- 1Department of Pharmacology and Therapeutics, Faculty of Basic Clinical Sciences, College of Health Sciences, Usmanu Danfodiyo University, Sokoto, Nigeria
- 2Nigerian COVID-19 Research Coalition, Nigerian Institute of Medical Research Institute, Abuja, Nigeria
- 3Centre for Advanced Medical Research and Training, Usmanu Danfodiyo University, Sokoto, Nigeria
- 4Department of Medical Biochemistry, Usmanu Danfodiyo University, Sokoto, Nigeria
- 5Department of veterinary Microbiology, Usmanu Danfodiyo University, Sokoto, Nigeria
- 6School of Public Health, University of the Western Cape, Cape Town, South Africa
- 7Department of Medicine, University of Abuja Teaching Hospital, Gwagwalada, Abuja, Nigeria
- 8Nigerian Centre for Disease Control and Prevention, Abuja, Nigeria
- 9Department of Clinical pharmacy and Pharmacy Practice, Faculty of Pharmaceutical sciences, Usmanu Danfodiyo University, Sokoto, Nigeria
- 10Department of Microbiology and Biotechnology, Federal University of Dutse, Dutse, Nigeria
- 11Centre for Environmental and Public Health Research and Development, Kano, Nigeria
- 12University of Nigeria Centre for Clinical Trials, University of Nigeria Teaching Hospital, Enugu, Ituku Ozalla, Nigeria
Several efforts to repurpose drugs for COVID-19 treatment have largely either failed to identify a suitable agent or agents identified did not translate to clinical use. Reasons that have been suggested to explain the failures include use of inappropriate doses, that are not clinically achievable, in the screening experiments, and the use of inappropriate pre-clinical laboratory surrogates to predict efficacy. In this study, we used an innovative algorithm, that incorporates dissemination and implementation considerations, to identify potential drugs for COVID-19 using iterative computational and wet laboratory methods. The drugs were screened at doses that are known to be achievable in humans. Furthermore, inhibition of viral induced cytopathic effect (CPE) was used as the laboratory surrogate to predict efficacy. Erythromycin, pyridoxine, folic acid and retapamulin were found to inhibit SARS-CoV-2 induced CPE in Vero cells at concentrations that are clinically achievable. Additional studies may be required to further characterize the inhibitions of CPE and the possible mechanisms.
1 Introduction
Coronavirus disease 2019 (COVID-19) pandemic was not a rude surprise because impending pandemics had long been predicted based on scientific data, and preparations existed for early detection and response (Platto et al., 2021). When SARS-CoV-2 finally crossed the species barrier and initiated a rapidly evolving worldwide morbidity and mortality events, even the most advanced health systems in terms of resources and scalability were easily overwhelmed (Shivalkar et al., 2021). Nonetheless, the rapid discovery, dissemination and implementation of effective vaccine is estimated to have saved millions of lives (Uttarilli et al., 2021) and can largely be traced to the availability of repurposed mRNA technology (Szabó et al., 2022) initially developed with therapeutics in view, though other vaccine platforms were also successful (Fathizadeh et al., 2021). A major drawback was the lack of equity in vaccine distribution with developed countries acquiring vaccine doses in multiples of their population needs while less developed economies that had neither the resources for competitive purchases nor capacity for unassisted vaccination campaigns were late in vaccine deployment (Evans et al., 2021; The Lancet Infectious Diseases, 2021). Efforts at identifying therapeutics for people who do develop COVID-19 have had only limited success with most early enthusiasm at discovery of effective repurposed drugs rapidly and consistently failing randomized clinical trials (Parvathaneni and Gupta, 2020). With the exception of Dexamethasone and related drugs, the few effective COVID-19 therapeutics like Nirmatrelvir/ritonavir (Paxlovid) and even less effective Remdesivir (Veklury) were priced out of the reach of most countries, especially developing economies (MacKenna et al., 2021). It is informative that both Nirmatrelvir and Remdesivir are essentially repurposed (Assmus et al., 2022).
Repurposing drugs appears to be an effective approach to rapidly discover therapeutics for emerging infectious diseases. What seems to be faulty is the lack of factoring dissemination and implementation into the initial repurposing inquiry. Such inclusion would involve reflection on potential hindrances to rapid worldwide use of the drug such as cost, inherent toxicity (Ngan et al., 2022), worldwide accessibility, ease of scalability and other factors that may together be described as off-label use likelihood (e.g., familiarity of clinicians with the drugs, background acceptability by the population, among others). Also, most of the effort at repurposing drugs have been conservative on molecular targets, by frequently restricting screening to inhibition of one target in the virus and/or disease evolution pathway (Hussien and Abdelaziz, 2020; Baby et al., 2021). However, lessons from HIV suggests that therapeutics with multiple targets may have higher effectiveness and tremendously lower the probability of resistance by the virus (Atta et al., 2019). It has been suggested that a new approach to drug repurposing is needed (Assmus et al., 2022) and that for emerging viral diseases, combination therapy or targets should be prioritized (Shyr et al., 2021). This so called one-drug-multiple-target approach is attractive and has been used successfully for non-viral diseases (Zhang, 2005). Fortunately, current in silico technology and supporting ecosystem (databases) enable highly cost-effective screening of multiple small molecules for potential activity at multiple targets and greatly reduces wet laboratory transition time and cost (Liang et al., 2021).
Methods of wet laboratory validation frequently used is inhibition of viral multiplication as measured by reduced viral number on quantitative PCR (qPCR) (Caly et al., 2020). However, qPCR is not optimized for rapid high throughput screening of large number of compounds. Furthermore, reduction in viral load may not be predictive of clinical efficacy, especially where the mechanism of viral disease involves cellular cytopathy and a low viral threshold for cytopathy exists (Doms et al., 2016). SARS-CoV-2 induces cellular cytopathy in its target cells and inhibition of cytopathic effect has been employed as a high throughput screening method for drug repurposing against COVID-19 (Severson et al., 2007).
This study was therefore designed as a cost-effective, easy to conduct, high throughput procedure to repurpose drugs using in silico screening against multiple SARS-COV-2 and COVID-19 targets, then select from the hits using criteria that will enhance dissemination and implantation, then conduct wet laboratory validation of the selected hits using inhibition of cytopathic effect (CPE) as a measure of efficacy.
2 Materials and methods
2.1 Theoretical framework
Although orthodox medicines are developed for narrow indications, by their nature, small molecules (drugs) have the capacity for activity at various sites (Kim et al., 2016). This phenomena is usually described as target and off-target effects, some of which may have clinical benefits (Kim et al., 2016; Shoshan and Linder, 2008; Rawls, Dougherty, Papin). Drugs with multiple sites of activity are essentially ‘combination therapies’ and therefore may be expected to show a high threshold for viruses to develop resistance. For example, although the rate of resistant mutation is unknown with SARS-CoV-2, it is estimated as 1 in 106 for HIV-1 virus (Riemenschneider and Heider, 2016; Haankuku and Njuho, 2021; Samizi et al., 2021). This suggests that a three-drug combination with different sites of action will acquire 1 in 1018 resistant viruses: a number much higher than the possible viral loads in the body. When repurposing drugs for viral infections, it is therefore prudent to screen drugs for potential activity at all known targets (confirmed and putative) in the viral cycle and select those drugs with the highest overall score (negative binding score) across the targets. It is also prudent that the minimum potential sites of activity of a candidate drug for selection will thus be three sites.
Although different virus families interact with their host cells in very different ways, they have historically been categorized as either generating abortive, persistent or cytolytic infections (Heaton, 2017). SARS-CoV-2 is known to cause CPE in infected cells and SARS-CoV-2 Spike variant has been shown to have enhanced cytopathic and fusogenic effects in infected cells (Rocheleau et al., 2021). We, therefore, propose that inhibition of virus induced CPE by drugs would be more rapid and predictive of clinical efficacy than inhibition of viral replication, especially where virus induced cellular death is a major pathway of disease, as with COVID-19. This is because viral load suppression alone may not go below the viral threshold for CPE though the suppression may be of multiple folds compared to control. Lack of inhibition of CPE may be one of the reasons many repurposed drugs fail. In addition, inhibition of CPE could be more cost-effective to rapidly screen multiple batches of candidate drugs than qPCR.
