
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 20 March 2023
Sec. Pharmacology of Anti-Cancer Drugs
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1136251
This article is part of the Research Topic Recent advances in molecular mechanisms involved in targeted therapy for cancer treatment View all 5 articles
As non-canonical nucleic acid secondary structures consisting of cytosine-rich nucleic acids, i-motifs can form under certain conditions. Several i-motif sequences have been identified in the human genome and play important roles in biological regulatory functions. Due to their physicochemical properties, these i-motif structures have attracted attention and are new targets for drug development. Herein, we reviewed the characteristics and mechanisms of i-motifs located in gene promoters (including c-myc, Bcl-2, VEGF, and telomeres), summarized various small molecule ligands that interact with them, and the possible binding modes between ligands and i-motifs, and described their effects on gene expression. Furthermore, we discussed diseases closely associated with i-motifs. Among these, cancer is closely associated with i-motifs since i-motifs can form in some regions of most oncogenes. Finally, we introduced recent advances in the applications of i-motifs in multiple areas.
DNA carries the genetic information necessary to synthesize RNA and proteins, which are essential for the development and maintaining the normal function of living organisms. Scientists have never stopped exploring the function and application of DNA. In 1953, Watson and Crick (1974) first put forward the molecular model of DNA double helix structure, namely, B-form DNA (B-DNA), which describes the DNA molecule as a right-handed twisted coil composed of a purine and pyrimidine inner core held together by hydrogen bonds, with a sugar-phosphate backbone extending from these paired bases. Further studies have revealed the conformation of A-DNA, Z-DNA, triplex DNA, hairpins, and cruciform, as well as tetraplex structures, including G4 (guanine-quadruplex) and i-motifs (intercalated-motif) (Dickerson et al., 1982; Shakked et al., 1983; Bochman et al., 2012). B-DNA is the major conformation of DNA in the physiological state, while the other configurations are uniformly referred to as non-B-form secondary structures (Bacolla and Wells, 2004) and have not been studied well compared to B-DNA. Therefore, researchers have placed particular emphasis on exploiting treatment strategies in connection with non-B-form secondary structures.
In recent decades, G4 structures have attracted increasing research interest. G4 is a non-B-form secondary structure with G-rich nucleic acid sequences under physiological pH. In G4s (guanine-quadruplexes), each guanine interacts with its two adjacent guanines to form a square planar configuration comprising four guanine residues (Figure 1). G4s may occur in one or several different ssDNA or even RNA molecules. Moreover, the extension direction between DNA strands can not only be the same but the opposite; thus, G4s play different roles in gene regulation (Burge et al., 2006; Pascale et al., 2006; Bochman et al., 2012). Thus, G4s are not only DNA sequences that block transcription (Robinson et al., 2021) but can also affect the activity and function of DNA repair (Linke et al., 2021). G4s may also be involved in the development of cutting-edge therapeutic strategies for the treatment of viral infections and cancer (Ruggiero et al., 2021).
Scientists have also discovered another DNA structure with gene regulation function, which follows the principle of complementary base pairing and is present in the complementary cytosine (C)-rich sequences in nucleic acid sequences forming G4s. These C-rich sequences form another type of non-B-form secondary structure known as an i-motif, which forms in slightly acidic microenvironments. Studies have reported that the influential factors of i-motif formation are similar to those of G4 structures. This review summarized researches on i-motifs, including their structure, biological function, mechanism, and application.
I-motif structures are four-stranded non-covalent complexes that form within certain C-rich sequences, where hemi-protonated cytosine-cytosine base pairs intercalate with each other (Day et al., 2014; Mir et al., 2017; Kretzmann et al., 2021; Serrano-Chacón et al., 2021) (Figure 1). These structures can form in both DNA and RNA (Collin and Gehring, 1998) and include intramolecular and intermolecular i-motif structures. The presence of i-motif structures was proven by simulating the normal human physiological state in vitro through circular dichroism and native gel electrophoresis (Wright et al., 2017). In 2018, scientists at the Garvan Medical Research Institute in Australia demonstrated the presence of i-motif structures in human cells using fluorescent antibodies against i-motifs, namely, iMabs, which specifically bind to i-motifs (Zeraati et al., 2018).
Huppert and Balasubramanian (2005) suggested that the human genome contains up to 3,776,000 DNA fragments forming G4s; thus, a considerable number of i-motifs are theoretically present. The promoter regions of oncogenes including Rb (Jakubowska et al., 2001; Xu and Sugiyama, 2005; Xu and Sugiyama, 2006), RET (Guo et al., 2007; Bielecka and Juskowiak, 2015), VEGF (Guo et al., 2008; Kimura et al., 2022), c-myb (Li et al., 2016), c-myc (Sun and Hurley, 2009; Reilly et al., 2014), and Bcl-2 (Panasiuk et al., 2006; Kendrick et al., 2009; Lin et al., 2013; Yang et al., 2020) contain i-motifs. In addition to those cancer-associated genomic regions, i-motifs also occur in telomeres, centromeres, and ribosomes (Bochman et al., 2012; Mondal et al., 2019).
The stabilizing ability of i-motif DNA conformations is also influenced by factors similar to those of G4 structures. The structural stability of G4 depends on many elements, including pH, the specific gene sequence, the size of rings between adjacent bases, temperature, cations, and small molecule ligands (Hardin et al., 2000; Bugaut and Balasubramanian, 2008; Sun and Hurley, 2009; Guédin et al., 2010; Abdelhamid et al., 2019). Many studies have reported factors affecting i-motif structural stability. I-motifs are more sensitive to pH compared to G4s and can fold/unfold swiftly with changes in environmental pH (Del Toro et al., 2009; Jin et al., 2009; Bucek et al., 2010; Lannes et al., 2015; Abou Assi et al., 2018). Moreover, the thermal stability of i-motifs depends on the length of the loop (Reilly et al., 2015); for example, [Ru(phen)2(dppz)]2+ (1, Figure 2) tends to interact with i-motif DNA with longer-length thymidine rings and stabilizes the structure (Pages et al., 2019). Cations such as Na+ and K+ tend to stabilize/destabilize i-motif structures in different buffer systems (Mathur et al., 2004; Gao and Hou, 2021). Some specific small molecule ligands can combine and stabilize i-motifs (Fedoroff et al., 2000). These effects are summarized in Table 1.
G4s and i-motifs transform dynamically with cell cycle progression. Consequently, there is a general consensus that both DNA structures are highly likely to participate in controlling gene expression by turning them on or off at the DNA level. Biffi et al. (2013) found that G4 structures mostly occurred during the S phase, while Zeraati et al. (2018) reported a maximum number of i-motif structures in the late G1 phase. Most i-motif sequences are located in the regulatory regions of the human genome (Huppert and Balasubramanian, 2005; Belmonte-Reche and Morales, 2020). Therefore, i-motifs are considered to be closely related to DNA replication, transcription, and translation of human genes, which are summarized in this section.
Promoters, just like switches, are important factors in controlling gene activity. Promoter function depends on their structure, which impacts their affinity with RNA polymerase and is associated with imbalanced gene expression and illnesses. In recent years, i-motifs located in promoters have been demonstrated to be closely connected with gene transcription. To date, many small molecule ligands have been reported, which may interact with these i-motifs and play important roles in regulating the biological activities of specific genes.
Bcl-2 is a proto-oncogene, the expression product of which is suspected as the motivator for cancers and autoimmune diseases by activating superfluous cells (Roy et al., 2014). The human Bcl-2 gene has two main promoters, P1 and P2, both of which regulate the initiation of gene transcription (Dexheimer et al., 2006). P1 is a GC-rich promoter in which i-motif structures have been reported in vitro (Amato et al., 2022). Previous studies proved that G4 structures provide a negative signal, resulting in the silencing of gene expression (Brooks and Hurley, 2009). In contrast, the i-motif in the Bcl-2 P1 promoter is a positive signal to activate gene transcription (Kang et al., 2014).
The small molecule ligand biproline amide derivative PBP1 (2, Figure 2) specifically binds the Bcl-2 i-motif, which promotes folding of the C-rich DNA sequence into i-motif structure at neutral pH and up-regulating Bcl-2 expression in both RNA and protein synthesis. Its isomer PBP2 (3, Figure 2) has a lower affinity for the Bcl-2 i-motif and reduces the level of active caspase enzymes 3/7 in HCT-116 cells in flow cytometry analysis (Debnath et al., 2017). Yang et al. (2020) reported that the natural flavonoids P5 and P6 (4, 5, Figure 2) had a higher affinity for the Bcl-2 i-motif compared to P1 (6, Figure 2). The acridone derivative A22 (7, Figure 2) (Li et al., 2020) stimulated Bcl-2 expression, reducing hepatocyte apoptosis and alleviating inflammation, endoplasmic reticulum stress, and cirrhosis in a NAFLD/NASH (non-alcoholic fatty liver disease/non-alcoholic steatohepatitis) model. Thus, A22 (7) may be a promising compound for the treatment of liver illnesses. In the quest for therapy for diffuse large B cell lymphoma (DLBCL), Kendrick et al. (2017) proposed an innovative strategy to simultaneously use GQC-05 and IMC-76 (8, Figure 2), which respectively recognize the Myc G4 and the Bcl-2 i-motif. This treatment not only regulated Myc and Bcl-2 gene expression but also significantly decelerated tumor growth in DLBCL xenografted mice.
hnRNP LL (heterogeneous nuclear ribonucleoprotein LL) protein, which shows tissue-specific distribution, activates T cells by transferring the transcriptional genome and then advances cell proliferation and inhibits cell death (Oberdoerffer et al., 2008). Roy et al. (2016) defined hnRNP LL as an active transcription factor for Bcl-2, which recognizes the i-motif by binding the preferred loop to its four RRM (RNA recognition motif). Among the four RRMs, RRM1 and RRM2 strongly combine with the Bcl-2 i-motif to drive the i-motif transformation into a more stable hairpin structure and finally upregulate Bcl-2 transcription. Furthermore, the small molecule IMC-48 (9, Figure 2) increased i-motif and hnRNP LL levels by binding the central ring of the i-motif. In contrast, IMC-76 (8) wrapped flexible hairpins to decrease the levels of this compound (Cui et al., 2014; Kendrick et al., 2014).
Located in the human chromosome at 4q12-13, the c-kit proto-oncogene is a key point in cancer occurrence and proliferation (Abdel-Magid, 2021). The transcriptional product of c-kit is a type III receptor tyrosine kinase, which participates in regulating hematopoietic stem cell proliferation and differentiation. Recent findings have shown that c-kit mutations are strongly related to the morbidity and prognosis of gastrointestinal stromal tumors, small-cell lung cancer, melanoma, and systemic mastocytosis (Pathania et al., 2021). Given the overexpression of c-kit gene in some diseases, targeting and inhibiting c-kit expression is considered a novel strategy.
Through spectroscopic analysis, Bucek and his team (2009; 2010) reported the presence of i-motif-forming sequences in the c-kit promoter, where G4s and i-motifs coexist with duplexes at pH values of 3 to 6.5. Additionally, compounds such as terpyridine derivatives (Wei and Gao, 2015), trisubstituted isoalloxazines (Bejugam et al., 2007), and benzo[a]phenoxazine derivatives (McLuckie et al., 2011) have high affinities for c-kit G4s.
