
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 20 January 2023
Sec. Experimental Pharmacology and Drug Discovery
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1110146
This article is part of the Research TopicCell fate and disease: The role of autophagy, apoptosis, ferroptosis, and necroptosis in human health and treatmentView all 7 articles
 Zhongqing Zhang1,2†
Zhongqing Zhang1,2† Tianlei Xie1,2†
Tianlei Xie1,2† Shun Zhang1,3†
Shun Zhang1,3† Haoli Yin1,3
Haoli Yin1,3 Xuyu Zhang1,2
Xuyu Zhang1,2 Siyuan Zhang4
Siyuan Zhang4 Wei Chen3
Wei Chen3 Ding Yu5
Ding Yu5 Xuefeng Qiu1,3*
Xuefeng Qiu1,3* Wei Zhao4*
Wei Zhao4* Hongqian Guo1,3*
Hongqian Guo1,3* Junlong Zhuang1,3*
Junlong Zhuang1,3*Purpose: Prostate cancer (PCa) poses a great threat to humans. The study aimed to evaluate the potential of TQB3720 in promoting ferroptosis to suppress prostate cancer, providing a theoretical basis for PCa therapy.
Methods: PCa cells and nude mice models were divided into TQB3720, enzalutamide (ENZ), and control groups. Sulforhodamine B assay, colony formation assessment, organoids culture system, and the CCK8 assay were used for detecting proliferation. Western blot assay was processed to detect the expression of androgen receptor (AR), ferroptosis, and apoptosis-related genes. Flow cytometry was applied to measure the intracellular ROS levels. ELISA was performed to determine the cellular oxidized glutathione (GSSG) and malondialdehyde (MDA) levels. RT-qPCR was conducted to detect the mRNA expression of genes in AR signaling. BODIPYTM™ 581/591 was processed for detection of intracellular lipid peroxidation levels. The interaction of AR with other translational factor complex proteins was explored using Co-immunoprecipitation (Co-IP), and the chromatin immunoprecipitation (ChIP) assay was performed to detect the binding of AR-involved translational complex to downstream genes promoter. Luciferase reporter assay was conducted to examine the translation activity of GPX4 promoter, and immunohistochemistry (IHC) was conducted to analyze the levels of c-MYC, Ki-67 and AR in TQB3720-treated cancer tissues.
Results: Here, we found TQB3720 inhibits the growth of prostate cancer in vitro and in vivo. TQB3720 treatment induced intracellular levels of GSSG and MDA significantly, by which hints AR antagonist caused ferroptosis-related cell death. Moreover, molecular evidence shown TQB3720 regulates downstream of AR signaling by binding AR resulting in inhibition of AR entry into the nucleus. Additional, we also proved that TQB3720 abrogates the interaction between AR and SP1 and leads to decrease GPX4 transcription.
Conclusion: TQB3720 promotes ferroptosis in prostate cancer cells by reducing the AR/SP1 transcriptional complex binding to GPX4 promoter. As a result, it is suggested to be a potential drug for clinic prostate cancer treatment.
Prostate cancer (PCa) is a malignant tumor of the genitourinary system, accounting for about 14.1% of malignant tumors in men (Sung et al., 2021). Early-stage patients mostly have no obvious symptoms, whereas late-stage patients may experience abnormal urination, abnormal urodynamics, blood in semen, erectile dysfunction, discomfort in the pelvic region, and bone pain (Litwin and Tan, 2017). Most patients are in the middle to the late stage at diagnosis. Therefore, it is important to develop novel drugs for countering against PCa (Sekhoacha et al., 2022).
In previous studies, AR was reported to transduce hormonal signals to regulates multiple prostate cancer events, proliferation, apoptosis, migration, invasion, and differentiation (Jacob et al., 2021; Westaby et al., 2022). As a translational factor, the transactivation function of AR can be regulated by Specificity Protein 1 (SP1) (Chen et al., 2008), the interaction between AR and SP1 stimulated the production of Vascular Endothelial Growth Factor (VEGF) (Eisermann et al., 2013) and Erythropoietin-Producing Hepatocellular carcinoma cell surface type-A receptor 3 (EPHA3) (Diao et al., 2018), which promotes PCa growth and progession. Thus, targeting the AR/SP1 translational complex showing therapeutic potential in prostate cancer (Wang and Ferrari, 2006; Yuan et al., 2010). In the past 30 years, huge number of anti-AR drugs have been developed and approved for different stages of prostate cancer, such as flutamide, bicalutamide, nilumet, and enzalutamide (Bassetto et al., 2016). First-generation AR antagonists have shortcomings, including shorter half-life and lower steady-state levels in the brain (Clement and Karen, 2022). Thus, the scientists develop the second-generation AR antagonists to address these shortcomings. Nowadays, TQB3720 has been considered as a second-generation AR antagonist (Chen et al., 2022). However, the molecular mechanisms of TQB3720 in PCa remains unexplored completely.
To uncover the potential mechanism of TQB3720 in PCa, we focused on ferroptosis. Emerging studies have shown that ferroptosis is involved in the development of prostate cancer. Ferroptosis is an iron-dependent and non-apoptotic form of cell death characterized by the accumulation of iron and ROS (Shen et al., 2021). However, iron homeostasis is essential in the development of prostate cancer, iron toxicity inhibits the growth of cancer cells (Raymonvil, 2017). Ferroptosis-mediating factors (SLC7A11, SLC3A2, GPX4) are aberrantly expressed in AR-resistant prostate cancer (Zaffaroni and Beretta, 2022). The induction of ferroptosis in tumor cells is expected to address drug resistance to conventional radiotherapy (Zhang et al., 2019; Zhou et al., 2019). TQB3720, a second-generation AR antagonist which is developed by Chia Tai Tianqing Pharmaceutical Group Co., Ltd., competes for binding to AR and causes its loss of function (Chen et al., 2022). Most importantly, this chemical is under the evaluation of the tolerance and pharmacokinetics in clinical trial phase 1 stage (ClinicalTrials.gov Identifier: NCT04853498). In this study, we investigated the potential molecular mechanisms of TQB3720 in ferroptosis of prostate cancer development using the in vivo and in vitro models.
The prostate cancer cell lines (LNCap95 and 22RV1) were purchased from American Type Culture Collection (ATCC). Cells were cultured with RPMI-1640 medium (Gibco) containing 10% fetal bovine serum and incubated in a cell incubator (Thermo Fisher Scientific) with 37°C and 5% CO2. Moreover, inhibitors of multiple death pathways, including Ferrostatin-1 (Sigma-Aldrich), Liprostatin-1 (Sigma-Aldrich), Necrosulfonamide (Selleckchem), ZVAD-FMK (Selleckchem), and Chloroquine (Selleckchem), were used to detect the function of TQB3720 (TQB) and enzalutamide (ENZ). Synbio Technologies Co. Ltd. (Suzhou, China) synthesized mixture siRNAs (target AR and GPX4) and HA-AR while their transfection was conducted using the lipofectamine 2000 reagent (Invitrogen) as our previous reports (Zhao et al., 2014; Huang et al., 2019).