Also, it has been shown that only about 12.5% of positive research findings are ever disseminated and implemented (DI) and that even these (DI) take over 10 years (Wilson et al., 2010). To improve this status, a theory-informed approach built into research design has been suggested (Wilson et al., 2010; McCormack et al., 2013; Morrato et al., 2013; McDavitt et al., 2016). We propose that DI of study results are more successful if designed into the research a priori by involving stakeholders like infectious disease clinicians, policymakers and clinical protocol developers in a needs assessment and other phases of the study: thus, making them, by their participation, aware of the rationale and thoroughness of the primary data generation and its analysis.
2.2 Repurposing algorithm
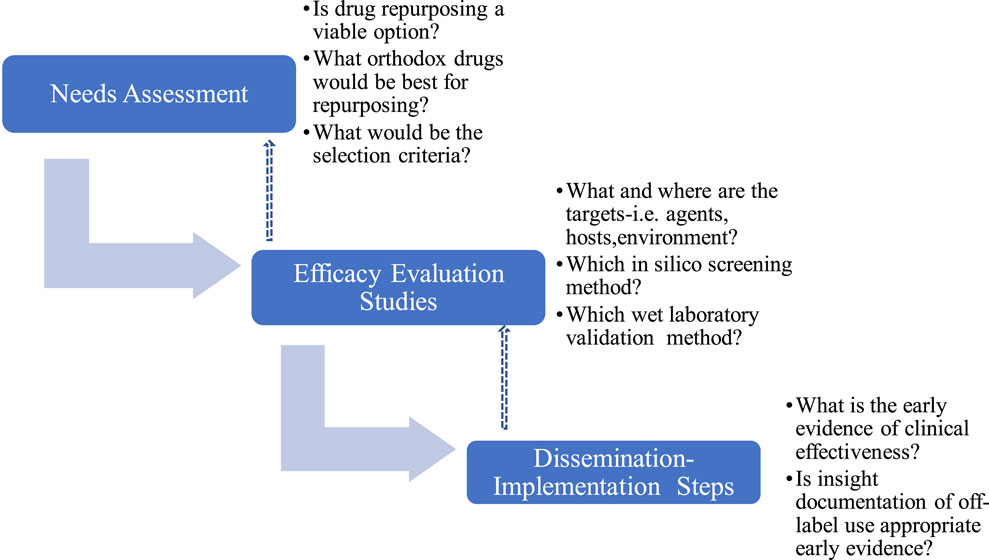
The repurposing algorithm used in this study includes i) Needs assessment, NA ii) Evaluation and Efficacy Studies, EES and iii) Dissemination and Implementation Steps (DIS), which were iterative with continuous review and enrichment as knowledge about SARS-CoV-2 and COVID-19 accumulated (Figure 1). Where required, consensus was reached by modified Delphi protocol (Yarrington et al., 2021) involving subject specialists and measurements were made using 4-points Likert’s scale to evoke forced choices and avoid neutrality.
2.2.1 Needs assessment
This involved developing a drug repurposing group that included those who will be directly involved in early phase laboratory studies (SOB, AY, AAA), later phase wet laboratory validation studies (SOB, AY, AAA, MUI, MBB, AS), clinical pharmacologist (SOB), policymakers(MAP,CLO), public health experts (EI,CLO, KE) and infectious disease clinicians (ZH) some of whom did not start the study but iteratively became involved as COVID-19 scenario evolved and needs became recognized. Consensus were reached using modified Delphi protocols involving relevant experts at each stage, with multiple iterations. The process involved questions and brainstorming on what drugs to select for repurposing efforts that will translate to feasible clinical implementation. In this regard, the opinions of infectious disease clinicians and clinical pharmacologists were considered particularly important because they may be the earliest advocates and champions of off-label use. For example, irrespective of potential for efficacy it will be inconceivable and a herculean task to attempt to repurpose digoxin for a viral infection because clinicians are unlikely to offer digoxin off-label irrespective of potential benefits (Holmes and Chen, 2012; Marino et al., 2017). The expected outcome of the needs assessment is to itemize a set of criteria for selecting drug candidates for repurposing.
2.2.2 Efficacy evaluation studies
2.2.2.1 Selection of drugs (ligands), SAR-CoV-2 protein (targets), and development of local database
The criteria for selecting drug candidates, previously developed, were applied to drugs with FDA-registration and whose structures was available on Drugbank (www.drugbank.com). One thousand four hundred and ninety drugs that met all the criteria were, therefore, downloaded.
Iterative scoping literature review was then conducted to understand the emerging knowledge of life cycle of SARS-CoV-2 virus as well as the host pathways involved in COVID-19. The review was conducted by a subgroup (SOB, AY, AAA) of the repurposing group and was continuously updated. The results were updated to all current members of the group in an iterative fashion.
The crystal structures of SARS-CoV-2 proteins critical in the life cycle of the virus, as well as their critical role in the pathogenesis of COVID-19, were identified based on a literature search and downloaded from the Protein Data Bank(www.rcsb.org) provided they also met the criteria as highlighted by Warren et al. (2012) for selection of crystal structure for molecular docking, namely, availability of experimental data for the protein, an R-free value of less than 0.45, the difference between R and R-free values should be equal to or less than 0.05, a density precision index of < 0.5, and structure with a higher resolution of less than 3.5 Å. A local database of both the ligands and targets was then created on a desktop computer.
2.2.2.2 Molecular docking using schrodinger maestro
The computer simulations were run on a 27 inches iMac desktop workstation using the Maestro graphical user interface of industry standard Schrodinger software (www.schrodinger.com). Hardware specifications include Graphics- AMD Radeon Pro 5700 8 GB, Memory- 24 GB 2133 MHz DDR4, Processor- 3.6 GHz 10-Core Intel Core i9 and 5K retina display all of which were optimized for structural visualization. All the molecular dockings were performed using Maestro Schrodinger version 9.2 on default settings and using protocols previously established in several studies (Gill et al., 2018; Leslie et al., 2021; Rahman et al., 2021; Nagasubramanian et al., 2022).
2.2.2.2.1 Protein preparation, SiteMap, and grid box generation
SARS-CoV-2 proteins (targets) stored in the local database were uploaded onto Maestro software and prepared using the Protein Preparation Wizard, keeping the settings at default. ‘Full’ protein preparation was done, which includes both H-bond network optimization and geometry minimization, as these have been shown to give significantly better results than the minimal preparation (Madhavi Sastry et al., 2013; Kontoyianni, 2017). Briefly, the structures were loaded into the Maestro workspace by clicking the Import and Process tab, of the wizard. Thereafter, unwanted chains and waters beyond 5 Å away from the binding pocket and Het groups were deleted by clicking the Review and Modify tab. Finally, the orientations of hydrogen-bonded groups and energy of the structures were optimized and minimised using optimized potentials for liquid simulations 2005 (OPLS-2005) force field by clicking the Refine tab. To check for errors due to automation, the prepared structures were reviewed manually for residues that have missing atoms, overlapping atoms, and atoms that are incorrectly typed. Also, a Ramachandran Plot was developed and inspected for stearic clashes.
The location of the primary binding site on the prepared proteins was predicted using the SiteMap module of the Schrödinger software suite. SiteMap uses the interaction energies to find the most energetic favourable areas. First, it traces the sites that include multiple site points on a grid. The number of site points for a site was set to 15 and the number of sites to be found was set to 5 and a more restrictive definition of hydrophobicity and OPLS force field were used for all the calculations (Pink et al., 1988). Binding pockets site-mapping is a robust method of identifying more potential binding sites than limiting to the binding pocket of the co-crystallized ligand (Madhavi Sastry et al., 2013). More so, in some cases, the location of a binding site for protein-ligand or protein-protein interactions is not resolved even though the protein structures are available (Kontoyianni, 2017).
Furthermore, receptor grid boxes were generated using Glide’s Receptor Grid Generation module. A binding site residue identified by the SiteMap tool was selected and served as the grid-defining area for all the respective targets. Default Van der Waals radius scaling parameters were used (scaling factor of 1, partial charge cut-off of 0.25) at the binding pocket with the radius of 20 Å around the sitemap, setting the dimension of the grid box at 10 Ǻ × 10 Ǻ × 10 Ǻ.
