- 1Department of Pathology, Xiangya Hospital, Central South University, Changsha, Hunan, China
- 2Department of Pharmacy, Shanghai Pudong New Area People’s Hospital, Shanghai, China
- 3Department of Pharmacy, Xiangya Hospital, Central South University, Changsha, Hunan, China
- 4National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, China
- 5Department of Orthopedic Surgery, The Second Hospital University of South China, Hengyang, Hunan, China
- 6Ben May Department for Cancer Research, Pritzker School of Molecular Engineering, University of Chicago, Chicago, IL, United States
- 7State Key Laboratory of Silkworm Genome Biology, Medical Research Institute, Southwest University, Chongqing, China
- 8Department of Gynecology, Xiangya Hospital, Central South University, Changsha, Hunan, China
Autophagy is a self-recycling and conserved process, in which the senescent cytoplasmic components are degraded in cells and then recycled to maintain homeostatic balance. Emerging evidence has suggested the involvement of autophagy in oncogenesis and progression of various cancers, such as ovarian cancer (OC). Meanwhile, the non-coding RNAs (ncRNAs) frequently regulate the mRNA transcription and other functional signaling pathways in cell autophagy, displaying promising roles in human cancer pathogenesis and therapeutic response. This article mainly reviews the cutting-edge research advances about the interactions between ncRNAs and autophagy in OC. This review not only summarizes the underlying mechanisms of dynamic ncRNA-autophagy association in OC, but also discusses their prognostic implications and therapeutic biomarkers. The aim of this review was to provide a more in-depth knowledge framework exploring the ncRNA-autophagy crosstalk and highlight the promising treatment strategies for OC patients.
Introduction
Ovarian cancer (OC) is a major threat to women’s health. With the deepening of research, the relationship between tumorigenesis and gene alterations has been gradually known and valued. It has been reported that nearly one-fifth of OC is mainly caused by BRCA1 and BRCA2 mutations (Norquist et al., 2016; Kuchenbaecker et al., 2017). Furthermore, OC is related to infertility, endometriosis, and the use of oral contraceptives and intrauterine devices (La Vecchia, 2017; Kralickova et al., 2020; Balayla et al., 2021). OC is characterized by strong concealment and high mortality (Kuroki and Guntupalli, 2020). It was estimated that there were more than 300,000 new OC cases and more than 200,000 deaths worldwide in 2020 (Cabasag et al., 2022). Most patients with OC have no symptoms in the early stage and are usually diagnosed with the metastatic and peritoneal spread in the late stage. In addition, the 5-year survival rate of OC patients is about 47% and has not improved in the past twenty years (Moufarrij et al., 2019). At present, the first-line treatment strategy for OC remains surgery, followed by chemotherapy using agents such as platinum and Taxus (Gogineni et al., 2021). Recently, bevacizumab and PARP inhibitors such as olaparib, rucaparib, and niraparib have received FDA approval for use in OC patients (O'Malley, 2019). Nevertheless, a high recurrence rate and chemotherapeutic resistance remain serious obstacles to treating OC (Norouzi-Barough et al., 2018). Hence, it is necessary to investigate the underlying mechanism of tumorigenesis and progression, as well as explore novel and effective therapeutic approaches. Recent studies have implied that non-coding RNAs (ncRNAs) play vital roles in the initiation and progression of OC through autophagy regulation.
The ncRNAs were originally regarded as “junk DNA” for a long time, possessing no functions (Sharma et al., 2022). However, with the breakthrough of sequencing technology, the roles of ncRNAs in humans were redefined (Costa, 2005; Chen et al., 2022a). Currently, it is well known that ncRNAs are unique molecules that could be used as protein transcription templates to generate functional biomarkers involved in physiological and pathological activities (Mattick and Makunin, 2006). They are mainly divided into three categories: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). MiRNAs, with a length of about 21 nucleotides, have the functions of inhibiting transcription and degrading messenger RNAs (mRNAs) by interacting with the 3′-untranslated region of the target genes (Filipowicz et al., 2008). LncRNAs, which are greater than 200 nucleotides in length, participate in a wide variety of biological processes. Their functions mainly include acting as sponges and scaffolds and signaling (Stackhouse et al., 2020). CircRNAs are single-stranded, covalently closed molecules and can serve as competing endogenous RNAs (ceRNAs) or miRNA sponges to regulate biological activities (Zhou et al., 2020). LncRNAs and circRNAs contain binding sites for miRNAs and bind to them to inhibit their activity and functions (Salmena et al., 2011; Huang et al., 2018a).
Recent studies have suggested the functional roles of ncRNAs in the regulation of cell autophagy and participating in many kinds of cancers, including OC. For example, miR-145-3p could downregulate the expression level of HDAC4 to activate autophagic cell death in multiple myeloma (Wu et al., 2020). lncRNA GBCDRlnc1 can reduce the chemotherapy sensitivity of gallbladder cancer cells by activating cell autophagy (Cai et al., 2019). Overexpression of circATG7 has stimulative effects on cell autophagy in pancreatic cancer cells, thus promoting cancer progression (He et al., 2022). Meanwhile, in OC, lncRNA XIST could trigger cell autophagy by inhibiting the downstream target miR-506-3p, leading to enhanced resistance to carboplatin (Xia et al., 2022). In addition, the aberrant lnc-CTSLP8 displayed crucial roles in regulating cell autophagy and metastasis in OC cells (Wang et al., 2021a).
This paper reviews the effect of ncRNAs-associated signaling on the occurrence and development of OC by regulating autophagic processes. Furthermore, the underlying mechanisms and functional status of ncRNAs-autophagy interaction in prognosis and drug resistance of OC patients are also discussed. These findings would further improve our understanding of the ncRNA-autophagy axis in the malignant progression of OC and offer promising targets to enhance the therapeutic effect for OC.
Functional roles of autophagy modulation
Autophagy is a highly conserved and complex process in all eukaryotes, which maintains the homeostasis of cells and organisms through degradation and recycling (Kim and Klionsky, 2000; Yu et al., 2018a; Peng et al., 2021). Autophagy is classified into three categories: micro-autophagy, macro-autophagy, and chaperone-mediated autophagy (CMA). In micro-autophagy, the lysosome membrane invaginates or protrudes to engulf cellular components directly (Mijaljica et al., 2011). Macro-autophagy refers to the isolation of cellular components from lysosomes. More specifically, double-membrane vesicles, namely autophagosomes, are newly synthesized to isolate cellular components and transport them to lysosomes (Yorimitsu and Klionsky, 2005). Dynamic membrane rearrangement is involved in both micro-autophagy and macro-autophagy (Wang and Klionsky, 2003). In addition, CMA uses molecular chaperones to recognize targeted proteins containing specific pentapeptide motifs (Massey et al., 2004; Yang et al., 2019). Three kinds of autophagy have their functions to maintain metabolism and internal balance together. Among the three types of autophagy, macro-autophagy (called ‘autophagy’ hereafter) is the most thoroughly studied and widely applied in human physiological and pathological processes.
