- 1Department of Respiratory Medicine, The First Hospital of Jilin University, Changchun, Jilin, China
- 2Tongji University Cancer Center, Shanghai Tenth People’s Hospital of Tongji University, School of Medicine, Tongji University, Shanghai, China
Lung cancer is the common malignant tumor with the highest mortality rate. Lung cancer patients have achieved benefits from immunotherapy, including immune checkpoint inhibitors (ICIs) therapy. Unfortunately, cancer patients acquire adaptive immune resistance, leading to poor prognosis. Tumor microenvironment (TME) has been demonstrated to play a critical role in participating in acquired adaptive immune resistance. TME is associated with molecular heterogeneity of immunotherapy efficacy in lung cancer. In this article, we discuss how immune cell types of TME are correlated with immunotherapy in lung cancer. Moreover, we describe the efficacy of immunotherapy in driven gene mutations in lung cancer, including KRAS, TP53, EGFR, ALK, ROS1, KEAP1, ZFHX3, PTCH1, PAK7, UBE3A, TNF-α, NOTCH, LRP1B, FBXW7, and STK11. We also emphasize that modulation of immune cell types of TME could be a promising strategy for improving adaptive immune resistance in lung cancer.
Introduction
Lung cancer is the common malignant tumor and displays the highest mortality rate (Chen P. et al., 2022; Choi and Mazzone, 2022). Lung cancer had 1.8 million deaths (18% of the total cancer-related deaths) worldwide in 2020 (Sung et al., 2021). Lung cancer is the most frequently occurring tumor in males and the third commonly diagnosed tumor in females. Lung cancer is the first cause of tumor death in males and the second leading cause of cancer mortality in women (Sung et al., 2021). In the United States, there are 2,36,740 new lung cancer cases and 1,30,180 lung cancer-related deaths (Siegel et al., 2022). The 5-year survival rate of lung cancer is only 10%–20% in some countries (Sung et al., 2021).
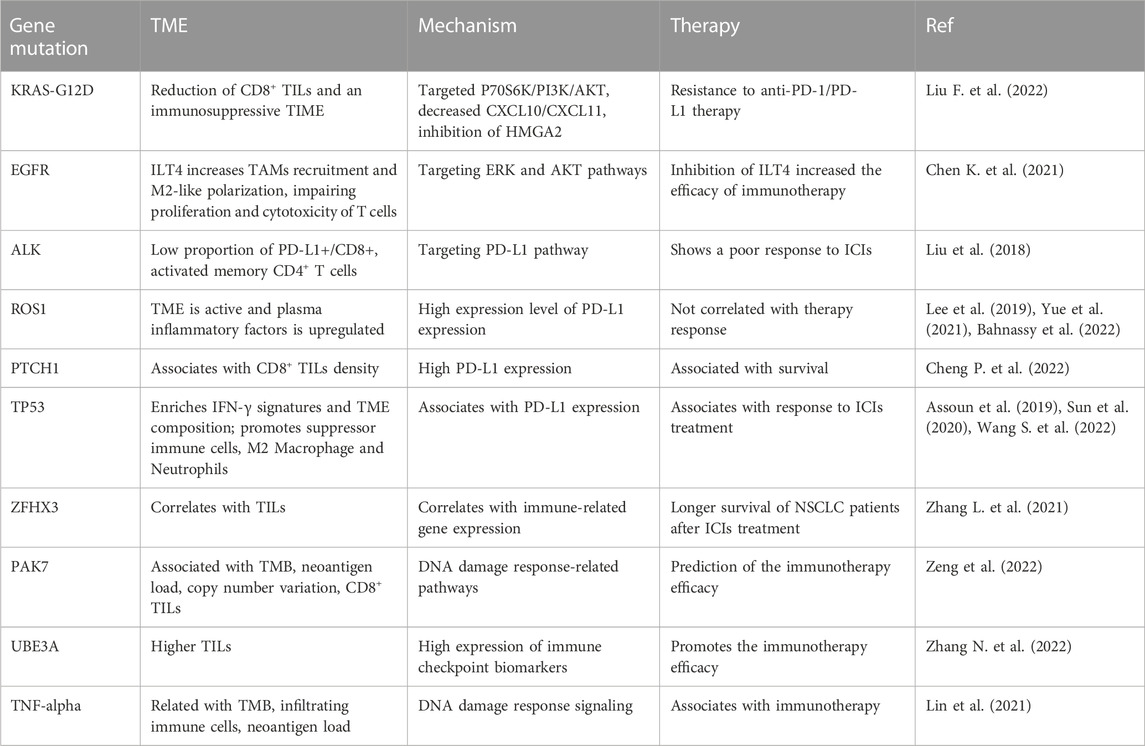
Lung cancer has three-type categories, including small cell lung cancer (SCLC, 14%), non-small cell lung cancer (NSCLC, 82%) and unspecified histology (3%) (Miller et al., 2022). The NSCLC includes large cell carcinoma, adenocarcinoma, and squamous cell carcinoma (Mengoli et al., 2018). The global lung cancer occurrence could be due to outdoor ambient PM2.5 and tobacco (Guo et al., 2020; Turner et al., 2020; Frazer et al., 2022). Multiple gene mutations have been found in NSCLC patient, including epidermal growth factor receptor (EGFR) (Zhao D. et al., 2022; Castaneda-Gonzalez et al., 2022), Kirsten rat sarcoma viral oncogene homolog (KRAS) (Desage et al., 2022; Garcia-Robledo et al., 2022), anaplastic lymphoma kinase (ALK) (Cognigni et al., 2022; Xiang et al., 2022), Erb-B2 Receptor Tyrosine Kinase 2 (ERBB2) (Ni and Zhang, 2021; Vathiotis et al., 2021; Yu X. et al., 2022), B-Raf proto-oncogene (BRAF) (Abdayem and Planchard, 2022; Riudavets et al., 2022; Sforza et al., 2022), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), AKT serine/threonine kinase 1 (AKT1), mitogen-activated protein kinase kinase 1 (MAP2K1) (Kim and Giaccone, 2018; Han et al., 2021), c-ros oncogene 1 (ROS1) (Guaitoli et al., 2021; Yu Z. Q. et al., 2022), neurotrophic tyrosine receptor kinase (NTRK) (Liu C. et al., 2022; Qin and Patel, 2022), and mesenchymal-epithelial transition factor (MET) (Pao and Girard, 2011; Olmedo et al., 2022) (Figure 1). In SCLC patients, gene mutations often include retinoblastoma (Rb), TP53, PTEN, FBXW7, VHL mutations (Cardona et al., 2019; Guan et al., 2022). In addition, targeted therapy, immunotherapy, antiangiogenic therapy and combination therapy have been used in the clinic for lung cancer patients (Luo et al., 2021; Wang et al., 2021; Guo et al., 2022) (Figure 1). NSCLC patients with KRAS mutation or EGFR often have a worse benefit from immunotherapy (Di Nicolantonio et al., 2021).
FIGURE 1. Gene Mutations and therapies are illustrated in lung cancer. Lung cancer has three-type categories, including SCLC, NSCLC, and unspecified histology. NSCLC patients have multiple gene mutations. Targeted therapy, immunotherapy, antiangiogenic therapy and combination therapy have been used in the clinic for lung cancer patients.
Immunotherapy has improved the therapeutic outcomes in lung cancer patients (Caliman et al., 2022; Catalano et al., 2022; Martin and Enrico, 2022; Tartarone et al., 2022; Yang et al., 2022). Immune checkpoint inhibitors (ICIs) have been used for the cancer therapy, including anti-PD-1, anti-PD-L1 and anti-CTLA-4. Anti-PD-1 drugs in NSCLC have cemiplimab, pembrolizumab, and nivolumab (Bote et al., 2022; Mussafi et al., 2022). The anti-PD-L1 monoclonal antibodies have durvalumab and atezolizumab in NSCLC. Anti-CTLA-4 (ipilimumab) is also be used in lung cancer because of CTLA-4 as a checkpoint on lymphocytes (Ackermann et al., 2019; Dawe et al., 2020; Peng et al., 2022). However, PD-1/PD-L1 monoclonal antibodies and anti-CTLA-3 treatment obtain a good response in a subgroup of lung cancer patients. Moreover, adaptive immune resistance is observed in lung cancer patients and attenuated the immunotherapeutic benefits (Gkountakos et al., 2021; Gemelli et al., 2022). Furthermore, immunotherapy often causes side-effects, such as endocrinopathy, colitis, pneumonitis, nephritis in lung cancer patients (Bredin and Naidoo, 2022; Hao et al., 2022).
Tumor microenvironment (TME) is a unique environment and composed of many other types of cells, such as stromal, endothelial and immune cells, which has shown to participate in tumor development, initiation and progression as well as metastasis (Eulberg et al., 2022; Nallasamy et al., 2022). The TME cellular components consist of MDSCs, Treg cells, M1 macrophages, M2 macrophages, cytotoxic CD8+ T cells and NK cells (Gajewski et al., 2013; Cao et al., 2022). It has been known that tumor cells inhibit the anticancer functions of TME and promote pro-tumorigenic functions of TME (Faraj et al., 2022; Tiwari et al., 2022). Tumor immune microenvironment (TIME) consists of tumor cells, different cell types of the immune system and their interactions in the TME niche (Binnewies et al., 2018).
In this review article, we described the association between immune cell types in TME and immunotherapy in lung cancer. Moreover, we discussed the efficacy of immunotherapy in driven gene mutations in lung cancer, including KRAS, TP53, EGFR, ALK, ROS1, KEAP1, ZFHX3, PTCH1, and STK11. Furthermore, we concluded that targeting TME could be helpful to overcome resistance to PD-1/PD-L1 blockade in lung cancer.
Immunotherapy, driven gene mutations, and TME
TME has been identified to take part in tumorigenesis and cancer metastasis. Several adverse conditions in TME, such as acidity, hypoxia, and nutrient restriction, have been unraveled to affect the responses of immunotherapy (Li and Qiao, 2022). Moreover, TME governs immune cell functions via regulation of immune cells activation (Ahluwalia et al., 2021; Genova et al., 2021). In this section, the relationship of TME, driven gene mutations and immunotherapy will be summarized. Evidence dissects that immune therapy benefit is associated with driven gene mutations and smoking status in NSCLC patients. These driven genes include EGFR, KRAS, ALK, and BRAF (Skoulidis and Heymach, 2019). One study identified that the most frequently mutated genes included TP53, KRAS, ERBB2, SMAD4, ERBB4, EGFR, BRAF, and MET (Cinausero et al., 2019). In the following sections, we will describe how these driven gene mutations modulate the TIME and affect the anti-PD-1/PD-L1 therapy in NSCLC patients (Table 1). We highlight that the efficacy of immunotherapy is modulated by these key gene mutations in NSCLC patients.
The efficacy of immunotherapy in KRAS-mutant NSCLC
KRAS oncogenic pathway affected TME via modulation of cancer-associated fibroblasts and immune cells (Dias Carvalho et al., 2018; Ceddia et al., 2022). KRAS-mutant cancer cells govern immune responses through regulation of immune cell recruitment, activation, and differentiation, leading to enhancement of protumorigenic ability and promotion of tumor cell evasion (Dias Carvalho et al., 2018). KRAS pathway controls populations of myeloid cells, T cells, fibroblasts, endothelial cells, ECM composition. In KRAS-mutant lung cancer patients, M2 macrophages, MDSCs, CD4+FoxP3+ Treg cells, IL-17-producing T helper cells displayed a pro-tumorigenic TME (Cullis et al., 2018). Studies have shown that the efficacy of anti-PD-1 and anti-PD-L1 immunotherapy was associated with promotion of immunogenicity and an inflammatory TME (Liu et al., 2020; Ceddia et al., 2022). Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations are linked to superior patient response to PD-1/PD-L1 inhibitors. KRAS mutations are associated with treatment efficacy and prognosis in NSCLC (Wood et al., 2016; Ferrer et al., 2018; Roman et al., 2018). Targeting KRAS variant has be shown to have potential treatment applications in NSCLC (Ricciuti et al., 2016; Uras et al., 2020; Li J. X. et al., 2022). Notably, KRAS mutations are linked to immune therapy resistance in NSCLC patients (Kim et al., 2017; Adderley et al., 2019).
In clinic study, NSCLC patients with KRAS mutation obtained treatment benefit from immunotherapy via anti-PD-1 and anti-PD-L1 approaches (Liu et al., 2020). Interestingly, suppression of PD-L1 in combination with docetaxel failed to enhance an anti-tumor response in a KRAS-mutant lung adenocarcinoma mouse model (Liu et al., 2020). This study indicated that the combination of immunotherapy and chemotherapy need to be revaluated in NSCLC patients with KRAS mutations (Liu et al., 2020). Moreover, evidence has demonstrated resistance to ICIs in NSCLC with KRAS mutation via modulation of tumor metabolism and TME functions (Li W. et al., 2022). KRAS-G12D mutation induced immune suppression and caused the resistance to anti-PD-1/PD-L1 therapy in NSCLC. KRAS-G12D point mutation was negatively associated with PD-L1 expression level and CXCL10/CXCL11, resulting in a reduction of CD8+ TILs and an immunosuppressive TIME (Liu F. et al., 2022). KRAS-G12D mutation reduced PD-L1 expression through P70S6K/PI3K/AKT pathway and decreased CXCL10/CXCL11 expression via inhibition of HMGA2 in lung cancer cells. Paclitaxel plus PD-L1 blockade treatments promoted CD8+ TILs recruitment due to CXCL10/CXCL11 upregulation (Liu J. et al., 2022). This study suggested that chemotherapy plus ICIs are effective in NSCLC patients with KRAS-G12D mutation (Liu C. et al., 2022).
