
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Pharmacol. , 16 August 2023
Sec. Pharmacology of Ion Channels and Channelopathies
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1210140
Missense variants in CALM genes encoding the Ca2+-binding protein calmodulin (CaM) cause severe cardiac arrhythmias. The disease mechanisms have been attributed to dysregulation of RyR2, for Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT) and/or CaV1.2, for Long-QT Syndrome (LQTS). Recently, a novel CALM2 variant, G114R, was identified in a mother and two of her four children, all of whom died suddenly while asleep at a young age. The G114R variant impairs closure of CaV1.2 and RyR2, consistent with a CPVT and/or mild LQTS phenotype. However, the children carrying the CALM2 G114R variant displayed a phenotype commonly observed with variants in NaV1.5, i.e., Brugada Syndrome (BrS) or LQT3, where death while asleep is a common feature. We therefore hypothesized that the G114R variant specifically would interfere with NaV1.5 binding. Here, we demonstrate that CaM binding to the NaV1.5 IQ-domain is severely impaired for two CaM variants G114R and G114W. The impact was most severe at low and intermediate Ca2+ concentrations (up to 4 µM) resulting in more than a 50-fold reduction in NaV1.5 binding affinity, and a smaller 1.5 to 11-fold reduction at high Ca2+ concentrations (25–400 µM). In contrast, the arrhythmogenic CaM-N98S variant only induced a 1.5-fold reduction in NaV1.5 binding and only at 4 µM Ca2+. A non-arrhythmogenic I10T variant in CaM did not impair NaV1.5 IQ binding. These data suggest that the interaction between NaV1.5 and CaM is decreased with certain CaM variants, which may alter the cardiac sodium current, INa. Overall, these results suggest that the phenotypic spectrum of calmodulinopathies may likely expand to include BrS- and/or LQT3-like traits.
The cytosolic calcium (Ca2+) binding protein calmodulin (CaM) serves as a critical mediator of intra-cellular Ca2+ signals in a multitude of physiological processes (Chin and Anthony, 2000; Xia and Storm, 2005; Clapham, 2007; Sorensen et al., 2013; Berchtold and Villalobo, 2014). The multifaceted nature of CaM comes from its ubiquitous expression and its ability to interact with, and relay information to, more than 350 cellular target proteins (Yap et al., 2000; Tidow and Nissen, 2013).
This extraordinary versatility is due to the two lobes of CaM, the N-lobe and C-lobe, each containing two Ca2+-binding EF hands (Figure 1A). The lobes differ in Ca2+-affinity and -kinetics, allowing CaM to respond to changes in Ca2+ over a broad range of concentration and time. This range is even further expanded by target-specific changes in Ca2+ binding-affinities and -kinetics upon CaM binding to protein targets (Villarroel et al., 2014; Søndergaard et al., 2015a; Søndergaard et al., 2015b).
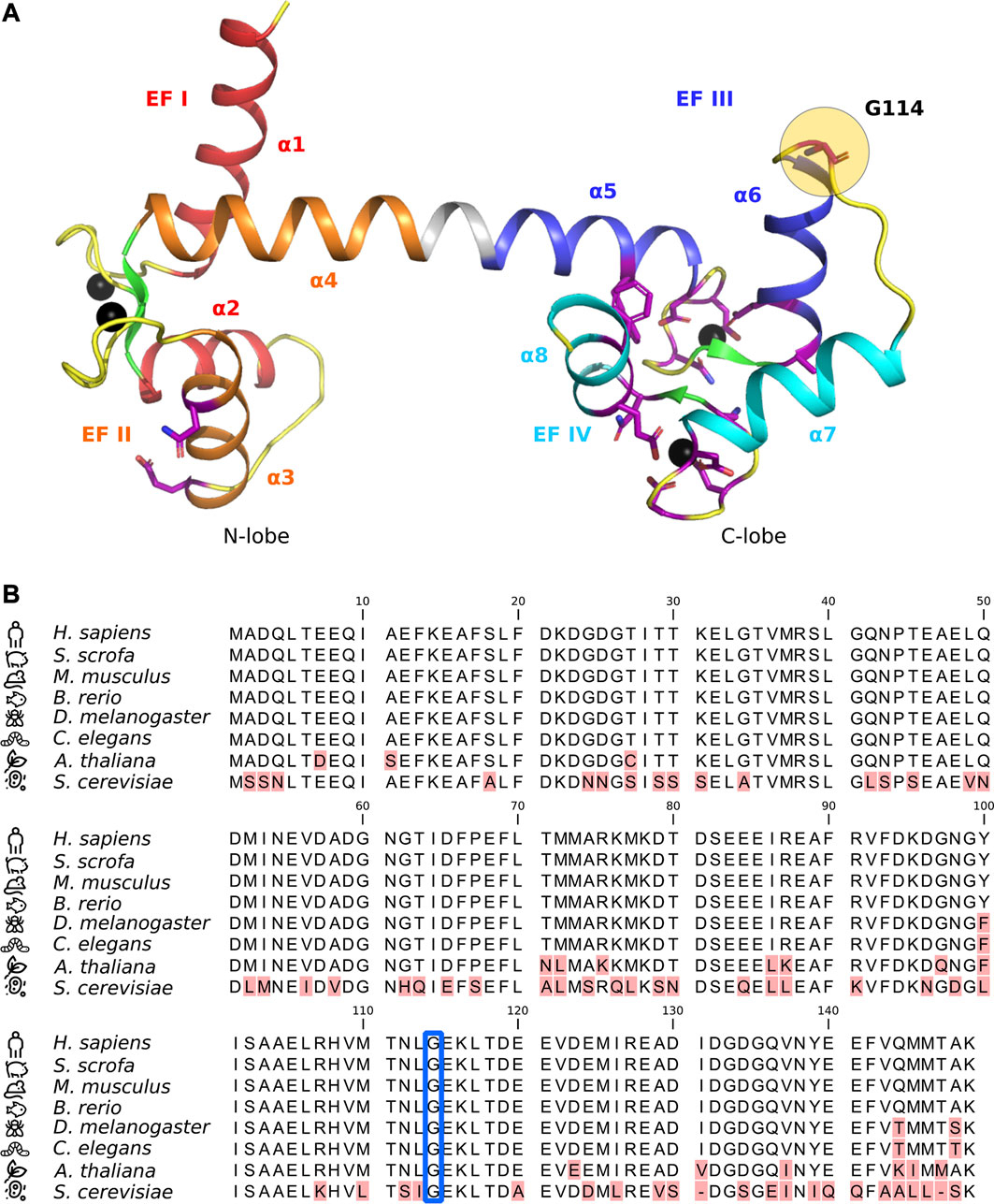
FIGURE 1. (A) Crystal structure of Ca2+-bound calmodulin (CaM) (PDB ID: 1CLL) with EF hands I (red), II (orange), III (blue), and IV (cyan) indicated along with their corresponding alpha-helices (α1-8). Beta-sheets are shown in green and flexible loops/unstructured regions in yellow. Amino acid residues known to harbor arrhythmogenic substitutions (Crotti et al., 2019) are shown in purple stick representation, with residue G114 highlighted with a yellow circle. Ca2+-ions are shown as black spheres. (B) CaM protein sequence alignment. Red highlights indicate amino acid differences or gaps in the alignment. Residue G114 is indicated by a blue frame. Numbering is according to immature human CaM (including the initial M).
The cellular importance of CaM Ca2+-sensing and integrity is highlighted by the protein’s unique genetic architecture and evolutionary conservation. Mammals have three independent genes (CALM1-3) that all encode an identical CaM protein. Moreover, the protein sequence is invariant in all vertebrates, underpinning the extreme selection pressure against any amino acid variation in this central Ca2+-sensor protein (Figure1B) (Berchtold et al., 1993; Toutenhoofd and Strehler, 2000; Friedberg and Rhoads, 2001).