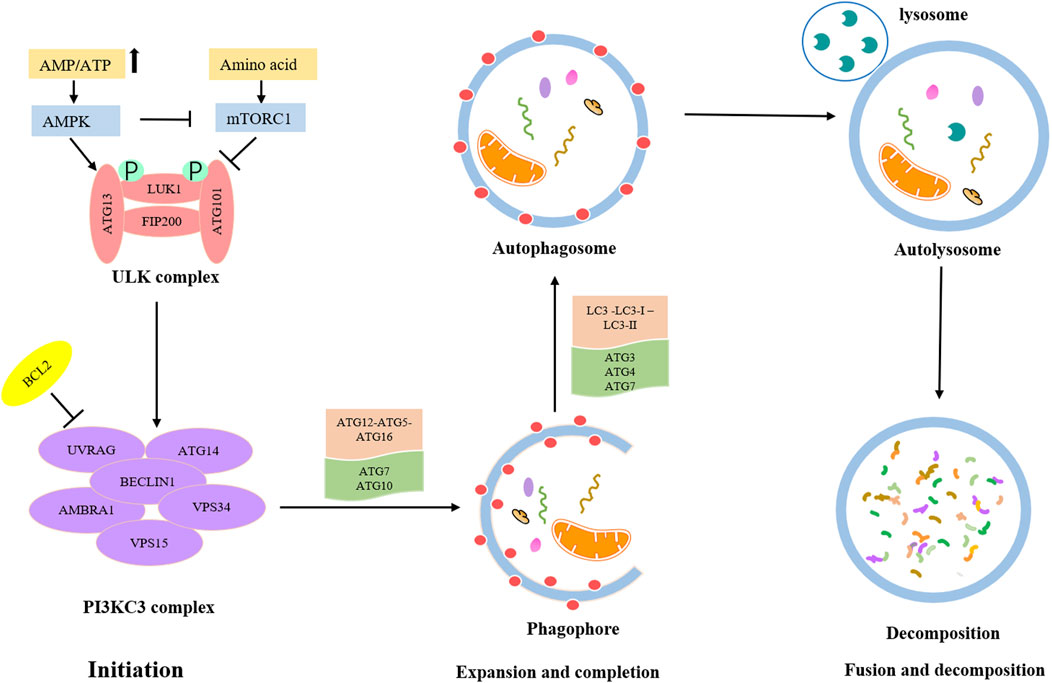
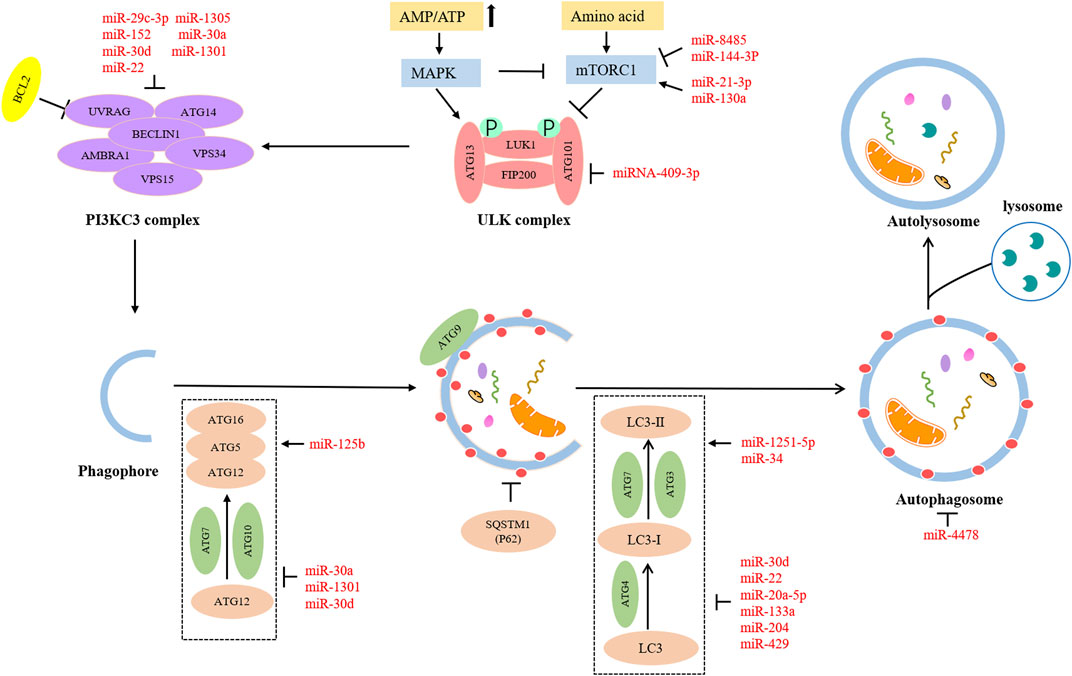
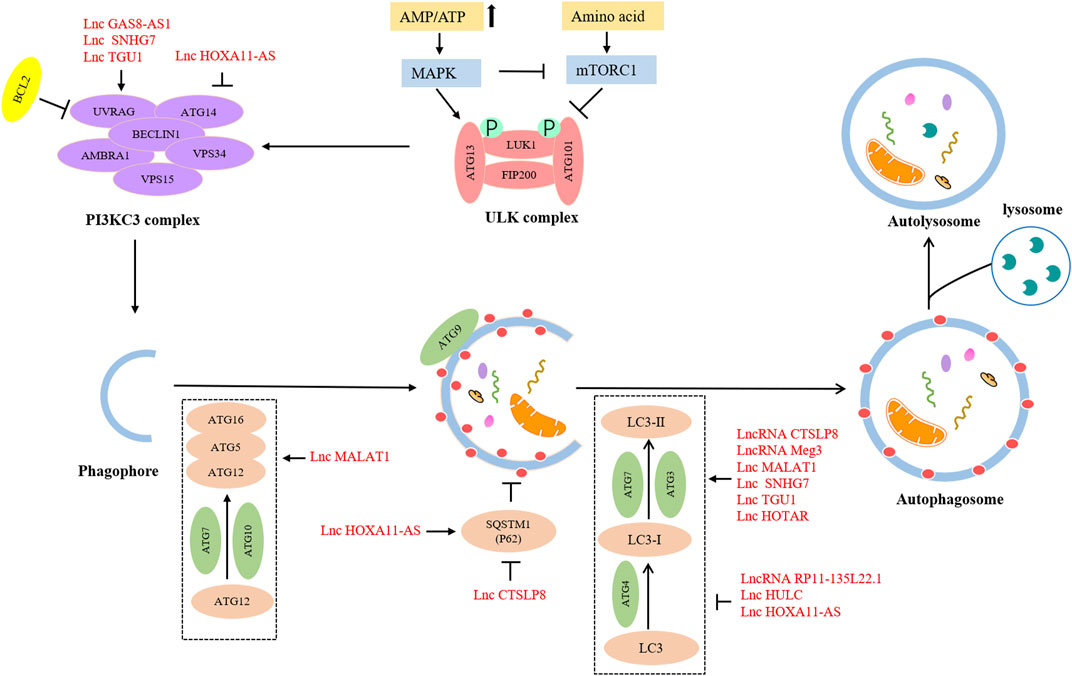
The detailed process of autophagy is shown in Figure 1. Autophagy is initially triggered by various stress conditions, including hypoxia, oxidative stress, infection, nutritional deficiencies, and growth factor reduction. These autophagy-trigger signals cause the activation of MAPK and mTOR pathways, promoting or inhibiting the progression of autophagy. The process of autophagy can be divided into the following three main phases: (Norquist et al., 2016) Initiation phase, wherein the pre-initiation complex, unc-51-like kinase (ULK), composed of ULK1, ATG13, ATG101, and FIP200 is activated, which leads to the phosphorylation of the class III PI3K (PI3KC3) complex, consisting of BECN-1, UVRAG, AMBRA1, VPS15, and VPS34 (Russell et al., 2013). This complex recruits ATG9-containing vesicles to trigger phagophore nucleation. Meanwhile, the activated PI3KC3 complex can be inhibited by BCL2; (Kuchenbaecker et al., 2017) Expansion and completion phase, wherein the most critical step is to recognize cargo and lengthen phagophore, which is conducive to forming the autophagosome. At this stage, there are two vital ubiquitin–like conjugation pathways (Klionsky and Schulman, 2014). The first one involves the combination of ATG12 and ATG5 to form ATG5-ATG12 conjugate and interacts with ATG16L1, leading to the formation of the ATG12-ATG5-ATG16L1 complex. This ubiquitin–like conjugation pathway requires the participation of ATG7 and ATG10. The second stage consists of the cleavage of LC3 into LC3-Ⅰ with the help of ATG4, and then LC3-Ⅰ is converted into LC3-II with the catalysis of ATG7 and ATG3. Consequently, LC3-II is coupled with phosphatidylethanolamine to promote the expansion of the phagophore and the formation of autophagosomes; (La Vecchia, 2017) Fusion and decomposition phase, in which autophagosomes further fuse with lysosomes to form autolysosomes, where activated lysosomal hydrolase decomposes the substrate. Eventually, the degraded substrate is transferred to the cytoplasm for cell recycling.