Cells were treated with different concentrations of TQB3720 (0, 0.5, 1, 2, and 5 μM, Chia Tai Tianqing Pharmaceutical Group Co., Ltd.), while similar concentrations of Enzalutamide (ENZ, 0, 0.5, 1, 2, and 5 μM, Selleckchem) were used as a reference to clarify the dose-time-effect of TQB3720. After incubation for 24, 48, and 72 h, the plates were taken out, and 50 μL of 10% trichloroacetic acid (TCA) was added to each well before fixing for 1 h at 4°C. After fixation, the liquid in the wells was discarded, and the cells were washed 5 times with deionized water to remove the TCA. 100 μL of 0.4% SRB reagent (LMAI Bio Co. Ltd., China) was added to each well and kept at room temperature for 20 min. The liquid in the wells was discarded, and the unbound reagent was removed using 1% acetic acid and dried at room temperature. 150 μL of dimethyl sulfoxide (DMSO, Sigma-Aldrich) was then added and shaken for 10 min on a microplate shaker to lyse the cells (QB-9001, Qilinbeier Co. Ltd., China). The absorbance value (OD570) of each well was measured by a Microplate reader (Thermo Fisher Scientific) and used to calculate the effect of the drug on cell proliferation.
LNCap95 and 22RV1 cells were inoculated in 6-well plates with a cell density of 500 cells/well. The experiments were divided into control, TQB3720 (5 µM), and GA+ENZ (5 µM) groups. After 48 h of treatment, the cells were washed 3 times with PBS buffer and stained with crystal violet (Sangon Biotech, China). Colonies formed in each group were observed, and the number of colonies with more than 10 cells was counted by photographing them under a microscope (Olympus). Colony formation rate was calculated as previous reports (Liu et al., 2021; Sun et al., 2022).
The prostate cancer tissue was collected from Department of Urology, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School with patients written informed consents. This study has been approved by Ethics Committee of Nanjing University Medical School. The collected prostate cancer tissues were clipped on ice and resuspended by adding collagenase (Invitrogen). After digestion, the cells were filtered and the filtrate was added to DMEM/F12 (Gibco) to terminate the digestion. Centrifugation was then used to separate the supernatant. Erythrocyte lysate was used to resuspend the cells. After centrifugation, the supernatant was removed and DMEM/F12 was added to resuspend cells. After centrifugation, the supernatant was removed, cells were counted, and matrigel (Gibco) was mixed. Samples were dropped in each well of the plate, culture dishes were placed, and martrigel was coagulated. 500 nM of A83-01 medium (STEMCELL technologies) was added to each well, and the cells were incubated. The sample plates were then digested for 15–25 min and uniformly distributed in microtiter plates. The sample plates were incubated for 22–26 h, then replaced with the drug-containing medium. To maintain the growth conditions after spreading, the plates were incubated for 24 h. This was done to mimic the conditions during in vivo drug delivery.
The cells were inoculated in 100 mm culture dishes and incubated for 48 h after drug treatment. After sufficient lysis of the cells, the supernatant was collected by centrifugation. The protein content was determined using the BCA kit (Sigma-Aldrich). SDS-PAGE electrophoresis was performed to separate proteins with different relative molecular masses, which were then transferred onto PVDF membranes (Thermo Fisher Scientific). The proteins were blocked with 5% skim milk for 2 h, washed 3 times with TBST, and incubated with primary antibodies (GPX4, AR, FKBP51, PSA, LK2, S100P, TMPRSS2, Bax, caspase-3, Bcl-2, GPX1, AR, PSA, 1:1000, all purchased from Abcam) overnight at 4°C. PVDF membranes were washed with TBST, and peroxidase-labeled secondary antibody (1:10000, Abcam) was added, incubated for 2 h at room temperature, and washed 4 times with PBST. The proteins were visualized by a DAB immunohistochemistry color development kit (Sangon Biotech, China) and photographed using a gel imaging analysis system (Thermo Fisher Scientifi). Relative protein levels were calculated by ImageJ as previous report (Sun et al., 2022).
The cells were cultured for 6 h after TQB3720 treatment. DCFH-DA staining solution and serum-free culture medium were diluted at 1:1000, and 2 mL of the working solution was added to cell-containing wells. The cells were then incubated for 20 min at 37°C. The cells were digested using trypsin, collected, and resuspended in PBS buffer after 3 washes with a serum-free culture medium. Intracellular ROS levels were measured with an Accuri C6 flow cytometer (BD Accuri C6) with an excitation wavelength of 488 nm and an emission wavelength of 525 nm.
Cellular GSSG and MDA levels were measured by ELISA assay. The standard solutions were diluted, and the standard curve was plotted following the ELISA kit (Sigma, Shanghai) manufacturer’s instructions. 40 μL of sample and 10 μL of biotin-labeled antibody were added to each ELISA plate well. 50 μL of standard enzyme reagent was added to each well except the blanks. The plate was sealed and incubated at 37°C for 30 min. The sealing membrane was removed, and the liquid was discarded and dried. Each well was filled with the washing solution and allowed to stand for 30 s. 50 μL of reagent A and 50 μL of reagent B were added to each well, gently shaken, and mixed. The cell mixture was then incubated, and the samples were developed at 37°C for 10 min in darkness. 50 μL of termination solution was used to terminate the reaction and the results were detected using a microplate reader (Bio-Rad).
RT-qPCR assay was conducted to detect the expression of AR signaling-related genes (AR, FKBP51, PSA, LK2, S100P, TMPRSS2), ferroptosis-related gene (GPX4), and apoptosis-related genes (Bax, caspase-3, Bcl-2). The cells were inoculated in cell culture flasks with 1 × 106 cells. After 24 h, TQB3720 was introduced in doses of 0, 2, and 5 μM, and the treatment was terminated after 48 h. The cells were collected, and total RNA was extracted by TRIzol reagent (Invitrogen) and reverse transcribed into cDNA using a reverse transcription kit (QIAGEN). The amplification conditions included 95°C for 3 min, 95°C for 15 s, 58°C for 30 s, 72°C for 30 s, and 35 cycles. A fluorescence real-time quantitative PCR kit (Thermo Fisher Scientific) was used for RT-qPCR detection with primers synthesized by Invitrogen (Table 1), and GAPDH was used as an internal reference. All the used primers were purchased from Sangon (Shanghai, China). Ct values were analyzed and processed as 2−ΔΔCT (Ren et al., 2021).
The cells were grown in 24-well plates, and when the cell density grew to about 60%–70%, the drug was added for 7 h. 1 μmol/L BODIPYTM 581/591 solution was added into the wells with the cells and incubated for 1 h at 37°C. Random photographic recordings were processed under fluorescence microscopy. The blue color represented the hochest-stained nuclei, while the red indicated the lipid peroxidation level.
The effect of TQB on cell proliferation was detected using the CCK8 kit (DOJINDO, Japan). The cells were inoculated in 96-well plates at a density of 8 × 103 cells per well and incubated at 37°C in a 5% CO2 incubator. After culture apposition, the culture medium was discarded, and the cells were treated with TBQ, HA-AR, and siGPX4 for 48 h. The original culture medium was discarded and 100 μL of culture medium containing 10% CCK8 reagent was added to each well and incubated for 2 h at 37°C. The absorbance value of each well was measured at 450 nm using a microplate reader (Bio-Rad, USA).
After 24 h of treatment, the cells were collected, lysed with 0.5 mL of lysis solution, and then centrifuged for 30 min in an ice bath to obtain the supernatant for immunoprecipitation assay. Briefly, the assay followed our previous report (Zhao et al., 2014). The antibodies (anti-AR, cat.no. 06-680, Sigma, Shanghai; anti-SP1, cat.no. ab231778, Abcam, Shanghai; anti-Tubulin-α, cat.no. ab52866, Abcam) for immunoprecipitation were added to the supernatant and incubated overnight at 4°C. Agarose beads protein A-binding antibodies were added and incubated at 4°C for 4 h. The precipitates were washed 4 times with lysate, each time for 10 min. The precipitate was added to 20 μL of 2×SDS protein buffer, denatured by boiling for 10 min, and subjected to SDS-PAGE electrophoresis.
