- 1Department of Pharmacology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Tehran Heart Center, Cardiovascular Diseases Research Institute, Tehran University of Medical Sciences, Tehran, Iran
- 3Afzalipour Faculty of Medicine, Kerman University of Medical Sciences, Kerman, Iran
- 4Student Research Committee, Guilan University of Medical Sciences, Rasht, Iran
- 5Department of Immunology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran
- 6Autophagy Research Center, Department of Clinical Biochemistry, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran
- 7Department of Clinical Biochemistry, School of Pharmacy and Pharmaceutical Sciences, Isfahan University of Medical Sciences, Isfahan, Iran
- 8School of Medicine, Shahrekord University of Medical Sciences, Shahrekord, Iran
- 9Department of Clinical Biochemistry, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
- 10Department of Life Science Engineering, Faculty of New Sciences and Technologies, University of Tehran, Tehran, Iran
- 11Department of Clinical Biochemistry, Mashhad University of Medical Sciences, Mashhad, Iran
- 12Student Research Committee, Department of Immunology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 13Department of Internal Medicine, Lorestan University of Medical Sciences, Khorramabad, Iran
- 14Department of Surgery, Kashan University of Medical Sciences, Kashan, Iran
- 15Research Center for Biochemistry and Nutrition in Metabolic Diseases, Kashan University of Medical Sciences, Kashan, Iran
- 16Nutrition and Food Security Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
Breast cancer (BC) is the most common malignancy among women worldwide. Like many other cancers, BC therapy is challenging and sometimes frustrating. In spite of the various therapeutic modalities applied to treat the cancer, drug resistance, also known as, chemoresistance, is very common in almost all BCs. Undesirably, a breast tumor might be resistant to different curative approaches (e.g., chemo- and immunotherapy) at the same period of time. Exosomes, as double membrane-bound extracellular vesicles 1) secreted from different cell species, can considerably transfer cell products and components through the bloodstream. In this context, non-coding RNAs (ncRNAs), including miRNAs, long ncRNAs (lncRNAs), and circular RNAs (circRNAs), are a chief group of exosomal constituents with amazing abilities to regulate the underlying pathogenic mechanisms of BC, such as cell proliferation, angiogenesis, invasion, metastasis, migration, and particularly drug resistance. Thereby, exosomal ncRNAs can be considered potential mediators of BC progression and drug resistance. Moreover, as the corresponding exosomal ncRNAs circulate in the bloodstream and are found in different body fluids, they can serve as foremost prognostic/diagnostic biomarkers. The current study aims to comprehensively review the most recent findings on BC-related molecular mechanisms and signaling pathways affected by exosomal miRNAs, lncRNAs, and circRNAs, with a focus on drug resistance. Also, the potential of the same exosomal ncRNAs in the diagnosis and prognosis of BC will be discussed in detail.
1 Introduction
Breast cancer (BC), the most common malignancy among women, is the second leading cause of cancer death for women worldwide (DeSantis et al., 2017). The National Breast Cancer Coalition (NBCC) has estimated the incidence of BC to be 12.9% in 2022, and unfortunately, a woman dies from BC every 13 min (Oza et al., 2021). BC survival rates are considerably different among the nations, as high-income countries have reported a 5-year survival of 80%, while low-income nations have reported a 5-year survival of less than 40% (Coleman et al., 2008). Age, family history of BC, reproductive and environmental parameters, and genetic predisposition are considered substantial risk factors for BC onset and progression (Shah et al., 2014). Almost 40% of recurrent BCs are detected in patients without particular symptoms in regular examinations, highlighting the significance of BC management and follow-up. Clinical assessments, such as physical examination, should be carried out every 4–6 months for at least 5 years, and afterward, for every 12 months, along with annual mammography evaluation (Shah et al., 2014; Rautalin et al., 2022).
Based upon immunohistochemical staining for key proteins’ expression, breast tumors are considered to contain at least 1% of the following genes: human epidermal growth factor receptor 2 (HER2, Erbb2 gene), estrogen receptor (ER), and progesterone receptor (PR, Pgr gene) (Hammond et al., 2010). Tumors that do not express the three mentioned proteins are referred to as “basal-like” or “triple-negative” breast cancer (TNBC) (Klinge, 2018). Primary breast tumors have predominantly been reported to be ER+/PR+/HER2-, which determine the basis of applying therapeutics, including radiation therapy, surgery, and endocrine therapies (e.g., anti-estrogen therapy) (Klinge, 2018). Nevertheless, 30%–40% of BC cases have been found to be resistant to endocrine therapies and develop metastatic conditions (Ring and Dowsett, 2004; Piggott et al., 2018). According to the PAM50 test that analyzes 50 genes expressed in primary breast tumors, more individualized therapeutics are planned to be used in clinical settings (Cejalvo et al., 2018).
Non-coding RNAs (ncRNAs), which comprise 99% of the total cellular RNAs in human cells (Consortium, 2012; Romano et al., 2017), have recently attracted much attention in relation to BC pathogenesis and development (Sobhani et al., 2022). In brief, ncRNAs are a large family of RNA molecules, classified into two subclasses based on their size: minor or short ncRNAs with less than 200 nucleotides in size (Krichevsky et al., 2003) and major or long ncRNAs with more than 200 nucleotides in size. Multiple ncRNAs are categorized in the aforementioned groups, in which miRNAs, lncRNAs, and circRNAs have been reported to be more essential in cancer pathophysiology (Cech and Steitz, 2014; Beermann et al., 2016). NcRNA molecules are involved in several biological processes, as well as protein coding/decoding, transcription regulation, and gene expression modulation, in both physiological and pathological conditions (Krichevsky et al., 2003; Gregory and Shiekhattar, 2005; Esquela-Kerscher and Slack, 2006; Huarte and Rinn, 2010; Kasinski and Slack, 2011; Rinn and Huarte, 2011; Tay et al., 2014; Anastasiadou et al., 2018). More interestingly, ncRNAs can be packaged into EVs, especially exosomes (Meldolesi, 2018), to be locally or systemically transmitted among the cells (Palazzo and Lee, 2015; Sun et al., 2018a); a characteristic that enables ncRNA transfer from tumor cells to normal cells, and vice versa. The specific structure of EVs (i.e., their bilayer membranes) supports the process of ncRNA transmission and protects them against circulatory nucleases and other possible threat factors (Teixeira et al., 2015; Mateescu et al., 2017).
Exosomal ncRNAs have previously been reported to be prominent in BC-related pathogenic mechanisms, such as cell proliferation, invasion, metastasis, migration, and especially drug resistance (Bullock et al., 2015; Xie et al., 2019). In drug resistance, exosomal ncRNAs interfere with various mechanisms, from drug absorption to its efflux. In addition, they can link resistant cells to sensitive ones through their exosome-dependent transmission (Ashekyan et al., 2022). In the current review, the crosstalk between exosomal ncRNAs, including exosomal miRNAs, lncRNAs, and circRNAs, and BC drug resistance is discussed in detail, and the possible roles of the corresponding RNAs in the deceleration or exacerbation of chemoresistance are comprehensively highlighted. Since exosomal ncRNAs are considered circulating agents with diagnostic and therapeutic potential, and regarding the limitations and disadvantages of the current diagnostic/therapeutic strategies, prognostic/diagnostic potentials of the aforestated RNA molecules will also be reviewed in brief.
2 BC drug resistance in brief: Underlying mechanisms
During the relapse abundance among BC patients, it seems necessary to elucidate the resistance-related mechanisms in detail (Harbeck and Gnant, 2017; Ji et al., 2019). There are two types of drug resistance; intrinsic resistance and acquired resistance. In intrinsic drug resistance, cancer cannot naturally be targeted by a specific agent, which can be the result of genetic mutations, tumor heterogeneity, or the absence of drug target expression. In the acquired type, therapeutic effectiveness is attenuated over time (Cosentino et al., 2021). Modifications of HER receptor signaling have been reported to have a substantial role in developing BC drug resistance. Lee et al. discovered that concentrations of heat shock protein 90 (HSP90) were highly elevated in drug-resistant BC cells, and a mixture of lapatinib and HSP90 inhibitors represented an advantageous therapeutic strategy for HER2+ patients (Lee et al., 2020). As a xenobiotic transporter, the BC-resistant protein (BCRP, also known as, ATP-binding cassette G2 (ABCG2)) contributes to multidrug resistance (MDR) and is responsible for the efflux of anticancer drugs (Szczygieł et al., 2022). In parenthesis, MDR1 is one of the most well-known ABC transporters that induces chemoresistance (Robey et al., 2018). Furthermore, BC heterogeneity in the tumor microenvironment can impact the response to the therapeutic approach, and thus tumor progression (Kim and Zhang, 2016). The increased glucose uptake and disrupted oxidative phosphorylation and glycolysis are linked to cancer progression toward advanced stages, as well as the development of drug resistance against approved chemotherapy drugs, such as paclitaxel, cisplatin, doxorubicin, and tamoxifen (Varghese et al., 2020). Intercellular communications between tumor cells and the surrounding cells, including immune cells, adipocytes, and fibroblasts, significantly affect the increase in resistance against anticancer drugs (Fontana et al., 2022). On the other hand, the enhanced levels of mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), EGFR, and phospho-ribosomal protein S6 kinase beta-1 (p-S6K1) are linked to BC radioresistance (Gray et al., 2019). Multiple evaluations have shown that a large number of miRNAs, as well as lncRNAs and circRNAs, contribute to the progression of BC drug resistance. However, tumor suppressor ncRNAs, which are silenced in chemoresistant BC, can suppress drug resistance if upregulated (Zhu et al., 2011; Gao et al., 2016).
3 Exosomes: Biogenesis and biological features
Exosomes, as small EVs (30–150 nm), are produced during endosomal maturation (Chahar and Casola, 2015). Typically, exosomes range from 1.13 g/mL (derived from B cells) to 1.19 g/m (derived from epithelial cells) in density (Zakharova and Fomina, 2007; Bobrie et al., 2011). Exosomal characteristics, as well as the type of cargos transferred by these EVs, closely depend on the cell of origin and the state involved in exosome generation. Exosomes can carry multiple cargoes, including different RNA molecules, DNA, peptides, and several proteins such as oncoproteins, transcriptional regulators, tumor suppressors, and splicing factors (Valadi and BossiosSjostrandLeeLotvall, 2007). The corresponding EVs facilitate intercellular communications by transferring the aforementioned biologically active molecules (Chahar and Casola, 2015). Structurally, exosomes are produced by endosomes through inward budding from the limited multivesicular body (MVB) membrane (Minciacchi and Di Vizio, 2015). Throughout the process, invagination of the endosomal membrane results in the formation of intraluminal vesicles (ILVs) in large MVBs (Huotari and Helenius, 2011). Through fusion, most ILVs will be released into the extracellular space in the form of exosomes (Yellon and Davidson, 2014). The biogenesis of exosomes involves a set of consecutive molecular machinery, of which the endosomal sorting complex, required for the ESCRT transport machinery, is the most substantial system, playing a crucial role in ILV formation (JH, 2008). ESCRT has four complexes, namely, ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, and the associated proteins, i.e., vacuolar protein sorting-associated protein 4A (VPS4A), tumor susceptibility gene 101 protein (TSG101), and ALG-2-interacting protein X (ALIX), which increase the generation rate of MVBs, vesicle budding (Henne and Emr, 2011), sorting, binding, and clustering of ubiquitinylated proteins and receptors in the late endosomes (Kalra and Mathivanan, 2016). The ESCRT-0 ubiquitin-binding subunits induce the sequestration and recognition of ubiquitinated cargo proteins into the endosomal membrane domains (Henne and Emr, 2011). Facilitating ILV budding, where cargo is transferred into the lumen, can be induced by ESCRT-I and ESCRT-II. ALIX recruits ESCRT-III for the acceleration of pulling, spiral generation, and full budding (Kalra and Mathivanan, 2016). Moreover, VPS4A and TSG101 have regulatory roles in exosome biogenesis through the ESCRT-dependent pathway (Baietti et al., 2012). Eventually, following ILV formation, the ESCRT-III complex is separated from the MVB membrane, while the sorting protein VPS4A supplies its energy (Henne and Emr, 2011). Despite the modulatory roles of ESCRT-associated mechanisms being controversial in exosome release, various ESCRT components, as well as the ubiquitinated proteins, are found in exosomes that are separated from multiple cells (Zhang et al., 2019). Recent studies have also clarified the possible role of the ESCRT-independent pathway in sorting exosomal cargos into MVBs and subsequent biogenesis of exosomes containing lipids and associated proteins, like tetraspanin (Babst, 2011; Stuffers et al., 2019). Unlike ESCRT-mediated protein sorting, it has been shown that RNA loading into exosomes is correlated with cargo domains and self-organizing lipids (Janas et al., 2015). Tetraspanins, as transmembrane proteins (e.g., CD9, CD63, and CD81) found in exosomes, are significantly involved in the ESCRT-independent pathway (Chairoungdua et al., 2010; van Niel and Raposo, 2018). Mechanistically, tetraspanins trigger the organization of membranous microdomains, called tetraspanin-enriched microdomains (TEMs), using several cytosolic and transmembrane signaling proteins (ME, 2003). Recently, both ESCRT-dependent and -independent mechanisms have been reported to cooperate to regulate exosome biogenesis (Maas and Weaver, 2017), which shows the corresponding pathways can work synergistically. The presence of different subpopulations of exosomes may be explained by the presence of different biogenic machineries, as well as different cell species and cellular homeostasis (Zhang et al., 2019). The small size and unified appearance of exosomes help them escape from mononuclear clearance, resulting in their prolonged circulation time to affect cell-to-cell interactions, more efficiently (Zhang et al., 2019). Regarding the role of exosomes as essential mediators of intercellular communications, they have been demonstrated to impact the pathogenesis of several disorders, such as cancers (Colombo and Thery, 2014). Meanwhile, the possible clinical applications of exosomes, especially as diagnostic biomarkers and therapeutic delivery vehicles, have attracted much attention recently (Jiang et al., 2019).
4 Non-coding RNAs: A summary of their structure and subclassifications
Coding RNAs (i.e., mRNAs) are known for their ability to encode proteins that can serve as enzymes, signal transductors, transcription factors, etc., while ncRNAs predominantly have regulatory effects (Li and Liu, 2019). However, about 98% of all transcriptional output is ncRNA, and only 2% is responsible for the formation of coding RNAs (Legnini et al., 2017). Although the term ncRNA refers to an RNA molecule without coding capacity, recent evaluations have demonstrated that a number of ncRNAs can surprisingly be translated into proteins (Legnini et al., 2017). NcRNAs are categorized into two major subclasses, according to the nucleotide length, long ncRNAs and small ncRNAs, which are classified into further subgroups (Costa, 2005). Small ncRNAs mostly contribute to post-transcriptional gene regulation, whereas long ncRNAs have roles in epigenetic modifications (Sana et al., 2012). MiRNAs, small nuclear RNAs (snoRNAs); small interfering RNAs (siRNAs); rRNAs; tRNAs; and Piwi-interacting RNAs (piRNAs) are key RNA molecules belonging to the small ncRNA subclass; on the other hand, pseudogenes; antisense RNAs (asRNAs); long intergenic ncRNAs (lincRNAs); and circRNAs belong to the long ncRNA subclass (Chan, 2018). Several investigations have shown that ncRNAs are essential molecules with the ability to affect a wide spectrum of cellular processes, including inflammation, oxidative stress, autophagy, fibrosis, and pathophysiological processes associated with malignancies such as cell proliferation, migration, angiogenesis, and especially drug resistance (Chan, 2018; Li et al., 2019). Moreover, ncRNAs have been found to serve as hallmarks of cancer cells, suggesting their possible role as prognostic and diagnostic biomarkers (Sana et al., 2012). Among multiple ncRNA molecules, miRNAs, lncRNAs, and circRNAs have been identified to be central to the regulation of cancer-related processes, such as drug resistance.
MiRNAs are small single-stranded RNA molecules (20–24 nucleotides) primarily contributing to post-transcriptional gene modulation through binding to the target gene’s 3′-untranslated region (3′UTR) to suppress the translation (Ruan and Ouyang, 2009; Sana et al., 2012; Bahmyari et al., 2021; Mafi et al., 2022). Subcellular localization of miRNAs, the affinity of miRNA-related interconnections, and the number of target mRNAs and miRNAs influence the miRNA–target gene interactions (O'Brien et al., 2018). Extracellular miRNAs, which can be transferred to target cells by vesicles (e.g., exosomes) or through binding to proteins, are key messengers in modulating cell-to-cell communications (Wang and Sen, 2016). Since miRNAs can affect multiple aspects of tumorigenesis such as angiogenesis, immune deregulation, metastasis, and drug resistance, research studies have recently introduced miRNAs as significant biologically active components of exosomes, affecting neoplastic conditions (Hayes and Lawler, 2014; Kozomara and Griffiths-Jones, 2018). Being inside the exosomes protects miRNAs, as well as the other ncRNAs, from RNase-dependent degradation (Lima et al., 2009).