As a human proto-oncogene, c-myb plays an important role in regulating cell proliferation and differentiation in the hematopoietic and gastrointestinal systems. The c-myb mutation or overexpression induces cancerous lesions including acute myeloid leukemia and breast, colon, and gastroesophageal cancers (Gonda, 1998; Ramsay et al., 2003). Hence, it is vital to regulate c-myb expression when treating these diseases.
The c-myb G4 DNA blocks the transcriptional activity of the T7 RNA polymerase (Mishra et al., 2019). Wang et al. (2020) reported that the natural compound hyoscine butylbromide specifically bound c-myb G4 DNA with a binding constant of 1.18 × 105 L/mol. There remains comparatively scarce researches on c-myb i-motifs, most of which focus on factors affecting the formation and structural stability of the c-myb i-motif. Intramolecular i-motif structures formed in a stretch of cytosine-rich sequence S6 in the transcription start site of c-myb at pH 7.0 and ions like H+ and K+ promoted the transformation of the double helix to G4/i-motif structures (Li et al., 2016). Moreover, Li et al. (2016) screened out a natural product, Fangchinoline (10, Figure 3), which combines the c-myb i-motif DNA mainly in a “1 + 1” mode with a low affinity.
The c-myc is a proto-oncogene, the protein of which widely regulates transcription (Dang et al., 2006). The c-myc expression has important implications for the metabolism and growth of cancers including adenomatous colorectal polyposis and lymphoma (Shim et al., 1997; He et al., 1998). Both c-myc G4 and i-motif structures control c-myc transcription, as demonstrated in footprinting analysis of the c-myc promoter region in vitro.
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is closely related to chromatin remodeling, transcription, splicing, and translation (Bomsztyk et al., 2004). For c-myc, it is also a significant transcriptional activator that may speed carcinogenesis (Sutherland et al., 2016) (E)-1-(4-methoxyphenyl)-3-(4-morpholino-6-nitroquinolin-2-yl) prop-2-en-1-one (11, Figure 3) is the first-reported ligand for the hnRNP K protein. When this compound binds to hnRNP K, it induces i-motifs unfolding, resulting in c-myc downregulation. It also shows different anti-proliferative effects on human cancer cell lines, with IC50 values ranging from 1.36 M to 3.59 M (Shu et al., 2019). TMPyP4 (12, Figure 3) specifically binds to the top of the i-motif structure of nuclease hypersensitivity element III1 (NHE III1) in c-myc, subsequently preventing hnRNP K from binding to c-myc and ultimately silencing c-myc (Bialis et al., 2007). According to ITC data, TMPyP4 (12) can embed itself in two adjacent C-C+ base pairs through π–π stacking (Figure 7). Qin et al. (2017) confirmed that TMPyP4 (12) can bind to the major external groove of the i-motif structure owing to its large steric hindrance and ionic strength (Figure 7). TMPyP4 (12) also combines with both ends of the i-motif in the presence of Van der Waals and electrostatic forces (Cashman et al., 2008) (Figure 7).
The double acridine derivative a9 (13, Figure 3), which stabilizes the c-myc promoter NHE III1 G4 and i-motif, inhibited the proliferation of lymphatic carcinoma cell lines, including Raji and CA46, and limited the clone formation and migration of the SiHa human cervical squamous cell line. And the IC50 of these two lymphatic carcinoma cell lines are 3.385 μM and more than 50 μM respectively (Kuang et al., 2020). ActD (Actinomycin D) (14, Figure 3) binds non-specifically to the duplex DNA and unwinds it, after which ActD (14) preferentially binds to G4 DNA by terminal stacking, with a binding constant of 1.34 × 105 M-1 at pH 7.2. However, the binding constant of ActD (14) and c-myc i-motif is only 9.3 × 104 M-1 at pH 5, which is the weakest constant among three non-B DNA conformations (Niknezhad et al., 2016).
The RAS gene family is one of the most widespread proto-oncogenes and includes KRAS, NRAS, and HRAS. HRAS transmits signals to the nucleus and stimulates cell proliferation (Lowy, 1993). Miglietta et al. (2015) proved that the lateral loops of the HRAS i-motif provide binding sites for the RRM domains of heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). After hnRNP A1 stably binds to the i-motif sequence, gene transcription is activated. Compounds 1 and 2 (15, 16, Figure 3) show sub-micromolar affinities for the HRAS i-motif (Table 1), which is a C-rich sequence located upstream of the HRAS oncogene transcription start site. Compound 1 (15) is assumed to directly bind to the i-motif core region, while compound 2 (16) binds to the minor loop region (Journey et al., 2018).
Among the RAS gene family, KRAS is a frequently mutated oncogene with specific frequencies of variation in different cancer types. KRAS hyperactivation often results in sustained tumor proliferation, mostly common in lung, colorectal, and pancreatic carcinomas (Friday and Adjei, 2005; Baines et al., 2011; Cox et al., 2014). Thus, researchers have identified small molecule inhibitors with anticancer activity to treat KRAS-driven cancers (Kumar and Priya Doss, 2021a; Kumar and Priya Doss, 2021b; Udhaya Kumar et al., 2022).
While studies on the KRAS i-motif are insufficient, it is clear that the C-rich nucleic acid sequences in the KRAS promoter can form i-motifs, which are in a state of dynamic transition with hairpin species (Manzini et al., 1994; Amato et al., 2022). HnRNP K can bind selectively to the KRAS i-motif and upregulate KRAS transcription. Kaiser et al. (2017) showed that nitidine (17, Figure 3), a benzophilite alkaloid, is preferred to bind hairpin species in the central loop region of KRAS i-motifs (Figure 7). The cross-link between hnRNP K and i-motif is then destroyed, following with refolding of the i-motif and increasing gene transcription. Nitidine (17) also stabilized KRAS G4s and microscale thermophoresis analysis showed that it decreased KRAS expression in pancreatic cancer cells (Morgan et al., 2016). Thus, nitidine (17) plays a dual role in i-motif and G4s, providing a distinct mechanism for drug development.
Brown et al. (2017) reported that the nuclease hypersensitivity element (NHE) in the human PDGFR promoter could form an i-motif structure. The R1 mutation (T-to-C) in the PDGFR i-motif, increased the thermal stability of the i-motif (ΔT = 13.2 °C). NSC309874 (18, Figure 3) preferentially bound to the R1-mutated i-motif in medium-flux screening and finally downregulated gene expression in neuroblastoma cells.
The acridone derivative B19 (19, Figure 3) selectively induced i-motif formation at the c-myc promoter and downregulated its transcription, eventually causing apoptosis in cancer cells (Shu et al., 2018). Since the compound showed insufficient anticancer activity, Zhang et al. (2021) modified it into the acridone naphthalide derivative WZZ02 (20, Figure 3). In vitro experiments showed that WZZ02 (20) exhibited the specificity of stabilizing the G4 located in the PDGFR promoter and the potential to disrupt the complementary i-motif structure, leading to PDGFR downregulation in a dose-dependent manner. WZZ02 (20) also showed excellent anticancer activity in the MCF-7 xenograft tumor model by inducing cell apoptosis and cycle arrest and inhibiting proliferation, likely due to its intricate interaction with PDGFR G4 and i-motif. Hence, WZZ02 (20) may be a potential agent for cancer therapy.
The cell cycle checkpoint protein RAD17 is directly involved in the cellular DNA damage and replication detection system, playing a critical role in maintaining genomic stability and carcinogenesis (Zhou et al., 2013). While limited work has assessed i-motifs located in the RAD17 promoter sequence, the structure of the RAD17 i-motif changes rapidly when the pH drops, without observable hysteresis (Rogers et al., 2018).
The RET proto-oncogene encodes the receptor tyrosine kinase and is crucial for neurodevelopment. Most commonly, RET mutations cause two neural crest disorders, Hirschsprung’s disease and multiple endocrine neoplasia type 2 (Manié et al., 2001). Stable G4 and i-motif structures can form within the subterminal promoter region of human RET (Huppert and Balasubramanian, 2007). A fluorescent cytosine analog, 1,3-diaza-2-oxophenothiazine used as a pH probe showed DNA forming double strands at higher pH while shaping i-motifs in acidic environments. The RET i-motif also showed a higher anisotropic signal; thus, this system can be used to monitor reversible pH changes in the design of molecular logic gates and intricate biosensors (Bielecka and Juskowiak, 2015; Bielecka et al., 2019).
Vascular endothelial growth factor (VEGF) is a diffusible endothelial cell-specific mitogen and angiogenic factor and the major regulator of physiological angiogenesis irreplaceable for embryonic development and disease progression. Additionally, VEGF mRNA upregulation is associated with angiogenesis in most common human tumors and proliferative retinopathy. Thus, resisting VEFG is considered an effective strategy for the treatment of neoplasms and retinopathy (Ferrara, 1995; Matsumoto and Ema, 2014).
Guo et al. (2008) reported the presence of intramolecular i-motif structures in the poly C region of the proximal end of the VEGF promoter. In the context of epigenetics, accompanied by the cytosine-phosphate-guanine (CpG) methylation, VEGF i-motif structures could be more stable (Kimura et al., 2022). With the help of a DNA methylation detection system, Yoshida et al. (2016) showed decreased DNA polymerase efficiency with increased DNA methylation in VEGF i-motif sequences, indicating that the i-motif formation sequence may inhibit gene expression by increasing methylation. Takahashi et al. (2020) showed that the plant flavonol fisetin (Fis) (21, Figure 3) preferred to combine with the VEGF i-motif and provoked i-motif unfolding. Moreover, the fluorescence emission spectra showed that Fis (21) bound to the central ring of the VEGF i-motif.
Telomerase maintains the integrity of chromosome ends and is important in cell immortalization and carcinogenesis. Human telomerase mainly consists of three different subunits, among which the catalytic subunit hTERT is the rate-limiting determinant of telomerase activity. The hTERT expression is sensitized when carcinogenesis occurs. However, this expression is regulated not only by various activators and inhibitors but also epigenetic pathways like DNA methylation and histone modification (Takakura et al., 1999; Daniel et al., 2012).
Initially, TMPyP4 (12) was identified as a specific ligand for hTERT G4s (Fedoroff et al., 2000). Later, NMR spectroscopy results from Fernández et al. (2011) showed that TMPyP4 (12) also advances the formation of the h-telo (human telomeric) i-motif DNA structure in a non-intercalation mode. Pagano and colleagues (2018) showed that existing G4 ligands, including BRACO-19, mitoxantrone, phen-DC3, pyridostatin, and RHPS4 (22-26, Figure 4) also interact with the h-telo i-motif. Moreover, BRACO-19 (22), mitoxantrone (23), and phen-DC3 (24) destabilize the h-telo i-motif. However, Wright and his team (2016) reported the opposite finding, likely due to the different experimental conditions, in which mitoxantrone (23) firmly bound to the i-motif and induced its formation at pH 5.5. As mitoxantrone (23) is a well-known topoisomerase II inhibitor for the treatment of non-Hodgkin lymphoma and metastatic breast cancer and slowing the progression of multiple sclerosis, the mechanisms by which mitoxantrone (23) combines with the i-motif require urgent study.
