
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 04 August 2023
Sec. Pharmacogenetics and Pharmacogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1244516
 Polina Petkova-Kirova1
Polina Petkova-Kirova1 Stephan Baas2
Stephan Baas2 Gudrun Wagenpfeil3
Gudrun Wagenpfeil3 Philip Hartz1
Philip Hartz1 Marcus Michael Unger4*
Marcus Michael Unger4* Rita Bernhardt1*
Rita Bernhardt1*Parkinson’s disease (PD) is one of the most frequent neurological diseases affecting millions of people worldwide. While the majority of PD cases are of unknown origin (idiopathic), about 5%–10% are familial and linked to mutations in different known genes. However, there are also people with a genetic predisposition to PD who do not develop the disease. To elucidate factors leading to the manifestation of PD we compared the occurrence of single nucleotide polymorphisms (SNPs) in various cytochrome P450 (P450) genes in people with a genetic predisposition and suffering from PD (GPD) to that of people, who are genetically predisposed, but show no symptoms of the disease (GUN). We used the PPMI (Parkinson’s Progression Markers Initiative) database and the gene sequences of all 57 P450s as well as their three redox partners. Corresponding odds ratios (OR) and confidence intervals (CI) were calculated to assess the incidence of the various SNPs in the two groups of individuals and consequently their relation to PD. We identified for the first time SNPs that are significantly (up to 10fold!) over- or under-represented in GPD patients compared to GUN. SNPs with OR > 5 were found in 10 P450s being involved in eicosanoid, vitamin A and D metabolism as well as cholesterol degradation pointing to an important role of endogenous factors for the manifestation of PD clinical symptoms. Moreover, 12 P450s belonging to all P450 substrate classes as well as POR have SNPs that are significantly under-represented (OR < 0.2) in GPD compared to GUN, indicating a protective role of those SNPs and the corresponding P450s regarding disease advancement. To the best of our knowledge our data for the first time demonstrate an association between known PD predisposition genes and SNPs in other genes, shown here for different P450 genes and for their redox partner POR, which promote the manifestation of the disease in familial PD. Our results thus shed light onto the pathogenesis of PD, especially the switch from GUN to GPD and might further help to advance novel strategies for preventing the development or progression of the disease.
Parkinson’s disease (PD) is the second most common neurodegenerative disorder worldwide. According to the World Health Organization, disease prevalence has doubled in the last 25 years. For the year 2019, over 8.5 million people have been shown to suffer from PD with estimated 329,000 deaths caused by the disease, an increase of over 100% since 2000 (https://www.who.int/news-room/fact-sheets/detail/parkinson-disease). The reason for the degeneration of Substantia nigra pars compacta and the related loss of dopaminergic neurons, main characteristics of the disease, is not yet completely known. Therefore, treatment of PD remains symptomatic and new biomarkers for early diagnostics and novel approaches as well as candidates for causative and effective treatment are pursued.
Very recently we published a novel approach analyzing a possible association between the pathogenesis of PD and single nucleotide polymorphisms (SNPs) in cytochrome P450 (P450) genes (Hartz et al., 2022). The Parkinson’s Progression Markers Initiative (PPMI) database was used to evaluate possible relations between variations in the genes of the 57 human P450s and their 3 redox partners, a cytochrome P450 reductase (POR), adrenodoxin (Adx) and adrenodoxin reductase (AdR), and the origin and development of PD. Statistical analysis of the data was applied to calculate corresponding odds ratios (OR) and confidence intervals (CI) to estimate the link between the occurrence of a particular SNP in the genes of the 60 proteins and the incidence of PD. It was shown that SNPs in 26 out of 57 P450s and their three redox partners were significantly over-represented (with OR values >5) in patients with a genetic predisposition to PD (GPD) compared to healthy subjects (HC), taken as controls. Three main groups of P450s were shown to contain those SNPs: 1) xenobiotics-metabolizing P450s demonstrating the biggest accumulation of highly over-represented SNPs, posing a major role for toxic compounds in the pathogenesis of PD, 2) P450s involved in eicosanoid metabolism, confirming the relation of PD to inflammation and 3) P450s involved in the degradation of cholesterol (CYP46A1, CY7B1, CYP39A1), indicating a prominent role of brain cholesterol metabolism for the risk to develop PD. Additionally, POR, required for electron transfer from NADPH to microsomal P450s, displayed nine SNPs with OR > 5 in GPD and thus also seems to play an important role in the etiology of PD (Hartz et al., 2022). In contrast, in idiopathic PD patients (IPD), SNPs with OR values >5 were only described for CYP46A1 as well as Adx and POR. However, SNPs with OR values between 2 and 5 are much more abundant, especially in the group of xenobiotics-converting enzymes like CYP2E1 and CYP2C8 (Hartz et al., 2022).
While the majority of PD cases worldwide are idiopathic, about 10%–15% of all patients and about 25% of early-onset patients have a family history (first degree relatives) of PD and approximately 5%–10% are ascertained with a defined genetic predisposition (Sellbach et al., 2006; Lesage and Brice, 2009; Nalls et al., 2019). Mutations in seven genes have been unambiguously linked to typical familial PD: SNCA, LRRK2, VPS35, PRKN, PINK1, GBA and DJ-1 (Bandres-Ciga et al., 2020), but most of the patients with a genetic predisposition studied so far show changes in the LRRK2 (leucine rich repeat kinase 2) gene leading to changes in kinase activity and a-synuclein and mitochondrial impairments (Di Maio et al., 2018). Interestingly, in the PPMI database there are also individuals who have a genetic predisposition to PD, but do not show any signs of the disease (designated in our study as GUN). It is not clear at the moment which factors are necessary to promote the progression of the disease at the background of an existing genetic predisposition.
Thus, the aim of our present study was to analyze the differences in the occurrence of SNPs in the P450 genes and the genes of their three redox partners in GPD and GUN individuals to further shed light on factors eliciting disease manifestation.
Information for the genetic variants of the 57 P450s and their 3 redox partners was obtained as previously described (Hartz et al., 2022). Briefly, whole genome sequencing (WGS) VCF files were extracted from the Parkinson’s Progression Markers Initiative database (PPMI; July 2018 release) (Marek et al., 2018) for P450s, POR, Adx and AdR and genetic variants were identified. The identified variants were annotated with SnpEff 5.0 (Cingolani et al., 2012b) and SnpSift 5.0 (Cingolani et al., 2012a), further processed with the R programming language and classified in groups (IPD, GPD, GUN and HC) based on clinical diagnosis. Altogether 317 GPD, 344 GUN and 193 HC were included into our analyses. In addition to the genetic data, information for the intake of fish oil supplements and vitamin D by some of the patients was also obtained from the PPMI database (https://ida.loni.usc.edu/login.jsp?project=ppmi). A comparison was made, using Pearson’s chi-square test, between GUN individuals and GPD patients, for whom information in the PPMI database was available stating that they take vitamin D (or vitamin D-based medicines like Dekristol) or fish oil/omega-3 fatty acids. We aimed to assess whether the intake of these supplements would protect the genetically predisposed individuals (GUN) from getting symptoms of the disease (GPD).
IBM-SPSS Version 26 and 27 were used for statistical analysis. Due to the explorative nature of the study, we did not account for the issue of multiple statistical testing. Thus, we report raw 2-sided p-values without adjustment. The significance level p is set at 0.05. Risk factors are assessed using logistic regression and reported as odds ratios (OR) with 95% confidence intervals (CI) (Hartz et al., 2022). The odds ratio is a statistical measure describing the strength and direction of an association between two variables, in our case a certain SNP in a PD patient compared with HC or GUN. Calculation of OR is performed as follows (number of PD patients with a SNP/number of HC (GUN) with a SNP)/(number of PD patients without a SNP/number of HC (GUN) without a SNP). If OR is <1, the SNP is under-represented in PD patients, if it is >1, then it appears to be over-represented in PD patients compared to HC (GUN). The CI gives a measure as to how precise the OR value could be calculated.
The P450s are subdivided into 6 groups according to their main function and substrate class as suggested by Guengerich (Guengerich, 2017) and this classification is applied consistently throughout the whole study.
Our first approach was to compare the number of SNPs in GUN (individuals with genetic predisposition to PD, but without symptoms of the disease) and GPD (individuals with genetic predisposition to PD suffering from the disease) with that of the healthy individuals in the PPMI database, taken as controls (GUN/HC and GPD/HC, Table 1). Surprisingly, it turned out that both, GUN and GPD, have many SNPs, which are strongly over-represented compared to their occurrence in controls (Table 1). As can be seen from Table 1, most of the 57 P450s show SNPs with OR values >5, i.e., being more than 5 times over-represented in GUN and GPD compared to HC. Only 26 out of 57 P450s as well as the three redox partners do not show SNPs with OR values >5 when comparing GUN and HC and 31 out of 57 P450s as well as AdR and Adx when comparing GPD and HC. As shown previously (Hartz et al., 2022), this value is much higher (56 P450s and AdR) when comparing PD patients with unknown origin of the disease (IPD) and HC, where only CYP46A1 as well as Adx and POR were demonstrated to display SNPs with OR values >5 compared to HC.