As a result, genetic variation in all three CALM genes is ultra-rare. Until the first human missense variant was discovered in 2012, and linked to a severe cardiac arrhythmia (Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT)) and sudden cardiac death (SCD) (Nyegaard et al., 2012), mutations in the CALM genes were considered incompatible with life (Jensen et al., 2018). Since the initial discovery of human missense variants linked to CPVT, mutations have also been identified in individuals affected by long QT syndrome (LQTS) (Crotti et al., 2013) and Idiopathic Ventricular Fibrillation (IVF) (Marsman et al., 2014). In 2019, Crotti and co-workers presented an extensive collection of 28 unique CaM variants, identified in 74 carriers, in the International Calmodulinopathy Registry (Crotti et al., 2019). CALM variant carriers in this registry present with cardiac arrhythmia phenotypes including LQTS (49%), CPVT (28%), overlap LQTS/CPVT (4%), and a few cases of IVF, sudden unexplained death (SUD), or atypical phenotypes (Crotti et al., 2019).
Clinical characteristics of the calmodulinopathies include an early age of onset (a mean of 1.5 years for LQTS and 6 years for CPVT) and a high risk of a major arrhythmic event (68%), such as cardiac arrest or SCD/SUD (Crotti et al., 2019). The molecular mechanisms underlying the two main phenotypes has largely been ascribed to specific dysregulation of the two primary cardiac Ca2+ channels, CaV1.2 for LQTS, and RyR2 for CPVT-like phenotypes (Limpitikul et al., 2014; Yin et al., 2014; Søndergaard et al., 2019; 2020; 2017; Nyegaard and Overgaard, 2019; Holt et al., 2020). The broad phenotypic spectrum caused by CaM variants, including mechanistically different cardiac arrhythmias, is likely a consequence of CaM serving as a key regulator of multiple cardiac ion-channels, besides CaV1.2 and RyR2, that control cardiac excitation-contraction coupling. Indeed, given the vast number of CaM-regulated proteins, the phenotypic spectrum of calmodulinopathies is likely to expand even further as more carriers are discovered (Jensen et al., 2018; Urrutia et al., 2019).
In 2019, a novel CALM2 variant, G114R (immature protein numbering including initial Met), was identified in an Australian mother and two of her four children. Over a 10-year period, all four children died suddenly and unexpectedly while asleep, at ages ranging from 19 days to 18 months (Brohus et al., 2021). Further, the two children carrying the G114R variant had infections at the time of death, implying a potential presence of fever. In 2021, we showed that the G114R variant impairs CaM’s ability to bind Ca2+-ions and to interact with and regulate CaV1.2 and RyR2, with an impact suggesting an arrhythmogenic potential consistent with CPVT, IVF, or mild LQTS (Brohus et al., 2021).
Death while asleep or at rest have only been observed in a small subset of CaM variant carriers, and mainly for CaM variants with a severe impairment of Ca2+ binding and/or Ca2+-dependent inactivation of CaV1.2, larger than the effect imposed by G114R (Brohus et al., 2021). Therefore, the phenotype of the children carrying the CALM2 G114R variant to some degree represents an expansion of the known clinical manifestations of CaM variant carriers. The phenotype more closely resembles that of carriers of missense variants in the cardiac sodium channel, NaV1.5, for whom major arrhythmic events or death while asleep is a common feature (Schwartz et al., 2001; Postema and Wilde, 2008; Takigawa et al., 2008). Given that CaM is critical for NaV1.5 function, we hypothesized that the G114R variant would specifically interfere with NaV1.5 binding.
The NaV1.5 channel is implicated in both Brugada Syndrome (BrS) and LQT3, arrhythmic diseases that result from divergent molecular mechanisms. Intriguingly, both phenotypes can be caused by NaV1.5 channel mutations that perturb the interaction with and modulation by CaM (Shah et al., 2006; Yan et al., 2017; Urrutia et al., 2019; Kang et al., 2021; Wu and Liang, 2021). In some cases, the same NaV1.5 variant causes both phenotypes, which alludes to the difficulty in variant genotype-phenotype interpretation (Supplementary Figure S1).
The gating of NaV1.5 is modulated by CaM in a bi-directional manner: Both channel activation (peak current), fast inactivation, and persistent current depend on CaM. Several CaM binding domains (CaMBDs) have been identified, but their individual roles in the bi-directional modulation by CaM is still unclear (Kang et al., 2021). While the primary CaM binding site is an IQ-motif located in the C-terminal domain (CTD) of NaV1.5 (Chagot and Chazin, 2011; C; Wang et al., 2014; Gabelli et al., 2014; C; Wang et al., 2012; Gabelli et al., 2016), CaM has also been shown to interact with a preIQ-domain in the CTD (Yoder et al., 2019), the “inactivation gate” in the DIII-DIV linker (Potet et al., 2009; Sarhan, Van Petegem, and Ahern, 2009; Sarhan et al., 2012; Johnson et al., 2018), and an N-terminal domain (NTD) (Wang et al., 2020) (Figure2A).
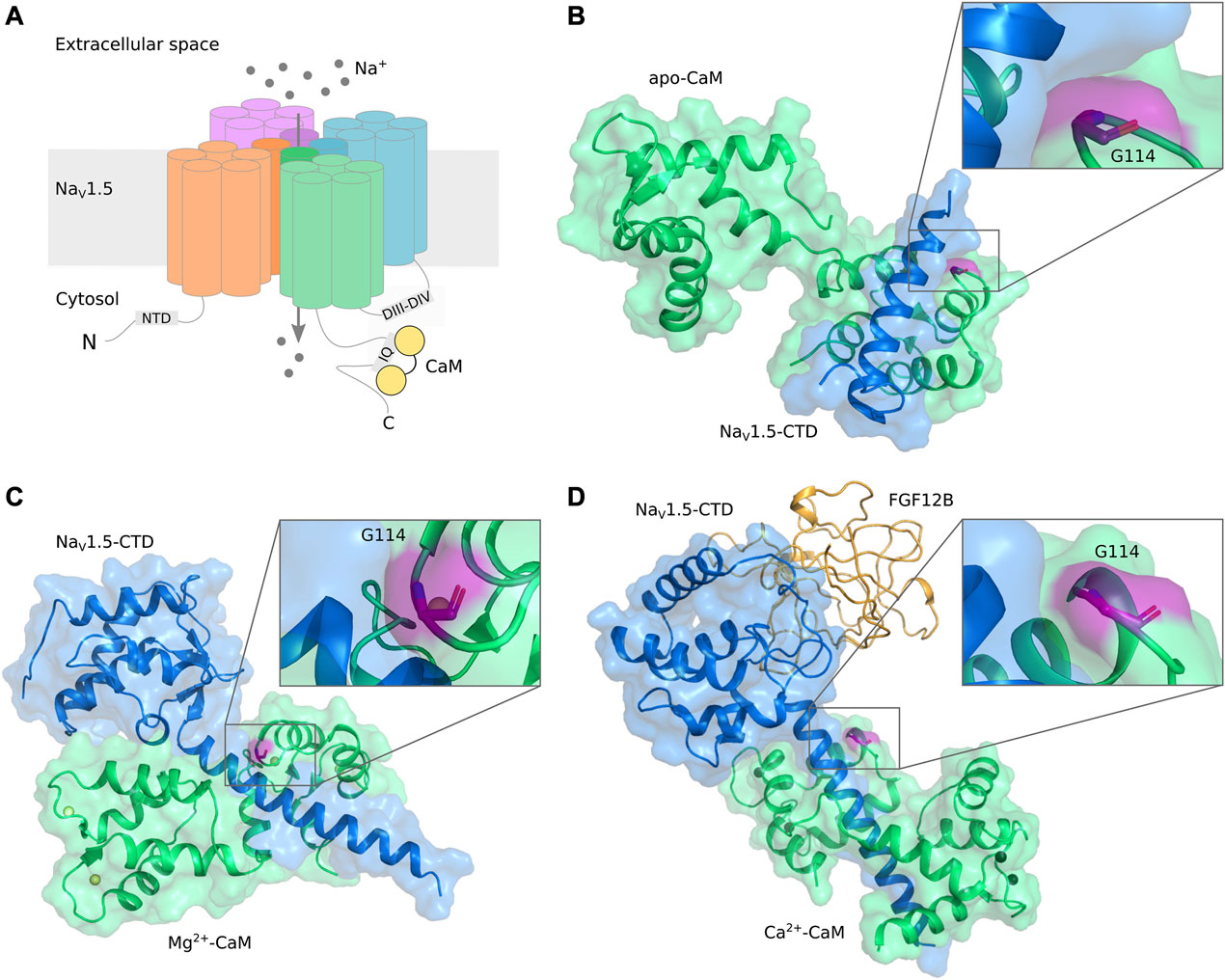
FIGURE 2. (A) Schematic representation of Nav1.5 structural elements and interaction with CaM. (B–D) Visualization of CaM-G114 in NaV1.5/CaM complexes. CaM (green) and the NaV1.5 CTD (blue) are displayed in cartoon and surface representation with CaM-G114 (purple) displayed in stick representation. The insets show that G114 is located at the interface between the CaM EF hand III-IV loop and the NaV1.5-IQ domain. (B) apo-CaM, PDB ID 2L53 (Chagot and Chazin, 2011); (C) Mg2+-bound CaM, PDB ID 4OVN (Gabelli et al., 2014); (D) Ca2+-bound CaM, PDB ID 4JQ0 (Wang et al., 2014). Ca2+ (black) and Mg2+ (yellow) ions are shown as spheres. CaM, calmodulin; NaV, voltage-gated sodium channel; CTD, C-terminal domain; FGF, fibroblast growth factor (orange).