The efficacy of immunotherapy in EGFR-mutant NSCLC
EGFR-mutant lung cancer patients exhibit therapy resistance (Passaro et al., 2021; Girard, 2022). Activation of EGFR has been reported to establish an immunosuppressive TME in NSCLC cells, including promotion of suppressive TAMs, Tregs, blockade of T cell infiltration and cytotoxicity, and induction of inhibitory cytokines, which impair the immunotherapy (Lin et al., 2019). About 50% of NSCLC patients with EGFR mutations acquired EGFR-tyrosine kinase inhibitor (TKI) resistance. EGFR pathway has been reported to regulate PD-L1 in NSCLC (Hsu et al., 2019). EGFR-TKI resistance upregulated PD-L1 expression and caused immune escape in lung cancer via activation of phosphatidylinositol-3 kinase (PI3K), mitogen-activated protein kinase (MAPK) and NF-kappa B (NF-κB) pathways (Peng et al., 2019). One study found that hypoxia-inducible factor 1α (HIF-1α) and NF-κB are critical to modulate the expression of PD-L1 in EGFR-mutant NSCLC cells (Guo et al., 2019). Another group suggested that TGF-β/Smad pathway participated in PD-L1-mediated EGFR-TKIs resistance in NSCLC with EGFR mutations (Zhang et al., 2019). Overexpression of PD-L1 increased gefitinib resistance in EGFR-mutant NSCLC cells, while depletion of PD-L1 reduced gefitinib resistance (Zhang et al., 2019). Activation of OPN/integrin αVβ3/FAK pathway is important for regulation of EGFR-TKI resistance in NSCLC with EGFR mutations (Fu et al., 2020). PD-L1 expression is correlated with TKIs response and prognosis in lung cancer patients with EGFR mutations (Lin et al., 2015).
Immunoglobulin-like transcript 4 (ILT4) belongs to the immunoglobulin superfamily and often expressed in myeloids, which can promote the proliferation, migration and invasion in human cancers. ILT4 induced immunosuppressive T cell infiltration and led to poor prognosis in lung cancer. ILT4 stimulated T cell senescence and reduced tumor immunity in the TME in human cancer (Gao et al., 2021; Yang et al., 2021). Moreover, ILT4 acts as a useful checkpoint molecule for immunotherapy (Gao et al., 2018). One group showed that ILT4 expression can be elevated after EGFR activation in NSCLC cells, which was mediated by activated ERK and AKT cellular signaling pathways (Chen K. et al., 2021). Moreover, ILT4 increased recruitment of TAMs and M2-like polarization in NSCLC cells with EGFR activation, leading to impairing proliferation and cytotoxicity of T cells (Chen X. et al., 2021). Furthermore, inhibition of ILT4 promoted the efficacy of PD-L1 inhibitors and abrogated TAMs- and T cell-involved immunosuppression in NSCLC cells with EGFR activation. In vivo study showed that knockdown of ILT4 and PD-L1 blockade synergistically retarded mouse tumor growth and inhibited immune escape (Chen K. et al., 2021). Animal study data further showed that inhibition of ILT4 alone repressed tumor progression and immune evasion in EGFR mutant NSCLC. This work implied that inhibition of ILT4 increased the efficacy of immunotherapy in EGFR-mutant NSCLC (Chen X. et al., 2021). One retrospective study determined the association between PD-L1, TILs and immunotherapy response in uncommon EGFR-mutant NSCLC patients (Chen et al., 2020). Among 600 NSCLC cases with EGFR mutations, 49 cases were borne with uncommon alterations, such as Ex20, L861Q, S7681, G719X. Uncommon EGFR-mutant NSCLC patients had a high PD-L1 expression and CD8+ TILs and displayed a favorable response to anti-PD-1 therapy (Chen et al., 2020). Therefore, like in common EGFR-mutant NSCLC patients, combination of CD8+ TILs and PD-L1 level in TME can determine the anti-PD-1/PD-L1 therapy efficacy for NSCLC patients with uncommon EGFR mutations (Chen et al., 2020). Anti-CD73 in combination with anti-PD-L1 therapy enhanced T cell response via upregulation of the number of CD8+ T cells and promotion of TNF-α and IFN-γ production in EGFR-mutant NSCLC, leading to inhibition of tumor growth (Tu et al., 2022).
ERBB-family genetic alterations and KRAS mutations regulated response to anti-PD-1 inhibitors in NSCLC with metastasis (Cinausero et al., 2019). NSCLC patients with KRAS mutations had a better anti-PD-1 therapy efficacy and a longer PFS and OS. NSCLC patients with EGFR mutation, ERBB2 mutation and ERBB4 mutations had a worse response to anti-PD-1 therapy (Cinausero et al., 2019). STK11/LKB1 mutations were linked to resistance of PD-1 blockade in KRAS-mutant lung cancer (Skoulidis et al., 2018). Biton et al. (2018) also reported that TP53, STK11, and EGFR mutations represented the anti-PD-1 treatment efficacy in lung adenocarcinoma. NSCLC patients with STK11 mutation displayed chemotherapy resistance, while co-mutations with KRAS or TP53 modulated TIME of STK11-mutant NSCLC tumors and affected immunotherapy response (Malhotra et al., 2022). Additionally, NSCLC patients with EGFR/HER2 exon 20 insertions had a higher expression of PD-L1 and exhibited the sensitive to anti-PD-1/PD-L1 therapy (Chen X. et al., 2021).
The efficacy of immunotherapy in ALK-rearranged NSCLC
ALK-rearranged tumors exhibited more resting memory CD4+ T cells and less activated memory CD4+ T cells and CD8+ T cells (Jin et al., 2020). Anti-PD-1/PD-L1 therapy is useful for the treatment of ALK-translocated NSCLC patients (Bylicki et al., 2017). ALK positivity and EGFR mutations have been reported to be adverse predictors for NSCLC patients (Bahnassy et al., 2022). A retrospective analysis showed that ALK rearrangement and EGFR mutations were involved in poor response to blockade of PD-1 pathway in NSCLC (Gainor et al., 2016). This could be due to low rates of PD-L1 expression and CD8+ TILS in the TME in NSCLC patients (Gainor et al., 2016). Similarly, PD-L1 expression and CD8+ T cells infiltration have a clinical relationship in lung cancer patients with ALK-rearranged and EGFR-mutated tumors (Liu et al., 2018). Lung cancer patients with ALK-rearrangement or EGFR mutations had lowest proportion of PD-L1+/CD8+ tumors and the shortest overall survival. Lung cancer patients with ALK-rearrangement or EGFR mutations showed a poor response to ICIs (Liu et al., 2018). Strikingly, PD-L1 expression and CD8 expression are biomarkers for prediction of prognosis with poor prognosis in patients with EGFR mutations or ALK rearrangement (Liu et al., 2018). Interestingly, a retrospectively study indicated that cytotoxic chemotherapy affected the TIME in NSCLC patients with wild type of ALK and EGFR (Sakai et al., 2019). Platinum-based adjuvant chemotherapy modulated PD-L1 expression, CD8+ TIL density and tumor mutation burden (TMB) in NSCLC patients (Sakai et al., 2019).
The efficacy of immunotherapy in ROS1-rearrangement NSCLC
One research group reported that expression of ROS1 and ROS1-rearrangement was observed in 18.57% and 15.71% of the 70 NSCLC patients, respectively (Bahnassy et al., 2022). ROS1 expression was not correlated with PD-1, PD-L1, survival and therapy response (Bahnassy et al., 2022). Another research simultaneous genotypic screening of three gene mutations, including ROS1, ALK and EGFR, to measure the prevalence and clinicopathologic features of ROS1 mutations and immunotherapy efficacy in NSCLC patients (Lee et al., 2019). This study found that among 407 NSCLC cause, there were 14 ROS1 and 19 ALK rearrangements and 106 EGFR mutations. Among 130 NSCLC tumors, 29 samples had high expression of PD-L1. Among 14 cases with ROS1 mutations, 12 samples exhibited PD-L1 expression and 5 cases displayed high expression level of PD-L1 expression (Lee et al., 2019). This work indicated that ROS1 rearrangement was overlapped with high expression of PD-L1 in NSCLC patients (Lee et al., 2019). Similarly, the correlation among oncogenic mutations in ROS1, ALK, EGFR and PD-L1 had been reported in lung adenocarcinoma (Rangachari et al., 2017). This retrospective work explored 71 cases of lung cancer and found that 29.6% cases had a PD-L1 TPS of high than 50%. Of 19 cases with ALK, ROS1, or EGFR mutations, 18 cases had a PD-L1 TPS less than 50%. Moreover, lung cancer with a PD-L1 TPS of high than 50% was correlated with smoking status (Rangachari et al., 2017). In addition, it has been compared with ALK, ROS1, EGFR, and PD-L1 between cytological tumors and surgical tumors in NSCLC to explore the adequacy of PD-L1 expression by a retrospective study (Ekin et al., 2021). Among 220 NSCLC cases, there were 64 small histology biopsies, 90 surgical biopsies and 66 cytology samples. However, there was no difference between two types of samples (154 histological plus surgical and 66 cytology samples) in cellular adequacy for EGFR, ROS1, ALK, and PD-L1. There was no change in the expression positivity rates for these four biomarkers between two types of samples (Ekin et al., 2021). ROS1-rearranged lung adenocarcinoma patient had active TME and increased plasma inflammatory factors when the patient received immune therapy and ceritinib chemotherapy. PD-L1 expression was elevated in tumor samples during treatment, suggesting that the patient obtained a limited benefit from combination therapies (Yue et al., 2021).
The efficacy of immunotherapy in TP53-mutant NSCLC
An immunohistochemical work illustrated that PD-L1 expression was associated with poor overall survival and PFS in NSCLC patients. CD8+ TILs were correlated with therapy response and a good PFS and overall survival. P53 expression was observed in most of NSCLC samples, but was not correlated with PD-L1 expression (Rashed et al., 2017). Serra et al. (2018) found that RAS/TP53 mutations were associated with PD-L1 expression in lung adenocarcinoma. Moreover, Dong et al. (2017) uncovered that TP53 mutation and KRAS mutation can predict the response to anti-PD-1 immunotherapy in lung adenocarcinoma. Zhang L. et al. (2021) reported that 219 cases from 350 NSCLC patients harbored TP53 mutations. Coexisting TP53 and ZFHX3 mutations were correlated with prognosis, indicating that TP53 and ZFHX3 mutations could be prognostic factors for late-stage NSCLC cases undergoing anti-PD-1/PD-L1 therapy. Another study also clarified that TP53 mutations were associated with response to ICIs treatment and a longer survival in advanced NSCLC patients (Assoun et al., 2019). Notably, NSCLC patients with TP53 plus KEAP1 mutations had a better PFS after treatment with PD-1/PD-L1 monotherapy (Wang S. et al., 2022). Strikingly, the TP53-missense mutation patients displayed enriched IFN-γ signatures and TME composition compared with TP53 wild-type patients (Sun et al., 2020). TP53 non-sense mutation patients exhibited promotion of suppressor immune cells, such as M2 Macrophage and Neutrophils. Upregulation of TMB and neoantigen levels were observed in both TP53 non-sense and missense mutations. TP53 missense was linked to better benefit of anti-PD-1/PD-L1 therapy (Sun et al., 2020).
The efficacy of immunotherapy in PTCH1-mutant NSCLC
Patched 1 (PTCH1) is one component of hedgehog pathway, which has been correlated with tumor malignancies (Sigafoos et al., 2021). In NSCLC patients, PTCH1 was underexpressed in the tumor specimens compared with normal lung samples (Herreros-Pomares et al., 2022). NSCLC patients with overexpression of PTCH1 displayed a better outcome (Herreros-Pomares et al., 2022). Moreover, PTCH1 expression was found to be correlated with NSCLC development (Barbirou et al., 2022). One circulating tumor cell NGS assay in early-stage lung cancer patients showed that more than 50% of lung cancer patients presented four common mutations, including Notch1, EGFR, IGF2, and PTCH1 (Wan et al., 2021). Genetic mutation analysis demonstrated that 147 mutant genes were discovered in small cell lung cancer patients, including TP53, RB1, KMT2D, PTCH1, APC, LRRK2, ARID2, and BRCA1 (Jin et al., 2021). In addition, elevated mutations of six genes were linked to advanced clinical stages II and III, such as SETD2, WT1, EPHA3, ACVR1B, NOTCH1 and KDM6A (Jin et al., 2021). Similarly, TP53 and RB1 mutations are two most frequently mutations in SCLC, while FGFR1, KIT, PTCH1, RICTOR, and RET mutations are low-frequency mutations (Dowlati et al., 2016). One retrospective study used the data from 180 lung squamous cell carcinoma and reported that patched receptor 1 (PTCH1) gene mutation was linked to CD8+ TILs density (Cheng et al., 2022). CD8+ TILs and high expression of PD-L1 were correlated with better disease-free survival in lung squamous cell carcinoma patients (Cheng et al., 2022). Serial sequencing of circulating tumor DNA (ctDNA) showed that PTCH1 mutation and β2 microglobulin (B2M) mutations were observed in NSCLC patients with anti-PD-1 treatment. Moreover, PTCH1 and B2M mutations were associated with distant metastasis in NSCLC patients (Li et al., 2019).
