., et al. (2009). Regulation of glucose-6-phosphatase gene expression by insulin and metformin. Horm. Metab. Res. 41 (10), 730–735. doi:10.1055/s-0029-1225360
Owen, M. R., Doran, E., and Halestrap, A. P. (2000). Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 348 (Pt 3), 607–614. doi:10.1042/bj3480607
Panni, S., Lovering, R. C., Porras, P., and Orchard, S. (2020). Non-coding RNA regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech. 1863 (6), 194417. doi:10.1016/j.bbagrm.2019.194417
Parvar, S. N., Mirzaei, A., Zare, A., Doustimotlagh, A. H., Nikooei, S., Arya, A., et al. (2023). Effect of metformin on the long non-coding RNA expression levels in type 2 diabetes: an in vitro and clinical trial study. Pharmacol. Rep. 75 (1), 189–198. doi:10.1007/s43440-022-00427-3
Pecinova, A., Brázdová, A., Drahota, Z., Houštěk, J., and Mráček, T. (2019). Mitochondrial targets of metformin-Are they physiologically relevant? Biofactors 45 (5), 703–711. doi:10.1002/biof.1548
Rovira-Llopis, S., Bañuls, C., Diaz-Morales, N., Hernandez-Mijares, A., Rocha, M., and Victor, V. M. (2017). Mitochondrial dynamics in type 2 diabetes: pathophysiological implications. Redox Biol. 11, 637–645. doi:10.1016/j.redox.2017.01.013
Saito, A., Kitayama, J., Horie, H., Koinuma, K., Ohzawa, H., Yamaguchi, H., et al. (2020). Metformin changes the immune microenvironment of colorectal cancer in patients with type 2 diabetes mellitus. Cancer Sci. 111 (11), 4012–4020. doi:10.1111/cas.14615
Sang, L., Ju, H. Q., Yang, Z., Ge, Q., Zhang, Z., Liu, F., et al. (2021). Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 3 (1), 90–106. doi:10.1038/s42255-020-00325-z
Sathishkumar, C., Prabu, P., Mohan, V., and Balasubramanyam, M. (2018). Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genomics 12 (1), 41. doi:10.1186/s40246-018-0173-3
Sebastian, D., Hernández-Alvarez, M. I., Segalés, J., Sorianello, E., Muñoz, J. P., Sala, D., et al. (2012). Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc. Natl. Acad. Sci. U. S. A. 109 (14), 5523–5528. doi:10.1073/pnas.1108220109
Shin, N. R., Gu, N., Choi, H. S., and Kim, H. (2020). Combined effects of Scutellaria baicalensis with metformin on glucose tolerance of patients with type 2 diabetes via gut microbiota modulation. Am. J. Physiol. Endocrinol. Metab. 318 (1), E52-E61–E61. doi:10.1152/ajpendo.00221.2019
Shu, C., Yan, D., Chen, C., Mo, Y., Wu, L., Gu, J., et al. (2019). Metformin exhibits its therapeutic effect in the treatment of pre-eclampsia via modulating the Met/H19/miR-148a-5p/P28 and Met/H19/miR-216-3p/EBI3 signaling pathways. Int. Immunopharmacol. 74, 105693. doi:10.1016/j.intimp.2019.105693
Shu, L., Hou, X., Song, G., Wang, C., and Ma, H. (2021). Comparative analysis of long non-coding RNA expression profiles induced by resveratrol and metformin treatment for hepatic insulin resistance. Int. J. Mol. Med. 48 (5), 206. doi:10.3892/ijmm.2021.5039
Solomon, W. L., Hector, S. B. E., Raghubeer, S., Erasmus, R. T., Kengne, A. P., and Matsha, T. E. (2020). Genome-wide DNA methylation and LncRNA-associated DNA methylation in metformin-treated and -untreated diabetes. Epigenomes 4 (3), 19. doi:10.3390/epigenomes4030019
Sugawara, K., and Ogawa, W. (2023). New mechanism of metformin action mediated by lysosomal presenilin enhancer 2. J. Diabetes Investig. 14 (1), 12–14. doi:10.1111/jdi.13925
Sultan, H. K., El-Ayat, W. M., AbouGhalia, A. H., Lasheen, N. N., and Moustafa, A. S. (2021). Study of long non-coding RNA and mitochondrial dysfunction in diabetic rats. Tissue Cell 71, 101516. doi:10.1016/j.tice.2021.101516