After 48 h of treatment, cells were collected and lysed. ChIP experiments were then performed to show DNA sequences (including GPX4 promoter) bound to AR or SP1 proteins. The experiments were performed using the ChIP kit (cat.no. ab185913, Abcam) following the manufacturer’s instructions. DNA-binding proteins were cross-linked to DNA in vivo using formaldehyde. After chromosomal separation, the sheared DNA fragments were associated with the binding proteins. The Specific antibodies (anti-AR, cat.no. 06-680, Sigma, Shanghai; anti-SP1, cat.no. ab231778, Abcam, Shanghai; anti-Tubulin-α, cat.no. ab52866, Abcam) were used to precipitate and to isolate the complex containing targeted proteins and its binding DNAs. The DNA was released by reverse cross-linking, and the protein was digested. RT-qPCR was performed to detect GPX4 promoter, and western blot was conducted to detect targeted proteins.
The promoter (−500 bp, 50 bp) of GPX4, recorded as RefSeq NM_00208 in NCBI database, was constructed into pGLO vector. After 12 h transfection with GPX4 promoter (pGLO system, cat.no.2920, Promega, Madison, USA), the cells were incubated with 5 μM TQB7320 or 5 μM ENZ for 48 h. Afterwards, the cells were lysed by using passive lysis buffer according to the protocols of Promega kit (cat.no. J3081, Promega). The luciferase activity was determined by using Promega GloMaxTM 20/20 instrument. Luciferase assay substrate was added into the supernatant of cell samples. After mixing completely and measuring immediately as fluorescence intensity I, they were added with the “stop & glo” solution to stop the reaction and the corresponding luciferase activity was recorded as fluorescence intensity II. The relative luciferase activity was calculated as the ratio of fluorescence value II to I.
LNCap95 and 22RV1 cells were cultured in 10% fetal bovine serum, RPMI 1640 medium at 37°C, and 5% CO2. The cells were then inoculated subcutaneously on the back shoulder of mice at the logarithmic growth stage, and then randomly divided into several groups: TQB3720 (0 mg/kg body weight), TQB3720 (20 mg/kg body weight), and TQB3720 (40 mg/kg body weight). The animal experiments were approved by the biomedical ethics committee of Chengdu Medical College. The initiating day was recorded as the 0 days. The tumor size of mice was measured every 3 days. Finally, the cancerous tissues were collected for further analysis.
The assay was conducted using the ready-to-use immunohistochemistry kit (Biolab, cat.no. GS4974, Bejing, China). Following the manufacturer’s instructions, the negative control group of the experiment was substituted with phosphate buffer solution (PBS) for the primary antibody. Tissue sections were incubated in an oven at 70°C for 2 h, dewaxed in xylene 3 times, and hydrated in alcohol. The sections were then soaked in a 3% H2O2 mixture, rinsed with PBS buffer and distilled water in turns, placed in 0.01 mol/L raffinate buffer, transferred to an autoclave (120°C∼140°C) and heat-repaired antigen for 10 min. The sections were incubated with primary antibody in a wet box for 20 min overnight at 4°C. The samples were kept at room temperature, rinsed, and a secondary antibody was added, and incubation was continued for 30 min in a wet box at 37°C. The sections were washed 3 times with PBS and developed with DAB. Mayer’s hematoxylin was re-stained for 5 s, and the films were sealed with neutral gum. Images were taken immediately using a LSM 780 microscope (Zeiss, Germany), LSM 880 Fast Ariyscan, or SP8 microscope (Leica, Germany).
All the data were analyzed using SPSS 20.0 statistical software (IBM, Armonk, New York, USA). Measurement data were expressed as mean ± standard deviation (
Sulforhodamine B assays were used to determine the effects of TQB3720 and ENZ on the proliferation of LNCap95 and 22RV1 cells to determine the dose-time effect of TQB3720. Here, both TQB3720 and positive control, ENZ inhibited the survival rate of LNCap and 22RV1 cells. Importantly, the effects of both drugs were dose-time dependent (Figures 1A–D). 5 µM TQB3720 and ENZ had the most significant effects and were used for the following experiments. Effects of TQB3720 on the proliferation of LNCap95, 22RV1 cells were determined by assessing colony formation. TQB3720 and ENZ significantly decreased the survival rate of both cell lines. Compared with the ENZ group, the inhibitory effect of TQB3720 was more evident, although there were no statistically significant differences (Figure 1E). Moreover, an organoids culture system was used to detect the effect of TQB3720 on organoid formation from prostate cancer tumors. It was observed that TQB3720 inhibited the growth of prostate tissue (Figure 1F), suggesting the inhibitory effects of TQB3720 on the growth of prostate cells.
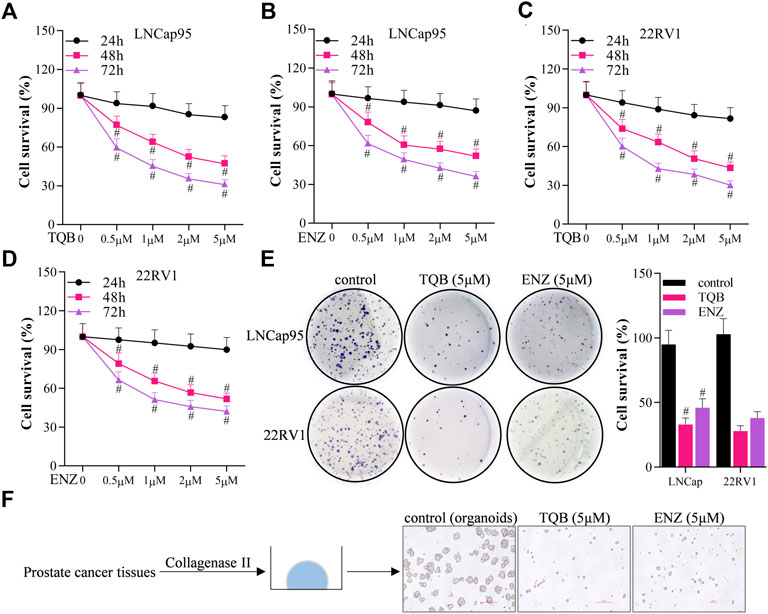
FIGURE 1. TQB3720 inhibited the growth of prostate cells (A–D) Sulforhodamine B assays were conducted to detect the effect of TQB3720 on the proliferation of LNCap95 and 22RV1 cells, #p < 0.01 vs. 24 h. (E) Effect of TQB3720 on colony formation of LNCap95, 22RV1 cells, #p < 0.01 vs. 24 h. (F) Organoid assay showing the effect of TQB3720 on organoid formation of clinic human prostate cancer primary cells.
Above data demonstrated that TQB3720 inhibits the growth of prostate cells. To uncover the detailed effect of 2nd generation of AR antagonist on PCa cells, Sulforhodamine B assays were conducted to determine the effect of TQB3720 and other inhibitors such as ferrostatub-1, liprostatin-1, necrosulfonamide, ZVAD-FAK, and chloroquine on the proliferation of LNCap95 and 22RV1 cells. After 48 h of TQB3720 treatment, the proliferation of LNCap95 and 22RV1 cells significantly decreased, while the inhibition was rescued by ferrostatin-1 and liprostatin-1, necrosulfonamide, ZVAD-FMK, and chloroquine failed to rescue PCa cells (Figures 2A, B). The results suggest that ferroptosis is a major cell death when TQB3720 treated PCa cells. Western blot assay indicated that 2 and 5 µM of TQB3720 inhibited GPX4 expression in LNCap95 and 22RV1 cells (Figures 2C, D). The results hint that TQB3720 induces iron-dependent death in PCa cells.