Autophagy is a flexible mechanism of cell self-protection (Hill and Wang, 2022). When human beings are in a state of stress, such as hunger, infection, and hypoxia, autophagy can be induced and provide energy for cellular activities (Parzych and Klionsky, 2014). Previous reports have summarized that autophagy dysfunction needs to be responsible for many diseases, such as neurodegenerative, cardiovascular, metabolic, and inflammatory disorders and cancers (Saha et al., 2018). Zhou et al. have proved that autophagy activation plays an important role in the pathogenic mechanism of lupus nephritis. It has a negative correlation with podocyte lesions, which is answerable to the progression of lupus nephritis (Zhou et al., 2019). Djajadikerta et al. have suggested that autophagy blockade is detrimental to the degradation of toxic protein aggregates involved in neurodegenerative diseases, such as amyotrophic lateral sclerosis, Alzheimer’s disease, and Parkinson’s disease. Autophagy activation may be beneficial in delaying the progression of these diseases by accelerating the breakdown of toxic protein aggregates (Djajadikerta et al., 2020). Accumulating evidence has also confirmed that autophagy is related to the occurrence, development, and treatment of various cancers. For example, autophagy can be used as an accelerator of monocarboxylate transporter 1 expression via the activation of the Wnt/β-catenin signaling pathway, promoting the metastasis of liver cancer (Fan et al., 2018). Autophagy can cooperate with TrkC/NT-3 axis to enhance the viability of hypoxic glioblastoma cells (Jawhari et al., 2017). The deletion of TTK protein kinase could inhibit cisplatin-induced autophagy via activating the mTOR signaling pathway, thus enhancing the sensitivity of OC cells to cisplatin (Qi et al., 2021). Collectively, autophagy plays a significant part in a wide variety of diseases. Targeting autophagy may provide a new strategy with wide application prospects for clinical management.
ncRNA-autophagy signaling interaction in OC
The correlations between ncRNAs and cell autophagy in human cancers have been well recognized by numerous investigators (Liang et al., 2021). Elevated miR106A-5p is dedicated to the suppression of autophagy, which is correlated with late-stage, easy relapse, and undesirable prognosis of patients with nasopharyngeal carcinoma (Zhu et al., 2021). lncRNA EIF3J-DT can directly bind to ATG14 and reduce the degradation of ATG14. Thus, autophagy is activated, and chemotherapy resistance is enhanced in gastric cancer patients (Luo et al., 2021). circRNA CDYL is significant in facilitating cell proliferation of breast cancer by enhancing autophagy, leading to poor clinical outcomes (Liang et al., 2020). Additionally, extensive research has proved that ncRNAs also take part in the regulation of autophagy in many aspects and affect the progression of OC (Huang et al., 2018b). Vescarelli et al. have shown that overexpressed miR-200c enhances the sensitivity of OC cells to olaparib via regulating the expression of neuropilin 1 (Vescarelli et al., 2020). Song et al. have demonstrated that circRNF144B can strengthen Beclin-1 ubiquitination levels by modulating miR-342-3p and FBXL11, which results in the inhibition of autophagy and the promotion of OC progression (Song et al., 2022). Many researchers have speculated that ncRNAs have a great possibility of being developed as the targets for prognosis and treatment of OC. Therefore, to gain a deeper understanding, we concluded the possible mechanism and biological effects of ncRNAs associated with autophagy in OC, aiming to search for possible and promising therapeutic strategies and targets.
Autophagy-associated miRNAs in ovarian cancers
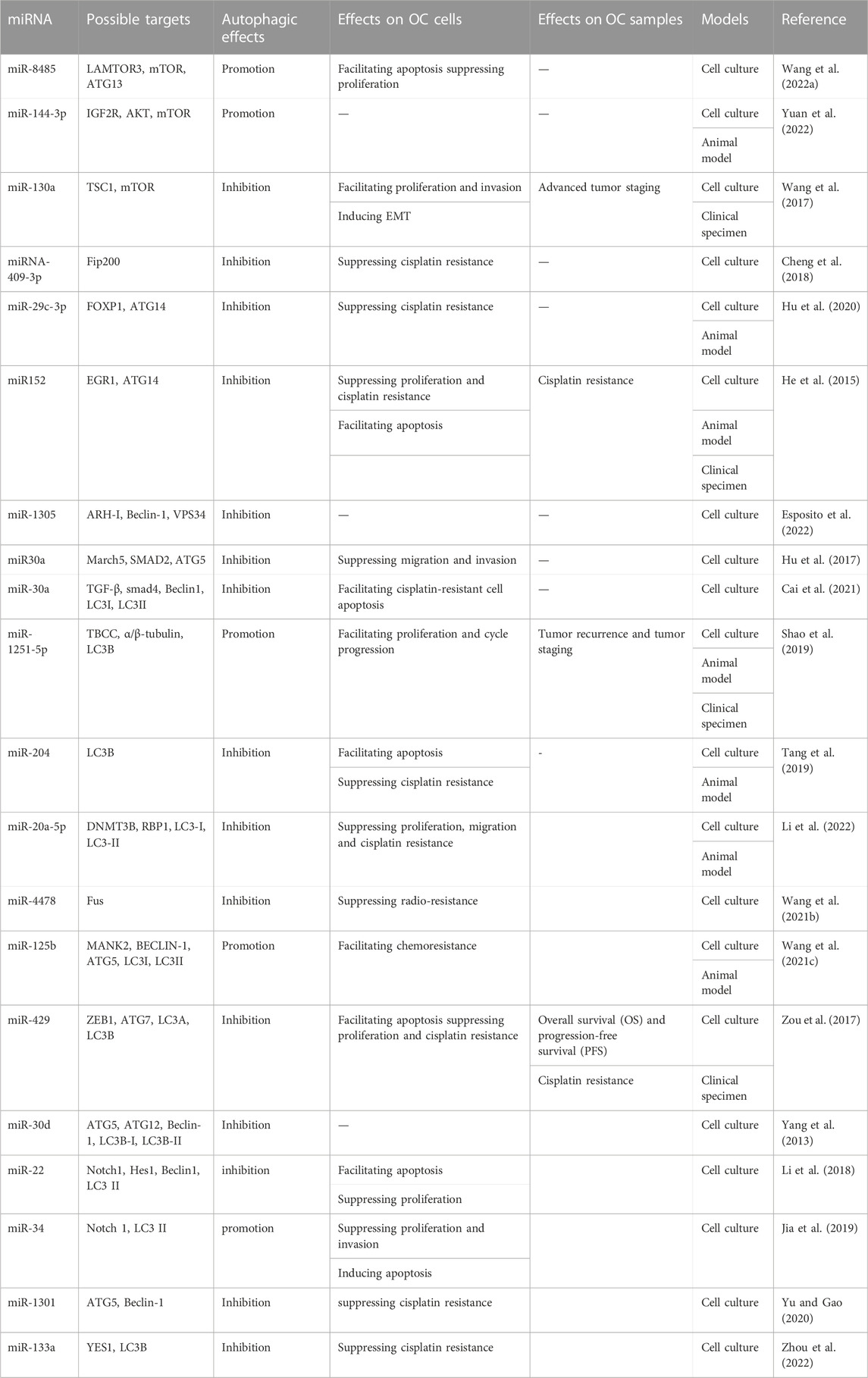
The miRNAs are an important participant in modulating gene expression, which affects cellular processes, and their aberrant expression contributes to the progression of cancers (Deb et al., 2018; Zhang et al., 2022a). Emerging evidence has indicated that aberrant miRNAs can affect the various stages of autophagy, thereby producing stimulative or suppressive effects on OC progression. Hence, we have summarized some miRNAs involved in autophagy in OC (Table 1; Figure 2).
Numerous miRNAs can take part in the initiation stage of autophagy. The mTOR is a major player in the anabolic and catabolic processes that regulate the growth and proliferation of cells (Saxton and Sabatini, 2017). Autophagy is a catabolic process. Moreover, mTOR is an essential component of mTORC1, and mTORC1 is a negative factor of the ULK1 complex involved in initiating autophagy. The activation of mTORC1 suppresses the phosphorylation of ULK1, leading to the inhibition of autophagy (Hosokawa et al., 2009). Therefore, several miRNAs may regulate autophagy via the mTOR pathway. MiR8485 could bind to LAMTOR3, which resulted in decreased mTOR expression level and increased ATG13 and LC3-Ⅱ expression level, indicating the encouraging effect on autophagy (Wang et al., 2022a). MiR144-3p negatively targeted IGF2R, reduced phosphorylation levels of AKT and mTOR in OC, and promoted autophagy (Yuan et al., 2022). MiR-130a downregulated TSC1 to activate the mTOR pathway and cause the inhibition of autophagy (Wang et al., 2017). Fip200 is the first discovered interactor of the ULK complex, which is a key kinase to initiate autophagy (Hara et al., 2008). A report suggested that the Fip200-mediated autophagy could be disturbed by miRNA-409-3p (Cheng et al., 2018). ATG14 is a distinct subunit of the autophagy-associated PI3KC3 complex. It can interact with Beclin-1 via coiled-coil structure and enlist PI3KC3 complex into the endoplasmic reticulum, thereby promoting autophagosome formation and autophagy process (Matsunaga et al., 2010). Furthermore, it has been mentioned in the literature that ATG14 may act as a fusion medium between autophagosomes and lysosomes (Diao et al., 2015). Researchers have found that overexpressed miR-29c-3p and miR152 have direct or indirect inhibitory effects on the expression of ATG14, thus blocking autophagy progress (He et al., 2015; Hu et al., 2020). As the homologous gene of mammalian yeast ATG6, Beclin-1 is an indispensable regulator of autophagy initiation. It can induce the activation of Vps34 to form the Beclin-1-Vp34 complex, which gives an impetus to phosphatidylinositol 3-phosphate generation and lipid membrane elongation (Kihara et al., 2001). A review has suggested the role of ARH-I in triggering autophagy via substituting bcl-2 and binding to Beclin-1 (Lu et al., 2014). MiR-1305 could target ARH-1 to decrease the amounts of Beclin-1 and Vp34. Thus, autophagy flux was reduced by miR-1305 (Esposito et al., 2022).
