- 1School of Integrated Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing, China
- 2Collaborative Innovation Center of Jiangsu Province of Cancer Prevention and Treatment of Chinese Medicine, Nanjing University of Chinese Medicine, Nanjing, China
- 3Research Center for Pathogenesis Theory of Cancerous Toxin and Application, Nanjing University of Chinese Medicine, Nanjing, China
- 4The First Clinical Medical College, Guizhou University of Traditional Chinese Medicine, Guiyang, China
Colorectal cancer (CRC) is the third most common malignancy in terms of global tumor incidence, and the rates of morbidity and mortality due to CRC are rising. Experimental models of CRC play a vital role in CRC research. Clinical studies aimed at investigating the evolution and mechanism underlying the formation of CRC are based on cellular and animal models with broad applications. The present review classifies the different experimental models used in CRC research, and describes the characteristics and limitations of these models by comparing the research models with the clinical symptoms. The review also discusses the future prospects of developing new experimental models of CRC.
1 Introduction
Colorectal cancer (CRC) is the most common malignancy worldwide, in terms of both morbidity and mortality (Sung et al., 2021). The understanding of the origin of CRC has increased dramatically over the past few decades. However, despite breakthroughs in diagnosis and treatment, CRC continues to be a major health concern worldwide. The morbidity and mortality due to CRC are on the rise owing to the overall low screening rates and changes in lifestyle, including poor diets, irregular lifestyles, smoking, and other factors (Minami et al., 2022). Strategies for the early screening and intervention of precancerous CRC lesions in developed countries have reduced the rates of incidence and mortality due to CRC (Zorzi and Urso, 2022). Similar to studies on other illnesses, research studies on CRC critically depend on experimental models with reliable and distinct characteristics. Although CRC tumors have heterogeneous characteristics, experimental models of CRC are established in such a manner that they represent the characteristics of CRC tumors. Selection of the appropriate model that reflects the tumor system is a crucial challenge in cancer screening. Therefore, experimental models of CRC have been extensively studied for determining the optimum model for studying the invasion, progression, and early detection of CRC. This review discusses the significance of CRC models as a platform for screening drugs and developing novel therapeutic approaches for CRC. The application of cellular and animal models of CRC were also summarized and discussed to aid further preclinical studies on CRC.
2 Cellular models based on intestinal cells and CRC cells
In vitro models of CRC established using intestinal cells and CRC cells are frequently employed for obtaining rapidly growing cellular models of CRC and for facilitating experimental control. In vitro models of CRC can simultaneously generate several populations of homogeneous cells. Specific cellular targets of macroscopic systems can be conveniently studied using these models by analyzing the experimental results (Saeidnia et al., 2015).
The first mammalian cell line was established in 1943, which served as a prelude to in vitro cell culture. The CoLo 205 CRC cell line was established in 1957, which promoted in vitro studies on CRC. Figure 1 depicts the history of development of in vitro models of CRC (Sanford et al., 1948; Ricci et al., 2007; Sharma et al., 2010; Jedrzejczak, 2017).
2.1 Two-dimensional (2D) cellular models of CRC
CRC cell lines are in vitro tumor models with different origins and types, and serve as fundamental tools for investigating the biomarkers of drug sensitivity, resistance, and toxicity. CRC cell lines are established by isolating CRC cells from patients or animals with CRC followed by culture on artificial media. The appropriate cell lines are selected based on the type of cancer or gene expression levels, according to the aims of the study. SW620, Caco-2, RKO, SW480, HT8, HT29, HT116, LoVo, and LS174 T cell lines are currently widely used in basic research studies on CRC (Akashi et al., 2000; Vécsey et al., 2002; Lind et al., 2004; Barretina et al., 2012; Ahmed et al., 2013; Gemei et al., 2013; Mouradov et al., 2014; Maletzki et al., 2015; Boot et al., 2016; Berg et al., 2017; Mooi et al., 2018; Kim et al., 2020; Bian et al., 2021).
Although the characteristics of CRC cell lines are highly consistent with those of human cancer models, they have certain limitations. CRC cell lines facilitate the investigation of the molecular and phenotypic characteristics of CRC. However, as only one side of the cells is in contact with the medium during culture, the majority of cells gradually flatten, undergo abnormal division, and lose their differentiation phenotype following isolation from tissues and plate culture. Additionally, CRC cells continue to proliferate in vitro, which may cause the cell lines to lose the characteristics of the original tumor. Another limitation of CRC cell lines is the scarcity of matrix ingredients in the tumor microenvironment (TME), including the cells and acellular components constituting the structural complexity of the in vivo environment. Altogether, these indicate that CRC cell lines fail to accurately mimic the in vivo growth characteristics of tumor cells.
2.2 Three-dimensional (3D) cellular models of CRC
Owing to the limitations of 2D cellular models of CRC, researchers are committed towards exploiting novel and physiologically representative models of CRC. In vitro 3D culture models, including spheroids and organoids, are therefore used for overcoming the limitations of 2D cellular models. Spheroids comprise a mixture of single-cell or multicellular systems, while organoids are generally formed of specific stem cells or ancestral cells from organs (Kimlin et al., 2013; Boucherit et al., 2020). Spheroids and organoids are superior at mimicking tumor cell heterogeneity and the complex interactions among different cells (Thoma et al., 2014).
2.2.1 Spheroids
Spheroids are one of the most commonly used models in CRC research. They are constructed by suspending cancer cell lines or isolated tumor tissues from patients in CRC. They have a convenient mode of production and application, and are particularly effective for studying micrometastases or avascular tumors. Spheroid models can be categorized into four types according to the origin and morphology of the cancer cells from which they are derived. These categories include multicellular tumor spheroids (MCTS), tumorospheres, tissue-derived tumor spheres (TDTS), and organotypic multicellular spheroids (OMS; Figure 2) (Weiswald et al., 2015).
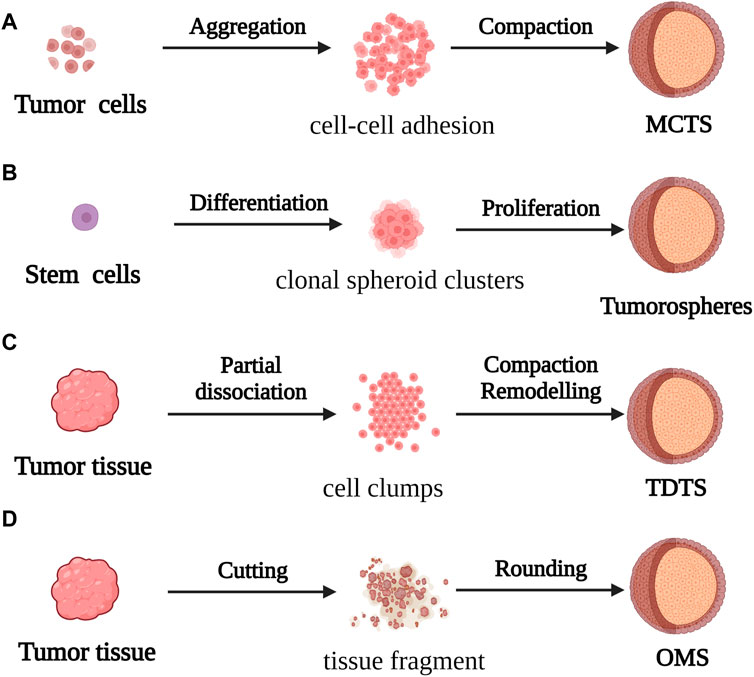
FIGURE 2. For the formation process of spherical cancer models (A) MCTS: Cell suspensions cultured under non-adherent conditions were aggregated and compacted to obtain MCTS; (B) Tumorospheres: Stem cells cultured under low-adherent conditions formed Tumorospheres by clonal proliferation (C) TDTS: Partial dissociation of tumor tissue and compaction/remodeling produced TDTS; (D) OMS: Cut tumor tissue aggregates formed OMS during culture under non-adherent conditions.
MCTS models, first constructed by Bauleth-Ramos, consist of colonic epithelia, human intestinal fiber cells, and human mononuclear cells, and are inoculated into hydrogel microwells to form the spheroid model (Inch et al., 1970; Bauleth-Ramos, T et al., 2020). MCTS models are similar to solid tumors in terms of the growth kinetics, metabolic rate, and resistance to chemotherapy and radiotherapy in vivo (Ivascu and Kubbies, 2006), and have been employed for screening and evaluating the efficacy of drugs. However, the variability of MCTS models makes it difficult to obtain repeatable and stable experimental data, which affects the use of these models in tumor research.
The tumorosphere model of CRC stem cells (CSCs) was used in the early 2000s for evaluating the differentiation capacity of tumors. However, because there are no morphological phenotypes associated with the phenotypic instability of CSCs, the tumorosphere model is unable to faithfully simulate the in vivo 3D framework and physiological condition of tumors (Valent et al., 2012).
The TDTS models consist of cancer and stromal cells, and are commonly used in studies on CRC. TDTS models of CRC tumors have a unique histological feature similar to the poorly differentiated globules produced by permanent cancer cell lines, and can fully simulate the characteristics of in vitro 3D cell culture models of CRC (Santini and Rainaldi, 1999; Weiswald et al., 2009).
OMS models are enriched in stem cells which can represent the complexity of parental tumor cells similar to in vivo tissues by forming an extracellular layer of epithelioid cells and an intracellular layer of mesenchymal cells, and thus maintaining the multicellular nature of CRC (Rajcevic et al., 2014). However, the difficulty of producing homogeneous spheres in a reproducible manner combined with the insufficiency of stable experimental data can prove to be a challenge during the application of the OMS model in CRC research and drug development.
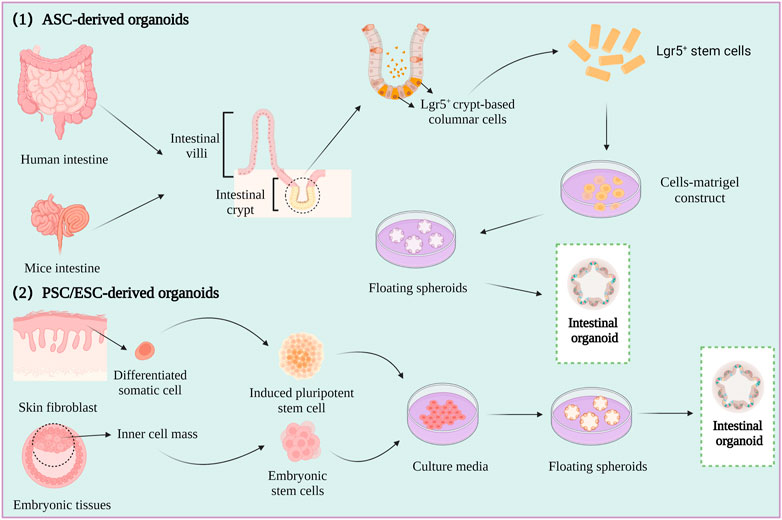
2.2.2 Organoids
Spheroids are a simple experimental model that only partly represent the in vivo characteristics of tumor tissues. However, organoids are relatively complex three-dimensional (3D) culture models that are frequently used in CRC research. Organoids are self-organizing organotypic cultures that are produced from various stem cells, including tissue specific adult stem cells (ASCs), embryonic stem cells (ESCs), or induced pluripotent stem cells (iPSCs) (Fujii et al., 2018; Fujii and Sato, 2021). The stem cells are grown in matrigel 3D culture conditions to mimic the in vivo growth environment, and to produce stable, near-physiological epithelial structures (Figure 3) (Lancaster and knoblich, 2014; Huch and Koo, 2015).
The first intestinal epithelial 3D organoids were constructed by growing leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5+) intestinal stem cells in a medium containing stem cell niche restatement factors and tissue-specific growth factors (Sato et al., 2011). An increasing number of studies have described the formation of patient-derived organoids (PDOs) by culturing minced human CRC tumors in human intestinal stem cell medium (HISC), and the phenotype and genotype of the PDOs have been reported to be highly similar to those of the original tumor (Van et al., 2015; Vlachogiannis et al., 2018).
Organoids are typically used for investigating the mechanism underlying the development of CRC, screening anti-CRC drugs, and determining the efficacy and mechanism of action of drugs. However, there are various limitations to the application of organoids in studies on CRC, which are described hereafter. First, the current methods for organoid culture lack the technological means for maintaining the blood vessels, immune system, and peripheral nervous system of tumor cells, and organoids lacking these characteristics cannot be used in CRC research (Bredenoord et al., 2017). Second, as PDO models lack the cellular and acellular components of the TME of the original tumor, they cannot equivalently represent the in vivo environment of the tumor (Li X. et al., 2020). Third, there are no specific media for culturing organoids to date. Furthermore, it is unclear whether organoids can represent the overall heterogeneity of the tumor and all cell types in the tumor. Organoids can be applied to relevant studies by optimizing the culture conditions for maintaining the expression of genes related to microsatellite instability, B-Raf proto-oncogene, serine/threonine kinase (BRAF) mutations, poor differentiation, or mucinous phenotypes related to CRC. The application of organoids to CRC research can be improved by employing the co-culture model of organoids in which immune cells and mesenchymal cells are co-cultured for simulating the in vivo TME.