The efficacy of immunotherapy in ZFHX3-mutant NSCLC
ZFHX3 was reported to suppress alpha-fetoprotein expression. ZFHX3 mRNA expression in tumor tissues was linked to overall survival rate in 140 NSCLC patients. Low expression of ZFHX3 in NSCLC patients was associated with LNM and poor prognosis (Minamiya et al., 2012). Song et al. (2021) reported genomic profiles and TIME of lung cancer with brain metastasis. High-frequent ZFHX3 was found in 40% lung tumors and 28% brain tumors. A majority of lung cancer patients had lesions-shared mutations, such as EGFR mutation. Zhou et al. (2020) reported that 19% ZFHX3 mutation frequency was identified in lung cancer patients by next-generation sequencing. Another study also identified that the mutation of ZFHX3 in NSCLC patients could have benefit from ICIs treatment (Principe, 2022). ZFHX3 was identified as a genomic mutation for prediction of immunotherapy in NSCLC patients (Wang Z. et al., 2022). ZFHX3 mutation in NSCLC patients was correlated with TILs, immune-related gene expression and tumor mutation burden. ZFHX3 mutation was also linked to longer overall survival of NSCLC patients after treatment with ICIs (Zhang J. et al., 2021).
The efficacy of immunotherapy in PAK7-mutant NSCLC
Evidence has shown that p21-activated kinase (PAK7) regulates carcinogenesis in a variety of malignancies (Gu et al., 2013; Han et al., 2014; Quan et al., 2020; Wang et al., 2020). Suppression of PAK7 increased radio-sensitivity in hepatocellular carcinoma (HCC) (Gu and Kong, 2021). Depletion of PAK7 by shRNA transfection induced apoptosis and G2/M phase arrest, decreased clone formation and elevated γ-H2AX expression in HCC cells (Gu and Kong, 2021). PAK7 expression was upregulated in breast tumor samples and associated with differentiation and TNM stage in breast cancer patients. PAK7 activated Wnt/β-Catenin pathway and caused promotion of proliferation and migration as well as inhibition of apoptosis in breast cancer (Li et al., 2018). In esophageal squamous cell cancers (ESCC), high expression of PAK7 was correlated with LNM (He et al., 2016). Moreover, PAK7 was regulated by Aurora-A via binding with E2F1 in ESCC cells. PAK7 induced cisplatin resistance of ESCC with Aurora-A overexpression (He et al., 2016). One group revealed that PAK7 could be related to gemcitabine resistance in NSCLC cells (Zhang et al., 2013). PAK7 mutations were found to be associated with tumor mutation burden, neoantigen load, copy number variation, CD8+ TILs, mutation rate in the DDR-related pathways, suggesting that PAK7 mutations could be a helpful biomarker for prediction of the immunotherapy efficacy in NSCLC patients (Zeng et al., 2022).
The efficacy of immunotherapy in UBE3A-mutant NSCLC
UBE3A, also known as E6AP, acts as an E3 ligase and critically involves in carcinogenesis (Owais et al., 2020; Zheng et al., 2021). For example, UBE3A promoted tumor progression via disruption of ZNF185 in ESCC (Zheng et al., 2021). UBE3A targeted SIRT6 and regulated liver tumorigenesis, which was dependent on ANXA2 (Kohli et al., 2018). Downregulation of E6AP led to decreased expression of p15, p16 and p19 in NSCLC. E6AP represses the expression of CDC6 via inhibiting its E2F1 transcription (Gamell et al., 2017). UBE3A deletion promoted the immunotherapy efficacy in NSCLC patients (Zhang L. X. et al., 2022). NSCLC patients with UBE3A deletion had higher TILs and higher expression of immune checkpoint biomarkers (Zhang N. et al., 2022).
The efficacy of immunotherapy in TNF-α-mutant NSCLC
A mutated human tumor necrosis factor alpha (TNF-α) has been reported to improve the therapeutic index in the mouse fibroblast cell line L929 and mice (Yan et al., 2006). Similarly, TNF-α mutant was also found to promote cytotoxicity and receptor binding affinity (Shin et al., 1998). Pharmacokinetics of the recombinant mutated human TNF-α (rmhTNF-α) displayed that rmhTNF-α has a low systemic toxicity and high anticancer ability (Li et al., 2010). Phase II multicenter, randomized, double-blind trial showed that rmhTNF-α plus chemotherapies displayed higher response rate compared with chemotherapy alone group in multiple types of cancers. 11.39% patients had a response in the chemotherapy alone, while 27.47% patients had a response in the chemotherapy plus rmhTNF-α treatment. In lung cancer patients, the combination treatment caused 48.89% patients a response (Li et al., 2012). Moreover, a randomized phase III trial in stage IIIB/IV NSCLC patients showed that rmhTNF-α potentiated the efficacy of chemotherapy in advanced NSCLC patients (Ma et al., 2015). TNF-α alternation was uncovered for prediction of survival of ICIs in NSCLC patients. TNF-α mutations were linked to prolonged overall survival in NSCLC patients undergoing immunotherapy (Lin et al., 2021). TNF-α mutations were also related with TMB, DDR mutations and neoantigen load, and infiltrating immune cells (Lin et al., 2021).
The efficacy of immunotherapy in NOTCH-mutant NSCLC
Notch signaling pathway is critically involved in tumorigenesis, which consists of four receptors, Notch1, Notch2, Notch 3, Notch4, and several ligands, such as delta-like proteins (DLL1, DLL3, DLL4), Jagged-1 and Jagged-2 (Gao et al., 2020; Majumder et al., 2021). In general, 20%–25% of SCLC patients exhibited Notch mutations with loss-of-function (LOF). Notch can act as a tumor suppressor in SCLC and also enhance non-neuroendocrine plasticity to facilitate tumor growth in SCLC (Hong et al., 2022). Mice with genetic loss of Nocth1 or Nocth2 facilitated SCLC tumorigenesis and formed non-neuroendocrine populations via regulation of RUNX2/REST pathway and STING (stimulator of interferon genes) (Hong et al., 2022). Li Y. et al. (2022) reported that Notch pathway was correlated with TIME in SCLC. Notch1 gene mutation was negatively linked to PD-L1 expression in SCLC patients. Higher expression of DLL3 was found in SCLC patients and associated with PD-L1 levels. Hence, SCLC patients with positive DLL3 expression and Notch1 wild type had PD-L1 overexpression, which could be likely to have good immunotherapy efficacy. Notch2 mutation was a prognostic factor in NSCLC patients and could be provide a new treatment option for patients without EGFR mutations (Niu et al., 2021).
The high-mutated NOTCH pathway could act as a biomarker for predicting the prognosis of ICIs-treated NSCLC patients because NSCLC patients with high-mutated NOTCH pathway had a better PFS and OS (Li et al., 2021). Zhang et al. (2020) also identified that Notch mutation acted as a new predictor for efficacious immunotherapy in NSCLC patients. Notch1/2/3 mutation had a correlation with better ICI treatment outcomes, including PFS and overall survival due to regulation of transcription of genes that were related to immune activation and DNA damage response (Zhang et al., 2020). Notch4 mutation was also a potential response biomarker for ICIs therapy in several cancer types, including NSCLC (Long et al., 2021). Cancer patients with Notch4 mutation displayed better responses for ICI therapy, including ORR, DCB, PFS and overall survival. Notch4 mutation was linked to increased immunogenicity, high TMB, anticancer immunity and activation of the antigen-processing machinery (Long et al., 2021).
The efficacy of immunotherapy in LRP1B-mutant NSCLC
LRP1B has been reported to be frequently mutated in numerous types of cancers, including lung cancer (Principe et al., 2021). The bioinformatics analysis showed that LRP1B mutation was linked to age and MUC16 and TP53 mutation status in gastric cancer patients (Hu et al., 2021). The next-generation sequencing (NGS) data showed that 13.98% of NSCLC patients had LRP1B mutation (Xu et al., 2023). LRP1B mutation was correlated with high TMB in NSCLC. Moreover, NSCLC patients with LRP1B mutation had a high infiltrating levels of immune cells and immune molecules. Additionally, LRP1B mutations were linked to several pathways in the immune system, including cell cycle, Notch, mTOR and insulin pathways (Xu et al., 2023). LRP1B mutation was associated with TMB and outcomes in NSCLC patients with immunotherapy (Chen et al., 2019). LRP1B mutation was correlated with a better survival in NSCLC patients. Moreover, LRP1B mutations was also associated with immunocytes and enriched pathways, such as cell cycle mitotic, antigen processing and presentation pathways (Chen et al., 2019). Another group reported that LRP1B mutation was correlated with better outcomes to ICIs in combination with chemotherapy in NSCLC patients (Zhou J. et al., 2022). Hence, LRP1B mutations could be critical in promoting immunotherapy and might be a biomarker for judgement of treatment responsiveness.
The efficacy of immunotherapy in FBXW7-mutant NSCLC
FBXW7, one of F-box proteins, has been identified to regulate carcinogenesis and progression (Wang et al., 2014; Yan et al., 2020; Liu F. et al., 2022). FBXW7 mutation caused drug resistance via targeting several downstream substrates for ubiquitination and degradation, including Mcl-1, mTOR, snail and CCDC6 in NSCLC (Peng and Chen, 2019). Several compounds displayed an effective treatment efficacy in NSCLC patients with FBXW7 mutation, such as rabdosia, MS-275 and rapamycin (Peng and Chen, 2019). By analysis of TCGA data, 30.9% of lung adenocarcinoma presents FBXW7 deletion, and 63.5% of lung squamous cell carcinoma exhibited FBXW7 deletion. FBXW7 deletion led to lung oncogenesis and contributed to gefitinib resistance (Xiao et al., 2018). One study revealed that 5.6% of NSCLC patients (7 cases) had FBXW7 truncating mutations in 125 NSCLC cases. In these seven patients with FBXW7 mutation after they obtained immunotherapy, four cases presented partial response, two cases showed stable disease, and one case displayed progressive disease (Liu J. et al., 2022). FBXW7-mutant NSCLC patients had 13 months for median progression-free survival (PFS), while FBXW7 wild type patients had 4 months for PFS. FBXW7-mutant patients had a higher TMB and the activation of T cells. Moreover, FBXW7 mutation was linked to upregulation of CD8+ T cell infiltration and M1 macrophages. FBXW7 gene mutation could predict the prognosis of immunotherapy in patients with NSCLC (Liu C. et al., 2022).
LncRNAs and circRNAs regulate TME and immunotherapy in lung cancer
Besides these gene mutations, non-coding RNAs have been reported to involve in regulating TME and immunotherapy in lung cancer. Non-coding RNAs have been reported to involved in human cancer development and progression (Ghafouri-Fard et al., 2020; Yan and Bu, 2021; Chen T. et al., 2022; Liu and Shang, 2022). Evidence has shown that lncRNAs play an essential role in NSCLC initiation, development and progression (Osielska and Jagodzinski, 2018; Wang et al., 2018; Hu et al., 2022). Moreover, non-coding RNAs are critically involved in cancer drug resistance in human cancers (Jiang et al., 2020; Zhou X. et al., 2022; Xie et al., 2022). LncRNAs and exosomal lncRNAs regulate tumor progression, drug sensitivity and TME remodeling in lung cancer (Entezari et al., 2022). The role of lncRNAs in the regulation of PD-1 and PD-L1 pathways and TME in cancer immunotherapy has been discussed (Jiang W. et al., 2021; Dai et al., 2021; Zhang P. et al., 2022). For example, lncRNA C5orf64 was characterized as a potential indicator for TME and mutation pattern remodeling in lung cancer (Pang et al., 2021). LncRNA C5orf64 expression was positively associated with neutrophils, monocytes, M2 macrophages and eosinophils, and negatively linked to Tregs and plasma cells (Pang et al., 2021). High expression of C5orf64 was linked to upregulation of PD-1, PD-L1 and CTLA-4 expression. Interestingly, lung adenocarcinoma patients with high expression of C5orf64 had a low frequency of TP53 mutation (Pang et al., 2021). Together, lncRNA C5orf64 could be a useful indicator for TME modulation and immunotherapy in lung cancer. Jiang Y. et al. (2021) found that cancer-associated fibroblasts (CAFs)-derived exosomes can regulate lncRNA OIP5-AS1 and modulate miR-142-5p and control the expression of PD-L1, leading to promotion of lung cancer progression. Moreover, N6-methyladenosin (m6A) related lncRNA signatures with TME have been defined to predict the immunotherapy in lung cancer (Weng et al., 2021; Zhao et al., 2021; Zhang W. et al., 2022; Yan et al., 2022). Recently, the circular RNA circHMGB2 was uncovered to promote immunosuppression and resistance to anti-PD-1 therapy via targeting miR-181a-5p and upregulating CARM1 in lung cancer, suggesting that circHMGB2 reshaped the TME and governed immunotherapy in lung cancer (Zhang L. X. et al., 2022).
Conclusion and perspectives
TME is critically involved in immunotherapy in lung cancer. The efficacy of immunotherapy was regulated by driven gene mutations in lung cancer, including KRAS, TP53, EGFR, ALK, ROS1, ZFHX3, and PTCH1 (Figure 2). Targeting TME could abolish immune resistance of anti-PD-1/PD-L1 treatment in lung cancer. It has been suggested that PD-1/PD-L1 blockade should be combined with other therapy such as chemotherapy to improve the anticancer efficacy in human cancer (Wu et al., 2022).