There are some miRNAs participating in the expansion and completion stage of autophagy. ATG5 is an essential ingredient in forming autophagic vesicles, and its downregulation can directly lead to restricting or blocking autophagy. On the other hand, the ATG5-ATG12-ATG16 complex is involved in the regulation of autophagy as a pivotal ubiquitin-like conjugation pathway (Ye et al., 2018). TGFB1-SMAD2/3 pathway could upregulate March5 expression level via downregulating miR30A expression level. In addition, increased March5 levels acted as a positive regulator of ATG5 to promote autophagy (Hu et al., 2017). Another study showed that miR30A also could contribute to lower autophagy flux by inhibiting the TGF-β/Smad4 pathway (Cai et al., 2021). LC3 family, including LC3A, LC3B, and LC3C, is a human homolog of Atg8 coding gene of yeast (Wild et al., 2014). It plays a core part in a ubiquitin-like conjugation pathway of autophagy. Highly expressed miR-1251-5p might suppress LC3B expression via inversely modulating the expression level of TBCC and α/β-tubulin, thus impairing autophagy (Shao et al., 2019). LC3B could be inhibited by miR-204 to reduce autophagy (Tang et al., 2019). MiR-20a-5p could upregulate RBP1 expression via the downregulation of DNMT3B expression. Moreover, the elevated RBP1 had a remarkable restraining effect on autophagy by reducing the conversion of LC3-I to LC3-II (Li et al., 2022). Fus is a gene encoding an RNA-binding protein. Fus-containing stress particles can co-locate with the cellular autophagosomes. Accordingly, overexpression of miR-4478 could impair autophagy by suppressing fus (Wang et al., 2021b).
Furthermore, many miRNAs can act on multiple steps of autophagy to produce accumulation effects. Beclin-1, ATG5, and the conversion rate of LC3-I to LC3-II could be increased by miR125 via attenuating MKNK2 expression, which synergistically promoted autophagy (Wang et al., 2021c). The overexpression of miR-429 caused a decrease in anti-ATG7 and anti-LC3A/B by directly targeting ZEB1, jointly resulting in a reduction in autophagy (Zou et al., 2017). MiR-30d could inhibit multiple autophagy-associated core proteins such as Beclin-1, ATG12, ATG5, BNIP3L, and ATG2 and restrict transformation from LC3B-I to LC3B-II, leading to autophagy damage (Yang et al., 2013). Overall, different miRNAs target different autophagy-related factors to produce different effects, affecting OC progression.
Autophagy-associated lncRNAs in OC
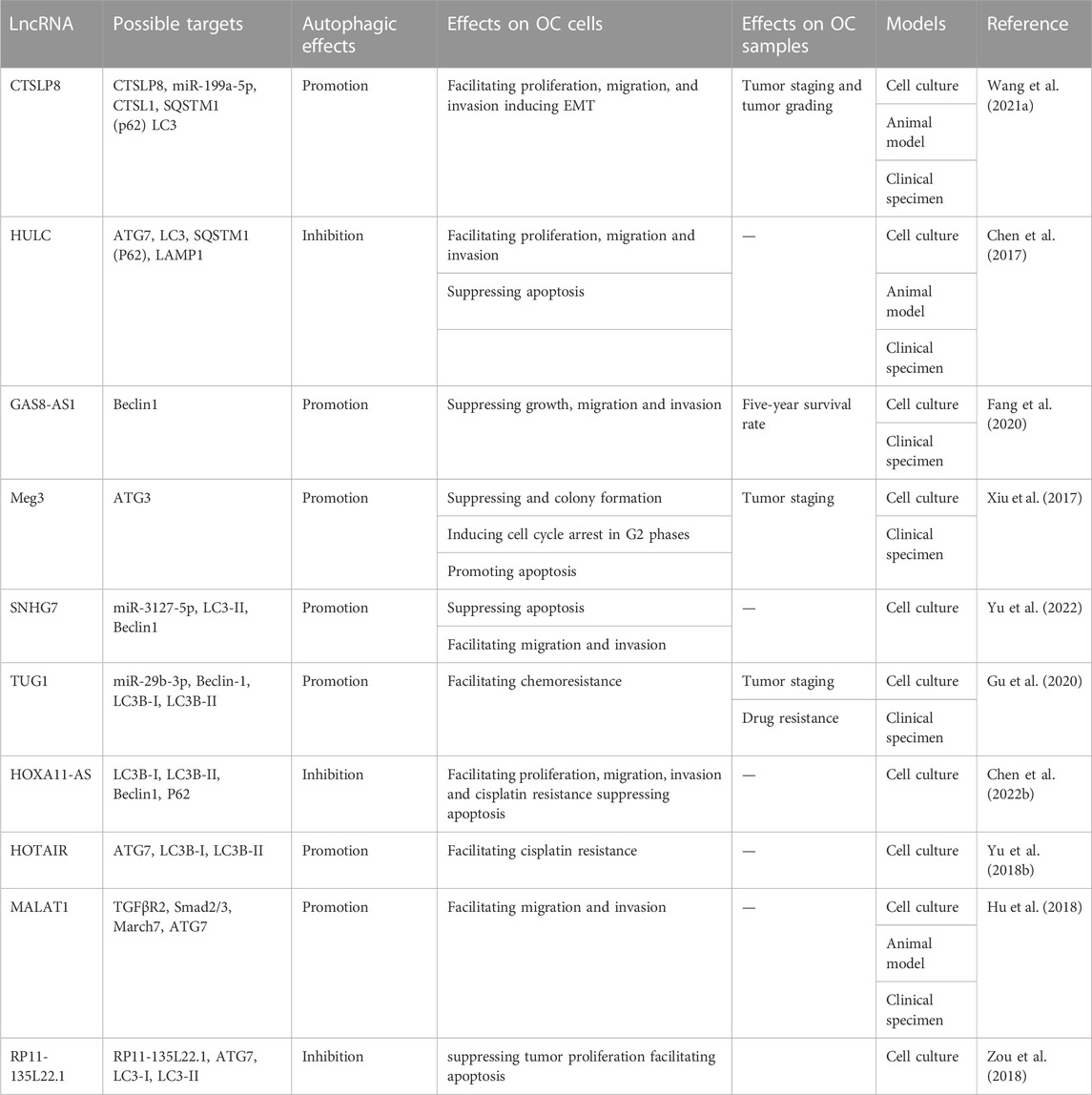
The lncRNAs are aberrantly expressed in many cancers and closely related to onset, development, metastasis, and drug resistance in many cancers via the regulation of autophagy (Chen et al., 2017; Xin et al., 2018; Wang et al., 2019). With advances in research, some scholars have deepened their understanding of lncRNAs in the modulation of autophagy (Table 2; Figure 3).