[Tb2(DL-Cys)4(H2O)8]Cl2 and [Tb2(DL-HVal)4(H2O)8]Cl6.2H2O (27, 28, Figure 4) are terbium amino acid complexes that can bind not only h-telo G4s but also i-motifs without conformational changes (Xu et al., 2006). The models for the interactions between the h-telo i-motif and a ruthenium (II) polypyridine compound (29, Figure 5) are complicated. Especially, the cis isoform (30, Figure 5) can bind the major groove of the h-telo i-motif more tightly due to its smaller spatial structure compared to the mer (31, Figure 5) and trans (32, Figure 5) isoforms. Three isoforms combine with the i-motif in different ways: the Λ-cis has 68 contacts with the i-motif core, only 52 in Δ-cis, and even fewer in the mer (31) and trans (32) isoforms. In addition, luminescence lifetime data supported that the cis (30) isoform can be used as a small molecule to detect death-associated protein (DAP) i-motifs (Spence et al., 2020).
Sheng et al. (2017) reported that thiazole orange (TO) (33, Figure 6) stabilized the h-telo i-motif no matter the sequence, therefore available as probes in i-motif DNA analysis. Moreover, they identified several novel i-motif-binding ligands, including tobramycin, alexidine, tilorone, chlorhexidine, phenazopyridine, amodiaquine, harmalol, quinalizarin, and minocycline tyrothricin (34-42, Figure 6). Recent research is lacking regarding how these compounds interact with i-motifs; thus, further exploration is needed. Slightly interacting with i-motif DNA, [Ru(bpy)2(dppz)]2+ (43, Figure 6) preferentially binds the h-telo G4 sequence and can act as a “photoswitch” to monitor the dynamic transition of G4 DNA structures (Shi et al., 2010).
Berberine (44, Figure 6) binds to the h-telo i-motif structure via electrostatic interactions, with a weak dissociation constant (Table 1) (Xu et al., 2016; Gargallo et al., 2021). Berberine (44) can also be used as an effective fluorescent probe to monitor the conformational conversion of i-motifs driven by pH (Xu et al., 2016). Absorption titration and thermodynamic analyses showed that phenanthroline compounds 1–3 (45-47, Figure 6) bind to the C-C+ base pairs of telomeric i-motifs in the presence of π-π stacking, intercalation, or hydrophobic interaction. These compounds show a higher affinity for G4 DNA compared to the i-motif, which is attributed to telomeric G4s having larger π-π stacking interactions and van der Waals contacts (Wang et al., 2013). The 1,8-, 1,4-, 1,5-, and 2,6-disubstituted anthraquinone monomers (48-51, Figure 6) were investigated to assess their effects on the thermal stability of the i-motif. When anthraquinones modify the TAA loop of the h-telo DNA sequence in the intercalation or external binding mode, the melting temperature increases significantly, accordingly enhancing the stability of the i-motif structure (Gouda et al., 2017).
In their analysis of structure-activity relationships, Wei et al. (2015) reported that the coumarin group may be the determinant of the specific affinities of terpyridine derivatives 1–3 (52-54, Figure 6) for the h-telo i-motif, among which terpyridine derivative 1 (52) reduced 88.7% of the telomerase activity and slightly restrained the bioactivity of topoisomerase I activity at 5 μmol/L. Moreover, macrocyclic bis-acridine (BisA) (55, Figure 6) shows a considerable affinity for the telomeric i-motif. While BisA (55) did not show obvious effects on the physiological function and conformation of telomeric i-motifs (Bonnet et al., 2022).
In addition, chemically modified cytosines have different effects on the formation of i-motif structures under physiological conditions. The telomeric i-motif became less stable at a neutral pH due to the methylated cytosine in MCF7 and MCF10A cell lines (Wright et al., 2020). Xu et al. (2015) also reported that the telomeric i-motif was stable with small amounts of cytosine methylation modification. However, numerous hydroxymethylation and methylation modifications could lead to structural destabilization. In plants, cytosine methylation is similarly meaningful in the epigenetic regulation of telomeric DNA (Školáková et al., 2020), which can also fine-tune the stability and the pH dependence of i-motifs. Balasubramaniyam et al. (2021) reported that the halogenation of cytosine at C5 accelerated i-motif folding, which eventually altered the pH dependence. Moreover, the hydrophilic hydroxyl group at C5 is more tolerant to immunostimulation compared to the hydrophobic methyl group (Kandimalla et al., 2001). Therefore, modifying the deoxycytidine or cytidine of i-motifs may be a new strategy to regulate immunity in vivo.
The synthetic C6T i-motif has two unequal broad grooves and is used to explore possible binding modes with compounds with long molecular structures such as polyamines. Putrescine (56, Figure 9) with the shortest molecular length, only binds to the loop region (Figure 7) and, thus, has the lowest affinity for i-motifs. In contrast, the longer molecule spermidine (57, Figure 9) can bind the loop region and the groove; therefore, it has two quite different binding constants. The longest molecule, spermine (58, Figure 9), can simultaneously bind two broad grooves of the C6T i-motif, with the highest binding affinity (Molnar et al., 2019) (Table 1). Based on mechanisms of polyamine binding to i-motifs, additional research is needed to identify additional ligands with greater affinity by altering the molecular length of the polyamines. Kinetic analysis has shown that the addition of 5′-terminal guanidino-i-clamp (Tsvetkov et al., 2019) or i-clamp (Tsvetkov et al., 2018) can decrease the unfolding rate of synthetic i-motifs, which guarantees the stability of i-motifs. It is also possible to become unstable given the steric hindrance.
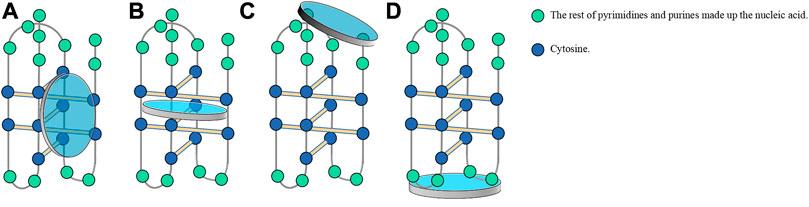
FIGURE 7. Usual modes of ligands binding to the i-motif. (A) Binding to the major external groove of the i-motif owing to large steric hindrance and ionic strength. (B) Embedding in C-C+ base pairs by π-π stacking. (C) Stacking at two ends of the i-motif in the presence of Van der Waals and electrostatic forces. (D) Binding to hairpin species in the central loop region of the i-motif due to steric hindrance and molecular length.
Additionally, BmPOUM2 has been reported to be a significant regulator of the wing disc cuticle protein gene, namely, BmWCP4. Silkworms cannot complete metamorphosis when the BmPOUM2 is expressed. An i-motif structure and the novel BmILF i-motif-binding protein have been identified. BmILF specifically binds the i-motif and activates BmPOUM2, which prevents silkworm metamorphosis into moths (Niu et al., 2018). For agriculturists, these findings provide new insights into the epigenetic mechanism for how to control the growth of silkworms to achieve a high silk yield.
The above summary showed that natural i-motif structures are mainly located in the promoter regions of various oncogenes. Hence, most current studies on the biological function of i-motifs have focused on how to build targeted and effective anticancer strategies through the combination of i-motif and other molecules. Therefore, attention to genes with very few or no drug studies, such as c-kit, c-myb, RAD17, and RET, may demonstrate the value and application of i-motifs in the medical field. We hold a strong belief that i-motifs in promoters can be subtly regulated by ligands under specific conditions to stabilize or disrupt their structures and, ultimately, regulate their biological functions.
The discussion above provides a preliminary conclusion that i-motif-forming sequences are crucial for regulating chromosome integrity, mitosis, and protein synthesis and are linked to challenges such as cancer and aging (Hampton, 2011; Armanios and Blackburn, 2012; von Figura et al., 2009). The following section discusses in detail diseases related to i-motifs.
DM is often accompanied by cardiovascular complications like coronary heart disease, which is a common reason for clinical death (Rogowicz-Frontczak et al., 2012). Therefore, it is particularly urgent to treat diabetes. The insulin minisatellite region is the insulin-linked polymorphic region (ILPR), the polymorphism length of which is strongly linked to the genetic susceptibility of insulin-dependent diabetes mellitus (IDDM) (Catasti et al., 1996). The C-rich duplex sequence in ILPR forms intermolecular/intramolecular i-motifs through pairing between C+ and C (Jolad et al., 2005; Dhakal et al., 2012). Catasti et al. (1997) suggested that the stable folding of the i-motif in C-rich sequences led to the loose structure upstream of the insulin gene, promoting insulin expression. Dhakal et al. (2010) indicated that, simply from a mechanical perspective, when insulin i-motifs interact with RNA polymerases, the unfolding force of the i-motif is greater than the stall force, interrupting gene transcription (Galburt et al., 2007).
Experimental results at the cellular and individual levels will reveal whether insulin i-motifs advance or inhibit insulin transcription and provide more therapeutic options for patients with DM. Thus, more studies are needed on i-motifs in the human insulin gene at these levels.
HIV-1 is a human immunodeficiency virus and a basic pathogen of acquired immunodeficiency syndrome (AIDS) (Radestock et al., 2013). The i-motif structure can also form in the promoter of the HIV-1 DNA genome. Moreover, the i-motif located in the long terminal repeat (LTR) of the HIV-1 promoter has a unique folding pattern that differs from that in the human genome. Ruggiero et al. (2019) reported that hnRNP K induces the formation of the HIV-1 LTR i-motif and subsequently represses the transcription, finally decreasing the virulence of HIV-1. These findings lay a theoretical foundation for innovative antiviral drug design based on the selective recognition of the HIV-1 i-motif.
Depression is a neuropsychiatric disorder strongly associated with the serotonin transporter (SERT). The linkage polymorphic region of SERT contains two C-rich allelic variants that regulate susceptibility to depression by altering SERT expression levels. Both variants can form i-motifs; however, the mechanism requires further exploration (Zhang et al., 2015; Thorne et al., 2021). As a key regulator of serotonin, i-motifs in the SERT-linkage polymorphic region will be an important pharmacological breakthrough for treating depression.
Fragile X syndrome (FXS) is the most common consequence of genetic intellectual disability caused by the CGG/CCG tandem repeat sequences of Fragile X Messenger Ribonucleoprotein 1 (FMR1) on the X chromosome. CCG repeats prefer to form stable intermolecular i-motif structures (Tekendo-Ngongang et al., 2021; Zhang et al., 2022). Chen et al. (2018) reported that when the CCG trinucleotide repeat region is bound to the CoII (Chro)2 dimer (59, Figure 9), the i-motif unfolds and is restored the double-helix structure, suggesting that CoII (Chro)2 (59) could be a new drug for the treatment or diagnosis of neurological diseases.
In addition, hexanucleotide repeat amplification sequence G4C2 in the C9orf72 gene is the most common single genetic factor for frontotemporal dementia and amyotrophic lateral sclerosis. G4C2 can fold into unusual secondary structures, including R-loops, i-motifs, and G4s (Kumar et al., 2016). These findings indicate that i-motifs can be a new target in gene therapy for neuropsychiatric disorders.
Tumor occurrence is often accompanied by gene mutations. Any gene abnormality, such as the inactivation of the anti-oncogene or the activation of the proto-oncogene can cause cancer. Recent reports have demonstrated the central role of Bcl-2 in orchestrating the interplay between apoptosis and senescence. Bcl-2 over-activation results in the abnormal proliferation of cancer cells (Nahta and Esteva, 2003; Kim et al., 2004). As one of the most important transcription factors, c-myc protein is particularly significant for the reprogramming of multiple types of cancer cells, as well as their proliferation and chemoresistance (Fatma et al., 2022). The c-myc inactivation can lead to sustained tumor regression, which may be a key therapy to reverse cancerous growth and restore antitumor immune responses in patients with high c-myc expression (Dhanasekaran et al., 2022). Additionally, telomeres and VEGF play vital roles in indefinite proliferation and nutrient supply, which are essential for tumor growth.