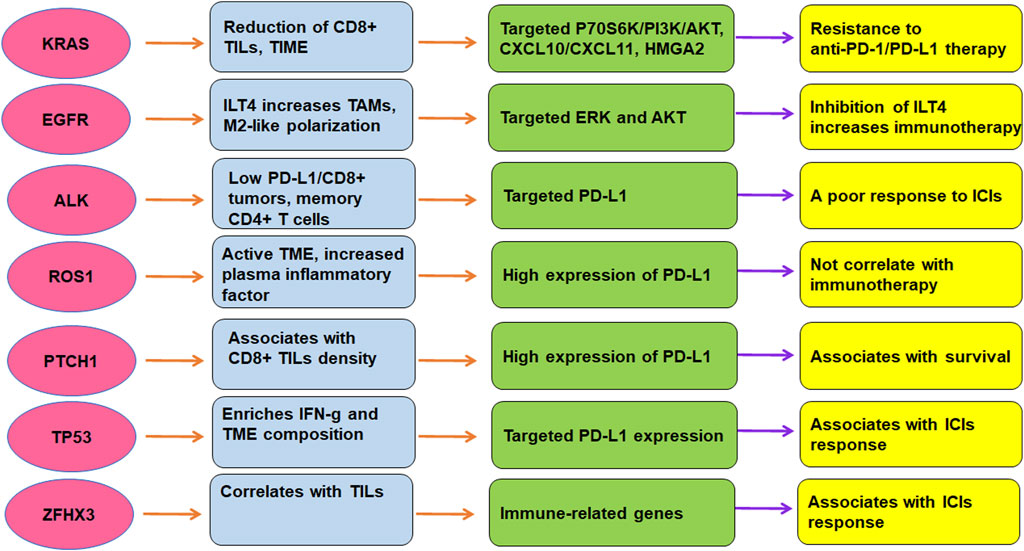
FIGURE 2. The role of gene mutations in regulation of TME and immunotherapy in lung cancer. The efficacy of immunotherapy was regulated by driven gene mutations in lung cancer, including KRAS, TP53, EGFR, ALK, ROS1, ZFHX3, and PTCH1.
Several issues need to be clarified regarding the TME and immunotherapy in NSCLC. For example, several reports showed that mRNA vaccine could be applied for cancer treatment via regulation of TME (Zhong et al., 2021; Zhao W. et al., 2022; Huang et al., 2022). Proteomics, genomics, and metabolomics might be good approaches to explore the mechanism of gene mutation-driven lung cancer and TME (Zhou et al., 2019; Bourbonne et al., 2022). Recently, several studies used the single-cell profiling of lung cancer to determine the TME and immunotherapy (Maynard et al., 2020; Wu et al., 2021; Hui et al., 2022). Hui et al. (2022) reported single-cell profiling of immune cells after chemotherapy and pembrolizumab in advanced NSCLC. This study found the synergistical increase of CD4+ cells and B cells were positively correlated with chemoimmunotherapy. Moreover, this work identified several positive outcomes, such as promotion of TNFRSF4+ Tregs, LAMP3+ DCs, intratumoral CD4+ T clones and CD8+ T clones (Hui et al., 2022). In addition, single-cell RNA sequencing was used to evaluate therapy-induced evolution in lung cancer patients, including TN (patients before initiating systemic therapy, TKI naive), RD (residual disease) and PD (on-therapy progressive disease) (Maynard et al., 2020). Transcriptional differences between RD and TN tumor cells suggested cell-state-specific programs, while transcriptional differences between PD and TN tumor cells indicated that immune modulation and invasion are critical for cancer progression. RD patients displayed active T-lymphocytes and reduced macrophages, while PD patients displayed immunosuppressive cell states (Maynard et al., 2020). Wu et al. also reported single-cell profiling of tumor heterogeneity and TME in advanced NSCLC. This work identified not only common cell types but also rare cell types in tumors including T helper 17 cells and follicular dendritic cells. Different NSCLC patients exhibited larger heterogeneity in chromosomal structure, intercellular signaling network and cellular composition and so on (194). Further investigations are necessary to determine the underlying molecular mechanisms of TME in regulation of immunotherapy resistance. In addition, besides TME, immunotherapy resistance could be caused by other factors in lung cancer, which should be explored in the future.
Author contributions
MW wrote the manuscript; LZ, XY, and JL searched the literatures, made the tables and figures. YT edited and revised the manuscript. All authors have approved for the final submission of the manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (grant No. 81900037).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ALK, anaplastic lymphoma kinase; BRAF, v-Raf murine sarcoma viral oncogene homolog B; CNV, copy number variation; CTLA-4, cytotoxic T-lymphocyte antigen 4; DDR, DNA damage response and repair; EGFR, epidermal growth factor receptor; FBXW7, F-box and WD-40 domain protein 7; ICIs, immune checkpoint inhibitors; KRAS, Kirsten rat sarcoma; LRP1B, low-density lipoprotein receptor-related protein 1B; mTOR, mammalian target of rapamycin; NSCLC, non-small cell lung carcinoma; OS, overall survival; PD-1, programmed cell death-1; PD-L1, programmed cell death ligand-1; PFS, progression-free survival; ROS1, receptor tyrosine kinase c-ros oncogene 1; TMB, tumor mutational burden; TME, tumor microenvironment; TIME, tumor immune microenvironment; TNFα, tumor necrosis factor α; ZFHX3, zinc finger homeobox 3.
References
Abdayem, P., and Planchard, D. (2022). Ongoing progress in BRAF-mutated non-small cell lung cancer. Clin. Adv. Hematol. Oncol. 20 (11), 662–672. Cited in: Pubmed; PMID 36331404.
Ackermann, C. J., Reck, M., Paz-Ares, L., Barlesi, F., and Califano, R. (2019). First-line immune checkpoint blockade for advanced non-small-cell lung cancer: Travelling at the speed of light. Lung Cancer 134, 245–253. Epub 20190619Cited in: Pubmed; PMID 31319988. doi:10.1016/j.lungcan.2019.06.007
Adderley, H., Blackhall, F. H., and Lindsay, C. R. (2019). KRAS-mutant non-small cell lung cancer: Converging small molecules and immune checkpoint inhibition. EBioMedicine 41, 711–716. Epub 20190307Cited in: Pubmed; PMID 30852159. doi:10.1016/j.ebiom.2019.02.049
Ahluwalia, P., Ahluwalia, M., Mondal, A. K., Sahajpal, N. S., Kota, V., Rojiani, M. V., et al. (2021). Natural killer cells and dendritic cells: Expanding clinical relevance in the non-small cell lung cancer (NSCLC) tumor microenvironment. Cancers (Basel) 13 (16), 4037. Epub 20210811. doi:10.3390/cancers13164037
Assoun, S., Theou-Anton, N., Nguenang, M., Cazes, A., Danel, C., Abbar, B., et al. (2019). Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer 132, 65–71. Epub 20190408Cited in: Pubmed; PMID 31097096. doi:10.1016/j.lungcan.2019.04.005
Bahnassy, A. A., Ismail, H., Mohanad, M., El-Bastawisy, A., and Yousef, H. F. (2022). The prognostic role of PD-1, PD-L1, ALK, and ROS1 proteins expression in non-small cell lung carcinoma patients from Egypt. J. Egypt Natl. Canc Inst. 34 (1), 23. Epub 20220530Cited in: Pubmed; PMID 35644823. doi:10.1186/s43046-022-00121-8
Barbirou, M., Miller, A., Manjunath, Y., Ramirez, A. B., Ericson, N. G., Staveley-O'Carroll, K. F., et al. (2022). Single circulating-tumor-cell-targeted sequencing to identify somatic variants in liquid biopsies in non-small-cell lung cancer patients. Curr. Issues Mol. Biol. 44 (2), 750–763. Epub 20220202. doi:10.3390/cimb44020052
Binnewies, M., Roberts, E. W., Kersten, K., Chan, V., Fearon, D. F., Merad, M., et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24 (5), 541–550. Epub 20180423Cited in: Pubmed; PMID 29686425. doi:10.1038/s41591-018-0014-x
Biton, J., Mansuet-Lupo, A., Pecuchet, N., Alifano, M., Ouakrim, H., Arrondeau, J., et al. (2018). TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin. Cancer Res. 24 (22), 5710–5723. Epub 20180515Cited in: Pubmed; PMID 29764856. doi:10.1158/1078-0432.CCR-18-0163
Bote, H., Mesas, A., Baena, J., Herrera, M., and Paz-Ares, L. (2022). Emerging immune checkpoint inhibitors for the treatment of non-small cell lung cancer. Expert Opin. Emerg. Drugs 27 (3), 289–300. Epub 20221006. doi:10.1080/14728214.2022.2113377
Bourbonne, V., Geier, M., Schick, U., and Lucia, F. (2022). Multi-omics approaches for the prediction of clinical endpoints after immunotherapy in non-small cell lung cancer: A comprehensive review. Biomedicines 10 (6), 1237. Epub 20220526. doi:10.3390/biomedicines10061237
Bredin, P., and Naidoo, J. (2022). The gut microbiome, immune check point inhibition and immune-related adverse events in non-small cell lung cancer. Cancer Metastasis Rev. 41 (2), 347–366. Epub 20220725Cited in: Pubmed; PMID 35876944. doi:10.1007/s10555-022-10039-1
Bylicki, O., Paleiron, N., Margery, J., Guisier, F., Vergnenegre, A., Robinet, G., et al. (2017). Targeting the PD-1/PD-L1 immune checkpoint in EGFR-mutated or ALK-translocated non-small-cell lung cancer. Target Oncol. 12 (5), 563–569. Cited in: Pubmed; PMID 28624922. doi:10.1007/s11523-017-0510-9
Caliman, E., Fancelli, S., Petroni, G., Gatta Michelet, M. R., Cosso, F., Ottanelli, C., et al. (2022). Challenges in the treatment of small cell lung cancer in the era of immunotherapy and molecular classification. Lung Cancer 175, 88–100. Epub 20221123Cited in: Pubmed; PMID 36493578. doi:10.1016/j.lungcan.2022.11.014
Cao, H., Gao, S., Jogani, R., and Sugimura, R. (2022). The tumor microenvironment reprograms immune cells. Cell Reprogr. 24 (6), 343–352. Epub 20221026. doi:10.1089/cell.2022.0047
Cardona, A. F., Rojas, L., Zatarain-Barron, Z. L., Ruiz-Patino, A., Ricaurte, L., Corrales, L., et al. (2019). Multigene mutation profiling and clinical characteristics of small-cell lung cancer in never-smokers vs. Heavy smokers (Geno1.3-CLICaP). Front. Oncol. 9, 254. Epub 20190417. doi:10.3389/fonc.2019.00254
Castaneda-Gonzalez, J. P., Chaves, J. J., and Parra-Medina, R. (2022). Multiple mutations in the EGFR gene in lung cancer: A systematic review. Transl. Lung Cancer Res. 11 (10), 2148–2163. Cited in: Pubmed; PMID 36386461. doi:10.21037/tlcr-22-235
Catalano, M., Shabani, S., Venturini, J., Ottanelli, C., Voltolini, L., and Roviello, G. (2022). Lung cancer immunotherapy: Beyond common immune checkpoints inhibitors. Cancers (Basel) 14 (24), 6145. Epub 20221213. doi:10.3390/cancers14246145
Ceddia, S., Landi, L., and Cappuzzo, F. (2022). KRAS-mutant non-small-cell lung cancer: From past efforts to future challenges. Int. J. Mol. Sci. 23 (16), 9391. Epub 20220820. doi:10.3390/ijms23169391
Chen, H., Chong, W., Wu, Q., Yao, Y., Mao, M., and Wang, X. (2019). Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front. Immunol. 10, 1113. Epub 20190521. doi:10.3389/fimmu.2019.01113
Chen, K., Cheng, G., Zhang, F., Zhu, G., Xu, Y., Yu, X., et al. (2020). PD-L1 expression and T cells infiltration in patients with uncommon EGFR-mutant non-small cell lung cancer and the response to immunotherapy. Lung Cancer 142, 98–105. Epub 20200219Cited in: Pubmed; PMID 32120230. doi:10.1016/j.lungcan.2020.02.010
Chen, K., Pan, G., Cheng, G., Zhang, F., Xu, Y., Huang, Z., et al. (2021). Immune microenvironment features and efficacy of PD-1/PD-L1 blockade in non-small cell lung cancer patients with EGFR or HER2 exon 20 insertions. Thorac. Cancer 12 (2), 218–226. Epub 20201118. doi:10.1111/1759-7714.13748
Chen, X., Gao, A., Zhang, F., Yang, Z., Wang, S., Fang, Y., et al. (2021). ILT4 inhibition prevents TAM- and dysfunctional T cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in NSCLC with EGFR activation. Theranostics 11 (7), 3392–3416. Epub 20210119. doi:10.7150/thno.52435
Chen, P., Liu, Y., Wen, Y., and Zhou, C. (2022). Non-small cell lung cancer in China. Cancer Commun. (Lond) 42 (10), 937–970. Epub 20220908. doi:10.1002/cac2.12359
Chen, T., Liu, J., Zhang, H., Li, J., and Shang, G. (2022). Long intergenic noncoding RNA00265 enhances cell viability and metastasis via targeting miR-485-5p/USP22 Axis in osteosarcoma. Front. Oncol. 12, 907472. Epub 20220526. doi:10.3389/fonc.2022.907472
Cheng, X., Wang, L., and Zhang, Z. (2022). Prognostic significance of PD-L1 expression and CD8(+) TILs density for disease-free survival in surgically resected lung squamous cell carcinoma: A retrospective study. J. Thorac. Dis. 14 (6), 2224–2234. Cited in: Pubmed; PMID 35813758. doi:10.21037/jtd-22-630
Choi, H. K., and Mazzone, P. J. (2022). Lung cancer screening. Med. Clin. North Am. 106 (6), 1041–1053. Epub 20221004Cited in: Pubmed; PMID 36280331. doi:10.1016/j.mcna.2022.07.007