In the initiation stage, overexpressed lncRNA GAS8-AS1 interacted with Beclin-1 to trigger autophagy (Fang et al., 2020). In the expansive and completion stage, lncRNA Meg3 might upregulate the level of ATG3 by forming Meg3-ATG3 complex and trigger ATG3-dependent autophagy (Xiu et al., 2017). Moreover, more lncRNAs can affect multiple procedures of autophagy. The lncRNA SNHG7 served as a sponge of miR-3127-5p to active autophagy via upregulating Beclin-1 and LC3-II expression levels (Yu et al., 2022). LncRNA TUG1 could directly bind to miR-29b-3p and increase the expression of Beclin-1 and convert LC3B-I to LC3B-II to promote autophagy (Gu et al., 2020). In addition to declining the expression of Beclin-1 and LC3-II, lncRNA HOXA11-AS also could increase the p62 expression level to suppress the autophagy process (Chen et al., 2022b). ATG7 expression could be upregulated to facilitate autophagy by lncRNA HOTAIR and lncRNA MALAT1. However, lncRNA HOTAIR could result in an increased ratio of LC3-II to LC3-I (Yu et al., 2018b; Hu et al., 2018). Although lncRNA HULC and lncRNA RP11-135L22.1 could downregulate ATG7 and LC3 -II expression to impair autophagy, lncRNA HULC-mediated ATG7 downregulation also could induce the expression of LAMP1 to suppress autophagy (Chen et al., 2017; Zou et al., 2018). Collectively, these studies present strong evidence that lncRNAs-associated signaling pathways have impacts on OC development by regulating cell autophagy.
Autophagy-associated circRNAs in OC
It is more than 40 years since the first discovery of circRNA in plant viroids (Sanger et al., 1976). After then, accumulating data indicate that circRNAs perform vital roles in the initiation and progression of OC through modulating autophagy. For example, the direct binding of circMUC16 and miR-199a-5p alleviated the expression inhibition of RUNX1, which, in turn, caused increased circMUC16 expression. Additionally, circMUC16 could combine with ATG13 and facilitate its expression, and overexpressed circMUC16 could promote autophagy (Gan et al., 2020). The interaction between circEEF2 with miR-6881-3p and ANXA2 has been implicated in autophagy. The former interaction upregulated the ATG5 and ATG7 expression, and the latter downregulated the expression of p-mTOR, resulting in the promotion of autophagy (Yong et al., 2020). Furthermore, circRAB11FIP1 regulated ATG14 and ATG7 by sponging miR-129 or binding with DSC1 protein to promote autophagy (Zhang et al., 2021). Taken together, circRNAs function as a sponge to interact with miRNAs, regulating the autophagy of OC.
Prognosis values of autophagy-associated ncRNAs in OC patients
A growing body of evidence has shown that several autophagy-associated ncRNAs may be crucial players in the prognosis of OC patients, and they have great potential to become prognostic biomarkers and treatment targets. Recently, based on abnormally expressed autophagy-related lncRNAs, Li et al. have proposed an autophagy-associated lncRNA risk model for the prediction of prognosis and therapeutic effect in OC patients (Li et al., 2021). However, there is no substantive data to prove that they are involved in the specific roles of autophagy regulation in the occurrence and development of ovarian cancer.
Up to now, only a small number of ncRNAs have been detailly revealed in the OC outcome. For instance, lncRNA GAS8-AS1 interacted with Beclin-1 to promote autophagy, and its deletion caused an enhanced ability of OC cells to metastasize, invade, and proliferate (Fang et al., 2020). An elevation of lncRNA RP11-135L22.1 showed a prohibitive role in cisplatin-induced autophagy, leading to the reduction of cell proliferation and increased cell apoptosis (Zou et al., 2018). Similarly, high expression of miR-22 and miR-34 could suppress cell proliferation and expedite cell apoptosis via the downregulation of the notch signaling pathway (Li et al., 2018; Jia et al., 2019). The suppressive effects of radiation on the growth of OC cells could be reinforced, and the response of DNA damage repair to radiation could be attenuated, thus improving the sensitivity of OC cells to radiotherapy by upregulating miR-4478 to downregulate Fus and reduce autophagy (Wang et al., 2021b). NF-κB signaling pathway could be inhibited by miR-1301 to attenuate cisplatin resistance and delay the development of EMT in OC, ultimately refraining the progression of OC (Yu and Gao, 2020). Additionally, autophagy could be suppressed by miR-133a and miR-1301, thereby suppressing cisplatin resistance (Yu and Gao, 2020; Zhou et al., 2022). More importantly, the imbalance of ncRNA homeostasis was also closely related to the clinical characteristics of OC patients. For example, miR-130a was upregulated and could promote OC progression through inhibiting TSC3 expression in OC samples. The aberrant miR-130a-TSC3 axis has also been proved to be associated with the patients’ tumor stage (Wang et al., 2017). He and colleague reported an autophagy-associated miRNA, miR-152, in cisplatin-resistant OC samples. This group found that of miR-152 overexpression could weaken cisplatin resistant by preventing cyto-protective autophagy and enhancing cell apoptosis (He et al., 2015). In addition, the expression of LncMeg3 was significantly downregulated in OC specimens, and was negatively correlated with stage of OC patients (Xiu et al., 2017). Based on these findings, clarifying the detailed roles of autophagy-associated ncRNAs may become a new direction for clinical management of OC patients.
Involvement of ncRNA-autophagy axis in immune regulation
Previous studies have suggested the involvement of autophagy in immune regulation (Jiang et al., 2019; Wang et al., 2022b; Xie et al., 2022). The autophagic decomposition of cyclic GMP-AMP synthase could be astricted by TRIM14, thus enhancing congenital immunity (Chen et al., 2016). Autophagy blocking has an intensive effect on programmed death ligand-1 (PD-L1)-associated immunological suppression in bladder cancer (Tsai et al., 2022). With progress in research, the findings have indicated that ncRNA-mediated autophagy is involved in immunomodulation (Eng et al., 2021). Mir223 could suppress autophagy flux by lowering ATG16L1 expression, causing aggravation of inflammation in the central nervous system (Li et al., 2019). The tumor-infiltrating lymphocyte of CD4 was expressed highly under the condition of autophagy suppression by modulating miR-155 and activating TNF-related apoptosis-inducing ligand in lung cancer (Zarogoulidis et al., 2016). Several autophagy-associated lncRNAs are related to the immune microenvironment. They may be candidates for predicting the prognosis of OC patients (Zhang et al., 2022b). Collectively, the double roles of ncRNAs in autophagy and immunomodulation determine their importance in cancers, providing a theoretical basis for future cancer immunotherapy.
Conclusion and perspectives
The ncRNAs participate in the oncogenesis and progression of cancers through regulating cell autophagy, although the specific mechanisms still need to be further explored. These autophagy-associated ncRNA might be conductive to suppress or facilitate cell growth, and affect the therapeutic response. Therefore, it is reasonable to speculate that ncRNA inhibitors or activators can be developed to improve therapeutic effect and prolong the survival of OC patients. In addition, some ncRNAs could be stably expressed in the biological fluids of tumors, and participate in the regulation of cell autophagy (Keller et al., 2011). Serving as the independent indicators of prognosis, these ncRNAs in liquid biopsy can predict the recurrence and disease-free survival rates of cancer patients. Thus, the strategies targeting autophagy-associated ncRNAs would provide novel therapeutic concepts and application prospects for OV cancers.