In this study, we illustrate that CaM-G114 is located exactly in the binding interface between CaM and the IQ-domain of NaV1.5 and demonstrate that CaM variants G114R and G114W impair the interaction with the IQ-domain in a Ca2+-dependent manner with the largest impact occurring at a free Ca2+-concentration range of 3 nM–4 µM. Thus, the apoCaM interaction with the NaV1.5-IQ domain is impaired for these CaM variants and their Ca2+-sensing ability in the CaM/NaV1.5-IQ complex has markedly changed.
Materials and methods can be found in the Supplementary Material.
CaM-G114 is the terminating residue of the second helix of EF hand III and it so far constitutes the only amino acid residue known to harbor a mutation in the loop between EF hands III and IV (L113-T118) (Figure 1A) (Bycroft et al., 2018; Crotti et al., 2019; Chen et al., 2022). A protein sequence alignment of CaM from different species shows that residue 114 is extremely evolutionarily conserved (Figure1B, blue square). It is a glycine in all species investigated, including yeast, emphasizing the universal importance of its integrity.
High resolution structures of CaM (green) in complex with the NaV1.5-CTD (blue) reveal that CaM-G114 (purple) is located at the interface between CaM and the channel (Figure 2). In the apo-form, only the C-lobe of CaM binds to the NaV1.5 IQ-domain (Figure 2B), whereas in the Mg2+- and Ca2+-bound forms, CaM wraps around the NaV1.5 CTD with both its lobes (Figure 2C, D). In all cases, the interaction brings CaM-G114 and the IQ-domain into close proximity (Figure 2, insets). We therefore hypothesized that substitution of residue G114 would affect the binding between CaM and the NaV1.5 channel.
To test the hypothesis that the integrity of CaM-G114 is essential for the interaction with the NaV1.5 IQ-domain, we monitored the fluorescence anisotropy (FA) signal of the TAMRA-labeled IQ-domain during titration with CaM at eight different Ca2+-concentrations, resulting in eight binding curves for each CaM variant (Figure 3A). The CaM variant N98S was included as an arrhythmogenic control, known to cause both LQTS and CPVT (Nyegaard et al., 2012; Makita et al., 2014; Jiménez-Jáimez et al., 2016). Another CaM variant, CaM-I10T, identified in the UK Biobank resource (Bycroft et al., 2018), was included as a non-arrhythmogenic control.
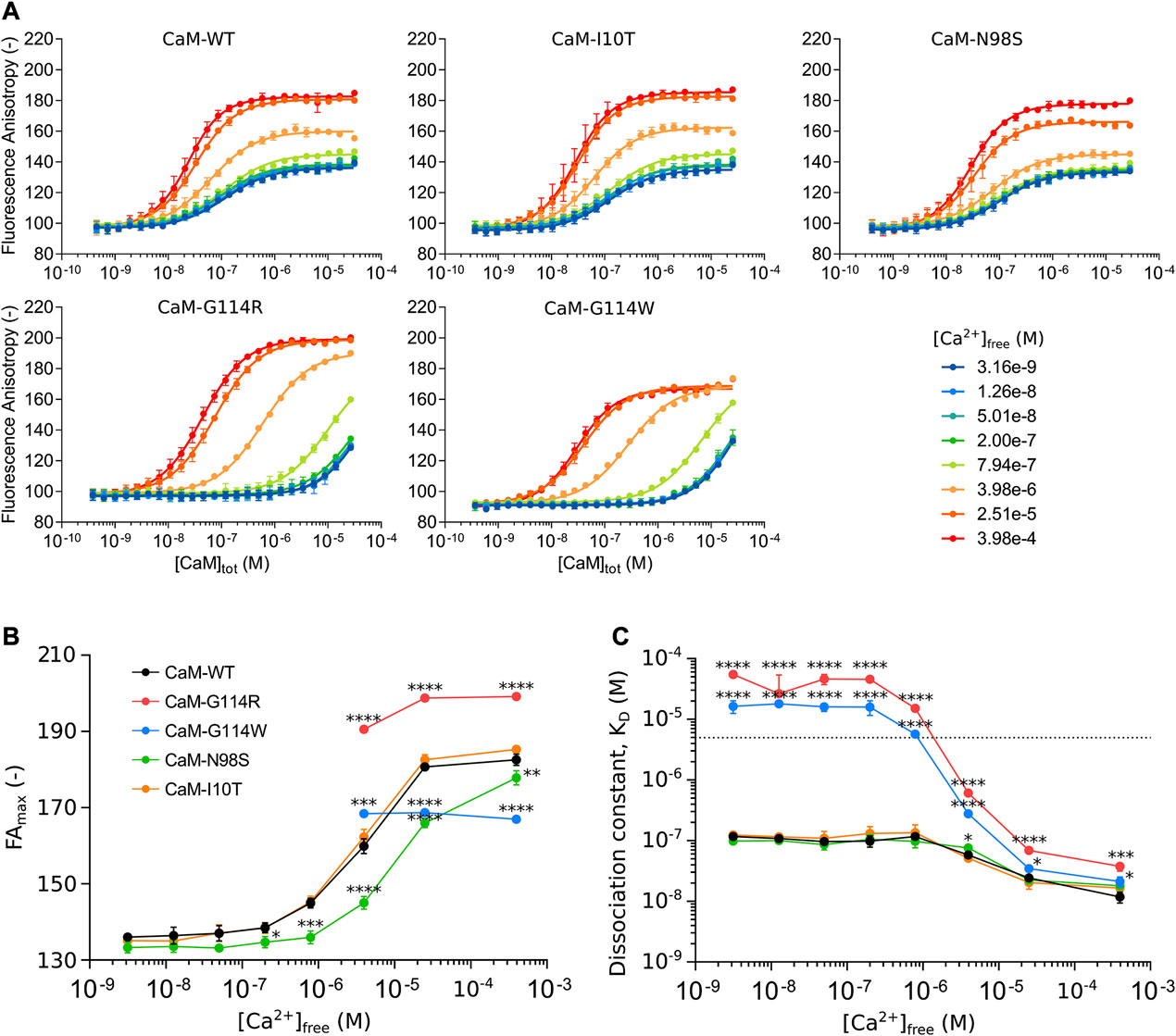
FIGURE 3. (A) CaM binding to the NaV1.5 IQ-domain monitored by fluorescence anisotropy (FA) as a function of total CaM-concentration ([CaM]tot) at eight free Ca2+ concentrations ([Ca2+]free). (B) Ca2+-dependent conformational changes of the CaM/NaV1.5 IQ-domain complex represented by changes in the maximum FA plateau (FAmax) as a function of [Ca2+]free. (C) Ca2+-dependent changes in the CaM affinity of the NaV1.5 IQ-domain represented by the dissociation constant (KD) as a function of [Ca2+]free. Color scheme as in panel B. Each data point represents the mean of three replicates with the standard deviation shown as error bars. For panel B and C, statistically significant differences between CaM-WT and other variants were determined by a 1-way ANOVA at each Ca2+-concentration with Dunnett’s multiple comparisons test: **** (p-value <0.0001), *** (p-value <0.001), * (p-value <0.05).