2.2.2.2.2 Ligand preparation
LigPrep wizard from the Maestro builder panel was used to prepare ligands and generate a 3D structure of the ligands in the ionization/tautomeric states using Epik. Hydrogen atoms were added while salts were removed at ionizing pH (7 ± 2). Energy minimization was performed using OPLS-2005 force field by using the standard energy function of molecular mechanics and RMSD cut-off 0.01 Ǻ to generate the low-energy ligand isomer. The top-ranking ligand conformation was selected for the subsequent molecular docking.
2.2.2.2.3 In-silico ligand to target interaction studies(docking)
Once the protein and ligand were prepared and receptor grid-generated, ligands were then docked to the proteins using Grid-based Ligand Docking with Energetics (GLIDE) docking protocol. The ligands were docked using Extra Precision (XP) mode. The docked conformers were evaluated using the Glide (G) Score. The G Score was calculated as follows (James and Ramanathan, 2018; Kurkinen et al., 2019):
Where, vdW means Van der Waals energy, Coul means Coulomb energy, Lipo means lipophilic contact, HBond designates hydrogen-bonding, Metal indicates metal-binding, BuryP means penalty for buried polar groups, RotB indicates a penalty for freezing rotatable bonds, Site denotes polar interactions in the active site and the a = 0.065 and b = 0.130 are coefficients of vdW and Coul. The types of ligands-protein interactions were first evaluated and provided they form hydrophilic, hydrophobic, and electrostatic interactions, the ligands were then ranked from the arithmetic sum of the docking scores in the Standard Precision method (SP-docking).
2.2.2.3 Wet laboratory validation of in silico studies by inhibition of SARS-CoV-2 induced CPE
Wet laboratory evaluation of drug’s capacity to inhibit SARS-CoV-2 CPE was performed using the methods described and validated by Severson et al. (2007).
Analytic standards of drugs that met the criteria for selection after the in silico screening were purchased from MedChem Express (United States); all were purchased already dissolved in DSMO.
Other materials used in this study include SARS-CoV-2 virus (P3), Glacial acetic acid, ethanol 96% 0.4% (wt./vol), Trypan blue in 0.9% NaCl solution, Vero cells, SARS-CoV-2 clinical sample (ID 1239), Minimal Essential Medium/Earls Balance Salts (MEM/EBSS) (HyClone laboratories, Utah, United States), Trypsin, Tissue cultureware, fetal bovine serum (FBS) (Gibco, Life Technologies Ltd., Paisley, United Kingdom), and Penicillin/streptomycin (PenStrep).
2.2.2.3.1 Culturing SARS-CoV-2 in Vero-6 cells
Vero cells were thawed and transferred into a 15-mL conical tube containing 10 mL of MEM/EBSS supplemented with 10% FBS and 1% Penstrep (this step was performed to remove the cryo-preservative-dimethyl sulfoxide (DMSO)). The 15 mL tube was then centrifuged at room temperature for 5 min at 200 g. The supernatant was discarded, and the cells were resuspended in 5 mL of MEM/EBSS containing 10% FBS and 1% Penstrep. The Vero cell suspension was transferred to a 25 cm2 (T25) tissue culture flask with a vented cap and incubated at 37°C in 5% CO2. The cells were monitored daily, and the media was changed every 3 days. At about 90% confluent monolayer, the cells were subcultured into a new tissue culture flask.
Confluent monolayer of Vero cells was detached using 0.25% trypsin-EDTA (Gibco) and resuspended in MEM/EBSS containing 10% FBS and with the use of hemocytometer and trypan blue exclusion method; about 2.1 million Vero cells were counted and seeded in a T-75 flask containing MEM/EBSS plus 1% FBS without Penstrep and incubated for about 24 h at 37°C in 5% CO2. Thereafter the flask was viewed under an inverted microscope for confluence. A confluence of 70%–80% was desirable. Meanwhile, a positive SARS-COV-2 sample (sample ID 1239) confirmed by PCR, with a Circle threshold (CT) value of 12, obtained from CAMRET Sokoto, was filtered with a 0.22 µm Membrane filter. Once the 70%–80% confluence monolayer was achieved, the medium was removed leaving about 2.5 mL. Then, 250 µL of the filtered virus sample was added to the flask and incubated for 1 h at 37°C in 5% CO2. Thereafter, the media was replenished up to a total volume of 10 mL with MEM/EBSS having 1% FBS without Penstrep. The flask was incubated for 96 h at 37°C in 5% CO2. CPE was checked daily with an inverted microscope until significant CPE was seen. The supernatant was then collected from the infected flask and centrifuged for 5 min at 500 × g, room temperature, to remove any cellular debris. The clarified supernatant was then stored in 1.5-mL screw-cap tubes at −80°C in aliquots of 1 mL until use.
2.2.2.3.2 Inhibition of SARS-CoV-2 induced CPE by selected drugs-rapid qualitative assay
This study was designed as a high-throughput qualitative assessment of CPE inhibition by candidate drugs. Drugs that show positive CPE inhibition will then be tested in dose ranges in a separate study; thus, reducing the cost and time of this initial screening. All candidate drugs were screened at the maximum achievable plasma concentration (Cmax) at the recommended dose, as identified in literature. This concentration will also be the maximum tested in subsequent experiments to avoid spurious efficacy at concentrations (in vitro) not achievable in vivo. Such non-physiological result was a major drawback of several Ivermectin repurposing studies. The controls were three, as follows: 1) vehicle control groups (DMSO), 2) a negative control (Vero cells infected with SARS-CoV-2 virus without any treatment) and 3) a Vero cells growth control. All experiments were run in triplicates.
Briefly, we seeded 96-well plates with 6 × 10 (Szabó et al., 2022) cells/mL of Vero E6 (200 μL per well), using MEM with 10% of fetal bovine serum (FBS) without antibiotics. Plates were incubated overnight at 37°C in a 5% CO2 atmosphere. The following day, the 96 wells plates were viewed under an inverted microscope for confluence of about 50%.
Before drug treatment, cell culture supernatant was removed from each well and the wells were washed with 150 μL phosphate buffer solution (PBS). Except for the negative and cell growth control wells where only PBS (50 μL) was added, each well was infected with 50 μL SARS-CoV-2 diluted in PBS at a multiplicity of infection (MOI) of 0.1. All were then incubated for 1 h at 37°C in 5% CO2 with intermittent shaking of the plates at 15 min interval to allow for viral adsorption. Thereafter, the supernatant was removed and 200 μL of the respective drugs diluted in MEM having 1% FBS without antibiotics were added to the different treatment groups and incubated at 37°C in 5% CO2. The cells were viewed using an inverted microscope at 48 h to check for CPE. Two investigators (MBB and AAA) jointly visually assessed the slides and agreed on the percentage inhibition of CPE demonstrated by the test drugs.
3 Results
3.1 Outcome of needs assessment
3.1.1 Selection criteria and rationale -drug targets
Thirteen drug targets selected after several iterations of modified Delphi protocol, extending to the end of wet laboratory studies, are shown in Table 1. Major targets are those assessed as critical for SARS-CoV-2 replication and are non-redundant because there are no alternative pathways to replication that exclude these enzymes. All other targets were classified as minor for the contrary reasons. Targets are considered validated if at least one publication exists that demonstrated that inhibition of the target resulted in reduction of viral replication.