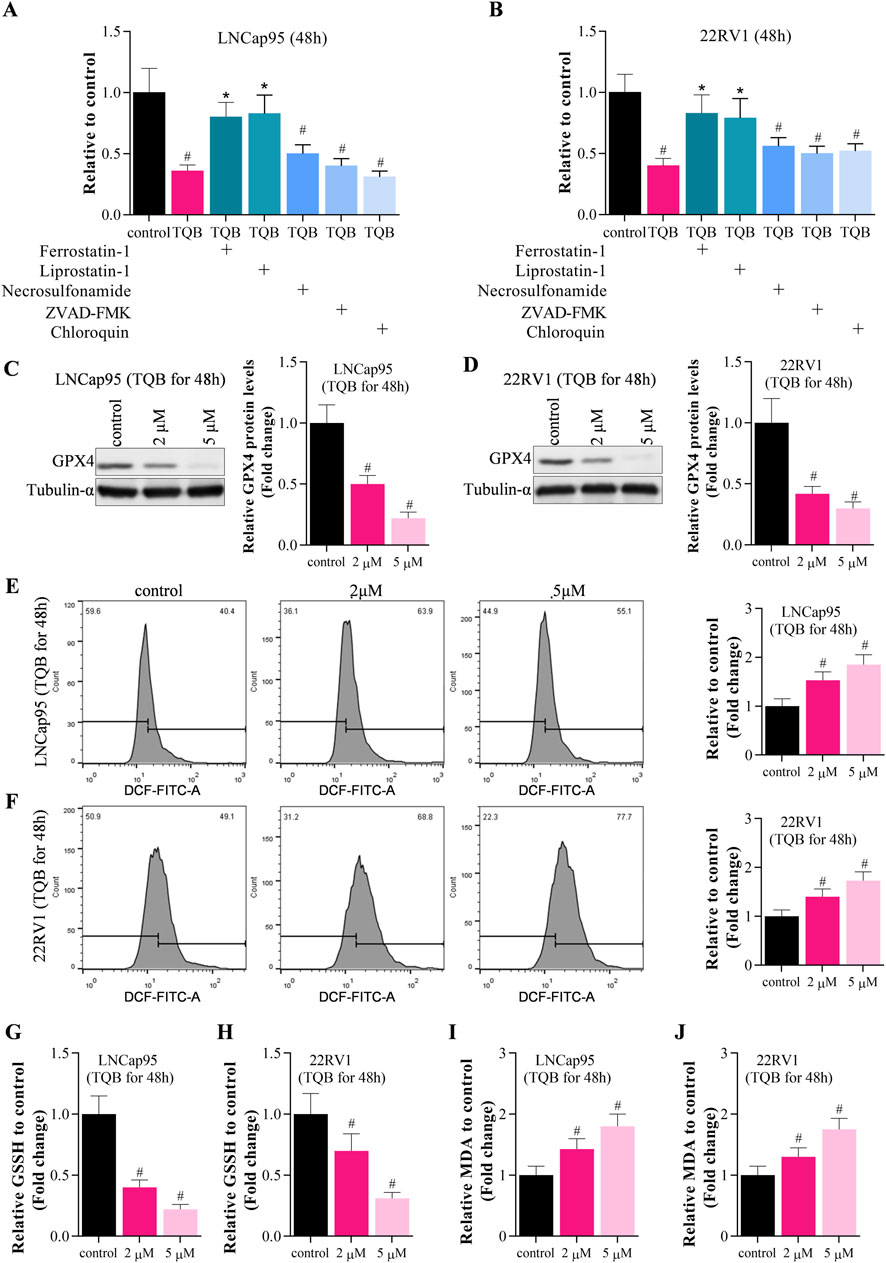
FIGURE 2. TQB3720 induced ferroptosis of prostate cancer cells. (A,B) The proliferation of LNCap95 and 22RV1 cells significantly decreased following TQB3720 incubation for 48 h, whereas ferrostatin-1 and liprostatin-1 abolished this effect, #p < 0.01 vs. control, *p < 0.01 vs. TQB3720 (5 μM) (C,D) Western blot showed effects of TQB3720 on expression of ferroptosis marker GPX4, #p < 0.01 vs. control. (E,F) Flow cytometry assay showed the effects of TQB3720 on ROS levels using DCFH-DA probes, #p < 0.01 vs. control. (G,H) ELISA results showed the effects of TQB3720 on GSSG levels in cancer cells, #p < 0.01 vs. control. (I–J) ELISA assay proved that the effects of TQB7320 on MDA levels in cancer cells, #p < 0.01 vs. control.
GPX4 impairs lipid peroxide toxicity through its catalytic activity and maintains membrane lipid bilayer homeostasis, and inactivation of GPX4 induces Reactive Oxygen Species (ROS) accumulation and triggers iron-dependent death (Kang et al., 2022; Yang W et al., 2022). In Figures 2E, F, data shown that 2 and 5 µM TQB3720 treatment increased the intracellular level of ROS significantly. Compared with the control, ELISA assay also indicated that cellular oxidized glutathione (GSSG) and malondialdehyde (MDA) levels were significantly upregulated by TQB3720 treatment in PCa cells (Figures 2G–J). Taken together, data in Figure 2 demonstrated that TQB3720 induces ferroptosis in prostate cancer cells.
To identify the underlying molecular mechanism of TQB3720 induced-ferroptosis, the expression of AR signaling-related genes (AR, FKBP51, PSA, LK2, S100P, TMPRSS2), ferroptosis-related genes (GPX4), and apoptosis-related genes (Bax, caspase-3, Bcl-2). The expression levels of FKBP51, PSA, LK2, S100P, TMPRSS2, and Bcl-2 GPX4 were downregulated by 2 and 5 µM of TQB3720. The apoptosis-related genes, caspase-3 was also downregulated, and Bax was up-related in TQB3720 groups compared with the controls (Figures 3A–D). Moreover, the knockdown of AR abrogates GPX4 expression, but no effect on GPX1 (Figures 3E, F). The BODIPY 581/591 assay revealed the knockdown of AR-induced lipid peroxidation (Figure 3G). In Figure 3H, TQB3720 induced PCa cells lipid peroxidation. Exogenous AR inhibited intracellular lipid peroxidation, while knockdown of GPX4 induced PCa cells lipid peroxidation (Figure 3H). When Furthermore, HA-AR increased cell viability, while si-GPX4 had inhibitory effects. Cell viability gradually decreased with increasing TQB3720 concentration (Figure 3I). Our results indicate that TQB3720 induces ferroptosis via the AR-GPX4 axis.