As i-motifs form in specific regions of most oncogenes, a link must exist between cancer development and therapy. As mentioned above, researchers often establish specialized models, screen and modify drugs with certain cytotoxicity, and use the effects between drugs and i-motif to verify the anticancer potential of the studied drugs. For example, in the anti-DLBCL model, IMC-76 (8) was found to bind the Bcl-2 i-motif and upregulate Bcl-2 expression, thus slowing cancer cell growth (Kendrick et al., 2017). In the anti-lymphoid Raji cell line model, a9 (13) interacted with the c-myc i-motif and showed high cytotoxic effects, with an IC50 reaching 3.385 μM (Kuang et al., 2020). In the cancer model associated with VEGF, Fis (21) was used as a probe to specifically recognize the VEGF i-motif; therefore, this system can be used to diagnose VEGF-associated cancers (Takahashi et al., 2020). In the cancer model associated with telomeres, terpyridine derivative 1 (52) binds to the h-telo i-motif to inhibit telomerase and topoisomerase activity (Wei and Gao, 2015). Table 1 summarizes studies on drugs. Although clinical studies on these potential anti-cancer drugs are lacking, those on the association of i-motifs with cancer are essential.
Since these oncogenes have been introduced above, the retinoblastoma gene Rb is highlighted here. Rb is the first tumor suppressor gene identified in humans, the functional incapacitation of which is related to retinoblastoma tumorigenesis. The product of Rb, pRB protein, controls the cell cycle transition from the G1 to the S phase, pausing the cell cycle in a static state. Clinical findings suggest that a dysregulation of the G1-S control pathway may occur in retinoblastoma and sporadic lung, breast, and bladder cancers (Dannenberg et al., 2000). Thus, restoring or enhancing Rb function may be a late-model strategy for these cancers. Lee et al. (2015) identified the Rb i-motif based on the pH, providing evidence for retinoblastoma diagnosis. Studies on the structural characteristics, biological functions, and targeted ligands of i-motifs in RB are scarce (Xu and Sugiyama, 2005) and require future exploration.
In addition to these diseases, i-motifs also have specific roles in fighting bacterial infections. Hemiprotonic phenanthroline-phenanthroline+ compounds are synthesized based on the unique structure of half-protonated nucleotide base pairs of the i-motif DNA from oncogenes. These compounds not only selectively resist tumors but also have broad-spectrum antibacterial activity, providing new therapeutic candidate drugs for patients with cancer accompanied by infection (Zhao et al., 2022).
The diseases discussed in this review are i-motif-related. Among these diseases, cancer is a primary focus. The study of these diseases should consider the following four aspects: the specific gene locus of the i-motif, the effect of i-motif formation on the disease, pathways involved in the i-motif regulation of the disease, and different drug design schemes for different signaling molecules (Figure 8). As described above, current research on these diseases is still lacking. Therefore, additional research on this topic is needed.
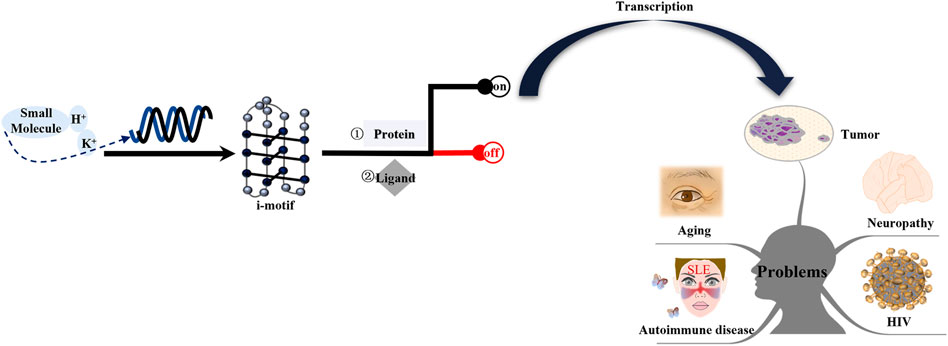
FIGURE 8. Mechanism of i-motifs in vivo. The i-motif DNA sequence folds into i-motif structures in specific conditions, including acid pH, K+, or some small molecules. I-motifs mainly have two effects: firstly, they directly bind to proteins such as hnRNP A1, hnRNP K, and hnRNP LL; second, they can directly interact with ligands. The result is switching gene expression on or off. Activated genes produce corresponding proteins after transcription. These proteins interact with other factors and form complexes that are transported to certain tissue. The release of these proteins can lead to tumors, neuropathy, aging, HIV, and autoimmune disease.
Applications of i-motifs have been reported in different fields. This section introduces and describes the applications of i-motifs in medicine.
In recent years, gene-based medicines wrapped in microparticles have been successfully delivered into the human body to concentrate in the reticuloendothelial system in vivo, thus realizing the targeted transportation of medicine. As a drug delivery system (DDS), the nanomaterial-i-motif DNA DDS has been widely applied due to its excellent biosafety and simplicity of synthesis.
Aptamers are usually short oligonucleotide sequences or polypeptides that can bind the corresponding ligands with high affinity and strong specificity and have become valuable molecular tools in the development of bioanalysis and targeted therapy. A structure-switchable aptamer (SW-Apt) with an i-motif is one reported tool. The i-motif forms under an acidic pH, thus making an SW-Apt with high binding capacity to target cells, which modulates the specific recognition of the aptamer with the help of the dependence of the i-motif on pH and is enlightening for aptamer construction (Li et al., 2018). The aptamer DNA-cyclodextrin makes the system freely slide along the polymer PEG chain and the assembly of drugs and the i-motif DNA can respond to pH change to release drugs under intracellular acidic conditions, enhancing cellular uptake and healing efficacy (Jang et al., 2017). A DNA nanocapsule with an acidic pH-responsive i-motif DNA and a tumor cell-specific aptamer is first triggered by an acidic pH and then targets cells via the specific aptamer recognition and releases the carried drug doxorubicin, successfully providing selective cytotoxicity to cancer cells (Yuan et al., 2022). Similarly, a nanocarrier based on bovine serum albumin and DNA including a pH-responsive i-motif and a cancer cell-targeted guanine-quadruplex-structured aptamer also showed accurate targeting and efficient therapeutic effect on cancer cells (Yu et al., 2021). In addition to targeted therapy, i-motifs are also useful in immunotherapy. As cyclic dinucleotides (CDNs) like c-di-GMP (CDG) are agonists for stimulator of interferon genes (STING), STING-activating DNA nanovaccines (STING-NVs) with i-motif DNA on the surface showed potential to improve immunosuppression in vitro and in vivo in a murine melanoma model. In acidic environments such as the cell endosome, the i-motif was formed and CDG was released, facilitating the cell delivery of CDG (Zhang et al., 2020). Nanotubes of the anti-inflammatory drug dexamethasone modified with the i-motif were used to target the reticuloendothelial system. The i-motifs manipulated the intracellular release of dexamethasone with pH changes, thereby regulating the anti-inflammatory activity of macrophages (Sellner et al., 2017).
Hydrogels equipped with functional nucleic acids can be used to construct molecules based on aptamers, DNA enzymes, i-motif nanostructures, siRNA, and CpG oligonucleotides, all of which provide additional recognition sites, catalytic activities, and therapeutic potential (Li et al., 2016). DNA nanogels containing gemcitabine enhance the anticancer activity, for the intramolecular i-motif structure forming under acidic conditions, which facilitates nanogel disintegration and gemcitabine release (Pan et al., 2019). An intelligent DNA nanosystem based on controllable DNA nanohydrogels consists of a pH-responsive i-motif sequence, doxorubicin, the CpG fusion sequence, and an aptamer for immunostimulation and chemotherapy. This nanosystem integrates targeting, immune response, and chemotherapy to fight malignancy (Wei et al., 2019).
In addition to triggering drug release under certain conditions, i-motifs can also act as diagnostic biomarkers in biomedical sciences. Heydari et al. (2016) used tamoxifen (Tam) (60, Figure 9) as the ligand and prepared an electrochemical biosensor with a nanosilica-modified carbon paste electrode to detect and differentiate human telomeric DNA. In acidic environments, the interaction between i-motifs and Tam (60) produced oxidation peaks; when the pH goes increased, the interaction decreased and the oxidation peak changed. Takahashi et al. (2020) reported the excited-state intramolecular proton transfer reaction was significantly induced and the intensity of the tautomer emission band of Fis (21) was enhanced when Fis (21) bound to the VEGF i-motif, which could be used for the diagnosis of cancers associated with VEGF. In addition, thioflavin T (61, Figure 9) can distinguish structural changes in RET and Rb i-motif sequences based on pH changes; thus, it can be used as a special probe for these two nucleotide sequences and may play a good supporting role in genetic diagnosis and treatment (Lee et al., 2015). A metal-organic framework (MOF)-shell-confined i-motif-based pH probe strategy can be used to distinguish metabolic behaviors of cancer and normal cells based on the pH, in which the labeled i-motif optimizes for pH sensing and the MOF shell limits endocrine acid diffusion (Yang et al., 2021). The dual-pyrene-functionalized i-motif can provide analytical results regarding pH changes; thus, it shows potential as a pH-sensitive fluorescent probe (Dembska et al., 2013). Likewise, i-motifs tethered on framework nucleic acids can act as a controlling unit and can be designed as logical sensors in response to extracellular changes in H+ and K+ (Peng et al., 2020).
Regarding the application of i-motifs, we mainly introduce those in nanomedicine. These studies demonstrated their important roles as detectors and in DDS made of i-motifs and nanomaterials. The application of i-motifs in the medical field is promising and warrants further exploration. In addition to the medical field, other practical applications of i-motifs are possible; however, the specific direction requires further exploration.
This review clarified the structure, ligands, related diseases, and applications of various i-motifs. As an emerging cytosine-rich non-B-DNA structure, i-motifs form in the promoters of many human oncogenes. As discussed above, i-motifs have significant biological functions. To elucidate the concrete regulation mechanisms of i-motifs, we extensively discussed the relevant ligand compounds. The analysis and summarization of previous research results showed that the same ligand compound can show different results for different experimental conditions (Table 1). The effects of ligands on the i-motif also vary among genes. Increasing numbers of ligands have been reported. Table 1 summarizes ligand compounds that interact with i-motifs under different experimental conditions. These ligands affect the stability of i-motifs, activate/inhibit the gene expression in many promoters, and have important implications for cell growth and development, which are closely related to cancers, mental disorders, aging, infection, and other diseases.
G4 DNA binds its ligands mainly through three modes; namely, end accumulation, intermolecular insertion, and non-specific binding; for example, porphyrinoids, telomestatin, and Se2SAP (Rezler et al., 2005; Baker et al., 2006). Figure 7 shows the major modes by which ligands bind to i-motifs. However, continuing research will reveal novel ligands and new modes. Therefore, this topic warrants additional research to explore i-motif ligand drugs, explain the regulatory mechanisms at micro and macro levels, and provide more new drugs and treatment methods for genetically-related human diseases.