Takahashi, N., Kimura, A. P., Otsuka, K., Ohmura, K., Naito, S., Yoshida, M., et al. (2019). Dreh, a long noncoding RNA repressed by metformin, regulates glucose transport in C2C12 skeletal muscle cells. Life Sci. 236, 116906. doi:10.1016/j.lfs.2019.116906
Veeranki, S., Givvimani, S., Kundu, S., Metreveli, N., Pushpakumar, S., and Tyagi, S. C. (2016). Moderate intensity exercise prevents diabetic cardiomyopathy associated contractile dysfunction through restoration of mitochondrial function and connexin 43 levels in db/db mice. J. Mol. Cell Cardiol. 92, 163–173. doi:10.1016/j.yjmcc.2016.01.023
Wada, J., and Nakatsuka, A. (2016). Mitochondrial dynamics and mitochondrial dysfunction in diabetes. Acta Med. Okayama 70 (3), 151–158. doi:10.18926/AMO/54413
Wang, G., Lin, F., Wan, Q., Wu, J., and Luo, M. (2021b). Mechanisms of action of metformin and its regulatory effect on microRNAs related to angiogenesis. Pharmacol. Res. 164, 105390. doi:10.1016/j.phrs.2020.105390
Wang, L., Zhao, D., Tang, L., Li, H., Liu, Z., Gao, J., et al. (2021a). Soluble epoxide hydrolase deficiency attenuates lipotoxic cardiomyopathy via upregulation of AMPK-mTORC mediated autophagy. J. Mol. Cell Cardiol. 154, 80–91. doi:10.1016/j.yjmcc.2020.12.013
Wang, Q., Shang, J., Zhang, Y., and Zhou, W. (2019b). Metformin and sitagliptin combination therapy ameliorates polycystic ovary syndrome with insulin resistance through upregulation of lncRNA H19. Cell Cycle 18 (19), 2538–2549. doi:10.1080/15384101.2019.1652036
Wang, Y., An, H., Liu, T., Qin, C., Sesaki, H., Guo, S., et al. (2019a). Metformin improves mitochondrial respiratory activity through activation of AMPK. Cell Rep. 29 (6), 1511–1523. doi:10.1016/j.celrep.2019.09.070
Wang, Y., Tang, H., Ji, X., Zhang, Y., Xu, W., Yang, X., et al. (2018). Expression profile analysis of long non-coding RNAs involved in the metformin-inhibited gluconeogenesis of primary mouse hepatocytes. Int. J. Mol. Med. 41 (1), 302–310. doi:10.3892/ijmm.2017.3243
Watanabe, T., Saotome, M., Nobuhara, M., Sakamoto, A., Urushida, T., Katoh, H., et al. (2014). Roles of mitochondrial fragmentation and reactive oxygen species in mitochondrial dysfunction and myocardial insulin resistance. Exp. Cell Res. 323 (2), 314–325. doi:10.1016/j.yexcr.2014.02.027
Wolf, P., Winhofer, Y., Anderwald, C. H., Krššák, M., and Krebs, M. (2014). Intracellular lipid accumulation and shift during diabetes progression. Wien Med. Wochenschr 164 (15-16), 320–329. doi:10.1007/s10354-014-0292-y
Wu, H., Esteve, E., Tremaroli, V., Khan, M. T., Caesar, R., Mannerås-Holm, L., et al. (2017). Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23 (7), 850–858. doi:10.1038/nm.4345
Wu, Z., Bai, Y., Qi, Y., Chang, C., Jiao, Y., Bai, Y., et al. (2023). Metformin ameliorates ferroptosis in cardiac ischemia and reperfusion by reducing NOX4 expression via promoting AMPKα. Pharm. Biol. 61 (1), 886–896. doi:10.1080/13880209.2023.2212700
Xia, C., Liang, S., He, Z., Zhu, X., Chen, R., and Chen, J. (2018). Metformin, a first-line drug for type 2 diabetes mellitus, disrupts the MALAT1/miR-142-3p sponge to decrease invasion and migration in cervical cancer cells. Eur. J. Pharmacol. 830, 59–67. doi:10.1016/j.ejphar.2018.04.027
Xu, J., Xiang, P., Liu, L., Sun, J., and Ye, S. (2020b). Metformin inhibits extracellular matrix accumulation, inflammation and proliferation of mesangial cells in diabetic nephropathy by regulating H19/miR-143-3p/TGF-β1 axis. J. Pharm. Pharmacol. 72 (8), 1101–1109. doi:10.1111/jphp.13280