2.3 Application of cellular models of CRC
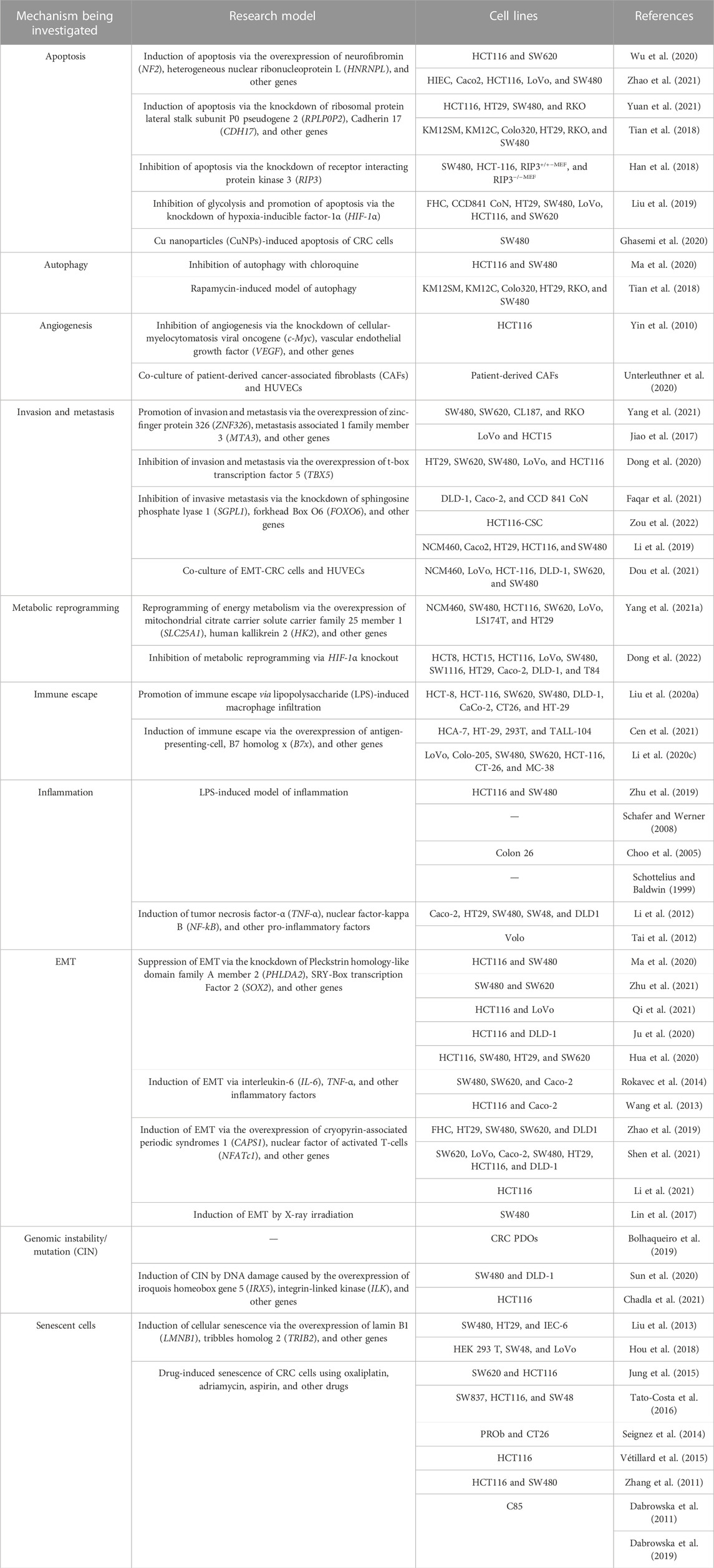
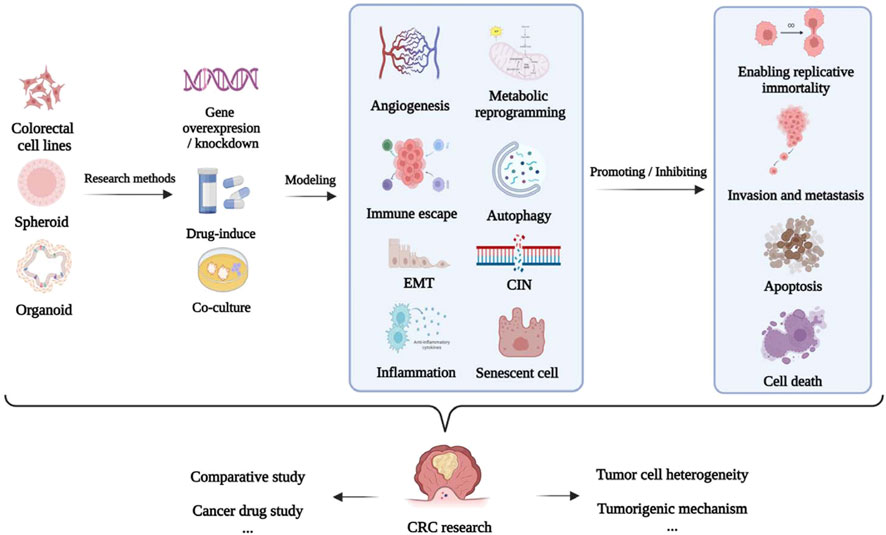
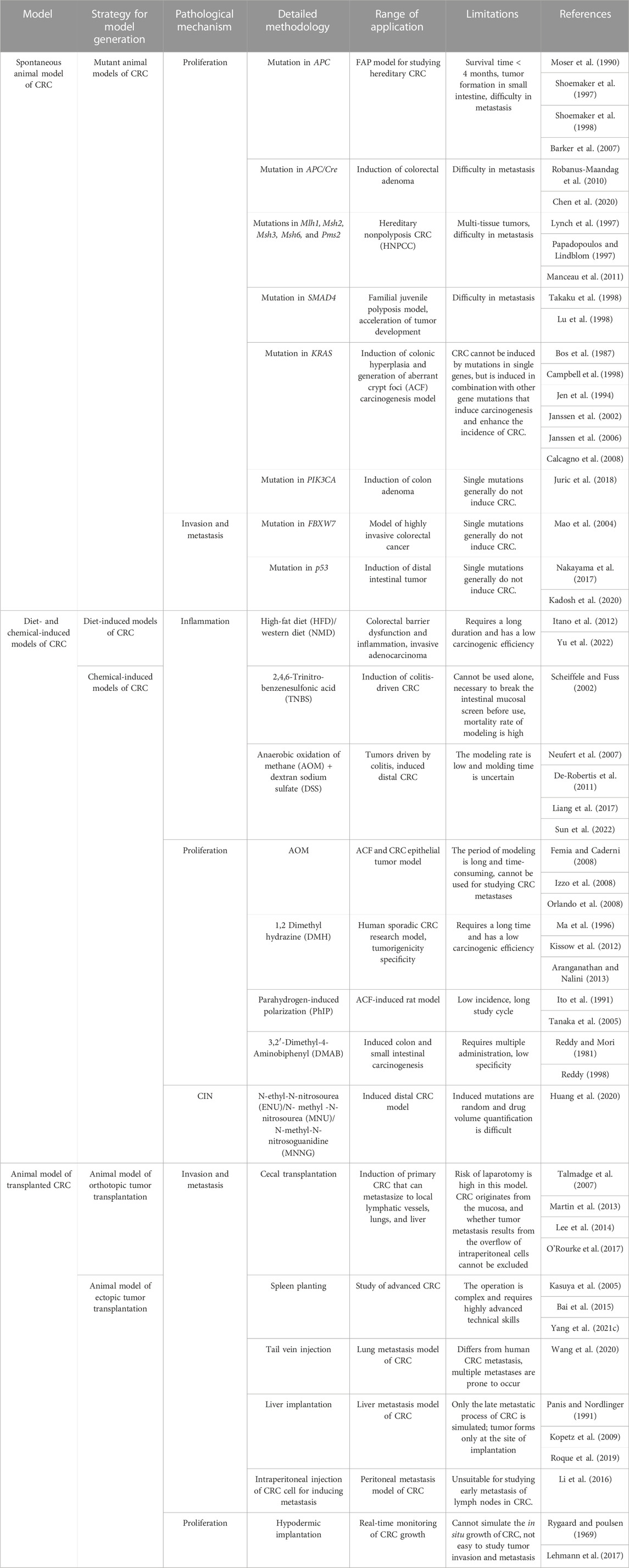
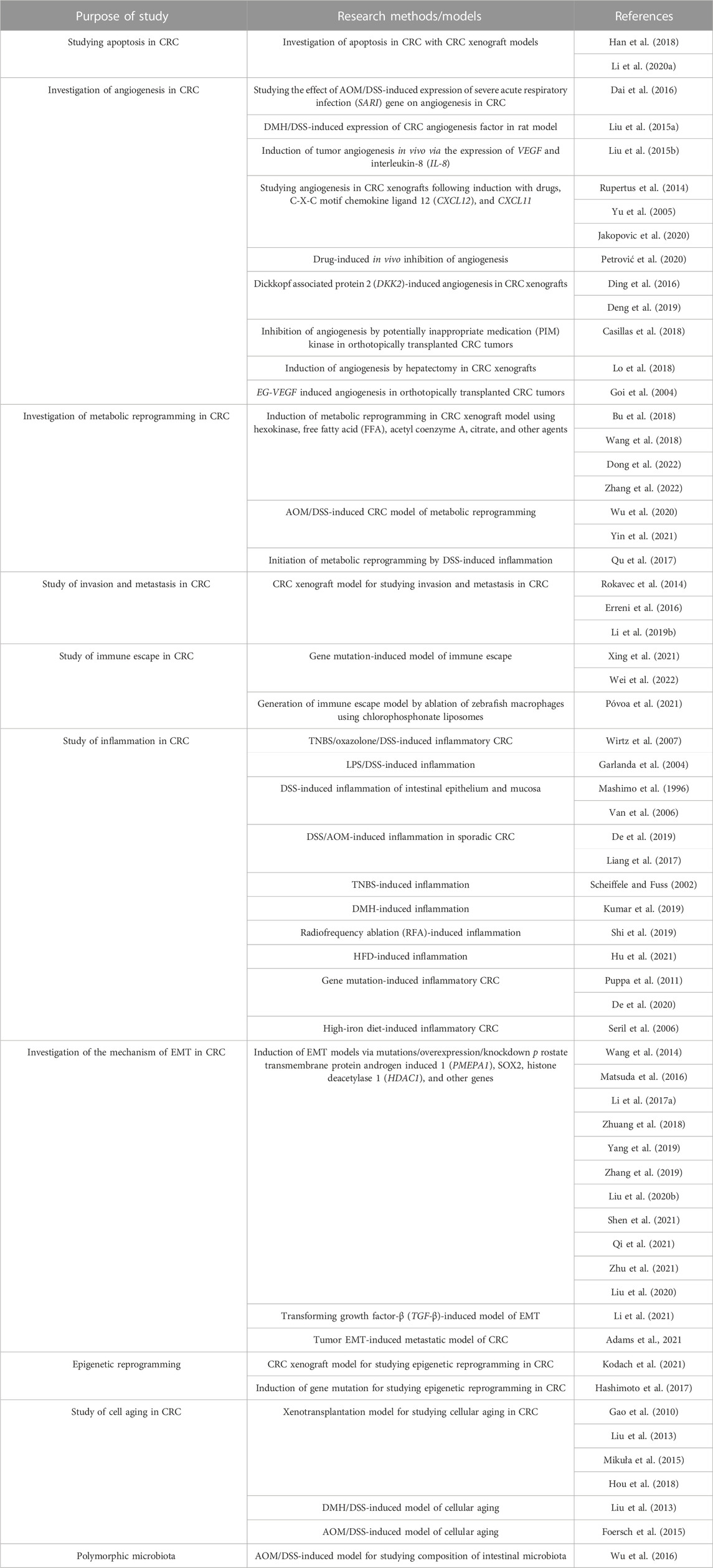
The establishment of models using the corresponding tumor cells is crucial for investigating the mechanism underlying the development of CRC and discovery of anti-CRC drugs (Senga and Grose, 2021). The applications of different cellular models of CRC according to the different molecular mechanisms underlying tumor formation, including epithelial–mesenchymal transition (EMT), apoptosis, invasion, metastasis, chromosome instability (CIN), and immune escape, are summarized in Table 1 and Figure 4.