Cinausero, M., Laprovitera, N., De Maglio, G., Gerratana, L., Riefolo, M., Macerelli, M., et al. (2019). KRAS and ERBB-family genetic alterations affect response to PD-1 inhibitors in metastatic nonsquamous NSCLC. Ther. Adv. Med. Oncol. 11, 1758835919885540. Epub 20191114. doi:10.1177/1758835919885540
Cognigni, V., Pecci, F., Lupi, A., Pinterpe, G., De Filippis, C., Felicetti, C., et al. (2022). The landscape of ALK-rearranged non-small cell lung cancer: A comprehensive review of clinicopathologic, genomic characteristics, and therapeutic perspectives. Cancers (Basel) 14 (19), 4765. Epub 20220929. doi:10.3390/cancers14194765
Cullis, J., Das, S., and Bar-Sagi, D. (2018). Kras and tumor immunity: Friend or foe? Cold Spring Harb. Perspect. Med. 8 (9), a031849. Epub 20180904. doi:10.1101/cshperspect.a031849
Dai, S., Liu, T., Liu, Y. Y., He, Y., Liu, T., Xu, Z., et al. (2021). Long non-coding RNAs in lung cancer: The role in tumor microenvironment. Front. Cell Dev. Biol. 9, 795874. Epub 20220103. doi:10.3389/fcell.2021.795874
Dawe, D. E., Harlos, C. H., and Juergens, R. A. (2020). Immuno-oncology-the new paradigm of lung cancer treatment. Curr. Oncol. 27, S78–S86. Epub 20200401. doi:10.3747/co.27.5183
Desage, A. L., Leonce, C., Swalduz, A., and Ortiz-Cuaran, S. (2022). Targeting KRAS mutant in non-small cell lung cancer: Novel insights into therapeutic strategies. Front. Oncol. 12, 796832. Epub 20220216. doi:10.3389/fonc.2022.796832
Di Nicolantonio, F., Vitiello, P. P., Marsoni, S., Siena, S., Tabernero, J., Trusolino, L., et al. (2021). Precision oncology in metastatic colorectal cancer - from biology to medicine. Nat. Rev. Clin. Oncol. 18 (8), 506–525. Epub 20210416Cited in: Pubmed; PMID 33864051. doi:10.1038/s41571-021-00495-z
Dias Carvalho, P., Guimaraes, C. F., Cardoso, A. P., Mendonca, S., Costa, A. M., Oliveira, M. J., et al. (2018). KRAS oncogenic signaling extends beyond cancer cells to orchestrate the microenvironment. Cancer Res. 78 (1), 7–14. Epub 20171220. doi:10.1158/0008-5472.CAN-17-2084
Dong, Z. Y., Zhong, W. Z., Zhang, X. C., Su, J., Xie, Z., Liu, S. Y., et al. (2017). Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 23 (12), 3012–3024. Epub 20161230. doi:10.1158/1078-0432.CCR-16-2554
Dowlati, A., Lipka, M. B., McColl, K., Dabir, S., Behtaj, M., Kresak, A., et al. (2016). Clinical correlation of extensive-stage small-cell lung cancer genomics. Ann. Oncol. 27 (4), 642–647. Epub 20160122. doi:10.1093/annonc/mdw005
Ekin, Z., Nart, D., Savas, P., and Veral, A. (2021). Comparison of PD-L1, EGFR, ALK, and ROS1 status between surgical samples and cytological samples in non-small cell lung carcinoma. Balk. Med. J. 38 (5), 287–295. in: Pubmed; PMID 34558414. doi:10.5152/balkanmedj.2021.20086
Entezari, M., Ghanbarirad, M., Taheriazam, A., Sadrkhanloo, M., Zabolian, A., Goharrizi, M., et al. (2022). Long non-coding RNAs and exosomal lncRNAs: Potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed. Pharmacother. 150, 112963. Epub 20220422. doi:10.1016/j.biopha.2022.112963
Eulberg, D., Fromming, A., Lapid, K., Mangasarian, A., and Barak, A. (2022). The prospect of tumor microenvironment-modulating therapeutical strategies. Front. Oncol. 12, 1070243. Epub 20221208. doi:10.3389/fonc.2022.1070243
Faraj, J. A., Al-Athari, A. J. H., Mohie, S. E. D., Kadhim, I. K., Jawad, N. M., Abbas, W. J., et al. (2022). Reprogramming the tumor microenvironment to improve the efficacy of cancer immunotherapies. Med. Oncol. 39 (12), 239. Epub 20220929Cited in: Pubmed; PMID 36175691. doi:10.1007/s12032-022-01842-5
Ferrer, I., Zugazagoitia, J., Herbertz, S., John, W., Paz-Ares, L., and Schmid-Bindert, G. (2018). KRAS-Mutant non-small cell lung cancer: From biology to therapy. Lung Cancer 124, 53–64. Epub 20180719Cited in: Pubmed; PMID 30268480. doi:10.1016/j.lungcan.2018.07.013
Frazer, K., Bhardwaj, N., Fox, P., Stokes, D., Niranjan, V., Quinn, S., et al. (2022). Systematic review of smoking cessation inventions for smokers diagnosed with cancer. Int. J. Environ. Res. Public Health 19 (24), 17010. Epub 20221218. doi:10.3390/ijerph192417010
Fu, Y., Zhang, Y., Lei, Z., Liu, T., Cai, T., Wang, A., et al. (2020). Abnormally activated OPN/integrin αVβ3/FAK signalling is responsible for EGFR-TKI resistance in EGFR mutant non-small-cell lung cancer. J. Hematol. Oncol. 13 (1), 169. Epub 20201207Cited in: Pubmed; PMID 33287873. doi:10.1186/s13045-020-01009-7
Gainor, J. F., Shaw, A. T., Sequist, L. V., Fu, X., Azzoli, C. G., Piotrowska, Z., et al. (2016). EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: A retrospective analysis. Clin. Cancer Res. 22 (18), 4585–4593. Epub 20160525. doi:10.1158/1078-0432.CCR-15-3101
Gajewski, T. F., Schreiber, H., and Fu, Y. X. (2013). Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14 (10), 1014–1022. . Cited in: Pubmed; PMID 24048123. doi:10.1038/ni.2703
Gamell, C., Gulati, T., Levav-Cohen, Y., Young, R. J., Do, H., Pilling, P., et al. (2017). Reduced abundance of the E3 ubiquitin ligase E6AP contributes to decreased expression of the INK4/ARF locus in non-small cell lung cancer. Sci. Signal 10 (461), eaaf8223. Epub 20170110. doi:10.1126/scisignal.aaf8223
Gao, A., Liu, X., Lin, W., Wang, J., Wang, S., Si, F., et al. (2021). Tumor-derived ILT4 induces T cell senescence and suppresses tumor immunity. J. Immunother. Cancer 9 (3), e001536. Cited in: Pubmed; PMID 33653799. doi:10.1136/jitc-2020-001536
Gao, A., Sun, Y., and Peng, G. (2018). ILT4 functions as a potential checkpoint molecule for tumor immunotherapy. Biochim. Biophys. Acta Rev. Cancer 1869 (2), 278–285. Epub 20180410Cited in: Pubmed; PMID 29649510. doi:10.1016/j.bbcan.2018.04.001
Gao, Y., Bai, L., and Shang, G. (2020). Notch-1 promotes the malignant progression of osteosarcoma through the activation of cell division cycle 20. Aging (Albany NY) 13 (2), 2668–2680. Epub 20201219. doi:10.18632/aging.202314
Garcia-Robledo, J. E., Rosell, R., Ruiz-Patino, A., Sotelo, C., Arrieta, O., Zatarain-Barron, L., et al. (2022). KRAS and MET in non-small-cell lung cancer: Two of the new kids on the 'drivers' block. Ther. Adv. Respir. Dis. 16, 17534666211066064. in: Pubmed; PMID 35098800. doi:10.1177/17534666211066064
Gemelli, M., Noonan, D. M., Carlini, V., Pelosi, G., Barberis, M., Ricotta, R., et al. (2022). Overcoming resistance to checkpoint inhibitors: Natural killer cells in non-small cell lung cancer. Front. Oncol. 12, 886440. Epub 20220531. doi:10.3389/fonc.2022.886440
Genova, C., Dellepiane, C., Carrega, P., Sommariva, S., Ferlazzo, G., Pronzato, P., et al. (2021). Therapeutic implications of tumor microenvironment in lung cancer: Focus on immune checkpoint blockade. Front. Immunol. 12, 799455. Epub 20220107. doi:10.3389/fimmu.2021.799455
Ghafouri-Fard, S., Shoorei, H., Branicki, W., and Taheri, M. (2020). Non-coding RNA profile in lung cancer. Exp. Mol. Pathol. 114, 104411. Epub 20200226. doi:10.1016/j.yexmp.2020.104411
Girard, N. (2022). New strategies and novel combinations in EGFR TKI-resistant non-small cell lung cancer. Curr. Treat. Options Oncol. 23 (11), 1626–1644. Epub 20221015Cited in: Pubmed; PMID 36242712. doi:10.1007/s11864-022-01022-7
Gkountakos, A., Delfino, P., Lawlor, R. T., Scarpa, A., Corbo, V., and Bria, E. (2021). Harnessing the epigenome to boost immunotherapy response in non-small cell lung cancer patients. Ther. Adv. Med. Oncol. 13, 17588359211006947. Epub 20210525. doi:10.1177/17588359211006947
Gu, J., Li, K., Li, M., Wu, X., Zhang, L., Ding, Q., et al. (2013). A role for p21-activated kinase 7 in the development of gastric cancer. FEBS J. 280 (1), 46–55. Epub 20121123. doi:10.1111/febs.12048
Gu, Y. F., and Kong, L. T. (2021). Inhibiting p21-activated kinase (PAK7) enhances radiosensitivity in hepatocellular carcinoma. Hum. Exp. Toxicol. 40 (12), 2202–2214. Epub 20210624. doi:10.1177/09603271211027948
Guaitoli, G., Bertolini, F., Bettelli, S., Manfredini, S., Maur, M., Trudu, L., et al. (2021). Deepening the knowledge of ROS1 rearrangements in non-small cell lung cancer: Diagnosis, treatment, resistance and concomitant alterations. Int. J. Mol. Sci. 22 (23), 12867. Epub 20211128. doi:10.3390/ijms222312867
Guan, X., Bao, G., Liang, J., Yao, Y., Xiang, Y., and Zhong, X. (2022). Evolution of small cell lung cancer tumor mutation: From molecular mechanisms to novel viewpoints. Semin. Cancer Biol. 86, 346–355. Epub 20220330Cited in: Pubmed; PMID 35367118. doi:10.1016/j.semcancer.2022.03.015
Guo, H., Li, W., and Wu, J. (2020). Ambient PM2.5 and annual lung cancer incidence: A nationwide study in 295 Chinese counties. Int. J. Environ. Res. Public Health 17 (5), 1481. Epub 20200225. doi:10.3390/ijerph17051481
Guo, Q., Liu, L., Chen, Z., Fan, Y., Zhou, Y., Yuan, Z., et al. (2022). Current treatments for non-small cell lung cancer. Front. Oncol. 12, 945102. Epub 20220811. doi:10.3389/fonc.2022.945102
Guo, R., Li, Y., Wang, Z., Bai, H., Duan, J., Wang, S., et al. (2019). Hypoxia-inducible factor-1α and nuclear factor-κB play important roles in regulating programmed cell death ligand 1 expression by epidermal growth factor receptor mutants in non-small-cell lung cancer cells. Cancer Sci. 110 (5), 1665–1675. Epub 20190323. doi:10.1111/cas.13989
Han, J., Liu, Y., Yang, S., Wu, X., Li, H., and Wang, Q. (2021). MEK inhibitors for the treatment of non-small cell lung cancer. J. Hematol. Oncol. 14 (1), 1. Epub 20210105Cited in: Pubmed; PMID 33402199. doi:10.1186/s13045-020-01025-7
Han, K., Zhou, Y., Gan, Z. H., Qi, W. X., Zhang, J. J., Fen, T., et al. (2014). p21-activated kinase 7 is an oncogene in human osteosarcoma. Cell Biol. Int. 38 (12), 1394–1402. Epub 20140806. doi:10.1002/cbin.10351
Hao, Y., Zhang, X., and Yu, L. (2022). Immune checkpoint inhibitor-related pneumonitis in non-small cell lung cancer: A review. Front. Oncol. 12, 911906. Epub 20220816Cited in: Pubmed; PMID 36052257. doi:10.3389/fonc.2022.911906
He, S., Liu, M., Zhang, W., Xu, N., and Zhu, H. (2016). Over expression of p21-activated kinase 7 associates with lymph node metastasis in esophageal squamous cell cancers. Cancer Biomark. 16 (2), 203–209. . Cited in: Pubmed; PMID 26682509. doi:10.3233/CBM-150557
Herreros-Pomares, A., Doria, P., Gallach, S., Meri-Abad, M., Guijarro, R., Calabuig-Farinas, S., et al. (2022). A sonic hedgehog pathway score to predict the outcome of resected non-small cell lung cancer patients. Ann. Surg. Oncol. 30, 1225–1235. Epub 20220921Cited in: Pubmed; PMID 36131117. doi:10.1245/s10434-022-12565-2
Hong, D., Knelson, E. H., Li, Y., Durmaz, Y. T., Gao, W., Walton, E., et al. (2022). Plasticity in the absence of NOTCH uncovers a RUNX2-dependent pathway in small cell lung cancer. Cancer Res. 82 (2), 248–263. Epub 20211122. doi:10.1158/0008-5472.CAN-21-1991
Hsu, P. C., Jablons, D. M., Yang, C. T., and You, L. (2019). Epidermal growth factor receptor (EGFR) pathway, yes-associated protein (YAP) and the regulation of programmed death-ligand 1 (PD-L1) in non-small cell lung cancer (NSCLC). Int. J. Mol. Sci. 20 (15), 3821. Epub 20190805. doi:10.3390/ijms20153821