LncRNAs, which are more than 200 nucleotides in length, are not translated into functional proteins but rarely encode small functional peptides. LncRNAs, first observed in eukaryotic cells, are located in the cytoplasm or nucleus (Dempsey and Cui, 2017). According to the chromosomal position, lncRNAs are categorized into aslncRNAs, divergent lncRNAs, enhancer RNAs (eRNAs), intronic lncRNAs, promoter-associated lncRNAs, intergenic lncRNAs, and transcription start site-associated lncRNAs (Hombach and Kretz, 2016). Abnormal expression of lncRNAs has been reported to accelerate cancer progression by interfering with tumorigenesis, cell survival and proliferation, metastasis and invasion, and drug resistance (Hung et al., 2014; Prensner et al., 2014). By being packaged inside the exosomes, lncRNAs can release into the tumor microenvironment and transferred to recipient cells, moderating the process of cancer metastasis and progression (Fan et al., 2018). Exosomal lncRNAs are also considered potential tumor markers based on their specificity and sensitivity (Yousefi et al., 2020).
Eventually, circRNAs are single-stranded covalently closed molecules originating from pre-mRNA back-splicing (Sanger et al., 1976; Jeck et al., 2013; Salami et al., 2022). CircRNAs lack 5’ to 3’ polarity or poly A tail, and thus are less sensitive to RNA exonuclease- or RNase R-related degradation (Jeck and Sharpless, 2014; Chen and Yang, 2015; Najafi et al., 2022); a prominent characteristic making circRNAs potential prognostic and diagnostic biomarkers and therapeutic agents (Zhou et al., 2020). CircRNAs can interfere with pathogenic processes of multiple diseases, including cancer (Li et al., 2020a), neurological disorders (Mehta and Vemuganti, 2020), cardiovascular diseases (ufiero et al., 2019), and autoimmune defects (Zhou et al., 2019). Nevertheless, the underlying mechanisms by which circRNAs affect physiological/pathological circumstances are not fully understood (Zhou et al., 2020). Some circRNAs can act as protein decoys, recruiters, and scaffolds (Xiao and Wilusz, 2020). In addition, circRNAs exert biological functions, like serving as miRNA sponges, transcriptional regulators, and protein templates (Kristensen et al., 2019; Yu and Kuo, 2019; Huang et al., 2020). Beyond the significance of circRNAs, alone, exosomal circRNAs are considerably involved in cancer-associated mechanisms, including cell proliferation, migration, invasion, metastasis, and drug resistance (Wang et al., 2018a).
5 Functional roles of exosomal ncRNAs in multiple diseases
As mentioned previously, exosomal ncRNAs can potentially upregulate or downregulate the processes that impact tumor expansion, like cell proliferation, tumor metastasis, invasion, immunomodulation, angiogenesis, and specifically drug resistance (Fan et al., 2018). Also, tumor-suppressed ncRNAs can be transferred by exosomes to other tumor or non-tumor cells (Li and Xu, 2019). In this context, exosomal ncRNAs have been shown to be correlated with various human cancers, such as BC, lung cancer, hepatocellular carcinoma (HCC), glioblastomas, and prostate cancer. (Li et al., 2018a; He et al., 2018; Hu et al., 2020a; Movahedpour et al., 2021; Movahedpour et al., 2022a; Taghvimi et al., 2022). For instance, in the case of HCC, exosomal ncRNAs interfere with the process of liver fibrosis and subsequent cirrhosis and consequently provoke tumorigenesis and HCC development (Quail and Joyce, 2013; Bukong et al., 2014). In addition to being involved in cancer progression, exosomal ncRNAs have been demonstrated to contribute to the onset of metabolic disorders, including osteoporosis, type 2 diabetes mellitus (T2DM) and diabetic nephropathy, and obesity (Li et al., 2021a; Mafi et al., 2021). NcRNAs are also involved in the pathogenesis and progression of neurodegenerative diseases, such as Parkinson’s and Alzheimer’s diseases (Jea et al., 2019; Dorostgou et al., 2022; Vakili et al., 2023). NcRNAs encapsulated in exosomes are even associated with infectious diseases (e.g., viral hepatitis) and autoimmune disorders, such as rheumatoid arthritis (RA) (Wang et al., 2018b; Jiao et al., 2021). The aforementioned pathological roles of exosomal ncRNAs can help researchers find the ambiguous aspects of disease pathophysiology and develop novel diagnostic and therapeutic targets to improve disease management (Dea et al., 2014; Lv et al., 2020a; Li et al., 2021a). Especially, in the case of cancer and related pathological mechanisms, drug resistance is of great significance, as it results in conditions where chemotherapy does not work. Regarding the global incidence of BC and the role of chemoresistance in its ineffective treatment, potential interactions between exosomal ncRNAs and BC drug resistance will be comprehensively discussed as follows.
6 Molecular mechanisms by which exosomal ncRNAs interfere with BC drug resistance
Exosomes transport biologically active molecules between various cell types to mediate the initiation and progression of BC. In this regard, exosomal ncRNAs have a remarkable impact on a variety of tumor biology-related processes, including tumor growth, metastasis, migration, and drug resistance. Recently, it has been discovered that exosomal ncRNAs, particularly exosomal miRNAs, lncRNAs, and circRNAs, are involved in BC regulatory mechanisms (Chen et al., 2021; Singh et al., 2022). Exclusively, the latest findings on the crosstalk between exosomal ncRNAs (miRNAs, lncRNAs, and circRNAs) and BC drug resistance will be reviewed in the upcoming sections (Figures 1, 2).
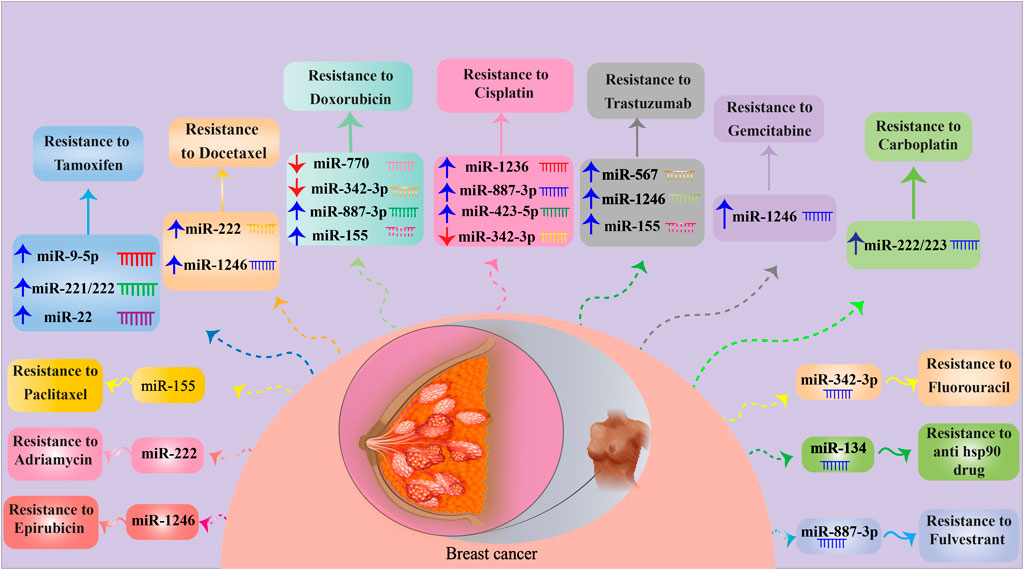
FIGURE 1. MiRNAs involved in BC drug resistance. For detailed information about signaling pathways targeted by miRNAs, see Table 1. ↑ and ↓ indicate up- and down-regulation, respectively.
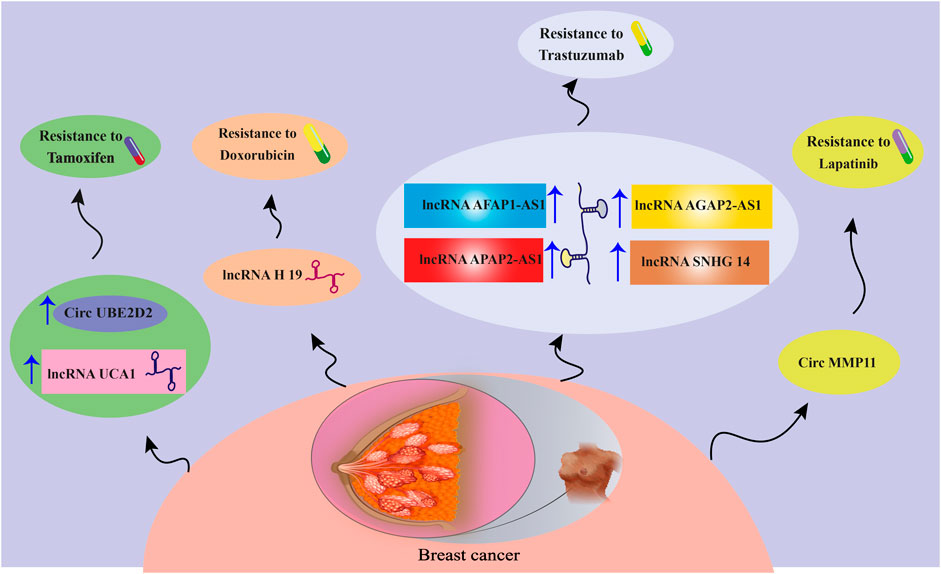
FIGURE 2. A schematic view of the effects of different lncRNAs and circRNAs on BC drug resistance. For more information about the relevant molecular mechanisms, visit Table 1.
6.1 Exosomal miRNAs and BC chemoresistance
MiRNAs are considered to be central to the regulation of genes linked to BC drug resistance (Fu and Tong, 2020; Purwanto et al., 2021; Liu et al., 2022). Several mechanisms modulated by miRNAs have been recognized in relation to BC chemoresistance; targeting genes involved in drug efflux and metabolism; cellular responses to chemotherapeutics (e.g., apoptosis, cell cycle arrest, and DNA repair); epigenetic alterations (e.g., DNA methylation and histone modifications); and deregulation of drug targets and receptors, which are the best examples in the field (Kutanzi et al., 2011). On the other hand, the miRNA-containing exosomes secreted by tumor cells can possibly be internalized by other cells to deliver gene modulatory characteristics to the recipient cells (Bach et al., 2017; Schwarzenbach, 2017; Salehi et al., 2022). As an example, it has been demonstrated that endothelial cell-derived exosomal miR-503, which is increased after neoadjuvant chemotherapy, can impair tumor growth, invasion, and proliferation in BC by inhibiting the expression of cyclin D2- and D3-encoding genes (CCND2 and CCND3) (Bovy et al., 2015). MiRNA expression profiles in BC cells were also reported to be either upregulated or downregulated in response to three chemotherapy drugs: docetaxel, epirubicin, and vinorelbine. In the same study, 12 of 22 upregulated miRNAs showed a significant upregulation, following pre-neoadjuvant chemotherapy, and thus play key roles in several pathways related to BC drug resistance, such as p53, Wnt, MAPK, and ErbB signaling pathways (Zhong et al., 2016). Here, the exosomal miRNA–drug resistance network in BC is highlighted with a focus on the chemotherapy drug subtypes.
6.1.1 Tamoxifen
Tamoxifen (TAM) is an effective FDA-approved chemotherapeutic agent to combat ERα-positive breast tumors, particularly those in premenopausal patients. However, it has many adverse effects attributed to its estrogenic activities in other tissues (Cuzick and Baum, 1985; Freedman et al., 2003; Lumachi et al., 2013; Cuzick et al., 2015). TAM efficacy is attenuated by the development of drug resistance through a wide range of underlying mechanisms orchestrated by exosomal miRNAs (Ali et al., 2016; Yao et al., 2020). Liu et al. declared that exosomes derived from TAM-resistant MCF-7 (ER-positive) cells could transfer miR-9-5p to TAM-sensitive MCF-7 cells, resulting in the inhibition of cell apoptosis and promotion of MCF-7 resistance to TAM through downregulating the expression of the adiponectin gene (ADIPOQ) (Liu et al., 2021). Furthermore, the expression of miR-9-5p was up-modulated in BC tissue more than in normal breast tissue, and the increased miR-9-5p was also found to be associated with reduced ER expression in BC (Barbano et al., 2017). ADIPOQ, located on chromosome 3q27 and encoding adiponectin, can be linked to BC-related cell invasion (Mantzoros et al., 2004; Falk Libby et al., 2016). In addition, it has been elucidated that ADIPOQ overexpression induces autophagy and apoptosis in BC cells by activating the serine/threonine protein kinase 1/liver kinase B1 (STK11/LKB1)-associated AMP-activated protein kinase-Unc-51-like kinase 1 (AMPK–ULK1) pathway to decrease BC growth and progression (Chung et al., 2017; Zhang et al., 2017).
TAM-resistant MCF-7 cells can also induce TAM resistance in sensitive cells by propagating the exosomes containing miR-221/222 in ER-positive BC. In the presence of TAM, TAM-resistant MCF-7 cell-derived exosomes suppressed apoptosis and stimulated the colony-forming ability in TAM-sensitive MCF-7 cells. Exosomal miR-221/222 significantly inhibited the expression of P27 and ERα, promoting TAM resistance in recipient cells (Wei et al., 2014). Consistently, previous studies also demonstrated that miR-221/222 was notably upregulated in TAM-resistant MCF-7 cells and possibly involved in TAM resistance by regulating the expression of p27/Kip1 (Miller et al., 2008). In contrast, a recent evaluation indicated that miR-221/222 suppression could abolish TAM resistance, mediated by restoring the expression of ERα and PTEN (Ouyang et al., 2021).
According to the study conducted by Gao et al., CD63+ cancer-associated fibroblasts (CAFs) secreted miR-22-enriched exosomes that could mitigate ERα expression and activate the PI3K/AKT pathway via PTEN downregulation to promote TAM resistance in BC cells (Gao et al., 2020). In addition, loss of ERα expression and CAFs is linked to a poor response of BC cells to TAM, with the suppression of the activity of CD63+ CAFs to enhance TAM sensitivity in BC cells, in vivo (Gao et al., 2020). CAFs originally constitute the major stromal components of the breast tumor microenvironment, indicating their effect on promoting cancer progression and mediating chemoresistance (Su et al., 2018; Ershaid et al., 2019). Activation of the PI3K/AKT signaling pathway can induce TAM resistance in BC cells, which may result in a decrease in ERα expression (Toska et al., 2017; Mills et al., 2018).
6.1.2 Docetaxel
Although docetaxel (DOC)-based chemotherapy is an effective neoadjuvant approach to improve survival outcomes in BC patients (Heys et al., 2002), chemoresistance mediated by ncRNAs is almost unavoidable (Brown et al., 2004; Huang et al., 2018). According to the evaluations of Chen et al., exosomes from DOC-resistant MCF-7 cells play substantial roles in transmitting DOC resistance to recipient DOC-sensitive cells through the delivery of responsible miRNAs. This study reported the top 20 most common miRNAs in DOC-resistant BC cell-derived exosomes involved in axon guidance, MAPK signaling pathway, cell cycle regulation, Wnt signaling, and TGF-β signaling cascade, which result in treatment failure when upregulated (Chen et al., 2014a). There are several exosomal miRNAs, being up- and down-modulated, secreted from adriamycin- and DOC-resistant BC cells. These BC cells can transfer miR-100, miR-30a, and miR-222 to drug-sensitive cells to modulate cell cycle distribution and drug-induced apoptosis. Exosomal miR-222 derived from DOC-resistant cells could also induce chemoresistance by decreasing PTEN expression in recipient cells (Chen et al., 2014b). In parenthesis, PTEN is an essential tumor suppressor and a pivotal component of the PI3K/PTEN/Akt signaling pathway associated with cellular processes (McCubrey et al., 2006; Steelman et al., 2008; Xia et al., 2020). PTEN underexpression was reported to serve as a predictive marker for poor outcomes in BC patients (Xu et al., 2017).