3 CRC animal models based on experimental animals
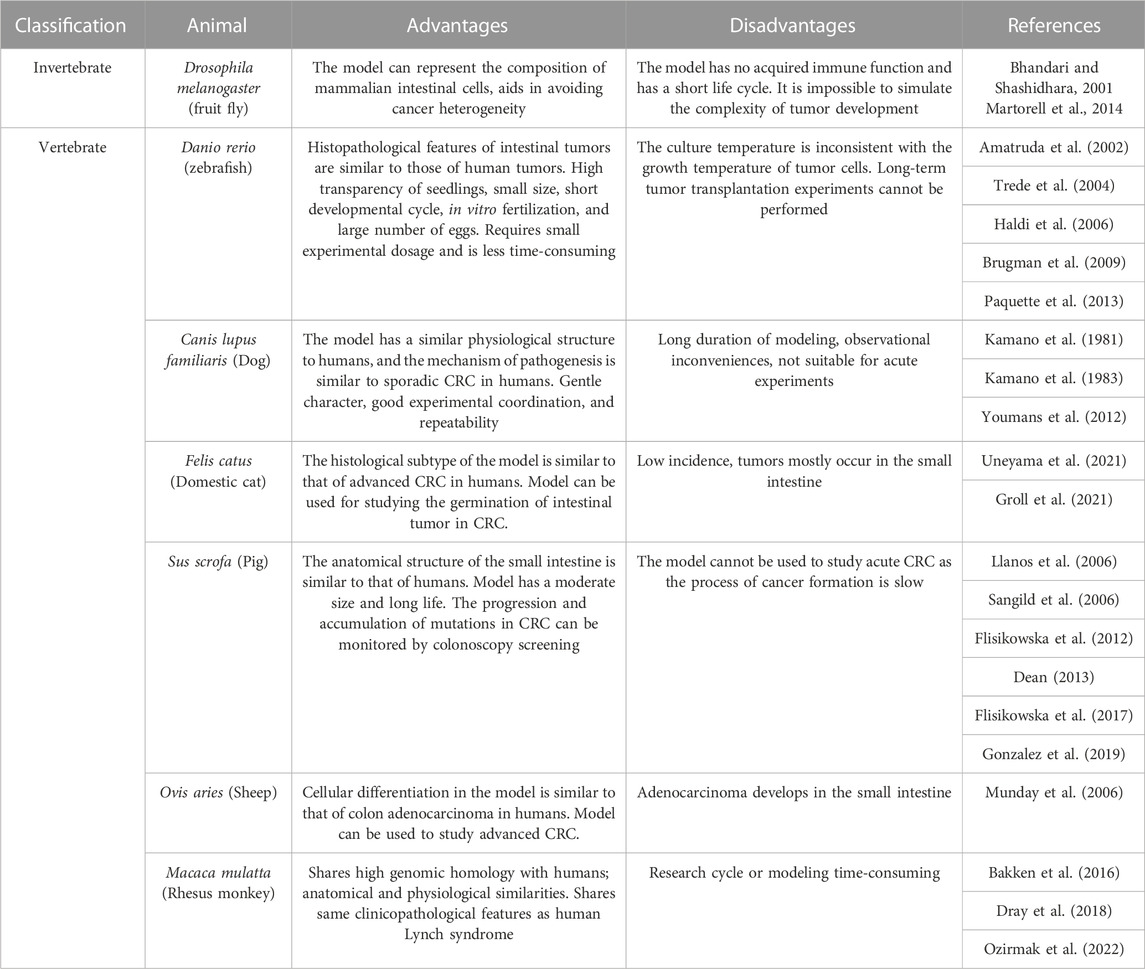
The occurrence of diseases such as cancer that occur spontaneously in animals is largely attributed to genetic diversity and immune functions. Therefore, studying the methods for generating animal models of CRC can aid in elucidating the mechanisms underlying the development of cancer (Marian, 2004). Animal models can compensate for the limitations of cellular models that are incapable of simulating the mechanism underlying the development of CRC. Rat and murine models are the most frequently used animal models of CRC, and other animal models of CRC, including fruit fly, zebrafish, and pigs, are also commonly used as sentinels and preclinical models in CRC research.
3.1 Rodent models
Rodent models are conducive tools for conducting cancer research, and are extensively used for elucidating the etiopathogenesis and molecular mechanisms underlying the development of CRC. Previous studies have demonstrated that the protein-coding genes of mice and humans share high homogeneity (Mouse Genome Sequencing Consortium, 2002). Additionally, the use of murine models is advantageous owing to the fact that mice have a short intergenerational interval, high reproducibility, and similar genetic background and formula as humans, compared to other animal models. Murine models of CRC can therefore be used as effective tools for studying the mechanism underlying the pathogenesis of CRC and determining novel strategies for the prevention and treatment of CRC (Doyle et al., 2012).
Transgenic mice models can serve as effective tools for preclinical evaluation and screening during the optimization and development of anticancer drugs. Mutations in APC (adenomatous polyposis coli) are commonly inherited in adenoma-carcinoma transitions observed during the development of CRC (Van et al., 2000). Additionally, the absence of mutations in DNA mismatch repair (MMR) genes increases deletion mutations in APC, which accelerates the formation of adenomas (Huang et al., 2004). It has been reported that mutations in tumor protein 53 (p53), Kirsten rats arcomaviral oncogene homolog (KRAS), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), F-box and WD repeat domain containing 7 (FBXW7), SMAD family member 4 (SMAD4), transcription factor 7-like 2 (TCF7L2), NRAS proto-oncogene (NRAS), AT-rich interaction domain 1 A (ARID1A), SRY-box transcription factor 9 (SOX9), and APC membrane recruitment protein 1 (FAM123B) can also increase the risk of CRC (Cancer Genome Atlas Network, 2012). Transgenic murine models are extensively used for studying the occurrence and elimination of tumors, underlying molecular pathways, and genomic regulation via gain-of-function or loss-of-function mutations in oncogenes and cancer suppressor genes.
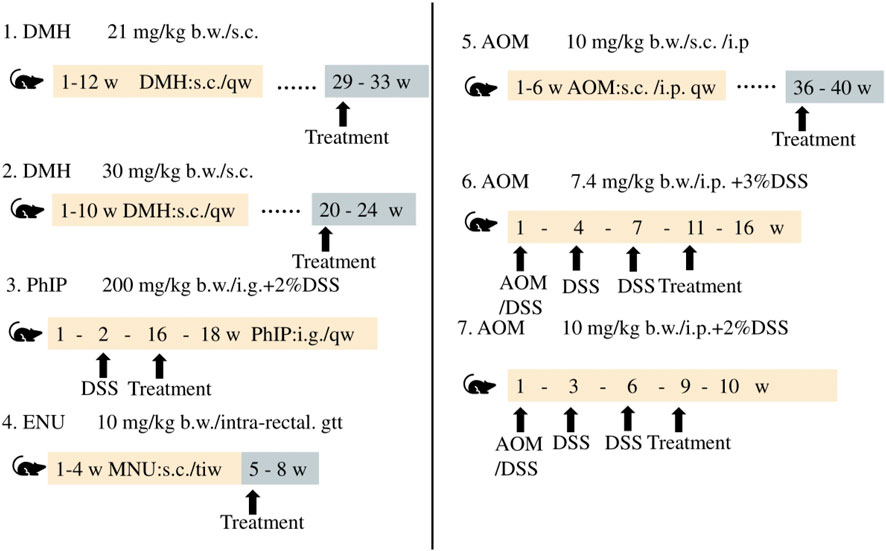
CRC is caused by various risk factors, including poor dietary habits, environment, exposure to carcinogenic chemicals, and other factors (Hecht, 2003; Mehta et al., 2017). Animal models of CRC generated by treatment with chemicals serve as effective models in studies aimed at determining novel therapeutic approaches and investigating the diagnosis, prognosis, and identification of predictive markers. The differences among the methods and duration of treatment for inducing CRC with different chemical agents are depicted in Figure 5.
The use of chemical agents for generating models of CRC requires a long duration and these models have longer experimental cycles. Mofikawa et al. established the first orthotopic transplantation model of CRC in 1986 by transplanting human CRC cells under the cecal wall of nude mice. This shortened the period of study using animal models of CRC, and initiated the establishment of tumor transplantation models. Table 2 summarizes the different murine models of CRC, and describes their scope of application and limitations in tumor research.
3.2 Other animal models of CRC
In addition to rodents, invertebrates such as fruit fly can be used for personalized diagnosis and developing potential therapeutic strategies for CRC. Vertebrates such as zebrafish, dogs, cats, pigs, and non-human primates are also used in studies on CRC. The advantages and disadvantages of the different animal models used in CRC research are summarized in Table 3.
3.3 Application of animal models of CRC
The carcinogenesis of CRC is affected by several contributing factors. The selection of the animal model of CRC depends on the purpose of the study, as summarized in Table 4.
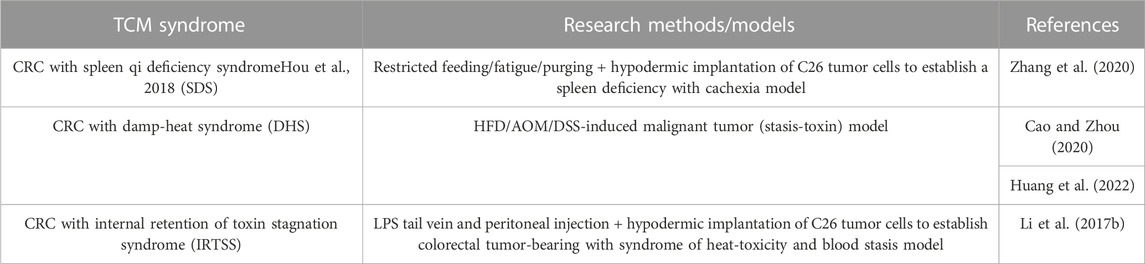
Traditional Chinese medicine (TCM) and western medicine are two different medical theoretical systems. The research model based on the etiological mechanism theory of TCM is applied to animal studies with TCM syndrome, as shown in Table 5.
4 Conclusions and future directions
Understanding the inherent advantages and limitations of the different models of CRC, and the appropriate application of these models in drug development and studies on the mechanism of tumor occurrence and development are important in CRC research.
Human cell lines and xenograft models have been extensively employed over the past few decades owing to their low cost and ease of application. However, these models are incapable of reproducing the heterogeneity of CRC tumors (Harma et al., 2010). The cell co-culture technique can overcome the limitations of monolayer cell culture, and enables the construction of in vitro physiological or pathological models that closely represent the in vivo condition, and can be used for studying the interactions between cells, and between cells and the culture environment. It has been reported that 3D models can mimic the physiological characteristics of parental tumors, including tumor heterogeneity (Li et al., 2019). However, the shape, size, and activity of organoids are different under the same culture conditions, and the matrix limits the penetration of drugs and hinders drug screening (Zhao et al., 2020). It is therefore imperative to construct a model that closely represents the characteristics of CRC in vivo.
The intestinal microarray platforms used in CRC research, which consist of intestinal organoids and organic chips, can summarize the important structural features and functions of the natural duodenum. This platform can be applied for studying drug conveyance, metabolism, and drug-drug interactions (Kasendra et al., 2018). Multi-locus transfer chips consist of multiple 3D organoids that connect the CRC-like organs, liver, lungs, and endothelial flow via recirculating fluid systems, and enables cell tracking by fluorescence imaging technology. The transfer sites of CRC cells are also included in multi-locus transfer chips (Aleman and Skardal, 2019).
Animal models of CRC have been widely used for studying the complexity of CRC. There are primarily two types of animal models, namely, in situ models and the cell and tissue transplantation models of CRC. Owing to the relatively simple modeling approach of human tumor xenotransplantation, this model is presently widely used for studying the efficacy of anti-CRC drugs. The effects of CRC xenotransplantation can be closely related to clinical activity via the rational application of these models. For instance, genetically engineered murine models have been used for studying the progression of tissue-specific molecular changes in CRC by determining the effect of specific molecular targets. Chemical induced-CRC animal model is one of the most commonly CRC models, in which CAC model is usually induced by AOM/DSS to study the mechanism of inflammation related-tumorigenesis and development (Zeng et al., 2022). The CRC model with TCM syndrome is an artificial disease and syndrome experimental animal model created by simulating and replicating characteristics of human disease prototype according to TCM theory. An animal model combining with CRC and TCM syndromes might be useful to mimic the clinical characteristics of CRC patients with TCM syndrome (Zhang et al., 2020). Mouse is the commonly used to the models mentioned above, however, it is increasingly accepted that the use of larger animal models, especially dogs and pigs, can provide deeper insights in cancer research (Croker et al., 2009).
The application of molecular tools and genetic strategies has aided the advancement of cancer research, and the cellular and animal models of CRC are being continually improved. Further understanding of the genetic and epigenetic events in CRC, including the alterations in molecular networks associated with the initial stages of development, are facilitated by high-resolution approaches.