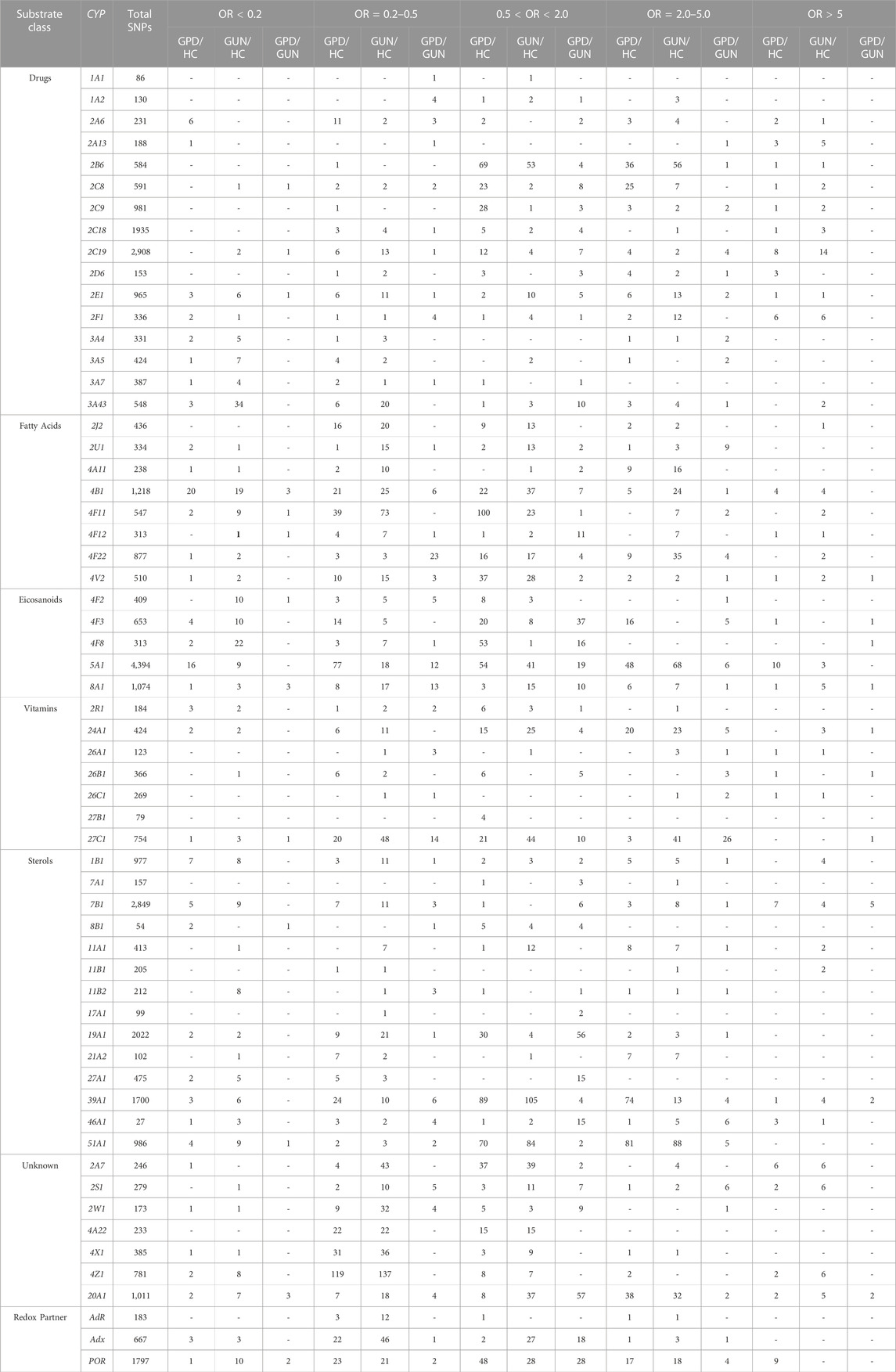
TABLE 1. Summary of the single nucleotide polymorphisms (SNPs) with statistically significant association (p-value < 0.05) to Parkinson´s Disease (PD) for the 57 human cytochrome P450 genes and their 3 redox partner genes, Adx, AdR and POR. Compared are GPD and HC (GPD/HC), GUN and HC (GUN/HC) and GPD and GUN (GPD/GUN) and SNPs for the various comparisons are subdivided into groups based on the strength of association to PD, represented by the odds ratio (OR). SNPs with OR > 5 and OR < 0.2 are most strongly associated to PD being more than 5 times over- or underrepresented, respectively, in GPD patients when comparing GPD and HC (GPD/HC), in GUN patients when comparing GUN and HC (GUN/HC) and in GPD patients when comparing GPD and GUN patients (GPD/GUN). The cytochrome P450s (CYPs) are subdivided into 6 groups according to their main function and substrate class (Guengerich, 2017).
Regarding P450s with SNPs displaying OR values <0.2 i.e., SNPs that are more than 5 times over-represented in HC compared to GPD or GUN, it was found that there are 23 P450s as well as AdR in GPD/HC and 18 P450s as well as AdR in GUN/HC present which do not have SNPs with OR < 0.2 (Table 1). Again, the number of P450s with OR < 0.2 is lower when comparing IPD patients with controls (37 P450s as well as AdR and Adx do not display SNPs with OR < 0.2) (Hartz et al., 2022).
To identify novel players leading to the differences between GPD and GUN individuals, i.e., to question why GUN individuals do not show symptoms of PD although they also have a genetic predisposition, while GPD individuals show such symptoms, we compared the distribution of SNPs between the two groups (GPD/GUN, Table 1; Table 2). It can be seen that the genes of 7 P450s as well as AdR do not show any significant differences between GPD patients and GUN (Table 2). In contrast, three P450 genes (CYP4V2, CYP24A1 and CYP39A1) display SNPs with OR values >10 when comparing the two groups. Most P450s fall into the group with marginal effects (OR values between 0.50 and 2.00). Differences between the two groups with OR values >5 or <0.2 are found in 10 and 12 P450s, respectively (Table 1). POR has also two SNPs that appear with OR values below 0.2 when comparing GPD and GUN individuals. Moreover, as can be seen in Table 1, considerably less SNPs are statistically different between GPD and GUN individuals (GPD/GUN) than between HC and GUN individuals (GUN/HC) or HC and GPD patients (GPD/HC). This is especially true for SNPs, which are more than five times over-represented in HC (OR < 0.2). More than 300 SNPs in 43 P450s are over-represented in HC compared with GUN and GPD patients (108 for GPD/HC and 227 for GUN/HC). In contrast, only 18 SNPs in 12 P450 genes are over-represented in GUN compared with GPD patients.
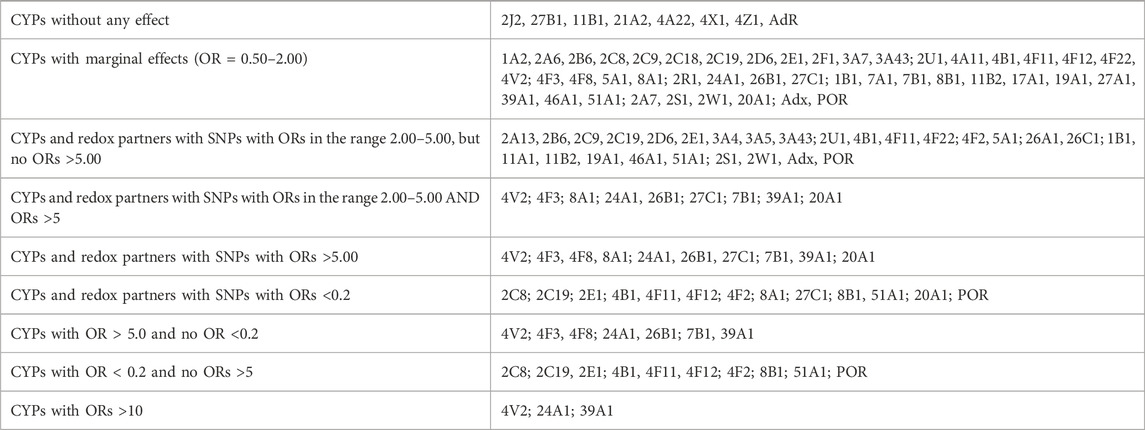
TABLE 2. Classification of cytochrome P450 genes (CYPs) and genes of their redox partners, Adx, AdR and POR according to the strength of association of their SNPs to Parkinson´s Disease based on the GPD/GUN comparison.
After that, we were looking at the association of SNPs in different P450 genes concerning their over- or under-representation between GPD and GUN in the various classes of P450s, divided according to their major substrate class (Guengerich, 2017). As shown in Figure 1, SNPs with OR > 5 are found in P450 genes involved in fatty acid and eicosanoid, in vitamin as well as in sterol metabolism. Considering SNPs with OR < 0.2, besides these 3 classes also P450s involved in the biotransformation of drugs and xenobiotics are affected. In addition, in both groups (OR > 5 and OR < 0.2) CYP20A1, an orphan P450, seems to play a prominent role.