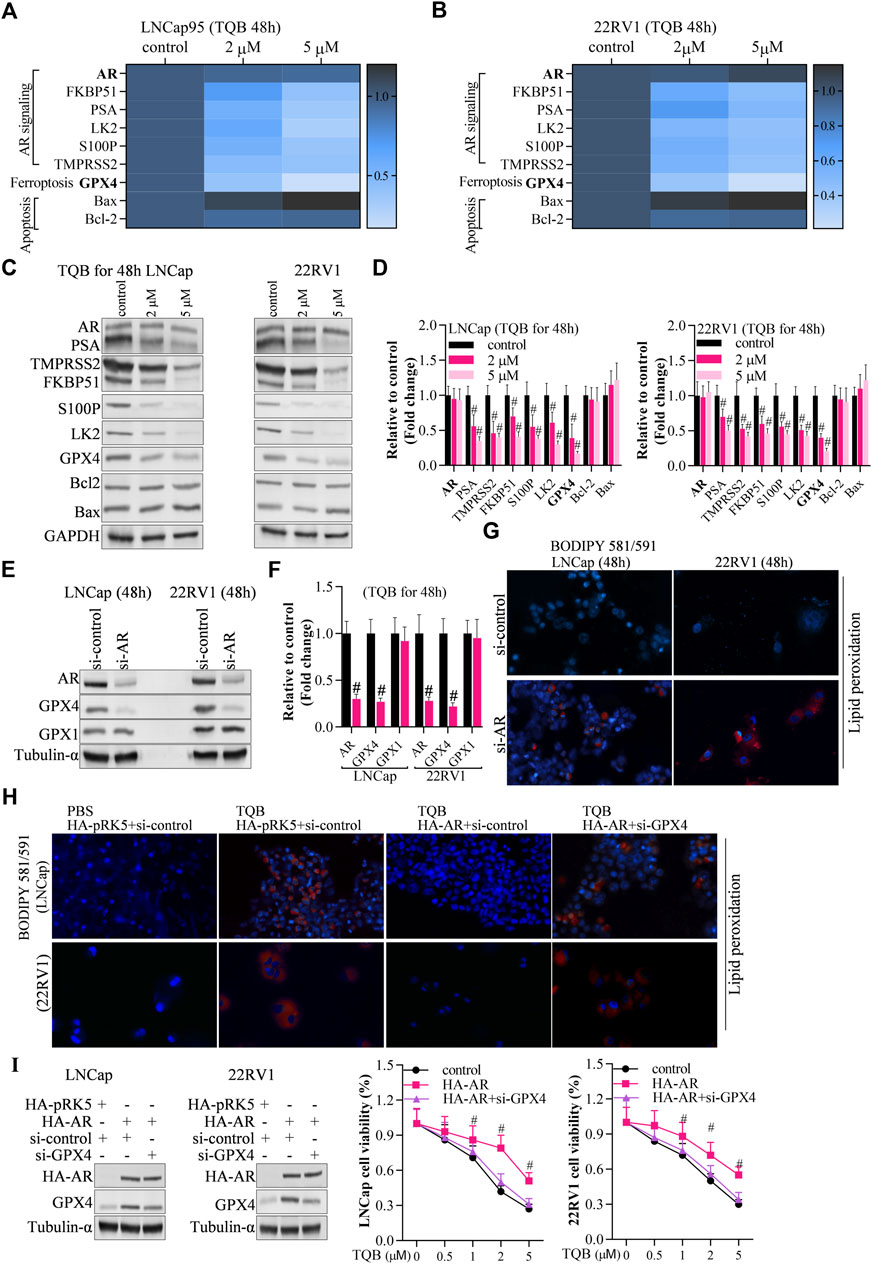
FIGURE 3. TQB3720 induced ferroptosis via AR-GPX4 axis (A,B) RT-qPCR detected the effects of on expression of genes in AR signaling pathway, ferroptosis pathway, and apoptotic pathway, #p < 0.01 vs. control. (C,D) Western blot examined the effects of on expression of genes in AR signaling pathway, ferroptosis pathway, and apoptotic pathway, #p < 0.01 vs. control. (E,F) Western blot determined the expression of AR and GPX4 after knockdown of AR, #p < 0.01 vs. si-control. (G) The BODIPY 581/591 assay revealed the effects of AR knockdown on lipid peroxidation. Blue indicates the hochest stained nuclei, and red indicates lipid peroxidation levels. (H) The cells were transfected with HA-AR for 24 h after knocking down GPX4 for 48 h, and then they were incubated with 5 μM TQB3720 for another 48 h. The BODIPY 581/591 assay was used to determine the lipid peroxidation levels indicated by hochest-stained nuclei in blue and lipid peroxidation in red. (I) The cells were transfected with HA-AR for 24 h after knocking GPX4 down for 48 h and then incubated with 5 μM TQB3720 for another 48 h, and then, MTT assay was used to detect the cell proliferation, #p < 0.01 vs. control.
As a second-generation AR antagonist, above data proved that TQB3720 alleviates AR downstream genes expression. Here, we found PSA was significantly downregulated by TQB3720 and ENZ treatment, while AR expression without change (Figure 4A). Then, the cells were incubated with DHT for 2h, followed by TQB3720 and ENZ stimulation for the next 48 h. They were collected for nuclei-cytoplasm separation, and the contents of AR in the nucleus and plasma of the cells were detected using western blot. TQB3720 and ENZ inhibited DHT-mediated promotion of AR entry into the nucleus (Figures 4B–D). Therefore, TQB3720 did not affect AR protein levels but it suppressed downstream of AR signaling by inhibiting AR entry into the nucleus (Figures 4C, D). This mechanism indicates that administration of TQB7320 does not develop drug resistance.

FIGURE 4. TQB3720 inhibited downstream of AR signaling by inhibiting AR entry into the nucleus (A, B) Western blot examined the effects of TQB3720 and ENZ on PSA expressions, and the protein band density was determined using ImageJ software, #p < 0.01 vs. control (absence of any indicated chemicals). (C,D) Western blot examined the effects of TQB3720 and ENZ on AR entry into nuclei from cytoplasm, and the protein band density was determined using ImageJ software, #p < 0.01 vs. control (absence of any indicated chemicals).
To identify whether TQB3720 disturb the AR/SP1 transcriptional factor complex in PCa cells, co-IP, ChIP and Luciferase reporter assays were performed. In both LNCap95 and 22RV1, an interaction between AR and SP1 was observed. However, this interaction was diminished after incubation of TQB3720 or ENZ (Figures 5A, B). By using anti-AR or anti-SP1, the ChIP data also confirmed that TQB3720 and ENZ groups showed less content of immunoprecipitated GPX4 promoter than in the controls (Figures 5C, D). Additionally, luciferase reporter assay showed that TQB3720 impaired GPX4 expression. The ChIP as well as RT-qPCR showed that TQB3720 inhibited the binding of AR/SP1 to the PSA promoter (Supplementary Figure S1). The highlighted results confirmed our speculation that TQB3720 decreases GPX4 expression by inhibiting the interaction between AR and SP1.

FIGURE 5. TQB3720 impaired GPX4 expression by inhibiting AR binding to SP1 (A, B) Co-IP assay showed the effect of TQB3720 on the interaction between AR and SP1 in LNcap95 and 22RV1. (C, D) RT-qPCR detected content of AR- or SP1-binding GPX4 promoter after performing ChIP assay, #p < 0.01 vs. control. (E) Luciferase reporter assay analyzed the impacts of TQB7320 on luciferase activity of GPX4 promoter, #p < 0.01 vs. control.
The effect of TQB3720 on prostate cell tumor growth in vivo was verified through xenograft experiments. Compared with control group (TQB3720, 0 mg/kg), 20 and 40 mg/kg of TQB3720 significantly inhibited tumor growth (Figures 6A, B). Additionally, 40 mg/kg TQB3720 downregulated Ki-67 and c-MYC expression (Figure 6C). Heterogeneous tumor tissues were collected for nucleoplasmic separation, and the levels of AR proteins in the nucleus and plasma were detected by western blot. The results showed that AR entry into the nucleus was inhibited by 40 mg/kg TQB3720 (Figure 6D). Moreover, GPX4 expression was downregulated by 40 mg/kg TQB3720 in vivo (Figure 6E). The in vivo results were similar with those obtained in vitro, showing that TQB3720 impairs GPX4 expression and inhibits AR entry into the nucleus.