Hu, Q., Ma, H., Chen, H., Zhang, Z., and Xue, Q. (2022). LncRNA in tumorigenesis of non-small-cell lung cancer: From bench to bedside. Cell Death Discov. 8 (1), 359. Epub 20220813Cited in: Pubmed; PMID 35963868. doi:10.1038/s41420-022-01157-4
Hu, S., Zhao, X., Qian, F., Jin, C., and Hou, K. (2021). Correlation between LRP1B mutations and tumor mutation burden in gastric cancer. Comput. Math. Methods Med. 2021, 1522250. Epub 20210921. doi:10.1155/2021/1522250
Huang, T., Peng, L., Han, Y., Wang, D., He, X., Wang, J., et al. (2022). Lipid nanoparticle-based mRNA vaccines in cancers: Current advances and future prospects. Front. Immunol. 13, 922301. Epub 20220826. doi:10.3389/fimmu.2022.922301
Hui, Z., Zhang, J., Ren, Y., Li, X., Yan, C., Yu, W., et al. (2022). Single-cell profiling of immune cells after neoadjuvant pembrolizumab and chemotherapy in IIIA non-small cell lung cancer (NSCLC). Cell Death Dis. 13 (7), 607. Epub 20220713Cited in: Pubmed; PMID 35831283. doi:10.1038/s41419-022-05057-4
Jiang, W., Pan, S., Chen, X., Wang, Z. W., and Zhu, X. (2021). The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol. Cancer 20 (1), 116. Epub 20210908Cited in: Pubmed; PMID 34496886. doi:10.1186/s12943-021-01406-7
Jiang, Y., Wang, K., Lu, X., Wang, Y., and Chen, J. (2021). Cancer-associated fibroblasts-derived exosomes promote lung cancer progression by OIP5-AS1/miR-142-5p/PD-L1 axis. Mol. Immunol. 140, 47–58. Epub 20211013Cited in: Pubmed; PMID 34653794. doi:10.1016/j.molimm.2021.10.002
Jiang, W., Xia, J., Xie, S., Zou, R., Pan, S., Wang, Z. W., et al. (2020). Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Updat 50, 100683. Epub 20200225. doi:10.1016/j.drup.2020.100683
Jin, R., Liu, C., Zheng, S., Wang, X., Feng, X., Li, H., et al. (2020). Molecular heterogeneity of anti-PD-1/PD-L1 immunotherapy efficacy is correlated with tumor immune microenvironment in East Asian patients with non-small cell lung cancer. Cancer Biol. Med. 17 (3), 768–781. Cited in: Pubmed; PMID 32944405. doi:10.20892/j.issn.2095-3941.2020.0121
Jin, W., Lei, Z., Xu, S., Fachen, Z., Yixiang, Z., Shilei, Z., et al. (2021). Genetic mutation analysis in small cell lung cancer by a novel NGS-based targeted resequencing gene panel and relation with clinical features. Biomed. Res. Int. 2021, 3609028. Epub 20210405. doi:10.1155/2021/3609028
Kim, C., and Giaccone, G. (2018). MEK inhibitors under development for treatment of non-small-cell lung cancer. Expert Opin. Investig. Drugs 27 (1), 17–30. Epub 20171213. doi:10.1080/13543784.2018.1415324
Kim, J. H., Kim, H. S., and Kim, B. J. (2017). Prognostic value of KRAS mutation in advanced non-small-cell lung cancer treated with immune checkpoint inhibitors: A meta-analysis and review. Oncotarget 8 (29), 48248–48252. in: Pubmed; PMID 28525386. doi:10.18632/oncotarget.17594
Kohli, S., Bhardwaj, A., Kumari, R., and Das, S. (2018). SIRT6 is a target of regulation by UBE3A that contributes to liver tumorigenesis in an ANXA2-dependent manner. Cancer Res. 78 (3), 645–658. Epub 20171207. doi:10.1158/0008-5472.CAN-17-1692
Lee, J., Park, C. K., Yoon, H. K., Sa, Y. J., Woo, I. S., Kim, H. R., et al. (2019). PD-L1 expression in ROS1-rearranged non-small cell lung cancer: A study using simultaneous genotypic screening of EGFR, ALK, and ROS1. Thorac. Cancer 10 (1), 103–110. Epub 20181126. doi:10.1111/1759-7714.12917
Li, W., Ye, L., Huang, Y., Zhou, F., Wu, C., Wu, F., et al. (2022). Characteristics of Notch signaling pathway and its correlation with immune microenvironment in SCLC. Lung Cancer 167, 25–33. Epub 20220329Cited in: Pubmed; PMID 35381444. doi:10.1016/j.lungcan.2022.03.019
Li, J. X., Li, R. Z., Ma, L. R., Wang, P., Xu, D. H., Huang, J., et al. (2022). Targeting mutant kirsten rat sarcoma viral oncogene homolog in non-small cell lung cancer: Current difficulties, integrative treatments and future perspectives. Front. Pharmacol. 13, 875330. Epub 20220420. doi:10.3389/fphar.2022.875330
Li, Y., Hu, L., Peng, X., Xu, H., Tang, B., and Xu, C. (2022). Resistance to immune checkpoint inhibitors in KRAS-mutant non-small cell lung cancer. Cancer Drug Resist 5 (1), 129–146. Epub 20220208. doi:10.20517/cdr.2021.102
Li, K., Xu, X., He, Y., Tian, Y., Pan, W., Xu, L., et al. (2018). P21-activated kinase 7 (PAK7) interacts with and activates Wnt/β-catenin signaling pathway in breast cancer. J. Cancer 9 (10), 1821–1835. Epub 20180422. doi:10.7150/jca.24934
Li, L., Wang, Y., Shi, W., Zhu, M., Liu, Z., Luo, N., et al. (2019). Serial ultra-deep sequencing of circulating tumor DNA reveals the clonal evolution in non-small cell lung cancer patients treated with anti-PD1 immunotherapy. Cancer Med. 8 (18), 7669–7678. Epub 20191106. doi:10.1002/cam4.2632
Li, M., Qin, X., Xue, X., Zhang, C., Yan, Z., Han, W., et al. (2010). Safety evaluation and pharmacokinetics of a novel human tumor necrosis factor-alpha exhibited a higher antitumor activity and a lower systemic toxicity. Anticancer Drugs 21 (3), 243–251. . Cited in: Pubmed; PMID 20166241. doi:10.1097/cad.0b013e328333d5ce
Li, M., Xu, T., Zhang, Z., Xue, X., Zhang, C., Qin, X., et al. (2012). Phase II multicenter, randomized, double-blind study of recombinant mutated human tumor necrosis factor-alpha in combination with chemotherapies in cancer patients. Cancer Sci. 103 (2), 288–295. Epub 20120109. doi:10.1111/j.1349-7006.2011.02153.x
Li, T., and Qiao, T. (2022). Unraveling tumor microenvironment of small-cell lung cancer: Implications for immunotherapy. Semin. Cancer Biol. 86 (2), 117–125. Epub 20220929Cited in: Pubmed; PMID 36183998. doi:10.1016/j.semcancer.2022.09.005
Li, X., Wang, Y., Li, X., Feng, G., Hu, S., and Bai, Y. (2021). The impact of NOTCH pathway alteration on tumor microenvironment and clinical survival of immune checkpoint inhibitors in NSCLC. Front. Immunol. 12, 638763. Epub 20210709. doi:10.3389/fimmu.2021.638763
Lin, A., Wei, T., Meng, H., Luo, P., and Zhang, J. (2019). Role of the dynamic tumor microenvironment in controversies regarding immune checkpoint inhibitors for the treatment of non-small cell lung cancer (NSCLC) with EGFR mutations. Mol. Cancer 18 (1), 139. Epub 20190916Cited in: Pubmed; PMID 31526368. doi:10.1186/s12943-019-1062-7
Lin, A., Zhang, H., Meng, H., Deng, Z., Gu, T., Luo, P., et al. (2021). TNF-alpha pathway alternation predicts survival of immune checkpoint inhibitors in non-small cell lung cancer. Front. Immunol. 12, 667875. Epub 20210916. doi:10.3389/fimmu.2021.667875
Lin, C., Chen, X., Li, M., Liu, J., Qi, X., Yang, W., et al. (2015). Programmed death-ligand 1 expression predicts tyrosine kinase inhibitor response and better prognosis in a cohort of patients with epidermal growth factor receptor mutation-positive lung adenocarcinoma. Clin. Lung Cancer 16 (5), e25–e35. Epub 20150219Cited in: Pubmed; PMID 25801750. doi:10.1016/j.cllc.2015.02.002
Liu, C., Zheng, S., Jin, R., Wang, X., Wang, F., Zang, R., et al. (2020). The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. 470, 95–105. Epub 20191020Cited in: Pubmed; PMID 31644929. doi:10.1016/j.canlet.2019.10.027
Liu, C., Zheng, S., Wang, Z., Wang, S., Wang, X., Yang, L., et al. (2022). KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun. (Lond) 42 (9), 828–847. Epub 20220711. doi:10.1002/cac2.12327
Liu, F., Wei, Y., Zhang, H., Jiang, J., Zhang, P., and Chu, Q. (2022). NTRK fusion in non-small cell lung cancer: Diagnosis, therapy, and TRK inhibitor resistance. Front. Oncol. 12, 864666. Epub 20220317. doi:10.3389/fonc.2022.864666
Liu, J., Chen, T., Li, S., Liu, W., Wang, P., and Shang, G. (2022). Targeting matrix metalloproteinases by E3 ubiquitin ligases as a way to regulate the tumor microenvironment for cancer therapy. Semin. Cancer Biol. 86 (2), 259–268. Epub 20220618Cited in: Pubmed; PMID 35724822. doi:10.1016/j.semcancer.2022.06.004
Liu, J., and Shang, G. (2022). The roles of noncoding RNAs in the development of osteosarcoma stem cells and potential therapeutic targets. Front. Cell Dev. Biol. 10, 773038. Epub 20220216. doi:10.3389/fcell.2022.773038
Liu, S. Y., Dong, Z. Y., Wu, S. P., Xie, Z., Yan, L. X., Li, Y. F., et al. (2018). Clinical relevance of PD-L1 expression and CD8+ T cells infiltration in patients with EGFR-mutated and ALK-rearranged lung cancer. Lung Cancer 125, 86–92. Epub 20180914Cited in: Pubmed; PMID 30429043. doi:10.1016/j.lungcan.2018.09.010
Liu, X. Y., Cui, Y. N., Li, J., Zhang, Z., and Guo, R. H. (2022). Effect of FBXW7 gene mutation on the prognosis of immunotherapy in patients with non-small cell lung cancer. Zhonghua Yi Xue Za Zhi 102 (13), 914–921. Cited in: Pubmed; PMID 35385962. doi:10.3760/cma.j.cn112137-20211021-02332
Long, J., Wang, D., Yang, X., Wang, A., Lin, Y., Zheng, M., et al. (2021). Identification of NOTCH4 mutation as a response biomarker for immune checkpoint inhibitor therapy. BMC Med. 19 (1), 154. Epub 20210721Cited in: Pubmed; PMID 34284787. doi:10.1186/s12916-021-02031-3
Luo, W., Wang, Z., Zhang, T., Yang, L., Xian, J., Li, Y., et al. (2021). Immunotherapy in non-small cell lung cancer: Rationale, recent advances and future perspectives. Precis. Clin. Med. 4 (4), 258–270. Epub 20211202. doi:10.1093/pcmedi/pbab027
Ma, X., Song, Y., Zhang, K., Shang, L., Gao, Y., Zhang, W., et al. (2015). Recombinant mutated human TNF in combination with chemotherapy for stage IIIB/IV non-small cell lung cancer: A randomized, phase III study. Sci. Rep. 4, 9918. Epub 20150421. doi:10.1038/srep09918
Majumder, S., Crabtree, J. S., Golde, T. E., Minter, L. M., Osborne, B. A., and Miele, L. (2021). Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 20 (2), 125–144. Epub 20201208Cited in: Pubmed; PMID 33293690. doi:10.1038/s41573-020-00091-3
Malhotra, J., Ryan, B., Patel, M., Chan, N., Guo, Y., Aisner, J., et al. (2022). Clinical outcomes and immune phenotypes associated with STK11 co-occurring mutations in non-small cell lung cancer. J. Thorac. Dis. 14 (6), 1772–1783. Cited in: Pubmed; PMID 35813711. doi:10.21037/jtd-21-1377
Martin, C., and Enrico, D. (2022). Current and novel therapeutic strategies for optimizing immunotherapy outcomes in advanced non-small cell lung cancer. Front. Oncol. 12, 962947. Epub 20221208. doi:10.3389/fonc.2022.962947
Maynard, A., McCoach, C. E., Rotow, J. K., Harris, L., Haderk, F., Kerr, D. L., et al. (2020). Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 182 (5), 1232–1251.e22. e22 Epub 20200820Cited in: Pubmed; PMID 32822576. doi:10.1016/j.cell.2020.07.017
Mengoli, M. C., Longo, F. R., Fraggetta, F., Cavazza, A., Dubini, A., Ali, G., et al. (2018). The 2015 world health organization classification of lung tumors: New entities since the 2004 classification. Pathologica 110 (1), 39–67. Cited in: Pubmed; PMID 30259912.