The interaction between CaM-WT and the NaV1.5 IQ-domain depends on the level of Ca2+, apparent as a change in FA values for the CaM-saturated IQ-domain from low (Figure 3A, blue) to high (Figure 3A, red) Ca2+ concentrations. The assay thus allowed us to explore the Ca2+-dependency of the interaction, by determining the maximum FA plateau (FAmax) (Figure 3B, Supplementary Table S1) and the binding affinity (Figure 3C, Supplementary Table S2) at each of the eight Ca2+-concentrations.
The FA signal is a measure of the tumbling rate of the TAMRA-labeled NaV1.5 IQ-domain (Rossi and Taylor, 2011). As more CaM is added, more CaM/NaV1.5 IQ-domain complex forms, and the FA signal will increase (due to a reduced tumbling rate of the IQ-domain) until reaching FAmax which represents the tumbling rate of the saturated complex (Figure 3A). The tumbling rate depends on the conformation of the complex, and a plot of the FAmax value as a function of [Ca2+]free reveals a Ca2+-dependent increase in FAmax of the CaM-WT/NaV1.5 IQ-domain complex, in turn reflecting a Ca2+-induced conformational change (Figure 3B, black, Supplementary Table S1). The Ca2+-dependent conformational change is accompanied by a 10-fold increase in the CaM-WT binding affinity of the NaV1.5 IQ-domain (Figure 3C, black, Supplementary Table S2).
Curiously, the Ca2+-dependent development in the FA signal for the CaM-G114 variants is very different from CaM-WT. First, no FAmax plateau is reached at free Ca2+ concentrations <4 µM (Figure 3A, B). Second, the FAmax values ≥4 µM free Ca2+ are significantly different from those of the CaM-WT/NaV1.5-IQ complex (Figure 3B, red and blue, Supplementary Table S1). Interestingly, the change in FAmax imposed by the two G114 substitutions occurs in opposite directions relative to CaM-WT at saturating Ca2+ (FAmax increases for CaM-G114R and decreases for CaM-G114W, Supplementary Table S3).
Moreover, the CaM-G114R and -G114W mutations cause a dramatic reduction in IQ-domain binding affinity compared to CaM-WT (Figure 3C, red and blue, Supplementary Table S2). At Ca2+-concentrations below 4 μM, the affinity is reduced to an extent where the dissociation constant could not be accurately determined (KD < 5 µM) (Figure 3C, stapled line). However, the data demonstrates that the affinity is reduced at least 47-fold compared to CaM-WT at these Ca2+-concentrations and estimates of KD-values could be determined by assuming an identical FAmax value at all Ca2+ concentrations (Figure 3C, see Methods section for details, Supplementary Table S2, S4). At free Ca2+-concentrations at and above 4 μM, the IQ-domain affinity of CaM-G114R and -G114W is still significantly reduced compared to CaM-WT, but to a smaller extent (1.4 to 11-fold) (Figure 3C; Supplementary Table S4).
In contrast to the CaM-G114 variants, the Ca2+-dependent increase in FAmax observed for the CaM-WT/IQ-domain complex is also apparent for the CaM-I10T/IQ-domain complex (Figure 3B, orange). Like for CaM-WT, the FAmax value for the CaM-N98S/IQ-domain complex increases with Ca2+, but the transition is shifted to higher Ca2+-concentrations (Figure 3B, green, Supplementary Tables S1, S3). Further, the arrhythmogenic CaM-N98S variant only reduced the NaV1.5 affinity significantly (1.5-fold compared to CaM-WT) at intermediate 4 uM free Ca2+ (Figure 3C; Supplementary Tables S2, S4). This effect is consistent with the observed 4-fold reduction in CaM C-lobe Ca2+ affinity imposed by the N98S substitution, but different from the G114R and G114W substitutions, although they have a similar impact on C-lobe Ca2+ binding (3- and 7-fold reduction compared to CaM-WT) (Brohus et al., 2021). The non-arrhythmogenic CaM-I10T variant displayed no difference in IQ-domain affinity compared to CaM-WT across any of the Ca2+ concentrations tested (Figure3C, orange). These results are consistent with the observation that both residue I10 and N98 are located away from the CaM/NaV1.5 IQ-domain binding interface (Supplementary Figure S2).
The dramatic effect of the G114 variants on NaV1.5 affinity at low Ca2+ concentrations (≤200 nM) appear specific to the IQ-domain, as the effect of CaM-G114R and G114W on the NaV1.5 NTD were much smaller at the corresponding Ca2+ concentrations and comparable in magnitude to the effects of arrhythmogenic N98S (within a 4-fold difference from CaM-WT) (Supplementary Figure S3, S4; Supplementary Table S5, S6). As observed for the IQ-domain, the interaction between the non-arrhythmogenic CaM-I10T and the NaV1.5 NTD did not differ from the CaM-WT/NTD interaction.
In this study, we use an FA-based assay to investigate the Ca2+-dependent interactions between CaM-WT and the NaV1.5 IQ-domain and the recently identified CaM binding domain in the NaV1.5 NTD, and how these are affected by mutations in CaM.
Intriguingly, the CaM/NaV1.5 IQ-domain interaction displays a very different Ca2+ sensitivity profile compared to the interactions between CaM and the NaV1.5 NTD, CaV1.2 IQ-domain, and RyR2-CaMBD2 (Wang et al., 2018; Brohus et al., 2019; Wang et al., 2020; Brohus et al., 2021). While the binding affinity increases 10-fold for the CaM-WT/NaV1.5 IQ-domain complex from low nM to high µM Ca2+-concentrations, the affinities of the CaM-WT/NaV1.5 NTD, CaM-WT/CaV1.2 IQ-domain, and CaM-WT/RyR2-CaMBD2 complexes increase more than 1000-fold across the same Ca2+-range (Supplementary Figure S5) (Brohus et al., 2021). Moreover, apoCaM binds to the NaV1.5 IQ-domain with high affinity (117 nM—in good agreement with affinities determined by others (Shah et al., 2006; Yan et al., 2017)), much higher than to the NaV1.5 NTD (825-fold), the CaV1.2 IQ-domain (26-fold), and RyR2 CaMBD2 (7-fold). For Ca2+-saturated CaM, the interaction with the NaV1.5 IQ-domain is more than 100-fold weaker than the interaction with the CaM binding domains from RyR2 and CaV1.2 (Brohus et al., 2021). These results corroborate an essential role of apoCaM in modulating the NaV1.5 channel via the IQ-domain (Kang et al., 2021).