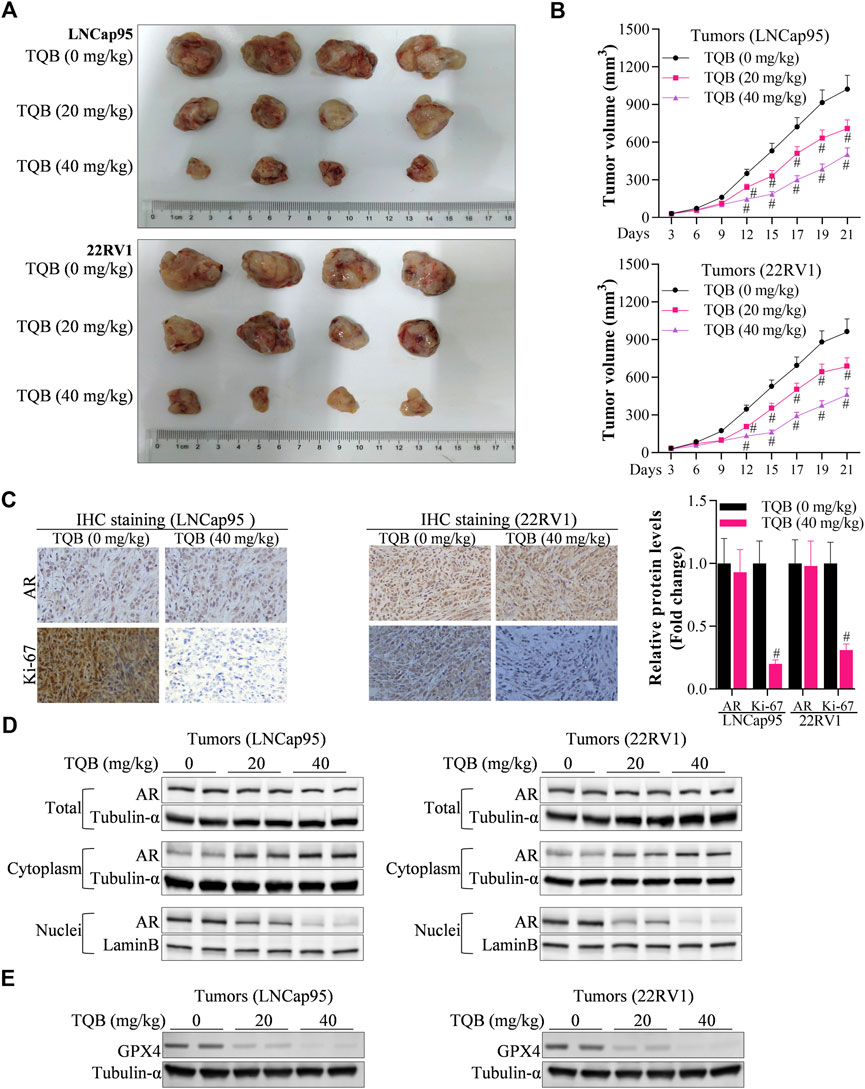
FIGURE 6. TQB3720 inhibited PCa carcinogenesis. (A, B) The tumor volume in each group of nude mice, #p < 0.01 vs. TQB3720 (0 mg/kg). (C) IHC examined the influences of TQB7320 on the expression levels of AR, Ki-67, #p < 0.01 vs. TQB3720 (0 mg/kg). (D) Western blot showed the impacts of TQB7320 on content of AR proteins in the nucleus or cytoplasm tumors from nude mice. (E) Western blot examined the effects of TQB7320 on expression of GPX4 protein in tumors.
Prostate cancer, especially metastatic PCa, poses a great threat to humans worldwide (Raju et al., 2022). Therefore, developing effective and safe anti-cancer agents is critical to prolong the life of patients, reduce death rates, and improve survival. Traditional androgen deprivation therapy (ADT), which includes surgical and pharmacologic deprivation, improves the prognosis of PCa patients by lowering serum testosterone levels and inhibiting the androgen pathway (Cho et al., 2003). However, after 18–24 months of ADT, almost all PCa patients evolve into destructive-resistant prostate cancer (Oudard et al., 2014). Therefore, AR antagonists that act directly on the androgen receptor (AR) are critical in PCa treatment. The first-generation AR antagonists, such as bicalutamide and flutamide, have shown a low affinity for the receptor, poor antimutagenic ability, and hepatotoxic adverse effects in clinical applications (Soloway et al., 2000). Although second-generation AR antagonists have partially solved these problems, they are associated with drug resistance (Semenas et al., 2012). In this study, TQB3720 promoted ferroptosis in prostate cancer cells by alleviating the AR/SP1 transcriptional complex. It is suggested to be a potential drug for prostate cancer treatment, with lower drug resistance than ENZ.
During androgen deficiency, the cells develop an adaptive response by either allowing AR mutation or proliferation (Wang, 2009). AR regulates other signaling pathways to achieve the self-activating process. The transcriptional activity of AR is usually activated by androgen receptor signaling (Cronauer et al., 2003). Thus in depot-resistance diseases, there is a strong selective pressure to maintain the AR control signaling pathway (Clement and Karen, 2022). In this study, the expressions of FKBP51, PSA, LK2, S100P, TMPRSS2, GPX4, and Bcl-2 were downregulated by TQB3720 as well as caspase-3. As a result, TQB3720 improved apoptosis and inhibited AR signaling. Moreover, TQB3720 regulates AR downstream signaling by inhibiting AR entry into the nucleus, which is unique compared with first and second-generation AR antagonists. Moreover, the results from this study verified that AR binds to SP1. As reported in a previous study, aberrant expression and activation of SP1 protein in tumor tissues regulate tumor proliferation, angiogenesis, and metastatic ability (Yuan et al., 2005). Our results suggest that by inhibiting AR binding to SP1, TQB3720 is effective in prostate cancer. Based on the data from the clinical trial database (https://www.clinicaltrials.gov), TQB3720 has been investigated in numerous advanced cancers. Moreover, data from the global data database (https://pharmacn.globaldata.com) indicate that TQB3720 has been used in cancers, such as non-small cell lung cancer, gastric cancer, and cancer of the gallbladder. TQB3720, therefore, exhibits potential as a clinical prostate cancer treatment.
To clarify the molecular mechanisms of TQB3720 in the suppression of prostate cancer progression, we examined the effects of TQB3720 on ferroptosis. Ferroptosis refers to iron-dependent cell death caused by the excessive accumulation of ROS and lipid peroxides (Latunde-Dada, 2017). Highly malignant cancer cells have an innate susceptibility to ferroptosis, and thus induction of ferroptosis may be a new approach to cancer therapy (Mga et al., 2022). In this study, to evaluate the potential of TQB3720 to promote ferroptosis in prostate cancer, molecular mechanisms of TQB3720 were studied using in vivo and in vitro models. TQB3720 inhibits the growth of prostate cells and induces ferroptosis in prostate cancer cells. The level of ROS was higher in TQB3720 groups. TQB3720 treatment also significantly increased the levels of GSSG and MDA.
Moreover, ENZ was used as a reference for TQB3720, and in vitro experiments confirmed the non-inferiority of TQB3720 compared with ENZ, suggesting that TQB3720 maybe be a potential drug for prostate cancer treatment. GPX4 is the most potent defense against ferroptosis (Chang et al., 2022). In this study, TQB3720 induced ferroptosis by impairing GPX4 expression. Several therapeutic oncology regimens are highly susceptible to GPX4 inhibition, highlighting the importance of GPX4 inhibitors in targeting ferroptosis in cancer (Zhan et al., 2022). However, most inhibitors are associated with poor pharmacological properties in animal models, limiting their potential for clinical translation. The development and optimization of GPX4-targeted drugs with improved pharmacokinetics and selectivity remain a major challenge to the use of GPX4 inhibitors for cancer treatment (Mtca et al.). Thus, available and in vivo stable anticancer drugs with GPX4 inhibitory activities may provide an alternative approach. Previously, it was found that inhibition of GPX4 activity and expression activated ferroptosis and effectively inhibited tumor progression (Zhan et al., 2022). Ferroptosis is regulated by GPX4 and iron transport regulatory proteins (Song et al., 2020). Consistent with these reports, our study showed that TQB3720 suppressed GPX4 expression and GSSG levels while increasing the MDA levels. The cellular redox homeostasis factor GSSG and the lipid peroxidation product MDA are essential in maintaining cellular redox homeostasis (Wong and Bruice, 2012). The main manifestations of ferroptosis in biochemical metabolism are ionized iron deposition leading to membrane lipid peroxidation and excessive oxidative stress that impairs intracellular redox homeostasis, reducing antioxidant capacity and increasing intracellular lipid reactive oxygen species, ultimately leading to oxidative cell death (Yang Z et al., 2022).