Miller, K. D., Nogueira, L., Devasia, T., Mariotto, A. B., Yabroff, K. R., Jemal, A., et al. (2022). Cancer treatment and survivorship statistics. CA Cancer J. Clin. 72 (5), 409–436. Epub 20220623. doi:10.3322/caac.21731
Minamiya, Y., Saito, H., Ito, M., Imai, K., Konno, H., Takahashi, N., et al. (2012). Suppression of Zinc Finger Homeobox 3 expression in tumor cells decreases the survival rate among non-small cell lung cancer patients. Cancer Biomark. 11 (4), 139–146. . Cited in: Pubmed; PMID 23144151. doi:10.3233/CBM-2012-00272
Mussafi, O., Mei, J., Mao, W., and Wan, Y. (2022). Immune checkpoint inhibitors for PD-1/PD-L1 axis in combination with other immunotherapies and targeted therapies for non-small cell lung cancer. Front. Oncol. 12, 948405. Epub 20220817. doi:10.3389/fonc.2022.948405
Nallasamy, P., Nimmakayala, R. K., Parte, S., Are, A. C., Batra, S. K., and Ponnusamy, M. P. (2022). Tumor microenvironment enriches the stemness features: The architectural event of therapy resistance and metastasis. Mol. Cancer 21 (1), 225. Epub 20221222Cited in: Pubmed; PMID 36550571. doi:10.1186/s12943-022-01682-x
Ni, J., and Zhang, L. (2021). Progress in treatment of non-small cell lung cancer harboring HER2 aberrations. Onco Targets Ther. 14, 4087–4098. Epub 20210706. doi:10.2147/OTT.S312820
Niu, L., Dang, C., Li, L., Guo, N., Xu, Y., Li, X., et al. (2021). Next-generation sequencing-based identification of EGFR and NOTCH2 complementary mutations in non-small cell lung cancer. Oncol. Lett. 22 (2), 594. Epub 20210607. doi:10.3892/ol.2021.12855
Olmedo, M. E., Cervera, R., Cabezon-Gutierrez, L., Lage, Y., Corral de la Fuente, E., Gomez Rueda, A., et al. (2022). New horizons for uncommon mutations in non-small cell lung cancer: BRAF, KRAS, RET, MET, NTRK, HER2. World J. Clin. Oncol. 13 (4), 276–286. in: Pubmed; PMID 35582653. doi:10.5306/wjco.v13.i4.276
Osielska, M. A., and Jagodzinski, P. P. (2018). Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomed. Pharmacother. 101, 322–333. Epub 20180322Cited in: Pubmed; PMID 29499406. doi:10.1016/j.biopha.2018.02.099
Owais, A., Mishra, R. K., and Kiyokawa, H. (2020). The HECT E3 ligase e6ap/ube3a as a therapeutic target in cancer and neurological disorders. Cancers (Basel) 12 (8), 2108. Epub 20200729. doi:10.3390/cancers12082108
Pang, Z., Chen, X., Wang, Y., Wang, Y., Yan, T., Wan, J., et al. (2021). Long non-coding RNA C5orf64 is a potential indicator for tumor microenvironment and mutation pattern remodeling in lung adenocarcinoma. Genomics 113, 291–304. Epub 20201210Cited in: Pubmed; PMID 33309768. doi:10.1016/j.ygeno.2020.12.010
Pao, W., and Girard, N. (2011). New driver mutations in non-small-cell lung cancer. Lancet Oncol. 12 (2), 175–180. Cited in: Pubmed; PMID 21277552. doi:10.1016/S1470-2045(10)70087-5
Passaro, A., Janne, P. A., Mok, T., and Peters, S. (2021). Overcoming therapy resistance in EGFR-mutant lung cancer. Nat. Cancer 2 (4), 377–391. Epub 20210415Cited in: Pubmed; PMID 35122001. doi:10.1038/s43018-021-00195-8
Peng, L., Wang, Z., Stebbing, J., and Yu, Z. (2022). Novel immunotherapeutic drugs for the treatment of lung cancer. Curr. Opin. Oncol. 34 (1), 89–94. . Cited in: Pubmed; PMID 34636350. doi:10.1097/CCO.0000000000000800
Peng, S., Wang, R., Zhang, X., Ma, Y., Zhong, L., Li, K., et al. (2019). EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol. Cancer 18 (1), 165. Epub 20191120Cited in: Pubmed; PMID 31747941. doi:10.1186/s12943-019-1073-4
Peng, Z., and Chen, Q. (2019). Research progress in the role of FBXW7 in drug resistance against non-small cell lung cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 44 (4), 444–448. Cited in: Pubmed; PMID 31113922. doi:10.11817/j.issn.1672-7347.2019.04.016
Principe, C., Dionisio de Sousa, I. J., Prazeres, H., Soares, P., and Lima, R. T. (2021). LRP1B: A giant lost in cancer translation. Pharm. (Basel) 14 (9), 836. Epub 20210824. doi:10.3390/ph14090836
Principe, D. R. (2022). Patients deriving long-term benefit from immune checkpoint inhibitors demonstrate conserved patterns of site-specific mutations. Sci. Rep. 12 (1), 11490. Epub 20220707Cited in: Pubmed; PMID 35798829. doi:10.1038/s41598-022-15714-5
Qin, H., and Patel, M. R. (2022). The challenge and opportunity of NTRK inhibitors in non-small cell lung cancer. Int. J. Mol. Sci. 23 (6), 2916. Epub 20220308. doi:10.3390/ijms23062916
Quan, L., Cheng, Z., Dai, Y., Jiao, Y., Shi, J., and Fu, L. (2020). Prognostic significance of PAK family kinases in acute myeloid leukemia. Cancer Gene Ther. 27 (1-2), 30–37. Epub 20190320Cited in: Pubmed; PMID 30890765. doi:10.1038/s41417-019-0090-1
Rangachari, D., VanderLaan, P. A., Shea, M., Le, X., Huberman, M. S., Kobayashi, S. S., et al. (2017). Correlation between classic driver oncogene mutations in EGFR, ALK, or ROS1 and 22C3-PD-L1 ≥50% expression in lung adenocarcinoma. J. Thorac. Oncol. 12 (5), 878–883. Epub 20170116. doi:10.1016/j.jtho.2016.12.026
Rashed, H. E., Abdelrahman, A. E., Abdelgawad, M., Balata, S., and Shabrawy, M. E. (2017). Prognostic significance of programmed cell death ligand 1 (PD-L1), CD8+ tumor-infiltrating lymphocytes and p53 in non-small cell lung cancer: An immunohistochemical study. Turk Patoloji Derg. 1 (1), 211–222. Cited in: Pubmed; PMID 28832075. doi:10.5146/tjpath.2017.01398
Ricciuti, B., Leonardi, G. C., Metro, G., Grignani, F., Paglialunga, L., Bellezza, G., et al. (2016). Targeting the KRAS variant for treatment of non-small cell lung cancer: Potential therapeutic applications. Expert Rev. Respir. Med. 10 (1), 53–68. Epub 20151117. doi:10.1586/17476348.2016.1115349
Riudavets, M., Cascetta, P., and Planchard, D. (2022). Targeting BRAF-mutant non-small cell lung cancer: Current status and future directions. Lung Cancer 169, 102–114. Epub 20220526Cited in: Pubmed; PMID 35696864. doi:10.1016/j.lungcan.2022.05.014
Roman, M., Baraibar, I., Lopez, I., Nadal, E., Rolfo, C., Vicent, S., et al. (2018). KRAS oncogene in non-small cell lung cancer: Clinical perspectives on the treatment of an old target. Mol. Cancer 17 (1), 33. Epub 20180219Cited in: Pubmed; PMID 29455666. doi:10.1186/s12943-018-0789-x
Sakai, H., Takeda, M., Sakai, K., Nakamura, Y., Ito, A., Hayashi, H., et al. (2019). Impact of cytotoxic chemotherapy on PD-L1 expression in patients with non-small cell lung cancer negative for EGFR mutation and ALK fusion. Lung Cancer 127, 59–65. Epub 20181123Cited in: Pubmed; PMID 30642552. doi:10.1016/j.lungcan.2018.11.025
Serra, P., Petat, A., Maury, J. M., Thivolet-Bejui, F., Chalabreysse, L., Barritault, M., et al. (2018). Programmed cell death-ligand 1 (PD-L1) expression is associated with RAS/TP53 mutations in lung adenocarcinoma. Lung Cancer 118, 62–68. Epub 20180206Cited in: Pubmed; PMID 29572005. doi:10.1016/j.lungcan.2018.02.005
Sforza, V., Palumbo, G., Cascetta, P., Carillio, G., Manzo, A., Montanino, A., et al. (2022). BRAF inhibitors in non-small cell lung cancer. Cancers (Basel) 14 (19), 4863. Epub 20221005. doi:10.3390/cancers14194863
Shin, N. K., Lee, I., Chang, S. G., and Shin, H. C. (1998). A novel tumor necrosis factor-alpha mutant with significantly enhanced cytotoxicity and receptor binding affinity. Biochem. Mol. Biol. Int. 44 (6), 1075–1082. in: Pubmed; PMID 9623760. doi:10.1080/15216549800202142
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. Epub 20220112. doi:10.3322/caac.21708
Sigafoos, A. N., Paradise, B. D., and Fernandez-Zapico, M. E. (2021). Hedgehog/GLI signaling pathway: Transduction, regulation, and implications for disease. Cancers (Basel) 13 (14), 3410. Epub 20210707. doi:10.3390/cancers13143410
Skoulidis, F., Goldberg, M. E., Greenawalt, D. M., Hellmann, M. D., Awad, M. M., Gainor, J. F., et al. (2018). STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8 (7), 822–835. Epub 20180517Cited in: Pubmed; PMID 29773717. doi:10.1158/2159-8290.CD-18-0099
Skoulidis, F., and Heymach, J. V. (2019). Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 19 (9), 495–509. Epub 20190812Cited in: Pubmed; PMID 31406302. doi:10.1038/s41568-019-0179-8
Song, Z., Yang, L., Zhou, Z., Li, P., Wang, W., Cheng, G., et al. (2021). Genomic profiles and tumor immune microenvironment of primary lung carcinoma and brain oligo-metastasis. Cell Death Dis. 12 (1), 106. Epub 20210121Cited in: Pubmed; PMID 33479213. doi:10.1038/s41419-021-03410-7
Sun, H., Liu, S. Y., Zhou, J. Y., Xu, J. T., Zhang, H. K., Yan, H. H., et al. (2020). Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine 60, 102990. Epub 20200911Cited in: Pubmed; PMID 32927274. doi:10.1016/j.ebiom.2020.102990
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. Epub 20210204. doi:10.3322/caac.21660
Tartarone, A., Lerose, R., Tartarone, M., and Aieta, M. (2022). Potential role of tumor-derived exosomes in non-small-cell lung cancer in the era of immunotherapy. Life (Basel) 12 (12), 2104. Epub 20221214. doi:10.3390/life12122104
Tiwari, A., Trivedi, R., and Lin, S. Y. (2022). Tumor microenvironment: Barrier or opportunity towards effective cancer therapy. J. Biomed. Sci. 29 (1), 83. Epub 20221017Cited in: Pubmed; PMID 36253762. doi:10.1186/s12929-022-00866-3
Tu, E., McGlinchey, K., Wang, J., Martin, P., Ching, S. L., Floc'h, N., et al. (2022). Anti-PD-L1 and anti-CD73 combination therapy promotes T cell response to EGFR-mutated NSCLC. JCI Insight 7 (3), e142843. Epub 20220208Cited in: Pubmed; PMID 35132961. doi:10.1172/jci.insight.142843
Turner, M. C., Andersen, Z. J., Baccarelli, A., Diver, W. R., Gapstur, S. M., Pope, C. A., et al. (2020). Outdoor air pollution and cancer: An overview of the current evidence and public health recommendations. CA Cancer J. Clin. 70, 460–479. Epub 20200825. doi:10.3322/caac.21632
Uras, I. Z., Moll, H. P., and Casanova, E. (2020). Targeting KRAS mutant non-small-cell lung cancer: Past, present and future. Int. J. Mol. Sci. 21 (12), 4325. Epub 20200617. doi:10.3390/ijms21124325
Vathiotis, I. A., Charpidou, A., Gavrielatou, N., and Syrigos, K. N. (2021). HER2 aberrations in non-small cell lung cancer: From pathophysiology to targeted therapy. Pharm. (Basel) 14 (12), 1300. Epub 20211214. doi:10.3390/ph14121300
Wan, L., Liu, Q., Liang, D., Guo, Y., Liu, G., Ren, J., et al. (2021). Circulating tumor cell and metabolites as novel biomarkers for early-stage lung cancer diagnosis. Front. Oncol. 11, 630672. Epub 20210531. doi:10.3389/fonc.2021.630672
Wang, L., Ma, L., Xu, F., Zhai, W., Dong, S., Yin, L., et al. (2018). Role of long non-coding RNA in drug resistance in non-small cell lung cancer. Thorac. Cancer 9 (7), 761–768. Epub 20180503. doi:10.1111/1759-7714.12652
Wang, M., Herbst, R. S., and Boshoff, C. (2021). Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 27 (8), 1345–1356. Epub 20210812Cited in: Pubmed; PMID 34385702. doi:10.1038/s41591-021-01450-2