Further bioinformatics analyses found several overexpressed miRNAs, such as let-7a, let-7c, miR-103a, let-7b, miR-16, miR-23a, miR-27a, miR-23b, and miR-30a, as well as underexpressed miRNAs, such as miR-25, miR-130a, miR-20b, miR-425, miR-4725-5p, miR-455-3p, miR-551, and miR-92, in exosomes originated from DOC-resistant MCF-7 cells (Chen et al., 2019a). The overexpressed miRNAs mostly target the signaling pathways involved in the regulation of stem cell pluripotency and the TGF-β, FOXO, MAPK, and Wnt signaling pathways. However, the MAPK, TGF-β, FOXO, mTOR, and PI3K/Akt signaling pathways were found to be major targets for underexpressed miRNAs (Sebolt-Leopold and Herrera, 2004; Wang et al., 2008; Loh et al., 2013; Coomans de Brachène and Demoulin, 2016; Dey et al., 2017; Chen et al., 2019a; Movahedpour et al., 2022b). CCND1 and PTEN are considered the most common exosomal miRNA targets with high and low expression levels, respectively (Chen et al., 2019a). Overexpression of CCND1, the pivotal factor for cell transition from the G1 to the S phase, was reported in 50% of human BCs (Elsheikh et al., 2008). Another investigation also demonstrated that CCND1 organized the miRNA signature that induced the Wnt/β-catenin signaling pathway, serving as downstream and/or upstream targets of the Wnt/β-catenin axis (Wang et al., 2018c).
Ccng2 was found to be downregulated in BC and served as a gene directly targeted by miR-1246. MiR-1246 has been reported to be overexpressed in human BC cells, particularly metastatic BC MDA-MB-231 cells (ER-negative). Exosomes derived from drug-resistant MDA-MB-231 cells can reduce apoptosis and promote the migration, invasion, and resistance to DOC, epirubicin (Liu et al., 2022), and gemcitabine (GEM) in non-malignant cells through transferring the aforementioned miRNA, i.e., miR-1246, leading to suppression of Ccng2 gene expression (Sakha et al., 2016; Zhong et al., 2016; Li et al., 2017a). Ccng2, as a tumor suppressor gene, has a close relationship with cell cycle, DNA damage, and p53-related pathways (Bates et al., 1996; Montagner et al., 2012; Chang et al., 2015; Zimmermann et al., 2016). MiR-1246-mediated targeting of Ccng2 also promotes cancer progression and chemoresistance in other cancers (Hasegawa et al., 2014; Lin et al., 2018).
6.1.3 Doxorubicin
TNBCs are usually reported with a poorer prognosis, rapid progression, early metastasis, and containing no effective molecular targets for chemotherapy (Haffty et al., 2006). Doxorubicin, sold under the brand name adriamycin (ADM), is an anthracycline agent, which is considered an effective chemotherapeutic to treat BC (Jones et al., 1987; Wang et al., 2015). Nevertheless, doxorubicin resistance results in unsuccessful BC chemotherapy through different mechanisms, and overcoming the resistance can be a promising achievement in BC management (Nabholtz et al., 2003; Koike Folgueira et al., 2005; Lovitt et al., 2018). In this context, miR-770 was demonstrated to be overexpressed in chemo-sensitive TNBC cells, while it was downmodulated in chemoresistant cells. Moreover, overexpression of miR-770 inhibited tumor metastasis and doxorubicin resistance by induction of apoptosis in vivo and in vitro. MiR-770 is transferred by exosomes and causes chemosensitivity in cancer cells via the downregulation of stathmin 1 (STMN1) and suppressed cell invasion and migration by modification of the epithelial–mesenchymal transition (EMT) pathway (Li et al., 2018b). STMN1 or oncoprotein 18 is an essential cytoplasmic phosphoprotein responsible for cellular microtubule dynamics and depolymerization, involving cell cycle progression (Marklund et al., 1996; Rubin and Atweh, 2004). STMN1 participates in metastasis and chemoresistance of BC (Kuang et al., 2015; Kuang et al., 2016; Obayashi et al., 2017).
Santos et al. (2018) declared that exosomes separated from breast cancer stem cells (CSCs) and doxorubicin- and paclitaxel-resistant cells (i.e., MDA-MB-231 and MCF-7) could be transferred to recipient-sensitive cells, resulting in stimulation of migration and drug resistance through miR-155 delivery. Functionally, miR-155 upregulation was linked to the increased levels of B cell-specific Moloney murine leukemia virus integration site 1 (BMI1), snail family transcriptional repressor 1 (SNAI1 or SNAIL), SNAI2 (SLUG), enhancer of zeste homolog 2 (EZH2), and SRY-box transcription factor 9 (SOX9) and repression of E-cadherin in resistant cells, demonstrating molecular changes in EMT. MiR-155 also decreased the C/EBP-β, TGF-β, and FOXO-3a expression in sensitive cells co-cultured with exosomes from CSCs and chemoresistant cells (Santos et al., 2018). CSCs have a self-renewal ability, which is in line with tumor recurrence, resistance, and metastasis against chemotherapeutic agents (Lee et al., 2016). These cells can arise from epithelial cells subjected to EMT, determined by E-cadherin loss of expression accompanied by the upregulation of transcription factors such as BMI1 and EZH2. Therefore, the epithelial cell transformation into a mesenchymal state was triggered, resulting in the development of aggressive cancerous cells (Hiscox et al., 2006; Kajiyama et al., 2007; Dave et al., 2012; Proctor et al., 2013). Consistently, accumulating evidence has revealed the effects of overexpressed miR-155, as an oncomiR, on the development of chemoresistance in BC (Johansson et al., 2013; Ouyang et al., 2014; Shen et al., 2015; Yu et al., 2015; Chiu et al., 2016; Li et al., 2017b).
Sprouty2 was found to be deregulated in BC through involvement in metastasis- and invasion-related pathways targeted by miRNAs (Li et al., 2013). On the other hand, cyclin-dependent kinase (CDK) inhibitor p27, an essential factor in cancer cell cycle arrest, autophagy, and angiogenesis, as well as the PTEN, is targeted by miRNAs in regulating BC drug resistance (Li et al., 2013; Zhong et al., 2013). Further investigations have detected that exosomes secreted by ADM-resistant MCF-7 BC cells could transfer drug-resistance characteristics to recipient-sensitive cells through miR-222 delivery (Yu et al., 2016). Activation of the PTEN/Akt/FOXO1 signaling pathway is believed to be responsible for the underlying mechanisms through which miR-222 affects ADM resistance in BC cells. MiR-222 causes PTEN to be suppressed and also results in Akt overexpression and FOXO1 downmodulation. The suppression of the PTEN/Akt/FOXO1 axis is linked to increased ADM sensitivity in BC cells (Shen et al., 2017). The upregulation of FOXO, following PTEN activation and Akt suppression, has represented tumor-suppressive effects such as the induction of cell cycle arrest and/or cancer cell apoptosis (Greer and Brunet, 2005). More recently, a study exhibited that lncRNA-GAS5 could alleviate ABCB1-mediated ADM resistance of BC cells by repressing miR-221-3p, which directly targeted Dickopf Wnt signaling pathway inhibitor 2 (DKK2). Subsequently, the Wnt/β-catenin signaling axis was activated, leading to the inhibition of ABCB1 expression and further alleviation of ADM resistance (Chen et al., 2020a).
6.1.4 Anti-Hsp90 drugs
Anti-Hsp90 drugs, such as 17-AAG or PU-H71, have shown promising anticancer effects in TNBC therapy during pre-clinical investigations (Eiseman et al., 2005; Caldas-Lopes et al., 2009; Proia et al., 2014). O’Brien et al. observed that EV-derived miR-134 was downmodulated in Hs578T TNBC cells compared to normal breast cells, in which it was shown to be underexpressed in TNBC aggressive clonal variant (Hs578 Ts(i)8) cells. Indeed, miR-134 loss is linked to elevated cellular aggressiveness. Also, miR-134 could act as a potential tumor suppressor by inhibiting STAT5B, which, in turn, reduced the Hsp90 and Bcl-2 expression levels. It has previously been revealed that STAT5B is involved in BC tumorigenesis and increases the transcription of Hsp90 (Perotti et al., 2008), which enhances the survival and apoptotic resistance of BC cells (Workman et al., 2007; Gallerne et al., 2013). Direct delivery of miR-134 caused suppression of TNBC cell proliferation and promotion of cisplatin-induced apoptosis, whereas delivery of exosomal miR-134 inhibited cell proliferation, migration, and invasion, as well as the enhancement of sensitivity to anti-Hsp90 drugs (O'Brien et al., 2015). MiR-134 originates from the 14q32 locus, a region usually deleted in tumor progression (O'Brien et al., 2015; Takayama et al., 1992; Kerangueven et al., 1997; Bando et al., 1999; Haller et al., 2010; Gattolliat et al., 2011). In line with this finding, other observations have reported that miR-134 has a tumor-suppressive role and its levels are inversely associated with tumor progression (Li et al., 2012; Sarver et al., 2013; Yin et al., 2013).
6.1.5 Cisplatin
Cisplatin (DDP) is another effective chemotherapeutic agent for BC therapy, especially for TNBCs that show an ineffective response to anti-HER-2 therapies (Silver et al., 2010). Although patients initially exhibit positive responses against DDP, drug resistance remains a major challenge that causes failure in treatment (Smith et al., 2007). Jia et al. have revealed that adipose mesenchymal stem cell (ADMSC)-derived exosomes, containing miR-1236, can decrease BC cell resistance against DDP by inhibiting the solute carrier family 9 member A1 (SLC9A1) Na+/H+ anti-porter and inactivating the Wnt/β-catenin signaling pathway (Jia et al., 2020). Indeed, miR-1236 directly binds to SLC9A1 and suppresses its expression, which is notably overexpressed in DDP-resistant BC cells. In addition, ADMSC-derived exosomes induce a further increase in caspase-3, promoting cell apoptosis in DDP-resistant BC cells (Jia et al., 2020; Asadi et al., 2022). Effective functions of ADMSC-derived exosomes in cancer control and treatment have been reported to be exerted by regulating various cellular behaviors, like proliferation, migration, and apoptosis, by means of miRNA cargos (Reza et al., 2016; Hong et al., 2019). It has been noted that SCL9A1 acts as an oncogene and is involved in the development of drug-resistant BC cells (Chen et al., 2019b). SLC9A1 was also discovered to be a Wnt/β-catenin signaling activator, participating in BC carcinogenesis, metastasis, and tumor progression (Prosperi and Goss, 2010; Sun et al., 2018b). Recently, it has been revealed that about 60 miRNAs, particularly miR-423-5p, miR-370-3p, and miR-373, are significantly upregulated in exosomes from DDP-resistant TNBC cells compared to DDP-sensitive ones. DDP-resistant cell-derived exosomes could induce DDP resistance in recipient cells by miR-423-5p delivery, which leads to P-gp overexpression, invasion, and migration, as well as apoptosis suppression (Wang et al., 2019a).
Yu et al. indicated the underexpression of miR-342-3p in samples collected from patients with metastatic and refractory BC types. Their investigations further revealed that the MSC-derived exosomes consisting of miR-342-3p could inhibit invasion, metastasis, and chemoresistance of BC cells to doxorubicin, fluorouracil, and cisplatin by suppressing the inhibitor of differentiation 4 (ID4). Also, ID4 silencing significantly reduced tumor growth and drug resistance and influenced the EMT by upregulating the E-cadherin and downregulating the N-cadherin and SNAIL (Yu et al., 2022). A similar investigation on TNBC patients declared that miR-342-3p loss of function contributed to metabolic carcinogenic pathways in TNBC by overexpression of MCT1 (Romero-Cordoba et al., 2018).
Recent in vivo and in vitro findings have shown that exosomes derived from MDA-MB-231 cells developed resistance-related mechanisms in BC cells by transferring miR-887-3p, which was overexpressed in BC cells. MiR-887-3p induced drug resistance through negative BTBD7 regulation, resulting in the activation of the Notch1/Hes1 signaling pathway (Wang et al., 2022a). In contrast, when miR-887-3p is inhibited in exosomes, drug resistance and tumor development are reduced, while BC cell apoptosis is increased (Wang et al., 2022a). Upregulation of miR-887-3p was observed in BC cell lines, and the suppressed miR-887-3p improved BC cell sensitivity to 5-fluorouracil therapy (Lv et al., 2020b). BTBD7 plays an important role in tumorigenesis, in which its expression can be declined in BC cells and tissues. BTBD7 expression is associated with low recurrence and repressed BC progression through inactivating Notch1 signaling (Chen et al., 2020b). Notch1 participates in cell growth, differentiation, and apoptosis. Notch1 activation was also proven in TNBC, as a facilitator of TNBC formation (Miao et al., 2020).
6.1.6 Trastuzumab
Currently, trastuzumab (TZB), an anti-HER2 monoclonal antibody, has been introduced as an effective agent to cure human HER-2-positive BC (Daniels et al., 2018), while its effectiveness can mostly be restricted by chemoresistance-associated mechanisms (Adamczyk et al., 2018). In this context, Han et al. showed significant downregulation of miR-567 in TZB-resistant BC cells compared to the sensitive ones. Overexpression of miR-567 reversed TZB resistance, but miR-567 downregulation could induce TZB resistance, in vivo and in vitro. Functionally, miR-567 can be transferred to recipient BC cells by packaging into exosomes and reverses the chemoresistance by suppressing the autophagic flux, which is mediated by inhibiting the expression of Atg5, post-transcriptionally (Han et al., 2020). Autophagy-related gene 5 (Atg5) has a key function in the early stages of autophagy, i.e., autophagosome formation, and is associated with cell differentiation and carcinogenesis (Le Bars et al., 2014; Rai et al., 2019). Growing evidence has demonstrated the essential role of autophagy in preserving the survival of tumor cells in adverse conditions (Degenhardt et al., 2006). Additionally, the inhibitory functions of miR-567 in the oncogenesis of BC have been proven (Bertoli et al., 2017). Another investigation elucidated that exosomal miR-155 and miR-1246 were the principal overexpressed miRNAs in HER2-positive BC with TZB resistance, suggesting a remarkable prognostic value for these miRNAs (Zhang et al., 2020).
6.2 Exosomal lncRNAs and BC chemoresistance
6.2.1 Tamoxifen
The overexpression of exosomal lncRNA urothelial carcinoma-associated 1 (UCA1), isolated from TAM-resistant LCC2 cells compared to TAM-sensitive MCF-7 cells, was an interesting finding in the field of exosomal lncRNA–BC drug resistance relationships. Exosome-transmitted lncRNA UCA1 was linked to TAM resistance in ER-positive BC cells, which might act by suppressing cleaved caspase-3 expression and cell apoptosis (Xu et al., 2016). The oncogenic function of UCA1 in BCs was identified through recruiting various mechanisms, such as inhibition of p27 (Huang et al., 2014) and acting as a miR-143 sponge (Tuo et al., 2015).
Chen et al. reported an association between the expression of myeloid-specific lncRNA and hypoxia-inducible factor 1α (HIF-1α)-stabilizing lncRNA (HISLA) in tumor-associated macrophages (TAMs) with poor chemotherapeutic responses and survival rates in BC patients (Chen et al., 2019c). The findings revealed that EV-mediated transmission of HISLA promoted apoptotic resistance and aerobic glycolysis in BC cells. Functionally, HISLA suppressed the hydroxylation and degradation of HIF-1α by blocking the interaction between HIF-1α and prolyl hydroxylase domain protein 2 (PHD2) (Chen et al., 2019c). Metabolic reprogramming is an indicator for developing cancer cells (Hay, 2016), with frequently reported deterioration of glucose metabolism in BC (Lloyd et al., 2016; Ibrahim-Hashim et al., 2017). HIF-1α, an oxygen-sensing transcription factor, is known to determine glucose metabolism by oxidation or glycolysis in cancer cells, which could be considered adaptive changes to TAM circumstances (Hsu and Sabatini, 2008; DeBerardinis and Chandel, 2016). Under physiological conditions, HIF-1α can be quickly hydroxylated and degraded by PHD2; however, multiple factors involved in TAM may modulate PHD2 activity to inhibit HIF-1α degradation, leading to the maintenance of HIF-1α protein, and promote aerobic glycolysis in cancer cells (Hsu and Sabatini, 2008; Semenza, 2012; Semenza, 2013).
6.2.2 Trastuzumab
LncRNA SNHG14 is a well-studied lncRNA promoted in TZB-resistant BC cells through overexpression of the poly (a)-binding protein cytoplasmic 1 (PABPC1) gene, leading to Nrf2 signaling activation (Dong et al., 2018). PABPs can participate in sequence-specific interactions with single-stranded poly (A) via an RNA recognition motif (RRM). In the cytoplasm, PABPs are categorized as PABPC, and inside the nucleus, they are known as PABPN1 (Burgess et al., 2011).