Author contributions
YY and BP: Conception and design. YL, QL, WL, AT, KZ and JW: Data curation. ZX, JL and FZ: Writing the manuscript and revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by grants from the China Postdoctoral Science Foundation (2021T140754, 2020M672521).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Balayla, J., Gil, Y., Lasry, A., and Mitric, C. (2021). Ever-use of the intra-uterine device and the risk of ovarian cancer. J. Obstet. Gynaecol. 41 (6), 848–853. doi:10.1080/01443615.2020.1789960
Cabasag, C. J., Fagan, P. J., Ferlay, J., Vignat, J., Laversanne, M., Liu, L., et al. (2022). Ovarian cancer today and tomorrow: A global assessment by world region and human development index using globocan 2020. Int. J. Cancer 151 (9), 1535–1541. doi:10.1002/ijc.34002
Cai, Q., Wang, S., Jin, L., Weng, M., Zhou, D., Wang, J., et al. (2019). Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 18 (1), 82. doi:10.1186/s12943-019-1016-0
Cai, Y., An, B., Yao, D., Zhou, H., and Zhu, J. (2021). MicroRNA miR-30a inhibits cisplatin resistance in ovarian cancer cells through autophagy. Bioengineered 12 (2), 10713–10722. doi:10.1080/21655979.2021.2001989
Chen, M., Meng, Q., Qin, Y., Liang, P., Tan, P., He, L., et al. (2016). TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell. 64 (1), 105–119. doi:10.1016/j.molcel.2016.08.025
Chen, S., Wu, D. D., Sang, X. B., Wang, L. L., Zong, Z. H., Sun, K. X., et al. (2017). The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell. Death Dis. 8 (10), e3118. doi:10.1038/cddis.2017.486
Chen, S., Zhang, C., Shen, L., Hu, J., Chen, X., and Yu, Y. (2022). Noncoding RNAs in cataract formation: Star molecules emerge in an endless stream. Pharmacol. Res. 184. doi:10.1016/j.phrs.2022.106417
Chen, Y., Cui, Z., Wu, Q., Wang, H., Xia, H., and Sun, Y. (2022). Long non-coding RNA HOXA11-AS knockout inhibits proliferation and overcomes drug resistance in ovarian cancer. Bioengineered 13 (5), 13893–13905. doi:10.1080/21655979.2022.2086377
Cheng, Y., Ban, R., Liu, W., Wang, H., Li, S., Yue, Z., et al. (2018). MiRNA-409-3p enhances cisplatin-sensitivity of ovarian cancer cells by blocking the autophagy mediated by Fip200. Oncol. Res. doi:10.3727/096504017X15138991620238
Costa, F. F. (2005). Non-coding RNAs: New players in eukaryotic biology. Gene 357 (2), 83–94. doi:10.1016/j.gene.2005.06.019
Deb, B., Uddin, A., and Chakraborty, S. (2018). miRNAs and ovarian cancer: An overview. J. Cell. Physiol. 233 (5), 3846–3854. doi:10.1002/jcp.26095
Diao, J., Liu, R., Rong, Y., Zhao, M., Zhang, J., Lai, Y., et al. (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520 (7548), 563–566. doi:10.1038/nature14147
Djajadikerta, A., Keshri, S., Pavel, M., Prestil, R., Ryan, L., and Rubinsztein, D. C. (2020). Autophagy induction as a therapeutic strategy for neurodegenerative diseases. J. Mol. Biol. 432 (8), 2799–2821. doi:10.1016/j.jmb.2019.12.035
Eng, G. W. L., Zheng, Y., Yap, D. W. T., Teo, A. Y. T., and Cheong, J. K. (2021). Autophagy and ncRNAs: Dangerous liaisons in the crosstalk between the tumor and its microenvironment. Cancers (Basel) 14 (1). doi:10.3390/cancers14010020
Esposito, A., Ferraresi, A., Salwa, A., Vidoni, C., Dhanasekaran, D. N., and Isidoro, C. (2022). Resveratrol contrasts IL-6 pro-growth effects and promotes autophagy-mediated cancer cell dormancy in 3D ovarian cancer: Role of miR-1305 and of its target ARH-I. Cancers (Basel) 14 (9). doi:10.3390/cancers14092142
Fan, Q., Yang, L., Zhang, X., Ma, Y., Li, Y., Dong, L., et al. (2018). Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/β-catenin signaling pathway activation in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 37 (1), 9. doi:10.1186/s13046-018-0673-y
Fang, Y. J., Jiang, P., Zhai, H., and Dong, J. S. (2020). LncRNA GAS8-AS1 inhibits ovarian cancer progression through activating beclin1-mediated autophagy. Onco Targets Ther. 13, 10431–10440. doi:10.2147/OTT.S266389
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9 (2), 102–114. doi:10.1038/nrg2290
Gan, X., Zhu, H., Jiang, X., Obiegbusi, S. C., Yong, M., Long, X., et al. (2020). CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer 19 (1), 45. doi:10.1186/s12943-020-01163-z
Gogineni, V., Morand, S., Staats, H., Royfman, R., Devanaboyina, M., Einloth, K., et al. (2021). Current ovarian cancer maintenance strategies and promising new developments. J. Cancer 12 (1), 38–53. doi:10.7150/jca.49406
Gu, L., Li, Q., Liu, H., Lu, X., and Zhu, M. (2020). Long noncoding RNA TUG1 promotes autophagy-associated paclitaxel resistance by sponging miR-29b-3p in ovarian cancer cells. Onco Targets Ther. 13, 2007–2019. doi:10.2147/OTT.S240434
Hara, T., Takamura, A., Kishi, C., Iemura, S., Natsume, T., Guan, J. L., et al. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell. Biol. 181 (3), 497–510. doi:10.1083/jcb.200712064
He, J., Yu, J. J., Xu, Q., Wang, L., Zheng, J. Z., Liu, L. Z., et al. (2015). Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy 11 (2), 373–384. doi:10.1080/15548627.2015.1009781
He, Z., Cai, K., Zeng, Z., Lei, S., Cao, W., and Li, X. (2022). Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell. Death Dis. 13 (3), 233. doi:10.1038/s41419-022-04677-0
Hill, C., and Wang, Y. (2022). Autophagy in pulmonary fibrosis: Friend or foe? Genes. Dis. 9 (6), 1594–1607. doi:10.1016/j.gendis.2021.09.008
Hosokawa, N., Hara, T., Kaizuka, T., Kishi, C., Takamura, A., Miura, Y., et al. (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20 (7), 1981–1991. doi:10.1091/mbc.e08-12-1248
Hu, J., Meng, Y., Zhang, Z., Yan, Q., Jiang, X., Lv, Z., et al. (2017). MARCH5 RNA promotes autophagy, migration, and invasion of ovarian cancer cells. Autophagy 13 (2), 333–344. doi:10.1080/15548627.2016.1256520
Hu, J., Zhang, L., Mei, Z., Jiang, Y., Yi, Y., Liu, L., et al. (2018). Interaction of E3 ubiquitin ligase MARCH7 with long noncoding RNA MALAT1 and autophagy-related protein ATG7 promotes autophagy and invasion in ovarian cancer. Cell. Physiol. Biochem. 47 (2), 654–666. doi:10.1159/000490020
Hu, Z., Cai, M., Zhang, Y., Tao, L., and Guo, R. (2020). miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. Cell. Cycle 19 (2), 193–206. doi:10.1080/15384101.2019.1704537
Huang, M. S., Zhu, T., Li, L., Xie, P., Li, X., Zhou, H. H., et al. (2018). LncRNAs and CircRNAs from the same gene: Masterpieces of RNA splicing. Cancer Lett. 415, 49–57. doi:10.1016/j.canlet.2017.11.034
Huang, Y., Shi, J., and Xu, Y. (2018). Long non-coding RNA NNT-AS1 contributes to cell proliferation, metastasis and apoptosis in human ovarian cancer. Oncol. Lett. 15 (6), 9264–9270. doi:10.3892/ol.2018.8492
Jawhari, S., Bessette, B., Hombourger, S., Durand, K., Lacroix, A., Labrousse, F., et al. (2017). Autophagy and TrkC/NT-3 signaling joined forces boost the hypoxic glioblastoma cell survival. Carcinogenesis 38 (6), 592–603. doi:10.1093/carcin/bgx029
Jia, Y., Lin, R., Jin, H., Si, L., Jian, W., Yu, Q., et al. (2019). MicroRNA-34 suppresses proliferation of human ovarian cancer cells by triggering autophagy and apoptosis and inhibits cell invasion by targeting Notch 1. Biochimie 160, 193–199. doi:10.1016/j.biochi.2019.03.011
Jiang, G. M., Tan, Y., Wang, H., Peng, L., Chen, H. T., Meng, X. J., et al. (2019). The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 18 (1), 17. doi:10.1186/s12943-019-0944-z