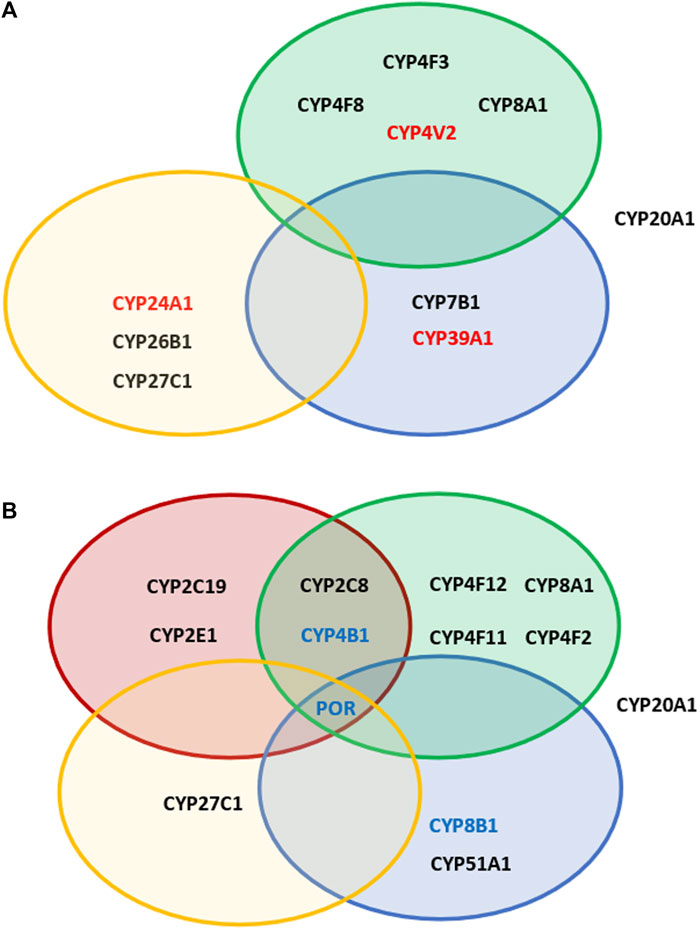
FIGURE 1. Overview of the effect of different SNPs in cytochromes P450 involved in various physiological pathways. Shown are P450s, which display SNPs with OR values >5 (A) and <0.2 (B) in GPD patients vs GUN individuals. Red circles: P450s participating in the biotransformation of xenobiotics; green circles: P450s involved in immune response and inflammation; blue circles: P450s involved in sterol metabolism or degradation of cholesterol; yellow circles: P450s involved in the metabolism of vitamins A and D. In addition, CYP20A1 is shown, which remains an „orphan“ P450, but is known to be involved in neurophysiological functions. P450s shown in red in (A) are those with OR values >10 (GPD/GUN). P450s shown in blue in (B) are those having SNPs found to be under-represented when considering both, GPD/HC and GPD/GUN.
As already mentioned, there are no SNPs with OR > 5 in the drug- and xenobiotics metabolizing P450s (Figure 1; Table 1; Table 2) which show an association with the manifestation of PD (GPD/GUN). However, except for seven P450s (CYP1A1, CYP1A2, CYP2A6, CYP2C8, CYP2C18, CYP2F1 and CYP3A), all the other enzymes in this group have SNPs which are 2-5fold over-represented in GPD compared to GUN individuals. Additionally, three P450s (CYP2C8, CYP2C19 and CYP2E1) have one SNP each, more than five times over-represented in GUN compared to GPD (OR < 0.2) (Figure 1).
With the exception of CYP4V2, the fatty acid converting P450s do not seem to possess SNPs that are highly over-represented in GPD patients compared to GUN individuals (OR > 5) (Table 1; Table 2). CYP4V2 shows one SNP that is ten times over-represented in GPD patients compared to GUN individuals (OR = 10.02). It is found in an intron region and described as a modifier of function. No SNPs with an OR value <0.2 have been found in CYP4V2 and only one SNP has been described with an OR value between 2.0 and 5.0. Looking further into the fatty acids metabolizing group of enzymes, there are five P450 genes (out of 8) with a total of 17 SNPs with OR values between 2 and 5, and three P450 genes with 5 SNPs displaying OR < 0.2. CYP2U1 with 9 SNPs having OR values between 2 and 5 and CYP4B1 displaying 3 SNPs with OR < 0.2 are especially remarkable.
In contrast to the drug and xenobiotics metabolizing P450s, P450s involved in eicosanoid metabolism display significant differences between GPD and GUN. The differences can be traced to SNPs in CYP4F3, CYP4F8 and CYP8A1 showing OR values >5 for GPD/GUN. The three SNPs with OR > 5 are all located in introns and identified as modifiers of function. In addition, differences in SNPs with OR values between 2.0 and 5.0 have been found for CYP4F2, CYP4F3, CYP5A1 and CYP8A1 and 82 SNPs are described in the eicosanoid group with OR values between 0.5 and 2.0. Additionally, the eicosanoid group is one of the groups with the biggest number of highly protective SNPs (OR < 0.2). The described data indicates substantial variations between GUN and GPD in genes coding eicosanoid metabolizing P450s.
Some of the P450s belonging to the vitamin group also show strong differences in their SNPs between GPD and GUN (Table 1; Table 2). CYP24A1 has one SNP with OR = 10.02 and CYP26B1 and CYP27C1 have one SNP each with OR > 5 (OR = 6.15 and 6.25, respectively). All three SNPs are located in intron regions and described as modifiers. Additionally, CYP27C1 has a SNP with OR < 0.2. The three P450s (CYP24A1, CYP26B1 and CYP27C1) also have 5, 3 and 26 SNPs, respectively, with OR values between 2.0 and 5.0. Altogether, this indicates an important contribution of this group of P450 genes for the development of PD.
When considering P450s involved in sterol metabolism, it becomes obvious that mainly SNPs in those P450s are different between GPD and GUN that are also different between HC and GPD: CYP7B1 and CYP39A1 displaying SNPs with OR values >5 as well as CYP46A1 with 6 SNPs showing OR values between 2.0 and 5.0. Once again, all SNPs with OR values >5 are located in intron regions and information on their exact effect on function is not available.
From the group of P450s with unknown function, CYP20A1 plays an outstanding role. It shows two SNPs with OR > 5, two SNPs with OR values between 2.0 and 5.0 and 57 SNPs with OR values between 0.5 and 2.0. The two SNPs with OR > 5 are located in introns and described as modifiers of function.
When comparing the occurrence of special SNPs in the three groups (HC, GPD, GUN), it turns out that all SNPs with OR > 5 discussed so far are represented in GPD as well as in HC, but under-represented in GUN. This means that they show up as “protective” when compared with HC. When looking at SNPs which are either over- or under-represented in GPD/GUN and in GPD/HC as well, the picture is different. It turns out that in many cases the effect is rather moderate with OR values between 1.4 and 3.3 (Table 3). Six P450s are identified in the xenobiotics metabolizing group, 3 each in the fatty acid and in the eicosanoid group, 2 in the vitamin group, 5 in the sterol group and 1 in the group of P450s with unknown function. CYP5A1 with 13 SNPs, CYP2C8 with 10 SNPs, CYP4F3 and CYP4B1 with 7 SNPs each as well as the redox partner POR with 8 SNPs seem to be of special importance. While in many cases the SNPs seem to have a moderate effect on the progression of PD, some of them seem to be more important. Thus, one of the SNPs in CYP5A1 which has an OR value of 2.13 in the GPD/GUN ratio, shows a value of 5.22 when looking at GPD and HC. This SNP is also over-represented in IPD patients compared to HC (OR = 4, data not shown). Moreover, one of the SNPs in CYP4F3 displaying an OR value of 2.99 in the GPD/GUN ratio, shows a value of 10.21 when comparing GPD and HC. Further on, except for CYP2D6, CYP3A4, CYP24A1 and CYP7A1, all 20 P450s in Table 3 as well as POR show SNPs with OR values <1 in GPD/GUN, some of which with OR values equal or below 0.2 (CYP2F1, CYP4B1 with three such SNPs; CYP5A1, CYP8B1, POR with 1 SNP each). It is certainly worth mentioning that out of the 75 SNPs in all P450s and POR, 26 are over-represented in GPD patients compared to HC and GUN individuals (OR > 1), but 49 are under-represented in GPD patients compared to HC and GUN individuals, showing the important role of protective SNPs in the appearance of PD.