The lncRNA AGAP2 antisense RNA 1 (AGAP2-AS1) is another lncRNA having high expression levels in TZB-resistant BC cells and promotes resistance by reducing TZB-induced apoptosis. Thus, silencing of AGAP2-AS1 could re-sensitize BC cells to TZB-induced cytotoxicity. AGAP2-AS1 could also disseminate TZB resistance through packaging into exosomes (Zheng et al., 2019). According to the study conducted by Qian et al., exosomal AGAP2-AS1 caused TZB resistance by inducing autophagy in HER2-positive BC cells. In detail, the corresponding exosomal lncRNA exerts its effects by promoting Atg10 expression. Mechanistically, the embryonic lethal abnormal visual system (ELAV)-like RNA-binding protein 1 (ELAVL1) interacted with and stabilized the AGAP2-AS1, and subsequently, the AGAP2-AS1–ELAVL1 complex directly attached to and promoted H3K4 trimethylation, as well as H3K27 acetylation at the Atg10’s promoter region, thus resulting in activation of Atg10 transcription (Qian et al., 2021). AGAP2-AS1-targeting antisense oligonucleotides (ASOs) promoted TZB-related cytotoxicity. The elevated serum levels of exosomal AGAP2-AS1 were linked to poor response against TZB treatment (Luo et al., 2020; Qian et al., 2021).
6.2.3 Doxorubicin
It has been found that lncRNA H19 is upregulated in doxorubicin-resistant BC cells, while its suppression significantly decreases doxorubicin resistance by reducing cell viability, colony-forming ability, and inducing apoptosis. H19 can disseminate doxorubicin resistance among sensitive cells by being packaged into exosomes (Wang et al., 2020). H19 levels have been reported to be elevated in serum samples from patients unresponsive to doxorubicin (Wang et al., 2020). H19 also participates in the onset and development of BC through multiple mechanisms (Collette et al., 2017). For instance, H19 results in the overexpression of transcriptional factor LIN28, which has a critical role in BC stem cell maintenance, by sponging miRNA let-7 (Peng et al., 2017). Moreover, a previous study has demonstrated that H19 suppresses apoptosis in ERα-positive BCs, thereby promoting paclitaxel (PTX) resistance by repressing transcription of the Bik pro-apoptotic gene (Si et al., 2016). H19 was also reported to mediate doxorubicin resistance in BC cells by regulating the cullin4A–MDR1 signaling pathway (Zhu et al., 2017).
6.3 Exosomal circRNAs and BC drug resistance
6.3.1 Tamoxifen
The upregulation of circRNA UBE2D2 (circ-UBE2D2) was elucidated by Hu et al. in TAM-resistant ERα-positive BC cells. TAM-resistant cells could release exosomes containing circ-UBE2D2, thus inducing TAM resistance in drug-sensitive BC cells. Mechanistically, circ-UBE2D2 interacts with miR-200a-3p, and subsequently decreases its function, thereby promoting metastasis, cancer cell viability, and TAM resistance (Hu et al., 2020b). It has been shown that miR-200a-3p is a target for LINC00894-002 to contribute to TAM resistance in MCF-7 BC cells (Zhang et al., 2018). Previously, circ-UBE2D2 was displayed to be overexpressed in BC; therefore, blocking the circ-UBE2D2 could lead to the inhibition of BC tumorigenesis through targeting miR-1236 or miR-1287 (Wang et al., 2019b).
6.3.2 Lapatinib
Despite lapatinib (LAP) being a small-molecule tyrosine kinase inhibitor with high levels of effectiveness and limited side effects for treating HER2-positive BC (Rusnak et al., 2001; Chan, 2006), innate or acquired LAP resistance has led to an obstacle for BC therapy (Liu et al., 2009; Wang et al., 2011). A recent assessment conducted by Wu et al. demonstrated the circ-MMP11 overexpression in LAP-resistant cells, which could be transported to sensitive cells through exosomes to induce cell viability, invasion, migration, and repressing apoptosis in LAP-sensitive cells. Regarding the underlying mechanism, circ-MMP11 serves as a potential sponge for miR-153-3p, increasing the expression of anillin (ANLN) (Wu et al., 2021). ANLN is upregulated in BC, whose up-regulation contributes to drug resistance in BC cells against doxorubicin (Zhou et al., 2015). Additionally, circ-MMP11 (hsa_circ_0062558) has been reported with carcinogenic effects by serving as a miR-1204 competitive endogenous RNA (ceRNA) (Li et al., 2020b). MiR-153-3p is another circRNA with critical inhibitory roles in BC progression and LAP resistance (Yu et al., 2018).
6.3.3 Non-exosomal circRNAs
Although limited evidence is available with respect to the effects of exosomal circRNAs on the development of BC drug resistance, a large number of investigations have reported the crucial functions of non-exosomal circRNAs in chemotherapy resistance during BC progression. For instance, Wang et al. demonstrated that circATXN7 could act as a miR-149-5p sponge to upregulate homeobox A11 (HOXA11), resulting in increased doxorubicin resistance in BC cells (Wang et al., 2022b). Moreover, Hsa_circ_0092276 promoted resistance to doxorubicin in BC cells by modulating the ATG7 via sponging miR-384, leading to activated autophagy (Wang et al., 2021a).
CircWAC is another cirRNA that can induce PTX resistance in TNBC cells by suppressing the miR-142, a tumor suppressor miRNA, inducing the overexpression of WWP1 and activation of the PI3K/Akt pathway (Wang et al., 2021b). Consistently, circ_0006528 was found to partially participate in PTX resistance in BC cells by stimulating the expression of CDK8 by sponging miR-1299 (Liu et al., 2020). Further studies showed that the upmodulated circ-RNF111 was associated with PTX resistance, as well as cancer cell viability, cell invasion, and glycolysis. Indeed, circ-RNF111 is negatively correlated with miR-140-5p expression, hence triggering E2F3 in BC tissues (Zang et al., 2020).
Recently, it has been shown that circ_0085495 causes ADM resistance in BC cells by triggering the inhibition of miR-873-5p, and consequently the overexpression of integrin ß1 (Xie and Zheng, 2022). Similarly, the role of circ_0001667 in developing ADM resistance in BC cells was observed in another study, in which circ_0001667 knockdown attenuated ADM resistance, decreased cancer cell proliferation, and enhanced apoptosis by depleting nuclear receptor co-activator 3 (NCOA3) via releasing miR-4453 (Cui et al., 2022). Zhu et al. revealed that circFBXL5 might enhance 5-FU resistance in BC cells by promoting cell migration and invasion and inhibiting apoptosis by regulating the miR-216b/HMGA2 axis (Zhu et al., 2021). Another investigation identified that circ_0001598 promoted TZB resistance by modulating the miR-1184/programmed death-ligand 1 (PD-L1) signaling pathway in HER2-positive BC (Huang et al., 2021). CircFAT1 also promoted oxaliplatin resistance in BC through the activation of Notch and Wnt signaling pathways, principally through regulation of the miR-525-5p/SKA1 axis (Yao et al., 2021). CircUBAP2, another circRNA involved in drug resistance, could increase DDP resistance in TNBC cells by serving as a ceRNA for miR-300 to overexpress the anti-silencing function 1B histone chaperone (ASF1B), resulting in activation of the PI3K/Akt/mTOR signaling pathway (Wang et al., 2022c).
On the other hand, several circRNAs have shown a tumor-suppressive function; in this regard, hsa_circ_0025202 could reduce cancer cell proliferation, hinder tumor growth, promote apoptosis, and enhance TAM sensitivity in BC cells. Also, it has been shown that hsa_circ_0025202 exerts its functions through binding to miR-197-3p, thereby increasing HIPK3 expression (Li et al., 2021b). Furthermore, circ_0025202 was found to improve TAM sensitivity in BC cells by targeting miR-182-5p, leading to the regulation of FOXO-3a expression and activity (Sang et al., 2019).
7 Conclusion and perspectives
Chemoresistance, whether inherent or acquired, contributes to poor prognosis and treatment failure in cancer patients. Elucidating the underlying mechanisms of drug resistance development might help researchers to suggest a more effective approach for treating patients. Considering drug resistance, BC therapy has proven to be complicated. Recently, exosomal ncRNAs have been surprisingly found to regulate drug resistance in BC cells and TAM. Beyond drug resistance, exosomal ncRNAs play a substantial role in the development of cancer-related behaviors, such as cell proliferation, angiogenesis, and migration, by regulating several molecular mechanisms (Figure 3). As exosomal ncRNAs can be transferred through the bloodstream, they seem to have tremendous potential as diagnostic markers in clinical settings. Moreover, loading ncRNAs, as well as ncRNA agonists or inhibitors, into particular exosomes, and then delivering the exosomes into the bloodstream to be distributed among cancer cells can provide an exosome-based approach to treat malignancies. However, a wide variety of molecular mechanisms directing exosomal ncRNAs to a certain human cell or organ have remained unclear. In the current study, the contribution of exosomal ncRNAs and their target mechanisms/signaling pathways to BC drug resistance was highlighted in detail. The majority of findings reviewed here were the outcomes of in vitro evaluations, as there are limited human studies in this field. Notwithstanding, new insights were provided here that might help cancer researchers overcome drug resistance by designing exosomal ncRNAs-based therapeutics. Further assessments are definitely needed to elucidate the unclear aspects of BC chemoresistance in association with exosomal ncRNAs.
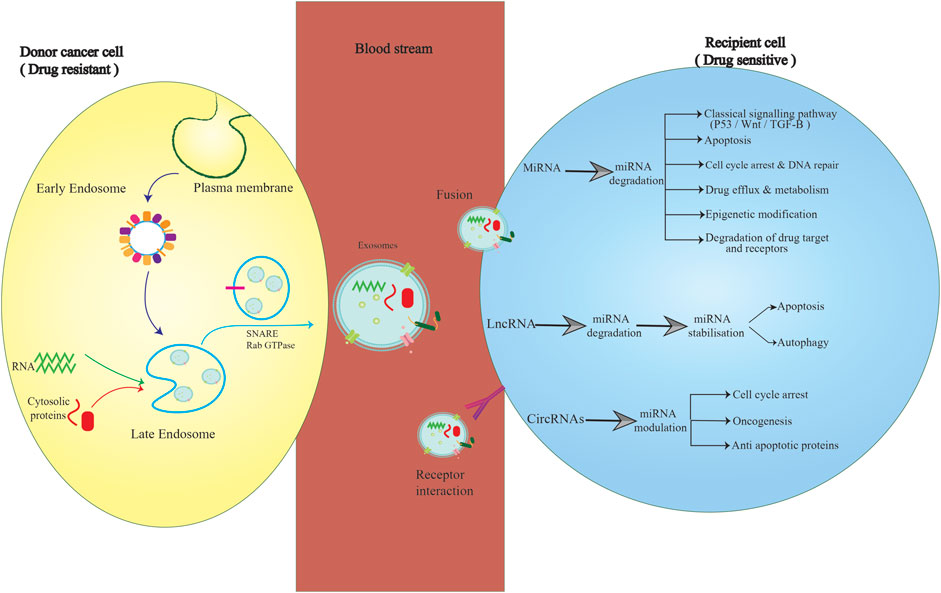
FIGURE 3. Exosomal delivery of ncRNAs between cancer and non-cancer cells. Exosomes derived from drug-resistant BC cells encapsulate and then deliver miRNAs, lncRNAs, and circRNAs to drug-sensitive cells, thus promoting multiple signaling pathways in recipient cells. This characteristic can be considered the basis for designing more effective chemotherapeutic approaches by attenuating drug resistance.
Author contributions
The authors confirm contributions to this paper as follows: Investigation and database searching, writing, and draft preparation: MaR, FM, AK, SY, OV, YM, VV, RD, SA, SG, MeR, NM, ZA, and AzA. Review, editing, and validation: OV, AM, HM, and MA. Supervision and project administration: MA, AbA, HM, and AM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamczyk, A., Kruczak, A., Harazin-Lechowska, A., Ambicka, A., Grela-Wojewoda, A., Domagała-Haduch, M., et al. (2018). Relationship between HER2 gene status and selected potential biological features related to trastuzumab resistance and its influence on survival of breast cancer patients undergoing trastuzumab adjuvant treatment. Onco Targets Ther. 11, 4525–4535. doi:10.2147/OTT.S166983
Ali, S., Rasool, M., Chaoudhry, H., Pushparaj, P. N., Jha, P., Hafiz, A., et al. (2016). Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation 12 (3), 135–139. doi:10.6026/97320630012135
Anastasiadou, E., Jacob, L. S., and Slack, F. J. (2018). Non-coding RNA networks in cancer. Nat. Rev. Cancer 18 (1), 5–18. doi:10.1038/nrc.2017.99
Asadi, M., Taghizadeh, S., Kaviani, E., Vakili, O., Taheri-Anganeh, M., Tahamtan, M., et al. (2022). Caspase-3: Structure, function, and biotechnological aspects. Biotechnol. Appl. Biochem. 69 (4), 1633–1645. doi:10.1002/bab.2233
Ashekyan, O., Abdallah, S., Shoukari, A. A., Chamandi, G., Choubassy, H., Itani, A. R. S., et al. (2022). Spotlight on exosomal non-coding RNAs in breast cancer: An in silico analysis to identify potential lncRNA/circRNA-miRNA-Target Axis. Int. J. Mol. Sci. 23 (15), 8351. doi:10.3390/ijms23158351
Babst, M. (2011). MVB vesicle formation: ESCRT-dependent, ESCRTindependent and everything in between. Curr. Opin. Cell Biol. 23, 452–457. doi:10.1016/j.ceb.2011.04.008
Bach, D. H., Hong, J. Y., Park, H. J., and Lee, S. K. (2017). The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. cancer 141 (2), 220–230. doi:10.1002/ijc.30669
Bahmyari, S., Jamali, Z., Khatami, S. H., Vakili, O., Roozitalab, M., Savardashtaki, A., et al. (2021). microRNAs in female infertility: An overview. Cell Biochem. Funct. 39 (8), 955–969. doi:10.1002/cbf.3671
Baietti, M. F. Z. Z., Mortier, E., Melchior, A., Degeest, G., Geeraerts, A., et al. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14 (7), 677–685. doi:10.1038/ncb2502
Bando, T., Kato, Y., Ihara, Y., Yamagishi, F., Tsukada, K., and Isobe, M. (1999). Loss of heterozygosity of 14q32 in colorectal carcinoma. Cancer Genet. Cytogenet. 111 (2), 161–165. doi:10.1016/s0165-4608(98)00242-8
Barbano, R., Pasculli, B., Rendina, M., Fontana, A., Fusilli, C., Copetti, M., et al. (2017). Stepwise analysis of MIR9 loci identifies miR-9-5p to be involved in Oestrogen regulated pathways in breast cancer patients. Sci. Rep. 7 (1), 45283–45312. doi:10.1038/srep45283
Bates, S., Rowan, S., and Vousden, K. H. (1996). Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13 (5), 1103–1109.
Beermann, J., Piccoli, M-T., Viereck, J., and Thum, T. (2016). Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol. Rev. 96, 1297–1325. doi:10.1152/physrev.00041.2015
Bertoli, G., Cava, C., Diceglie, C., Martelli, C., Rizzo, G., Piccotti, F., et al. (2017). MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res. Treat. 161 (3), 605–616. doi:10.1007/s10549-016-4079-2
Bliss, S. A., Sinha, G., Sandiford, O. A., Williams, L. M., Engelberth, D. J., Guiro, K., et al. (2016). Mesenchymal stem cell–derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Res. 76 (19), 5832–5844. doi:10.1158/0008-5472.CAN-16-1092
Bobrie, A. C. M., Raposo, G., and Thery, C. (2011). Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic (Copenhagen, Den. 12 (12), 1659–1668. doi:10.1111/j.1600-0854.2011.01225.x
Bovy, N., Blomme, B., Frères, P., Dederen, S., Nivelles, O., Lion, M., et al. (2015). Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget 6 (12), 10253–10266. doi:10.18632/oncotarget.3520
Brown, I., Shalli, K., McDonald, S. L., Moir, S. E., Hutcheon, A. W., Heys, S. D., et al. (2004). Reduced expression of p27 is a novel mechanism of docetaxel resistance in breast cancer cells. Breast cancer Res. 6 (5), R601–R607. doi:10.1186/bcr918
Bukong, T. N. M-H. F., Kodys, K., Bala, S., and Szabo, G. (2014). Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 10 (10), e1004424. doi:10.1371/journal.ppat.1004424
Bullock, M. D., Silva, A. M., Kanlikilicer-Unaldi, P., Filant, J., Rashed, M. H., Sood, A. K., et al. (2015). Exosomal non-coding RNAs: Diagnostic, prognostic and therapeutic applications in cancer. Noncoding RNA 1 (1), 53–68. doi:10.3390/ncrna1010053
Burgess, H. M., Richardson, W. A., Anderson, R. C., Salaun, C., Graham, S. V., and Gray, N. K. (2011). Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J. Cell Sci. 124 (19), 3344–3355. doi:10.1242/jcs.087692
Caldas-Lopes, E., Cerchietti, L., Ahn, J. H., Clement, C. C., Robles, A. I., Rodina, A., et al. (2009). Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc. Natl. Acad. Sci. 106 (20), 8368–8373. doi:10.1073/pnas.0903392106
Cech, T. R., and Steitz, J. A. (2014). The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 157 (1), 77–94. doi:10.1016/j.cell.2014.03.008
Cejalvo, J. M., Pascual, T., Fernández-Martínez, A., Brasó-Maristany, F., Gomis, R. R., Perou, C. M., et al. (2018). Clinical implications of the non-luminal intrinsic subtypes in hormone receptor-positive breast cancer. Cancer Treat. Rev. 67, 63–70. doi:10.1016/j.ctrv.2018.04.015
Chahar, H. S. B. X., and Casola, A. (2015). Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses 19 (7), 3204–3225. doi:10.3390/v7062770
Chairoungdua, A. S. D., Pochard, P., Hull, M., and Caplan, M. J. (2010). Exosome release of beta-: A novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 190 (6), 1079–1091. doi:10.1083/jcb.201002049
Chan, A. (2006). Lapatinib - overview and current role in metastatic breast cancer. Cancer Res. Treat. 38 (4), 198–200. doi:10.4143/crt.2006.38.4.198
Chan, J. J. T. Y. (2018). Noncoding RNA:RNA regulatory networks in cancer. Int. J. Mol. Sci. 19 (5), 1310.