Although CRC research has advanced immensely in recent years, several clinical issues remain to be resolved to date, which is partly attributed to the absence of suitable preclinical research models. The application of in vivo and in vitro models in CRC research, combined with advanced scientific techniques for simulating a more realistic tumor environment in vivo and in vitro, can help replicate the complex scenarios of tumor occurrence and development, identify novel therapeutic approaches for inhibiting tumor growth, and elucidate the molecular mechanisms underlying tumor formation.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (82074318, 81930117 and 82004310), Natural Science Foundation Youth Project of Jiangsu Province (BK 20200846), Natural Science Research of Jiangsu Higher Education Institutions of China (19KJA310007), Qinglan Project of Jiangsu Province, College Students’ Innovative Entrepreneurial Training Plan Program (202010315023Z and 202010315025), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Acknowledgments
The authors must be grateful to the BioRender (www.biorender.com), as the figures in this review were drawn by using the BioRender platform.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, D., Eide, P. W., Eilertsen, I. A., Danielsen, S. A., Eknæs, M., Hektoen, M., et al. (2013). Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2 (9), e71. doi:10.1038/oncsis.2013.35
Akashi, H., Han, H. J., Iizaka, M., and Nakamura, Y. (2000). Growth-suppressive effect of non-steroidal anti-inflammatory drugs on 11 colon-cancer cell lines and fluorescence differential display of genes whose expression is influenced by sulindac. Int. J. Cancer 88 (6), 873–880. doi:10.1002/1097-0215(20001215)88:6<873:aid-ijc6>3.0.co;2-b
Aleman, J., and Skardal, A. (2019). A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol. Bioeng. 116 (4), 936–944. doi:10.1002/bit.26871
Amatruda, J. F., Shepard, J. L., Stern, H. M., and Zon, L. I. (2002). Zebrafish as a cancer model system. Cancer Cell. 1 (3), 229–231. doi:10.1016/s1535-6108(02)00052-1
Aranganathan, S., and Nalini, N. (2013). Antiproliferative efficacy of hesperetin (citrus flavanoid) in 1,2-dimethylhydrazine-induced colon cancer. Phytotherapy Res. PTR 27 (7), 999–1005. doi:10.1002/ptr.4826
Bai, J. S., Wang, J., and Zhao, X. F. (2015). Nude mice hemispleen method in hepatic metastases of colon cancer model. 37, 447–450. doi:10.11724/jdmu.2015.05.08
Bakken, T. E., Miller, J. A., Ding, S. L., Sunkin, S. M., Smith, K. A., Ng, L., et al. (2016). A comprehensive transcriptional map of primate brain development. Nature 535 (7612), 367–375. doi:10.1038/nature18637
Barker, N., Van Es, J. H., Kuipers, J., Kujala, P., Van den Born, M., Cozijnsen, M., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449 (7165), 1003–1007. doi:10.1038/nature06196
Barretina, J., Caponigro, G., Stransky, N., Venkatesan, K., Margolin, A. A., Kim, S., et al. (2012). The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483 (7391), 603–607. doi:10.1038/nature11003
Berg, K. C. G., Eide, P. W., Eilertsen, I. A., Johannessen, B., Bruun, J., Danielsen, S. A., et al. (2017). Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol. Cancer 16 (1), 116. doi:10.1186/s12943-017-0691-y
Bhandari, P., and Shashidhara, L. S. (2001). Studies on human colon cancer gene APC by targeted expression in Drosophila. Oncogene 20 (47), 6871–6880. doi:10.1038/sj.onc.1204849
Bian, X., Cao, F., Wang, X., Hou, Y., Zhao, H., and Liu, Y. (2021). Establishment and characterization of a new human colon cancer cell line, PUMC-CRC1. Sci. Rep. 11 (1), 13122. doi:10.1038/s41598-021-92491-7
Bolhaqueiro, A. C. F., Ponsioen, B., Bakker, B., Klaasen, S. J., Kucukkose, E., Van Jaarsveld, R. H., et al. (2019). Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 51 (5), 824–834. doi:10.1038/s41588-019-0399-6
Boot, A., Van Eendenburg, J., Crobach, S., Ruano, D., Speetjens, F., Calame, J., et al. (2016). Characterization of novel low passage primary and metastatic colorectal cancer cell lines. Oncotarget 7 (12), 14499–14509. doi:10.18632/oncotarget.7391
Bos, J. L., Fearon, E. R., Hamilton, S. R., Verlaan-de Vries, M., van Boom, J. H., van der Eb, A. J., et al. (1987). Prevalence of Ras gene mutations in human colorectal cancers. Nature 327 (6120), 293–297. doi:10.1038/327293a0
Boucherit, N., Gorvel, L., and Olive, D. (2020). 3D tumor models and their use for the testing of immunotherapies. Front. Immunol. 11, 603640–640. doi:10.3389/fimmu.2020.603640
Bredenoord, A. L., Clevers, H., and Knoblich, J. A. (2017). Human tissues in a dish: The research and ethical implications of organoid technology. Science 355 (6322), eaaf9414. doi:10.1126/science.aaf9414
Brugman, S., Liu, K. Y., Lindenbergh-Kortleve, D., Samsom, J. N., Furuta, G. T., Renshaw, S. A., et al. (2009). Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology 137 (5), 1757–1767. doi:10.1053/j.gastro.2009.07.069
Bu, P., Chen, K. Y., Xiang, K., Johnson, C., Crown, S. B., Rakhilin, N., et al. (2018). Aldolase B mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell. Metab. 27 (6), 1249–1262. doi:10.1016/j.cmet.2018.04.003
Calcagno, S. R., Li, S., Colon, M., Kreinest, P. A., Thompson, E. A., Fields, A. P., et al. (2008). Oncogenic K-ras promotes early carcinogenesis in the mouse proximal colon. Int. J. Cancer 122, 2462–2470. doi:10.1002/ijc.23383
Campbell, S. L., Khosravi-Far, R., Rossman, K. L., Clark, G. J., and Der, C. J. (1998). Increasing complexity of Ras signaling. Oncogene 17 (11), 1395–1413. doi:10.1038/sj.onc.1202174
Cancer Genome Atlas Network (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 (7407), 330–337. doi:10.1038/nature11252
Cao, W., and Zhou, X. (2020). Establishment of intestinal cancer model in mice with damp-heat, phlegm-stagnation and stasis-toxin. J. Hunan Univ. Chin. Med. 40 (01), 38–41. doi:10.3969/j.issn.1674-070X.2020.01.009
Casali, A., and Batlle, E. (2009). Intestinal stem cells in mammals and drosophila. Cell. Stem Cell. 4 (2), 124–127. doi:10.1016/j.stem.2009.01.009
Casillas, A. L., Toth, R. K., Sainz, A. G., Singh, N., Desai, A. A., Kraft, A. S., et al. (2018). Hypoxia-inducible PIM kinase expression promotes resistance to antiangiogenic agents. Clin. Cancer Res. 24 (1), 169–180. doi:10.1158/1078-0432.CCR-17-1318
Cen, B., Wei, J., Wang, D., Xiong, Y., Shay, J. W., and DuBois, R. N. (2021). Mutant APC promotes tumor immune evasion via PD-L1 in colorectal cancer. Oncogene 40 (41), 5984–5992. doi:10.1038/s41388-021-01972-6
Chadla, P., Arbi, M., Nikou, S., Kalliakoudas, T., Papadaki, H., Taraviras, S., et al. (2021). Integrin-linked-kinase overexpression is implicated in mechanisms of genomic instability in human colorectal cancer. Dig. Dis. Sci. 66 (5), 1510–1523. doi:10.1007/s10620-020-06364-6
Chen, L., Vasoya, R. P., Toke, N. H., Parthasarathy, A., Luo, S., Chiles, E., et al. (2020). HNF4 regulates fatty acid oxidation and is required for renewal of intestinal stem cells in mice. Gastroenterology 158 (4), 985–999. doi:10.1053/j.gastro.2019.11.031
Choo, M. K., Sakurai, H., Koizumi, K., and Saiki, I. (2005). Stimulation of cultured colon 26 cells with TNF-alpha promotes lung metastasis through the extracellular signal-regulated kinase pathway. Cancer Lett. 230 (1), 47–56. doi:10.1016/j.canlet.2004.12.027
Croker, A. K., Goodale, D., Chu, J., Postenka, C., Hedley, B. D., Hess, D. A., et al. (2009). High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell. Mol. Med. 13 (8B), 2236–2252. doi:10.1111/j.1582-4934.2008.00455.x
Dabrowska, M., Skoneczny, M., and Rode, W. (2011). Functional gene expression profile underlying methotrexate-induced senescence in human colon cancer cells. Tumour Biol. 32 (5), 965–976. doi:10.1007/s13277-011-0198-x
Dabrowska, M., Skoneczny, M., Uram, L., and Rode, W. (2019). Methotrexate-induced senescence of human colon cancer cells depends on p53 acetylation, but not genomic aberrations. Anticancer Drugs 30 (4), 374–382. doi:10.1097/CAD.0000000000000731
Dai, L., Cui, X., Zhang, X., Cheng, L., Liu, Y., Yang, Y., et al. (2016). SARI inhibits angiogenesis and tumour growth of human colon cancer through directly targeting ceruloplasmin. Nat. Commun. 7, 11996. doi:10.1038/ncomms11996
De Oliveira, T., Ramakrishnan, M., Diamanti, M. A., Ziegler, P. K., Brombacher, F., and Greten, F. R. (2019). Loss of Stat6 affects chromatin condensation in intestinal epithelial cells causing diverse outcome in murine models of inflammation-associated and sporadic colon carcinogenesis. Oncogene 38 (11), 1787–1801. doi:10.1038/s41388-018-0551-2
De Robertis, M., Massi, E., Poeta, M. L., Carotti, S., Morini, S., Cecchetelli, L., et al. (2011). The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J. Carcinog. 10, 9. doi:10.4103/1477-3163.78279
De Santis, S., Verna, G., Serino, G., Armentano, R., Cavalcanti, E., Liso, M., et al. (2020). Winnie-APCMin/+ mice: A spontaneous model of colitis-associated colorectal cancer combining genetics and inflammation. Int. J. Mol. Sci. 21 (8), 2972. doi:10.3390/ijms21082972
Dean, P. G. (2013). Commentary on "The pig as a preclinical model for intestinal ischemia-reperfusion and transplantation studies. J. Surg. Res. 185 (2), 541–542. doi:10.1016/j.jss.2012.10.014
Deng, F., Zhou, R., Lin, C., Yang, S., Wang, H., Li, W., et al. (2019). Tumor-secreted dickkopf-2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics 9 (4), 1001–1014. doi:10.7150/thno.30056
Ding, C., Li, L., Yang, T., Fan, X., and Wu, G. (2016). Combined application of anti-VEGF and anti-EGFR attenuates the growth and angiogenesis of colorectal cancer mainly through suppressing AKT and ERK signaling in mice model. BMC Cancer 16 (1), 791. doi:10.1186/s12885-016-2834-8
Dolara, P., Luceri, C., De Filippo, C., Femia, A. P., Giovannelli, L., Caderni, G., et al. (2005). Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat. Res. 591 (1-2), 237–246. doi:10.1016/j.mrfmmm.2005.04.022
Dong, M. J., Zhou, Y., Duan, M., Gao, Q. M., and Zhao, J. H. (2020). Clinical significance and mechanism of TBX5 gene in colorectal cancer. Zhonghua Zhong Liu Za Zhi 42 (5), 383–390. doi:10.3760/cma.j.cn112152-112152-20190829-00560
Dong, S., Liang, S., Cheng, Z., Zhang, X., Luo, L., Li, L., et al. (2022). ROS/PI3K/AKT and Wnt/β-catenin signalings activate HIF-1α-induced metabolic reprogramming to impart 5-fluorouracil resistance in colorectal cancer. J. Exp. Clin. Cancer Res. 41 (1), 15. doi:10.1186/s13046-021-02229-6
Dou, R., Liu, K., Yang, C., Zheng, J., Shi, D., Lin, X., et al. (2021). EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clin. Transl. Med. 11 (12), 595. doi:10.1002/ctm2.595
Doyle, A., McGarry, M. P., Lee, N. A., and Lee, J. J. (2012). The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 21 (2), 327–349. doi:10.1007/s11248-011-9537-3
Dray, B. K., Raveendran, M., Harris, R. A., Benavides, F., Gray, S. B., Perez, C. J., et al. (2018). Mismatch repair gene mutations lead to lynch syndrome colorectal cancer in rhesus macaques. Genes. Cancer 9 (3-4), 142–152. doi:10.18632/genesandcancer.170
Erreni, M., Siddiqui, I., Marelli, G., Grizzi, F., Bianchi, P., Morone, D., et al. (2016). The fractalkine-receptor Axis improves human colorectal cancer prognosis by limiting tumor metastatic dissemination. J. Immunol. 196 (2), 902–914. doi:10.4049/jimmunol.1501335
Faqar-Uz-Zaman, W. F., Schmidt, K. G., Thomas, D., Pfeilschifter, J. M., Radeke, H. H., and Schwiebs, A. (2021). S1P lyase siRNA dampens malignancy of DLD-1 colorectal cancer Cells. Lipids 56 (2), 155–166. doi:10.1002/lipd.12282
Femia, A. P., and Caderni, G. (2008). Rodent models of colon carcinogenesis for the study of chemopreventive activity of natural products. Planta Med. 74 (13), 1602–1607. doi:10.1055/s-2008-1074577
