
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Pharmacol., 13 June 2023
Sec. Pharmacogenetics and Pharmacogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1234219
This article is part of the Research TopicThe Utilization of Bench-to-Bedside Approaches in PharmacogenomicsView all 5 articles
 Eric Rytkin1*†
Eric Rytkin1*† Kseniia Kriukova2,3†
Kseniia Kriukova2,3† Natalia Denisenko4†
Natalia Denisenko4† Dmitriy Ivashchenko4†
Dmitriy Ivashchenko4† Michael Zastrozhin5
Michael Zastrozhin5 Karin Mirzaev4‡
Karin Mirzaev4‡ Dmitry Sychev4‡
Dmitry Sychev4‡Editorial on the Research Topic
The utilization of bench-to-bedside approaches in pharmacogenomics
Pharmacogenetic testing is gaining prominence in clinical practice as evidence-based recommendations, and prospective studies demonstrate the advantages of a personalized genotype-based approach over standard therapy regimens (Weinshilboum and Wang, 2017; Luzum et al., 2021). In recent years, pharmacogenomics has emerged as a promising field that combines pharmacology and genomics to determine how an individual’s genetic makeup influences their response to medications. This personalized medicine approach can potentially revolutionize patient care by optimizing drug selection and dosage based on individual genetic profiles. A bench-to-bedside approach is crucial to translate pharmacogenomic research findings into clinical practice. This editorial explores the utilization of bench-to-bedside approaches in pharmacogenomics and features different aspects of them—from organizing pharmacogenetic laboratories within the academic infrastructure of educational and research institutions to the selection of Predictive Biomarkers.
Today, pharmacogenetic testing is becoming more and more affordable. The price of genotyping is decreasing every year. There is a growing demand from patients for effective and safe pharmacotherapy. But doctors’ awareness of the possibilities of pharmacogenetic testing remains low: according to various estimates, only up to 30% of specialists are familiar with this technology (Rahawi et al., 2020; Muflih et al., 2021). Consequently, the work on the implementation of pharmacogenetic testing is in demand. It is important not only to educate physicians about the possibilities of personalized pharmacotherapy but also to campaign among patients. Although there are genetic tests on the market that have no proven benefit, it is important to introduce people to new technologies. It is necessary to overcome the psychological barrier that slows down the use of genetic tests. Typically, patients are wary of genome analysis (McCarthy et al., 2020). It is important to include pharmacogenetic testing in clinical guidelines. This will lead to more active prescribing of single nucleotide polymorphism analysis. To date, CPIC and DPWG have made numerous suggestions for personalized pharmacotherapy selection. But the practical application will be limited until pharmacogenetic testing is included in clinical guidelines for the treatment of diseases.
The Organization of Pharmacogenetic Laboratories in Academic Institutions:
While commercial laboratories offer accessibility and scalability, they often lack expertise in marker selection, test prescription strategies, and result interpretation. This results in a lack of credibility among physicians, clinic leaders, health system representatives, and patients (Luzum et al., 2021). To overcome these limitations, organizing pharmacogenetic laboratories within the academic infrastructure of educational and research institutions is a promising approach. This organizational form provides expert support, including marker selection and result interpretation, and offers extensive training programs for healthcare professionals and patients. Academic laboratories can also establish well-described biobanks, collect pharmacogenetic and clinical data, and secure multiple funding sources through research and grant projects.
In academic institutions, Pharmacogenomics laboratories of such a kind are usually organized in the form of Core Facilities. These core facilities usually include equipment for nucleic acid extraction and quantitative and qualitative analysis and may employ various methods such as polymerase chain reaction or high-throughput sequencing (Henslee and Telgenhoff, 2019). Additionally, access to pharmacokinetic laboratory and bioinformatics services can be incorporated into the laboratory structure. Providing access to the pharmacogenetic core facility for other university departments allows collaborative research studies and grant programs (Table 1). Effective laboratory operation relies on financial statements, close interaction with the finance department, and a well-organized procurement system.