Chang, Y-Y., Kuo, W-H., Hung, J-H., Lee, C-Y., Lee, Y-H., Chang, Y-C., et al. (2015). Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol. cancer 14 (1), 36–13. doi:10.1186/s12943-015-0301-9
Chen, F., Chen, J., Yang, L., Liu, J., Zhang, X., Zhang, Y., et al. (2019). Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 21 (4), 498–510. doi:10.1038/s41556-019-0299-0
Chen, J., Lai, Y-H., Ooi, S., Song, Y., Li, L., and Liu, T-Y. (2020). BTB domain-containing 7 predicts low recurrence and suppresses tumor progression by deactivating Notch1 signaling in breast cancer. Breast Cancer Res. Treat. 184 (2), 287–300. doi:10.1007/s10549-020-05857-2
Chen, L. L. Y. L., and Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. doi:10.1080/15476286.2015.1020271
Chen, Q., Li, Y., Liu, Y., Xu, W., and Zhu, X. (2021). Exosomal non-coding RNAs-mediated crosstalk in the tumor microenvironment. Front. Cell Dev. Biol. 9, 646864. doi:10.3389/fcell.2021.646864
Chen, Q., Liu, Y., Zhu, X. L., Feng, F., Yang, H., and Xu, W. (2019). Increased NHE1 expression is targeted by specific inhibitor cariporide to sensitize resistant breast cancer cells to doxorubicin in vitro and in vivo. BMC Cancer 19 (1), 211. doi:10.1186/s12885-019-5397-7
Chen, W-X., Cai, Y-Q., Lv, M-M., Chen, L., Zhong, S., Ma, T-F., et al. (2014). Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumor Biol. 35 (10), 9649–9659. doi:10.1007/s13277-014-2242-0
Chen, W-X., Liu, X-M., Lv, M-M., Chen, L., Zhao, J-H., Zhong, S., et al. (2014). Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PloS one 9 (4), e95240. doi:10.1371/journal.pone.0095240
Chen, W. X., Xu, L. Y., Cheng, L., Qian, Q., He, X., Peng, W. T., et al. (2019). Bioinformatics analysis of dysregulated microRNAs in exosomes from docetaxel-resistant and parental human breast cancer cells. Cancer Manag. Res. 11, 5425–5435. doi:10.2147/CMAR.S201335
Chen, Z., Pan, T., Jiang, D., Jin, L., Geng, Y., Feng, X., et al. (2020). The lncRNA-GAS5/miR-221-3p/DKK2 Axis modulates ABCB1-mediated adriamycin resistance of breast cancer via the wnt/β-catenin signaling pathway. Mol. Ther. Nucleic Acids 19, 1434–1448. doi:10.1016/j.omtn.2020.01.030
Chiu, C. F., Chang, Y. W., Kuo, K. T., Shen, Y. S., Liu, C. Y., Yu, Y. H., et al. (2016). NF-κB-driven suppression of FOXO3a contributes to EGFR mutation-independent gefitinib resistance. Proc. Natl. Acad. Sci. U. S. A. 113 (18), E2526–E2535. doi:10.1073/pnas.1522612113
Chung, S. J., Nagaraju, G. P., Nagalingam, A., Muniraj, N., Kuppusamy, P., Walker, A., et al. (2017). ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy 13 (8), 1386–1403. doi:10.1080/15548627.2017.1332565
Coleman, M., Quaresma, M., Berrino, F., Lutz, J., De Angelis, R., Capocaccia, R., et al. (2008). Cancer survival in five continents: A worldwide population-based study (CONCORD). Lancet Oncol. 9 (8), 730–756. doi:10.1016/S1470-2045(08)70179-7
Collette, J., Le Bourhis, X., and Adriaenssens, E. (2017). Regulation of human breast cancer by the long non-coding RNA H19. Int. J. Mol. Sci. 18 (11), 2319. doi:10.3390/ijms18112319
Colombo, M. R. G., and Thery, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. doi:10.1146/annurev-cellbio-101512-122326
Consortium, E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489 (7414), 57–74. doi:10.1038/nature11247
Coomans de Brachène, A., and Demoulin, J. B. (2016). FOXO transcription factors in cancer development and therapy. Cell Mol. Life Sci. 73 (6), 1159–1172. doi:10.1007/s00018-015-2112-y
Cosentino, G., Plantamura, I., Tagliabue, E., Iorio, M. V., and Cataldo, A. (2021). Breast cancer drug resistance: Overcoming the challenge by capitalizing on MicroRNA and tumor microenvironment interplay. Cancers (Basel). 13 (15), 3691. doi:10.3390/cancers13153691
Costa, F. F. (2005). Non-coding RNAs: New players in eukaryotic biology. Gene 357, 83–94. doi:10.1016/j.gene.2005.06.019
Cui, Y., Fan, J., Shi, W., and Zhou, Z. (2022). Circ_0001667 knockdown blocks cancer progression and attenuates adriamycin resistance by depleting NCOA3 via releasing miR-4458 in breast cancer. Drug Dev. Res. 83 (1), 75–87. doi:10.1002/ddr.21845
Cuzick, J., and Baum, M. (1985). Tamoxifen and contralateral breast cancer. Lancet 326 (8449), 282. doi:10.1016/s0140-6736(85)90338-1
Cuzick, J., Sestak, I., Cawthorn, S., Hamed, H., Holli, K., Howell, A., et al. (2015). Tamoxifen for prevention of breast cancer: Extended long-term follow-up of the IBIS-I breast cancer prevention trial. lancet Oncol. 16 (1), 67–75. doi:10.1016/S1470-2045(14)71171-4
Daniels, B., Kiely, B. E., Lord, S. J., Houssami, N., Lu, C. Y., Ward, R. L., et al. (2018). Long-term survival in trastuzumab-treated patients with HER2-positive metastatic breast cancer: Real-world outcomes and treatment patterns in a whole-of-population Australian cohort (2001-2016). Breast Cancer Res. Treat. 171 (1), 151–159. doi:10.1007/s10549-018-4804-0
Dave, B., Mittal, V., Tan, N. M., and Chang, J. C. (2012). Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res. 14 (1), 202. doi:10.1186/bcr2938
Dea, S., De Nardis, V., Marcantonio, P., Mandolini, C., Paganelli, C., Vitale, E., et al. (2014). Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: Effect of glycemic control. J. Clin. Endocrinol. Metab. 99, E1681–E1685. doi:10.1210/jc.2013-3843
DeBerardinis, R. J., and Chandel, N. S. (2016). Fundamentals of cancer metabolism. Sci. Adv. 2 (5), e1600200. doi:10.1126/sciadv.1600200
Degenhardt, K., Mathew, R., Beaudoin, B., Bray, K., Anderson, D., Chen, G., et al. (2006). Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10 (1), 51–64. doi:10.1016/j.ccr.2006.06.001
Dempsey, J. L. C. J., and Cui, J. Y. (2017). Long non-coding RNAs: A novel paradigm for toxicology. Toxicol. Sci. 155 (1), 3–21. doi:10.1093/toxsci/kfw203
DeSantis, C. E., Ma, J., Goding Sauer, A., Newman, L. A., and Jemal, A. (2017). Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 67 (6), 439–448. doi:10.3322/caac.21412
Dey, N., De, P., and Leyland-Jones, B. (2017). PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell signaling to clinical trials. Pharmacol. Ther. 175, 91–106. doi:10.1016/j.pharmthera.2017.02.037
Dong, H., Wang, W., Mo, S., Liu, Q., Chen, X., Chen, R., et al. (2018). Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J. Cell Mol. Med. 22 (10), 4935–4947. doi:10.1111/jcmm.13758
Dorostgou, Z., Yadegar, N., Dorostgou, Z., Khorvash, F., and Vakili, O. (2022). Novel insights into the role of circular RNAs in Parkinson disease: An emerging renaissance in the management of neurodegenerative diseases. J. Neurosci. Res. 100 (9), 1775–1790. doi:10.1002/jnr.25094
Eiseman, J. L., Lan, J., Lagattuta, T. F., Hamburger, D. R., Joseph, E., Covey, J. M., et al. (2005). Pharmacokinetics and pharmacodynamics of 17-demethoxy 17-[[(2-dimethylamino) ethyl] amino] geldanamycin (17DMAG, NSC 707545) in CB-17 SCID mice bearing MDA-MB-231 human breast cancer xenografts. Cancer Chemother. Pharmacol. 55 (1), 21–32. doi:10.1007/s00280-004-0865-3
Elsheikh, S., Green, A. R., Aleskandarany, M. A., Grainge, M., Paish, C. E., Lambros, M. B., et al. (2008). CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res. Treat. 109 (2), 325–335. doi:10.1007/s10549-007-9659-8
Ershaid, N., Sharon, Y., Doron, H., Raz, Y., Shani, O., Cohen, N., et al. (2019). NLRP3 inflammasome in fibroblasts links tissue damage with inflammation in breast cancer progression and metastasis. Nat. Commun. 10 (1), 4375. doi:10.1038/s41467-019-12370-8
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncomirs—microRNAs with a role in cancer. Nat. Rev. cancer 6 (4), 259–269. doi:10.1038/nrc1840
Falk Libby, E., Liu, J., Li, Y. I., Lewis, M. J., Demark-Wahnefried, W., and Hurst, D. R. (2016). Globular adiponectin enhances invasion in human breast cancer cells. Oncol. Lett. 11 (1), 633–641. doi:10.3892/ol.2015.3965
Fan, Q. Y. L., Zhang, X., Peng, X., Wei, S., Su, D., et al. (2018). The emerging role of exosomederived non-coding RNAs in cancer biology. Cancer Lett. 414, 107–115. doi:10.1016/j.canlet.2017.10.040
Fontana, F., Anselmi, M., and Limonta, P. (2022). Molecular mechanisms of cancer drug resistance: Emerging biomarkers and promising targets to overcome tumor progression. Cancers (Basel). 14 (7), 1614. doi:10.3390/cancers14071614
Freedman, A. N., Graubard, B. I., Rao, S. R., McCaskill-Stevens, W., Ballard-Barbash, R., and Gail, M. H. (2003). Estimates of the number of US women who could benefit from tamoxifen for breast cancer chemoprevention. J. Natl. Cancer Inst. 95 (7), 526–532. doi:10.1093/jnci/95.7.526
Fu, R., and Tong, J. S. (2020). miR-126 reduces trastuzumab resistance by targeting PIK3R2 and regulating AKT/mTOR pathway in breast cancer cells. J. Cell. Mol. Med. 24 (13), 7600–7608. doi:10.1111/jcmm.15396
Gallerne, C., Prola, A., and Lemaire, C. (2013). Hsp90 inhibition by PU-H71 induces apoptosis through endoplasmic reticulum stress and mitochondrial pathway in cancer cells and overcomes the resistance conferred by Bcl-2. Biochimica Biophysica Acta (BBA)-Molecular Cell Res. 1833 (6), 1356–1366. doi:10.1016/j.bbamcr.2013.02.014
Gao, M., Miao, L., Liu, M., Li, C., Yu, C., Yan, H., et al. (2016). miR-145 sensitizes breast cancer to doxorubicin by targeting multidrug resistance-associated protein-1. Oncotarget 7 (37), 59714–59726. doi:10.18632/oncotarget.10845
Gao, Y., Li, X., Zeng, C., Liu, C., Hao, Q., Li, W., et al. (2020). CD63(+) cancer-associated fibroblasts confer tamoxifen resistance to breast cancer cells through exosomal miR-22. Adv. Sci. (Weinh) 7 (21), 2002518. doi:10.1002/advs.202002518
Gattolliat, C., Thomas, L., Ciafre, S., Meurice, G., Le Teuff, G., Job, B., et al. (2011). Expression of miR-487b and miR-410 encoded by 14q32. 31 locus is a prognostic marker in neuroblastoma. Br. J. cancer 105 (9), 1352–1361. doi:10.1038/bjc.2011.388
Gray, M., Turnbull, A. K., Ward, C., Meehan, J., Martínez-Pérez, C., Bonello, M., et al. (2019). Development and characterisation of acquired radioresistant breast cancer cell lines. Radiat. Oncol. 14 (1), 64. doi:10.1186/s13014-019-1268-2
Greer, E. L., and Brunet, A. (2005). FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24 (50), 7410–7425. doi:10.1038/sj.onc.1209086
Gregory, R. I., and Shiekhattar, R. (2005). MicroRNA biogenesis and cancer. Cancer Res. 65 (9), 3509–3512. doi:10.1158/0008-5472.CAN-05-0298
Haffty, B. G., Yang, Q., Reiss, M., Kearney, T., Higgins, S. A., Weidhaas, J., et al. (2006). Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 24 (36), 5652–5657. doi:10.1200/JCO.2006.06.5664
Haller, F., Von Heydebreck, A., Zhang, J. D., Gunawan, B., Langer, C., Ramadori, G., et al. (2010). Localization-and mutation-dependent microRNA (miRNA) expression signatures in gastrointestinal stromal tumours (GISTs), with a cluster of co-expressed miRNAs located at 14q32. 31. J. Pathology A J. Pathological Soc. G. B. Irel. 220 (1), 71–86. doi:10.1002/path.2610
Hammond, M. E., Hayes, D. F., Dowsett, M., Allred, D. C., Hagerty, K. L., Badve, S., et al. (2010). American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 134 (7), e48–e72. doi:10.5858/134.7.e48
Han, M., Hu, J., Lu, P., Cao, H., Yu, C., Li, X., et al. (2020). Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 11 (1), 43. doi:10.1038/s41419-020-2250-5
Harbeck, N., and Gnant, M. (2017). Breast cancer. Lancet 389 (10074), 1134–1150. doi:10.1016/S0140-6736(16)31891-8
Hasegawa, S., Eguchi, H., Nagano, H., Konno, M., Tomimaru, Y., Wada, H., et al. (2014). MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br. J. cancer 111 (8), 1572–1580. doi:10.1038/bjc.2014.454
Hay, N. (2016). Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 16 (10), 635–649. doi:10.1038/nrc.2016.77
Hayes, J. P. P., and Lawler, S. (2014). MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 20 (8), 460–469. doi:10.1016/j.molmed.2014.06.005
He, Y., Deng, F., Yang, S., Wang, D., Chen, X., Zhong, S., et al. (2018). Exosomal microRNA: A novel biomarker for breast cancer. Biomark. Med. 12, 177–188. doi:10.2217/bmm-2017-0305
Henne, W. M. B. N., and Emr, S. D. (2011). The ESCRT pathway. Dev. Cell 21 (1), 77–91. doi:10.1016/j.devcel.2011.05.015
Heys, S. D., Hutcheon, A. W., Sarkar, T. K., Ogston, K. N., Miller, I. D., Payne, S., et al. (2002). Neoadjuvant docetaxel in breast cancer: 3-year survival results from the aberdeen trial. Clin. breast cancer 3, S69–S74. doi:10.3816/cbc.2002.s.015
Hiscox, S., Jiang, W. G., Obermeier, K., Taylor, K., Morgan, L., Burmi, R., et al. (2006). Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int. J. Cancer 118 (2), 290–301. doi:10.1002/ijc.21355
Hombach, S. K. M., and Kretz, M. (2016). Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 937, 3–17. doi:10.1007/978-3-319-42059-2_1
Hong, P., Yang, H., Wu, Y., Li, K., and Tang, Z. (2019). The functions and clinical application potential of exosomes derived from adipose mesenchymal stem cells: A comprehensive review. Stem Cell Res. Ther. 10 (1), 242. doi:10.1186/s13287-019-1358-y
Hsu, P. P., and Sabatini, D. M. (2008). Cancer cell metabolism: Warburg and beyond. Cell 134 (5), 703–707. doi:10.1016/j.cell.2008.08.021
Hu, C., Meiners, S., Lukas, C., Stathopoulos, G. T., and Chen, J. (2020). Role of exosomal microRNAs in lung cancer biology and clinical applications. Cell Prolif. 53, e12828. doi:10.1111/cpr.12828
Hu, K., Liu, X., Li, Y., Li, Q., Xu, Y., Zeng, W., et al. (2020). Exosomes mediated transfer of Circ_UBE2D2 enhances the resistance of breast cancer to tamoxifen by binding to MiR-200a-3p. Med. Sci. Monit. 26, e922253. doi:10.12659/MSM.922253
Huang, A. Z. H., Wu, Z., Chen, M., and Huang, Y. (2020). Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 10 (8), 3503–3517. doi:10.7150/thno.42174
Huang, J., Zhou, N., Watabe, K., Lu, Z., Wu, F., Xu, M., et al. (2014). Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 5 (1), e1008. doi:10.1038/cddis.2013.541
Huang, L., Ma, J., and Cui, M. (2021). Circular RNA hsa_circ_0001598 promotes programmed death-ligand-1-mediated immune escape and trastuzumab resistance via sponging miR-1184 in breast cancer cells. Immunol. Res. 69 (6), 558–567. doi:10.1007/s12026-021-09237-w
Huang, P., Li, F., Li, L., You, Y., Luo, S., Dong, Z., et al. (2018). lncRNA profile study reveals the mRNAs and lncRNAs associated with docetaxel resistance in breast cancer cells. Sci. Rep. 8 (1), 17970–18015. doi:10.1038/s41598-018-36231-4
Huarte, M., and Rinn, J. L. (2010). Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet. 19 (R2), R152–R161. doi:10.1093/hmg/ddq353
Hung, C. L. W. L., Yu, Y. L., Chen, H. W., Srivastava, S., Petrovics, G., et al. (2014). A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. U. S. A. 111 (52), 18697–18702. doi:10.1073/pnas.1415669112
Huotari, J. H. A., and Helenius, A. (2011). Endosome maturation. EMBO J. 30 (17), 3481–3500. doi:10.1038/emboj.2011.286
Ibrahim-Hashim, A., Robertson-Tessi, M., Enriquez-Navas, P. M., Damaghi, M., Balagurunathan, Y., Wojtkowiak, J. W., et al. (2017). Defining cancer subpopulations by adaptive strategies rather than molecular properties provides novel insights into intratumoral evolution. Cancer Res. 77 (9), 2242–2254. doi:10.1158/0008-5472.CAN-16-2844