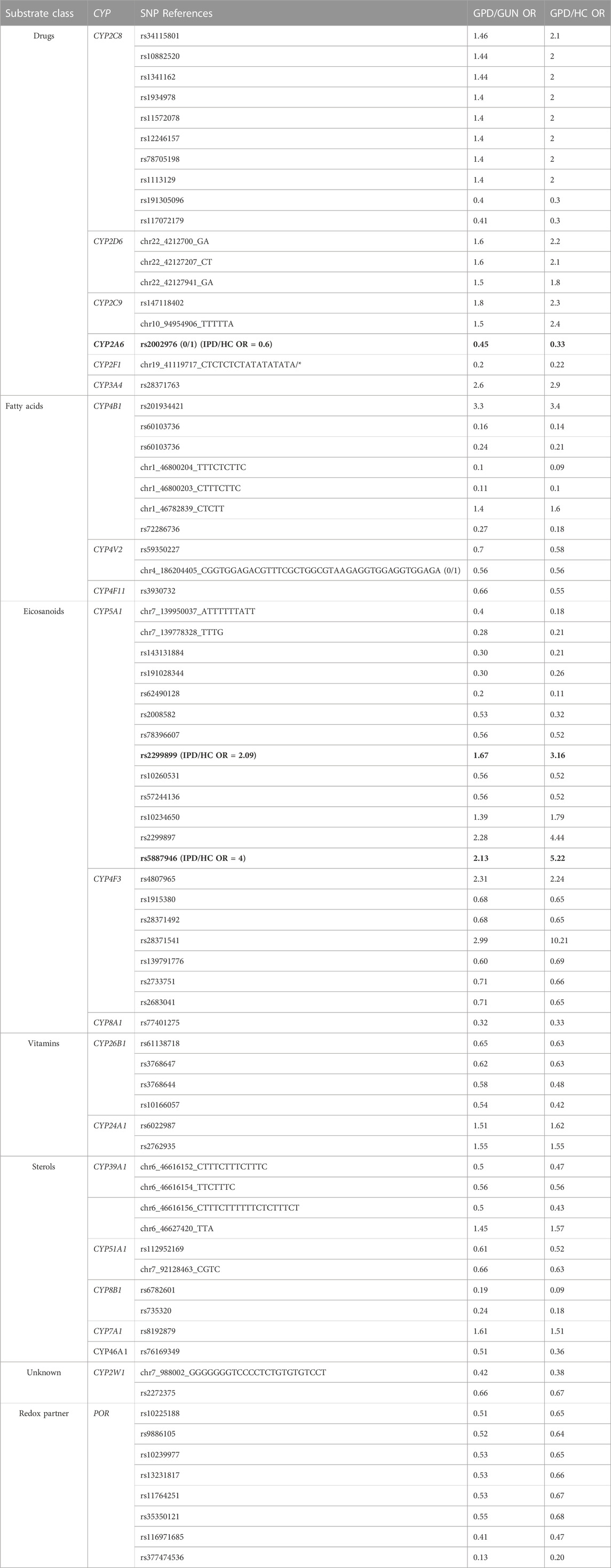
TABLE 3. Summary of all significant SNPs that are over- or under-represented in GPD patients when compared to both, HC (GPD/HC) and GUN (GPD/GUN) individuals. Marked in bold are SNPs, that are over- (OR > 1) or under-represented (OR < 1) also in IPD patients.
Intense worldwide investigations are carried out to identify novel markers and targets to better diagnose, understand and treat PD. There is a special lack in deciphering the detailed biochemical basis of PD to derive directions for a causative treatment of the disease. Our very recent study identified various P450 genes as novel potential players in the pathogenesis of PD (Hartz et al., 2022). A special subgroup of PD patients are those with familial PD caused by monogenetic mutations in various genes such as LRRK2, SNCA, GBA and others (Nalls et al., 2019; Bandres-Ciga et al., 2020).
It is well known, however, that not all individuals who have such a genetic predisposition to PD develop the disease and there are people, who, although genetically predisposed, do not show symptoms of the disease. Using the PPMI database and considering genetic differences in the 57 human P450 genes and their three redox partners, AdR, Adx and POR, we aimed to disclose the classes of P450s, the individual P450s and the specific SNPs that are important in turning genetic predisposition into symptomatic disease. We based our discussion and conclusions on two types of SNPs. First, SNPs that are over- or under-represented when comparing GPD and GUN (GPD/GUN, Table 1; Table 2) were identified regardless of whether these SNPs were also differently presented when comparing GPD/HC. Special attention was paid to strongly over- or under-represented SNPs, i.e., those with OR > 5, being more than 5 times over-represented in GPD patients compared to GUN, and those with OR < 0.2, being more than 5 times under-represented in GPD compared to GUN. The list of SNPs with OR > 5 is shown in detail (including designation of the SNP, OR values and confidence intervals) in Table 4. Second, SNPs over-represented (or under-represented) in GPD patients when compared to both, HC and GUN, i.e., which display significant OR values in both, the GPD/HC and GPD/GUN comparisons (Table 3), are considered. Possible consequences of associations between the SNPs in P450 genes and the switch from GUN to GPD are being discussed paying attention to the affected P450s and their potential metabolic impact.
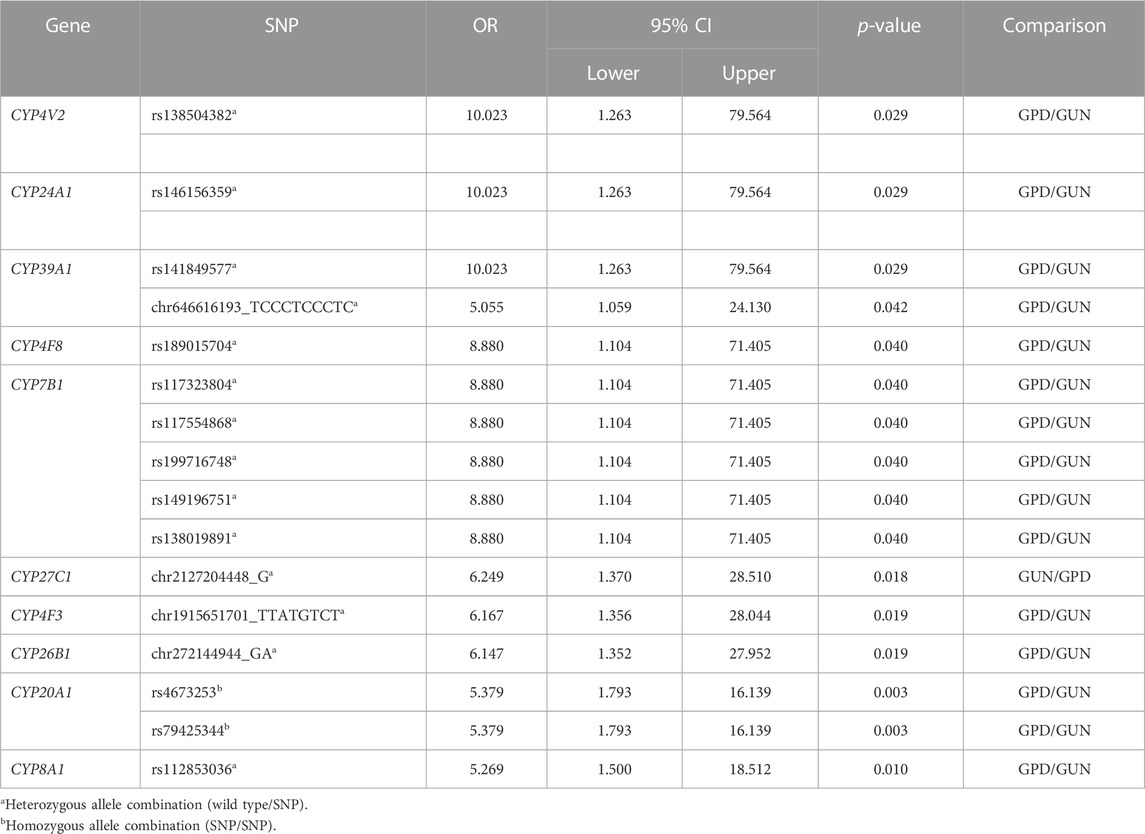
TABLE 4. Summary of all significant single nucleotide polymorphisms (SNP) with OR > 5 when comparing GPD and GUN patients. The confidence interval (CI) of the OR is indicated by the lower and upper limit for a confidence level of 95%.
When analyzing the data from the PPMI database, we were first of all surprised that GPD as well as GUN individuals showed much more highly significant SNPs (OR > 5 and OR < 0.2) in their P450 genes than IPD patients when compared with healthy individuals (Hartz et al., 2022). Whether people with genetic predisposition generally have more SNPs in their genes than other individuals or whether the higher number of SNPs is directly related to the genetic predisposition is unclear and needs further investigation.
When comparing GPD and GUN, 7 P450 genes as well as AdR did not show any differences in their SNPs between GPD and GUN (Table 2), suggesting that they do not play a significant role for the progression of the disease from GUN to GPD. Ten P450 genes have SNPs with OR > 5 and 12 P450s as well as POR have SNPs with OR < 0.2. Taking a closer look at the SNPs with OR values >5 for GPD/GUN, it is interesting to note that most of those SNPs are over-represented in GPD and abundant in HC as well, but are barely found in GUN. This suggests that the SNPs are especially unfavorable when combined with a genetic predisposition (mostly mutations in LRRK2 or GBA are found in these groups in the PPMI database). Interestingly, the number of these SNPs in IPD patients is also slightly (but not statistically significantly) increased compared with HC (data not shown). Taking into account that PD is a multifactorial disease and that mutations in different genes (which are not identified yet) can also be expected to contribute to the occurrence of IPD, this observation seems to be very reasonable. Interestingly, GUN individuals display a lower number of the corresponding SNP this way decreasing their disposition to the disease. The underlying mechanisms for this negative selection (in GUN, SNPs which are present in HC, GPD and even IPD, are under-represented) is not yet clear. Concerning the consequences of the SNPs with OR > 5 (Table 1), it has to be stated that all of them are found in intron regions of corresponding P450 genes and designated as modifiers of function so that differential regulation of gene expression (through binding of transcription factors, mRNAs, etc.) rather than a direct effect on P450s function and expression seems a plausible scenario. The finding that the SNPs are found in non-coding regions and have not been detected in previous genome-wide association studies coincides very well with observations of other laboratories (Ohnmacht et al., 2020).