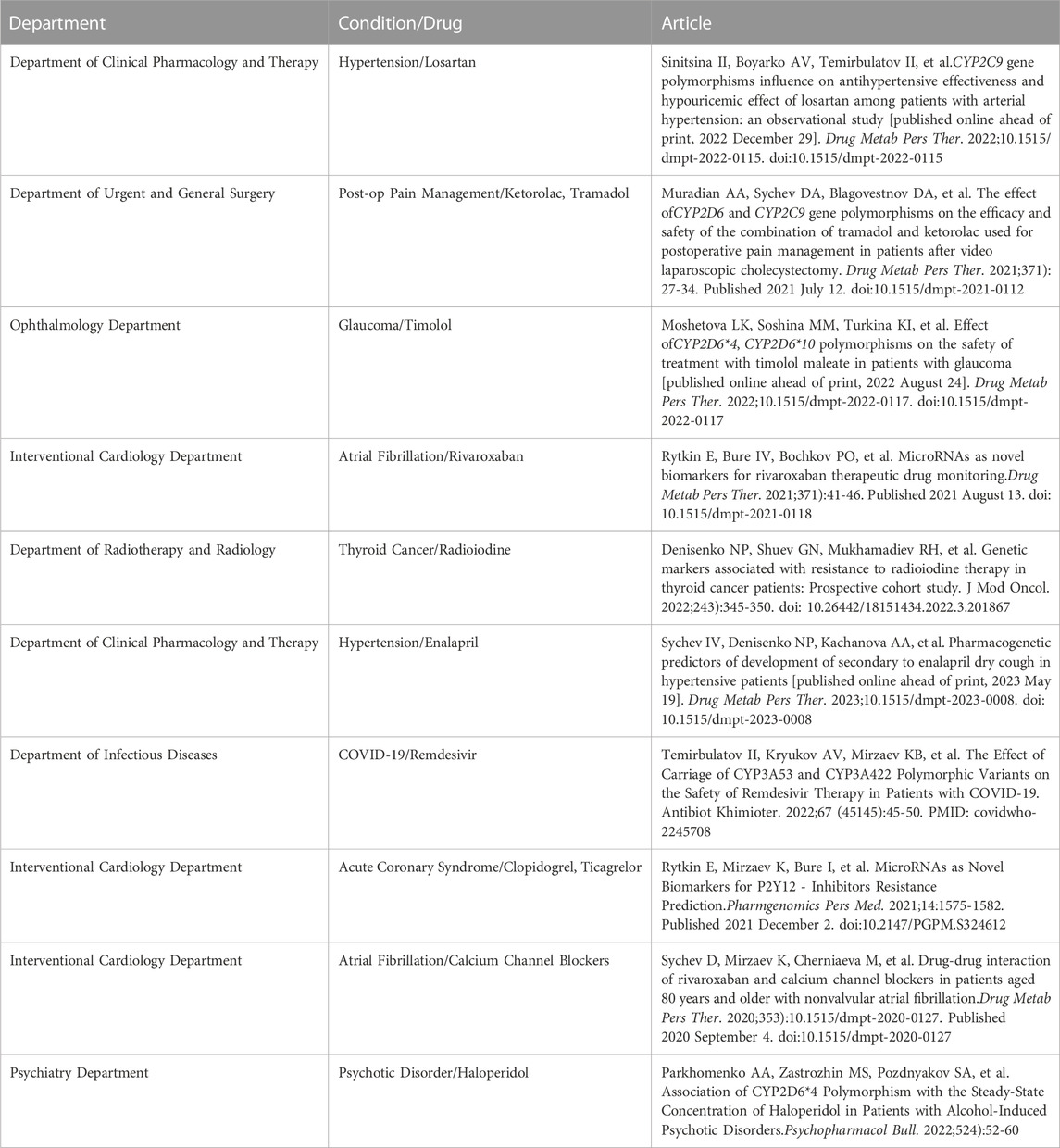
TABLE 1. The summary of the collaborative studies conducted in the Russian Medical Academy of Continuous Professional Education. This features the collaborations between Clinical Departments and the Pharmacogenomics Center.
Preventive medicine and personalized medicine aim to provide better-individualized healthcare based on unique patient factors and biomarkers, including genetic factors. The research on predictive biomarkers may vary from pharmacogenomics of antidepressant drugs (Kee et al.) to acute coronary syndrome management (Azzahhafi et al.) and antithrombotic therapy (Asiimwe et al.). (Asiimwe et al.; Azzahhafi et al.; Kee et al.) By identifying genetic variations as predictive biomarkers, pharmacogenomic information on the optimal medication can be obtained, potentially improving patient outcomes. Similar approaches can be applied to other areas, such as neurodegenerative and neuroinflammatory disorders.