Keller, A., Leidinger, P., Gislefoss, R., Haugen, A., Langseth, H., Staehler, P., et al. (2011). Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 8 (3), 506–516. doi:10.4161/rna.8.3.14994
Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell. Biol. 152 (3), 519–530. doi:10.1083/jcb.152.3.519
Kim, J., and Klionsky, D. J. (2000). Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69, 303–342. doi:10.1146/annurev.biochem.69.1.303
Klionsky, D. J., and Schulman, B. A. (2014). Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 21 (4), 336–345. doi:10.1038/nsmb.2787
Kralickova, M., Lagana, A. S., Ghezzi, F., and Vetvicka, V. (2020). Endometriosis and risk of ovarian cancer: What do we know? Arch. Gynecol. Obstet. 301 (1), 1–10. doi:10.1007/s00404-019-05358-8
Kuchenbaecker, K. B., Hopper, J. L., Barnes, D. R., Phillips, K. A., Mooij, T. M., Roos-Blom, M. J., et al. (2017). Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317 (23), 2402–2416. doi:10.1001/jama.2017.7112
Kuroki, L., and Guntupalli, S. R. (2020). Treatment of epithelial ovarian cancer. BMJ 371, m3773. doi:10.1136/bmj.m3773
La Vecchia, C. (2017). Ovarian cancer: Epidemiology and risk factors. Eur. J. Cancer Prev. 26 (1), 55–62. doi:10.1097/CEJ.0000000000000217
Li, H., Lei, Y., Li, S., Li, F., and Lei, J. (2022). MicroRNA-20a-5p inhibits the autophagy and cisplatin resistance in ovarian cancer via regulating DNMT3B-mediated DNA methylation of RBP1. Reprod. Toxicol. 109, 93–100. doi:10.1016/j.reprotox.2021.12.011
Li, Y., Gu, Y., Tang, N., Liu, Y., and Zhao, Z. (2018). miR-22-Notch signaling pathway is involved in the regulation of the apoptosis and autophagy in human ovarian cancer cells. Biol. Pharm. Bull. 41 (8), 1237–1242. doi:10.1248/bpb.b18-00084
Li, Y., Wang, J., Wang, F., Gao, C., Cao, Y., and Wang, J. (2021). Development and verification of an autophagy-related lncRNA signature to predict clinical outcomes and therapeutic responses in ovarian cancer. Front. Med. (Lausanne) 8. doi:10.3389/fmed.2021.715250
Li, Y., Zhou, D., Ren, Y., Zhang, Z., Guo, X., Ma, M., et al. (2019). Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy 15 (3), 478–492. doi:10.1080/15548627.2018.1522467
Liang, G., Ling, Y., Mehrpour, M., Saw, P. E., Liu, Z., Tan, W., et al. (2020). Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 19 (1), 65. doi:10.1186/s12943-020-01152-2
Liang, J., Zhang, L., and Cheng, W. (2021). Non-coding RNA-mediated autophagy in cancer: A protumor or antitumor factor? Biochim. Biophys. Acta Rev. Cancer 1876 (2). doi:10.1016/j.bbcan.2021.188642
Lu, Z., Baquero, M. T., Yang, H., Yang, M., Reger, A. S., Kim, C., et al. (2014). DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy 10 (6), 1071–1092. doi:10.4161/auto.28577
Luo, Y., Zheng, S., Wu, Q., Wu, J., Zhou, R., Wang, C., et al. (2021). Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 17 (12), 4083–4101. doi:10.1080/15548627.2021.1901204
Massey, A., Kiffin, R., and Cuervo, A. M. (2004). Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell. Biol. 36 (12), 2420–2434. doi:10.1016/j.biocel.2004.04.010
Matsunaga, K., Morita, E., Saitoh, T., Akira, S., Ktistakis, N. T., Izumi, T., et al. (2010). Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell. Biol. 190 (4), 511–521. doi:10.1083/jcb.200911141
Mattick, J. S., and Makunin, I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15 (1), R17–R29. doi:10.1093/hmg/ddl046
Mijaljica, D., Prescott, M., and Devenish, R. J. (2011). Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 7 (7), 673–682. doi:10.4161/auto.7.7.14733
Moufarrij, S., Dandapani, M., Arthofer, E., Gomez, S., Srivastava, A., Lopez-Acevedo, M., et al. (2019). Epigenetic therapy for ovarian cancer: Promise and progress. Clin. Epigenetics 11 (1), 7. doi:10.1186/s13148-018-0602-0
Norouzi-Barough, L., Sarookhani, M. R., Sharifi, M., Moghbelinejad, S., Jangjoo, S., and Salehi, R. (2018). Molecular mechanisms of drug resistance in ovarian cancer. J. Cell. Physiol. 233 (6), 4546–4562. doi:10.1002/jcp.26289
Norquist, B. M., Harrell, M. I., Brady, M. F., Walsh, T., Lee, M. K., Gulsuner, S., et al. (2016). Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2 (4), 482–490. doi:10.1001/jamaoncol.2015.5495
O'Malley, D. M. (2019). New therapies for ovarian cancer. J. Natl. Compr. Canc Netw. 17(5):619–621. doi:10.6004/jnccn.2019.5018
Parzych, K. R., and Klionsky, D. J. (2014). An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal 20 (3), 460–473. doi:10.1089/ars.2013.5371
Peng, Z., Yuan, L., XuHong, J., Tian, H., Zhang, Y., Deng, J., et al. (2021). Chiral nanomaterials for tumor therapy: Autophagy, apoptosis, and photothermal ablation. J. Nanobiotechnology 19 (1), 220. doi:10.1186/s12951-021-00965-7
Qi, G., Ma, H., Li, Y., Peng, J., Chen, J., and Kong, B. (2021). TTK inhibition increases cisplatin sensitivity in high-grade serous ovarian carcinoma through the mTOR/autophagy pathway. Cell. Death Dis. 12 (12), 1135. doi:10.1038/s41419-021-04429-6
Russell, R. C., Tian, Y., Yuan, H., Park, H. W., Chang, Y. Y., Kim, J., et al. (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell. Biol. 15 (7), 741–750. doi:10.1038/ncb2757
Saha, S., Panigrahi, D. P., Patil, S., and Bhutia, S. K. (2018). Autophagy in health and disease: A comprehensive review. Biomed. Pharmacother. 104, 485–495. doi:10.1016/j.biopha.2018.05.007
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 146 (3), 353–358. doi:10.1016/j.cell.2011.07.014
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Saxton, R. A., and Sabatini, D. M. (2017). mTOR signaling in growth, metabolism, and disease. Cell. 168 (6), 960–976. doi:10.1016/j.cell.2017.02.004
Shao, Y., Liu, X., Meng, J., Zhang, X., Ma, Z., and Yang, G. (2019). MicroRNA-1251-5p promotes carcinogenesis and autophagy via targeting the tumor suppressor TBCC in ovarian cancer cells. Mol. Ther. 27 (9), 1653–1664. doi:10.1016/j.ymthe.2019.06.005
Sharma, U., Tuli, H. S., Uttam, V., Choudhary, R., Sharma, B., Sharma, U., et al. (2022). Role of hedgehog and hippo signaling pathways in cancer: A special focus on non-coding RNAs. Pharmacol. Res. 186. doi:10.1016/j.phrs.2022.106523
Song, W., Zeng, Z., Zhang, Y., Li, H., Cheng, H., Wang, J., et al. (2022). CircRNF144B/miR-342-3p/FBXL11 axis reduced autophagy and promoted the progression of ovarian cancer by increasing the ubiquitination of Beclin-1. Cell. Death Dis. 13 (10), 857. doi:10.1038/s41419-022-05286-7
Stackhouse, C. T., Gillespie, G. Y., and Willey, C. D. (2020). Exploring the roles of lncRNAs in GBM pathophysiology and their therapeutic potential. Cells 9 (11). doi:10.3390/cells9112369
Tang, J., Zhu, J., Ye, Y., Liu, Y., He, Y., Zhang, L., et al. (2019). Inhibition LC3B can increase chemosensitivity of ovarian cancer cells. Cancer Cell. Int. 19, 199. doi:10.1186/s12935-019-0921-z
Tsai, T. F., Chang, A. C., Chen, P. C., Ho, C. Y., Chen, H. E., Chou, K. Y., et al. (2022). Autophagy blockade potentiates cancer-associated immunosuppression through programmed death ligand-1 upregulation in bladder cancer. J. Cell. Physiol. 237 (9), 3587–3597. doi:10.1002/jcp.30817
Vescarelli, E., Gerini, G., Megiorni, F., Anastasiadou, E., Pontecorvi, P., Solito, L., et al. (2020). MiR-200c sensitizes Olaparib-resistant ovarian cancer cells by targeting Neuropilin 1. J. Exp. Clin. Cancer Res. 39 (1), 3. doi:10.1186/s13046-019-1490-7
Wang, C. W., and Klionsky, D. J. (2003). The molecular mechanism of autophagy. Mol. Med. 9 (3-4), 65–76. doi:10.1007/bf03402040