Wang, S., Jiang, M., Yang, Z., Huang, X., and Li, N. (2022). The role of distinct co-mutation patterns with TP53 mutation in immunotherapy for NSCLC. Genes Dis. 9 (1), 245–251. Epub 20200409Cited in: Pubmed; PMID 35005121. doi:10.1016/j.gendis.2020.04.001
Wang, Z., Ge, Y., Li, H., Fei, G., Wang, S., and Wei, P. (2022). Identification and validation of a genomic mutation signature as a predictor for immunotherapy in NSCLC. Biosci. Rep. (11), 42. in: Pubmed; PMID 36305643. doi:10.1042/BSR20220892
Wang, X., Liu, S., Shao, Z., and Zhang, P. (2020). Bioinformatic analysis of the potential molecular mechanism of PAK7 expression in glioblastoma. Mol. Med. Rep. 22 (2), 1362–1372. Epub 20200603. doi:10.3892/mmr.2020.11206
Wang, Z., Liu, P., Inuzuka, H., and Wei, W. (2014). Roles of F-box proteins in cancer. Nat. Rev. Cancer 14 (4), 233–247. in: Pubmed; PMID 24658274. doi:10.1038/nrc3700
Weng, C., Wang, L., Liu, G., Guan, M., and Lu, L. (2021). Identification of a N6-methyladenosine (m6A)-Related lncRNA signature for predicting the prognosis and immune landscape of lung squamous cell carcinoma. Front. Oncol. 11, 763027. Epub 20211118. doi:10.3389/fonc.2021.763027
Wood, K., Hensing, T., Malik, R., and Salgia, R. (2016). Prognostic and predictive value in KRAS in non-small-cell lung cancer: A review. JAMA Oncol. 2 (6), 805–812. Cited in: Pubmed; PMID 27100819. doi:10.1001/jamaoncol.2016.0405
Wu, F., Fan, J., He, Y., Xiong, A., Yu, J., Li, Y., et al. (2021). Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 12 (1), 2540. Epub 20210505Cited in: Pubmed; PMID 33953163. doi:10.1038/s41467-021-22801-0
Wu, M., Huang, Q., Xie, Y., Wu, X., Ma, H., Zhang, Y., et al. (2022). Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 15 (1), 24. Epub 20220312Cited in: Pubmed; PMID 35279217. doi:10.1186/s13045-022-01242-2
Xiang, Y., Zhang, S., Fang, X., Jiang, Y., Fang, T., Liu, J., et al. (2022). Therapeutic advances of rare ALK fusions in non-small cell lung cancer. Curr. Oncol. 29 (10), 7816–7831. Epub 20221016. doi:10.3390/curroncol29100618
Xiao, Y., Yin, C., Wang, Y., Lv, H., Wang, W., Huang, Y., et al. (2018). FBXW7 deletion contributes to lung tumor development and confers resistance to gefitinib therapy. Mol. Oncol. 12 (6), 883–895. Epub 20180509. doi:10.1002/1878-0261.12200
Xie, W., Chu, M., Song, G., Zuo, Z., Han, Z., Chen, C., et al. (2022). Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin. Cancer Biol. 83, 303–318. Epub 20201115Cited in: Pubmed; PMID 33207266. doi:10.1016/j.semcancer.2020.11.004
Xu, F., Cui, W. Q., Liu, C., Feng, F., Liu, R., Zhang, J., et al. (2023). Prognostic biomarkers correlated with immune infiltration in non-small cell lung cancer. FEBS Open Bio 13 (1), 72–88. Epub 20221128. doi:10.1002/2211-5463.13501
Yan, H., and Bu, P. (2021). Non-coding RNA in cancer. Essays Biochem. 65 (4), 625–639. in: Pubmed; PMID 33860799. doi:10.1042/ebc20200032
Yan, L., Lin, M., Pan, S., Assaraf, Y. G., Wang, Z. W., and Zhu, X. (2020). Emerging roles of F-box proteins in cancer drug resistance. Drug Resist Updat 49, 100673. Epub 20191217. doi:10.1016/j.drup.2019.100673
Yan, Q., Hu, B., Chen, H., Zhu, L., Lyu, Y., Qian, D., et al. (2022). A novel algorithm for lung adenocarcinoma based on N6 methyladenosine-related immune long noncoding RNAs as a reliable biomarker for predicting survival outcomes and selecting sensitive anti-tumor therapies. J. Clin. Lab. Anal. 36 (9), e24636. Epub 20220810. doi:10.1002/jcla.24636
Yan, Z., Zhao, N., Wang, Z., Li, B., Bao, C., Shi, J., et al. (2006). A mutated human tumor necrosis factor-alpha improves the therapeutic index in vitro and in vivo. Cytotherapy 8 (4), 415–423. in: Pubmed; PMID 16923618. doi:10.1080/14653240600845278
Yang, T., Xiong, Y., Zeng, Y., Wang, Y., Zeng, J., Liu, J., et al. (2022). Current status of immunotherapy for non-small cell lung cancer. Front. Pharmacol. 13, 989461. Epub 20221013. doi:10.3389/fphar.2022.989461
Yang, Z., Gao, A., Shi, W., Wang, J., Zhang, X., Xu, Z., et al. (2021). ILT4 in colorectal cancer cells induces suppressive T cell contexture and disease progression. Onco Targets Ther. 14, 4239–4254. Epub 20210720. doi:10.2147/OTT.S290348
Yu, X., Ji, X., and Su, C. (2022). HER2-Altered non-small cell lung cancer: Biology, clinicopathologic features, and emerging therapies. Front. Oncol. 12, 860313. Epub 20220329. doi:10.3389/fonc.2022.860313
Yu, Z. Q., Wang, M., Zhou, W., Mao, M. X., Chen, Y. Y., Li, N., et al. (2022). ROS1-positive non-small cell lung cancer (NSCLC): Biology, diagnostics, therapeutics and resistance. J. Drug Target 30 (8), 845–857. Epub 20220614. doi:10.1080/1061186X.2022.2085730
Yue, D., Qian, J., Chen, Z., Zhang, B., Chen, P., Zhang, L., et al. (2021). Short-term response to immune-chemotherapy and immune features of a ceritinib-resistant patient with ROS1-rearranged lung adenocarcinoma. J. Immunother. Cancer 9 (2), e001967. Cited in: Pubmed; PMID 33558279. doi:10.1136/jitc-2020-001967
Zeng, H., Tong, F., Bin, Y., Peng, L., Gao, X., Xia, X., et al. (2022). The predictive value of PAK7 mutation for immune checkpoint inhibitors therapy in non-small cell cancer. Front. Immunol. 13, 834142. Epub 20220203. doi:10.3389/fimmu.2022.834142
Zhang, H. H., Zhang, Z. Y., Che, C. L., Mei, Y. F., and Shi, Y. Z. (2013). Array analysis for potential biomarker of gemcitabine identification in non-small cell lung cancer cell lines. Int. J. Clin. Exp. Pathol. 6 (9), 1734–1746. Epub 20130815. Cited in: Pubmed; PMID 24040438.
Zhang, J., Zhou, N., Lin, A., Luo, P., Chen, X., Deng, H., et al. (2021). ZFHX3 mutation as a protective biomarker for immune checkpoint blockade in non-small cell lung cancer. Cancer Immunol. Immunother. 70 (1), 137–151. Epub 20200711Cited in: Pubmed; PMID 32653938. doi:10.1007/s00262-020-02668-8
Zhang, L., Zhang, T., Shang, B., Li, Y., Cao, Z., and Wang, H. (2021). Prognostic effect of coexisting TP53 and ZFHX3 mutations in non-small cell lung cancer patients treated with immune checkpoint inhibitors. Scand. J. Immunol. 94 (3), e13087. Epub 20210613. doi:10.1111/sji.13087
Zhang, K., Hong, X., Song, Z., Xu, Y., Li, C., Wang, G., et al. (2020). Identification of deleterious NOTCH mutation as novel predictor to efficacious immunotherapy in NSCLC. Clin. Cancer Res. 26 (14), 3649–3661. Epub 20200402Cited in: Pubmed; PMID 32241817. doi:10.1158/1078-0432.CCR-19-3976
Zhang, L. X., Gao, J., Long, X., Zhang, P. F., Yang, X., Zhu, S. Q., et al. (2022). The circular RNA circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas via the miR-181a-5p/CARM1 axis. Mol. Cancer 21 (1), 110. Epub 20220507Cited in: Pubmed; PMID 35525959. doi:10.1186/s12943-022-01586-w
Zhang, N., Shen, J., Gou, L., Cao, M., Ding, W., Luo, P., et al. (2022). UBE3A deletion enhances the efficiency of immunotherapy in non-small-cell lung cancer. Bioengineered 13 (5), 11577–11592. in: Pubmed; PMID 35531878. doi:10.1080/21655979.2022.2069328
Zhang, P., Li, H., Cheng, X., and Wang, W. (2022). Comprehensive analysis of immune cell infiltration of m6a-related lncRNA in lung squamous cell carcinoma and construction of relevant prognostic models. Biomed. Res. Int. 2022, 9139823. Epub 20220714. doi:10.1155/2022/9139823
Zhang, W., Zhang, Q., Xie, Z., Che, L., Xia, T., Cai, X., et al. (2022). N6 -Methyladenosine-Related long non-coding RNAs are identified as a potential prognostic biomarker for lung squamous cell carcinoma and validated by real-time PCR. Front. Genet. 13, 839957. Epub 20220603. doi:10.3389/fgene.2022.839957
Zhang, Y., Zeng, Y., Liu, T., Du, W., Zhu, J., Liu, Z., et al. (2019). The canonical TGF-β/Smad signalling pathway is involved in PD-L1-induced primary resistance to EGFR-TKIs in EGFR-mutant non-small-cell lung cancer. Respir. Res. 20 (1), 164. Epub 20190722Cited in: Pubmed; PMID 31331328. doi:10.1186/s12931-019-1137-4
Zhao, D., Liu, X., Shan, Y., Li, J., Cui, W., Wang, J., et al. (2022). Recognition of immune-related tumor antigens and immune subtypes for mRNA vaccine development in lung adenocarcinoma. Comput. Struct. Biotechnol. J. 20, 5001–5013. Epub 20220905Cited in: Pubmed; PMID 36187916. doi:10.1016/j.csbj.2022.08.066
Zhao, W., Zhou, W., Rong, L., Sun, M., Lin, X., Wang, L., et al. (2022). Epidermal growth factor receptor mutations and brain metastases in non-small cell lung cancer. Front. Oncol. 12, 912505. Epub 20221115. doi:10.3389/fonc.2022.912505
Zhao, J., Lin, X., Zhuang, J., and He, F. (2021). Relationships of N6-methyladenosine-related long non-coding RNAs with tumor immune microenvironment and clinical prognosis in lung adenocarcinoma. Front. Genet. 12, 714697. Epub 20211020. doi:10.3389/fgene.2021.714697
Zheng, Z., Zhang, B., Yu, H., Li, S., Song, N., Jin, X., et al. (2021). UBE3A activates the NOTCH pathway and promotes esophageal cancer progression by degradation of ZNF185. Int. J. Biol. Sci. 17 (12), 3024–3035. Epub 20210713. doi:10.7150/ijbs.61117
Zhong, H., Liu, S., Cao, F., Zhao, Y., Zhou, J., Tang, F., et al. (2021). Dissecting tumor antigens and immune subtypes of glioma to develop mRNA vaccine. Front. Immunol. 12, 709986. Epub 20210827. doi:10.3389/fimmu.2021.709986
Zhou, J., Bao, M., Gao, G., Cai, Y., Wu, L., Lei, L., et al. (2022). Increased blood-based intratumor heterogeneity (bITH) is associated with unfavorable outcomes of immune checkpoint inhibitors plus chemotherapy in non-small cell lung cancer. BMC Med. 20 (1), 256. Epub 20220729Cited in: Pubmed; PMID 35902848. doi:10.1186/s12916-022-02444-8
Zhou, X., Ao, X., Jia, Z., Li, Y., Kuang, S., Du, C., et al. (2022). Non-coding RNA in cancer drug resistance: Underlying mechanisms and clinical applications. Front. Oncol. 12, 951864. Epub 20220817. doi:10.3389/fonc.2022.951864
Zhou, J. G., Zhong, H., Zhang, J., Jin, S. H., Roudi, R., and Ma, H. (2019). Development and validation of a prognostic signature for malignant pleural mesothelioma. Front. Oncol. 9, 78. Epub 20190215. doi:10.3389/fonc.2019.00078
Keywords: TME, PD-L1, PD-1, time, immunotherapy, resistance
Citation: Wang M, Zhu L, Yang X, Li J, Liu Y and Tang Y (2023) Targeting immune cell types of tumor microenvironment to overcome resistance to PD-1/PD-L1 blockade in lung cancer. Front. Pharmacol. 14:1132158. doi: 10.3389/fphar.2023.1132158
Received: 26 December 2022; Accepted: 06 February 2023;
Published: 15 February 2023.
Edited by:
Lin Qi, Second Xiangya Hospital, Central South University, ChinaReviewed by:
Xin Cheng, Dana–Farber Cancer Institute, United StatesChangli Wang, Tianjin Medical University Cancer Institute and Hospital, China
Copyright © 2023 Wang, Zhu, Yang, Li, Liu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Tang, dGFuZ195aW5nQGpsdS5lZHUuY24=
 Man Wang1
Man Wang1 Yu’e Liu
Yu’e Liu Ying Tang
Ying Tang