Janas, T. J. M., Sapoń, K., and Janas, T. (2015). Mechanisms of RNA loading into exosomes. FEBS Lett. 589, 1391–1398. doi:10.1016/j.febslet.2015.04.036
Jea, C. J., Yang, G., Yan, Q. q., Zhao, J., and Li, S. (2019). Exosome-encapsulated microRNAs as promising biomarkers for Alzheimer’s disease. Rev. Neurosci. 31, 77–87. doi:10.1515/revneuro-2019-0001
Jeck, W. R. S. J., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi:10.1261/rna.035667.112
Jeck, W. R. S. N., and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 (5), 453–461. doi:10.1038/nbt.2890
Jh, H. (2008). ESCRT complexes and the biogenesis of multivesicular bodies. Cur Opin. Cell Biol. 20 (1), 4–11. doi:10.1016/j.ceb.2007.12.002
Ji, X., Lu, Y., Tian, H., Meng, X., Wei, M., and Cho, W. C. (2019). Chemoresistance mechanisms of breast cancer and their countermeasures. Biomed. Pharmacother. 114, 108800. doi:10.1016/j.biopha.2019.108800
Jia, Z., Zhu, H., Sun, H., Hua, Y., Zhang, G., Jiang, J., et al. (2020). Adipose mesenchymal stem cell-derived exosomal microRNA-1236 reduces resistance of breast cancer cells to cisplatin by suppressing SLC9A1 and the wnt/β-catenin signaling. Cancer Manag. Res. 12, 8733–8744. doi:10.2147/CMAR.S270200
Jiang, L. G. Y., Du, Y., and Liu, J. (2019). Exosomes: Diagnostic biomarkers and therapeutic delivery vehicles for cancer. Mol. Pharm. 16 (8), 3333–3349. doi:10.1021/acs.molpharmaceut.9b00409
Jiao, Y., Xu, P., Shi, H., Chen, D., and Shi, H. (2021). Advances on liver cell-derived exosomes in liver diseases. J. Cell Mol. Med. 25, 15–26. doi:10.1111/jcmm.16123
Johansson, J., Berg, T., Kurzejamska, E., Pang, M. F., Tabor, V., Jansson, M., et al. (2013). MiR-155-mediated loss of C/EBPβ shifts the TGF-β response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene 32 (50), 5614–5624. doi:10.1038/onc.2013.322
Jones, R. B., Holland, J. F., Bhardwaj, S., Norton, L., Wilfinger, C., and Strashun, A. (1987). A phase I-II study of intensive-dose adriamycin for advanced breast cancer. J. Clin. Oncol. 5 (2), 172–177. doi:10.1200/JCO.1987.5.2.172
Kajiyama, H., Shibata, K., Terauchi, M., Yamashita, M., Ino, K., Nawa, A., et al. (2007). Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int. J. Oncol. 31 (2), 277–283. doi:10.3892/ijo.31.2.277
Kalra, H. D. G., and Mathivanan, S. (2016). Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 17 (2), 170. doi:10.3390/ijms17020170
Kasinski, A. L., and Slack, F. J. (2011). Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 11 (12), 849–864. doi:10.1038/nrc3166
Kerangueven, F., Noguchi, T., Coulier, F., Allione, F., Wargniez, V., Simony-Lafontaine, J., et al. (1997). Genome-wide search for loss of heterozygosity shows extensive genetic diversity of human breast carcinomas. Cancer Res. 57 (24), 5469–5474.
Kim, I. S., and Zhang, X. H. (2016). One microenvironment does not fit all: Heterogeneity beyond cancer cells. Cancer Metastasis Rev. 35 (4), 601–629. doi:10.1007/s10555-016-9643-z
Klinge, C. M. (2018). Non-coding RNAs in breast cancer: Intracellular and intercellular communication. Noncoding RNA 4 (4), 40. doi:10.3390/ncrna4040040
Koike Folgueira, M. A. A., Carraro, D. M., Brentani, H., da Costa Patrão, D. F., Barbosa, E. M., Netto, M. M., et al. (2005). Gene expression profile associated with response to doxorubicin-based therapy in breast cancer. Clin. Cancer Res. 11 (20), 7434–7443. doi:10.1158/1078-0432.CCR-04-0548
Kozomara, A. B. M., and Griffiths-Jones, S. (2018). Mirbase: From microRNA sequences to function. Nucleic Acids Res. 47 (D1), D155–D162. doi:10.1093/nar/gky1141
Krichevsky, A. M., King, K. S., Donahue, C. P., Khrapko, K., and Kosik, K. S. (2003). A microRNA array reveals extensive regulation of microRNAs during brain development. Rna 9 (10), 1274–1281. doi:10.1261/rna.5980303
Kristensen, L. S. A. M., Stagsted, L., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Kuang, X-Y., Chen, L., Zhang, Z-J., Liu, Y-R., Zheng, Y-Z., Ling, H., et al. (2015). Stathmin and phospho-stathmin protein signature is associated with survival outcomes of breast cancer patients. Oncotarget 6 (26), 22227–22238. doi:10.18632/oncotarget.4276
Kuang, X-Y., Jiang, H-S., Li, K., Zheng, Y-Z., Liu, Y-R., Qiao, F., et al. (2016). The phosphorylation-specific association of STMN1 with GRP78 promotes breast cancer metastasis. Cancer Lett. 377 (1), 87–96. doi:10.1016/j.canlet.2016.04.035
Kutanzi, K. R., Yurchenko, O. V., Beland, F. A., Checkhun, V. F., and Pogribny, I. P. (2011). MicroRNA-mediated drug resistance in breast cancer. Clin. epigenetics 2 (2), 171–185. doi:10.1007/s13148-011-0040-8
Le Bars, R., Marion, J., Le Borgne, R., Satiat-Jeunemaitre, B., and Bianchi, M. W. (2014). ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5, 4121. doi:10.1038/ncomms5121
Lee, G., Hall, R. R., and Ahmed, A. U. (2016). Cancer stem cells: Cellular plasticity, niche, and its clinical relevance. J. Stem Cell Res. Ther. 6 (10), 363. doi:10.4172/2157-7633.1000363
Lee, H. J., Shin, S., Kang, J., Han, K. C., Kim, Y. H., Bae, J. W., et al. (2020). HSP90 inhibitor, 17-DMAG, alone and in combination with lapatinib attenuates acquired lapatinib-resistance in ER-positive, HER2-overexpressing breast cancer cell line. Cancers (Basel) 12 (9), 2630. doi:10.3390/cancers12092630
Legnini, I. D. T. G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9. doi:10.1016/j.molcel.2017.02.017
Li, B. H. J., Hong, M., Wang, Y., Yu, T., Zang, S., et al. (2019). piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene 38 (26), 5227–5238. doi:10.1038/s41388-019-0788-4
Li, C., Ni, Y. Q., Xu, H., Xiang, Q. Y., Zhao, Y., Zhan, J. K., et al. (2021). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 6 (1), 383. doi:10.1038/s41392-021-00779-x
Li, C., and Xu, X. (2019). Biological functions and clinical applications of exosomal non-coding RNAs in hepatocellular carcinoma. Cell. Mol. life Sci. CMLS 76 (21), 4203–4219. doi:10.1007/s00018-019-03215-0
Li, D. P., Fan, J., Wu, Y. J., Xie, Y. F., Zha, J. M., and Zhou, X. M. (2017). MiR-155 regulated by TGF-β promotes epithelial-mesenchymal transition, invasion and metastasis of human hepatocellular carcinoma cells in vitro. Am. J. Transl. Res. 9 (6), 2956–2965.
Li, H., Li, Q., and He, S. (2021). Hsa_circ_0025202 suppresses cell tumorigenesis and tamoxifen resistance via miR-197-3p/HIPK3 axis in breast cancer. World J. Surg. Oncol. 19 (1), 39–12. doi:10.1186/s12957-021-02149-x
Li, J. L. C., and Liu, C. (2019). Coding or noncoding, the converging concepts of RNAs. Front. Genet. 22 (10), 496. doi:10.3389/fgene.2019.00496
Li, J. S. D., Pu, W., Wang, J., and Peng, Y. (2020). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 6 (4), 319–336. doi:10.1016/j.trecan.2020.01.012
Li, J., Wang, Y., Luo, J., Fu, Z., Ying, J., Yu, Y., et al. (2012). miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 586 (20), 3761–3765. doi:10.1016/j.febslet.2012.09.016
Li, S., Yao, J., Xie, M., Liu, Y., and Zheng, M. (2018). Exosomal miRNAs in hepatocellular carcinoma development and clinical responses. J. Hematol. Oncol. 54 (11), 54. doi:10.1186/s13045-018-0579-3
Li, X., Liu, X., Xu, W., Zhou, P., Gao, P., Jiang, S., et al. (2013). c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 288 (25), 18121–18133. doi:10.1074/jbc.M113.478560
Li, X. J., Ren, Z. J., Tang, J. H., and Yu, Q. (2017). Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell. Physiology Biochem. 44 (5), 1741–1748. doi:10.1159/000485780
Li, Y., Liang, Y., Sang, Y., Song, X., Zhang, H., Liu, Y., et al. (2018). MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell death Dis. 9 (1), 14–12. doi:10.1038/s41419-017-0030-7
Li, Z., Chen, Z., Feng, Y., Hu, G., and Jiang, Y. (2020). CircMMP11 acts as a ce-circRNA in breast cancer progression by regulating miR-1204. Am. J. Transl. Res. 12 (6), 2585–2599.
Lima, L. G. C. R., Monteiro, R. Q., Moreira, M. E., and Barcinski, M. A. (2009). TumorDerived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 283, 168–175. doi:10.1016/j.canlet.2009.03.041
Lin, S-S., Peng, C-Y., Liao, Y-W., Chou, M-Y., Hsieh, P-L., and Yu, C-C. (2018). miR-1246 targets CCNG2 to enhance cancer stemness and chemoresistance in oral carcinomas. Cancers 10 (8), 272. doi:10.3390/cancers10080272
Liu, G., Zhang, Z., Song, Q., Guo, Y., Bao, P., and Shui, H. (2020). Circ_0006528 contributes to paclitaxel resistance of breast cancer cells by regulating miR-1299/CDK8 Axis. Onco Targets Ther. 13, 9497–9511. doi:10.2147/OTT.S252886
Liu, J., Zhu, S., Tang, W., Huang, Q., Mei, Y., and Yang, H. (2021). Exosomes from tamoxifen-resistant breast cancer cells transmit drug resistance partly by delivering miR-9-5p. Cancer Cell Int. 21 (1), 55–15. doi:10.1186/s12935-020-01659-0
Liu, L., Greger, J., Shi, H., Liu, Y., Greshock, J., Annan, R., et al. (2009). Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: Activation of AXL. Cancer Res. 69 (17), 6871–6878. doi:10.1158/0008-5472.CAN-08-4490
Liu, M., Yang, J., Lv, W., Wang, S., Du, T., Zhang, K., et al. (2022). Down-regulating GRP78 reverses pirarubicin resistance of triple negative breast cancer by miR-495-3p mimics and involves the p-AKT/mTOR pathway. Biosci. Rep. 42 (1), BSR20210245. doi:10.1042/BSR20210245
Lloyd, M. C., Cunningham, J. J., Bui, M. M., Gillies, R. J., Brown, J. S., and Gatenby, R. A. (2016). Darwinian dynamics of intratumoral heterogeneity: Not solely random mutations but also variable environmental selection forces. Cancer Res. 76 (11), 3136–3144. doi:10.1158/0008-5472.CAN-15-2962
Loh, Y. N., Hedditch, E. L., Baker, L. A., Jary, E., Ward, R. L., and Ford, C. E. (2013). The Wnt signalling pathway is upregulated in an in vitro model of acquired tamoxifen resistant breast cancer. BMC Cancer 13, 174. doi:10.1186/1471-2407-13-174
Lovitt, C. J., Shelper, T. B., and Avery, V. M. (2018). Doxorubicin resistance in breast cancer cells is mediated by extracellular matrix proteins. BMC cancer 18 (1), 41–11. doi:10.1186/s12885-017-3953-6
Lumachi, F., Brunello, A., Maruzzo, M., Basso, U., and Mm Basso, S. (2013). Treatment of estrogen receptor-positive breast cancer. Curr. Med. Chem. 20 (5), 596–604. doi:10.2174/092986713804999303
Luo, N., Zhang, K., Li, X., and Hu, Y. (2020). ZEB1 induced-upregulation of long noncoding RNA ZEB1-AS1 facilitates the progression of triple negative breast cancer by binding with ELAVL1 to maintain the stability of ZEB1 mRNA. J. Cell. Biochem. 121 (10), 4176–4187. doi:10.1002/jcb.29572
Lv, Q., Deng, J., Chen, Y., Wang, Y., Liu, B., and Liu, J. (2020). Engineered human adipose stem-cell-derived exosomes loaded with miR-21-5p to promote diabetic cutaneous wound healing. Mol. Pharm. 17, 1723–1733. doi:10.1021/acs.molpharmaceut.0c00177
Lv, Z., Wang, S., Zhao, W., and He, N. (2020). MicroRNA analysis of NCI-60 human cancer cells indicates that miR-720 and miR-887 are potential therapeutic biomarkers for breast cancer. Drug Discov. Ther. 14 (4), 197–203. doi:10.5582/ddt.2020.03058
Maas, S. L. B. X., and Weaver, A. M. (2017). Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 27 (3), 172–188. doi:10.1016/j.tcb.2016.11.003
Mafi, A., Rahmati, A., Babaei Aghdam, Z., Salami, R., Salami, M., Vakili, O., et al. (2022). Recent insights into the microRNA-dependent modulation of gliomas from pathogenesis to diagnosis and treatment. Cell. Mol. Biol. Lett. 27 (1), 65–32. doi:10.1186/s11658-022-00354-4
Mafi, A., Yadegar, N., Salami, M., Salami, R., Vakili, O., and Aghadavod, E. (2021). Circular RNAs; powerful microRNA sponges to overcome diabetic nephropathy. Pathology-Research Pract. 227, 153618. doi:10.1016/j.prp.2021.153618
Mantzoros, C., Petridou, E., Dessypris, N., Chavelas, C., Dalamaga, M., Alexe, D. M., et al. (2004). Adiponectin and breast cancer risk. J. Clin. Endocrinol. Metab. 89 (3), 1102–1107. doi:10.1210/jc.2003-031804
Marklund, U., Larsson, N., Gradin, H. M., Brattsand, G., and Gullberg, M. (1996). Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J. 15 (19), 5290–5298. doi:10.1002/j.1460-2075.1996.tb00914.x
Mateescu, B., Kowal, E. J., van Balkom, B. W., Bartel, S., Bhattacharyya, S. N., Buzás, E. I., et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA–an ISEV position paper. J. Extracell. vesicles 6 (1), 1286095. doi:10.1080/20013078.2017.1286095
McCubrey, J. A., Steelman, L. S., Abrams, S. L., Lee, J. T., Chang, F., Bertrand, F. E., et al. (2006). Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv. enzyme Regul. 46 (1), 249–279. doi:10.1016/j.advenzreg.2006.01.004
Me, H. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422. doi:10.1146/annurev.cellbio.19.111301.153609
Mehta, S. L. D. R., and Vemuganti, R. (2020). Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 186, 101746. doi:10.1016/j.pneurobio.2020.101746
Meldolesi, J. (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol. 28 (8), R435–R444. doi:10.1016/j.cub.2018.01.059
Miao, K., Lei, J. H., Valecha, M. V., Zhang, A., Xu, J., Wang, L., et al. (2020). NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nat. Commun. 11 (1), 3256. doi:10.1038/s41467-020-16936-9
Miller, T. E., Ghoshal, K., Ramaswamy, B., Roy, S., Datta, J., Shapiro, C. L., et al. (2008). MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 283 (44), 29897–29903. doi:10.1074/jbc.M804612200
Mills, J. N., Rutkovsky, A. C., and Giordano, A. (2018). Mechanisms of resistance in estrogen receptor positive breast cancer: Overcoming resistance to tamoxifen/aromatase inhibitors. Curr. Opin. Pharmacol. 41, 59–65. doi:10.1016/j.coph.2018.04.009
Minciacchi, V. R. F. M., and Di Vizio, D. (2015). Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 40, 41–51. doi:10.1016/j.semcdb.2015.02.010
Montagner, M., Enzo, E., Forcato, M., Zanconato, F., Parenti, A., Rampazzo, E., et al. (2012). SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487 (7407), 380–384. doi:10.1038/nature11207
Movahedpour, A., Khatami, S. H., Karami, N., Vakili, O., Naeli, P., Jamali, Z., et al. (2022). Exosomal noncoding RNAs in prostate cancer. Clin. Chim. Acta 537, 127–132. doi:10.1016/j.cca.2022.10.018
Movahedpour, A., Khatami, S. H., Khorsand, M., Salehi, M., Savardashtaki, A., Mirmajidi, S. H., et al. (2021). Exosomal noncoding RNAs: Key players in glioblastoma drug resistance. Mol. Cell. Biochem. 476, 4081–4092. doi:10.1007/s11010-021-04221-2
Movahedpour, A., Vakili, O., Khalifeh, M., Mousavi, P., Mahmoodzadeh, A., Taheri-Anganeh, M., et al. (2022). Mammalian target of rapamycin (mTOR) signaling pathway and traumatic brain injury: A novel insight into targeted therapy. Cell Biochem. Funct. 40 (3), 232–247. doi:10.1002/cbf.3692
Nabholtz, J-M., Falkson, C., Campos, D., Szanto, J., Martin, M., Chan, S., et al. (2003). Docetaxel and doxorubicin compared with doxorubicin and cyclophosphamide as first-line chemotherapy for metastatic breast cancer: Results of a randomized, multicenter, phase III trial. J. Clin. Oncol. 21 (6), 968–975. doi:10.1200/JCO.2003.04.040
Najafi, S., Zarch, S. M. A., Majidpoor, J., Pordel, S., Aghamiri, S., Rasul, M. F., et al. (2022). Recent insights into the roles of circular RNAs in human brain development and neurologic diseases. Int. J. Biol. Macromol.