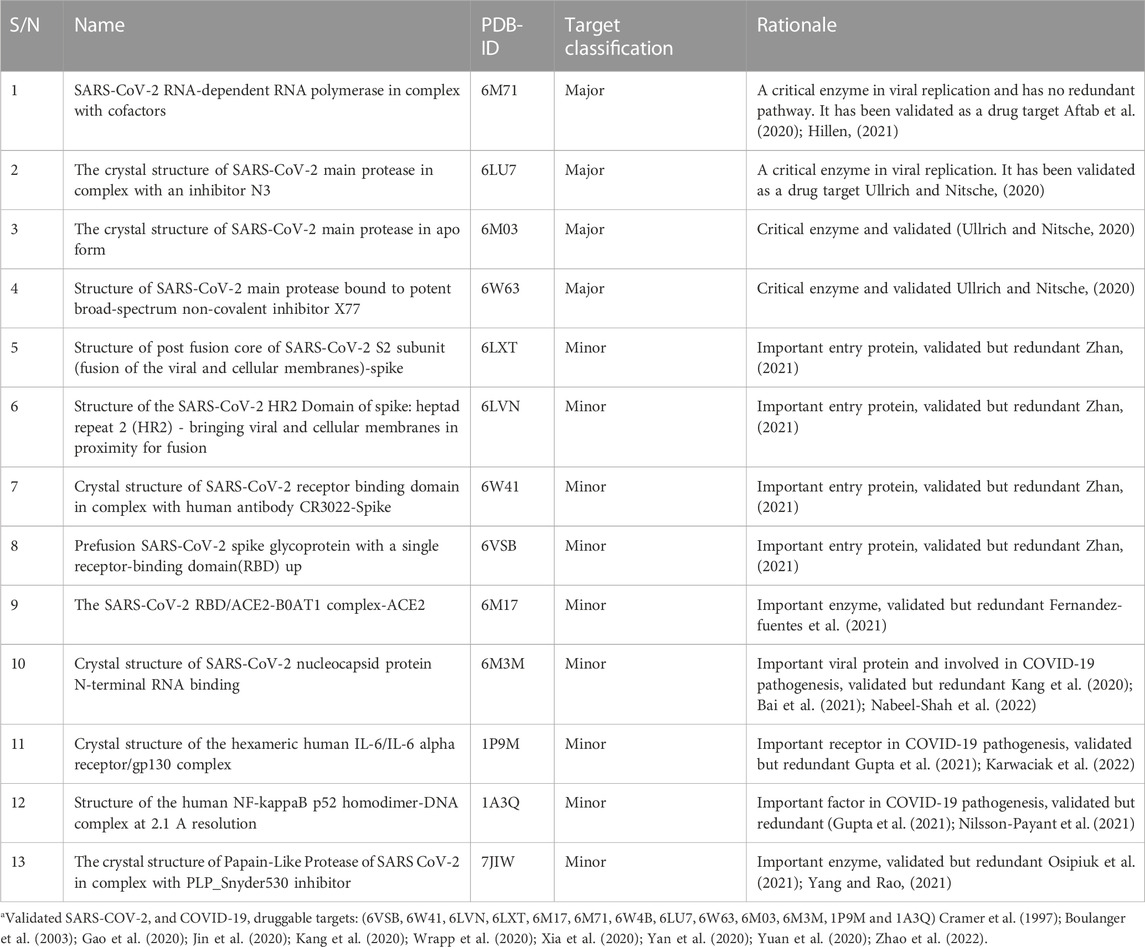
TABLE 1. Drug targets and rationalea
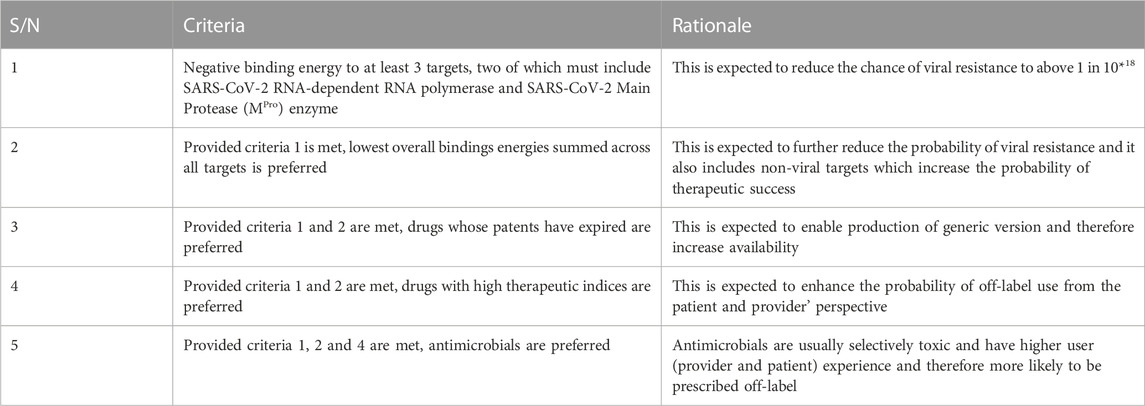
3.1.2 Selection criteria and rationale -drugs for repurposing for COVID-19
Five criteria developed for selecting drugs for repurposing after modified Delphi protocol iterations extending to the end of wet laboratory studies are shown in Table 2. All criteria must be met for a drug to be selected. The group of experts expect that drugs that meet these criteria should be rapidly available in all parts of the world if the drugs are found to be effective and will have a high threshold before resistance would emerge.
The rationales are expected outcomes based on consensus between subject experts in a modified Delphi process.
3.2 Outcome of molecular docking and final selection for wet laboratory screening
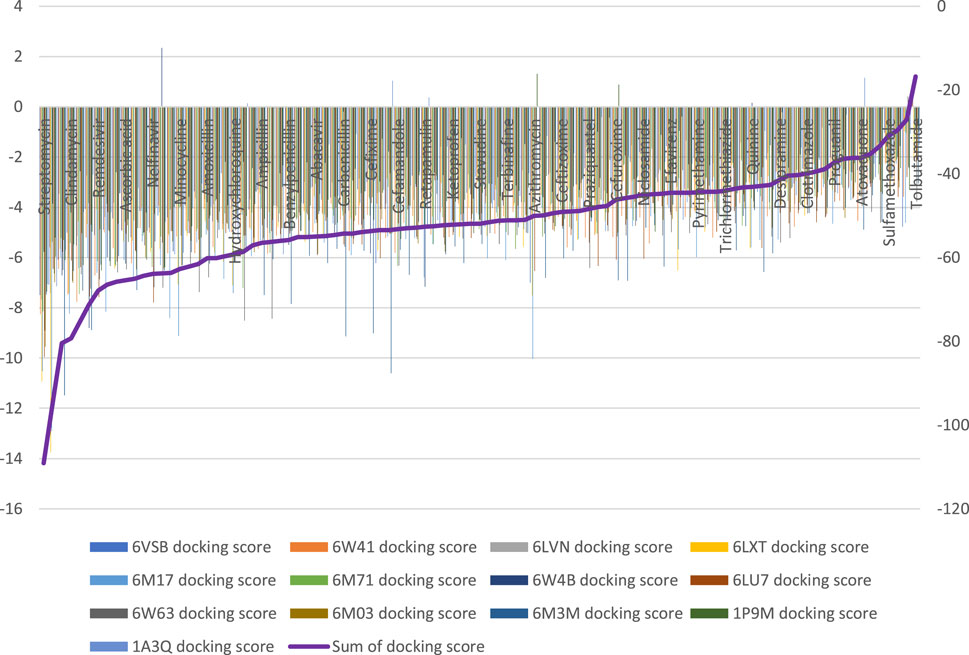
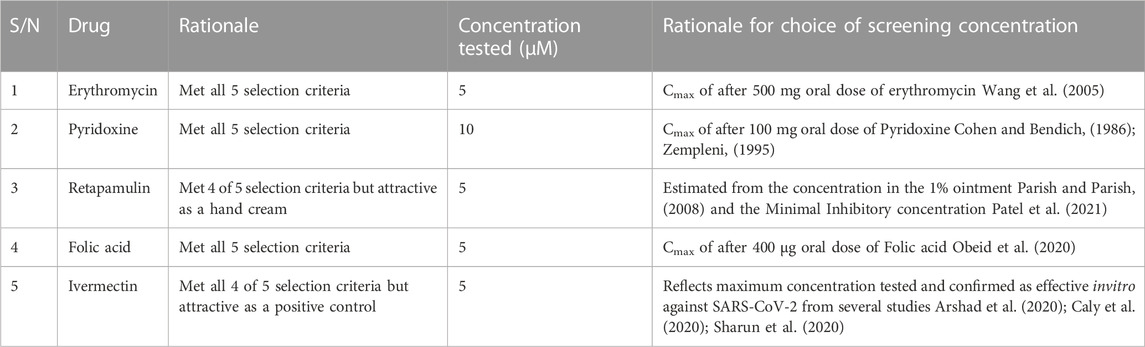
Ninety-four drugs met the criteria for selection based on the docking result, Figure 2; Supplementary Table S1. Of these ninety-four, Streptomycin had the most favorable sum of scores across the targets (−109.1) while Tolbutamide had the least favorable docking across targets (−16.8). Nonetheless, after applying other criteria, Table 3 were the drugs selected for wet laboratory validation based on ability to inhibit SARS-CoV-2 induced CPE in Vero cells.