Regarding cancer treatment, previous studies hypothesized that the inhibition of i-motif formation or decreasing the structural stability to fight cancer. Researchers often target a regulatory factor, which can be an enzyme, protein, or receptor molecule. Interactions between i-motifs and ligand molecules change the function of regulatory factors to downregulate or upregulate gene expression and inhibit cancer cell growth and proliferation to ultimately alleviate or even cure cancer. This field of research requires more corresponding experiments to improve the therapy choices for patients. In addition, i-motifs can function as DNA carriers or probe molecules, playing important roles in transmitting gene drugs in the nano-drug delivery system with the rapid development of nanomedicine.
Conceptualization and supervision, YW, CW, and JZ; original manuscript preparation, XiL, WP, XuL, YG, YY, and LC; manuscript review and editing, YW, CW, and JZ. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Natural Science Foundation of China (81903467), the Fund of Guangzhou Science and Technology Program (202002030410), the Fund of the Guangdong Education Department (2020KTSCX102), a High-level University Construction Fund of Guangdong Province (06-410-2107212), a University Student Laboratory Open Fund from Guangzhou Medical University (PX-1020418), and the Undergraduates Research Project of Guangzhou Medical University (02-410-2206290, 02-410-2206305, 2022A102, and 2022A118).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Magid, A. F. (2021). The potential of c-KIT kinase inhibitors in cancer treatment. ACS Med. Chem. Lett. 12, 1191–1192. doi:10.1021/acsmedchemlett.1c00332
Abdelhamid, M. A. S., Gates, A. J., and Waller, Z. A. E. (2019). Destabilization of i-motif DNA at neutral pH by G-quadruplex ligands. Biochemistry 58, 245–249. doi:10.1021/acs.biochem.8b00968
Abou Assi, H., GaravíS, M., GonzáLEZ, C., and Damha, M. J. (2018). i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 46, 8038–8056. doi:10.1093/nar/gky735
Amato, J., Iaccarino, N., D'Aria, F., D'Amico, F., Randazzo, A., Giancola, C., et al. (2022). Conformational plasticity of DNA secondary structures: Probing the conversion between i-motif and hairpin species by circular dichroism and ultraviolet resonance Raman spectroscopies. Phys. Chem. Chem. Phys. PCCP 24, 7028–7044. doi:10.1039/d2cp00058j
Armanios, M., and Blackburn, E. H. (2012). The telomere syndromes. Nat. Rev. Genet. 13, 693–704. doi:10.1038/nrg3246
Bacolla, A., and Wells, R. D. (2004). Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 279, 47411–47414. doi:10.1074/jbc.R400028200
Baines, A. T., Xu, D., and Der, C. J. (2011). Inhibition of ras for cancer treatment: The search continues. Future Med. Chem. 3, 1787–1808. doi:10.4155/fmc.11.121
Baker, E. S., Lee, J. T., Sessler, J. L., and Bowers, M. T. (2006). Cyclo[n]pyrroles: Size and site-specific binding to G-quadruplexes. J. Am. Chem. Soc. 128, 2641–2648. doi:10.1021/ja0564968
Balasubramaniyam, T., Oh, K. I., Jin, H. S., Ahn, H. B., Kim, B. S., and Lee, J. H. (2021). Non-canonical helical structure of nucleic acids containing base-modified nucleotides. Int. J. Mol. Sci. 22, 9552. doi:10.3390/ijms22179552
Bejugam, M., Sewitz, S., Shirude, P. S., Rodriguez, R., Shahid, R., and Balasubramanian, S. (2007). Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: Small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 129, 12926–12927. doi:10.1021/ja075881p
Belmonte-Reche, E., and Morales, J. C. (2020). G4-iM grinder: When size and frequency matter. G-quadruplex, i-motif and higher order structure search and analysis tool. NAR genomics Bioinforma. 2, lqz005. doi:10.1093/nargab/lqz005
Bialis, T., Dexheimer, T., Gleasonguzman, M., Yang, D., and Hurley, L. (2007). Transcriptional consequences of targeting the i-motif structure of the c-Myc promoter with TMPyP4. Cancer Res. 67, 3169.
Bielecka, P., Dembska, A., and Juskowiak, B. (2019). Monitoring of pH using an i-motif-forming sequence containing a fluorescent cytosine analogue, tC. Mol. (Basel, Switz. 24, 952. doi:10.3390/molecules24050952
Bielecka, P., and Juskowiak, B. (2015). Fluorescent sensor for PH monitoring based on an i-motif-switching aptamer containing a tricyclic cytosine analogue (tC). Mol. (Basel, Switz. 20, 18511–18525. doi:10.3390/molecules201018511
Biffi, G., Tannahill, D., Mccafferty, J., and Balasubramanian, S. (2013). Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186. doi:10.1038/nchem.1548
Bochman, M. L., Paeschke, K. K., and Zakian, V. A. (2012). DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780. doi:10.1038/nrg3296
Bomsztyk, K., Denisenko, O., and Ostrowski, J. (2004). hnRNP K: one protein multiple processes. BioEssays news Rev. Mol. Cell. Dev. Biol. 26, 629–638. doi:10.1002/bies.20048
Bonnet, H., Morel, M., Devaux, A., Boissieras, J., Granzhan, A., Elias, B., et al. (2022). Assessment of presumed small-molecule ligands of telomeric i-DNA by biolayer interferometry (BLI). Chem. Commun. Camb. Engl. 58, 5116–5119. doi:10.1039/d2cc00836j
Brooks, T. A., and Hurley, L. H. (2009). The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer 9, 849–861. doi:10.1038/nrc2733
Brown, R. V., Wang, T., Chappeta, V. R., Wu, G., Onel, B., Chawla, R., et al. (2017). The consequences of overlapping G-quadruplexes and i-motifs in the platelet-derived growth factor receptor β core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J. Am. Chem. Soc. 139, 7456–7475. doi:10.1021/jacs.6b10028
Bucek, P., Gargallo, R., and Kudrev, A. (2010). Spectrometric study of the folding process of i-motif-forming DNA sequences upstream of the c-kit transcription initiation site. Anal. Chim. Acta 683, 69–77. doi:10.1016/j.aca.2010.10.008
Bucek, P., Jaumot, J., Aviñó, A., Eritja, R., and Gargallo, R. (2009). pH-Modulated Watson-Crick duplex-quadruplex equilibria of guanine-rich and cytosine-rich DNA sequences 140 base pairs upstream of the c-kit transcription initiation site. Chem. (Weinheim der Bergstrasse, Ger. 15, 12663–12671. doi:10.1002/chem.200901631
Bugaut, A., and Balasubramanian, S. (2008). A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 47, 689–697. doi:10.1021/bi701873c
Burge, S., Parkinson, G. N., Hazel, P., Todd, A. K., and Neidle, S. (2006). Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415. doi:10.1093/nar/gkl655
Cashman, D. J., Buscaglia, R., Freyer, M. W., Dettler, J., Hurley, L. H., and Lewis, E. A. (2008). Molecular modeling and biophysical analysis of the c-MYC NHE-III1 silencer element. J. Mol. Model. 14, 93–101. doi:10.1007/s00894-007-0254-z
Catasti, P., Chen, X., Deaven, L. L., Moyzis, R. K., Bradbury, E. M., and Gupta, G. (1997). Cystosine-rich strands of the insulin minisatellite adopt hairpins with intercalated cytosine+.cytosine pairs. J. Mol. Biol. 272, 369–382. doi:10.1006/jmbi.1997.1248
Catasti, P., Chen, X., Moyzis, R. K., Bradbury, E. M., and Gupta, G. (1996). Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 264, 534–545. doi:10.1006/jmbi.1996.0659
Chen, Y. W., Satange, R., Wu, P. C., Jhan, C. R., Chang, C. K., Chung, K. R., et al. (2018). Co(II)(Chromomycin)₂ complex induces a conformational change of CCG repeats from i-motif to base-extruded DNA duplex. Int. J. Mol. Sci. 19, 2796. doi:10.3390/ijms19092796
Collin, D., and Gehring, K. (1998). Stability of chimeric DNA/RNA cytosine tetrads: Implications for i-motif formation by RNA. J. Am. Chem. Soc. 120, 4069–4072. doi:10.1021/ja973346r
Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J., and Der, C. J. (2014). Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851. doi:10.1038/nrd4389
Cui, Y., Koirala, D., Kang, H., Dhakal, S., Yangyuoru, P., Hurley, L. H., et al. (2014). Molecular population dynamics of DNA structures in a bcl-2 promoter sequence is regulated by small molecules and the transcription factor hnRNP LL. Nucleic Acids Res. 42, 5755–5764. doi:10.1093/nar/gku185
Dang, C. V., O'Donnell, K. A., Zeller, K. I., Nguyen, T., Osthus, R. C., and Li, F. (2006). The c-Myc target gene network. Seminars cancer Biol. 16, 253–264. doi:10.1016/j.semcancer.2006.07.014
Daniel, M., Peek, G. W., and Tollefsbol, T. O. (2012). Regulation of the human catalytic subunit of telomerase (hTERT). Gene 498, 135–146. doi:10.1016/j.gene.2012.01.095
Dannenberg, J. H., Van Rossum, A., Schuijff, L., and Te Riele, H. (2000). Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14, 3051–3064. doi:10.1101/gad.847700
Day, H. A., Pavlou, P., and Waller, Z. A. (2014). i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 22, 4407–4418. doi:10.1016/j.bmc.2014.05.047
Debnath, M., Ghosh, S., Chauhan, A., Paul, R., Bhattacharyya, K., and Dash, J. (2017). Preferential targeting of i-motifs and G-quadruplexes by small molecules. Chem. Sci. 8, 7448–7456. doi:10.1039/c7sc02693e
Del Toro, M., Bucek, P., Aviñó, A., Jaumot, J., GonzáLEZ, C., Eritja, R., et al. (2009). Targeting the G-quadruplex-forming region near the P1 promoter in the human BCL-2 gene with the cationic porphyrin TMPyP4 and with the complementary C-rich strand. Biochimie 91, 894–902. doi:10.1016/j.biochi.2009.04.012
Dembska, A., Rzepecka, P., and Juskowiak, B. (2013). Spectroscopic characterization of i-motif forming c-myc derived sequences double-labeled with pyrene. J. Fluoresc. 23, 807–812. doi:10.1007/s10895-013-1184-z
Dexheimer, T., Sun, D., and Hurley, L. (2006). Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 128, 5404–5415. doi:10.1021/ja0563861
Dhakal, S., Lafontaine, J. L., Yu, Z., Koirala, D., and Mao, H. (2012). Intramolecular folding in human ILPR fragment with three C-rich repeats. PLoS One 7, e39271. doi:10.1371/journal.pone.0039271
Dhakal, S., Schonhoft, J. D., Koirala, D., Yu, Z., Basu, S., and Mao, H. (2010). Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J. Am. Chem. Soc. 132, 8991–8997. doi:10.1021/ja100944j
Dhanasekaran, R., Deutzmann, A., Mahauad-Fernandez, W. D., Hansen, A. S., Gouw, A. M., and Felsher, D. W. (2022). The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 19, 23–36. doi:10.1038/s41571-021-00549-2
Dickerson, R., Drew, H., Conner, B., Wing, R., Fratini, A., and Kopka, M. (1982). The anatomy of A-B-and Z-DNA. Science 216, 475–485. doi:10.1126/science.7071593