O'Brien, J. H. H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 402 (9).
O'Brien, K., Lowry, M. C., Corcoran, C., Martinez, V. G., Daly, M., Rani, S., et al. (2015). miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 6 (32), 32774–32789. doi:10.18632/oncotarget.5192
Obayashi, S., Horiguchi, J., Higuchi, T., Katayama, A., Handa, T., Altan, B., et al. (2017). Stathmin1 expression is associated with aggressive phenotypes and cancer stem cell marker expression in breast cancer patients. Int. J. Oncol. 51 (3), 781–790. doi:10.3892/ijo.2017.4085
Ouyang, M., Li, Y., Ye, S., Ma, J., Lu, L., Lv, W., et al. (2014). MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One 9 (5), e96228. doi:10.1371/journal.pone.0096228
Ouyang, Y. X., Feng, J., Wang, Z., Zhang, G. J., and Chen, M. (2021). miR-221/222 sponge abrogates tamoxifen resistance in ER-positive breast cancer cells through restoring the expression of ERα. Mol. Biomed. 2 (1), 20–11. doi:10.1186/s43556-021-00045-0
Oza, P., Sharma, P., and Patel, S. (2021). A drive through computer-aided diagnosis of breast cancer: A comprehensive study of clinical and technical aspects. Recent innovations in computing. Proc. ICRIC 1, 233–249.
Palazzo, A. F., and Lee, E. S. (2015). Non-coding RNA: What is functional and what is junk? Front. Genet. 6, 2. doi:10.3389/fgene.2015.00002
Peng, F., Li, T. T., Wang, K. L., Xiao, G. Q., Wang, J. H., Zhao, H. D., et al. (2017). H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 8 (1), e2569. doi:10.1038/cddis.2016.438
Perotti, C., Liu, R., Parusel, C. T., Böcher, N., Schultz, J., Bork, P., et al. (2008). Heat shock protein-90-alpha, a prolactin-STAT5 target gene identified in breast cancer cells, is involved in apoptosis regulation. Breast Cancer Res. 10 (6), R94–R18. doi:10.1186/bcr2193
Piggott, L., Silva, A., Robinson, T., Santiago-Gómez, A., Simões, B. M., Becker, M., et al. (2018). Acquired resistance of ER-positive breast cancer to endocrine treatment confers an adaptive sensitivity to TRAIL through posttranslational downregulation of c-FLIP. Clin. Cancer Res. 24 (10), 2452–2463. doi:10.1158/1078-0432.CCR-17-1381
Prensner, J. R. Z. S., Erho, N., Schipper, M., Iyer, M. K., Dhanasekaran, S. M., et al. (2014). RNA biomarkers associated with metastatic progression in prostate cancer: A multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15 (13), 1469–1480. doi:10.1016/S1470-2045(14)71113-1
Proctor, E., Waghray, M., Lee, C. J., Heidt, D. G., Yalamanchili, M., Li, C., et al. (2013). Bmi1 enhances tumorigenicity and cancer stem cell function in pancreatic adenocarcinoma. PLoS One 8 (2), e55820. doi:10.1371/journal.pone.0055820
Proia, D. A., Zhang, C., Sequeira, M., Jimenez, J-P., He, S., Spector, N., et al. (2014). Preclinical activity profile and therapeutic efficacy of the HSP90 inhibitor ganetespib in triple-negative breast cancer. Clin. Cancer Res. 20 (2), 413–424. doi:10.1158/1078-0432.CCR-13-2166
Prosperi, J. R., and Goss, K. H. (2010). A wnt-ow of opportunity: Targeting the wnt/beta-catenin pathway in breast cancer. Curr. Drug Targets 11 (9), 1074–1088. doi:10.2174/138945010792006780
Purwanto, I., Heriyanto, D. S., Widodo, I., Hakimi, M., Hardianti, M. S., Aryandono, T., et al. (2021). MicroRNA-223 is associated with resistance towards platinum-based chemotherapy and worse prognosis in Indonesian triple-negative breast cancer patients. Breast Cancer Targets Ther. 13, 1–7. doi:10.2147/BCTT.S291014
Qian, X., Qu, H., Zhang, F., Peng, S., Dou, D., Yang, Y., et al. (2021). Exosomal long noncoding RNA AGAP2-AS1 regulates trastuzumab resistance via inducing autophagy in breast cancer. Am. J. Cancer Res. 11 (5), 1962–1981.
Quail, D. F. J. J., and Joyce, J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19 (11), 1423–1437. doi:10.1038/nm.3394
Rai, S., Arasteh, M., Jefferson, M., Pearson, T., Wang, Y., Zhang, W., et al. (2019). The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy 15 (4), 599–612. doi:10.1080/15548627.2018.1534507
Rautalin, M., Jahkola, T., and Roine, R. P. (2022). Breast reconstruction–prospective follow up on breast cancer patients’ health-related quality of life. World J. Surg. 46, 836–844. doi:10.1007/s00268-021-06426-4
Reza, A., Choi, Y. J., Yasuda, H., and Kim, J. H. (2016). Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci. Rep. 6, 38498. doi:10.1038/srep38498
Ring, A., and Dowsett, M. (2004). Mechanisms of tamoxifen resistance. Endocr. Relat. Cancer 11 (4), 643–658. doi:10.1677/erc.1.00776
Rinn, J. L., and Huarte, M. (2011). To repress or not to repress: This is the guardian's question. Trends Cell Biol. 21 (6), 344–353. doi:10.1016/j.tcb.2011.04.002
Robey, R. W., Pluchino, K. M., Hall, M. D., Fojo, A. T., Bates, S. E., and Gottesman, M. M. (2018). Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18 (7), 452–464. doi:10.1038/s41568-018-0005-8
Romano, G., Veneziano, D., Acunzo, M., and Croce, C. M. (2017). Small non-coding RNA and cancer. Carcinogenesis 38 (5), 485–491. doi:10.1093/carcin/bgx026
Romero-Cordoba, S. L., Rodriguez-Cuevas, S., Bautista-Pina, V., Maffuz-Aziz, A., D'Ippolito, E., Cosentino, G., et al. (2018). Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer. Sci. Rep. 8 (1), 12252. doi:10.1038/s41598-018-29708-9
Ruan, K. F. X., and Ouyang, G. (2009). MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 285 (2), 116–126. doi:10.1016/j.canlet.2009.04.031
Rubin, C. I., and Atweh, G. F. (2004). The role of stathmin in the regulation of the cell cycle. J. Cell. Biochem. 93 (2), 242–250. doi:10.1002/jcb.20187
Rusnak, D. W., Lackey, K., Affleck, K., Wood, E. R., Alligood, K. J., Rhodes, N., et al. (2001). The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol. Cancer Ther. 1 (2), 85–94.
Sakha, S., Muramatsu, T., Ueda, K., and Inazawa, J. (2016). Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 6 (1), 38750–38811. doi:10.1038/srep38750
Salami, R., Salami, M., Mafi, A., Vakili, O., and Asemi, Z. (2022). Circular RNAs and glioblastoma multiforme: Focus on molecular mechanisms. Cell Commun. Signal. 20 (1), 13. doi:10.1186/s12964-021-00809-9
Salehi, M., Vafadar, A., Khatami, S. H., Taheri-Anganeh, M., Vakili, O., Savardashtaki, A., et al. (2022). Gastrointestinal cancer drug resistance: The role of exosomal miRNAs. Mol. Biol. Rep. 49 (3), 2421–2432. doi:10.1007/s11033-021-07007-3
Sana, J., Faltejskova, P., Svoboda, M., and Slaby, O. (2012). Novel classes of non-coding RNAs and cancer. J. Transl. Med. 103 (10), 103. doi:10.1186/1479-5876-10-103
Sang, Y., Chen, B., Song, X., Li, Y., Liang, Y., Han, D., et al. (2019). circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a Axis in breast cancer. Mol. Ther. 27 (9), 1638–1652. doi:10.1016/j.ymthe.2019.05.011
Sanger, H. L. K. G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Santos, J. C., Lima, N. D. S., Sarian, L. O., Matheu, A., Ribeiro, M. L., and Derchain, S. F. M. (2018). Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 8 (1), 829. doi:10.1038/s41598-018-19339-5
Sarver, A. L., Thayanithy, V., Scott, M. C., Cleton-Jansen, A-M., Hogendoorn, P. C., Modiano, J. F., et al. (2013). MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J. rare Dis. 8 (1), 7–11. doi:10.1186/1750-1172-8-7
Schwarzenbach, H. (2017). Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol. Res. Treat. 40 (7-8), 423–429. doi:10.1159/000478019
Sebolt-Leopold, J. S., and Herrera, R. (2004). Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer 4 (12), 937–947. doi:10.1038/nrc1503
Semenza, G. L. (2013). HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. investigation 123 (9), 3664–3671. doi:10.1172/JCI67230
Semenza, G. L. (2012). Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 33 (4), 207–214. doi:10.1016/j.tips.2012.01.005
Shah, R., Rosso, K., and Nathanson, S. D. (2014). Pathogenesis, prevention, diagnosis and treatment of breast cancer. World J. Clin. Oncol. 5 (3), 283–298. doi:10.5306/wjco.v5.i3.283
Shen, H., Wang, D., Li, L., Yang, S., Chen, X., Zhou, S., et al. (2017). MiR-222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway. Gene 596, 110–118. doi:10.1016/j.gene.2016.10.016
Shen, R., Wang, Y., Wang, C. X., Yin, M., Liu, H. L., Chen, J. P., et al. (2015). MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 7 (10), 2115–2126.
Si, X., Zang, R., Zhang, E., Liu, Y., Shi, X., Zhang, E., et al. (2016). LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 7 (49), 81452–81462. doi:10.18632/oncotarget.13263
Silver, D. P., Richardson, A. L., Eklund, A. C., Wang, Z. C., Szallasi, Z., Li, Q., et al. (2010). Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J. Clin. Oncol. 28 (7), 1145–1153. doi:10.1200/JCO.2009.22.4725
Singh, D., Assaraf, Y. G., and Gacche, R. N. (2022). Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist. Updat. 63, 100851. doi:10.1016/j.drup.2022.100851
Smith, L., Welham, K. J., Watson, M. B., Drew, P. J., Lind, M. J., and Cawkwell, L. (2007). The proteomic analysis of cisplatin resistance in breast cancer cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 16 (11), 497–506. doi:10.3727/096504007783438358
Sobhani, N., Chahwan, R., Roudi, R., Morris, R., Volinia, S., Chai, D., et al. (2022). Predictive and prognostic value of non-coding RNA in breast cancer. Cancers 14 (12), 2952. doi:10.3390/cancers14122952
Steelman, L. S., Navolanic, P. M., Sokolosky, M. L., Taylor, J. R., Lehmann, B. D., Chappell, W. H., et al. (2008). Suppression of PTEN function increases breast cancer chemotherapeutic drug resistance while conferring sensitivity to mTOR inhibitors. Oncogene 27 (29), 4086–4095. doi:10.1038/onc.2008.49
Stuffers, S. S. W. C., Stenmark, H., and Brech, A. (2019). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10 (7), 925–937. doi:10.1111/j.1600-0854.2009.00920.x
Su, S., Chen, J., Yao, H., Liu, J., Yu, S., Lao, L., et al. (2018). CD10(+)GPR77(+) cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 172 (4), 841–856.e16. doi:10.1016/j.cell.2018.01.009
Sun, D. I., Tasca, A., Haas, M., Baltazar, G., Harland, R. M., Finkbeiner, W. E., et al. (2018). Na+/H+ exchangers are required for the development and function of vertebrate mucociliary epithelia. Cells Tissues Organs 205 (5-6), 279–292. doi:10.1159/000492973
Sun, Z., Yang, S., Zhou, Q., Wang, G., Song, J., Li, Z., et al. (2018). Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer 17 (1), 82. doi:10.1186/s12943-018-0831-z
Szczygieł, M., Markiewicz, M., Szafraniec, M. J., Hojda, A., Fiedor, L., and Urbanska, K. (2022). Systemic mobilization of breast cancer resistance protein in response to oncogenic stress. Cancers (Basel) 14 (2), 313. doi:10.3390/cancers14020313
Taghvimi, S., Vakili, O., Soltani Fard, E., Khatami, S. H., Karami, N., Taheri-Anganeh, M., et al. (2022). Exosomal microRNAs and long noncoding RNAs: Novel mediators of drug resistance in lung cancer. J. Cell. physiology 237 (4), 2095–2106. doi:10.1002/jcp.30697
Takayama, H., Suzuki, T., Mugishima, H., Fujisawa, T., Ookuni, M., Schwab, M., et al. (1992). Deletion mapping of chromosomes 14q and 1p in human neuroblastoma. Oncogene 7 (6), 1185–1189.