In conclusion, TQB3720 promotes ferroptosis in prostate cancer cells by alleviating the AR/SP1 transcriptional complex. TQB3720-based cancer therapy is expected to bypass the drawbacks of conventional therapies mediated by apoptosis due to its ability to induce natural non-apoptotic forms. It is, therefore, a potential drug for prostate cancer treatment, with equivalent efficacy as ENZ. In future, we shall explore the clinical application of TQB3720 and identify strategies for improving the effectiveness and safety of the drug.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Ethics Committee of Nanjing University Medical School. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Biomedical Ethics Committee of Chengdu Medical College.
XQ, WZ, HG, and JZ conceived and designed the work. ZZ, TX, ShZ, HY, XZ, SiZ, WC, and DY performed experiments. ZZ, TX, and ShZ collected and analyzed the data. WZ and JZ checked the raw data and results. JZ, HG, and SiZ obtained fundings. WZ, JZ, and SiZ drafted the manuscript. All authors revised and agree to be accountable for the content of the work.
This work was funded by the National Natural Science Foundation of China (81974394, 82273301 and 81972388), Natural Science Foundation of Jiangsu Province for Excellent Young Scholars (BK20200051) and the Sichuan Province Science and Technology Program (2022072).
Author DY was employed by the company Chia Tai Tianqing Pharmaceutical Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1110146/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | RT-qPCR determined the effects of TQB3720 on the binding of AR/SP1 to PSA promoter after conducting ChIP assay. (A,B) The cell lines were incubated with TQB7320 and ENZ for 48 h, we used RT-qPCR to detect content of AR- or SP1-binding PSA promoter after performing ChIP assay, #p < 0.01 vs. control.
Bassetto, M., Ferla, S., Pertusati, F., Kandil, S., Westwell, A. D., Brancale, A., et al. (2016). Design and synthesis of novel bicalutamide and enzalutamide derivatives as antiproliferative agents for the treatment of prostate cancer. Eur. J. Med. Chem. 118, 230–243. doi:10.1016/j.ejmech.2016.04.052
Chang, M. T., Tsai, L-C., Goto, K. N., Lee, K. H., and Shyur, L. F. (2022) Phyto-sesquiterpene lactones DET and DETD-35 induce ferroptosis in vemurafenib sensitive and resistant melanoma via GPX4 inhibition and metabolic reprogramming. Pharmacol. Res. 178, 106148. doi:10.1016/j.phrs.2022.106148
Chen, P. H., Tsao, Y. P., Wang, C. C., and Chen, S. L. (2008). Nuclear receptor interaction protein, a coactivator of androgen receptors (AR), is regulated by AR and Sp1 to feed forward and activate its own gene expression through AR protein stability. Nucleic Acids Res. 36 (1), 51–66. doi:10.1093/nar/gkm942
Chen, Y., Zhou, Q., Hankey, W., Fang, X., and Yuan, F. (2022). Second generation androgen receptor antagonists and challenges in prostate cancer treatment. Cell Death Dis. 13 (7), 632. doi:10.1038/s41419-022-05084-1
Cho, D., Blasio, C. J. D., Rhee, A. C., and Kattan, M. W. (2003). Prognostic factors for survival in patients with hormone-refractory prostate cancer (HRPC) after initial androgen deprivation therapy (ADT). Urol. Oncol. 21, 282. doi:10.1016/s1078-1439(03)00057-7
Clement, C., and Karen, A. (2022). Targeting the androgen receptor signaling pathway in advanced prostate cancer. Am. J. Health-System Pharm. 79 (15), 1224–1235. doi:10.1093/ajhp/zxac105
Cronauer, M. V., Schulz, W. A., Ackermann, R., and Burchardt, M. (2003). Inhibition of p53 function diminishes androgen receptor-mediated signaling in prostate cancer cell lines. Oncogene 23, 3541. doi:10.1038/sj.onc.1207346
Diao, X., Chen, X., Pi, Y., Zhang, Y., Wang, F., Liu, P., et al. (2018). Androgen receptor induces EPHA3 expression by interacting with transcription factor SP1. Oncol. Rep. 40 (2), 1174–1184. doi:10.3892/or.2018.6503
Eisermann, K., Broderick, C. J., Bazarov, A., Moazam, M. M., and Fraizer, G. C. (2013). Androgen up-regulates vascular endothelial growth factor expression in prostate cancer cells via an Sp1 binding site. Mol. Cancer 12, 7. doi:10.1186/1476-4598-12-7
Huang, N., Guo, W., Ren, K., Li, W., Jiang, Y., Sun, J., et al. (2019). LncRNA AFAP1-AS1 supresses miR-139-5p and promotes cell proliferation and chemotherapy resistance of non-small cell lung cancer by competitively upregulating RRM2. Front. Oncol. 9, 1103. doi:10.3389/fonc.2019.01103
Jacob, A., Raj, R., Allison, D. B., and Myint, Z. W. (2021). Androgen receptor signaling in prostate cancer and therapeutic strategies. Cancers (Basel) 13 (21), 5417. doi:10.3390/cancers13215417
Kang, X., Huo, Y., Jia, S., He, F., Li, H., Zhou, Q., et al. (2022). Silenced LINC01134 enhances oxaliplatin sensitivity by facilitating ferroptosis through GPX4 in hepatocarcinoma. Front. Oncol. 12, 939605. doi:10.3389/fonc.2022.939605
Latunde-Dada, G. O. (2017). Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochimica Biophysica Acta (BBA) - General Subj. 1861, 1893–1900. doi:10.1016/j.bbagen.2017.05.019
Litwin, M. S., and Tan, H. J. (2017). The diagnosis and treatment of prostate cancer: A review. JAMA 317 (24), 2532–2542. doi:10.1001/jama.2017.7248
Liu, Y., Yang, Y., Zhang, L., Lin, J., Li, B., Yang, M., et al. (2021). LncRNA ASAP1-IT1 enhances cancer cell stemness via regulating miR-509-3p/YAP1 axis in NSCLC. Cancer Cell Int. 21 (1), 572. doi:10.1186/s12935-021-02270-7
Mga, B., Hrh, B., and Aykb, C. (2022). Molecular mechanisms of reactive oxygen species in regulated cell deaths: Impact of ferroptosis in cancer therapy. Gene Rep. 27, 101614. doi:10.1016/j.genrep.2022.101614
Oudard, D. L. M. R., Delanoy, F., Ozgüroglu, B., Ozguroglu, M., Mercier, F., Guillot, A., et al. (2014). Prostate-specific antigen flare induced by cabazitaxel-based chemotherapy in patients with metastatic castration-resistant prostate cancer. Eur. J. cancer official J. Eur. Organ. Res. Treat. Cancer (EORTC) Eur. Assoc. Cancer Res. (EACR) 50 (9), 1602–1609. doi:10.1016/j.ejca.2014.03.015
Raju, G. N. L., Bhat, P. P., and Nagini, S. (2022). Utility of prostate-specific antigen isoforms and prostate health index in the diagnosis of metastatic prostate cancer. J. Laboratory Physicians doi:10.1055/s-0042-1757723
Raymonvil, A. D. (2017). Serum iron concentration and prostate cancer in the United States. Minneapolis: Walden University ScholarWorks.