Flisikowska, T., Merkl, C., Landmann, M., Eser, S., Rezaei, N., Cui, X., et al. (2012). A porcine model of familial adenomatous polyposis. Gastroenterology 143 (5), 1173–1175. doi:10.1053/j.gastro.2012.07.110
Flisikowska, T., Stachowiak, M., Xu, H., Wagner, A., Hernandez-Caceres, A., Wurmser, C., et al. (2017). Porcine familial adenomatous polyposis model enables systematic analysis of early events in adenoma progression. Sci. Rep. 7 (1), 6613. doi:10.1038/s41598-017-06741-8
Foersch, S., Sperka, T., Lindner, C., Taut, A., Rudolph, K. L., Breier, G., et al. (2015). VEGFR2 signaling prevents colorectal cancer cell senescence to promote tumorigenesis in mice with colitis. Gastroenterology 149 (1), 177–189. doi:10.1053/j.gastro.2015.03.016
Fujii, M., Matano, M., Toshimitsu, K., Takano, A., Mikami, Y., Nishikori, S., et al. (2018). Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell. Stem Cell. 23 (6), 787–793. doi:10.1016/j.stem.2018.11.016
Fujii, M., and Sato, T. (2021). Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mat. 20, 20156–20169. doi:10.1038/s41563-020-0754-0
Gao, F. H., Hu, X. H., Li, W., Liu, H., Zhang, Y. J., Guo, Z. Y., et al. (2010). Oridonin induces apoptosis and senescence in colorectal cancer cells by increasing histone hyperacetylation and regulation of p16, p21, p27 and c-Myc. BMC Cancer 10, 610. doi:10.1186/1471-2407-10-610
Garlanda, C., Riva, F., Polentarutti, N., Buracchi, C., Sironi, M., De Bortoli, M., et al. (2004). Intestinal inflammation in mice deficient in Tir8, an inhibitory member of the IL-1 receptor family. Proc. Natl. Acad. Sci. U. S. A. 101 (10), 3522–3526. doi:10.1073/pnas.0308680101
Ghasemi, P., Shafiee, G., Ziamajidi, N., and Abbasalipourkabir, R. (2020). Copper nanoparticles induce apoptosis and oxidative stress in SW480 human colon cancer cell line. Biol. Trace Elem. Res. doi:10.1007/s12011-022-03458-2
Goi, T., Fujioka, M., Satoh, Y., Tabata, S., Koneri, K., Nagano, H., et al. (2004). Angiogenesis and tumor proliferation/metastasis of human colorectal cancer cell line SW620 transfected with endocrine glands-derived-vascular endothelial growth factor, as a new angiogenic factor. Cancer Res. 64 (6), 1906–1910. doi:10.1158/0008-5472.can-3696-2
Gonzalez, L. M., Stewart, A. S., Freund, J., Kucera, C. R., Dekaney, C. M., Magness, S. T., et al. (2019). Preservation of reserve intestinal epithelial stem cells following severe ischemic injury. Am. J. Physiol. Gastrointest. Liver Physiol. 316 (4), G482-G494–G494. doi:10.1152/ajpgi.00262.2018
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y., and Wrana, J. L. (2015). Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526 (7575), 715–718. doi:10.1038/nature15382
Groll, T., Schopf, F., Denk, D., Mogler, C., Schwittlick, U., Aupperle-Lellbach, H., et al. (2021). Bridging the species gap: Morphological and molecular comparison of feline and human intestinal carcinomas. Cancers (Basel). 13 (23), 5941. doi:10.3390/cancers13235941
Haldi, M., Ton, C., Seng, W. L., and McGrath, P. (2006). Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis 9, 9139–9151. doi:10.1007/s10456-006-9040-2
Han, Q., Ma, Y., Wang, H., Dai, Y., Chen, C., Liu, Y., et al. (2018). Resibufogenin suppresses colorectal cancer growth and metastasis through RIP3-mediated necroptosis. J. Transl. Med. 16 (1), 201. doi:10.1186/s12967-018-1580-x
Harma, S., Haber, D., and Settleman, J. (2010). Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer 10 (4), 241–253. doi:10.1038/nrc2820
Hashimoto, K., Yamada, Y., Semi, K., Yagi, M., Tanaka, A., Itakura, F., et al. (2017). Cellular context-dependent consequences of APC mutations on gene regulation and cellular behavior. Proc. Natl. Acad. Sci. U. S. A. 114 (4), 758–763. doi:10.1073/pnas.1614197114
Hason, M., and Bartůněk, P. (2019). Zebrafish models of cancer-new insights on modeling human cancer in a non-mammalian vertebrate. Genes. (Basel). 10 (11), 935. doi:10.3390/genes10110935
Hecht, S. S. (2003). Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer 3 (10), 733–744. doi:10.1038/nrc1190
Hou, Z., Guo, K., Sun, X., Hu, F., Chen, Q., Luo, X., et al. (2018). TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol. Cancer 17 (1), 172. doi:10.1186/s12943-018-0922-x
Hu, X., Fatima, S., Chen, M., Xu, K., Huang, C., Gong, R. H., et al. (2021). Toll-like receptor 4 is a master regulator for colorectal cancer growth under high-fat diet by programming cancer metabolism. Cell. Death Dis. 12 (8), 791. doi:10.1038/s41419-021-04076-x
Hua, R., Yu, J., Yan, X., Ni, Q., Zhi, X., Li, X., et al. (2020). Syndecan-2 in colorectal cancer plays oncogenic role via epithelial-mesenchymal transition and MAPK pathway. Biomed. Pharmacother. 121, 109630. doi:10.1016/j.biopha.2019.109630
Huang, J., Jiang, T., Kang, J., Xu, J., Dengzhang, Y., Zhao, Z., et al. (2022). Synergistic effect of Huangqin decoction combined treatment with Radix Actinidiae chinensis on DSS and AOM-induced colorectal cancer. Front. Pharmacol. 13, 933070. doi:10.3389/fphar.2022.933070
Huang, J., Zheng, S., Jin, S. H., and Zhang, S. Z. (2004). Somatic mutations of APC gene in carcinomas from hereditary non-polyposis colorectal cancer patients. World J. Gastroenterol. 10 (6), 834–836. doi:10.3748/wjg.v10.i6.834
Huang, Z., Liu, C. A., Cai, P. Z., Xu, F. P., Zhu, W. J., Wang, W. W., et al. (2020). Omega-3PUFA Attenuates MNU-induced colorectal cancer in rats by blocking PI3K/AKT/Bcl-2 signaling. Onco Targets Ther. 13, 1953–1965. doi:10.2147/OTT.S241298
Huch, M., and Koo, B. K. (2015). Modeling mouse and human development using organoid cultures. Development 142 (18), 3113–3125. doi:10.1242/dev.118570
Inch, W. R., McCredie, J. A., and Sutherland, R. M. (1970). Growth of nodular carcinomas in rodents compared with multi-cell spheroids in tissue culture. Growth 34 (3), 271–282.
Itano, O., Fan, K., Yang, K., Suzuki, K., Quimby, F., Dong, Z., et al. (2012). Effect of caloric intake on Western-style diet-induced intestinal tumors in a mouse model for hereditary colon cancer. Nutr. Cancer 64 (3), 401–408. doi:10.1080/01635581.2012.660672
Ito, N., Hasegawa, R., Sano, M., Tamano, S., Esumi, H., Takayama, S., et al. (1991). A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (PhIP). Carcinogenesis 12 (8), 1503–1506. doi:10.1093/carcin/12.8.1503
Ivascu, A., and Kubbies, M. (2006). Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J. Biomol. Screen 11 (8), 922–932. doi:10.1177/1087057106292763
Izzo, A. A., Aviello, G., Petrosino, S., Orlando, P., Marsicano, G., Lutz, B., et al. (2008). Increased endocannabinoid levels reduce the development of precancerous lesions in the mouse colon. J. Mol. Med. Berl. 86 (1), 89–98. doi:10.1007/s00109-007-0248-4
Jakopovic, B., Oršolić, N., and Kraljević, P. S. (2020). Antitumor, immunomodulatory and antiangiogenic efficacy of medicinal mushroom extract mixtures in advanced colorectal cancer animal model. Molecules 25 (21), 5005. doi:10.3390/molecules25215005
Janssen, K. P., Alberici, P., Fsihi, H., Gaspar, C., Breukel, C., Franken, P., et al. (2006). APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology 131 (4), 1096–1109. doi:10.1053/j.gastro.2006.08.011
Janssen, K. P., El-Marjou, F., Pinto, D., Sastre, X., Rouillard, D., Fouquet, C., et al. (2002). Targeted expression of oncogenic KRAS in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology 123 (2), 492–504. doi:10.1053/gast.2002.34786
Jedrzejczak, S. M. (2017). “History of cell culture,” in In new insights into cell culture technology (Rijeka, Croatia: InTech). doi:10.5772/66905
Jen, J., Powell, S. M., Papadopoulos, N., Smith, K. J., Hamilton, S. R., Vogelstein, B., et al. (1994). Molecular determinants of dysplasia in colorectal lesions. Cancer Res. 54 (21), 5523–5526.
Jiao, T., Li, Y., Gao, T., Zhang, Y., Feng, M., Liu, M., et al. (2017). MTA3 regulates malignant progression of colorectal cancer through Wnt signaling pathway. Tumour Biol. 39 (3), 1010428317695027. doi:10.1177/1010428317695027
Ju, S., Wang, F., Wang, Y., and Ju, S. (2020). CSN8 is a key regulator in hypoxia-induced epithelial-mesenchymal transition and dormancy of colorectal cancer cells. Mol. Cancer 19 (1), 168. doi:10.1186/s12943-020-01285-4
Jung, Y. R., Kim, E. J., Choi, H. J., Park, J. J., Kim, H. S., Lee, Y. J., et al. (2015). Aspirin targets SIRT1 and AMPK to induce senescence of colorectal carcinoma cells. Mol. Pharmacol. 88 (4), 708–719. doi:10.1124/mol.115.098616
Juric, D., Rodon, J., Tabernero, J., Janku, F., Burris, H. A., Schellens, J. H. M., et al. (2018). Phosphatidylinositol 3-kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: Results from the first-in-human study. J. Clin. Oncol. 36 (13), 1291–1299. doi:10.1200/JCO.2017.72.7107
Kadosh, E., Snir-Alkalay, I., Venkatachalam, A., May, S., Lasry, A., Elyada, E., et al. (2020). The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 586 (7827), 133–138. doi:10.1038/s41586-020-2541-0
Kamano, T., Kishino, H., Mizukami, K., Azuma, N., Tamura, J., Katami, A., et al. (1983). Histopathological study on N-ethyl-N'-nitro-N-nitrosoguanidine-induced colon cancer in dogs. Int. J. Cancer 32 (2), 255–258. doi:10.1002/ijc.2910320219
Kamano, T., Kurihara, M., Kishino, H., Mizukami, K., Kidokoro, T., Wakabayashi, K., et al. (1981). Experimental colonic cancer in a dog. Jpn. J. Surg. 11 (3), 214–218. doi:10.1007/BF02468841
Kasendra, M., Tovaglieri, A., Sontheimer-Phelps, A., Jalili-Firoozinezhad, S., Bein, A., Chalkiadaki, A., et al. (2018). Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 8 (1), 2871. doi:10.1038/s41598-018-21201-7
Kasuya, H., Kuruppu, D. K., Donahue, J. M., Choi, E. W., Kawasaki, H., Tanabe, K. K., et al. (2020). Establishment and characterization of 18 human colorectal cancer cell lines. Sci. Rep. 10 (1), 6801. doi:10.1038/s41598-020-63812-z
Kasuya, H., Kuruppu, D. K., Donahue, J. M., Choi, E. W., Kawasaki, H., and Tanabe, K. K. (2005). Mouse models of subcutaneous spleen reservoir for multiple portal venous injections to treat liver malignancies. Cancer Res. 65 (9), 3823–3827. doi:10.1158/0008-5472.CAN-04-2631
Kim, S. C., Kim, H. S., Kim, J. H., Jeong, N., Shin, Y. K., Kim, M. J., et al. (2020). Establishment and characterization of 18 human colorectal cancer cell lines. Sci. Rep. 10 (1), 6801. doi:10.1038/s41598-020-63812-z
Kimlin, L. C., Casagrande, G., and Virador, V. M. (2013). In vitro three-dimensional (3D) models in cancer research: An update. Mol. Carcinog. 52 (3), 167–182. doi:10.1002/mc.21844
Kissow, H., Hartmann, B., Holst, J. J., Viby, N. E., Hansen, L. S., Rosenkilde, M. M., et al. (2012). Glucagon-like peptide-1 (GLP-1) receptor agonism or DPP-4 inhibition does not accelerate neoplasia in carcinogen treated mice. Regul. Pept. 179 (1-3), 91–100. doi:10.1016/j.regpep.2012.08.016
Kodach, L. L., Jacobs, R. J., Voorneveld, P. W., Wildenberg, M. E., Verspaget, H. W., Van Wezel, T., et al. (2011). Statins augment the chemosensitivity of colorectal cancer cells inducing epigenetic reprogramming and reducing colorectal cancer cell 'stemness' via the bone morphogenetic protein pathway. Gut 60 (11), 1544–1553. doi:10.1136/gut.2011.237495
Kopetz, S., Lesslie, D. P., Dallas, N. A., Park, S. I., Johnson, M., Parikh, N. U., et al. (2009). Synergistic activity of the SRC family kinase inhibitor dasatinib and oxaliplatin in colon carcinoma cells is mediated by oxidative stress. Cancer Res. 69 (9), 3842–3849. doi:10.1158/0008-5472.CAN-08-2246
Kumar, V. L., Verma, S., and Das, P. (2019). Artesunate suppresses inflammation and oxidative stress in a rat model of colorectal cancer. Drug Dev. Res. 80 (8), 1089–1097. doi:10.1002/ddr.21590
Lancaster, M. A., and Knoblich, J. A. (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345 (6194), 1247125. doi:10.1126/science.1247125
Lee, W. Y., Hong, H. K., Ham, S. K., Kim, C. I., and Cho, Y. B. (2014). Comparison of colorectal cancer in differentially established liver metastasis models. Anticancer Res. 34 (7), 3321–3328.