A potential dysregulation of NaV1.5, caused by human CaM mutations, has previously been investigated for a handful of LQTS-causing CaM variants (D96V, D130G, F142L, E141G) (Yin et al., 2014; Boczek et al., 2016; Rocchetti et al., 2017; Tarasov et al., 2023). However, the results for these variants have been largely unremarkable. Co-expression of CaM and human NaV1.5 in tsA102 cells, and subsequent whole-cell patch clamp recordings, showed no effect on channel function for any of the CaM variants investigated. Only when expressing a fetal NaV1.5 splice variant, CaM-D130G caused a 7.5-fold increase in persistent Na+ current, and only at 1 µM free Ca2+. Moreover, native Na+ currents from fetal mouse cardiomyocytes were not affected by CaM-D130G (Yin et al., 2014). Along the same lines, co-expression of CaM-E141G and NaV1.5 in tsA102 cells, and subsequent whole-cell patch clamp recordings, caused a 1.7-fold increase in persistent Na+ current, but the effect was no longer apparent when co-expressed with CaM-WT (Boczek et al., 2016). Since these studies, the NaV1.5 channel has been under the radar in terms of studying its implication in calmodulinopathies. However, and interestingly, Tarasov and co-workers recently demonstrated that CaM-D96V specifically increased the late current of the NaV1.6 isoform, but not of NaV1.5, speculating that this was due to a reduced CaM affinity for the NaV1.6 IQ-domain (Tarasov et al., 2023).
We have previously shown that CaM-G114R and -G114W reduce the affinity of CaM for both the CaV1.2 IQ-domain and for RyR2-CaMBD2 at low to medium Ca2+-concentrations (Brohus et al., 2021). Here we show that both mutations also reduce CaM’s affinity for the NaV1.5 IQ-domain, but the effect is dramatically larger than for the CaMBDs of CaV1.2 and RyR2, particularly at free Ca2+-concentrations ≤200 nM, the physiological Ca2+ concentration in the cardiomyocytes at rest, where the CaM/NaV1.5 IQ-domain interaction is essentially abolished.
The interaction between apoCaM and NaV1.5 is critical for channel function, by tuning channel activity. ApoCaM binding to the NaV1.5 CTD causes an increase in peak channel open probability as well as a decrease in persistent channel open probability, effects that have divergent implications in disease (Kang et al., 2021). Disruption or weakening of apoCaM binding reduces peak open probability of the channel, corresponding to a loss-of-function effect, such as that observed with the BrS phenotype. However, impaired apoCaM binding can also lead to an increase in persistent NaV1.5 late current, corresponding to a gain-of-function effect, such as that observed with the LQT3 phenotype (Yan et al., 2017; Kang et al., 2021). The dramatic reduction in the affinity of apoCaM for the NaV1.5 IQ-domain, caused by substitution of CaM-G114 could thus result in similar divergent effects, and may provide a mechanistic explanation for the mixed phenotypic pattern observed for carriers of CaM-G114 mutations, and potentially other residues affecting NaV1.5 binding.
But how can a mutation in one of six CaM-encoding alleles display a dominant effect through an ion-channel if the affinity for this channel is dramatically reduced? Several points provide hints towards a possible explanation. The intracellular pool of CaM is limited in cardiomyocytes, suggesting a dynamic competition among CaM target binding sites (Persechini and Stemmer, 2002; Wu et al., 2007). In the GTEx transcript database (GTEx Consortium, 2013), CALM2 represents ∼50% of the total CALM transcript pool in ventricular tissue, and a heterozygous CALM2 missense mutation may thus be present in 25% of the CaM protein pool. Since the impact of a specific CaM mutation differs between targets, the available CaM protein will be redistributed accordingly. Targets affected by a large reduction in CaM binding affinity may experience a larger deficit in CaM saturation than expected from the CaM-mutant/CaM-WT protein ratio, thereby increasing the risk of experiencing a “haploinsufficiency-like” phenotype. This notion is corroborated by a study investigating LQT3 mutations within the NaV1.5 IQ-domain (Yan et al., 2017). The study demonstrated that these variants increase the persistent Na+ current amplitude of NaV1.5 in whole-cell patch clamp recordings in HEK-cells, and that IQ-domains containing these mutations reduce the CaM binding affinity. Overexpression of CaM-WT rescued the increased current for these LQT3-NaV1.5 channels (Yan et al., 2017).
Another possible explanation for a dominant effect of the CaM-G114 variants is that CaM binds to other parts of the NaV1.5 channel than the IQ-domain. One example is binding of apoCaM to the preIQ-domain with high affinity (∼40 nM) (Yoder et al., 2019). Such binding may anchor the CaM-G114 variant to the channel and mediate a pathogenic effect through a compromised IQ-domain binding, induced by the C-lobe mutation.
Intrinsic mutations in NaV1.5 are responsible for BrS and LQTS3, arrhythmogenic conditions both known for cardiac events to frequently occur during rest/sleep (Schwartz et al., 2001; Postema and Wilde, 2008; Takigawa et al., 2008). Additionally, for BrS, fever has been established as a trigger of these events (Adler et al., 2013; Michowitz et al., 2018). Some of these arrhythmogenic channel mutations occur in the CaM-binding IQ-domain of the NaV1.5 CTD and perturb the CaM/NaV1.5 interaction (Supplementary Figure S1) (Shah et al., 2006; Yan et al., 2017; Kang et al., 2021; Wu and Liang, 2021). This, together with the impaired apoCaM/NaV1.5 IQ-domain interaction presented in this work, opens a possible mechanistic explanation for the clinical presentation observed for the CaM-G114R carriers, who died suddenly at a very young age while asleep, potentially triggered by a fever from the infections they each had at the time of death (Brohus et al., 2021). Other intrinsic NaV1.5 BrS/LQTS mutations occur in the NTD of the channel, a domain for which the role of CaM has only recently been explored (Wang et al., 2020). Wang and others demonstrated the ability of CaM to interact with the NTD of NaV1.5 and discussed the potential implication of altered CaM binding in the presence of intrinsic channel disease mutations. We find that the binding of CaM to the NaV1.5-NTD depends dramatically on Ca2+-concentration (Supplementary Figures S3, S4). When the Ca2+-concentration approaches µM range, the CaM/NaV1.5-NTD affinity increases and is comparable to that of the IQ-domain, supporting a potential Ca2+-triggered role of the NaV1.5 NTD in CaM regulation of channel activity.
It is not surprising that the phenotypic range of calmodulinopathies may not yet be fully mapped, as CaM interacts with a myriad of cardiac target proteins, that may or may not be affected by specific CaM missense mutations. In addition to CaM mutation effects on NaV1.5, evidence of KV7.1 effects are accumulating, further expanding the phenotypic spectrum of calmodulinopathies. Kato and others described a family of 14 CaM-N138K carriers who displayed a variably expressed LQTS phenotype from asymptomatic carriers to carriers experiencing sudden death as children (Kato et al., 2022). In support of the LQTS phenotype, the CaM-N138K variant caused an impairment of CaV1.2 inactivation by whole-cell patch clamp recordings of HEK-cells. However, the variant also caused an unexpected potentiation of the KV7.1 current by the same technique in CHO cells, providing a possible explanation for the variably expressed LQTS phenotype, by countering the CaV1.2 effects (Kato et al., 2022). Another comprehensive study, involving 13 arrhythmogenic CaM variants, revealed differential effects of the CaM variants on KV7.1 binding affinity, channel trafficking, and channel gating (activation) (Kang et al., 2023). Interestingly, as the only one of the 13 variants, CaM-G114W diminished the interaction with the KV7.1 channel, both at resting and elevated Ca2+ concentrations, and induced an increase in KV7.1 trafficking to the cell membrane (Kang et al., 2023).