Ren, K., Sun, J., Liu, L., Yang, Y., Li, H., Wang, Z., et al. (2021). TP53-Activated lncRNA GHRLOS regulates cell proliferation, invasion, and apoptosis of non-small cell lung cancer by modulating the miR-346/APC Axis. Front. Oncol. 11, 676202. doi:10.3389/fonc.2021.676202
Sekhoacha, M., Riet, K., Motloung, P., Gumenku, L., Adegoke, A., and Mashele, S. (2022). Prostate cancer review: Genetics, diagnosis, treatment options, and alternative approaches. Molecules 27 (17), 5730. doi:10.3390/molecules27175730
Semenas, A., Boorjian, S., A., Mongan, N., P., and Persson, J. L. (2012). Overcoming drug resistance and treating advanced prostate cancer. Curr. drug targets-The Int. J. timely in-depth Rev. drug targets 13, 1308–1323. doi:10.2174/138945012802429615
Shen, L. D., Qi, W. H., Bai, J. J., Zuo, C. Y., Bai, D. L., Gao, W. D., et al. (2021). Resibufogenin inhibited colorectal cancer cell growth and tumorigenesis through triggering ferroptosis and ROS production mediated by GPX4 inactivation. anatomical Rec. Adv. Integr. Anat. Evol. Biol. 304 (12), 313–322. doi:10.1002/ar.24378
Soloway, M. S., Schellhammer, P. F., Sharifi, R., Block, N. L., Venner, P. M., Patterson, A. L., et al. (2000). Bicalutamide and flutamide, each in combination with luteinizing hormone-releasing hormone analogs, in advanced prostate cancer: Exploratory analysis of impact of extent of disease by bone scan on outcome. Adv. Prostate Cancer Explor. Analysis Impact Extent Dis. by Bone Scan onOutcome 2 (3), 137–145. doi:10.1046/j.1525-1411.2000.23003.x
Song, X., Wang, X., Liu, Z., and Yu, Z. (2020). Role of GPX4-mediated ferroptosis in the sensitivity of triple negative breast cancer cells to gefitinib. Front. Oncol. 10, 597434. doi:10.3389/fonc.2020.597434
Sun, Q., Gui, Z., Zhao, Z., Xu, W., Zhu, J., Gao, C., et al. (2022). Overexpression of LncRNA MNX1-AS1/PPFIA4 activates AKT/HIF-1α signal pathway to promote stemness of colorectal adenocarcinoma cells. J. Oncol. 2022, 8303409. doi:10.1155/2022/8303409
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Wang, H. (2009). Essential and specific roles of Skp2 in Rb-deficiency induced tumorigenesis and hormone-sensitive prostate cancer cells. New York: Yeshiva University.
Wang, L. G., and Ferrari, A. C. (2006). Mithramycin targets sp1 and the androgen receptor transcription level-potential therapeutic role in advanced prostate cancer. Transl. Oncogenomics 1, 19–31.
Westaby, D., Fenor de La Maza, M. L. D., Paschalis, A., Jimenez-Vacas, J. M., Welti, J., de Bono, J., et al. (2022). A new old target: Androgen receptor signaling and advanced prostate cancer. Annu. Rev. Pharmacol. Toxicol. 62, 131–153. doi:10.1146/annurev-pharmtox-052220-015912
Wong, C. G., and Bruice, T. C. (2012). Utility of dual electrochemical gsh and gssh evaluation and other redox/detoxification quantitation as functional parameters in assessment of retinal toxicity during in vivo drug studies with large animal eyes. Invest. Ophthalmol. Vis. 53, 5557.
Yang, W., Wang, Y., Zhang, C., Huang, Y., Yu, J., Shi, L., et al. (2022). Maresin1 protect against ferroptosis-induced liver injury through ROS inhibition and Nrf2/HO-1/GPX4 activation. Front. Pharmacol. 13, 865689. doi:10.3389/fphar.2022.865689
Yang, Z., Huang, S., Liu, Y., Chang, X., Liang, Y., Li, X., et al. (2022). Biotin-targeted Au(I) radiosensitizer for cancer synergistic therapy by intervening with redox homeostasis and inducing ferroptosis. J.Med. Chem. 65, 8401.doi:10.1021/acs.jmedchem.2c00300
Yuan, H. Q., Yang, L. L., Jiang, A. L., Kong, F., Zhang, J. Y., and Young, C. (2005). Inhibitory effects of quercetin on proliferation of prostate cancer cells and activity of transcription factor Sp1. Chin. J. Biochem. Mol. Biol. 21, 672–677.
Yuan, H., Young, C. Y., Tian, Y., Liu, Z., Zhang, M., and Lou, H. (2010). Suppression of the androgen receptor function by quercetin through protein-protein interactions of Sp1, c-Jun, and the androgen receptor in human prostate cancer cells. Mol. Cell Biochem. 339 (1-2), 253–262. doi:10.1007/s11010-010-0388-7
Zaffaroni, N., and Beretta, G. (2022). Ferroptosis inducers for prostate cancer therapy. Curr. Med. Chem. 29 (24), 4185–4201. doi:10.2174/0929867329666220111120924
Zhan, S., Lu, L., Pan, S. S., Wei, X. Q., Miao, R. R., Liu, X. H., et al. (2022). Targeting NQO1/GPX4-mediated ferroptosis by plumbagin suppresses in vitro and in vivo glioma growth. Br. J. cancer 127 (2), 364–376. doi:10.1038/s41416-022-01800-y
Zhang, Z., Jiaying, Q., Li, Q., and Kai, Y. (2019). The valuable role of holo-lactoferrin in ferroptosis and its application in radiotherapy of triple negative breast cancer (P05-014-19). Curr. Dev. Nutr. 3, nzz030. doi:10.1093/cdn/nzz030.P05-014-19
Zhao, W., Chang, C., Cui, Y., Zhao, X., Yang, J., Shen, L., et al. (2014). Steroid receptor coactivator-3 regulates glucose metabolism in bladder cancer cells through coactivation of hypoxia inducible factor 1α. J. Biol. Chem. 289 (16), 11219–11229. doi:10.1074/jbc.M113.535989
Keywords: prostate cancer, androgen receptor, TQB3720, ferroptosis, GPx4
Citation: Zhang Z, Xie T, Zhang S, Yin H, Zhang X, Zhang S, Chen W, Yu D, Qiu X, Zhao W, Guo H and Zhuang J (2023) Second generation androgen receptor antagonist, TQB3720 abrogates prostate cancer growth via AR/GPX4 axis activated ferroptosis. Front. Pharmacol. 14:1110146. doi: 10.3389/fphar.2023.1110146
Received: 28 November 2022; Accepted: 09 January 2023;
Published: 20 January 2023.
Edited by:
Xiawei Cheng, East China University of Science and Technology, ChinaReviewed by:
Kaijie Wu, The First Affiliated Hospital of Xi’an Jiaotong University, ChinaCopyright © 2023 Zhang, Xie, Zhang, Yin, Zhang, Zhang, Chen, Yu, Qiu, Zhao, Guo and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuefeng Qiu, aHlkZXdvb2RzQDE2My5jb20=; Wei Zhao, encxOTg2MjY1MjBAMTI2LmNvbQ==; Hongqian Guo, ZHIuZ2hxQG5qdS5lZHUuY24=; Junlong Zhuang, emh1YW5namxAbmpnbHl5LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.