Fatma, H., Maurya, S. K., and Siddique, H. R. (2022). Epigenetic modifications of c-MYC: Role in cancer cell reprogramming, progression and chemoresistance. Seminars cancer Biol. 83, 166–176. doi:10.1016/j.semcancer.2020.11.008
Fedoroff, O. Y., Rangan, A., Chemeris, V. V., and Hurley, L. H. (2000). Cationic porphyrins promote the formation of i-motif DNA and bind peripherally by a nonintercalative mechanism. Biochemistry 39, 15083–15090. doi:10.1021/bi001528j
FernáNDEZ, S., Eritja, R., Aviñó, A., Jaumot, J., and Gargallo, R. (2011). Influence of pH, temperature and the cationic porphyrin TMPyP4 on the stability of the i-motif formed by the 5'-(C3TA2)4-3' sequence of the human telomere. Int. J. Biol. Macromol. 49, 729–736. doi:10.1016/j.ijbiomac.2011.07.004
Ferrara, N. (1995). The role of vascular endothelial growth factor in pathological angiogenesis. Breast cancer Res. Treat. 36, 127–137. doi:10.1007/bf00666035
Friday, B. B., and Adjei, A. A. (2005). K-ras as a target for cancer therapy. Biochimica biophysica acta 1756, 127–144. doi:10.1016/j.bbcan.2005.08.001
Galburt, E. A., Grill, S. W., Wiedmann, A., Lubkowska, L., Choy, J., Nogales, E., et al. (2007). Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature 446, 820–823. doi:10.1038/nature05701
Gao, B., and Hou, X. M. (2021). Opposite effects of potassium ions on the thermal stability of i-motif DNA in different buffer systems. ACS Omega 6, 8976–8985. doi:10.1021/acsomega.0c06350
Gargallo, R., Aviñó, A., Eritja, R., Jarosova, P., Mazzini, S., Scaglioni, L., et al. (2021). Study of alkaloid berberine and its interaction with the human telomeric i-motif DNA structure. Spectrochimica acta. Part A, Mol. Biomol. Spectrosc. 248, 119185. doi:10.1016/j.saa.2020.119185
Gonda, T. J. (1998). The c-Myb oncoprotein. Int. J. Biochem. Cell Biol. 30, 547–551. doi:10.1016/s1357-2725(98)00003-x
Gouda, A. S., Amine, M. S., and Pedersen, E. B. (2017). Improved i-motif thermal stability by insertion of anthraquinone monomers. Org. Biomol. Chem. 15, 6613–6621. doi:10.1039/c7ob01393k
GuéDIN, A., Gros, J., Alberti, P., and Mergny, J. L. (2010). How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 38, 7858–7868. doi:10.1093/nar/gkq639
Guo, K., Gokhale, V., Hurley, L. H., and Sun, D. (2008). Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 36, 4598–4608. doi:10.1093/nar/gkn380
Guo, K., Pourpak, A., Beetz-Rogers, K., Gokhale, V., Sun, D., and Hurley, L. H. (2007). Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 129, 10220–10228. doi:10.1021/ja072185g
Hampton, T. (2011). Studies probe role of telomere length in predicting, modulating cancer risk. JAMA 305, 2278–2279. doi:10.1001/jama.2011.772
Hardin, C. C., Perry, A. G., and White, K. (2000). Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers 56, 147–194. doi:10.1002/1097-0282(2000/2001)56:3<147:Aid-bip10011>3.0.Co;2-n
He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., Da Costa, L. T., et al. (1998). Identification of c-MYC as a target of the APC pathway. Sci. (New York, N.Y.) 281, 1509–1512. doi:10.1126/science.281.5382.1509
Heydari, E., Raoof, J. B., Ojani, R., and Bagheryan, Z. (2016). SiO2 nanoparticles modified CPE as a biosensor for determination of i-motif DNA/Tamoxifen interaction. Int. J. Biol. Macromol. 89, 421–427. doi:10.1016/j.ijbiomac.2016.05.001
Huppert, J. L., and Balasubramanian, S. (2007). G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413. doi:10.1093/nar/gkl1057
Huppert, J. L., and Balasubramanian, S. (2005). Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916. doi:10.1093/nar/gki609
Jakubowska, A., Zajaczek, S., Haus, O., Limon, J., Kostyk, E., Krzystolik, Z., et al. (2001). Novel RB1 gene constitutional mutations found in Polish patients with familial and/or bilateral retinoblastoma. Hum. Mutat. 18, 459. doi:10.1002/humu.1220
Jang, D., Lee, Y. M., Lee, J., Doh, J., and Kim, W. J. (2017). Remission of lymphoblastic leukaemia in an intravascular fluidic environment by pliable drug carrier with a sliding target ligand. Sci. Rep. 7, 40739. doi:10.1038/srep40739
Jin, K. S., Shin, S. R., Ahn, B., Rho, Y., Kim, S. J., and Ree, M. (2009). pH-dependent structures of an i-motif DNA in solution. J. Phys. Chem. B 113, 1852–1856. doi:10.1021/jp808186z
Jolad, V. V., Murad, F. K., Arnold, J. R., and Fisher, J. (2005). Solution conformation of d(C(4)ACAC(4)TGT)(2); an intramolecularly folded i-motif from the insulin minisatellite. Org. Biomol. Chem. 3, 2234–2236. doi:10.1039/b504606h
Journey, S. N., Alden, S. L., Hewitt, W. M., Peach, M. L., Nicklaus, M. C., and Schneekloth, J. S. (2018). Probing the hras-1(Y) i-motif with small molecules. MedChemComm 9, 2000–2007. doi:10.1039/c8md00311d
Kaiser, C. E., Van Ert, N. A., Agrawal, P., Chawla, R., Yang, D., and Hurley, L. H. (2017). Insight into the complexity of the i-motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J. Am. Chem. Soc. 139, 8522–8536. doi:10.1021/jacs.7b02046
Kandimalla, E. R., Yu, D., Zhao, Q., and Agrawal, S. (2001). Effect of chemical modifications of cytosine and guanine in a CpG-motif of oligonucleotides: Structure-immunostimulatory activity relationships. Bioorg. Med. Chem. 9, 807–813. doi:10.1016/s0968-0896(00)00316-3
Kang, H. J., Kendrick, S., Hecht, S. M., and Hurley, L. H. (2014). The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 136, 4172–4185. doi:10.1021/ja4109352
Kendrick, S., Akiyama, Y., Hecht, S. M., and Hurley, L. H. (2009). The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 131, 17667–17676. doi:10.1021/ja9076292
Kendrick, S., Kang, H. J., Alam, M. P., Madathil, M. M., Agrawal, P., Gokhale, V., et al. (2014). The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 136, 4161–4171. doi:10.1021/ja410934b
Kendrick, S., Muranyi, A., Gokhale, V., Hurley, L. H., and Rimsza, L. M. (2017). Simultaneous drug targeting of the promoter MYC G-quadruplex and BCL2 i-motif in diffuse large B-cell lymphoma delays tumor growth. J. Med. Chem. 60, 6587–6597. doi:10.1021/acs.jmedchem.7b00298
Kim, R., Emi, M., Tanabe, K., and Toge, T. (2004). Therapeutic potential of antisense Bcl-2 as a chemosensitizer for cancer therapy. Cancer 101, 2491–2502. doi:10.1002/cncr.20696
Kimura, K., Oshikawa, D., Ikebukuro, K., and Yoshida, W. (2022). Stabilization of VEGF i-motif structure by CpG methylation. Biochem. Biophysical Res. Commun. 594, 88–92. doi:10.1016/j.bbrc.2022.01.054
Kretzmann, J. A., Irving, K. L., Smith, N. M., and Evans, C. W. (2021). Modulating gene expression in breast cancer via DNA secondary structure and the CRISPR toolbox. Nar. Cancer 3, zcab048. doi:10.1093/narcan/zcab048
Kuang, G., Zhang, M., Kang, S., Hu, D., Li, X., Wei, Z., et al. (2020). Syntheses and evaluation of new bisacridine derivatives for dual binding of G-quadruplex and i-motif in regulating oncogene c-myc expression. J. Med. Chem. 63, 9136–9153. doi:10.1021/acs.jmedchem.9b01917
Kumar, S. U., and Priya Doss, C. G. (2021a). Computational investigation to identify potent inhibitors of the GTPase-Kirsten RAt sarcoma virus (K-Ras) mutants G12C and G12D. Comput. Biol. Med. 139, 104946. doi:10.1016/j.compbiomed.2021.104946
Kumar, S. U., and Priya Doss, C. G. (2021b). Residue interaction networks of K-Ras protein with water molecules identifies the potential role of switch II and P-loop. Comput. Biol. Med. 135, 104597. doi:10.1016/j.compbiomed.2021.104597
Kumar, V., Kashav, T., Islam, A., Ahmad, F., and Hassan, M. I. (2016). Structural insight into C9orf72 hexanucleotide repeat expansions: Towards new therapeutic targets in FTD-ALS. Neurochem. Int. 100, 11–20. doi:10.1016/j.neuint.2016.08.008
Lannes, L., Halder, S., Krishnan, Y., and Schwalbe, H. (2015). Tuning the pH response of i-motif DNA oligonucleotides. Chembiochem a Eur. J. Chem. Biol. 16, 1647–1656. doi:10.1002/cbic.201500182
Lee, I. J., Patil, S. P., Fhayli, K., Alsaiari, S., and Khashab, N. M. (2015). Probing structural changes of self assembled i-motif DNA. Chem. Commun. Camb. Engl. 51, 3747–3749. doi:10.1039/c4cc06824f
Li, H., Hai, J., Zhou, J., and Yuan, G. (2016). The formation and characteristics of the i-motif structure within the promoter of the c-myb proto-oncogene. J. Photochem. Photobiol. B, Biol. 162, 625–632. doi:10.1016/j.jphotobiol.2016.07.035
Li, L., Jiang, Y., Cui, C., Yang, Y., Zhang, P., Stewart, K., et al. (2018). Modulating aptamer specificity with pH-responsive DNA bonds. J. Am. Chem. Soc. 140, 13335–13339. doi:10.1021/jacs.8b08047
Li, X., Wang, J., Gong, X., Zhang, M., Kang, S., Shu, B., et al. (2020). Upregulation of BCL-2 by acridone derivative through gene promoter i-motif for alleviating liver damage of NAFLD/NASH. Nucleic Acids Res. 48, 8255–8268. doi:10.1093/nar/gkaa615
Lin, J., Hou, J. Q., Xiang, H. D., Yan, Y. Y., Gu, Y. C., Tan, J. H., et al. (2013). Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: Implications for epigenetic regulation. Biochem. Biophysical Res. Commun. 433, 368–373. doi:10.1016/j.bbrc.2012.12.040
Linke, R., Limmer, M., Juranek, S. A., Heine, A., and Paeschke, K. (2021). The relevance of G-quadruplexes for DNA repair. Int. J. Mol. Sci. 22, 12599. doi:10.3390/ijms222212599
Lowy, D. R., and Willumsen, B. M. (1993). Function and regulation of RAS. Annu. Rev. Biochem. 62, 851–891. doi:10.1146/annurev.bi.62.070193.004223
Manié, S., Santoro, M., Fusco, A., and Billaud, M. (2001). The RET receptor: Function in development and dysfunction in congenital malformation. Trends Genet. 17, 580–589. doi:10.1016/s0168-9525(01)02420-9
Manzini, G., Yathindra, N., and Xodo, L. E. (1994). Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 22, 4634–4640. doi:10.1093/nar/22.22.4634
Mathur, V., Verma, A., Maiti, S., and Chowdhury, S. (2004). Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem. Biophysical Res. Commun. 320, 1220–1227. doi:10.1016/j.bbrc.2004.06.074
Matsumoto, K., and Ema, M. (2014). Roles of VEGF-A signalling in development, regeneration, and tumours. J. Biochem. 156, 1–10. doi:10.1093/jb/mvu031
Mcluckie, K. I., Waller, Z. A., Sanders, D. A., Alves, D., Rodriguez, R., Dash, J., et al. (2011). G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J. Am. Chem. Soc. 133, 2658–2663. doi:10.1021/ja109474c