To effectively utilize bench-to-bedside approaches in pharmacogenomics, it is crucial to acquire knowledge of predictive biomarkers and study them as both predictive biomarkers and pharmacogenomic tools. Cross et al. discussed the concept of polygenic risk scores (PRS) as an overview of predictive biomarkers in personalized medicine (Cross et al.). PRS is a statistical approach that combines information from multiple genetic variants to estimate an individual’s genetic risk for a particular condition or response to treatment. The integration of PRS into pharmacogenomics has the potential to enhance the predictive power and precision of personalized medicine approaches.
Cross et al. highlighted that PRS can be derived from genome-wide association studies (GWAS) and applied to various medical conditions, including cardiovascular diseases, psychiatric disorders, and cancer (Cross et al.). By considering a broader set of genetic markers, PRS can capture the cumulative effect of multiple genetic variants, allowing for more accurate risk assessment and treatment predictions. Additionally, incorporating PRS into pharmacogenomics can help identify individuals who are at a higher risk of adverse drug reactions or poor treatment response, enabling clinicians to personalize medication choices accordingly.
The utilization of bench-to-bedside approaches in pharmacogenomics holds great promise for advancing personalized medicine. Organizing pharmacogenetic laboratories within academic institutions provides several advantages, including expert support, extensive training programs, and the establishment of well-described biobanks. Integrating predictive biomarkers, such as PRS, into pharmacogenomics enhances the ability to predict treatment response and optimize therapeutic interventions. By embracing these approaches, we can pave the way for a deeper understanding of diseases and the development of targeted therapies that improve patient outcomes.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cross, B., Turner, R., and Pirmohamed, M. (2022). Polygenic risk scores: An overview from bench to bedside for personalised medicine. Front. Genet. 13, 1000667. doi:10.3389/fgene.2022.1000667
Henslee, C. R., and Telgenhoff, D. (2019). Molecular genetic testing laboratory management: Emerging challenges for quality assurance. J. Histotechnol. 42, 240–244. doi:10.1080/01478885.2019.1630083
Luzum, J. A., Petry, N., Taylor, A. K., Van Driest, S. L., Dunnenberger, H. M., and Cavallari, L. H. (2021). Moving pharmacogenetics into practice: It’s all about the evidence. Clin. Pharmacol. Ther. 110, 649–661. doi:10.1002/cpt.2327
McCarthy, M. J., Chen, Y., Demodena, A., Fisher, E., Golshan, S., Suppes, T., et al. (2020). Attitudes on pharmacogenetic testing in psychiatric patients with treatment-resistant depression. Depress Anxiety 37, 842–850. doi:10.1002/da.23074
Muflih, S., Alshogran, O. Y., Al-Azzam, S., Al-Taani, G., and Khader, Y. S. (2021). Physicians’ knowledge and attitudes regarding point-of-care pharmacogenetic testing: A hospital-based cross-sectional study. Pharmgenomics Pers. Med. 14, 655–665. doi:10.2147/PGPM.S307694
Rahawi, S., Naik, H., Blake, K. V., Owusu Obeng, A., Wasserman, R. M., Seki, Y., et al. (2020). Knowledge and attitudes on pharmacogenetics among pediatricians. J. Hum. Genet. 65, 437–444. doi:10.1038/s10038-020-0723-0
Keywords: pharmacogenomics, core facilities, bench to bedside translation, clinical application, biomarkers
Citation: Rytkin E, Kriukova K, Denisenko N, Ivashchenko D, Zastrozhin M, Mirzaev K and Sychev D (2023) Editorial: The utilization of bench-to-bedside approaches in pharmacogenomics. Front. Pharmacol. 14:1234219. doi: 10.3389/fphar.2023.1234219
Received: 03 June 2023; Accepted: 05 June 2023;
Published: 13 June 2023.
Edited and reviewed by:
José A. G. Agúndez, University of Extremadura, SpainCopyright © 2023 Rytkin, Kriukova, Denisenko, Ivashchenko, Zastrozhin, Mirzaev and Sychev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eric Rytkin, ZXJ5dGtpbkBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.