Lehmann, B., Biburger, M., Brückner, C., Ipsen-Escobedo, A., Gordan, S., Lehmann, C., et al. (2017). Tumor location determines tissue-specific recruitment of tumor-associated macrophages and antibody-dependent immunotherapy response. Sci. Immunol. 2 (7), 6413. doi:10.1126/sciimmunol.aah6413
Li, M., and Izpisua Belmonte, J. C. (2019a). Organoids-preclinical models of human disease. N. Engl. J. Med. 380 (6), 569–579. doi:10.1056/NEJMra1806175
Li, Q., Tang, H., Hu, F., and Qin, C. (2019b). Silencing of FOXO6 inhibits the proliferation, invasion, and glycolysis in colorectal cancer cells. J. Cell. Biochem. 120 (3), 3853–3860. doi:10.1002/jcb.27667
Li, Q., Zhang, S., Hu, M., Xu, M., and Jiang, X. (2020a). Silencing of synaptotagmin 13 inhibits tumor growth through suppressing proliferation and promoting apoptosis of colorectal cancer cells. Int. J. Mol. Med. 45 (1), 234–244. doi:10.3892/ijmm.2019.4412
Li, S., Shi, X., Chen, M., Xu, N., Sun, D., Bai, R., et al. (2019c). Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int. J. Cancer 145 (5), 1395–1407. doi:10.1002/ijc.32245
Li, S., Zhang, J., Qian, S., Wu, X., Sun, L., Ling, T., et al. (2021). S100A8 promotes epithelial-mesenchymal transition and metastasis under TGF-β/USF2 axis in colorectal cancer. Cancer Commun. 41 (2), 154–170. doi:10.1002/cac2.12130
Li, X., Larsson, P., Ljuslinder, I., Öhlund, D., Myte, R., Löfgren-Burström, A., et al. (2020b). Ex vivo organoid cultures reveal the importance of the tumor microenvironment for maintenance of colorectal cancer stem cells. Cancers (Basel) 12 (4), 923. doi:10.3390/cancers12040923
Li, Y., Deuring, J., Peppelenbosch, M. P., Kuipers, E. J., De Haar, C., and Van der Woude, C. J. (2012). IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinogenesis 33 (10), 1889–1896. doi:10.1093/carcin/bgs214
Li, Y., Liu, Y., Zhao, N., Yang, X., Li, Y., Zhai, F., et al. (2020c). Checkpoint regulator B7x is epigenetically regulated by HDAC3 and mediates resistance to HDAC inhibitors by reprogramming the tumor immune environment in colorectal cancer. Cell. Death Dis. 11 (9), 753. doi:10.1038/s41419-020-02968-y
Li, Y., Qian, L. Y., Tang, P. L., Guo, Y., Men, C., Cui, Y., et al. (2017b). Cathelicidin LL37 promotes epithelial and smooth-muscle-like differentiation of adipose-derived stem cells through the wnt/β-catenin and NF-κB pathways. Chin. J. Traditional Chin. Med. 82 (03), 1336–1345. doi:10.1134/S0006297917110116
Li, Y., Yang, Y., Li, J., Liu, H., Chen, F., Li, B., et al. (2017a). USP22 drives colorectal cancer invasion and metastasis via epithelial-mesenchymal transition by activating AP4. Oncotarget 8 (20), 32683–32695. doi:10.18632/oncotarget.15950
Li, Z., Wang, J., Zhou, T., and Ye, X. (2016). Establishment of a colorectal cancer nude mouse visualization model of HIF-1α overexpression. Oncol. Lett. 11 (4), 2725–2732. doi:10.3892/ol.2016.4287
Liang, X., Xie, R., Su, J., Ye, B., Wei, S., Liang, Z., et al. (2017). Inhibition of RNA polymerase III transcription by triptolide attenuates colorectal tumorigenesis. J. Exp. Clin. Cancer Res. 38 (1), 217. doi:10.1186/s13046-019-1232-x
Lin, S., Chen, S., Chen, Z., Dai, Q., and Ke, C. (2017). X-ray-induced epithelial-mesenchymal transition in SW480 colorectal cancer cells and its potential mechanisms. J. BUON 22 (6), 1457–1462.
Lind, G. E., Thorstensen, L., Løvig, T., Meling, G. I., Hamelin, R., Rognum, T. O., et al. (2004). A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol. Cancer 3, 28. doi:10.1186/1476-4598-3-28
Liu, C., Yao, Z., Wang, J., Zhang, W., Yang, Y., Zhang, Y., et al. (2020a). Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell. Death Differ. 27 (6), 1765–1781. doi:10.1038/s41418-019-0460-0
Liu, J., Deng, G. H., Zhang, J., Wang, Y., Xia, X. Y., Luo, X. M., et al. (2015a). The effect of chronic stress on anti-angiogenesis of sunitinib in colorectal cancer models. Psychoneuroendocrinology 52, 130–142. doi:10.1016/j.psyneuen.2014.11.008
Liu, L., Wang, J., Shi, L., Zhang, W., Du, X., Wang, Z., et al. (2013). β-Asarone induces senescence in colorectal cancer cells by inducing lamin B1 expression. Phytomedicine 20 (6), 512–520. doi:10.1016/j.phymed.2012.12.008
Liu, M., Xiao, Y., Tang, W., Li, J., Hong, L., Dai, W., et al. (2020b). HOXD9 promote epithelial-mesenchymal transition and metastasis in colorectal carcinoma. Cancer Med. 9 (11), 3932–3943. doi:10.1002/cam4.2967
Liu, W., Li, W., Liu, H., and Yu, X. (2019). Xanthohumol inhibits colorectal cancer cells via downregulation of Hexokinases II-mediated glycolysis. Int. J. Biol. Sci. 15 (11), 2497–2508. doi:10.7150/ijbs.37481
Liu, W. X., Gu, S. Z., Zhang, S., Ren, Y., Sang, L. X., and Dai, C. (2015b). Angiopoietin and vascular endothelial growth factor expression in colorectal disease models. World J. Gastroenterol. 21 (9), 2645–2650. doi:10.3748/wjgv21i9.2645
Llanos, J. C., Bakonyi Neto, A., Lerco, M. M., Clark, R. M., Polachini do Valle, A., and Sousa, M. M. (2006). Induction of short gut syndrome and transplantation in a porcine model. Transpl. Proc. 38 (6), 1855–1856. doi:10.1016/j.transproceed.2006.06.085
Lo Dico, R., Tijeras-Raballand, A., Bonnin, P., Launay, J. M., Kaci, R., Pimpie, C., et al. (2018). Hepatectomy increases metastatic graft and growth in an immunocompetent murine model of peritoneal metastases. Eur. J. Surg. Oncol. 44 (6), 784–791. doi:10.1016/j.ejso.2018.01.096
Lu, S. L., Kawabata, M., Imamura, T., Akiyama, Y., Nomizu, T., Miyazono, K., et al. (1998). HNPCC associated with germline mutation in the TGF-beta type II receptor gene. Nat. Genet. 19 (1), 17–18. doi:10.1038/ng0598-17
Lynch, H. T., Smyrk, T., and Lynch, J. (1997). An update of HNPCC (Lynch syndrome). Cancer Genet. Cytogenet 93 (1), 84–99. doi:10.1016/s0165-4608(96)00290-7
Ma, Q., Hoper, M., Anderson, N., and Rowlands, B. J. (1996). Effect of supplemental L-arginine in a chemical-induced model of colorectal cancer. World J. Surg. 20 (8), 1087–1091. doi:10.1007/s002689900165
Ma, Z., Lou, S., and Jiang, Z. (2020). PHLDA2 regulates EMT and autophagy in colorectal cancer via the PI3K/AKT signaling pathway. Aging (Albany NY) 12 (9), 7985–8000. doi:10.18632/aging.103117
Maletzki, C., Gock, M., Randow, M., Klar, E., Huehns, M., Prall, F., et al. (2015). Establishment and characterization of cell lines from chromosomal instable colorectal cancer. World J. Gastroenterol. 21 (1), 164–176. doi:10.3748/wjgv21.i1.164
Manceau, G., Karoui, M., Charachon, A., Delchier, J. C., and Sobhani, I. (2011). HNPCC (hereditary non-polyposis colorectal cancer) or lynch syndrome: A syndrome related to a failure of DNA repair system. Bull. Cancer 98 (3), 323–336. doi:10.1684/bdc.2011.1328
Mao, J. H., Perez-Losada, J., Wu, D., Delrosario, R., Tsunematsu, R., Nakayama, K. I., et al. (2004). Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432, 775–779. doi:10.1038/nature03155
Marian, B. (2004). Colorectal cancer: Modeling causes, prevention and therapy. Drug Discov. Today Dis. Models 1 (1), 11–17. doi:10.1016/jddmod.2004.07.006
Martin, E. S., Belmont, P. J., Sinnamon, M. J., Richard, L. G., Yuan, J., Coffee, E. M., et al. (2013). Development of a colon cancer GEMM-derived orthotopic transplant model for drug discovery and validation. Clin. Cancer Res. 19 (11), 2929–2940. doi:10.1158/1078-0432.CCR-12-2307
Martorell, Ò., Merlos-Suárez, A., Campbell, K., Barriga, F. M., Christov, C. P., Miguel-Aliaga, I., et al. (2014). Conserved mechanisms of tumorigenesis in the Drosophila adult midgut. PLoS ONE 9, 88413. doi:10.1371/journal.pone.0088413
Mashimo, H., Wu, D. C., Podolsky, D. K., and Fishman, M. C. (1996). Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science 274 (5285), 262–265. doi:10.1126/science.274.5285.262
Matsuda, Y., Miura, K., Yamane, J., Shima, H., Fujibuchi, W., Ishida, K., et al. (2016). SERPINI1 regulates epithelial-mesenchymal transition in an orthotopic implantation model of colorectal cancer. Cancer Sci. 07 (5), 619–628. doi:10.1111/cas.12909
Mehta, G., Hsiao, A. Y., Ingram, M., Luker, G. D., and Takayama, S. (2012). Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control Release 164 (2), 192–204. doi:10.1016/j.jconrel.2012.04.045
Mehta, R. S., Song, M., Nishihara, R., Drew, D. A., Wu, K., Qian, Z. R., et al. (2017). Dietary patterns and risk of colorectal cancer: Analysis by tumor location and molecular subtypes. Gastroenterology 152 (8), 1944–1953.e1. doi:10.1053/j.gastro.2017.02.015
Mikuła-Pietrasik, J., Sosińska, P., Maksin, K., Kucińska, M. G., Piotrowska, H., Murias, M., et al. (2015). Colorectal cancer-promoting activity of the senescent peritoneal mesothelium. Oncotarget 6 (30), 29178–29195. doi:10.18632/oncotarget.4932
Minami, Y., Kanemura, S., Kusaka, J., Kinouchi, M., Suzuki, S., Nishino, Y., et al. (2022). Associations of cigarette smoking, alcohol drinking and body mass index with survival after colorectal cancer diagnosis by anatomic subsite: A prospective patient cohort study in Japan. Jpn. J. Clin. Oncol. 52 (12), 1375–1388. doi:10.1093/jjco/hyac140
Mooi, J. K., Luk, I. Y., and Mariadason, J. M. (2018). Cell line models of molecular subtypes of colorectal cancer. Methods Mol. Biol. 1765, 3–26. doi:10.1007/978-1-4939-7765-9_1
Moser, A. R., Pitot, H. C., and Dove, W. F. (1990). A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247 (4940), 322–324. doi:10.1126/science.2296722
Mouradov, D., Sloggett, C., Jorissen, R. N., Love, C. G., Li, S., Burgess, A. W., et al. (2014). Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 74 (12), 3238–3247. doi:10.1158/0008-5472.CAN-14-0013
Mouse Genome Sequencing Consortium Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420 (6915), 520–562. doi:10.1038/nature01262
Munday, J. S., Brennan, M. M., Jaber, A. M., and Kiupel, M. (2006). Ovine intestinal adenocarcinomas: Histologic and phenotypic comparison with human colon cancer. Comp. Med. 56 (2), 136–141.
Nakayama, M., Sakai, E., Echizen, K., Yamada, Y., Oshima, H., Han, T. S., et al. (2017). Intestinal cancer progression by mutant p53 through the acquisition of invasiveness associated with complex glandular formation. Oncogene 36 (42), 5885–5896. doi:10.1038/onc.2017.194
Neufert, C., Becker, C., and Neurath, M. F. (2007). An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2, 1998–2004. doi:10.1038/nprot.2007.279
O'Rourke, K. P., Loizou, E., Livshits, G., Schatoff, E. M., Baslan, T., Manchado, E., et al. (2017). Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 35 (6), 577–582. doi:10.1038/nbt.3837
Orlando, F. A., Tan, D., Baltodano, J. D., Khoury, T., Gibbs, J. F., Hassid, V. J., et al. (2008). Aberrant crypt foci as precursors in colorectal cancer progression. J. Surg. Oncol. 98 (3), 207–213. doi:10.1002/jso.21106
Ozirmak Lermi, N., Gray, S. B., Bowen, C. M., Reyes-Uribe, L., Dray, B. K., Deng, N., et al. (2022). Comparative molecular genomic analyses of a spontaneous rhesus macaque model of mismatch repair-deficient colorectal cancer. PLoS Genet. 18 (4), 1010163. doi:10.1371/journal.pgen.1010163
Panis, Y., and Nordlinger, B. (1991). Experimental models for hepatic metastases from colorectal tumors. Ann. Chir. 45 (3), 222–228.