Miglietta, G., Cogoi, S., Pedersen, E. B., and Xodo, L. E. (2015). GC-elements controlling HRAS transcription form i-motif structures unfolded by heterogeneous ribonucleoprotein particle A1. Sci. Rep. 5, 18097. doi:10.1038/srep18097
Mir, B., Serrano, I., Buitrago, D., Orozco, M., Escaja, N., and GonzáLEZ, C. (2017). Prevalent sequences in the human genome can form mini i-motif structures at physiological pH. J. Am. Chem. Soc. 139, 13985–13988. doi:10.1021/jacs.7b07383
Mishra, S., Chaudhary, R., Singh, S., Kota, S., and Misra, H. S. (2019). Guanine quadruplex DNA regulates gamma radiation response of genome functions in the radioresistant bacterium deinococcus radiodurans. J. Bacteriol. 201, 001544–e219. doi:10.1128/jb.00154-19
Molnar, M. M., Liddell, S. C., and Wadkins, R. M. (2019). Effects of polyamine binding on the stability of DNA i-motif structures. ACS omega 4, 8967–8973. doi:10.1021/acsomega.9b00784
Mondal, M., Bhattacharyya, D., and Gao, Y. Q. (2019). Structural properties and influence of solvent on the stability of telomeric four-stranded i-motif DNA. Phys. Chem. Chem. Phys. PCCP 21, 21549–21560. doi:10.1039/c9cp03253c
Morgan, R. K., Batra, H., Gaerig, V. C., Hockings, J., and Brooks, T. A. (2016). Identification and characterization of a new G-quadruplex forming region within the kRAS promoter as a transcriptional regulator. Biochimica biophysica acta 1859, 235–245. doi:10.1016/j.bbagrm.2015.11.004
Nahta, R., and Esteva, F. J. (2003). Bcl-2 antisense oligonucleotides: A potential novel strategy for the treatment of breast cancer. Seminars Oncol. 30, 143–149. doi:10.1053/j.seminoncol.2003.08.016
Niknezhad, Z., Hassani, L., and Norouzi, D. (2016). Investigating actinomycin D binding to G-quadruplex, i-motif and double-stranded DNA in 27-nt segment of c-MYC gene promoter. Mater. Sci. Eng. C, Mater. Biol. Appl. 58, 1188–1193. doi:10.1016/j.msec.2015.09.072
Niu, K., Zhang, X., Deng, H., Wu, F., Ren, Y., Xiang, H., et al. (2018). BmILF and i-motif structure are involved in transcriptional regulation of BmPOUM2 in Bombyx mori. Nucleic Acids Res. 46, 1710–1723. doi:10.1093/nar/gkx1207
Oberdoerffer, S., Moita, L. F., Neems, D., Rui, P. F., Hacohen, N., and Rao, A. (2008). Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321, 686–691. doi:10.1126/science.1157610
Pagano, A., Iaccarino, N., Abdelhamid, M. A. S., Brancaccio, D., Garzarella, E. U., Di Porzio, A., et al. (2018). Common G-quadruplex binding agents found to interact with i-motif-forming DNA: Unexpected multi-target-directed compounds. Front. Chem. 6, 281. doi:10.3389/fchem.2018.00281
Pages, B. J., Gurung, S. P., Mcquaid, K., Hall, J. P., Cardin, C. J., and Brazier, J. A. (2019). Stabilization of long-looped i-motif DNA by polypyridyl ruthenium complexes. Front. Chem. 7, 744. doi:10.3389/fchem.2019.00744
Pan, G., Mou, Q., Ma, Y., Ding, F., Zhang, J., Guo, Y., et al. (2019). pH-responsive and gemcitabine-containing DNA nanogel to facilitate the chemodrug delivery. ACS Appl. Mater. Interfaces 11, 41082–41090. doi:10.1021/acsami.9b14892
Panasiuk, A., Dzieciol, J., Panasiuk, B., and Prokopowicz, D. (2006). Expression of p53, Bax and Bcl-2 proteins in hepatocytes in non-alcoholic fatty liver disease. World J. gastroenterology 12, 6198–6202. doi:10.3748/wjg.v12.i38.6198
Pascale, H., Parkinson, G. N., and Stephen, N. (2006). Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures. Nucleic Acids Res. 34, 2117–2127. doi:10.1093/nar/gkl182
Pathania, S., PentikäINEN, O. T., and Singh, P. K. (2021). A holistic view on c-Kit in cancer: Structure, signaling, pathophysiology and its inhibitors. Biochimica biophysica acta. Rev. cancer 1876, 188631. doi:10.1016/j.bbcan.2021.188631
Peng, P., Wang, Q., Du, Y., Wang, H., Shi, L., and Li, T. (2020). Extracellular ion-responsive logic sensors utilizing DNA dimeric nanoassemblies on cell surface and application to boosting AS1411 internalization. Anal. Chem. 92, 9273–9280. doi:10.1021/acs.analchem.0c01612
Qin, T., Liu, K., Song, D., Yang, C., and Su, H. (2017). Porphyrin bound to i-motifs: Intercalation versus external groove binding. Chem. Asian J. 12, 1578–1586. doi:10.1002/asia.201700398
Radestock, B., Morales, I., Rahman, S. A., Radau, S., Glass, B., Zahedi, R. P., et al. (2013). Comprehensive mutational analysis reveals p6Gag phosphorylation to be dispensable for HIV-1 morphogenesis and replication. J. virology 87, 724–734. doi:10.1128/jvi.02162-12
Ramsay, R. G., Barton, A. L., and Gonda, T. J. (2003). Targeting c-Myb expression in human disease. Expert Opin. Ther. targets 7, 235–248. doi:10.1517/14728222.7.2.235
Reilly, S. M., Lyons, D. F., Wingate, S. E., Wright, R. T., Correia, J. J., Jameson, D. M., et al. (2014). Folding and hydrodynamics of a DNA i-motif from the c-MYC promoter determined by fluorescent cytidine analogs. Biophysical J. 107, 1703–1711. doi:10.1016/j.bpj.2014.08.014
Reilly, S. M., Morgan, R. K., Brooks, T. A., and Wadkins, R. M. (2015). Effect of interior loop length on the thermal stability and pK(a) of i-motif DNA. Biochemistry 54, 1364–1370. doi:10.1021/bi5014722
Rezler, E., Seenisamy, J., Bashyam, S., Kim, M., White, E., Wilson, W., et al. (2005). Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 127, 9439–9447. doi:10.1021/ja0505088
Robinson, J., Raguseo, F., Nuccio, S. P., Liano, D., and Di Antonio, M. (2021). DNA G-quadruplex structures: More than simple roadblocks to transcription? Nucleic Acids Res. 49, 8419–8431. doi:10.1093/nar/gkab609
Rogers, R. A., Fleming, A. M., and Burrows, C. J. (2018). Unusual isothermal hysteresis in DNA i-motif pH transitions: A study of the RAD17 promoter sequence. Biophysical J. 114, 1804–1815. doi:10.1016/j.bpj.2018.03.012
Rogowicz-Frontczak, A., Araszkiewicz, A., Pilacinski, S., Zozulinska-Ziolkiewicz, D., Wykretowicz, A., and Wierusz-Wysocka, B. (2012). Carotid intima-media thickness and arterial stiffness in type 1 diabetic patients with and without microangiopathy. Archives Med. Sci. AMS 8, 484–490. doi:10.5114/aoms.2012.29404
Roy, B., Talukder, P., Kang, H. J., Tsuen, S. S., Alam, M. P., Hurley, L. H., et al. (2016). Interaction of individual structural domains of hnRNP LL with the BCL2 promoter i-motif DNA. J. Am. Chem. Soc. 138, 10950–10962. doi:10.1021/jacs.6b05036
Roy, M. J., Vom, A., Czabotar, P. E., and Lessene, G. (2014). Cell death and the mitochondria: Therapeutic targeting of the BCL-2 family-driven pathway. Br. J. Pharmacol. 171, 1973–1987. doi:10.1111/bph.12431
Ruggiero, E., Lago, S., Šket, P., Nadai, M., Frasson, I., Plavec, J., et al. (2019). A dynamic i-motif with a duplex stem-loop in the long terminal repeat promoter of the HIV-1 proviral genome modulates viral transcription. Nucleic Acids Res. 47, 11057–11068. doi:10.1093/nar/gkz937
Ruggiero, E., Zanin, I., Terreri, M., and Richter, S. N. (2021). G-quadruplex targeting in the fight against viruses: An update. Int. J. Mol. Sci. 22, 10984. doi:10.3390/ijms222010984
Sellner, S., Kocabey, S., Zhang, T., Nekolla, K., Hutten, S., Krombach, F., et al. (2017). Dexamethasone-conjugated DNA nanotubes as anti-inflammatory agents in vivo. Biomaterials 134, 78–90. doi:10.1016/j.biomaterials.2017.04.031
Serrano-ChacóN, I., Mir, B., Escaja, N., and GonzáLEZ, C. (2021). Structure of i-motif/duplex junctions at neutral pH. J. Am. Chem. Soc. 143, 12919–12923. doi:10.1021/jacs.1c04679
Shakked, Z., Rabinovich, D., Kennard, O., Cruse, W., Salisbury, S. A., and Viswamitra, M. A. (1983). Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J. Mol. Biol. 166, 183–201. doi:10.1016/s0022-2836(83)80005-9
Sheng, Q., Neaverson, J. C., Mahmoud, T., Stevenson, C. E. M., Matthews, S. E., and Waller, Z. A. E. (2017). Identification of new DNA i-motif binding ligands through a fluorescent intercalator displacement assay. Org. Biomol. Chem. 15, 5669–5673. doi:10.1039/c7ob00710h
Shi, S., Geng, X., Zhao, J., Yao, T., Wang, C., Yang, D., et al. (2010). Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: Preferential binding to G-quadruplexes over i-motif. Biochimie 92, 370–377. doi:10.1016/j.biochi.2010.01.003
Shim, H., Dolde, C., Lewis, B. C., Wu, C. S., Dang, G., Jungmann, R. A., et al. (1997). c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U. S. A. 94, 6658–6663. doi:10.1073/pnas.94.13.6658
Shu, B., Cao, J., Kuang, G., Qiu, J., Zhang, M., Zhang, Y., et al. (2018). Syntheses and evaluation of new acridone derivatives for selective binding of oncogene c-myc promoter i-motifs in gene transcriptional regulation. Chem. Commun. Camb. Engl. 54, 2036–2039. doi:10.1039/c8cc00328a
Shu, B., Zeng, P., Kang, S., Li, P. H., Hu, D., Kuang, G., et al. (2019). Syntheses and evaluation of new Quinoline derivatives for inhibition of hnRNP K in regulating oncogene c-myc transcription. Bioorg. Chem. 85, 1–17. doi:10.1016/j.bioorg.2018.12.020
ŠkoláKOVá, P., Badri, Z., Foldynová-TrantíRKOVá, S., Ryneš, J., Šponer, J., Fojtová, M., et al. (2020). Composite 5-methylations of cytosines modulate i-motif stability in a sequence-specific manner: Implications for DNA nanotechnology and epigenetic regulation of plant telomeric DNA. Biochimica biophysica acta. General Subj. 1864, 129651. doi:10.1016/j.bbagen.2020.129651
Spence, P., Fielden, J., and Waller, Z. A. E. (2020). Beyond solvent exclusion: I-motif detecting capability and an alternative DNA light-switching mechanism in a ruthenium(II) polypyridyl complex. J. Am. Chem. Soc. 142, 13856–13866. doi:10.1021/jacs.0c04789
Sun, D., and Hurley, L. H. (2009). The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 52, 2863–2874. doi:10.1021/jm900055s
Sutherland, C., Cui, Y., Mao, H., and Hurley, L. H. (2016). A mechanosensor mechanism controls the G-quadruplex/i-motif molecular switch in the MYC promoter NHE III(1). J. Am. Chem. Soc. 138, 14138–14151. doi:10.1021/jacs.6b09196
Takahashi, S., Bhattacharjee, S., Ghosh, S., Sugimoto, N., and Bhowmik, S. (2020). Preferential targeting cancer-related i-motif DNAs by the plant flavonol fisetin for theranostics applications. Sci. Rep. 10, 2504. doi:10.1038/s41598-020-59343-2
Takakura, M., Kyo, S., Kanaya, T., Hirano, H., Takeda, J., Yutsudo, M., et al. (1999). Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 59, 551–557.