3.3 Inhibition of SARS-CoV-2 induced CPE in vero cells
In this rapid qualitative assay, at the concentrations tested (Cmax of the drugs), all the drugs inhibited SARS-CoV-2 induced CPE in Vero cells (Figure 3) with percentage inhibition ranging from 60% to 80%, when compared to SARS-CoV-2 infected controls. Erythromycin caused the highest inhibition of CPE compared to other drugs tested, all at their Cmax routine dose schedules.
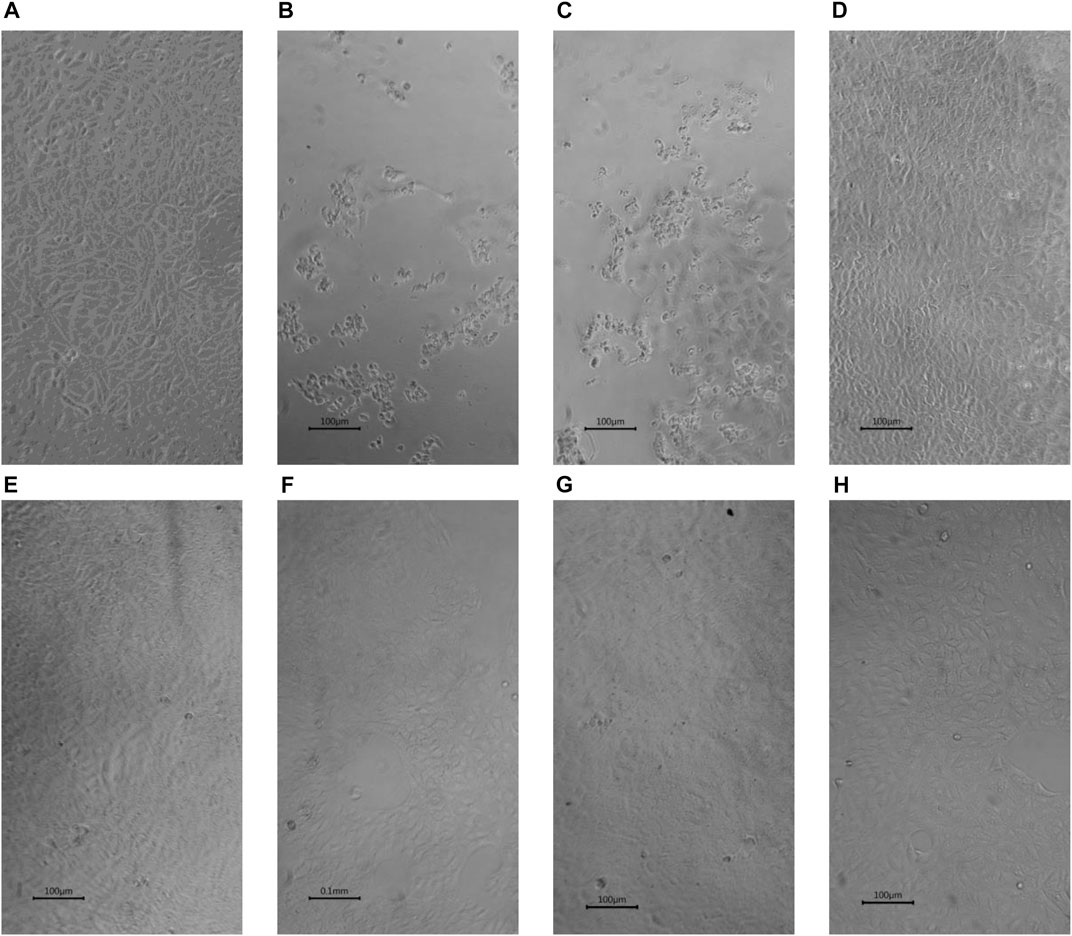
FIGURE 3. Inhibition by selected drugs of SARS-CoV-2 induced CPE in Vero cells (x10) (A) Vero cells. (B) SARS-CoV-2 infected Vero showing ≥ 80% Cytopathic effects (CPE) (C) SARS-CoV-2 infected Vero cells with 0.1% DMSO showing ≥ 80% CPE. (D) SARS-CoV-2 infected Vero cells with 5 µM Erythromycin in 0.1% DMSO showing ≥ 80% inhibition of CPE. (E) SARS-CoV-2 infected Vero cells with 5 µM Retapamulin in 0.1% DMSO showing ≥ 60% inhibition of CPE .(F) SARS-CoV-2 infected Vero cells with 10 µM Pyridoxine in 0.1% DMSO showing ≥ 60% inhibition of CPE. (G) SARS-CoV-2 infected Vero cells with 5 µM Folic acid in 0.1% DMSO showing ≥ 60% inhibition of CPE. (H) SARS-CoV-2 infected Vero cells with 5 µM Ivermectin in 0.1% DMSO showing ≥ 80% inhibition of CPE.All images were taken and prepared by AAA, then reviewed by MBB.
4 Discussion and conclusion
In this study, a novel algorithm was developed and implemented for repurposing drugs for emerging viral diseases using SARS-CoV-2 and COVID-19 as case studies. At each step, scenarios sought to maximize the probability of identifying an effective agent that may easily translate to clinical application through off-label principles while awaiting further studies. Also, the protocol was designed to be easily executed even in LMIC using CPE. These objectives appear successful because erythromycin, pyridoxine and folic acid that this study found effective in inhibiting SARS-CoV-2 induced CPE are almost ubiquitously available in the world and are accepted to be of low safety concerns (Butterworth and Tamura, 1989; Ericson et al., 2015; AlSaad et al., 2017; Field and Stover, 2018), especially when life time dosing is not under consideration. Indeed, Folic acid has previously been suggested to be a potential inhibitor of SARS-CoV-2 MPro (Serseg et al., 2020; Lokhande et al., 2021) and SARS-CoV-2 nucleocapsid protein (Chenmeng et al., 2022), but these studies stopped at in silico docking and focused on a single target. In a separate study, folic acid was shown to inhibit SARS-CoV-2 invasion of cells by methylating ACE2 and reducing the transmissibility in mice (Zhang et al., 2022), altogether suggesting a multitarget effect. In COVID-19 patients, serum levels of folic acid have been correlated with clinical outcome (Acosta-Elias and Espinosa-Tanguma, 2020)and high dose folic acid has been suggested as possible add on therapy or supplement for COVID-19 (Asad and Shuja, 2021). Paradoxically folic acid has been used as inert placebo in clinical trials of COVID-19 infection (Kaur et al., 2021) instead of as an active comparator thereby possibly biasing the result. Supplements, with pyridoxine 4.9 g daily and folic acid 400 μg daily, as a component, have been shown to improve COVID-19 clinical outcome (Beigmohammadi et al., 2020). Erythromycin, the prototype macrolide antibiotic, has not been much studied for SARS-CoV-2 except in computational docking (Anwar et al., 2021; Oliver and Hinks, 2021), which suggested activity against SARS-CoV-2 MPro, but these studies also mainly have single target.
Retapamulin is less ubiquitous and will probably be difficult to access in LMIC, nevertheless, it is attractive as a potential hand or surface ointment solution. Ivermectin has previously been established to inhibit SARS-CoV-2 replication but at physiologically impossible dose (Schmith et al., 2020). In this study ivermectin served as a positive control and the result supports that inhibition of CPE is its additional or consequential effect and further reassures that the overall study design could be predictive. Use of qualitative assessment of CPE inhibition is one of the drawbacks of this study but it is conceivably more useful for rapid screening of large library of compounds compared to quantitative methods. Further studies of the drugs identified are needed and may include quantification of CPE inhibition, graded dose response of CPE inhibition, insight into mechanisms of actions and clinical trials. Dose response studies would be particularly important if doses beyond posology are under consideration; a likely situation where current clinical doses are not expected to be effective (like ivermectin) or have been shown not to be effective in previous clinical trials. Meanwhile, we propose that the result of the present study should be sufficient for off-label prescription of erythromycin, pyridoxine, and folic acid in COVID-19, and is particularly important during rapidly evolving, high mortality epidemics.