Papadopoulos, N., and Lindblom, A. (1997). Molecular basis of HNPCC: Mutations of MMR genes. Hum. Mutat. 10 (2), 89–99. doi:10.1002/(SICI)1098-1004(1997)10:2<89:AID-HUMU1>3.0.CO;2-H
Paquette, C. E., Kent, M. L., Buchner, C., Tanguay, R. L., Guillemin, K., Mason, T. J., et al. (2013). A retrospective study of the prevalence and classification of intestinal neoplasia in zebrafish (Danio rerio). Zebrafish 10 (2), 228–236. doi:10.1089/zeb.2012.0828
Petrović, J., Glamočlija, J., Ilić-Tomić, T., Soković, M., Robajac, D., Nedić, O., et al. (2020). Lectin from Laetiporus sulphureus effectively inhibits angiogenesis and tumor development in the zebrafish xenograft models of colorectal carcinoma and melanoma. Int. J. Biol. Macromol. 148, 129–139. doi:10.1016/j.ijbiomac.2020.01.033
Póvoa, V., Rebelo de Almeida, C., Maia-Gil, M., Sobral, D., Domingues, M., Martinez-Lopez, M., et al. (2021). Innate immune evasion revealed in a colorectal zebrafish xenograft model. Nat. Commun. 12 (1), 1156. doi:10.1038/s41467-021-21421-y
Puppa, M. J., White, J. P., Sato, S., Cairns, M., Baynes, J. W., and Carson, J. A. (2011). Gut barrier dysfunction in the APC(Min/+) mouse model of colon cancer cachexia. Biochim. Biophys. Acta 1812 (12), 1601–1606. doi:10.1016/j.bbadis.2011.08.010
Qi, Z. P., Yalikong, A., Zhang, J. W., Cai, S. L., Li, B., Di, S., et al. (2021). HDAC2 promotes the EMT of colorectal cancer cells and via the modular scaffold function of ENSG00000274093.1. J. Cell. Mol. Med. 25 (2), 1190–1197. doi:10.1111/jcmm.16186
Qu, D., Shen, L., Liu, S., Li, H., Ma, Y., Zhang, R., et al. (2017). Chronic inflammation confers to the metabolic reprogramming associated with tumorigenesis of colorectal cancer. Cancer Biol. Ther. 18 (4), 237–244. doi:10.1080/15384047.2017.1294292
Rajcevic, U., Knol, J. C., Piersma, S., Bougnaud, S., Fack, F., Sundlisaeter, E., et al. (2014). Colorectal cancer derived organotypic spheroids maintain essential tissue characteristics but adapt their metabolism in culture. Proteome Sci. 12, 39. doi:10.1186/1477-5956-12-39
Reddy, B. S. (1998). Colon carcinogenesis models for chemoprevention studies. Hematol. Oncol. Clin. North Am. 12 (5), 963–973. doi:10.1016/s0889-8588(05)70036-8
Reddy, B. S., and Mori, H. (1981). Effect of dietary wheat bran and dehydrated citrus fiber on 3,2'-dimethyl-4-aminobiphenyl-induced intestinal carcinogenesis in F344 rats. Carcinogenesis 2 (1), 21–25. doi:10.1093/carcin/2.1.21
Ricci-Vitiani, L., Lombardi, D. G., Pilozzi, E., Biffoni, M., Todaro, M., Peschle, C., et al. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445 (7123), 111–115. doi:10.1038/nature05384
Robanus-Maandag, E. C., Koelink, P. J., Breukel, C., Salvatori, D. C., Jagmohan-Changur, S. C., Bosch, C. A., et al. (2010). A new conditional APC-mutant mouse model for colorectal cancer. Carcinogenesis 31 (5), 946–952. doi:10.1093/carcin/bgq046
Rokavec, M., Öner, M. G., Li, H., Jackstadt, R., Jiang, L., Lodygin, D., et al. (2014). IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 124 (4), 1853–1867. doi:10.1172/JCI73531
Roque-Lima, B., Roque, C. C. T. A., Begnami, M. D., Peresi, P., Lima, E. N. P., Mello, C. A. L., et al. (2019). Development of patient-derived orthotopic xenografts from metastatic colorectal cancer in nude mice. J. Drug Target 27 (9), 943–949. doi:10.1080/1061186X.2018.1509983
Rupertus, K., Sinistra, J., Scheuer, C., Nickels, R. M., Schilling, M. K., Menger, M. D., et al. (2014). Interaction of the chemokines I-TAC (CXCL11) and SDF-1 (CXCL12) in the regulation of tumor angiogenesis of colorectal cancer. Clin. Exp. Metastasis 31 (4), 447–459. doi:10.1007/s10585-014-9639-4
Rygaard, J., and Poulsen, C. O. (1969). Heterotransplantation of a human malignant tumour to “nude”mice. Acta Pathol. Microbiol. Scand. 77, 758–760. doi:10.1111/j.1699-0463.1969.tb04520.x
Saeidnia, S., Manayi, A., and Abdollahi, M. (2015). From in vitro experiments to in vivo and clinical studies; pros and cons. Curr. Drug Discov. Technol. 12 (4), 218–224. doi:10.2174/1570163813666160114093140
Sanford, K. K., Earle, W. R., and Likely, G. D. (1948). The growth in vitro of single isolated tissue cells. J. Natl. Cancer Inst. 9 (3), 229–246.
Sangild, P. T., Siggers, R. H., Schmidt, M., Elnif, J., Bjornvad, C. R., Thymann, T., et al. (2006). Diet and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology 130 (6), 1776–1792. doi:10.1053/j.gastro.2006.02.026
Santini, M. T., and Rainaldi, G. (1999). Three-dimensional spheroid model in tumor biology. Pathobiology 67 (3), 148–157. doi:10.1159/000028065
Sato, T., Stange, D. E., Ferrante, M., Vries, R. G., Van Es, J. H., Van den Brink, S., et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141 (5), 1762–1772. doi:10.1053/j.gastro.2011.07.050
Schafer, M., and Werner, S. (2008). Cancer as an overhealing wound: An old hypothesis revisited. Nat. Rev. Mol. Cell. Biol. 9 (8), 628–638. doi:10.1038/nrm2455
Scheiffele, F., and Fuss, I. J. (2002). Induction of TNBS colitis in mice. Curr. Protoc. Immunol. 15, 15.19. doi:10.1002/0471142735.im1519s49
Schottelius, A., and Baldwin, A. (1999). A role for transcription factor NF-κB in intestinal inflammation. Int. J. Color. Dis. 14, 18–28. doi:10.1007/s003840050178
Seignez, C., Martin, A., Rollet, C. E., Racoeur, C., Scagliarini, A., Jeannin, J. F., et al. (2014). Senescence of tumor cells induced by oxaliplatin increases the efficiency of a lipid A immunotherapy via the recruitment of neutrophils. Oncotarget 5 (22), 11442–11451. doi:10.18632/oncotarget.2556
Senga, S. S., and Grose, R. P. (2021). Hallmarks of cancer-the new testament. Open Biol. 11 (1), 200358. doi:10.1098/rsob.200358
Seril, D. N., Liao, J., West, A. B., and Yang, G. Y. (2006). High-iron diet: Foe or feat in ulcerative colitis and ulcerative colitis-associated carcinogenesis. J. Clin. Gastroenterol. 40 (5), 391–397. doi:10.1097/00004836-200605000-00006
Sharma, S. V., Haber, D. A., and Settleman, J. (2010). Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer 10 (4), 241–253. doi:10.1038/nrc2820
Shen, T., Yue, C., Wang, X., Wang, Z., Wu, Y., Zhao, C., et al. (2021). NFATc1 promotes epithelial-mesenchymal transition and facilitates colorectal cancer metastasis by targeting SNAI1. Exp. Cell. Res. 408 (1), 112854. doi:10.1016/j.yexcr.2021.112854
Shi, L., Wang, J., Ding, N., Zhang, Y., Zhu, Y., Dong, S., et al. (2019). Inflammation induced by incomplete radiofrequency ablation accelerates tumor progression and hinders PD-1 immunotherapy. Nat. Commun. 10 (1), 5421. doi:10.1038/s41467-019-13204-3
Shoemaker, A. R., Gould, K. A., Luongo, C., Moser, A. R., and Dove, W. F. (1997). Studies of neoplasia in the Min mouse. Biochim. Biophys. Acta 1332 (2), 25–48. doi:10.1016/s0304-419x(96)00041-8
Shoemaker, A. R., Moser, A. R., Midgley, C. A., Clipson, L., Newton, M. A., and Dove, W. F. (1998). A resistant genetic background leading to incomplete penetrance of intestinal neoplasia and reduced loss of heterozygosity in APCMin/+ mice. Proc. Natl. Acad. Sci. U. S. A. 95 (18), 10826–10831. doi:10.1073/pnas.95.18.10826
Smits, R., Kartheuser, A., Jagmohan-Changur, S., Leblanc, V., Breukel, C., De Vries, A., et al. (1997). Loss of APC and the entire chromosome 18 but absence of mutations at the Ras and Tp53 genes in intestinal tumors from APC1638N, a mouse model for APC-driven carcinogenesis. Carcinogenesis 18, 321–327. doi:10.1093/carcin/18.2.321
Sun, Q., Yang, H., Liu, M., Ren, S., Zhao, H., Ming, T., et al. (2022). Berberine suppresses colorectal cancer by regulation of Hedgehog signaling pathway activity and gut microbiota. Phytomedicine 103, 154227. doi:10.1016/j.phymed.2022.154227
Sun, X., Jiang, X., Wu, J., Ma, R., Wu, Y., Cao, H., et al. (2020). IRX5 prompts genomic instability in colorectal cancer cells. J. Cell. Biochem. 121 (11), 4680–4689. doi:10.1002/jcb.29693
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tai, J., Wang, G., Liu, T., Wang, L., Lin, C., and Li, F. (2012). Effects of siRNA targeting c-Myc and VEGF on human colorectal cancer Volo cells. J. Biochem. Mol. Toxicol. 26 (12), 499–505. doi:10.1002/jbt.21455
Takaku, K., Oshima, M., Miyoshi, H., Matsui, M., Seldin, M. F., and Taketo, M. M. (1998). Intestinal tumorigenesis in compound mutant mice of both Dpc4 (SMAD4) and APC genes. Cell. 92, 645–656. doi:10.1016/s0092-8674(00)81132-0
Talmadge, J. E., Singh, R. K., Fidler, I. J., and Raz, A. (2007). Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am. J. Pathol. 170 (3), 793–804. doi:10.2353/ajpath.2007.060929
Tanaka, T., Suzuki, R., Kohno, H., Sugie, S., Takahashi, M., and Wakabayashi, K. (2005). Colonic adenocarcinomas rapidly induced by the combined treatment with 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine and dextran sodium sulfate in male ICR mice possess β-catenin gene mutations and increases immunoreactivity for β-catenin, cyclooxygenase-2 and inducible nitric oxide synthase. Carcinogenesis 26, 229–238. doi:10.1093/carcin/bgh292
Tato-Costa, J., Casimiro, S., Pacheco, T., Pires, R., Fernandes, A., Alho, I., et al. (2016). Therapy-induced cellular senescence induces epithelial-to-mesenchymal transition and increases invasiveness in rectal cancer. Clin. Colorectal Cancer 15 (2), 170–178. doi:10.1016/j.clcc.2015.09.003
Thoma, C. R., Zimmermann, M., Agarkova, I., Kelm, J. M., and Krek, W. (2014). 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 69-70, 29–41. doi:10.1016/j.addr.2014.03.001
Tian, X., Han, Z., Zhu, Q., Tan, J., Liu, W., Wang, Y., et al. (2018). Silencing of cadherin-17 enhances apoptosis and inhibits autophagy in colorectal cancer cells. Biomed. Pharmacother. 108, 331–337. doi:10.1016/j.biopha.2018.09.020
Trede, N. S., Langenau, D. M., Traver, D., Look, A. T., and Zon, L. I. (2004). The use of zebrafish to understand immunity. Immunity 20 (4), 367–379. doi:10.1016/s1074-7613(04)00084-6
Uneyama, M., Chambers, J. K., Nakashima, K., Uchida, K., and Nakayama, H. (2021). Histological classification and immunohistochemical study of feline colorectal epithelial tumors. Vet. Pathol. 58 (2), 305–314. doi:10.1177/0300985820974279
Unterleuthner, D., Neuhold, P., Schwarz, K., Janker, L., Neuditschko, B., Nivarthi, H., et al. (2020). Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis 23 (2), 159–177. doi:10.1007/s10456-019-09688-8
Valent, P., Bonnet, D., De Maria, R., Lapidot, T., Copland, M., Melo, J. V., et al. (2012). Cancer stem cell definitions and terminology: The devil is in the details. Nat. Rev. Cancer 12 (11), 767–775. doi:10.1038/nrc3368
Van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161 (4), 933–945. doi:10.1016/j.cell.2015.03.053
Van der Sluis, M., De Koning, B. A., De Bruijn, A. C., Velcich, A., Meijerink, J. P., Van Goudoever, J. B., et al. (2006). MUC2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131 (1), 117–129. doi:10.1053/j.gastro.2006.04.020
Van, R. B., Tops, C. M., and Vasen, H. F. (2000). From gene to disease; the APC gene and familial adenomatous polyposis coli. Ned. Tijdschr. Geneeskd. 144 (42), 2007–2009.