Tekendo-Ngongang, C., Grochowsky, A., Solomon, B. D., and Yano, S. T. (2021). Beyond trinucleotide repeat expansion in fragile X syndrome: Rare coding and noncoding variants in FMR1 and associated phenotypes. Genes (Basel) 12, 1669. doi:10.3390/genes12111669
Thorne, B. N., Ellenbroek, B. A., and Day, D. J. (2021). Evaluation of i-motif formation in the serotonin transporter-linked polymorphic region. Chembiochem a Eur. J. Chem. Biol. 22, 349–353. doi:10.1002/cbic.202000513
Tsvetkov, V. B., Zatsepin, T. S., Belyaev, E. S., Kostyukevich, Y. I., Shpakovski, G. V., Podgorsky, V. V., et al. (2018). i-Clamp phenoxazine for the fine tuning of DNA i-motif stability. Nucleic Acids Res. 46, 2751–2764. doi:10.1093/nar/gky121
Tsvetkov, V. B., Zatsepin, T. S., Turaev, A. V., Farzan, V. M., Pozmogova, G. E., Aralov, A. V., et al. (2019). DNA i-motifs with guanidino-i-clamp residues: The counterplay between kinetics and thermodynamics and implications for the design of pH sensors. Comput. Struct. Biotechnol. J. 17, 527–536. doi:10.1016/j.csbj.2019.04.006
Udhaya Kumar, S., Bithia, B., Thirumal Kumar, D., George Priya, D., and Zayed, H. (2022). Mutational landscape of K-Ras substitutions at 12th position-a systematic molecular dynamics approach. J. Biomol. Struct. Dyn. 40, 1571–1585. doi:10.1080/07391102.2020.1830177
Von Figura, G., Hartmann, D., Song, Z., and Rudolph, K. L. (2009). Role of telomere dysfunction in aging and its detection by biomarkers. J. Mol. Med. (Berlin, Ger. 87, 1165–1171. doi:10.1007/s00109-009-0509-5
Wang, L., Wu, Y., Chen, T., and Wei, C. (2013). The interactions of phenanthroline compounds with DNAs: Preferential binding to telomeric quadruplex over duplex. Int. J. Biol. Macromol. 52, 1–8. doi:10.1016/j.ijbiomac.2012.08.015
Wang, S., Yang, Y., Fan, S., Li, H., and David, D. Y. C. (2020). Determination of the binding of natural products to the human c-myb oncogene promoter G-quadruplex DNA by capillary electrophoresis and electrospray ionization mass spectrometry. Se Pu 38, 1069–1077. doi:10.3724/sp.J.1123.2020.03001
Watson, J. D., and Crick, F. H. (1974). Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 248, 765. doi:10.1038/248765a0
Wei, C., and Gao, N. (2015). Cytotoxicities, telomerase and topoisomerases I inhibitory activities and interactions of terpyridine derivatives with DNAs. Chem. Res. Chin. Univ. 31, 970–975. doi:10.1007/s40242-015-5256-2
Wei, H., Zhao, Z., Wang, Y., Zou, J., Lin, Q., and Duan, Y. (2019). One-step self-assembly of multifunctional DNA nanohydrogels: An enhanced and harmless strategy for guiding combined antitumor therapy. ACS Appl. Mater. interfaces 11, 46479–46489. doi:10.1021/acsami.9b15874
Wright, E. P., Abdelhamid, M. A. S., Ehiabor, M. O., Grigg, M. C., Irving, K., Smith, N. M., et al. (2020). Epigenetic modification of cytosines fine tunes the stability of i-motif DNA. Nucleic Acids Res. 48, 55–62. doi:10.1093/nar/gkz1082
Wright, E. P., Day, H. A., Ibrahim, A. M., Kumar, J., Boswell, L. J., Huguin, C., et al. (2016). Mitoxantrone and analogues bind and stabilize i-motif forming DNA sequences. Sci. Rep. 6, 39456. doi:10.1038/srep39456
Wright, E. P., Huppert, J. L., and Zoë, A. E., W. (2017). Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 45, 2951–2959. doi:10.1093/nar/gkx090
Xu, B., Devi, G., and Shao, F. (2015). Regulation of telomeric i-motif stability by 5-methylcytosine and 5-hydroxymethylcytosine modification. Org. Biomol. Chem. 13, 5646–5651. doi:10.1039/c4ob02646b
Xu, H., Zhang, H., and Qu, X. (2006). Interactions of the human telomeric DNA with terbium-amino acid complexes. J. Inorg. Biochem. 100, 1646–1652. doi:10.1016/j.jinorgbio.2006.05.015
Xu, L., Hong, S., Sun, N., Wang, K., Zhou, L., Ji, L., et al. (2016). Berberine as a novel light-up i-motif fluorescence ligand and its application in designing molecular logic systems. Chem. Commun. Camb. Engl. 52, 179–182. doi:10.1039/c5cc08242k
Xu, Y., and Sugiyama, H. (2006). Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 34, 949–954. doi:10.1093/nar/gkj485
Xu, Y., and Sugiyama, H. (2005). Structural and functional characterizations of the G-quartet and i-motif elements in retinoblastoma susceptibility genes (Rb). Nucleic acids symp. Ser. 49, 177–178. doi:10.1093/nass/49.1.177
Yang, H., Chen, J., Liang, Y., Zhang, Y., Yin, W., Xu, Y., et al. (2021). A MOF-shell-confined I-Motif-Based pH probe (MOFC-i) strategy for sensitive and dynamic imaging of cell surface pH. ACS Appl. Mater. interfacesInterfaces 13, 45291–45299. doi:10.1021/acsami.1c13720
Yang, Y., Fu, H., Qian, C., Li, H., and Chen, D. D. Y. (2020). Characterization of interaction between Bcl-2 oncogene promoter I-Motif DNA and flavonoids using electrospray ionization mass spectrometry and pressure-assisted capillary electrophoresis frontal analysis. Talanta 215, 120885. doi:10.1016/j.talanta.2020.120885
Yoshida, W., Yoshioka, H., Bay, D. H., Iida, K., Ikebukuro, K., Nagasawa, K., et al. (2016). Detection of DNA methylation of G-quadruplex and i-motif-forming sequences by measuring the initial elongation efficiency of polymerase chain reaction. Anal. Chem. 88, 7101–7107. doi:10.1021/acs.analchem.6b00982
Yu, J., Zhang, J., Jin, J., and Jiang, W. (2021). Microenvironment-responsive DNA-conjugated albumin nanocarriers for targeted therapy. J. Mater. Chem. B 9, 8424–8436. doi:10.1039/d1tb01022k
Yuan, B., XI, Y., Qi, C., Zhao, M., Zhu, X., and Tang, J. (2022). A sequentially triggered DNA nanocapsule for targeted drug delivery based on pH-responsive i-motif and tumor cell-specific aptamer. Front. Bioeng. Biotechnol. 10, 965337. doi:10.3389/fbioe.2022.965337
Zeraati, M., Langley, D. B., Schofield, P., Moye, A. L., Rouet, R., Hughes, W. E., et al. (2018). I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 10, 631–637. doi:10.1038/s41557-018-0046-3
Zhang, A., Sokolova, I., Domissy, A., Davis, J., Rao, L., Hana Utami, K., et al. (2022). Maturation delay of human GABAergic neurogenesis in fragile X syndrome pluripotent stem cells. Stem cells Transl. Med. 11, 613–629. doi:10.1093/stcltm/szac022
Zhang, M., Wei, Z., Gong, X., Li, X., Kang, S., Wang, J., et al. (2021). Syntheses and evaluation of acridone-naphthalimide derivatives for regulating oncogene PDGFR-β expression. Bioorg. Med. Chem. 34, 116042. doi:10.1016/j.bmc.2021.116042
Zhang, X., Wang, L., Huang, F., Li, J., Xiong, L., Xue, H., et al. (2015). Evaluation of the promoter region polymorphism (5-HTTLPR) in the serotonin transporter gene in females with postpartum depression. Exp. Ther. Med. 9, 245–249. doi:10.3892/etm.2014.2043
Zhang, Y., Shen, T., Zhou, S., Wang, W., Lin, S., and Zhu, G. (2020). pH-responsive STING-activating DNA nanovaccines for cancer immunotherapy. Adv. Ther. 3, 2000083. doi:10.1002/adtp.202000083
Zhao, Z., Li, X., Cui, Z., Tong, T., Zhang, Y., Zhang, Y., et al. (2022). Synthesis of hemiprotonic phenanthroline-phenanthroline(+) compounds with both antitumor and antimicrobial activity. J. Med. Chem. 65, 2532–2547. doi:10.1021/acs.jmedchem.1c01982
Keywords: i-motif, biological activities, molecular mechanism, ligand compounds, oncogene, telomere, gene-targeted therapy
Citation: Luo X, Zhang J, Gao Y, Pan W, Yang Y, Li X, Chen L, Wang C and Wang Y (2023) Emerging roles of i-motif in gene expression and disease treatment. Front. Pharmacol. 14:1136251. doi: 10.3389/fphar.2023.1136251
Received: 02 January 2023; Accepted: 27 February 2023;
Published: 20 March 2023.
Edited by:
Ning Ji, China Academy of Chinese Medical Sciences, ChinaReviewed by:
Udhaya Kumar. S., Baylor College of Medicine, United StatesCopyright © 2023 Luo, Zhang, Gao, Pan, Yang, Li, Chen, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Wang, d2FuZ2NoYW5nMDQ3QGhvdG1haWwuY29t; Yuqing Wang, d2FuZ3l1cUBnemhtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.