In conclusion, drug repurposing efforts need to be purposeful to optimize yields and avoid identifying drugs whose translation to clinical use may be compromised due to toxicity, availability, or inappropriate dosing requirements. This study has presented results that suggest that erythromycin, pyridoxine, and folic acid could be useful in the treatment of COVID-19.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SB was the principal investigator. MI was a co-PI. SB, AY, AA, MI, MB, and AS conceived the study and drafted the initial design. EI, ZH, MP, CO, AB, YD, and IF contributed to the final design. SB, AY, and AA conducted the docking and simulation studies. SB, MI, MB, AY, AA, and AS conducted wet laboratory experiments. All authors contribute to the drafting of the final manuscript.
Funding
This work was funded by the Tertiary Education Trust Fund (TETFund) under The TETFund COVID-19 Special Intervention Research grant (grant number TETFund/DR&D/CE/SI/COVID-19/UDUS/VOL 1). The views stated in this manuscript are solely those of the authors and do not necessarily reflect those of the TETFund. The funding agency did not contribute to the design of this work nor the preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1130828/full#supplementary-material
References
Acosta-Elias, J., and Espinosa-Tanguma, R. (2020). The folate concentration and/or folic acid metabolites in plasma as factor for COVID-19 infection. Front. Pharmacol. 11, 1062. doi:10.3389/fphar.2020.01062
Aftab, S. O., Ghouri, M. Z., Masood, M. U., Haider, Z., Khan, Z., Ahmad, A., et al. (2020). Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 18, 275–315. doi:10.1186/s12967-020-02439-0
AlSaad, D., Awaisu, A., Elsalem, S., Abdulrouf, P. V., Thomas, B., and AlHail, M. (2017). Is pyridoxine effective and safe for post-partum lactation inhibition? A systematic review. J. Clin. Pharm. Ther. 42, 373–382. doi:10.1111/jcpt.12526
Anwar, F., Altayb, H. N., Al-Abbasi, F. A., Kumar, V., and Kamal, M. A. (2021). The computational intervention of macrolide antibiotics in the treatment of COVID-19. Curr. Pharm. Des. 27, 1202–1210. doi:10.2174/1381612827666210125121954
Arshad, U., Pertinez, H., Box, H., Tatham, L., Rajoli, R. K. R., Curley, P., et al. (2020). Prioritization of anti-SARS-cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 108, 775–790. doi:10.1002/cpt.1909
Asad, D., and Shuja, S. H. (2021). Role of folate, cobalamin, and probiotics in COVID-19 disease management [letter]. Drug Des. Dev. Ther. 15, 3709–3710. doi:10.2147/DDDT.S333295
Assmus, F., Driouich, J. S., Abdelnabi, R., Vangeel, L., Touret, F., Adehin, A., et al. (2022). Need for a standardized translational drug development platform: Lessons learned from the repurposing of drugs for COVID-19. Microorganisms 10, 1639. doi:10.3390/microorganisms10081639
Atta, M. G., De Seigneux, S., and Lucas, G. M. (2019). Clinical pharmacology in HIV therapy. Clin. J. Am. Soc. Nephrol. 14, 435–444. doi:10.2215/CJN.02240218
Baby, K., Maity, S., Mehta, C. H., Suresh, A., Nayak, U. Y., and Nayak, Y. (2021). Targeting SARS-CoV-2 main protease: A computational drug repurposing study. A Comput. Drug Repurposing Study 52, 38–47. doi:10.1016/j.arcmed.2020.09.013
Bai, Z., Cao, Y., Liu, W., and Li, J. (2021). The sars-cov-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses 13, 1115. doi:10.3390/v13061115
Beigmohammadi, M. T., Bitarafan, S., Hoseindokht, A., Abdollahi, A., Amoozadeh, L., Mahmoodi Ali Abadi, M., et al. (2020). Impact of vitamins A, B, C, D, and e supplementation on improvement and mortality rate in ICU patients with coronavirus-19: A structured summary of a study protocol for a randomized controlled trial. Trials 21, 614. doi:10.1186/s13063-020-04547-0
Boulanger, M. J., Chow, D., Brevnova, E. E., and Garcia, K. C. (2003). Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science 300, 2101–2104. doi:10.1126/science.1083901
Butterworth, C. E., and Tamura, T. (1989). Folic acid safety and toxicity: A brief review. Am. J. Clin. Nutr. 50, 353–358. doi:10.1093/ajcn/50.2.353
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., and Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 178, 104787. doi:10.1016/j.antiviral.2020.104787
Chenmeng, Y., Wei, J. L., Qin, R. S., Hou, J. P., Zang, G. C., Zhang, G. Y., et al. (2022). Folic acid: A potential inhibitor against SARS-CoV-2 nucleocapsid protein. Pharm. Biol. 60, 862–878. doi:10.1080/13880209.2022.2063341
Cohen, M., and Bendich, A. (1986). Safety of pyridoxine - a review of human and animal studies. Toxicol. Lett. 34, 129–139. doi:10.1016/0378-4274(86)90202-x
Cramer, P., Larson, C. J., Verdine, G. L., and Müller, C. W. (1997). Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution. EMBO J. 16, 7078–7090. doi:10.1093/emboj/16.23.7078
Doms, R. W. (2016). “Basic concepts: A step-by-step guide to viral infection,” in Viral pathogenesis: From basics to systems biology. Editors M. G. Katze, M. J. Korth, G. L. Law, and N. B. T. V. P. Nathanson (London: Elselvier Ltd.) doi:10.1016/B978-0-12-800964-2.00003-3
Ericson, J. E., Arnold, C., Cheeseman, J., Cho, J., Kaneko, S., Wilson, E., et al. (2015). Use and safety of erythromycin and metoclopramide in hospitalized infants. J. Pediatr. Gastroenterol. Nutr. 61, 334–339. doi:10.1097/MPG.0000000000000792
Evans, N. G., Berger, Z. D., Phelan, A. L., and Silverman, R. D. (2021). Covid-19, equity, and inclusiveness. BMJ 373, n1631. doi:10.1136/bmj.n1631
Fathizadeh, H., Afshar, S., Masoudi, M. R., Gholizadeh, P., Asgharzadeh, M., Ganbarov, K., et al. (2021). SARS-CoV-2 (Covid-19) vaccines structure, mechanisms and effectiveness: A review. Int. J. Biol. Macromol. 188, 740–750. doi:10.1016/j.ijbiomac.2021.08.076
Fernandez-fuentes, N., Molina, R., and Oliva, B. (2021). A collection of designed peptides to target sars-cov-2 spike rbd—ace2 interaction. Int. J. Mol. Sci. 22, 11627. doi:10.3390/ijms222111627
Field, M. S., and Stover, P. J. (2018). Safety of folic acid. Ann. N. Y. Acad. Sci. 1414, 59–71. doi:10.1111/nyas.13499
Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., et al. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368, 779–782. doi:10.1126/science.abb7498
Gill, B. S., Kumar, S., and Navgeet, R. (2018). Ganoderic acid A targeting β-catenin in wnt signaling pathway: In silico and in vitro study. Interdiscip. Sci. – Comput. Life Sci. 10, 233–243. doi:10.1007/s12539-016-0182-7
Gupta, A., Pradhan, A., Maurya, V. K., Kumar, S., Theengh, A., Puri, B., et al. (2021). Therapeutic approaches for SARS-CoV-2 infection. Methods 195, 29–43. doi:10.1016/j.ymeth.2021.04.026
Haankuku, U., and Njuho, P. (2021). The estimation of transmitted drug resistance mutation strains probability in the treatment of HIV using the beta-binomial model. AIDS Res. Hum. Retroviruses 37, 468–477. doi:10.1089/AID.2020.0166