Vécsey-Semjén, B., Becker, K. F., Sinski, A., Blennow, E., Vietor, I., Zatloukal, K., et al. (2002). Novel colon cancer cell lines leading to better understanding of the diversity of respective primary cancers. Oncogene 21 (30), 4646–4662. doi:10.1038/sj.onc.1205577
Vétillard, A., Jonchère, B., Moreau, M., Toutain, B., Henry, C., Fontanel, S., et al. (2015). Akt inhibition improves irinotecan treatment and prevents cell emergence by switching the senescence response to apoptosis. Oncotarget 6 (41), 43342–43362. doi:10.18632/oncotarget.6126
Vlachogiannis, G., Hedayat, S., Vatsiou, A., Jamin, Y., Fernández-Mateos, J., Khan, K., et al. (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359 (6378), 920–926. doi:10.1126/scienceaao2774
Wang, G., Yang, X., Li, C., Cao, X., Luo, X., and Hu, J. (2014). PIK3R3 induces epithelial-to-mesenchymal transition and promotes metastasis in colorectal cancer. Mol. Cancer Ther. 13 (7), 1837–1847. doi:10.1158/1535-7163.MCT-14-0049
Wang, H., Wang, H. S., Zhou, B. H., Li, C. L., Zhang, F., Wang, X. F., et al. (2013). Epithelial-mesenchymal transition (EMT) induced by TNF-α requires AKT/GSK-3β-mediated stabilization of snail in colorectal cancer. PLoS One 8 (2), e56664. doi:10.1371/journal.pone.0056664
Wang, J., Chen, D., Song, W., Liu, Z., Ma, W., Li, X., et al. (2020). ATP6L promotes metastasis of colorectal cancer by inducing epithelial-mesenchymal transition. Cancer Sci. 111 (2), 477–488. doi:10.1111/cas.14283
Wang, L., Zuo, X., Xie, K., and Wei, D. (2018). The role of CD44 and cancer stem cells. Methods Mol. Biol. 1692, 31–42. doi:10.1007/978-1-4939-7401-6_3
Wei, J., Zhang, J., Wang, D., Cen, B., Lang, J. D., and DuBois, R. N. (2022). The COX-2-PGE2 pathway promotes tumor evasion in colorectal adenomas. Cancer Prev. Res. (Phila). 15 (5), 285–296. doi:10.1158/1940-6207.CAPR-21-0572
Weiswald, L. B., Bellet, D., and Dangles, M. V. (2015). Spherical cancer models in tumor biology. Neoplasia 17 (1), 1–15. doi:10.1016/j.neo.2014.12.004
Weiswald, L. B., Richon, S., Validire, P., Briffod, M., Lai-Kuen, R., Cordelières, F. P., et al. (2009). Newly characterised ex vivo colospheres as a three-dimensional colon cancer cell model of tumour aggressiveness. Br. J. Cancer 101 (3), 473–482. doi:10.1038/sj.bjc.6605173
Wirtz, S., Neufert, C., Weigmann, B., and Neurath, M. F. (2007). Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2 (3), 541–546. doi:10.1038/nprot.2007.41
Wu, M., Wu, Y., Deng, B., Li, J., Cao, H., Qu, Y., et al. (2016). Isoliquiritigenin decreases the incidence of colitis-associated colorectal cancer by modulating the intestinal microbiota. Oncotarget 7 (51), 85318–85331. doi:10.18632/oncotarget.13347
Wu, X., Mao, F., Li, N., Li, W., Luo, Y., Shi, W., et al. (2020). NF2/Merlin suppresses proliferation and induces apoptosis in colorectal cancer cells. Front. Biosci. (Landmark Ed. 25 (3), 513–525. doi:10.2741/4817
Wu, Z., Zuo, M., Zeng, L., Cui, K., Liu, B., Yan, C., et al. (2021). OMA1 reprograms metabolism under hypoxia to promote colorectal cancer development. EMBO Rep. 22 (1), 50827. doi:10.15252/embr.202050827
Xing, C., Wang, M., Ajibade, A. A., Tan, P., Fu, C., Chen, L., et al. (2021). Microbiota regulate innate immune signaling and protective immunity against cancer. Cell. Host Microbe 29 (6), 959–974.e7. doi:10.1016/j.chom.2021.03.016
Yang, Y., He, J., Zhang, B., Zhang, Z., Jia, G., Liu, S., et al. (2021a). SLC25A1 promotes tumor growth and survival by reprogramming energy metabolism in colorectal cancer. Cell. Death Dis. 12 (12), 1108. doi:10.1038/s41419-021-04411-2
Yang, Y. S., Wen, D., and Zhao, X. F. (2021c). Sophocarpine can enhance the inhibiting effect of oxaliplatin on colon cancer liver metastasis-in vitro and in vivo. Naunyn Schmiedeb. Arch. Pharmacol. 394 (6), 1263–1274. doi:10.1007/s00210-020-02032-8
Yang, Y., Yan, T., Han, Q., Zhang, M., Zhang, Y., Luo, Y., et al. (2021b). ZNF326 promotes colorectal cancer epithelial-mesenchymal transition. Pathol. Res. Pract. 225, 153554. doi:10.1016/j.prp.2021.153554
Yang, Z., Wu, D., Chen, Y., Min, Z., and Quan, Y. (2019). GRHL2 inhibits colorectal cancer progression and metastasis via oppressing epithelial-mesenchymal transition. Cancer Biol. Ther. 20 (9), 1195–1205. doi:10.1080/15384047.2019.1599664
Yin, K., Lee, J., Liu, Z., Kim, H., Martin, D. R., Wu, D., et al. (2021). Mitophagy protein PINK1 suppresses colon tumor growth by metabolic reprogramming via p53 activation and reducing acetyl-CoA production. Cell. Death Differ. 28 (8), 2421–2435. doi:10.1038/s41418-021-00760-9
Yin, Y., Cao, L. Y., Wu, W. Q., Li, H., Jiang, Y., and Zhang, H. F ( (2010). Blocking effects of siRNA on VEGF expression in human colorectal cancer cells. World J. Gastroenterol. 16 (9), 1086–1092. doi:10.3748/wjg.v16.i9.1086
Youmans, L., Taylor, C., Shin, E., Harrell, A., Ellis, A. E., Séguin, B., et al. (2012). Frequent alteration of the tumor suppressor gene APC in sporadic canine colorectal tumors. PLoS One 7 (12), 50813. doi:10.1371/journal.pone.0050813
Yu, H. K., Ahn, J. H., Lee, H. J., Lee, S. K., Hong, S. W., Yoon, Y., et al. (2005). Expression of human apolipoprotein(a) kringles in colon cancer cells suppresses angiogenesis-dependent tumor growth and peritoneal dissemination. J. Gene Med. 7 (1), 39–49. doi:10.1002/jgm.638
Yu, Y., Cai, Y., Yang, B., Xie, S., Shen, W., Wu, Y., et al. (2022). High-fat diet enhances the liver metastasis potential of colorectal cancer through microbiota dysbiosis. Cancers (Basel). 14 (11), 2573. doi:10.3390/cancers14112573
Yuan, H., Tu, S., Ma, Y., and Sun, Y. (2021). Downregulation of lncRNA RPLP0P2 inhibits cell proliferation, invasion and migration, and promotes apoptosis in colorectal cancer. Mol. Med. Rep. 23 (5), 309. doi:10.3892/mmr.2021.11948
Zeng, S., Tan, L., Sun, Q., Chen, L., Zhao, H., Liu, M., et al. (2022). Suppression of colitis-associated colorectal cancer by scutellarin through inhibiting Hedgehog signaling pathway activity. Phytomedicine 98, 153972. doi:10.1016/j.phymed.2022.153972
Zhang, C., Wang, X. Y., Zhang, P., He, T. C., Han, J. H., Zhang, R., et al. (2022). Cancer-derived exosomal HSPC111 promotes colorectal cancer liver metastasis by reprogramming lipid metabolism in cancer-associated fibroblasts. Cell. Death Dis. 13 (1), 57. doi:10.1038/s41419-022-04506-4
Zhang, L., Wang, X., Lai, C., Zhang, H., and Lai, M. (2019). PMEPA1 induces EMT via a non-canonical TGF-β signalling in colorectal cancer. J. Cell. Mol. Med. 23 (5), 3603–3615. doi:10.1111/jcmm.14261
Zhang, W. L., Li, N., Shen, Q., Fan, M., Guo, X. D., Zhang, X. W., et al. (2020). Establishment of a mouse model of cancer cachexia with spleen deficiency syndrome and the effects of atractylenolide I. Acta Pharmacol. Sin. 41 (2), 237–248. doi:10.1038/s41401-019-0275-z
Zhang, Y., Gao, Y., Zhang, G., Huang, S., Dong, Z., Kong, C., et al. (2011). DNMT3a plays a role in switches between doxorubicin-induced senescence and apoptosis of colorectal cancer cells. Int. J. Cancer 128 (3), 551–561. doi:10.1002/ijc.25365
Zhao, G. X., Xu, Y. Y., Weng, S. Q., Zhang, S., Chen, Y., Shen, X. Z., et al. (2019). CAPS1 promotes colorectal cancer metastasis via Snail mediated epithelial mesenchymal transformation. Oncogene 38 (23), 4574–4589. doi:10.1038/s41388-019-0740-7
Zhao, Q., Guan, J., and Wang, X. (2020). Intestinal stem cells and intestinal organoids. J. Genet. Genomics 47 (6), 289–299. doi:10.1016/j.jgg.2020.06.005
Zhao, Y., Wang, Y., and Wang, Q. (2021). HNRNPL affects the proliferation and apoptosis of colorectal cancer cells by regulating PD-L1. Pathol. Res. Pract. 218, 153320. doi:10.1016/j.prp.2020.153320
Zhao, Y., Zhang, B., Ma, Y., Zhao, F., Chen, J., Wang, B., et al. (2022). Colorectal cancer patient-derived 2d and 3d models efficiently recapitulate inter- and intratumoral heterogeneity. Adv. Sci. (Weinheim, Baden-Wurttemberg, Ger. 9 (22), e2201539. doi:10.1002/advs.202201539
Zhu, G., Cheng, Z., Lin, C., Hoffman, R. M., Huang, Y., Singh, S. R., et al. (2019). MyD88 regulates LPS-induced NF-ĸB/MAPK cytokines and promotes inflammation and malignancy in colorectal cancer cells. Cancer Genomics Proteomics 16 (6), 409–419. doi:10.21873/cgp.20145
Zhu, Y., Huang, S., Chen, S., Chen, J., Wang, Z., Wang, Y., et al. (2021). SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell. Death Dis. 12 (5), 449. doi:10.1038/s41419-021-03733-5
Zhuang, Y. W., Wu, C. E., Zhou, J. Y., Zhao, Z. M., Liu, C. L., Shen, J. Y., et al. (2018). Solasodine reverses stemness and epithelial-mesenchymal transition in human colorectal cancer. Biochem. Biophys. Res. Commun. 505 (2), 485–491. doi:10.1016/j.bbrc.2018.09.094
Zorzi, M., and Urso, E. D. L. (2022). Impact of colorectal cancer screening on incidence, mortality and surgery rates: Evidences from programs based on the fecal immunochemical test in Italy. Dig. Liver Dis. S1590-8658 (22), 336–341. doi:10.1016/j.dld.2022.08.013
Zou, W., Zhang, Y., Bai, G., Zhuang, J., Wei, L., Wang, Z., et al. (2022). siRNA-induced CD44 knockdown suppresses the proliferation and invasion of colorectal cancer stem cells through inhibiting epithelial-mesenchymal transition. J. Cell. Mol. Med. 26 (7), 1969–1978. doi:10.1111/jcmm.17221
Glossary
Keywords: colorectal cancer, cellular models, animal models, preclinical studies, drug development
Citation: Liu L, Yan Q, Chen Z, Wei X, Li L, Tang D, Tan J, Xu C, Yu C, Lai Y, Fan M, Tao L, Shen W, Li L, Wu M, Cheng H and Sun D (2023) Overview of research progress and application of experimental models of colorectal cancer. Front. Pharmacol. 14:1193213. doi: 10.3389/fphar.2023.1193213
Received: 18 April 2023; Accepted: 05 June 2023;
Published: 04 July 2023.
Edited by:
Li Li, The University of Queensland, AustraliaReviewed by:
Haibo Xu, Chengdu University of Traditional Chinese Medicine, ChinaLihong Zhou, Shanghai University of Traditional Chinese Medicine, China
Copyright © 2023 Liu, Yan, Chen, Wei, Li, Tang, Tan, Xu, Yu, Lai, Fan, Tao, Shen, Li, Wu, Cheng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haibo Cheng, aGJjaGVuZ19uanVjbUAxNjMuY29t; Dongdong Sun, c3VuZGRAbmp1Y20uZWR1LmNu
†These authors have contributed equally to this work
 Li Liu
Li Liu Qiuying Yan2,3†
Qiuying Yan2,3† Yueyang Lai
Yueyang Lai Dongdong Sun
Dongdong Sun