In conclusion, the data presented in this study warrants a potential expansion of the phenotypic spectrum of calmodulinopathies. Moreover, these results emphasize our incomplete understanding of the molecular mechanisms possible for calmodulinopathy-related diseases and point to the complexity in variant interpreting due to the mixed phenotypes caused by individual CaM mutations. Molecularly, the multifaceted effects of CaM mutations may act additively or synergistically, thereby contributing to compound and mixed phenotypic expressions. Also, given the high number of CaM-binding targets in cardiomyocytes, the likelihood of observing variably expressed phenotypes in CaM mutation carriers increases, compared to carriers of single ion-channel (or single pathway effecting protein) mutations with pure ‘classical’ phenotypes (Figure 4). This brings the calmodulinopathies to the forefront of scientific research into personalized medicine. Careful interrogation of the CALM genes in large cohorts of sequenced individuals with unexplained BrS-like or atypical heart arrhythmia phenotypes should be performed to confirm an expansion of the phenotypic spectrum of calmodulinopathies to include CALM-BrS. Such knowledge will allow for earlier and more accurate diagnosis and treatment of individuals with calmodulinopathies.
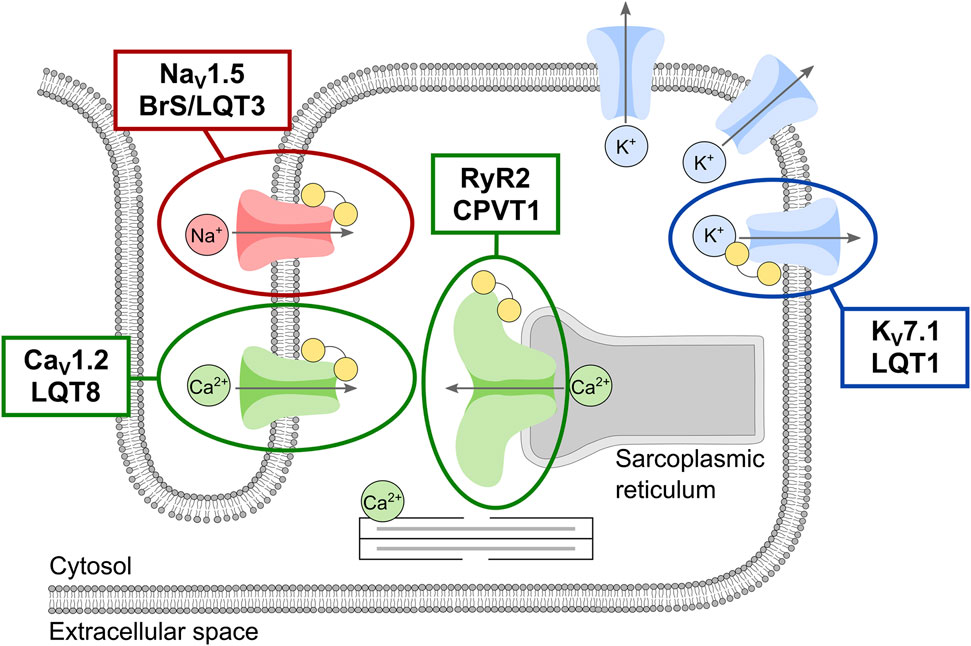
FIGURE 4. Schematic overview of the phenotypic spectrum of the main calmodulin (CaM)-regulated cardiac ion-channels responsible for the cardiac action potential and excitation-contraction coupling. Ion-channels are shown and colored according to the permeating ion: the ryanodine receptor (RyR2, green) and the voltage-gated calcium (CaV1.2, green), sodium (NaV1.5, red), and potassium (KV7.1, blue) channels. CaM is shown as yellow dumbbells. The involvement of multiple potassium channels in shaping the cardiac action potential is represented by multiple copies of this channel. Clinical phenotypes associated with intrinsic channel mutations are given for the CaM-regulated channels.
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author.
Written informed consent was obtained from the individuals, and minors’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
MN, RW, and MO contributed to conception and design of the study. MB and MO organized the laboratory work, which was carried out by A-OB. MB wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a research grant from the Lundbeck Foundation (no. R324-2019-1933), and a research grant from the Independent Danish Research Foundation, Council for Natural Sciences (no. 2032-00333B).
MN and MO acted as expert witnesses in the 2022 New South Wales Inquiry into the convictions of the Australian woman carrying the CaM-G114R variant.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1210140/full#supplementary-material
Adler, A., Guy, T., Heller, K., Zeltser, D., Ohayon, T., Rozovski, U., et al. (2013). Fever-induced Brugada pattern: how common is it and what does it mean? Heart rhythm. 10 (9), 1375–1382. doi:10.1016/j.hrthm.2013.07.030
Berchtold, M. W., and Villalobo., A. (2014). The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochimica Biophysica Acta 1843 (2), 398–435. doi:10.1016/j.bbamcr.2013.10.021
Berchtold, M. W., Egli, R., Rhyner, J. A., Hameister, H., and Strehler, E. E. (1993). Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3, 4611-P21.3, and 19q13.2-Q13.3. Genomics 16 (2), 461–465. doi:10.1006/geno.1993.1211
Boczek, N. J., Gomez-Hurtado, N., Ye, D., Calvert, M. L., Tester, D. J., Kryshtal, D., et al. (2016). Spectrum and prevalence of CALM1-CALM2-and CALM3-encoded calmodulin variants in long QT syndrome and functional characterization of a novel long QT syndrome-associated calmodulin missense variant, E141G. Circ. Cardiovasc. Genet. 9 (2), 136–146. doi:10.1161/CIRCGENETICS.115.001323
Brohus, M., Arsov, T., Wallace, D. A., Jensen, H. H., Nyegaard, M., Crotti, L., et al. (2021). Infanticide vs. Inherited cardiac arrhythmias. Europace 23 (3), 441–450. doi:10.1093/europace/euaa272
Brohus, M., Søndergaard, M. T., Michael, T., van Petegem, F., and Overgaard, M. T. (2019). Ca2+-Dependent calmodulin binding to cardiac ryanodine receptor (RyR2) calmodulin-binding domains. Biochem. J. 476 (2), 193–209. doi:10.1042/BCJ20180545
Bycroft, C., Freeman, C., Petkova, D., Gavin, B., Elliott, L. T., Sharp, K., et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562 (7726), 203–209. doi:10.1038/s41586-018-0579-z
Chagot, B., and Chazin, W. J. (2011). Solution NMR structure of apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 406 (1), 106–119. doi:10.1016/j.jmb.2010.11.046
Chen, S., Francioli, L. C., Goodrich, J. K., Collins, R. L., Wang, Q., Alföldi, J., et al. (2022). A genome-wide mutational constraint map quantified from variation in 76,156 human genomes. BioRxiv. doi:10.1101/2022.03.20.485034
Chin, D., and Anthony, R. (2000). Calmodulin: A prototypical calcium sensor. Trends Cell. Biol. 10 (8), 322–328. doi:10.1016/s0962-8924(00)01800-6
Crotti, L., Johnson, C. N., Graf, E., De Ferrari, G. M., Cuneo, B. F., Ovadia, M., et al. (2013). Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 127 (9), 1009–1017. doi:10.1161/CIRCULATIONAHA.112.001216
Crotti, L., Spazzolini, C., Tester, D. J., Ghidoni, A., Beckmann, B. M., Behr, E. R., et al. (2019). Calmodulin mutations and life-threatening cardiac arrhythmias: insights from the international calmodulinopathy registry. Eur. Heart J. 40 (35), 2964–2975. doi:10.1093/eurheartj/ehz311
Friedberg, F., and Rhoads, A. R. (2001). Evolutionary aspects of calmodulin. IUBMB Life 51 (4), 215–221. doi:10.1080/152165401753311753
Gabelli, S. B., Jesse, B., Tomaselli, G. F., and Amzel, L. M. (2016). Calmodulin and Ca(2+) control of voltage gated Na(+) channels. Channels (Austin, Tex.) 10 (1), 45–54. doi:10.1080/19336950.2015.1075677