When the impact of SNPs for the manifestation of GPD vs. GUN is considered concerning the class of P450 affected, it turns out that, in contrast to the GPD/HC group described previously (Hartz et al., 2022), P450s involved in the metabolism of drugs and xenobiotics, although playing some role, are not of high importance in turning GUN into GPD (Table 1). However, 6 out of 16 drug and xenobiotics metabolizing P450s show SNPs being present in GPD/HC as well as GPD/GUN. CYP2D6, CYP2C9 and CYP3A4 show OR values between 1.4 and 2.9 in both, GPD/HC and GPD/GUN, while CYP2A6 and CYP2F1 display protective SNPs with OR < 1. Special attention should be given to CYP2C8, which displays 10 SNPs, though with moderate OR values, which are found in GPD/GUN and also in GPD/HC (Table 3). Summarizing, this suggests that toxic compounds, e.g., pesticides, seem to be of rather moderate importance for the manifestation of PD in genetically predisposed individuals.
In contrast to the drug and xenobiotics metabolizing P450s, CYP4V2 belonging to the “fatty acid” family and CYP20A1 designated as an “orphan” P450 (Guengerich, 2017) as well as three P450s involved in eicosanoid, three involved in vitamin and three involved in sterol metabolism display SNPs that are highly over-represented in GPD patients compared to GUN individuals (OR > 5). This indicates that endogenous factors such as variations in the metabolism of eicosanoids, vitamins and sterols rather than exogenous factors such as variations in the degradation of external substances like xenobiotics, may play a more important role in the manifestation of PD symptoms when comparing GPD and GUN individuals.
CYP4V2 shows a SNP with OR = 10.02 in GPD/GUN (Table 1). It also shows two protective SNPs, that are under-represented in GPD/GUN and GPD/HC (Table 3). CYP4V2 is widely expressed in different organs and tissues, also in brain (Li et al., 2004), and is described to show ω-hydroxylase activity on saturated and polyunsaturated fatty acids (PUFAs) of medium and long chain length (Nakano et al., 2009; Nakano et al., 2012). Mutations in CYP4V2 are the main cause for Bietti’s crystalline dystrophy (Li et al., 2004), an inherited disease of the retina. It was demonstrated that functional impairment of CYP4V2 leads to a malfunction of global lipid metabolism. Patients show a marked reduction in the transformation of fatty acid precursors into ω-3 PUFAs, and a decreased synthesis of eicosapentaenoic acids (20:5ω-3) as well as docosahexaenoic acids (for review see (García-García et al., 2019). Lipidomic analyses of the cells of those patients also demonstrate an accumulation of cholesterol (Hata et al., 2018). Moreover, a role of CYP4V2 in ischemic stroke due to venous thromboembolism has been shown (Yue et al., 2019; Long et al., 2022).
In view of the involvement of CYP4V2 in the metabolism of saturated and polyunsaturated fatty acids, we checked whether the consumption of fish oil and omega fatty acid supplements has an effect on the manifestation of GPD since for some individuals in the PPMI database corresponding information is available. Analysis of the data, however, did not show any statistically significant differences in the number of GUN and GPD in dependence on the intake of fish (or krill) oil (21.2% (73 out of 344) GUN vs. 15.5% (49 out of 317) GPD, p = 0.056), thus demonstrating no protective role of the oil on the manifestation of the disease. However, it cannot be excluded that a sub-group of individuals, e.g., people showing defects in the biosynthesis of anti-inflammatory compounds derived from fatty acids, will profit from the intake of fish or krill oil.
Analyzing the role of P450s of the “eicosanoid” group, our results show that P450s involved in eicosanoid metabolism and inflammation seem to have a key function in the manifestation of GPD. Indeed, 3 out of 5 P450s involved in eicosanoid metabolism also have SNPs with OR values >5 (CYP4F3, CYP4F8 and CYP8A1) suggesting an important effect of eicosanoids in differentiating between GPD and GUN. A similar importance of eicosanoids was described by (Hartz et al., 2022) for the association between SNPs in these genes and PD. In addition, 4 P450s (CYP4F2, CYP4F3, CYP5A1, CYP8A1) of this group have SNPs with OR values between 2 and 5 (Table 1). Moreover, CYP5A1 displays 13 SNPs when considering simultaneously GPD/GUN and GPD/HC with OR values up to 5.2 in GPD/HC and >2 in GPD/GUN (Table 3). CYP5A1 is a thromboxane synthase and its function is related to the action of CYP8A1, a prostacyclin synthase. While both P450s use prostaglandin H2 (PGH2) as a substrate, prostacyclin (prostaglandin I2, PGI2), resulting from the catalytic action of CYP8A1, is a potent vasodilator and platelet antiaggregatory eicosanoid, while thromboxane A2 (TXA2), resulting from the catalytic action of CYP5A1, is a potent inducer of platelet aggregation and vasoconstriction. The balance of the two products affects important physiological processes such as blood pressure regulation, blood coagulation and inflammation (Rendic and Peter Guengerich, 2018). PGI2 is neuroprotective in neuronal cultures and in the ischemic brain with pronounced effect on microglia (Lin et al., 2002; Tsai et al., 2005). Moreover, its enhanced synthesis decreases glial activation and alleviates motor impairments in hemiparkinsonian rats (Tsai et al., 2013). Increased TXA2, in turn, has been demonstrated to be involved in amyloid beta protein (Abeta)-induced dysfunction of the dopaminergic nigrostriatal pathway and motor function deficits in Abeta-injected rats (Yagami et al., 2004). In addition, CYP4B1 and CYP4F3, also involved in fatty acid and eicosanoid metabolism, show differences in the occurrence of SNPs when comparing GPD/GUN as well as GPD/HC (Table 3). CYP4B1 has 7 SNPs, one of them with OR > 3 in both cases. CYP4F3 has also 7 such SNPs, one of them with OR = 10.21 for GPD/HC and OR = 2.99 for GPD/GUN. Interestingly, CYP2C8 with 10 SNPs, besides playing an important role in drug and xenobiotics metabolism, possesses also epoxygenase activity and metabolizes long-chain polyunsaturated fatty acids such as arachidonic acid, eicosapentaenoic acid, docosahexaenoic acid, and linoleic acid to epoxide products (Rifkind et al., 1995). Taken together, this data underlines the remarkable importance of inflammation on the manifestation of the disease based on a genetic predisposition to PD.
When analyzing P450s involved in the metabolism of vitamins concerning differences in SNPs between GPD and GUN, it becomes obvious that again three P450s play an important role (Table 1; Table 4). CYP24A1 is one of the three P450s displaying a SNP with OR = 10.02 for GPD/GUN. The SNP is located in an intron region and suggested to behave as a modifier of function. It has also two SNPs that are over-represented in GPD patients compared to HC and GUN with OR values in the range 1.5–1.6 (Table 3). CYP24A1 is responsible for the degradation of 25OH- as well as 1α, 25-dihydroxy vitamin D3 to 24-hydroxylated products (Chen et al., 1993; Sakaki et al., 2000). It is a central player in vitamin D homeostasis and in this way in vitamin D function. Since the precursor of vitamin D3, 7-dehydrocholesterol, is also the precursor of cholesterol, the biosynthesis of vitamin D is closely related to sterol metabolism and steroid hormone biosynthesis (Schuster, 2011; Saponaro et al., 2020). Changes in the expression and functionality of CYP24A1 may thus have broad consequences. First, malfunction could lead to an accumulation of the precursor 7-dehydrocholesterol and in this way to increased levels of cholesterol. On the other hand, an unbalanced high expression of CYP24A1 may lead to vitamin D deficiency thereby causing various disorders connected to bone metabolism, immunomodulation and proliferative disorders like cancer (Schuster et al., 2006). Very recently it was found that vitamin D activation in astrocytes could play an important role in PD pathogenesis and a protective role of vitamin D was suggested (Mazzetti et al., 2022). Moreover, a pathogenic role for CYP24A1 in multiple sclerosis and ischemic stroke has been proposed. It was shown that the risk allele rs2248359_C, which is associated with an increased expression of CYP24A1 in frontal cortex, increases the risk of multiple sclerosis, possibly by changing immunomodulation in brain (Ramasamy et al., 2014). CYP24A1 genetic polymorphisms were also shown to be significantly associated with the occurrence of an ischemic stroke (Yang et al., 2020). In this relation, it should be mentioned that overexpression of CYP24A1 could serve as a possible drug target as successful attempts for selective inhibition of CYP24A1 have already been made (Schuster et al., 2006; Alshabrawy et al., 2022). Due to the importance of CYP24A1 in vitamin D metabolism, we checked whether there is a difference in the manifestation of the disease in dependence on the intake of vitamin D as a supplement, since data for some individuals are available in the PPMI database. However, we found no statistically significant difference in the total number of GPD vs. GUN taking vitamin D (39.5% (136 out of 344) GUN vs. 33.1% (105 out of 317) GPD, p = 0.087) suggesting that there is no effect of nutritional supplementation with vitamin D on the manifestation of PD in genetically pre-disposed individuals. Besides CYP24A1, also CYP27C1 and CYP26B1 from the vitamin group display SNPs with OR > 5 in GPD patients compared with GUN individuals (Table 1). CYP27C1 so far has not been extensively studied. It is known that it does not catalyze the oxidation of vitamin D3, 1α- or 25-hydroxy vitamin D3 or cholesterol at detectable levels as do its relatives CYP27A1 and CYP27B1 (Kakimoto et al., 2022), but it was shown to catalyze the 3,4-desaturation of retinoids (Wu et al., 2006; Kramlinger et al., 2016). CYP27C1 is highly expressed in skin (Johnson et al., 2017), but was also shown to be expressed in the brain (https://www.proteinatlas.org). More information is available on CYP26B1. Human CYP26B1 was first described in 1999 (Nelson, 1999) and shown to be responsible for the generation of several hydroxylated forms of retinoic acid in the human brain thus playing an important role in protecting it from exposure to retinoids (White et al., 2000). Retinoic acid, which is synthesized from vitamin A, is an essential regulator of gene expression and its homeostasis is of great importance for a balanced metabolism and for neuronal differentiation. In human neurons, this homeostasis is maintained by anabolic as well as catabolic enzymes such as the catabolic P450s CYP26A1 and CYP26B1 (Stoney et al., 2016). Knock-out of the two enzymes in mice is embryonic lethal while postnatal global deletion results in severe dermatitis, blepharitis, splenomegaly, lymphadenomegaly, systemic inflammation and reduced life span (Snyder et al., 2020). The authors further concluded that CYP26B1, but not CYP26A1, is the main post-natal enzyme clearing all-trans-retinoic acid, which coincides well with our results showing that many SNPs in CYP26B1 (but not CYP26A1) display differences (under- or over-representation) in GPD vs. GUN and thus in the manifestation of PD.