Heaton, N. S. (2017). Revisiting the concept of a cytopathic viral infection. PLoS Pathog. 13, e1006409. doi:10.1371/journal.ppat.1006409
Hillen, H. S. (2021). Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 48, 82–90. doi:10.1016/j.coviro.2021.03.010
Holmes, M. D., and Chen, W. Y. (2012). Hiding in plain view: The potential for commonly used drugs to reduce breast cancer mortality. Breast Cancer Res. 14, 216. doi:10.1186/bcr3336
Hussien, M. A., and Abdelaziz, A. E. M. (2020). Molecular docking suggests repurposing of brincidofovir as a potential drug targeting SARS-CoV-2 ACE2 receptor and main protease. Netw. Model. Anal. Heal. Inf. Bioinforma. 9, 56. doi:10.1007/s13721-020-00263-6
James, N., and Ramanathan, K. (2018). Discovery of potent ALK inhibitors using pharmacophore-informatics strategy. Cell. biochem. Biophys. 76, 111–124. doi:10.1007/s12013-017-0800-y
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. doi:10.1038/s41586-020-2223-y
Kang, S., Yang, M., Hong, Z., Zhang, L., Huang, Z., Chen, X., et al. (2020). Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 10, 1228–1238. doi:10.1016/j.apsb.2020.04.009
Karwaciak, I., Karaś, K., Sałkowska, A., Pastwińska, J., and Ratajewski, M. (2022). Chlorpromazine, a clinically approved drug, inhibits SARS-CoV-2 nucleocapsid-mediated induction of IL-6 in human monocytes. Molecules 27, 3651. doi:10.3390/molecules27123651
Kaur, H., Sarma, P., Bhattacharyya, A., Prajapat, M., Kumar, S., Prakash, A., et al. (2021). Folic acid as placebo in controlled clinical trials of hydroxychloroquine prophylaxis in COVID-19: Is it scientifically justifiable? Med. Hypotheses 149, 110539. doi:10.1016/j.mehy.2021.110539
Kim, D., Lee, J., Lee, S., Park, J., and Lee, D. (2016). Predicting unintended effects of drugs based on off-target tissue effects. Biochem. Biophys. Res. Commun. 469, 399–404. doi:10.1016/j.bbrc.2015.11.095
Kontoyianni, M. (2017). Docking and virtual screening in drug discovery. Methods Mol. Biol. 1647, 255–266. doi:10.1007/978-1-4939-7201-2_18
Kurkinen, S. T., Lätti, S., Pentikäinen, O. T., and Postila, P. A. (2019). Getting docking into shape using negative image-based rescoring. J. Chem. Inf. Model. 59, 3584–3599. doi:10.1021/acs.jcim.9b00383
Leslie, V. A., Mohammed Alarjani, K., Malaisamy, A., and Balasubramanian, B. (2021). Bacteriocin producing microbes with bactericidal activity against multidrug resistant pathogens. J. Infect. Public Health 14, 1802–1809. doi:10.1016/j.jiph.2021.09.029
Liang, H., Zhao, L., Gong, X., Hu, M., and Wang, H. (2021). Virtual screening FDA approved drugs against multiple targets of SARS-CoV-2. Clin. Transl. Sci. 14, 1123–1132. doi:10.1111/cts.13007
Lokhande, K. B., Doiphode, S., Vyas, R., and Swamy, K. V. (2021). Molecular docking and simulation studies on SARS-CoV-2 Mpro reveals Mitoxantrone, Leucovorin, Birinapant, and Dynasore as potent drugs against COVID-19. J. Biomol. Struct. Dyn. 39, 7294–7305. doi:10.1080/07391102.2020.1805019
MacKenna, B., Bates, C., Hulme, W., Evans, D., Davy, S., A Kennedy, N., et al. (2021). A comprehensive high cost drugs dataset from the NHS in England - an OpenSAFELY-TPP Short Data Report. Wellcome Open Res. 6, 360. doi:10.12688/wellcomeopenres.17360.1
Madhavi Sastry, G., Adzhigirey, M., Day, T., Annabhimoju, R., and Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 27, 221–234. doi:10.1007/s10822-013-9644-8
Marino, A., Giordano, L., and Ardu, F. (2017). Digoxin poisoning: New prospects for therapy. G. Ital. Nefrol. 34, 82–87.
McCormack, L., Sheridan, S., Lewis, M., Boudewyns, V., Melvin, C. L., Kistler, C., et al. (2013). Communication and dissemination strategies to facilitate the use of health-related evidence. Evid. Rep. Technol. Assess. Full. Rep. 213, 1–520. doi:10.23970/ahrqepcerta213
McDavitt, B., Bogart, L. M., Mutchler, M. G., Wagner, G. J., Green, H. D., Lawrence, S. J., et al. (2016). Dissemination as dialogue: Building trust and sharing research findings through community engagement. Prev. Chronic Dis. 13, E38. doi:10.5888/pcd13.150473
Morrato, E. H., Concannon, T. W., Meissner, P., Shah, N. D., and Turner, B. J. (2013). Dissemination and implementation of comparative effectiveness evidence: Key informant interviews with Clinical and Translational Science Award institutions. J. Comp. Eff. Res. 2, 185–194. doi:10.2217/cer.13.10
Nabeel-Shah, S., Lee, H., Ahmed, N., Burke, G. L., Farhangmehr, S., Ashraf, K., et al. (2022). SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience 25, 103562. doi:10.1016/j.isci.2021.103562
Nagasubramanian, K., Jha, S., Rathore, A. S., and Gupta, K. (2022). Identification of small molecule modulators of class II transactivator-I using computational approaches. J. Biomol. Struct. Dyn. 11, 13. doi:10.1080/07391102.2022.2133011
Ngan, D. K., Xu, T., Xia, M., Zheng, W., and Huang, R. (2022). Repurposing drugs as COVID-19 therapies: A toxicity evaluation. Drug Discov. Today 27, 1983–1993. doi:10.1016/j.drudis.2022.04.001
Nilsson-Payant, B. E., Uhl, S., Grimont, A., Doane, A. S., Cohen, P., Patel, R. S., et al. (2021). The NF-κB transcriptional footprint is essential for SARS-CoV-2 replication. J. Virol. 95, e0125721. doi:10.1128/JVI.01257-21
Obeid, R., Schon, C., Pietrzik, K., Menzel, D., Wilhelm, M., Smulders, Y., et al. (2020). Pharmacokinetics of sodium and calcium salts of (6S)-5-methyltetrahydrofolic acid compared to folic acid and indirect comparison of the two salts. Nutrients 12, 3623–3716. doi:10.3390/nu12123623
Oliver, M. E., and Hinks, T. S. C. (2021). Azithromycin in viral infections. Rev. Med. Virol. 31, e2163. doi:10.1002/rmv.2163
Osipiuk, J., Azizi, S. A., Dvorkin, S., Endres, M., Jedrzejczak, R., Jones, K. A., et al. (2021). Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 12, 743–749. doi:10.1038/s41467-021-21060-3
Parish, L. C., and Parish, J. L. (2008). Retapamulin: A new topical antibiotic for the treatment of uncomplicated skin infections. Drugs Today 44, 91–102. doi:10.1358/dot.2008.44.2.1153446
Parvathaneni, V., and Gupta, V. (2020). Utilizing drug repurposing against COVID-19 – efficacy, limitations, and challenges. Life Sci. 259, 118275. doi:10.1016/j.lfs.2020.118275
Patel, A. B., Lighter, J., Fulmer, Y., Copin, R., Ratner, A. J., and Shopsin, B. (2021). Retapamulin activity against pediatric strains of mupirocin-resistant methicillin-resistant Staphylococcus aureus. Pediatr. Infect. Dis. J. 40, 637–638. doi:10.1097/INF.0000000000003123
Pink, D. A., Chisholm, D. M., and Chapman, D. (1988). Models of protein lateral arrangements in lipid bilayer membranes. Application to electron spin resonance studies of cytochrome c oxidase. Chem. Phys. Lipids 46, 267–277. doi:10.1016/0009-3084(88)90041-2
Platto, S., Wang, Y., Zhou, J., and Carafoli, E. (2021). History of the COVID-19 pandemic: Origin, explosion, worldwide spreading. Biochem. Biophys. Res. Commun. 538, 14–23. doi:10.1016/j.bbrc.2020.10.087