Gabelli, S. B., Bianchet, M. A., Farinelli, F., Srinivas, A., Yoder, J., Jean, J., et al. (2014). Regulation of the NaV1.5 cytoplasmic domain by calmodulin. Nat. Commun. 5, 5126. doi:10.1038/ncomms6126
GTEx Consortium (2013). The genotype-tissue expression (GTEx) project. Nat. Genet. 45 (6), 580–585. doi:10.1038/ng.2653
Holt, C., Hamborg, L., Lau, K., Brohus, M., Larsen, K. T., Sommer, C., et al. (2020). The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor. J. Biol. Chem. 295 (22), 7620–7634. doi:10.1074/jbc.RA120.013430
Jensen, H. H., Brohus, M., Nyegaard, M., and Overgaard, M. T. (2018). Human calmodulin mutations. Front. Mol. Neurosci. 11, 396. doi:10.3389/fnmol.2018.00396
Jiménez-Jáimez, J., Ortega, Á., Macías-Ruiz, R., Francesca Perin, M., Perin, F., Rodríguez-Vázquez del Rey, M. M., et al. (2016). Calmodulin 2 mutation N98S is associated with unexplained cardiac arrest in infants due to low clinical penetrance electrical disorders. PloS One 11 (4), e0153851. doi:10.1371/journal.pone.0153851
Johnson, C. N., Potet, F., Thompson, B., Chazin, W. J., Glazer, A. M., Voehler, M. W., et al. (2018). A mechanism of calmodulin modulation of the human cardiac sodium channel. Struct. Lond. Engl. 26 (5), 683–694. doi:10.1016/j.str.2018.03.005
Kang, P. W., Chakouri, N., Diaz, J., Tomaselli, G. F., Yue, D. T., and Ben-Johny, M. (2021). Elementary mechanisms of calmodulin regulation of NaV1.5 producing divergent arrhythmogenic phenotypes. Proc. Natl. Acad. Sci. U. S. A. 118 (21), e2025085118. doi:10.1073/pnas.2025085118
Kang, P. W., Woodbury, L., Angsutararux, P., Sambare, N., Shi, J., Marras, M., et al. (2023). Arrhythmia-associated calmodulin variants interact with KCNQ1 to confer aberrant membrane trafficking and function. Prepr. Serv. Biol. 2023, 526031. doi:10.1101/2023.01.28.526031
Kato, K., Isbell, H. M., Fressart, V., Denjoy, I., Amal, D., Itoh, H., et al. (2022). Novel CALM3 variant causing calmodulinopathy with variable expressivity in a 4-generation family. Circulation. Arrhythmia Electrophysiol. 15 (3), e010572. doi:10.1161/CIRCEP.121.010572
Limpitikul, W. B., Dick, I. E., Yue, D. T., Overgaard, M. T., and George, A. L. (2014). Calmodulin mutations associated with long QT syndrome prevent inactivation of cardiac L-type Ca(2+) currents and promote proarrhythmic behavior in ventricular myocytes. J. Mol. Cell. Cardiol. 74, 115–124. doi:10.1016/j.yjmcc.2014.04.022
Makita, N., Yagihara, N., Crotti, L., Johnson, C. N., Beckmann, B-M., Roh, M. S., et al. (2014). Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ. Cardiovasc. Genet. 7 (4), 466–474. doi:10.1161/CIRCGENETICS.113.000459
Marsman, R. F., Barc, J., Beekman, L., Alders, M., Dooijes, D., Arthur van den, Wijngaard, et al. (2014). A mutation in CALM1 encoding calmodulin in familial idiopathic ventricular fibrillation in childhood and adolescence. J. Am. Coll. Cardiol. 63 (3), 259–266. doi:10.1016/j.jacc.2013.07.091
Michowitz, Y., Milman, A., Sarquella-Brugada, G., Andorin, A., Champagne, J., Postema, P. G., et al. (2018). Fever-related arrhythmic events in the multicenter survey on arrhythmic events in Brugada syndrome. Heart rhythm. 15 (9), 1394–1401. doi:10.1016/j.hrthm.2018.04.007
Nyegaard, M., Overgaard, M. T., Søndergaard, M. T., Vranas, M., Behr, E. R., Hildebrandt, L. L., et al. (2012). Mutations in calmodulin cause ventricular Tachycardia and sudden cardiac death. Am. J. Hum. Genet. 91 (4), 703–712. doi:10.1016/j.ajhg.2012.08.015
Nyegaard, M., and Overgaard, M. T. (2019). The international calmodulinopathy registry: recording the diverse phenotypic spectrum of un-CALM hearts. Eur. Heart J. 40 (35), 2976–2978. doi:10.1093/eurheartj/ehz463
Persechini, A., and Stemmer, P. M. (2002). Calmodulin is a limiting factor in the cell. Trends Cardiovasc. Med. 12 (1), 32–37. doi:10.1016/s1050-1738(01)00144-x
Postema, P. G., and Wilde, A. A. M. (2008). Arrhythmias in Brugada syndrome: changing throughout day and season? Heart rhythm. 5 (11), 1528–1529. doi:10.1016/J.HRTHM.2008.08.033
Potet, F., Chagot, B., Anghelescu, M., Viswanathan, P. C., Stepanovic, S. Z., Kupershmidt, S., et al. (2009). Functional interactions between distinct sodium channel cytoplasmic domains through the action of calmodulin. J. Biol. Chem. 284 (13), 8846–8854. doi:10.1074/jbc.M806871200
Rocchetti, M., Sala, L., Dreizehnter, L., Crotti, L., Sinnecker, D., Mura, M., et al. (2017). Elucidating arrhythmogenic mechanisms of long-QT syndrome CALM1-F142L mutation in patient-specific induced pluripotent stem cell-derived cardiomyocytes. Cardiovasc. Res. 113 (5), 531–541. doi:10.1093/cvr/cvx006
Rossi, A. M., and Taylor, C. W. (2011). Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 6 (3), 365–387. doi:10.1038/nprot.2011.305
Sarhan, M. F., and Ahern, C. A. (2009). A double tyrosine motif in the cardiac sodium channel domain III-IV linker couples calcium-dependent calmodulin binding to inactivation gating. J. Biol. Chem. 284 (48), 33265–33274. doi:10.1074/jbc.M109.052910
Sarhan, M. F., Tung, C., and Ahern, C. A. (2012). Crystallographic basis for calcium regulation of sodium channels. Proc. Natl. Acad. Sci. U. S. A. 109 (9), 3558–3563. doi:10.1073/pnas.1114748109
Schwartz, P. J., Priori, S. G., Spazzolini, C., Moss, A. J., Vincent, G. M., Napolitano, C., et al. (2001). Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation 103 (1), 89–95. doi:10.1161/01.cir.103.1.89
Shah, V. N., Tammy, L., Chazin, W. J., Williams, C. K., and Balser, J. R. (2006). Calcium-dependent regulation of the voltage-gated sodium channel HH1: intrinsic and extrinsic sensors use a common molecular switch. Proc. Natl. Acad. Sci. U. S. A. 103 (10), 3592–3597. doi:10.1073/pnas.0507397103
Søndergaard, M. T., Liu, Y., Brohus, M., Guo, W., Nani, A., Carvajal, C., et al. (2019). Diminished inhibition and facilitated activation of RyR2-mediated Ca2+ release is a common defect of arrhythmogenic calmodulin mutations. FEBS J. 286 (22), 4554–4578. doi:10.1111/febs.14969