Sterol-converting P450s also show a strong association of SNPs in corresponding P450 genes with the manifestation of PD in GPD vs. GUN. CYP39A1 is the third P450 found in our studies with an OR = 10.02 (GPD/GUN). Moreover, it possesses one SNP with OR = 1.5 for GPD/GUN and 1.6 for GPD/HC. This P450 plays a prominent role in the degradation of cholesterol. It converts 24S-hydroxycholesterol, formed by CYP46A1 from cholesterol, into 7α,24S-dihydroxycholesterol (Li-Hawkins et al., 2000). Further hydroxylation by CYP8B1 and CYP27A1 leads to formation of bile acids. CYP39A1 is mainly localized in liver, but also found in the brain and in the eye (Baloni et al., 2020; Li et al., 2021). It was shown to be directly involved in neural cholesterol clearance and changed bile acid metabolism in Alzheimer’s disease cohorts (Baloni et al., 2020). Increasing the expression of CYP39A1 could be beneficial to reduce brain 24S-hydroxycholesterol, whose accumulation has been shown to be related to oxidative stress and reported in the brain of patients with dementia and Alzheimer’s disease (Matsuoka et al., 2020). Interestingly, using both metabolomic as well as epigenetic approaches, bile acid metabolism, being closely related to cholesterol degradation, was demonstrated to be the top perturbed metabolic pathway in the brain of PD patients (Vishweswaraiah et al., 2022) underlining the role of cholesterol degradation for the manifestation of PD as described here. Finally, using GWAS investigations it was reported that four intron variants of CYP39A1 (rs6907129, rs6905960, rs7749491, rs16874881) are strongly associated with the occurrence of levodopa-induced dyskinesia in patients aged over 50 years at the onset of PD (Ryu et al., 2020). CYP7B1 is another P450 gene having SNPs with OR values >5 in GPD/GUN. It shows five such SNPs, all of which with the same OR value of 8.88 and all of which clustered in the same patients. CYP7B1 is also involved in the degradation of cholesterol leading to 7α-hydroxylation of 27-hydroxycholesterol, which is formed from cholesterol with the participation of CYP27A1. The latter one is found ubiquitously while CYP7B1 mRNA transcripts are mostly found in tissues involved in steroid and bile acid biosynthesis (brain, testes, ovaries, prostate, liver) as well as reabsorption (colon, kidney, and small intestines) (Wu et al., 1999). It has been shown that the blood-brain barrier is permeable to hydroxylated cholesterol metabolites and that 24-hydroxycholesterol, which is the substrate of CYP39A1, is formed almost exclusively in the brain, while 27-hydroxycholesterol, the substrate of CYP7B1, is mainly supplied to the brain (Björkhem et al., 2018). Interestingly, Björkhem and coworkers found that the level of oxysterols in the cerebrospinal fluid may reflect neurodegenerative changes occurring in PD patients (Björkhem et al., 2013), which is in line with the association between SNPs in CYP39A1 and CYP7B1 and the manifestation of PD symptoms found in our study. Moreover, defects in CYP7B1 have been associated with hereditary spastic paraplegia type 5, a group of neurodegenerative disorders characterized by progressive neurodegeneration of the corticospinal tract motor neurons (Schöls et al., 2017; Chou et al., 2020; Prestsæter et al., 2020). Hence, defects in CYP7B1 might make an individual more vulnerable for neurodegeneration in general, i.e., also for disorders besides PD.
Finally, CYP20A1, belonging to the group of “orphan” P450s, displays SNPs with OR > 5 (GPD/GUN). It has two SNPs with OR = 5.38 that are both found in the same patients. CYP20A1 is especially abundant in S. nigra and the hippocampus of human brain (Stark et al., 2008) and was very recently shown to have important neurophysiological functions in zebra fish (Lemaire et al., 2016; Brun et al., 2021). Unfortunately, so far only aniline and some luminogenic substances have been shown to be weakly active with CYP20A1 expressed in yeast (Durairaj et al., 2019) indicating on possible endogenic substrates (Brun et al., 2021).
Interestingly, POR, which did not show SNPs with OR > 5 when comparing all SNPs between GPD and GUN, displayed 4 SNPs with OR values between 2 and 5 (Table 1) as well as 8 SNPs with OR values <1 when analysing SNPs in GPD/HC and GPD/GUN (Table 3). This points to a possible modulatory effect of this redox partner on the manifestation of PD in GPD patients.
Taken together, we were able, to the best of our knowledge for the first time, to identify SNPs in the genes of proteins, which are associated with the switch from GUN individuals (with a predisposition to PD, but without symptoms of the disease) to GPD (with disease symptoms). Most impressive, some of the SNPs are up to 10 fold over-represented in GPD patients vs. GUN individuals indicating a significant role of these SNPs for the manifestation of the disease. Out of the 57 human P450s and their three redox partners (Adx, AdR and POR), 10 P450s show SNPs with OR values >5 when comparing GPD and GUN. Since those SNPs occur under-represented in GUN compared with GPD and even HC, it seems that they are not sufficient to cause PD, when occurring alone. They seem to exert their effect only when combined with other SNPs, e.g., of the predisposition genes as found here, or with SNPs in other so far unidentified genes as proposed for IPD patients. This defines important novel players in the pathogenesis of PD, especially when looking at patients with a genetic predisposition. Altogether, this underlines the well-accepted multivariant basis for the development of PD. Limitations of the study are the lack of an analysis of potential combinations of various SNPs in the different P450s of individual GPD patients and GUN individuals as well as the lack of experimental studies. However, those were outside the focus of this paper and common to most of the communications on identified PD risk factors reported so far. Comparison of the age of GPD patients at the time of PD diagnosis with GUN individuals at the time when listed in the PPMI database, shows no statistically significant difference between the two groups (58.2 ± 1.6 (GPD) vs. 57.6 ± 0.7 (GUN) years, p = 0.78). Nevertheless, it cannot be excluded that GUN individuals might develop symptoms of the disease at a later stage. However, taking into account that a genetic predisposition is mostly related to an early onset of the disease (Ylikotila et al., 2015), we believe that the average age of 57.61 years for GUN (at which they were enrolled in the PPMI study), is already the age at which the first PD symptoms, if any, would have developed. Interestingly, the P450s with the greatest effect on the switch from GUN to GPD do not belong to the group involved in drug and xenobiotics metabolism, like in the case of GPD/HC, but to fatty acid/eicosanoid, vitamin and sterol metabolism (Figure 1), which is further confirmed when examining also the SNPs occurring with similar OR values in GPD patients compared to both, HC and GUN individuals. Thus, endogenous rather than exogenous factors seem to be responsible for the differentiation between GPD and GUN and inflammation, vitamin A and D metabolism, cholesterol degradation and pathways affected by CYP20A1 seem to be key players in eliciting PD manifestation. Additional to deleterious SNPs that are highly over-represented in GPD patients, protective SNPs are equally important. These SNPs are largely missing in GPD but are present in GUN (Figure 1). Twelve P450s have SNPs with OR values below 0.2 when comparing GPD and GUN individuals and two P450s have SNPs with OR values below 0.2 when comparing GPD both with GUN and HC. The CYPs with protective SNPs belong to all substrate classes. Moreover, also POR shows SNPs with OR < 0.2 in both approaches indicating on a contribution of this protein, which is the redox partner of all microsomal P450s, for the transformation from GUN to GPD. Further studies, especially experimental investigations are needed to examine the consequences of the SNPs found in this study, characterize in detail biochemical pathways being affected and identify possible novel potential targets for PD early diagnostics, intervention and treatment.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
RB and MU designed the project. RB and PP-K performed analyses and discussion of the results and wrote the paper. PP-K, SB, and PH performed statistical analyses of the data and participated in the data analyses. MU participated in analyses of the data and writing of the manuscript. GW performed statistical analyses of the data. All authors contributed to the article and approved the submitted version.
The work was supported by a research grant from the “Rolf M. Schwiete Stiftung” Mannheim/Germany. The work was made possible by accession to the PPMI database.