Rahman, Z., Bazaz, M. R., Devabattula, G., Khan, M. A., and Godugu, C. (2021). Targeting H3K9 methyltransferase G9a and its related molecule GLP as a potential therapeutic strategy for cancer. J. Biochem. Mol. Toxicol. 35, e22674. doi:10.1002/jbt.22674
Rawls, K., Dougherty, B. V., and Papin, J. (2020). Metabolic network reconstructions to predict drug targets and off-target effects. Methods Mol. Biol. 2088, 315–330. doi:10.1007/978-1-0716-0159-4_14
Riemenschneider, M., and Heider, D. (2016). Current approaches in computational drug resistance prediction in HIV. Curr. HIV Res. 14, 307–315. doi:10.2174/1570162x14666160321120232
Rocheleau, L., Laroche, G., Fu, K., Stewart, C. M., Mohamud, A. O., Cote, M., et al. (2021). Identification of a high-frequency intrahost SARS-CoV-2 spike variant with enhanced cytopathic and fusogenic effects. MBio 12, e0078821. doi:10.1128/mBio.00788-21
Samizi, F. G., Panga, O. D., Malugu, S. S., Gitige, C. G., and Mmbaga, E. J. (2021). Rate and predictors of HIV virological failure among adults on first-line antiretroviral treatment in Dar Es Salaam, Tanzania. J. Infect. Dev. Ctries. 15, 853–860. doi:10.3855/jidc.13603
Schmith, V. D., Zhou, J., and Lohmer, L. R. L. (2020). The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther. 108, 762–765. doi:10.1002/cpt.1889
Serseg, T., Benarous, K., Yousfi, M. H., and Lepidine, E. (2020). Hispidin and lepidine E: Two natural compounds and folic acid as potential inhibitors of 2019-novel coronavirus main protease (2019- nCoVMpro), molecular docking and SAR study. Curr. Comput. Aided. Drug Des. 17, 469–479. doi:10.2174/1573409916666200422075440
Severson, W. E., Shindo, N., Sosa, M., Fletcher, T., White, E. L., Ananthan, S., et al. (2007). Development and validation of a high-throughput screen for inhibitors of SARS CoV and its application in screening of a 100,000-compound library. J. Biomol. Screen. 12, 33–40. doi:10.1177/1087057106296688
Sharun, K., Dhama, K., Patel, S. K., Pathak, M., Tiwari, R., Singh, B. R., et al. (2020). Ivermectin, a new candidate therapeutic against SARS-CoV-2/COVID-19. Ann. Clin. Microbiol. Antimicrob. 19, 23. doi:10.1186/s12941-020-00368-w
Shivalkar, S., Pingali, M. S., Verma, A., Singh, A., Singh, V., Paital, B., et al. (2021). Outbreak of COVID-19: A detailed overview and its consequences. Adv. Exp. Med. Biol. 1353, 23–45. doi:10.1007/978-3-030-85113-2_2
Shoshan, M. C., and Linder, S. (2008). Target specificity and off-target effects as determinants of cancer drug efficacy. Expert Opin. Drug Metab. Toxicol. 4, 273–280. doi:10.1517/17425255.4.3.273
Shyr, Z. A., Cheng, Y. S., Lo, D. C., and Zheng, W. (2021). Drug combination therapy for emerging viral diseases. Drug Discov. Today 26, 2367–2376. doi:10.1016/j.drudis.2021.05.008
Szabó, G. T., Mahiny, A. J., and Vlatkovic, I. (2022). COVID-19 mRNA vaccines: Platforms and current developments. Mol. Ther. 30, 1850–1868. doi:10.1016/j.ymthe.2022.02.016
The Lancet Infectious Diseases (2021). COVID-19 vaccine equity and booster doses. Lancet Infect. Dis. 21, 1193. doi:10.1016/S1473-3099(21)00486-2
Ullrich, S., and Nitsche, C. (2020). The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 30, 127377. doi:10.1016/j.bmcl.2020.127377
Uttarilli, A., Amalakanti, S., Kommoju, P. R., Sharma, S., Goyal, P., Manjunath, G. K., et al. (2021). Super-rapid race for saving lives by developing COVID-19 vaccines. J. Integr. Bioinform. 18, 27–43. doi:10.1515/jib-2021-0002
Wang, L. Q., Hu, Z. y., Yu, Q., Guo, X., Xiong, J., Huang, Z. z., et al. (2005). Pharmacokinetics of erythromycin stinoprate capsule. J. Cent. South Univ. Med. Sci. 30, 197–201.
Warren, G. L., Do, T. D., Kelley, B. P., Nicholls, A., and Warren, S. D. (2012). Essential considerations for using protein-ligand structures in drug discovery. Drug Discov. Today 17, 1270–1281. doi:10.1016/j.drudis.2012.06.011
Wilson, P. M., Petticrew, M., Calnan, M. W., and Nazareth, I. (2010). Disseminating research findings: What should researchers do? A systematic scoping review of conceptual frameworks. Implement. Sci. 5, 91. doi:10.1186/1748-5908-5-91
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi:10.1126/science.abb2507
Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., et al. (2020). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell. Res. 30, 343–355. doi:10.1038/s41422-020-0305-x
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi:10.1126/science.abb2762
Yang, H., and Rao, Z. (2021). Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 19, 685–700. doi:10.1038/s41579-021-00630-8
Yarrington, M. E., Moehring, R. W., David, M. Z., Hamilton, K. W., Klompas, M., Rhee, C., et al. (2021). A modified Delphi approach to develop a trial protocol for antibiotic de-escalation in patients with suspected sepsis. Antimicrob. Steward. Healthc. Epidemiol. 1, e44. doi:10.1017/ash.2021.205
Yuan, M., Wu, N. C., Zhu, X., Lee, C. C. D., So, R. T. Y., Lv, H., et al. (2020). A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633. doi:10.1126/science.abb7269
Zempleni, J. (1995). Pharmacokinetics of vitamin b6 supplements in humans. J. Am. Coll. Nutr. 14, 579–586. doi:10.1080/07315724.1995.10718546
Zhan, Y. (2021). Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 82, 1124–1130. doi:10.1002/ddr.21815
Zhang, H. Y. (2005). One-compound-multiple-targets strategy to combat Alzheimer’s disease. FEBS Lett. 579, 5260–5264. doi:10.1016/j.febslet.2005.09.006
Zhang, Y., Pang, Y., Xu, B., Chen, X., Liang, S., Hu, J., et al. (2022). Folic acid restricts SARS-CoV-2 invasion by methylating ACE2. Front. Microbiol. 13, 980903. doi:10.3389/fmicb.2022.980903
Keywords: SARS-CoV-2, COVID-19 treatment, repurpose, cytopathic action, docking & simulation
Citation: Bello SO, Yunusa A, Adamu AA, Imam MU, Bello MB, Shuaibu A, Igumbor EU, Habib ZG, Popoola MA, Ochu CL, Bello AY, Deeni YY and Okoye I (2023) Innovative, rapid, high-throughput method for drug repurposing in a pandemic—A case study of SARS-CoV-2 and COVID-19. Front. Pharmacol. 14:1130828. doi: 10.3389/fphar.2023.1130828
Received: 23 December 2022; Accepted: 20 February 2023;
Published: 01 March 2023.
Edited by:
Miguel Sierra-Hoffman, Texas A&M University, United StatesReviewed by:
Mithun Rudrapal, Technology and Research, IndiaAnoop Kumar, Delhi Pharmaceutical Sciences and Research University, India
Copyright © 2023 Bello, Yunusa, Adamu, Imam, Bello, Shuaibu, Igumbor, Habib, Popoola, Ochu, Bello, Deeni and Okoye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaibu Oricha Bello, b3JpY2hhLmJlbGxvQHVkdXNvay5lZHUubmc=
 Shaibu Oricha Bello
Shaibu Oricha Bello Abdulmajeed Yunusa1,3
Abdulmajeed Yunusa1,3 Mustapha Umar Imam
Mustapha Umar Imam Muhammad Bashir Bello
Muhammad Bashir Bello Ehimario Uche Igumbor
Ehimario Uche Igumbor