Søndergaard, M. T., Liu, Y., Guo, W., Wei, J., Wang, R., Brohus, M., et al. (2020). Role of cardiac ryanodine receptor calmodulin-binding domains in mediating the action of arrhythmogenic calmodulin N-domain mutation N54I. FEBS J. 287 (11), 2256–2280. doi:10.1111/febs.15147
Søndergaard, M. T., Liu, Y., Larsen, K. T., Nani, A., Tian, X., Holt, C., et al. (2017). The arrhythmogenic calmodulin p.Phe142Leu mutation impairs C-domain Ca2+ binding but not calmodulin-dependent inhibition of the cardiac ryanodine receptor. J. Biol. Chem. 292 (4), 1385–1395. doi:10.1074/jbc.M116.766253
Søndergaard, M. T., Sorensen, A. B., Skov, L. L., Kjaer-Sorensen, K., Bauer, M. C., Nyegaard, M., et al. (2015a). Calmodulin mutations causing catecholaminergic polymorphic ventricular Tachycardia confer opposing functional and biophysical molecular changes. FEBS J. 282 (4), 803–816. doi:10.1111/febs.13184
Søndergaard, M. T., Tian, X., Liu, Y., Wang, R., Chazin, W. J., Chen, S. R. W., et al. (2015b). Arrhythmogenic calmodulin mutations affect the activation and termination of cardiac ryanodine receptor-mediated Ca2+ release. J. Biol. Chem. 290 (43), 26151–26162. doi:10.1074/jbc.M115.676627
Sorensen, A. B., Søndergaard, M. T., and Overgaard, M. T. (2013). Calmodulin in a heartbeat. FEBS J. 280 (21), 5511–5532. doi:10.1111/febs.12337
Takigawa, M., Noda, T., Shimizu, W., Miyamoto, K., Okamura, H., Satomi, K., et al. (2008). Seasonal and circadian distributions of ventricular fibrillation in patients with Brugada syndrome. Heart rhythm. 5 (11), 1523–1527. doi:10.1016/j.hrthm.2008.08.022
Tarasov, M., Struckman, H. L., Olgar, Y., Miller, A., Demirtas, M., Bogdanov, V., et al. (2023). NaV1.6 dysregulation within myocardial T-tubules by D96V calmodulin enhances proarrhythmic sodium and calcium mishandling. J. Clin. Investigation 133 (7), e152071. doi:10.1172/JCI152071
Tidow, H., and Nissen, P. (2013). Structural diversity of calmodulin binding to its target sites. FEBS J. 280 (21), 5551–5565. doi:10.1111/febs.12296
Toutenhoofd, S. L., and Strehler, E. E. (2000). The calmodulin multigene family as a unique case of genetic redundancy: multiple levels of regulation to provide spatial and temporal control of calmodulin pools? Cell. Calcium 28 (2), 83–96. doi:10.1054/ceca.2000.0136
Urrutia, J., Aguado, A., Muguruza-Montero, A., Núñez, E., Malo, C., Casis, O., et al. (2019). The crossroad of ion channels and calmodulin in disease. Int. J. Mol. Sci. 20 (2), 400. doi:10.3390/ijms20020400
Villarroel, A., Taglialatela, M., Bernardo-Seisdedos, G., Alaimo, A., Agirre, J., Alberdi, A., et al. (2014). The ever changing moods of calmodulin: how structural plasticity entails transductional adaptability. J. Mol. Biol. 426 (15), 2717–2735. doi:10.1016/j.jmb.2014.05.016
Wang, C., Chung, B. C., Yan, H., Wang, H. G., Lee, S. Y., and Pitt, G. S. (2014). Structural analyses of Ca2+/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation. Nat. Commun. 5 (1), 4896. doi:10.1038/ncomms5896
Wang, C., Chung, B. C., Yan, H., Lee, S. Y., and Pitt, G. S. (2012). Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Struct. Lond. Engl. 20 (7), 1167–1176. doi:10.1016/j.str.2012.05.001
Wang, K., Brohus, M., Holt, C., Overgaard, M. T., Wimmer, R., and Filip Van Petegem, (2020a). Arrhythmia mutations in calmodulin can disrupt cooperativity of Ca2+ binding and cause misfolding. J. Physiology 598 (6), 1169–1186. doi:10.1113/JP279307
Wang, K., Holt, C., Lu, J., Brohus, M., Larsen, K. T., Overgaard, M. T., et al. (2018). Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel. Proc. Natl. Acad. Sci. U. S. A. 115 (45), E10556–E10565. doi:10.1073/pnas.1808733115
Wang, Z., Vermij, S. H., Sottas, V., Anna, S., Ross-Kaschitza, D., Zaklyazminskaya, E. V., et al. (2020b). Calmodulin binds to the N-terminal domain of the cardiac sodium channel Nav1.5. Channels (Austin, Tex) 14 (1), 268–286. doi:10.1080/19336950.2020.1805999
Wu, X., and Donald, M. (2007). Free and bound intracellular calmodulin measurements in cardiac myocytes. Cell. Calcium 41 (4), 353–364. doi:10.1016/j.ceca.2006.07.011
Wu, X., and Liang, H. (2021). Calmodulin interactions with voltage-gated sodium channels. Int. J. Mol. Sci. 22 (18), 9798. doi:10.3390/ijms22189798
Xia, Z., and Storm, D. R. (2005). The role of calmodulin as a signal integrator for synaptic plasticity. Nat. Rev. Neurosci. 6 (4), 267–276. doi:10.1038/nrn1647
Yan, H., Wang, C., Marx, S. O., and Pitt, G. S. (2017). Calmodulin limits pathogenic Na+ channel persistent current. J. General Physiology 149 (2), 277–293. doi:10.1085/jgp.201611721
Yap, K. L., Kim, J., Truong, K., Sherman, M., Yuan, T., and Ikura, M. (2000). Calmodulin target database. J. Struct. Funct. Genomics 1 (1), 8–14. doi:10.1023/a:1011320027914
Yin, G., Hassan, F., Ayman, R. H., Murphy, L. L., Crotti, L., Peter, J., et al. (2014). Arrhythmogenic calmodulin mutations disrupt intracellular cardiomyocyte Ca2+ regulation by distinct mechanisms. J. Am. Heart Assoc. 3 (3), e000996. doi:10.1161/JAHA.114.000996
Keywords: calmodulin, calmodulinopathy, arrhythmogenic, cardiac ion-channel regulation, calmodulin target binding, experimental variant interpretation, SCN5A, NaV1.5
Citation: Brohus M, Busuioc A-O, Wimmer R, Nyegaard M and Overgaard MT (2023) Calmodulin mutations affecting Gly114 impair binding to the NaV1.5 IQ-domain. Front. Pharmacol. 14:1210140. doi: 10.3389/fphar.2023.1210140
Received: 24 April 2023; Accepted: 25 July 2023;
Published: 16 August 2023.
Edited by:
Mohamed Chahine, Laval University, CanadaReviewed by:
Jon Silva, Washington University in St. Louis, United StatesCopyright © 2023 Brohus, Busuioc, Wimmer, Nyegaard and Overgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Toft Overgaard, bXRvQGJpby5hYXUuZGs=
†ORCID: Malene Brohus, 0000-0001-9247-8551; Reinhard Wimmer, orcid.org/0000-0001-6942-5056; Mette Nyegaard, orcid.org/0000-0003-4973-8543; Michael Toft Overgaard, orcid.org/0000-0002-1423-2481
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.