Data used for analyses presented in this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (http://www.ppmi-info.org/data). The authors thank Prof. Dr. Andreas Keller and Dr. Tobias Fehlmann for their contribution in the extraction and classification of the data from the PPMI database.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CYP, cytochrome P450; P450, cytochrome P450; Adx, adrenodoxin; POR, NADPH-dependent cytochrome P450 reductase; CYP, POR, Adx, AdR designate the corresponding genes; PD, Parkinson’s disease; IPD, idiopathic (sporadic) PD; GPD, PD with a genetic predisposition; GUN, persons with genetic predisposition for PD, but without symptoms of disease; PPMI, The Parkinson’s Progression Markers Initiative; OR, odds ratio; CI, confidence interval.
Alshabrawy, A. K., Cui, Y., Sylvester, C., Yang, D., Petito, E. S., Barratt, K. R., et al. (2022). Therapeutic potential of a novel vitamin D3 oxime analogue, VD1-6, with CYP24A1 enzyme inhibitory activity and negligible vitamin D receptor binding. Biomolecules 12, 960. doi:10.3390/biom12070960
Baloni, P., Funk, C. C., Yan, J., Yurkovich, J. T., Kueider-Paisley, A., Nho, K., et al. (2020). Metabolic network analysis reveals altered bile acid synthesis and metabolism in Alzheimer’s disease. Cell. Rep. Med. 1, 100138. doi:10.1016/j.xcrm.2020.100138
Bandres-Ciga, S., Diez-Fairen, M., Kim, J. J., and Singleton, A. B. (2020). Genetics of Parkinson’s disease: An introspection of its journey towards precision medicine. Neurobiol. Dis. 137, 104782. doi:10.1016/j.nbd.2020.104782
Björkhem, I., Lövgren-Sandblom, A., Leoni, V., Meaney, S., Brodin, L., Salveson, L., et al. (2013). Oxysterols and Parkinson’s disease: Evidence that levels of 24S-hydroxycholesterol in cerebrospinal fluid correlates with the duration of the disease. Neurosci. Lett. 555, 102–105. doi:10.1016/j.neulet.2013.09.003
Björkhem, I., Patra, K., Boxer, A. L., and Svenningsson, P. (2018). 24S-Hydroxycholesterol correlates with tau and is increased in cerebrospinal fluid in Parkinson’s disease and corticobasal syndrome. Front. neurology 9, 756. doi:10.3389/fneur.2018.00756
Brun, N. R., Salanga, M. C., Mora-Zamorano, F. X., Lamb, D. C., Goldstone, J. V., and Stegeman, J. J. (2021). Orphan cytochrome P450 20a1 CRISPR/Cas9 mutants and neurobehavioral phenotypes in zebrafish. Sci. Rep. 11, 23892. doi:10.1038/s41598-021-03068-3
Chen, K. S., Prahl, J. M., and DeLuca, H. F. (1993). Isolation and expression of human 1,25-dihydroxyvitamin D3 24-hydroxylase cDNA. Proc. Natl. Acad. Sci. U. S. A. 90, 4543–4547. doi:10.1073/pnas.90.10.4543
Chou, C.-T., Soong, B.-W., Lin, K.-P., Tsai, Y.-S., Jih, K.-Y., Liao, Y.-C., et al. (2020). Clinical characteristics of Taiwanese patients with Hereditary spastic paraplegia type 5. Ann. Clin. Transl. neurology 7, 486–496. doi:10.1002/acn3.51019
Cingolani, P., Patel, V. M., Coon, M., Nguyen, T., Land, S. J., Ruden, D. M., et al. (2012a). Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 3, 35. doi:10.3389/fgene.2012.00035
Cingolani, P., Platts, A., Le Wang, L., Coon, M., Nguyen, T., Wang, L., et al. (2012b). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92. doi:10.4161/fly.19695
Di Maio, R., Hoffman, E. K., Rocha, E. M., Keeney, M. T., Sanders, L. H., Miranda, B. R. de, et al. (2018). LRRK2 activation in idiopathic Parkinson’s disease. Sci. Transl. Med. 10, eaar5429. doi:10.1126/scitranslmed.aar5429
Durairaj, P., Fan, L., Du, W., Ahmad, S., Mebrahtu, D., Sharma, S., et al. (2019). Functional expression and activity screening of all human cytochrome P450 enzymes in fission yeast. FEBS Lett. 593, 1372–1380. doi:10.1002/1873-3468.13441
García-García, G. P., Martínez-Rubio, M., Moya-Moya, M. A., Pérez-Santonja, J. J., and Escribano, J. (2019). Current perspectives in Bietti crystalline dystrophy. Clin. Ophthalmol. Auckl. N.Z.) 13, 1379–1399. doi:10.2147/OPTH.S185744
Guengerich, F. P. (2017). Intersection of the roles of cytochrome P450 enzymes with xenobiotic and endogenous substrates: Relevance to toxicity and drug interactions. Chem. Res. Toxicol. 30, 2–12. doi:10.1021/acs.chemrestox.6b00226
Hartz, P., Fehlmann, T., Wagenpfeil, G., Unger, M. M., and Bernhardt, R. (2022). A CYPome-wide study reveals new potential players in the pathogenesis of Parkinson’s disease. Front. Pharmacol. 13, 1094265. doi:10.3389/fphar.2022.1094265
Hata, M., Ikeda, H. O., Iwai, S., Iida, Y., Gotoh, N., Asaka, I., et al. (2018). Reduction of lipid accumulation rescues Bietti’s crystalline dystrophy phenotypes. Proc. Natl. Acad. Sci. U. S. A. 115, 3936–3941. doi:10.1073/pnas.1717338115
Johnson, K. M., Phan, T. T. N., Albertolle, M. E., and Guengerich, F. P. (2017). Human mitochondrial cytochrome P450 27C1 is localized in skin and preferentially desaturates trans-retinol to 3,4-dehydroretinol. J. Biol. Chem. 292, 13672–13687. doi:10.1074/jbc.M116.773937
Kakimoto, A., Ogura, H., Suenaga, M., Mishima, T., Fujioka, S., Ouma, S., et al. (2022). Role of cytochrome P450 for vitamin D metabolisms in patients with neurodegenerative disorders. Clin. Park. Relat. Disord. 7, 100162. doi:10.1016/j.prdoa.2022.100162
Kramlinger, V. M., Nagy, L. D., Fujiwara, R., Johnson, K. M., Phan, T. T. N., Xiao, Y., et al. (2016). Human cytochrome P450 27C1 catalyzes 3,4-desaturation of retinoids. FEBS Lett. 590, 1304–1312. doi:10.1002/1873-3468.12167
Lemaire, B., Kubota, A., O’Meara, C. M., Lamb, D. C., Tanguay, R. L., Goldstone, J. V., et al. (2016). Cytochrome P450 20A1 in zebrafish: Cloning, regulation and potential involvement in hyperactivity disorders. Toxicol. Appl. Pharmacol. 296, 73–84. doi:10.1016/j.taap.2016.02.001
Lesage, S., and Brice, A. (2009). Parkinson’s disease: From monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 18, R48–R59. doi:10.1093/hmg/ddp012
Li, A., Jiao, X., Munier, F. L., Schorderet, D. F., Yao, W., Iwata, F., et al. (2004). Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. Am. J. Hum. Genet. 74, 817–826. doi:10.1086/383228
Li, Z., Wang, Z., Lee, M. C., Zenkel, M., Peh, E., Ozaki, M., et al. (2021). Association of rare CYP39A1 variants with exfoliation syndrome involving the anterior chamber of the eye. JAMA 325, 753–764. doi:10.1001/jama.2021.0507
Li-Hawkins, J., Lund, E. G., Bronson, A. D., and Russell, D. W. (2000). Expression cloning of an oxysterol 7alpha-hydroxylase selective for 24-hydroxycholesterol. J. Biol. Chem. 275, 16543–16549. doi:10.1074/jbc.M001810200
Lin, H., Lin, T.-N., Cheung, W.-M., Nian, G.-M., Tseng, P.-H., Chen, S.-F., et al. (2002). Cyclooxygenase-1 and bicistronic cyclooxygenase-1/prostacyclin synthase gene transfer protect against ischemic cerebral infarction. Circulation 105, 1962–1969. doi:10.1161/01.cir.0000015365.49180.05
Long, F., Wang, D., Su, Q., Zhang, Y., Li, J., Xia, S., et al. (2022). CYP4 subfamily V member 2 (CYP4V2) polymorphisms were associated with ischemic stroke in Chinese Han population. BMC Med. genomics 15, 246. doi:10.1186/s12920-022-01393-8
Marek, K., Chowdhury, S., Siderowf, A., Lasch, S., Coffey, C. S., Caspell-Garcia, C., et al. (2018). The Parkinson’s progression markers initiative (PPMI) – establishing a PD biomarker cohort. Ann. Clin. Transl. Neurol. 5, 1460–1477. doi:10.1002/acn3.644
Matsuoka, H., Katayama, M., Ohishi, A., Miya, K., Tokunaga, R., Kobayashi, S., et al. (2020). Orphan nuclear receptor RORα regulates enzymatic metabolism of cerebral 24S-hydroxycholesterol through CYP39A1 intronic response element activation. Int. J. Mol. Sci. 21, 3309. doi:10.3390/ijms21093309
Mazzetti, S., Barichella, M., Giampietro, F., Giana, A., Calogero, A. M., Amadeo, A., et al. (2022). Astrocytes expressing Vitamin D-activating enzyme identify Parkinson’s disease. CNS Neurosci. Ther. 28, 703–713. doi:10.1111/cns.13801