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 (7483), 344–352. doi:10.1038/nature12986
Teixeira, F., Castro, H., Cruz, T., Tse, E., Koldewey, P., Southworth, D. R., et al. (2015). Mitochondrial peroxiredoxin functions as crucial chaperone reservoir in Leishmania infantum. Proc. Natl. Acad. Sci. 112 (7), E616–E624. doi:10.1073/pnas.1419682112
Toska, E., Osmanbeyoglu, H. U., Castel, P., Chan, C., Hendrickson, R. C., Elkabets, M., et al. (2017). PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 355 (6331), 1324–1330. doi:10.1126/science.aah6893
Tuo, Y. L., Li, X. M., and Luo, J. (2015). Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharmacol. Sci. 19 (18), 3403–3411.
ufiero, S. R. Y., Pinto, Y. M., and Creemers, E. E. (2019). Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 16 (8), 503–514. doi:10.1038/s41569-019-0185-2
Vakili, O., Asili, P., Babaei, Z., Mirahmad, M., Keshavarzmotamed, A., Asemi, Z., et al. (2023). Circular RNAs in Alzheimer’s disease: A new perspective of diagnostic and therapeutic targets. CNS Neurological Disorders-Drug Targets. doi:10.2174/1871527321666220829164211
Valadi, H. E. K., and BossiosSjostrandLeeLotvall, A. M. J. J. J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9 (6), 654–659. doi:10.1038/ncb1596
van Niel, G. D. A. G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19 (4), 213–228. doi:10.1038/nrm.2017.125
Varghese, E., Samuel, S. M., Líšková, A., Samec, M., Kubatka, P., and Büsselberg, D. (2020). Targeting glucose metabolism to overcome resistance to anticancer chemotherapy in breast cancer. Cancers (Basel) 12 (8), 2252. doi:10.3390/cancers12082252
Wang, B., Wang, Y., Wang, X., Gu, J., Wu, W., Wu, H., et al. (2022). Extracellular vesicles carrying miR-887-3p promote breast cancer cell drug resistance by targeting BTBD7 and activating the notch1/hes1 signaling pathway. Dis. Markers 2022, 5762686. doi:10.1155/2022/5762686
Wang, B., Zhang, Y., Ye, M., Wu, J., Ma, L., and Chen, H. (2019). Cisplatin-resistant MDA-MB-231 cell-derived exosomes increase the resistance of recipient cells in an exosomal miR-423-5p-dependent manner. Curr. drug Metab. 20 (10), 804–814. doi:10.2174/1389200220666190819151946
Wang, G., Gormley, M., Qiao, J., Zhao, Q., Wang, M., Di Sante, G., et al. (2018). Cyclin D1-mediated microRNA expression signature predicts breast cancer outcome. Theranostics 8 (8), 2251–2263. doi:10.7150/thno.23877
Wang, H., Shan, S., Wang, H., and Wang, X. (2022). CircATXN7 contributes to the progression and doxorubicin resistance of breast cancer via modulating miR-149-5p/HOXA11 pathway. Anticancer Drugs 33 (1), e700–e710. doi:10.1097/CAD.0000000000001243
Wang, J. C. J., and Sen, S. (2016). MicroRNA as biomarkers and diagnostics. J. Cell Physiol. 231, 25–30. doi:10.1002/jcp.25056
Wang, L., Yang, X., Zhou, F., Sun, X., and Li, S. (2022). Circular RNA UBAP2 facilitates the cisplatin resistance of triple-negative breast cancer via microRNA-300/anti-silencing function 1B histone chaperone/PI3K/AKT/mTOR axis. Bioengineered 13 (3), 7197–7208. doi:10.1080/21655979.2022.2036894
Wang, L., Zhang, Q., Zhang, J., Sun, S., Guo, H., Jia, Z., et al. (2011). PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer 11, 248. doi:10.1186/1471-2407-11-248
Wang, L., Zhou, Y., Jiang, L., Lu, L., Dai, T., Li, A., et al. (2021). CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway. Mol. cancer 20 (1), 43–15. doi:10.1186/s12943-021-01332-8
Wang, Q., Liang, D., Shen, P., Yu, Y., Yan, Y., and You, W. (2021). Hsa_circ_0092276 promotes doxorubicin resistance in breast cancer cells by regulating autophagy via miR-348/ATG7 axis. Transl. Oncol. 14 (8), 101045. doi:10.1016/j.tranon.2021.101045
Wang, S. E., Xiang, B., Guix, M., Olivares, M. G., Parker, J., Chung, C. H., et al. (2008). Transforming growth factor beta engages TACE and ErbB3 to activate phosphatidylinositol-3 kinase/Akt in ErbB2-overexpressing breast cancer and desensitizes cells to trastuzumab. Mol. Cell Biol. 28 (18), 5605–5620. doi:10.1128/MCB.00787-08
Wang, X., Pei, X., Guo, G., Qian, X., Dou, D., Zhang, Z., et al. (2020). Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell Physiol. 235 (10), 6896–6904. doi:10.1002/jcp.29585
Wang, Y., Li, J., Du, C., Zhang, L., Zhang, Y., Zhang, J., et al. (2019). Upregulated circular RNA circ-ube2d2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl. Oncol. 12 (10), 1305–1313. doi:10.1016/j.tranon.2019.05.016
Wang, Y. L. J., Ma, J., Sun, T., Zhou, Q., Wang, W., et al. (2018). Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 18 (1), 116. doi:10.1186/s12943-019-1041-z
Wang, Y., Zheng, F., Gao, G., Yan, S., Zhang, L., Wang, L., et al. (2018). MiR-548a-3p regulates inflammatory response via TLR4/NF-κB signaling pathway in rheumatoid arthritis. J. Cell Biochem. 120, 1133–1140. doi:10.1002/jcb.26659
Wang, Z., Liang, S., Lian, X., Liu, L., Zhao, S., Xuan, Q., et al. (2015). Identification of proteins responsible for adriamycin resistance in breast cancer cells using proteomics analysis. Sci. Rep. 5 (1), 9301–9311. doi:10.1038/srep09301
Wei, Y., Lai, X., Yu, S., Chen, S., Ma, Y., Zhang, Y., et al. (2014). Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast cancer Res. Treat. 147 (2), 423–431. doi:10.1007/s10549-014-3037-0
Workman, P., Burrows, F., Neckers, L., and Rosen, N. (2007). Drugging the cancer chaperone HSP90: Combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann. N. Y. Acad. Sci. 1113 (1), 202–216. doi:10.1196/annals.1391.012
Wu, X., Ren, Y., Yao, R., Zhou, L., and Fan, R. (2021). Circular RNA circ-MMP11 contributes to lapatinib resistance of breast cancer cells by regulating the miR-153-3p/ANLN Axis. Front. Oncol. 11, 639961. doi:10.3389/fonc.2021.639961
Xia, Q., Ali, S., Liu, L., Li, Y., Liu, X., Zhang, L., et al. (2020). Role of ubiquitination in PTEN cellular homeostasis and its implications in GB drug resistance. Front. Oncol. 10, 1569. doi:10.3389/fonc.2020.01569
Xiao, M. S. A. Y., and Wilusz, J. E. (2020). Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 30 (3), 226–240. doi:10.1016/j.tcb.2019.12.004
Xie, H., and Zheng, R. (2022). Circ_0085495 knockdown reduces adriamycin resistance in breast cancer through miR-873-5p/integrin β1 axis. Anticancer Drugs 33 (1), e166–e177. doi:10.1097/CAD.0000000000001174
Xie, Y., Dang, W., Zhang, S., Yue, W., Yang, L., Zhai, X., et al. (2019). The role of exosomal noncoding RNAs in cancer. Mol. Cancer 18 (1), 37. doi:10.1186/s12943-019-0984-4
Xu, C. G., Yang, M. F., Ren, Y. Q., Wu, C. H., and Wang, L. Q. (2016). Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 20 (20), 4362–4368.
Xu, F., Zhang, C., Cui, J., Liu, J., Li, J., and Jiang, H. (2017). The prognostic value and potential drug target of phosphatase and tensin homolog in breast cancer patients: A meta-analysis. Med. Baltim. 96 (36), e8000. doi:10.1097/MD.0000000000008000
Yao, J., Deng, K., Huang, J., Zeng, R., and Zuo, J. (2020). Progress in the understanding of the mechanism of tamoxifen resistance in breast cancer. Front. Pharmacol. 11, 592912. doi:10.3389/fphar.2020.592912
Yao, Y., Li, X., Cheng, L., Wu, X., and Wu, B. (2021). Circular RNA FAT atypical cadherin 1 (circFAT1)/microRNA-525-5p/spindle and kinetochore-associated complex subunit 1 (SKA1) axis regulates oxaliplatin resistance in breast cancer by activating the notch and Wnt signaling pathway. Bioengineered 12 (1), 4032–4043. doi:10.1080/21655979.2021.1951929
Yellon, D. M. D. S., and Davidson, S. M. (2014). Exosomes: Nanoparticles involved in cardioprotection? Circ. Res. 114 (2), 325–332. doi:10.1161/CIRCRESAHA.113.300636
Yin, C., Wang, P. Q., Xu, W. P., Yang, Y., Zhang, Q., Ning, B. F., et al. (2013). Hepatocyte nuclear factor-4α reverses malignancy of hepatocellular carcinoma through regulating miR-134 in the DLK1-DIO3 region. Hepatology 58 (6), 1964–1976. doi:10.1002/hep.26573
Yousefi, H. M. M., Molaei, F., Mashouri, L., Reza Aref, A., Momeny, M., et al. (2020). Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 39 (5), 953–974. doi:10.1038/s41388-019-1040-y
Yu, C., and Kuo, H. C. (2019). The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 29 (26), 29. doi:10.1186/s12929-019-0523-z
Yu, D-D., Wu, Y., Zhang, X-H., Lv, M-M., Chen, W-X., Chen, X., et al. (2016). Exosomes from adriamycin-resistant breast cancer cells transmit drug resistance partly by delivering miR-222. Tumor Biol. 37 (3), 3227–3235. doi:10.1007/s13277-015-4161-0
Yu, D. D., Lv, M. M., Chen, W. X., Zhong, S. L., Zhang, X. H., Chen, L., et al. (2015). Role of miR-155 in drug resistance of breast cancer. Tumour Biol. 36 (3), 1395–1401. doi:10.1007/s13277-015-3263-z
Yu, L., Xu, Q., Yu, W., Duan, J., and Dai, G. (2018). LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop. Biochem. Biophys. Res. Commun. 506 (4), 819–825. doi:10.1016/j.bbrc.2018.10.131
Yu, S., Zhou, Y., Niu, L., Qiao, Y., and Yan, Y. (2022). Mesenchymal stem cell-derived exosome mir-342-3p inhibits metastasis and chemo-resistance of breast cancer through regulating ID4. Genes Genomics 44 (5), 539–550. doi:10.1007/s13258-021-01200-1
Zakharova, L. S. M., and Fomina, A. F. (2007). T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J. Cell Physiol. 212 (1), 174–181. doi:10.1002/jcp.21013
Zang, H., Li, Y., Zhang, X., and Huang, G. (2020). Circ-RNF111 contributes to paclitaxel resistance in breast cancer by elevating E2F3 expression via miR-140-5p. Thorac. Cancer 11 (7), 1891–1903. doi:10.1111/1759-7714.13475
Zhang, K., Wang, Y. W., and Ma, R. (2017). Bioinformatics analysis of dysregulated microRNAs in the nipple discharge of patients with breast cancer. Oncol. Lett. 13 (5), 3100–3108. doi:10.3892/ol.2017.5801
Zhang, X., Wang, M., Sun, H., Zhu, T., and Wang, X. (2018). Downregulation of linc00894-002 contributes to tamoxifen resistance by enhancing the TGF-β signaling pathway. Biochem. (Mosc) 83 (5), 603–611. doi:10.1134/S0006297918050139
Zhang, Y., Liu, Y., Liu, H., and Tang, W. H. (2019). Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 19 (9), 19. doi:10.1186/s13578-019-0282-2
Zhang, Z., Zhang, L., Yu, G., Sun, Z., Wang, T., Tian, X., et al. (2020). Exosomal miR-1246 and miR-155 as predictive and prognostic biomarkers for trastuzumab-based therapy resistance in HER2-positive breast cancer. Cancer Chemother. Pharmacol. 86 (6), 761–772. doi:10.1007/s00280-020-04168-z
Zheng, Z., Chen, M., Xing, P., Yan, X., and Xie, B. (2019). Increased expression of exosomal AGAP2-AS1 (AGAP2 antisense RNA 1) in breast cancer cells inhibits trastuzumab-induced cell cytotoxicity. Med. Sci. Monit. 25, 2211–2220. doi:10.12659/MSM.915419
Zhong, S., Chen, X., Wang, D., Zhang, X., Shen, H., Yang, S., et al. (2016). MicroRNA expression profiles of drug-resistance breast cancer cells and their exosomes. Oncotarget 7 (15), 19601–19609. doi:10.18632/oncotarget.7481
Zhong, S., Li, W., Chen, Z., Xu, J., and Zhao, J. (2013). MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene 531 (1), 8–14. doi:10.1016/j.gene.2013.08.062
Zhou, W., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 172 (19), 172. doi:10.1186/s12943-020-01286-3
Zhou, W., Wang, Z., Shen, N., Pi, W., Jiang, W., Huang, J., et al. (2015). Knockdown of ANLN by lentivirus inhibits cell growth and migration in human breast cancer. Mol. Cell Biochem. 398 (1-2), 11–19. doi:10.1007/s11010-014-2200-6
Zhou, Z. S. B., Huang, S., and Zhao, L. (2019). Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis. 10 (7), 503. doi:10.1038/s41419-019-1744-5
Zhu, M., Wang, Y., Wang, F., Li, L., and Qiu, X. (2021). CircFBXL5 promotes the 5-FU resistance of breast cancer via modulating miR-216b/HMGA2 axis. Cancer Cell Int. 21 (1), 384. doi:10.1186/s12935-021-02088-3
Zhu, Q. N., Wang, G., Guo, Y., Peng, Y., Zhang, R., Deng, J. L., et al. (2017). LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget 8 (54), 91990–92003. doi:10.18632/oncotarget.21121
Zhu, Y., Yu, F., Jiao, Y., Feng, J., Tang, W., Yao, H., et al. (2011). Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 17 (22), 7105–7115. doi:10.1158/1078-0432.CCR-11-0071
Zimmermann, M., Arachchige-Don, A. P., Donaldson, M. S., Patriarchi, T., and Horne, M. C. (2016). Cyclin G2 promotes cell cycle arrest in breast cancer cells responding to fulvestrant and metformin and correlates with patient survival. Cell Cycle 15 (23), 3278–3295. doi:10.1080/15384101.2016.1243189
Keywords: breast neoplasms, drug resistance, exosomes, microRNAs, circular RNA, long non-coding RNA
Citation: Rezaee M, Mohammadi F, Keshavarzmotamed A, Yahyazadeh S, Vakili O, Milasi YE, Veisi V, Dehmordi RM, Asadi S, Ghorbanhosseini SS, Rostami M, Alimohammadi M, Azadi A, Moussavi N, Asemi Z, Aminianfar A, Mirzaei H and Mafi A (2023) The landscape of exosomal non-coding RNAs in breast cancer drug resistance, focusing on underlying molecular mechanisms. Front. Pharmacol. 14:1152672. doi: 10.3389/fphar.2023.1152672
Received: 28 January 2023; Accepted: 29 March 2023;
Published: 19 April 2023.
Edited by:
Sujit Nair, Viridis Biopharma Pvt. Ltd., IndiaReviewed by:
Mihir Khambete, Yale University, United StatesJessian Munoz, Texas Children’s Hospital, United States
Copyright © 2023 Rezaee, Mohammadi, Keshavarzmotamed, Yahyazadeh, Vakili, Milasi, Veisi, Dehmordi, Asadi, Ghorbanhosseini, Rostami, Alimohammadi, Azadi, Moussavi, Asemi, Aminianfar, Mirzaei and Mafi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mina Alimohammadi, bWluYS5hbGltb2hhbW1hZGkxMUBnbWFpbC5jb20=; Abbas Azadi, YWJhc2F6YWRpNDdAZ21haWwuY29t; Hamed Mirzaei, aC5taXJ6YWVpMjAwMkBnbWFpbC5jb20=; Alireza Mafi, YXJtYWZpLm1AZ21haWwuY29t
 Malihe Rezaee
Malihe Rezaee Fatemeh Mohammadi
Fatemeh Mohammadi Atoosa Keshavarzmotamed4
Atoosa Keshavarzmotamed4 Sheida Yahyazadeh
Sheida Yahyazadeh Omid Vakili
Omid Vakili Mehdi Rostami
Mehdi Rostami Mina Alimohammadi
Mina Alimohammadi Hamed Mirzaei
Hamed Mirzaei Alireza Mafi
Alireza Mafi