Wang, J., Da, C., Su, Y., Song, R., and Bai, Z. (2021). MKNK2 enhances chemoresistance of ovarian cancer by suppressing autophagy via miR-125b. Biochem. Biophys. Res. Commun. 556, 31–38. doi:10.1016/j.bbrc.2021.02.084
Wang, J., Xie, S., Yang, J., Xiong, H., Jia, Y., Zhou, Y., et al. (2019). The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12 (1), 81. doi:10.1186/s13045-019-0747-0
Wang, L., Liu, Y., Li, H., Zhang, C., Wang, H., Dai, S., et al. (2021). miR-4478 sensitizes ovarian cancer cells to irradiation by inhibiting Fus and attenuating autophagy. Mol. Ther. Nucleic Acids 23, 1110–1119. doi:10.1016/j.omtn.2020.11.024
Wang, X., Li, X., Lin, F., Sun, H., Lin, Y., Wang, Z., et al. (2021). The lnc-CTSLP8 upregulates CTSL1 as a competitive endogenous RNA and promotes ovarian cancer metastasis. J. Exp. Clin. Cancer Res. 40 (1), 151. doi:10.1186/s13046-021-01957-z
Wang, X. R., Jiang, Z. B., Xu, C., Meng, W. Y., Liu, P., Zhang, Y. Z., et al. (2022). Andrographolide suppresses non-small-cell lung cancer progression through induction of autophagy and antitumor immune response. Pharmacol. Res. 179. doi:10.1016/j.phrs.2022.106198
Wang, Y., Chen, B., Xiao, M., Wang, X., and Peng, Y. (2022). Brucea javanica oil emulsion promotes autophagy in ovarian cancer cells through the miR-8485/LAMTOR3/mTOR/ATG13 signaling Axis. Front. Pharmacol. 13. doi:10.3389/fphar.2022.935155
Wang, Y., Zhang, X., Tang, W., Lin, Z., Xu, L., Dong, R., et al. (2017). miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-κB in high-grade serous ovarian carcinoma. Cell. Death Differ. 24 (12), 2089–2100. doi:10.1038/cdd.2017.129
Wild, P., McEwan, D. G., and Dikic, I. (2014). The LC3 interactome at a glance. J. Cell. Sci. 127 (1), 3–9. doi:10.1242/jcs.140426
Wu, H., Liu, C., Yang, Q., Xin, C., Du, J., Sun, F., et al. (2020). MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy 16 (4), 683–697. doi:10.1080/15548627.2019.1635380
Xia, X., Li, Z., Li, Y., Ye, F., and Zhou, X. (2022). LncRNA XIST promotes carboplatin resistance of ovarian cancer through activating autophagy via targeting miR-506-3p/FOXP1 axis. J. Gynecol. Oncol. 33 (6), e81. doi:10.3802/jgo.2022.33.e81
Xie, Y., Yu, L., Cheng, Z., Peng, Y., Cao, Z., Chen, B., et al. (2022). SHED-derived exosomes promote LPS-induced wound healing with less itching by stimulating macrophage autophagy. J. Nanobiotechnology 20 (1), 239. doi:10.1186/s12951-022-01446-1
Xin, X., Wu, M., Meng, Q., Wang, C., Lu, Y., Yang, Y., et al. (2018). Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 17 (1), 94. doi:10.1186/s12943-018-0843-8
Xiu, Y. L., Sun, K. X., Chen, X., Chen, S., Zhao, Y., Guo, Q. G., et al. (2017). Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 8 (19), 31714–31725. doi:10.18632/oncotarget.15955
Yang, Q., Wang, R., and Zhu, L. (2019). Chaperone-mediated autophagy. Adv. Exp. Med. Biol. 1206, 435–452. doi:10.1007/978-981-15-0602-4_20
Yang, X., Zhong, X., Tanyi, J. L., Shen, J., Xu, C., Gao, P., et al. (2013). mir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem. Biophys. Res. Commun. 431 (3), 617–622. doi:10.1016/j.bbrc.2012.12.083
Ye, X., Zhou, X. J., and Zhang, H. (2018). Exploring the role of autophagy-related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front. Immunol. 9, 2334. doi:10.3389/fimmu.2018.02334
Yong, M., Hu, J., Zhu, H., Jiang, X., Gan, X., and Hu, L. (2020). Circ-EEF2 facilitated autophagy via interaction with mir-6881-3p and ANXA2 in EOC. Am. J. Cancer Res. 10 (11), 3737–3751.
Yorimitsu, T., and Klionsky, D. J. (2005). Autophagy: Molecular machinery for self-eating. Cell. Death Differ. 12(2):1542–1552. doi:10.1038/sj.cdd.4401765
Yu, J. L., and Gao, X. (2020). MicroRNA 1301 inhibits cisplatin resistance in human ovarian cancer cells by regulating EMT and autophagy. Eur. Rev. Med. Pharmacol. Sci. 24 (4), 1688–1696. doi:10.26355/eurrev_202002_20343
Yu, L., Chen, Y., and Tooze, S. A. (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14 (2), 207–215. doi:10.1080/15548627.2017.1378838
Yu, Y., Zhang, X., Tian, H., Zhang, Z., and Tian, Y. (2018). Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J. BUON 23 (5), 1396–1401.
Yu, Z., Wang, Y., Wang, B., and Zhai, J. (2022). Metformin affects paclitaxel sensitivity of ovarian cancer cells through autophagy mediated by long noncoding RNASNHG7/miR-3127-5p Axis. Cancer Biother Radiopharm. 37 (9), 792–801. doi:10.1089/cbr.2019.3390
Yuan, D., Guo, T., Qian, H., Ge, H., Zhao, Y., Huang, A., et al. (2022). Icariside II suppresses the tumorigenesis and development of ovarian cancer by regulating miR-144-3p/IGF2R axis. Drug Dev. Res. 83 (6), 1383–1393. doi:10.1002/ddr.21967
Zarogoulidis, P., Petanidis, S., Domvri, K., Kioseoglou, E., Anestakis, D., Freitag, L., et al. (2016). Autophagy inhibition upregulates CD4(+) tumor infiltrating lymphocyte expression via miR-155 regulation and TRAIL activation. Mol. Oncol. 10 (10), 1516–1531. doi:10.1016/j.molonc.2016.08.005
Zhang, J., Yan, H., and Fu, Y. (2022). Effects of autophagy-related genes on the prognosis and immune microenvironment of ovarian cancer. Biomed. Res. Int. 2022. doi:10.1155/2022/6609195
Zhang, Y., Liu, Q., Zhang, X., Huang, H., Tang, S., Chai, Y., et al. (2022). Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J. Nanobiotechnology 20 (1), 279. doi:10.1186/s12951-022-01472-z
Zhang, Z., Zhu, H., and Hu, J. (2021). CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell. Death Dis. 12 (2), 219. doi:10.1038/s41419-021-03486-1
Zhou, W. Y., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 19 (1), 172. doi:10.1186/s12943-020-01286-3
Zhou, X. J., Klionsky, D. J., and Zhang, H. (2019). Podocytes and autophagy: A potential therapeutic target in lupus nephritis. Autophagy 15 (5), 908–912. doi:10.1080/15548627.2019.1580512
Zhou, Y., Wang, C., Ding, J., Chen, Y., Sun, Y., and Cheng, Z. (2022). miR-133a targets YES1 to reduce cisplatin resistance in ovarian cancer by regulating cell autophagy. Cancer Cell. Int. 22 (1), 15. doi:10.1186/s12935-021-02412-x
Zhu, Q., Zhang, Q., Gu, M., Zhang, K., Xia, T., Zhang, S., et al. (2021). MIR106A-5p upregulation suppresses autophagy and accelerates malignant phenotype in nasopharyngeal carcinoma. Autophagy 17 (7), 1667–1683. doi:10.1080/15548627.2020.1781368
Zou, J., Liu, L., Wang, Q., Yin, F., Yang, Z., Zhang, W., et al. (2017). Downregulation of miR-429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. Am. J. Transl. Res. 9 (3), 1357–1368.
Keywords: non-coding RNAs, cell autophagy, ovarian cancer, cancer pathogenesis, prognosis
Citation: Peng B, Li J, Yan Y, Liu Y, Liang Q, Liu W, Thakur A, Zhang K, Xu Z, Wang J and Zhang F (2023) Non-coding RNAs: The recently accentuated molecules in the regulation of cell autophagy for ovarian cancer pathogenesis and therapeutic response. Front. Pharmacol. 14:1162045. doi: 10.3389/fphar.2023.1162045
Received: 09 February 2023; Accepted: 20 March 2023;
Published: 31 March 2023.
Edited by:
Zhiyun Wei, Tongji University, ChinaReviewed by:
Ruifang Liu, University at Buffalo, United StatesWenrui Wang, Bengbu Medical College, China
Copyright © 2023 Peng, Li, Yan, Liu, Liang, Liu, Thakur, Zhang, Xu, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan Zhang, a2VsbHl6Zmh1YW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Bi Peng
Bi Peng Jing Li
Jing Li Yuanliang Yan3
Yuanliang Yan3 Yuanhong Liu
Yuanhong Liu Qiuju Liang
Qiuju Liang Wei Liu
Wei Liu Abhimanyu Thakur
Abhimanyu Thakur Kui Zhang
Kui Zhang