Nakano, M., Kelly, E. J., and Rettie, A. E. (2009). Expression and characterization of CYP4V2 as a fatty acid omega-hydroxylase. Drug metabolism Dispos. Biol. fate Chem. 37, 2119–2122. doi:10.1124/dmd.109.028530
Nakano, M., Kelly, E. J., Wiek, C., Hanenberg, H., and Rettie, A. E. (2012). CYP4V2 in Bietti’s crystalline dystrophy: Ocular localization, metabolism of ω-3-polyunsaturated fatty acids, and functional deficit of the p.H331P variant. Mol. Pharmacol. 82, 679–686. doi:10.1124/mol.112.080085
Nalls, M. A., Blauwendraat, C., Vallerga, C. L., Heilbron, K., Bandres-Ciga, S., Chang, D., et al. (2019). Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurology 18, 1091–1102. doi:10.1016/S1474-4422(19)30320-5
Nelson, D. R. (1999). A second CYP26 P450 in humans and zebrafish: CYP26B1. Archives Biochem. biophysics 371, 345–347. doi:10.1006/abbi.1999.1438
Ohnmacht, J., May, P., Sinkkonen, L., and Krüger, R. (2020). Missing heritability in Parkinson’s disease: The emerging role of non-coding genetic variation. J. neural Transm. 127, 729–748. doi:10.1007/s00702-020-02184-0
Prestsæter, S., Koht, J., Lamari, F., Tallaksen, C. M. E., Hoven, S. T. J., Vigeland, M. D., et al. (2020). Elevated hydroxycholesterols in Norwegian patients with hereditary spastic paraplegia SPG5. J. neurological Sci. 419, 117211. doi:10.1016/j.jns.2020.117211
Ramasamy, A., Trabzuni, D., Forabosco, P., Smith, C., Walker, R., Dillman, A., et al. (2014). Genetic evidence for a pathogenic role for the vitamin D3 metabolizing enzyme CYP24A1 in multiple sclerosis. Multiple Scler. Relat. Disord. 3, 211–219. doi:10.1016/j.msard.2013.08.009
Rendic, S. P., and Peter Guengerich, F. (2018). Human cytochrome P450 enzymes 5-51 as targets of drugs and natural and environmental compounds: Mechanisms, induction, and inhibition - toxic effects and benefits. Drug metab. Rev. 50, 256–342. doi:10.1080/03602532.2018.1483401
Rifkind, A. B., Lee, C., Chang, T. K., and Waxman, D. J. (1995). Arachidonic acid metabolism by human cytochrome P450s 2C8, 2C9, 2E1, and 1A2: Regioselective oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Archives Biochem. biophysics 320, 380–389. doi:10.1016/0003-9861(95)90023-3
Ryu, H.-S., Park, K. W., Choi, N., Kim, J., Park, Y.-M., Jo, S., et al. (2020). Genomic analysis identifies new loci associated with motor complications in Parkinson’s disease. Front. neurology 11, 570. doi:10.3389/fneur.2020.00570
Sakaki, T., Sawada, N., Komai, K., Shiozawa, S., Yamada, S., Yamamoto, K., et al. (2000). Dual metabolic pathway of 25-hydroxyvitamin D3 catalyzed by human CYP24. Eur. J. Biochem. 267, 6158–6165. doi:10.1046/j.1432-1327.2000.01680.x
Saponaro, F., Saba, A., and Zucchi, R. (2020). An update on vitamin D metabolism. Int. J. Mol. Sci. 21, 8672. doi:10.3390/ijms21228672
Schöls, L., Rattay, T. W., Martus, P., Meisner, C., Baets, J., Fischer, I., et al. (2017). Hereditary spastic paraplegia type 5: Natural history, biomarkers and a randomized controlled trial. Brain a J. neurology 140, 3112–3127. doi:10.1093/brain/awx273
Schuster, I. (2011). Cytochromes P450 are essential players in the vitamin D signaling system. Biochimica biophysica acta 1814, 186–199. doi:10.1016/j.bbapap.2010.06.022
Schuster, I., Egger, H., Herzig, G., Reddy, G. S., Schmid, J. A., Schüssler, M., et al. (2006). Selective inhibitors of vitamin D metabolism—New concepts and perspectives. Anticancer Res. 26, 2653–2668.
Sellbach, A. N., Boyle, R. S., Silburn, P. A., and Mellick, G. D. (2006). Parkinson’s disease and family history. Park. Relat. Disord. 12, 399–409. doi:10.1016/j.parkreldis.2006.03.002
Snyder, J. M., Zhong, G., Hogarth, C., Huang, W., Topping, T., LaFrance, J., et al. (2020). Knockout of Cyp26a1 and Cyp26b1 during postnatal life causes reduced lifespan, dermatitis, splenomegaly, and systemic inflammation in mice. FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 34, 15788–15804. doi:10.1096/fj.202001734R
Stark, K., Wu, Z.-L., Bartleson, C. J., and Guengerich, F. P. (2008). mRNA distribution and heterologous expression of orphan cytochrome P450 20A1. Drug metabolism Dispos. Biol. fate Chem. 36, 1930–1937. doi:10.1124/dmd.108.022020
Stoney, P. N., Fragoso, Y. D., Saeed, R. B., Ashton, A., Goodman, T., Simons, C., et al. (2016). Expression of the retinoic acid catabolic enzyme CYP26B1 in the human brain to maintain signaling homeostasis. Brain Struct. Funct. 221, 3315–3326. doi:10.1007/s00429-015-1102-z
Tsai, M.-J., Shyue, S.-K., Weng, C.-F., Chung, Y., Liou, D.-Y., Huang, C.-T., et al. (2005). Effect of enhanced prostacyclin synthesis by adenovirus-mediated transfer on lipopolysaccharide stimulation in neuron-glia cultures. Ann. N. Y. Acad. Sci. 1042, 338–348. doi:10.1196/annals.1338.031
Tsai, M.-J., Weng, C.-F., Yu, N.-C., Liou, D.-Y., Kuo, F.-S., Huang, M.-C., et al. (2013). Enhanced prostacyclin synthesis by adenoviral gene transfer reduced glial activation and ameliorated dopaminergic dysfunction in hemiparkinsonian rats. Oxidative Med. Cell. Longev. 2013, 649809. doi:10.1155/2013/649809
Vishweswaraiah, S., Akyol, S., Yilmaz, A., Ugur, Z., Gordevičius, J., Oh, K. J., et al. (2022). Methylated cytochrome P450 and the solute carrier family of genes correlate with perturbations in bile acid metabolism in Parkinson’s disease. Front. Neurosci. 16, 804261. doi:10.3389/fnins.2022.804261
White, J. A., Ramshaw, H., Taimi, M., Stangle, W., Zhang, A., Everingham, S., et al. (2000). Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc. Natl. Acad. Sci. U. S. A. 97, 6403–6408. doi:10.1073/pnas.120161397
Wu, Z.-L., Bartleson, C. J., Ham, A.-J. L., and Guengerich, F. P. (2006). Heterologous expression, purification, and properties of human cytochrome P450 27C1. Archives Biochem. biophysics 445, 138–146. doi:10.1016/j.abb.2005.11.002
Wu, Z., Martin, K. O., Javitt, N. B., and Chiang, J. Y. (1999). Structure and functions of human oxysterol 7α-hydroxylase cDNAs and gene CYP7B1. J. lipid Res. 40, 2195–2203. doi:10.1016/s0022-2275(20)32094-0
Yagami, T., Takahara, Y., Ishibashi, C., Sakaguchi, G., Itoh, N., Ueda, K., et al. (2004). Amyloid beta protein impairs motor function via thromboxane A2 in the rat striatum. Neurobiol. Dis. 16, 481–489. doi:10.1016/j.nbd.2004.04.013
Yang, W., Ma, F., Wang, L., He, X., Zhang, H., Zheng, J., et al. (2020). The association analysis between CYP24A1 genetic polymorphisms and the risk of ischemic stroke in Chinese Han population. Brain Behav. 10, e01503. doi:10.1002/brb3.1503
Ylikotila, P., Tiirikka, T., Moilanen, J. S., Kääriäinen, H., Marttila, R., and Majamaa, K. (2015). Epidemiology of early-onset Parkinson's disease in Finland. Park. Relat. Disord. 21, 938–942. doi:10.1016/j.parkreldis.2015.06.003
Keywords: Parkinson’s disease, cytochromes P450, POR, Adx, eicosanoids, cholesterol, vitamin D, retinoic acid
Citation: Petkova-Kirova P, Baas S, Wagenpfeil G, Hartz P, Unger MM and Bernhardt R (2023) SNPs in cytochrome P450 genes decide on the fate of individuals with genetic predisposition to Parkinson’s disease. Front. Pharmacol. 14:1244516. doi: 10.3389/fphar.2023.1244516
Received: 22 June 2023; Accepted: 21 July 2023;
Published: 04 August 2023.
Edited by:
Amit V. Pandey, University of Bern, SwitzerlandReviewed by:
D. Fernando Estrada, University at Buffalo, United StatesCopyright © 2023 Petkova-Kirova, Baas, Wagenpfeil, Hartz, Unger and Bernhardt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rita Bernhardt, cml0YWJlcm5AbXgudW5pLXNhYXJsYW5kLmRl; Marcus Michael Unger, bS51bmdlckBzYi5zaGcta2xpbmlrZW4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.