Xu, Y., Fang, H., Xu, Q., Xu, C., Yang, L., and Huang, C. (2020a). LncRNA GAS5 inhibits NLRP3 inflammasome activation-mediated pyroptosis in diabetic cardiomyopathy by targeting miR-34b-3p/AHR. Cell Cycle 19 (22), 3054–3065. doi:10.1080/15384101.2020.1831245
Yan, L., Zhou, J., Gao, Y., Ghazal, S., Lu, L., Bellone, S., et al. (2015). Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 34 (23), 3076–3084. doi:10.1038/onc.2014.236
Yang, F., Qin, Y., Lv, J., Wang, Y., Che, H., Chen, X., et al. (2018a). Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis. 9 (10), 1000. doi:10.1038/s41419-018-1029-4
Yang, F., Qin, Y., Wang, Y., Meng, S., Xian, H., Che, H., et al. (2019). Metformin inhibits the NLRP3 inflammasome via AMPK/mTOR-dependent effects in diabetic cardiomyopathy. Int. J. Biol. Sci. 15 (5), 1010–1019. doi:10.7150/ijbs.29680
Yang, Q., Vijayakumar, A., and Kahn, B. B. (2018b). Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 19 (10), 654–672. doi:10.1038/s41580-018-0044-8
Yang, Q., Wang, G., Fang, D., Gao, X., Liang, Y., Wang, L., et al. (2022). The role of MicroRNA networks in tissue-specific direct and indirect effects of metformin and its application. Biomed. Pharmacother. 151, 113130. doi:10.1016/j.biopha.2022.113130
You, G., Long, X., Song, F., Huang, J., Tian, M., Xiao, Y., et al. (2020). Metformin activates the AMPK-mTOR pathway by modulating lncRNA TUG1 to induce autophagy and inhibit atherosclerosis. Drug Des. Devel Ther. 14, 457–468. doi:10.2147/DDDT.S233932
Zhang, Y., Liu, Y., Liu, X., Yuan, X., Xiang, M., Liu, J., et al. (2022a). Exercise and metformin intervention prevents lipotoxicity-induced hepatocyte apoptosis by alleviating oxidative and ER stress and activating the AMPK/Nrf2/HO-1 signaling pathway in db/db mice. Oxid. Med. Cell Longev. 2022, 2297268. doi:10.1155/2022/2297268
Zhang, Z. M., Liu, Z. H., Nie, Q., Zhang, X. M., Yang, L. Q., Wang, C., et al. (2022b). Metformin improves high-fat diet-induced insulin resistance in mice by downregulating the expression of long noncoding RNA NONMMUT031874.2. Exp. Ther. Med. 23 (5), 332. doi:10.3892/etm.2022.11261
Zhou, G., Myers, R., Li, Y., Chen, Y., Shen, X., Fenyk-Melody, J., et al. (2001). Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108 (8), 1167–1174. doi:10.1172/JCI13505
Zhou, X., Ao, X., Jia, Z., Li, Y., Kuang, S., Du, C., et al. (2022). Non-coding RNA in cancer drug resistance: underlying mechanisms and clinical applications. Front. Oncol. 12, 951864. doi:10.3389/fonc.2022.951864
Zhou, X., Wang, Q., Nie, L., Zhang, P., Zhao, P., Yuan, Q., et al. (2020). Metformin ameliorates the NLPP3 inflammasome mediated pyroptosis by inhibiting the expression of NEK7 in diabetic periodontitis. Arch. Oral Biol. 116, 104763. doi:10.1016/j.archoralbio.2020.104763
Glossary
Keywords: long non-coding RNA, T2DM, metformin, mechanism, AMPK
Citation: Chang W, Li W and Li P (2023) The anti-diabetic effects of metformin are mediated by regulating long non-coding RNA. Front. Pharmacol. 14:1256705. doi: 10.3389/fphar.2023.1256705
Received: 11 July 2023; Accepted: 02 November 2023;
Published: 20 November 2023.
Edited by:
Ahmed Esmat Abdel Moneim, Helwan University, EgyptReviewed by:
Tarun Pant, Medical College of Wisconsin, United StatesMaria Theresa Montales, University of Arkansas for Medical Sciences, United States
Ting Yuan, Goethe-Universität Frankfurt, Germany
Copyright © 2023 Chang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenguang Chang, Y2hhbmdzdWJtaXRAMTI2LmNvbQ==; Peifeng Li, cGVpZmxpQGhvdG1haWwuY29t
 Wenguang Chang
Wenguang Chang Wei Li2
Wei Li2